Abstract
Protein structure prediction and design can be regarded as two inverse processes governed by the same folding principle. Although progress remained stagnant over the last two decades, the recent application of deep neural networks to spatial constraint prediction and end-to-end model training has significantly improved the accuracy of protein structure prediction, largely solving the problem at the fold level for single-domain proteins. The field of protein design has also witnessed dramatic improvement, where noticeable examples have shown that information stored in neural-network models can be used to advance functional protein design. Thus, incorporation of deep learning techniques into different steps of protein folding and design approaches represents an exciting future direction and should continue to have a transformative impact on both fields.
Keywords: Deep learning, protein structure prediction, protein design, template-based modeling, template-free modeling, end-to-end training
Introduction
The diverse physiological functions performed by proteins across all organisms are mediated by the unique three-dimensional structures adopted by specific amino acid sequences. Given the cost, both financially and timewise, associated with experimentally determining a protein’s structure and function, extensive effort has been made to develop computational methods capable of modeling the structures of natural protein sequences and/or designing new sequences with novel structures and functions beyond proteins observed in nature. The technique of deep machine learning [1], which has revolutionized many fields of research, including computer vision, speech recognition, strategy games and medical diagnosis, has recently made a significant impact on protein structure prediction and design. In this review, we will highlight methods used for protein structure prediction and protein design, as well as the impact brought about by deep learning on these fields, where a particular emphasis will be put on developments that have occurred within the past few years.
Protein Structure Prediction and Impact Brought About by Deep Learning
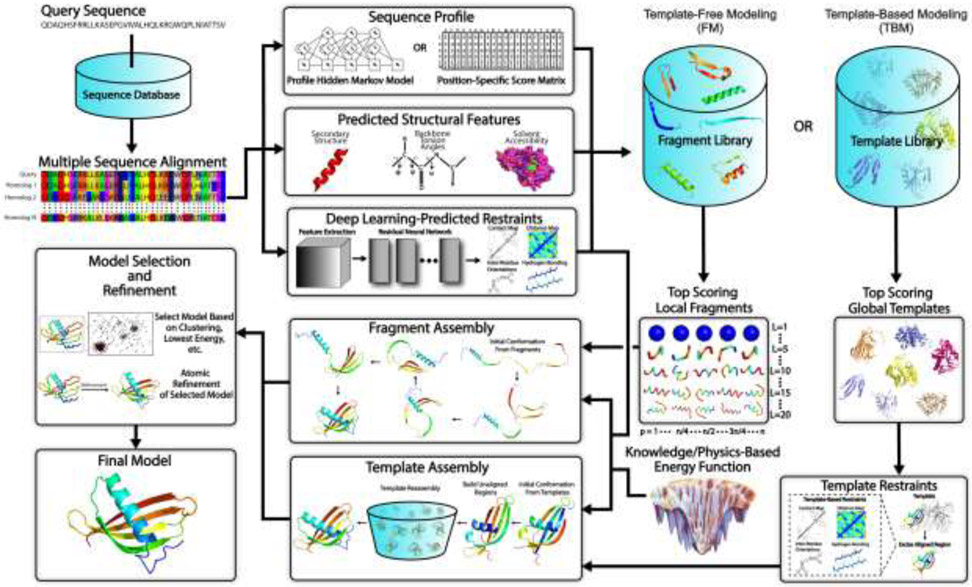
The goal of protein structure prediction is to use computational methods to determine the spatial location of every atom in a protein molecule starting from its amino acid sequence. Depending on whether a template structure is used, protein structure prediction approaches can be generally categorized as either template-based modeling (TBM) or template-free modeling (FM) methods. While TBM constructs models by copying and refining structural frameworks of other related proteins, called templates, identified from the PDB, FM aims to predict protein structures without using global template structures. FM methods have also been referred to as ab initio or de novo modeling approaches. A general pipeline that illustrates the key steps involved in traditional TBM and FM methods is depicted in Fig. 1.
Fig 1.
Typical steps involved in template-free and template-based protein structure prediction approaches. Starting from a query sequence, an MSA is generated by identifying homologous sequences from a sequence database. The MSA is then converted into a sequence profile and used to predict structural features such as the secondary structure, backbone torsion angles and solvent accessibility. For fragment assembly-based FM methods, these structural features together with the sequence profile are used to search a fragment library to identify high scoring local fragments. For TBM methods, they are used by threading protocols to identify global template structures. Meanwhile, co-evolutionary information is extracted from the MSA and fed into a deep residual neural network to predict spatial restraints such as inter-residue long-range contacts, distances, hydrogen bonds and torsion angles. For full-length model construction, structure assembly simulations are performed under the guidance of a composite force field which usually combines the generic knowledge- and/or physics-based energy function with deep neural network feature prediction (plus template-based restraints in the case of TBM). Finally, representative models are typically selected from the lowest energy conformations or based on structural clustering, followed by atomic-level refinement to generate the final model.
Classical Approaches to Template Based Modeling
There are four key steps involved in TBM methods: (1) identification of experimentally solved proteins (templates) structurally related to the protein to be modeled (query), (2) alignment of the query and the template proteins, (3) construction of the initial structure frameworks by copying the aligned regions of the template structure, and (4) construction of the unaligned regions and refinement of the global structure. The first two steps are intertwined and usually done in a single procedure called template recognition, while the last two steps are often accomplished in the template structure refinement procedure.
Depending on the evolutionary distance between the query and template, TBM has been historically divided into comparative modeling (CM), which is designed for targets with close homologous templates where the templates can typically be identified by sequence-based alignment, and threading, which is designed for detecting more distantly homologous templates by combining sequence profiles and/or Hidden Markov Model (HMM) alignment with local structure feature prediction [2,3]. Examples of predicted local structural features include torsion angles, secondary structure, and solvent accessibility [2]. With the progress of the field, the difference between CM and threading has become increasingly blurred and most of the TBM approaches nowadays start with templates identified by advanced threading programs. Since different threading programs are trained with different scoring function and alignment algorithms, the template recognition and alignment results are often diverse for the same query sequence. This has resulted in the prevalence of meta-threading programs [4], which collect and combine template alignments from a set of complementary threading algorithms. Since there are many more ways for a threading program to get incorrect alignments than to get a correct alignment [5], the consensus template selected from the meta-threading templates often has a higher accuracy on average than any of the individual threading programs.
Since threading templates only provide gapped Cα traces, which have no practical use for detailed protein function annotation and/or virtual ligand screening, many programs have been developed to assemble and refine full-length atomic structural models starting from the template alignments. Among them, MODELLER [6] is one of earliest programs which builds atomic models by optimally satisfying spatial restraints derived from a threading alignment, where the restraints are expressed as probability density functions for the restrained features. While TBM approaches based solely on restraint satisfaction often constrain the models close to the template, TASSER represents one of the first approaches that showed a consistent ability to draw the templates closer to the native structure [7]. The most successful TBM method is probably I-TASSER [8], which is an extension of TASSER and has been consistently ranked as the top automated method in the community-wide Critical Assessment of Structure Prediction (CASP) experiment, whose goal is to benchmark the state of the art in protein structure prediction [9]. In the I-TASSER pipeline, continuous fragments are excised from the template alignments and reassembled through replica-exchange Monte Carlo (REMC) simulations, where the unaligned regions (mainly loops) are built ab initio using a lattice-based system injunction with the aligned fragments. One of the key reasons for the success of I-TASSER, especially on template refinement, is its effective combination of multiple threading templates (often more than 20-50) under the guidance of an optimal knowledge-based force field whose parameters were extensively optimized using large-scale structural decoys. Following a similar idea, RosettaCM was developed which assembles global structural folds by recombining aligned segments of threading templates and building unaligned regions de novo in torsion space using gradient-based minimization [10].
Classical Approaches to Template-Free Modeling (FM)
Unlike TBM, FM approaches predict protein structures without the use of global template information (Fig 1). One of the most effective methods for constructing FM models is fragment assembly, an idea originally pioneered by Bowie and Eisenberg in 1994 [11]. More modern fragment assembly approaches include Rosetta [12] and QUARK [13], which first identify local structural fragments, with either discrete (3 and 9 AA long) or continuous lengths (1-20 AA), from other unrelated proteins based on the profile-profile similarity and comparison of the local structural features such as secondary structure, solvent accessibility and torsion angles, either predicted for the query or extracted from the templates. In the next step of fragment assembly simulations, the backbone torsion angles for a specific region of the simulated structure are replaced with those from a selected fragment, either assuming ideal bond lengths and angles [12], or directly taking these from the fragments themselves [13]. Loop closure may also be used, which adjusts the torsion angles around the substitution site in order to prevent large conformational changes downstream [14]. The rationale for constructing models through fragment assembly is two-fold: it reduces the entropy of the conformational search space, while ensuring the local structures of models are well formed as the fragments are selected from experimental structures of other proteins, which can help compensate for inaccuracies in the energy functions used for modeling. To improve the efficiency of conformational sampling, Rosetta [12] uses simulated annealing Monte Carlo simulations, while QUARK [13] uses REMC simulations with as many as 11 different conformational moves and extracts distance-profile–based contact maps from the generated fragments in order to guide the simulations towards the native structure [15].
Both QUARK and Rosetta have demonstrated excellent performance in the FM section of the CASP experiment by successfully folding protein targets that lack identifiable homology templates [16-18]. Despite the success, the Monte Carlo simulation-based fragment assembly process can be time-consuming compared to TBM approaches, since FM methods need to create models starting from random conformations. Encouraging progress has been recently made by rapid optimization techniques such as gradient descent to accurately fold protein sequences [19,20]. One condition for the success is that a significantly high number of long-range spatial constraints are required to reshape and smoothen the energy landscape so that the gradient descent-based optimization search is not overly trapped in local minima. Meanwhile, many repeated simulations must be performed in order to ensure the identification of the global minimum energy state [19].
Early Effort in Inter-residue Contact Prediction to Assist FM
Since the structural fold of a protein can be specified by the inter-residue contact map, considerable effort has been devoted to contact prediction. One of the earliest sequence-based contact prediction methods used correlated mutations observed in multiple sequence alignments (MSAs) to predict inter-residue contact maps [21]. The hypothesis behind the approach was that if mutations that occur at two positions in an MSA are correlated, these positions are more likely to form a contact in 3D space. This is because there is evolutionary pressure to conserve the structures of proteins and a mutation at one position may be rescued by a corresponding mutation at a nearby residue. The accuracy of co-evolution-based contact map prediction remained low for many years due to the inability to distinguish between direct and indirect interactions, where indirect interactions occur when residues appear to co-evolve but do not actually form contacts. For example, if Residues A and B are both in contact with Residue C, A and B often appear as if they co-evolve even when there is no physical contact between them. There is evidence showing that such co-evolution may have a functional cause [22] rather than a structural one, which resulted in the failure of structure-based contact derivation.
Progress in contact prediction remained stagnant for some time. However, a leap in contact prediction accuracy took place when algorithms started utilizing global prediction approaches. Early methods mainly predicted contacts between residue pairs one-at-a-time using techniques such as mutual information, thus ignoring the interactions with other residue pairs and the global context in which the interactions took place; this is largely why it was difficult for these local methods to distinguish between direct and indirect interactions. The introduction of global statistical models determined through the use of direct coupling analysis (DCA) was more successfully able to distinguish between these direct and indirect interactions [23,24]. The goal of such global statistical models is to determine the set of direct interactions that most harmoniously accounts for the observed sequence co-variation by simultaneously considering the entire set of pairwise interactions. Since all pairwise interactions are simultaneously considered, instead of just considering one interaction at a time and ignoring the global context in which the interactions take place, DCA was able to significantly improve the contact prediction accuracy.
Many DCA techniques fit a Markov random field (MRF), or more specifically a Potts model, to an MSA. An MRF is a graphical model that represents each column of an MSA as a node that describes the distribution of amino acids at a given position, where the edges between nodes indicate the joint distributions of amino acids between each pair of positions. The couplings or co-evolutionary parameters can be determined from the edge weights. Since fitting an MRF model using its actual likelihood function is computationally intractable due to the need to calculate the partition function, various approximations have been developed including those based on message passing [23], Gaussian approximation [25], mean-field approximation [24], and pseudo-likelihood maximization [26]. Another popular method was introduced by PSICOV [27], which determines the coupling parameters by estimating the inverse covariance matrix or precision matrix using a graphical LASSO penalty (L1 regularization) instead of directly fitting an MRF model to an MSA. This was later extended by ResPRE [28], where the inverse covariance matrix is estimated using L2 regularization instead of L1 regularization. Network deconvolution has also been used to distinguish direct from indirect interactions determined from co-evolutionary data [29].
Accurate Structural Feature Prediction by Deep Learning Techniques
The field of protein structure prediction has been considerably transformed by the recent use of deep machine learning techniques to generate high quality geometric feature prediction. In addition to the high accuracy of model training enabled by multi-layer neural networks [1], another important advantage of deep learning is its ability to predict multiple structural features, including contacts, distances, inter-residue torsion angles and hydrogen bonds. The combination of these structural features with the classical folding simulation methods has significantly improved the modeling accuracy of protein structure prediction, especially for FM protein targets which lack homologous templates [16,19,20].
The early focus of deep learning in protein structure prediction was on contact map prediction following the long history of contact prediction in the field. Along this line, RaptorX-Contact [30] reformulated the pair-wise contact prediction problem as an image segmentation task where the whole contact map is regarded as the image and each residue pair corresponds to a pixel in the image. The success of this approach can be partially attributed to the ability of deep learning to simultaneously consider the global set of pair-wise interactions instead of considering only one interaction at a time, thereby leading to more accurate discrimination between direct and indirect contacts [30]. The approach introduced by RaptorX-Contact was adapted by methods such as ResPRE [28] and TripletRes [31], which use a similar deep learning architecture but with a unique set of features that include multiple co-evolutionary coupling matrices directly deduced from deep MSAs without post-processing.
A similar residual neural network was later extended to predict the probability that the distance between two residues falls within a given distance range instead of predicting a binary contact map [32]. The power of distance map-guided folding was convincingly demonstrated by AlphaFold in the CASP13 experiment, in which the program utilized an ultra-deep neural network composed of 220 residual blocks to predict distance maps for a query sequence [19]. The distance maps were then used to guide their fragment assembly and gradient descent-based folding simulations for full-length structure construction. AlphaFold also used a unique fragment generation strategy where they leveraged deep learning to produce short structural fragments de novo. To accomplish this, they trained a generative network to create fragments based on prediction of the torsional angles for a selected region of a protein. This approach allows for the generation of fragments conditioned on the input features and eliminates the need to identify near native fragments from a library of existing fragment structures.
The success of deep learning-based contact and distance map prediction has raised the question of what other constraints can be accurately predicted using deep learning. As protein structure modelers have known for years that knowledge-based energy functions that are dependent only on residue-residue distances are often not as accurate as those that use both distances and orientations [33], a natural extension of distance prediction is inter-residue torsion angle orientation prediction. Orientation-dependent energy functions are important as certain types of inter-residue interactions require not only distance proximity but also specific orientations between the residue pairs, e.g., β-strand pairing. Furthermore, the geometry of a structure cannot be uniquely determined without torsional angle orientation information, as distance information alone cannot differentiate between a pair of mirrored structures. Recently, trRosetta has advanced the idea of inter-residue torsion angle prediction by simultaneously predicting both pairwise residue distances and inter-residue torsion angles from co-evolutionary features using a unified deep ResNet [20]. More recently, Li et al. extended the deep learning predictor TripletRes to DeepPotential and predicted the ensemble of contact, distance, torsion angle and hydrogen bonding maps, which were found to be highly effective at modeling non-homologous protein targets in the CASP14 experiment [34].
End-to-End Training with Attention Networks Has Nearly Solved the Single-Domain Protein Structure Prediction Problem
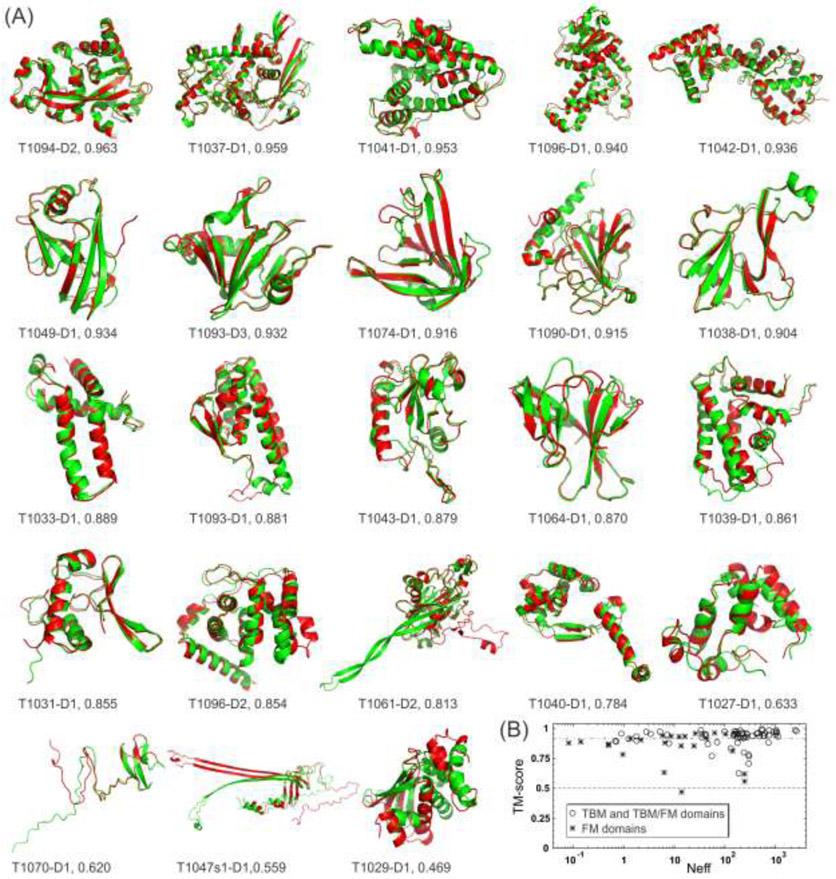
The most exciting progress in the history of protein structure prediction was recently brought about by AlphaFold2, the second iteration of AlphaFold developed by the Google DeepMind team [35], which achieved an unprecedented modeling accuracy in the CASP14 experiment. Out of the 89 domains with experimentally released structures, AlphaFold2 generated first-rank models with TM-scores >0.5 for 88 domains, where 59 of them had TM-scores >0.914. Here, TM-score is a sequence length-independent metric that measures protein structural similarity and takes a value in the range of (0, 1] [36], where PDB statistics show that a TM-score >0.5 indicates that two structures share approximately the same SCOP/CATH fold [37]. Moreover, we collected a set of 112 single-domain proteins whose structures were solved by both NMR and X-ray crystallography and had sequence identities >95% and alignment gaps < 10 AA, where we found the average TM-score was 0.807±0.107 between the NMR and X-ray structures. Thus, AlphaFold2 could fold nearly all individual domains in CASP14, with around 2/3 (=59/89) of the cases having accuracy comparable to low-to-medium resolution experimental models if we use a cutoff TM-score of 0.914 (=0.807+0.107). Fig. 2A lists a comparison of the AlphaFold2 first-rank models (green) overlaid on the experimentally solved structures (red) for all 23 free-modeling (FM) targets, which are the hardest targets to model due to the lack of templates in the PDB. AlphaFold2 created the correct fold for the core regions for all but one target (T1029-D1), which was a small single-domain protein (125 AA) whose structure was solved by NMR. For the other two targets with the lowest TM-scores (T1047d1-D1 and T1070-D1), which came from one chain of a heterodimer and the N-terminal domain of a 4-domain protein, respectively, the error of the AlphaFold2 models was mainly at the disordered tail regions but, again, the core regions were correctly folded. These data suggest that AlphaFold2 nearly solved the problem of single-domain protein structure prediction, at least at the fold level.
Fig 2.
Domain-level protein structure prediction results for AlphaFold2 in the CASP14 experiment. (A) The first-rank models by AlphaFold2 (green) superposed on the experimental structures (red) for the 23 FM domains, together with the domain ID and TM-score values. The pictures are listed in descending order of the TM-scores of the AlphaFold2 models. (B) TM-score versus Neff, the number of effective sequences in the multiple sequence alignments collected by DeepMSA, for all 89 FM (stars) and TBM and TBM/FM (circles) domains. Dashed and dashed-dotted lines mark the two TM-score cutoffs at 0.5 and 0.914, respectively.
Although most of the top participating groups in CASP14 achieved quite a significant improvement over CASP13 [38], AlphaFold2 outperformed the second-best group by a large margin with the average TM-score differing by 23% (0.903 vs 0.732). For the FM targets, the gap increased to 38% (0.840 vs 0.608). Interestingly, there was nearly no correlation (with a PCC =0.145) between the TM-score of the AlphaFold2 models and the logarithm of the Neff (the number of effective sequences) of the multiple sequence alignments collected by the DeepMSA program [39] searched through the metagenome sequence databases (Fig. 2B). For the other top ten groups, such correlation is obvious with PCCs ranging from 0.491 to 0.637. Although different groups use different strategies to collect MSAs and some may involve manual MSA search, the data shown in Fig. 2B is encouraging as the modeling accuracy by AlphaFold2 likely depends less on the availability of evolutionarily homologous sequences in the sequence databases.
Compared to the first iteration of AlphaFold in CASP13, which was driven by convolutional neural network-based distance map prediction, one of the major new developments of AlphaFold2 is the attention-based neural network architecture that attends arbitrarily over the full MSA, which allows the system to select relevant sequences from the MSAs and extract richer input information. Moreover, instead of using gradient descent optimization to construct models based on the predicted distance restraints, as AlphaFold did in CASP13, AlphaFold2 utilizes a full end-to-end training system from sequence to structure models using iterative structural refinement based on local structural error estimation. As part of this, the system replaces traditional folding simulations with a structure module composed of 3D equivariant transformer neural networks, which treat each amino acid as a gas of 3D rigid bodies and directly builds the protein backbone and sidechains. All these advantages, together with the extensive computing resources which are beyond what are accessible to most of the academic research laboratories, contribute to the significant improvement of the state of the art of deep learning-based protein structure prediction [35]. As an unprecedented achievement made by an industrial research company, however, the scientific impact will critically depend on whether and how far the method and technique, including the source codes of the programs, are made publicly available to the community.
Advances in Functional De Novo Protein Design
De Novo Protein Design
Protein design can be conceptually regarded as the inverse of protein structure prediction in that protein structure prediction aims to model unknown 3D structures from known sequences, while protein design attempts to identify new amino acid sequences that fold into given structural frameworks. De novo protein design usually contains two steps, the construction of a structural framework (or fold) and the identification/optimization of new amino acid sequences for that framework.
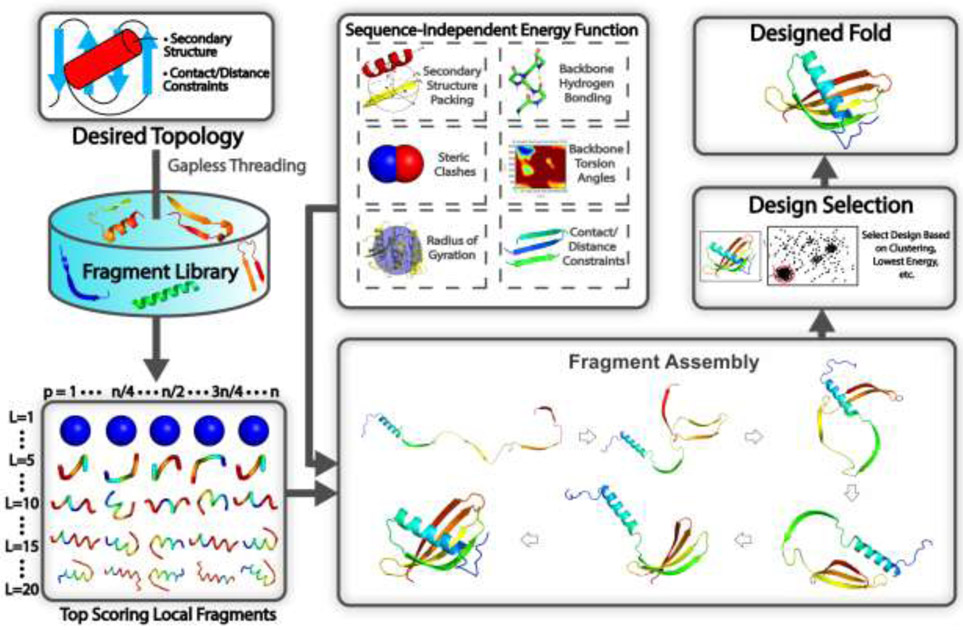
In addition to its use in protein structure prediction, the idea of fragment assembly has been successfully used to address the first step in de novo protein design, which is the construction of new protein folds beyond those observed in nature. One of the landmark achievements in de novo protein design was the design of Top7 in 2003 [40], which was one of the few proteins designed without a natural structural analog. The design of Top7 and other more recent de novo designed proteins have expanded on the strategies used by fragment assembly-based structure prediction methods, where a generic pipeline for such approaches is highlighted in Fig. 3. Instead of starting from an amino acid sequence, popular structure design methods such as RosettaRemodel [41] start from a predefined secondary structure and other user-defined constraints such as inter-residue distances, which define a target fold. Fragments are then picked with secondary structures and backbone torsion angles that are compatible with the predefined secondary structure. The simulation strategy is slightly altered as the amino acid-specific energy function is replaced with an energy function that is independent of the amino acid sequence and generic side-chain centers of mass are used to avoid steric clashes [41]. Another popular method for designing backbone structures is to generate them using idealized parametric models [42], although this approach is typically more useful for designing helical bundle proteins and is not as effective at designing proteins with more complex topologies or hydrogen bonding networks.
Fig 3.
Typical steps involved in a fragment assembly-based approach to design new protein structures. Starting from the desired secondary structure together with user-defined packing restraints, such as residue-residue contact/distance restraints, the query is searched through a non-redundant PDB structure library using gapless threading to generate position-specific fragment structures. High scoring fragments, which may range from 1-20 residues long, are identified based on the complementarity between the desired secondary structure and a fragment’s secondary structure and backbone torsion angles. Then during the folding simulations, the top scoring local fragments are assembled under the guidance of a sequence-independent energy function, which accounts for fundamental rules that govern protein folding such as secondary structure packing, backbone hydrogen bonding, favorable backbone torsion angles, steric clashes, radius of gyration, as well as the artificial contact/distance restraints supplied by the user. As the method is sequence independent, generic side-chain centers of mass, typically those for valine, are used to evaluate energy terms such as steric clashes. Following the folding simulations, the final design may be selected based on clustering of the simulation decoys, by selecting the lowest energy structure, or through whatever filter the user deems appropriate.
Following the generation of the initial target folds based on the input constraints, iterative rounds of sequence and structure optimization are performed [41] for amino acid sequence design. Here, sequence design and structure optimization can be performed using combined physics and knowledge-based energy functions such as Rosetta [43] or EvoEF2 [44]. These approaches typically start from a fixed protein backbone, where the amino acid side-chain conformation or rotamer of a randomly selected position is substituted for another rotamer randomly selected from a rotamer library. The corresponding energy changes caused by the mutation are then calculated using the physical energy function, where mutations are accepted or rejected based on the Metropolis criterion. Following sequence design, local structure optimization is performed and the sequence design/backbone optimization is iteratively repeated [41].
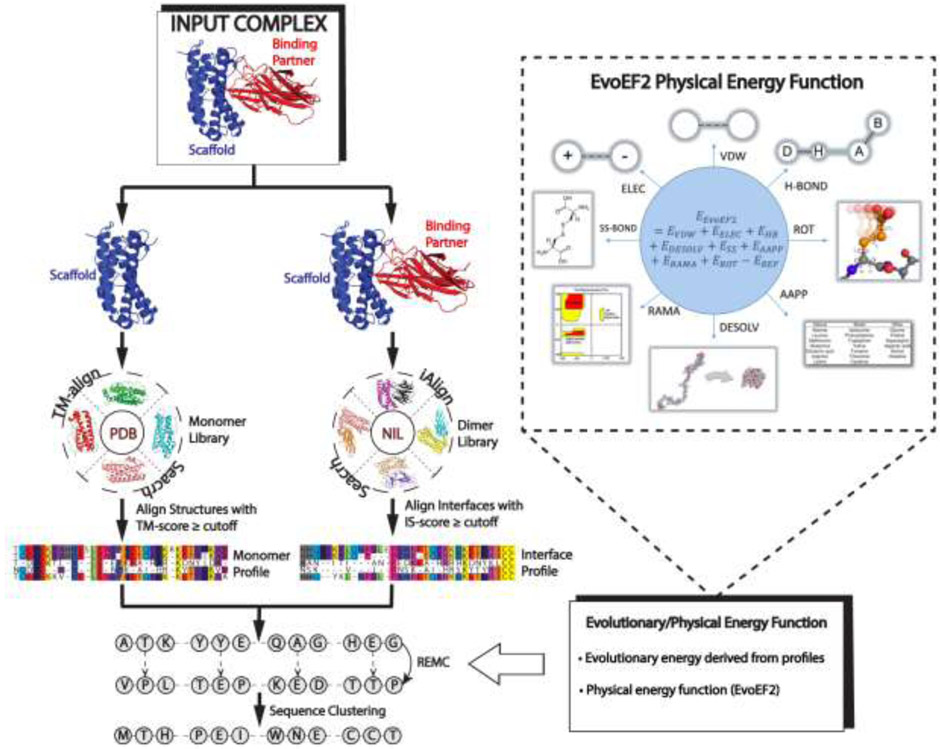
Most recently, Pearce et al. proposed an automated protein design pipeline, EvoDesign (Fig. 4) [45], which incorporates evolutionary profiles derived from natural structural analogs in the force field in order to enhance the folding stability of the designed sequences. For protein-protein interaction (PPI) design, EvoDesign starts from an input complex structure and identifies both monomeric and interface structural analogs from databases of solved protein structures. These structural analogs are converted into PPI evolutionary profiles, which are then combined with a physical energy function for PPI design, EvoEF2, to guide the REMC sequence design simulations.
Fig 4.
A protocol for evolution-based protein-protein interaction design used by EvoDesign. The procedure starts from an input complex, for which monomer/interface structural homologs are identified from the PDB library through TM-align and iAlign searches, respectively. Structural profiles are then constructed from the alignments of the monomer/interface analogs and used in conjunction with a physics-based potential, EvoEF2, to guide the REMC simulations to design novel protein sequences. The final designs are selected from the center of the largest cluster of designed sequence decoys.
De Novo Design of Proteins with Complex Structures and Functions
The past few years have seen rapid progress in de novo protein design, where proteins with increasingly complex structural characteristics and functions have been created [46-56]. Earlier de novo designed proteins had highly idealized structures without functional sites and with a single low energy conformation. However, recent work by Wei et al. demonstrated that it is possible to design proteins that adopt multiple low energy states that assume significantly different conformations [46]. In the study, the authors used Rosetta to design a helical bundle that either adopted a short (~66 Å height) or long (~100 Å height) state based on the environmental conditions, which mimicked the action of membrane fusion proteins. Additionally, new studies have focused on designing proteins with more complex logical functions for use in synthetic biology. In this regard, Chen et al. was able to design logic gates that controlled transcription and enzymatic activity via the association of different designed coiled-coil heterodimers [47]. The backbone structures of each coiled coil were designed in a previous study using parametric modeling to generate the helices and loop fragments to connect them into a single chain [48]. The association between different heterodimers was achieved using the Rosetta HBNet protocol [49], which can be used to exhaustively enumerate all of the hydrogen bond networks available for a given design space in order to design highly specific protein-protein interactions.
Rosetta has also been applied to the classical problem of designing proteins with significant β-sheet content, which have enriched hydrogen bonding patterns. For example, Dou et al. designed fluorescence-activated β-barrel proteins using either ideal parametric models or fragment assembly [50]. Interestingly, the authors found that the ideal backbones generated by the parametric models had unfavorable steric strain and hydrogen bonding interactions. These problems were alleviated by building backbones using fragment assembly and introducing kinks and bulges into the structures, producing a stable and functional protein. Another challenging problem in protein design is the ability to create proteins that can bind to highly functionalized small molecules. Polizzi et al. addressed this problem by creating a unit of protein structure called the van der Mer, which directly maps the backbone of each amino acid to preferred positions of interacting chemical groups [56]. They then used their method to design proteins capable of binding the complex drug apixaban, which has implications for the de novo design of customized biosensors and enzymes, among other applications.
De Novo Design of Therapeutic Proteins
Other studies have focused on designing proteins for therapeutic applications. One strategy to accomplish this goal is to design proteins that are capable of binding natural proteins with high affinity. For instance, Chevalier et al. described a protocol for generating large pools of mini-proteins with different backbone scaffolds composed of ~40 residues produced by fragment assembly [51]. The authors demonstrated that given advances in high throughput experimental techniques and computational modeling, an unprecedented number of designed proteins could be tested. This resulted in the production of highly stable designs that could bind to influenza hemagglutinin and provide prophylactic protection without eliciting an adverse immune response [51]. Another study by Silva et al. used parametric modeling to design mimics of IL-2 and IL-15 capable of binding the IL-2 receptor βγc heterodimer but without binding sites for CD25 and CD215, producing a potent anti-cancer effect without the toxicity of natural IL-2 therapeutics [52]. Another strategy is to use de novo design methods such as Rosetta, TopoBuilder, and EvoDesign to generate computationally designed immunogens with topologies designed to stabilize functional motifs that are capable of inducing the production of virus-neutralizing antibodies [53-55,57]. These successes highlight the potential for de novo protein design to create therapeutics with tailor-made characteristics and superior efficacy compared to those produced by traditional approaches.
Given the havoc caused by the ongoing COVID-19 pandemic, researchers are seeking to develop new proteins that can serve as therapeutic treatments against the epidemic. Along this line, Huang et al. proposed the design of de novo peptides to inhibit the association of the SARS-CoV-2 Spike protein, which is the pathogen behind COVID-19, with the human ACE2 receptor [58]. The in silico assay experiments showed that the peptide inhibitors designed by EvoEF2 and EvoDesign had a significantly higher affinity for the binding domain of the Spike protein than the wildtype hACE2 receptor did. With a similar goal, Cao et al. applied Rosetta’s fragment assembly design method to design protein inhibitors for the SARS-CoV-2 Spike protein [59]. The authors used two design strategies, either incorporating the native helical interface between ACE2 and the Spike protein or generating novel interfaces de novo by optimizing the rotamer interaction field. After affinity maturation, they found the second approach was able to create proteins capable of potently inhibiting SARS-CoV-2 with picomolar affinity.
Improving the Accuracy of De Novo Fold Design without User-Defined Constraints
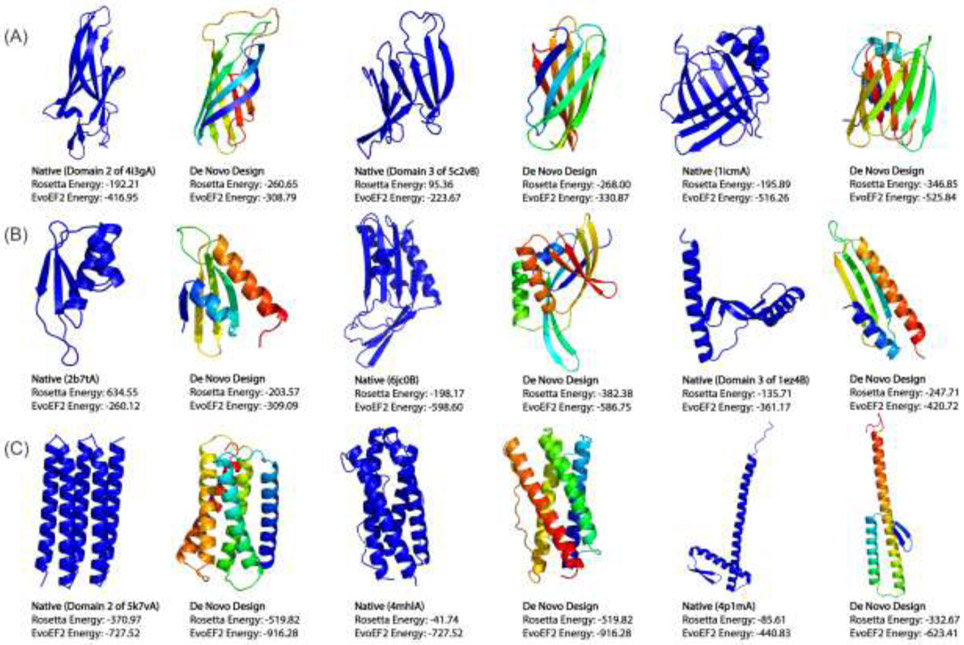
One prominent challenge associated with designing proteins with novel structures and functions is that de novo protein design remains somewhat of an art form, as designers are often required to manually specify how the different secondary structure elements (SSEs) should pack in order to produce a well-folded protein [50,51,60]. Furthermore, the design success rate is quite low in some studies, which may be improved by altering the design procedure in an iterative fashion based on the experimental results [51]. To improve the success rate of de novo protein design, Pearce and Zhang recently developed a new method called FoldDesign (https://zhanglab.ccmb.med.umich.edu/FoldDesign/). The method combines fragment assembly with multiple conformational movements and an optimized physics- and knowledge-based energy function to design protein-like scaffolds. Fig. 5 presents scaffolds designed using FoldDesign starting from 9 unique secondary structure topologies (α, β and αβ folds) obtained from native proteins without any pre-defined contact or distance restraints, where the sequences for each scaffold were designed using EvoDesign [45]. Notably, even in the absence of user-defined packing constraints, the method is able to produce well-folded scaffolds with complex tertiary structures, such as those composed of curved β-sheets, which required extensive pre-definition of packing rules in previous studies [50]. To assess the quality of the designed scaffolds, the designs along with the native proteins whose secondary structure were used as input to FoldDesign were scored using the Rosetta ref2015 [43] and EvoEF2 energy functions [44]. All of the designs had lower Rosetta energies and 7 out of 9 had lower EvoEF2 energies than their native counterparts, demonstrating the ability of using computational simulations to produce protein-like structures with new folds starting only from loose constraints such as the desired secondary structure composition.
Fig 5.
Protein folds designed de novo starting from 9 unique secondary structures. The designed folds and corresponding wildtype native proteins (with denoted PDB IDs) whose secondary structures were used as input are shown side-by-side for (A) 3 β proteins, (B) 3 α/β and α+β proteins, and (C) 3 a proteins. Even in the absence of pre-defined packing rules, such as inter-residue distance restraints, the designed new folds have well-packed topologies with lower or comparable Rosetta and EvoEF2 energies.
Deep Learning Applied to Protein Design
Deep learning has recently been successfully employed to various protein design strategies. One such strategy is to design a sequence given a known protein structure, where the native sequence recapitulation rate, or the percentage of native amino acids recovered at each position, is typically used as one of the key validation criterions. As an example of such methods, SPIN combines a neural network composed of two hidden layers with features such as the backbone torsion angles for a selected residue, fragment-derived sequence profiles, and rotamer-based energy profiles to design favorable sequences for a given structure, where the method achieved a native sequence recapitulation rate of 30.7% [61]. SPIN was later extended to SPIN2, which added an additional hidden layer and extra local/nonlocal features, thereby obtaining a native sequence recovery rate of 34.4% [62]. Using a different approach, Anand et al. recently extracted features around a local, voxelized environment for each residue, which were then fed into a convolutional neural network with six 3D convolutional layers [63]. The authors found that their method was able to achieve designs with greater sequence diversity than Rosetta. Using a different strategy, Greener et al. utilized variational autoencoders to add metal binding sites to existing protein sequences and to design new sequences conditioned on the desired topology of a protein [64]. This approach removes the constraint of starting from a known protein structure, directly allowing the generation of sequences conditioned on the desired fold of a protein.
Most recently, Anishchenko et al. set out to answer the question if the information stored in deep neural networks used to predict inter-residue distances and orientations could be applied to design new protein sequences and structures [65]. To address this, they used deep network hallucination, where they performed Monte Carlo sampling in sequence space, at each step feeding the sequences into the trRosetta deep neural network architecture in order to predict their distance maps and comparing them against a background distance map distribution. Mutations were accepted or rejected based on the Metropolis criterion, where the objective of the simulations was to maximize the information gain (Kullback-Leibler divergence) between the predicted distance maps and the background distribution. The method was able to produce diverse sequences that adopted stable, monomeric folds as assessed by circular dichroism. The developed method was then extended in two additional studies, where the hallucination was either completely constrained to design sequences for a fixed fold [66] or to design sequences that recapitulated native interfaces [67], while allowing the remainder of the protein to be hallucinated freely. The ability to constrain the design simulation to recapitulate native structural motifs is particularly impactful when considering functional inhibitor design, where the native interface from a known binder could be incorporated into the hallucinated proteins. However, the ability to freely hallucinate interfaces that can bind to therapeutic targets would remove such limitations and would address a long-standing problem in the field which is generating high affinity binders to arbitrary protein targets. Nevertheless, these studies demonstrate that it is possible to utilize the information stored in the deep neural networks used for protein structure prediction to design new protein sequences and structures.
Conclusion and Future Directions
The prediction of protein structures from amino acid sequences alone has remained an outstanding problem in structural biology since Anfisen first demonstrated that the information encoded in a protein sequence determines its structure more than 60 years ago. Now more than ever, there is an urgent need to develop high accuracy protein structure prediction methods, as advancements in high-throughput sequencing technology have greatly exacerbated the gap between the number of known sequences and the number of experimentally determined protein structures. For some time since the development of profile-based threading methods and fragment assembly approaches, progress in the field has remained slow and only incremental gains have been achieved. Nevertheless, recent advancements in co-evolution-based contact map prediction and especially the more recent deep learning-based spatial restraint prediction and end-to-end model training have revolutionized the field of protein structure prediction, greatly improving its accuracy and the ability to fold proteins, in particular those that lack homologous templates in the PDB.
The success of inter-residue contact- and distance-guided folding approaches raises the question of what other constraints can be predicted using deep learning and incorporated into structure assembly simulations. The most recent studies demonstrated that prediction of inter-residue torsional angles [20] and hydrogen-bonding networks [34] may represent a future direction of the field, where the use of other restraints should also be investigated. In addition to specific spatial feature predictions, the AlphaFold2 team [35] recently demonstrated that an end-to-end training system powered by attention-based neural networks could self-learn the feature derivation process and refine models based on the estimated local structure errors. They generated models with a TM-score above 0.5 for all domains (except for one whose structure was solved by NMR) in the CASP14 experiment, marking the solution of the single-domain protein structure prediction problem at a fold level [68]. Nevertheless, protein structure prediction is multifaced, including single-domain, multi-domain and quaternary complex structure modeling, the latter two of which were not assessed in CASP14. Even for single-domain structures, there were nearly 1/3 of cases for which the AlphaFold2 models were below the level of experimental accuracy. Given that most proteins perform their functions through interaction with other domains and chain partners in cells and that function annotation and drug discovery studies often requires atomic resolution models, all these problems must be carefully addressed before the convincing claim of a complete solution to the protein structure prediction problem. Another important dilemma raised by the successful use of deep learning is the difficulty in understanding what information is being learned by such approaches. Traditional energy force fields used for protein folding are easily interpretable as they include explicit terms that account for various physically important constraints that guide protein folding. However, deep learning is essentially a black box that does not provide any easily interpretable information on the physical principles that underlie protein folding. Thus, the ability to fold proteins based on first principles or physical principles, which is essential to understand the dynamics of protein folding, remains elusive. Nevertheless, while there certainly are many challenges in the field, the progress witnessed within the last few years provides hope that one of the most difficult and meaningful biological problems, predicting structures of proteins at their equilibrium state starting from the amino acid sequences alone, could be solved through the use of deep learning within the foreseeable future.
As the reverse procedure of ab initio folding, protein design has by far witnessed much less involvement of deep machine learning models. Given that the same physical principle governs both procedures, one can expect that more accurately modeled sequence and structure relationships obtained from deep neural network learning should help increase the accuracy and success rate of de novo protein design. Indeed, the use of deep network hallucination confirmed that it is possible to use the information stored in neural networks utilized for protein structure prediction to design novel protein sequences and structures. The extension of such networks to functional protein design should have dramatic implications as current de novo design approaches require users to pre-specify the length and composition of secondary structure elements. The ability to allow deep learning to select the most favorable composition of the designed scaffolds for a particular application would simplify the design process and allow for a more comprehensive exploration of viable solutions. Nevertheless, one drawback to such approaches is that most of the sophisticated deep learning models in structural bioinformatics are trained on MSAs, where the MSA construction often involves lengthy and time-consuming genome database searching; this may render it infeasible to incorporate these deep learning models with extensive sequence design simulations because each step of the sequence design iterations generates a new sequence and therefore requires new MSA construction and model training. In this regard, development of accurate and single sequence-based deep learning models might be important to overcome this barrier, which has in fact been demonstrated through the use of transformer neural networks by AlphaFold2. Overall, the integration of advanced deep learning algorithms with traditional structural folding approaches represents an exciting future avenue for both protein structure prediction and protein design and should continue to enable the next wave of innovation in both fields.
Acknowledgements
This work is supported in part by the National Institute of General Medical Sciences (GM136422, S10OD026825), the National Institute of Allergy and Infectious Diseases (AI134678), and the National Science Foundation (IIS1901191, DBI2030790, MTM2025426).
Footnotes
Declaration of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 2015, 521:436–444. [DOI] [PubMed] [Google Scholar]
- 2.Wu ST, Zhang Y: MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins-Structure Function and Bioinformatics 2008, 72:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soding J: Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21:951–960. [DOI] [PubMed] [Google Scholar]
- 4.Zheng W, Zhang C, Wuyun Q, Pearce R, Li Y, Zhang Y: LOMETS2: improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic Acids Res 2019, 47:W429–W436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y: Progress and challenges in protein structure prediction. Current Opinion in Structural Biology 2008, 18:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sali A, Blundell TL: Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 1993, 234:779–815. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Skolnick J: Automated structure prediction of weakly homologous proteins on a genomic scale'. Proceedings of the National Academy of Sciences of the United States of America 2004, 101:7594–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy A, Kucukural A, Zhang Y: I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010, 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J: Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins 2019, 87:1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song YF, DiMaio F, Wang RYR, Kim D, Miles C, Brunette TJ, Thompson J, Baker D: High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21:1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowie JU, Eisenberg D: An Evolutionary Approach to Folding Small Alpha-Helical Proteins That Uses Sequence Information and an Empirical Guiding Fitness Function. Proceedings of the National Academy of Sciences of the United States of America 1994, 91:4436–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohl C, Strauss C, Misura K, Baker D: Protein structure prediction using Rosetta. Methods in enzymology 2004, 383:66–93. [DOI] [PubMed] [Google Scholar]
- 13.Xu D, Zhang Y: Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins-Structure Function and Bioinformatics 2012, 80:1715–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canutescu AA, Dunbrack RL: Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Science 2003, 12:963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Zhang Y: Toward optimal fragment generations for ab initio protein structure assembly. Proteins-Structure Function and Bioinformatics 2013, 81:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W, Li Y, Zhang CX, Pearce R, Mortuza SM, Zhang Y: Deep-learning contact-map guided protein structure prediction in CASP13. Proteins-Structure Function and Bioinformatics 2019, 87:1149–1164.** The authors incorporated predicted contact maps from deep learning into the classic I-TASSER and QUARK frameworks, where the newly developed contact map-based folding approaches, C-I-TASSER (as 'Zhang-Server') and C-QUARK (as 'QUARK'), placed as the first and second best automated servers in CASP13, respectively.
- 17.Zhang CX, Mortuza SM, He BJ, Wang YT, Zhang Y: Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins-Structure Function and Bioinformatics 2018, 86:136–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovchinnikov S, Park H, Kim DE, DiMaio F, Baker D: Protein structure prediction using Rosetta in CASP12. Proteins-Structure Function and Bioinformatics 2018, 86:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin CL, Zidek A, Nelson AWR, Bridgland A, et al. : Improved protein structure prediction using potentials from deep learning. Nature 2020, 577:706-+.** Description of the top CASP13 human group, AlphaFold, which convincingly demonstrated the power of deep learning-based distance prediction to accurately fold proteins. AlphaFold also introduced deep learning to directly generate fragments de novo, instead of relying on identifying near native fragment structures from the PDB.
- 20.Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D: Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences 2020, 10.1073/pnas.1914677117:201914677.** The authors developed trRosetta, which is the first method to use deep learning to predict inter-residue orientations to assist 3D structure constructions. The source codes are publicly released.
- 21.Gobel U, Sander C, Schneider R, Valencia A: Correlated mutations and residue contacts in proteins. Proteins 1994, 18:309–317. [DOI] [PubMed] [Google Scholar]
- 22.Kass I, Horovitz A: Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins 2002, 48:611–617. [DOI] [PubMed] [Google Scholar]
- 23.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T: Identification of direct residue contacts in protein-protein interaction by message passing. Proceedings of the National Academy of Sciences of the United States of America 2009, 106:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M: Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proceedings of the National Academy of Sciences of the United States of America 2011, 108:E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldassi C, Zamparo M, Feinauer C, Procaccini A, Zecchina R, Weigt M, Pagnani A: Fast and Accurate Multivariate Gaussian Modeling of Protein Families: Predicting Residue Contacts and Protein-Interaction Partners. Plos One 2014, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekeberg M, Lovkvist C, Lan YH, Weigt M, Aurell E: Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Physical Review E 2013, 87. [DOI] [PubMed] [Google Scholar]
- 27.Jones DT, Buchan DWA, Cozzetto D, Pontil M: PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 2012, 28:184–190. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Hu J, Zhang CX, Yu DJ, Zhang Y: ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics 2019, 35:4647–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun HP, Huang Y, Wang XF, Zhang Y, Shen HB: Improving accuracy of protein contact prediction using balanced network deconvolution. Proteins-Structure Function and Bioinformatics 2015, 83:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Sun SQ, Li Z, Zhang RY, Xu JB: Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. Plos Computational Biology 2017, 13.** In this study, the authors proposed the use of deep residual neural networks for inter-residue contact map prediction, thus dramatically improving the prediction accuracy.
- 31.Li Y, Zhang C, Bell EW, Zheng W, Zhou X, Yu DJ, Zhang Y: Deducing high-accuracy protein contact-maps from a triplet of coevolutionary matrices through deep residual convolutional networks. bioRxiv 2020:326140.* In this study, the authors developed TripleRes for deep learning-based contact map prediction, which was ranked as one of the best methods in CASP13 and CASP14. The method extends on previous deep learning-based contact prediction approaches, including multiple co-evolutionary feature matrices.
- 32.Xu J: Distance-based protein folding powered by deep learning. Proc Natl Acad Sci U S A 2019, 116:16856–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Zhang Y: A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PLoS One 2010, 5:e15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Zheng W, Zhang C, Bell E, Huang X, Pearce R, Zhou X, Zhang Y: Protein 3D Structure Prediction by Zhang Human Group in CASP14. In 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction: 2020. [Google Scholar]
- 35.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Tunyasuvunakool K, Ronneberger O, Bates R, Žídek A, Bridgland A, et al. : High Accuracy Protein Structure Prediction Using Deep Learning. In 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction: 2020.** The authors introduce AlphaFold2, which uses end-to-end training based on attention neural networks to fold proteins. The authors were able to achieve unprecedented levels of accuracy, marking a fold-level solution of the single-domain protein structure prediction problem.
- 36.Zhang Y, Skolnick J: Scoring function for automated assessment of protein structure template quality. Proteins-Structure Function and Bioinformatics 2004, 57:702–710. [DOI] [PubMed] [Google Scholar]
- 37.Xu JR, Zhang Y: How significant is a protein structure similarity with TM-score=0.5? Bioinformatics 2010, 26:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishin N: 3D Assessment. In 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction: 2020. [Google Scholar]
- 39.Zhang C, Zheng W, Mortuza SM, Li Y, Zhang Y: DeepMSA: constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. Bioinformatics 2020, 36:2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D: Design of a novel globular protein fold with atomic-level accuracy. Science 2003, 302:1364–1368. [DOI] [PubMed] [Google Scholar]
- 41.Huang PS, Ban YEA, Richter F, Andre I, Vernon R, Schief WR, Baker D: RosettaRemodel: A Generalized Framework for Flexible Backbone Protein Design. Plos One 2011, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang PS, Oberdorfer G, Xu CF, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, Baker D: High thermodynamic stability of parametrically designed helical bundles. Science 2014, 346:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alford RF, Leaver-Fay A, Jeliazkov JR, O'Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, et al. : The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput 2017, 13:3031–3048.* Description of the Rosetta ref2015 energy function, including a detailed breakdown of each energy term. The ref2015 energy function achieved state-of-the-art results on various modeling and design tasks.
- 44.Huang X, Pearce R, Zhang Y: EvoEF2: accurate and fast energy function for computational protein design. Bioinformatics 2020, 36:1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearce R, Huang X, Setiawan D, Zhang Y: EvoDesign: Designing Protein-Protein Binding Interactions Using Evolutionary Interface Profiles in Conjunction with an Optimized Physical Energy Function. J Mol Biol 2019, 431:2467–2476.* The authors introduce EvoDesign for PPI design by extending evolutionary profile-guided design to include interface profiles and a newly developed physical energy function to improve the ability to design native-like protein sequences.
- 46.Wei KY, Moschidi D, Bick MJ, Nerli S, McShan AC, Carter LP, Huang PS, Fletcher DA, Sgourakis NG, Boyken SE, et al. : Computational design of closely related proteins that adopt two well-defined but structurally divergent folds. Proceedings of the National Academy of Sciences of the United States of America 2020, 117:7208–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen ZB, Kibler RD, Hunt A, Busch F, Pearl J, Jia MX, VanAernum ZL, Wicky BIM, Dods G, Liao H, et al. : De novo design of protein logic gates. Science 2020, 368:78-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen ZB, Boyken SE, Jia MX, Busch F, Flores-Solis D, Bick MJ, Lu PL, VanAernum ZL, Sahasrabuddhe A, Langan RA, et al. : Programmable design of orthogonal protein heterodimers. Nature 2019, 565:106-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyken SE: De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity (vol 352, aag1318, 2016). Science 2016, 353:879–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dou JY, Vorobieva AA, Sheffler W, Doyle LA, Park H, Bick MJ, Mao BC, Foight GW, Lee MY, Gagnon LA, et al. : De novo design of a fluorescence-activating beta-barrel. Nature 2018, 561:485-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam KH, Yao GR, et al. : Massively parallel de novo protein design for targeted therapeutics. Nature 2017, 550:74-+.* The authors describe a combined computational and experimental protocol that allows for the testing of an unprecedented number of designed proteins. The experiments resulted in the design of potent inhibitors for haemagglutinin and botulinum neurotoxin B.
- 52.Silva DA, Yu S, Ulge UY, Spangler JB, Jude KM, Labao-Almeida C, Ali LR, Quijano-Rubio A, Ruterbusch M, Leung I, et al. : De novo design of potent and selective mimics of IL-2 and IL-15. Nature 2019, 565:186-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sesterhenn F, Yang C, Bonet J, Cramer JT, Wen X, Wang Y, Chiang CI, Abriata LA, Kucharska I, Castoro G, et al. : De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 2020, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. : Proof of principle for epitope-focused vaccine design. Nature 2014, 507:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sesterhenn F, Galloux M, Vollers SS, Csepregi L, Yang C, Descamps D, Bonet J, Friedensohn S, Gainza P, Corthesy P, et al. : Boosting subdominant neutralizing antibody responses with a computationally designed epitope-focused immunogen. Plos Biology 2019, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polizzi NF, DeGrado WF: A defined structural unit enables de novo design of small-molecule-binding proteins. Science 2020, 369:1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ong E, Huang X, Pearce R, Zhang Y, He Y: Computational Design of SARS-CoV-2 Spike Glycoproteins to Increase Immunogenicity by T Cell Epitope Engineering. Comput Struct Biotechnol J 2020, 10.1016/j.csbj.2020.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang X, Pearce R, Zhang Y: De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging 2020, 12:11263–11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park Y-J, et al. : De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 2020, 10.1126/science.abd9909:eabd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang PS, Feldmeier K, Parmeggiani F, Velasco DAF, Hocker B, Baker D: De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy. Nature Chemical Biology 2016, 12:29-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li ZX, Yang YD, Faraggi E, Zhan J, Zhou YQ: Direct prediction of profiles of sequences compatible with a protein structure by neural networks with fragment-based local and energy-based nonlocal profiles. Proteins-Structure Function and Bioinformatics 2014, 82:2565–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Connell J, Li ZX, Hanson J, Heffernan R, Lyons J, Paliwal K, Dehzangi A, Yang YD, Zhou YQ: SPIN2: Predicting sequence profiles from protein structures using deep neural networks. Proteins-Structure Function and Bioinformatics 2018, 86:629–633. [DOI] [PubMed] [Google Scholar]
- 63.Anand N, Eguchi RR, Derry A, Altman RB, Huang P-S: Protein Sequence Design with a Learned Potential. bioRxiv 2020, 10.1101/2020.01.06.895466:2020.2001.2006.895466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greener JG, Moffat L, Jones DT: Design of metalloproteins and novel protein folds using variational autoencoders. Scientific Reports 2018, 8.* The authors extend deep learning to generate novel protein sequences conditioned on protein topologies and to add metal binding sites to existing protein sequences. This eliminates the need to start from an existing structure and allows users to generate sequences directly using deep learning.
- 65.Anishchenko I, Chidyausiku TM, Ovchinnikov S, Pellock SJ, Baker D: De novo protein design by deep network hallucination. bioRxiv 2020, 10.1101/2020.07.22.211482:2020.2007.2022.211482.** The authors demonstrate that deep neural networks used for protein structure prediction can be applied to protein design. The method also allows for the generation of stable proteins without predefinition of the length and composition of the secondary structure elements, thereby allowing deep learning to more fully sample the total range of favorable scaffolds.
- 66.Norn C, Wicky BIM, Juergens D, Liu S, Kim D, Koepnick B, Anishchenko I, Players F, Baker D, Ovchinnikov S: Protein sequence design by explicit energy landscape optimization. bioRxiv 2020, 10.1101/2020.07.23.218917:2020.2007.2023.218917. [DOI] [Google Scholar]
- 67.Tischer D, Lisanza S, Wang J, Dong R, Anishchenko I, Milles LF, Ovchinnikov S, Baker D: Design of proteins presenting discontinuous functional sites using deep learning. bioRxiv 2020, 10.1101/2020.11.29.402743:2020.2011.2029.402743. [DOI] [Google Scholar]
- 68.Callaway E: 'It will change everything': DeepMind's AI makes gigantic leap in solving protein structures. Nature 2020, 588:203–204. [DOI] [PubMed] [Google Scholar]