ABSTRACT
Lanthipeptides are ribosomally synthesized and posttranslationally modified peptides, with modifications that are incorporated during biosynthesis by dedicated enzymes. Various modifications of the peptides are possible, resulting in a highly diverse group of bioactive peptides that offer a potential reservoir for use in the fight against a plethora of diseases. Their activities range from the antimicrobial properties of lantibiotics, especially against antibiotic-resistant strains, to antiviral activity, immunomodulatory properties, antiallodynic effects, and the potential to alleviate cystic fibrosis symptoms. Lanthipeptide biosynthetic genes are widespread within bacterial genomes, providing a substantial repository for novel bioactive peptides. Using genome mining tools, novel bioactive lanthipeptides can be identified, and coupled with rapid screening and heterologous expression technologies, the lanthipeptide drug discovery pipeline can be significantly sped up. Lanthipeptides represent a group of bioactive peptides that hold great potential as biotherapeutics, especially at a time when novel and more effective therapies are required. With this review, we provide insight into the latest developments made toward the therapeutic applications and production of lanthipeptides, specifically looking at heterologous expression systems.
KEYWORDS: antimicrobial agents, antimicrobial peptides, antiviral agents, drug resistance, heterologous expression systems, lantibiotics, lanthipeptides
INTRODUCTION
Lantibiotics, first described in 1988, are ribosomally synthesized and posttranslationally modified peptides (RiPPs) with antimicrobial activity that contain meso-lanthionine and 3-methyl-lanthionine (1, 2). While most lanthionine-containing RiPPs are lantibiotics, there are some that lack antimicrobial activity. To account for the few lanthionine-containing RiPPs without antimicrobial activity the overall term “lanthipeptides” is used (3). Modification occurs at the precursor peptide, which consists of an N-terminal leader peptide, important for recognition by modification enzymes, and a C-terminal-modified core peptide (1, 2, 4, 5) (Fig. 1; see Fig. S1A, S2A, S3, and S4 in the supplemental material). Lanthionine cross-links form as result of a sequence of dehydration and cyclization reactions catalyzed by specific (dedicated) lanthionine synthetases, such as LanB and LanC in class I lantibiotics, or by single multifunctional synthetases, such as LanM, LanKC, and LanL described for classes II, III, and IV, respectively (Fig. 1) (reviewed in references 6 and 7). The class I LanB dehydratases are aminoacyl-tRNA dependent, with initial glutamylation of Ser/Thr, followed by glutamate elimination and generation of Dha/Dhb (8, 9). The multifunctional synthetases (LanM, LanKC, and LanL) require (d)NTPs for dehydration to phosphorylate Ser/Thr residues, followed by elimination and generation of Dha/Dhb (10–13). The class II LanMs consist of two domains (an N-terminal dehydratase and a C-terminal cyclase domain), whereas the multifunctional synthetases in classes III and IV have three domains consisting of a N-terminal lyase domain, central kinase domain, and C-terminal cyclase domain (5, 10–13). The N-terminal dehydratase domain of class II synthetases is responsible for the phosphorylation and elimination steps, and in classes III and IV, this is carried out by two different domains, namely, the kinase and lyase domains, respectively. Dehydrated Ser/Thr (i.e., Dha/Dhb) are subsequently cyclized by the addition of a Cys thiol through a Michael-type addition: the resulting enolate intermediate undergoes protonation to form either Lan (Dha-Cys) or MeLan (Dhb-Cys) cross-links (14). The main differentiating factor between class III and class IV synthetases is in their C-terminal cyclase domains, with class IV featuring the conserved zinc-binding domain, also present in class I and II cyclases, while this feature is lacking in class III cyclase representatives (5, 11, 15). Furthermore, in the case of some class III lanthipeptides, the enolate intermediate formed during the first nucleophilic attack is not protonated and undergoes an additional Michael addition, with a second Dha yielding a bicyclic structure termed labionin (Lab) (15, 16). Based on these synthetase differences, lanthipeptides are grouped into four classes, with further division into subclasses based on differences in the amino acid sequences and tertiary structures of the mature lanthipeptides (Fig. 1; Fig. S1 to S4) (3, 17). Lantibiotics may undergo additional posttranslational modifications (PTMs), such as the introduction of d-amino acids, oxidative decarboxylation of the C terminus, formation of a lysinoalanine ring, and formation of an N-terminal lactate group (Fig. 1) (18–21). This results in the formation of a diverse number of lantibiotics and lanthipeptides. In most cases, the structural genes of lantibiotics form part of a biosynthetic gene cluster and contain the biosynthetic machinery for modification, export, leader processing, and regulation. This was first shown for the lantibiotics nisin (22), epidermin (23), and subtilin (24) and has subsequently been illustrated for numerous other lanthipeptides.
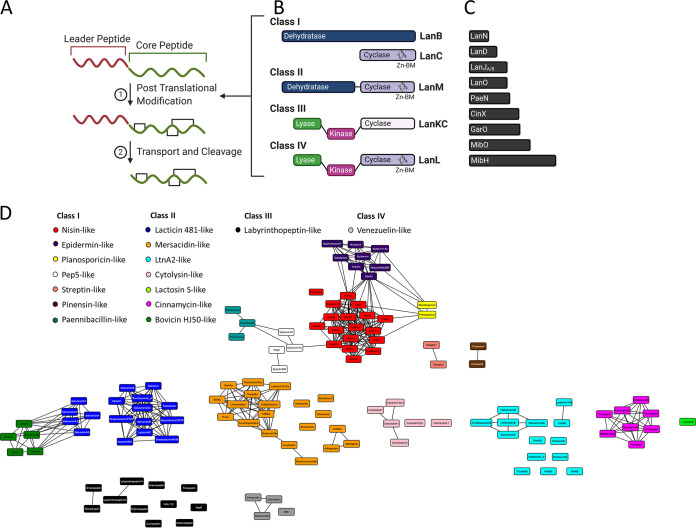
FIG 1.
Biosynthesis and classification of lanthipeptides. (A) Generalized scheme of lanthipeptide biosynthesis (precursor peptide made up of leader and core peptides). (B and C) Four main classes of synthetases (B) and additional modification enzymes (C) involved in PTM of lanthipeptides. (D) Sequence similarity network of lanthipeptide core peptides generated with the Enzyme Function Initiative-Enzyme Similarity Tool (EST-EFI [https://efi.igb.illinois.edu/efi-est/]) (E value cutoff, 10−3) and visualized in Cytoscape (v.3.8.0). Abbreviations: LanN, lysinoalanine synthase; LanD, flavin-containing Cys decarboxylase; LanJA/B, dehydrogenase (A indicates Zn2+ dependent and B indicates flavin dependent); LanO, oxidoreductase; PaeN, acetylation (paenibacillin); CinX, α-ketoglutarate (α-KG)/iron(II)-dependent hydroxylase (cinnamycin); GarO, flavin-dependent monooxygenase (actagardine); MibO, hydroxylase (microbisporicin); MibH, Trp halogenase (microbisporicin); and Zn-BM, zinc binding motif.
Since the first description of lantibiotics, numerous lantibiotics have been identified and characterized, with nisin, first reported in 1928, being the most well known: nisin has been used as a food preservative for over 50 years, which is currently the only industrial application of a lantibiotic (1, 25, 26). Although the antimicrobial properties of lantibiotics, especially against antibiotic-resistant and clinically relevant strains of Staphylococcus (27–35), Enterococcus (27–30, 33, 35), and Clostridium (28, 36–40) spp., have been reported in several studies (Table 1), very few clinical studies have been published (reviewed in reference 41). This may be changing, as more recent studies have shown that some lantibiotics have broader applications than initially appreciated due to activities in addition to their antibacterial properties, such as antiviral activity (42–45), immunomodulatory properties (46, 47), antiallodynic effects (16, 48), and the ability to alleviate cystic fibrosis (49–54) (Table 1). Lanthipeptide biosynthetic genes are widespread within the genomes of taxonomically distinct bacterial species, providing a substantial repository for peptides with a wide range of potentially novel structures and bioactivities (26). More recent interrogation of the genomes of understudied phyla suggests that lanthipeptides are likely much more diverse than is currently appreciated, and many novel posttranslational modification mechanisms have yet to be described (55). Such genome mining efforts have indicated that lanthipeptide synthetases have been repurposed for production of natural products other than lanthipeptides, thereby expanding natural product diversity. The development of rapid screening methods of large lantibiotic/lanthipeptide libraries further adds to the discovery pipeline (56–58). With current advances in heterologous gene expression, these peptides may be produced by laboratory strains at high levels (Table 2). Heterologous expression of unusual biosynthetic systems identified through (meta)genome mining efforts could result in the discovery of natural products with new scaffolds that potentially have interesting biological activities.
TABLE 1.
Lanthipeptide producers and applications
| Lanthipeptide(s) | Producer strain | Class, subclass | Type testeda | Bioactivity (reference[s]) |
|---|---|---|---|---|
| Nisin | Lactococcus lactis | I, nisin-like | In vivo, in vitro, TC | Antimicrobial (160), anticancer (129), immunomodulatory (46) |
| Subtilin | Bacillus subtilis | I, nisin-like | In vitro, in vivo | Antimicrobial (161, 162) |
| Ericin | Bacillus subtilis | I, nisin-like | In vitro | Antimicrobial (163) |
| CMB001 | Paenibacillus kyungheensis | I, nisin-like | In vitro | Antimicrobial (33) |
| Gallidermin/epidermin | Staphylococcus gallinarum/S. epidermidis | I, epidermin-like | In vitro, TC | Antimicrobial (164), immunomodulatory (47) |
| Clausin | Bacillus clausii | I, epidermin-like | In vitro, in vivo | Antimicrobial (27) |
| Mutacin 1140 | Streptococcus mutans | I, epidermin-like | In vitro, in vivo, TC | Antimicrobial (40, 165) |
| Mutacin B-Ny266 | Streptococcus mutans | I, epidermin-like | In vitro, in vivo | Antimicrobial (98, 166) |
| Planosporicin | Planispora alba | I, planosporicin-like | In vivo, in vitro | Antimicrobial (28, 82) |
| NAI-107 (microbisporicin) | Microspora corallina | I, planosporicin-like | In vivo, in vitro | Antimicrobial (28, 29) |
| Mersacidin | Bacillus amyloliquefaciens | II, mersacidin-like | In vitro, in vivo | Antimicrobial (31, 34) |
| Actagardine | Actinoplanes liguraie | II, mersacidin-like | In vivo, in vitro | Antimicrobial (79, 167) |
| Duramycin/cinnamycin | Streptomyces cinnamoneus | II, cinnamycin-like | In vitro, in vivo, TC | Antimicrobial (168), antiviral (141, 168), anticancer (135, 136), ion channel regulator (50, 51), immunomodulatory (121) |
| Lacticin-3147 | Lactococcus lactis | II, mersacidin-like; II, Ltn2-like | In vivo, in vitro | Antimicrobial (32, 169) |
| Amyloliquecidin | Bacillus amyloliquefaciens | II, mersacidin-like; II, Ltn2-like | In vivo, in vitro | Antimicrobial (27) |
| Pinensins | Chitinophaga pinensins | I, pinensin-like | In vitro | Antimicrobial (170), antifungal (170) |
| Labyrinthopeptins | Actinomadura namibiensis | III, labyrinthopeptin-like | In vivo, in vitro, TC | Antiviral (44), antiallodynic (16) |
| NAI-112 | Actinoplanes sp. strain DSM 24059 | III, labyrinthopeptin-like | In vivo, in vitro | Antimicrobial (mild) (48), antiallodynic (48), antinociceptive (48) |
TC, tissue culture.
TABLE 2.
Examples of lanthipeptide heterologous expression systems
| Lanthipeptide | Wild typea | Class, subclass | Heterologous producer | Vector(s) usedb | Cleavagec | Intracellular/secretedd | Yielde | Culture conditionsf | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| Nisin | L. lactis | I, nisin-like | E. coli BL21(DE3) | pRSFnisA-nisB (HispLanA and LanB), pACYCnisC (LanC) | In vitro, trypsin | IC (SF, ISF) | 24 mg/liter (precursor) | TB, induced with IPTG, expressed 15 h at 18°C | 149 |
| L. lactis | I, nisin-like | E. coli BL21(DE3) | pRSFGFPnisA-nisB (HisGFPpLanA and LanB), pACYCnisC (LanC) | In vitro, thrombin, hNisP | IC (SF) | 1.99 mg/liter (core) | TB, induced with IPTG, expressed 24–48 h at 26°C | 151 | |
| L. lactis | I, nisin-like | L. lactis NZ9000 | pIL3EryBTC (LanB, LanT and LanC), pNZnisA (pLanA) | In vitro, hNisP | Secreted | 1.7–3.5 mg/liter (core) | MEM, induced with nisin, expressed 3–24 h at 30°C | 146, 147, 171 | |
| Pep5 | Sta. epidermidis 5 | I, Pep5-like | E. coli BL21(DE3) | pRSFGFPpep5-nisB (HisGFPpLanA and LanB), pACYCnisC (LanC) | In vitro, thrombin, hNisP | IC (SF) | ND | TB, induced with IPTG, expressed 24–48 h at 26°C | 151 |
| Gallidermin | Sta. gallinarum Tu3928 | I, epidermin-like | L. lactis NZ9000 | pIL3EryBTC (LanB, LanT and LanC), pNZEgdmA (pLanA), pNZEgdmD (LanD) | In vitro, trypsin | Secreted | ND | MEM, induced with nisin, expressed 2 h at 30°C | 147 |
| NAI-107 (microbisporicin) | Microbispora sp. strain 107891 | I, planosporicin-like | E. coli BL21(DE3) | pETmibA-mibB (HispLanA and LanB), pCDFmibC (LanC), pACYCtRNAGluX4-mgltX (tRNAGluCUC-1 iso-acceptor and GluRS) | In vitro, LysC | IC (SF) | ND | TB, induced with IPTG, expressed 18–20 h at 25°C | 9 |
| Ala(0)-actagardine | A. liguriae ATCC 31048 | II, mersacidin-like | E. coli BL21(DE3) | pRSFgarA-garM (HispLanA and LanM), pCDFgarO(2x) (GarO) | In vitro, trypsin aminopeptidase A8200 | IC (SF) | 4.2 mg/liter (precursor) | LB, induced with IPTG, expressed 20 h at 18°C | 172 |
| A. garbadinensis ATCC 31049 | II, cinnamycin-like | S. lividans | Cosmid CosAG14 (contains garA BGC) | In vivo | Secreted | 50–80 mg/liter (core) | GM1, not induced, expressed 72 h at 30°C | 159 | |
| Bovicin | Str. bovis HJ50 | II, bovicin HJ50-like | E. coli BL21(DE3) and C43(DE3) | pET28abovA (HispLanA), pET28abovA-bovM (HispLanA and LanM), pACYCbovT150M (truncated HisLanT fused to LanM), pACYCbovT150 (truncated HisLanT) | In vitro and in vivo, BovT, BovT150, BovT50M | IC (SF, ISF), secreted | 0.2–0.91 μg/ml (core) in cell lysates | LB, induced with IPTG, expressed 20 h at 16°C | 155 |
| Carnolysin | C. maltaromaticum C2 | II, cytolysin-like | E. coli BL21(DE3) | pRSFcrnA1-crnM (HispLan and LanM), pRSFcrnA2-crnM (HispLan and LanM), pETcrnJ (LanJB) | In vitro, CrnT150, GluC | IC (SF, ISF) | 0.5 mg/liter (precursor) | LB, induced with IPTG, expressed 20 h at 18°C | 173 |
| Cinnamycin | St. cinnamoneus DSM 40005 | II, cinnamycin-like | E. coli BL21(DE3) | pRSFcinA-cinM (HispLanA and LanM), pACYCcinX (CinX) | In vitro, LysC | 2 mg/liter (precursor) | LB, induced with IPTG, expressed 20 h at 18°C | 20 | |
| Haloduracin | B. halodurans C-125 | II, mersacidin-like (Halα); II, LtnA2-like (Halβ) | E. coli BL21(DE3) | pRSFhalA-halM1 (HispLanA and LanM), pRSFhalB-halM2 (HispLanA and LanM) | In vitro, factor Xa | IC (SF) | 1–2 mg/liter (precursors) | LB, induced with IPTG, expressed 18 h at 18°C | 149 |
| Lichenicidin | B. licheniformis 189 | II, mersacidin-like (Bliα); II, LtnA2-like (Bliβ) | E. coli BL21 Gold(DE3) | pRSFTM1A1 (pLanA, LanT, and LanM), pRSFTPM2A2 (pLanA, LanT, LanP, and LanM) | In vivo, LicT, LicP | Secreted | 4 mg/liter Bliα (core), 6 mg/liter Bliβ (core) | Medium M, induced by autoinduction/IPTG, expressed 16 h at 30°C | 154 |
| Roseocin | Streptomyces roseosporus NRRL 11379 | II, mersacidin-like (Rosα); II, LtnA2-like (Rosβ) | E. coli BL21 | pRSFRosA1-rosM (HispLanA and LanM), pRSFRosA2-rosM (HispLanA and LanM) | In vitro, GluC | IC (SF) | 4 mg/liter (both α and β; precursor) | LB, induced with IPTG, expressed 24 h at 18°C | 174 |
| Nukacin ISK-1 | Sta. warneri ISK-1 | II, lacticin 481-like | E. coli BL21(DE3) | pET-nukAM (HispLanA and LanM) | In vitro, LysC | IC (SF) | 1.5 mg/liter (precursor) | 2× YT, induced with IPTG, expressed 20 h at 20°C | 150 |
| Prochlorosins (1.7, 2.11, and 3.3) | Prochlorococcus sp. strain MIT 9313 | II, mersacidin-like | E. coli BL21(DE3) | pRSFprocA-procM (HispLanA and LanM) | In vitro, trypsin, LysC, TEV | IC (SF, ISF) | 10–35 mg/liter (precursors) | LB, induced with IPTG, expressed 20 h at 18°C | 149 |
| Ruminococcin A | Ruminococcus gnavus E1 | II, lacticin 481-like | E. coli BL21(DE3) | pWLEOgrv4-grvM (HisGFPpLanA and LanM) | In vitro, TEV | IC (SF) | 6 mg/liter (precursor) | TB, induced with IPTG, expressed 18–24 h at 30°C | 152 |
| NAI-112 | Actinoplanes DSM24059 | III, labyrinthopeptin-like | E. coli BL21(DE3) | pRSFAplA (HispLanA), pACYCAplKC (LanKC) | In vitro, metalloproteases (AplP) | IC (ISF) | 1.25 mg/liter (precursor) | LB, induced with IPTG, expressed 20 h at 16°C | 175 |
| Labyrinthopeptins | Actinomadura namibiensis DSM 6313 | III, labyrinthopeptin-like | S. lividans | pUWLabSG2 (pLabyA1; LanKC and ABC transporters),g pUWLabSG6 (pLabyA2; LanKC and ABC transporters)g | In vivo | Secreted | 86 mg/liter LabyA1 (core), 14 mg/liter LabyA2 (core) | YEME, R2YE (LabyA1), and NZ amine (LabyA2), not induced, expressed 384 h at 28°C | 158 |
| Venezuelin | Streptomyces venezuelae ATCC 10712 | IV, venezuelin-like | E. coli BL21(DE3) | pET28MBPVenA (MBPHispVenA), pET28VenL (HisVenL and mutants) | In vitro, TEV | IC (SF)h | ND | LB, induced with IPTG, expressed 2.5–3 h at 37°C | 12 |
| Globisporin | St. globisporus NRRL B2293 | IV, venezuelin-like | E. coli BL21(DE3) | pET28asgbA-sgbL (LanL and either HisMBPpLanA or HispLanA) | In vitro, thrombin, TEV | IC (SF, ISF) | 0.3–0.7 mg/liter (precursor) | LB, induced with IPTG, expressed 1 h at 37°C | 5 |
| SflA | Streptomyces sp. strain NRRL S-1022 | IV, venezuelin-like | E. coli BL21(DE3) | Plasmid coexpressing SflA (HispLanA) and SflL (LanL) | In vitro, metalloproteases (StrS1022-P2 and -P4) | IC (SF) | 10 μg/liter (precursor) | TB, induced with IPTG, expressed 18–20 h at 18°C | 176 |
| Streptocollin | St. collinus Tu 365 | IV, venezuelin-like | St. coelicolor M1146 and M1152 | Cosmid A12-1 (contains stcA BGC) | In vivo | Secreted | 5.4–10 mg/liter (core) | SFM agar, not induced, expressed 240 h at 29°C | 124 |
Strain in which the biosynthetic gene cluster is found.
Vectors used for expression are given, and peptides/proteins expressed from vectors are indicated. pLanA, precursor LanA; HispLanA, His-tagged pLanA; HisGFPpLan, His-tagged GFP fused to pLanA.
hNisP, heterologously produced nisin protease; GluC, glutamyl endopeptidase; TEV, tobacco etch virus protease; LysC, endoproteinase LysC.
IC, intracellular; SF, soluble fraction; ISF, insoluble fraction.
ND, not determined; core, core peptide; precursor, precursor peptide.
LB, Luria-Bertani; TB, Terrific Broth; IPTG, isopropyl-β-d-thiogalactopyranoside; MEM, minimal expression medium; TSB, tryptic soy broth; 2× YT, 2× yeast extract-tryptone; SFM, soya flour-mannitol; YEME, yeast extract-malt extract; R2YE, R2 medium containing yeast extract.
Labyrinthopeptin A1 leader peptide modified (Arg-1 to Met-1) and used for pLabyA1 and to replace the pLabyA2 leader peptide.
Modification performed in vitro after cleavage of His-tagged precursor peptide from MBP.
In this review, we evaluate advances made in heterologous gene expression and report on progress made in the medical application of lanthipeptides.
APPLICATIONS OF LANTHIPEPTIDES
Due to the diversity and complex nature of lantibiotics, they have been explored for use in various medical applications, with some in clinical trials (Table 1). Lantibiotics have mainly been studied for their application as antimicrobials. However, they have bioactivities that extend beyond antimicrobial. Furthermore, their modification machinery can be used to stabilize peptides, improving their in vivo efficacy under a variety of conditions, such as stroke and diabetic nephropathy (59, 60). An increase in the number of sequenced genomes, including data from unculturable organisms, has led to an increase in the in silico analyses of genomic data that may yet reveal lantibiotics/lanthipeptides with more diverse bioactivities (26). With the help of in vitro/in vivo engineering, these putative peptides can then be functionally expressed, increasing the arsenal of candidates with potential medical applications. The development of efficient in vitro/in vivo strategies for screening and expressing these peptides makes large-scale production a realistic possibility.
TREATMENT OF INFECTIONS
Although a substantial arsenal of antibiotics is currently available for the treatment of a wide range of infections, antibiotics are losing efficacy against once treatable infections—a phenomenon accelerated by the incorrect use of antibiotics and the resulting microbial multidrug resistance (61, 62). A pessimistic view may be that we are treating ourselves back into an era without antibiotics. The World Health Organization (WHO) reports the high rates of resistance to antibiotics commonly used to treat serious bacterial infections, and the Centers for Disease Control and Prevention (CDC) estimates that in the United States more than 2.8 million people contract infections that are caused by microorganisms resistant to one or more of the prescribed antibiotics (61, 62). Although antibiotic resistance is on the increase, challenges faced by drug discovery programs have led to an antibiotic discovery void, with the introduction of very few new antibiotic classes in the last 2 decades (63). However, new technologies in bioinformatics, structural and chemical biology, and high-throughput screening techniques can aid in novel antibiotics making it into the drug discovery pipeline.
Lantibiotics are attractive antimicrobials as they are active at low concentrations and mostly target high-value targets, such as lipid II (Fig. 2). The majority of lantibiotics bind to the cell wall precursor lipid II, preventing cell wall biosynthesis and facilitating the disruption of the bacterial membrane (64). As an example, the prototypical lantibiotic nisin binds to the pyrophosphate moiety of lipid II, with its two N-terminal rings crucial for this interaction (65). Formation of the pore complex results in cell membrane permeabilization and dissipation of the proton motive force (64). The ability of certain globular lantibiotics such as epidermin-like lantibiotics to form pores is dependent on membrane thickness. These peptides are much shorter than nisin-like lantibiotics and cannot form pores in cell membranes exceeding 40 Å (66). However, due to their ability to bind to lipid II, they are still able to disrupt cell wall biosynthesis.
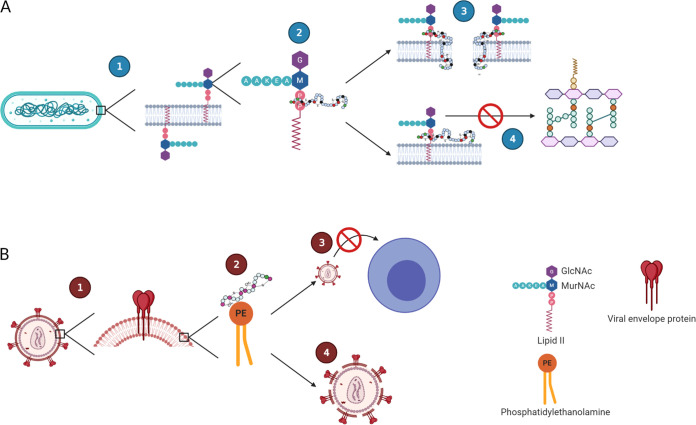
FIG 2.
Lanthipeptide antimicrobial and antiviral modes of action. (A) Antimicrobial mode of action of lipid II binding lantibiotics. The peptidoglycan precursor lipid II (1) is the binding site for lipid II binding lantibiotics, which bind to the pyrophosphate cage of lipid II (2). Once bound, the lantibiotics can undergo a conformational change, resulting in insertion into the membrane and subsequent pore formation (3). Additionally, binding of lantibiotics to lipid II can result in inhibition of cell wall biosynthesis (4). (B) Proposed antiviral mechanism of lanthipeptides. Envelope viruses harbor lipids in their virus envelope derived from the host cell membranes, including phosphatidylethanolamine (PE) (1). Lanthipeptides effective against envelope virus bind to PE distributed on the outer virus envelope (2), which can interfere with viral entry into host cells (3) and virolysis (4). The figure was created with BioRender.
Lantibiotics are mainly produced by Gram-positive organisms and as a result are very effective against closely related Gram-positive bacteria, with limited activity against Gram-negative bacteria. This is due to the structural design of the of Gram-negative bacterial outer membrane, which prevents access to the peptidoglycan layer (home of lipid II) and cytoplasmic membrane. Furthermore, the anionic cell surface of Gram-negative bacteria results in binding of the cationic lantibiotics, where such an interaction potentially increases the stability of the outer membrane through electrostatic interactions (67). This is not surprising considering that most lantibiotics are produced by Gram-positive bacteria. Despite this, lantibiotics have shown promising activity against several of the pathogens on the WHO priority list (63). As such, lantibiotics have been exhaustively tested in vitro against various bacteria, with several also tested for their in vivo efficacy (Table 1). This review discusses some of these examples regarding lantibiotic antimicrobial activity and other potential bioactivities to illustrate the versatility of lantibiotics/lanthipeptides in their application for human health.
LANTIBIOTICS AGAINST GRAM-POSITIVE BACTERIA
Staphylococci and streptococci.
Some of the most promising in vivo antimicrobial results for lantibiotics have been obtained against staphylococci. Staphylococci represent one of the main genera represented on the skin, either as commensals or pathogens, with Staphylococcus aureus most commonly presenting as a pathogen. Unsurprisingly, the vast majority of skin and soft tissue infections (SSTIs) are caused by S. aureus and are usually associated with boils, abscesses, carbuncles and localized wound sepsis (68).
Several lantibiotics are effective against even antibiotic-resistant S. aureus in vitro and in vivo (27–34, 69). However, there are limited published studies evaluating lantibiotics as topical treatments. A relatively recent study used a promising system comprising the incorporation of nisin into nanofibers (69). This approach proved to be effective in the treatment and prevention of S. aureus in mice. After 7 days (with dressings changed on days 2 and 4), the nisin-eluting nanofibers reduced the amount of viable S. aureus in wounds to 4.3 × 102 CFU/wound, compared to 2.2 × 107 CFU/wound for the control nanofibers. The nisin-eluting and control nanofibers also had a positive effect on wound healing and closure compared to the gauze control, with up to 90% wound closure compared to 74% closure, respectively. The use of nanofibers as a delivery vehicle for lantibiotics has potential as it provides controlled release over a prolonged period as well as provides a scaffold for improved wound healing. In another study, nisin, clausin, and the two-component lantibiotic amyloliquecidin were shown to be just as effective as a commercially available antimicrobial (mupirocin) in the treatment of S. aureus-induced skin infections in vivo (27). Lantibiotics were administered as polyvinyl alcohol formulations and along with the mupirocin control were able to significantly reduce the viable S. aureus numbers in wounds to 2.1 × 104 CFU/wound for mupirocin, 6.5 × 104 CFU/wound for amyloliquecidin, and 1.6 × 105 CFU/wound for nisin and clausin after 7 days (treatments on days 2, 4, and 6) compared to the control treatment (1.4 × 107 CFU/wound). In this study, the positive effect on wound closure was also observed, indicating a beneficial effect, potentially involving the activation of the hosts’ immune system (46, 47). An alternative mechanism for the delivery of lantibiotics may be via administration of an organism producing them as a probiotic. It is hypothesized that lantibiotics produced by commensal bacteria may play a role in maintaining microbial balance, through direct antimicrobial activity (reviewed in reference 70). This was demonstrated, to some extent, using lantibiotic-producing staphylococcal commensals to provide resistance against colonization by S. aureus (71). The authors identified antimicrobial-producing coagulase-negative Staphylococcus (CoNS) isolates collected from healthy human skin. These isolates produced known lantibiotics, including epidermin and Pep5, as well as the novel lantibiotics Sh-lantibiotic-α and Sh-lantibiotic-β produced by Staphylococcus hominis A9. Using this lantibiotic-producing strain, complete eradication of S. aureus colonized on the backs of mice could be achieved after twice-daily applications for 1 week. Furthermore, these lantibiotics were shown to synergize with the human cationic peptide LL-37, increasing activity against S. aureus 16- and 32-fold for Sh-lantibiotic-α and Sh-lantibiotic-β, respectively. To further investigate the potential of CoNS to treat skin diseases associated with S. aureus, such as atopic dermatitis (AD), the authors used formulations of antimicrobial-producing CoNS in autologous microbiome skin transplants. Using their formulation, the amount of S. aureus that could be isolated from the skin of AD patients was significantly reduced after 24 h. These results illustrate that the use of lantibiotic-producing strains for autologous microbiome skin transplants holds potential and should be investigated further. However, although the lantibiotic-producing capability of commensal staphylococci may provide a host benefit, the opposite may also be true for pathogenic strains. An example is growth attenuation of S. aureus in a mouse abscess model through the disruption of the lantibiotic gene, indicating a potential role in pathogenesis (72). In both of these cases, antimicrobial activity undoubtedly plays a role in terms of competitive exclusion: their potential immune-regulatory roles cannot be overlooked.
In addition to their application in the treatment of topical S. aureus infections, several lantibiotics have shown potential for use in other applications related to staphylococcal infections. This includes both preventative and therapeutic approaches to protect against Staphylococcus colonization and formation of biofilms on medical devices, such as catheters, cardiac devices, and prosthetic implants, which can complicate treatment (73–75).
Lantibiotics are susceptible to low bioavailability when used systemically: perhaps due to the ability of some to activate the immune system, resulting in rapid degradation/inactivation in vivo, or the binding to host components (29, 40, 47, 76). Despite this, some success has been reported for the systemic use of lantibiotics for the treatment of infections. For example, the class I lantibiotic microbisporicin (NAI-107) has shown promise, with equivalent or superior activity compared to reference treatments (e.g., penicillin, vancomycin, and linezolid) in methicillin-resistant S. aureus (MRSA) and glycopeptide-intermediate S. aureus infections (28, 29). In a rat granuloma pouch model, two 20-mg/kg doses (at 12 or 24 h) of microbisporicin, administered directly after infection with MRSA, resulted in a 3-log reduction in bacterial cell counts after 72 h, with no regrowth up to 96 h (29). Similar results were also obtained in a rat endocarditis model, and in both models, microbisporicin performed similar to or better than treatment with 100 mg/kg vancomycin (29). Interestingly, microbisporicin was more effective when administered intravenously compared to subcutaneously (1.5 to 2.5 times higher), suggesting that not all lantibiotics are equally capable of crossing host membrane or tissue barriers (29, 76). Similarly, the class II lantibiotic mersacidin has also shown potential with superior activity over vancomycin against MRSA in an in vivo infection model (50% effective doses [ED50s] of 2.59 to 10.81 mg/kg of body weight and 7.20 to 18.98 mg/kg, respectively) where mice were injected intraperitoneally with lethal amounts of S. aureus (MRSA and methicillin-sensitive S. aureus) and treated subcutaneously (34). Mersacidin also has reduced bioavailability when injected subcutaneously, which only improves after using the more water-soluble potassium mersacidin (34). This supports our interpretation that there may be an interaction between the peptide and host components when administered into tissue rather than intravenously. A combination of mutacin 1140 analogs have recently been used to improve the efficacy of lantibiotics in the treatment of systemic S. aureus infection (77). The two analogs had substitutions at positions 2 (K2A) and 13 (R13A) resulting in improved activity and stability (77). The two analogs were more active than native mutacin, with 2- and 8-fold increases in activity (MIC) against MRSA (S. aureus ATCC 33591) for K2A and R13A, respectively, compared to the native peptide. In vivo stability was also increased, with up to 4.1- and 5.7-fold higher peak plasma concentrations (K2A and R13A variants, respectively) compared to the native mutacin 1140 after 60 min (administered intravenously at 10 mg/kg in mice). Further investigation revealed that the K2A mutacin exhibited the lowest clearance levels and highest AUC (area under the concentration-time curve), while R13A had the longest half-life and highest Vss (volume of distribution at steady state) in vivo. Efficacy against MRSA in a murine systemic infection model also proved promising, with intravenous administration of the combined analogs (1:1 ratio at 10 mg/kg, with 5 mg/kg of each analog), resulting in 100% survival of animals after 5 days, compared to 100% mortality in the vehicle group. Furthermore, bacterial loads in the liver and kidneys were significantly reduced compared to the vehicle group. Using a combination of either different lantibiotics or analogs with distinct pharmacokinetic and activity profiles can be advantageous, as this can increase both efficacy as well as antimicrobial spectrum in vivo.
Several lantibiotics have also proved effective against pathogenic streptococci. Streptococci include several pathogenic strains and are divided into alpha- and beta-hemolytic streptococci. Alpha-hemolytic streptococci includes Streptococcus pneumoniae, which is the cause of pneumococcal infections, including otitis media, sinusitis, pneumonia, and meningitis. Mice infected intraperitoneally with S. pneumoniae, at concentrations sufficient to result in death, were treated with either nisin or vancomycin (78). Two intravenous treatments with nisin (0.16 mg/kg) resulted in survival of all animals, whereas the survival of mice treated with vancomycin was only 83% when treated with 1.25 mg/kg, and 100% survival was only achieved after treatment with 2.5 or 5.0 mg/kg. Nisin had low blood and tissue levels (serum half-life of 0.9 h), but this was still sufficient to prevent death of the mice (78). Like microbisporicin, nisin was also more effective when administered intravenously (0.16 versus 2.5 mg/kg for 100% survival). In another study, carboxamides of actagardine were generated (79). The monocarboxamides were more active than other variants in vitro, and a more water-soluble derivative was effective in a murine septicemia model, with effective dose values (ED50, 0.23 to 3.5 mg/kg/day) comparable to those of the reference antibiotics used (0.03 to 26 mg/kg/day) (79). Compared to nisin, the actagardine derivative was, however, eliminated faster, with a serum half-life of 0.3 h (78). Microbisporicin also proved highly effective against S. pneumonia, with ED50 values lower than those of linezolid (0.51 versus 15.9 mg/kg, respectively) (29).
Beta-hemolytic streptococci are divided into groups A and B. Group A streptococci are found on the skin and inside the throat and are responsible for most beta-hemolytic streptococcal infections. Common infections caused by group A beta-hemolytic streptococci (GAS [Streptococcus pyogenes]) include impetigo, cellulitis, pharyngitis, and scarlet fever. Several lantibiotics, including microbisporicin, mersacidin, actagardine, and lantibiotics produced by streptococci (e.g., salivaricin 9 and streptin) are active against GAS (80, 81). The same water-soluble derivatives of mersacidin and actagardine successfully used against S. aureus and S. pneumoniae also proved effective in the treatment of S. pyogenes in vivo (34, 79). Planosporicin (ED50, 3.75 mg/kg) has also been reported to be effective in preventing septicemia caused by S. pyogenes in mice when administered intravenously or subcutaneously (82).
Enterococci.
Enterococcal infections, and specifically those caused by vancomycin-resistant enterococci (VRE), are characterized as a serious threat by the CDC and resulted in 54,500 cases (5,400 deaths) in the United States in 2017 (61). Lantibiotics, including lacticin 3147, nisin, mersacidin, epidermin, haloduracin, amyloliquecidin, clausin, and microbisporicin, have promising in vitro activity against enterococci, including drug-resistant strains (27–30, 33, 35). In vivo efficacy in mice has also been illustrated for microbisporicin, where intravenous and subcutaneous administration exhibited the lowest ED50 values (2.3 and 2.8 mg/kg) against two VRE strains compared to linezolid (5.1 and 22.4 mg/kg) (29). Recently, a novel approach using synthetic biology was used to generate a peptide with two lipid II binding motifs (N-terminal domain of nisin and C-terminal domain of haloduracin α) (83). This variant, termed TL19, displayed increased activity (MIC, 0.9 to 15 μM) against several multidrug-resistant (MDR) Enterococcus faecium strains in vitro compared to nisin (MIC, 1.9 to 3.8 μM) and the C-terminally truncated nisin(1–22) variant (MIC, 30 to 60 μM). Although this variant does not form pores, the additional lipid II binding motif is sufficient to counteract this. Using synthetic biology approaches such as this can be invaluable to generate more effective and stable lantibiotics.
Similar to lantibiotic-producing strains effective against S. aureus on skin, lantibiotic-producing probiotics have also shown promise in inhibiting VRE colonization of the gastrointestinal tract (GIT) (84). Kim et al. (84) used a four-strain formulation consisting of Clostridium bolteae, Blautia producta (nisin-like lantibiotic producer), Bacteroides sartorii, and Parabacteroides distasonis and reported significant reductions in VRE growth in the gastrointestinal tract of mice compared to the controls (phosphate-buffered saline [PBS] and consortia without a lantibiotic producer). Interestingly, the use of consortia was found to be essential for colonization of the lantibiotic-producing Blautia producta strain (85). In the same study, Kim et al. also showed that microbiomes with a high abundance of lantibiotic genes were associated with a lower abundance of Enterococcus faecium (84). Furthermore, colonization of germfree mice with microbiomes containing a great abundance of lantibiotic genes resulted in significantly smaller amounts of VRE compared to microbiomes with a low abundance of lantibiotic genes. These results further illustrate the potential of using lantibiotic-producing strains as a delivery vehicle for the treatment or prevention of diseases caused by pathogenic bacteria.
Although VRE is a threat, the emergence of carbapenem-resistant enterococci (CRE)—which are resistant to all treatments evaluated to date—is even more serious (61). To our knowledge, lantibiotics have not been specifically tested against CRE, but given the beneficial effects reported in the context of VRE, investigation of lantibiotic activity against these strains is warranted.
Clostridia.
An important foodborne pathogen, involved in severe GIT infections, is Clostridium difficile. Clostridium difficile-associated diarrhea (CDAD) is one of the major causes of hospital-associated diarrhea, with more than 220,000 hospitalizations in 2017 in the United States alone, with at least $1 billion in excess medical costs (61). Current treatment of CDAD includes oral administration of vancomycin and metronidazole; however, vancomycin treatment can lead to secondary colonization of VRE in the GIT or even the spread of vancomycin resistance within a hospital environment. Several lantibiotics are effective against C. difficile, with some preventing spore outgrowth (28, 36–40). Prevention of spore outgrowth can help in curbing the growth and spread of C. difficile, which may contribute to the successful treatment of CDAD. Actagardine (NVB-302) is currently being developed for treatment of C. difficile and has successfully completed phase I clinical trials. In an in vitro GIT infection model, actagardine compared well with vancomycin in the treatment of C. difficile, with less deleterious effects on Bacteriodes fragilis (a GIT commensal) (38). Combination treatment with actagardine and ramoplanin was especially effective against multiple C. difficile strains (39). The two-component lantibiotic lacticin 3147 has also shown in vitro potential for use as a treatment for CDAD (36). In a fecal fermentation model, it completely eliminates C. difficile (36). However, in a porcine model, neither of the lacticin 3147 peptides could be detected in digesta of pigs 2 h after oral administration (86). Use of the producing strain has also been investigated (87). While the strain was capable of surviving passage through the GIT, no lacticin 3147 or antimicrobial activity could be detected in the feces of pigs, with the producer strain also incapable of preventing Listeria monocytogenes infection in mice. This further illustrates a potential pitfall of using a peptide antibiotic, which may be prone to proteolytic degradation. Degradation can potentially be addressed by encapsulation of the peptides or by using a genetically tailored probiotic strain overexpressing the peptide (69, 88, 89). Additionally, stability is an important factor to consider when evaluating two-component lantibiotics for therapeutic use, as this can be different for the respective peptides.
More recently, promising in vivo results have been reported for various variants of mutacin 1140 (40, 89, 90). Of specific interest is variant OG716 (amino acid substitutions Phe1Val and Arg13Asn), showing superior activity against C. difficile in hamster models of CDAD (90, 91). Oral administration of OG716 three times a day (days 2 to 5) resulted in 100% survival of animals, with C. difficile spore and toxin levels near or below detection limits. Furthermore, considering the sizes of OG716 (2.2 kDa) and vancomycin (1.4 kDa), the ED50 values of OG716 are very promising (10.97 and 13.3 μmol/kg/day for OG716 and vancomycin, respectively) (91). Importantly, the various mutacin 1140 variants had low toxicity against human cell lines and in animal models (40, 89, 91). As mentioned earlier, the stability of these peptides in the GIT is an important aspect to consider, and although amino acid substitutions resulted in increased stability of mutacin 1140 variants, they are still susceptible to proteolytic degradation (40, 90). To address this, the use of target-specific enteric-coated capsules is currently being investigated (89).
Mycobacteria.
Mycobacterium tuberculosis is the causative agent of the respiratory tract infection known as tuberculosis. Worldwide an estimated 10 million people have contracted the disease, with a fatality rate of 11 to 15% (92). Cases of multiple- and extensively-drug-resistant M. tuberculosis place an immense burden on the efforts to try and control the spread of M. tuberculosis, especially in developing countries (92). The unique cell wall and slow-growth nature of M. tuberculosis may make it difficult for lantibiotics (and other treatments) to exert antimicrobial activity. However, the ability of lantibiotics to bind to lipid II gives them an advantage over treatments such as rifampin, which need to be transported across the plasma membrane. The lipid II structure of mycobacteria does differ from those of other bacteria, due to modifications on both N-acetylmuramic acid (MurNAc) and the peptide side chain (93). Despite these differences, nisin has activity against the nonpathogenic mycobacteria M. smegmatis and M. bovis, with intracellular ATP leakage and dissipation of the proton motive force: additionally, hinge mutants were shown to have enhanced activity (94, 95). Nisin, CMB001, and lacticin 3147 are also active against clinical mycobacterial isolates in vitro, with lacticin 3147 (MIC90, 7.5 μg/ml) and CMB001 (MIC, 0.3 μg/ml) showing the best activity against M. tuberculosis (33, 96). Although these lantibiotics have potential, in vivo studies are still required, and an appropriate delivery system still needs to be developed to reach M. tuberculosis residing within tissues. For example, in the context of M. tuberculosis-infected macrophages in the distal lung, promising results have been reported for the in vivo efficacy of class IIa bacteriocins complexed with phosphatidylcholine-cardiolipin liposomes (97). Given the rise of drug-resistant mycobacteria, further research is warranted to establish the feasibility and use of lantibiotics as an antimycobacterial treatment.
LANTIBIOTIC ACTIVITY AGAINST GRAM-NEGATIVE BACTERIA
Lantibiotics are not particularly active against Gram-negative bacteria, mainly due to the inability of the peptides to cross the outer membrane of these organisms, but several reports of their limited antimicrobial activity are available. This includes activity against Helicobacter pylori, Neisseria meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae, Campylobacter jejuni, Serratia marcescens, Proteus vulgaris, and Escherichia coli (98–100). Activity against Gram-negative bacteria has been shown to be improved by combination with colistin (polymyxin E) or addition of chelating agents such as EDTA (30, 99). Recently a nisin-like lantibiotic (CMB001) with a similar structure to subtilin was shown to have in vitro activity (10 to 15 μg/ml) against MDR Acinetobacter baumannii (33). Furthermore, studies have also shown that lantibiotic (specifically nisin) fusion with peptides capable of penetrating Gram-negative outer membranes results in increased potency (100). Addition of these penetrating peptides to the C-terminal domain of nisin resulted in a significant improvement (up to 12-fold) in activity against several MDR Gram-negative pathogenic bacteria (100). The potential of bioengineering lantibiotics with increased activity against Gram-negative pathogens may further expand the antimicrobial arsenal of lantibiotics. Additionally, the immunomodulatory properties of some lantibiotics can also result in their indirect activity against Gram-negative pathogens (47).
RESISTANCE AGAINST LANTIBIOTICS
The discovery of novel antibiotics is limited by several steps, including target selection, which is important if a novel antibiotic is to remain effective for a prolonged period after administration. In this respect, several lantibiotics target cell wall components, including lipid II, and/or possess dual modes of action. Despite this, lantibiotics, as with any antibiotic, are still susceptible to the development of resistance, and this is an important aspect to consider when evaluating their application as antimicrobial therapies. Changes in the cell wall or alterations in membrane composition are some of the major mechanisms employed by target organisms to protect themselves against lantibiotics.
The cationic nature of lantibiotics is a crucial element in their initial interaction with susceptible bacteria; as such, any change in their charge or the charge of their membrane targets would result in altered attraction. Bacterial membranes are overall negatively charged, and in the case of Gram-positive bacteria, this is because of teichoic acids in their cell wall (101). Due to this anionic nature, cationic antimicrobial peptides (ctAMPs), including lantibiotics, are attracted to the bacterial cell wall. The dltA operon, found in numerous bacteria, has been identified as an innate defense against lantibiotics (102–104). The different genes in this operon are responsible for the d-alanylation of teichoic acids, resulting in the incorporation of positive charges and consequentially alternating the electrostatic interaction of ctAMPs with the cells (102, 104). Additionally, it has been proposed that d-alanylation of lipoteichoic acids decreases the flexibility and permeability of the cell wall, protecting the host from ctAMPs (105).
Alterations in the lipid composition of the bacterial cell membrane can also affect the efficacy of lantibiotics. Nisin has been shown to have high affinity for anionic phospholipids (106), such as phosphatidylglycerol and diphosphatidylglycerol, which coincidentally are the most common phospholipids in bacterial membranes (reviewed in reference 107). Changes in the anionic nature of phospholipids in the bacterial membrane would influence resistance. The phosphatidylglycerol lysyltransferase MprF is a mechanism employed by bacteria to alter their membrane charge through alteration of phosphatidylglycerol (108). This protein catalyzes the transfer of a lysine residue from lysyl-tRNAs to phosphatidylglycerol resulting in lysylphosphatidylglycerol. The addition of positively charged l-lysine can subsequently result in the repulsion of ctAMPs, including lantibiotics (108).
Another interesting mode of resistance is that of immune mimicry, whereby resistant target strains harbor an immunity protein (LanI) or immune-specific ABC transporter [LanFE(G)] that confers immunity to a specific lantibiotic (109). Immunity proteins (LanI) and immune-specific ABC transporters [LanFE(G)] are present in most lantibiotic operons and provide protection to the host from its own mature lantibiotic. The immunity protein LanI is a cell-associated lipoprotein and acts by intercepting the mature lantibiotic, rendering it inert (109, 110). The ABC transporters LanFE(G) act through transporting mature lantibiotics from the membrane, thereby preventing pore formation (109). Usually cross-immunity is rare as these immunity elements are specific to a certain lantibiotic. However, it has been shown that cross-immunity is possible, even in strains that do not harbor lantibiotic biosynthetic genes (111). Additionally, more nonspecific ABC transporters have also been identified that are capable of conferring resistance (112, 113). In general, these transporters can be grouped as either CprABC- or BceAB-type transporter families (reviewed in reference 114). These observations are concerning as these immunity elements can also potentially be horizontally transferred, resulting in the increased resistance to lantibiotics.
Resistance mechanisms are especially important to consider as lantibiotics migrate into the clinical setting, where such resistance can be devastating. For additional information on these and other lantibiotic resistance mechanisms, the reader is referred to the comprehensive reviews by Clemens et al. from 2018 (114) and Draper et al. from 2015 (115).
OTHER THERAPEUTIC APPLICATIONS OF LANTIBIOTICS AND LANTHIPEPTIDES
In addition to the antimicrobial potential of lantibiotics, lanthipeptides (including lantibiotics) have shown a diverse range of bioactivities. This is likely due to the diverse nature of these peptides and the unique properties that the various PTMs can confer. Examples are presented below to illustrate the diversity of lantibiotic bioactivity.
Immune modulation.
Several ctAMPs (such as LL-37 and α- and β-defensins) play a crucial role in modulating the immune system during infection and injury (116). Most complex species have ctAMPs, which interact with the innate immune system and are generally short overall and positively charged, with a large proportion of hydrophobic residues, making them very similar to lanthipeptides.
Lantibiotics have shown immune-regulatory properties, with nisin, gallidermin, and Pep5 being capable of inducing the release of multiple chemokines at levels similar to that of LL-37, with nisin modulating multiple signaling pathways (47). The protective effect as a result of the immune-modulatory properties of nisin pretreatment provides effective protection for mice infected with Salmonella enterica serovar Typhimurium and E. coli (47). In line with this interpretation, nisin has also been shown to activate neutrophil release of neutrophil extracellular traps (NETs), a mechanism used by neutrophils to trap, contain, and kill pathogenic organisms and which is thought to be particularly important in the immune response to pathogens too large to be destroyed by phagocytosis alone (46), as reviewed in reference 117. The formation of NETs may thus—apart from contributing to the reduced loads of S. Typhimurium and E. coli reported in mice pretreated with nisin—also contribute to immune activity against larger pathogens, such as fungi (118). Although the antimicrobial properties associated with NET formation can be advantageous, chronic NET formation, which entails cellular release of large amounts of free radicals as well as nuclear material such as histones, is also associated with chronic inflammation and increased risk for autoimmune diseases, such as rheumatoid arthritis (reviewed in reference 119). This should be considered when evaluating nisin (or potentially other lanthipeptides) as a chronic therapeutic strategy. In addition to effects on the innate immune system, nisin has also been shown to have effects on the adaptive response in mice (120). This was demonstrated through the addition of nisaplin (a commercially available nisin preparation) to the feed of mice, resulting in the short-term increase of CD4+ and CD8+ cells and T lymphocytes. The nature and significance of this modulation remain to be further elucidated.
Other immune-regulatory properties of lantibiotics come in the form of indirect modulation of phospholipase A2. Phospholipase A2 plays an important role in inflammatory responses, resulting from its role in the release of arachidonic acid. The oxidative metabolism of arachidonic acid results in eicosanoids, such as prostaglandins and leukotrienes, which are strong mediators of the immune system. Cinnamycin-like lantibiotics can indirectly inactivate phospholipase A2 by sequestering phosphatidylethanolamine (PE; substrate for phospholipase A2), thereby having the potential to indirectly mediate inflammatory responses (121). The cinnamycin-like lantibiotic ancovenin is also an inhibitor of angiotensin-converting enzyme (ACE) (122), which is responsible for the poor stability of angiotensin II in circulation and which has been implicated in hypertension and diabetic inflammation and fibrosis (123). On the topic of ACE inhibition and maintenance of angiotensin II function, peptidase-resistant lanthionine-stabilized angiotensin-(1–7), was recently shown to confer benefit in the context of experimental diabetic nephropathy and cerebral stroke (59, 60). Interestingly, streptocollin, which has a similar structure to cinnamycin-like lantibiotics (although forming part of the venezuelin-like class IV lanthipeptides) (Fig. S2G and S4) is unable to inhibit phospholipase A2, although its PE binding has yet to be evaluated (124). This peptide is, however, able to partially inhibit protein tyrosine phosphatase 1B (PTP1B). Protein tyrosine phosphatase 1B is a regulator of various signaling pathways and is best known for its role in insulin signaling, but it also has roles in immune cell signaling (125). Inhibition of PTP1B has been identified as a potential strategy to improve insulin sensitivity and also has therapeutic potential in the treatment of Alzheimer’s disease and diabetes (126).
These results suggest that lantibiotics can interact and modulate the immune system, potentially using similar mechanisms employed by human and other ctAMPs. Additional cell biology research is required to fully understand how lantibiotics/lanthipeptides interact with cells of the immune system. This will give further insight into the roles these peptides might play in host-microbe interaction and potential therapeutic application.
Neuropathic pain relief.
Labyrinthopeptin-like lantibiotics have limited antibacterial activity. However, labyrinthopeptin A2 and NAI-112 have been shown to have antiallodynic and antinociceptive properties in mice (16, 48). Labyrinthopeptin A2 administered intravenously at concentrations ranging from 0.01 to 3.0 mg/kg resulted in significant attenuation of tactile allodynia (ED50, 50 μg/kg). Efficacy remained stable over 6 h posttreatment, with loss of efficacy after 24 h (16). Similarly, NAI-112 was also able to significantly reduce allodynia and hyperalgesia 2 h after administration, albeit at much higher doses (>10 mg/kg) (48). Significant antinociceptive effects could be observed at somewhat lower doses (from 3 mg/kg). At this point, due to differences in experimental procedures, the efficacies of these two lanthipeptides cannot be directly compared. More research is warranted in this context, as the mechanisms of action of these peptides have not been elucidated, although a potential interaction with the vanilloid pathway has been suggested for NAI-112 (48).
Ion channel regulation.
Duramycin has potential in the treatment of cystic fibrosis, which is caused by abnormal chloride ion transport into cells. It has been demonstrated in tissue culture that the efflux of chloride observed after duramycin treatment is associated with a decrease in intracellular calcium levels (49). It was therefore proposed that the efflux of chloride from epithelial cells is likely due to the interaction duramycin has with cell membranes. This is supported by the interaction that duramycin has with PE, whereby it can be deposited into the cell membrane and indirectly affect ion channel function (49–51). Duramycin has undergone phase I and II clinical trials, with phase II clinical trials reporting it to be safe, with overall positive results on the pulmonary function of cystic fibrosis patients (52–54). This effect of duramycin to lower cellular calcium levels may also have broader application in health, given the known association between intracellular accumulation of calcium and a variety of diseases linked to cumulative oxidative damage and chronic inflammation, such as neurodegenerative disease, cancer, accelerated aging, and type II diabetes (reviewed in reference 127).
Anticancer treatment.
Nisin (nisins A and Z) has been shown to be effective in vitro and in vivo against head and neck squamous cell carcinoma (HNSCC) (128, 129). Nisin appears to preferentially induce apoptosis in HNSCC cells in a dose-dependent manner, with minimal effect on primary keratinocytes. This may be due to the structural differences in the plasma membrane, specifically the phospholipid content, of the different cell types. This is supported by the observation that nisin binds to phosphatidylcholine, which is known to be increased in cancer cells (along with PE) (reviewed in references 130, to ,132). The mechanism by which nisin induces apoptosis has been proposed to be calcium dependent (influx of calcium) (129). The subsequent influx of calcium results in the activation of calpain-1, resulting in caspase 3-independent apoptosis (128). This is further supported by the observation that nisin affects plasma membrane integrity through the release of lactose dehydrogenase (LDH) (76). Additionally, the potency of nisin in vitro and in vivo can be further increased in combination with 5-fluorouracil or doxorubicin (133, 134). The involvement of calcium in this context and the increased efficacy by addition of doxorubicin, which is known to exert anticancer effects via induction of free radical damage in cancer cells, again suggest that lantibiotics may have a role in the modulation of redox status, although the nature of this modulation varies between lantibiotics.
Duramycin has also shown potential for the treatment of cancer and has been shown to induce apoptosis and reduce proliferation in tumor cells (135, 136). Furthermore, due to its high affinity for PE, duramycin can be more selective toward cancerous cells. An interesting application of duramycin as an anticancer treatment is its fusion to IgG, generating a new duramycin-IgG variant (137, 138). Fusion of IgG to duramycin does not influence its PE binding capability and has the advantage of reducing duramycin cytotoxicity. The IgG fused to duramycin helps guide the host immune cells to apoptotic cells, resulting in enhanced phagocytosis. Furthermore, tumor growth (MethA tumors) is inhibited in mice after treatment with duramycin-IgG (138). Since duramycin binds to PE and the Fc region on (fused) IgG antibodies interacts with phagocytic cells to enhance phagocytosis, duramycin is likely cleared from the site effectively soon after inducing apoptosis in cancer cells, via phagocytosis, which would explain its lower cytotoxicity to surrounding normal cells.
The urokinase plasminogen activator (uPA) is a serine protease responsible for the conversion of plasminogen to plasmin. The urokinase plasminogen activator system has been implicated in activities associated with tumor progression and metastasis and has been identified as a potential target for anticancer therapy (reviewed in reference 139). Using a phage display system, Urban et al. screened a lanthipeptide library for peptides capable of binding to uPA (57). Using this system, they identified several novel lanthipeptides capable of inhibiting the catalytic ability of uPA. The application of these peptides was not specifically evaluated for their anticancer capabilities, but they do show potential. More importantly, this study has highlighted techniques that could vastly increase the efficiency with which potential candidates may be screened for anticancer activity or, in fact, other bioactivities.
Antiviral capacity.
Various lantibiotics, including nisin, labyrinthopeptin, and duramycin, have been evaluated for their antiviral properties (42–45, 140, 141). Of interest are the antiviral properties of labyrinthopeptin A1 and duramycin, which show antiviral activity through their ability to bind PE (Fig. 2). The PE binding capability of duramycin has proven useful once again, with duramycin being able to inhibit the entry of filo- and flaviviruses into host cells (42, 141). Phosphatidylethanolamine is a ligand for the T-cell Ig mucin domain (TIM) protein TIM1, and together they are involved in phosphatidylserine (PS)-dependent phagocytosis of apoptotic cells (42). Additionally, TIM family proteins have also been shown to promote infection of enveloped viruses as a result of virion lipid content, specifically PS and PE (42). Duramycin was therefore evaluated for its ability to inhibit TIM1-mediated virus entry through blocking virus attachment to TIM1 (42). Duramycin was reported to be most effective at the entry phase of the viral infection, with no effect observed when duramycin was administered postinfection, and was effective in inhibiting viral entry into human TIM1-expressing cells (hTIM1-293T) as well as cells naturally expressing TIM1 (Vero and A549 cells). This inhibitory effect was shown to be specific for TIM1-mediated entry of viruses through interaction of duramycin with PE present in the viral membrane (42). Through inhibition of Zika virus binding to TIM1, duramycin has also been shown to be effective in reducing infection of placental cells and explants (141).
Labyrinthopeptin A1 has shown promising antiviral activity against several viruses, including human immunodeficiency virus (HIV) and herpes simplex virus (HSV), with activity against laboratory-adapted strains and clinical isolates (including drug-resistant strains) (43). Labyrinthopeptin was able to inhibit cell-free viral infection as well as inhibit cell-to-cell spread of HIV in vitro. This inhibitory activity was dependent on time of drug administration and was only effective if the drug was administered within 1 h after infection. Like duramycin, these results suggest that labyrinthopeptin A1 likely also interferes with the viral entry process. Labyrinthopeptin A1 was shown to interact with the virus (interaction with envelope protein gp120) and not receptors on the host cell. However, binding to the virus is highly likely to be via interaction with lipids (specifically PE) in the viral membrane (44). A moderate degree of synergism when combined with other commonly used antiretroviral therapies was seen (43). An advantage of not having significant antibacterial activity is that labyrinthopeptin A1 does not have a negative effect on host microbiota, such as vaginal lactobacilli, reducing the risk of dysbiosis (43). Importantly, labyrinthopeptin A1 did not stimulate targeted immune cells (peripheral blood mononuclear cells [PBMCs]), as expression of CD69 and CD25 remained unchanged and did not result in significant induction of inflammatory cytokine secretion from these cells (43). Additionally, labyrinthopeptin A1 was not cytotoxic against vaginal epithelial cells or other nonepithelial cells at effective antiviral concentrations (43).
Labyrinthopeptin A1 and A2 have also been tested against a variety of other enveloped viruses, with labyrinthopeptin A1 being the most effective, conferring broad-spectrum antiviral activity (44). Of interest is the observation that labyrinthopeptins bind to PE and may be responsible for labyrinthopeptin binding to viral membranes. Furthermore, it was shown that the antiviral effect was a result of virolysis (viral membrane disruption), although similar effects on TIM1-mediated entry to those reported for duramycin cannot be excluded (42, 44). Additionally, labyrinthopeptins are effective against respiratory syncytial virus (RSV) in vitro and have shown promising results in vivo (45). Moreover, the labyrinthopeptins are not affected by resistance mutations usually detrimental toward RSV entry inhibitors. The mode of action against RSV is similar to that reported for other viruses (i.e., interaction with the virus-associated PE) (45). Although promising results were reported using an in vivo murine model, treatment was not as effective compared to in vitro models and requires additional research (45).
Efforts at generating lanthipeptides new to nature have also shown promise for generating lanthipeptides capable of inhibiting HIV budding from cells (142). A bacterial reverse two-hybrid (BRTH) system was used to screen potential lanthipeptide analogues for their ability to inhibit the interaction of the HIV p6 protein with the ubiquitin E2 variant (UEV) domain of human TSG101 (important for budding of HIV from infected cells). Using prochloricin A2.8 as a backbone, the amino acids between the two rings were randomized and modification performed by ProcM (LanM). In vitro testing using the BRTH system resulted in the identification of one peptide, XY3-3, capable of disrupting the interaction between HIV p6 and UEV. Further in vitro testing revealed that the lanthipeptide had more than 10-fold increase in activity compared to a previously identified inhibitor and specifically binds to UEV (142, 143). Both lanthionine bridges were also shown to be crucial for activity. To access the peptides’ ability to prevent Gag-mediated budding of virus-like particles in cell-based assays, the cell-penetrating Tat peptide was fused to the N terminus of XY3-3. The newly generated XY3-3-Tat was not toxic to cells at concentrations up to 500 nM and inhibited viral budding by 65% at 100 nM. The peptide interfered with the degradation of the epidermal growth factor receptor (at 500 nM), which is mediated by the UEV domain of TSG101, further supporting binding of XY3-3-Tat to UEV. Although further testing is required, lanthipeptides such as XY3-3-Tat and labyrinthopeptins may prove useful in antiviral therapy. Furthermore, methods based on BRTH and phage display systems provide a platform for identifying and testing novel lanthipeptides (57, 142).
HETEROLOGOUS EXPRESSION OF LANTHIPEPTIDES
Most studies investigating lanthipeptides require pure peptide at relativity high yields. This can be troublesome as some of the producing strains suffer from low production yields and production of other contaminating peptides. Furthermore, for a lanthipeptide to be commercially viable, industrial-scale production would need to be feasible. Additionally, lanthipeptides that are identified by genome mining are not always readily produced by the native producer or the native producer is not available (144). Furthermore, regulation of lanthipeptide and associated gene expression can be very complex and, in some cases, has not been fully elucidated. The design and expression of new-to-nature lanthipeptides are also not possible without the use of a heterologous expression system or chemical synthesis. Several heterologous expression systems have been investigated to produce lanthipeptides (Table 2). An important feature of any lanthipeptide expression system is the inclusion of the appropriate modification enzymes. In most cases, the precursor peptide is expressed to neutralize the bioactivity of the core peptide, reducing potential toxic effects of the core peptide against the expressing host. Furthermore, addition of affinity tags, such as multiple histidines, aids in purification via affinity chromatography.
As the native producer of nisin, it is not surprising that Lactococcus lactis has been used as a heterologous host to produce lantibiotics (Table 2). Additionally, the genes involved in the regulation of nisin expression have also been incorporated into a commercially available expression system, namely, the nisin-controlled gene expression system (NICE). In the native nisin-producing bacterium L. lactis, nisin biosynthesis is autoregulated by a two-component regulatory system made up of NisK (histidine sensor kinase) and NisR (transcriptional activator) (145). Mature nisin acts as its own inducing peptide, with NisK acting as its receptor. Once nisin is bound to NisK, a signal transduction cascade is initiated that results in the autophosphorylation of NisK. This subsequently results in the transfer of phosphate to NisR, which binds to the promoter regions in the nisin operon resulting in induced expression. This regulatory machinery is extremely sensitive and tightly controlled, which makes it ideal for use in a heterologous expression system. Using L. lactis as the heterologous host, yields of ∼6.0 mg/liter (precursor peptide) have been reported (146). L. lactis peptides have been expressed from class I and II by using modifications of enzymes from both classes (147). A rapid screening system (nanoFlemming) using L. lactis has also been developed capable of assessing peptide libraries at nanoliter scale (58). Systems like these allow for the rapid screening of large peptide libraries that can significantly streamline the discovery pipeline.
Escherichia coli is a molecular workhorse, and a wealth of resources are available, ranging from different expression strains, cloning tools, and expression systems reviewed in reference 148. Due to these advantages, E. coli has been utilized as a heterologous expression host to produce lanthipeptides from all four classes (Table 2). From these studies, it seems that the multifunctional synthetases (LanM, LanKC, and LanL) are simpler to express in E. coli than the dedicated class I synthetases (i.e., LanB and LanC). This may be due to the increased complexity of modification, specifically the requirement for two dedicated modification enzymes. Shi et al. (149) were the first to report successful expression of a class I lantibiotic in E. coli with in vivo modification of precursor nisin by LanB and LanC. Using this system, a yield of 24 mg/liter modified precursor nisin (13.8 mg/liter core peptide theoretical) was obtained. To further improve on this yield, the inclusion of optimized tRNAGlu sequences and glutamyl tRNA synthetase can increase the efficiency of class I modification enzymes (9). Shi et al. (149) also successfully expressed the class II lanthipeptides haloduracin (Halα and Halβ) and prochlorosins (1.7, 2.11, 3.2, and 3.3), modified using their respective LanM synthetases. Yields of the various precursor prochlorosins ranged from 10 to 35 mg/liter and fully modified haloduracin precursor peptides from 1 to 2 mg/liter (149). Previously, precursor nukacin ISK was produced in E. coli at 1.5 mg/liter, which is significantly less than prochlorosins but similar to haloduracin (150). This seems to indicate that the type of lanthipeptide as well as the modification enzymes used can influence production yields. Interestingly, prochlorosins were obtained from the soluble fraction when modified, but are mostly in the insoluble fraction when ProcM (LanM) is absent (149). Sequestration of heterologously expressed peptides/proteins to inclusion bodies is a potential limitation (151). To address this, fusion of precursor peptides to “solubility enhancers” has been investigated, including green fluorescent protein (GFP) and mannose-binding protein (MBP) (56, 151, 152). Using these larger fusion tags has been shown to improve stability and solubility of heterologously expressed proteins and can reduce toxicity to the heterologous host (56, 151–153). Importantly, fusion to these larger proteins does not interfere with the modification and may improve the contact time of the precursor peptides with their respective synthetases. An additional advantage of using GFP is its fluorescence, which can help in optimization of expression through real-time in vivo monitoring of expression and evaluation of purification (151–153). Although using these larger fusion proteins can result in reductions of final lanthipeptide yield, further optimization of these systems is required to unleash their full potential. Another possibility is to secrete the lanthipeptides outside the cell. This has been done successfully for the two-component lantibiotic lichenicidin, with yields of 4 and 6 mg/liter for the alpha- and beta-peptides, respectively (154). In this example, secretion and cleavage are performed by the bifunctional LanT (LicT), with additional cleavage performed by an extracellular protease LanP (LicP). This system has the advantage of not having to cleave the precursor peptides in vitro after purification, but removes the ability to utilize affinity chromatography during initial purification. Secretion can, however, still be performed without leader peptide cleavage, using E. coli secretion systems (e.g., the twin-arginine translocation [TAT] pathway) or by removing/disrupting the protease domain of the bifunctional LanT (if using a bifunctional LanT) (155).
Streptomyces spp. are known for their production of secondary metabolites and RiPPs, making them an intriguing platform for heterologous lanthipeptide production (156, 157). Streptomyces spp. have been used for the heterologous production of lanthipeptides represented in three of the four classes (II to IV), including cinnamycin, labyrinthopeptins, and streptocollin (124, 158, 159). Using Streptomyces lividans, fully modified labyrinthopeptides could be produced at yields of 86 and 14 mg/liter for labyrinthopeptin A1 and A2, respectively (158). These values are lower than those produced by the wild type (90 and 36 mg/liter for A1 and A2, respectively), but it should be noted that this process was not optimized, and more importantly, copurification of closely related peptides is eliminated when the peptides are heterologously expressed (43, 158). Similarly, actagardine was also successfully expressed in S. lividans, with yields of 50 to 80 mg/liter (159). Expression of streptocollin in Streptomyces coelicolor also proved fruitful, yielding a 5.5-fold increase in production over the wild-type strain (from 1.8 mg/liter to 10 mg/liter) (124).
In addition to the use of these expression systems to produce lanthipeptides, they can also be used for fundamental studies and as tools in the drug discovery pipeline. To this end, heterologous expression has been used in several studies to investigate fundamentals of lanthipeptides, including structure-function relationships, gene function, and regulation of expression. This is, in part, made possible by genetic tools available for heterologous hosts, such as E. coli and L. lactis, which allow for specific manipulation of genes involved in the modification and processing of the peptides or to easily make changes to peptide structure and study the effect on bioactivity. The refactoring of promoters and transcriptional units for target gene clusters can be used to improve expression in heterologous hosts and facilitate the expression of pathways that are silent in the producing strain. Furthermore, rapid screening methods have also been developed and are essential for testing of large libraries of potential lanthipeptides (56–58).
CONCLUDING REMARKS
Lantibiotics (and lanthipeptides in general) represent a diverse range of peptides and make up the largest group of RiPPs. Due to this diversity, a plethora of bioactive peptides have been discovered, with activities as diverse as the peptides themselves. Recent studies illustrating the widespread nature of lanthipeptides in bacterial genomes are promising, signifying an untapped source of potential biotherapeutics with novel mechanisms of action. This is especially important in current times, given the rise in resistance toward available therapeutic interventions. It is therefore promising to see the significant advances in the lantibiotic/lanthipeptide discovery pipeline over the past few years. These include methods for rapid evaluation of large lanthipeptide libraries and development of more effective production systems.
However, there remains an innovation chasm between academic research and commercialization of lanthipeptides. To help bridge this gap, future studies should focus on identifying potential applications of novel peptides and evaluating their modes of action. Additionally, increased focus should be placed on in vivo assessment to help identify and address shortcomings, such as low bioavailability. Another aspect that requires further innovation is the production of lanthipeptides. Despite significant steps being made in the heterologous expression of lanthipeptides, the complex nature of their PTMs and low production yields remain a hurdle. Future research needs to focus on the fine-tuning of expression systems to produce lanthipeptides at feasible yields and see lanthipeptides enter the commercial market.
Footnotes
Supplemental material is available online only.
Contributor Information
Anton Du Preez van Staden, Email: advstaden@outlook.com.
Robert M. Kelly, North Carolina State University
REFERENCES
- 1.Schnell N, Entian K-D, Schneider U, Götz F, Zähner H, Kellner R, Jung G. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278. 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 2.Weil HP, Beck-Sickinger AG, Metzger J, Stevanovic S, Jung G, Josten M, Sahl HG. 1990. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur J Biochem 194:217–223. 10.1111/j.1432-1033.1990.tb19446.x. [DOI] [PubMed] [Google Scholar]
- 3.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khusainov R, Moll GN, Kuipers OP. 2013. Identification of distinct nisin leader peptide regions that determine interactions with the modification enzymes NisB and NisC. FEBS Open Bio 3:237–242. 10.1016/j.fob.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegemann JD, van der Donk WA. 2018. Investigation of substrate recognition and biosynthesis in class IV lanthipeptide systems. J Am Chem Soc 140:5743–5754. 10.1021/jacs.8b01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repka LM, Chekan JR, Nair SK, van der Donk WA. 2017. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev 117:5457–5520. 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegemann JD, Süssmuth RD. 2020. Matters of class: coming of age of class III and IV lanthipeptides. RSC Chem Biol 1:110–127. 10.1039/D0CB00073F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega MA, Hao Y, Walker MC, Donadio S, Sosio M, Nair SK, van der Donk WA. 2016. Structure and tRNA specificity of MibB, a lantibiotic dehydratase from actinobacteria involved in NAI-107 biosynthesis. Cell Chem Biol 23:370–380. 10.1016/j.chembiol.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krawczyk B, Völler GH, Völler J, Ensle P, Süssmuth RD. 2012. Curvopeptin: a new lanthionine-containing class III lantibiotic and its co-substrate promiscuous synthetase. ChemBioChem 13:2065–2071. 10.1002/cbic.201200417. [DOI] [PubMed] [Google Scholar]
- 11.Müller WM, Schmiederer T, Ensle P, Süssmuth RD. 2010. In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew Chem Int Ed Engl 49:2436–2440. 10.1002/anie.200905909. [DOI] [PubMed] [Google Scholar]
- 12.Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. 2010. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol 8:e1000339. 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimafuji C, Noguchi M, Nishie M, Nagao J, Shioya K, Zendo T, Nakayama J, Sonomoto K. 2015. In vitro catalytic activity of N-terminal and C-terminal domains in NukM, the post-translational modification enzyme of nukacin ISK-1. J Biosci Bioeng 120:624–629. 10.1016/j.jbiosc.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, van der Donk WA. 2015. Michael-type cyclizations in lantibiotic biosynthesis are reversible. ACS Chem Biol 10:1234–1238. 10.1021/acschembio.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, van der Donk WA. 2012. Biosynthesis of the class III lantipeptide catenulipeptin. ACS Chem Biol 7:1529–1535. 10.1021/cb3002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Gühring H, Vértesy L, Wink J, Hoffmann H, Brönstrup M, Sheldrick GM, Süssmuth RD. 2010. Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew Chem Int Ed Engl 49:1151–1154. 10.1002/anie.200905773. [DOI] [PubMed] [Google Scholar]
- 17.Pag U, Sahl H-G. 2002. Multiple activities in lantibiotics—models for the design of novel antibiotics? Curr Pharm Des 8:815–833. 10.2174/1381612023395439. [DOI] [PubMed] [Google Scholar]
- 18.Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Inger C, Abildgaard M, Nes IF. 1994. In vivo conversion of l-serine to d-alanine in a ribosomally synthesized polypeptide. J Biol Chem 269:27183–27185. 10.1016/S0021-9258(18)46966-9. [DOI] [PubMed] [Google Scholar]
- 19.Kupke T, Kempter C, Gnau V, Jung G, Götz F. 1994. Mass spectroscopic analysis of a novel enzymatic reaction. Oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J Biol Chem 269:5653–5659. 10.1016/S0021-9258(17)37510-5. [DOI] [PubMed] [Google Scholar]
- 20.Ökesli A, Cooper LE, Fogle EJ, van der Donk WA. 2011. Nine post-translational modifications during the biosynthesis of cinnamycin. J Am Chem Soc 133:13753–13760. 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velásquez JE, Zhang X, van der Donk WA. 2011. Biosynthesis of the antimicrobial peptide epilancin 15X and its N-terminal lactate. Chem Biol 18:857–867. 10.1016/j.chembiol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian KD. 1992. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol 58:3730–3743. 10.1128/AEM.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnell N, Engelke G, Augustin J, Rosenstein R, Ungermann V, Götz F, Entian KD. 1992. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem 204:57–68. 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- 24.Klein C, Kaletta C, Schnell N, Entian KD. 1992. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol 58:132–142. 10.1128/AEM.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers LA. 1928. The inhibiting effect of Streptococcus lactis on Lactobacillus bulgaricus. J Bacteriol 16:321–325. 10.1128/JB.16.5.321-325.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker MC, Eslami SM, Hetrick KJ, Ackenhusen SE, Mitchell DA, van der Donk WA. 2020. Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genomics 21:387. 10.1186/s12864-020-06785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Staden ADP, Heunis T, Smith C, Deane S, Dicks LMT. 2016. Efficacy of lantibiotic treatment of Staphylococcus aureus-induced skin infections, monitored by in vivo bioluminescent imaging. Antimicrob Agents Chemother 60:3948–3955. 10.1128/AAC.02938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, Parenti F. 2008. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15:22–31. 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Jabés D, Brunati C, Candiani G, Riva S, Romanó G, Donadio S. 2011. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother 55:1671–1676. 10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunati C, Thomsen TT, Gaspari E, Maffioli S, Sosio M, Jabes D, Løbner-Olesen A, Donadio S. 2018. Expanding the potential of NAI-107 for treating serious ESKAPE pathogens: synergistic combinations against Gram-negatives and bactericidal activity against non-dividing cells. J Antimicrob Chemother 73:414–424. 10.1093/jac/dkx395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruszewska D, Sahl H-G, Bierbaum G, Pag U, Hynes SO, Ljungh Å. 2004. Mersacidin eradicates methicillin-resistant Staphylococcus aureus (MRSA) in a mouse rhinitis model. J Antimicrob Chemother 54:648–653. 10.1093/jac/dkh387. [DOI] [PubMed] [Google Scholar]
- 32.Piper C, Casey PG, Hill C, Cotter PD, Ross RP. 2012. The lantibiotic lacticin 3147 prevents systemic spread of Staphylococcus aureus in a murine infection model. Int J Microbiol 2012:806230–806236. 10.1155/2012/806230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karczewski J, Krasucki SP, Asare-Okai PN, Diehl C, Friedman A, Brown CM, Maezato Y, Streatfield SJ. 2020. Isolation, characterization and structure elucidation of a novel lantibiotic from Paenibacillus sp. Front Microbiol 11:2905. 10.3389/fmicb.2020.598789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee S, Chatterjee DK, Jani RH, Blumbach J, Ganguli BN, Klesel N, Limbert M, Seibert G. 1992. Mersacidin, a new antibiotic from Bacillus. In vitro and in vivo antibacterial activity. J Antibiot (Tokyo) 45:839–845. 10.7164/antibiotics.45.839. [DOI] [PubMed] [Google Scholar]
- 35.Piper C, Draper LA, Cotter PD, Ross RP, Hill C. 2009. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J Antimicrob Chemother 64:546–551. 10.1093/jac/dkp221. [DOI] [PubMed] [Google Scholar]
- 36.Rea MC, Clayton E, O'Connor PM, Shanahan F, Kiely B, Ross RP, Hill C. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J Med Microbiol 56:940–946. 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- 37.Gut IM, Blanke SR, van der Donk WA. 2011. Mechanism of inhibition of Bacillus anthracis spore outgrowth by the lantibiotic nisin. ACS Chem Biol 6:744–752. 10.1021/cb1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowther GS, Baines SD, Todhunter SL, Freeman J, Chilton CH, Wilcox MH. 2013. Evaluation of NVB302 versus vancomycin activity in an in vitro human gut model of Clostridium difficile infection. J Antimicrob Chemother 68:168–176. 10.1093/jac/dks359. [DOI] [PubMed] [Google Scholar]
- 39.Mathur H, O'Connor PM, Hill C, Cotter PD, Ross RP. 2013. Analysis of anti-Clostridium difficile activity of thuricin CD, vancomycin, metronidazole, ramoplanin, and actagardine, both singly and in paired combinations. Antimicrob Agents Chemother 57:2882–2886. 10.1128/AAC.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kers JA, Sharp RE, Defusco AW, Park JH, Xu J, Pulse ME, Weiss WJ, Handfield M. 2018. Mutacin 1140 lantibiotic variants are efficacious against Clostridium difficile infection. Front Microbiol 9:415. 10.3389/fmicb.2018.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ongey EL, Yassi H, Pflugmacher S, Neubauer P. 2017. Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol Lett 39:473–482. 10.1007/s10529-016-2279-9. [DOI] [PubMed] [Google Scholar]
- 42.Richard AS, Zhang A, Park S-J, Farzan M, Zong M, Choe H. 2015. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc Natl Acad Sci U S A 112:14682–14687. 10.1073/pnas.1508095112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Férir G, Petrova MI, Andrei G, Huskens D, Hoorelbeke B, Snoeck R, Vanderleyden J, Balzarini J, Bartoschek S, Brönstrup M, Süssmuth RD, Schols D. 2013. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. PLoS One 8:e64010. 10.1371/journal.pone.0064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochnow H, Rox K, Birudukota NVS, Weichert L, Hotop S-K, Klahn P, Mohr K, Franz S, Banda DH, Blockus S, Schreiber J, Haid S, Oeyen M, Martinez JP, Süssmuth RD, Wink J, Meyerhans A, Goffinet C, Messerle M, Schulz TF, Kröger A, Schols D, Pietschmann T, Brönstrup M. 2019. Labyrinthopeptins exert broad-spectrum antiviral activity through lipid-binding-mediated virolysis. J Virol 94:e01471-19. 10.1128/JVI.01471-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blockus S, Sake SM, Wetzke M, Grethe C, Graalmann T, Pils M, Le Goffic R, Galloux M, Prochnow H, Rox K, Hüttel S, Rupcic Z, Wiegmann B, Dijkman R, Rameix-Welti M-A, Eléouët J-F, Duprex WP, Thiel V, Hansen G, Brönstrup M, Haid S, Pietschmann T. 2020. Labyrinthopeptins as virolytic inhibitors of respiratory syncytial virus cell entry. Antiviral Res 177:104774. 10.1016/j.antiviral.2020.104774. [DOI] [PubMed] [Google Scholar]
- 46.Begde D, Bundale S, Mashitha P, Rudra J, Nashikkar N, Upadhyay A. 2011. Immunomodulatory efficacy of nisin—a bacterial lantibiotic peptide. J Pept Sci 17:438–444. 10.1002/psc.1341. [DOI] [PubMed] [Google Scholar]
- 47.Kindrachuk J, Jenssen H, Elliott M, Nijnik A, Magrangeas-Janot L, Pasupuleti M, Thorson L, Ma S, Easton DM, Bains M, Finlay B, Breukink EJ, Georg-Sahl H, Hancock RE. 2013. Manipulation of innate immunity by a bacterial secreted peptide: lantibiotic nisin Z is selectively immunomodulatory. Innate Immun 19:315–327. 10.1177/1753425912461456. [DOI] [PubMed] [Google Scholar]
- 48.Iorio M, Sasso O, Maffioli SI, Bertorelli R, Monciardini P, Sosio M, Bonezzi F, Summa M, Brunati C, Bordoni R, Corti G, Tarozzo G, Piomelli D, Reggiani A, Donadio SA. 2014. Glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol 9:398–404. 10.1021/cb400692w. [DOI] [PubMed] [Google Scholar]
- 49.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. 2010. Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. APMIS 118:982–990. 10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 50.Zebedin E, Koenig X, Radenkovic M, Pankevych H, Todt H, Freissmuth M, Hilber K. 2008. Effects of duramycin on cardiac voltage-gated ion channels. Naunyn Schmied Arch Pharmacol 377:87–100. 10.1007/s00210-007-0248-5. [DOI] [PubMed] [Google Scholar]
- 51.Sheth TR, Henderson RM, Hladky SB, Cuthbert AW. 1992. Ion channel formation by duramycin. Biochim Biophys Acta Biomembr 1107:179–185. 10.1016/0005-2736(92)90345-M. [DOI] [PubMed] [Google Scholar]
- 52.Grasemann H, Stehling F, Brunar H, Widmann R, Laliberte TW, Molina L, Döring G, Ratjen F. 2007. Inhalation of Moli1901 in patients with cystic fibrosis. Chest 131:1461–1466. 10.1378/chest.06-2085. [DOI] [PubMed] [Google Scholar]
- 53.Steiner I, Errhalt P, Kubesch K, Hubner M, Holy M, Bauer M, Müller M, Hinterberger S, Widmann R, Mascher D, Freissmuth M, Kneussl M. 2008. Pulmonary pharmacokinetics and safety of nebulized duramycin in healthy male volunteers. Naunyn Schmied Arch Pharmacol 378:323–333. 10.1007/s00210-008-0293-8. [DOI] [PubMed] [Google Scholar]
- 54.Zeitlin PL, Boyle MP, Guggino WB, Molina L. 2004. A phase I trial of intranasal Moli1901 for cystic fibrosis. Chest 125:143–149. 10.1378/chest.125.1.143. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Doroghazi JR, Zhao X, Walker MC, van der Donk WA. 2015. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria. Appl Environ Microbiol 81:4339–4350. 10.1128/AEM.00635-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Si T, Tian Q, Min Y, Zhang L, Sweedler JV, van der Donk WA, Zhao H. 2018. Rapid screening of lanthipeptide analogs via in-colony removal of leader peptides in Escherichia coli. J Am Chem Soc 140:11884–11888. 10.1021/jacs.8b05544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urban JH, Moosmeier MA, Aumüller T, Thein M, Bosma T, Rink R, Groth K, Zulley M, Siegers K, Tissot K, Moll GN, Prassler J. 2017. Phage display and selection of lanthipeptides on the carboxy-terminus of the gene-3 minor coat protein. Nat Commun 8:1500. 10.1038/s41467-017-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt S, Montalbán-López M, Peterhoff D, Deng J, Wagner R, Held M, Kuipers OP, Panke S. 2019. Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat Chem Biol 15:437–443. 10.1038/s41589-019-0250-5. [DOI] [PubMed] [Google Scholar]
- 59.Cassis P, Locatelli M, Corna D, Villa S, Rottoli D, Cerullo D, Abbate M, Remuzzi G, Benigni A, Zoja C. 2019. Addition of cyclic angiotensin-(1–7) to angiotensin-converting enzyme inhibitor therapy has a positive add-on effect in experimental diabetic nephropathy. Kidney Int 96:906–917. 10.1016/j.kint.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 60.Kuipers A, Moll GN, Levy A, Krakovsky M, Franklin R. 2020. Cyclic angiotensin-(1–7) contributes to rehabilitation of animal performance in a rat model of cerebral stroke. Peptides 123:170193. 10.1016/j.peptides.2019.170193. [DOI] [PubMed] [Google Scholar]
- 61.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. 10.15620/cdc:82532. [DOI]
- 62.World Health Organization. 2017. Global Antimicrobial Resistance Surveillance System (GLASS) report: early implementation 2016–2017. World Health Organization, Geneva Switzerland. [Google Scholar]
- 63.World Health Organization. 2019. 2019 antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline. https://apps.who.int/iris/bitstream/handle/10665/330420/9789240000193-eng.pdf.
- 64.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl H-G. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 65.Hsu S-TD, Breukink E, Tischenko E, Lutters MAG, de Kruijff B, Kaptein R, Bonvin AMJJ, van Nuland NAJ. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 66.Bonelli RR, Schneider T, Sahl H-G, Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob Agents Chemother 50:1449–1457. 10.1128/AAC.50.4.1449-1457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helander IM, Mattila-Sandholm T. 2000. Permeability barrier of the Gram-negative bacterial outer membrane with special reference to nisin. Int J Food Microbiol 60:153–161. 10.1016/S0168-1605(00)00307-X. [DOI] [PubMed] [Google Scholar]
- 68.Marra F, Patrick DM, Chong M, McKay R, Hoang L, Bowie WR. 2012. Population-based study of the increased incidence of skin and soft tissue infections and associated antimicrobial use. Antimicrob Agents Chemother 56:6243–6249. 10.1128/AAC.00649-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heunis TDJ, Smith C, Dicks LMT. 2013. Evaluation of a nisin-eluting nanofiber scaffold to treat Staphylococcus aureus-induced skin infections in mice. Antimicrob Agents Chemother 57:3928–3935. 10.1128/AAC.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christensen GJM, Brüggemann H. 2014. Bacterial skin commensals and their role as host guardians. Benef Microbes 5:201–215. 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 71.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim J-N, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DYM, Gallo RL. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9:eaah4680. 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coulter SN, Schwan WR, Ng EYW, Langhorne MH, Ritchie HD, Westbrock-Wadman S, Hufnagle WO, Folger KR, Bayer AS, Stover CK. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol 30:393–404. 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 73.van Staden AD, Brand AM, Dicks LMT. 2012. Nisin F-loaded brushite bone cement prevented the growth of Staphylococcus aureus in vivo. J Appl Microbiol 112:831–840. 10.1111/j.1365-2672.2012.05241.x. [DOI] [PubMed] [Google Scholar]
- 74.Ghiselli R, Giacometti A, Cirioni O, Dell'Acqua G, Mocchegiani F, Orlando F, D'Amato G, Rocchi M, Scalise G, Saba V. 2004. RNAIII-inhibiting peptide and/or nisin inhibit experimental vascular graft infection with methicillin-susceptible and methicillin-resistant Staphylococcus epidermidis. Eur J Vasc Endovasc Surg 27:603–607. 10.1016/j.ejvs.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 75.Saising J, Dube L, Ziebandt A-K, Voravuthikunchai SP, Nega M, Götz F. 2012. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 56:5804–5810. 10.1128/AAC.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dreyer L, Smith C, Deane SM, Dicks LMT, van Staden AD. 2019. Migration of bacteriocins across gastrointestinal epithelial and vascular endothelial cells, as determined using in vitro simulations. Sci Rep 9:11481. 10.1038/s41598-019-47843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geng M, Ravichandran A, Escano J, Smith L. 2018. Efficacious analogs of the lantibiotic mutacin 1140 against a systemic methicillin-resistant Staphylococcus aureus infection. Antimicrob Agents Chemother 62:e01626-18. 10.1128/AAC.01626-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldstein B. 1998. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J Antimicrob Chemother 42:277–278. 10.1093/jac/42.2.277. [DOI] [PubMed] [Google Scholar]
- 79.Malabarba A, Pallanza R, Berti M, Cavalleri B. 1990. Synthesis and biological activity of some amide derivatives of the lantibiotic actagardine. J Antibiot 43:1089–1097. 10.7164/antibiotics.43.1089. [DOI] [PubMed] [Google Scholar]
- 80.Wescombe PA, Tagg JR. 2003. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl Environ Microbiol 69:2737–2747. 10.1128/AEM.69.5.2737-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wescombe PA, Upton M, Renault P, Wirawan RE, Power D, Burton JP, Chilcott CN, Tagg JR. 2011. Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology (Reading) 157:1290–1299. 10.1099/mic.0.044719-0. [DOI] [PubMed] [Google Scholar]
- 82.Castiglione F, Cavaletti L, Losi D, Lazzarini A, Carrano L, Feroggio M, Ciciliato I, Corti E, Candiani G, Marinelli F, Selva E. 2007. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry 46:5884–5895. 10.1021/bi700131x. [DOI] [PubMed] [Google Scholar]
- 83.Zhao X, Yin Z, Breukink E, Moll GN, Kuipers OP. 2020. An engineered double lipid II binding motifs-containing lantibiotic displays potent and selective antimicrobial activity against Enterococcus faecium. Antimicrob Agents Chemother 64:e02050-19. 10.1128/AAC.02050-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, Gjonbalaj M, Eaton V, Fontana E, Amoretti L, Wright R, Caballero S, Wang Z-MX, Jung H-J, Morjaria SM, Leiner IM, Qin W, Ramos RJJF, Cross JR, Narushima S, Honda K, Peled JU, Hendrickson RC, Taur Y, van den Brink MRM, Pamer EG. 2019. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572:665–669. 10.1038/s41586-019-1501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG. 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21:592–602.e4. 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gardiner GE, Rea MC, O'Riordan B, O'Connor P, Morgan SM, Lawlor PG, Lynch PB, Cronin M, Ross RP, Hill C. 2007. Fate of the two-component lantibiotic lacticin 3147 in the gastrointestinal tract. Appl Environ Microbiol 73:7103–7109. 10.1128/AEM.01117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobson A, Crispie F, Rea MC, O'Sullivan O, Casey PG, Lawlor PG, Cotter PD, Ross P, Gardiner GE, Hill C. 2011. Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol Ecol 76:602–614. 10.1111/j.1574-6941.2011.01069.x. [DOI] [PubMed] [Google Scholar]
- 88.Heunis TDJ, Botes M, Dicks LMT. 2010. Encapsulation of Lactobacillus plantarum 423 and its bacteriocin in nanofibers. Probiotics Antimicrob Proteins 2:46–51. 10.1007/s12602-009-9024-9. [DOI] [PubMed] [Google Scholar]
- 89.Rajeshkumar NV, Kers JA, Moncrief S, Defusco AW, Park JH, Handfield M. 2019. Preclinical evaluation of the maximum tolerated dose and toxicokinetics of enteric-coated lantibiotic OG253 capsules. Toxicol Appl Pharmacol 374:32–40. 10.1016/j.taap.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 90.Kers JA, DeFusco AW, Park JH, Xu J, Pulse ME, Weiss WJ, Handfield M. 2018. OG716: designing a fit-for-purpose lantibiotic for the treatment of Clostridium difficile infections. PLoS One 13:e0197467. 10.1371/journal.pone.0197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pulse ME, Weiss WJ, Kers JA, DeFusco AW, Park JH, Handfield M. 2019. Pharmacological, toxicological and dose range assessment of OG716, a novel lantibiotic for the treatment of Clostridium difficile-associated infection. Antimicrob Agents Chemother 63:e01904-18. 10.1128/AAC.01904-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.World Health Organization. 2019. Global tuberculosis report 2019. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-report-2019.
- 93.Mahapatra S, Yagi T, Belisle JT, Espinosa BJ, Hill PJ, McNeil MR, Brennan PJ, Crick DC. 2005. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J Bacteriol 187:2747–2757. 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Montville TJ, Chung HJ, Chikindas ML, Chen Y. 1999. Nisin A depletes intracellular ATP and acts in bactericidal manner against Mycobacterium smegmatis. Lett Appl Microbiol 28:189–193. 10.1046/j.1365-2672.1999.00511.x. [DOI] [PubMed] [Google Scholar]
- 95.Healy B, Field D, O'Connor PM, Hill C, Cotter PD, Ross RP. 2013. Intensive mutagenesis of the nisin hinge leads to the rational design of enhanced derivatives. PLoS One 8:e79563. 10.1371/journal.pone.0079563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carroll J, Draper LA, O'Connor PM, Coffey A, Hill C, Ross RP, Cotter PD, O'Mahony J. 2010. Comparison of the activities of the lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int J Antimicrob Agents 36:132–136. 10.1016/j.ijantimicag.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 97.Sosunov V, Mischenko V, Eruslanov B, Svetoch E, Shakina Y, Stern N, Majorov K, Sorokoumova G, Selishcheva A, Apt A. 2007. Antimycobacterial activity of bacteriocins and their complexes with liposomes. J Antimicrob Chemother 59:919–925. 10.1093/jac/dkm053. [DOI] [PubMed] [Google Scholar]
- 98.Mota-Meira M, Lapointe G, Lacroix C, Lavoie MC. 2000. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob Agents Chemother 44:24–29. 10.1128/AAC.44.1.24-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naghmouchi K, Baah J, Hober D, Jouy E, Rubrecht C, Sané F, Drider D. 2013. Synergistic effect between colistin and bacteriocins in controlling Gram-negative pathogens and their potential to reduce antibiotic toxicity in mammalian epithelial cells. Antimicrob Agents Chemother 57:2719–2725. 10.1128/AAC.02328-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Q, Montalban-Lopez M, Kuipers OP. 2018. Increasing the antimicrobial activity of nisin-based lantibiotics against Gram-negative pathogens. Appl Environ Microbiol 84:e00052-18. 10.1128/AEM.00052-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Armstrong JJ, Baddiley J, Buchanan JG, Carss B, Greenberg GR. 1958. Isolation and structure of ribitol phosphate derivatives (teichoic acids) from bacterial cell walls. J Chem Soc 1958:4344–4354. 10.1039/jr9580004344. [DOI] [Google Scholar]
- 102.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol 187:6719–6725. 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kovács M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Brückner R. 2006. A functional Dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188:5797–5805. 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the Dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 105.Saar-Dover R, Bitler A, Nezer R, Shmuel-Galia L, Firon A, Shimoni E, Trieu-Cuot P, Shai Y. 2012. d-Alanylation of lipoteichoic acids confers resistance to cationic peptides in group B Streptococcus by increasing the cell wall density. PLoS Pathog 8:e1002891. 10.1371/journal.ppat.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Demel RA, Peelen T, Siezen RJ, Kruijff B, Kuipers OP. 1996. Nisin Z, mutant nisin Z, and lacticin 481 interactions with anionic lipids correlate with antimicrobial activity. A monolayer study. Eur J Biochem 235:267–274. 10.1111/j.1432-1033.1996.00267.x. [DOI] [PubMed] [Google Scholar]
- 107.Sohlenkamp C, Geiger O. 2016. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 40:133–159. 10.1093/femsre/fuv008. [DOI] [PubMed] [Google Scholar]
- 108.Ernst CM, Staubitz P, Mishra NN, Yang S-J, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e1000660. 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stein T, Heinzmann S, Solovieva I, Entian K-D. 2003. Function of Lactococcus lactis nisin immunity genes NisI and NisFEG after coordinated expression in the surrogate host Bacillus subtilis. J Biol Chem 278:89–94. 10.1074/jbc.M207237200. [DOI] [PubMed] [Google Scholar]
- 110.Hacker C, Christ NA, Duchardt-Ferner E, Korn S, Göbl C, Berninger L, Düsterhus S, Hellmich UA, Madl T, Kötter P, Entian K-D, Wöhnert J. 2015. The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J Biol Chem 290:28869–28886. 10.1074/jbc.M115.679969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Draper LA, Grainger K, Deegan LH, Cotter PD, Hill C, Ross RP. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol Microbiol 71:1043–1054. 10.1111/j.1365-2958.2008.06590.x. [DOI] [PubMed] [Google Scholar]
- 112.Suarez JM, Edwards AN, McBride SM. 2013. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J Bacteriol 195:2621–2631. 10.1128/JB.00166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kawada-Matsuo M, Yoshida Y, Zendo T, Nagao J, Oogai Y, Nakamura Y, Sonomoto K, Nakamura N, Komatsuzawa H. 2013. Three distinct two-component systems are involved in resistance to the class I bacteriocins, nukacin ISK-1 and nisin A, in Staphylococcus aureus. PLoS One 8:e69455. 10.1371/journal.pone.0069455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clemens R, Zaschke-Kriesche J, Khosa S, Smits SHJ. 2018. Insight into two ABC transporter families involved in lantibiotic resistance. Front Mol Biosci 4:91. 10.3389/fmolb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Draper LA, Cotter PD, Hill C, Ross RP. 2015. Lantibiotic resistance. Microbiol Mol Biol Rev 79:171–191. 10.1128/MMBR.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scott MG, Davidson DJ, Gold MR, Bowdish D, Hancock REW. 2002. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J Immunol 169:3883–3891. 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- 117.Driouich A, Smith C, Ropitaux M, Chambard M, Boulogne I, Bernard S, Follet‐Gueye M, Vicré M, Moore J. 2019. Root extracellular traps versus neutrophil extracellular traps in host defence, a case of functional convergence? Biol Rev Camb Rev 94:1685–1700. 10.1111/brv.12522. [DOI] [PubMed] [Google Scholar]
- 118.Urban CF, Nett JE. 2019. Neutrophil extracellular traps in fungal infection. Semin Cell Dev Biol 89:47–57. 10.1016/j.semcdb.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Apel F, Zychlinsky A, Kenny EF. 2018. The role of neutrophil extracellular traps in rheumatic diseases. Nat Rev Rheumatol 14:467–475. 10.1038/s41584-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 120.Pablo MA, Gaforio JJ, Gallego AM, Ortega E, Gálvez AM, Alvarez de Cienfuegos López G. 1999. Evaluation of immunomodulatory effects of nisin-containing diets on mice. FEMS Immunol Med Microbiol 24:35–42. 10.1111/j.1574-695X.1999.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 121.Märki F, Hänni E, Fredenhagen A, van Oostrum J. 1991. Mode of action of the lanthionine-containing peptide antibiotics duramycin, duramycin B and C, and cinnamycin as indirect inhibitors of phospholipase A2. Biochem Pharmacol 42:2027–2035. 10.1016/0006-2952(91)90604-4. [DOI] [PubMed] [Google Scholar]
- 122.Kido Y, Hamakado T, Yoshida T, Anno M, Motoki Y, Wakamiya T, Shiba T. 1983. Isolation and characterization of ancovenin, a new inhibitor of angiotensin I converting enzyme, produced by actinomycetes. J Antibiot 36:1295–1299. 10.7164/antibiotics.36.1295. [DOI] [PubMed] [Google Scholar]
- 123.Bernstein KE, Khan Z, Giani JF, Cao D-Y, Bernstein EA, Shen XZ. 2018. Angiotensin-converting enzyme in innate and adaptive immunity. Nat Rev Nephrol 14:325–336. 10.1038/nrneph.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Iftime D, Jasyk M, Kulik A, Imhoff JF, Stegmann E, Wohlleben W, Süssmuth RD, Weber T. 2015. Streptocollin, a type IV lanthipeptide produced by Streptomyces collinus Tü 365. Chembiochem 16:2615–2623. 10.1002/cbic.201500377. [DOI] [PubMed] [Google Scholar]
- 125.Través PG, Pardo V, Pimentel-Santillana M, González-Rodríguez Á, Mojena M, Rico D, Montenegro Y, Calés C, Martín-Sanz P, Valverde AM, Boscá L. 2014. Pivotal role of protein tyrosine phosphatase 1B (PTP1B) in the macrophage response to pro-inflammatory and anti-inflammatory challenge. Cell Death Dis 5:e1125. 10.1038/cddis.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ricke KM, Cruz SA, Qin Z, Farrokhi K, Sharmin F, Zhang L, Zasloff MA, Stewart AFR, Chen H-H. 2020. Neuronal protein tyrosine phosphatase 1B hastens amyloid β-associated Alzheimer’s disease in mice. J Neurosci 40:1581–1593. 10.1523/JNEUROSCI.2120-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith C. 2018. Natural antioxidants in prevention of accelerated ageing: a departure from conventional paradigms required. J Physiol Biochem 74:549–558. 10.1007/s13105-018-0621-5. [DOI] [PubMed] [Google Scholar]
- 128.Kamarajan P, Hayami T, Matte B, Liu Y, Danciu T, Ramamoorthy A, Worden F, Kapila S, Kapila Y. 2015. Nisin ZP, a bacteriocin and food preservative, inhibits head and neck cancer tumorigenesis and prolongs survival. PLoS One 10:e0131008. 10.1371/journal.pone.0131008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Joo NE, Ritchie K, Kamarajan P, Miao D, Kapila YL. 2012. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med 1:295–305. 10.1002/cam4.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng M, Bhujwalla ZM, Glunde K. 2016. Targeting phospholipid metabolism in cancer. Front Oncol 6:e0131008. 10.3389/fonc.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.El Jastimi R, Edwards K, Lafleur M. 1999. Characterization of permeability and morphological perturbations induced by nisin on phosphatidylcholine membranes. Biophys J 77:842–852. 10.1016/S0006-3495(99)76936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.El Jastimi R, Lafleur M. 1999. Nisin promotes the formation of non-lamellar inverted phases in unsaturated phosphatidylethanolamines. Biochim Biophys Acta Biomembr 1418:97–105. 10.1016/S0005-2736(99)00027-9. [DOI] [PubMed] [Google Scholar]
- 133.Rana K, Sharma R, Preet S. 2019. Augmented therapeutic efficacy of 5-fluorouracil in conjunction with lantibiotic nisin against skin cancer. Biochem Biophys Res Commun 520:551–559. 10.1016/j.bbrc.2019.10.058. [DOI] [PubMed] [Google Scholar]
- 134.Preet S, Bharati S, Panjeta A, Tewari R, Rishi P. 2015. Effect of nisin and doxorubicin on DMBA-induced skin carcinogenesis—a possible adjunct therapy. Tumor Biol 36:8301–8308. 10.1007/s13277-015-3571-3. [DOI] [PubMed] [Google Scholar]
- 135.Broughton LJ, Crow C, Maraveyas A, Madden LA. 2016. Duramycin-induced calcium release in cancer cells. Anticancer Drugs 27:173–182. 10.1097/CAD.0000000000000313. [DOI] [PubMed] [Google Scholar]
- 136.Yates KR, Welsh J, Udegbunam NO, Greenman J, Maraveyas A, Madden LA. 2012. Duramycin exhibits antiproliferative properties and induces apoptosis in tumour cells. Blood Coagul Fibrinolysis 23:396–401. 10.1097/MBC.0b013e3283538875. [DOI] [PubMed] [Google Scholar]
- 137.Thorpe PE, Soares MIM, He J. March 2016. Selected antibodies and duramycin peptides binding to anionic phospholipids and aminophospholipids and their use in the treatment of viral infections and cancer. European patent EP2283868B1. https://patents.google.com/patent/EP2283868B1/en. Accessed 16 April 2020.
- 138.He J, Thorpe PE. 2004. Anti-tumor effects of duramycin-IgG conjugate. Proceedings of the American Association for Cancer Research, abstr 4561. Cancer Res 64(Suppl 7):1053. [Google Scholar]
- 139.Mahmood N, Mihalcioiu C, Rabbani SA. 2018. Multifaceted role of the urokinase-type plasminogen activator (UPA) and its receptor (UPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol 8:24. 10.3389/fonc.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Małaczewska J, Kaczorek-Łukowska E, Wójcik R, Siwicki AK. 2019. Antiviral effects of nisin, lysozyme, lactoferrin and their mixtures against bovine viral diarrhoea virus. BMC Vet Res 15:318. 10.1186/s12917-019-2067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, Pereira L. 2016. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 20:155–166. 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang X, Lennard KR, He C, Walker MC, Ball AT, Doigneaux C, Tavassoli A, van der Donk WA. 2018. A lanthipeptide library used to identify a protein-protein interaction inhibitor. Nat Chem Biol 14:375–380. 10.1038/s41589-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. 2008. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem Biol 3:757–764. 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 144.Velásquez JE, van der Donk WA. 2011. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol 15:11–21. 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kuipers OP, Beerthuyzen MM, de Ruyter PGGA, Luesink EJ, de Vos WM. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–27304. 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 146.Lagedroste M, Reiners J, Smits SHJ, Schmitt L. 2019. Systematic characterization of position one variants within the lantibiotic nisin. Sci Rep 9:935. 10.1038/s41598-018-37532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Heel AJ, Mu D, Montalbán-López M, Hendriks D, Kuipers OP. 2013. Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth Biol 2:397–404. 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- 148.Mesa-Pereira B, Rea MC, Cotter PD, Hill C, Ross RP. 2018. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front Microbiol 9:1654. 10.3389/fmicb.2018.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shi Y, Yang X, Garg N, van der Donk WA. 2011. Production of lantipeptides in Escherichia coli. J Am Chem Soc 133:2338–2341. 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nagao J, Harada Y, Shioya K, Aso Y, Zendo T, Nakayama J, Sonomoto K. 2005. Lanthionine introduction into nukacin ISK-1 prepeptide by co-expression with modification enzyme NukM in Escherichia coli. Biochem Biophys Res Commun 336:507–513. 10.1016/j.bbrc.2005.08.125. [DOI] [PubMed] [Google Scholar]
- 151.Van Staden ADP, Faure LM, Vermeulen RR, Dicks LMT, Smith C. 2019. Functional expression of GFP-fused class I lanthipeptides in Escherichia coli. ACS Synth Biol 8:2220–2227. 10.1021/acssynbio.9b00167. [DOI] [PubMed] [Google Scholar]
- 152.Ongey EL, Giessmann RT, Fons M, Rappsilber J, Adrian L, Neubauer P. 2018. Heterologous biosynthesis, modifications and structural characterization of ruminococcin-A, a lanthipeptide from the gut bacterium Ruminococcus gnavus E1, in Escherichia coli. Front Microbiol 9:1688. 10.3389/fmicb.2018.01688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vermeulen RR, Van Staden ADP, Dicks L. 2020. Heterologous expression of the class IIa bacteriocins, plantaricin 423 and mundticin ST4SA, in Escherichia coli using green fluorescent protein as a fusion partner. Front Microbiol 11:1634. 10.3389/fmicb.2020.01634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kuthning A, Mösker E, Süssmuth RD. 2015. Engineering the heterologous expression of lanthipeptides in Escherichia coli by multigene assembly. Appl Microbiol Biotechnol 99:6351–6361. 10.1007/s00253-015-6557-6. [DOI] [PubMed] [Google Scholar]
- 155.Wang J, Ge X, Zhang L, Teng K, Zhong J. 2016. One-pot synthesis of class II lanthipeptide bovicin HJ50 via an engineered lanthipeptide synthetase. Sci Rep 6:38630. 10.1038/srep38630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ahmed Y, Rebets Y, Estévez MR, Zapp J, Myronovskyi M, Luzhetskyy A. 2020. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb Cell Fact 19:5. 10.1186/s12934-020-1277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Myronovskyi M, Luzhetskyy A. 2019. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat Prod Rep 36:1281–1294. 10.1039/C9NP00023B. [DOI] [PubMed] [Google Scholar]
- 158.Krawczyk JM, Völler GH, Krawczyk B, Kretz J, Brönstrup M, Süssmuth RD. 2013. Heterologous expression and engineering studies of labyrinthopeptins, class III lantibiotics from Actinomadura namibiensis. Chem Biol 20:111–122. 10.1016/j.chembiol.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 159.Boakes S, Cortés J, Appleyard AN, Rudd BAM, Dawson MJ. 2009. Organization of the genes encoding the biosynthesis of actagardine and engineering of a variant generation system. Mol Microbiol 72:1126–1136. 10.1111/j.1365-2958.2009.06708.x. [DOI] [PubMed] [Google Scholar]
- 160.Mattick ATR, Hirsch A, Berridge NJ. 1947. Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet 250:5–8. 10.1016/S0140-6736(47)90004-4. [DOI] [PubMed] [Google Scholar]
- 161.Liu W, Hansen JN. 1992. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem 267:25078–25085. 10.1016/S0021-9258(19)74008-3. [DOI] [PubMed] [Google Scholar]
- 162.Steenken W, Wolinsky E. 1949. The tuberculostatic effect of subtilin in vitro and in vivo. J Bacteriol 57:453–457. 10.1128/JB.57.4.453-457.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian K. 2002. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J Bacteriol 184:1703–1711. 10.1128/JB.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kellner R, Jung G, Hörner T, Zähner H, Schnell N, Entian KD, Götz F. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem 177:53–59. 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- 165.Hillman JD, Johnson KP, Yaphe BI. 1984. Isolation of a Streptococcus mutans strain producing a novel bacteriocin. Infect Immun 44:141–144. 10.1128/IAI.44.1.141-144.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mota-Meira M, Morency H, Lavoie MC. 2005. In vivo activity of mutacin B-Ny266. J Antimicrob Chemother 56:869–871. 10.1093/jac/dki295. [DOI] [PubMed] [Google Scholar]
- 167.Arioli V, Berti M, Silvestri LG. 1976. Gardimycin, a new antibiotic from actinoplanes. III. Biological properties. J Antibiot 29:511–515. 10.7164/antibiotics.29.511. [DOI] [PubMed] [Google Scholar]
- 168.Naruse N, Tenmyo O, Tomita K, Konishi M, Miyaki T, Kawaguchi H, Fukase K, Wakamiya T, Shiba T. 1989. Lanthiopeptin, a new peptide antibiotic. Production, isolation and properties of lanthiopeptin. J Antibiot 42:837–845. 10.7164/antibiotics.42.837. [DOI] [PubMed] [Google Scholar]
- 169.Ryan MP, Rea MC, Hill C, Ross RP. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62:612–619. 10.1128/AEM.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mohr KI, Volz C, Jansen R, Wray V, Hoffmann J, Bernecker S, Wink J, Gerth K, Stadler M, Müller R. 2015. Pinensins: the first antifungal lantibiotics. Angew Chem Int Ed Engl 54:11254–11258. 10.1002/anie.201500927. [DOI] [PubMed] [Google Scholar]
- 171.Montalbán-López M, Deng J, van Heel AJ, Kuipers O. 2018. P specificity and application of the lantibiotic protease NisP. Front Microbiol 9:160. 10.3389/fmicb.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shi Y, Bueno A, van der Donk WA. 2012. Heterologous production of the lantibiotic Ala(0)actagardine in Escherichia coli. Chem Commun (Camb) 48:10966–10968. 10.1039/c2cc36336d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lohans CT, Li JL, Vederas JC. 2014. Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with d-amino acids. J Am Chem Soc 136:13150–13153. 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 174.Singh M, Chaudhary S, Sareen D. 2020. Roseocin, a novel two‐component lantibiotic from an actinomycete. Mol Microbiol 113:326–337. 10.1111/mmi.14419. [DOI] [PubMed] [Google Scholar]
- 175.Chen S, Xu B, Chen E, Wang J, Lu J, Donadio S, Ge H, Wang H. 2019. Zn-dependent bifunctional proteases are responsible for leader peptide processing of class III lanthipeptides. Proc Natl Acad Sci U S A 116:2533–2538. 10.1073/pnas.1815594116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ren H, Shi C, Bothwell IR, van der Donk WA, Zhao H. 2020. Discovery and characterization of a class IV lanthipeptide with a nonoverlapping ring pattern. ACS Chem Biol 15:1642–1649. 10.1021/acschembio.0c00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Fig. S1 to S4. Download AEM00186-21_Supp_1_seq5.pdf, PDF file, 0.9 MB (937.2KB, pdf)