ABSTRACT
Negative-stranded RNA (NSR) viruses include both animal- and plant-infecting viruses that often cause serious diseases in humans and livestock and in agronomic crops. Rice stripe tenuivirus (RSV), a plant NSR virus with four negative-stranded/ambisense RNA segments, is one of the most destructive rice pathogens in many Asian countries. Due to the lack of a reliable reverse-genetics technology, molecular studies of RSV gene functions and its interaction with host plants are severely hampered. To overcome this obstacle, we developed a mini-replicon-based reverse-genetics system for RSV gene functional analysis in Nicotiana benthamiana. We first developed a mini-replicon system expressing an RSV genomic RNA3 enhanced green fluorescent protein (eGFP) reporter [MR3(−)eGFP], a nucleocapsid (NP), and a codon usage-optimized RNA-dependent RNA polymerase (RdRpopt). Using this mini-replicon system, we determined that RSV NP and RdRpopt are indispensable for the eGFP expression from MR3(−)eGFP. The expression of eGFP from MR3(−)eGFP can be significantly enhanced in the presence of four viral suppressors of RNA silencing (VSRs), NSs, and P19-HcPro-γb. In addition, NSvc4, the movement protein of RSV, facilitated eGFP trafficking between cells. We also developed an antigenomic RNA3-based replicon in N. benthamiana. However, we found that the RSV NS3 coding sequence acts as a cis element to regulate viral RNA expression. Finally, we made mini-replicons representing all four RSV genomic RNAs. This is the first mini-replicon-based reverse-genetics system for monocot-infecting tenuivirus. We believe that the mini-replicon system described here will allow studies of the RSV replication, transcription, cell-to-cell movement, and host machinery underpinning RSV infection in plants.
IMPORTANCE Plant-infecting segmented negative-stranded RNA (NSR) viruses are grouped into three genera: Orthotospovirus, Tenuivirus, and Emaravirus. Reverse-genetics systems have been established for members of the genera Orthotospovirus and Emaravirus. However, there is still no reverse-genetics system available for Tenuivirus. Rice stripe virus (RSV) is a monocot-infecting tenuivirus with four negative-stranded/ambisense RNA segments. It is one of the most destructive rice pathogens and causes significant damage to the rice industry in Asian countries. Due to the lack of a reliable reverse-genetics system, molecular characterizations of RSV gene functions and the host machinery underpinning RSV infection in plants are extremely difficult. To overcome this obstacle, we developed a mini-replicon-based reverse-genetics system for RSV in Nicotiana benthamiana. This is the first mini-replicon-based reverse-genetics system for tenuivirus. We consider that this system will provide researchers a new working platform to elucidate the molecular mechanisms dictating segmented tenuivirus infections in plants.
KEYWORDS: rice stripe tenuivirus, reverse-genetics system, mini-replicon, negative-sense/ambisense RNA virus
INTRODUCTION
Negative-sense RNA (NSR) viruses include well-known members of medical importance such as Ebola virus (EBOV), vesicular stomatitis virus (VSV), influenza A virus (FLUAV), and Rift Valley fever virus (RVFV) (1, 2) and include serious plant pathogens of agronomic importance such as tomato spotted wilt virus (TSWV), rice stripe virus (RSV), and rose rosette virus (RRV) (3–5). There are three genera of segmented NSR viruses infecting plants: Orthotospovirus, Tenuivirus, and Emaravirus. TSWV and RSV are the representative viruses for Orthotospovirus and Tenuivirus, respectively (6, 7). RRV and European mountain ash ringspot-associated virus (EMAraV) are important members of the genus Emaravirus (8, 9).
Tenuiviruses are classified in the genus Tenuivirus, family Phenuiviridae, within the order Bunyavirales. Most viruses in the family Phenuiviridae infect animals. RSV is one of most devastating causal agents of rice and often causes severe damage to rice production in China and many other Asian countries (6, 10, 11). RSV is transmitted by the small brown planthopper (Laodelphax striatellus) in a persistent and circulative-propagative manner (6, 12–14). The RSV genome consists of four RNA segments and encodes seven proteins through an antisense or ambisense coding strategy (15–18). RSV RNA1 is of negative polarity and encodes the RNA-dependent RNA polymerase (RdRp) (17). RSV RNA2 encodes the NS2 protein in the viral (v) strand and the NSvc2 protein from the viral complementary (vc) strand (19). The NS2 protein is a weak viral suppressor of RNA silencing (VSR) and is required for RSV systemic infection in plants. The NSvc2 protein is a putative glycoprotein that targets the endoplasmic reticulum (ER) and Golgi apparatus and functions as a helper factor to conquer insect midgut barriers after being cleaved into two mature proteins (i.e., NSvc2-N, the amino-terminal half, and NSvc2-C, the carboxy-terminal half) (12, 20). RSV RNA3 encodes the NS3 protein in the v strand, which is known as a VSR that binds single- and double-stranded RNAs to suppress RNA silencing (21, 22), and the NP protein in the vc strand, which interacts with viral genomic RNAs to form viral ribonucleoprotein complexes (RNPs) (23). RSV RNA4 encodes the SP protein in the v strand, a nonstructural and disease-specific protein that interacts with a host oxygen-evolving complex protein to interfere with host photosynthesis, and the NSvc4 protein in the vc strand, a protein involved in RSV cell-to-cell and long-distance movement in plants (4, 21). The four RSV genomic RNAs all contain highly conserved 5′ and 3′ untranslated regions (UTR), important for the initiation of viral RNA transcriptions. RSV RNA2, -3, and -4 all have a noncoding intergenic region (IGR) with multiple AU-rich regions that form secondary hairpin-like structures to act as transcription termination signals (24, 25).
Viral reverse-genetics systems are important tools for studies of viral gene functions, disease inductions, and host factors involved in virus infection in plants (26–32). Although reverse-genetics systems were first reported for segmented animal negative-stranded RNA viruses over 20 years ago (26, 33–42), establishing similar systems for segmented plant negative-stranded/ambisense RNA viruses has turned out to be very challenging. However, just recently, reverse-genetics systems have been established for a few nonsegmented and segmented plant NSR viruses. The first reverse-genetics system of the nonsegmented plant NSR viruses was established for sonchus yellow net virus (SYNV; a nucleorhabdovirus), followed by barley yellow striate mosaic virus (BYSMV) (a cytorhabdovirus) (27, 43–46). Recently, we established the first reverse-genetics system of segmented plant NSR viruses for TSWV (47). Soon after, the reverse-genetics system for RRV was also established, allowing for studies of emaravirus gene function and disease pathology in whole plants (9).
Of the three genera of segmented NSR viruses infecting plants, a reverse-genetics model has been established for two, Orthotospovirus and Emaravirus (9, 47, 48). There is still no reverse-genetics system available for the genus Tenuivirus. Recent progress on a TSWV and RRV reverse-genetics system encouraged us to establish a reverse-genetics system for RSV. In this study, we developed a mini-replicon-based reverse-genetics system for RSV gene function analyses in Nicotiana benthamiana. This represents the first mini-replicon-based reverse-genetics system for monocot-infecting tenuivirus. The developed mini-replicon reverse-genetics system will provide researchers with a novel platform for studies of RSV replication, transcription, movement, and host factors involved in interactions between the virus and the host plant. This system also provides a useful basis for the development of infectious RSV clones for assays in plants, including rice.
RESULTS
Development of a genomic RNA3 mini-replicon-based reverse-genetics system for RSV.
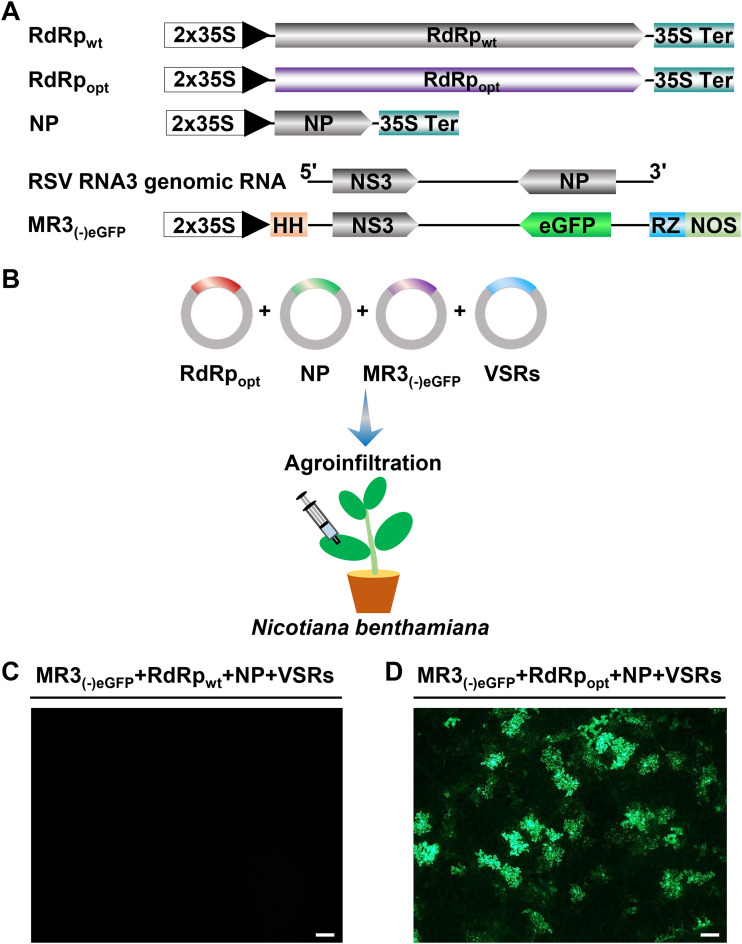
To establish a mini-replicon-based reverse-genetics system to investigate RSV infection in plants, we amplified the full-length RSV RNA3 sequence by reverse transcription PCR (RT-PCR) and inserted it between a hammerhead (HH) sequence and a hepatitis delta virus ribozyme (RZ) sequence in the pCB301-2 × 35S-RZ-NOS vector to produce RNA3(−). We then replaced the NP gene in RNA3(−) with an enhanced green fluorescent protein (eGFP) gene to produce an MR3(−)eGFP mini-replicon reporter (Fig. 1A). The expression of RNA3(−) and MR3(−)eGFP from these two vectors is driven by a doubled Cauliflower mosaic virus (CaMV) 35S promoter (2 × 35S) (Fig. 1A).
FIG 1.
Construction of an RSV RNA3(−)-based mini-replicon. (A) Schematics representing the RdRpwt, RdRpopt, NP, RSV RNA3(−), and MR3(−)eGFP mini-replicon. For MR3(−)eGFP, we replaced the NP gene in the gRNA3 with an eGFP gene. The 5′ untranslated, the 3′ untranslated, and the intergenic regions in RNA3(−) are indicated by a thin black line. 2 × 35S, doubled 35S promoter; HH, hammerhead ribozyme; RZ, hepatitis delta virus (HDV) ribozyme; 35S Ter, 35S terminator; NOS, nopaline synthase terminator. The minus sign and 5′ to 3′ designations represent the viral (genomic) strand of RNA3 in the figure. (B) Illustration of agroinfiltration using a mixed Agrobacterium culture carrying various mini-replicons into N. benthamiana leaves. VSRs, NSs plus P19-HcPro-γb. (C) An N. benthamiana leaf infiltrated with a mixed Agrobacterium culture carrying MR3(−)eGFP, RdRpwt, NP, and VSRs (NSs and P19-HcPro-γb). (D) An N. benthamiana leaf infiltrated with a mixed Agrobacterium culture carrying MR3(−)eGFP, RdRpopt, NP, and VSRs (NSs and P19-HcPro-γb) and showing eGFP fluorescence at 5 dpi under an inverted fluorescence microscope. Bar, 200 μm.
We constructed p2300-RdRpwt, pBIN-NS3, p2300-NP, and pCXSN-NSvc4 to express wild-type RSV RdRp (RdRpwt), NS3, NP, and NSvc4, respectively, in cells through agroinfiltration. After infiltration of a mixed Agrobacterium culture carrying the plasmids for MR3(−)eGFP, RdRpwt, NP, and four viral suppressors of RNA silencing (VSRs) (NSs and P19-HcPro-γb) into N. benthamiana leaves (Fig. 1B), no eGFP fluorescence was observed in the infiltrated leaves by 5 days post agroinfiltration (dpi) (Fig. 1C). As reported for TSWV RdRp (47), wild-type RSV RdRp was also predicted to have numerous putative intron splicing sites. This prediction prompted us to optimize its codon usage and to remove the predicted intron splicing sites. The optimized RdRp open reading frame (ORF) was then inserted into the p2300 vector to produce p2300-RdRpopt (RdRpopt). After infiltration of N. benthamiana leaves with a mixed Agrobacterium culture carrying plasmids for MR3(−)eGFP, RdRpopt, NP, and four VSRs, we observed eGFP fluorescence with a microscope at 5 dpi (Fig. 1D), suggesting that the sequence-optimized RdRpopt was now functional in N. benthamiana cells.
RSV NP and RdRpopt are required for MR3(−)eGFP expression.
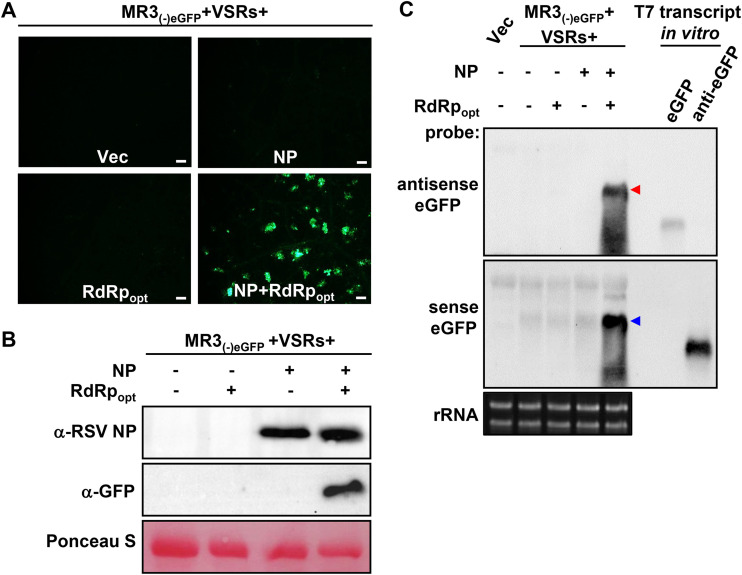
To investigate the roles of NP and RdRpopt in RSV replication in plants, we coexpressed MR3(−)eGFP and VSRs with empty vector (Vec), NP, RdRpopt, or both NP and RdRpopt in N. benthamiana leaves through agroinfiltration. The eGFP fluorescence was again detected in the leaves coexpressing MR3(−)eGFP with NP and RdRpopt. In contrast, the control leaves expressing MR3(−)eGFP alone, or coexpressing MR3(−)eGFP with NP or RdRpopt, did not show any eGFP fluorescence (Fig. 2A), indicating that the presence of both NP and RdRpopt is necessary for MR3(−)eGFP expression from the mini-replicon.
FIG 2.
RSV NP and RdRpopt are required for MR3(−)eGFP expression in N. benthamiana leaves. (A) N. benthamiana leaves were infiltrated with mixed Agrobacterium cultures carrying MR3(−)eGFP and four VSRs (NSs and P19-HcPro-γb) with empty vector (Vec), NP, RdRpopt, or both NP and RdRpopt. The infiltrated leaves were examined and photographed under an inverted fluorescence microscope at 5 dpi. Bars, 200 μm. (B) Western blot analyses using the samples described for panel A and an NP- or eGFP-specific antibody. The Ponceau S-stained RubisCO large-subunit gel was used to show sample loadings. (C) Northern blot analyses using the samples described for panel A and a DIG-labeled sense or an antisense eGFP probe. The red and blue arrowheads indicate the antigenomic and genomic RNA3 expressed in the infiltrated leaves, respectively. An ethidium bromide-stained rRNA gel was used to show sample loadings.
To confirm this observation, we analyzed the accumulation levels of eGFP protein, eGFP mRNA, and MR3(−)eGFP genomic RNA (gRNA) and anti-genomic RNA (agRNA) in the infiltrated N. benthamiana leaf tissues. Western blot analysis showed high levels of eGFP in the leaves coexpressing MR3(−)eGFP, NP, RdRpopt, and four VSRs (NSs and P19-HcPro-γb). In contrast, eGFP protein was not detected in the control leaves coexpressing MR3(−)eGFP with Vec, NP, or RdRpopt only by 5 dpi (Fig. 2B). Northern blot results showed that the gRNA and agRNA were detected in the leaf tissues coexpressing MR3(−)eGFP, NP, RdRpopt, and VSRs by 5 dpi (Fig. 2C). In contrast, no amplified gRNA or agRNA was detected in the control leaves coexpressing MR3(−)eGFP with Vec, NP, or RdRp only, respectively (Fig. 2C). Only primary transcripts of agRNA from MR3(−)eGFP were detected in these leaves. These results indicate that the presence of RSV NP and RdRpopt is required for the replication of both gRNA and agRNA from the MR3(−)eGFP mini-replicon. Intrinsically, the eGFP mRNA was not detected in the leaf tissues coexpressing MR3(−)eGFP, NP, RdRpopt, and VSRs.
Effect of viral suppressors of RNA silencing on MR3(−)eGFP expression.
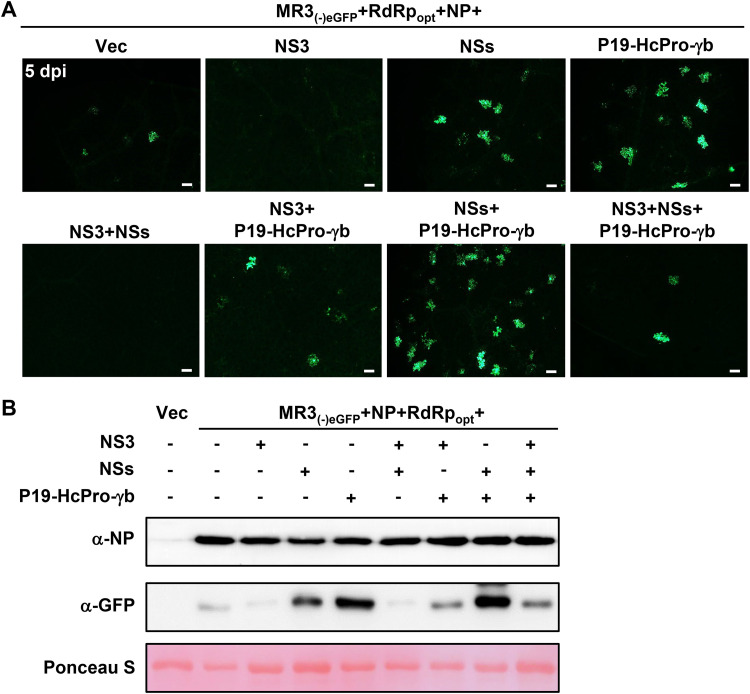
VSRs inhibit host RNA interference (RNAi) machinery and enhance nonviral gene expression in plants (43, 45). To investigate the roles of different VSRs on MR3(−)eGFP expression in plants, we infiltrated N. benthamiana leaves with mixed Agrobacterium cultures carrying MR3(−)eGFP, RdRpopt, NP, and VSRs, including NS3, NSs, and P19-HcPro-γb. By 5 dpi, the leaves coexpressing MR3(−)eGFP, RdRpopt, and NP with empty vector (Vec) showed a few cells with eGFP fluorescence. Many cells with eGFP fluorescence were observed in the leaves coexpressing MR3(−)eGFP, RdRpopt, and NP with three VSRs (P19-HcPro-γb) or four VSRs (NSs and P19-HcPro-γb) (Fig. 3A and B). Although RSV NS3 is a VSR (21, 22), coexpression of MR3(−)eGFP, RdRpopt, and NP with NS3 or with both NS3 and NSs resulted in almost no cells with eGFP fluorescence. Moreover, coexpression of MR3(−)eGFP, RdRpopt, and NP with both NS3 and P19-HcPro-γb resulted in some cells with eGFP fluorescence. These findings indicate that RSV NS3 can suppress eGFP expression from the MR3(−)eGFP mini-replicon and are supported by the Western blotting result (Fig. 3B). Based on these results, we decided to use VSRs, including NSs, P19, HcPro, and γb but not NS3, in the following experiments.
FIG 3.
Effects of VSRs on MR3(−)eGFP expression. (A) N. benthamiana leaves were infiltrated with mixed Agrobacterium cultures as indicated in the figure. The infiltrated N. benthamiana leaves were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (B) Western blot analyses using the samples described for panel A and NP- and eGFP-specific antibodies. Proteins in the leaves shown in panel A were detected using NP- and GFP-specific antibodies. A Ponceau S-stained RubisCO large-subunit gel was used to show sample loadings.
Dosage effects of NP and RdRpopt on MR3(−)eGFP expression.
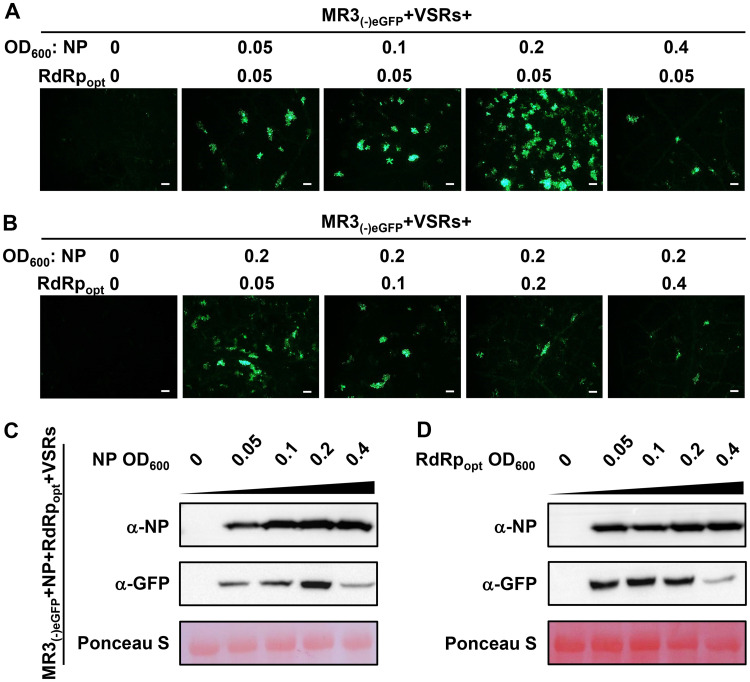
To optimize the expression of the MR3(−)eGFP mini-replicon in plant cells, we mixed the Agrobacterium culture carrying MR3(−)eGFP and four VSRs (NSs+P19-HcPro-γb) with the Agrobacterium cultures carrying NP (optical density at 600 nm [OD600] of 0, 0.05, 0.1, 0.2, or 0.4) or RdRpopt (OD600 of 0, 0.05, 0.1, 0.2, or 0.4) and then infiltrated them individually into N. benthamiana leaves. When the concentration of Agrobacterium culture carrying RdRp was fixed at an OD600 of 0.05 and the concentration of Agrobacterium culture carrying NP was increased from an OD600 of 0.05 to an OD600 of 0.4, the results showed that the strongest eGFP fluorescence was observed in the leaves coexpressing MR3(−)eGFP with NP at an OD600 of 0.2 and RdRpopt at an OD600 of 0.05 (Fig. 4A). When the concentration of Agrobacterium culture carrying NP was maintained at an OD600 of 0.2 while the concentration of Agrobacterium culture carrying RdRpopt was increased from an OD600 of 0.05 to an OD600 of 0.4, the number of cells with eGFP fluorescence decreased as the concentration of RdRpopt increased (Fig. 4B). These results were supported by the results from Western blot assays (Fig. 4C and D).
FIG 4.
Concentrations of NP and RdRpopt needed for maximum expression MR3(−)eGFP. (A and B) N. benthamiana leaves were infiltrated with various mixed Agrobacterium cultures as indicated in the figure. The concentrations of Agrobacterium cultures carrying NP or RdRpopt ranged from an OD600 of 0 to 0.4, as indicated. The infiltrated leaves were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (C and D) Western blot analyses of NP and eGFP expression in the infiltrated leaves described for panels A and B by using NP- and eGFP-specific antibodies. A Ponceau S-stained RubisCO large-subunit gel was used to show sample loadings.
RSV NSvc4 supports MR3(−)eGFP cell-to-cell movement.
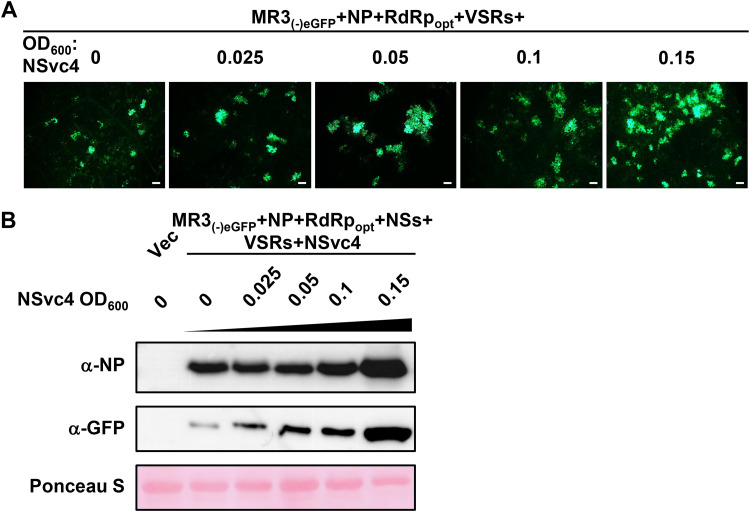
NSvc4 is the movement protein of RSV (18, 49). To investigate whether NSvc4 can also influence MR3(−)eGFP expression, we infiltrated N. benthamiana leaves with the mixed Agrobacterium cultures carrying MR3(−)eGFP (OD600, 0.2), NP (OD600, 0.2), RdRpopt (OD600, 0.05), NSs (OD600, 0.05), P19-HcPro-γb (OD600, 0.05), and NSvc4 (OD600, 0.025, 0.05, 0.1, or 0.15). In this experiment, the MR3(−)eGFP started to move out of the original cell with the addition of NSvc4 at an OD600 of 0.025, compared with that in the leaves coexpressing MR3(−)eGFP, NP, and RdRpopt without NSvc4 (Fig. 5A and Table 1). When the concentration of Agrobacterium culture carrying NSvc4 was further increased, stronger cell-to-cell movement of MR3(−)eGFP was observed (Fig. 5A and Table 1). Results of the Western blot assays agreed with the microscopic observations (Fig. 5B).
FIG 5.
Effect of RSV NSvc4 on eGFP expression from MR3(−)eGFP in cells. (A) N. benthamiana leaves were infiltrated with various mixed Agrobacterium cultures as indicated in the figure. The concentrations of Agrobacterium culture carrying NSvc4 ranged from an OD600 of 0 to 0.15, as indicated. The infiltrated leaves were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (B) Western blot analyses using the infiltrated leaf samples described for panel A and NP- and eGFP-specific antibodies. A Ponceau S-stained RubisCO large-subunit gel was used to show sample loadings.
TABLE 1.
RSV NSvc4 enhances cell-to-cell movement of the MR3(−)eGFP mini-replicon in the presence of NP, RdRpopt, and four VSRs
| NSvc4a OD600 | Total no. of eGFP fluorescent cells | No. of clustersb (% of total) with |
||
|---|---|---|---|---|
| 1 cell | 2 cells | ≥3 cells | ||
| 0 | 18 | 18 (100) | 0 (0) | 0 (0) |
| 0.025 | 54 | 15 (27.8) | 6 (22.2) | 5 (50) |
| 0.05 | 76 | 12 (15.8) | 7 (18.4) | 10 (65.8) |
| 0.1 | 95 | 8 (8.4) | 11 (23.2) | 13 (68.4) |
| 0.15 | 140 | 17 (12.1) | 9 (12.9) | 18 (75) |
Agrobacterium cultures harboring the plasmid encoding NSvc4 (final concentrations [OD600], 0, 0.025, 0.05, 0.1, and 0.15) were used to infiltrate N. benthamiana leaves.
eGFP fluorescent foci of RSV MR3(−)eGFP at 5 dpi.
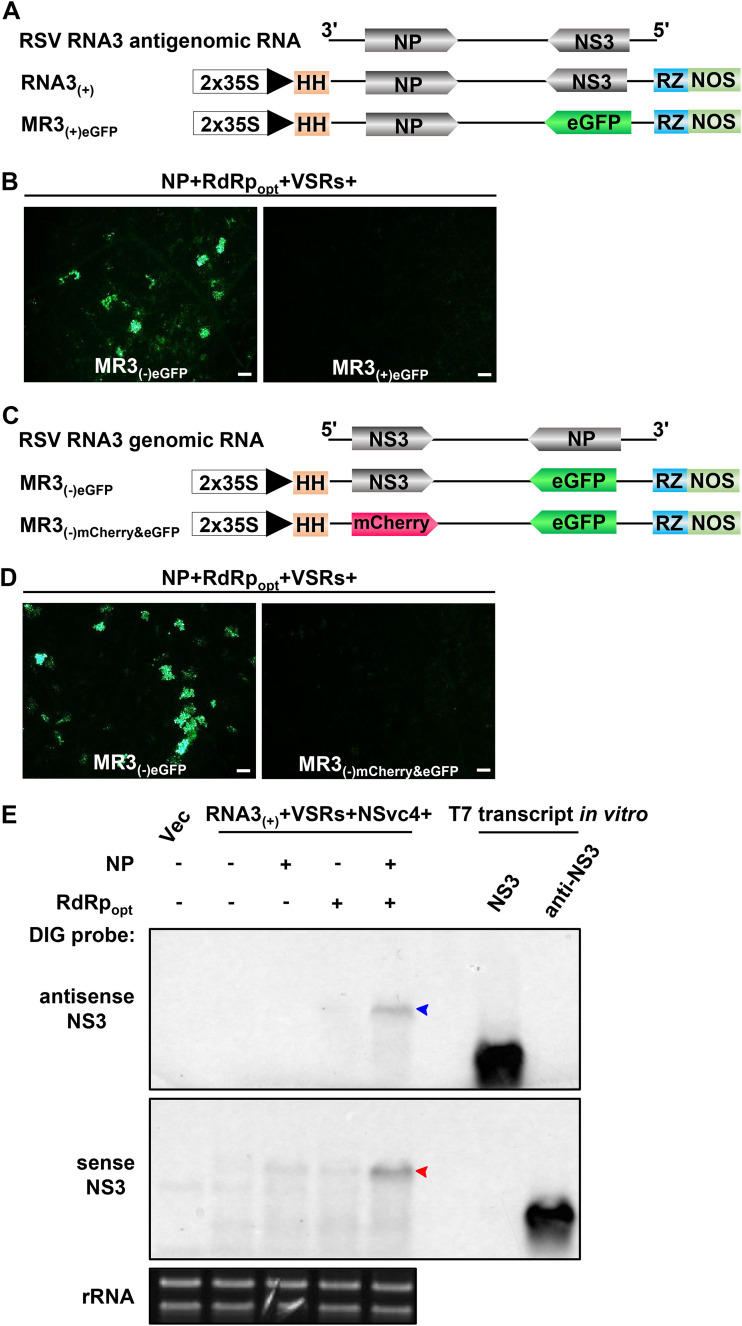
Development of an RSV antigenomic RNA3-based replicon system.
To develop an RSV antigenomic (ag) RNA-based mini-replicon, we replaced the NS3 gene in the RNA3(+)-agRNA vector [RNA3(+)] with an eGFP gene to produce MR3(+)eGFP (Fig. 6A). We then transiently coexpressed MR3(+)eGFP, NP, RdRpopt, and four VSRs (NSs plus P19-HcPro-γb) in N. benthamiana leaves through agroinfiltration. By 5 dpi, no eGFP fluorescence was observed in the infiltrated leaves. In contrast, strong eGFP fluorescence was observed in the leaves coexpressing MR3(−)eGFP, NP, RdRpopt, and four VSRs (Fig. 6B), indicating that the MR3(+)eGFP mini-replicon is incapable of expressing eGFP in plant cells.
FIG 6.
Construction and test of RNA3(+)-based mini-replicon in N. benthamiana leaves. (A) Schematics representing RNA3(+) and MR3(+)eGFP mini-replicons. The MR3(+)eGFP mini-replicon was made by replacing the NS3 gene in RNA3(+) with an eGFP gene. A plus sign and 3′ to 5′ designations represent the viral complementary (antigenomic) strand of RNA3. (B) N. benthamiana leaves were infiltrated with various mixed Agrobacterium cultures as indicated in the figure. The infiltrated leaves were observed at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (C) Schematics representing MR3(−)eGFP and MR3(−)mCherry&eGFP mini-replicons. The MR3(−)mCherry&eGFP mini-replicon was constructed by replacing the NS3 gene with an mCherry gene. A plus sign and 5′ to 3′ designations represent the viral (genomic) strand of RNA3. (D) The infiltrated N. benthamiana leaves were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (E) Northern blot analyses of RNA3 expression from RNA3(+) in the infiltrated leaves as described for panel C. After electrophoresis, the blots were probed with DIG-labeled antisense and sense RNA3 probes. The red and blue arrowheads indicate the RNA3 bands probed with DIG-labeled probes. An ethidium bromide-stained rRNA gel was used to show sample loadings.
Next, we replaced the NS3 gene in MR3(−)eGFP with an mCherry gene (MR3(−)mCherry&eGFP) (Fig. 6C) and coexpressed MR3(−)mCherry&eGFP or MR3(−)eGFP with NP, RdRpopt, and four VSRs in N. benthamiana leaves through agroinfiltration. By 5 dpi, in contrast to the leaves coexpressing MR3(−)eGFP, NP, RdRpopt, and VSRs, no eGFP fluorescence was observed for MR3(−)mCherry&eGFP (Fig. 6D), indicating that without the NS3 gene, this MR3(−)mCherry&eGFP mini-replicon is unable to express eGFP in N. benthamiana leaf cells. To confirm this result, we generated RNA3(+) to express full-length RSV agRNA3 (Fig. 6A) and transiently coexpressed RNA3(+), VSRs, and NSvc4 with one or two of the three plasmids (i.e., Vec, NP, and RdRpopt), respectively, in N. benthamiana leaves via agroinfiltration. The infiltrated leaves were harvested at 5 dpi and analyzed for the accumulation of RNA3(+)-derived gRNA3 and agRNA3 through Northern blot assays using digoxigenin (DIG)-labeled antisense and sense probes, respectively. The results showed that high levels of gRNA3 and agRNA3 were detected in the leaves coexpressing RNA3(+), NP, RdRpopt, VSRs, and NSvc4 (Fig. 6E). In contrast, no amplified gRNA3 or agRNA3 was detected in the leaves coexpressing RNA3(+), four VSRs, and NSvc4 with Vec, NP, or RdRpopt only (Fig. 6E). Only primary transcripts of agRNA3 from RNA3(+) was detected in these leaves. Based on these results, we conclude that the RNA3(+) replicon is functional in N. benthamiana in the presence of NP, RdRpopt, NSs, P19-HcPro-γb, and NSvc4.
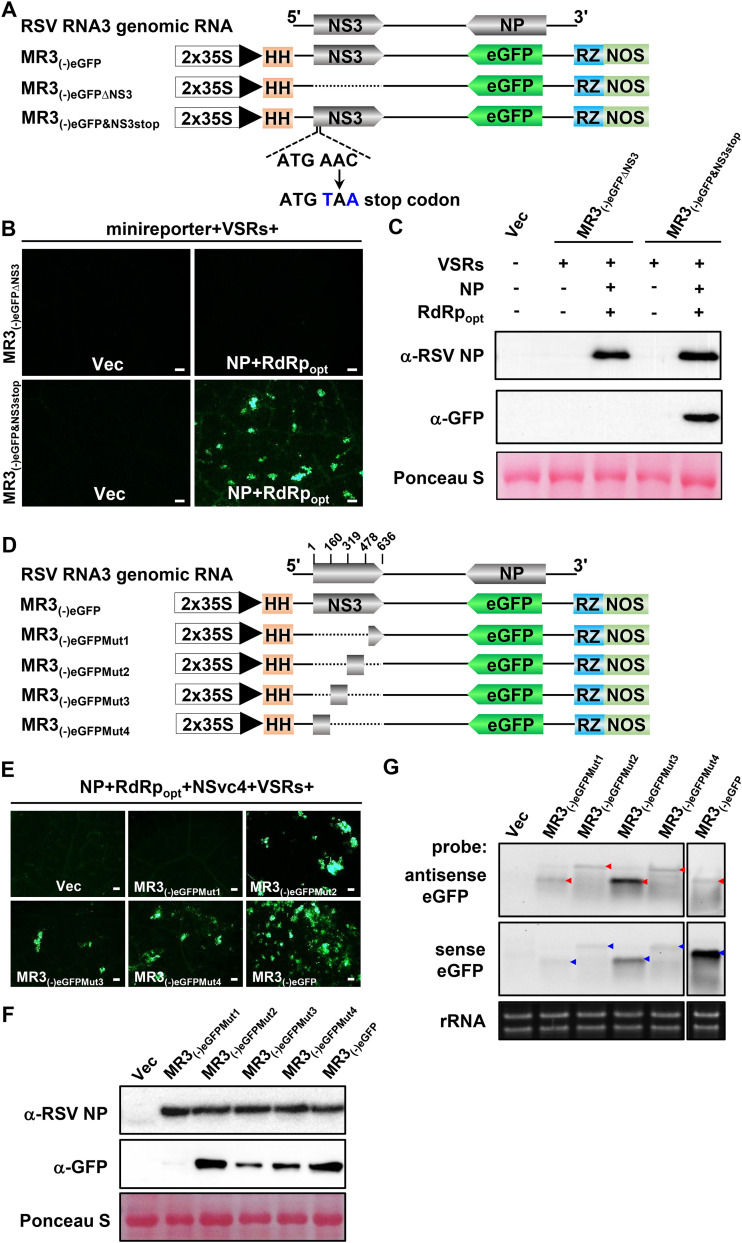
The NS3 gene is required for eGFP expression from MR3(−)eGFP.
To investigate the function of NS3 in eGFP expression from the MR3(−)eGFP mini-replicon, we introduced a stop codon (TAA) downstream of the start codon of the NS3 ORF [MR3(−)eGFP&NS3stop] (Fig. 7A) and coexpressed it with NP, RdRpopt, and VSRs in N. benthamiana leaves through agroinfiltration. By 5 dpi, strong eGFP fluorescence was observed in the infiltrated leaves, but not in leaves coexpressing MR3(−)eGFPΔNS3, NP, RdRpopt, and VSRs (Fig. 7B). Western blot results showed that eGFP was expressed in the leaves coexpressing MR3(−)eGFP&NS3stop, NP, RdRpopt, and VSRs, while no eGFP was accumulated in the leaves coexpressing MR3(−)eGFPΔNS3, NP, RdRpopt, and VSRs (Fig. 7C). This finding indicates that the NS3 is dispensable for eGFP expression from the MR3(−)eGFP mini-replicon. Deletion of the NS3 sequence from MR3(−)eGFP [MR3(−)eGFPΔNS3] abolished the expression of eGFP.
FIG 7.
Effect of NS3 on eGFP gene expression from the mini-replicons. (A and D) Schematics representing MR3(−)eGFP and its mutants. For MR3(−)eGFPΔNS3, the NS3 gene was deleted from MR3(−)eGFP. For MR3(−)eGFP&NS3stop, a stop codon was introduced downstream of the start codon of the NS3 gene in MR3(−)eGFP. For MR3(−)eGFPMut1, MR3(−)eGFPMut2, MR3(−)eGFPMut3, and MR3(−)eGFPMut4 mutant mini-replicons, a quarter of the NS3 gene in MR3(−)eGFP was deleted as shown in the figure. A minus sign and 5′ to 3′ designations represent the viral (genomic) strand of RNA3. (B and E) N. benthamiana leaves were infiltrated with various mixed Agrobacterium cultures as shown in the figure. The infiltrated leaves were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm. (C and F) Western blot analyses using the leaf samples described for panels B and E and NP- and eGFP-specific antibodies. A Ponceau S-stained RubisCO large-subunit gels was used to show sample loading. (G) Northern blot analyses of antigenomic and genomic RNA expressions from MR3(−)eGFP using a DIG-labeled antisense eGFP probe and a sense eGFP probe. An ethidium bromide-stained rRNA gel was used to show sample loadings.
To further confirm the role of the NS3 ORF in MR3(−)eGFP expression, we divided the NS3 ORF into four segments and generated four truncated MR3(−)eGFP mutant constructs: MR3(−)eGFPMut1, MR3(−)eGFPMut2, MR3(−)eGFPMut3, and MR3(−)eGFPMut4 (Fig. 7D). Each mutant was coexpressed with NP, RdRpopt, NSvc4, and VSRs in N. benthamiana leaves. By 5 dpi, eGFP fluorescence was observed in the leaves coexpressing MR3(−)eGFPMut2, MR3(−)eGFPMut3, or MR3(−)eGFPMut4 with NP, RdRpopt, NSvc4, and VSRs. However, the leaves coexpressing MR3(−)eGFPMut1, NP, RdRpopt, NSvc4, and VSRs did not show eGFP fluorescence (Fig. 7E). Western blot results agreed with the microscopic observations and showed that eGFP was not expressed from MR3(−)eGFPMut1 in cells (Fig. 7F), indicating that deletion of the first 159 nucleotides (nt) of NS3 significantly affects eGFP expression of MR3(−)eGFP mini-replicon. It is noteworthy that the Northern blot results showed that expression of gRNA and agRNA from MR3(−)eGFPMut1 was not affected (Fig. 7G). We also noticed that the mobility of gRNA and agRNA from MR3(−)eGFPMut2 and MR3(−)eGFPMut4 was altered compared with that from MR3(−)eGFPMut1, MR3(−)eGFPMut3, or MR3(−)eGFP. Therefore, we conclude that the NS3 ORF sequence is not necessary for viral replication but is required for the expression of eGFP from the RNA3(−)-derived mini-replicons in cells.
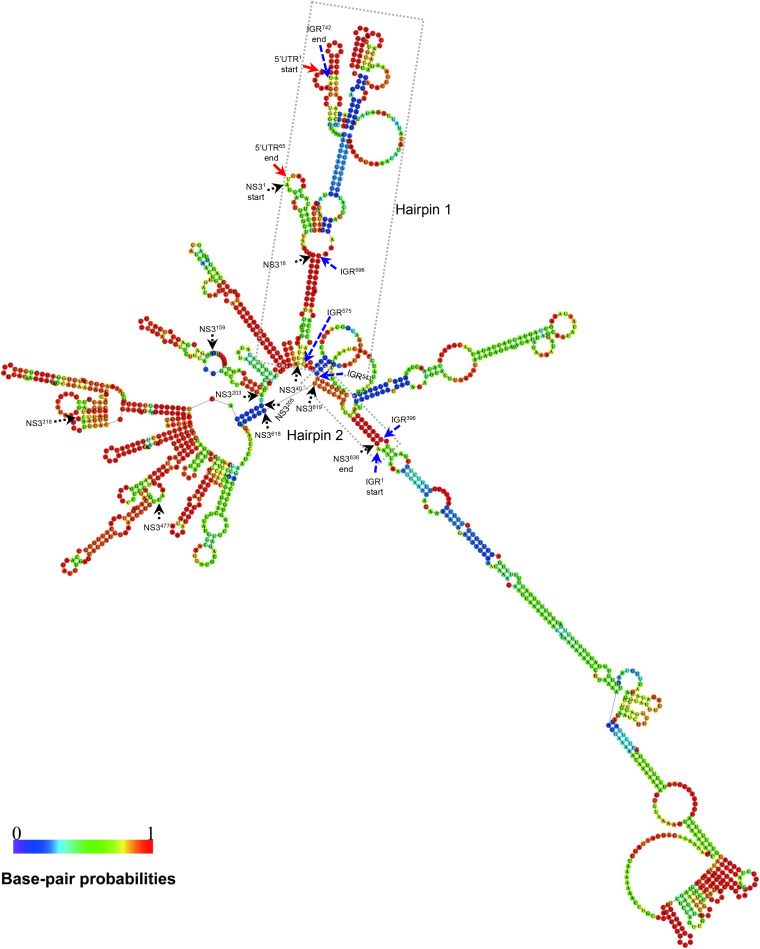
RSV RNA2, -3, and -4 IGRs can form secondary hairpin-like structures which are postulated to act as transcription termination signals (24, 25). To further investigate the possible role of RSV NS3 in viral transcription regulation, we predicted the RNA secondary structure and examined the possible RNA-RNA interaction between the NS3 coding sequence, IGR, and 5′ UTR of RSV RNA3. The 5′ UTR, NS3, and IGR of RSV RNA3 have 65, 636, and 742 nt, respectively. The secondary structure analysis showed that the nt 1 to 295 (nt 1–295) sequence of IGR formed a very long hairpin structure (Fig. 8). Strikingly, the nt 1–40 coding region sequence of NS3 base-paired with the nt 575–596 sequence of IGR and the nt 1–65 sequence of 5′ UTR and formed a long hairpin structure (Fig. 8). The nt 619–636 coding region sequence of NS3 base-paired with the nt 396–403 and the nt 537–543 sequences of IGR and formed a small hairpin-like structure. The nt 41–203 coding region sequence of NS3 itself formed four long and short hairpin structures. The nt 205–618 coding region sequence of NS3 formed a sector structure containing at least 9 hairpins (Fig. 8). The secondary structure analysis suggested that the coding sequence of NS3 likely interacts with the IGR and 5′ UTR of RNA3 in forming hairpin-like structures.
FIG 8.
RNA secondary structures of the 5′ UTR, NS3 ORF, and IGR of RSV RNA3 segment. Secondary structure was predicted using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) based on a thermodynamic prediction of minimal free energy (MFE). Red, black, and blue arrows indicate the nucleotide positions in the 5′ UTR, NS3, and IGR regions, respectively. Dotted boxes indicate that hairpins 1 and 2 were formed by the IGR, NS3, and 5′ UTR of RSV RNA3.
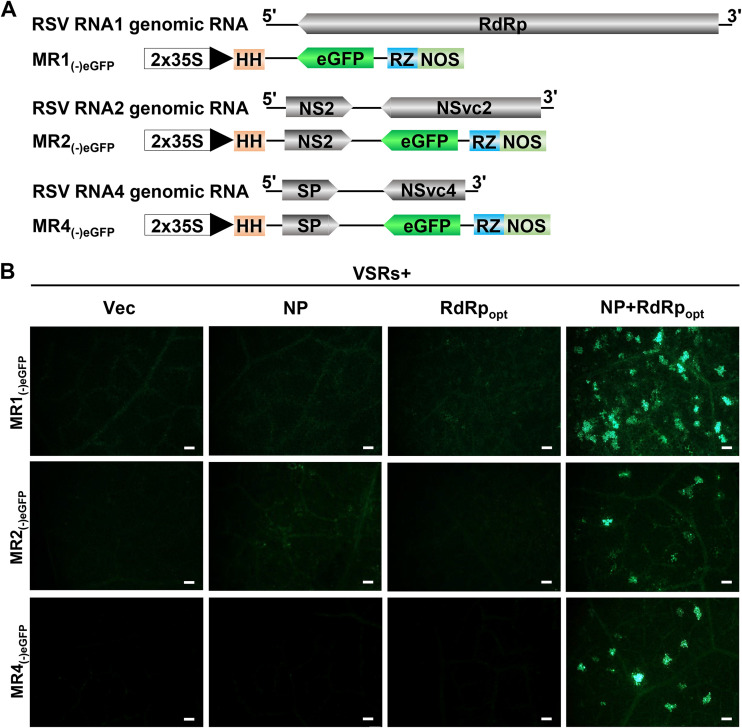
Development of RSV RNA1, RNA2, and RNA4 mini-replicons.
The RSV genome consists of four RNA segments encoding seven proteins by using an antisense or ambisense coding strategy (15–18). Because the MR3(−)eGFP mini-replicon is functional in N. benthamiana leaf cells (Fig. 1 and 2), we decided to develop mini-replicons for three other RSV genomic RNAs. We first produced full-length RNA1 and then replaced the RdRp ORF with the eGFP gene to produce MR1(−)eGFP (Fig. 9A). For RSV genomic RNA2 and RNA4, we first cloned the full-length RNA2 or RNA4 segments individually into the vector and then replaced the NSvc2 ORF with the eGFP gene to produce MR2(−)eGFP or replaced the NSvc4 ORF with the eGFP gene to produce MR4(−)eGFP (Fig. 9A). N. benthamiana leaves were then infiltrated with the mixed Agrobacterium cultures carrying various combinations of plasmids (Fig. 9B). By 5 dpi, strong eGFP fluorescence was observed in the leaves coexpressing MR1(−)eGFP, MR2(−)eGFP, or MR4(−)eGFP with NP, RdRpopt, and four VSRs (Fig. 9B). As expected, the leaves infiltrated with the mixed Agrobacterium cultures lacking NP, RdRpopt, or NP+RdRpopt did not show eGFP fluorescence.
FIG 9.
Establishment of mini-replicon systems for RSV RNA1, RNA2, and RNA4 genomic RNA segments in N. benthamiana leaves. (A) Schematics representing MR1(−)eGFP, MR2(−)eGFP, and MR4(−)eGFP mini-replicons. The RSV RdRp gene in MR1(−)eGFP, the NSvc2 gene in MR2(−)eGFP, and the NSvc4 gene in MR4(−)eGFP were replaced with an eGFP gene to produce MR1(−)eGFP, MR2(−)eGFP, and MR4(−)eGFP, respectively. The minus sign and 5′ to 3′ designations represent the viral (genomic) strand of RNA1, RNA2, and RNA4. (B) N. benthamiana leaves were infiltrated with various mixed Agrobacterium cultures as shown in the figure. The infiltrated leaf tissues were harvested at 5 dpi and examined and photographed under an inverted fluorescence microscope. Bars, 200 μm.
Collectively, the mini-replicon-based reverse-genetics system, representing all four RSV genomic RNAs, was created.
DISCUSSION
There are three genera of segmented NSR viruses infecting plants: Orthotospovirus, Tenuivirus, and Emaravirus. Reverse-genetics systems have been established for TSWV and RRV in the genera Orthotospovirus and Emaravirus (9, 47, 48), respectively. Here, we established a mini-replicon-based reverse-genetics system for RSV, the representative virus for the genus Tenuivirus. RSV is an important rice virus and poses a significant threat to rice production in China and many other Asian countries (6, 10, 11). During the past 20 years, the lack of a reliable reverse-genetics system significantly hampered studies of RSV gene functions and disease induction in plants. To overcome this obstacle, we launched a multiyear research that finally yielded a functional mini-replicon-based reverse-genetics system for RSV studies. We first developed a mini-replicon system to express RSV MR3(−)eGFP, NP, and codon usage-optimized RdRp (RdRpopt). Using this mini-replicon system, we determined that RSV NP and RdRpopt are indispensable for eGFP expression from MR3(−)eGFP. The expression of eGFP from MR3(−)eGFP was significantly enhanced in the presence of NSs and P19-HcPro-γb. In addition, NSvc4, the movement protein of RSV, facilitated eGFP trafficking between cells. Interestingly, coexpression of RSV NS3 inhibited eGFP expression from MR3(−)eGFP. We also found that the RSV NS3 gene sequence is not necessary for viral replication but regulates viral RNA expression. Secondary structure analysis showed that the coding sequence of NS3 base-paired with the sequence of the IGR and 5′ UTR of RNA3 to form a long hairpin structure. The phenomenon of coding sequence as a cis element in regulating viral RNA expression has not been reported previously for the negative-stranded/ambisense RNA viruses. Finally, based on the system of RNA3(−), we have also produced mini-replicons representing all RSV RNA genomic segments, allowing RSV functional studies in plants.
The choice of promoter for RNA transcription is critical for the development of reverse-genetics systems for plant NSR viruses. The genomic and antigenomic RNAs generated from the negative-stranded RNA virus clones were not infectious because their infectious ribonucleoprotein complexes (RNPs) contain not only viral gRNA but also NP and RdRp proteins (36, 50). Genomic RNAs of the same segmented NSV contain highly conserved 5′- and 3′-terminal untranslated sequences that have eight complementary nucleotides, capable of forming panhandle-like structures. These structures are known to play critical roles in viral gRNA and agRNA replications (50). Moreover, the NSR virus RNAs do not possess 5′ cap structures and 3′ poly(A) tails. The classical bacteriophage T7 promoter can produce accurate viral RNA 5′-end sequences without a cap. We initially produced an RSV mini-replicon system by using the T7 promoter. This mini-replicon system, however, did not express the eGFP gene from an RNA3-based mini-replicon in plant cells. In our recent report, we also reported that the T7 promoter-based system was unable to generate infectious TSWV RNA transcripts in plant cells (47). It is possible that the synthesis of viral genomic RNA transcripts through the T7 promoter and T7 RNA polymerase is incomplete or is inefficient in planta.
In a recent report, we described an expression vector with a double CaMV 35S promoter (an RNA Pol II promoter), a hammerhead (HH) ribozyme, and a hepatitis delta virus (HDV) ribozyme to produce infectious TSWV viral RNAs in plant cells (47). The mini-replicon system produced in this study also has an HH ribozyme and an HDV ribozyme before and after the viral sequence to ensure the correct ends. The results shown in Fig. 1 and 2 suggest that the vector-transcribed RSV genomic RNA did bind to viral NP and RdRp to form functional RNPs, which is needed for the synthesis of functional viral gRNA and agRNA. This finding supports earlier reports that the Pol II promoter can replicate not only nonsegmented plant NSR viruses (43, 45) but also segmented plant NSR viruses (9, 47).
RSV RdRp is one of the major components needed for the initiation of viral genomic RNA replication (17). RSV RdRp is a 337-kDa protein. When this protein was coexpressed with the mini-replicon in plant cells, no eGFP fluorescence was observed in the infiltrated leaves (Fig. 1A to C). Our computer prediction suggested that the RSV RdRp sequence contained many putative cryptic intron splicing sites. Because the segmented plant NSR viruses replicate in cytoplasm, their RdRp gene sequences should not have evolved to remove the cryptic intron splicing sites that could be spliced into the cell nucleus. We speculated that after the wild-type RSV RdRp sequences were expressed through the 2 × 35S promoter in the nucleus, they were quickly spliced, resulting in nonfunctional RdRp fragments. After the putative intron splicing sites were removed and the codon usage was optimized, the expressed RdRpopt was capable of supporting eGFP expression from the mini-replicon (Fig. 1D). In this study, we also determined that the less concentrated Agrobacterium culture carrying RdRpopt (OD600, 0.05) caused higher eGFP expression in cells. In contrast, an increase in OD600 of Agrobacterium culture carrying RdRpopt from 0.1 to 0.4 decreased eGFP expression from the mini-replicon (Fig. 4B and C), suggesting that an optimum concentration of RdRpopt is required during RSV infection in plants.
Analyses of the five different VSRs have indicated that in the presence of NSs or P19-HcPro-γb, the eGFP expression from the mini-replicon was significantly enhanced (Fig. 3B and C). These VSRs are known to function at different steps in the host RNA interference (RNAi) pathway during virus infection in plant (43, 51). Consequently, we conclude that these steps in the RNAi pathway can all affect eGFP expression from the mini-replicon. It is noteworthy that the presence of RSV NS3 alone or NS3 plus one or three of the four VSRs (i.e., NSs or P19-HcPro-γb) suppressed eGFP expression from the RNA3 mini-replicon (Fig. 3B and C), suggesting that RSV NS3 is a negative regulator of RSV RNA3 mini-replicon expression. A similar phenomenon was also reported for TSWV NSs during virus rescue assays using cDNA clones (47). We hypothesize that this negative regulation is caused by the cosuppression of NS3 gene expression. In this study, addition of NSvc4 significantly increased the number of cells with eGFP fluorescence (Fig. 5A and B), further confirming its role in cell-to-cell trafficking.
When the NS3 gene was replaced with an mCherry gene [MR3(−)mCherry&eGFP] or with an eGFP gene in MR3(+)eGFP, no mCherry or eGFP fluorescence was observed in the leaf tissues coexpressing NP, RdRpopt, and VSRs (Fig. 6C and D). Deletion of the NS3 gene sequence abolished the function of MR3(−)eGFP. However, gRNA and agRNA were detected in the leaves coexpressing RNA3(+), NP, RdRpopt, NSvc4, and VSRs (Fig. 6A and E). The MR3(−)eGFP&NS3stop mini-replicon contains a translation stop codon immediately after the start codon of NS3 and is still functional in N. benthamiana leaves (Fig. 7A to C). This finding suggests that the NS3 coding sequence is required for eGFP expression of the mini-replicon, probably required for viral transcription. Through nucleotide deletion assays, we determined that the region encompassing nt 1 to 477 in the NS3 ORF is not sufficient to express eGFP from the MR3(−)eGFP mini-replicon (Fig. 7D to G), even though the mutant mini-replicon [MR3(−)eGFPMut1] is capable of expressing RNA in cells, based on the Northern blot results. Because the positions of the gRNA and agRNA bands from MR3(−)eGFPMut2 and MR3(−)eGFPMut4 were altered, we speculate that the NS3 ORF sequence may act as a cis regulatory element during RSV RNA3 viral transcription. Importantly, the secondary structure analysis suggested that the coding sequence of NS3 interacts with the IGR and 5′ UTR to form a hairpin-like structure and that NS3 itself also forms a sophisticated hairpin-like structure (Fig. 8). Hairpin structures of TSWV and RSV IGR have been shown to regulate viral transcription termination (52). This is consistent with our findings that the NS3 coding sequence is involved in regulation of viral RNA transcription. In our earlier studies, deletion of the NSs coding sequence from the TSWV S-based mini-replicon system had no clear effect on RNA synthesis (47). Although the genome structure of RSV RNA3 is similar to that of TSWV S RNA, this is the first evidence showing that the NS3 coding region can act as a cis element to regulate the synthesis of viral RNA transcripts of segmented NSR viruses in plants.
Based on the established mini-replicon system for MR3(−)eGFP, we have also produced mini-replicons to express MR1(−)eGFP, MR2(−)eGFP, and MR4(−)eGFP, as described in the legend to Fig. 9A. In the presence of NP, RdRpopt, and VSRs, eGFP was expressed from MR1(−)eGFP, MR2(−)eGFP, and MR4(−)eGFP mini-replicons, respectively (Fig. 9B). We also constructed full-length infectious cDNA clones representing RSV genomic RNA1, RNA2, and RNA4. Infiltration of N. benthamiana leaves with the mixed Agrobacterium culture carrying RNA1(−), RNA2(−), RNA3(−), RNA4(−), and NP plus RdRpopt plus VSRs did not yield a systemic infection. Because N. benthamiana is an experimental host of RSV and the accumulations of RSV RNAs and proteins are lower than those in the rice plants, it is possible that the low level of RSV RNAs and proteins in the infiltrated N. benthamiana leaves fails to rescue RSV systemic infection. It is also possible that the difficulty of delivering four RSV RNA segments into the same cells prevents systemic infection. We have also infiltrated rice callus tissues with the mixed Agrobacterium culture carrying MR1(−)eGFP, MR2(−)eGFP, MR3(−)eGFP, MR4(−)eGFP, and NP plus RdRpopt plus VSRs; none of them work in rice callus tissues.
In conclusion, there are three genera of segmented NSR viruses infecting plants, and a replicon-based reverse-genetics system has been established for two of them. We have now established a mini-replicon-based reverse-genetics system for a tenuivirus in N. benthamiana. This is the first mini-replicon-based reverse-genetics system for the segmented monocot-infecting tenuivirus and will provide a useful platform for studies of RSV gene functions during viral replication, cell-to-cell movement, and interactions between RSV and host factors in plants. Knowledge gained from this study also benefits future construction of full-length multisegmented infectious clones for tenuivirus in plants.
MATERIALS AND METHODS
Plant growth and virus source.
Nicotiana benthamiana plants were grown inside a growth chamber maintained at 25°C, with a 16/8 h light/dark photoperiod, and were used for assays at about the 7-leaf stage. RSV was originally isolated from an RSV-infected rice plant as reported previously (12). Optimization of RSV RdRp ORF codon usage and deletion of putative intron splicing sites were performed based on the predictions using the GeneArt project manager online software (Thermo Fisher Scientific).
Plasmid construction. (i) Construction of RSV RdRp, RdRpopt, NP, NSvc4, NS3, and VSR mini-replicons.
Complementary DNAs (cDNAs) of RSV, NP, RdRpwt, NS3, and NSvc4 genes were individually amplified from a total RNA sample isolated from an RSV-infected rice plant through reverse transcription PCR (RT-PCR) using gene-specific primers. The resulting RT-PCR products were inserted individually into expression vector pCambia2300 (referred to as p2300 herein), pBINplus, or pCXSN to generate p2300-RdRpwt, pBIN-NS3, p2300-NP, and pCXSN-NSvc4, respectively. The pCXSV-NSs vector was constructed by inserting an NSs fragment amplified from a cDNA from a TSWV-infected N. benthamiana plant into the pCXSN vector. Plasmid pCB301-P19-HcPro-γb (P19-HcPro-γb), which can simultaneously express the tomato bushy stunt virus P19 protein, the tobacco etch virus HcPro protein, and the barley stripe mosaic virus γb protein, is from a previously published source (45). To construct p2300-RdRpopt, we first optimized RdRpwt codon usage and deleted the putative intron splicing sites in it at the GenScript Biotech Corp. (Nanjing, China), followed by inserting the synthesized sequence into the p2300 vector to produce p2300-RdRpopt (RdRpopt).
(ii) Construction of MR3(−)eGFP, MR3(-)mCherry&eGFP, and MR3(+)eGFP mini-replicons.
To generate the MR3(−)eGFP and MR3(+)eGFP mini-replicons, we first prepared cDNAs from a total RNA sample isolated from an RSV-infected rice plant by using a reverse transcription kit as instructed by the manufacturer (Promega, Madison, WI, USA). We then amplified the full-length RSV RNA3(−) and RNA3(+) sequences from this cDNA by PCR using RSV RNA3-specific primers (see Table S1 in the supplemental material) and a Phanta super-fidelity DNA polymerase (Vazyme Biotech, Nanjing, China). Both PCR products contained a hammerhead (HH) ribozyme (53) before the 5′ end, which was then cloned individually behind the 2 × 35S promoter in the pCB301-2 × 35S-RZ-NOS vector to generate pCB301-2 × 35S-HH-RNA3(−)-RZ-NOS [referred to as RNA3(−)] or pCB301-2 × 35S-HH-RNA3(+)-RZ-NOS [RNA3(+)]. The presence of an HH and a hepatitis delta virus ribozyme (RZ) in these two vectors allows the production of RNA3(−) and RNA3(+) with near-perfect ends. To produce an MR3(−)eGFP and an MR3(+)eGFP mini-replicon, we first amplified the eGFP gene from SR(+)eGFP (47) using primer FMF37 and FMF38 and then used it to replace the NP gene in RNA3(−) and RNA3(+) through in vitro homologous recombination using a ClonExpress II one-step cloning kit (Vazyme Biotech, Nanjing, China). The resulting plasmids are referred to as MR3(−)eGFP and MR3(+)eGFP, respectively. To produce an MR3(−)mCherry&eGFP mini-replicon with both the mCherry and eGFP genes, we PCR amplified the mCherry gene from the TSWV SR(−)mCherry&eGFP mini-replicon as reported previously (47) by using primers FMF267 and FMF269 and used it to replace the NS3 gene in MR3(−)eGFP through homologous recombination as described above. All primers used in this study are listed in Table S1.
(iii) Construction of mutant MR3(−)eGFP mini-replicons.
To produce the mutant MR3(−)eGFP mini-replicons, we first introduced a stop codon (TAA) after the original start codon of the NS3 gene by PCR using MR3(−)eGFP as the template and primers FMF528 and FMF36. This NS3stop fragment was then used to replace the NS3 gene in MR3(−)eGFP through homologous recombination using primers FMF40 and FMF529. The resulting plasmid was named MR3(−)eGFP&NS3stop. We then deleted a 636-nt fragment from the NS3 gene in MR3(−)eGFP via PCR using primers LLY40 and FMF42 and then inserted it into the pCB301 vector by homologous recombination using primers FMF37 and LLY105 to produce MR3(−)eGFPΔNS3.
To further investigate the role of the RSV NS3 gene in viral RNA replication and transcription from RNA3(−), we divided the NS3 ORF into four fragments and amplified them individually via PCR using specific primers. These four fragments comprise nucleotide positions 1 to 159, 160 to 318, 319 to 477, and 478 to 636. These fragments were then used to generate MR3(−)eGFPMut1, MR3(−)eGFPMut2, MR3(−)eGFPMut3, and MR3(−)eGFPMut4, respectively. Primers used in this study are listed in Table S1. The plasmids were transformed individually into Agrobacterium tumefaciens strain GV3101 through electroporation, and the transformants were maintained at −80°C until use.
Agrobacterium infiltration.
A. tumefaciens cultures were prepared as described previously (43, 44). Briefly, Agrobacterium cultures carrying specific plasmids were grown individually in culture medium and then diluted to an OD600 of 1.0, or as indicated in the text, in an infiltration buffer (10 mM morpholineethanesulfonic acid [MES] and 10 mM MgCl2, pH 5.6, supplemented with 100 μM acetosyringone). After 2 to 3 h of incubation in the dark and at room temperature (RT), Agrobacterium cultures harboring p2300-NP (OD600, 0.2), p2300-RdRp (OD600, 0.05), p2300-NSs (OD600, 0.1), pCB301-P19-HcPro-γb (OD600, 0.1), or one of the vectors (OD600, 0.2 each) with an eGFP and/or an mCherry reporter gene were mixed in equal volumes, or as indicated in the text. The mixed cultures were then infiltrated individually into leaves of six- to seven-leaf-stage N. benthamiana plants using 1-ml needleless syringes. The infiltrated plants were grown inside a growth chamber under the same growth conditions as described above.
Western blot assay.
The agroinfiltrated N. benthamiana leaves were harvested at 5 days post agroinfiltration (dpi) and homogenized (1 mg/sample) individually in 1 ml extraction buffer (150 mM NaCl, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 2% polyvinylpolypyrrolidone, 10 mM dithiothreitol, 10% glycerol, 0.5% Triton X-100, and 1× protease inhibitor cocktail reagent). The crude extract from each sample was mixed with a 5×loading buffer at a 1:4 ratio (vol/vol). All the samples were boiled for 10 min and then incubated on ice for 5 min, followed by electrophoresis in 12% SDS-PAGE gels. After the protein bands were transferred onto polyvinylidene difluoride (PVDF) membranes (GE Healthcare, UK), the blots were probed with an RSV NP-specific (1:5,000 diluted) or an eGFP-specific (1:3,000 diluted) antibody, followed by a horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit secondary antibody (1:10,000 diluted). The detection signal was visualized using an ECL substrate kit as instructed by the manufacturer (Thermo Fisher Scientific, Rockford, IL, USA). The Ponceau S-stained RubisCO large-subunit gels were used to show sample loadings.
Northern blot assay.
To detect the expression of RSV gRNAs, agRNAs, and eGFP mRNA, we isolated total RNA from the agroinfiltrated N. benthamiana leaf tissues by using an RNAprep pure plant kit (Tiangen Biotech, Beijing, China). The isolated total RNA samples were separated in 1% formaldehyde agarose gels by electrophoresis and then transferred onto Hybond-N+ membranes (GE Healthcare, UK) (54). DIG-labeled RNA probes specific for sense or antisense eGFP mRNA were in vitro synthesized using a DIG high prime RNA labeling kit (Roche, Basel, Switzerland). The blotted membranes were probed with DIG-labeled RNA probes specific for sense or antisense eGFP mRNA. The detection signal was visualized using a DIG high prime detection starter kit II as instructed by the manufacturer (Roche).
Fluorescence microscopy.
The agroinfiltrated N. benthamiana leaf tissues were harvested at 5 dpi and examined under an inverted fluorescence microscope (IX71-F22FL/DIC; Olympus, Tokyo, Japan) equipped with a green barrier filter. The captured images were processed using the ImagePro system (Olympus, Tokyo, Japan) and then Adobe Photoshop CS4 (San Jose, CA, USA).
Data availability.
All data produced in this study are presented in this article or in the supplemental material.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31630062, 31925032, and 31870143), the Fundamental Research Funds for the Central Universities (grant no. JCQY202104 and KYXK202012), the Youth Science and Technology Innovation Program, and a project funded by the China Postdoctoral Science Foundation (grant no. 2020M681643).
M.F., L.L., and X.T. conceived and designed the experiments, and M.Y., Y.Z., J.W., and Y.X. provided input. M.F., L.L., R.C., Y.Y., Y.D., M.C., and G.R. performed the experiments. M.F., X.D., X.Z., and X.T. wrote the manuscript.
We declare that no competing interests exist.
Footnotes
Supplemental material is available online only.
Contributor Information
Xueping Zhou, Email: zzhou@zju.edu.cn.
Xiaorong Tao, Email: taoxiaorong@njau.edu.cn.
Anne E. Simon, University of Maryland, College Park
REFERENCES
- 1.Fields BN, Knipe DM, Howley PM. 1996. Fields virology. Lippincott-Raven, New York, NY. [Google Scholar]
- 2.Elliott RM, Blakqori G. 2011. Molecular biology of orthobunyaviruses, p 1–39. In Plyusnin A, Elliott RM (ed), The Bunyaviridae: molecular and cellular biology. Horizon Scientific Press, Norwich, United Kingdom. [Google Scholar]
- 3.Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD. 2011. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12:938–954. 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong LF, Wu JX, Lu LN, Xu Y, Zhou XP. 2014. Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant 7:691–708. 10.1093/mp/sst158. [DOI] [PubMed] [Google Scholar]
- 5.Laney AG, Keller KE, Martin RR, Tzanetakis IE. 2011. A discovery 70 years in the making: characterization of the rose rosette virus. J Gen Virol 92:1727–1732. 10.1099/vir.0.031146-0. [DOI] [PubMed] [Google Scholar]
- 6.Falk BW, Tsai JH. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. Annu Rev Phytopathol 36:139–163. 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- 7.Zhu M, Jiang L, Bai BH, Zhao WY, Chen XJ, Li J, Liu Y, Chen ZQ, Wang BT, Wang CL, Wu Q, Shen QH, Dinesh-Kumar SP, Tao XR. 2017. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 29:2214–2232. 10.1105/tpc.17.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mielke N, Muehlbach HP. 2007. A novel, multipartite, negative-strand RNA virus is associated with the ringspot disease of European mountain ash (Sorbus aucuparia L.). J Gen Virol 88:1337–1346. 10.1099/vir.0.82715-0. [DOI] [PubMed] [Google Scholar]
- 9.Verchot J, Herath V, Urrutia CD, Gayral M, Lyle K, Shires MK, Ong K, Byrne D. 2020. Development of a reverse genetic system for studying rose rosette virus in whole plants. Mol Plant Microbe Interact 33:1209–1221. 10.1094/MPMI-04-20-0094-R. [DOI] [PubMed] [Google Scholar]
- 10.Otuka A, Matsumura M, Sanada-Morimura S, Takeuchi H, Watanabe T, Ohtsu R, Inoue H. 2010. The 2008 overseas mass migration of the small brown planthopper, Laodelphax striatellus, and subsequent outbreak of rice stripe disease in western Japan. Appl Entomol Zool 45:259–266. 10.1303/aez.2010.259. [DOI] [Google Scholar]
- 11.Wang HD, Chen JP, Zhang HM, Sun XL, Zhu JL, Wang AG, Sheng WX, Adams MJ. 2008. Recent rice stripe virus epidemics in Zhejiang province, China, and experiments on sowing date, disease-yield loss relationships, and seedling susceptibility. Plant Dis 92:1190–1196. 10.1094/PDIS-92-8-1190. [DOI] [PubMed] [Google Scholar]
- 12.Lu G, Li S, Zhou CW, Qian X, Xiang Q, Yang TQ, Wu JX, Zhou XP, Zhou YJ, Ding XS, Tao XR. 2019. Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog 15:e1007655. 10.1371/journal.ppat.1007655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Yang PC, Kang L, Cui F. 2016. Different pathogenicities of rice stripe virus from the insect vector and from viruliferous plants. New Phytol 210:196–207. 10.1111/nph.13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toriyama S. 1986. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiol Sci 3:347–351. [PubMed] [Google Scholar]
- 15.Zhu Y, Hayakawa T, Toriyama S, Takahashi M. 1991. Complete nucleotide sequence of RNA 3 of rice stripe virus: an ambisense coding strategy. J Gen Virol 72(Pt 4):763–767. 10.1099/0022-1317-72-4-763. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi M, Toriyama S, Hamamatsu C, Ishihama A. 1993. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J Gen Virol 74:769–773. 10.1099/0022-1317-74-4-769. [DOI] [PubMed] [Google Scholar]
- 17.Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A. 1994. Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol 75:3569–3579. 10.1099/0022-1317-75-12-3569. [DOI] [PubMed] [Google Scholar]
- 18.Xiong RY, Wu JX, Zhou YJ, Zhou XP. 2008. Identification of a movement protein of the tenuivirus rice stripe virus. J Virol 82:12304–12311. 10.1128/JVI.01696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du ZG, Xiao DL, Wu JG, Jia DS, Yuan ZJ, Liu Y, Hu LY, Han Z, Wei TY, Lin QY, Wu ZJ, Xie LH. 2011. p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol 12:808–814. 10.1111/j.1364-3703.2011.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao M, Liu XF, Li S, Xu Y, Zhou YJ, Zhou XP, Tao XR. 2014. Rice stripe tenuivirus NSvc2 glycoproteins targeted to the Golgi body by the N-terminal transmembrane domain and adjacent cytosolic 24 amino acids via the COP I- and COP II-dependent secretion pathway. J Virol 88:3223–3234. 10.1128/JVI.03006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong RY, Wu JX, Zhou YJ, Zhou XP. 2009. Characterization and subcellular localization of an RNA silencing suppressor encoded by rice stripe tenuivirus. Virology 387:29–40. 10.1016/j.virol.2009.01.045. [DOI] [PubMed] [Google Scholar]
- 22.Shen M, Xu Y, Jia R, Zhou X, Ye KQ. 2010. Size-independent and noncooperative recognition of dsRNA by the rice stripe virus RNA silencing suppressor NS3. J Mol Biol 404:665–679. 10.1016/j.jmb.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Lu G, Li J, Zhou YJ, Zhou XP, Tao XR. 2017. Model-based structural and functional characterization of the rice stripe tenuivirus nucleocapsid protein interacting with viral genomic RNA. Virology 506:73–83. 10.1016/j.virol.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Muhlberger E, Lotfering B, Klenk HD, Becker S. 1998. Three of four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol 72:8756–8764. 10.1128/JVI.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GT, Lu YW, Zheng HM, Lin L, Yan F, Chen JP. 2013. Transcription of ORFs on RNA2 and RNA4 of rice stripe virus terminate at an AUCCGGAU sequence that is conserved in the genus Tenuivirus. Virus Res 175:71–77. 10.1016/j.virusres.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Neumann G, Whitt MA, Kawaoka Y. 2002. A decade after the generation of a negative-sense RNA virus from cloned cDNA—what have we learned? J Gen Virol 83:2635–2662. 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AO, Li ZH. 2016. Developments in plant negative-strand RNA virus reverse genetics. Annu Rev Phytopathol 54:469–498. 10.1146/annurev-phyto-080615-095909. [DOI] [PubMed] [Google Scholar]
- 28.Jackson AO, Dietzgen RG, Goodin MM, Li Z. 2018. Development of model systems for plant rhabdovirus research. Adv Virus Res 102:23–57. 10.1016/bs.aivir.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Chen YT, Dessau M, Rotenberg D, Rasmussen DA, Whitfield AE. 2019. Entry of bunyaviruses into plants and vectors. Adv Virus Res 104:65–96. 10.1016/bs.aivir.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Feng MF, Feng ZK, Li ZH, Wang XB, Tao XB. 2020. Advances in reverse genetics system of plant negative-strand RNA viruses. Chin Sci Bull 65:4073–4083. (In Chinese.) 10.1360/TB-2020-0671. [DOI] [Google Scholar]
- 31.German TL, Lorenzen MD, Grubbs N, Whitfield AE. 2020. New technologies for studying negative-strand RNA viruses in plant and arthropod hosts. Mol Plant Microbe Interact 33:382–393. 10.1094/MPMI-10-19-0281-FI. [DOI] [PubMed] [Google Scholar]
- 32.Zang Y, Fang XD, Qiao JH, Gao Q, Wang XB. 2020. Reverse genetics systems of plant negative-strand RNA viruses are difficult to be developed but powerful for virus-host interaction studies and virus-based vector applications. Phytopathol Res 2:29–37. 10.1186/s42483-020-00068-5. [DOI] [Google Scholar]
- 33.Dunn EF, Pritlove DC, Jin H, Elliott RM. 1995. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology 211:133–143. 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- 34.Bridgen A, Elliott RM. 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci U S A 93:15400–15404. 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pekosz A, He B, Lamb RA. 1999. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc Natl Acad Sci U S A 96:8804–8806. 10.1073/pnas.96.16.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KJ, Novella IS, Teng MN, Oldstone MBA, de la Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMW) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flick R, Pettersson RF. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol 75:1643–1655. 10.1128/JVI.75.4.1643-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. 2006. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A 103:4663–4668. 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107–1113. 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 41.Conzelmann KK, Schnell M. 1994. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol 68:713–719. 10.1128/JVI.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volchkov VE, Volchkova VA, Muhlberger E, Kolesnikova LV, Weik M, Dolnik O, Klenk HD. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965–1969. 10.1126/science.1057269. [DOI] [PubMed] [Google Scholar]
- 43.Ganesan U, Bragg JN, Deng M, Marr S, Lee MY, Qian S, Shi M, Kappel J, Peters C, Lee Y, Goodin MM, Dietzgen RG, Li Z, Jackson AO. 2013. Construction of a sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. J Virol 87:10598–10611. 10.1128/JVI.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Ma XL, Qian SS, Zhou X, Sun K, Chen XL, Zhou XP, Jackson AO, Li ZH. 2015. Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog 11:e1005223. 10.1371/journal.ppat.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang XD, Yan T, Gao Q, Cao Q, Gao DM, Xu WY, Zhang ZJ, Ding ZH, Wang XB. 2019. A cytorhabdovirus phosphoprotein forms mobile inclusions trafficked on the actin/ER network for viral RNA synthesis. J Exp Bot 70:4049–4062. 10.1093/jxb/erz195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Q, Xu WY, Yan T, Fang XD, Cao Q, Zhang ZJ, Ding ZH, Wang Y, Wang XB. 2019. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol 223:2120–2133. 10.1111/nph.15889. [DOI] [PubMed] [Google Scholar]
- 47.Feng MF, Cheng RX, Chen ML, Guo R, Li LY, Feng ZK, Wu JY, Xie L, Hong J, Zhang ZK, Kormelink R, Tao XR. 2020. Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proc Natl Acad Sci U S A 117:1181–1190. 10.1073/pnas.1910787117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi K, Matsumoto-Yokoyama E, Ishikawa M. 2017. A tomato spotted wilt virus S RNA-based replicon system in yeast. Sci Rep 7:12647. 10.1038/s41598-017-12687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu S, Xu Y, Li CY, Li Y, Wu JX, Zhou XP. 2018. Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol Plant 11:269–287. 10.1016/j.molp.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Ferron F, Weber F, de la Torre JC, Reguera J. 2017. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res 234:118–134. 10.1016/j.virusres.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgyan J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci 16:265–272. 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 52.van Knippenberg I, Goldbach R, Kormelink R. 2005. Tomato spotted wilt virus S-segment mRNAs have overlapping 3'-ends containing a predicted stem-loop structure and conserved sequence motif. Virus Res 110:125–131. 10.1016/j.virusres.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Herold J, Andino R. 2000. Poliovirus requires a precise 5' end for efficient positive-strand RNA synthesis. J Virol 74:6394–6400. 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng MF, Zuo DP, Jiang XZ, Li S, Chen J, Jiang L, Zhou XP, Jiang T. 2018. Identification of strawberry vein banding virus encoded P6 as an RNA silencing suppressor. Virology 520:103–110. 10.1016/j.virol.2018.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download JVI.00589-21-s0001.pdf, PDF file, 124 KB (123.4KB, pdf)
Data Availability Statement
All data produced in this study are presented in this article or in the supplemental material.