Abstract
Exosomes are extracellular vesicles that mediate intercellular communication. They contain different molecules, such as DNA, RNA, lipid, and protein, playing essential roles in the pathogenesis of colorectal cancer (CRC). Exosomes derived from CRC are implicated in tumorigenesis, chemotherapy resistance, and metastasis. Besides, they can enhance CRC progression by increasing tumor cell proliferation, reducing apoptosis mechanistically through altering particular essential regulatory genes, or controlling several signaling pathways. Therefore, exosomes derived from CRC are essential biomarkers and can be used in the diagnosis. Indeed, it is crucial to understand the role of exosomes in CRC, which is necessary to develop diagnostic and therapeutic strategies for early detection and treatment. In the present review, we discuss the roles of exosomes in the diagnosis and treatment of CRC.
Keywords: Colorectal cancer, Exosomes, Biomarker, Extracellular vesicle, Diagnosis
Core Tip: Exosomes are extracellular vesicles that mediate intercellular communication. They contain molecules such as RNAs, DNA, lipids, and proteins, whose role is essential in the pathogenesis of colorectal cancer (CRC). Exosomes derived from CRC are implicated in tumorigenesis, chemotherapy resistance, and metastasis. Besides, they can enhance CRC progression by increasing tumor cell proliferation, reducing apoptosis mechanistically through upregulation or downregulation of particular essential regulatory genes, or controlling several signaling pathways. Therefore, exosomes derived from CRCs are essential biomarkers and can be used in the diagnosis.
INTRODUCTION
As a global health issue, colorectal cancer (CRC) is ranked as the 3rd most commonly diagnosed malignancy worldwide. Data indicate that CRC affects about one million people annually[1] and causes nearly 694000 related deaths in males and females with almost an equal gender distribution[2]. Lack of specific CRC symptoms imposes a challenge towards clinicians, as the symptoms may overlap with other non-cancerous diseases. Most clinicians rely on clinical evaluation, which has limited diagnostic accuracy. Conventional diagnoses using radiological imaging or histopathological analysis lack the responsiveness to detect the systematic spread of CRC at an early stage[3]. CRC markers, like carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), are commonly utilized to identify CRC. However, these markers demonstrate a less-than-desirable sensitivity and specificity[4]. In most CRC cases, surgery is the optimal treatment of choice, while most patients are diagnosed at the advanced stages[5]. Therefore, it is urgently necessary to develop new molecular markers for effective diagnosis[3]. The detection of biomarkers using patients’ peripheral blood is now considered a prospective diagnostic tool.
Liquid biopsy is a non-invasive procedure for the analysis of exosomes, circulating tumor cells, and also the circulating tumor DNA fragments in early stages by taking blood samples or other body fluids[6]. They have advantages in monitoring treatment efficacy and progression of the tumor, therapy resistance mechanisms, and tumor heterogeneity and evolution in real time[7,8]. Since many cancers are discovered at advanced stages, biomarkers are crucial for earlier detection and reduction of cancer mortality[9].
Exosomes are extracellular nano-sized vesicles that contain DNA, RNA, lipid, and protein species. Exosomes can be locally and systemically transferred while carrying their contents to the recipient cells, thus playing critical roles in intercellular communication. Exosomes originating from CRC cells are linked to tumorigenesis, tumor cell survival, chemotherapy resistance, and metastasis. In this process, proteins, RNAs, or mutant versions of proto-oncogenes are transferred to the exosomes’ target cells[10]. Some of the bioactive molecules that are released/carried by exosomes can/might be essential biomarkers for CRC. These markers have been previously mentioned as mRNA, non-coding RNA, DNA, and secretory protein. The use of exosomal markers as potential diagnostic and prognostic molecules may eventually become necessary. In the present review, we focus on the roles of exosomes in the early diagnosis and treatment of CRC.
COMPONENTS AND BIOGENESIS OF EXOSOMES
Initially, exosomes were thought to act only as cellular waste disposal[11]. In the 1980s, exosomes have been revealed to be derived from the endosomes and are secreted by reticulocytes[12,13]. Raposo et al[14] have reported more than a decade later that exosomes isolated from Epstein-Barr virus-transformed B lymphocytes are antigen-presenting particles and may induce T cell responses. It is now believed that exosomes are a crucial part of many cellular processes[15]. Exosomes belong to a large family of membrane vesicles known as extracellular vesicles (EVs). Generally speaking, the exosome is considered to be the smallest member among the other remaining EVs (namely, apoptotic bodies and microvesicles) with a diameter of 30-100 nm in size[16]. In particular, exosomes are of significant interest to the cancer research community. The components of exosomes include proteins and RNAs, which can be horizontally transmitted between adjacent or distant cells[17].
Exosomes usually contain some cellular contents that can be used as markers for exosome recognition. In this sense, exosomes can help determine the type of cell, from which an exosome originated, as well as cell health. Markers change because of viral vs bacterial infections and vary when a cell is cancerous as well. These usual markers are mRNA, microRNA (miRNA), protein, and lipid[18]. More recently, however, much attention has been paid to the use of circulating miRNAs as diagnostic/prognostic biomarkers of infectious diseases, such as human tuberculosis (TB) caused by Mycobacterium tuberculosis infection[19]. Fu et al[20] have identified 92 differentially expressed miRNAs in serum from patients with active pulmonary TB using a human miRNA microarray platform (Exiqon miRCURY™ LNA).
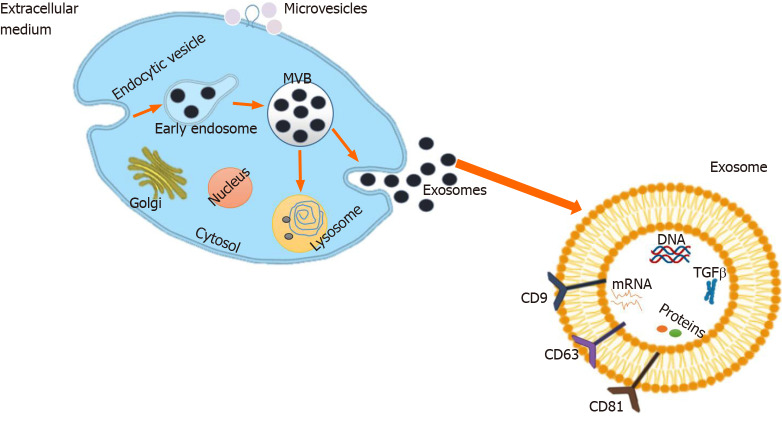
Different steps are involved in exosome biogenesis[21]. Figure 1 illustrates that exosome biogenesis starts with cell membrane internalization, which contributes to the early endosomal formation[22]. Several intraluminal vesicles (ILVs) are formed by endosomal membrane inward invagination, resulting in multivesicular body (MVB) formation. During this process, cytosolic constituents, namely, nucleic acids, lipids, and proteins, can be sorted into ILVs. MVBs are known as late endosomal structures containing dozens of ILVs that are ultimately delivered to lysosomes for degradation, or fused with the plasma membrane to release exosomes[23]. In this process, the ESCRT (endosomal sorting complexes required for transport) machinery is a crucial regulator of MVB formation. It consists of four different proteins, namely, ESCRT-0, -I, -II, and -III[24]. Other vital regulators required during exosome biogenesis are sphingolipid ceramide, and Rab GTPase (guanosine triphosphate) families, such as Rab27 and Rab11[25]. The proteins within the exosomes are highly conserved across species, and some exosome-enriched proteins, such as tetraspanins (CD63 and CD81), histocompatibility complex (MHC) molecules, ESCRT-III binding protein ALG2 interacting protein X (Alix), tumor susceptibility gene 101 protein (TSG101), and heat shock protein 70 (HSP70), are commonly used as markers for exosome recognition[26,27]. By analyzing data, common exosome markers (such as CD63, CD9, ALIX, TSG101, and HSP70) are enriched in urinary exosome-like vesicles compared with urine protein microvesicles[28]. Nevertheless, more proteomic studies are highly required to verify the specificity of these protein markers.
Figure 1.
Exosome biogenesis. Exosome biogenesis starts with budding into early endosome and further matures into the late endosome, collectively known as multivesicular bodies. Early endosome is formed from the plasma membrane via the endocytic pathway. Multivesicular body (MVB) can be formed by the invagination of the endosomal membrane. Dependent on the function and content, MVB then can be directed to fuse with plasma membrane and release to the extracellular space as exosomes. During the biogenesis of exosomes and prior to their secretion, proteins (e.g., tetraspanin, cytosolic proteins, and receptor), nucleic acids (e.g., mRNA, miRNA, and DNA), and lipids (e.g., sphingomyelin and cholesterol) are uploaded to exosomes. MVB: Multivesicular body; TGF: Transforming growth factor.
Exosomes are believed to exist in body fluids, including blood, urine, breast milk, and semen. Because of their ability to move from one place to another, transferring some essential molecules, such as DNA, RNA, mRNA, miRNA, lipid, and protein, plays a crucial role in intercellular communication for the diagnosis and prognosis of many diseases[29].
LEADING ROLES OF EXOSOMES IN CRC
Involvement of exosomes in proliferation, metastasis, epithelial-to-mesenchymal transition, and angiogenesis of CRC cells
Exosomes are highly involved in cell proliferation, metastasis, epithelial-to-mesenchymal transition (EMT), and angiogenesis in CRC. It is well known that cell proliferation increases the number of cells. It is characterized by disrupting the balance between cell division and cell loss through cell death or differentiation. Cell proliferation is increased in different cancers, including CRC. For example, it has been reported that miR1273g3p promotes the proliferation, migration, and invasion of LoVo cells via CNR1. This can occur by activating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway[30].
Moreover, Ren et al[31] have reported that self-growth or proliferation through exosome secretion, especially under hypoxic conditions, by reduction of the mitosis and activation duration can be promoted by CRC cells. In a recent study, the CRC cell-derived exosomes are separated from human colon cancer cell line SW480 and HCT116. When SW480 cells are incubated with exosomes marked with PKH67 fluorescent markers, they can absorb exosomes. Moreover, CRC cells are promoted by hypoxic conditions to release more exosomes. And, these exosomes promote the proliferation of CRC cells via shortening the mitosis duration and activating STAT3 signaling[31].
Metastasis is the spread of cancer cells through the lymphatic system or the bloodstream to new body areas. Exosomes have been reported to promote metastasis in CRC. For instance, a study has shown that the expression of serum exosomal miR-106b-3p in CRC patients with metastasis is dramatically higher compared with that of non-metastatic patients. Exosomal miR-106b-3p facilitates lung metastasis of CRC cells in vivo by targeting deleted liver cancer-1 (DLC-1). In that study, the authors have examined the miRNA expression profiles of five paired serum exosomal samples from metastatic CRC (mCRC) and non-mCRC patients via RNA sequencing. After evaluating the differentially expressed miRNAs in 80 CRC patients, they have chosen miR-106b-3p as a metastasis-associated miRNA of CRC. It shows that the expression of serum exosomal miR-106b-3p is dramatically advanced in mCRC patients compared with non-mCRC patients. Besides, high expression of serum exosomal miR-106b-3p in patients is interrelated with a poor prognosis. Cell migration, invasion, and EMT are promoted by coculture of low-mCRC cells and highly metastatic colon-cancer-cell-derived exosomes (CDEs), which are caused by the transport and transduction of miR-106b-3p in vitro. Furthermore, exosomal miR-106b-3p promotes lung metastasis of CRC cells in vivo. They have also shown that miR-106b-3p-triggered metastasis is regulated by DLC-1 in the liver. There is a negative correlation between miR-106b-3p and DLC-1 expression in human CRC tissues and mouse lung metastatic lesions. Generally speaking, their study has revealed that metastasis-associated miR-106b-3p from serum exosomes would be used as a potential prognostic biomarker and therapeutic target for CRC patients[32].
Moreover, a study aiming to explore the clinical and biological significance of miR-224 expression in CRC metastasis has also noted that miR-224 promotes CRC metastasis[33].
EMT is also a momentous event in the progression of CRC. Cumulative evidence shows that exosomes play a significant role in the EMT in CRC. For example, a study has shown that exosomes delivered by carcinoma-associated fibroblasts (CAFs) promote different features, including EMT[34]. Another study has also reported that the Wnt/β-catenin pathway transactivates miRNA-150 and enhances CRC cell EMT by hindering CREB signaling[35]. The chemosensitivity of metastatic cells is dramatically reduced compared with adherent HCT-8 cells. Of note, adherent new colonies experiencing EMT are not sensitive to both strategies of chemotherapy. Electron microscopy has shown that adherently developing HCT-8 cells produce exosomes, and exosomes are also taken up by metastatic cells. EMT is remarkably inhibited when exosomes produced by adherently developing HCT-8 cells are administered to metastatic cells. Exosomal miR-210 plays an important role in EMT that preserves the local cancer-development permissive milieu and also guides metastatic cells to avail new spread sites[36].
Angiogenesis is characterized by the formation of new blood vessels. This process includes the migration, growth, and differentiation of endothelial cells lining the inner walls of blood vessels. As mentioned earlier, exosomes play an essential role in promoting angiogenesis in different types of cancer, including CRC.
Research has shown the potential influence of exosomes in cancer evolution through a worldwide benchmark transcriptomic sequence of CRC cells. At first, 11327 microvesicular mRNAs are identified, which are linked to the physiology of donor CRC cells. Among them, 241 mRNAs are found in the microvesicles released by donor CRC cells, of which 27 are cell cycle-related. And these mRNAs promote the proliferation of the endothelium, which can induce tumor progression and metastasis[37].
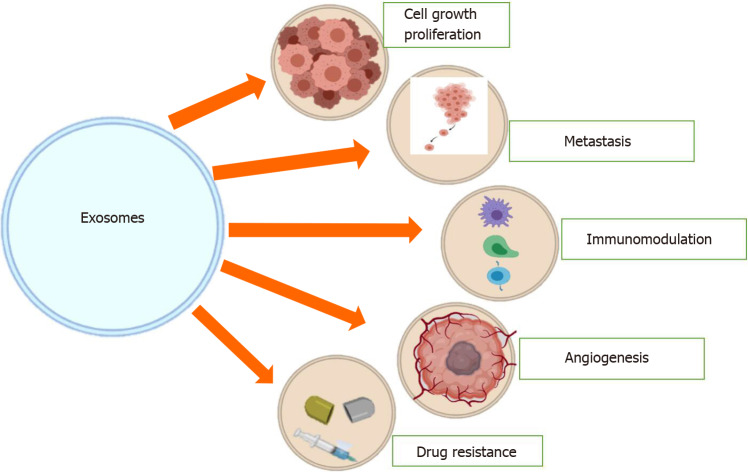
It has also been shown that by targeting promyelocytic leukemia mRNA in recipient endothelial cells, the miR-1246-containing exosomes from CRC cells can initiate Smad 1/5/8 signaling pathways, thus promoting angiogenic activities[38]. Based on the various studies mentioned above, it is quite clear that exosomes play an important role in different aspects of CRC, such as angiogenesis, EMT, metastasis, and cell proliferation (Figure 2)[39].
Figure 2.
Roles of tumor cell-derived exosomes. Exosomes are involved in tumor growth, tumorigenesis, angiogenesis, tumor immune escape, drug resistance, and metastasis.
Exosomes play an essential role in the CRC tumor microenvironment
The tumor microenvironment (TME) around a tumor contains adjacent blood vessels, fibroblasts, signaling molecules, immune cells, and extracellular matrix. The TME is an essential component in tumor growth or suppression. Exosomes partially form the microenvironment of multiple cancers, including CRC[40,41]. Exosomes enhance the expression of negative regulators of the immune system, including monocytes, regulatory T-cells (Tregs), and myeloid-derived suppressor cells (MDSCs), hence causing the tumor cells to escape from immune surveillance[42].
Exosomes derived from tumor cells can inhibit immune cell function. T cells do not internalize exosomes. Nonetheless, exosomes transmit signals to cell membrane receptors that modulate gene expression and human T lymphocyte functions[43]. Studies have demonstrated that EVs derived from CRC cells are enriched in transforming growth factor-beta 1 (TGF-β1) and can activate TGF-β/Smad signaling and inactivate SAPK signaling. Therefore, EVs from CRC can induce phenotypic modification of the T cells to Treg-like cells, the tumor-growth supporting cells. The CRC-EVs-induced-Treg-like cells have an unusual tumor-growth stimulating activity in vitro and in vivo[44].
CRC CDEs have significantly higher levels of miR-10b compared with healthy colorectal epithelial cells. Exosomal miR-10b substantially inhibits the expression of PIK3CA and decreases the PI3K/Akt/mTOR pathway activity. Exosomal miR-10b reduces the proliferation of fibroblasts but encourages the expression of TGF-β and SM α-actin, suggesting that exosomal miR-10b can cause fibroblasts to become CAFs. Activated fibroblasts can enhance CRC development in vitro and in vivo[45]. In most solid cancers, CAFs are the important cellular components of TME[46]. Exosomes derived from CAFs can promote neoplastic angiogenesis and tumor growth in CRC. They can also trigger cancer cell dedifferentiation via the Wnt signaling pathway, thus enhancing the chemical resistance of CRC[47,48].
There is also another study revealing that CAF exosomes can upsurge the number of cancer stem cells (CSCs) and trigger chemoresistance by activating the Wnt signaling pathway. CAFs are very well involved in tumor recurrence, and targeting them increases chemo-sensitivity. This research has examined whether fibroblasts can intensify CSCs to mediate chemoresistance. At first, CSCs are innately unaffected by chemotherapy. And, exosomes isolated from conditioned medium of fibroblasts promote the growth of CSCs under 5-Fu or oxaliplatin treatment. However, the above-mentioned effects are not observed when exosome secretion has been inhibited. In summary, their research findings recommend that blocking the secretion of CAF might be a new therapeutic strategy for advanced CRC treated with chemotherapy[49].
Tumor-associated macrophages (TAMs) play an immunosuppressive role in CRC. It has been found that miR-203 promotes the expression of M2 makers, therefore enhancing the differentiation of monocytes to TAMs[50]. M2 macrophages promote invasion and migration of CRC cells and provide significant BRG1 expression plasticity in response to TME[51]. Additionally, miR-1246 from CRC cells can reprogram macrophages to improve anti-inflammatory environment generation through up-regulating TGF-β[52]. The above facts highlight that exosomes play an essential role in the TME of CRC.
Exosomes are involved in the treatment of CRC
Managing CRC requires good therapeutic agents that are very accurate to inhibit CRC progression and increase patients’ survival rates. One of those agents is exosomes. Exosomes play a role in different cancer treatments by acting as vehicles that can transfer drugs to the target cells due to their small size (nano-scale dimension). Indeed, exosomes can carry interfering RNA (siRNA) or active pharmaceutical substances to the target cells or sites[53,54]. For example, during the prevention of CRC cell growth, in some cases, exosomes containing doxorubicin have been used to deliver the drugs to the targeted organs[55,56]. Different studies have shown that engineering exosomes play a significant role in the treatment of CRC by significantly decreasing cancer progression. It has been found that in CRC CDEs, miR-379 down-regulates the migration of CRC cells, and the transfer of these engineered miR-379-overexpressing exosomes to recipient cells decreases their migration[57]. It has also been reported that exosomal miRNA-375 prevents the spread of tumor cells by blocking Bcl-2 in CRC, suggesting that exosomal miRNA-375 can be regarded as a possible therapeutic target[58].
Another study has reported a pilot study for ascites-derived exosomes (Aexs) in conjunction with the granulocyte-macrophage colony-stimulating factor (GM-CSF) in the immunotherapy of CRC. Aexs isolated by sucrose/D2O density gradient ultracentrifugation are 60-90-nm vesicles that contain the diverse immunomodulatory indicators of exosomes and CEA associated with the tumor. Advanced CRC is used in a study consisting of 40 patients (HLA-A0201+CEA+). Besides, those patients are arbitrarily given medication, either Aex alone or Aex plus GM-CSF. Respondents from the two groups are administered with a set of four immunizations on a subcutaneous layer at a regular interval of 1 wk. The research discovers that both treatments are harmless, and they are well tolerated. Moreover, Aex plus GM-CSF, but not Aex alone, can trigger valuable tumor-specific antitumor cytotoxic T lymphocyte response. Consequently, the research shows that the immunotherapy of CRC with Aex together with GM-CSF is practicable and not dangerous. It can, therefore, be used as a substitute option in advanced CRC immunotherapy[59].
In patients with CRC, 5-Fu is generally prescribed. However, resistance to 5-Fu is one of the critical reasons for CRC treatment failure. To extricate from this problem, exosomes containing miRNAs have recently been established to regulate signaling pathways involved in the initiation and progression. In comparison with the single treatment with either miR-21i or 5-Fu, drug resistance can effectively be reversed. The cytotoxicity in 5-Fu-resistant CRC cells is significantly enhanced by the combinational delivery of miR-21i (miR-21 inhibitor oligonucleotide) and 5-Fu engineered exosomes. It is a potential approach to deliver the small functional RNA together with an anti-cancer drug using exosomes to reverse the drug resistance in CRC, thus enhancing the efficacy of the cancer treatment[60].
Moreover, exosomes containing miRNAs can help overcome many other forms of resistance to cancer therapy. For instance, by inhibiting autophagy in CRC cells, miR-214 enhances CRC radiosensitivity[61]. The resistance of CRC cells to OXA can be circumvented by the overexpression of miR-143, which is related to oxidative stress and cell death in CRC cells[62].
MiRNAs are non-coding RNAs that control the expression of mRNAs in a target cell[63]. This property also makes them suitable for a new approach to tumor therapy. When the expression of miRNAs is abnormal, tumor progression is affected[64], and restoring the expression of these miRNAs can prevent such disease progression. The tumor growth or metastasis can be restrained by the delivery of exosomal miRNA inhibitors, which interfere with tumor-promoting miRNAs[65]. By targeting KLF2 and KLF4, exosomal miR-25-3p from CRC cells may promote CRC-induced vascular permeability and angiogenesis and accelerate CRC metastasis. The development of pre-metastatic niches and CRC metastases can be significantly inhibited by exosomes loaded with the miR-25-3p inhibitor[66]. Briefly, using exosomes is of great significance in the treatment of tumors. Both anti-tumor and tumor-promoting miRNAs affect the tumor progression and balance the expression levels of these miRNAs.
It has also been reported that miR-128 targets the SIRT1/ROS/DR5 pathway to sensitize CRC cells to TRAIL-induced apoptosis. This makes exosomes together with TRAIL be considered a novel therapeutic for CRC [67]. Another study has also revealed that miR-128-3p transmitted by exosome increases the chemosensitivity of OXA-resistant CRC, altering the expression of Bmi1 and MRP5[68]. Besides, exosome-enclosed miR-140-3p demonstrates its suppressive effects on the development and metastasis of CRC cells through hindering BCL9 and BCL2[69]. Another trial has shown that the liver metastasis (LM) of CRC both in vitro and in vivo can be remarkably reduced[70] by using miR-20a-loading nanoparticles to target liver sinusoidal endothelial cells.
Exosomes can also decrease cancer cell proliferation, which is another momentous event in CRC progression. Researchers have reported that the overexpression of miR-194 significantly inhibits cell proliferation in HTC-116 cells, making it an excellent therapeutic agent for CRC[71].
Another study has shown that the production of exosomes with antiproliferative properties can be induced by treating cells with FF/CAP18 (analog of cathelicidin LL-37), which is a peptide limiting cancer cell proliferation[72]. This result would be caused by the expression of exosomal miRNAs, miR-584-5p, -1202, and -3162-5p. Some studies have also revealed the possibility of adjusting exosomes for therapeutic purposes by transfection with tumor-suppressor miRNAs[73]. Moreover, it has been shown that exosomes can inhibit tumor growth when coated with high-density antibodies to target specific ligands in CRC by using the doxorubicin (Dox) as cargo[74].
CRC continues to be one of the leading causes of cancer-related death worldwide, primarily due to resistance to treatment[75]. Many studies have demonstrated that this problem can be overcome. For example, a study has indicated that exosomal miR-46146 acts as a critical mediator of OXA resistance by targeting PDCD10, while increased expression of PDCD10 can reverse the effect of chemoresistance induced by exo-miR-46146[76]. Another study has found six up-regulated exosomes from the culture medium of resistant cells, namely, miR-96-5p, miR-1246, miR-135b, miR-21-5p, miR-425, and miR-1229-5p, and the expression levels of these six exosomal miRNAs in serum of CRC chemoresistant patients are markedly higher compared with chemosensitive controls, which may clearly distinguish the chemotherapy-resistant group from advanced CRC patients. However, targeting these miRNAs increases OXA and 5-Fu chemosensitivity[77].
Based on the above-mentioned facts, it is evident that exosomes play an essential role in the treatment of CRC. Nevertheless, it is also evident that in-depth research is highly required to let exosomes be used more effectively as routine treatments for CRC.
Exosomes are potential biomarkers for CRC
A biomarker may be a molecule that is secreted by a tumor or a specific response of the body to the presence of cancer. Given that early diagnosis of CRC is minimal, exosomes are now considered the source of diagnostic markers in CRC due to the significant role of exosomes in tumor formation, metastasis, chemoresistance, and invasion[78]. For example, a study has reported seven miRNAs, which are strongly expressed in the serum of CRC patients compared with healthy controls, and these miRNAs are miR-1229, miR-23a, let-7a, miR-223, miR-150, miR-1246, and miR-21[79], making these exosomes as potential biomarkers for CRC. A study has identified a novel biomarker to track CRC metastasis to the liver, showing that for early diagnosis and prognosis in CRC patients with LM, serum exosomal miR-122 can be a novel potential biomarker. To assess the diagnostic potential of serum exosomal miR-1222, a total of 135 serum samples, including those from CRC patients with LM (n = 35), CRC patients without LM (n = 50), and healthy controls (n = 50), are examined, and exosomes are isolated from the culture medium or serum using the Exosome Isolation Reagent (Invitrogen)[80].
Research has shown that flowing exosomes embraces significant effects as a biomarker for identification and forecast of cancers in human. Earlier, small RNA sequencing is used in the diagnosis of irrationally expressed exosomal miRNAs as nominees for analytical indicators in CRC patients. In this validation cohort, exosomal miR-125a-3p and miR-320c were found to be increased in plasma of CRC patients. And, plasma exosomal miR-125a-3p is readily reachable as an analytical bio-indicator for the initial phase of CRC. If jointed together with standard analytical indicators, miR-125a-3p can advance the indicative power[81].
Knowing patients’ outcomes is very important in diagnosing different types of cancer, including CRC, and exosomes are excellent tools to be used to track patients’ outcomes. For example, it has been revealed that increased expression miR-429 is correlated with enhanced malignant potential and poor prognosis of CRC patients, making miR-429 a biomarker in CRC diagnosis[82]. Another study has also shown that the expression of serum exosomal miR-874 may serve as a reliable marker for CRC diagnosis and prognosis prediction. Furthermore, serum miR-193b may serve as a promising novel prognostic biomarker for CRC[83]. Moreover, another study has found that serum miR-199a is an independent prognostic marker. Reduced expression of serum miR-199a is associated with poor prognosis in CRC. It may be a useful marker for diagnosis and prognosis in CRC[84]. Min et al[85] have shown that reduced expression of exosome-derived miR-92b in plasma is a promising biomarker for early CRC detection. Another study has found that the plasma exosomal miR-27a and miR-130a panel can serve as a non-invasive biomarker for early detection and prognosis prediction of CRC.
The above-mentioned studies all conclude that exosomal miRNAs play a significant role in the diagnosis of CRC and provide evidence for the routine application of specific exosomal components in the early detection of CRC. Additional information is presented in Table 1.
Table 1.
Exosomes as prognostic and diagnostic biomarkers for colorectal cancer
| Exosomes | Used samples | Expression | Function | Ref. |
| miR-23a, miR-301a | Serum | Increased | Diagnosis and prognosis | Karimi et al[86] |
| miR-193a, let-7g | Plasma | Increased, decreased | Diagnosis | Cho et al[87] |
| miR-548c-5p | Serum | Decreased | Diagnosis | Peng et al[88] |
| miR-92b | Plasma | Decreased | Diagnosis | Min et al[85] |
| miRNA-320d | Serum | Increased | Diagnosis | Tang et al[89] |
| let-7b-3p + miR-139-3p + miR-145-3p | Plasm | Increased | Diagnosis | Min et al[90] |
| miR-6803-5p | Serum | Increased | Diagnosis and prognosis | Yan et al[91] |
| miR-27a and miR-130a | Plasma | Increased | Diagnosis | Liu et al[92] |
| miR-122 | Serum and cultured media | Increased | Diagnosis | Sun et al[80] |
| miR-150-5p + CEA | Serum | Decreased | Used for early diagnosis | Peng et al[88] |
| miR-99b-5p | Serum | Decreased | Diagnosis | Zhao et al[93] |
CEA: Carcinoembryonic antigen.
CONCLUSION
This review shows that exosomes originating from CRC cells can be taken as a new research area for cancer diagnosis and medical treatment because these exosomes are linked to tumorigenesis, chemotherapy resistance, and tumor cell survival metastasis. Existing research has shown that the number of exosomes in the body fluid of CRC patients is exceptionally elevated compared with the healthy controls. Patients with a high number of exosomes mostly display poor prognoses. Therefore, exosomes from CRC can predict the prognosis for CRC patients.
The research on CRC exosomes has to be encouraged and continued because CRC exosomes are interesting small membrane-vesicles that can be used during cancer treatment and in many fields of our daily lives. It is also necessary to continue the research about comprehensive mechanisms for the function of exosomes in CRC to improve diagnostic accuracy and then reduce cancer mortality.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interests for this article.
Manuscript source: Invited manuscript
Peer-review started: January 22, 2021
First decision: February 28, 2021
Article in press: May 6, 2021
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Norčič G, Suzuki H S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Liu JH
Contributor Information
Yvette Umwali, Department of Clinical Laboratory, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, Shandong Province, China.
Cong-Bo Yue, Department of Clinical Laboratory, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, Shandong Province, China.
Abakundana Nsenga Ariston Gabriel, Department of Clinical Laboratory, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, Shandong Province, China.
Yi Zhang, Department of Clinical Laboratory, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, Shandong Province, China.
Xin Zhang, Department of Clinical Laboratory, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, Shandong Province, China. xinzhang@sdu.edu.cn.
References
- 1.Alrubaie A, Alkhalidi N, Abd-Alhusain S. A clinical study of newly-diagnosed colorectal cancer over 2 years in a gastroenterology center in Iraq. J Coloproctology. 2019;39:217–22. [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Norcic G. Liquid Biopsy in Colorectal Cancer-Current Status and Potential Clinical Applications. Micromachines (Basel) 2018;9 doi: 10.3390/mi9060300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–1360. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 5.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokunaga T, Chiba J, Ohnishi K. [Attempts to improve hybridoma technology for the production of human monoclonal antibodies] Gan To Kagaku Ryoho. 1987;14:2198–2204. [PubMed] [Google Scholar]
- 7.Hench IB, Hench J, Tolnay M. Liquid Biopsy in Clinical Management of Breast, Lung, and Colorectal Cancer. Front Med (Lausanne) 2018;5:9. doi: 10.3389/fmed.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 9.Martin KJ, Fournier MV, Reddy GP, Pardee AB. A need for basic research on fluid-based early detection biomarkers. Cancer Res. 2010;70:5203–5206. doi: 10.1158/0008-5472.CAN-10-0987. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-López L, Blancas I, Garrido JM, Mut-Salud N, Moya-Jódar M, Osuna A, Rodríguez-Serrano F. The role of exosomes on colorectal cancer: A review. J Gastroenterol Hepatol. 2018;33:792–799. doi: 10.1111/jgh.14049. [DOI] [PubMed] [Google Scholar]
- 11.Johnstone RM. The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem Cell Biol. 1992;70:179–190. doi: 10.1139/o92-028. [DOI] [PubMed] [Google Scholar]
- 12.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 14.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Chen G, Lin X, Xing X, Cai Z, Liu X, Liu J. Role of exosomes in hepatocellular carcinoma cell mobility alteration. Oncol Lett. 2017;14:8122–8131. doi: 10.3892/ol.2017.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahebi R, Langari H, Fathinezhad Z, Bahari Sani Z, Avan A, Ghayour Mobarhan M, Rezayi M. Exosomes: New insights into cancer mechanisms. J Cell Biochem. 2020;121:7–16. doi: 10.1002/jcb.29120. [DOI] [PubMed] [Google Scholar]
- 19.Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, Shaughnessy RG. Circulating microRNAs as Potential Biomarkers of Infectious Disease. Front Immunol. 2017;8:118. doi: 10.3389/fimmu.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1–11. doi: 10.1016/j.pharmthera.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 24.Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30; sup pp 1. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 26.Budnik V, Ruiz-Cañada C, Wendler F. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci. 2016;17:160–172. doi: 10.1038/nrn.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Chutipongtanate S, Greis KD. Multiplex Biomarker Screening Assay for Urinary Extracellular Vesicles Study: A Targeted Label-Free Proteomic Approach. Sci Rep. 2018;8:15039. doi: 10.1038/s41598-018-33280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu K, Zhang C, Du T, Gabriel ANA, Wang X, Li X, Sun L, Wang N, Jiang X, Zhang Y. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed Pharmacother. 2021;134:111111. doi: 10.1016/j.biopha.2020.111111. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Qian X, Zhu M, Li A, Fang M, Zhu Y, Zhang J. miR1273g3p promotes proliferation, migration and invasion of LoVo cells via cannabinoid receptor 1 through activation of ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Mol Med Rep. 2018;17:4619–4626. doi: 10.3892/mmr.2018.8397. [DOI] [PubMed] [Google Scholar]
- 31.Ren R, Sun H, Ma C, Liu J, Wang H. Colon cancer cells secrete exosomes to promote self-proliferation by shortening mitosis duration and activation of STAT3 in a hypoxic environment. Cell Biosci. 2019;9:62. doi: 10.1186/s13578-019-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, Han P, Zhang B, Yao K, Li C, Bai J, Cui B. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci (Lond) 2020;134:419–434. doi: 10.1042/CS20191087. [DOI] [PubMed] [Google Scholar]
- 33.Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y, Lee TJ, Luo Z, Liu L, Xu D, Tili E, Jin V, Middleton J, Chakravarti A, Lautenschlaeger T, Croce CM. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2015;112:E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo YH, Wang LQ, Li B, Xu H, Yang JH, Zheng LS, Yu P, Zhou AD, Zhang Y, Xie SJ, Liang ZR, Zhang CM, Zhou H, Qu LH. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7:42513–42526. doi: 10.18632/oncotarget.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Ther. 2016;17:1062–1069. doi: 10.1080/15384047.2016.1219815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D, Gho YS. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada N, Tsujimura N, Kumazaki M, Shinohara H, Taniguchi K, Nakagawa Y, Naoe T, Akao Y. Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim Biophys Acta. 2014;1839:1256–1272. doi: 10.1016/j.bbagrm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Guo W, Gao Y, Li N, Shao F, Wang C, Wang P, Yang Z, Li R, He J. Exosomes: New players in cancer (Review) Oncol Rep. 2017;38:665–675. doi: 10.3892/or.2017.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. 2017;189:259–267. doi: 10.1111/cei.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol. 2009;183:3720–3730. doi: 10.4049/jimmunol.0900970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tucci M, Mannavola F, Passarelli A, Stucci LS, Cives M, Silvestris F. Exosomes in melanoma: a role in tumor progression, metastasis and impaired immune system activity. Oncotarget. 2018;9:20826–20837. doi: 10.18632/oncotarget.24846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep. 2016;6:20254. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-β1-mediated suppression. Oncotarget. 2016;7:27033–27043. doi: 10.18632/oncotarget.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F, Zhang J. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer. 2018;105:336–349. doi: 10.1016/j.bulcan.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A, Nagrath D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H, Li XL, Tao DD, Wu YQ, Gong JP, Qin JC. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. 2019;38:1951–1965. doi: 10.1038/s41388-018-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H, Hamblin MR. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics. 2019;11:1627–1645. doi: 10.2217/epi-2019-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grossman JG, Nywening TM, Belt BA, Panni RZ, Krasnick BA, DeNardo DG, Hawkins WG, Goedegebuure SP, Linehan DC, Fields RC. Recruitment of CCR2+ tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology. 2018;7:e1470729. doi: 10.1080/2162402X.2018.1470729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J, Yuan X, Hu J, Wang G. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019;79:146–158. doi: 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 52.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, Chen Z, Zhu H. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 54.Xu P, Wang J, Sun B, Xiao Z. Integrated analysis of miRNA and mRNA expression data identifies multiple miRNAs regulatory networks for the tumorigenesis of colorectal cancer. Gene. 2018;659:44–51. doi: 10.1016/j.gene.2018.03.050. [DOI] [PubMed] [Google Scholar]
- 55.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 56.Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lötvall J, Kim YK, Gho YS. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7:7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 57.Clancy C, Khan S, Glynn CL, Holian E, Dockery P, Lalor P, Brown JA, Joyce MR, Kerin MJ, Dwyer RM. Screening of exosomal microRNAs from colorectal cancer cells. Cancer Biomark. 2016;17:427–435. doi: 10.3233/CBM-160659. [DOI] [PubMed] [Google Scholar]
- 58.Zaharie F, Muresan MS, Petrushev B, Berce C, Gafencu GA, Selicean S, Jurj A, Cojocneanu-Petric R, Lisencu CI, Pop LA, Pileczki V, Eniu D, Muresan MA, Zaharie R, Berindan-Neagoe I, Tomuleasa C, Irimie A. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J Gastrointestin Liver Dis. 2015;24:435–443. doi: 10.15403/jgld.2014.1121.244.375. [DOI] [PubMed] [Google Scholar]
- 59.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18:10. doi: 10.1186/s12951-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, Qian XL, Liao WT, Ding YQ, Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16. doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes SE, Pereira DM, Roma-Rodrigues C, Fernandes AR, Borralho PM, Rodrigues CMP. Convergence of miR-143 overexpression, oxidative stress and cell death in HCT116 human colon cancer cells. PLoS One. 2018;13:e0191607. doi: 10.1371/journal.pone.0191607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15:1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24:542–556. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 66.Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lian B, Yang D, Liu Y, Shi G, Li J, Yan X, Jin K, Liu X, Zhao J, Shang W, Zhang R. miR-128 Targets the SIRT1/ROS/DR5 Pathway to Sensitize Colorectal Cancer to TRAIL-Induced Apoptosis. Cell Physiol Biochem. 2018;49:2151–2162. doi: 10.1159/000493818. [DOI] [PubMed] [Google Scholar]
- 68.Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, Xie Y, Sun Z, Wang C. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021 doi: 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marquez J, Fernandez-Piñeiro I, Araúzo-Bravo MJ, Poschmann G, Stühler K, Khatib AM, Sanchez A, Unda F, Ibarretxe G, Bernales I, Badiola I. Targeting liver sinusoidal endothelial cells with miR-20a-loaded nanoparticles reduces murine colon cancer metastasis to the liver. Int J Cancer. 2018;143:709–719. doi: 10.1002/ijc.31343. [DOI] [PubMed] [Google Scholar]
- 71.Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3:560–566. doi: 10.3892/etm.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayashi M, Kuroda K, Ihara K, Iwaya T, Isogai E. Suppressive effect of an analog of the antimicrobial peptide of LL37 on colon cancer cells via exosomeencapsulated miRNAs. Int J Mol Med. 2018;42:3009–3016. doi: 10.3892/ijmm.2018.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kyuno D, Zhao K, Bauer N, Ryschich E, Zöller M. Therapeutic Targeting Cancer-Initiating Cell Markers by Exosome miRNA: Efficacy and Functional Consequences Exemplified for claudin7 and EpCAM. Transl Oncol. 2019;12:191–199. doi: 10.1016/j.tranon.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Gao Y, Gong C, Wang Z, Xia Q, Gu F, Hu C, Zhang L, Guo H, Gao S. A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine. 2018;14:1973–1985. doi: 10.1016/j.nano.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 75.Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8:57–84. doi: 10.1177/1758834015614530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y, Zhu M. Novel exosomal miR-46146 transfer oxaliplatin chemoresistance in colorectal cancer. Clin Transl Oncol. 2020;22:1105–1116. doi: 10.1007/s12094-019-02237-1. [DOI] [PubMed] [Google Scholar]
- 77.Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei B, Zhou L, Yin Y, Huang Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother Pharmacol. 2019;84:315–325. doi: 10.1007/s00280-019-03867-6. [DOI] [PubMed] [Google Scholar]
- 78.Zhou J, Li XL, Chen ZR, Chng WJ. Tumor-derived exosomes in colorectal cancer progression and their clinical applications. Oncotarget. 2017;8:100781–100790. doi: 10.18632/oncotarget.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, Guo Z, Zhang G, Xu M, Xu X, Sun H, Wang S. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer. 2020;11:630–637. doi: 10.7150/jca.33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. doi: 10.1038/s41598-017-04386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong SJ, Cai XJ, Li SJ. The Clinical Significance of MiR-429 as a Predictive Biomarker in Colorectal Cancer Patients Receiving 5-Fluorouracil Treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu J, Zhao J, Zhang R. Prognostic significance of serum miR-193b in colorectal cancer. Int J Clin Exp Pathol. 2017;10:9509–9514. [PMC free article] [PubMed] [Google Scholar]
- 84.Tan HY, Zheng YB, Liu J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:8657–8663. doi: 10.26355/eurrev_201812_16630. [DOI] [PubMed] [Google Scholar]
- 85.Min L, Chen L, Liu S, Yu Y, Guo Q, Li P, Zhu S. Loss of Circulating Exosomal miR-92b is a Novel Biomarker of Colorectal Cancer at Early Stage. Int J Med Sci. 2019;16:1231–1237. doi: 10.7150/ijms.34540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karimi N, Ali Hosseinpour Feizi M, Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B, Teimourian S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc. 2019;82:215–220. doi: 10.1097/JCMA.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 87.Cho WC, Kim M, Park JW, Jeong SY, Ku JL. Exosomal miR-193a and let-7g accelerate cancer progression on primary colorectal cancer and paired peritoneal metastatic cancer. Transl Oncol. 2021;14:101000. doi: 10.1016/j.tranon.2020.101000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peng ZY, Gu RH, Yan B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. 2018 doi: 10.1002/jcb.27291. [DOI] [PubMed] [Google Scholar]
- 89.Tang Y, Zhao Y, Song X, Niu L, Xie L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal. 2019;33:e23004. doi: 10.1002/jcla.23004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Min L, Zhu S, Chen L, Liu X, Wei R, Zhao L, Yang Y, Zhang Z, Kong G, Li P, Zhang S. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles. 2019;8:1643670. doi: 10.1080/20013078.2019.1643670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan S, Jiang Y, Liang C, Cheng M, Jin C, Duan Q, Xu D, Yang L, Zhang X, Ren B, Jin P. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem. 2018;119:4113–4119. doi: 10.1002/jcb.26609. [DOI] [PubMed] [Google Scholar]
- 92.Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, Xu T, Xu M, Wang S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 93.Zhao YJ, Song X, Niu L, Tang Y, Xie L. Circulating Exosomal miR-150-5p and miR-99b-5p as Diagnostic Biomarkers for Colorectal Cancer. Front Oncol. 2019;9:1129. doi: 10.3389/fonc.2019.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]