Abstract
SARS-CoV-2 virus, the causative agent of COVID-19 pandemic, has a genomic organization consisting of 16 nonstructural proteins (nsps), 4 structural proteins, and 9 accessory proteins. Relative of SARS-CoV-2, SARS-CoV, has genomic organization, which is very similar. In this article, the function and structure of the proteins of SARS-CoV-2 and SARS-CoV are described in great detail. The nsps are expressed as a single or two polyproteins, which are then cleaved into individual proteins using two proteases of the virus, a chymotrypsin-like protease and a papain-like protease. The released proteins serve as centers of virus replication and transcription. Some of these nsps modulate the host’s translation and immune systems, while others help the virus evade the host immune system. Some of the nsps help form replication-transcription complex at double-membrane vesicles. Others, including one RNA-dependent RNA polymerase and one exonuclease, help in the polymerization of newly synthesized RNA of the virus and help minimize the mutation rate by proofreading. After synthesis of the viral RNA, it gets capped. The capping consists of adding GMP and a methylation mark, called cap 0 and additionally adding a methyl group to the terminal ribose called cap1. Capping is accomplished with the help of a helicase, which also helps remove a phosphate, two methyltransferases, and a scaffolding factor. Among the structural proteins, S protein forms the receptor of the virus, which latches on the angiotensin-converting enzyme 2 receptor of the host and N protein binds and protects the genomic RNA of the virus. The accessory proteins found in these viruses are small proteins with immune modulatory roles. Besides functions of these proteins, solved X-ray and cryogenic electron microscopy structures related to the function of the proteins along with comparisons to other coronavirus homologs have been described in the article. Finally, the rate of mutation of SARS-CoV-2 residues of the proteome during the 2020 pandemic has been described. Some proteins are mutated more often than other proteins, but the significance of these mutation rates is not fully understood.
Keywords: SARS-CoV-2, SARS-CoV, structure, function, proteins
Introduction
A novel coronavirus infection was first identified in Wuhan, China, in December 2019. The virus was found to cause a respiratory illness, which was later named COVID-19. The disease quickly spread across the world and in March 2020 World Health Organization declared it a pandemic. The causative agent for the disease was first identified as a novel coronavirus using metagenomic RNA sequencing of bronchoalveolar lavage of a patient suffering from the disease in Wuhan, China. 1 The sequencing revealed that the novel virus had most proteins homologous to SARS-CoV, which caused the SARS outbreak in 2003, and thus was named SARS-CoV-2 by International Classification of Diseases.
SARS-CoV-2 is a type of coronavirus, which are spherical, enveloped viruses with surface projections that give rise to the corona appearance. Coronaviruses contain a positive-sense RNA genome, which is wrapped up in helical nucleocapsid. The genome size of SARS-CoV-2 is about 30 kb. Among the RNA viruses, coronaviruses have the largest genome size. Coronaviruses are one of the two genera of classification under the family Coronaviridae. Coronaviridae, along with Arteriviridae and Roniviridae, fall under the order Nidoviridae. 2
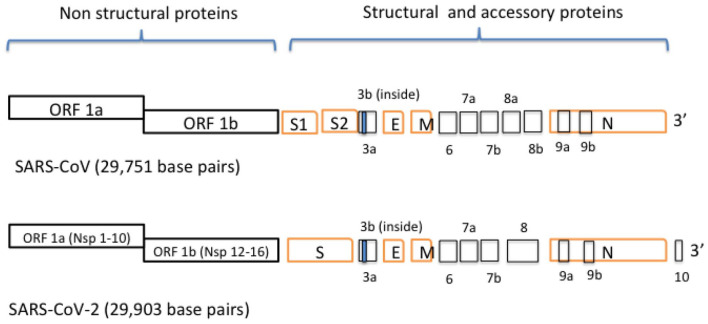
SARS-CoV-2 has a similar genome organization as other coronaviruses. The 5′ two thirds of the genome encodes for gene 1 proteins associated with the synthesis of viral RNA and the 3′ one third is responsible for encoding all of the structural and accessory proteins.3-5 In SARS-CoV-2, first two third of the genome consists of the replicase genes encoding for large polyproteins, pp1a and pp1ab, which are later converted into 16 nonstructural proteins by the process of proteolytic cleavage using multiple proteases: a virally encoded chymotrypsin-like protease and two papain-like proteases. Open Reading Frames (ORFs) for structural proteins like spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins occupy the remaining one third of the genome. 6 In addition to these components of the genomes shared by other coronaviruses, SARS-CoV genome also has eight ORFs which codes for accessory proteins named as ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b. 6 Accessory proteins differ from nonstructural proteins in that they do not have homology with proteins from other groups of viruses. They are generally small and have indirect roles in the function of a virus. Eight accessory genes are arranged in such a way that two are located between S and E genes (ORFs 3a and 3b), five are found between M and N genes, and one is arranged within the N gene. In this article, we have discussed in detail what is known about all the nonstructural, structural, and accessory proteins (Figure 1) (Table 1). Several review articles have already discussed structural and functional insights into these proteins.8,9 In this article, major structures of these proteins solved until December 15, 2020, are included.
Figure 1.
Genome organization of SARS-CoV and SARS-CoV-2. The upper panel shows the genomic organization of SARS-CoV. It contains two polyproteins pp1a and pp1b synthesized from ORF1a and 1b. These polypeptides undergo a series of proteolytic cleavages to form 16 nonstructural proteins. These 16 nonstructural proteins are encoded by the first two thirds of the genome (figure not drawn to scale). The second one third of the genome encodes for four structural proteins S, E, M, and N. Interspersed within these genes are genes for the expression accessory proteins. SARS-CoV-2, a more recently discovered virus, has genomic organization almost identical to SARS-CoV. Accessory protein 8 for SARS-CoV-2 is not divided into 8a and 8b as in SARS-CoV and two ORF for spike proteins are not present in this virus. In addition, SARS-CoV-2 protein contains accessory protein 10 potentially not present in SARS-CoV.
ORF indicates Open Reading Frames.
Table 1.
Tabulation of amino acid lengths of SARS-CoV and SARS-CoV-2 proteins and percentage identity for common proteins of the two species.
| Gene | Length of amino acids in SARS-CoV-2 | Length of amino acids in SARS-CoV | Percentage identity between homologous proteins | Number of amino acid residues with mutation rate greater than 0.01 during the pandemic (N) | N/amino acid length |
|---|---|---|---|---|---|
| Nsp1 | 180 | 180 | 84.44 | 0 | 0.000 |
| Nsp2 | 628 | 628 | 68.34 | 5 | 0.008 |
| Nsp3 | 1922 | 1922 | 75.82 | 10 | 0.005 |
| Nsp4 | 500 | 500 | 80.00 | 1 | 0.002 |
| Nsp5 | 306 | 306 | 96.08 | 5 | 0.016 |
| Nsp6 | 390 | 390 | 88.15 | 6 | 0.015 |
| Nsp7 | 83 | 83 | 98.80 | 1 | 0.012 |
| Nsp8 | 198 | 198 | 97.47 | 0 | 0.000 |
| Nsp9 | 113 | 113 | 97.35 | 1 | 0.009 |
| Nsp10 | 139 | 139 | 97.12 | 0 | 0.000 |
| Nsp11 | 13 | 13 | 84.60 | 0 | 0.000 |
| Nsp12 | 932 | 932 | 96.14 | 7 | 0.008 |
| Nsp13 | 601 | 601 | 99.83 | 5 | 0.008 |
| Nsp14 | 527 | 527 | 95.07 | 2 | 0.004 |
| Nsp15 | 346 | 346 | 88.73 | 4 | 0.012 |
| Nsp16 | 298 | 298 | 93.29 | 1 | 0.003 |
| S | 1273 | 1255 | 75.96 | 21 | 0.016 |
| ORF3a | 275 | 274 | 72.36 | 9 | 0.033 |
| ORF3b | 151 | 154 | No significant similarity | – | – |
| E | 75 | 76 | 94.74 | 0 | 0.000 |
| M | 222 | 221 | 90.54 | 0 | 0.000 |
| ORF6 | 61 | 63 | 68.85 | 0 | 0.000 |
| ORF7a | 121 | 122 | 85.35 | 1 | 0.008 |
| ORF7b | 43 | 44 | 81.40 | 1 | 0.023 |
| ORF8a | 121 (only ORF8) | 39 | 31.71 | 2 | 0.017 |
| ORF8b | 121 (only ORF8) | 84 | 40.48 | ||
| N | 419 | 422 | 90.52 | 16 | 0.038 |
| ORF9a | 97 | 98 | 72.45 | – | – |
| ORF9b | 73 | 70 | 77.14 | 4 | 0.055 |
| ORF10 | 38 | – | – |
Abbreviation: ORF, open reading frames.
Open reading frames of SARS-CoV-2 proteins were detected from GenBank accession number NC_045512.2. Similarly, open reading frames of SARS-CoV Tor2 were detected from GenBank accession number AY274119. Pairwise alignment of the proteins was conducted using NCBI blast and percentage identity was tabulated. 7 In the pblast algorithm, max target sequence of 100, short queries automatically adjusted to parameters for short input sequences, expect threshold of 0.05 and world size of 6 was set. BLOSUM62 matrix with gap costs of 11 for existence and 1 for extension was set. Number of amino acids with mutation rate higher than 0.01 and mutational frequency rate was tabulated in the last two columns.
The polyproteins 1a and 1b encode for nonstructural proteins that form replication/transcription complex (RTC) in double-membrane vesicles (DMVs). Using discontinuous transcription, through the RTC a nested set of subgenomic messenger RNAs are synthesized. These subgenomic mRNAs possess a common 5′ leader and 3′ terminal sequence and are translated into viral proteins. 10
Nonstructural Proteins (Nsps)
The enzymatic activities and functional domains for many of the essential nsps are predicted to be conserved between the different genera of Coronaviruses, highlighting their importance in the viral replication. In addition to the nsps described in this article with defined functions, there are several other nsps whose biological functions and roles in coronavirus life cycle remain to be characterized.
The synthesis of RNA of coronaviruses is accomplished by the replication-transcription complex (RTC) in conjunction with a complex vesicular network where the nsps have extensive interactions among each other. The viral replication machinery is harbored to the convoluted membranes by the transmembrane proteins nsp3, nsp4, and nsp6 preventing the dsRNAs from immune degradation.11-13 To enhance the whole machinery, nsp1 suppresses the host gene expression and induces the blockage of innate immune response in infected cells. 14 Nsp5, also known as 3C-like protease (3CLpro), is first produced automatically by cleavage, which then cleaves downstream nsps at 11 sites to release nsp4 to nsp16. 15 Nsp12 and nsp8 have a major role in the assembly of the entire RNA polymerase replicative machinery. It requires the presence of nsp7 along with nsp8 and nsp12 to bind nucleic acid and perform efficient RNA synthesis. N-terminal of nsp14 is an exonuclease domain (ExoN),16,17 which prevents lethal mutagenesis as the domain is involved in the role of proofreading (Figure 13).
Figure 13.
Cartoon representation of crystal structure of SARS-CoV-2 nsp9 dimer structure showing loops (green), β-sheets (yellow), and helices (red) (PDB 6WXD). The inter-subunit interactions to form a dimer are due to van der Waals interactions between the interfacing copies of α1 helix C-terminal as a result of self-association of GxxxG protein-protein binding motif. 77
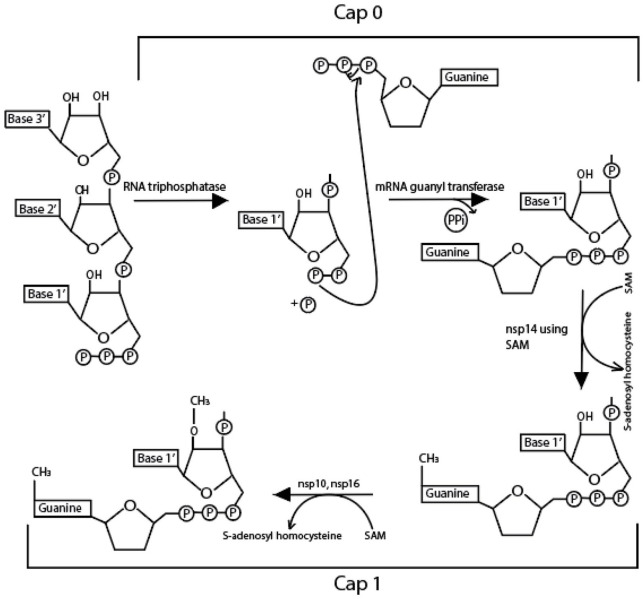
On the other hand, various nsps—10, 13, 14, and 16—play significant roles in mRNA capping. The mRNA cap has many significant biological roles in the virus as it is critical for the stability, translation, and evasion of the host immune response to mRNAs. Uncapped RNA molecules in cytoplasmic granular compartments are degraded as they can trigger innate immune response. Initially, 5’γ-phosphate of the nascent RNA chains (pppN-RNA) is hydrolyzed by the nsp13 helicase, an RNA 5’-triphosphatase. 18 Then, GpppN-RNA is formed after a yet unidentified GTase transfers a GMP molecule to the 5’-diphosphate of the RNA chains (ppN-RNA). Furthermore, the domain nsp14C-terminal N7-MTase methylates the cap structure at the guanosine’s N7 position, generating cap-0 (m7GpppN-RNA), using SAM (S-Adenosyl Methionine) as a methyl donor (Figure 2). Finally, nsp16 (SAM)-dependent 2’-O-MTase activity promotes the methylation of the ribose 2’-O position to form cap-1 (m7GpppNm-RNA) in the first transcribed nucleotide (Figure 2). In the final steps, nsp10 serves as an allosteric activator.19,20 The known structure and function of each nsp is described below (Table 2) (Figure 3).
Figure 2.
Cap0 and Cap1 activity. Methylation is carried out at the 5ʹ end of newly synthesized RNA using nsp14 (cap0). Methylation is then again carried out at the 2ʹ oxygen of terminal ribose using nsp10 and 14.
Table 2.
Structure and function of nonstructural proteins of SARS-CoV and SARS-CoV-2.
| Name | Functional name | Structure solved (SARS-CoV-2) | Structure solved (SARS-CoV) | Structure description | Function |
|---|---|---|---|---|---|
| Nsp1 | Virulent factor | Cryo-EM Structure and X-Ray Crystallography structure PDB:7K5I, 7K3 N, 7K7 P | NMR Structure PDB: 2GDT |
The SARS-CoV-2 nsp113-127 like that of SARS-CoV hosts a unique topological arrangement, which gives to the formation of a six-stranded (n = 6) beta-barrel. In addition, there is an alpha1 helix which is positioned as a cap along one opening of the beta-barrel, two 310 helices that run parallel to each other and the beta5 strand which is though not a part of the beta-barrel but forms a beta-sheet interaction with the beta4 strand. As evident in the crystal structure of nsp113 127, nsp1 of SARS-CoV-2 has large number of flexible loops. | It inhibits host translation, causes invasion from host immune response and leads to efficient viral gene expression in infected cells.14,21 |
| Nsp2 | Endosome-associated protein | – | – | N/A | It is entirely unknown. In SARS-CoV-2 as well, the other proteins nsp2 attaches to may offer some clues. Nsp2 interacts with PHB1 and PHB2 host protein complexes, which are involved in mitochondrial biogenesis. 22 |
| Nsp3 | Cutting and untagging protein | X-Ray Crystallography PDB: 6YWL, 6WEY, 6WOJ, 7CZ4, 7CJD, 7C33, 7LLZ, 7LOS, 7CMD, 7JIW,7LLZ | X-Ray Crystallography PDB: 4MM3, | It contains two transmembrane domains, which is released from pp1a/1ab by the papain-like protease domain, which is a part of nsp3 itself. | It releases nsp1 and nsp2 from polyprotein, interacts with other viral nsps as well as RNA to form replication/transcription complex 23 and removes tags from old proteins set for destruction. 24 |
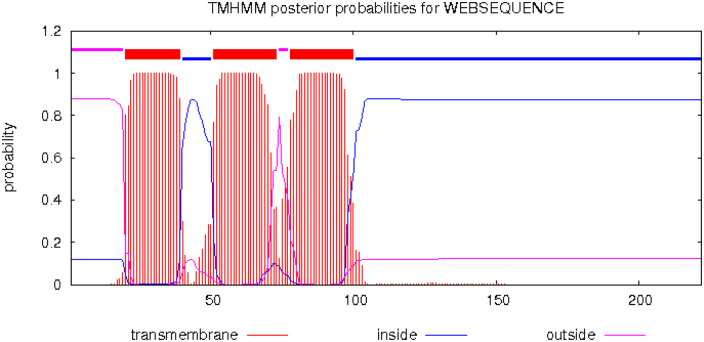
| Nsp4 | Double-membrane vesicle maker | – | – | It is predicted to contain four transmembrane domains, both termini projecting at the cytoplasmic side of the membrane, and three loop regions. | Nsp3, 4, and 6 are predicted to function to nucleate and anchor viral replication complexes on double-membrane vesicles in the cytoplasm.11-13 |
| Nsp5 | Protease (3CLpro) | X-Ray Crystallography PDB: 6M2N, 2M2N, 7L0D, 6M2Q, 7JKV, 7JQ3, 7JPY, 7JPZ, 7JQ0, 7JQ1, 7JQ4, 7JQ5, 7JQ2 | X-Ray Crystallography PDB: 2HOB, 3SN8 | 3CLpro monomer has 3 domains, domain I, domain II, domain III and a long loop. The active site of 3CLpro is located in the gap between domains I and II, and has a CysHis catalytic dyad. | 3CLpro is first automatically cleaved from polyproteins to produce mature enzyme, which then cleaves downstream nsps at 11 sites to release nsp4-nsp16. 15 |
| Nsp6 | Double-membrane vesicle factory | – | – | Nsp6 protein possesses 7 putative transmembrane helices located in endoplasmic reticulum (ER). | Nsp3, 4 and 6 are predicted to function to nucleate and anchor viral replication complexes on double-membrane vesicles in the cytoplasm.11-13 |
| Nsp7 | Copy assistant | Nsp7-nsp8-nsp12 structure solved (X-Ray Crystallography) PDB: 7JLT, 6YHU, 7DCD, 7BW4, 6M71 | Nsp7-nsp8 structure solved (X-Ray Crystallography) PDB: 2AHM | It has a hexadecameric structure with 8 nsp7 and nsp8s that encircles double-stranded RNA. | SARS-CoV nsp7 dimerizes and interacts with other proteins such as nsp5, nsp8, nsp9, and nsp13. 14 |
| Nsp8 | Primase | Nsp7-nsp8-nsp12 structure solved (X-Ray Crystallography) PDB: 7JLT, 6YHU, 7DCD, 7BW4, 6M71 | Nsp7-nsp8 structure solved (X-Ray Crystallography) PDB: 2AHM | It has a hexadecameric structure with 8 nsp7 and nsp8s that encircles double-stranded RNA. | Nsp8 enzyme is able of de novo initiate replication and has been proposed to operate as primase
25
. Nsp8 is known to colocalize with RdRp to copy the SARS-CoV genome. 25 |
| Nsp9 | RNA-binding protein | X-Ray Crystallography PDB: 6WXD | X-Ray Crystallography PDB: 3EE7 | It consists of an unusual fold and its core is made up of 6-stranded enclosed β-barrel and a series of extended loops projects outward from it. | It is a single-stranded RNA-binding protein, which displays an oligosaccharide/oligonucleotide binding fold. 26 |
| Nsp10 | Methyltransferase stimulator | Solved as nsp10-nsp16-SAM complex (X-Ray Crystallography) PDB: 7BQ7, 7JYY | Solved as nsp10-nsp16-SAM complex (X-Ray Crystallography) PDB:3R24 | It comprises a central anti-parallel pair of β-strands, surrounded by a broad crossover loop on one side. On the other side, a helical domain with loops is present, which generates 2 zinc fingers. | It stimulates nsp16 to execute S-adenosyl-L-methionine (SAM)-dependent methyltransferase (MTase) activity 20 |
| Nsp12 | RNA-dependent RNA polymerase | Solved as nsp7-nsp8-nsp12 (Electron Microscopy) PDB: 6M71, 7JLT, 6YHU, 7DCD, 7BW4, 7AAP | Solved as nsp7-nsp8-nsp12 (Electron Microscopy) PDB: 6NUR Solved as nsp7-nsp8 complex (Electron Microscopy) PDB: 6NUS |
It consists of N-terminal and polymerase domain which resembles a cupped “right hand” consisting a finger, a palm, and a thumb subdomain | Nsp12, in association with nsp7, nsp8, and other essential components of the RNA synthesis machinery, forms a viral replication complex. 27 |
| Nsp13 | Helicase | X-Ray Crystallography PDB: 6ZSL, 7NI0, 7NN0, 7NNG | – | Nsp13 adopts a triangular pyramid shape comprising five domains: two “RecA-like” domains (1A and 2A), and 1B domain, N-terminal zinc-binding domain (ZBD) and stalk domain, which connects ZBD and 1B domain. | It unwinds dsRNA or DNA with a 5′→3′ polarity, using energy from nucleotide hydrolysis. 28 |
| Nsp14 | Proofreading exonuclease | – | Nsp14-nsp10 complex solved (X-Ray Crystallography) PDB: 5C8U | The ExoN domain features a core, twisted β-sheet consisting of five β-strands with one Mg2+ ion at its active site. The N7-MTase domain features a MTase fold with central β-sheet consisting of five β-strands. β1 and β2 sheets have a ligand-binding cavity in-between. | Its N-terminal exoribonuclease domain has a proofreading role, which prevents lethal mutagenesis, whereas the C-terminal domain functions as a (guanine-N7) methyltransferase (N7-MTase) for mRNA capping. 29 |
| Nsp15 | Endonuclease | X-Ray Crystallography PDB: 7KEG, 7KEH, 7KF4 | Catalytically inactive mutant version of Nsp15 solved (X-Ray Crystallography) PDB: 2RHB | Nsp15 forms dimers of trimers, which finally assembles into a hexamer. Each subunit consists of N-terminal domain, a middle domain and C-terminal catalytic endonuclease domain. | Nsp15 preferentially cleaves 3′ of uridines in a manganese dependent manner. This is thought to be an important way for the virus to hide from antiviral defense. 30 |
| Nsp16 | Methyltransferase | Solved as nsp10-nsp16-SAM complex (X-Ray Crystallography) PDB: 7BQ7, 7JYY | Solved as nsp10-nsp16-SAM complex (X-Ray Crystallography) PDB:3R24 | It consists of Rossmann-like β-sheet fold surrounded by 11 α-helices, 7 β-strands, and loops in the 2′-O-MTase catalytic core. | Nsp16 recruits N7-methylated capped RNA and SAM which promotes the assembly of the enzymatically active nsp10/nsp16 complex. This complex converts 7mGpppG (cap-0) into 7mGpppG2′Om (cap-1) RNA by 2′-OH methylation of N1. 31 |
Abbreviations: Cryo-EM, cryogenic electron microscopy; DNA, deoxyribonucleic acid; NMR = nuclear magnetic resonance; RdRp, RNA-dependent RNA polymerase; RNA, ribonucleic acid.
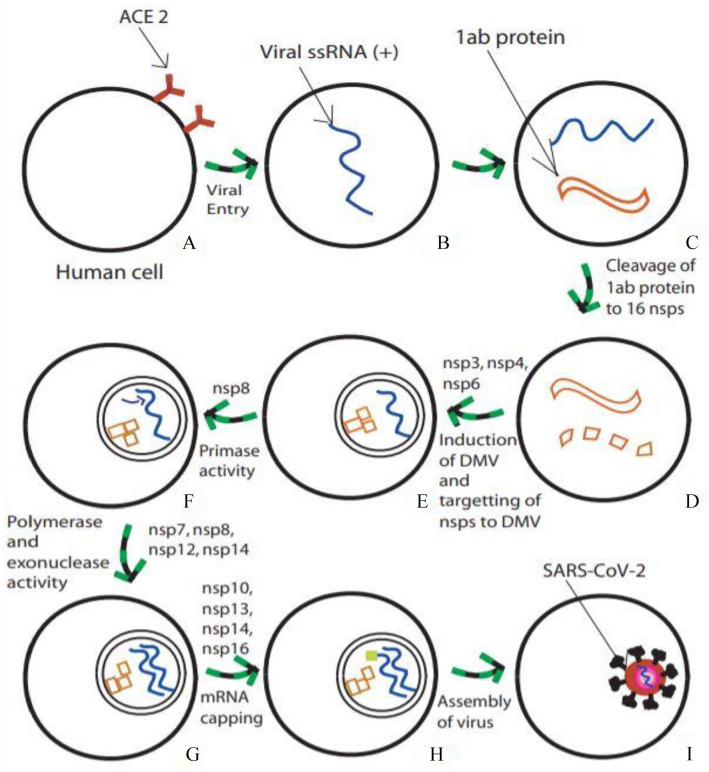
Figure 3.
Hypothetical SARS-CoV-2 entry and replication inside human host cell. (A) Human host cell with angiotensin-converting enzyme 2 (ACE 2) receptors, which attaches the virus and aids its entry into the cell. (B) Positive-sense single-stranded viral RNA within the cell. (C) Translation of the RNA into 1ab proteins (pp1a and pp1b). (D) Cleavage of pp1a and pp1b into 16 nonstructural proteins by virally encoded chymotrypsin-like protease and two papain-like proteases. (E) Induction of double-membraned vesicles (DMV) and localization of cleaved nsps with the help of nsp3, nsp4 and nsp6. (F) nsp8 acts as a primase for RNA replication. (G) nsp7, nsp8, nsp12, and nsp14 assist polymerase and exonuclease activities. (H) mRNA capping is assisted by nsp10, nsp13, nsp14 and nsp16. (I) Finally, replicated RNA and other translated viral proteins assembled into a new virus.
Fifteen nsps have been described in the order of their functions. First nsp1 and nsp2, which act to suppress the immune system of the host, have been described. This is followed by the proteases nsp3 and nsp5, which are used to cleave the polyprotein into individual nsps. Thereafter, nsp4 and nsp5, which help anchor the nsps to double-membrane vesicles, are described. Following these proteins, nsp7, nsp8, and nsp12, which help polymerize RNA of the virus, are described. Thereafter, proofreading exonuclease nsp14 is described. After that, nsps involved in cap0 and cap1 activity, nsps 13, 10, and 16 are described. Finally endonuclease nsp15 and RNA-binding protein nsp9 are described.
At the beginning of the year 2021, an analysis on how much the virus has changed during the year 2020 was carried out. Using more than 290 000 SARS-CoV-2 viral proteomes collected throughout the world, hotspots of mutation in the proteome were detected. Mutation rate for a given residue X was calculated as the ratio of the number of sequences with mutated residue compared with original Wuhan virus at position X over the total number of sequences. 32 This mutation rate of different proteins and different sites was compared to get a sense on which part of the proteome mutated the fastest and which part of the proteome remained stable.
Virulent factor:·nsp1
The size of Coronavirus nsp1 differs among the different lineages of the virus within this genus. While nsp3-nsp16 from different Coronavirus genera share conserved functional domains, the N-terminal region of the ORF 1 polyprotein, especially nsp1 sequence, remains highly divergent among Coronaviruses. SARS-CoV belonging to the lineage B has nsp1 of 180 amino acids, which is translated from the farther 5’ coding region. However, only alpha and beta genera encode nsp1 and the gene 1 of delta and gamma genera encode only 15 nsps (nsp2-nsp16).5,17,33 Bioinformatics analysis of the primary amino acid sequence has not revealed any known cellular or viral homologs of nsp1 making it unique. 33 The biological functions of alpha and beta Coronaviruses demonstrate remarkable similarity, despite the lack of overall sequence similarity and known protein motifs, indicating its significance in the life cycle of these different lineages of Coronaviruses. 34
Nsp 1 downregulates the host translation by interacting with the 40S ribosomal subunit. This interaction on the other hand induces an endonucleolytic cleavage near the 5’UTR of host mRNAs, targeting them for degradation. Besides, viral mRNAs are protected from cleavage due to the presence of a 5’-end leader sequence. SARS-CoV breaches the host immune response via inhibiting type I interferon expression 14 as well as host antiviral signaling pathway 21 in the infected cells. Nsp1, thus, expedites efficient viral gene expression in infected cells.
The SARS-CoV-2 Nsp1 35 , like that of SARS-CoV hosts a unique topological arrangement, which gives rise to the formation of a six-stranded (n = 6) beta-barrel. The beta-barrel is primarily antiparallel with the exception of beta1 and beta2. In addition, there is an alpha1 helix which is positioned as a cap along one opening of the beta-barrel, two 310 helices that run parallel to each other and the beta5 strand, which though not a part of the beta-barrel, forms a beta-sheet interaction with the beta4 strand. As evident in the crystal structure of nsp1 35 , nsp1 of SARS-CoV-2 has large number of flexible loops. 35
The alignment of the primary amino acid sequence of nsp1 of SARS-CoV with that of SARS-CoV-2 has revealed the sequence identity of 84.4% (Table 1). Similar to SARS-CoV, the nonstructural protein 1 (nsp1) is the first protein synthesized by SARS-CoV-2 in infected cells to inhibit innate immune system of host. The combined cryo-electron microscopic and biochemical experiments showed that SARS-CoV2 nsp1 binds to the human 40S subunit in ribosomal complex, including the pre-initiation complex 43S. 36 However, nsp1 protein from SARS-CoV-2 binds to both 40S subunit and 80S ribosome. The protein then inserts its C-terminal domain at the gateway to the mRNA channel as a hairpin of alpha helices where it obstructs mRNA binding. The binding in this channel strictly relies on two specific amino acid side chains of nsp1. Potent inhibition in translation has also been observed in the presence of nsp1 in lysates from human cells. On the basis of the high-resolution structure of 40S-nsp1 complex for SARS-CoV-2, residues of nsp1 crucial for mediating translation inhibition have been singled out. 36
Nsp1 is one of the least mutated proteins during the SARS-CoV-2 pandemic. Mutation rate was lower than 0.01 for all amino acid residues of the protein (Figure 4). 32
Figure 4.
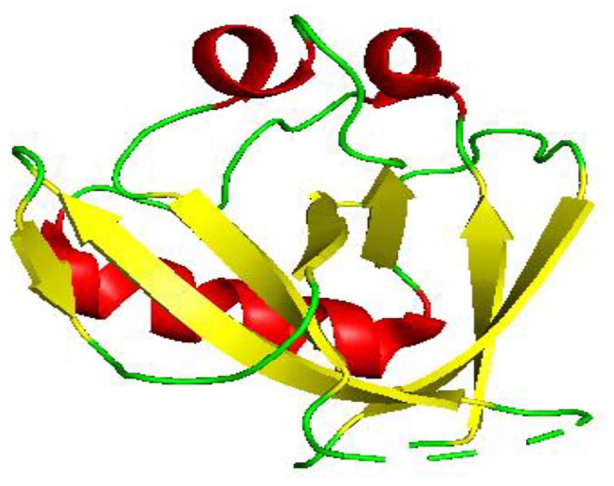
Crystal structure of SARS-CoV-2 nsp1 globular domain (Cartoon representation) comprising of residues 13 to 127 showing red helix, yellow sheet and green loop (PDB 7K3N). The structure of SARS-CoV-2 nsp1, like that of SARS-CoV, has six-stranded beta-barrel (yellow) and has additionally an alpha1 helix (red) and large number of flexible loops (green).
Endosome associated protein: nsp2
The function of nsp2 is not entirely known. It is thought to associate with the endosome of the host and disrupt host cell environment. 37 It is also a very conserved protein among coronaviruses. Nsp2 and nsp3 of SARS-CoV are detected not only as matured processed proteins but also as nsp2 and nsp3 precursors,38-41 suggesting the role of precursors in replication. These results suggest that nsp2 may be involved in regulating functions of nsp1 and nsp3. It has been found that the resulting mutant had decreased but not delayed growth, when the entire coding sequence of nsp2 was deleted in SARS-CoV, indicating the indispensable role of nsp2 for replication in culture. Similarly, in SARS-CoV-2, the other proteins nsp2 attaches to may provide some insights. Nsp2 interacts with PHB1 and PHB2 host protein complexes, which are involved in mitochondrial biogenesis. 22 During the pandemic of 2020, the protein did not mutate significantly. Five residues, T85, I120, A318, V381, and L550, demonstrated mutation rate higher than 0.01. 32
Protease: nsp5
Nsp5, also known as 3CLpro, is a 33.8 kDa cysteine protease, which processes two replicase polyproteins, pp1a (486 kDa) and pp1ab (790 kDa). 3CLpro monomer has 3 domains; domain I (residues 8-101), domain II (residues 101-184) and domain III (residues 201-303), and a long loop (residues 185-200) which connects domains II and III. The active site of 3CLpro is located in between the gaps of domains I and II and has a CysHis catalytic dyad (Cys145 and His41). 42 The sulfur of cysteine serves as a nucleophile and the imidazole ring of the histidine as a general base.
3CLpro is first automatically cleaved from polyproteins to produce a mature enzyme, which then cleaves downstream nsps at 11 sites to release nsp4-nsp16. 15 It is able to cleave a peptide bond between a glutamine at position P1 and a small amino acid (serine, alanine, or glycine) at position P2. 3CLpro expedites the maturation of nsps directly, which is crucial in the life cycle of the virus. The detailed investigation on the structure and catalytic mechanism of 3CLpro makes it an attractive target for anti-corona virus drug development. Inhibitors targeting at SARS-CoV 3CLpro comprises mainly peptide inhibitors and small-molecule inhibitors.
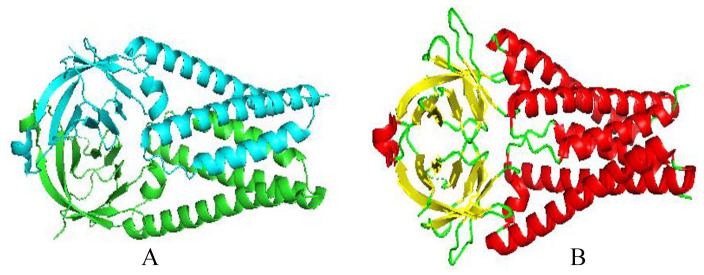
The amino acid sequence alignment of nsp5 for SARS-CoV and SARS-CoV-2 revealed the sequence identity of 96.06 (Table 1). The three dimensional structure of SARS-CoV-2 is highly similar to that of the SARS-CoV as expected from high sequence identity. 43 Domain I (residues 10-99) and domain II (residues 100-182) which harbor the binding site between them are six-stranded antiparallel B barrels. The domain III (198-303) is a globular cluster of five helices which is involved in regulating dimerization of the 3CLpro through a salt-bridge interaction between E290 of one promoter and R4 of the other. 44 The tight dimer formed by two molecules of 3CLpro oriented at right angles to each other has a contact interface, predominantly between the domain II of molecules A and the NH2-terminal residues (N-finger) of molecule B (Figure 5). Dimerization is crucial for the catalytic activity because the N-finger of each of the two protomers interacts with E290 of the other protomers and thereby shape the S1 pocket of substrate binding site. 45 To reach this interaction site, the N-finger is squeezed in between domains II and III of the parent monomer and domain II of the other monomer. Interestingly, there is a polar interaction between the two domains in the SARS-CoV but not in the SARS-CoV-2. 3CLpro dimer involves a 2.60-Å hydrogen bond between the T285 residue side chain groups of each protomers and favored by a hydrophobic contact between the I286 side chain and T285 Cγ2 side chain. In SARS-CoV-2, threonine is replaced by alanine and isoleucine by leucine. It has been shown previously that replacing S284, T285, and I286 with alanine residues in SARS-CoV 3CLpro results in a 3.6-fold enhancement of the protease’s catalytic activity, resulting in a slightly closer packing of the dimer’s two domains III against each other. 46
Figure 5.
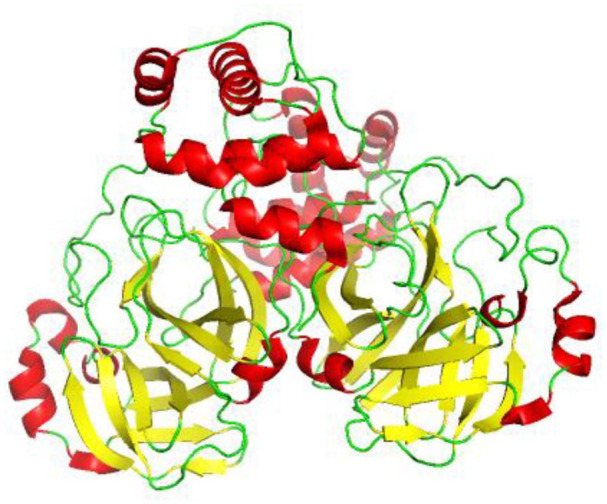
SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor in cartoon representation showing red helix, yellow sheet, and green loop (PDB 2M2 N). 3CL pro has two chains (A and B) with three domains (I, II, III). A long loop connects domains II and III. The B barrels of each domain I and II are composed of six-stranded B-sheets (yellow) and domain III is composed of mainly alpha helices (red).
During the pandemic of 2020, nsp5 was lowly mutated. Only five residues, K90, L89, P132, G71 and G15, showed mutation rate between 0.01 and 0.02. It is interesting to note that domain I shows the highest mutation rate. 32
Cutting and untagging protein: nsp3
Nsp3, also known as papain-like protease, is one of the first nsp encoded by ORFab. Nsp3 is the largest nonstructural protein of SARS-CoV-2 comprising various functional domains. It has different domain organization in different Coronavirus genera. The individual corona viruses can possess 10 to 16 domains of which eight domains the ubiquitin-like domain 1 (Ubl1), the glutamic acid rich acidic domain (also called “hypervariable region”), a macrodomain (also named X-domain), the ubiquitin-like domain 2(Ubl2), the papain-like protease 2(PL2(pro)) domain, the nsp3 ectodomain (3Ecto, also called “zinc finger domain”), as well as domains Y1 and CoV-Y of unknown function and two transmembrane regions (TM1 and TM2) are conserved according to recent bioinformatics analysis (Figure 6). 47 In SARS-CoV and SARS-CoV-2, the macro X domain, which is suspected to bind to ADP ribose has most of the variations. The amino acid sequences of this macro X domain differ by 26% between SARS-CoV and SARS-CoV-2, but the ability to bind to ADP ribose is retained. SARS-CoV nsp3, with a molecular weight of 215 kDa, is a transmembrane, glycosylated, multidomain protein that has been shown to interact with other proteins involved in replication and transcription. It may also serve as a scaffolding protein for these processes.48-50 The two transmembrane domains are released from pp1a/1ab by the papain-like protease domain, which is a part of nsp3 itself. 23 Likewise, it also releases nsp1 and nsp2 from polyproteins and interacts with the other viral nsps as well as RNA to form a replication/transcription complex. It acts on post-translational modifications of the host proteins to block the innate immunity of the host and to promote cytokine expression. It can also interact with host proteins to support virus survival. Nsp3 is supposed to be the second most promising vaccine candidate besides S protein. In SARS-CoV-2, nsp3 has significantly two jobs—the first is cutting other viral proteins to free them to do their own tasks and the second is removing tags from old proteins which are set for destruction. The removal of tags from old and damaged proteins by nsp3 changes the balance of proteins, thus possibly compromising the cell’s ability to fight the virus. 24
Figure 6.
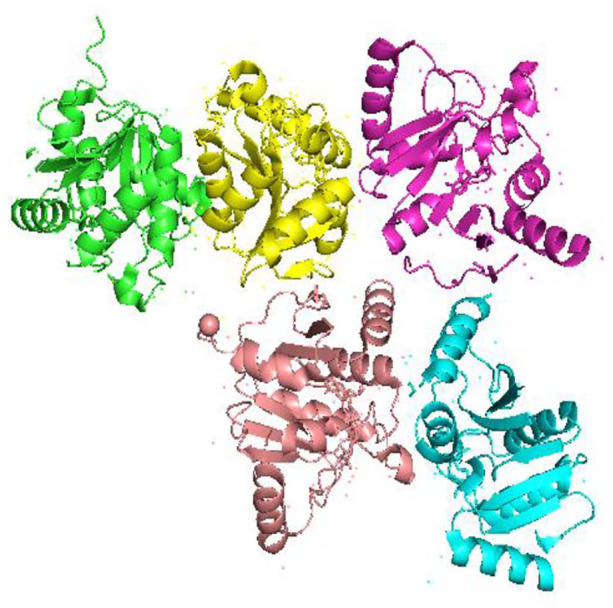
Crystal structure of SARS-CoV-2 nsp3 macrodomain (Cartoon representation) in complex with ADP ribose and showing five (A, B, C, D, E) different chains in different colors where ligands and water are shown in ball and stick representation (PDB 6YWL). SARS-CoV-2 encodes a large, multidomain nsp3 with an ADP ribose phosphate (ADRP) domain (also known as macrodomain), which is thought to interfere with the host immune response.
The papain-like protease domain recognizes the sequence LXGGX for proteolysis. The cleavage occurs between G and X residues. Proteins ubiquitin and ISG15 contain this consensus sequence at the C-terminus. Thus, papain-like protease can deubiquitinate and deISG15ylate proteins. These two covalent modifications are known to activate immune response. Removing this tag from proteins may suppress the immune system. Papain-like proteases in both SARS-CoV and SARS-CoV-2 have the catalytic core domain separated from N-terminal ubiquitin-like domain. The catalytic core domain takes the conformation of an open right hand with fingers, thumb, and palm. The finger is formed from four-stranded antiparallel beta sheets, the thumb is formed from four alpha helices, and the palm is formed from six-stranded beta sheets. Papain protease is a cysteine protease with catalytic triad consisting of cysteine, histidine, and aspartic acid. From experiments and from structural analysis, it has been found that SARS-CoV and SARS-CoV-2 papain proteases have similar catalytic efficiency. 8
Certain residues within the protein nsp3 demonstrated high level of mutational rate during the pandemic. T183, I1412, and A890 showed a mutation rate as high as 0.075. Other residues that showed mutational rates higher than 0.01 are I1683, A1736, T1363, H295, P968, T1189, and M1788. These mutational hotspot lie throughout the protein scattered in different domains. 32
Molecular anchor: nsp4
The nsp4 protein of SARS-CoV is about 500 amino acids long and has a calculated molecular mass of approximately 56 kDa. It is predicted to contain four transmembrane domains, 51 with both termini bulging out at the cytoplasmic side of the membrane, and three loop regions. Out of 16 nonstructural proteins, only nsp3, nsp4, and nsp6 have transmembrane domains. 52 The first transmembrane domain might display a cleavage signal sequence and is located after amino acid 32. 51 The other three predicted transmembrane domains are located between the residues 280 and 400, approximately leaving the cytoplasmic C-terminal tail of about 100 amino acids. In addition, an atypical glycosylation motif (NXC) occurs at position 131 between the first and second putative transmembrane domains. Loops 1 and 3 are located in the lumen of endoplasmic reticulum (ER), whereas loop 2 and the N- and C-termini are cytosolic.51,53 Nsp4 is the N-terminal-most ORF1ab protein, which shares greater than 50% similarity and identity with other group of corona viruses, suggesting the importance of nsp4 protein function for viral replication.
Nsp3, 4, and 6 are predicted to function in the nucleation and anchorage of viral replication complexes on the double-membrane vesicles in the cytoplasm in SARS-CoV.11-13 The interaction of nsp3 with nsp4 is very crucial in the replication of SARS-CoV. 52 In SARS-CoV-2 as well, nsp4, along with other proteins like nsp3 and nsp6, assists to build double-membrane vesicles for the favorable replication inside infected cells. Parts of new viruses’ copies are assembled inside these vesicles. 24
Nsp4 demonstrated one of the lowest rates of mutations among proteins during the pandemic of 2020, with only one residue, M324, with mutation rate higher than 0.01. 32
Double-membrane vesicle factory:·nsp6
SARS-CoV nsp6 is approximately 34 kDa membrane protein comprising predicted six transmembrane helices with a highly conserved C-terminus. 54 It is a common component of both alpha and beta corona viruses and resides in the ER of the host cells. It has been found that the presence of multiple phenylalanine residues in the outer membrane region of nsp6 favors the affinity between this region and the ER membrane, satisfying a more stable binding of the protein to the ER. 55
All three proteins viz nsp3, nsp4, and nsp6 of SARS-CoV are required for the formation of double membranes. 56 The nsp3 induces membrane proliferation and disordering when expressed alone, and nsp4 induces single-membrane vesicles. Nsp3 and nsp4, however, have the ability to pair membranes when co-expressed. Nsp6 also possesses membrane proliferation capability, inducing perinuclear vesicles localized around the organizing center of the microtubule. This activity appears to require the full-length form of nsp3 for action, as double-membrane vesicles with nsp4 and nsp6 were not seen in cells co-expressing the C-terminal truncation nsp3.
Although nsp6 stimulated internal cellular rearrangement is observed in the presence of nsp3 and nsp4, nsp6 alone can also cause membrane proliferation. It was observed that nsp6 induces autophagy including induction of vesicles that contain Atg5 and LC3-II. 57
During the pandemic of 2020, nsp6 demonstrated a fair amount of mutations along the length of the protein. Six mutations—L37, L142, V149, M86, M143, and K270—demonstrated mutation rate higher than the threshold of 0.01. 32
Copy assistants: nsp7 and nsp8
Nsp7 is an alpha helical protein of about 10 kDa,14,58 which has a single domain with a novel fold that consists of five helical secondary structures. Nsp7 localizes to the cytoplasmic membrane. 58 The central core consists of an N-terminal helical bundle (HB), which contains helices HB1, HB2, and HB3. There are hydrophobic inter-helical side chain interactions that stabilize and hold the helices together. SARS-CoV nsp7 dimerizes and interacts with other proteins such as nsp5, nsp8, nsp9, and nsp13. 14 The HB region of nsp7 is conserved and is known to interact with nsp8.
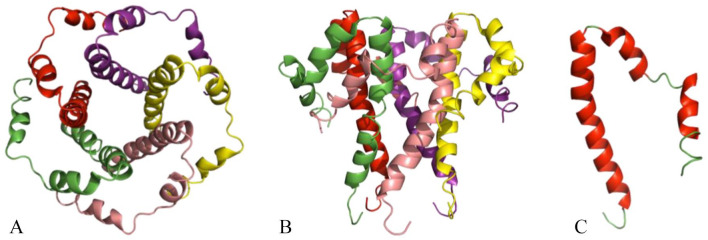
Nsp8 has a molecular mass of about 22 kDa and is unique for corona viruses. 14 The four monomers of nsp8 have different conformations: nsp8I and nsp8II. 58 Nsp8I is portrayed as a “golf club” like structure harboring an N-terminal “shaft” domain, which further contains three helices (NH1-3) and a C-terminal “head” domain, while nsp8II has a similar head domain, but the shaft helix NH3 bends into two shorter helices (Figure 7). The N-terminal is known to be highly conserved, suggesting that this domain may have a crucial role in the interaction with other molecules or complexes. 58 Nsp8 is unique to coronaviruses and was reported to be capable of synthesizing RNA only de novo with a low fidelity on ssRNA templates. There are stable interconnections of the golf club-like nsp8 molecules within the hexadecameric structure, which is unique in that it does not simply involve the stacking of its protein subunits. 25
Figure 7.
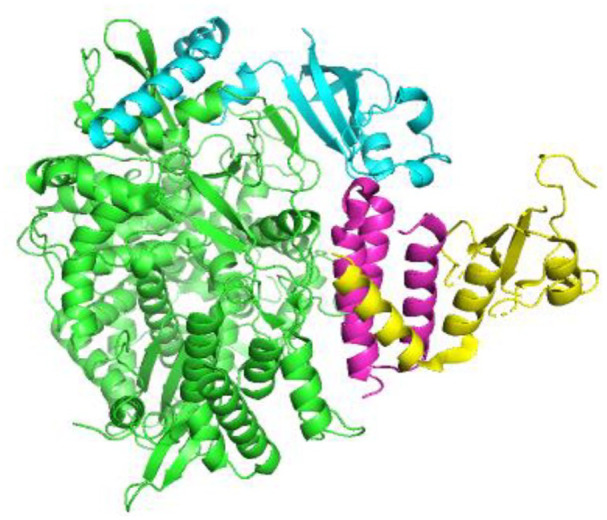
Crystal structure of RNA-dependent RNA polymerase of SARS-CoV-2 consisting of four chains represented by nsp12 (chain A), nsp8 (chain B, D), and nsp7 (chain C) in four different colors green, sky-blue, yellow, and purple, respectively (PDB 7W4Y).
The genome of SARS-CoV is assumed to contain two RNA-dependent RNA polymerase (RdRp) activities. The primary RdRp activity is associated with nsp12, whereas the secondary activity may reside in nsp8. The enzyme nsp8 is capable of initiating de novo and has been proposed to operate as a primase. Surprisingly, this protein was crystallized, only with the nsp7, forming a hexadecameric, dsRNA-encircling ring structure. The ring comprises eight nsp7 and eight nsp8 proteins. From this supercomplex, one can get an insight about the SARS-CoV transcription and replication machinery. Nsp8 is known to colocate with RdRp while copying the SARS-CoV genome. The crystal structure of the SARS-CoV hexadecameric nsp7-nsp8 supercomplex was solved to 2.4-A resolution by Yujia Zhai et al. This structure is believed to be the first to exhibit interactions between the coronavirus replication proteins. The supercomplex structure looks like a hollow cylinder with a central channel, which has two handles protruding from opposite sides. 59
The amino acid sequence alignment performed recently revealed that the nsp12 of SARS-CoV-2 shares 96.14% sequence identity with the nsp12 of SARS-CoV (Table 1). The comparative analysis of deduced amino acid sequences further reveals that nsp7 and nsp8 of SARS-CoV 2 shares 98.8% and 97.47% sequence identity with that of SARS-CoV, respectively.
Nsp7 and nsp8 remained largely unmutated during the pandemic of SARS-CoV-2 in 2020. Only one residue of Nsp7, M75, showed mutation rate higher than 0.01. 32
RNA-dependent RNA polymerase: nsp12
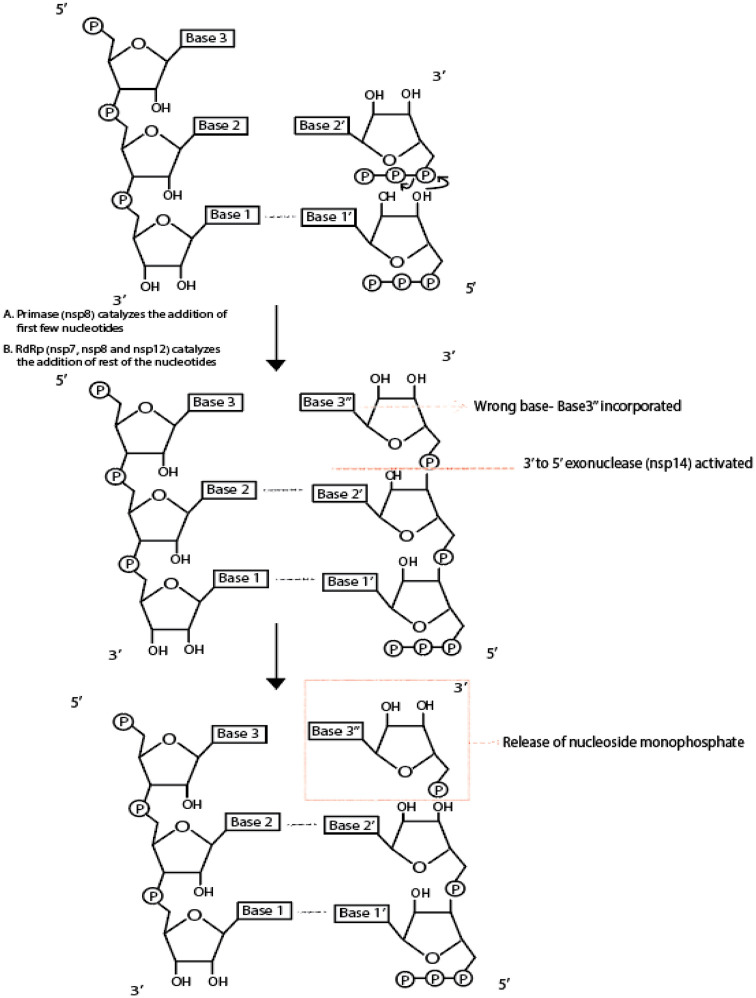
Made up of 932 amino acids, SARS-CoV-2 nsp12, an RNA-dependent RNA polymerase (RdRp), consists of polymerase domain resembling “cupped right hand” and an N-terminal domain. 60 Likewise, the SARS-CoV nsp12 has a “right hand” shaped fold that consists of characteristic fingers, palm, and thumb subdomains. The finger domain comprises three helices, one RdRp characteristic helix-loop-helix, and two-stranded β-sheets, whereas the palm domain consists of two helices and a β-hairpin that contains catalytic aspartates responsible for the nucleotide transfer reaction. 61 The index finger–thumb interaction site forms the nsp7-nsp8 heterodimer-binding site where most of the contacts between nsp12 and nsp7 are made. 27 The template entry site, template-primer exit site, nucleoside triphosphate (NTP) tunnels, and the polymerase-active site are found to be highly conserved across the Coronavirus family. 27 As shown in Figure 8, RdRp polymerizes ribonucleotides based on RNA template.
Figure 8.
RNA-dependent RNA polymerase activity and exonuclease proofreading activity. The figure shows, at a biochemical level, what reactions take place due to RNA-dependent RNA polymerase and 3ʹ to 5ʹ exonuclease.
Addition of the nsp7 and nsp8 co-factors greatly stimulates nsp12 activity. 62 Although other viral factors are necessary as well, the nsp12-nsp7-nsp8 complex is essential for nucleotide polymerization. 27 In the SARS-CoV-2 nsp12-nsp7-nsp8 complex crystal structure studied, nsp7-nsp8 heterodimer was found to interact with nsp12 on the polymerase thumb domain facing the NTP entry channel. The nsp12 polymerase index finger loop is present between the polymerase thumb domain and nsp7-nsp8. The binding is supposed to facilitate the interaction of nsp12 with other essential components of the RNA synthesis machinery for incorporation into viral replication complex. 27
The polymerase complex of SARS-CoV-2 highly resembles to that of SARS-CoV. It consists of an nsp12 core catalytic subunit bound together with an nsp7-nsp8 (nsp8.1) heterodimer with an additional nsp8, nsp8.2, subunit present at a different binding site. As in SARS-CoV polymerase complex, the nsp7-nsp8 heterodimer interacts above the thumb subdomain of nsp12. This interaction is predominantly mediated by nsp7, while nsp8 (nsp8I) contributes only a few interactions to the polymerase subunit nsp12. The other subunit nsp8 (nsp8II) grips the finger subdomain’s top region, and forms additional interactions with the interface domain. However, the SARS-CoV-2 nsp12-nsp7-nsp8 complex has a lesser efficiency (about 35%) for RNA synthesis particularly in comparison with its SARS-CoV counterpart, due to variations in the nsp8 subunit. The thermostability of nsp8 and nsp12 is also relatively lower owing to residue substitutions in the SARS-CoV-2 nsps. 63
During the pandemic of 2020, nsp12 was one of the most mutated proteins. One of the residues P323 was mutated at the rate of 0.996. Other mutated residues with mutation rate higher than 0.01 were V776, A185, E254, A656, T739, and V720. The most commonly mutated site P323 lies outside of the active site of the protein. It may, however, have a role to play in protein folding and interaction with nsp7 and nsp8. 32
Proofreading exonuclease: nsp14
Nsp14’s N-terminal contains an exonuclease (ExoN) domain,16,17 whereas the C-terminal carries an S-Adenosyl methionine-dependent guanosine N7-methyltransferase (N7-MTase) activity. Nsp14 of CoV is essential for viral replication and transcription (Figure 8).
Nsp14 MTase and nsp16 methylates the cap on a GTP guanine and C2ʹ hydroxyl of the nucleotide, respectively. 64 Moreover, one molecule of nsp10 interacts with nsp14 to stabilize and significantly enhance the enzymatic activities of nsp14. 29
ExoN domain has a proofreading role, which prevents lethal mutagenesis, whereas the C-terminal domain functions as a methyltransferase for mRNA capping. 29 While ExoN knockout SARS-CoV and mouse hepatitis virus were viable with an increased rate of mutation, ExoN knockout mutants of five other beta coronavirus, including SARS-CoV-2, were found to be nonviable. 65 Thus, in case of SARS-CoV-2, ExoN domain of nsp14 is believed to have an additional crucial function besides boosting the replication fidelity of the virus. ExoN is believed to be important in RNA synthesis, resistance of antiviral nucleoside analogues, fitness, immune antagonism, and virulence besides high fidelity replication. It has also been implicated to increase recombination, which is important in the evolution of viruses. 66
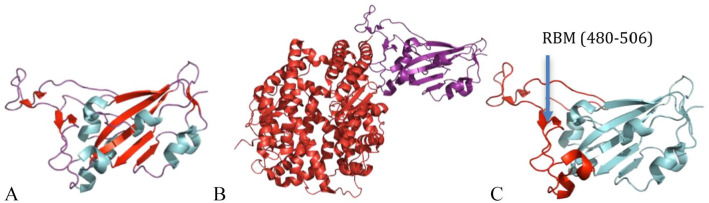
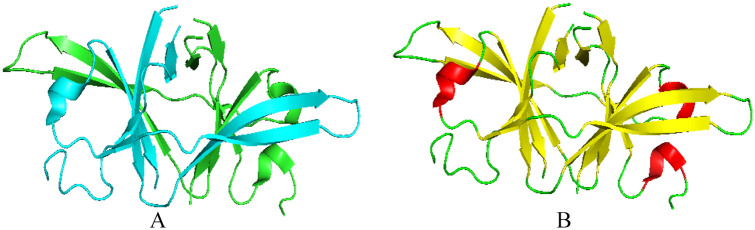
The crystal structure for the nsp10-nsp14 complex has been solved in SARS-CoV (Figure 9). The ExoN domain features a core and a twisted β-sheet consisting of five β-strands. Except for β3, the remaining strands form a parallel β-sheet that is flanked by α-helices along each side. One Mg2+ ion is observed in the active site. The N7-MTase domain of nsp14 exhibits an MTase fold in which the central β-sheet consists of five β-strands. β2′, β1′, β3′, and β4′ are parallelly positioned, whereas β8′ runs in an antiparallel position. Between strands β5 and β6 of the central sheet, a three-stranded antiparallel β2-sheet is present. A cavity between the β1 and β2 sheets serves as a ligand-binding pocket. Two small helices are present in the β1-sheet’s connecting loops, whereas α1′-helix is placed against the central β1-sheet opposite face. A long α2′-helix lies behind the α1′-helix. Nsp14’s zinc finger is located at the tip of this helix, which protrudes at its C terminal from the protein. 29
Figure 9.
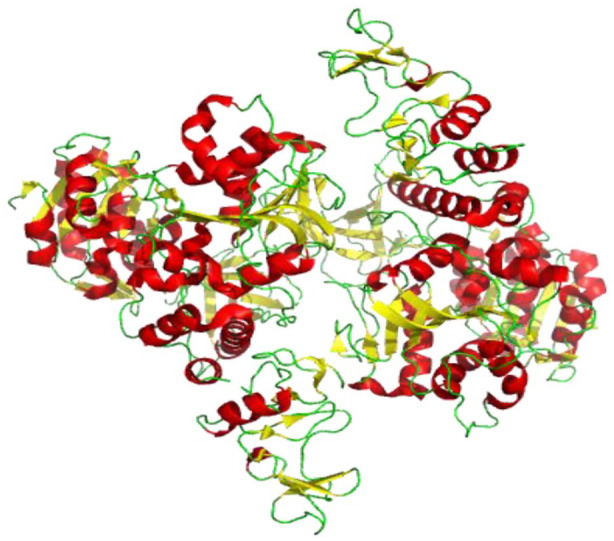
Cartoon representation of SARS-CoV nsp10-nsp14 complex structure showing loops (green), β-sheets (yellow), and helices (red) (PDB 5C8U). The nsp14 ExoN domain is stabilized by nsp10. The ExoN domain features a core, twisted β-sheet consisting of five β-strands and the N7-MTase domain consists of five β-strands.
Nsp14 showed very low rate of mutational variation during the pandemic of 2020. Only two residues, M501 and N129, showed mutational rates higher than 0.01. 32
Helicase: nsp13
The SARS-CoV-2 nsp13 (Figure 10), consisting of 596 amino acids, has a triangular pyramid shape comprising five domains: two “RecA-like” domains (1A and 2A), 1B domain, N-terminal Zinc binding domain (ZBD), and stalk domain, which connects ZBD and 1B domain. 1A, 2A, and 1B domains form the base of the pyramid, whereas ZBD and stalk domains form the apex. The SARS-CoV-2 nsp13 structure reported similar NTPase active site residues conservation as present in SARS-CoV nsp13, which contains the residues K288, S289, D374, E375, Q404, and R567. All of these residues were found to be concentrated in the cleft between domain 1A and 2B located at the base (Figure 10). 60
Figure 10.
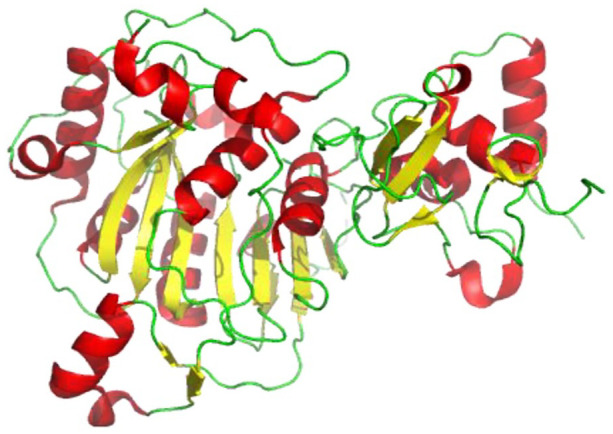
Cartoon representation of SARS-CoV-2 nsp13 structure showing loops (green), β-sheets (yellow), and helices (red) (PDB 6ZSL).
Twelve Cys/His conserved residues capable of binding at least three Zn2+ ions are present in the Zn2+ binding domain. The helicase domain has six conserved motifs among which two motifs are called the Walker A motif (GXXXXGK(T/S) containing a conserved Lys residue and the Walker B motif (R/K)XXXXGXXXXLhhhhDE) containing a Asp and a Glu residue. Lys in Walker A and Asp/Glu in Walker B participate in DNA/RNA helicases based ATP-hydrolysis. 67
The nsp13 helicase separates double-stranded RNA or DNA with a 5′→3′ polarity. SARS-CoV RdRp, nsp12, enhances the catalytic efficiency of nsp13 twofold by increasing the step size of nucleic acid unwinding. 28 Besides the helicase activity on double-stranded DNA and RNA, it is also capable of unwinding RNA/DNA duplex. Moreover, it has NTPase activity as well as 5′ mRNA capping activity. 8
An analysis of the mutation rate of nsp13 amino acid residues during the 2020 pandemic revealed that five residues, E261, K218, H290, K460, and A598, showed mutation rate greater than 0.01. 32
Methyltransferase stimulator: nsp10
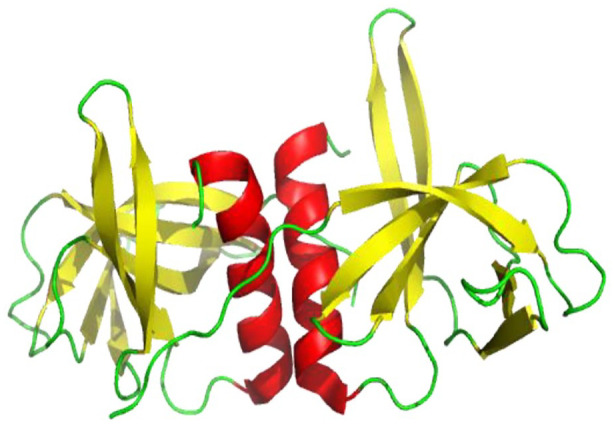
SARS-CoV-2 nsp10-nsp16 heterodimer crystal structure in complex with S-adenosylmethionine (SAM) has been solved. Nsp10’s hydrophobic surface that is positively charged interacts with a negatively charged, hydrophobic pocket at the nsp16 surface, thus stabilizing the SAM binding site. An antiparallel pair of β-strands surrounded by a crossover loop on one side and a helical domain with loops generating two zinc fingers on the other side is present in the center. In other coronaviruses, these structures are found to be involved in nonspecific RNA-binding. 68
SARS-CoV nsp10’s fold comprises 12 identical subunits giving rise to a spherical and hollow dodecameric architecture (Figure 11). From nsp10 monomer structure, two zinc fingers having sequence motifs C-(X)2-C-(X)5-H-(X)6-C and C-(X)2-C-(X)7-C-(X)-C have been identified. The dodecameric assembly of nsp10 has an outer diameter of 84 Å from where 12 C-terminal zinc fingers stick outward. On the other hand, the inner diameter is 36 Å and consists of the remaining 12 zinc fingers. 59
Figure 11.
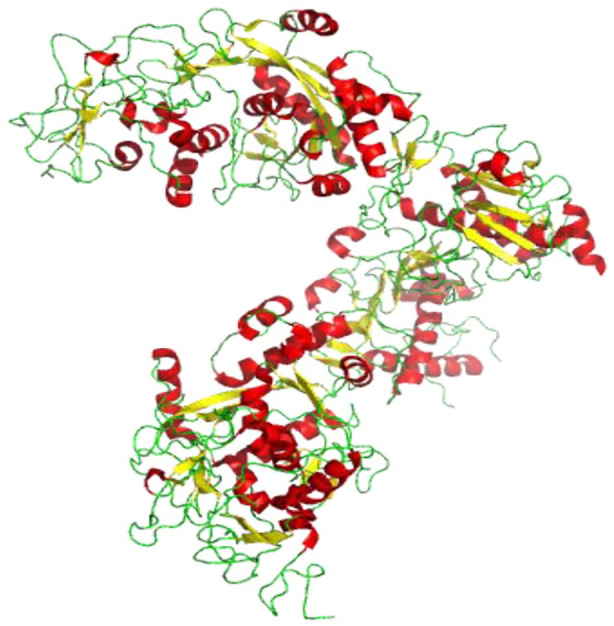
Cartoon representation of SARS-CoV-2 nsp10-nsp16 complex structure showing loops (green), β-sheets (yellow), and helices (red) (PDB 6W4 H). Nsp10’s positively charged and hydrophobic surface interacts with a hydrophobic pocket and a negatively charged nsp16 surface, which helps to stabilize the SAM binding site.
From sequence analysis, nsp10 is found to be related to the HIT-type zinc finger family. These are often found in nuclear proteins involved in gene regulation and chromatin remodeling. 59 Nsp10 interacts with nsp14 ExoN besides nsp16 to stabilize as well as stimulate its exoribonuclease activity. 29 In SARS-CoV, nsp10 acts as a stimulatory factor to execute nsp16’s S-adenosyl-L-methionine (SAM)-dependent methyltransferase (MTase) activity in which ribose 2′-O is methylated. Ribose 2′-O-methylation in the cap structure of viral RNAs is an integral step in viral escape from innate immune recognition. 20
Likewise, MERS-CoV nsp16, an S-adenosyl-L-methionine (SAM)-dependent 2′-O-methyltransferase (MTase), is believed to methylate the ribose 2′-OH of the first transcribed nucleotide of viral RNA cap structures, which is regulated by nsp10. 69 In SARS-CoV, nsp10 surface interacting with nsp14 and nsp16 was found to overlap with each other. Specific “hot spot” residues (F19, M44, G69, S72, H80, and Y96), within and around the nsp10 core, are highly conserved across coronaviruses, including MERS CoV. These residues can be targeted to inhibit nsp10 function. 70
Interestingly, none of the amino acids of nsp10 demonstrated mutation rate higher than 0.01 during the pandemic of 2020. 32
Methyltransferase: nsp16
As nsp16 is unstable in a variety of buffers and precipitates under various storage conditions, it has not been crystallized yet. 71 However, crystal structure of nsp10-nsp16 (Figure 10) heterodimer complexed with SAM has been solved in SARS-CoV-2. Nsp16 structure in the complex comprises 298 residues containing Rossmann-like β-sheet fold surrounded by 11 α-helices, 7 β-strands, and loops in the 2ʹ-O-MTase catalytic core. The SARS-CoV-2 nsp16 fold is constructed by a β-sheet featuring the canonical arrangement 3-2-1-4-5-7-6, in which β7 is the only antiparallel strand. Loops and α-helices sandwich this β-sheet. The catalytic core interacts with one SAM molecule near the Rossmann-like fold’s β1 and β2 strands, and various other loops form the negatively charged binding SAM binding pocket. 68
SARS-CoV nsp16, despite having SAM-dependent methyltransferase fold, does not exhibit this enzymatic activity alone.72,73 SARS-CoV nsp16 is only active in the presence of its stimulating partner nsp10. 19 The 2′-O-MTase encoded by SARS-CoV is composed of two subunits; the catalytic subunit nsp16 and the activating subunit nsp10. Nsp10 helps nsp16 to bind capped RNA substrate as well as the methyl donor SAM. 20
Likewise, MERS-CoV nsp16 is believed to methylate the ribose 2′-OH of the first transcribed nucleotide (N1) of viral RNA cap structures. Nsp16 recruits N7-methylated capped RNA and SAM which promotes the assembly of the enzymatically active nsp10/nsp16 complex. This complex converts 7mGpppG (cap-0) into 7mGpppG2′Om (cap-1) RNA by 2′-OH methylation of N1. 31
Only one amino acid residue, R216, of nsp16 demonstrated mutation rate higher than 0.01 during the 2020 SARS-CoV-2 pandemic. 32
Endonuclease: nsp15
Nsp15 cleaves 3ʹ of uridines in a manganese dependent manner. 30 This is believed to be an important way for the virus to hide from antiviral defense. Nsp15 cleaves polyuridines from the 5ʹ terminus of the negative strand of viral RNA. These polyuridines serve as pathogen associated molecular patterns for recognition by the host defense system. By getting rid of such polyuridines, the virus can evade the host immune system. 8
Nsp15 can be crystallized as a hexamer and was found to be competent for RNA binding (Figure 12). SARS-CoV-2 nsp15 generates dimers of trimers, which finally assembles into a hexamer. In SARS-CoV-2, each subunit of nsp15 contains 10 α-helices and 21 β-strands. The N-terminal domain is composed of an antiparallel β-sheet wrapped around two α-helices (α1 and α2). The middle domain is formed by 10 β-strands organized in three β-hairpins, a mixed β-sheet and three short helices, two α and one 310. The C-terminal catalytic endonuclease domain contains two antiparallel β-sheets and consists of a catalytic site.
Figure 12.
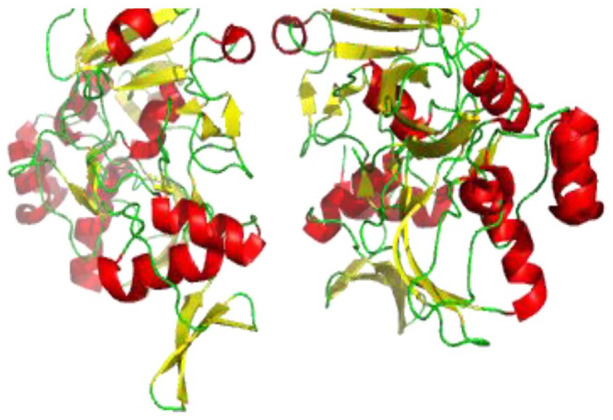
Cartoon representation of SARS-CoV-2 nsp15 structure showing loops (green), β-sheets (yellow), and helices (red)(PDB 6VWW). SARS-CoV-2 nsp15 generates dimers of trimmers, which finally assembles into a hexamer where each subunit of nsp15 contains 10 α-helices and 21 β-strands.
The SARS-CoV-2 nsp15 endonuclease oligomers were found to resemble those of SARS-CoV, H-CoV-229E, and MERS-CoV homologs. Monomeric folds of SARS-CoV-2 hexamer show higher (88%) similarity to that of SARS-CoV than to H-CoV-229E and MERS-CoV.
SARS-CoV-2 enzyme most likely operates in a manner identical to their SARS-CoV, H-CoV-229E, and MERS CoV homologs. Nevertheless, differential catalytic properties and potentially altered substrate specificity might be present in case of homologs. The hexamer is stabilized by the interaction of N-terminal oligomerization domains, but each subunit domain also contributes to the oligomer interactions. 74
In SARS-CoV, each subunit of nsp15 contains 9 α-helices and 21 β-strands. Every subunit is further organized into three domains; an N-terminal domain, a middle domain, and a C-terminal domain. 75 From alanine substitutions of highly conserved residues, it is interpreted that the C-terminal domain contains the active site, 76 which faces away from the center of the hexamer and contains the extreme C-terminal residues. From the top of the hexamer, a pore through the trimer is present, where N-terminal domains of the trimer line the bottom of the pore. This pore was found to have an inner diameter of ∼12 Å and did not interact with the substrate RNA. 75
During the 2020 pandemic, this protein was found to mutate at a moderate frequency. Four amino acid residues, K13, T34, R207, and T115, demonstrated mutation rate higher than 0.01. 32
RNA-binding protein: nsp9
SARS-CoV nsp9 is an ssRNA-binding protein which features an oligosaccharide or oligonucleotide binding fold. 26 It is implicated in virulence of the virus. 8
Like any other nsp9 homolog, the crystal structure of SARS-CoV-2 nsp9 (Figure 13) also consists of a fold unique to coronaviruses, the core of which is made up of a six-stranded enclosed β-barrel and a series of extended loops projecting outward from it. The individual β-strands of the barrel, C-terminal α helix, and N-terminal β-strand are linked by the loops. The inter-subunit interactions that form a dimer is due to van der Waals interactions between the interfacing copies of α1 helix C-terminal as a result of self-association of GxxxG protein-protein binding motif. 77 In SARS-CoV-2, nsp9 was observed to change the behavior of nsp8. 78
Similarly, crystals of SARS-CoV nsp9 also contain a dimer in the asymmetric unit. In each monomer, there are seven strands and one helix arranged into a single compact domain, which form a cone-shaped-barrel flanked by the C-terminal helix. The helix has two hydrophobic sites, one of which faces the barrel and the other interacts with the helix of the second monomer. This dimer is assembled by hydrophobic interactions and further stabilized by four long hydrogen bonds involving main-chain atoms. 26 The sequence identity of HCoV-229E nsp9 and SARS-CoV nsp9 is 45%. HCoV-229E nsp9 dimerization occurs via disulfide formation. Nevertheless, C69, the residue responsible for disulfide formation in HCoV-229E nsp9, is conserved in SARS-CoV nsp9. 79 Furthermore, HCoV-229E nsp9 and tRNA interacts nonspecifically. 80
During the pandemic of 2020, nsp9 of SARS-CoV-2 showed very little mutation. Only one residue, M101, underwent mutation higher than 0.01. 32
Structural Proteins
There are four structural proteins of SARS-CoV: Spike (S) protein, Envelope (E) protein, Nucleocapsid (N) protein, and Membrane (M) protein. They form the structural components of virions. The structure and function of these proteins are discussed in detail in the following section (Table 3).
Table 3.
Structure and function of structural proteins.
| Name of the protein | Structure solved (SARS-CoV-2) | Structure solved (SARS-CoV) | Description of the structure | Function |
|---|---|---|---|---|
| Spike (S) protein | SOLVED: S1 and S2 subunits of the S protein. Solved for: SARS-CoV-2 via Cryo-EM |
Solved for: SARS-CoV (Cryo-EM) | It forms homotrimer protruding from the viral surface. There are two subunits: S1 and S2. The S1 subunit houses the receptor-binding domains at the distal end. | It causes anchorage with a host cell receptor before merging. It is also involved in viral entry into the host cell. 81 |
| Nucleocapsid (N) protein | Solved for SARS-CoV-2-RNA binding domain PDB:6M3M (X-Ray Crystallography) |
Solved for SARS-CoV PDB:2OFZ PDB:2OG3 (X-Ray Crystallography) |
SARS-CoV-2-N-NTD crystal shows orthorhombic crystal packaging mode in which there are four monomers in one asymmetry unit. Each monomer in one asymmetric unit has the same type of right handed (loop)-(β-sheet core)-(loop) forming a sandwich structure. It consists of five antiparallel β-strands with a single short 310 helix and a sticking out β-hairpin between β2 and β5 strands, this structure as a whole contributes to form a β-sheet core. The structure of SARS-CoV-2-N-NTD looks similar to the hand shape (fingers, palm and wrist). | Nucleocapsid Protein (N) is believed to have multifunctional activities like forming helical ribonucleoprotein (RNP) complex during packaging the RNA genome, during replication and regulating viral RNA synthesis, transcription and regulating infected cell metabolism.82-85 The N protein protects the RNA of the virus by keeping it intact and stable within the virus. N proteins wrap and coil the RNA in long helical structure. |
| Membrane(M) protein | Not solved | Not solved | The M protein consists of three transmembrane domain which are flanked by a short glycosylated amino-terminal domain and a long carboxy-terminal tail (Cytoplasmic domain) outward and inward within the viral envelope, respectively | M protein plays a vital role in the assembly of viruses through protein-protein interactions: M-nucleocapsid (N), M-M and M-spike(S) interaction. 86 |
| Envelope (E) protein | NOT SOLVED: For SARS-CoV-2 | SOLVED: For another SARS-CoV via NMR | For SARS-CoV, two distinct subunits have been identified: hydrophobic domain with a transmembrane domain (TMD) and a charged cytoplasmic tail. | It reaches heavy localization at sites of intracellular transport, Viral assembly and budding at ER, Golgi complex ERGIC.87,88 It leads to formation of ion conductive pore in the membrane of the virus.89-92 |
Abbreviations: ER, endoplasmic reticulum; ERGIC, endoplasmic reticulum-Golgi body intermediate compartment; NMR = nuclear magnetic resonance; RNA, ribonucleic acid; RNP, ribonucleoprotein; TMD, transmembrane domain.
S protein
S or Spike proteins are structural glycoproteins that are involved in host-virus interactions during the viral entry into the host cell. 93 They facilitate the attachment of the viral particle to the host cells to mediate membrane fusion and viral entry into the host cell. The S proteins are exposed on the surface of the virus, making them one of the main targets of drug design and neutralization antibodies. 94 Thus, they are under extensive study for therapeutics and vaccine design.
S proteins consist of two subunits called S1 and S2. S1 binds with host cellular receptors while the S2 subunit facilitates the membrane fusion of the virus with the host cell. Most coronaviruses have S1 and S2 cleaved with only noncovalent bonding between them. SARS-CoV-2 shows an exception to this via having a noncleaved furin cleavage site between the S1 and S2 subunits that are then cleaved during the biosynthesis of the virus.95,96 All Coronaviruses activate membrane fusion when the S is cleaved by a host enzyme at S2-prime region via extensive irreversible changes in their structural conformation. 97 Different coronaviruses use different distinct domains in the S1 region for host receptor binding. For example, in case of the MERS-CoV the A domain is used for recognition of host receptors, in this case the human nonacetylated sialoside attachment receptors, which promotes the S-B domain allowing viral entry. 81 SARS-CoV and most SARS-related viruses show direct interaction of their S-B domain with angiotensin-converting enzyme 2 (ACE2). 98
Different coronaviruses interact to their respective host receptors using different domains with the S1 subunit. SARS-CoVs are known to interact directly with the human ACE2 or (hACE2) using their S-B domain. Ongoing research on SARS-CoV-2 suggests that it also uses similar mechanisms. Studies have shown that SARS-CoV-2 uses its S-B domain to interact with human ACE2 (hACE2) in a similar manner to the S protein of SARS-CoV seen during the 2002-2003 SARS outbreak. 99 S glycoproteins of SARS-CoV-2 have shown 76%, 80%, and 80% sequence identity with SARS-CoV S Urbani, Rinolophus sinicus (Chinese horseshoe bats) SARSr CoV ZXC21 S, and Rinolophus sinicus ZC45 S glycoproteins, respectively. 100 SARS-CoV-2 was also shown to have high sequence identity of S glycoprotein to another bat coronavirus called SARSr CoV RaTG13 at 97% sequence identity. 101 ACE2 was identified as potential SARS-CoV-2 receptor via comparison of samples of the virus that were cultured in HeLa cell line with and without ACE2 expression. 94
The SARS-CoV-2 S protein receptor-binding domain (RBD) is composed of a receptor-binding motif (RBM) which interacts with the hACE2 and a core made of interconnected loops and helixes (Figure 14). 102 Within the S proteins, SARS-CoV and CoV-2 share about 75% identity in S-B domain and 50% identity in RBMs within the B domain. 103 The RBM of SARS-CoV-2 S protein S-B domain is stretched from amino acids 438 to 506. 102 A peculiar feature seen in the S protein of SARS-CoV-2 is the presence of four amino acids (P,R,R,A) in the gap between S1 and S2 subunits. These amino acids form a furin cleavage site, which is not present in other SARS-related CoVs, not even in the closely related RaTG3 strain. So far, experimental results have not shown any advantage of this site during viral entry. Possible advantage of this site could possibly be in expanding the SARS-CoV-2’s tropism and transmissibility. 96
Figure 14.
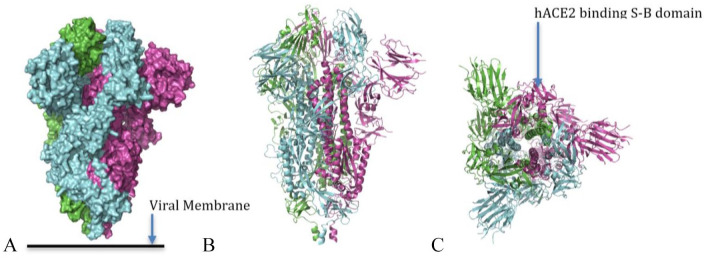
Surface representation of a closed trimer of SARS-CoV-2 S protein (A) composed of three chains shown in purple, green, and cyan. A cartoon representation of the side view of SARS-CoV-2 S trimer, (B) cartoon representation of top view of the SARS-CoV-2 trimer (C) showing the (closed) hACE2-binding S-B domain (pdb 6VXX) 99
S proteins in coronavirus exist as homotrimers protruding from the viral surface giving it the characteristic “crown” from which its name is derived (Figures 14 to 16). The S1 subunit of the S protein houses the RBDs at the distal end (Figure 14). It stabilizes the S2 subunit in its prefusion state. The S2 subunit houses the membrane fusion machinery and exists as a trimer (Figure 15). 105 Immediately upstream of the region involved in fusion, Coronaviruses have a region called S2-prime that is cleaved by the host protease. This cleavage leads to the activation of S2 protein activity leading to fusion between viral capsid and the host cell membrane. 106 Cryo-EM studies on the prefusion region stabilized in ectodomain trimer constructs for the SARS-CoV-2 S glycoprotein, with an abrogated furin S1/S2 cleavage site, show that the S-B domain in the S1 subunit can exist in different distinct organizations resulting in S proteins with multiple conformational states. The furin site may also help to explain the greater transmissibility of SARS-CoV-2 compared with SARS-CoV due to the fact that expression of furin-like proteases is quite common. This also explains the increase of tissue tropism in SARS-CoV-2 and its differences in pathogenicity. 107 The CoV-2 ectodomain, the parts extending outward of the viral particle, was determined to be a 160 Angstrom long trimer with a triangular cross section, closely resembling the structural binding of the SARS-CoV ectodomain. Superimposing CoV-2 and CoV S2 subunit structures gives a 1.2 degree-A root mean square deviation (rmsd) over 417 aligned C-alpha positions. The SARS-CoV 2 structure was constructed only for 27 to 1147 residues segment. This result combined with the 88% sequence similarity shows that S2 subunits of both SARS-CoV and CoV2 have significant structural similarity. Given the importance of S proteins in both the coronaviruses, this level of structural similarity of S2 subunit between the two viruses shows that antibodies targeting the SARS-CoV S2 subunit can also be expected to neutralize SARS-CoV-2 virus S proteins as well. 99 S glycoprotein’s trimers show densely protruding heterogeneous N-linked oligosaccharides on the trimer surface. They participate in S protein folding and priming of S protein. It may also be important in antibody recognition. 108 The SARS-CoV-2 S protein shows 22 such glycosylated protuberances per protomer. Comparisons with S proteins of CoV show that in S1, 9 out of 13 glycosylation sites are conserved, while in S2, all 9 are conserved for SARS-CoV-2. 109
Figure 16.
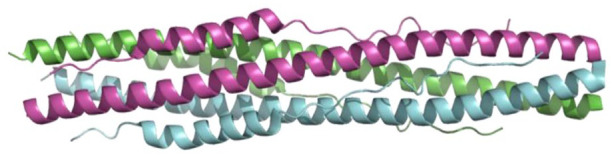
A cartoon representation showing a trimer of the S2 subunit of SARS-CoV-2 S protein (PDB 6LXT). 104
Figure 15.
A cartoon representation of hACE2-binding S-B domain (A) of SARS-CoV-2 S protein. A cartoon representation of SARS-CoV-2 S-B domain (purple) bound with an hACE2 (red) (B). A SARS-CoV-2 S-B domain (C) showing receptor-binding motif (RBM) comprising amino acids 438 to 506 in red and the core in cyan (pdb 6M0J). 102
Research on SARS-CoV S proteins have identified 14 positions in S-B domain which are important for viral binding to hACE2: T402, R426, Y436, Y440, Y442, L472, N473, Y475, N479, Y484, T486, T487, G488, and Y491. 110 Sequence analysis shows us that only 8 out of these 14 positions are fully conserved in SARS-CoV-2 S-B domain, with the other 6 positions showing substitutions. 99 In a study that used flow cytometry to measure the binding of SARS-CoV-2 and SARS-CoV’s RBD with hACE2 protein, it was found that the binding affinity between SARS-CoV-2 RBD and the hACE2 expressed in 293 T cells was greater than that of SARS-CoV RBD. 111 Another study by Yan et al 112 also found that the amino acid sequence for the RBD of SARS-CoV-2 shows some variations that may lead to greater binding affinity with hACE2 when compared with SARS-CoV RBD. These facts make the SARS-CoV-2 RBD an important target protein for vaccine and therapeutics design against COVID-19 and may help to explain its greater transmissibility.
HEK 293 T cells expressing hACE2 were studied for viral infection mechanism, and it was shown that the entry mechanism of SARS-CoV-2 into the host cell is via endocytosis. Phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) inhibitor studies have shown significant reduction of SARS-CoV-2 entry in cells proportional to the dose of the inhibitor applied. PIKfyve is the main enzyme synthesizing phosphatidylinositol-3,5-bisphosphate (PI(3,5)P2), an important phosphoinositides compound for endocytosis This highlights PIKfyve as another potential drug target for COVID-19. PI (3,5)P2 has two major downstream effector proteins: two pore channel subtype 2 (TPC2) and TRPML1. Inhibitor studies on both have shown different results: While TPC2 inhibition shows reduction in CoV-2 entry, inhibition of TRPML1 does not.
Another important step in coronavirus entry is the host protease activation of S protein. Previous studies during the SARS and MERS outbreaks have identified cathepsins as critical for viral entry. Studies using cathepsin inhibitors have shown that they are equally important for S priming for SARS-CoV-2 as in the SARS-CoV and MERS virus. Studies with HEK 293T cells expressing the SARS-CoV S protein show that expression of type II membrane serine proteases (TMPRSS) 2, 4, 11 A, 11D, and 11E enhance the cell-cell fusion of the S protein expressing cells and the control 293T cells. Thus, TMPRSS, being an important protein in viral entry of host cells for SARS-CoV, makes it another class of host protease that is a suitable target against COVID-19 due its possible involvement in activation of S protein in SARS-CoV-2.
During the pandemic of 2020, S protein of SARS-CoV-2 was the most frequently mutated protein. Amino acid positions, with mutation rates higher than 0.2, were D614, A222, and L18. There were a total of 21 amino acids positions with mutation rate higher than the threshold of 0.01. The residue with the highest mutation rate, D614 is located at the surface of the protomer. It is used to establish hydrogen bond with the adjacent protomer. This mutation could provide the protein with higher flexibility. As the residue changes ionizable property of the protein, it could also affect the pH response of the virus. Both D614 and A222 residues are potential sites of B cell epitope recognition. Changing these sites could help the virus evade the B cell response. N439, Y453, and N501 residues are located in the receptor-binding domain and their alteration could alter binding to the receptor ACE2. 32
N protein
Corona virus nucleocapsid protein (N) is important in forming a helical ribonucleoprotein (RNP) complex with the RNA genome. It may also have a role to play in replication and regulation of viral RNA synthesis, transcription, and regulation of infected cell metabolism.82-85 The N protein shields the viral ribonucleotides and helps the RNA be stable inside the virus.
N protein is a capsid protein whose primary function is to protect the genomic RNA by packaging it. N protein does so by first recognizing the genomic RNA, combining itself to genomic RNA and forming capsid by self-associating into the oligomer. 113 N protein has multifarious roles. It can manipulate the host cellular machinery, which is very important in corona virus life cycle. N protein of coronavirus can also deregulate the host cell cycle by inhibiting the S-phase processes. 114 - 116 It can downregulate the gene products such as cyclin E and cyclin dependent kinase (CDK2) which are expressed during the S phase of the cell cycle. 113 In addition, N protein can inhibit the production of interferon (IFN). 113 Although SARS-CoV infection does not produce INF, study suggests that cells which have pre-induced INF could prevent the SARS-CoV infection.117,118 In addition, N protein can upregulate the production of cyclooxygenase-2 (COX2) protein, which is the most important proinflammatory element induced during the infection of corona virus. 119
Various studies have demonstrated that the N protein combines to the leader RNA and is indispensable for maintaining extremely organized RNA conformation for replication and transcription of the viral genome.84,120 Further experimental studies demonstrated that N protein is involved in controlling host-pathogen interactions, progression of host cell cycle and apoptosis.116,121,122 Indeed N protein is extremely immunogenic and expressed in large amount that can trigger defensive immune responses against SARS-CoV-2 and other corona viruses.123-126
The nucleocapsid (N) protein is encoded from the 3ʹ end of structural ORF of SARS-CoV-2. The gene sequence that encodes this protein lies between the 28 274 and 29 533 nucleotides and is 1260 nucleotides in length with Transcription Regulatory Sequence (TRS) located from 28 254 to the putative start codon (AUG). 1 The protein is composed of 419 amino acids, which is 3 amino acids less than SARS-CoV nucleocapsid protein (422 amino acids long). 4
The crystal structure of SARS-CoV-2-N-NTD (N-terminal RNA-binding domain) was solved using X-ray crystallography at 2.7 Angstrom. 127 The position of N protein from 47 to 173 amino acid residues, which is mainly the region of RNA-binding domain, was cloned, expressed, and purified. The SARS-CoV-N-NTD structure was used as an exemplar in order to determine the structure of SARS-CoV-2-N-NTD.
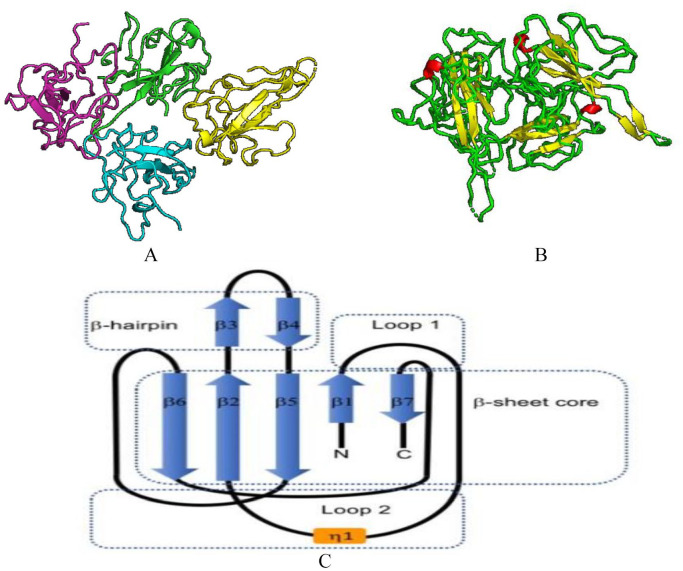
SARS-CoV-N-NTD crystal structure shows a monoclinic form of packing mode (PDB:2OFZ) and a cubic form of packing mode (PDB: 2OG3). 128 But the SARS-CoV-2-N-NTD crystal structure is somehow different; it shows an orthorhombic crystal packaging mode in which there are four monomers in one asymmetry unit. The difference in the crystal packing may contribute to other different contacts in the formation of ribonucleoprotein complex in SARS-CoV-2 from SARS-CoV. 127 In SARS-CoV-2-N-NTD crystal structure (Figure 17A), each monomer in one asymmetric unit has the same type of right handed structure (loop)-(β-sheet core)-(loop) forming a sandwich structure which is mostly conserved in corona viruses 127 (Figure 17B and C). 127 As shown in the figure (Figure 17C) SARS-CoV-2-N-NTD forms five antiparallel β-strands with a single short 310 helix and a β-hairpin sticking out between β2 and β5 strands. This structure as a whole contributes to the formation of a β-sheet core. Through mutational analysis, the hairpin was shown to be involved in RNA binding. 129 Structure of SARS-CoV-2-N-NTD looks similar to the hand shape (finger, palm and wrist). The finger region is basic and extends far beyond the β-sheet core, while the palm is basic and the wrist acidic. These structures form by folding of many aromatic and basic residues. 127
Figure 17.
Structure of SARS-CoV-2 nucleocapsid protein: (A) Structure of SARS-CoV-2-N-NTD (Cartoon representation), monomers in one asymmetric unit. There are a total of four monomers, which are represented by different colors (PDB 6M3M). (B) structure of SARS-CoV-2-N-NTD (Cartoon representation) showing green loops, yellow β sheets, and red 310 helices (η) (PDB 6M3M). (C) Structure of SARS-CoV-2-N-NTD showing (loop)-(β-sheet core)-(loop) with β-hairpin sticking out from β2 and β5 regions. 127 Here η1 represent the 310 helix.
The RNA-binding domain of SARS-CoV-2-N-NTD region was compared with HCoV-OC43-N-NTD region. HCoV-OC43 is a coronavirus that causes mild cold symptoms. Ribonucleoside 5ʹ-monophosphate binding mechanism in N protein is only thought to be present for HCoV-OC43-N-NTD. 130 HCoV-OC43-N-NTD structure suggests that corona viruses N protein consist of AMP, GMP, CMP, and UMP binding site alongside the middle of two β-strand of its β-sheet core. 130 This site in HCoV-OC43-N-NTD consists of phosphate group binding region, nitrogenous base binding region, and ribose pentose sugar 2’-hydroxyl group binding region. The phosphate group is bound by R112 and G68 via ionic interactions. Hydrophobic residues, consisting of F57, P61, Y63, Y102, Y124 and Y126, form a pocket, which recognizes and fits the nitrogenous bases. Y124 is mostly involved in interaction via the π-π stacking forces. S64 and R164 interacts with the pentose ribose sugar 2ʹ-hydroxyl group. 127
There are several differences in the structure of N-NTD in SARS-CoV-2 and HCoV-OC43. Kang et al tried to elucidate the contradictory mechanism of RNA binding by superimposition analysis that might be essential to discover the SARS-CoV-2-N-NTD target agents. By superimposing SARS-CoV-2-N-NTD with HCoV-OC43-N-NTD-AMP, complex the structural information of SARS-CoV-2-N-NTD ribonucleotide-binding site was revealed. This superposition analysis showed three distinct differences between these two structures. There is a difference in N-terminal tail of N-NTD with sequence variation; SARS-CoV-2 N-terminal tail residues are N48, N49, T50, and A51, whereas HCoV-OC43 N-terminal tail residues are V60, P61, Y62, and Y63. Furthermore, SARS-CoV-2 N-terminal tail region seems to be extended outward. This extension favors the movement of the N-terminal tail, which indeed changes the surface charge distribution, which makes the nucleotide-binding cavity easily available. On the other hand, this tail region is folded up to occupy a nitrogenous base-binding site in HCoV-OC43. There are also differences between the phosphate group binding sites. SARS-CoV-2-N-NTD has a larger side chain with residues T55 and A56, compared with HCoV-OC43-N-NTD side chain with S67 and G68 residues. T55, A56 residues cause additional polar properties on phosphate group binding site. Due to these differences, SARS-CoV-2-N-NTD has increased steric clash with ribonucleotide phosphate moiety. The last difference is that there is R89 residue on the edge of nitrogenous base recognizing the hydrophobic pocket in SARS-CoV-2-N-NTD but in the case of HCoV-OC43-N-NTD, Y102 is present. It is believed that there is an increase in polar properties and decrease in nonpolar properties in the nitrogenous base-binding site of SARS-CoV-2-N-NTD. 127
These distinct differences in the structure of SARS-CoV-2-N-NTD might be the reason why specific inhibitors are required to target SARS-CoV-2 N protein.
During the pandemic of 2020, N protein was the second most mutated protein in SARS-CoV-2 proteome after S protein. Even though the protein is only 419 amino acids long, there are 16 mutations with rate higher than the threshold of 0.01. The most common mutations—R203, G204, A220, D3, S235, S194, M234 and A376—do not lie in the crystallized region of the N protein. 32
E protein
E proteins of coronaviruses are another class of the structural proteins of the virus that form its outer capsid. They are short proteins of 76 to 109 amino acids in the size range of 8.4 to 12 kDa. 131 E proteins play important roles in the coronaviruses during infection like morphogenesis and pathogenesis and have been shown to be important for the virulence factor of the virus in mouse model studies. 132 They are also known to form ion channels on the virus surface. Given their importance in viral assembly and infectivity, their study is equally important among other structural proteins. 133
The structure of SARS-CoV E protein has been solved and well studied. The structure of SARS-CoV-2 E protein is yet to be solved but given its similarity in function, we can learn much from the E protein of the SARS-CoV. In the SARS-CoV two distinct structural subunits have been identified: a large hydrophobic domain and a charged cytoplasmic tail. The hydrophobic domain consists of a transmembrane domain (TMD) of 25 amino acids. The ion channels are predicted to form via oligomerization of alpha helixes contained in the TMD. The hydrophobicity of TMD comes from an abundance of valine and leucine residues, both nonpolar and neutral amino acids, in its sequence (Figure 18).89-92
Figure 18.
Top and side view, repectively, of a SARS-CoV E protein pentameric ion channel (A and B) in ribbon diagram represntation. Ribbon diagram representation of a single monomer of E protein (C) that forms the ion channel pentamer (pdb 5X29).
Coronavirus, during its assembly, has a distinct localization within the host cell in the ER-Golgi body intermediate compartment (ERGIC). 87 They bud into the ERGIC from where the host secretory system is expected to release the newly formed virus from the cell. 88 The localization of E protein in the virus assembly is mainly in the ER and the Golgi body. No concrete results have been found on which region of the E protein is responsible for the targeting of ERGIC for localization. 87 Studies indicate that it might be present in both the N-terminus and C-terminus for the SARS-CoV E protein. C-terminus is expected to contain the most information on Golgi targeting with additional information in the N-terminus of the protein.134,135 Most of these localization studies were conducted using epitope tagged E proteins. Concerns over the effect the tag has on the localization of the protein in the cell have been raised regarding these results. Till now no significant evidence has emerged supporting this concern.136,137
E protein also undergoes several post-translational modifications, which affect their subcellular trafficking and protein-protein interaction. One such modification is palmitoylation, which has also been seen in SARS-CoV. This modification occurs at the cysteine of the TMD in the protein. Experiments with mutated E protein that block this modification show protein instability and reduction of viral load indicating that palmitoylation is important for viral assembly. 138 Uniquely, SARS-CoV E protein, unlike other coronavirus proteins, also undergoes ubiquitination. CoV nsp3 interacts with E protein via ubiquitination of the N-terminal of nsp3. This modification has negative correlation with the protein’s stability and half-life.139,140
One of the most well characterized protein-protein interactions among coronaviruses proteins are the interactions between E and M proteins 141 Occurring in the ERGIC between the C-termini of the two proteins, the reduction in virus-like proteins (VLPs) when the C-terminus is removed shows the importance of this interaction for viral assembly.132,134,141,142 Ion channel formation via homo-oligomerization of E proteins due to interaction with itself is another known coronavirus protein-protein interation. 143 We can infer the importance of the TMD in oligomerization of E proteins via experimental results of research on synthetically created TMD sequences of E protein which are capable of forming dimers, pentamers, and so on which may help in formation of the ion channels. 144 Various residues within the TMD have been suggested to be important for these interactions. Some of them are from arginine 15 to the next alanine and valine 25 to the next phenylalanine. Mutations in these residues show disruption in oligomerization and channel formation by the E proteins. 145 C-terminus interaction between E protein and the N protein is also known to occur. But this interaction is not well understood and their effect on viral assembly is still largely unknown, though research with co-expression of E, N, and M proteins have shown to cause higher viral loads.146,147 In SARS-CoV, the E protein also interacts with the S protein via the TMD of the E protein which has three cysteine residues that interact with three cysteines in the C-terminus of the S protein.91,93
Five host cell proteins have been found to interact with the E protein: e. Bcl-xL, PALS1, syntenin, sodium/potassium (Na+/K+) ATPase α-1 subunit, and stomatin. Interaction of these proteins to the E protein may be the origin of various symptoms observed in the patients during the SARS outbreak like lymphopenia, inflammatory cytokines, breach of alveolar wall, and disruption of epithelial sodium channels.136,148-151.
During the 2020 pandemic, E protein showed very low mutation frequency with no residues with higher than 0.01 mutation rate. 32
M protein
Membrane (M) proteins of SARS-CoV-2 and other corona viruses play vital roles in the assembly of viruses through protein-protein interactions—M-nucleocapsid (N), M-M, and M-spike (S) interactions. 86 During viral infection, it has been suggested that the M protein binds to the viral S protein and the host surface receptor(s), which promote membrane fusion.152,153 This protein results in antigenicity displayed by the virus to the host’s immune response. As the most abundantly found protein in the coronavirus virions, the M protein may also be one of the vital components of viral assembly and morphogenesis, contributing to the regulation of replication and packaging of genomic RNA into viral particles.154,155
Membrane M glycoprotein is a transmembrane protein with three N-terminal domains. This protein is glycosylated in the Golgi apparatus after its expression and combines with other envelope protein like Spike (S) protein and Nucleocapsid (N) protein and also with each other during the virus assembly. 156 N protein creates a complex with the genomic RNA and M protein that activates the development of interacting virions in the intermediate of ER and Golgi interface compartment with this complex156,157
SARS-CoV-2 M protein consists of 222 amino acids encoded by 669 nucleotides. 1 SARS-CoV-2 M protein sequence is more than 90% identical to the SARS-CoV membrane M protein (221 amino acids). 158 Almost 60% of the residues are neutral and nonpolar, indicating that the protein is highly hydrophobic. The overall M protein is theoretically basic, that is, it has a pI value of 9.51 and it is positively charged.
M is a transmembrane protein as predicted by TMHMM web server. 159 It is predicted to have three TM domains between codons 20 and 100, which contribute to about one third of the M protein region. First of the three putative TM domains (TMI) is from 20 to 39 codons (19 amino acid residues), second domain (TMII) is from 51 to 73 codons (23 amino acid residues), and the third domain (TMIII) is from 78 to 100 codons (23 amino acid residues). The residues from 1 to 19 codons N-terminal lie outside the viral envelope. TMI and TMII are linked by a chain of 11 residues (40-50 codons) that probably lie interior to the viral envelope. TMII and TMIII are linked by a very short chain of four residues (74-77 codons) that probably lies outside the viral envelope. In addition, there is 122 residues (101-222 codons) long carboxy-terminal tail that lies inside the viral envelope (Figure 19). Hydrophobic region predicted by ProtScale web server tool from ExPASy 160 suggest that the transmembrane region from 20 to 100 codons are highly hydrophobic and the TMIII region seems to be less hydrophobic than the other two regions.
Figure 19.
Transmembrane domains predicted by TMHMM web server in which there are three domains connected by two linker peptides.
Although the amino acid composition of CoVs M proteins is heterogeneous, all M proteins share a common structural characteristic. The M protein consists of three transmembrane domains which are flanked by a short glycosylated amino-terminal domain and a long carboxy-terminal tail.86,132
Similar to E protein, M protein showed very low mutational rate in the SARS-CoV-2 proteome during the pandemic. None of the residues of the protein demonstrated mutation rate higher than 0.01. 32
Accessory Proteins
A group of accessory proteins that play an important role in changing the environment that favors the replication of virus inside the infected host cell are encoded by the genome of SARS-CoV-2. The major accessory proteins encoded by SARS-CoV-2 are ORF3a, 3b, 6, 7a, 7b, 8, 9b, and 10 (Table 4). 161 The structure and function of these proteins are still confounding and unsolved. Based on the research of other coronaviruses, scientists have gained a good understanding of this novel virus and have hypothesized about the structure and function of these accessory proteins. As SARS-CoV-2 has shown nucleotide similarity of 89.1% with SARS-CoV, the structure and function of accessory proteins of SARS-CoV-2 has been predicted based on the available information of SARS-CoV. 1 Important accessory proteins of SARS-CoV-2 are the following:
Table 4.
Structure and function of accessory proteins.
| Structure solved (SARS-CoV-2) | Structure solved (SARS-CoV) | Description of the structure | Function | Functional name | |
|---|---|---|---|---|---|
| 3a | Solved for SARS-CoV-2157
(Cryo-EM Struture) PDB: 6XDC |
Not solved | It is an ion channel with 275 amino acid residues (274 amino acid residues in SARS-CoV). It is an O-glycosylated protein with three transmembrane helices followed by cytosolic domains with multiple β-strands per protomer chain and an N-terminal ectodomain and a C-terminal endodomain in both intracellular and plasma membranes.162,163 | Interactions with some structural proteins (S, M and E) and observed inducing apoptosis in vitro 164 - 166 | Hole borer |
| 3b | Not solved | Not solved | It is 154 amino acids long for SARS-CoV (151 for SARS-CoV-2). 1 | It induces apoptosis and necrosis, and hinders antiviral innate immune response. 167 - 169 | Immune modulator |
| 6 | Not solved | Not solved | It is 63 amino acids long in SARS-CoV (61 for SARS-CoV-2). 1 | It suppress IFN induction and IFN signaling pathway.169,170 | Type 1 interferon antagonist |
| 7a | Solved for SARS-CoV-2 (X-Ray Crystallography) PDB:7Ci3 |
Solved for SARS-CoV 171 (X-Ray Diffraction) PDB: 1XAK | It is a type I transmembrane protein with 121 amino acids (122 for SARS-CoV that contain 15-amino-acid N-terminal signal peptide sequence, an 81-amino-acid luminal domain, a 21-amino-acid transmembrane domain and a short C-terminal tail. 171 | It induces apoptosis, inhibits cellular protein synthesis and arrests cell cycle at the GO/G1 phase. 172 - 174 | Immune evador |
| 7b | Not solved | Not solved | It is an integral membrane protein with 44 amino acids (43 for SARS-CoV-2). 1 | Immune evador | |
| 8 | Solved for SARS-CoV-2 (X-Ray Diffraction) PDB: 7JTL | 8a—Not solved | It is a 39-amino-acid-long polypeptide in which residues 1-35 are identical to the N-terminal of 8ab. | It is found to enhance replication in some studies and shows interaction with other structural proteins.175,176 ORF8 is not splitted into ORF 8a and 8b and have amino acids length of 121 in SARS-CoV-2. 1 | Immune modulator |
| 8b—Not solved | It is a 84-amino-acid-long polypeptide and residues 9-84 are identical to the C terminal of 8ab. | ||||
| 9b | Solved for SARS-CoV-2 (X-Ray Crystallography) PDB: 6Z4U. | Solved for SARS-CoV 177 (X-Ray Diffraction) PDB: 2CME | It is 70 amino acids long protein (73 in case of SARS-CoV-2). It contains a novel dimeric fold having β strands like structure with an amphipathic surface and a central hydrophobic cavity. 177 | It shows interaction with some nonstructural proteins, and incorporates into mature virions. 178 - 180 | Internal N protein |
| 10 | Not solved | Not found | It is a 38-amino-acid-long protein. It has one potential TM domain. Presence of one α-helix and two β-strands and β-α-β motif with a β-molecular recognition feature occurring in the first β-strand is revealed through the prediction of secondary structural elements of this protein | It has potential immune modulatory role. | Mystery protein |
Abbreviations: IFN, interferon; ORF, Open Reading Frames; TM, transmembrane.
Hole borer ORF3a protein
ORF3a protein is the largest accessory protein of SARS-CoV-2 with 275 amino acid residues (Figure 20) and is assumed to create holes in the membrane of the infected cells to facilitate the escape of the virus. 1 This protein is encoded by the ORF3a gene located between S and E genes in the genome. One of the symptoms of COVID-19, inflammation, is found to be triggered by this protein. The structure and function of this protein can be predicted by comparing it with its homolog of SARS-CoV. SARS-CoV also possesses this protein which is 274 amino acids long. 3 ORF3a is found to be highly conserved within Betacoronavirus subgenus Sarbecovirus that includes SARS-CoV and other related bat coronaviruses, which are assumed to be potential sources of SARS coronaviruses that are harmful to humans. 181 Study of interaction of SARS-CoV ORF3a protein with other proteins of the virus uncovered that it interacts with structural proteins like S and M in the Golgi apparatus, which is close to site of virus assembly and budding, as well as with another structural protein, E, and accessory protein ORF7a. 164 Incorporation of ORF3a into virus particles through the interaction with S protein has been observed in infected cells and SARS patients. 165 However, for the formation of virus-like particles (VLPs), ORF3a protein is not essential as VLPs are produced even in the absence of 3a protein. 182 In the majority of the SARS patients, this protein was found to be effectively expressed in cell surface and was detected as a result of the triggering of a humoral and cellular immune response. 183 In addition, it was observed that ORF3a protein can cause activation of NF-kB (nuclear factor kappa B) and JNK (c-jun N-terminal kinase) which in turn can upregulate the activity of interleukin 8 (IL-8) and RANTES (CCL5). 184 Study of in vitro expression in cell culture models showed that ORF3a protein causes G1 phase cell cycle arrest by lowering cyclin D3 expression and inhibiting retinoblastoma protein (Rb) phosphorylation 185 and incites apoptosis in Vero E6 cells. 166 Some studies suggested that virus-induced immune pathology and the disease outcome is likely to be decided by a dysregulated immune response with an intense upregulation of proinflammatory cytokines.166,186 In a study, ORF3a protein was found to regulated the expression and secretion of fibrinogen in human ling epithelial cell line, A549 and virus-infected Vero E6 cells which might be responsible for SARS pathogenesis by increasing cytokine production if produced in excessive amount. 187 Deletion of the gene encoding this protein reduces viral titer and morbidity in infected animal models. 188 ORF3a protein of SARS-CoV-2 is expected to perform similar functions as in SARS-CoV. So, this protein has been considered as a potential target to develop therapeutics against the disease caused by SARS. The accurate role of the ORF3a protein and its functions in infected cells and viral pathogenesis is still unclear and needs further elucidation.
Figure 20.
Structure of SARS-CoV-2 ORF3a protein (Cartoon representation). (A) Showing green chain A, blue chain B. (B) Showing green loops, yellow β sheets, and red alpha helices (PDB 6XDC).
ORF indicates Open Reading Frames.
ORF3a dimers and tetramers obtain a new fold having a large polar cavity that extend halfway across the membrane and is easily approachable to the cytosol portion as well as to the surrounding bilayer through different water- and lipid-filled openings as shown by the structure in lipid nanodiscs. Transmembrane region of this protein consists of three helices per promoter. N-termini are aligned on the extracellular side while C-termini toward cytosolic side of the bilayer. Transmembrane helices are present which are connected by short intracellular and extracellular linkers. A pair of opposing β-sheets packed against one another in an eight-stranded β-sandwich is formed from each promoter chain. Strands β1, β2, β6, and N-terminal half of β7 form the outer sheet while strands β3, β4, β5, β8, and C-terminal part of β7 contribute to inner sheet of the protein. Strong and stable link is formed in the dimer as a result of interaction between β-sandwiches from each promoter. 162
During the SARS-CoV-2 pandemic of 2020, this protein was highly mutated. There were nine sites that showed mutation rate higher than 0.01. These sites are Q57, G172, T223, V202, Q38, R122, K75, S166, and H182. 32
Immune modulator ORF3b protein
SARS-CoV-2 ORF3b protein, 151 amino acids polypeptide, is translated from ORF3b found between S and E genes. 1 ORF3b intersects the stretch of RNA of ORF3a, but it is still unclear whether the virus utilizes this gene to make proteins. SARS-CoV ORF3b is 154 amino acids long and is reported to be present in the nucleolus and mitochondria.167,189 Interestingly, it does not share any homology with any other known proteins. 3 ORF3b is not essential for SARS-CoV replication as observed in cell culture. 182 Serum samples of the infected SARS patients showed the presence of anti-p3b antibodies. 190 In addition to this, p3b was found to be involved in inducing apoptosis and necrosis independent of its subcellular localization.167,168 Likewise, immune-histochemical analysis of SARS-CoV infected Vero E6 cells displayed the expression of ORF3b protein in infected cells through the regulation of host innate immune response and hindering antiviral innate response by inhibiting both interferon production and signaling pathways. 169 This protein was observed inhibiting antiviral response by downregulating type-I interferon (IFN-β) and also restraining mitochondrial antiviral response.117,191 ORF3b protein was found to induce cell growth arrest at the G0/G1 phase using flow cytometry. 192 From the study of SARS-CoV ORF3b protein, the functions of SARS-CoV-2 ORF3b protein could be predicted.
Type I interferon agonist ORF6 protein
SARS-CoV-2 protein ORF6 is a 61-amino-acid residues long membrane-associated protein translated from ORF6 of the viral genome. 1 When the cell is attacked or infected by any foreign threat like a virus, it sends signals to the immune system of the body to defend itself. In the case of SARS-CoV-2 infection, ORF6 protein was involved in blocking the transmission of such signal to immune system and inactivating some of the virus-fighting proteins present in the cell. But SARS-CoV ORF6 protein is a 63-amino-acids-long protein localized in the ER and Golgi apparatus and was detected in the lungs and intestine tissues (ileum) of SARS patients as well as in virus-infected Vero E6 cells.193,194 Virus capture assay showed the presence of ORF6 protein in mature virus particles. 195 One study have shown that this protein is not required for viral replication as observed in cultured cells of BALB/c mice. 182 However, other studies have hinted that ORF 6 protein might be involved in SARS-CoV replication and acts as type I interferon antagonist that targets various steps of the IFN pathway and suppresses IFN induction and IFN signaling pathway.169,170 The nature of protein from ORF6 found in SARS-CoV-2 could be hypothesized from these findings of SARS-CoV. During the pandemic of 2020, none of the residues of this protein demonstrated mutation rate higher than 0.01. 32
Immune evaders ORF7a and 7b proteins
SARS-CoV-2 ORF7a is a 121-amino-acid-long transmembrane protein translated from ORF7 (Figure 21). 1 It is found to decrease the supply of tetherin in an infected cell, a substance responsible for trapping new viruses, and preventing their escape outside the cell. Studies have also found that this protein can induce apoptosis in infected cells, which could have toxic effects in the lung cells of the patients. In case of SARS-CoV, bicistronic subgenomic RNA 7 translates both these accessory proteins.3,4 SARS-CoV ORF7a is a protein with 122-amino-acid residues that contain 15-amino-acid signal peptide sequence, an 81-amino-acid luminal domain, a 21-amino-acid transmembrane domain, and a short C-terminal tail. 171 It was detected in the lung tissue of SARS patients and virus infected Vero E6 cells.171,196 Accurate subcellular localization of this protein is still not clear. Some studies have reported its presence within the ER 197 while others have observed it in the Golgi compartment. 172 SARS-CoV-2 ORF7a is found to show a significantly higher binding efficiency to CD14+ monocytes compared with SARS-CoV-2 ORF7a via viral protein based flow cytometry and cell surface marker analysis, suggesting a difference in their ability to bind with CD14+ monocytes. 198 ORF7a protein is found to be incorporated into mature virions. 199 The deletion of ORF7a from the viral genome does not affect replication and RNA synthesis in cell culture. 182 Several studies confirmed that ORF7a is not essential for viral replication both in vitro and in vivo. One study showed that recombinant viruses with deletion of ORF 7a, 7b, 8a, 8b, and 9b produced viral particles having similar morphology and replication pattern as the wild type SARS-CoV in transgenic mice. 200 However, this protein was found inducing apoptosis in several cell lines through a caspase-dependent pathway, 173 inhibiting cellular protein synthesis, activating p38 mitogen-activated protein kinase, 172 and arresting cell cycle at the GO/G1 phase. 174
Figure 21.

Structure of SARS-CoV-2 ORF7a protein (cartoon representation) showing green loops, yellow β sheets (PDB 7Ci3).
ORF indicates Open Reading Frames.
SARS-CoV-2 7b protein is a 43-amino-acid poly-peptide. 1 ORF7b overlaps the same stretch of RNA as ORF7a in the case of SARS-CoV. SARS-CoV ORF7b protein is an integral membrane protein with 44 amino acids. When expressed in virus-infected Vero cells, it was observed to be localized in the Golgi compartment as well as associated with intracellular viruses and purified virions. 201 The presence of an anti-7b antibody in SARS-infected patient serum samples indicated in vivo expression of this protein. 190 But the actual function of this protein in both SARS-CoV and SARS-CoV-2 is still unknown.
Like SARS-CoV ORF7a, SARS-CoV-2 ORF7a is a type I transmembrane protein consisting of an N-terminal signaling region (residues 1–15), an Ig-like ectodomain (residues 16–96), a hydrophobic transmembrane domain (residues 97–116), and a typical ER retention motif (residues 117–121). Seven β-strands which is divided into two tightly packed β-sheets comprises the overall structure of the SARS-CoV-2 ORF7a. Four strands form the larger β-sheet while the other three form smaller β-sheet. The “sandwich” β-sheet structure is stabilized by two disulfide bonds (formed by residues Cys23-Cys58and Cys35-Cys67) by connecting different strands of large and small β-sheets. A conserved Ig-like β-sandwich fold with seven β-strands is observed through the sequence alignment of SARS-CoV-2 ORF7a with SARS-CoV ORF7a. Comparing the structure of SARS-CoV-2 ORF7a with that of SARS-CoV, the varied residues were found to be present on the amphipathic side of the larger β-sheet only which suggests that major residues are present on the major functional interface. Majority of the variations in the sequence are dispersed on the ectodomain portion with the variations in eight residues, suggesting potentially distinct protein functions. The structure of this protein explains the molecular mechanism that governs the various pathological processes of SARS strains and works as an immunomodulating factor for immune cell binding as well as triggers inflammatory responses. Thus, identifying the biological significance of SARS-CoV-2 ORF7a-leukocyte interactions and resulting immune responses could provide an insight as a potential therapeutical targets. 198
During the pandemic of 2020, one sites each of 7a and 7b proteins showed mutation rates higher than 0.01. For 7a protein it was T14 and for 7b protein it was S5. 32
Immune modulator ORF8 protein
This protein is 121-amino-acid long and is translated from ORF8, but not separated into ORF 8a and 8b in SARS-CoV-2. 1 The gene for this protein is unusual in SARS-CoV-2 as compared with other coronaviruses. Its function in novel coronavirus can be predicted by studying the protein in SARS-CoV. Subgenomic RNA 8 of SARS-CoV translates 8a and 8b proteins, which differs from ORF8 in its conformation. Their expression was found in virus-infected cells. 175 It shows sequence similarity of less that 20% with SARS-CoV ORF8 indicating its noteworthy divergence. ORF8 present in both strains of SARS have a signal sequence for ER import. 202 This protein is shown to interrupt signaling when exogenously overexpressed in cells and downregulates MHC-I in cells.203,204 It was confirmed from epidemiological studies that early human and animal SARS-CoV isolates possessed only one intact ORF8. But, during the peak of the SARS epidemic in 2003, the virus isolated from humans showed deletion in the middle of ORF8, resulting in the separation of ORF8 into two smaller ORFs, 8a and 8b.205,206 During the late phase of the SARS pandemic, larger deletions (82-nucleotide and 415-nucleotide deletion) were additionally found in some virus-infected human samples.205,207 This finding led to the hypothesis that this genomic change might be responsible for the zoonotic transition of the virus from animal to human during the outbreak and the deletion of 29-nucleotide could have some connection with the adaptation of SARS-CoV to humans. But the addition of 29-nucleotide sequence into the human sample utilizing reverse genetics showed very little effect on the growth of the virus as well as RNA replication in cell culture. 182 This result also suggested that 29-nucleotide deletion might not be the reason for the adaptation of the virus to humans with enhanced pathogenicity. The reason for this mutation is still not clear and needs to be investigated. This protein, with an N-terminal hydrophobic signal sequence, is encoded by the undeleted ORF 8 or ORF8ab. ORF8a gives a 39-amino-acid polypeptide in which residues 1 to 35 are identical to the N-terminal of 8ab. Likewise, ORF8b produces a 84-amino-acid polypeptide and residues 9 to 84 are identical to the C terminal of 8ab. 208 Studies have shown that ORF8a protein interacts with S protein, 8b interacts with M, E, 3a, and 7a proteins, while ORF8 protein showed interaction with S, 3a, and 7a protein. The protein level of E was downregulated by the expression of ORF8b protein, but the expression of E gene mRNA was not affected. 175 Some studies suggest that ORF8 of SARS-CoV is not essential for viral replication both in vitro and in vivo.182,200 But one study showed that the integrity of ORF8 facilitates replication in reservoir bat cell, non-host cell lines, and in the human epithelial cultures. 176
During the SARs-CoV-2 pandemic of 2020, protein residues of ORF8 showed mutation rate higher than 0.01 at two sites, A65 and S24. 32
Three sets of intramolecular disulfide bonds per monomer and a single intermolecular disulfide bond formed by Cys20 of each monomer crystallizes ORF8 as a covalent dimer (Figure 22). Two antiparallel β-sheets is found at the core of each ORF8 monomer, smaller sheet formed by β1, β2, β5, and β6, and larger one consists of β3, β4, β7, and β8. Two novel intermolecular interfaces layered onto an ORF7 fold, one intervened by a disulfide bond and by noncovalent bond, is uncovered by the structure of the SARS-CoV-2 ORF8 protein. This structural characteristic is novel in case of SARS-CoV-2. SARS-CoV-2 ORF8 shows a sequence similarity of 16% with ORF7a. ORF8 monomer aligns with the Ig-like fold of ORF7a and both of them share two sets of structural disulfide linkages considered central to the Ig-like fold. The analysis of structure set up a molecular framework useful to understand the rapid evolution, pathogenicity of the virus, and its antibody neutralizing ability. 202
Figure 22.
Structure of SARS-CoV-2 ORF8 protein (cartoon representation) showing green loops, yellow β sheets (PDB 7JTL).
ORF indicates Open Reading Frames.
Internal N ORF 9b protein
Accessory protein from ORF9b of SARS-CoV-2 is 97 amino acids long found overlapping the stretch of N protein (Figure 23). 1 Expression of this protein in SARS-CoV-2 is still not clear. In the case of SARS-CoV, ORF9b protein is a 98-amino-acid polypeptide translated from the second ORF of RNA9 that shows no sequence homology with any known proteins.6,178 RNA9 is also found to encode N protein of SARS-CoV and ORF9b is observed as a complete internal ORF within the N gene in an alternate reading frame. 178 The study of the crystal structure of 9b protein revealed a novel dimeric fold having β strands like structure with an amphipathic surface and a central hydrophobic cavity, that binds lipid to stabilize the molecule allowing its probable association with intracellular vesicles. 177 It is involved in hindering interferon, a key element in the protection against viral infection. SARS-CoV infected cells and the lung and ileum tissues from SARS patients showed the expression of ORF9b protein. 137 Likewise, the expression of this protein in vitro and in vivo is shown by the presence of anti-9b antibodies in the serum of SARS patients. 209 Co-expression of this protein along with E and M proteins shows its incorporation in mature virions which are packaged into VLPs indicating 9b as a potential structural component of virions. 178 SARS-CoV ORF9b protein shows self-interaction and interaction with nsp5, nsp14, and one accessory protein ORF6 protein but the significance behind these interactions is not well understood.179,180 ORF9b protein is not essential for viral replication in vitro and in vivo as demonstrated by several reverse genetic studies.179,200 It may have a function during virus assembly as a membrane-attachment point for other viral proteins. 177 This protein is predicted to exhibit similar action in SARS-CoV-2.
Figure 23.
Structure of SARS-CoV-2 ORF9b protein (Cartoon representation). (A) Showing green chain A, blue chain B. (B) Showing green loops, yellow β sheets, and red alpha helices (PDB 6Z4U).
ORF indicates Open Reading Frames.
During the SARS-CoV-2 pandemic of 2020, this protein showed one of the highest rates of mutation. Residue L67 and G50 were mutated at a rate higher than 0.2. In addition, S6 and H9 were mutated at a rate higher than 0.01. 32
Mystery ORF 10
The genome annotation of SARS-CoV-2 uncovers the presence of 10 open reading frames (ORFs). Out of which, ORF10 is the last one with 117 nucleotides and is positioned upstream of the 3ʹ-untranslated region (3ʹ-UTR) and downstream of N gene encoding a protein with the length of 38 amino acids.210-212 The function of this protein is still unclear and it does not share similarity in sequence with any other known protein. Biochemical and functional characterization of this protein using different bioinformatics tools indicated this protein as highly ordered, thermally stable, hydrophobic protein possessing at least one transmembrane region. 213 Presence of one α-helix and two β-strands and β-α-β motif with a β-molecular recognition feature occurring in the first β-strand is revealed through the prediction of secondary structural elements of this protein.214,215 The primary α-helix region of this protein is shown to possess large numbers of cytotoxic T lymphocytes (CTL) epitopes which could elevate immune response including potential mortal immunopathological response toward SARS-CoV-2.214,216 This protein is found to contain 11 CTL epitopes of nine amino acids length each, across various human leukocyte antigen (HLA) subtypes which is highest number of immunogenic epitopes of all putative ORF proteins and makes it a potential target for vaccine development.214,217 ORF 10 is shown to interact with multiple members of the Cullin-ubiquitin-ligase complex and controls the host-ubiquitin machinery for viral pathogenesis using different bioinformatics tools.213,215,218,219 In a study, however, SARS-CoV-2 ORF 10 is found to be not essential in vitro or in vivo in humans. 211
Future directions
Solving the structures of proteins in SARS-CoV-2 has a critical role to play in vaccine and drug development. Solving of the structure of S protein led to proper use of viral epitopes for vaccination in different platforms. Similarly, solving the structure of essential proteins can lead to better structure-based drug design. While most of the structures of proteins have been solved, some accessory, structural and nonstructural protein structures have not been solved. This will probably be the focus of the international scientific community in the future. Solving structure helps not only in structure-based drug design but also in finding out the function of the protein. If the structure of the protein is homologous to already known protein, that function can be predicted.
A number of nonstructural proteins, accessory proteins, and even some structural proteins have undefined function. It would make sense to figure out the function of these proteins through reverse genetics and protein-protein interaction studies. Reverse genetics is critical to determining the importance of every protein in pathogenesis and survival of the virus. Such reverse genetics method of assembling SARS-CoV-2 virus with desired mutation has been designed by Xie et al. 220
There appears to be strong evidences that nonstructural proteins of SARS-CoV-2 work in tandem. They are all generally thought to work in replication-transcription of the virus. But in what order to the protein function and how they work in concert is not clear. Therefore, development of a cell free system or a cell culture-based system, where all the purified proteins will be provided with substrates they can act on would test the ability of the system to work in concert. Such system has been developed for the functioning of replication machinery and other DNA repair systems before.
In addition, several proteins have known enzymatic activity. The enzymatic activity can be biochemically probed for cofactor and substrate specificity. Additional precise enzyme kinetics can be determined.
Vilar et al have determined the hotspots of mutation in the SARS-CoV-2 proteome during the pandemic of 2020. Whether those hotspots occur because those regions allow greater amino acid flexibility or because there is mechanism in the virus to cause mutations at greater frequency in the hotspot is not clear. This can be investigated further.
Acknowledgments
We thank Anusa Thapa for proofreading the article.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: HKB initiated the write up. RG wrote on S and E proteins. PK wrote up the section on accessory proteins. Section on non structural proteins 1 to 8 was written by AM. SR wrote up on non structural proteins 9 to 16 and drew Figures 2, 3 and 8. AB wrote the section on N and M protein. HKB wrote on the mutation data from SARS-CoV-2 2020 pandemic and also wrote introduction, future directions and abstract sections.
ORCID iD: Hitesh Kumar Bhattarai  https://orcid.org/0000-0002-7147-1411
https://orcid.org/0000-0002-7147-1411
References
- 1. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. doi:10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193-292. doi:10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399-1404. doi:10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4. Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394-1399. doi:10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 5. Thiel V, Ivanov KA, Putics Á, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305-2315. doi:10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 6. Liu DX, Fung TS, Chong KKL, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97-109. doi:10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-410. [DOI] [PubMed] [Google Scholar]
- 8. Arya R, Kumari S, Pandey B, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2020;433:166725. doi:10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi:10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hussain S, Pan J, Chen Y, et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288-5295. doi:10.1128/jvi.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697-3708. doi:10.1128/jvi.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136-10141. doi:10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Meer Y, Snijder EJ, Dobbe JC, et al. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641-7657. doi:10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Narayanan K, Huang C, Lokugamage K, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82:4471-4479. doi:10.1128/jvi.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang H, Xie W, Xue X, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi:10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108-5113. doi:10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991-1004. doi:10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619-5632. doi:10.1128/jvi.78.11.5619-5632.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Decroly E, Debarnot C, Ferron F, et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi:10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Su C, Ke M, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi:10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81:11620-11633. doi:10.1128/jvi.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar R, Verma H, Singhvi N, et al. Comparative genomic analysis of rapidly evolving SARS-CoV-2 reveals mosaic pattern of phylogeographical distribution. mSystems. 2020;5:e00505-20. doi:10.1128/mSystems.00505-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853-879. doi:10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 24. Corum J, Zimmer C. Bad news wrapped in protein: inside the coronavirus genome A string of RNA. The New York Times. April 3, 2020. https://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html.
- 25. Zhai Y, Sun F, Li X, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980-986. doi:10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Egloff MP, Ferron F, Campanacci V, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792-3796. doi:310.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi:10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adedeji AO, Marchand B, Te Velthuis AJ, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi:10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci USA. 2015;112:9436-9441. doi:10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78:12218-12224. doi:10.1128/jvi.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aouadi W, Blanjoie A, Vasseur J-J, Debart F, Canard B, Decroly E. Binding of the methyl donor S-adenosyl-l-methionine to middle east respiratory syndrome coronavirus 2′-O-methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J Virol. 2017;91:e02217-16. doi:10.1128/jvi.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vilar S, Isom DG. One year of SARS-CoV-2: how much has the virus changed? Biology (Basel). 2021;10:91. doi:10.3390/biology10020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Connor RF, Roper RL. Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol. 2007;15:51-53. doi:10.1016/j.tim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narayanan K, Ramirez SI, Lokugamage KG, Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89-100. doi:10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clark LK, Green TJ, Petit CM. Structure of nonstructural protein 1 from SARS-CoV-2. J Virol. 2021;95:e02019-20. doi:10.1128/jvi.02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schubert K, Karousis ED, Jomaa A, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959-966. [DOI] [PubMed] [Google Scholar]
- 37. Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020;92:584-588. doi:10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol. 2005;79:13399-13411. doi:10.1128/jvi.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harcourt BH, Jukneliene D, Kanjanahaluethai A, et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600-13612. doi:10.1128/jvi.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schiller JJ, Kanjanahaluethai A, Baker SC. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288-302. doi:10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Denison MR, Zoltick PW, Hughes SA, et al. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274-284. doi:10.1016/0042-6822(92)90703-r. http://huji-primo.hosted.exlibrisgroup.com/openurl/972HUJI/972HUJI_SP?sid=EMBASE&sid=EMBASE&issn=00426822&id=doi:10.1016%2F0042-6822%2892%2990703-R&atitle=Intracellular+processing+of+the+N-terminal+ORF+1a+proteins+of+the+coronavirus+MHV-A59+requires+multiple+proteolytic+events&stitle=VIROLOGY&title=Virology&volume=189&issue=1&spage=274&epage=284&aulast=Denison&aufirst=M.R.&auinit=M.R.&aufull=Denison+M.R.&coden=VIRLA&isbn=&pages=274-284&date=1992&auinit1=M&auini. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang H, Yang M, Ding Y, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA. 2003;100:13190-13195. doi:10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan J, Verschueren KHG, Anand K, et al. pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J Mol Biol. 2005;354:25-40. doi:10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi J, Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035-1045. doi:10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213-3224. doi:10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lim L, Shi J, Mu Y, Song J. Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS ONE. 2014;9:e101941. doi:10.1371/journal.pone.0101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neuman BW. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016;135:97-107. doi:10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189-15198. doi:10.1128/jvi.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanjanahaluethai A, Chen Z, Jukneliene D, Baker SC. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391-401. doi:10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Neuman BW, Joseph JS, Saikatendu KS, et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279-5294. doi:10.1128/jvi.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJ, de Haan CA. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol. 2007;81:12323-12336. doi:10.1128/jvi.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakai Y, Kawachi K, Terada Y, Omori H, Matsuura Y, Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165-174. doi:10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oostra M, Hagemeijer MC, van Gent M, et al. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol. 2008;82:12392-12405. doi:10.1128/jvi.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baliji S, Cammer SA, Sobral B, Baker SC. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J Virol. 2009;83:6957-6962. doi:10.1128/jvi.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35-48. doi:10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524-13. doi:10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cottam EM, Maier HJ, Manifava M, et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335-1347. doi:10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clementz MA, Kanjanahaluethai A, O’Brien TE, Baker SC. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118-129. doi:10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Su D, Lou Z, Sun F, et al. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol. 2006;80:7902-7908. doi:10.1128/jvi.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mirza MU, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020;10:320-328. doi:10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Azzi A, Lin SX. Human SARS-coronavirus RNA-dependent RNA polymerase: activity determinants and nucleoside analogue inhibitors. Proteins. 2004;57:12-14. doi:10.1002/prot.20194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111:E3900-E3909. doi:10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peng Q, Peng R, Yuan B, et al. Structural and biochemical characterization of nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi:10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. 2020;11:3717. doi:10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ogando NS, Zevenhoven-Dobbe JC, van der Meer Y, Bredenbeek PJ, Posthuma CC, Snijder EJ. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94:e01246-20. doi:10.1128/jvi.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gribble J, Stevens LJ, Agostini ML, et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17:e1009226. doi:10.1371/journal.ppat.1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adedeji AO, Singh K, Sarafianos SG. Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Cell Mol Biol. 2012;58:114-121. doi:10.1170/T929. [PMC free article] [PubMed] [Google Scholar]
- 68. Lemus MR, Minasov G, Shuvalova L, et al. The crystal structure of nsp10-nsp16 heterodimer from SARS CoV-2 in complex with S-adenosylmethionine [published online ahead of print April 26, 2020]. bioRxiv. doi:10.1101/2020.04.17.047498. [Google Scholar]
- 69. Aouadi W, Blanjoie A, Vasseur J-J, Debart F, Canard B, Decroly E. Binding of the methyl donor S-adenosyl-l-methionine to middle east respiratory syndrome coronavirus 2’-O-methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J Virol. 2017;91:e02217-16. doi:10.1128/jvi.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bouvet M, Lugari A, Posthuma CC, et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem. 2014;289:25783-25796. doi:10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Debarnot C, Imbert I, Ferron F, et al. Crystallization and diffraction analysis of the SARS coronavirus nsp10-nsp16 complex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:404-408. doi:10.1107/S1744309111002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Von Grotthuss M, Wyrwicz LS, Rychlewski L. mRNA Cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701-702. doi:10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Decroly E, Imbert I, Coutard B, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol. 2008;82:8071-8084. doi:10.1128/jvi.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596-1605. doi:10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhardwaj K, Palaninathan S, Alcantara JMO, et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283:3655-3664. doi:10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106-1117. doi:10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Littler DR, Gully BS, Colson RN, Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23:101258. doi:10.1101/2020.03.28.013920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sutton G, Fry E, Carter L, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341-353. doi:10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ponnusamy R, Moll R, Weimar T, Mesters JR, Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J Mol Biol. 2008;383:1081-1096. doi:10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ponnusamy R, Mesters JR, Ziebuhr J, Moll R, Hilgenfeld R. Non structural proteins 8 and 9 of human coronavirus 229E. Adv Exp Med Biol. 2006;581:49-54. doi:10.1007/978-0-387-33012-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Park YJ, Walls AC, Wang Z, et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26:1151-1157. doi:10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nelson GW, Stohlman SA, Tahara SM. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181-188. doi:10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 83. Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2019;94:e01925-19. doi:10.1128/jvi.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288-4295. doi:10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang Q, Yu L, Petros AM, et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059-6063. doi:10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 86. Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J Virol. 2010;84:11418-11428. doi:10.1128/jvi.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363-382. doi:10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Westerbeck JW, Machamer CE. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J Virol. 2015;89:9313-9323. doi:10.1128/jvi.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li Y, Surya W, Claudine S, Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289:12535-12549. doi:10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10:e1004077. doi:10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wu Q, Zhang Y, Lü H, et al. The E protein is a multifunctional membrane protein of SARS-CoV. Genomics Proteomics Bioinformatics. 2003;1:131-144. doi:10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Du Y, Zuckermann FA, Yoo D. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth. Virus Res. 2010;147:294-299. doi:10.1016/j.virusres.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93-116. doi:10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi:10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871-5876. doi:10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39-43. doi:10.1016/0966-842X(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 97. Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120-134. doi:10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Song W, Gui M, Wang X, Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi:10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281-292.e6. doi:10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535-538. doi:10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. doi:10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-220. doi:10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 103. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127-20. doi:10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343-355. doi:10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119-129. doi:10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Walls AC, Tortorici MA, Snijder J, et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017;114:11157-11162. doi:10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Walls A, Tortorici MA, Bosch B-J, et al. Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo-electron microscopy. Protein Sci. 2017;26:113-121. doi:10.1002/pro.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xiong X, Tortorici MA, Snijder J, et al. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J Virol. 2017;92: e01628-17. doi:10.1128/jvi.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Walls AC, Xiong X, Park YJ, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li F, Li W, Farzan M, Harrison SC. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864-1868. doi:10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 111. Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613-620. doi:10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444-1448. doi:10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Surjit M, Lal SK. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol. 2008;8:397-405. doi:10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li YH, Li J, Liu XE, et al. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J Virol Methods. 2005;130:45-50. doi:10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387-2396. doi:10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Surjit M, Liu B, Chow VTK, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669-10681. doi:10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Spiegel M, Pichlmair A, Martínez-Sobrido L, et al. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079-2086. doi:10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zheng B, He ML, Wong KL, et al. Potent inhibition of SARS-associated coronavirus (SCoV) infection and replication by type I interferons (IFN-α/β) but not by type II interferon (IFN-γ). J Interf Cytokine Res. 2004;24:388-390. doi:10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- 119. Yan X, Hao Q, Mu Y, et al. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int J Biochem Cell Biol. 2006;38:1417-1428. doi:10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120. Tang TK, Wu MPJ, Chen ST, et al. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925-937. doi:10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Du L, Zhao G, Lin Y, et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644-1651. doi:10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hsieh P-K, Chang SC, Huang C-C, et al. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79:13848-13855. doi:10.1128/jvi.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi:10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Liu SJ, Leng CH, Lien SP, et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100-3108. doi:10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shang B, Wang XY, Yuan JW, et al. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem Biophys Res Commun. 2005;336:110-117. doi:10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lin Y, Shen X, Yang RF, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141-145. doi:10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kang S, Yang M, Hong Z, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228-1238. doi:10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Saikatendu KS, Joseph JS, Subramanian V, et al. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81:3913-3921. doi:10.1128/jvi.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tan YW, Fang S, Fan H, Lescar J, Liu DX. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816-4825. doi:10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lin SY, Liu CL, Chang YM, Zhao J, Perlman S, Hou MH. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247-2257. doi:10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J Virol. 2007;81:2249-2262. doi:10.1128/jvi.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hogue BG, Machamer CE. Coronavirus structural proteins and virus assembly. In: Perlman S, Gallagher T, Snijder EJ, eds. Nidoviruses. Washington, DC: American Society of Microbiology; 2014:179-200. doi:10.1128/9781555815790.ch12. [Google Scholar]
- 133. Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, Dediego ML, Enjuanes L, Aguilella VM. Analysis of SARS-CoV e protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta. 2013;1828:2026-2031. doi:10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Corse E, Machamer CE. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J Virol. 2002;76:1273-1284. doi:10.1128/jvi.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Cohen JR, Lin LD, Machamer CE. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J Virol. 2011;85:5794-5803. doi:10.1128/jvi.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Nieto-Torres JL, DeDiego ML, Álvarez E, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69-82. doi:10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nal B, Chan C, Kien F, et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol. 2005;86:1423-1434. doi:10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 138. Lopez LA, Riffle AJ, Pike SL, Gardner D, Hogue BG. Importance of conserved cysteine residues in the coronavirus envelope protein. J Virol. 2008;82:3000-3010. doi:10.1128/jvi.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Álvarez E, DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Marcos-Villar L, Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281-291. doi:10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Keng CT, Åkerström S, Leung CS, et al. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. Microbes Infect. 2011;13:179-188. doi:10.1016/j.micinf.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lim KP, Liu DX. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J Biol Chem. 2001;276:17515-17523. doi:10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mortola E, Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174-178. doi:10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pervushin K, Tan E, Parthasarathy K, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi:10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Torres J, Wang J, Parthasarathy K, Liu DX. The transmembrane oligomers of coronavirus protein E. Biophys J. 2005;88:1283-1290. doi:10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX, Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065-2071. doi:10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Tseng YT, Wang SM, Huang KJ, Wang CT. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J Biomed Sci. 2014;21:34. doi:10.1186/1423-0127-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Siu YL, Teoh KT, Lo J, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008;82:11318-11330. doi:10.1128/jvi.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi:10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, et al. The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis. PLoS Pathog. 2014;10:e1004320. doi:10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Teoh KT, Siu YL, Chan WL, et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838-3852. doi:10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yang Y, Xiong Z, Zhang S, et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. 2005;392:135-143. doi:10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fleming JO, Shubin RA, Sussman MA, Casteel N, Stohlman SA. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology. 1989;168:162-167. doi:10.1016/0042-6822(89)90415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751-752. doi:10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74:8127-8134. doi:10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838-6850. doi:10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Escors D, Ortego J, Laude H, Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J Virol. 2001;75:1312-1324. doi:10.1128/jvi.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hu Y, Wen J, Tang L, et al. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinformatics. 2003;1:118-130. doi:10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. TMHMM server. [Google Scholar]
- 160. ExPASy-ProtScale server. [Google Scholar]
- 161. Pascual MR. Coronavirus SARS-CoV-2: analysis of subgenomic mRNA transcription, 3CLpro and PL2pro protease cleavage sites and protein synthesis. https://arxiv.org/ftp/arxiv/papers/2004/2004.00746.pdf. Published 2020.
- 162. Kern DM, Sorum B, Hoel CM, et al. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs [published online ahead of print June 18, 2020]. bioRxiv. doi:10.1101/2020.06.17.156554. [Google Scholar]
- 163. Lu W, Zheng BJ, Xu K, et al. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA. 2006;103:12540-12545. doi:10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tan Y, Teng E, Shen S, et al. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol. 2004;78:6723-6734. doi:10.1128/JVI.78.13.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Zeng R, Yang R, Shi M, et al. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J Mol Biol. 2004;341:271-279. doi:10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Law HKW, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366-2374. doi:10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yuan X, Yao Z, Shan Y, et al. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 2005;114:70-79. doi:10.1016/j.virusres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Khan S, Fielding BC, Tan THP, et al. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20-27. doi:10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548-557. doi:10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81:9812-9824. doi:10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Nelson CA, Pekosz A, Lee CA, Diamond MS, Fremont DH. Structure and intracellular targeting of the SARS-coronavirus orf7a accessory protein. Structure. 2005;13:75-85. doi:10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kopecky-Bromberg SA, Martinez-Sobrido L, Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol. 2006;80:785-793. doi:10.1128/jvi.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tan Y, Fielding BC, Goh P, et al. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol. 2004;78:14043-14047. doi:10.1128/jvi.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Yuan X, Wu J, Shan Y, et al. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74-85. doi:10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Keng CT, Choi YW, Welkers MRA, et al. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132-142. doi:10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Muth D, Corman VM, Roth H, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep. 2018;8:15177. doi:10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Meier C, Aricescu AR, Assenberg R, et al. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157-1165. doi:10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Xu K, Zheng BJ, Zeng R, et al. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279-285. doi:10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. von Brunn A, Teepe C, Simpson JC, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi:10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Calvo E, DeDiego ML, Garcíac P, López JA, Pérez-Brena P, Falcónc A. Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo. Virus Res. 2012;169:282-288. doi:10.1016/j.virusres.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450-452. doi:10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yount B, Roberts RS, Sims AC, et al. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol. 2005;79:14909-14922. doi:10.1128/JVI.79.23.14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Lu B, Tao L, Wang T, et al. Humoral and cellular immune responses induced by 3a DNA vaccines against severe acute respiratory syndrome (SARS) or SARS-like coronavirus in mice. Clin Vaccine Immunol. 2009;16:73-77. doi:10.1128/CVI.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kanzawa N, Nishigaki K, Hayashi T, Ishii Y, Furukawa S. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580:6807-6812. doi:10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yuan X, Yao Z, Wu J, et al. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am J Respir Cell Mol Biol. 2007;37:9-19. doi:10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- 186. Reghunathan R, Jayapal M, Hsu L, et al. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi:10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Tan Y, Tham P, Chan D, et al. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J Virol. 2005;79:10083-10087. doi:10.1128/JVI.79.15.10083-10087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9:e02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Yuan X, Shan Y, Yao Z, et al. Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol Cells. 2006;21:186-191. [PubMed] [Google Scholar]
- 190. Guo J, Petric M, Campbell W, McGeer PL. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251-256. doi:10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Freundt EC, Yu L, Park E, Lenardo MJ, Xu X. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631-6640. doi:10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Yuan X, Shan Y, Zhao Z, Chen J, Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J. 2005;2:66. doi:10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Geng H, Liu YM, Chan WS, et al. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 2005;579:6763-6768. doi:10.1016/j.febslet.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Pewe L, Zhou H, Netland J, et al. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J Virol. 2005;79:11335-11342. doi:10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Huang C, Peters CJ, Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J Virol. 2007;81:5423-5426. doi:10.1128/jvi.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Fielding BC, Tan Y, Shuo S, et al. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78:7311-7318. doi:10.1128/JVI.78.14.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Fielding BC, Gunalan V, Tan THP, et al. Severe acute respiratory syndrome coronavirus protein 7a interacts with hSGT. Biochem Biophys Res Commun. 2006;343:1201-1208. doi:10.1016/j.bbrc.2006.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Zhou Z, Huang C, Zhou Z, et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14+ monocytes. iScience. 2021;24:102187. doi:10.1016/j.isci.2021.102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Huang C, Ito N, Tseng CK, Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J Virol. 2006;80:7287-7294. doi:10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Dediego ML, Pewe L, Alvarez E, Teresa M, Perlman S, Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379-389. doi:10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81:718-731. doi:10.1128/jvi.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion [published online ahead of print August 27, 2020]. bioRxiv. doi:10.1101/2020.08.27.270637. [Google Scholar]
- 203. Li J, Liao C, Wang Q, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi:10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Zhang Y, Zhang J, Chen Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I [published online ahead of print May 24, 2020]. bioRxiv. doi:10.1101/2020.05.24.111823. [Google Scholar]
- 205. He JF, Peng GW, Min J, et al. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666-1669. doi:10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 206. Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276-278. doi:10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 207. Chiu R, Chim S, Tong Y, et al. Tracing SARS-coronavirus variant with large genomic deletion. Emerg Infect Dis. 2005;11:168-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Oostra M, de Haan CAM, Rottier PJM. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol. 2007;81:13876-13888. doi:10.1128/jvi.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Qiu M, Shi Y, Guo Z, et al. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882-889. doi:10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi:10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Pancer K, Milewska A, Owczarek K, et al. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020;16:e1008959. doi:10.1371/journal.ppat.1008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Schuster NA. Characterization and structural prediction of the putative ORF10 protein in SARS-CoV-2 [published online ahead of print January 4, 2021]. bioRxiv. doi:10.1101/2020.10.26.355784. [Google Scholar]
- 213. Cagliani R, Forni D, Clerici M, Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect Genet Evol. 2020;83:104353. doi:10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Hassan SS, Attrish D, Ghosh S, et al. Notable sequence homology of the ORF10 protein introspects the architecture of SARS-COV-2. Int J Biol Macromol. 2021;181:801-809. doi:10.1101/2020.09.06.284976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459-468. doi:10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Mishra S. Designing of cytotoxic and helper T cell epitope map provides insights into the highly contagious nature of the pandemic novel coronavirus SARS-CoV2. R Soc Open Sci. 2020;7:201141. doi:10.26434/chemrxiv.12253463.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569-575. doi:10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Díaz J. SARS-CoV-2 molecular network structure. Front Physiol. 2020;11:870. doi:10.3389/fphys.2020.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Li J, Guo M, Tian X, et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Medicine (New York, NY). 2021;2:99-112.e7. doi:10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Xie X, Lokugamage KG, Zhang X, et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat Protoc. 2021;16:1761-1784. doi:10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]