Abstract
Wheat (Triticum aestivum) has a large allohexaploid genome. Subgenome-divergent regulation contributed to genome plasticity and the domestication of polyploid wheat. However, the specificity encoded in the wheat genome determining subgenome-divergent spatio-temporal regulation has been largely unexplored. The considerable size and complexity of the genome are major obstacles to dissecting the regulatory specificity. Here, we compared the epigenomes and transcriptomes from a large set of samples under diverse developmental and environmental conditions. Thousands of distal epigenetic regulatory elements (distal-epiREs) were specifically linked to their target promoters with coordinated epigenomic changes. We revealed that subgenome-divergent activity of homologous regulatory elements is affected by specific epigenetic signatures. Subgenome-divergent epiRE regulation of tissue specificity is associated with dynamic modulation of H3K27me3 mediated by Polycomb complex and demethylases. Furthermore, quantitative epigenomic approaches detected key stress responsive cis- and trans-acting factors validated by DNA Affinity Purification and sequencing, and demonstrated the coordinated interplay between epiRE sequence contexts, epigenetic factors, and transcription factors in regulating subgenome divergent transcriptional responses to external changes. Together, this study provides a wealth of resources for elucidating the epiRE regulomics and subgenome-divergent regulation in hexaploid wheat, and gives new clues for interpreting genetic and epigenetic interplay in regulating the benefits of polyploid wheat.
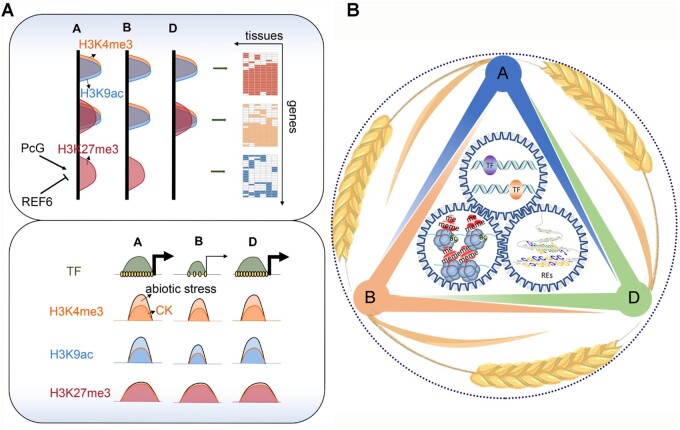
This epigenomic atlas reveals the interplay between regulatory elements, transcription factors, and epigenetic factors in subgenome-divergent spatio-temporal regulation.
Introduction
Common wheat (Triticum aestivum, 2n = 6x = 42, AABBDD) is one of the most commonly cultivated crops worldwide. It has a large (16 Gb) and complex allohexaploid genome (International Wheat Genome Sequencing et al., 2018). Compared to its diploid and tetraploid progenitors, common wheat has much broader adaptability to a wide range of environmental conditions. This has been ascribed to the convergence of individual genomes adapted to different environments, as well as the fast generation of new diversity in hexaploid wheat (Dubcovsky and Dvorak, 2007; Soltis and Soltis, 2009; Feldman et al., 2012; Nieto Feliner et al., 2020). Recent studies revealed extensive subgenome-divergent transcription in multiple tissues and subgenome-biased histone marks in leaves (Pfeifer et al., 2014; Jiao et al., 2018; Ramirez-Gonzalez et al., 2018; Li et al., 2019b), while how the subgenome divergence is dynamically and precisely regulated is still largely unknown.
Accurate responses to developmental and environmental cues depend on precise spatio-temporal transcription, which is orchestrated by the interplay between regulatory elements (REs) and the synergistic binding of transcription factors (TFs) and chromatin modifying complexes (Long et al., 2016). Investigating RE specificity regarding TFs is hampered by the large number of TFs encoded by a genome. Given that RE activities are generally accompanied by prompt biochemical changes to nearby chromatin marks, quantitative epigenomic approaches can be leveraged to study transcriptional regulation. First, on the basis of the principle that functionally related REs have a similar epigenetic environment, studies in animals have efficiently linked epigenetically regulated elements (epiREs) and their target genes according to coordinated epigenetic features (Ernst et al., 2011; Delaneau et al., 2019). Second, the changes to specific epigenomic architecture are synchronized with TF binding, and characterizing epigenomic changes may help identify cis- and trans-acting factors driving RE turnover (Rada-Iglesias et al., 2012; Huang et al., 2016; Wang et al., 2016). Third, some epigenetic factors can directly determine developmental specificity via tissue-specific expression and the interplay with cognate REs (Mozgova and Hennig, 2015; Cheng et al., 2018). Thus, quantitative epigenomic analysis is an effective approach for investigating the interplay between REs, TFs, and epigenetic factors in specific transcriptional regulation.
To apply quantitative epigenomic approaches, the epigenetic modifications involved in transcriptional regulation must be elucidated. In human studies, H3K4me3 is mostly enriched in promoter regions (Guenther et al., 2007), while H3K4me1 and H3K27ac are typical markers for active enhancers (Long et al., 2016). But in plants, H3K4me1 is not over-represented in enhancers (Li et al., 2019b; Lu et al., 2019). Instead, H3K4me3, H3K9ac, and H3K27ac are present in active REs (Oka et al., 2017; Li et al., 2019a, 2019b; Lu et al., 2019), which could be employed for linking the functionally related REs. In addition to these active markers, the presence of the repressive mark H3K27me3 reflects the poised or repressed state of REs (Rada-Iglesias et al., 2011). These REs may be activated in specific stages, as reported for both animals and plants (Mozgova and Hennig, 2015; Xiao and Wagner, 2015; Xiao et al., 2017). Recent studies also reported the regulation of distal accessible chromatin regions by H3K4me3 and H3K27me3 Hi-ChIP loops (Ricci et al., 2019). Thus, for studies on wheat, the active marks, including H3K4me3 and H3K9ac/H3K27ac, and the repressive mark H3K27me3 are the primarily enriched marks associated with gene activity. Moreover, the advantage of using histone marks for investigating RE specificity lies in the fact that the induction or repression can be distinguished, facilitating subsequent mechanism research.
In this study, we characterized the regulatory chromatin marks across diverse samples, based on which the epiRE functional interactions with cognate target genes were systematically detected. The specific epigenomic architecture involved in subgenome-divergent epiRE regulation was characterized, and the developmental fate and stress responses regulated by the interplay between epiRE sequence contexts, epigenetic factors, and TFs were further clarified. The results presented herein highlight the power of integrating quantitative epigenomic approaches for deciphering the subgenome-divergent spatio-temporal regulation.
Results
Systematic mapping of the epigenetic architecture in multiple tissues and in response to diverse external stimuli
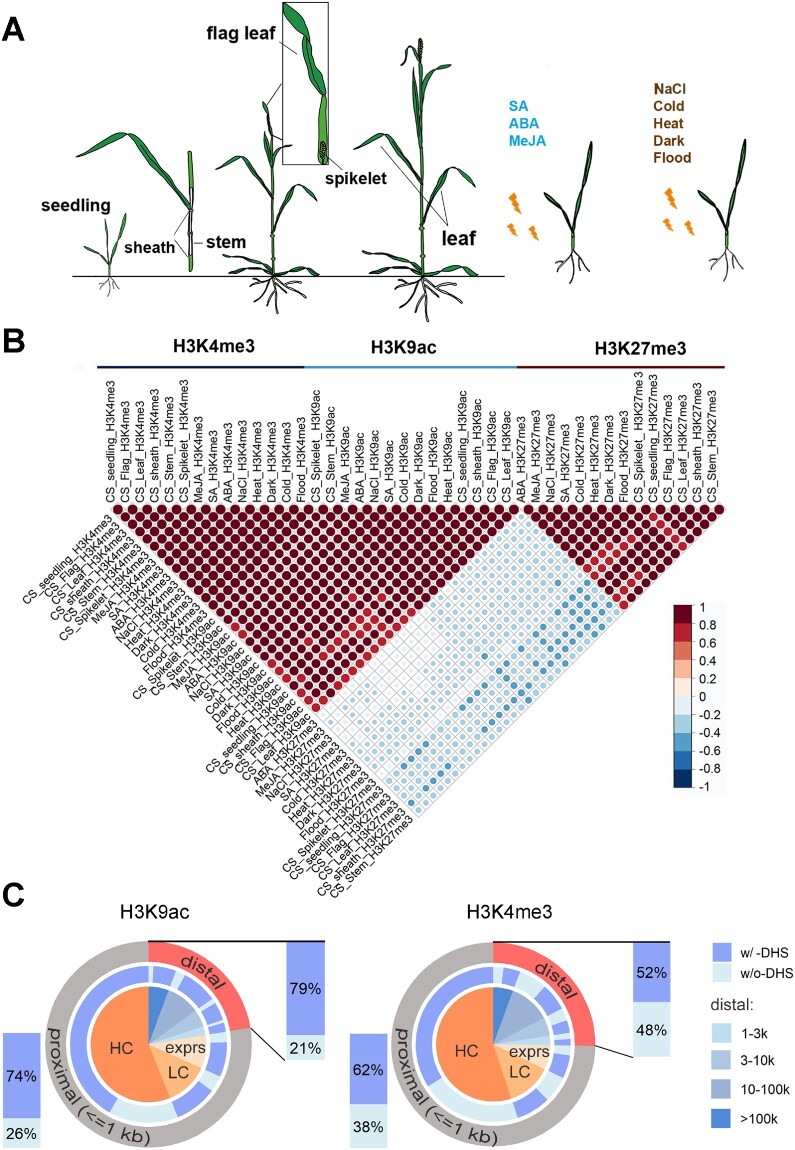
To systematically characterize the dynamic usage of REs and the mechanism determining RE specificity in common wheat, we investigated chromatin activity in six tissues and in response to eight external stimuli (Figure 1A). We applied chromatin immunoprecipitation coupled to massively parallel DNA sequencing (ChIP-seq) to examine the following three histone modifications that capture regulatory activities in wheat (Li et al., 2019b): the acetylation of histone 3 at lysine 9 (H3K9ac) and the tri-methylation of histone 3 at lysine 4 (H3K4me3) associated with active regulatory regions as well as the tri-methylation of histone 3 at lysine 27 (H3K27me3) associated with Polycomb-repressed regions. The gene expression profiles across 14 samples were also examined. All samples were prepared in biological replicates.
Figure 1.
Profiles of epigenomic modifications across typical developmental stages and in response to various external stimuli. A, ChIP-seq assay was performed for H3K9ac, H3K4me3, and H3K27me3 marks during six developmental stages, including seedling, sheath, stem, flag leaf, and boot spikelet (Feekes 10) and following exposures to eight external stimuli, including phytohormones and abiotic stresses. B, Scatter plots presenting the correlations among the densities of epigenetic marks across samples. C, Overlap between H3K9ac and H3K4me3 and DHS in gene proximal and distal regions. HC: high confidence genes; LC: low confidence genes; exprs: expressed coding genes predicted based on RNA-seq data (read count ≥ 3). Please also refer to the heatmap in Supplemental Figure 1.
An analysis of correlation coefficients revealed that the densities of the active marks H3K9ac and H3K4me3 were highly correlated (r = 0.71–0.95), but were negatively correlated with the density of H3K27me3 (Figure 1B). A genome-wide analysis detected 326,062 genomic regions with at least one of these three major regulatory marks, accounting for 2.2% of the whole genome and representing an expansive set of potentially functional REs across the common wheat genome. All data were visualized with a customized genome browser (http://bioinfo.sibs.ac.cn/dynamic_epigenome), and the raw data and peak files were retrieved from the Gene Expression Omnibus database [GSE139019].
A recent study characterizing histone marks and accessible chromatin regions in 13 plant species reported limited association between accessible chromatin and histone marks in distal regions (Lu et al., 2019). We determined the overlap between H3K9ac and H3K4me3 and accessible chromatin characterized by DNase I-hypersensitive sites (DHSs) in wheat seedlings, in both proximal genic regions (promoters) and distal regions (putative enhancers) (Figure 1C). To avoid misclassification of promoters as putative enhancers due to incomplete gene annotation, we integrated the gene annotation file and RNA-seq data. All potential coding regions were collected and classified as high confidence genes, low confidence genes and transcribed coding regions. Excluding peaks proximal to these potential coding regions resulted in 22% H3K9ac and 25% H3K4me3 peaks. Among these distal peaks, 79% (19,279) H3K9ac and 52% (20,622) H3K4me3 peaks overlap with DHS. These ratios are much higher as compared to those observed in maize and other species characterized previously (Oka et al., 2017; Lu et al., 2019). In addition, heatmap in Supplemental Figure 1 displayed high H3K9ac and H3K4me3 read densities surrounding DHS summit in both proximal and distal regions. Together, enrichment of H3K9ac and H3K4me3 reflect active regulatory activity in both proximal and distal regions in wheat.
Genome-wide assignment of distal epiRE to cognate target genes
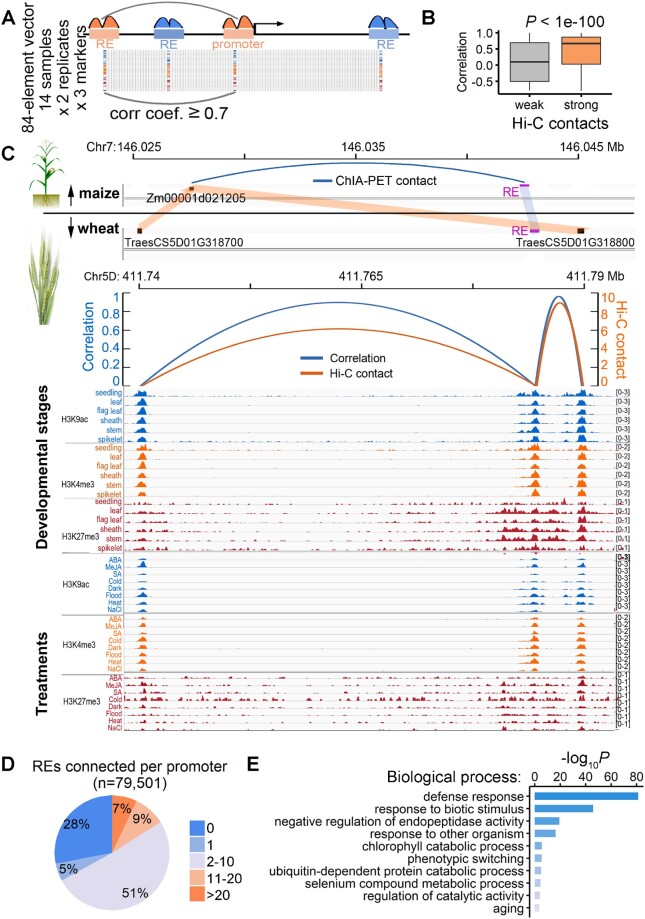
To investigate how REs regulate the spatio-temporal specificity of gene expression, linking the REs to their functional target genes is necessary. However, REs may affect the activity of cognate genes independent of the relative distance, location, or orientation, and assigning REs to target genes and further dissecting their specific regulatory mechanism is nontrivial, especially in the large and complex wheat genome. Previous studies in animals revealed that the epigenetic activities of distal REs are highly synchronized with those at the promoters of target genes (Delaneau et al., 2019). Accordingly, we correlated the density of three epigenetic markers in all samples at distal epigenetic peak regions with the density in all promoters within ± 500 kb, as previously described (Delaneau et al., 2019; Figure 2A). A total of 79,501 genes were targeted by 223,976 distal epiREs that were highly correlated (r ≥ 0.7, P < 0.05) with at least one promoter (Supplemental Data Set 1), reflecting the coordinated activities of distal epiRE–promoter pairings and providing an extensive map of candidate distal REs controlling specific genes. To validate these predicted distal epiRE–promoter interactions, we first compared their correlation coefficients with the strength of the chromosomal interactions mapped by Hi-C recently published (Concia et al., 2020), because physical proximity is generally a prerequisite for RE regulation (Schoenfelder and Fraser, 2019). We observed that epiRE pairs with strong physical interactions detected by Hi-C have highly correlated epigenetic activities (Figure 2B).
Figure 2.
Genome-wide map of functional distal epiRE–promoter interactions. A, Workflow diagram to link distal REs to cognate promoters by correlating epigenomic patterns. B, Distribution of the correlation coefficients of epigenetic activity between distal RE–promoter pairs with 1% weakest (left) and 1% strongest (right) Hi-C contacts. C, Genomic tracts illustrating a conserved distal RE–promoter interaction between maize and wheat. The top panel depicts the chromatin contact in maize detected by ChIA-PET. The bottom panels indicate the homologous regions (shaded in light blue and orange) connected both functionally, as reflected by parallel epigenetic activities (orange arcs, left y-axis), and physically, as measured by Hi-C (blue arcs, right y-axis). D, Proportion of 79,501 promoters correlated (r ≥ 0.7 and P < 0.05) with distal epiREs within 500 kb. E, Significantly enriched functional categories of genes regulated by multiple epiREs (result for top 2,000 genes). An unfiltered list of over-represented GO categories is provided (P < 10−4).
We subsequently assessed whether the pairs could be verified by independent experiments. There is currently a lack of experimental evidence of distal regulation in wheat, but the evolutionarily conserved RE interactions may reflect the functional significance of a particular regulatory architecture. Therefore, we compared the RE interactions detected in this study with those reported for maize (Zea mays), which were recently profiled based on analyses of chromatin interactions with paired-end tags (ChIA-PET; Peng et al., 2019). Of the previously described 9,152 distal epiRE–promoter pairs, we identified conserved sequences in common wheat for 65 pairs, of which 29 pairs (45%) were highly correlated in terms of chromatin activity (r ≥ 0.7 and P < 0.05). An example of the conserved RE–promoter pairs between maize and wheat is presented in Figure 2C. Interestingly, the maize gene was duplicated in wheat, but the interaction is “remembered” by both copies. Additionally, the correlations between the distal epiREs and the promoters closely paralleled the frequency of long-range chromatin interactions determined by Hi-C in common wheat. We further compared the performance in predicting gene expression based on RE epigenome between epigenome correlation method and the traditional way of target assignment based on proximity, i.e. assigning REs to the nearest genes. The predictability is better when using epigenome correlation, especially for genes regulated by multiple REs (Supplemental Figure 2). Together, the results validated the distal epiRE–promoter pairings from aspects of both high-throughput interactions and expression predictability.
A further examination of distal epiRE–promoter pairs revealed that 67% of the promoters were connected to more than one distal regulatory region marked by the histone marks, indicating the combinatorial regulation of gene activity by multiple REs (Figure 2D), which is similar to the results of a study on humans (Thurman et al., 2012). The number of distal REs assigned to a particular promoter represents a quantitative measure of the complexity of the cis-regulation. To examine the functional features of genes regulated by a highly complex mechanism, we ranked all common wheat genes based on the number of distal epiREs paired with their promoter. We then searched for the over-represented functional terms associated with the top 2,000 genes using GO terms curated by GOMAP (Lawrence-Dill, 2019). The common wheat genes associated with a highly complex cis-regulation were markedly enriched for defense responses (Figure 2E; Supplemental Figure 3). This result is consistent with the findings of a similar study on humans showing abundant RE regulation of immunity-related genes (Thurman et al., 2012). Thus, the complexity of defense responses is potentially encoded by the complex cis-regulatory circuit.
Subgenome-divergent epigenetic architecture of REs is indicative of target tissue specificity
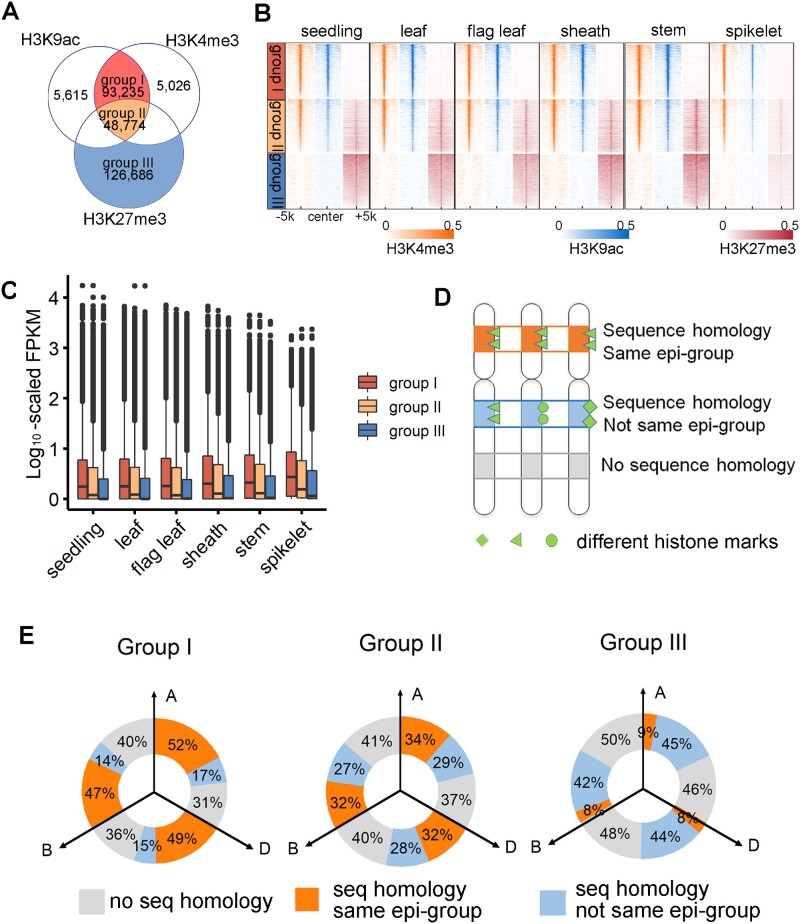
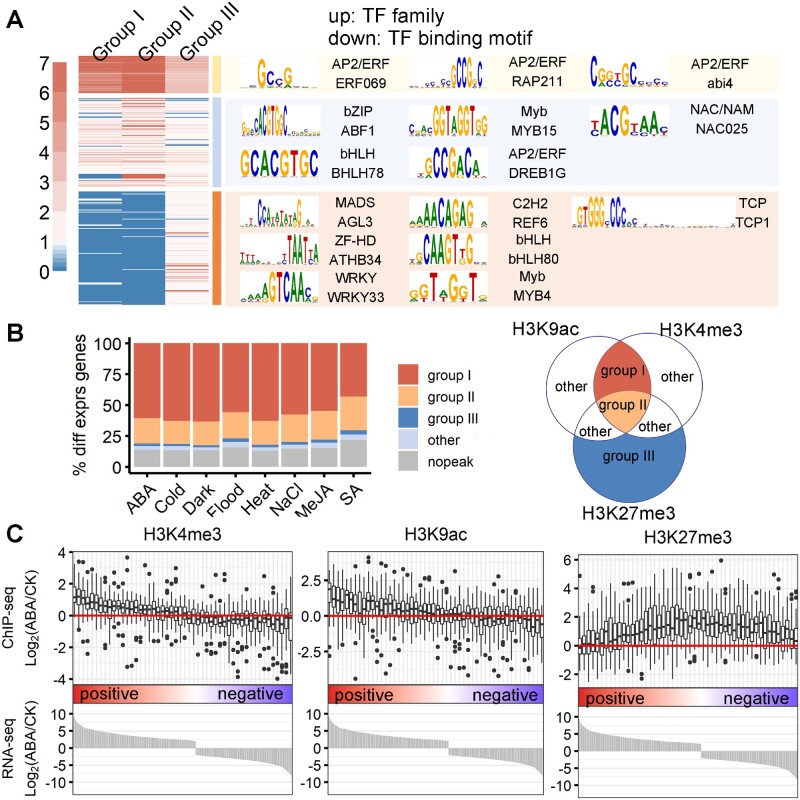
With the genes targeted by epiREs defined, we next investigated the relationship between subgenome-divergent regulation and tissue specificity modulated by epiREs. Firstly, all of the detected peaks for six tissues were compared, which revealed a significant overlap between the active histone marks H3K9ac (96%) and H3K4me3 (97%) (Figure 3, A and B; Supplemental Data Set 2). The overlapping peak regions represent active epiREs with relatively high confidence. Around 34% of these active epiREs were also overlapping with H3K27me3 peak in at least one tissue. In addition, 126,686 regions had H3K27me3 peak only.
Figure 3.
Subgenome-divergent epigenetic architecture of epiREs. A, Venn diagram presenting the overlap of the H3K9ac, H3K4me3, and H3K27me3 peaks in all six tissues. B, Heatmap showing read density of each histone modification across tissues surrounding REs from the three groups. C, Normalized expression levels of genes targeted by different epiRE groups. D, Comparison of epiRE sequences and epigenetic groups across subgenomes lead to three types of result. E, Donut plot presenting the sequence homology and similarity of the epigenetic features across subgenomes between three epiRE groups. The grey area indicates the fraction of regions with no sequence homology between subgenomes. The light blue area indicates the fraction of regions with homologous sequences between subgenomes, but marked by different epigenetic features. The orange area represents the homologous regions with epigenetic architecture from the same group.
Genes targeted by different epiRE groups displayed different levels of expression. Group I targets were highly expressed in all the tissues examined, while group III targets were lowly expressed in most tissues (Figure 3C). Previous studies reported that H3K27me3 is mediated by Polycomb repressive complex 2 (PRC2), which is responsible for tissue-specific gene repression and developmental patterning (Mozgova and Hennig, 2015; Xiao and Wagner, 2015).
We next compared the sequence features and epigenetic architecture of the three groups across subgenomes (see “Methods” section). Figure 3D illustrated the three types of comparison results. The sequence similarity of three epiRE groups is comparable across subgenomes, while the consistency of epigenetic group is different. For example, 47%–52% of group I epiREs belong to the same epi-group across subgenomes, while the number is only 8%–9% for group III epiREs (Figure 3E). Genes targeted by group III are mostly nonconserved (Figure 4A) with only 12% have 1:1:1 correspondence (triplet) across three subgenomes.
Figure 4.
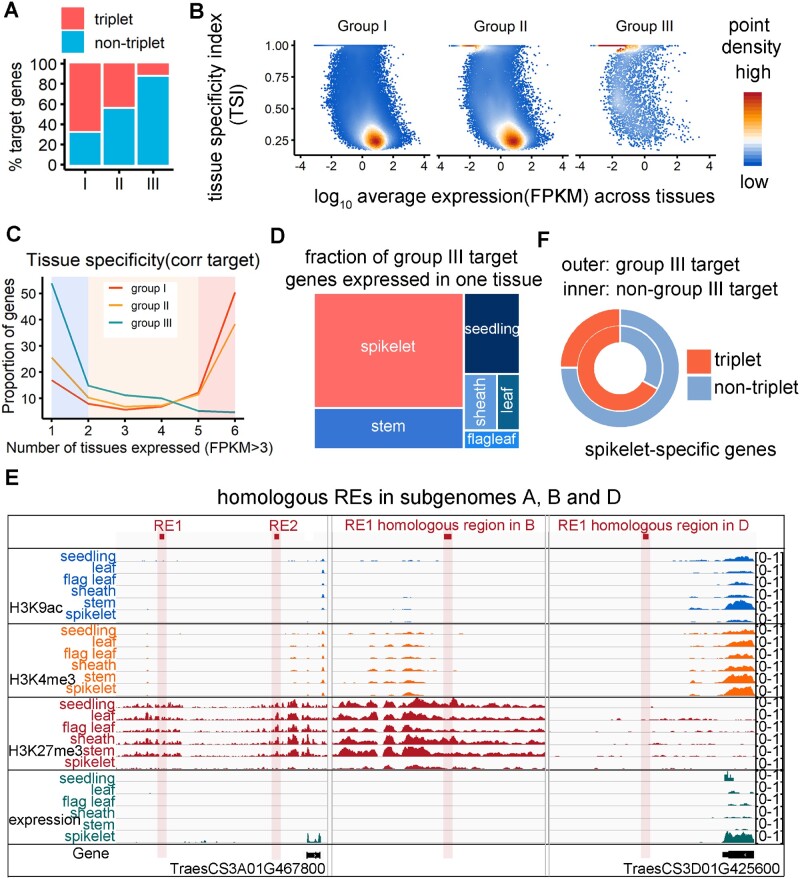
Subgenome unbalanced epigenetic feature of REs is indicative of target tissue specificity. A, Fraction of groups I, II, and III target genes belonging to triplets (1.1:1 correspondence across the three sub-genomes) and nontriplets. B, Density plot showing the distribution of tissue specificity and expression level of genes targeted by three epiRE groups. The tissue specificity is measured by the tissue specificity index (TSI) across tissues. C, Proportion of genes expressed specifically or universally across tissues. Genes targeted by three epi-RE groups are plotted. D, Fraction of group III target genes higher expressed (FPKM>3) in only one tissue. The area of the colormap represents the fraction of targets. E, Genomic tracks presenting subgenome-divergent epigenetic regulation of group III epiRE and expression profile of target genes across tissues. F, Fraction of spikelet-specific genes belonging to triplets. The outer circle represents group III targets, whereas the inner circle represents spikelet-specific genes not targeted by group III.
To further dissect the functional difference between these epiRE groups, we examined their target gene expression across diverse tissues. Notably, the group III targets have apparently higher tissue specificity (Figure 4B), 54% of which were expressed in only one tissue (i.e. FPKM > 3 in only one tissue; Figure 4C; Supplemental Data Set 3), most commonly the spikelet (Figure 4D; statistical test in Supplemental Figure 4). GO enrichment analysis using group III epiREs targets with spikelet-specific expression revealed the enrichment of floral development-related terms (Supplemental Figure 5). Genomic tracks in Figure 4E illustrated a distal epiRE from group III, which has sequence homology across subgenomes, but had different epigenetic architecture and linked to genes with different expression profiles across tissues.
We next wondered whether the subgenome-divergent regulation of tissue specificity is dependent on group III epiREs, in other words, whether spikelet specific genes are preferentially divergent across subgenome, no matter targeted by group III epiREs or not. We thus examined the subgenome conservation of spikelet-specific genes targeted or not targeted by group III epiREs. It is clear that only group III epiRE-targeted spikelet-specific genes are highly diverse across subgenomes (Figure 4F). Thus, the subgenome-divergent regulation of tissue specificity is preferentially targeted by group III epiREs which are preferentially marked by H3K27me3.
Extensive elimination of the H3K27me3 surrounding spikelet-specific epiREs associated with spikelet-specific H3K27 demethylase
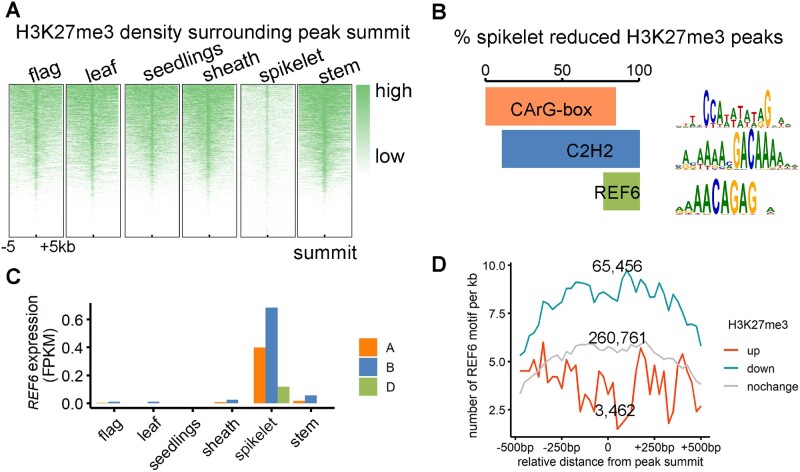
Genome-wide profiling of the H3K27me3 pattern across tissues detected extensive decrease of H3K27me3 in spikelet, which potentially contributed to the derepression of spikelet-specific epiREs and genes (Figure 5A). We next assessed how the H3K27me3 marks are specifically removed. The interplay between RE sequence contexts, epigenetic factors, and TFs determines the specific addition or removal of epigenetic modifications (Wang et al., 2016). We first examined the enriched DNA sequence features surrounding the group III epiREs specifically activated in the spikelet (Supplemental Data Set 4). The most abundant motifs included the CArG-box and C2H2 TF-binding site (Figure 5B). The CArG-box is bound by MADS-box TFs mostly involved in regulating flowering and floral development. This is consistent with the results of earlier investigation on Arabidopsis thaliana, in which flower-specific TFs were confirmed as responsible for removing the H3K27me3 surrounding flower-specific REs (Derkacheva and Hennig, 2014; Mozgova and Hennig, 2015; Wang et al., 2016). Among the enriched C2H2 TF-binding sequences, the RELATIVE OF EARLY FLOWERING 6 (REF6)-binding motif CTCTGYTY was significantly over-represented, present in 24% of the epiREs with decreased H3K27me3 levels.
Figure 5.
Spikelet-specific H3K27me3 elimination associated with spikelet-specific expression of H3K27 demethylase. A, H3K27me3 ChIP-seq density 5kb surrounding the peak summit in each tissue. The peak summits from all tissues were collected. B, Percentage of spikelet reduced H3K27me3 peaks carrying the enriched motifs. The overlapping area represents the coexistence of given motifs in one H3K27me3 peak region. C, REF6 expression level in each tissue. D, Distribution of REF6 motifs surrounding H3K27me3 peaks up- or downregulated in boot spikelets (Feekes 10). The number of epiREs for each group were labeled.
REF6 is a major plant-specific H3K27 demethylase (Cui et al., 2016). Examination of REF6 expression across tissues revealed the gene was much higher expressed in the spikelet as compared to other tissues (Figure 5C). Its homolog, Early Flowering 6 (ELF6), ubiquitously expressed in all tissues, also displayed a considerable upregulated expression in the spikelet (Supplemental Figure 6A). This phenomenon is highly conserved among Triticeae species, but not in other well-established plant model species such as Arabidopsis, rice (Oryza sativa), and maize, in which REF6 is ubiquitously expressed (Supplemental Figure 6, B–E), suggesting that the H3K27 demethylases are sub-functionalized in Triticeae species related to developmental transition. We further examined the distribution of REF6 binding motifs surrounding H3K27me3 peaks, and found that peaks downregulated in spikelets were enriched for REF6 motifs (Fisher’s exact test P = 1.59–26; Figure 5D), indicating spikelet-specific REF6 potentially involved in extensive reduction of H3K27me3 and derepression of spikelet specific REs.
Prediction and validation of key cis- and trans-factors responsive to external changes based on quantitative epigenomic alterations
To further investigate the genome-scale interplay between the epigenetic architecture and RE sequence context, we detected the enriched TF-binding motifs for epiREs from all three groups (Figure 6A; Supplemental Figures 7–9). Motif enrichment score in each of the three groups was calculated for all JASPAR plant motifs (see “Methods” section and Supplemental Data Set 5). The TF-binding sequences CArG-box and C2H2 were specifically enriched in group III. In contrast, groups I and II, which were preferentially marked by H3K4me3 and H3K9ac, were highly enriched for AP2/ERF TF binding motifs, which are mostly related to external and developmental programs (Xie et al., 2019). Further comparison to expression data revealed that 70%–83% of the phytohormone and abiotic stress responsive genes were targeted by the group I and group II epiREs (Figure 6B). We next examined the relationship between epigenetic changes and gene expression changes following stress and phytohormone treatments. All genes responsive to the treatments were ranked according to their expression-level fold-change, and the corresponding epigenetic changes were plotted for each mark (Figure 6C; Supplemental Figure 10). The H3K4me3 and H3K9ac changes were positively associated with gene expression changes, but the H3K27me3 changes were not. Thus, H3K4me3 and H3K9ac are likely involved in responses to external stimuli.
Figure 6.
H3K4me3 and H3K9ac alterations synchronized with transcriptional changes in response to phytohormones and abiotic stresses. A, Cluster of TF-binding motifs enriched in three RE groups. Color scale represents the enrichment score of different motifs in three RE groups. The consensus sequence LOGO of the top enriched motifs is provided on the right. Please refer to Supplemental Figures 7–9 for details regarding the clustering. B, Bar plot presenting the fraction of genes responsive to stress and phytohormone treatments enriched for epiREs from different groups. C, Relationship between epigenetic changes and gene expression changes in response to different treatments. The ABA treatment is presented here, whereas the results for the other treatments are presented in Supplemental Figure 10. Genes responsive to ABA are ranked by expression fold-changes (x-axis), and the fold-changes of the read density of histone marks were plotted (y-axis). CK, control.
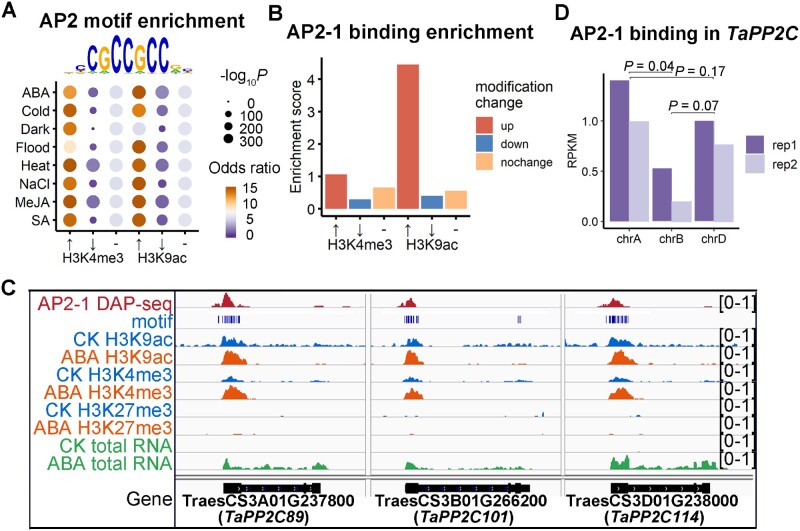
To determine the mechanisms underlying specific alteration of H3K4me3 and H3K9ac, we examined the related epigenetic factors and transcription factors. There were no significant expression-level changes to the enzymes associated with H3K4me3 and H3K9ac (Supplemental Figure 11), and the rapid epigenetic changes following these external treatments are likely associated with specific TFs. Given the close association between the H3K9ac and H3K4me3 changes and target gene expression changes, the regulatory regions with dynamic H3K4me3 and H3K9ac alterations may provide insightful clues regarding the key cis- and trans-acting factors controlling responses to environmental cues. Motif enrichment analysis revealed that AP2/ERF TF-binding sites are top enriched in epiREs with increased H3K4me3 and H3K9ac levels triggered by these treatments (Figure 7A; Supplemental Figure 12). This is consistent with previous reports that some AP2/ERF TFs are key regulators of various stress responses (Xie et al., 2019).
Figure 7.
Prediction and validation of key trans-acting factors in response to external changes. A, Enrichment analysis of AP2 motif surrounding epiREs with stress-induced (↑) or repressed (↓) H3K4me3 and H3K9ac marks. (-) represents no significant epigenetic change. B, Enrichment of AP2-1 binding as determined by DAP-seq surrounding ABA-induced H3K4me3 and H3K9ac REs. C, Genomic tracks presenting AP2-1 binding at AP2 motifs in promoter region of ABA-induced PP2C. The changes of H3K4me3 and H3K9ac densities and PP2C expression change in response to the ABA treatment were also shown. D, Quantification of AP2-1 binding surrounding PP2C. Student’s t test was applied.
We clarified the quantitative epigenetic strategy using ABA-triggered changes. There are 10 AP2/ERF TFs induced by ABA, among which AP2-1 exhibited the highest upregulated expression. We successfully obtained the genome-wide binding of AP2-1 in subgenome A via DNA affinity purification and sequencing (DAP-seq; Bartlett et al., 2017). The binding of AP2-1 was significantly enriched in epiREs with apparently increased H3K4me3 and H3K9ac levels (Figure 7B). The genomic tracks in Figure 7C illustrated the binding of AP2-1 in densely distributed AP2 motifs in promoter of ABA highly induced PP2C. Together, the orchestrated epigenetic change and specific TF binding ensured prediction of responsive cis- and trans-acting factors by quantitative epigenomic alterations.
Coordinated subgenome divergence between epigenomic profile, TF binding, and cis-element density
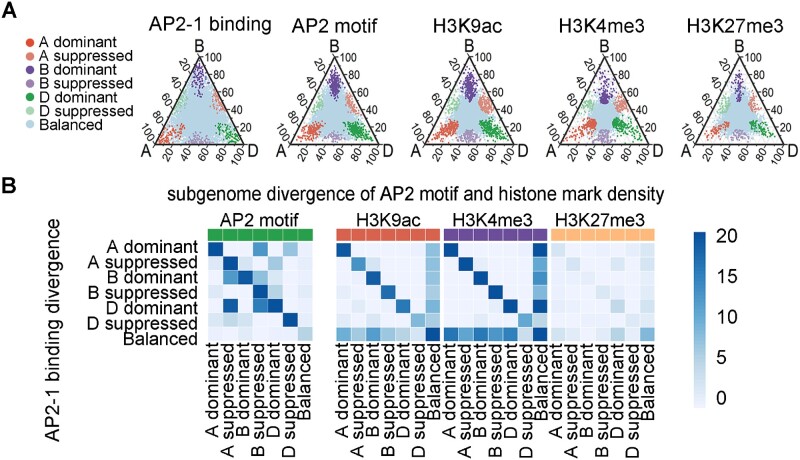
A closer examination of the genomic tracks in Figure 7C lead to the observation that AP2-1 displayed stronger binding surrounding PP2C in subgenomes A and D. The quantitative difference is statistically significant (Figure 7D). Accordingly, the active epigenetic marks and AP2 motifs also had higher densities in subgenomes A and D. We thus wondered whether this quantitative association of the subgenome divergence is a general feature on genome-wide level, which may reflect regulatory mechanisms regarding subgenome divergence. We quantitatively compared the AP2-1 binding regions, epigenetic modifications, as well as AP2 motif densities across subgenomes, respectively. Each data set was divided to seven categories based on the relative density in A, B, and D subgenomes. A ternary plot previously proposed (Ramirez-Gonzalez et al., 2018) was employed for visualization (Figure 8A). Further enrichment analysis revealed that the subgenome-biased AP2-1 binding was apparently correlated with the subgenome-biased H3K9ac and H3K4me3 read densities, and also positively correlated with the AP2 motif density (Figure 8B). In summary, these findings revealed the subgenome-divergent regulation is likely orchestrated by the interplay between genetic and epigenetic heterogeneity (Figure 9).
Figure 8.
Relationship between subgenome-divergent AP2-1 binding and subgenome-divergent densities of AP2 motif and epigenetic marks. A, Ternary plots presenting the relative binding densities of AP2-1 and three epigenetic marks, as well as the relative AP2 motif density across three subgenomes. Each circle represents a homologous region present in all three subgenomes. The distance for each region was determined based on the ratio of the normalized read (or motif) density for one sub-genome to the read density for all sub-genomes. B, Enrichment of the overlap between the biased binding of AP2-1 and the biased densities of epigenetic marks and motifs. Dark blue represents a significant overlap.
Figure 9.
Model illustrating the specificity of subgenome-divergent development and stress responses encoded in wheat REs. (A) Top panel: subgenome-divergent RE regulation by H3K27me3 contribute to tissue specificity. Bottom panel: coordinated subgenome divergence between epigenomic profile, TF binding, and cis-element density during stress responses. (B) Schematic diagram illustrating the interplay among REs, epigenetic factors, and TFs in regulating subgenome divergence.
Discussion
Subgenome-divergent regulation contributed to genome plasticity and success of polyploid wheat under domestication (Dubcovsky and Dvorak, 2007; Soltis and Soltis, 2009; Feldman et al., 2012; Nieto Feliner et al., 2020). The study of regulatory specificity in hexaploid wheat is challenged by the large and complex genome. We herein characterized the epigenomic architecture in a large spectrum of samples, and applied quantitative epigenomic approaches to reveal the interplay between RE sequence contexts, epigenetic factors, and TFs in subgenome-divergent spatio-temporal regulation.
Selection of epigenetic markers for dissecting the specific RE regulome
The epigenome (i.e. second dimension of the genome) harbors essential cell-specific information (Pikaard and Mittelsten Scheid, 2014). In addition to examining the three histone marks closely associated with gene activity included in this study, there are other epigenetic strategies useful for elucidating transcriptional regulation. One popular method is based on chromatin openness, which is generally characterized by DNase I hypersensitive site sequencing or Assay for Transposase-Accessible Chromatin using sequencing (Boyle et al., 2008; Thurman et al., 2012). Both types of data were recently reported in wheat leaves (Li et al., 2019b; Jordan et al., 2020; Lu et al., 2020). Previous studies on humans revealed that rather than being fully nucleosome free, open chromatin regions are generally associated or flanked by specific histone modifications (e.g. H3K4me1 and H3K27ac) related to enhancer deployment (Shlyueva et al., 2014). Thus, the high-throughput data characterizing the open chromatin state are generally highly correlated with the data for typical enhancer markers. This has been confirmed in common wheat and maize (Oka et al., 2017; Li et al., 2019a). Here, we observed that large proportions of H3K9ac and H3K4me3 marks overlapping with DHS in both proximal and distal regions, which are much higher as compared to those reported in other plant species, indicating that these two marks are good predictor of active regulatory activity of REs. In addition, the fact that proximal and distal REs enriched with similar epigenetic modifications also makes it feasible to assign epiRE targets via correlating of epigenetic patterns. In the present study, we applied both active and repressive histone markers to clarify RE specificity, of which the repressive REs are not necessarily detectable based on the open chromatin data. Considering the advantages and limitations of high-throughput data, the further application of chromatin openness to connect functionally related REs may enrich the present RE–target map.
Specific linkage between distal REs and targets: physical versus functional RE interactions
The interaction between distal REs and their cognate promoters can be detected based either on the correlated epigenetic activities between functionally related REs or on the physical proximity between REs and promoters as determined by chromosome conformation capture methods. A recent investigation involving humans (Delaneau et al., 2019) and the current study (Figure 2) indicate that these approaches produce consistent results. However, the resolution required for detecting physical interactions is highly dependent on the sequencing depth, resulting in increasing costs proportional to genome size. Accordingly, characterizing RE interactions across tissues and under various conditions in wheat is not practical. Additionally, studies on animals revealed the relative consistency of Hi-C data among tissues, suggesting Hi-C experiments are not sensitive enough to analyze the dynamics among tissues or under different conditions (Dixon et al., 2012). Another advantage of using epigenetic correlations to link functionally relevant REs and target genes is that the induction or repression can be distinguished based on the epigenetic features. Because incorporating chromosome proximity information may help distinguish between direct and indirect regulation, we included the Hi-C pair information for the RE pairs detected based on coordinated epigenetic activities (Supplemental Data Set 1).
Specific spatio-temporal regulation: the epigenetic machinery and the interplay with cis- and trans-acting factors
The expansion and diversification of epigenetic complexes contributed significantly to the complexity of transcriptional regulation. The specificity of the epigenetic machinery is mainly realized via co-factors and/or the sub-functionalization of epigenetic factors (Wang et al., 2016; Xiao et al., 2017; Cheng et al., 2018), both of which were uncovered in the present study. First, the spikelet-specific expression of REF6 potentially contributes to the elimination of H3K27me3 surrounding spikelet-specific REs, which is a phenomenon unique to Triticeae species (Supplemental Figure 6). The REF6 gene is universally expressed in other well-studied monocots and dicots (Supplemental Figure 6). The epigenetic machinery in wheat has expanded extensively, especially in hexaploid wheat. The relaxation of selective constraints may lead to the substantial sub-functionalization of epigenetic factors, whose functions and underlying mechanisms should be investigated in future studies. Second, the coordinated binding of TFs and epigenetic markers was detected near stress responsive REs, which provides the foundation for employing epigenomic changes to detect cis- and trans-acting factors driving responses. Given the large non-coding regions of wheat and other crops with large genomes, focusing on functional loci with significant epigenomic changes may help to rapidly narrow down the potential master stress responsive REs and TFs.
Subgenome-divergent RE regulation by H3K27me3 and the benefits of polyploidy
One intriguing finding in this study is that H3K27me3 in hexaploid wheat is closely associated with subgenome divergent RE regulation. The interplay between H3K27me3 and REs regulates the activity of a large proportion of subgenome-divergent tissue specific genes, particularly the spikelet-specific genes. In addition to the increased expression of H3K27 demethylase genes REF6 and ELF6, the genes encoding specific enzymes responsible for the addition of H3K27me3 marks were also highly expressed in the spikelet (Supplemental Figure 13). This may help explain the moderate recovery of the H3K27me3 level in the late spikelet developmental stage (Supplemental Figure 14). These findings are indicative of a complex feedback loop between H3K27 demethylase and PcGs in subgenome-divergent transcriptional regulation of reproductive development.
Heterosis and gene redundancy are the major benefits of polyploidy (Panchy et al., 2016). How the activities of subgenome divergent gene copies are finely regulated to contribute to the benefits of polyploidy is a fundamental issue. In addition to H3K27me3 regulation of subgenome-divergent tissue specific genes, the nontriplet stress responsive genes were also preferentially targeted by REs marked by H3K27me3 (Supplemental Figure 15), despite that their transcriptional changes are not correlated with H3K27me3 alterations (Figure 6C). These result indicated that H3K27me3 may have a diverse role in subgenome-divergent RE regulation. It was recently reported that H3K27me3 pattern is associated with higher order chromosomal interactions in both animals and plants (Cheutin and Cavalli, 2014; Baker et al., 2015). Thus, H3K27me3 may regulate subgenome-divergent gene copies both in cis and in trans. In summary, all of the findings described herein not only provide useful resources for elucidating subgenome-divergent regulation in hexaploid wheat, but also give new clues regarding the generic and epigenetic interplay in regulating the benefits of polyploid wheat.
Materials and methods
Plant materials and growth conditions
Common wheat (Triticum aestivum cultivar “Chinese Spring”) seeds were surface-sterilized via a 10-min incubation in 30% H2O2 and then thoroughly washed five times with distilled water. The seeds were germinated in water for 3 days at 22°C. The germinated seeds with residual endosperm were transferred to soil (1:1:3 mixture of vermiculite: perlite: peat soil) or Hoagland solution and grown under 16 h light/8 h dark condition at 22°C in greenhouse. The seedlings (above-ground parts) in soil were harvested after 9-day growth. The stems, stem leaves, flag leaves, sheaths, spikelets at booting stage (Feeke 10) and spikelets at flowering stage (Feekes 10.5) were harvested and either frozen in liquid nitrogen for an RNA isolation or directly vacuum-infiltrated with a formaldehyde cross-linking solution for use in the ChIP-seq assay. Regarding the cold and heat stress treatments, 7-day-old seedlings grown in soil were transferred to 4°C or 40°C, respectively, and grown under 16 h light/8 h dark conditions for 7 days. For the flooding treatment, 7-day-old seedlings grown in soil (9 × 9 × 9 cm pot) were submerged in the water with the water level at ∼5 cm above the bottom of the pot, which was incubated under 16 h light/8 h dark condition for 7 days. To assess the effects of darkness, 7-day-old seedlings grown in soil were incubated at 22°C in darkness for 7 days. For the hormone and NaCl treatments, germinated seeds were grown in Hoagland solution at 22°C under 16 h light/8 h dark conditions for 7 days, after which they were treated with 100 μm ABA, 100 μm MeJA, 500 μm SA, or 150 mM NaCl and incubated for another 7 days. The aerial parts of the treated and control plants were harvested and either frozen in liquid nitrogen for an RNA isolation or directly vacuum-infiltrated with a formaldehyde cross-linking solution for use in the ChIP-seq assay.
ChIP-seq assay and RNA sample preparation and sequencing
A total of 90 ChIP-seq and 30 RNA-seq data sets were generated with biological duplicates. ChIP-seq assay was completed as previously described (Wang et al., 2016), with antibodies specific for H3 trimethyl-Lys 27 (Millipore, Upstate, USA), H3 trimethyl-Lys 4 (Abcam, Cambridge, England), and H3 acetyl-Lys 9 (Millipore). For each ChIP-seq assay, approximately 30 seedlings were pooled and ground to a powder. More than 10 ng ChIP DNA or 2 μg total RNA was used to prepare each sequencing sample. Libraries were constructed and sequenced by Genenergy Biotechnology Co. Ltd. (Shanghai, China) and Novogene (Beijing, China). The libraries were sequenced with the HiSeq X Ten system (illumina, San Diego, CA, USA) to produce 150-bp paired-end reads.
Processing of ChIP-seq and RNA-seq data
Trimmomatic (version 0.36; Bolger et al., 2014) was used to trim adaptor, with parameters “PE ILLUMINACLIP: Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:'true' LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36”. Next, the program Sickle (version 1.33) with parameters “pe -t sanger -q 20 -l 20 -n -g” was used to eliminate bases with low quality scores (<20) and short reads (length < 20). The remaining clean reads were mapped to the International Wheat Genome Sequencing Consortium (IWGSC) reference sequence (version 1.0) with the Burrows–Wheeler Aligner (version 0.7.5a-r405; Li and Durbin, 2010) for the ChIP-sequencing data. The HISAT2 program (version 2.1.0; Kim et al., 2015) was used for mapping the RNA sequencing (RNA-seq) reads to the reference sequences and gene models from the IWGSC RefSeq genome assembly (version 1.0). High-confidence genes from this gene model version were used throughout this study.
The MACS (version 1.3.7) program (Zhang et al., 2008) with parameters “–nolambda –nomodel” was used to identify the read-enriched regions (peaks) of the ChIP-seq data based on the following criteria: P < 1e − 5 and fold-change > 32. To quantify gene expression levels, the featureCount program of the Subread package (version 1.6.5; Liao et al., 2013) with parameters “-s 2 -p -t exon” was used to determine the RNA-seq read density for the high-confidence genes in the IWGSC RefSeq genome assembly (version 1.0). To compare expression levels across samples and genes, the RNA-seq read density of each gene was normalized based on the exon length in the gene and the sequencing depth (i.e. fragments per kilobase of exon model per million mapped reads). To quantify histone markers across genes for the figure prepared with Integrative Genomics Viewer (Robinson et al., 2011), the number of reads at each position was normalized against the total number of reads (reads per million mapped reads). The DEseq program (Anders and Huber, 2010) was used for detecting differentially expressed genes based on the following criteria: |log2 fold-change| > 1 and P < 0.05. The MAnorm package(Shao et al., 2012) was used for the quantitative comparison of ChIP-seq signals between samples with the following criteria: |M value| > 1 and P < 0.05.
Epi-group classification and comparison across subgenomes
We first sampled 12,000,000 aligned ChIP-seq reads from every BAM file such that the total number of read is the same across samples. Next, the overall peak regions were detected using MACS (version 1.3.7) with parameters “–nolambda –nomodel” and with all sampled BAM files as input, resulted in 329,675 peaks in total. The peak file for each sample was also detected in the same way. “intersect” function in BEDtools (version 2.27.1) was used to determine if there were overlaps between the overall peak regions and the peak summit for each sample. Blastn was used to compare epiRE sequences across subgenomes. epiREs with sequence identity > 85%, E-value < 0.01 and query coverage >55% were defined as homologous regions between subgenomes.
Detection of transcription factor-binding motifs
To detect enriched TF-binding motifs in the regions of interest, we downloaded the position weight matrices of 501 plant motifs from the JASPAR database (Khan et al., 2018). The motifs were then used to screen specific regions with the Find Individual Motif Occurrences program (Grant et al., 2011) of the MEME software toolkit (version 5.0.2) using default settings. To calculate the enrichment of a given motif in a tested region list, we shuffled regions with the same length distribution with the region list under test. The enrichment score of a given motif in the tested regions was calculated as the ratio between the number of the tested regions harboring the given motif and the number of shuffled regions harboring the given motif. Fisher’s exact test was used to measure statistical significance.
Connectivity between promoters and distal epiREs
The connectivity between promoters and distal epiREs was calculated as previously described (Thurman et al., 2012). For the full repertoire comprising 326,062 peak regions of all three markers in six tissues and in response to eight treatments, we compiled 79,501 high-confidence genes with at least one peak localized within promoter regions (3 kb upstream of a transcription start site). For the promoter of each compiled gene, an associated distal epiRE was identified if the Pearson correlation coefficient of the epigenetic activities (a 84-element vector; i.e. density of three epigenetic marks under 14 conditions) between the promoter peak and distal peaks was at least 0.7 with P < 0.05. The Pearson correlation of histone modifications between epiRE and promoters was performed with corr function in R (version 3.6.3).
Analysis of Hi-C data
The Hi-C sequencing data, which were previously published (International Wheat Genome Sequencing et al., 2018), were retrieved from the Gene Expression Omnibus database (accession number PRJEB25248). The Trimmomatic program was used to trim adapters with the same parameters as mentioned above. The remaining reads were aligned to the IWGSC reference sequence (version 1.0) with the default settings of Hi-C-Pro (Servant et al., 2015). Briefly, a two-step filtering process was used to ensure the chimeric reads were accurately aligned. After the reads were mapped, the low-quality reads and singletons were discarded. The aligned read pairs were assigned to DpnII restriction fragments, and the invalid pairs with an overhanging end or those that self-ligated or re-ligated were discarded. Finally, 41,879,083 valid pairs were obtained. These valid pairs were further converted to the .hic format with the script hicpro2juicebox.sh implemented by HiC-Pro. To quantity the Hi-C interaction between two given regions, juicer_tools (Durand et al., 2016) was used to extract the interaction matrix, with the resolution set to 50 kb and the Knight–Ruiz algorithm adopted for data normalization (Knight and Ruiz, 2013). The frequency of the interaction between two regions was measured based on the normalized read pair density bridging the constituent regions.
DAP-seq assay and data processing
DAP-seq was performed as previously described (Bartlett et al., 2017). Genomic DNA was extracted from wheat leaves using Plant DNAzol Reagent (Invitrogen) and fragmented. DNA was then end repaired using the End-It kit (Lucigen) and A-tailed using Klenow (3′–5′ exo-; NEB). Truncated Illumina Y-adapter was ligated to DNA using T4 DNA Ligase (Promega). Full length TF was cloned into pIX-Halo vector (Forward primer: ggctgtcgacgctcgagATGGACTTCGGCGCCGACAT; Reverse primer: gagtgcggccgcaagctgGTTCACGGTCACCTCCGGGA). Halo-tagged TF was expressed in vitro using TNT SP6 Coupled Wheat Germ Extract System (Promega). Halo-TF was immobilized by Magne HaloTag Beads (Promega) and then incubated with the DNA library. TF specific binding DNA was eluted for 10 min at 98°C and amplified with indexed Illumina primer using Phanta Max Super-Fidelity DNA Polymerase (Vazyme). Meanwhile, to capture background DNA which captured by Halo, pIX-Halo vector without TF cloned was expressed and incubated with the DNA library as well. The PCR product was purified using VAHTS DNA Clean Beads (Vazyme) and then sequenced by Novogene (Beijing, China) with the Illumina NovaSeq 6000 system to produce 150-bp paired-end reads. For data processing, the cleaning, mapping and peak calling steps were the same as described above for ChIP-seq data analysis. The peaks detected from samples introduced with Halo tag only were considered as nonspecific bindings, and TF peaks overlapping with peaks detected from Halo samples were removed for subsequent analysis.
Accession numbers
The ChIP-seq and RNA-seq data were deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139019).
Tracks for all sequencing data can be visualized through our local genome browser (http://bioinfo.sibs.ac.cn/dynamic_epigenome).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure 1. H3K9ac and H3K4me3 density surrounding DHS summit. DHS regions were classified to proximal and distal groups.
Supplemental Figure 2. The performance of predicting target gene expression by RE epigenome based on different target definition methods.
Supplemental Figure 3. Significantly enriched functional categories of genes regulated by different numbers of REs.
Supplemental Figure 4. Enrichment significance of group III target genes with tissue specific expression.
Supplemental Figure 5. GO enrichment analysis of spikelet-specific genes targeted by group III epiREs.
Supplemental Figure 6. Expression of REF6 and ELF6 in Triticeae species and other plant species.
Supplemental Figure 7. Motif clusters enriched in RE group I.
Supplemental Figure 8. Motif clusters enriched in RE group II.
Supplemental Figure 9. Motif clusters enriched in RE group III.
Supplemental Figure 10. Relationship between epigenetic changes and gene expression changes in response to stress and phytohormone treatments.
Supplemental Figure 11. Expression-level changes to the enzymes associated with H3K4me3 and H3K9ac in response to stress and phytohormone treatments.
Supplemental Figure 12. Enriched motifs at phytohormones and stresses induced H3K4me3 and H3K9ac REs.
Supplemental Figure 13. Expression levels of the enzymes associated with addition and removal of H3K27me3 in different tissues.
Supplemental Figure 14. Average H3K27me3 ChIP-seq density surrounding the peak summit in each tissue. The peak summits from all tissues were collected.
Supplemental Figure 15. Proportion of stresses and phytohormone regulated genes belonging to triplets and non-triplets.
Supplemental Data Set 1. Genomic coordinates of distal epiREs and cognate promoters.
Supplemental Data Set 2. Genomic coordinates of three epiRE groups.
Supplemental Data Set 3. List of group III targeted tissue-specific genes. For each gene, different metrics depicting tissue-specificity and functional annotation was provided.
Supplemental Data Set 4. List of peaks with altered H3K27me3 level during spikelet development.
Supplemental Data Set 5. Enrichment statistics for motifs shown in Fig. 6A.
Supplementary Material
Acknowledgment
We thank Huang Tao for his help in maintaining the high-performance computing server. We thank Dr. Jizeng Jia from Chinese Academy of Agricultural Sciences for insightful and constructive comments.
Funding
This study was supported by the National Natural Science Foundation of China (31921005 and 31770285), the National Science Fund for Excellent Young Scholars (32022012) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010302).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Meiyue Wang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Zijuan Li, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Yu’e Zhang, University of the Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, and The Innovation Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Yuyun Zhang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Yilin Xie, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Luhuan Ye, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Yili Zhuang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Kande Lin, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Fei Zhao, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Jingyu Guo, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; Henan University, School of Life Science, Kaifeng, Henan 457000, China.
Wan Teng, University of the Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, and The Innovation Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Wenli Zhang, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Yiping Tong, University of the Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, and The Innovation Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Yongbiao Xue, University of the Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, and The Innovation Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute of Genomics, Chinese Academy of Sciences, and China National Centre for Bioinformation, Beijing 100101, China; Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, Yangzhou University, Yangzhou 225009, China.
Yijing Zhang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Yongbiao Xue (ybxue@genetics.ac.cn) and Yijing Zhang (zhangyijing@cemps.ac.cn).
Y.J.Z., Y.B.X., and Y.P.T. conceived and designed the experiments. Z.J.L., Y.Z., L.H.Y., K.D.L, Y.L.Z., W.T., and W.L.Z performed the experiments. M.Y.W., Y.Y.Z., Y.L.X., F.Z., J.Y.G., and Y.J.Z. analyzed the data. Y.J.Z. wrote the manuscript with input from all authors.
References
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K, Dhillon T, Colas I, Cook N, Milne I, Milne L, Bayer M, Flavell AJ (2015) Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance. Plant J 84: 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A, O'Malley RC, Huang SSC, Galli M, Nery JR, Gallavotti A, Ecker JR (2017) Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Niu Q, Zhang B, Chen K, Yang R, Zhu JK, Zhang Y, Lang Z (2018) Downregulation of RdDM during strawberry fruit ripening. Genome Biol 19: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G (2014) Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev 25: 30–37 [DOI] [PubMed] [Google Scholar]
- Concia L, Veluchamy A, Ramirez-Prado JS, Martin-Ramirez A, Huang Y, Perez M, Domenichini S, Rodriguez Granados NY, Kim S, Blein T, et al. (2020) Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biol 21: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lu F, Qiu Q, Zhou B, Gu L, Zhang S, Kang Y, Cui X, Ma X, Yao Q, et al. (2016) REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat Genet 48: 694–699 [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zazhytska M, Borel C, Giannuzzi G, Rey G, Howald C, Kumar S, Ongen H, Popadin K, Marbach D, et al. (2019). Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science 364 [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Hennig L (2014) Variations on a theme: Polycomb group proteins in plants. J Exp Bot 65: 2769–2784 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL (2016) Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy A, Chalhoub B, Kashkush K (2012) Genomic plasticity in polyploid wheat. In Soltis PS, Soltis DE, eds, Polyploidy and Genome Evolution. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 109–135 [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, et al. (2016). Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell 36: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium, IWGSC RefSeq principal investigators, Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N IWGSC whole genome assembly principal investigators,Pozniak CJ, et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361(6403): eaar7191 [DOI] [PubMed] [Google Scholar]
- Jiao W, Yuan J, Jiang S, Liu Y, Wang L, Liu M, Zheng D, Ye W, Wang X, Chen ZJ (2018). Asymmetrical changes of gene expression, small RNAs and chromatin in two resynthesized wheat allotetraploids. Plant J 93: 828–842 [DOI] [PubMed] [Google Scholar]
- Jordan KW, He F, de Soto MF, Akhunova A, Akhunov E (2020) Differential chromatin accessibility landscape reveals structural and functional features of the allopolyploid wheat chromosomes. Genome Biol 21: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, Tan G, et al. (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46: D1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PA, Ruiz D (2013) A fast algorithm for matrix balancing. IMA J Numer Anal 33: 1029–1047 [Google Scholar]
- Lawrence-Dill C (2019). GOMAP wheat reference sequences 1.1 (CyVerse Data Commons).
- Li E, Liu H, Huang L, Zhang X, Dong X, Song W, Zhao H, Lai J (2019a) Long-range interactions between proximal and distal regulatory regions in maize. Nat Commun 10: 2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang M, Lin K, Xie Y, Guo J, Ye L, Zhuang Y, Teng W, Ran X, Tong Y, et al. (2019b). The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J (2016) Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FH, McKenzie N, Gardiner LJ, Luo MC, Hall A, Bevan MW (2020) Reduced chromatin accessibility underlies gene expression differences in homologous chromosome arms of diploid Aegilops tauschii and hexaploid wheat. GigaScience 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Marand AP, Ricci WA, Ethridge CL, Zhang X, Schmitz RJ (2019) The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat Plants 5: 1250–1259 [DOI] [PubMed] [Google Scholar]
- Mozgova I, Hennig L (2015) The polycomb group protein regulatory network. Annu Rev Plant Biol 66: 269–296 [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Casacuberta J, Wendel JF (2020) Genomics of evolutionary novelty in hybrids and polyploids. Front Genet 11: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka R, Zicola J, Weber B, Anderson SN, Hodgman C, Gent JI, Wesselink JJ, Springer NM, Hoefsloot HCJ, Turck F, et al. (2017). Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biol 18: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH (2016) Evolution of gene duplication in plants. Plant Physiol 171: 2294–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Xiong D., Zhao L., Ouyang W., Wang S., Sun J., Zhang Q., Guan P., Xie L., Li W., et al. (2019). Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nat Commun 10: 2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR International Wheat Genome Sequencing CMayer KF, Olsen OA (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Mittelsten Scheid O (2014) Epigenetic regulation in plants. Cold Spring Harbor Perspect Biol 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J (2012) Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, et al. (2018) The transcriptional landscape of polyploid wheat. Science 361: 662. [DOI] [PubMed] [Google Scholar]
- Ricci WA, Lu ZF, Ji LX, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejia-Guerra MK, Colome-Tatche M, et al. (2019). Widespread long-range cis-regulatory elements in the maize genome. Nat Plants 5: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Fraser P (2019) Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 20: 437–455 [DOI] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E (2015) HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan G-C, Orkin SH, Waxman DJ (2012) MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15: 272–286 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. (2012). The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen WH, Jin H, Xu L, Zhang Y (2016). Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet 12: e1005771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wagner D (2015) Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol 23: 15–24 [DOI] [PubMed] [Google Scholar]
- Xiao J, Jin R, Yu X, Shen M, Wagner JD, Pai A, Song C, Zhuang M, Klasfeld S, He C, et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat Genet 49: 1546–1552 [DOI] [PubMed] [Google Scholar]
- Xie ZL, Nolan TM, Jiang H, Yin YH (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.