Abstract
Extant species have wildly different numbers of chromosomes, even among taxa with relatively similar genome sizes (for example, insects)1,2. This is likely to reflect accidents of genome history, such as telomere–telomere fusions and genome duplication events3–5. Humans have 23 pairs of chromosomes, whereas other apes have 24. One human chromosome is a fusion product of the ancestral state6. This raises the question: how well can species tolerate a change in chromosome numbers without substantial changes to genome content? Many tools are used in chromosome engineering in Saccharomyces cerevisiae7–10, but CRISPR–Cas9-mediated genome editing facilitates the most aggressive engineering strategies. Here we successfully fused yeast chromosomes using CRISPR–Cas9, generating a near-isogenic series of strains with progressively fewer chromosomes ranging from sixteen to two. A strain carrying only two chromosomes of about six megabases each exhibited modest transcriptomic changes and grew without major defects. When we crossed a sixteen-chromosome strain with strains with fewer chromosomes, we noted two trends. As the number of chromosomes dropped below sixteen, spore viability decreased markedly, reaching less than 10% for twelve chromosomes. As the number of chromosomes decreased further, yeast sporulation was arrested: a cross between a sixteen-chromosome strain and an eight-chromosome strain showed greatly reduced full tetrad formation and less than 1% sporulation, from which no viable spores could be recovered. However, homotypic crosses between pairs of strains with eight, four or two chromosomes produced excellent sporulation and spore viability. These results indicate that eight chromosome–chromosome fusion events suffice to isolate strains reproductively. Overall, budding yeast tolerates a reduction in chromosome number unexpectedly well, providing a striking example of the robustness of genomes to change.
Chromosome engineering in S. cerevisiae is driven by technological advances7–10. A haploid yeast strain with 33 chromosomes has been generated by splitting natural chromosomes into smaller chromosomes7. On the other hand, the two largest S. cerevisiae chromosomes, IV and XII, were fused by homologous recombination8, producing yeast with a 3.2-Mb compound chromosome that grew well. Further fusions (chromosomes VII–V–XV–IV) generated a 4.3-Mb compound chromosome with the longest yeast chromosome arm engineered previously (3.7 Mb), and a haploid chromosome number of n = 12, with no observed effect on fitness9. CRISPR–Cas9 expression in S. cerevisiae10 permits efficient engineering without selection. We used this to push the lower limit of chromosome numbers in S. cerevisiae by fusing chromosomes, producing a series of strains with progressively fewer chromosomes without affecting gene content.
At least three potential biological obstacles might hinder engineering of karyotype. First, studies of the field bean Vicia faba have suggested that the length limit to chromosome arms is half the average spindle axis at telophase for normal development11. Longer arms might yield incomplete sister chromatid separation, lagging chromosomes and micronucleus formation, impairing fertility and development. Second, as centromeres are deleted, excess kinetochore proteins may cause problems. Finally, mitotic mechanics may be affected—centromeric force may not suffice to pull very long chromosomes poleward. The latter is a particular concern for S. cerevisiae, in which point centromeres are bound by single microtubules; most organisms with larger chromosomes have regional centromeres bound by multiple microtubules12. Karyotype engineering in S. cerevisiae, therefore, investigates whether point centromeres can segregate large chromosomes.
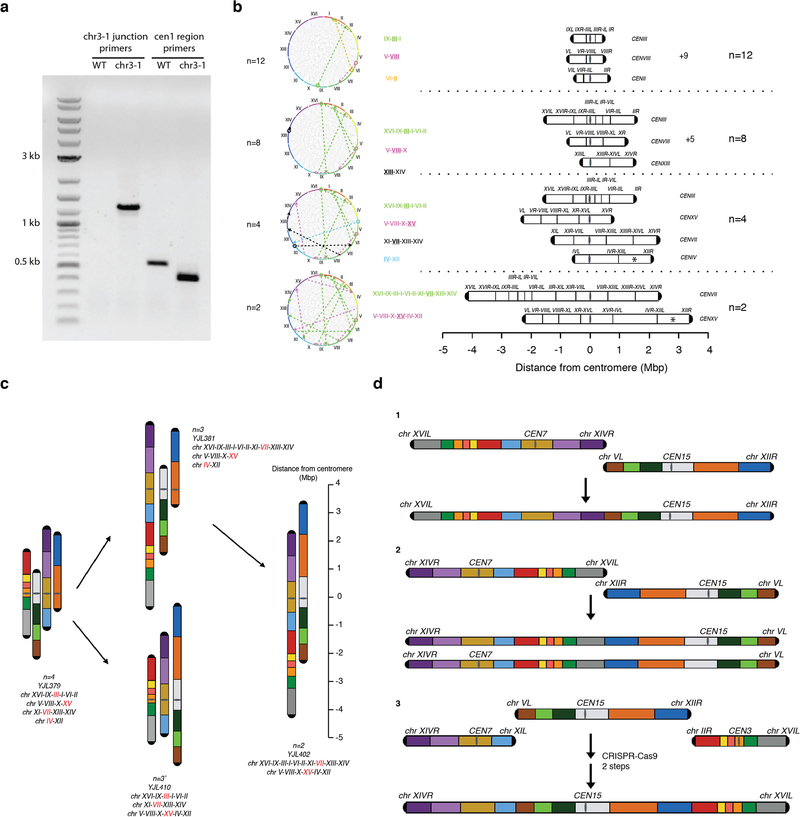
We devised specific paths to evaluate minimization of the number of chromosomes (n). We first fused all small chromosomes to maximize the number of chromosomes fused before hitting a potential chromosome arm length limit. Specifically, we fused chromosomes IX, III and I, then V and VIII, and finally II and VI to generate an n = 12 strain (Fig. 1).
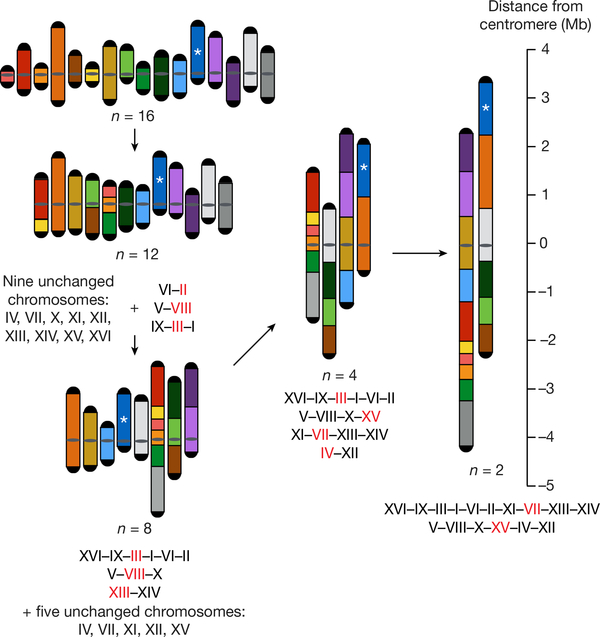
Fig. 1 |. Fusion chromosome paths and strategy.
This diagram shows how we fused chromosomes together from the wild type (n = 16) to n = 12, n = 8, n = 4 and finally n = 2. The 16 chromosomes are coloured uniquely and arranged by number. A ruler indicates the distance from the centromere. The centromeres of n = 4 and n = 2 are aligned to the 0 position. Please note that the length of the rDNA array, whose position is indicated with an asterisk, is omitted. Red lettering indicates the chromosome that has an active centromere in the compound chromosome. All the chromosomes are oriented from left to right (bottom to top in the figure) in the fusion chromosomes. For more details see Extended Data Fig. 1b.
We developed a CRISPR–Cas9-based strategy to fuse any two chromosomes while largely preserving isogenicity (Fig. 2a, Extended Data Fig. 1a and Extended Data Table 1; see Methods). Pulsed field gel electrophoresis, used for karyotyping S. cerevisiae, confirmed the presence of progressively fewer and larger chromosomes (Fig. 2b) with all chromosomes shorter than 600 kb merged in the n = 12 strain.
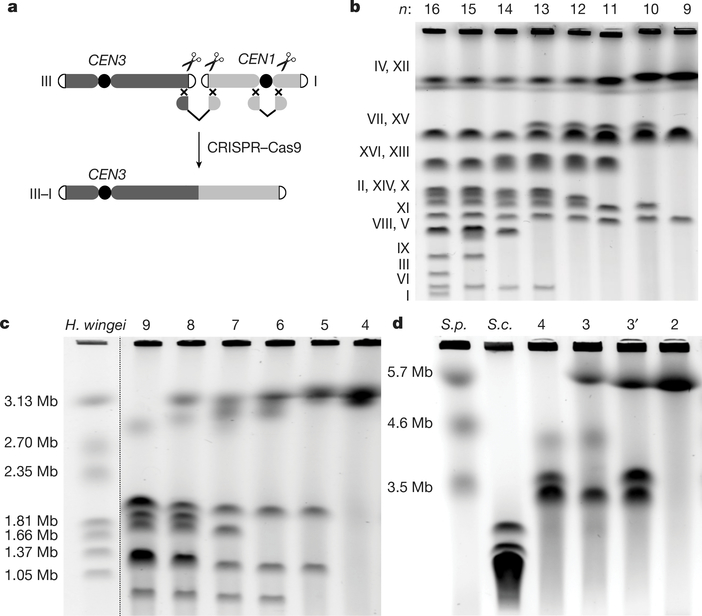
Fig. 2 |. Construction and characterization of fusion chromosomes.
a, A schematic showing a CRISPR–Cas9 based method to fuse any two chromosomes together. CEN3 and CEN1, centromeres of chromosomes III and I, respectively. b, Pulsed-field gel electrophoresis with a standard protocol for Saccharomyces cerevisiae. c, Pulsed-field gel electrophoresis with Hansenula wingei (also known as Wickerhamomyces canadensis) chromosomal DNA as a marker. d, Pulsed-field gel electrophoresis with S. pombe (S.p.) chromosomal DNA as a marker. S.c., S. cerevisiae. n = 3 strain is yJL381; n = 3′ strain is yJL410.
Given that 3.2-Mb and 4.3-Mb compound acrocentric chromosomes have been engineered previously8,9, we next produced a strain with four long chromosomes, each about 3 Mb long (for simplicity, the length of the rDNA array is omitted from lengths reported herein; Fig. 1). The serial disappearance of chromosomes confirmed that sequential fusion events occurred as planned (Fig. 2c). The n = 4 strain had four chromosomes, each around 3 Mb (Fig. 2d). At n = 4, the total chromosome number drops substantially below the smallest number previously achieved in engineered S. cerevisiae strains. Moreover, n = 4 is also lower than is seen in any extant Saccharomycetaceae species with point centromeres3.
Finally, to generate a strain with only two metacentric chromosomes, each about 6 Mb long, we planned to fuse the final four chromosomes in any viable order or orientation (Extended Data Fig. 1c), generating two versions of n = 3 strains by fusing different chromosomes in the n = 4 strain (Fig. 2d). To distinguish between these, we refer to them as n = 3 (yJL381) and n = 3′ (yJL410) (Extended Data Table 2). A further fusion cycle in n = 3 generated an n = 2 strain, with two metacentric chromosomes each about 6 Mb long (Fig. 2d). Each chromosome in the n = 2 strain now carries about half of the genomic content, which is unchanged compared to the n = 16 strain, except for the deletion of 14 centromeres and 28 telomeres.
We tried to generate an n = 1 strain by multiple strategies, for example, changing arm length (balanced versus dissimilar), position of centromere, and remaining telomeres (Extended Data Fig. 1d), but were unable to produce one. Has the limit of chromosome arm length been reached? Given that anaphase spindle axis length could extend up to 10 μm13, along with extrapolation from previous measurements (0.85 μm per 1 Mb)8, we estimate maximum arm length at about 5.9 Mb (see Methods), or slightly less than half of the full genome length if rDNA is included. This may make it more difficult to produce an n = 1 strain compared to other fusion steps, because the arm length of a metacentric single chromosome may approach the maximum. However, as we sampled only a few chromosome fusion paths (of 1019 possible), other paths might lead to an n = 1 strain. An accompanying study succeeded in producing an n = 1 strain using related methods, in combination with deleting repetitive regions14.
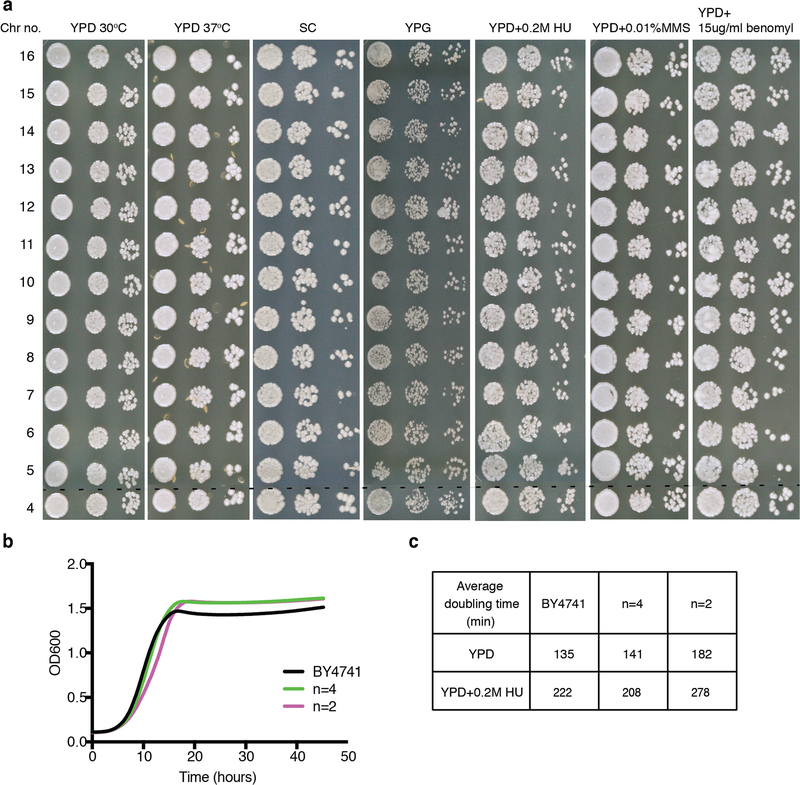
We analysed growth and resistance to various stresses for different n strains. Unexpectedly, strains with between four and sixteen chromosomes grew well in different media and stress conditions (Extended Data Fig. 2). Like n = 4, the n = 3 and n = 2 strains lacked obvious fitness defects (Fig. 3a). These strains are healthy, indicating that S. cerevisiae can handle large chromosomes and therefore, a regional centromere is not required to segregate S. pombe-sized chromosomes. To quantify small differences in fitness, we performed quantitative competitive growth assays by co-culturing differentially tagged BY4741 cells and n = 4 or n = 2 cells, and measuring changes in fluorescence ratios with time. The n = 4 strain grew similarly to the wild type (98.7 ± 0.3%), whereas the n = 2 strain grew slightly more slowly (91.3 ± 0.4%; Fig. 3b).
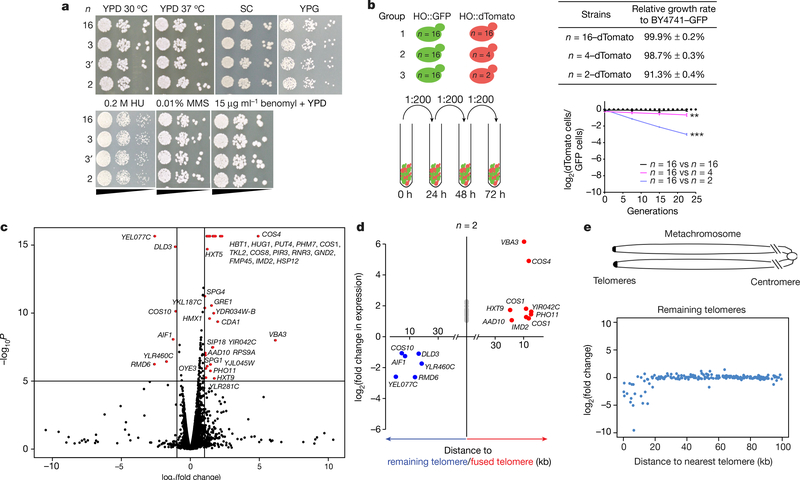
Fig. 3 |. Characterization of fusion chromosome strains.
a, Serial dilution assays under seven different conditions with n = 16, n = 3 and n = 2 strains. HU, hydroxyurea; MMS, methyl methanesulfonate; SC, synthetic complete medium; YPG, yeast extract peptone with 3% glycerol. b, Competitive growth assays. Each experimental group was tested in biological triplicate. Group 2 (n = 16 vs n = 4), P = 0.0048; group 3 (n = 16 vs n = 2), P = 0.00013 (one-sided t-test). *0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001. c, A volcano plot showing RNA-seq data by comparing the transcriptomes of the n = 2 and n = 16 strains. Red dots indicate genes whose expression was significantly different in the n = 2 strain compared to the n = 16 strain (P < 10−5, |fold change| > 2). Some DNA replication stress response genes, including RNR3 and HUG1, are also upregulated in the n = 2 strain. P values derived from two-sided t-test. YKL187C also known as FAT3; YLR281C also known as RSO55. d, Comparison of transcriptomes of n = 2 and n = 16 strains. Genes located within 20 kb of remaining telomeres are shown in blue; those within 20 kb of fused telomeres are shown in red; others are shown in grey. e, ‘Metachromosome’ of remaining telomere plots. Top, schematic diagram showing in the metachromosome view, all telomeres are aligned on the left; we show expression changes of genes within 100 kb of the nearest telomere. y-axis, log2(fold change in gene expression comparing transcriptome of n = 2 to n = 16); x-axis, distance of genes from the closest telomere.
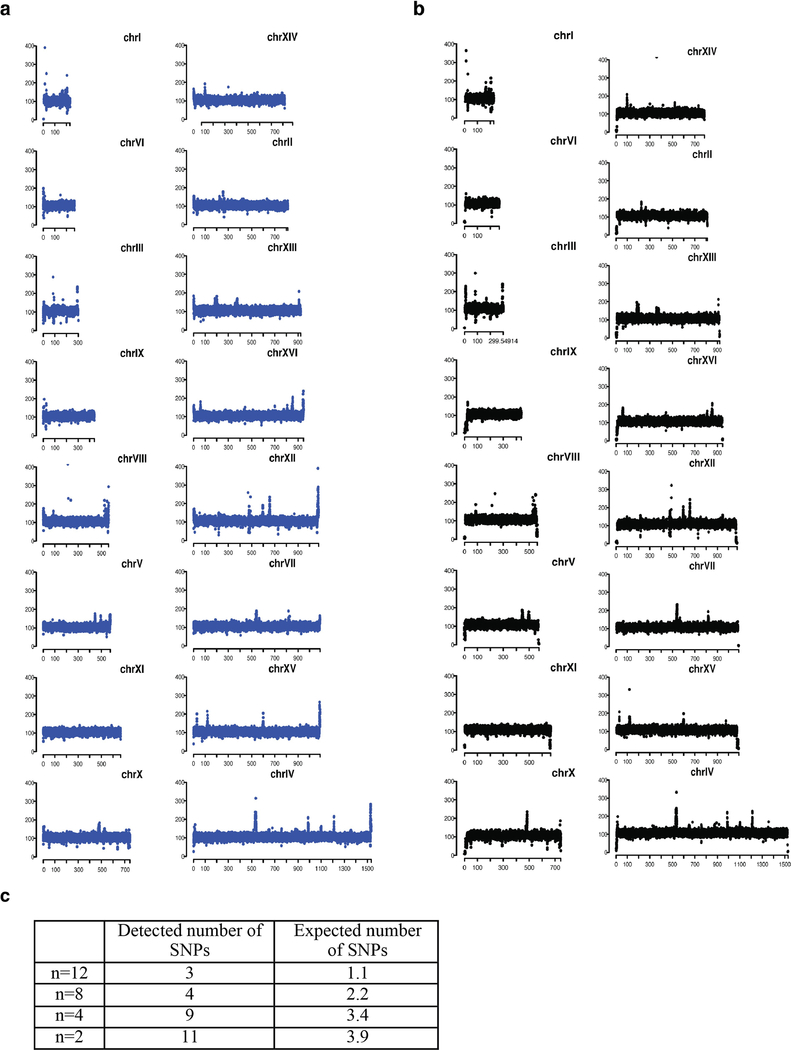
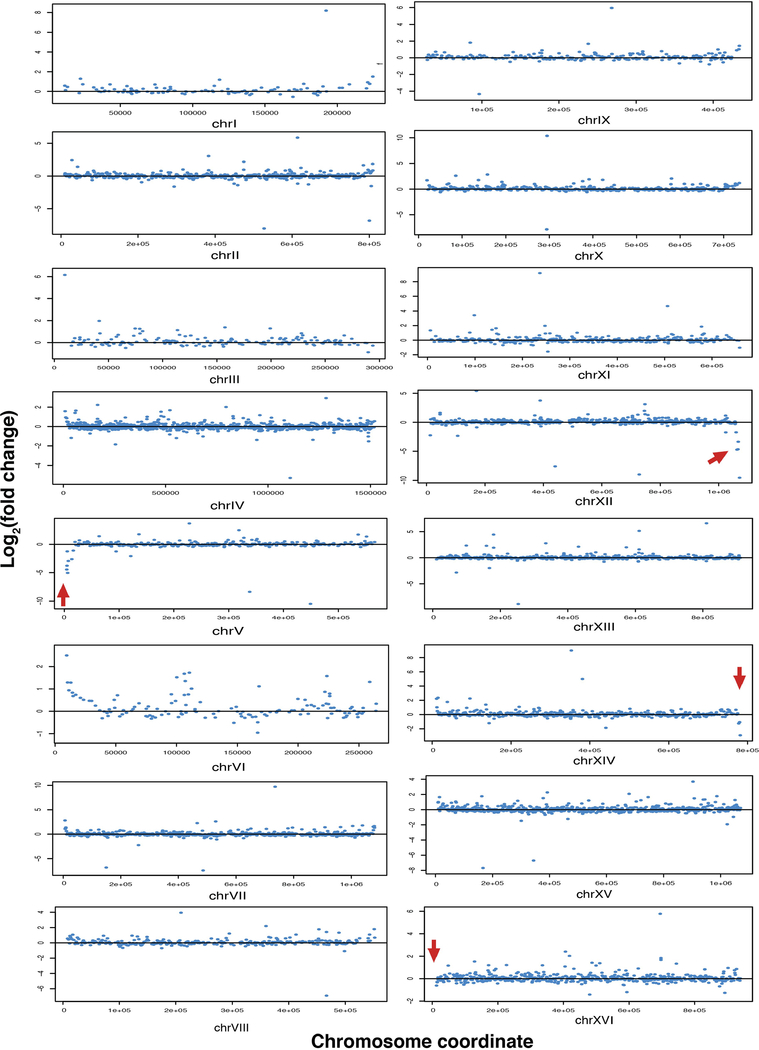
Fusion of chromosomes might trigger secondary genome or transcriptome changes. Sequencing of genomes from the n = 2, 4, 8 and 12 strains revealed no evidence of aneuploidy or regional copy number differences, and few new single nucleotide polymorphisms (SNPs) or small insertions/deletions (indels) (Extended Data Fig. 3 and Extended Data Table 3). The numbers of SNPs detected were within threefold of expected numbers based on reported error rates15 (Extended Data Fig. 3c). Two out of eleven SNPs (AMN1 L317F and QDR3 P586S) were predicted to be deleterious by the Sorting Intolerant From Tolerant (SIFT) online tool (http://sift.jcvi.org/), which assumes that important positions in coding sequences of proteins are conserved during evolution. AMN1 L317F is a potential suppressor of fusion chromosome state, because AMN1 controls mitotic exit16. Fusion efficiency did not drop markedly as n was reduced (Extended Data Table 1). RNA sequencing (RNA-seq) analysis revealed only minimal perturbations in the n = 8 and n = 4 strains (Extended Data Table 4). Because n = 2 does have a growth defect, we were unsurprised to find a few substantial transcriptome changes, including six downregulated genes (Fig. 3c and Extended Data Table 4). Strikingly, all these genes are positioned near the four remaining telomeres (Fig. 3d). As expression of the silencing genes SIR2, SIR3 and SIR4 was similar between n = 2 and wild-type strains, we hypothesize that downregulation of these six genes was due to enhanced telomere position effect (TPE). Consistent with this, further analysis of gene expression near the remaining telomeres showed reduced expression for genes within 20 kb of the telomeric repeats (Fig. 3e). The n = 2 strain also showed some upregulated genes lying close to newly fused telomeres; after fusion, when these genes are no longer subject to TPE, they are likely to be transcriptionally de-repressed (for example, genes in chromosomes IXR, VIL, VIIL and VIIIR; Extended Data Fig. 4). For the n = 4 and n = 8 strains, the few genes with substantially altered expression were also telomere-proximal (Extended Data Table 4). The significantly expressed subtelomeric genes overlapped extensively with subtelomeric genes upregulated in sir deletion strains (Extended Data Table 4). Finally, subtelomeric genes were highly enriched in genes whose expression was significantly affected in the n = 2, n = 4 and n = 8 strains (Extended Data Table 4).
Karyotype engineering to reduce chromosome number without affecting gene content revealed only mild changes in gross phenotypic and global gene expression. It is well known that karyotypic changes, including chromosomal rearrangement, polyploidy and hybridization, contribute to post-zygotic isolation, especially in plants17. About 100 million years ago, a whole-genome duplication (WGD) event4,5 resulted in a modal yeast chromosome number shift from n = 8 to n = 16. The major mechanism of chromosome number reduction in post-WGD species with n < 16 and non-WGD species with n < 8 was inferred to be via telomere fusion and loss of one centromere3, resembling the process intentionally engineered here.
Translocation events have been associated with reproductive isolation in Saccharomyces species18–21. A single naturally occurring reciprocal translocation was estimated to reduce spore viability to 50% when backcrossed to the wild type, owing to unbalanced segregation of essential genes on those translocation regions, while non-reciprocal translocation was 75%20. For chromosome–chromosome fusion, it is more difficult to model the effects on spore viability. The question remains, how much variation in chromosome number results in reproductive isolation (operationally defined here as <1% viability). Here, we are able to empirically assess the impact of chromosome number in isolation, because the strains represent an isogenic series (see Methods for definition of isogenic as used here) with the parent strain, BY4741.
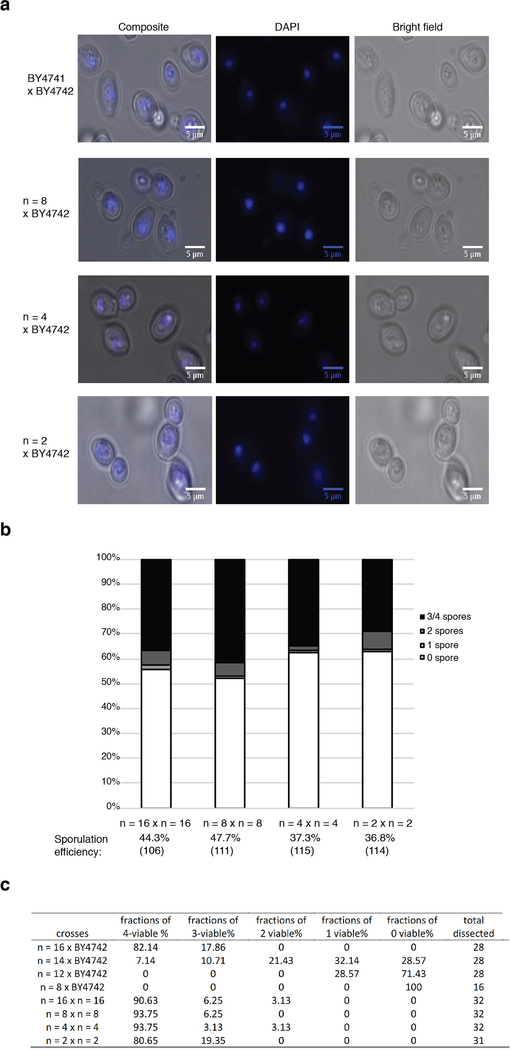
To explore reproductive isolation in this context, we mated each BY4741-derived strain from the fusion chromosome series (n = 16 to n = 2) to SK1 (n = 16), an efficient sporulating strain, and sporulated the diploids (Fig. 4a). All crosses readily produced viable zygotes, showing no barrier to zygote formation. We confirmed that nuclear fusion had occurred (Extended Data Fig. 5a), ruling out a block in karyogamy. We examined both success rate for meiosis, that is, sporulation efficiency, and the successful segregation of one genome’s worth of information into each spore, monitored by germination efficiency. For n = 16 × n = 16 crosses, more than 80% of asci developed 3–4 spores, and overall spore efficiency (per cent of asci with at least one spore) was 97.2%. For the ‘asymmetric’ (n < 16 × n = 16) crosses, as n decreased, sporulation efficiency dropped markedly. For n = 8 × n = 16 matings, fewer than 5% of asci developed 3 or 4 spores. For n = 2 or n = 4 × n = 16 matings, fewer than 1% of asci produced 3 or 4 spores (Fig. 4b).
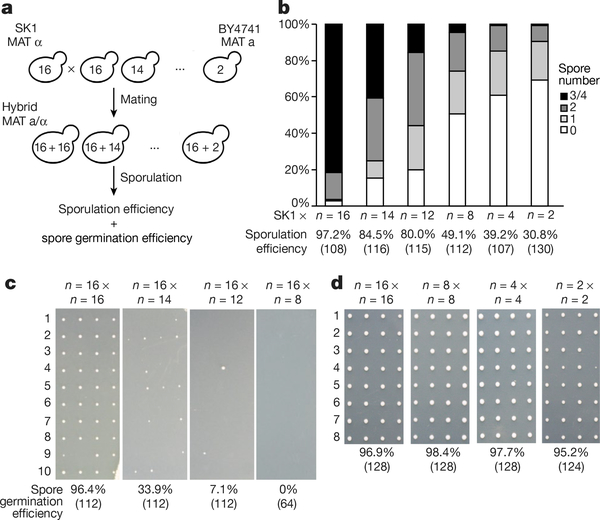
Fig. 4 |. Karyotype engineering leads to reproductive isolation.
a, Schematic of the experimental approach. The number inside the yeast is the number of chromosomes. b, Sporulation efficiency of hybrid strains. Y-axis, percentage of asci with 0–4 spores; x-axis, diploid strains. The number of asci counted is shown in parentheses. c, Spore clone germination rates for hybrid strain. Representative images are shown. The number of spores counted is shown in parentheses. d, Spore viability for wild-type diploid BY4743 2n = 32, isogenic diploids: 2n = 16 strain, 2n = 8 strain and 2n = 4 strain. Representative images are shown. The number of spores counted is shown in parentheses.
To test germination of offspring spore clones, we used a fully isogenic configuration, because spore clones that arise from n = 16 × n = 16 BY × SK1 hybrid diploids showed differences in spore clone colony size, reflecting differences between the BY and SK1 backgrounds. We crossed BY4741 strains (n = 16–8) to BY4742 (the isogenic n = 16 MATα partner of BY4741), and performed dissections of four-spored tetrads. Whereas n = 16 × n = 16 gave rise to more than 90% viable spore clones, n = 16 × n = 14 produced only 33.9% viable spores. In n = 16 × n = 12, survival dropped markedly to below 10% (Fig. 4c, Extended Data Fig. 5c). In n = 16 × n = 8, only very few tetrads formed after 15 days’ sporulation. We dissected 16 such tetrads and no spores (of 64) were viable. This did not reflect defects in chromosome fusion strains per se, as isogenic n = 8 × n = 8 diploids traverse meiosis as well as n = 16 diploids (Extended Data Fig. 5b), giving rise to 98.4% viable spores (Fig. 4d). Indeed, isogenic n = 4 × n = 4 and n = 2 × n = 2 diploids also sporulated well and generated viable spores (Fig. 4d, Extended Data Fig. 5b). We observed 5.6% small colonies in spores from the n = 2 × n = 2 cross only. Thus, the n = 8, 4 and 2 strains are fully capable of performing meiosis and generating viable progeny, but are incapable of generating viable progeny with the n = 16 strain. These results indicate that eight chromosome–chromosome fusion events suffice to result in virtually complete reproductive isolation. Reproductive isolation comes from at least three sources. 1) Owing to problems with nondisjunction in meiosis I, there is a steady rise in probability that a spore will inherit a genome missing at least one chromosome’s worth of genetic information as n drops. 2) Even if a full complement of genes is inherited, the probability of a lethal dosage imbalance is elevated in asymmetric crosses. 3) Recombination between the concatenated and native chromosomes is predicted to lead to the formation of deleterious and genetically unstable dicentric (or multicentric) chromosomes22.
We have shown that S. cerevisiae can survive with two chromosomes and grow relatively well with four chromosomes, at least under laboratory conditions. Why does it have sixteen small chromosomes whereas S. pombe can get by with only three larger ones?
We consider three possibilities for phylogenetic distribution of n. First, the extant pattern of values of n in yeast could be a historical product of a WGD event(s) and/or spontaneous chromosome fusions and breaks3. Saccharomyces (n = 16) and related genera arose as a WGD event relative to a large cluster of ‘preduplication’ yeasts3–5 (mostly n = 8). Second, a genome with 16 small chromosomes readily becomes aneuploid, allowing rapid and reversible adaptation to severe environmental changes23. However, aneuploidy can also result in deleterious imbalances24,25. With few large chromosomes, aneuploidy is more likely to be selected against, limiting potential for adaptation to environmental changes. Finally, sub-telomeres are enriched in ‘contingency’ genes that encode functions related to the cell wall and metabolism of different nutrients26. These genes are transcriptionally repressed under normal conditions but can be specifically expressed in appropriate environments or conditions27. A higher number of telomeres may allow more elaborate fine-tuning of these properties, improving the ability of a generalist species to adapt more rapidly to diverse environments and stresses.
We have efficiently generated a series of chromosome fusions, building a collection of strains with progressively fewer chromosomes, including n = 2. Unexpectedly, yeast growth was robust, and mostly indistinguishable from normal karyotype strains, even with chromosomes up to four times the maximum size (excluding rDNA) of wild-type strains, and with greatly reduced numbers of telomeres and centromeres. The n = 2 strain grew slightly more slowly than the wild type, perhaps owing to its large chromosomes or altered TPE. Meiosis was strongly affected in matings between strains with different numbers of chromosomes, suggesting that chromosome fusion and concomitant reduction in chromosome number suffices as a reproductive barrier. The strains described here may be used to probe aspects of meiotic recombination, replication origin timing, the role of 3D nuclear structure in transcriptional regulation in yeast, or recombination donor preference, to name a few. Reproductive isolation may also prove to be useful for future studies involving field release of engineered yeast or other cases in which genetic isolation is desirable.
The n = 2 strain is reproductively isolated from the n = 16 strain, but is it a new species? By classical biological species definitions (post-zygotic reproductive isolation)28,29, it could qualify. However, by phylogenetic species definitions (a group of organisms sharing an ancestor), n = 2 is not a new species because it has near-zero sequence divergence from n = 16, and its morphology is similar30. Eight fusions suffice to cause practically complete reproductive isolation, which, with further neutral evolution, would allow such pairs of strains to accumulate diverse mutations over time, leading to speciation by any definition.
Online content
Any Methods, including any statements of data availability and Nature Research reporting summaries, along with any additional references and Source Data files, are available in the online version of the paper at https://doi.org/10.1038/s41586-018-0374-x.
MEthodS
Strains.
All strains that we constructed in this study are listed in Extended Data Table 2. They are all derived from BY4741 (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0) by transformation events. For sporulation efficiency measurement experiments, the SK1 strain (MATα ho::LYS2, lys2, ura3, leu2::hisG, his3::hisG, trp1::hisG) was used to mate with strains of different n. For the tetrad dissection experiments, BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used to mate to strains with different values of n. 17 loxPsym sites were added at the sites of centromere deletion and telomere fusion anticipating future genome rearrangement experiments.
Calculation for potential paths to fuse 16 chromosomes into one chromosome.
The equation for this calculation is as follows: (32 × 30 × 28 × … × 2 × 16)/2 = 1.097 × 1019, if we take into consideration final centromere choice (16 conditions). In addition, we count the same configuration twice by arbitrarily defining left arm and right arm. To give a simple example to explain this point, suppose there are only two chromosomes: chr I and chr II, the final fusion chromosome chr IL-chr IR-chr IIL-chr IIR is the same as chr IIR-chr IIL-chr IR-chr IL, thus we divide by 2 to account for this.
CRISPR–Cas9 method.
The CRISPR–Cas9 method for genome editing in S. cerevisiae dramatically stimulates homologous recombination (HR) by co-expressing Cas9 and site-specific guide RNAs, together with transient linear PCR products as HR donors10. Here, we optimized this method to produce chromosome fusions. To fuse chrs I and III, for example, we targeted breaks adjacent to the chr III right telomere and chr I left telomere, and near CEN1 by Cas9 co-expression with three specific gRNAs (Fig. 2a). We synthesized two donor molecules, the first encoding homology to bridge the termini of these chromosomes, and the second a fragment of DNA spanning CEN1 to delete it. Through successful HR at both sites, we generated a compound chromosome III-I, with deletion of two telomeres, chr IIIR_tel and chr IL_tel, and one centromere, CEN1 in a single shot. The choice of gRNA telomere target site was chosen to optimize targeting specificity while minimizing deletion of sub-telomeric sequences to preserve genome content. At each stage, we picked plasmid-transformed colonies and then screened by PCR for a) presence of appropriate telomere-telomere junctions and b) deletion of appropriate centromeric DNA (Extended Data Fig. 1a).
The CRISPR–Cas9 method was performed as described10,31. In addition, another gRNA acceptor vector was constructed from gRNA-ura-HYB (Addgene plasmid #64330) by substituting the URA3 gRNA with a NotI restriction site. pRS426 gRNA acceptor vectors have two gRNA insertion sites, one defined by a NotI restriction site and another one by a HindIII restriction site. By using these two gRNAs acceptor vectors, up to 3 gRNAs could be co-expressed in yeast cells when co-transformed. To insert 20 nt gRNAs, Gibson assembly32 was used to assemble a restriction enzyme digested (for example, NotI or HindIII) and gel purified vector together with a 60 bp double-stranded oligonucleotides, which consists of a 20nt gRNA, and homologous sequences to the left and right of vectors (for example, for the NotI vector, 5′-GCAGTGAAAGATAAATGATC-20nt gRNA -GTTTTAGAGCTAGAAATAGC-3′; for the HindIII vector, 5′-CTGGGAGCTGCGATTGGCAG-20nt gRNA-GTTTTAGAGCTAGAAATAGC-3′). Single-stranded oligonucleotides were ordered from Integrated DNA Technologies and annealed10 by first denaturing the mixture at 95 °C for 5 min and then cooling to 10 °C with a ramp of 0.17° s−1. The donor DNAs linking two different chromosome arms together are made by two-step fusion PCR33 amplified from the wild type yeast genome or they were synthesized. These ‘linker donors’ usually have ~ 400bp sequences homologous to each chromosome arm (sometimes shorter sequences were used to avoid repetitive terminal sequences that would not target uniquely). The same rule applies for the centromere deletion donors, ~ 400bp flanking centromere regions were designed on each side. The centromere deletion donors were made by Polymerase Chain Assembly34,35. Donor DNA amplicons were about 800 bp long. The CRISPR–Cas9 method was performed stepwise: 1. We transformed a Cas9 expressing plasmid. 2. We co-transformed 50 ng gRNA expression plasmids and ~400 ng PCR amplicon donor DNAs into the strain already expressing Cas9.
Both pRS426 and pRS42H carrying 3 gRNAs in total were used, because we found that using 3 gRNAs gave a higher yield of correct colonies than only using 2 gRNAs. Then after co-transformation, yeast cells were plated on SC–Ura–Leu (Synthetic Complete medium lacking leucine and uracil) with 300 μg/ml hygromycin B plates for more than 2 days. Yeast cells were usually grown for 4 h in YPD after transformation to allow expression of hygromycin resistance, before plating on selective plates with 300 μg/ml hygromycin B. The yeast clones with correct fusion chromosome configurations were verified by PCR and pulse field electrophoresis. Sometimes the SC–Ura–Leu + Hyg plate selection is too stringent in that very few transformant colonies were obtained. In that case, SC–Ura–Leu plates were used.
PCR verification of fusion chromosomes.
To verify the fusion of two chromosomes, primers were designed to bind outside of homology sequences to the linker donors. Only clones bearing successfully reorganized fusion karyotypes generated appropriately sized PCR amplicons, which were absent from failed clones or wild type control colonies. To verify the deletion of a centromere, primers were designed to bind sequences flanking centromere regions. Typically, the successful deletion of centromeres produced amplicons ~120 bp shorter than wild type control samples. All verification primer information is included in Extended Data Table 5.
Pulsed-field gel electrophoresis.
For n = 16 to n = 9 strains (Fig. 2b), yeast chromosomes plugs were prepared and separated by clamped homogeneous electric field (CHEF) gel electrophoresis using the CHEF-DR III Pulsed-Field Electrophoresis System (Bio-Rad), as previously described36. The CHEF gel running condition was 6 V/cm, switch time: 60 s to 120 s, run time: 24 h, 14 °C, with a 0.5 × Tris-Borate-EDTA buffer and a 1% gel with low melting point agarose. For n = 9 to n = 2 strains (Figs. 2c and 2d), a 7 ml culture of yeast was grown over 48 h or until it reached stationary phase. Then yeast cell pellets were collected by centrifugation and zymolyase and agarose were added proportion to cell pellet weight. For 60 mg of cell pellet, 24 μl 25 mg/ml zymolyase 20T in 10 mM KPO4 (pH 7.5) and 540 μl 0.5% low melting point agarose in 100 mM EDTA (pH 7.5) were added. Low melting point agarose was fully dissolved in the EDTA solution by microwave heating and then kept at 42 °C. Cell pellets were resuspended with zymolyase and agarose solutions by pipetting with a wide bore pipette and transferred to BioRad molds (BioRad No.1703713). The molds were cooled in a 4 °C cold room for 30 m. The plugs were released from the molds, and added to 1 ml 500 mM EDTA, 10 mM Tris, pH 7.5 and incubated at 37 °C overnight. 5% sarcosyl and 5 mg/ml proteinase K in 500 mM EDTA pH 7.5 was added to the plug the next day and incubated at 50 °C overnight. The plugs were washed with 1 ml 2mM Tris-1mM EDTA buffer four times, for 1 h each. Such plugs could be used immediately or stored at 4 °C for at least a year. To separate 1 Mb to 3 Mb chromosomes, we used H. wingei chromosomal DNA as a marker. H. wingei has 7 chromosomes, varying from 1.05 Mb to 3.13 Mb. For n = 9 to n = 4 strain (Fig. 2c), the CHEF gel electrophoresis condition was that recommended for H. wingei chromosomes (BioRad No.170–3667). To separate chromosomes longer than 3 Mb, we adopted a different electrophoretic protocol, using S. pombe chromosomal DNA as a marker. For n = 4 to n = 2 strain (Fig. 2d), the CHEF gel electrophoresis condition was that recommended for S. pombe chromosomes (BioRad No.170–3633).
Calculation of maximum arm length.
The condensation of a fusion chromosome, chr IV-XII, in budding yeast has been studied by labelling TRP1 with red fluorescence and LYS4 with green fluorescence8. In mother cells, the distance between TRP1 and LYS4 was ~0.4 μM during anaphase. With ~470 kb between these two loci, extrapolating from this distance predicts that each 1 Mb of condensed chromosome during anaphase will extend ~0.85 μm (0.4 μm/0.4686 Mb). Anaphase spindle axis length can extend up to 10 μm13, suggesting the maximum distance a chromosome arm could extend is 5 μm. The maximum arm length limit, therefore, should be about 5.9 Mb (5 μm/0.85 μm per 1Mb).
Serial dilution assays.
Yeast strains were grown from single colonies in liquid YPD culture until they reached the stationary phase at 30 °C with rotation. Then culture was diluted to A600 = 0.01, and serially diluted (1:10) in water and plated on different media. YPG plates were prepared by adding 3% glycerol as carbon source to yeast extract peptone. All other compounds (HU, MMS, benomyl) were added to YPD medium. Plates were incubated at 30 °C for 2 days except for YPG plates, HU plates and MMS plates, which were incubated for 3 days.
Growth curve measurement.
Yeast strains were grown from single colonies in liquid YPD culture until they reached stationary phase at 30 °C with rotation. Then culture was diluted to A600 = 0.05 and grown in 96 well plates with 100 μl YPD or YPD + 0.2 M hydroxyurea for 48 h. Every 10 m, the plate would be shaken and a measurement of OD600 was taken by a BioTek Eon microplate spectrophotometer. Doubling time was measured by calculating the slope of the growth curve at exponential stage.
Competitive growth assays.
Competitive growth assays were carried out as described37. The GFP or dTomato cassette was integrated into the HO locus through selection of nourseothricin-resistant cells. The GFP-labelled BY4741 was co-cultured with dTomato-labelled BY4741 or n = 4 in 1:1 ratio, while the GFP-labelled BY4741 was mixed with dTomato-labelled n = 2 in 1:2.5 ratio in YPD. The next day, 30,000 total cells were sorted on a Sony SH800S Cell Sorter as T0. Cells were diluted 200-fold in fresh YPD medium and sorted every 24 h three times. Flowjo v.10 was used to analyse the data.
Genome sequencing.
Whole-genome DNA samples for sequencing were prepared using a Norgen Biotek fungi/yeast genomic DNA isolation kit (Cat No. 27300). Whole-genome shotgun libraries were made on a Beckman FXP automation workstation and prepared as follows: 500 ng genomic DNA, as input, was amplified by 2 cycles of PCR and sheared to 500 bp on a Covaris LE220. A library was prepared using KAPA High Throughput Library Preparation Kit (KK8234), and sequenced as 75 bp paired-end reads on an Illumina NextSeq 500. All raw reads were trimmed to remove adaptor sequence using Trimmomatic, and subsequently mapped to UCSC sacCer2 reference genome (S288C_reference_genome_R61–1-1_20080605) from Saccharomyces Genome Database using BWA-MEM standard options. BWA, Picard, GATK and SAMtools software were applied to align reads to transcriptome reference and call SNPs/Indels. The filtering criteria for SNPs using GATK were as follows: QD <2.0; FS >60.0; MQ <40.0; MQRankSum <−12.5; ReadPosRankSum <−8.0; SOR >4.0. The filtering criteria for Indels using GATK are as follows: QD <2.0; FS >200.0; ReadPosRankSum <−20.0; SOR >10.0. The variants common to the fused chromosome and the laboratory stock wild-type strain BY4741 were removed and considered as starting spontaneous mutations. The remaining variants were manually curated, by browsing through the bam files in IGV. Native 2-μm plasmids were absent from those sequenced fusion chromosome strains n = 12 to n = 2, according to the WGS results.
RNA sequencing.
For each strain, three independent colonies were grown in YPD liquid medium at 30 °C with rotation to saturation. The next day, an overnight culture was diluted to A600 = 0.1 and regrown. Cells were harvested at A600 = 0.4–0.6. Total RNA was extracted from three independent log-phase cultures using a QIAGEN RNeasy mini kit (Cat No.74106). The library was prepared with 500 ng total RNA as input, using a TruSeq RNA sample Preparation v2 kit (set A RS-122–2001 and set B RS-122–2002) with 13 cycles of amplification. The library was sequenced as 150 bp single end reads on an Illumina NextSeq 500. Reads were mapped to S288C reference genome (sacCer2) and differential gene expression analysis was performed with TopHat and Cuffdiff according to a standard pipeline38. Briefly, trimmed reads were mapped to the reference genome using TopHat, and aligned reads with more than 2 mismatches were excluded from the downstream analysis. RPKM were calculated using cufflinks and differentially expressed genes were analysed using cuffdiff. Genes that have been deleted in the fusion chromosomes were excluded in the analysis. The thresholds we used for differentially expressed genes are P < 10−5 and |fold change|>2.
Definition of Isogenic strain.
Isogenic, as used here, refers to genomic regions not deleted together with centromeres and telomeres. The deleted regions contain a maximum of 21 verified genes (n = 2 strain) as defined by SGD, and a maximum of 1.6% of the genome in the most extreme case, that is, comparing n = 2 and n = 16 strains, and consist primarily of the centromeres and telomeres themselves, and some adjacent repetitive subtelomeric elements, including X elements, Y’ elements, and highly repeated PAU and COS genes.
Mating type switching.
Mating type switch of fusion chromosome strains were carried out by transforming a CEN plasmid, which expresses endonuclease HO with its endogenous promoter, as described39.
DAPI staining of fixed yeast cells.
107 cells were spun down from an overnight culture. Pelleted cells were fixed by re-suspending in 70% EtOH and incubated for 2 h in room temperature. Cells were spun down, washed with water, re-suspended in 500 μl RNase A solution (2 mg/ml RNase A (Qiagen, 19101), 50 mM Tris pH 7.5 and 15 mM NaCl) and incubated for 2 h at 37 °C. Proteinase K (Invitrogen, 25530049) was added to a final concentration 1 mg/ml and incubated for 45 min at 37 °C. Cells were collected by centrifugation, washed with 1 ml of 50 mM Tris, and stored in 1ml of 50 mM Tris at 4 °C.
100–200 μl of fixed cells were pelleted by centrifugation, stained by re-suspending in 100 μl of DAPI solution (300 nM DAPI in 1× PBS) and incubated for 15 min. Cells were spun down and pellet was washed with 1× PBS, spun down again and re-suspended in 1× PBS for imaging.
Images were collected with NIS-Elements acquisition software, with 100× oil objective lens using Nikon Eclipse Ti microscope in both DAPI channel and bright field channel. Images were cropped in NIH ImageJ software with Bio-Formats plugin.
Sporulation and tetrad dissection.
For BY4743 diploid background, a diploid clone was inoculated in YPD at 30 °C with rotation overnight. The next day culture was diluted in YPD medium to grow at 30 °C for at least two generations to A600 = 4–8. The cells were pelleted by centrifugation (2,000g for 2 m) and washed three times with reagent-grade water. Cell pellets were re-suspended in sporulation medium (1% potassium acetate and 0.005% zinc acetate) with 0.1% (w/v) yeast extract and amino acid supplements (0.3 mM histidine, 2 mM leucine and 0.2 mM uracil), at a final A600 = 1.0, and incubated at 25 °C with rotation for 3–15 days before tetrad dissection. To digest asci, 50 μl cell cultures were pelleted by centrifugation (3,600 rpm × 3 m), re-suspended in 25 μl 0.5 mg/ml 20T zymolyase in 1 M sorbitol at 37 °C for 6–7 m, and diluted with 300 μl 1M sorbitol. Tetrads were dissected on a SINGER Instruments dissection microscope and germinated on YPD plates at 30 °C for 2 days. yJL346 was used in this experiment as the n = 8 strain rather than yJL342.
For crosses of the hybrid SK1 × BY4741 background, diploid strains were patched onto a YPG plate from −80 °C glycerol stocks and grown at 30 °C overnight (O/N), then patched onto a YPD plate containing 4% glucose and grown at 30 °C O/N, inoculated into YPD liquid culture and grown at 25 °C O/N, diluted to A600 = 0.8 and then grown in BYTA pre-sporulation medium (buffered yeast extract tryptone acetate, 1% yeast extract, 2% bacto tryptone, 1% potassium acetate and 50mM potassium phtalate) at 30 °C with rotation for ~16.5 h. Cell pellets were collected by centrifugation (3000 rpm × 5 min), washed twice with 25 ml water, re-suspended to A600 = 2.0 in sporulation medium (0.3% potassium acetate, 5% acetic acid, pH ~ 6.5, with 0.3mM histidine, 2 mM leucine and 0.2 mM uracil), incubated at 30 °C with shaking. The sporulation efficiency of each strain was measured at 48 h on a ZEISS phase contrast microscope under 60× magnification. For each strain, > 100 cells were counted to obtain the sporulation efficiency. We scored asci with 0 spore and cells that did not go through meiosis in the same category (asci with 0 spore). We calculated sporulation efficiency by dividing the number of asci with at least 1 spore to total cells. yJL346 was used in this experiment as n = 8 strain rather than yJL342.
Statistics and reproducibility.
Figures 2b–d, 3a and 4c, d and Extended Data Figs. 1a, 2a and 5a were repeated twice with similar results. Extended Data Fig. 2b was done with biological quadruplicates.
Code availability.
All codes used in this study are available at https://github.com/sunnysun515/fusion_yeast_chromosomes.
Data availability.
We submitted DNA sequences and RNA sequencing data to NCBI BioProject PRJNA471518. All other data are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1 |. Fusion chromosome paths and characterization.
a, PCR verification of fusion chromosomes. Two pairs of primers are used to verify the fusion of two chromosomes, here presenting the fusion of chromosomes I and III. Only with successful fusion would an amplicon show in fusion chromosome junction PCR. In addition, the centromere region PCR amplicon is shorter, owing to the deletion of a centromere. Marker: 2-log DNA marker. b, Circos diagrams for different values of n are shown in the upper panel, with 16 chromosomes laid out in circles in a clockwise manner. Each grey line connecting each pair of chromosomes represents a possible intertelomeric link. The dashed coloured lines represent the path we chose. (Open circle indicates the starting chromosome, arrows show direction of fusion chromosomes.) Each fusion chromosome is labelled in the same colour, providing detailed information on fusion chromosome paths. The underlined chromosome has the active centromere for the fusion chromosome. Unchanged chromosomes are not shown, but the number of unchanged chromosome is indicated after +, for example, +9 means nine unchanged chromosomes. Fusion chromosome lengths are shown below. Please note that the length of the rDNA array (normally 1–2 Mb)is omitted here. *Chromosome containing rDNA array. In addition, telomeric ends are clearly labelled with the original chromosomes; L/R stands for left/right telomere. Active centromeres are written on the right of each fusion chromosome diagram. When we deleted CEN15 and made 4 bp of mutations in gRNA sites in BY4741 to fuse chromosomes X and XV together, the fusion strain grew more slowly than the wild type. The growth defect could be due to altered expression of the gene neighbouring CEN15 (YOR001W; RRP6). For this reason, CEN15 was maintained in the remaining centromere. c, Fusion chromosome paths from n = 4 to n = 2. Red lettering indicates the chromosome that has an active centromere in the compound chromosome. d, Multiple strategies that we attempted to construct an n = 1 strain. We tried changing chromosome arm length (strategy 3), centromere position (strategy 2), and which telomeres remained (strategy 1 and 2). In strategy 2, we attempted both versions of n = 1 strains, keeping either CEN7 or CEN15 as the active centromere. In addition, we also attempted the strategy 1 in both SIR2+ and sir2Δ backgrounds.
Extended Data Fig. 2 |. Growth fitness assays.
a, Serial dilution assays in seven different conditions with n = 16 through n = 4 strains. b, Growth curve for BY4741, the n = 4 strain and the n = 2 strain in YPD medium at 30 °C. c, Doubling time calculation for these three strains. Each experimental group was tested in biological quadruplicate.
Extended Data Fig. 3 |. Whole-genome coverage maps and SNPs comparison.
a, Whole-genome coverage map of BY4741. Chromosomes are ranged according to their size. x-axis shows chromosome coordinate, and y-axis shows the coverage of reads that map to the reference genome of S288C. b, Whole-genome coverage map of n = 2 strain. For comparison, we mapped the reads back to the S288C genome. The main difference is telomere reads. Owing to deletion of telomeres in n = 2, either no reads or only few reads align back to fused telomere ends. c, Each round of CRISPR–Cas9 experiments takes at least 70 generations. The nucleotide mutation rate is 0.33 × 10−9 per base per generation15. If we multiply the number of rounds of CRISPR–Cas9 (16 – n) × 70 generations × yeast genome size (bp) 12 × 106 × 0.33 × 10−9, it gives the expected number of SNPs, as indicated in the table.
Extended Data Fig. 4 |. Chromosome plots of gene expression change in n = 2 vs n = 16 strains.
y-axis, log2 (fold change of gene expression (n = 2/n = 16)); x-axis, chromosome coordinates. Red arrows point to the four remaining telomeres in n = 2 strains.
Extended Data Fig. 5 |. DAPI staining of nucleus and sporulation efficiency for diploids.
a, DAPI staining of nuclei in diploids. b, Sporulation efficiencies for 2n = 32, 2n = 16, 2n = 8 and 2n = 4 homotypic diploids. y-axis: percentage of asci that have 0–4 spores. x-axis: different diploid strains. Sporulation efficiency was measured after shifting diploid strains in the sporulation medium for 10 days in room temperature. c, Fractions of asci with 0–4 viable spores in heterotypic and homotypic crosses.
Extended Data Table 1 |.
gRNA sequences and fusion efficiency of each fusion step
| Fusion step | Deletion | 20 nt gRNA (5’–>3’) | Deletion coordinate | Number of colonies | PCR verification |
|---|---|---|---|---|---|
| 1 | chr III_right_ telomere | GAGCTACTATCTTTGTCGGG | 299737 – 316617 | 10* | 4 |
| chr I_left_ telomere | CTCAATGTACGCGCCAGGCA | 1 – 6770 | |||
| CEN1 | AAGAAAGTTATATGTGTGAC | 151467 – 151578 | |||
| 2 | chr IX_right_ telomere | GTTGAGAGACAGGATGGTTA | 438945 – 439885 | NA | NA |
| chr III_left_ telomere | CCATTTGGAGTCTGCTCGGC | 1 – 3582 | |||
| CEN9 | TGAATAAGTTGAAGGACAAC | 355626 – 355853 | |||
| 3 | chr V_right_ telomere | TGCTAATCGTCTCTGCGGTT | 569244 – 576869 | 1 | 1 |
| chr VIII_left_ telomere | CCATTTTTTATCCTTATGCT | 1 – 6182 | |||
| CEN5 | TTTAGTTGAAACGCCAACAG | 151813 – 152187 | |||
| 4 | chr VI_right_ telomere | GAACTGTGCATCCACTCGTT | 269599–270148 | 3* | 2 |
| chr II_left_ telomere | GGGTTAACTACGCTATAGAC | 1–8703 | |||
| CEN6 | CTAAAACTGTCTTTTCGTGT | 148507–148624 | |||
| 5 | chr XIII_right_ telomere | CTGATAAAAGCAGGTGCCCT | 916969–924429 | 1* | 1 |
| chr XIV_left_ telomere | TCTGATCGGTCATACGTACA | 1–7493 | |||
| CEN14 | CTTCATCAAGATCGGGGAGC | 628758–628876 | |||
| 6 | chr XVI_right_ telomere | TGGTGTTATATAGTGGCACC | 941911–948062 | 1 | 1 |
| chr IX_left_ telomere | TTTGCCACCACCTGGGCGGG | 1–20857 | |||
| CEN16 | TTAGAATTACGACAACATAA | 555959–556070 | |||
| 7 | chr VIII_right_ telomere | CATCCGTGTGCGTATGCCAT | 556465–562643 | 10 | 2 |
| chr X_left_ telomere | TAGTGGCTCGCACTCATGCG | 1–7960 | |||
| CEN10 | TTGTTATACACAACGCGTCT | 436301–436421 | |||
| 8 | chr I_right_ telomere | ACCTGCGCGGCGCGGCGGTT | 227349–230208 | 48* | 2 |
| chr VI_left_ telomere | CTTTGTATGAGGGTACATCA | 1–6185 | |||
| CEN2 | CTTTGCGTGGTCTAGTGCAT | 238211–238325 | |||
| 9 | chr VII_right_ telomere | CTTTTACAACAACCGCCATA | 1083059–1090947 | 20* | 17 |
| chr XIII_left_ telomere | ATAGCGGCGGTACGTTTGTT | 1–8585 | |||
| CEN13 | CACTATTTATTTTACTATGG | 268036–268410 | |||
| 10 | chr IV_right_ telomere | TTGTGGTAGCAACACTATCA | 1524527–1531919 | 8 | 2 |
| chrXII_left_ telomere | GTTGTGTTGAGGTACTGTGT | 1–11385 | |||
| CEN12 | AGGAGAAAACTTGTAGTACG | 150818–150946 | |||
| 11 | chr XI_right_ telomere | CTGTTTAGACACTTGCGTCA | 664993–666454 | 16 | 9 |
| chr VII_left_ telomere | AAAAGGATCTATCTCCCGCT | 1–4700 | |||
| CEN11 | TATTTAGTATTGGACCATTG | 439774–439891 | |||
| 12 | chr X_right_ telomere | CATCCAGATCGGAAACGCTA | 743832–745742 | 2 | 1 |
| chr XV_left_ telomere | AGGTAAGGATACGGGGATAT | 1–5056 | |||
| CEN8 | TATTATACTAAATCGTTTTG | 105580–105700 | |||
| 13 | chr II_right_ telomere | CGTAACGTTGGTTCACATTT | 811675–813178 | 5* | 1 |
| chr XI_left_ telomere | AAATGAAGAAGTGCCATGGG | 1–2951 | |||
| CEN3 | TAAGCGGAAGGGGAAGGGTT | 114487–114489 | |||
| 14 | chr XV_right_ telomere | CTGCATCGTCTCGCTTTGCC | 1073916–1091289 | 135* | 2 out of 95 |
| chr IV_left_ telomere | CTTGCAACATCGCGACAGGT | 1–5566 | |||
| CEN4 | TCGGCATTTTGGCCGCTCCT | 449709–449819 | |||
Selection plates are SC–Ura–Leu, otherwise are SC–Ura–Leu+Hyg. The same CRISPR–Cas9 experiment as fusion step 14 was also carried out in an n = 15 strain background. As a result, 71 colonies grew and 1 out of 71 passed the PCR verification, suggesting that the efficiency does not change much as n drops.
Extended Data Table 2 |.
Summary table of fusion chromosome strains
| 1 | yJL373 | Chr03R was fused to chr01L, with cen1 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 III:I (cen3) | 15 |
| 2 | yJL374 | Chr09R was fused to chr03L, with cen9 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 IX:III:I (cen3) | 14 |
| 3 | yJL282 | Chr05R was fused to chr08L, with cen5 deleted | MATa his3Δ0 leu2Δ0 met 15Δ0 ura3Δ0 IX:III:I (cen3) V:VIII(cen8) | 13 |
| 4 | yJL303 | Chr06R was fused to chr02L, with cen6 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 IX:III:I (cen3) V:VIII(cen8) VI:II(cen2) | 12 |
| 5 | yJL310 | Chr13R was fused to chr14L, with cen14 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 IX:III:I (cen3) V:VIII(cen8) VI:II(cen2) XIII:XIV(cen13) | 11 |
| 6 | yJL320 | Chr16R was fused to chr09L, with cen16 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I(cen3) V:VIII(cen8) VI:II(cen2) XIII:XIV(cen13) | 10 |
| 7 | yJL336 | Chr08R was fused to chr10L, with cen10 deleted | MATa his3Δ0 leu2Δ0 met 15Δ0 ura3Δ0 XVI:IX:III:I(cen3) V:VIII:X(cen8) VI:II(cen2) XIII:XIV(cen13) | 9 |
| 8 | yJL342 | Chr01R was fused to chr06L, with cen2 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVl:IX:III:I:VI:II (cen3) V:VIII:X(cen8) XIII:XIV(cen13) | 8 |
| 8 | yJL346 | Chr07R was fused to chr13L, with cen13 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I(cen3) VI:II(cen2) V:VIII:X(cen8) VII:XIII:XIV(cen7) | 8 |
| 9 | yJL358 | Chr07R was fused to chr13L, with cen13 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I:VI:II (cen3) V:VIII:X(cen8) VII:XIII:XIV(cen7) | 7 |
| 10 | yJL369 | Chr04R was fused to chr12L, with cen12 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I:VI:II (cen3) IV:XII(cen4) V:VIII:X(cen8) VII:XIII:XIV(cen 7) | 6 |
| 11 | yJL375 | Chr11R was fused to chr07L, with cen11 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I:VI:II (cen3) IV:XII(cen4) V:VIII:X(cen8) XI:VII:XIII:XIV(cen7) | 5 |
| 12 | yJL379 | Chr10R was fused to chr15L, with cen8 deleted | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I: VI:II (cen3) IV:XII(cen4) V:VIII:X:XV(cen15) XI:VII:XIII:XIV(cen7) | 4 |
| 13 | yJL381 | Chr02R was fused to chr11L, with cen3 inactived | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I:VI:II:XI:VII:XIII:XIV (cen7) IV:XII(cen4) V:VIII:X:XV(cen15) | 3 |
| 13 | yJL410 | Chr15R was fused to chr04L, with cen4 deleted with parental strain: yJL379 | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 V:VIII:X:XV:IV:XII(cen15) XVI:IX:III:I:VI:II (cen3) XI:VII:XIII:XIV(cen7) | 3’ |
| 14 | yJL402 | Chr15R was fused to chr04L, with cen4 deleted with parental strain yJL381 | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 XVI:IX:III:I:VI:II:XI:VII:XIII:XIV (cen7) V:VIII:X:XV:IV:XII(cen15) | 2 |
Extended Data Table 3 |.
Variants identified from genome sequencing of different n strains
| N | Chromosome | Position | Reference | Mutation | ORF (amino acids mutation) |
|---|---|---|---|---|---|
| 12 | Chr II | 8972 | G | A | Non-coding |
| 8975 | T | C | Non-coding | ||
| 557493 | A | C | AMN1 (YBR158W, L317F) | ||
| Chr XIV | 577042 | ATCGGTGGTTAAACAA | A | Non-coding | |
| 8 | Chr II | 8972 | G | A | Non-coding |
| 8975 | T | C | Non-coding | ||
| 322190 | G | A | QDR3(YBR043C, P586S) | ||
| 557493 | A | C | AMN1 (YBR158W, L317F) | ||
| Chr XIV | 577042 | ATCGGTGGTTAAACAA | A | Non-coding | |
| 4 | Chr II | 8972 | G | A | Non-coding |
| 8975 | T | C | Non-coding | ||
| 9022 | GAAA | G | Non-coding (poly A region) | ||
| 322190 | G | A | QDR3(YBR043C, P586S) | ||
| 557493 | A | C | AMN1 (YBR158W, L317F) | ||
| Chr IV | 449785 | A | C | Non-coding | |
| 979062 | C | T | EXG2(YDR261C, A49T) | ||
| Mitochondria | 2483 | CTAATAAT | C | Non-coding (Tandem repeat region) | |
| 3916 | C | CT | Non-coding (poly T region) | ||
| 54569 | A | AG | Non-coding (poly G region) | ||
| Chr VII | 614706 | A | C | ADE6(YGR061C, C422G) | |
| Chr X | 648070 | C | A | ATP2(YJR121W, A158E) | |
| Chr XIV | 8046 | CCGTGA | C | Non-coding | |
| 8052 | T | TCTCACA | Non-coding | ||
| 246906 | C | T | RRG9(YNL213C, R67K) | ||
| 577042 | ATCGGTGGTTAAACAA | A | Non-coding | ||
| Chr XV | 976987 | AT | A | Non-coding (poly T region) | |
| 2 | Chr II | 8972 | G | A | Non-coding |
| 8975 | T | C | Non-coding | ||
| 322190 | G | A | QDR3(YBR043C, P586S) | ||
| 557493 | A | C | AMN1 (YBR158W, L317F) | ||
| Chr III | 114916 | AT | A | Non-coding | |
| Chr IV | 979062 | C | T | EXG2(YDR261C, A49T) | |
| Mitochondria | 2483 | CTAATAAT | C | Non-coding (Tandem repeat region) | |
| 3916 | C | CT | Non-coding (poly T region) | ||
| 29767 | T | A | Non-coding (in a AT rich region) | ||
| 54569 | A | AG | Non-coding (poly G region) | ||
| Chr V | 193248 | G | T | ISC1 (YER019W, synonymous mutation) | |
| Chr VII | 614706 | A | C | ADE6(YGR061C, C422G) | |
| Chr VIII | 133864 | A | T | Non-coding | |
| Chr X | 648070 | C | A | ATP2(YJR121W, A158E) | |
| Chr XIV | 8046 | CCGTGA | C | Non-coding | |
| 8052 | T | TCTCACA | Non-coding | ||
| 246906 | C | T | RRG9(YNL213C, R67K) | ||
| 577042 | ATCGGTGGTTAAACAA | A | Non-coding | ||
| Chr XV | 976987 | AT | A | Non-coding (poly T region) | |
Mutations inherited from previous steps are shaded.
Extended Data Table 4 |.
Gene expression changes in n = 2, 4 and 8 strains
| a | |||||
| N=2 | N=4 | N=8 | |||
| Expression decreases | RMD6*, YEL077C*, YLR460C*, AIF1*, DLD3*, COS10* | GEX2*, YRF1–4*, DLD3*, YLR460C*, AIF1*, YEL073C*, YEL077C*, RMD6*, COS7* | YLR460C*, YDR541C*, COS7* | ||
| Expression increases | YKL187C, SPG4, AAD10*, RPS9A, YJL045W, YLR281C, COS8*, OYE3, HXT5, IMD2*, HSP12, HMX1, HBT1, PHQ11*, SPG1, GRE1, SIP18, Y1R042C*, PIR3, FMP45, GND2, RNR3, YDR034W-B, HXT9*, PHM7, PUT4, COS1*, CDA1, TKL2, HUG1, COS4*, VBA3* | YER188W*, COS8*, IMD2*, YIR042C*, COS1*, COS4*, VBA3* | YIR042C*, COS1*, COS8* YER188W*, IMD2*, COS4* | ||
| b | |||||
| Gene Systematic name | N=2 | N=4 | N=8 | ||
| COS4/YFL062W | significant | significant | significant | ||
| YFR057W/YFR057W | significant | significant | significant | ||
| YPS5/YGL259W | significant | significant | significant | ||
| IMD2/YHR216W | significant | significant | significant | ||
| YGL258W-A /YGL258W-A | significant | not significant | significant | ||
| GEX1/YCL073C | significant | significant | not significant | ||
| VBA3/YCL069W | significant | significant | not significant | ||
| PAU4/YLR461W | significant | not significant | not significant | ||
| FDH1/YOR388C | significant | not significant * | not significant * | ||
| THI5/YFL058W | not significant | not significant | not significant | ||
| AAD15/YOL165C | Removed by design | Removed by design | not significant * | ||
| Fisher’s Exact test | p-value= 9.34 × e−12 | p-value= 1.16 × e−6 | p-value= 8.66 × e−7 | ||
| c | |||||
| Significantly changed in n=2 strain | Significantly changed in n=4 strain | Significantly changed in n=8 strain | All genes | ||
| Subtelomeric genes | 15 | 16 | 9 | 325 | |
| Non-subtelomeric genes | 24 | 0 | 0 | 6360 | |
| P value (Fisher’s Exact test) | 2.212 × e−10 | 2.20 × e−16 | 1.729 × e−12 | ||
a, Under- or overexpressed genes in n = 2, 4 and 8 strains. Genes in the fused chromosome arm are labelled as red. Genes in remaining unfused chromosome arms are in black.
Genes located near a telomere in n = 16 strain (within 20 kb).
b, Comparison of the expression changes of 21 subtelomeric genes, with changes in Sir2Δ, Sir3Δ and Sir4Δ strains in ref. 40. Twenty-one subtelomeric genes that are significantly expressed in ref. 40 are listed in the first column. Whether they are significantly expressed in our strains (n = 2, n = 4 and n = 8) or not is listed in the second, third and fourth columns. The last row shows the correlation between fusion chromosome strains and Sir protein depletion strains. IMD1, YAR075W, YCL076W, YCL075W, YCL074W, YFL063W, YNL337W are removed by design. YCL068C, YCL065W and YCL064C are part of the silent HML locus.
Unfused telomeres. The P value was calculated by a one-sided Fisher’s exact test.
c, Transcriptome change enrichments in sub-telomeres are significant in n = 2, 4 and 8 cells, calculated by one-sided Fisher’s exact test.
Extended Data table 5 |.
Primers for PCR verification of fusion chromosomes
| Fusion step | Purpose | Forward primer (5’–3’) | Reverse primer (5’–3’) |
|---|---|---|---|
| 1 | chr III-I junction | TGAAACGCACTGCAACATCTG | GGCTCAATCATCTACCGCATGG |
| CEN 1 deletion | CAACCAAACGTCCTCTTCTCTC | ACGATACATGGACTGACTCAAG | |
| 2 | chr IX-III junction | TTAAGGTGCGACCGGCAATG | CCTGTTTCGGCACTTGAGTC |
| CEN 9 deletion | CAACGAATTTCTCTCCGCCAGG | CACTTCAACAGTGCCAAAGACTCT AC | |
| 3 | chr V-VIII junction | TACGCCAAGTCGGTCAGGTC | TCCGAACTTGGTGTGTCTTCAG |
| CEN 5 deletion | ACCTCCTAGCACTTCGTAATG | GCTATTTATGTGCGGCTTTGTC | |
| 4 | chr VI-II junction | CAGATCCTTTCGCATTCCTACTTG | TCGTCGATGGTATTGGTGTAGAG |
| CEN 6 deletion | TTGGGCGATGGAAGAGGTAAAG | ACTTTCAACGCAAGAGCAAGAC | |
| 5 | chr XIII-XIV junction | CCAAGACTCTCACCTGCGAC | GCAATGGCTCAGTAACCTCG |
| CEN 14 deletion | CTGATGGACTCCGTAGAGAGC | AGGGTAGCATAAACCTGCTG | |
| 6 | chr XVI-IX junction | ACATTTGGGCCGTTGCTAGAAG | TATAGTCGGGCCTAGTTGCACTC |
| CEN 16 deletion | GGTTGAAGGAGTTAGTTTGTCG | GCCGCTTTGATGATTCTGCTTTAG | |
| 7 | chr VIII-X junction | GAACCGCTTCTGCTCAACTAG | CAATGACGGTGTTCGTGAAGC |
| CEN 10 deletion | CTCAGAAGGGAATTTCGTAAGC | CCAGTTTAGTTGTTGTGGATGC | |
| 8 | chr I-VI junction | CGTCAGCAGCGTCAGTAACTC | GCTGCAACAACTTCCCAATCATG |
| CEN 2 deletion | GGACTGAAAGCCAGTAACAAGC | TTCTCGTACCAAGCCGGTTC | |
| 9 | chr VII-XIII junction | AAAGTTTCCACCAGACGCTAAG | CT ACACTCGAACTCTGTTTCTCTC |
| CEN 13 deletion | AGGCTTTCGATTACCATGTGC | CTAAGGTAGCCAGAACTTCTCATC | |
| 10 | chr IV-XII junction | GCGTCACTTCTAAGAACAAGACTGC | ATGGTGAGAGATGGGTGATGGAG |
| CEN 12 deletion | GACAACCAAACTGGTGTATGC | TGCCATCATCTACTTCCTTTCC | |
| 11 | chr XI-VII junction | GCGAAAGCGAAGCCAATGTG | GCCATTAGCCTTCTATGTGTTC |
| CEN 11 deletion | GAACGACATTAACGGATACGCAAC | TGAAGAAGGTCAACATGAGGATGG | |
| 12 | chr X-XV junction | AATGCTGTGACACGCAGATAC | GGTACGCTCACCTCGTAAGTC |
| CEN 8 deletion | ACCCTCAGGTTGCTATGACG | ACGCACGAGCGAATTAACATTCC | |
| 13* | chr II-XI junction | GCTGCAACAACTTCCCAATCATG | TTTGCCAACACGAAAGGAACTC |
| 14 | chr XV-IV junction | GGTAGTAACCAACTCGTATCCCTG | TGGCATTCCTCTTTCACTTTCGTC |
| CEN 4 deletion | AGTGGTTGACATGCTGGCTAG | GGCCTCAAGAAAGAAACCTCTATG | |
Centromere 3 was inactivated by the deletion of only 3 bp CGG. Sanger sequencing was used to verify the deletion of CGG rather than PCR amplification.
Supplementary Material
Acknowledgements
We thank L. A. Mitchell, N. Agmon, and D. M. Truong for discussion and comments on this manuscript; L. A. Vale Silva and A. Hochwagen for sharing strains, advice and reagents; The NYU Langone Health Genome Technology Center, especially A. Heguy and P. Zappile, for deep sequencing libraries; M. S. Hogan and M. T. Maurano for help with the NextSeq 500 sequencer; Z. Kuang, X. Wang and Z. Tang for advice on bioinformatics analysis; and M. Delarue and L. Holt for help and access to spinning disk confocal microscopy. This work was supported by NSF grant MCB-1616111 to J.D.B.
Reviewer information Nature thanks G. Liti, K. Wolfe and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Footnotes
Competing interests J.D.B is a founder and Director of Neochromosome, Inc., the Center of Excellence for Engineering Biology, and CDI Labs, Inc. and serves on the Scientific Advisory Board of Modern Meadow, Inc., Recombinetics, Inc., and Sample6, Inc. All other authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41586-018-0374-x.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-018-0374-x.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kandul NP, Lukhtanov VA & Pierce NE Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 61, 546–559 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Taylor RW Ants with Attitude: Australian Jack-jumpers of the Myrmecia pilosula species complex, with descriptions of four new species (Hymenoptera: Formicidae: Myrmeciinae). Zootaxa 3911, 493–520 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Gordon JL, Byrne KP & Wolfe KH Mechanisms of chromosome number evolution in yeast. PLoS Genet. 7, e1002190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellis M, Birren BW & Lander ES Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Wolfe KH & Shields DC Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 (1997). [DOI] [PubMed] [Google Scholar]
- 6.IJdo JW, Baldini A, Ward DC, Reeders ST & Wells RA; JW. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc. Natl Acad. Sci. USA 88, 9051–9055 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda Y et al. Large-scale genome reorganization in Saccharomyces cerevisiae through combinatorial loss of mini-chromosomes. J. Biosci. Bioeng. 113, 675–682 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Neurohr G et al. A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science 332, 465–468 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Titos I, Ivanova T & Mendoza M Chromosome length and perinuclear attachment constrain resolution of DNA intertwines. J. Cell Biol. 206, 719–733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiCarlo JE et al. Genome engineering in Saccharomyces cerevisiae using CRISPR–Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert I & Oud JL There is an upper limit of chromosome size for normal development of an organism. Cell 88, 515–520 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Verdaasdonk JS & Bloom K Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12, 320–332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winey M & Bloom K Mitotic spindle form and function. Genetics 190, 1197–1224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Y et al. Creating a functional single chromosome yeast. Nature 10.1038/s41586-018-0382-x (2018). [DOI] [PubMed] [Google Scholar]
- 15.Lynch M et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl Acad. Sci. USA 105, 9272–9277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Shirogane T, Liu D, Harper JW & Elledge SJ Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell 112, 697–709 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Baack E, Melo MC, Rieseberg LH & Ortiz-Barrientos D The origins of reproductive isolation in plants. New Phytol. 207, 968–984 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Hou J, Friedrich A, de Montigny J & Schacherer J Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr. Biol. 24, 1153–1159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delneri D et al. Engineering evolution to study speciation in yeasts. Nature 422, 68–72 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Liti G, Barton DB & Louis EJ Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leducq JB et al. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat. Microbiol. 1, 15003 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Haber JE, Thorburn PC & Rogers D Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics 106, 185–205 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yona AH et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl Acad. Sci. USA 109, 21010–21015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheltzer JM et al. Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonney ME, Moriya H & Amon A Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev. 29, 898–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown CA, Murray AW & Verstrepen KJ Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20, 895–903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ai W, Bertram PG, Tsang CK, Chan TF & Zheng XF Regulation of subtelomeric silencing during stress response. Mol. Cell 10, 1295–1305 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Mayr E Systematics and the Origin of Species (Columbia Univ. Press, New York, 1942). [Google Scholar]
- 29.Dobzhansky T Genetics and the Origin of Species (Columbia Univ. Press, New York, 1937). [Google Scholar]
- 30.De Queiroz K Species concepts and species delimitation. Syst. Biol. 56, 879–886 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Neta Agmon JT et al. Human to yeast pathway transplantation: cross-species dissection of the adenine de novo pathway regulatory node. Preprint at https://www.biorxiv.org/content/early/2017/06/14/147579 (2017).
- 32.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Yon J & Fried M Precise gene fusion by PCR. Nucleic Acids Res. 17, 4895 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stemmer WP, Crameri A, Ha KD, Brennan TM & Heyneker HL Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Richardson SM, Wheelan SJ, Yarrington RM & Boeke JD GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16, 550–556 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell LA et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science 355, eaaf4831 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Breslow DK et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711–718 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science 355, eaaf4706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellahi A, Thurtle DM & Rine J The chromatin and transcriptional landscape of native Saccharomyces cerevisiae telomeres and subtelomeric domains. Genetics 200, 505–521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We submitted DNA sequences and RNA sequencing data to NCBI BioProject PRJNA471518. All other data are available from the corresponding author upon reasonable request.