Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected 78 million individuals and is responsible for over 1.7 million deaths to date. Infection is associated with the development of variable levels of antibodies with neutralizing activity, which can protect against infection in animal models1,2. Antibody levels decrease with time, but, to our knowledge, the nature and quality of the memory B cells that would be required to produce antibodies upon reinfection has not been examined. Here we report on the humoral memory response in a cohort of 87 individuals assessed at 1.3 and 6.2 months after infection with SARS-CoV-2. We find that titres of IgM and IgG antibodies against the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 decrease significantly over this time period, with IgA being less affected. Concurrently, neutralizing activity in plasma decreases by fivefold in pseudotype virus assays. By contrast, the number of RBD-specific memory B cells remains unchanged at 6.2 months after infection. Memory B cells display clonal turnover after 6.2 months, and the antibodies that they express have greater somatic hypermutation, resistance to RBD mutations and increased potency, indicative of continued evolution of the humoral response. Immunofluorescence and PCR analyses of intestinal biopsies obtained from asymptomatic individuals at 4 months after the onset of coronavirus disease 2019 (COVID-19) revealed the persistence of SARS-CoV-2 nucleic acids and immunoreactivity in the small bowel of 7 out of 14 individuals. We conclude that the memory B cell response to SARS-CoV-2 evolves between 1.3 and 6.2 months after infection in a manner that is consistent with antigen persistence.
Antibody responses to SARS-CoV-2 were initially characterized in a (6.2 months; range of 165 to 223 days) after the onset of symptoms. cohort of individuals convalescing from COVID-19 at approximately In this cohort, symptoms lasted for a median of 12 days (range of 0 to 40 days (1.3 months) after infection1. Between 31 August and 16 44 days) during the acute phase, and 10 (11%) of the participants were October 2020, 100 participants returned for a 6-month follow-up hospitalized. Consistent with other reports3,4, 38 (44%) of the particistudy visit. Although the initial criteria allowed enrolment of the pants reported persistent long-term symptoms attributable to COVIDclose contacts of individuals with SARS-CoV-2 infection confirmed 19 (Methods, Supplementary Tables 1, 2). The duration and severity by reverse-transcription PCR (RT–PCR)1, 13 of the contacts did not of symptoms during acute disease was significantly greater among seroconvert and were excluded from further analyses. The remaining participants with persistent post-acute symptoms at the second study 87 participants with RT–PCR-confirmed SARS-CoV-2 infection and/ visit (Extended Data Fig. 1m–q). Importantly, all 87 participants tested or seroconversion returned for analysis at approximately 191 days negative for SARS-CoV-2 at the 6-month follow-up study visit using an approved saliva-based PCR assay (Methods). Participant demographics and clinical characteristics are shown in Supplementary Tables 1, 2.
Plasma SARS-CoV-2 antibody reactivity
Antibody reactivity in plasma to the RBD and nucleoprotein (N) of SARS-CoV-2 was measured by enzyme-linked immunosorbent assay (ELISA) and automated serological assays1,5,6. Anti-RBD assays were strongly correlated (anti-RBD IgG ELISA and Pylon–IgG, anti-RBD IgM ELISA and Pylon–IgM at 1.3 months, r = 0.9200 and r = 0.7543, P < 0.0001, respectively) (Extended Data Fig. 2a–d), and anti-N assays showed a moderate correlation (anti-N IgG ELISA and Roche anti-N total immunoglobulin at 1.3 months, r = 0.3596, P = 0.0012) (Extended Data Fig. 2e, f). The anti-RBD and ELISA anti-N antibodies in plasma decreased significantly between 1.3 and 6.2 months (Fig. 1a–d). Notably, the decreased binding activity differed substantially by isotype and target. IgM showed the greatest decrease in anti-RBD reactivity (53%), followed by IgG (32%); anti-RBD IgA decreased by only 15% and anti-N IgG levels by 22% (Fig. 1e). By contrast, the Roche anti-N assay6 showed a small but significant increase (19%) in reactivity between the two time points (Extended Data Fig. 2g), which might be explained by the use of an antigen bridging approach7. In all cases, the magnitude of the decrease was inversely proportional to and directly correlated with the initial antibody levels, such that individuals with higher initial levels showed greater relative changes (Fig. 1f–i, Extended Data Fig. 2h). All measurements were strongly correlated between the two time points (Extended Data Fig. 2i–m) and anti-N IgG correlated with respective IgM, IgG and IgA anti-RBD reactivity at 1.3 months (Extended Data Fig. 2n–p). Notably, individuals with persistent post-acute symptoms had significantly higher levels of anti-RBD IgG and anti-N total antibody at both study visits (Extended Data Fig. 1a–l).
Fig. 1 |. Plasma antibody dynamics against SARS-CoV-2.
a–d, Results of ELISAs measuring plasma reactivity to RBD (a, b, c) and N protein (d) at the initial 1.3- and 6.2-month follow-up visit, respectively. a, Anti-RBD IgM. b, Anti-RBD IgG. c, Anti-RBD IgA d, Anti-N IgG. The normalized area under the curve (AUC) values for 87 individuals are shown in a–d for both time points. Positive and negative controls were included for validation1. e, Relative change in plasma antibody levels between 1.3 and 6.2 months for anti-RBD IgM, IgG, IgA and anti-N IgG in all 87 individuals. f–i, Relative change in antibody levels between 1.3 and 6.2 months plotted against the corresponding antibody levels at 1.3 months. f, Anti-RBD IgM. r = −0.83, P < 0.0001. g, Anti-RBD IgG. r = −0.76, 1.3 months 6.2 months 100 101 102 103 104 105 NT50 at 1.3 months P < 0.0001. h, Anti-RBD IgA. r = −0.67, P < 0.0001. i, Anti-N IgG. r = −0.87, P < 0.0001. j, Ranked average NT50 at 1.3 months (blue) and 6.2 months (red) for the 87 individuals studied. k, Graph shows NT50 for plasma from all 87 individuals collected at 1.3 and 6.2 months. P < 0.0001. l, Relative change in plasma neutralizing titres between 1.3 and 6.2 months plotted against the corresponding titres at 1.3 months. For a–e, k plotted values and horizontal bars indicate geometric mean. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test in a–d, k, and Friedman with Dunn’s multiple comparison test in e. The r and P values in f–i, l were determined by two-tailed Spearman’s correlations.
We measured plasma neutralizing activity using an HIV-1 virus pseudotyped with the SARS-CoV-2 spike protein1,8. Consistent with other reports9,10, the geometric mean half-maximal neutralizing titre (NT50) in this group of 87 participants was 401 and 78 at 1.3 and 6.2 months, respectively—representing a fivefold decrease (Fig. 1j, k). Neutralizing activity was directly correlated with the IgG anti-RBD ELISA measurements (Extended Data Fig. 2q, r). Moreover, the absolute magnitude of the decrease in neutralizing activity was inversely proportional to and directly correlated with the neutralizing activity at the earlier time point (Fig. 1l). We conclude that antibodies to RBD and plasma neutralizing activity decrease significantly, but remain detectable, 6 months after infection with SARS-CoV-2 in the majority of individuals.
To examine the phenotypic landscape of circulating B cells, we performed high-dimensional flow cytometry on 41 randomly selected individuals at both time points and compared the results to pre-COVID-19 samples from healthy individuals (n = 20). Global high-dimensional mapping with t-distributed stochastic neighbour embedding revealed significant persistent alterations in individuals who had recovered from COVID-19 (Extended Data Fig. 3a). The relative representation of clusters 2, 7, 8 and 10 (corresponding to naive, memory, plasmablast and plasma cells, respectively) was decreased at 1.3 months and remained so at the later time point. Metacluster 15—which corresponds to immature B cells that are recent immigrants from the bone marrow—was increased at the early time point but returned to control levels at the end of the observation period (Extended Data Fig. 3b–d).
SARS-CoV-2 memory B cell repertoire
Whereas plasma cells are the source of circulating antibodies, memory B cells contribute to recall responses. To identify and enumerate the circulating SARS-CoV-2-specific memory B cell compartment, we used flow cytometry to isolate individual B lymphocytes with receptors that bound to RBD1 (Fig. 2a, Extended Data Fig. 4). Notably, the percentage of RBD-binding memory B cells increased marginally between 1.3 and 6.2 months in 21 randomly selected individuals (Fig. 2b).
Fig. 2 |. Sequences of anti-SARS-CoV-2 RBD antibodies.
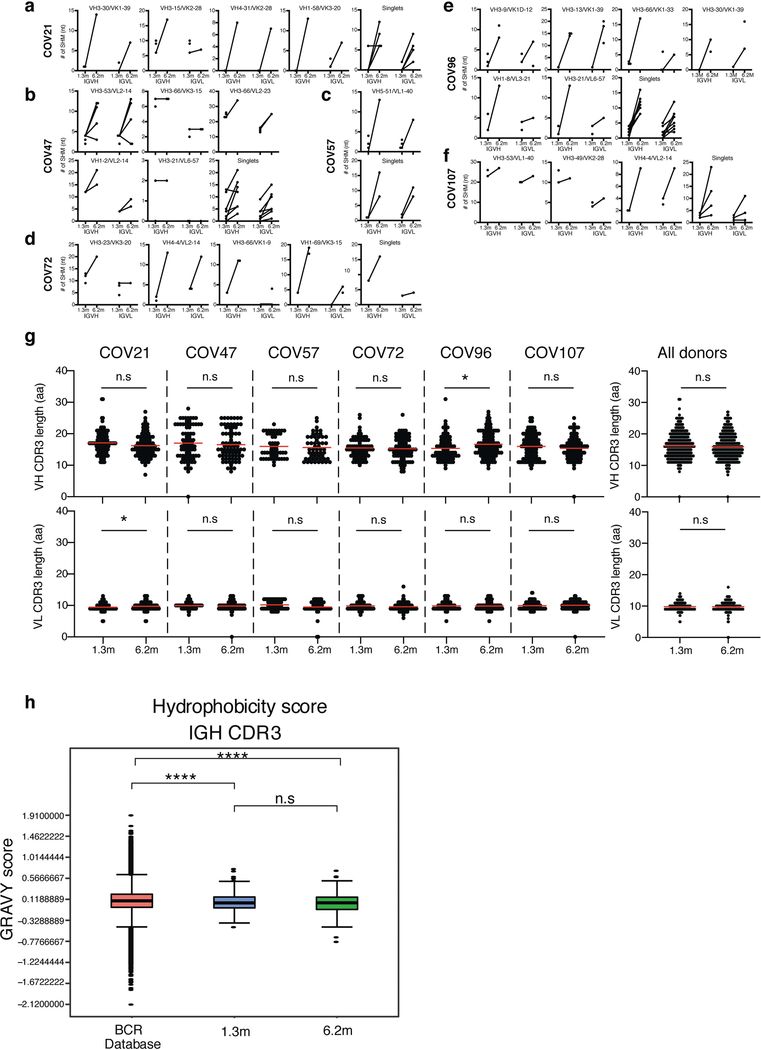
a, Representative flow cytometry plots showing dual AlexaFluor-647–RBD- and PE–RBD-binding B cells for six study individuals (designated COV21, COV47, COV57, COV72, COV96 and COV107) (the gating strategy is shown in Extended Data Fig. 5). The percentage of antigen-specific B cells is indicated. b, As in a. Graph summarizes the percentage of RBD-binding memory B cells in samples obtained at 1.3 and 6.2 months from 21 randomly selected individuals. Red horizontal bars indicate geometric mean values. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. c, Number of somatic nucleotide mutations in the IGVH (top) and IGVL (bottom) genes in antibodies obtained after 1.3 or 6.2 months from the indicated individual (left) or all six donors (right). Statistical significance was determined using two-tailed Mann–Whitney U-tests. Horizontal bars indicate median values. d, Pie charts show the distribution of antibody sequences from six individuals after 1.31 (top) or 6.2 months (bottom). The number in the inner circle indicates the number of sequences analysed for the individual denoted above the circle. Pie-slice size is proportional to the number of clonally related sequences. The black outline indicates the frequency of clonally expanded sequences detected in each participant. Coloured slices indicate persisting clones (same IGHV and IGLV genes and highly similar CDR3) found at both time points in the same participant. Grey slices indicate clones unique to the time point. White slices indicate singlets found at both time points, and the remaining white area indicates sequences that were isolated once. e, Graph shows relative clonality at both time points for all six donors. Red horizontal bars indicate mean values. Statistical significance was determined using two-tailed t-test.
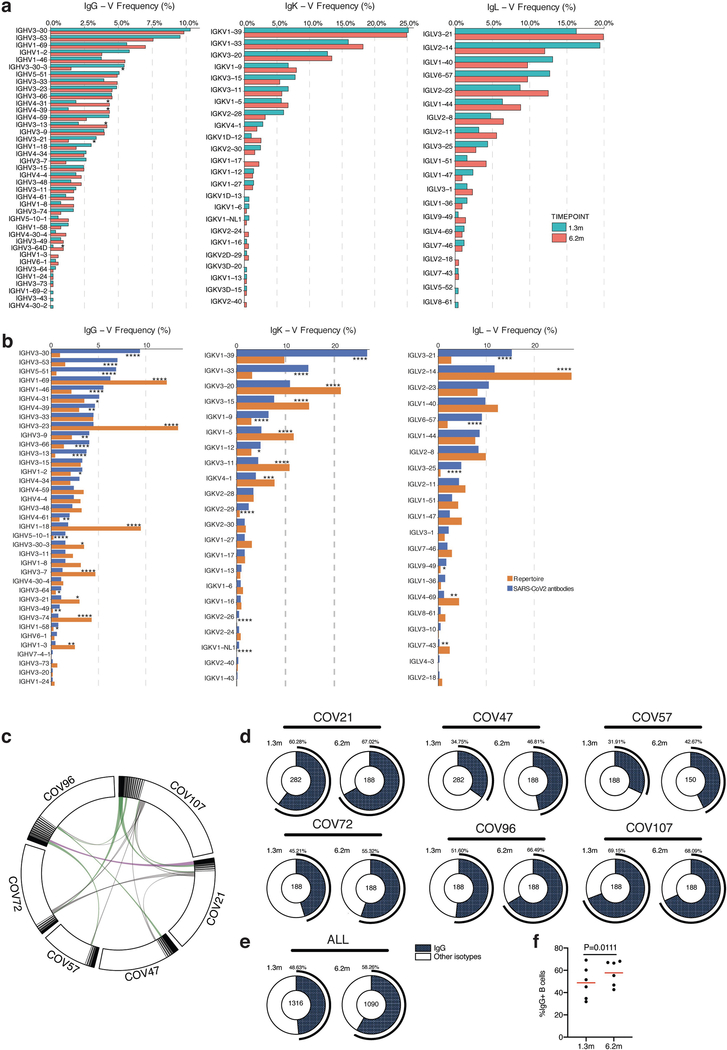
To determine whether there were changes in the antibodies produced by memory B cells after 6.2 months, we obtained 532 paired antibody heavy and light chains from the same 6 individuals who were examined at the earlier time point1 (Supplementary Table 3). There was no significant difference in the representation of IGV genes at the two time points, including the over-representation of the IGHV3–30 and IGHV3–53 gene segments1,11–16 (Extended Data Fig. 5a, b). In keeping with this observation (and similar to the earlier time point), antibodies that shared the same IGHV and IGLV genes comprised 8.6% of all sequences in different individuals (Extended Data Fig. 5c). As might be expected, there was a small—but significant—overall increase in the percentage of IgG-expressing anti-RBD memory cells, from 49% to 58% (P = 0.011) (Extended Data Fig. 5d–f). Consistent with the fractional increase in IgG memory cells, the extent of somatic hypermutation for both IGH and IGL differed significantly in all six individuals between the two time points. Whereas the average number of nucleotide mutations in IGH and IGL was only 4.2 and 2.8, respectively, at the first time point, these values were increased to 11.7 and 6.5, respectively, at the second time point (P < 0.0001) (Fig. 2c, Extended Data Fig. 6a–f). By contrast, the overall average length and hydrophobicity of complementarity-determining region 3 (CDR3) of IGH and IGL were unchanged (Extended Data Fig. 6g, h).
Similar to the earlier time point, we found expanded clones of memory B cells at 6.2 months (including 23 clones that appeared at both time points). However, expanded clones accounted for only 12.4% of all antibody sequences after 6.2 months, compared to 32% after 1.3 months (P = 0.0225) (Fig. 2d, e). In addition, the overall clonal composition of the memory compartment differed at the two time points in all of the individuals we examined (Fig. 2d). Forty-three expanded clones that were present at the earlier time point were not detectable after 6.2 months, and 22 new expanded clones appeared. In addition, the relative distribution of clones that appeared at both time points also varied. For example, the dominant clones in the individuals designated COV21 and COV57—in whom they represented 9.0% and 16.7% of all sequences, respectively, after 1.3 months —were reduced to 1.1% and 1.9%, respectively, of all sequences after 6.2 months (Fig. 2d, Supplementary Table 3). We conclude that, although the magnitude of the RBD-specific memory B cell compartment is conserved between 1.3 and 6.2 months after infection with SARS-CoV-2, there is extensive clonal turnover and antibody sequence evolution that is consistent with prolonged germinal centre reactions.
SARS-CoV-2 monoclonal antibodies
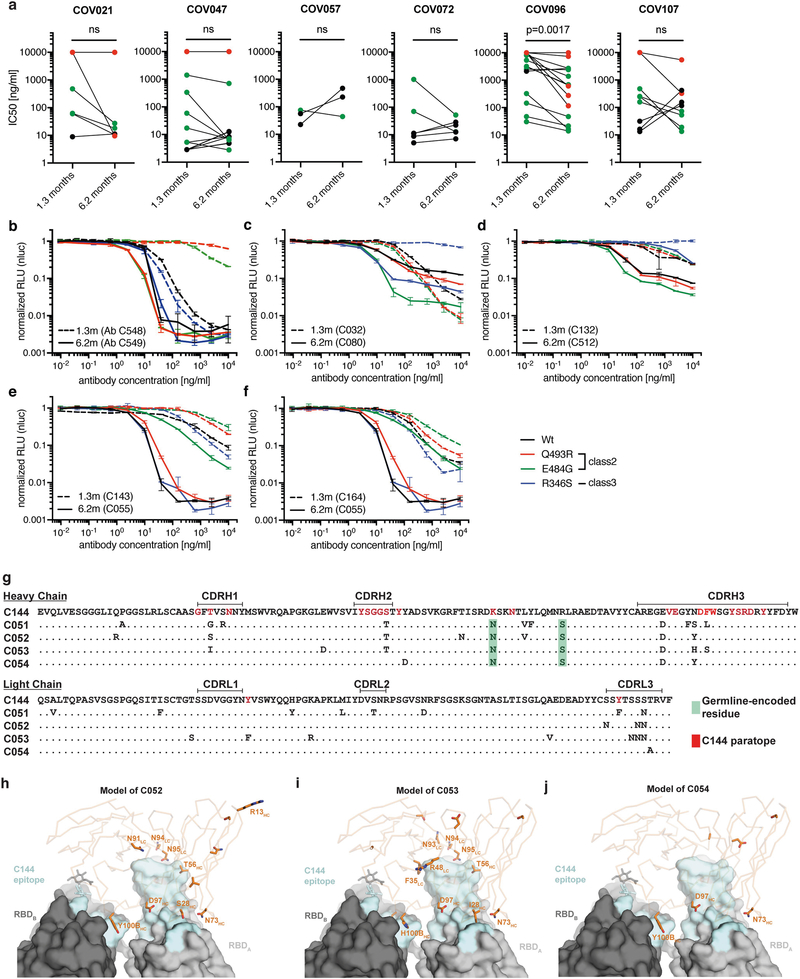
We tested 122 representative antibodies from the 6.2-month time point for reactivity to the RBD (Supplementary Table 4). The antibodies that we evaluated included: (1) 49 antibodies that were randomly selected from those antibodies that appeared only once; (2) 23 antibodies that appeared as singles at both 1.3 and 6.2 months; (3) 23 representatives of newly appearing expanded clones; and (4) 27 representatives of expanded clones appearing at both time points. One hundred and fifteen out of the 122 antibodies bound to RBD, which indicates that flow cytometry efficiently identified B cells that produce anti-RBD antibodies (Fig. 3a, Supplementary Tables 4, 5). Taking all antibodies together, the mean ELISA half-maximal effective concentration (EC50) was not significantly different at the two time points1 (Fig. 3a, Supplementary Table 4). However, comparison of the antibodies that were present at both time points revealed a significant improvement of the EC50 after 6.2 months (P = 0.0227) (Fig. 3b, Extended Data Fig. 7a).
Fig. 3 |. Reactivity of anti-SARS-CoV-2 RBD monoclonal antibodies.
a, Graph shows anti-SARS-CoV-2 RBD antibody reactivity. ELISA EC50 values for all antibodies measured at 1.3 months1, and 122 selected monoclonal antibodies measured at 6.2 months. Horizontal bars indicate geometric mean. Statistical significance was determined using two-tailed Mann–Whitney U-test. b, EC50 values for all 52 antibodies that appear at 1.3 and 6.2 months. Average of two or more experiments. Horizontal bars indicate geometric mean. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. c, Surface representation of the RBD with the ACE2-binding footprint indicated as a dotted line and selected residues found in circulating strains (grey) and residues that mediate resistance to class-2 (red) (C144) and −3 (green) (C135) antibodies highlighted as sticks. d, Graphs show ELISA binding curves for C144 (black dashed line) and its clonal relatives obtained after 6.2 months (C050, C051, C052, C053 and C054) (solid lines) binding to wild-type (WT), Q493R, R346S and E484K RBDs. e. Heat map shows log2-transformed relative fold change in EC50 against indicated RBD mutants for 26 antibody clonal pairs obtained at 1.3 and 6.2 months with the most pronounced changes in reactivity. The participant of origin for each antibody pair is indicated above. All experiments were performed at least twice.
To determine whether the antibodies expressed by memory B cells at the late time point also showed altered breadth, we compared them to earlier clonal relatives in binding assays using control and mutant RBDs. The substitutions E484K and Q493R17 were selected for resistance to class-2 antibodies (such as C144 and C121) that bind directly to the ACE2-interaction ridge in the RBD1,18–20, and R346S, N439K, and N440K were selected for resistance to class-3 antibodies (such as C135) that do not directly interfere with ACE2 binding1,17–20 (Fig. 3c). In addition, we also tested the V367F, A475V, S477N and V483A mutants of the RBD, which represent circulating variants that confer complete or partial resistance to class-1 and −2 antibodies17,18,21 (Fig. 3c). Out of 52 antibody clonal pairs that appeared at both time points, 43 (83%) showed overall increased binding to mutant RBDs at the 6.2-month time point (Extended Data Fig. 7b–k, Supplementary Table 5). For example, C144 (an antibody recovered at the 1.3-month time point) was unable to bind to RBD(Q493R) or RBD(E484K), but all five of its clonal derivatives collected at 6.2 months bound to RBD(Q493R) and one also showed binding to RBD(E484K) (Fig. 3d). Overall, the most pronounced increase in binding occurred for mutations affecting the RBD in amino acid positions such as E484, Q493, N439, N440 and R346, which are critical for the binding of class-2 and −3 antibodies17,18 (Fig. 3e, Extended Data Fig. 7b–k, Supplementary Table 5).
Next, we tested all 122 antibodies from the 6.2-month time point for activity in a pseudotyped SARS-CoV-2 neutralization assay1,8 (Fig. 4a, Supplementary Table 6). Consistent with RBD-binding assays, the mean neutralization half-maximal inhibitory concentrations (IC50) values were not significantly different at the two time points when all antibodies were compared1 (Fig. 4a). However, comparison of the antibodies that were present at both time points revealed a significant improvement of the IC50 values at 6.2 months (P = 0.0003) (Fig. 4b, Extended Data Fig. 8a).
Fig. 4 |. Neutralizing activity of anti-SARS-CoV-2 RBD monoclonal antibodies.
a, SARS-CoV-2 pseudovirus neutralization assay. IC50 values for all antibodies measured at 1.3 months1, and 122 selected antibodies measured at 6.2 months. Antibodies with IC50 values above 1 μg ml−1 were plotted at 1 μg ml−1. Mean of two independent experiments. Red bar indicates geometric mean. Statistical significance was determined using two-tailed Mann–Whitney U-test. b, IC50 values for 52 antibodies appearing at 1.3 and 6.2 months. Red bar indicates geometric mean. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. c, IC50 values for 5 pairs of monoclonal antibody clonal relatives obtained after 1.3 or 6.2 months for neutralization of wild-type and mutant SARS-CoV-2 pseudovirus. Antibody identifiers of the 1.3-month–6.2-month monoclonal antibody pairs as indicated. Bold styling denotes antibody pairs with substantial increase in neutralizing activity after 6.2 months. d, Graph shows the normalized relative luminescence units (RLU) for cell lysates of 293T cells expressing ACE2, 48 h after infection with SARS-CoV-2 pseudovirus containing wild-type RBD or one of three mutant RBDs (Q493R, E484G and R346S) in the presence of increasing concentrations of one of two monoclonal antibodies C144 (1.3 months) (dashed lines) or C051 (6.2 months) (solid lines). Experiments were performed at least twice. e, C051 binding model. Surface representation of two adjacent ‘down’ RBDs (RBDA and RBDB) on a spike trimer, with the C144 epitope on the RBDs highlighted in cyan and positions of amino acid mutations that accumulated in C051 compared to the parent antibody C144 highlighted as stick side chains on a Cα atom representation of C051 VHVL binding to adjacent RBDs. The C051 interaction with two RBDs was modelled on the basis of a cryo-electron microscopy structure of C144 Fab bound to spike trimer18. HC, heavy chain; LC, light chain.
To determine whether the antibodies exhibiting altered RBD binding also show increased neutralizing breadth, we tested five representative antibody pairs recovered at the two time points against HIV-1 viruses pseudotyped with E484G, Q493R, and R346S mutant spike proteins (Fig. 4c, Supplementary Table 6). Notably, the Q493R and E484G pseudotyped viruses were resistant to neutralization by C144; by contrast, C051 (a 6.2-month clonal derivative of C144) neutralized both variants, with IC50 values of 4.7 and 3.1 ng ml−1, respectively (Fig. 4c, d). Similarly, R346S pseudotyped viruses were resistant to C032, but C080 (a 6.2-month clonal derivative of C032) neutralized this variant with an IC50 of 5.3 ng ml−1 (Fig. 4c, Extended Data Fig. 8b–f). Consistent with the observed changes in binding and neutralizing activity, several late-appearing antibodies (for example, C051) had acquired mutations directly in or adjacent to the RBD-binding paratope (Fig. 4e, Extended Data Fig. 8g–j). We conclude that memory B cells that evolved during the observation period express antibodies with increased neutralizing potency and breadth.
SARS-CoV-2 antigen persistence
Antibody evolution occurs by somatic mutation and selection in germinal centres in which antigen can be retained in the form of immune complexes on the surface of follicular dendritic cells for prolonged periods of time. Residual protein in tissues represents another potential source of antigen. SARS-CoV-2 replicates in ACE2-expressing cells in the lungs, nasopharynx and small intestine22–25, and viral RNA has been detected in stool samples even after the virus is cleared from the nasopharynx26–28.
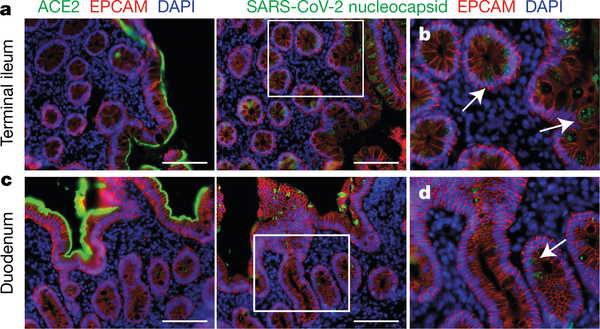
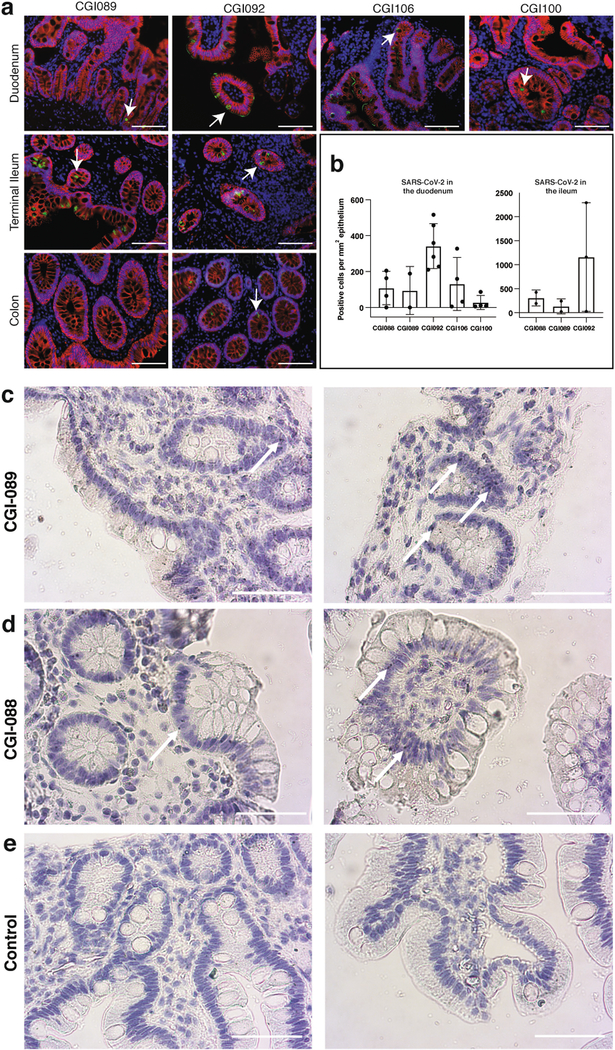
To determine whether there might be antigen persistence in the intestine after resolution of clinical illness, we obtained biopsies from the upper and lower gastrointestinal tract of 14 individuals at an average of 4 months (range of 2.8 to 5.7 months) after initial COVID-19 diagnosis (Supplementary Table 7). Immunostaining was performed to determine whether viral protein was also detectable in upper and lower gastrointestinal tract, with de-identified biopsies from individuals pre-dating the pandemic (n = 10) serving as controls. ACE2 and SARS-CoV-2 N protein was detected in intestinal enterocytes in 5 of 14 individuals (Fig. 5a–d, Extended Data Figs. 9a–h, 10a, b, Supplementary Table 7) but not in control samples (Extended Data Fig. 9i–l). When detected, immunostaining was sporadic, patchy, exclusive to the intestinal epithelium and not associated with inflammatory infiltrates (Extended Data Figs. 9a–h, 10a, b). Clinically approved nasopharyngeal-swab PCR assays were negative in all 14 individuals at the time of biopsy. However, biopsy samples from 3 of the 14 participants produced PCR amplicons that were sequence-verified as SARS-CoV-2 (Methods, Supplementary Table 7). In addition, viral RNA was detected by in situ hybridization in biopsy samples from the two participants who were tested (Extended Data Fig. 10c, d) but not in control samples (Extended Data Fig. 10e). Sampling variability during the endoscopic procedure probably contributed to incomplete concordance between detection of viral RNA and protein assays.
Fig. 5 |. Immunofluorescence imaging of intestinal biopsies.
a, Immunofluorescence images of human enterocytes stained for EPCAM (red), DAPI (blue) and either ACE2 (green in a, c) or SARS-CoV-2 N (green in b, d) in intestinal biopsies taken 92 d after onset of COVID-19 symptoms in participant CGI-088, in the terminal ileum (a, b) or duodenum (c, d). Regions in white boxes in the right panels of a, c are shown expanded in b, d, respectively. Arrows indicate enterocytes with detectable SARS-CoV-2 antigen. Scale bars, 100 μm. The experiments were repeated independently at least twice with similar results.
Neutralizing antibodies to SARS-CoV-2 develop in most individuals after infection but decay with time7,9,10,29–32. These antibodies are effective in prevention and therapy in animal models and are likely to have a role in protection from reinfection in humans2. Although there is a significant decrease in plasma neutralizing activity between 1.3 and 6.2 months, antibody titres remain measurable in most individu-als9,10,29–36.
Neutralizing monoclonal antibodies obtained from individuals during the early convalescence period showed notably low levels of somatic mutations, which some previous reports have attributed to defects in germinal centre formation11,12,14,37–40. Our data indicate that the anti-SARS-CoV-2 memory B cell response evolves during the first six months after infection, with accumulation of immunoglobulin somatic mutations, and production of antibodies with increased neutralizing breadth and potency. Persistent antibody evolution occurs in germinal centres and requires that B cells are exposed to antigen trapped in the form of immune complexes on follicular dendritic cells41. This form of antigen can be long-lived, because follicular dendritic cells do not internalize immune complexes. In addition, even small amounts of persistent viral antigen could fuel antibody evolution. The observation that SARS-CoV-2 mRNA and protein remains detectable in the small intestinal epithelium in some individuals at months after infection is consistent with the relative persistence of anti-RBD IgA antibodies and continued antibody evolution34–36.
Memory responses are responsible for protection from reinfection and are essential for effective vaccination. The observation that memory B cell responses do not decay after 6.2 months34,35,42, but instead continue to evolve, is strongly suggestive that individuals who are infected with SARS-CoV-2 could mount a rapid and effective response to the virus upon re-exposure.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03207-w.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Study participants
Previously enrolled study participants1 were asked to return for a 6-month follow-up visit at the Rockefeller University Hospital from 31 August through to 16 October 2020. Eligible participants were adults aged 18–76 years and were either diagnosed with SARS-CoV-2 infection by RT–PCR (cases) or were close contacts (for example, members of the same household, coworkers or members of same religious community) of someone who had been diagnosed with SARS-CoV-2 infection by RT–PCR (contacts). Close contacts without seroconversion against SARS-CoV-2 as assessed by serological assays (described in ‘High-throughput automated serology assays’) were not included in the subsequent analysis. Most study participants were residents of the greater New York city tri-state region and were asked to return approximately six months after the time of onset of COVID-19 symptoms. Participants presented to the Rockefeller University Hospital for blood sample collection and were asked to recall the symptoms and severity of clinical presentation during the acute (first six weeks) and the convalescent (seven weeks until second study visit) phase of COVID-19, respectively. The severity of acute infection was assessed by the World Health Organization (WHO) ‘Ordinal Clinical Progression/Improvement Scale’ (https://www.who.int/publications/i/item/covid-19-therapeutic-trial-synopsis). Shortness of breath was assessed through the modified Medical Research Council dyspnoea scale43. Participants who presented with persistent symptoms attributable to COVID-19 were identified on the basis of chronic shortness of breath or fatigue, deficit in athletic ability and/or three or more additional long-term symptoms such as persistent unexplained fevers, chest pain, new-onset cardiac sequalae, arthralgias, impairment of concentration or mental acuity, impairment of sense of smell or taste, neuropathy or cutaneous findings3,4. All participants at Rockefeller University provided written informed consent before participation in the study and the study was conducted in accordance with good clinical practice. Clinical data collection and management were carried out using the software iRIS by iMedRIS. The study was performed in compliance with all relevant ethical regulations and the protocol for studies with human participants was approved by the Institutional Review Board (IRB) of the Rockefeller University.
Gastrointestinal biopsy cohort
To determine whether SARS-CoV-2 can persist in the gastrointestinal tract, we recruited a cohort of 14 individuals with prior diagnosis of and recovery from COVID-19 illness. Eligible participants included adults, 18–76 years of age who were previously diagnosed with SARS-CoV-2 by RT–PCR or through a combination of clinical symptoms consistent with COVID-19 plus evidence of seroconversion, and presented to the gastroenterology clinics of Mount Sinai Hospital. Endoscopic procedures were performed for clinically indicated conditions as detailed in Supplementary Table 7. All participants were asymptomatic at the time of the endoscopic procedures and negative for SARS-CoV-2 by nasal-swab PCR (cycle threshold (Ct) cut-off < 38).
The CLIA-certified laboratory of the Mount Sinai Health System validated the laboratory-developed nasopharyngeal-swab real-time RT–PCR test according to the New York State Department of Health Wadsworth Center validation procedure for SARS-CoV-244. Informed consent was obtained from all participants. The biopsy-related studies were approved by the Mount Sinai Ethics Committee/IRB (IRB 16–0583, ‘The impact of viral infections and their treatment on gastrointestinal immune cells’).
SARS-CoV-2 saliva PCR test
The SARS-CoV-2 PCR method for saliva samples was developed and its performance characteristics determined by the Rockefeller University Clinical Genomics Laboratory. This laboratory-developed test has been authorized by New York state under an emergency use authorization for use by authorized laboratories. Saliva was collected into guanidine thiocyanate buffer as previously described45. RNA was extracted using either a column-based (Qiagen QIAmp DSP Viral RNA Mini Kit, 61904) or a magnetic-bead-based method as previously described46. Reverse-transcribed cDNA was amplified using primers and probes validated by the Centers for Disease Control and Prevention or by Columbia University Personalized Medicine Genomics Laboratory, respectively, and approved by the US Food and Drug Administration under the emergency use authorization. Viral RNA was considered detected if the Ct for two viral primer and probe combinations was < 40.
Blood sample processing and storage
Peripheral blood mononuclear cells were obtained by gradient centrifugation and stored in liquid nitrogen in the presence of FCS and DMSO. Heparinized plasma and serum samples were aliquoted and stored at −20 °C or below. Before experiments, aliquots of plasma samples were heat-inactivated (56 °C for 1 h) and then stored at 4 °C.
High-throughput automated serology assays
Plasma samples from 80 out of 87 participants were tested by high-throughput automated serology assays. The Roche Elecsys anti-SARS-CoV-2 assay was performed on Roche Cobas e411 (Roche Diagnostics). The Elecsys anti-SARS-CoV-2 assay uses a recombinant protein representing the N antigen for the determination of antibodies against SARS-CoV-2. This assay received emergency use authorization approval from the US Food and Drug Administration6. The Pylon COVID19 IgG and IgM assays were used to measure plasma IgG and IgM antibodies against SARS-CoV-2, respectively. Plasma samples were assayed on the Pylon 3D analyser (ET HealthCare) as previously described5. This assay was implemented clinically as a laboratory-developed test under New York State Department of Health regulations. In brief, the assay was performed using a unitized test strip containing wells with predispensed reagents. The COVID-19 reagent contains biotinylated recombinant versions of the SARS-CoV-2 S-protein RBD and trace amounts of N protein as antigens that bind IgG and IgM, respectively. The cut-off values for both Pylon assays were determined using the mean of non-COVID-19 samples plus 6 s.d. The results of a sample are reported in the form of a cut-off index or an index value, which were determined by the instrument readout of the test sample divided by instrument readout at cut-off.
ELISAs
ELISAs47,48 to evaluate antibodies binding to SARS-CoV-2 N (Sino Biological 40588-V08B), RBD and additional RBDs were performed by coating of high-binding 96-half-well plates (Corning 3690) with 50 μl per well of a 1 μg ml−1 protein solution in phosphate-buffered saline (PBS) overnight at 4 °C. Plates were washed 6 times with washing buffer (1× PBS with 0.05% Tween-20 (Sigma-Aldrich)) and incubated with 170 μl per well blocking buffer (1× PBS with 2% BSA and 0.05% Tween-20 (Sigma)) for 1 h at room temperature. Immediately after blocking, monoclonal antibodies or plasma samples were added in PBS and incubated for 1 h at room temperature. Plasma samples were assayed at a 1:67 starting dilution and 7 additional threefold serial dilutions. Monoclonal antibodies were tested at 10 μg ml−1 starting concentration and 10 additional fourfold serial dilutions. Plates were washed 6 times with washing buffer and then incubated with anti-human IgG, IgM or IgA secondary antibody conjugated to horseradish peroxidase (HRP) (Jackson Immuno Research 109–036-088 109–035-129 and Sigma A0295) in blocking buffer at a 1:5,000 dilution (IgM and IgG) or 1:3,000 dilution (IgA). Plates were developed by addition of the HRP substrate, TMB (ThermoFisher) for 10 min (plasma samples) or 4 min (monoclonal antibodies), then the developing reaction was stopped by adding 50 μl 1 M H2SO4 and absorbance was measured at 450 nm with an ELISA microplate reader (FluoStar Omega, BMG Labtech) with Omega and Omega MARS software for analysis. For plasma samples, a positive control (plasma from participant COV72, diluted 66.6-fold and 7 additional threefold serial dilutions in PBS) was added to every assay plate for validation. The average of its signal was used for normalization of all of the other values on the same plate with Excel software before calculating the AUC using Prism v.8.4 (GraphPad). For monoclonal antibodies, the EC50 was determined using four-parameter nonlinear regression (GraphPad Prism v.8.4).
Expression of RBD proteins
Mammalian expression vectors encoding the RBDs of SARS-CoV-2 (GenBank MN985325.1; S protein residues 319–539) and eight additional mutant RBD proteins (E484K, Q493R, R346S, N493K, N440K, V367F, A475V, S477N and V483A) with an N-terminal human IL-2 or Mu phosphatase signal peptide were previously described49.
SARS-CoV-2 pseudotyped reporter virus
SARS-CoV-2 pseudotyped particles were generated as previously described1,8. In brief, 293T cells were transfected with pNL 4–3ΔEnv-nanoluc and pSARS-CoV-2-SΔ19. For generation of RBD-mutant pseudoviruses, pSARS-CoV-2-SΔ19 carrying either of the following spike mutations was used instead of its wild-type counterpart: Q493R, R346S or E484G50. Particles were collected at 48 h after transfection, filtered and stored at −80 °C.
Pseudotyped virus neutralization assay
Fourfold serially diluted plasma from individuals convalescent from COVID-19, or monoclonal antibodies, were incubated with SARS-CoV-2 pseudotyped virus for 1 h at 37 °C. The mixture was subsequently incubated with 293TACE2 cells for 48 h, after which cells were washed with PBS and lysed with Luciferase Cell Culture Lysis 5× reagent (Promega). Nanoluc luciferase activity in lysates was measured using the Nano- Glo Luciferase Assay System (Promega) with the Glomax Navigator (Promega). The obtained relative luminescence units were normalized to those derived from cells infected with SARS-CoV-2 pseudotyped virus in the absence of plasma or monoclonal antibodies. The half-maximal inhibitory concentration for plasma (NT50) or monoclonal antibodies (IC50) was determined using four-parameter nonlinear regression (least squares regression method without weighting; constraints: top = 1, bottom = 0) (GraphPad Prism).
High-dimensional data analysis of flow cytometry data
High-dimensional viSNE and FlowSOM data analysis and visualization of flow cytometry data were performed on B cells using the Cytobank platform (https://cytobank.org). viSNE analysis was performed using equal sampling of 4,893 cells from each FCS file, with 75,00 iterations, a perplexity of 30 and a theta of 0.5. The following markers were used to generate viSNE maps: IgA, CD305, TGFb-RII, CD138, CD10, CD272, IgD, CD24, CD21, CD95, HLA-DR, IgG, CD279, CD38, IgM, CD274, CD27, CD23, CXCR5, CD32, CD86, CD40, CD85j, CD11c and CXCR3. Resulting viSNE maps were fed into the FlowSOM clustering algorithm51. The self-organizing map was generated using hierarchical consensus clustering on the t-distributed stochastic neighbour embedding axes.
Heat map visualization
Heat maps to display column-scaled z-scores of mean fluorescence intensity for individual FlowSOM clusters according to marker expression were created using the R function pheatmap.
Biotinylation of viral protein for use in flow cytometry
Purified and Avi-tagged SARS-CoV-2 RBD was biotinylated using the Biotin-Protein Ligase-BIRA kit according to manufacturer’s instructions (Avidity) as previously described1. Ovalbumin (Sigma, A5503–1G) was biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation kit according to the manufacturer’s instructions (Thermo Scientific). Biotinylated ovalbumin was conjugated to streptavidin–BV711 (BD biosciences, 563262) and RBD to streptavidin–PE (BD Biosciences, 554061) and streptavidin–AF647 (Biolegend, 405237)1.
Single-cell sorting by flow cytometry
Single-cell sorting by flow cytometry was previously described1. In brief, peripheral blood mononuclear cells were enriched for B cells by negative selection using a pan-B-cell isolation kit according to the manufacturer’s instructions (Miltenyi Biotec, 130–101-638). The enriched B cells were incubated in FACS buffer (1× PBS, 2% FCS, 1 mM EDTA) with the following anti-human antibodies (all at 1:200 dilution): anti-CD20–PECy7 (BD Biosciences, 335793), anti-CD3–APC–eFluro 780 (Invitrogen, 47–0037-41), anti-CD8–APC–eFluor 780 (Invitrogen, 47–0086-42), anti-CD16–APC–eFluor 780 (Invitrogen, 47–0168-41), anti-CD14–APC–eFluor 780 (Invitrogen, 47–0149-42), as well as Zombie NIR (BioLegend, 423105) and fluorophore-labelled RBD and ovalbumin (Ova) for 30 min on ice. Single CD3−CD8−CD14−CD16−CD20+Ova−RBD– PE+RBD–AF647+ B cells were sorted into individual wells of 96-well plates containing 4 μl of lysis buffer (0.5× PBS, 10 mM DTT, 3,000 units per ml RNasin ribonuclease inhibitors (Promega, N2615)) per well using a FACS Aria III and FACSDiva software (Becton Dickinson) for acquisition and FlowJo for analysis. The sorted cells were frozen on dry ice, and then stored at −80 °C or immediately used for subsequent RNA reverse transcription.
Antibody sequencing, cloning and expression
Antibodies were identified and sequenced as previously described1. In brief, RNA from single cells was reverse-transcribed (SuperScript III Reverse Transcriptase, Invitrogen, 18080–044) and the cDNA stored at −20 °C or used for subsequent amplification of the variable IGH, IGL and IGK genes by nested PCR and Sanger sequencing. Sequence analysis was performed using MacVector. Amplicons from the first PCR reaction were used as templates for sequence- and ligation-independent cloning into antibody expression vectors. Recombinant monoclonal antibodies and Fabs were produced and purified as previously described1.
Computational analyses of antibody sequences
Antibody sequences were trimmed on the basis of quality and annotated using Igblastn v.1.14. with IMGT domain delineation system. Annotation was performed systematically using Change-O toolkit v.0.4.54052. Heavy and light chains derived from the same cell were paired, and clonotypes were assigned on the basis of their V and J genes using in-house R and Perl scripts (Extended Data Fig. 5). All scripts and the data used to process antibody sequences are publicly available on GitHub (https://github.com/stratust/igpipeline).
The frequency distributions of human V genes in anti-SARS-CoV-2 antibodies from this study were compared to 131,284,220 previously generated IgH and IgL sequences53 and downloaded from cAb-Rep54 (a database of human shared BCR clonotypes available at https://cab-rep.c2b2.columbia.edu/). On the basis of the 82 distinct V genes that make up the 1,703 analysed sequences from immunoglobulin repertoire of the three participants present in this study, we selected the IgH and IgL sequences from the database that are partially coded by the same V genes and counted them according to the constant region. The frequencies shown in (Extended Data Fig. 5) are relative to the source and isotype analysed. We used the two-sided binomial test to check whether the number of sequences belonging to a specific IGHV or IGLV gene in the repertoire is different according to the frequency of the same IGV gene in the database. Adjusted P values were calculated using the false-discovery rate correction. In Extended Data Figs. 5, 6, significant differences are denoted with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Nucleotide somatic hypermutation and CDR3 length were determined using in-house R and Perl scripts. For somatic hypermutations, IGHV and IGLV nucleotide sequences were aligned against their closest germlines using Igblastn and the number of differences were considered nucleotide mutations. The average mutations for V genes was calculated by dividing the sum of all nucleotide mutations across all participants by the number of sequences used for the analysis. To calculate the GRAVY scores of hydrophobicity55 we used the Guy H. R. Hydrophobicity scale based on free energy of transfer (kcal per mole)56 implemented by the R package Peptides (https://CRAN.R-project.org/package=Peptides). We used 532 heavy-chain CDR3 amino acid sequences from this study and 22,654,256 IGH CDR3 sequences from the public database of memory B cell receptor sequences57. The Shapiro–Wilk test was used to determine whether the GRAVY scores were normally distributed. The GRAVY scores from all 532 IGH CDR3 amino acid sequences from this study were used to perform the test and 5,000 GRAVY scores of the sequences from the public database were randomly selected. The Shapiro–Wilk P values were 6.896 × 10−3 and 2.217 × 10−6 for sequences from this study and the public database, respectively, indicating that the data were not normally distributed. Therefore, we used the two-tailed Wilcoxon nonparametric test to compare the samples, which indicated a difference in hydrophobicity distribution (P = 5 × 10−6) (Extended Data Fig. 6h).
A heat map of log2-transformed relative fold change in EC50 against the indicated RBD mutants for antibody clonal pairs obtained at 1.3 and 6.2 months (Fig. 3e, Extended Data Fig. 7k) was created with R pheatmap package (https://github.com/raivokolde/pheatmap) using Euclidean distance and Ward.2 clustering method.
Biopsies and immunofluorescence
Endoscopically obtained mucosal biopsies were formalin-fixed and paraffin-embedded. Sections (5 μm) were cut, dewaxed in xylene, and rehydrated in graded alcohol and PBS. Heat-induced epitope retrieval was performed in target retrieval solution (DAKO, S1699) using a commercial pressure cooker. Slides were then cooled to room temperature, washed in PBS and permeabilized for 30 min in 0.1% Triton X-100 in PBS. Nonspecific binding was blocked with 10% goat serum (Invitrogen, 50062Z) for 1 h at room temperature. Sections were then incubated with a combination of primary antibodies diluted in blocking solution overnight at 4 °C. Slides were washed 3 times in PBS and then incubated in secondary antibody and DAPI (1 μg ml−1) for 1 h at room temperature. Sections were washed in PBS three times and then mounted with Fluoromount-G (Electron Microscopy Sciences, 1798425). Controls included omitting primary antibody (no primary 995 control) or substituting primary antibodies with nonreactive antibodies of the same isotype (isotype control). A Nikon Eclipse Ni microscope and digital SLR camera (Nikon, DS-Qi2) was used to visualize and image the tissue.
The antibody used to stain sections for N protein was raised in rabbits against SARS-CoV N and is cross-reactive with SARS-CoV-2 N protein58 (Supplementary Table 8).
SARS-CoV-2 PCR from intestinal biopsies
To determine whether SARS-CoV-2 RNA is present in the gastrointestinal tract, we isolated RNA from endoscopically obtained mucosal biopsies using Direct-zol miniprep kit (Zymo research, R2050). Reverse-transcribed cDNA was amplified using 2019-nCov Ruo Kit (IDT) to detect viral nucleocapsid genomic RNA. Amplification of subgenomic nucleocapsid RNA was done using following primers and probe: sgLeadSARSCov2_F 5′-CGATCTCTTGTAGATCTGTTCTC-3′28, wtN_R4 5′-GGTGAACCAAGACGCAGTAT-3′, wtN_P4 5′-/56-FAM/TAACCAGAA/ ZEN/TGGAGAACGCAGTGGG/3IABkFQ/−3′.
Quantitative PCR was performed using QuantTect probe PCR kit (Qiagen, 204345) under following conditions: 95 °C 15 s, 95 °C 15 s and 60 °C 1 min using the Applied Biosystem QuantStudio 6 Flex Real-Time PCR System. Viral RNA was considered detected if the Ct for viral primer and probe combinations was < 40. Samples from positive wells were column-purified and presence of N1 sequences additionally verified by Sanger sequencing.
SARS-CoV-2 RNA detection by probe proximity ligation
Probes were designed with a 20–25 nucleotide homology to SARS-CoV-2 genomic RNA. Probes were assessed by NCBI BLAST to exclude off target binding to other cellular transcripts. IDT OligoAnalyzer (Integrated DNA Technologies) was used to identify probe pairs with similar thermodynamic properties; melting temperature 45–60 °C, GC content of 40–55% and low self-complementary. The 3′ end of each one of the probes used for proximity ligation signal amplification is designed with a partially complementary sequence to the 61-bp-long backbone and partially to the 21-bp insert (Supplementary Table 8). Single-molecule fluorescence in situ hybridization (smFISH) probes were designed with a complementary 3′ end to the biotin detection probe (Supplementary Table 8).
Paraffin-embedded samples were sectioned at 10 μm. Sections were deparaffinized using 100% xylene, 5 min at room temperature, repeated twice. Slides were rinsed in 100% ethanol, 1 min at room temperature, twice, and air-dried. Endogenous peroxidase activity was eliminated by treating the samples with 0.3% hydrogen peroxide, 10 min at room temperature followed by washing with DEPC-treated water. Samples were incubated 15 min at 95–100 °C in antigen retrieval solution (ACDBio) rinsed in DEPC-treated water and dehydrated in 100% ethanol, 3 min at room temperature and air-dried. Tissue sections were permeabilized 30 min at 40 °C using RNAscope protease plus solution (ACDBio) and rinsed in DEPC-treated water.
Hybridization was performed overnight at 40 °C in a buffer based on DEPC-treated water containing 2× SSC, 20% formamide (Thermo Fischer Scientific), 2.5% (v/v) polyvinylsulfonic acid, 20 mM ribonucleoside vanadyl complex (New England Biolabs), 40 U ml−1 RNasin (Promega), 0.1% (v/v) Tween 20 (Sigma Aldrich), 100 μg ml−1 salmon sperm DNA (Thermo Fisher Scientific), 100 μg ml−1 yeast RNA (Thermo Fisher Scientific). DNA probes dissolved in DEPC-treated water were added at a final concentration of 100 nM (Integrated DNA Technologies). Samples were washed briefly and incubated in a buffer containing 2× SSC, 20% formamide, 40 U ml−1 RNasin at 40 °C and then washed four times (5 min each) in wash buffer, PBS, 0.1% (v/v) Tween 20, and 4 U ml−1 RNasin (Promega). Slides were then incubated with 100 nM insert and backbone oligonucleotides in PBS, 1× SSC, 0.1% (v/v) Tween 20, 100 μg ml−1 salmon sperm DNA (Thermo Fisher Scientific), 100 μg ml−1 yeast RNA (Thermo Fisher Scientific), 40 U ml−1 RNasin at 37 °C. After four washes, tissues were incubated at 37 °C with 0.1 U μl−1 T4 DNA ligase (New England Biolabs) in 50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 250 μg ml−1 BSA, 0.05% Tween 20, 40 U ml−1 RNasin, followed by incubation with 0.1 U μl−1 phi29 DNA polymerase in 50 mM Tris–HCl, 10 mM MgCl2, 10 mM (NH4)2SO4, 250 μM dNTPs, 1 mM DTT, 0.05% Tween 20, 40 U ml−1 RNasin pH 7.5 at 30 °C. Slides were washed and endogenous biotin was blocked using Avidin/Biotin blocking kit (Vector Laboratories) according to the manufacturer’s instructions. Rolling cycle amplicons were identified using a biotin-labelled DNA probe at a concentration of 5 nM at 37 °C in PBS, 1× SSC, 0.1% Tween 20, 100 μg ml−1 salmon sperm DNA, 100 μg ml−1 yeast RNA. After washing, samples were incubated with 1:100 diluted streptavidin–HRP (Thermo Fisher Scientific) in PBS, 60 min at room temperature followed by washing. Fluorescent labelling was accomplished using Alexa Fluor 647 Tyramide SuperBoostKit (Thermo Fischer Scientific) according to the manufacturer’s instructions. Hoechst 33342 was used for nuclear counterstaining (Thermo Fischer Scientific) and samples were mounted in ProLong gold antifade (Thermo Fischer Scientific).
SARS-CoV-2 RNA detection by smFISH
Hybridization was performed overnight at 40 °C in a buffer based on DEPC-treated water containing 2× SSC, 20% formamide (Thermo Fischer Scientific), 2.5% (v/v) polyvinylsulfonic acid, 20 mM ribonucleoside vanadyl complex (New England Biolabs), 40 U ml−1 RNasin (Promega), 0.1% (v/v) Tween 20 (Sigma Aldrich), 100 μg ml−1 salmon sperm DNA (Thermo Fisher Scientific), 100 μg ml−1 yeast RNA (Thermo Fisher Scientific). DNA probes dissolved in DEPC-treated water were added at a final concentration of 10 nM (Integrated DNA Technologies). Samples were washed briefly and incubated in a buffer containing 2× SSC, 20% formamide, 40 U ml−1 RNasin at 40 °C and then washed four times in wash buffer, PBS, 0.1% (v/v) Tween 20, and 4 U ml−1 RNasin (Promega). Samples were washed and endogenous biotin was blocked using Avidin/Biotin blocking kit (Vector Laboratories) according to the manufacturer’s instructions. Slides were incubated with a biotin-labelled DNA probe at a concentration of 10 nM at 37 °C in PBS, 1× SSC, 0.1% Tween 20, 100 μg ml−1 salmon sperm DNA, 100 μg ml−1 yeast RNA. After washing, samples were incubated with 1:100 diluted streptavidin–HRP (Thermo Fisher Scientific) in PBS, 60 min at room temperature followed by washing. Samples were labelled using ImmPACT-DAB substrate, counterstained using haematoxylin QS and imbedded in VectaMount AQ mounting medium (Vector Laboratories) according to the manufacturer’s instructions.
Extended Data
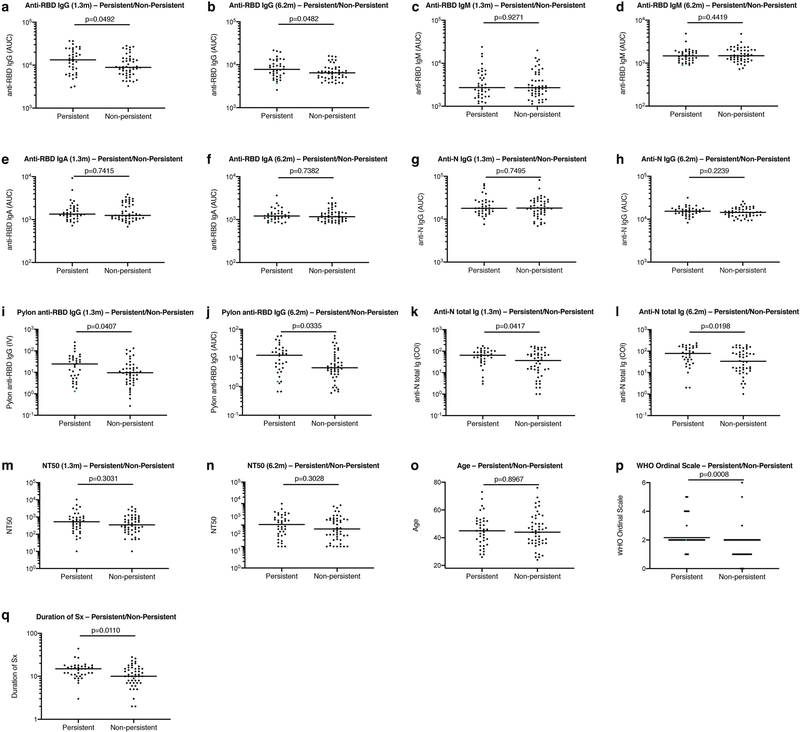
Extended Data Fig. 1 |. Clinical correlates of plasma antibody titres.
a, Normalized AUC anti-RBD IgG titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. b, Normalized AUC anti-RBD IgG titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. c, Normalized AUC anti-RBD IgM titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. d, Normalized AUC anti-RBD IgM titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. e, Normalized AUC anti-RBD IgA titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. f, Normalized AUC anti-RBD IgA titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. g, Normalized AUC anti-N IgG titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. h, Normalized AUC anti-N IgG titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. i, Index values (IV) of anti-RBD IgG titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. j, Index values of anti-RBD IgG titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. k, Cut-off index (COI) values of anti-N total Ig titres at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. l, COI values of anti-N total Ig titres at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. m, NT50 values at 1.3 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. n, NT50 values at 6.2 months for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. o, Age in years for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. p, Severity of acute infection as assessed by the WHO ‘Ordinal Clinical Progression/Improvement Scale’ for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. q, Duration of symptoms during acute infection for participants with (n = 38) or without (n = 49) persistent post-acute symptoms. Horizontal bars indicate median values. Statistical significance was determined using two-tailed Mann–Whitney U-tests.
Extended Data Fig. 2 |. Correlations of plasma antibody measurements.
a, Normalized AUC for IgG anti-RBD plotted against Pylon IgG anti-RBD index values at 1.3 months. b, Normalized AUC for IgG anti-RBD plotted against Pylon IgG anti-RBD index values at 6.2 months. c, Normalized AUC for IgM anti-RBD plotted against Pylon IgM anti-RBD index values at 1.3 months. d, Normalized AUC for IgM anti-RBD plotted against Pylon IgM anti-RBD index values at 6.2 months. e, Normalized AUC for IgG anti-N plotted against Roche COI values for anti-N total Ig titres at 1.3 months. f, Normalized AUC for IgG anti-N plotted against Roche COI values for anti-N total Ig titres at 6.2 months. g, Anti-N total Ig COI values for 80 individuals at the initial 1.3- and 6.2-month follow-up visit. h, Relative change in anti-N total Ig levels between 1.3 and 6.2 months plotted against the anti-N total Ig levels at 1.3 months. i, Normalized AUC for IgM anti-RBD at 6.2 months plotted against IgM anti-RBD at 1.3 months. j, Normalized AUC for IgG anti-RBD at 6.2 months plotted against IgG anti-RBD at 1.3 months. k, Normalized AUC for IgA anti-RBD at 6.2 months plotted against IgA anti-RBD at 1.3 months. l, Normalized AUC for IgG anti-N at 6.2 months plotted against IgG anti-N at 1.3 months. m, COI values for anti-N total Ig titres at 6.2 months plotted against anti-N total Ig titres at 1.3 months. n, Anti-RBD IgM titres at 1.3 months plotted against anti-N IgG titres at 1.3 months. o, Anti-RBD IgG titres at 1.3 months plotted against anti-N IgG titres at 1.3 months. p, Anti-RBD IgA titres at 1.3 months plotted against anti-N IgG titres at 1.3 months. q, NT50 values at 1.3 months plotted against anti-RBD IgG titres at 1.3 months. r, NT50 values at 6.2 months plotted against anti-RBD IgG titres at 6.2 months. The r and P values were determined by two-tailed Spearman’s correlations.
Extended Data Fig. 3 |. Persistent longitudinal changes in the phenotypic landscape of B cells in individuals recovered from COVID-19.
a, Global viSNE projection of pooled B cells for all participants pooled shown in background contour plots, with overlaid projections of concatenated controls, and convalescent participants at 1.3 and 6.2 months. b, viSNE projection of pooled B cells for all participants of B cell clusters identified by FlowSOM clustering. Column-scaled z-scores of median fluorescence intensity (MFI) as indicated by cluster and marker. c, Frequency of B cells from each group in FlowSOM clusters indicated. Each circle represents an individual control individual (n = 20) (grey), convalescent participant at 1.3-month post-infection (n = 41) (red) or convalescent participant at 6.2 months post-infection (n = 41) (green). Horizontal bars indicate mean values. Significance determined by two-tailed paired t-test for comparisons between time points within individuals and two-tailed unpaired t-test for comparison between controls and convalescent individuals. d, Individual viSNE projections of indicated protein expression.
Extended Data Fig. 4 |. Flow cytometry.
a, Gating strategy used for cell sorting. Gating was on singlets that were CD20+ and CD3−CD8−CD16−Ova−. Sorted cells were RBD–PE+ and RBD–AF647+. b, Flow cytometry showing the percentage of RBD-double-positive memory B cells from month 1.3 or month 6 post-infection in 21 randomly selected participants.
Extended Data Fig. 5 |. Frequency distributions of human V genes.
a, Two-sided binomial tests were used to compare the frequency distributions of human V genes of anti-SARS-CoV-2 antibodies from donors at 1.3 months and 6.2 months1. *P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001. b, Two-sided binomial tests were used to compare the frequency distributions of human V genes of anti-SARS-CoV-2 antibodies from this study to sequences from ref. 53. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. c, Sequences from all six individuals with clonal relationships depicted as circos plots as in Fig. 2d. Interconnecting lines indicate the relationship between antibodies that share V and J gene-segment sequences at both IGH and IGL. Purple, green and grey lines connect related clones, clones and singles, and singles to each other, respectively. d, For each participant, the number of IgG heavy-chain sequences (black) analysed at month 1.3 (left) or month 6.2 post-infection (right). The number in the inner circle indicates the number of cells that was sorted for each individual denoted above the circle. e, The same as d but showing combined data for all six participants. f, Comparison of the percentage of IgG-positive B cells from all six individuals at month 1.3 or month 6.2 post-infection. The horizontal bars indicate the mean. Statistical significance was determined using two-tailed t-test.
Extended Data Fig. 6 |. Analysis of antibody somatic hypermutation of persisting clones, CDR3 length and hydrophobicity.
a–f, Number of somatic nucleotide mutations in both the IGVH and IGVL of persisting clones found at month-1.3 and month-6.2 time points in six participants: COV21 (a), COV47 (b), COV57 (c), COV72 (d), COV96 (e) and COV107 (f). The VH and VL gene usage of each clonal expansion is indicated above the graphs, or are indicated as ‘singlets’ if the persisting sequence was isolated only once at both time points. Connecting line indicates the somatic hypermutation of the clonal pairs that were expressed as a recombinant monoclonal antibodies. g, For each individual, the amino acid length of the CDR3 at IGVH and IGVL is shown. The horizontal bars indicate the mean. The number of antibody sequences (IGVH and IGVL) evaluated for each participant are n = 90 (COV21), n = 78 (COV47), n = 53 (COV57), n = 87 (COV72), n = 104 (COV96), n = 120 (COV107). Right, all antibodies combined (n = 532 for both IGVH and IGVL). Statistical significance was determined using two-tailed Mann–Whitney U-tests and horizontal bars indicate median values. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. h, Distribution of the hydrophobicity GRAVY scores at the IGH CDR3 in 532 antibody sequences from this study compared to a public database: statistical analysis is provided in Methods. The box limits are at the lower and upper quartiles, the centre line indicates the median, the whiskers are 1.5× interquartile range and the dots represent outliers. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Extended Data Fig. 7 |. ELISA of wild-type or mutant RBD for monoclonal antibodies.
a, EC50 values for binding to wild-type RBD of shared singlets and shared clones of monoclonal antibodies obtained at the initial 1.3- and 6.2-month follow-up visit, divided by participant (n = 6 (COV21), n = 13 (COV47), n = 3 (COV57), n = 6 (COV72), n = 15 (COV96), n = 9 (COV107)). Lines connect shared singlets or clones. Monoclonal antibodies with improved EC50 at the 6.2-month follow-up visit are highlighted in green; remaining monoclonal antibodies are shown in black. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. b–j, Graphs show ELISA binding curves for different antibodies obtained at 1.3 months (dashed lines) and their clonal relatives obtained after 6.2 months (solid lines) binding to wild type, R346S, E484K, Q493R, N439K, N440K, A475V, S477N, V483A and V367F RBDs (colours as indicated). Antibody identifiers of pairs are as indicated on top of panels (1.3 months/6.2 months). k, Heat map shows log2-transformed relative fold change in EC50 against the indicated RBD mutants for 52 antibody clonal pairs obtained at 1.3 (black) and 6.2 months (red). The clonal and participant origin for each antibody pair is indicated above.
Extended Data Fig. 8 |. Neutralization of wild-type and mutant RBDs, C51 alignment and binding projection.
a, IC50 values of shared singlets and shared clones of monoclonal antibodies obtained at the initial 1.3- and 6.2-months follow-up visit, divided by participant (n = 6 (COV21), n = 13 (COV47), n = 3 (COV57), n = 6 (COV72), n = 15 (COV96), n = 9 (COV107)). Lines connect shared singlets or clones. Monoclonal antibodies with undetectable IC50 at 1.3 months are plotted at 10 μg ml−1 and are highlighted in red; monoclobal antibodies with improved IC50 at the 6.2-month follow-up visit are highlighted in green; remaining monoclonal antibodies are shown in black. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. b–f, The normalized relative luminescence values for cell lysates of 293TACE2 cells 48 h after infection with SARS-CoV-2 pseudovirus containing wild-type RBD or RBD mutants (wild-type, Q493R, E484G and R346S RBD shown in black, red, green and blue, respectively) in the presence of increasing concentrations of monoclonal antibodies obtained at the 1.3-month initial visit (1.3m, dashed lines) and their shared clones or singlets at the 6.2-month follow-up visit (6.2m, continuous lines). Antibody identifiers are as indicated. g, VH and VL amino acid sequence alignment of C144 and derivative antibodies C051, C052, C053 and C054. Germline-encoded residues are highlighted in green. Residues in the proximity of RBD-binding C144 paratope are highlighted in red. h–j, Surface representation of two adjacent ‘down’ RBDs (RBDA and RBDB) on a spike trimer with the C144 epitope on the RBDs highlighted in cyan and positions of amino acid mutations that accumulated in C052 (h), C053 (i) and C054 (j), compared to the parent antibody C144, highlighted as stick side chains on a Cα atom representation. The C052, C053 and C054 interactions with two RBDs was modelled on the basis of a cryo-electron microscopy structure of C144 Fab bound to spike trimer18.
Extended Data Fig. 9 |. SARS-CoV-2 antigen in human enterocytes in the gastrointestinal tract at three months after COVID-19 diagnosis, and pre-COVID-19 control individuals without detectable SARS-CoV-2 antigen.
a–j, Immunofluorescence images of human gastrointestinal tissue are shown. Staining is for EPCAM (red), DAPI (blue) and SARS-CoV-2 nucleocapsid (green) Samples are derived from intestinal biopsies in the gastrointestinal tract as indicated. a–h, Biopsies from participant CGI-088 (Supplementary Table 7) taken 92 d after the onset of COVID-19 symptoms. i, j, Biopsy taken 27 months before the onset of COVID-19 symptoms from participant CGI-088. Arrows indicate enterocytes with detectable SARS-CoV-2 antigen. Isotype and no-primary controls for each tissue are shown in the right two columns. Scale bars, 100 μm. k, l, Immunofluorescence images of biopsy samples in the gastrointestinal tract (ileal (k) and duodenal (l)) obtained from ten preCOVID-19 control individuals between January 2018 and October 2019 are shown. Staining is for EPCAM (red), DAPI (blue) and SARS-CoV-2 nucleocapsid (green). Scale bars, 100 μm. All experiments were repeated independently at least twice with similar results.
Extended Data Fig. 10 |. SARS-CoV-2 antigen and RNA is detectable in different intestinal segments in several individuals convalescent from COVID-19.
a, Immunofluorescence images of biopsy samples in the gastrointestinal tract in different individuals are shown. Staining is for EPCAM (red), DAPI (blue) and SARS-CoV-2 nucleocapsid (green). Samples are derived from intestinal biopsies from four participants (CGI-089, CGI-092, CGI-100 and CGI-106) taken at least three months after COVID-19 infection. Arrows indicate enterocytes with detectable SARS-CoV-2 antigen. Scale bars, 100 μm. The experiments were repeated independently at least twice with similar results. b, Quantification of SARS-CoV-2-positive cells by immunofluorescence. The number of cells staining positive for the N protein of SARS-CoV-2 per mm2 of intestinal epithelium is shown. The graphs show biopsy samples from the indicated individuals of the duodenum (left) and terminal ileum (right). Black dots represent the number of available biopsy specimen for each individual from the respective intestinal segment (CGI-088, n = 4 duodenal and n = 2 ileal; CGI-089, n = 2 duodenal and n = 2 ileal; CGI-092, n = 6 duodenal and n = 3 ileal; CGI-106, n = 4; CGI-100, n = 4). Boxes represent median values and whiskers the 95% confidence interval. c, d, SARS-CoV-2 viral RNA was visualized in intestinal biopsies of participant CGI-089 (c) and CGI-088 (d) using in situ hybridization. SARS-CoV-2 genomic RNA (black) and haematoxylin and eosin staining by smFISH–immunohistochemistry technique in the duodenum (left) or terminal ileum (right). Arrows indicate enterocytes with detectable SARS-CoV-2 RNA. e, Pre-COVID-19 control individuals show no detectable SARS-CoV-2 viral RNA in duodenal (left) or ileal (right) biopsies. Scale bars, 100 μm. The experiments were repeated independently three times with similar results.
Supplementary Material
Acknowledgements
We thank all study participants who devoted time to our research; B. Coller and S. Schlesinger, the Rockefeller University Hospital Clinical Research Support Office and nursing staff; M. Okawa Frank and R. B. Darnell for SARS-CoV-2 saliva PCR testing; C. M. Rice and all members of the M.C.N. laboratory for helpful discussions; M. Jankovic for laboratory support; and J. Vielmetter and the Protein Expression Center in the Beckman Institute at Caltech for expression assistance. This work was supported by NIH grant P01-AI138398-S1 (M.C.N. and P.J.B.) and 2U19AI111825 (M.C.N.).; the Caltech Merkin Institute for Translational Research and P50 AI150464-13 (P.J.B.), George Mason University Fast Grant (D.F.R. and P.J.B.) and the European ATAC grant EC 101003650 (D.F.R.); R37-AI64003 to P.D.B.; R01AI78788 to T. Hatziioannou; NCI R01CA234614, NIAID 2R01AI107301, NIDDK R01DK121072 and 1RO3DK117252 to R.E.S., NIH NIDDK R01 DK123749 01S1 to S.M. C.O.B. is supported by the HHMI Hanna Gray and Burroughs Wellcome PDEP fellowships. R.E.S. is an Irma Hirschl Trust Research Award Scholar. M.T. is supported by the Digestive Disease Research Foundation (DDRF). C.G. was supported by the Robert S. Wennett Post-Doctoral Fellowship, in part by the National Center for Advancing Translational Sciences (National Institutes of Health Clinical and Translational Science Award programme, grant UL1 TR001866), and by the Shapiro-Silverberg Fund for the Advancement of Translational Research. P.D.B. and M.C.N. are Howard Hughes Medical Institute Investigators.
Footnotes
Competing interests The Rockefeller University has filed a provisional patent application in connection with this work on which D.F.R. and M.C.N. are inventors (US patent 63/021,387). R.E.S. is on the scientific advisory board of Miromatrix Inc and is a consultant and speaker for Alnylam Inc. S.M. has served as a consultant for Takeda Pharmaceuticals, Morphic and Glaxo Smith Kline. Z.Z. received seed instruments and sponsored travel from ET Healthcare.
Reprints and permissions information is available at http://www.nature.com/reprints.
Data presentation
Extended Data Figures were arranged in Adobe Illustrator 2020.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Code availability
Computer code to process the antibody sequences is available at GitHub (https://github.com/stratust/igpipeline).
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03207-w.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Michael Laue, Stanley Perlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Data are provided in Supplementary Tables 1–8. The raw sequencing data and computer scripts associated with Fig. 2 have been deposited at https://github.com/stratust/igpipeline. This study also uses data from a DRYAD accession linked to ref. 57 (https://doi.org/10.5061/dryad.35ks2), the Protein Data Bank (accession codes 6VYB and 6NB6) and ref. 53.
References
- 1.Robbiani DF et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schäfer A et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med 218, e20201993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carfì A, Bernabei R & Landi F Persistent symptoms in patients after acute COVID-19. J. Am. Med. Assoc 324, 603–605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Rio C., Collins LF. & Malani P. Long-term Health Consequences of COVID-19. JAMA 324, 1723–1724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang HS et al. SARS-CoV-2 antibody characterization in emergency department, hospitalized and convalescent patients by two semi-quantitative immunoassays. Clin. Chim. Acta 509, 117–125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diagnostics Roche. Roche Diagnostics Elecsys Anti-SARS-CoV-2, https://www.fda.gov/media/137605 (2020).
- 7.Muecksch F et al. Longitudinal analysis of serology and neutralizing antibody levels in COVID19 convalescents. J. Infect. Dis jiaa659 (2020). [Google Scholar]
- 8.Schmidt F et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med 217, e20201181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wajnberg A et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370, 1227–1230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seow J et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol 5, 1598–1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouwer PJM et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju B et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Kreer C et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 182, 843–854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seydoux E et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 53, 98–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan M et al. Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisblum Y et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnes CO et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tortorici MA et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baum A et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182, 1284–1294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamers MM et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livanos AE et al. Gastrointestinal involvement attenuates COVID-19 severity and mortality. Preprint at 10.1101/2020.09.07.20187666 (2020). [DOI]
- 24.Qian Q et al. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis ciaa925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morone G et al. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front. Med 7, 562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SK et al. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol 10.1016/j.cgh.2020.06.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wölfel R et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Beaudoin-Bussières G et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. MBio 11, e02590–20398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford KHD et al. Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection. J. Infect. Dis jiaa618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer AS et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol 5, eabe0367–0313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin. Infect. Dis 579, 270–279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ripperger TJ et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity 53, 925–933.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan JM et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 10.1126/science.abf4063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodda LB et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 184, 169–183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Secchi M et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J. Clin. Invest 130, 6366–6378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Zost SJ et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen SCA et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microb 28, 516–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko N et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 183, 143–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Victora GD & Nussenzweig MC Germinal centers. Annu. Rev. Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Sokal A et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Preprint at 10.1101/2020.11.17.385252 (2020). [DOI] [PMC free article] [PubMed]
- 43.Mahler DA & Wells CK Evaluation of clinical methods for rating dyspnea. Chest 93, 580–586 (1988). [DOI] [PubMed] [Google Scholar]
- 44.Pujadas E et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med 8, e70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chomczynski P & Sacchi N Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem 162, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
- 46.DeAngelis MM, Wang DG & Hawkins TL Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 23, 4742–4743 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grifoni A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amanat F et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnes CO et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weisblum Y et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Gassen S et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Gupta NT et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soto C et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature 566, 398–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Y, Chen K, Kwong PD, Shapiro L & Sheng Z cAb-Rep: a database of curated antibody repertoires for exploring antibody diversity and predicting antibody prevalence. Front. Immunol 10, 2365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kyte J & Doolittle RF A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
- 56.Guy HR Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys. J 47, 61–70 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeWitt WS et al. A public database of memory and naive B-cell receptor sequences. PLoS ONE 11, e0160853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spiegel M et al. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol 79, 2079–2086 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are provided in Supplementary Tables 1–8. The raw sequencing data and computer scripts associated with Fig. 2 have been deposited at https://github.com/stratust/igpipeline. This study also uses data from a DRYAD accession linked to ref. 57 (https://doi.org/10.5061/dryad.35ks2), the Protein Data Bank (accession codes 6VYB and 6NB6) and ref. 53.