Abstract
KPC-producing Escherichia coli (KPC-Ec) remains uncommon, being mainly reported as the cause of sporadic episodes of infection rather than outbreak events. Here we retrospectively describe the dynamics of a large hospital outbreak sustained by KPC-Ec, involving 106 patients and 25 hospital wards, during a six-month period. Twenty-nine representative KPC-Ec isolates (8/29 from rectal swabs; 21/29 from other clinical specimens) have been investigated by Whole-Genome Sequencing (WGS). Outbreak isolates showed a multidrug-resistant profile and harbored several resistance determinants, including blaCTX-M-27, aadA5, dfrA17, sulI, gyrA1AB and parC1aAB. Phylogenomic analysis identified the ST131 cluster 1 (23/29 isolates), H30Rx clade C, as responsible for the epidemic event. A further two KPC-Ec ST131 clusters were identified: cluster 2 (n = 2/29) and cluster 3 (n = 1/29). The remaining KPC-Ec resulted in ST978 (n = 2/29) and ST1193 (n = 1/29), and were blaKPC-3 associated. The KPC-Ec ST131 cluster 1, originated in a previous KPC-Kp endemic context probably by plasmid transfer, and showed a clonal dissemination strategy. Transmission of the blaKPC gene to the globally disseminated high-risk ST131 clone represents a serious cause of concern. Application of WGS in outbreak investigations could be useful to better understand the evolution of epidemic events in order to address infection control and contrast interventions, especially when high-risk epidemic clones are involved.
Keywords: KPC-producing, Escherichia coli, ST131, WGS, outbreak, H30Rx
1. Introduction
The ongoing rise of carbapenemase-producing Enterobacterales (CPE) represents an important threat to public health worldwide, in both healthcare and community settings. The pervasive dissemination of CPE substantially impacts on patient safety since few therapeutic alternatives remain. Overall, the worldwide predominant carbapenemase is the Ambler class A Klebsiella pneumoniae carbapenemase (KPC) enzyme, encoded by alleles of the blaKPC gene, with KPC-producing K. pneumoniae (KPC-Kp) being the most common among KPC-producing CPE. Although less prevalent than in K. pneumoniae, KPC production in other species belonging to Enterobacterales is increasingly reported [1]. The presence of this highly transferable carbapenemase in Escherichia coli is of particular concern, being the most common community- and hospital-acquired pathogen, that can be transmitted among humans, animals and in the environment [2]. Moreover, the recent detection of the blaKPC gene in E. coli belonging to sequence type (ST) 131, a globally disseminated successful clone, represents a cause of serious concern [3]. In fact, the ST131 clone emerged as the most common extraintestinal pathogen that, in association with fluoroquinolone and extended-spectrum cephalosporin resistance, was responsible for the worldwide spread of the extended-spectrum beta-lactamase (ESBL) blaCTX-M-15 gene. Due to its ability to asymptomatically colonize the gastrointestinal tract of both community and healthcare-associated infections, a stable association of ST131 lineage with blaKPC could have relevant consequences for the management of E. coli infections [4].
KPC-producing E. coli (KPC-Ec) isolates were mainly reported in countries showing a high prevalence of KPC-Kp, probably reflecting a spill-over of resistance genes from the K. pneumoniae reservoir [5]. However, acquired carbapenem resistance in E. coli is still considered a rare and recent event. The first cases of KPC-Ec were observed in 2004–2005 in the USA (Cleveland, n = 1; New York City, n = 2; New Jersey, n = 1), and in Israel (Tel Aviv, n = 4) [3]. After the first detection in Europe—reported in 2008 in France from a patient initially hospitalized in Israel [6]—KPC-Ec has been sporadically reported in the USA [7], Israel [8] and some European countries [9,10].
In Italy, KPC-Kp has been endemic since 2013, but the presence of KPC-Ec isolates remains limited [11]. Results of the most recent Italian Nationwide survey highlighted that KPC was the most frequent carbapenemase from bloodstream infections, mainly in K. pneumoniae (95.2%). A recent epidemiological study showed that KPC-Ec were only 1.3% of KPC-producing invasive isolates, but they accounted for 81.4% of cabapenem-resistant E. coli, thus highlighting the propensity of these strains to cause invasive infections [12].
Outbreaks caused by KPC-Ec have been described in the USA, Greece, Canada, Israel, Italy and the UK [2,13,14,15,16,17]. Despite this, KPC-Ec has been more frequently reported in sporadic cases than as the cause of outbreaks [18]. In fact, unlike K. pneumoniae, in which the KPC determinant is often associated to predominant plasmids and epidemic clones (e.g., IncFII, -FIA, -I2 and ST258), the sporadic appearance of KPC-producing E. coli belonging to different genetic backgrounds is largely due to horizontal transfer of different blaKPC-harboring plasmid groups, thus suggesting a “less successful” association enzyme-pathogen than in KPC-Kp [19].
It is of particular concern that ST131 KPC-Ec strains able to cause outbreaks are emerging, as suggested by the recent reports from Israel, Italy and UK [13,14].
The aim of our study was to report an epidemiological and genomic (Whole-Genome Sequencing, WGS) investigation of a large nosocomial outbreak caused by KPC-Ec, which occurred in Northern Italy in 2016.
2. Results
2.1. Bacterial Isolates and Phenotypic Characterization
From February to July 2016, 123 KPC-Escherichia coli (KPC-Ec) isolates were collected from 106 patients. Twenty-five hospital wards were involved in the outbreak, with those of Internal Medicine (n = 34 patients), Cardiology Rehabilitation (n = 17), Cardiology (n = 10) and Nephrology (n = 9) being the most represented. From August 2016 to June 2017, eight additional KPC-Ec were isolated from rectal swabs (n = 7) and respiratory secretions (n = 1) of seven patients. Four of the above isolates were included in the WGS analysis for comparison. Twenty out of 106 patients belonging to the outbreak period were previously colonized by KPC-Klebsiella pneumoniae (KPC-Kp), while in 23 cases the detection of both species was associated, indicating a co-presence at the intestinal level. KPC-Ec was isolated prior to KPC-Kp in seven patients. Six KPC-Kp included in the study were isolated from different wards. Among them, four were from rectal swabs whereas the remaining two were from urine samples. According to both the MicroScan autoSCAN-4 semi-automated system and Sensititre broth microdilution results, representative isolates were consistently resistant to amoxicillin/clavulanate, piperacillin/tazobactam, cefotaxime, ceftazidime, aztreonam, ertapenem, and ceftolozane/tazobactam, but susceptible to colistin, tigecycline and ceftazidime-avibactam. Detailed data concerning the susceptibility results of the 29 KPC-Ec representative isolates are reported in Table 1. Of note, MIC values for imipenem ranged from 4 to >8 mg/L, whereas MICs of meropenem ranged from 4 to 8 mg/L.
Table 1.
Antimicrobial susceptibility profiles of selected KPC-positive E. coli strains.
| ID Strain | Antimicrobial Susceptibility (MIC, µg/mL) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | PTZ | CTX | CAZ | CZA | C/T | MER | IMI | ERT | AMK | GNT | AZT | CIP | TBR | TIG | SXT | COL | |
| sk46y46t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 4 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk36y36t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk35y35t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | >4 (R) | >4 (R) | >1 (R) | >4 (R) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk39y39t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk37y37t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 2/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk38y38t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk42y42t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | 16 (I) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk40y40t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk47y47t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | >8 (R) | >1 (R) | 16 (I) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk41y41t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 16/4 (R) | 8 (I) | 8 (I) | >1 (R) | >16 (R) | 4 (I) | >4 (R) | ≤0,06 (S) | >4 (R) | ≤1 (S) | ≤2/38 (S) | <0,25 (S) |
| sk43y43t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 32/4 (R) | 8 (I) | 8 (I) | >1 (R) | >16 (R) | 4 (I) | >4 (R) | ≤0,06 (S) | >4 (R) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk54y54t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | >8 (R) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk44y44t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk45y45t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk48y48t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk49y49t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | >8 (R) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | ≤2/38 (S) | <0,25 (S) |
| sk51y51t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk50y50t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk52y52t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk53y53t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk56y56t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk55y55t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 16/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk57y57t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk58y58t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 2/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk59y59t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk60y60t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 32/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 1 (S) |
| sk185y185t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk136y136t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk137y137t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
MIC: minimum inhibitory concentration; AMC: amoxicillin/clavulanate; PTZ: piperacillin/tazobactam; CTX: cefotaxime; CAZ: ceftazidime; MER: meropenem; IMI: imipenem; ERT: ertapenem; AMK: amikacin; GNT: gentamicin; AZT: aztreonam; CIP: ciprofloxacin; TBR: tobramycin; TIG: tigecycline; SXT: trimethoprim-sulfamethoxazole; CZA: ceftazidime/avibactam; C/T: ceftolozane/tazobactam; COL: colistin; S: susceptible; I: intermediate; R: resistant. Susceptibility results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2016) criteria. The “index strain” is indicated in bold.
2.2. Epidemiological Context
Epidemiological surveillance data showed that the epidemic event followed a previous KPC-Kp outbreak, starting in October 2015 and partially overlapping the KPC-Ec outbreak (Supplementary Figure S1). The prevalence of KPC-Ec increased from 0% in 2014 and 2015 to 2.8% in 2016. This trend seems to be related to the increase of KPC-Kp prevalence, from 12.4% in 2014, to 17.3% in 2015 and to 23.1% in 2016 (Supplementary Figure S1). Notably, 50 (47.2%) out of 106 patients’ results were positive to both KPC-Ec and KPC-Kp during previous hospitalizations. In particular, 20 out of 50 patients (40%) were previously colonized by KPC-Kp, while in 23 cases (46%) the isolation of both species was concomitant, indicating the co-presence at the intestinal level. Only in seven patients (14%), however, was KPC-Ec isolated previously with respect to KPC-Kp.
2.3. Whole-Genome Sequencing Characterization
Twenty-nine KPC-Ec isolates were further investigated by whole-genome sequencing. They were from urine (n = 11), rectal swabs (n = 8), blood (n = 2), purulent exudate (n = 2), respiratory secretions (n = 3), drainage fluid (n = 1), peritoneal fluid (n = 1) and a surgical wound swab (n = 1) (Table 2).
Table 2.
Main metadata of the 35 clinical samples included in the study and subjected to whole-genome sequencing. The “index strain” is indicated in bold.
| ID Sample | Microorganism | Ward | Isolation Date | Material |
|---|---|---|---|---|
| sk35y35t | Escherichia coli | Infectious Diseases | 15 February 2016 | Rectal swab |
| sk36y36t | Escherichia coli | Nephrology | 16 February 2016 | Rectal swab |
| sk37y37t | Escherichia coli | Medicine | 26 February 2016 | Urine |
| sk38y38t | Escherichia coli | Nephrology | 29 February 2016 | Rectal swab |
| sk39y39t | Escherichia coli | Cardiology Rehabilitation | 1 March 2016 | Urine |
| sk40y40t | Escherichia coli | Neurointensive Care | 3 March 2016 | Purulent exudate |
| sk42y42t | Escherichia coli | Dermatology | 3 March 2016 | Urine |
| sk41y41t | Escherichia coli | Medicine I Soap | 8 March 2016 | Blood |
| sk43y43t | Escherichia coli | Medicine | 17 March 2016 | Rectal swab |
| sk44y44t | Escherichia coli | Nephrology | 23 March 2016 | Blood |
| sk45y45t | Escherichia coli | Cardiology Rehabilitation | 5 April 2016 | Urine |
| sk46y46t | Escherichia coli | Cardiosurgery | 18 April 2016 | Bronchial aspirate |
| sk47y47t | Escherichia coli | Medicine I Soap | 18 April 2016 | Urine |
| sk48y48t | Escherichia coli | Nephrology | 21 April 2016 | Surgical wound swab |
| sk49y49t | Escherichia coli | Long-term Surgery II | 24 April 2016 | Peritoneal fluid |
| sk50y50t | Escherichia coli | Medicine II | 10 May 2016 | Respiratory secretion |
| sk51y51t | Escherichia coli | Cardiology Rehabilitation | 13 May 2016 | Urine |
| sk52y52t | Escherichia coli | Medicine I Soap | 22 May 2016 | Urine |
| sk53y53t | Escherichia coli | Medicine II men | 11 June 2016 | Urine |
| sk54y54t | Escherichia coli | Medicine II men | 25 June 2016 | Urine |
| sk55y55t | Escherichia coli | Surgery | 29 June 2016 | Drainage fluid |
| sk56y56t | Escherichia coli | Cardiology | 30 June 2016 | Urine |
| sk57y57t | Escherichia coli | Medicine soap | 13 July 2016 | Urine |
| sk58y58t | Escherichia coli | Medicine soap | 13 July 2016 | Rectal swab |
| sk59y59t | Escherichia coli | Medicine soap | 14 July 2016 | Purulent exudate |
| sk60y60t | Escherichia coli | Intensive care | 30 September 2016 | Rectal swab |
| sk185y185t | Escherichia coli | Infectious Diseases | 14 November 2016 | Respiratory secretion |
| sk136y136t | Escherichia coli | Nephrology | 23 January 2017 | Rectal swab |
| sk137y137t | Escherichia coli | Surgery | 5 June 2017 | Rectal swab |
| sk138y138t | Klebsiella pneumoniae | Nephrology | 22 January 2016 | Urine |
| sk139y139t | Klebsiella pneumoniae | Cardiology | 16 February 2016 | Rectal swab |
| sk140y140t | Klebsiella pneumoniae | Rehabilitation Medicine | 25 February 2016 | Rectal swab |
| sk141y141t | Klebsiella pneumoniae | Urology | 25 February 2016 | Urine |
| sk142y142t | Klebsiella pneumoniae | Infectious Diseases | 22 March 2016 | Rectal swab |
| sk143y143t | Klebsiella pneumoniae | Medicine | 31 March 2016 | Rectal swab |
Twenty-six out of 29 isolates belonged to ST131 while the remaining three isolates were of ST978 (n = 2) and ST1193 (n = 1). ST131 isolates carried the following resistance determinants: aadA5, aph(6)-Id, aph(3’’)-Ib, blaCTX-M-type, blaKPC-type, blaOXA-9, blaTEM-1A, mph(A), sul1, sul2, tet(A) and dfrA17 (Table 3). According to the observed quinolone resistance phenotype (see Table 3), ST131 isolates harbored the parC1aAB and gyrA1AB gene variants. Twenty-four ST131 strains harbored the blaKPC-2 gene, while only two strains’ results were blaKPC-3 positive. As shown in Table 3, all but one ST131 strain harbored the blaCTX-M-27, whereas the remaining one was positive for blaCTX-M-15. All the ST131 strains showed the O25b:H4 serotype and the fimbrial variant fimH30. Both the isolates belonging to ST978 carried the blaKPC-3 determinant, and showed the serotype O83:H27, and the fimH2 variant. The ST1193 isolate harbored the blaKPC-3 gene, and showed the serotype O75:H5, and the fimH64 variant.
Table 3.
Genomic characteristics of the sequenced isolates. The “index strain” is indicated in bold.
| ID Sample | Isolation Date | MLST | Cluster | Serotype | fimH | Resistance Genes | Virulence Genes | Plasmid Incompatibility Groups |
|---|---|---|---|---|---|---|---|---|
| sk35y35t | 15 February 2016 | 131 | ST131 cluster 3 | O25b:H4 | 30 | ampH, bla TEM-90 , bla KPC-2, blaOXA-9, bla CTX-M-15 , ampC2, catB4, bla OXA-1 , aac(3)-IIa, parC1aAB, gyrA1AB | iha, sat, iss, cnf1, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk36y36t | 16 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-122 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk37y37t | 26 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-90 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk38y38t | 29 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-122 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk39y39t | 1 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-141 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk40y40t | 3 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-122 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk42y42t | 3 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-79 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk41y41t | 8 March 2016 | 978 | ST978 | O83:H27 | 2 | aac(6)-Ib, ampH, ampC2, bla KPC-3 | vat, pic, gad | IncX3_1__JN247852 |
| sk43y43t | 17 March 2016 | 978 | ST978 | O83:H27 | 2 | aac(6)-Ib, ampH, ampC2, bla KPC-3 | vat, pic, gad | IncX3_1__JN247852 |
| sk44y44t | 23 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-122 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk45y45t | 5 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-79 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk46y46t | 18 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-156 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk47y47t | 18 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-54 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk48y48t | 21 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-192 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk49y49t | 24 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, bla TEM-122 , bla KPC-2 , bla OXA-9 , bla CTX-M-27 , ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk50y50t | 10 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-122 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk51y51t | 13 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-79 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk52y52t | 22 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-150 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk53y53t | 11 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-168 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk54y54t | 25 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, blaTEM-122, blaKPC-2, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk55y55t | 29 June 2016 | 131 | ST131 cluster 2 | O25b:H4 | 30 | ampH, bla TEM-79 , bla KPC-3 , bla OXA-9 , bla CTX-M-27 , ampC2, parC1aAB, gyrA1AB | sat, iss, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk56y56t | 30 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-79 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781, Col(BS512)_1_NC_010656 |
| sk57y57t | 13 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-168 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk58y58t | 13 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-141 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk59y59t | 14 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk60y60t | 30 September 2016 | 131 | ST131 cluster 2 | O25b:H4 | 30 | ampH, bla TEM-79 , bla KPC-3 , bla OXA-9 , bla CTX-M-27 , ampC2, parC1aAB, gyrA1AB | sat, iss, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk185y185t | 14 November 2016 | 1193 | ST1193 | O75:H5 | 64 | qnrB1, bla TEM-198 , bla KPC-3 , ampC2, aac(3)-IId, dfrA14 | iha, sat, vat | IncFIB(pQil)_1_pQil_JN233705, Col(BS512)_1_NC_010656 |
| sk136y136t | 23 January 2017 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, bla TEM-168 , bla KPC-2 , sulII, bla OXA-9 , bla CTX-M-27 , ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk137y137t | 5 June 2017 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, blaTEM-150, blaKPC-2, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk138y138t | 22 January 2016 | 35 | oqxBgb, bla KPC-2 , bla TEM-79 , bla OXA-9 , oqxA, tetD, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk139y139t | 16 February 2016 | 3033 | strA, strB, oqxBgb, catB4, bla KPC-2 , bla TEM-198 , bla OXA-1 , bla OXA-9 , bla CTX-M-15 , oqxA, dfrA14, sulII, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk140y140t | 25 February 2016 | 17 | oqxBgb, bla KPC-2 , bla TEM-122 , bla OXA-9 , oqxA, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk141y141t | 25 February 2016 | 35 | oqxBgb, bla KPC-2 , bla TEM-122 , bla OXA-9 , oqxA, tetD, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk142y142t | 22 March 2016 | 2279 | strA, strB, oqxBgb, bla KPC-2 , bla TEM-198 , bla OXA-9 , aac(3)-Iia, bla CTX-M-15 , oqxA, dfrA14, sulII, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk143y143t | 31 March 2016 | 17 | oqxBgb, bla KPC-2 , bla TEM-122 , bla OXA-9 , oqxA, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 |
The virulence genes sat (secret autotransporter toxin), iss (increased serum survival) and gad (glutamate decarboxylase) were found in all ST131 strains, while the iha (adherence protein) determinant was found in all isolates but one ST131. The senB (enterotoxin) gene was present in the majority of ST131 isolates, while the cnf1 (cytotoxic necrotizing factor type 1) was only identified in the isolate sk35y35t, which was the first KPC-Ec that emerged in the outbreak period. A different pattern of virulence genes was detected in the two ST978 strains, consisting of vat (vacuolating autotransporter toxin), pic (serine protease) and gad determinants. Lastly, the ST1193 isolate harbored iha, sat and vat as virulence genes (Table 3).
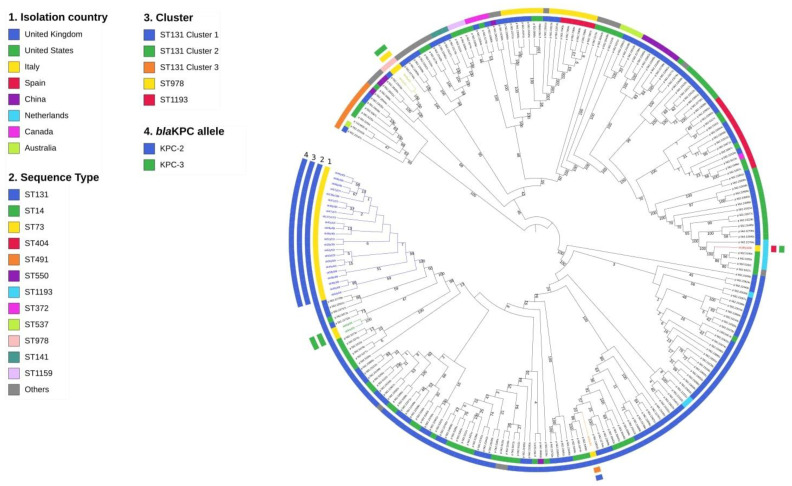
The comparison of the 29 KPC-Ec isolates with other E. coli genomes deposited in the PATRIC database is shown in Figure 1. Notably, most of them showed a major similarity with KPC-Ec strains collected in the UK.
Figure 1.
Maximum likelihood phylogenetic tree including the 29 KPC-producing Escherichia coli isolates and background strains retrieved from the PATRIC database. The clusters were identified as monophyletic highly supported groups and are reported on the colored inner ring. The geographic origin of all the strains is reported on the second level colored ring. The Sequence Type of the strains (Achtman scheme) is reported on the third colored ring, while the KPC variant is on the outer ring.
The phylogenetic reconstruction showed the presence of five distinct clusters, three of which belonged to ST131 (“ST131 cluster 1”, “ST131 cluster 2” and “ST131 cluster 3”), one to ST978 and another to ST1193 (Figure 1). The ST131 cluster 1, characteristic of the majority of isolates (n = 26), was found in all wards with the exception of the Intensive Care and the Infectious Diseases Units (Figure 2), thus representing the outbreak epidemic clone.
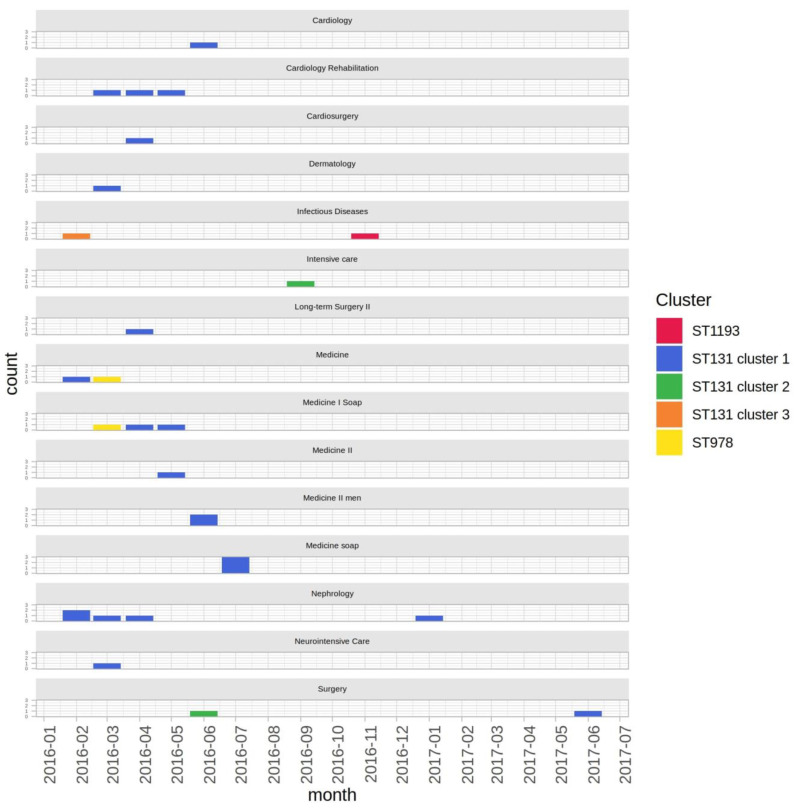
Figure 2.
Barplot of the number of isolates per month per ward. Bars were colored on the basis of the phylogenetic clusters shown in Figure 1.
Notably, sporadic isolates belonging to the ST131 cluster 1 were observed till June 2017. Based on phylogenetic data, the first isolate belonging to the ST131 cluster 1 (i.e., the outbreak index strain, coded sk36y36t) was isolated in the Nephrology Ward. The patient had been previously hospitalized for long periods in the past years in both Nephrology and ICU. Of note, the index patient was previously colonized by KPC-Kp (strain code, sk138y138t) (Table 2). The ST131 cluster 2 included two isolates, collected in Surgery and ICU in June and September 2016, respectively. The ST131 cluster 3 was represented by a single isolate, obtained in February 2016 from the Infectious Diseases Ward. ST978 strains were recovered in March 2016 from the blood culture and rectal swab of the same patient (Table 2 and Figure 2). Lastly, the ST1193 strain was isolated in November 2016 in the Infectious Diseases Ward. Figure 2 shows the ward distribution of the KPC-Ec strains belonging to different clusters and STs.
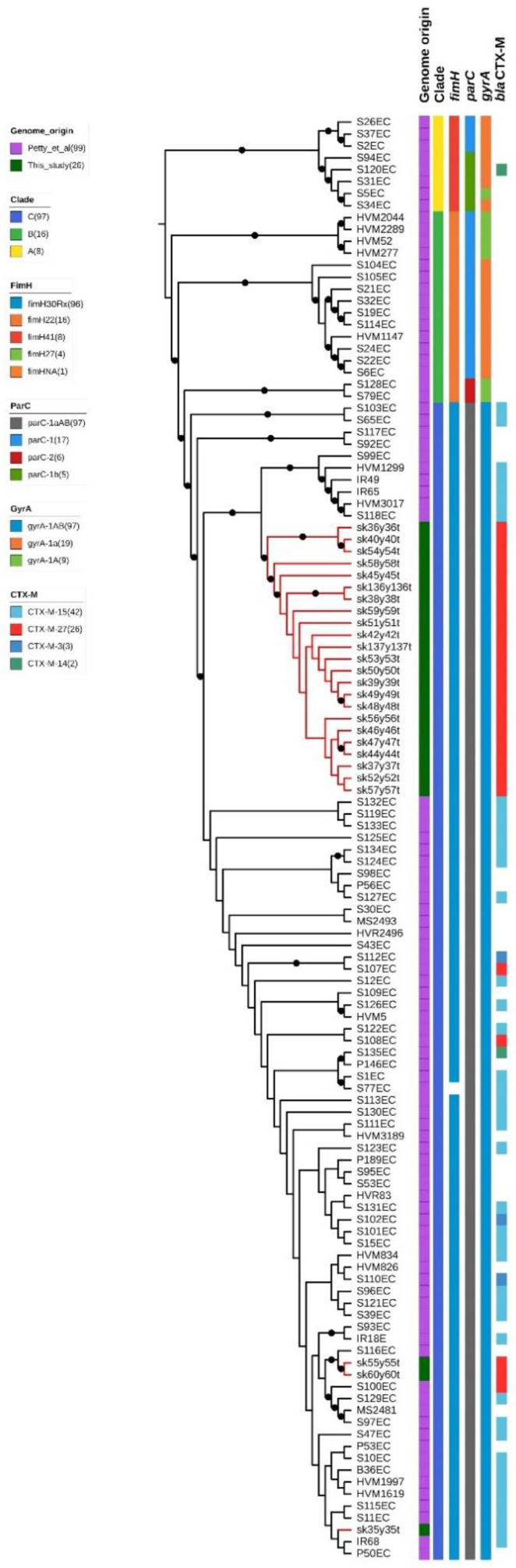
Recent studies have focused on deciphering the genomic evolution and diversity within the ST131 lineage [4,20,21]. Since three different ST131 clusters were identified, we compared their genomes with those published by Petty and colleagues [20]. As shown in Figure 3, all the studied strains could be assigned to the ST131 clade C, characterized by the fimbrial variant fimH30Rx, and the gyrA1AB and parC1aAB alleles, associated with fluoroquinolone resistance. Phylogeny highlighted that the only difference was the presence of the blaCTX-M-27 gene variant instead of blaCTX-M-15 for ST131 clusters 1 and 3.
Figure 3.
Maximum likelihood phylogenetic tree including the here studied ST131 strains and the ones from Petty et al. [20]. The branches corresponding to the strains of this study are highlighted in red. Metadata have been represented as follows: the first column indicates the origin of the strains; the second one the ST131 clades as called by Petty et al. [20]. The subsequent four columns report the allele variants of the fimH, parC, gyrA and blaCTX-M genes.
The six KPC-Kp strains, isolated during the early stages of the outbreak, were investigated for ST and plasmid incompatibility group arrangement. KPC-Kp strains belonged to ST35 (n = 2, from Nephrology and Urology), ST17 (n = 2, from the Rehabilitation Unit and Internal Medicine), ST3033 (n = 1, from Cardiology) and ST2279 (n = 1, from Infectious Diseases).
The positive colonization date (and co-isolation date, when it occurred) and related clinical samples of KPC-Ec and KPC-Kp isolates involved in WGS analyses are shown in Supplemental Tables S1 and S2.
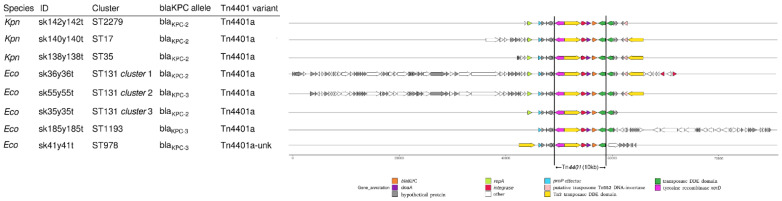
Regarding plasmids, IncFIBpQil was found in all the ST131 and ST1193 KPC-Ec strains, as well as in all the KPC-Kp strains. IncFII was found in all the ST131 cluster 1 and cluster 2 KPC-Ec strains; Col156 was found in all the ST131 cluster 1 isolates but one. IncX3 was detected in both the ST978 isolates. Lastly, all the KPC-Kp strains harbored IncFIB(K)_Kpn3. The KPC-Ec strains belonging to the epidemic ST131 of both cluster 1 and cluster 3 produced the KPC-2 enzyme, as well as the KPC-Kp isolates. The blaKPC-3 allele was instead found in the ST131 cluster 2, ST1193 and ST978 strains. Analysis of the blaKPC surrounding genetic environment showed that all clusters, except ST978 and ST1193, were characterized by a conserved blaKPC scaffold (Figure 4). ST978 and ST1193 showed two different scaffolds. All the clusters harbored the Tn4401a, a structural variant of the Tn4401 transposon.
Figure 4.
Arrow plot of the different blaKPC gene contigs aligned on the Tn4401 transposon (shown inside the vertical lines). The plot includes one representative strain for each Escherichia coli cluster and one for each Klebsiella pneumoniae ST.
Given the presence of the IncFIBpQil, a well-known KPC-harboring plasmid, the detection of the KPC-2 enzyme, and evaluating the extreme similarity of the blaKPC genomic environment in KPC-Kp and KPC-Ec isolates belonging to the ST131 cluster 1; a plasmid-mediated transmission of the KPC-2 determinant from KPC-Kp to KPC-Ec isolates could reasonably have happened.
Although different temperature conditions were tested, conjugation assay results were negative, thus indicating that the resistance determinants were not located on a conjugative plasmid.
3. Discussion
Our study describes a large intrahospital outbreak caused by KPC-Ec, involving a total of 106 inpatients in 25 wards. To the best of our knowledge, this is the largest outbreak caused by KPC-Ec reported worldwide. Genomic analyses allowed us to ascertain that five different KPC-Ec clusters were involved, three of which belonged to the high-risk ST131 clone. Notably, only the “cluster 1” was responsible for the epidemic event and was associated to the H30Rx clade C sub-clone. The ST131 cluster 1 strains co-harbored blaCTX-M-27 and blaKPC determinants and were the only ones provided with the senB virulence gene, coding for the secreted enterotoxin TieB.
Although its relevant ability to spread among patients in different wards, ST131 KPC-Ec showed a low propensity to cause infections. In fact, most samples were collected from rectal swabs, whereas only 20 isolates (16.2%) were obtained from other biological specimens. Of note, only two isolates (1.6%) were from blood cultures. Overall, no deaths were attributable to infection caused by the ST131 KPC-Ec cluster 1 outbreak clone. Conversely, the treatment of infections caused by ST131 KPC-Ec was a challenge. In fact, isolates were mostly nonsusceptible to beta-lactams, including carbapenems and ceftolozane/tazobactam, due to the presence of the blaKPC and blaCTX-M determinants. KPC-Ec were also mostly resistant to ciprofloxacin and trimethoprim-sulphamethoxazole, another typical feature related to the pandemic ST131 lineage. Therapeutic options were aminoglycosides, tigecycline, colistin and the ceftazidime/avibactam combination.
The dissemination of the blaKPC determinant in E. coli, largely due to horizontal transfer of plasmids or other mobile elements into diverse genetic backgrounds, has been previously described [19]. Since the acquisition of blaKPC by E. coli is a very uncommon event, no data are to date available in the literature about a different prevalence between blaKPC-2 and blaKPC-3 variants. Nonetheless, in our experience, the blaKPC-2 gene seems to be the most represented in Italy, mainly associated with ST131 (unpublished data from a multicentric clinical study). The blaKPC genetic background was conserved among both the KPC-Ec ST131 cluster 1 and the KPC-Kp strains of the same period, suggesting that transmission events of plasmid/mobile elements occurred. More importantly, the index patient was colonized during the same time period by both KPC-Kp (strain code sk138y138t) and KPC-Ec (strain code sk36y36t) strains (Table 2), the last one reasonably representing the origin of the outbreak. On the other hand, conjugation assays showed that the blaKPC gene was not located on a conjugative plasmid, highlighting that the outbreak was caused by the spread of a dominant clone that had acquired the plasmid, rather than by the dissemination of a resistance plasmid to unrelated strains. Transmissions were drastically interrupted in May 2016, thanks to the adoption of a strict cohorting of both colonized and infected patients, that was assisted by dedicated healthcare staff. During the outbreak period, infection prevention and control measures were implemented. Training courses for the staff based on infection control, contact precautions and hand washing campaigns were promoted. Only sporadic cases related to KPC-Ec (ST131 cluster 1) were observed till June 2017, representing the tail of the outbreak. After this period, no other cases related to KPC-Ec were observed.
Taking together epidemiological and WGS data, we can speculate that the KPC-Ec outbreak clone developed in the previous context of the KPC-Kp outbreak. Furthermore, the outbreak appeared to be caused by the diffusion of a dominant clone (that probably acquired a blaKPC-harboring plasmid), and not by the dissemination of a resistance plasmid among the strains of the different clusters.
Rapid application of WGS in outbreak investigations could be useful to better understand the dynamics of epidemic events in order to address infection control and contrast interventions.
4. Materials and Methods
4.1. Epidemiological Context and Characterization of Bacterial Isolates
We retrospectively studied an outbreak caused by KPC-Ec that occurred at the Hospital of Lecco (Northern Italy, close to Milan) across a six-month period (February to July 2016). The hospital accounts for about one thousand beds, and has a catchment area of about 340,000 inhabitants. During the outbreak period, as a part of the surveillance activity of the hospital team for infection control, 123 KPC-Ec nonrepetitive isolates were collected from 106 patients. A total of 103 isolates were from colonization surveillance rectal swabs. Rectal swabs have been used to screen intestinal colonization by KPC-Ec, as indicated by international guidelines (Centers for Diseases Control and Prevention, CDC, 2015; https://www.cdc.gov/hai/pdfs/cre/cre-guidance-508.pdf, accessed on 14 June 2021). Twenty strains from other sites were isolated from symptomatic patients with suspected infection. These isolates were from urine (n = 11), blood (n = 2), purulent exudate (n = 2), respiratory secretions (n = 2), drainage fluid (n = 1), peritoneal fluid (n = 1) and a surgical wound swab (n = 1). Strains from the same patients were included only when isolated from different sites. A further four KPC-Ec isolates were sporadically collected after the epidemic event and until June 2017 and were also investigated to verify their clonal relationship to outbreak strains. To better clarify the origin of the blaKPC gene in the KPC-Ec strains, six KPC-Kp isolates were collected from patients cocolonized by KPC-Kp and KPC-Ec and were included in the study. Of them, two were obtained at the beginning of the episode (including those isolated from the index patient), while the remaining were collected during the outbreak from patients admitted to those wards that were mainly involved in the episode. Isolates from rectal swabs were screened for carbapenemase production using chromogenic Brilliance CRE agar (Thermo Fisher Scientific). Bacterial isolates were identified to the species level using MALDI-TOF mass spectrometry (Vitek MS, bioMérieux), while susceptibility testing was routinely determined by the Vitek 2 system (bioMérieux). Isolates suspected of carbapenemase production (MIC values for ertapenem and/or meropenem >0.125 mg/L) were evaluated to assess the presence of specific carbapenem resistance determinants using the immunochromatographic technique (RESIST-4 O.K.N.V., Coris BioConcept) and/or a molecular dedicated assay (Xpert Carba-R, Cepheid).
To characterize epidemic isolates and better understand the dynamics of the outbreak, a total of 29 KPC-Ec strains (four of which were sporadically isolated in the post-outbreak period) were selected as representatives based on the site of infection or colonization, date and ward of admission (Table 2). MIC values of these isolates were determined by the MicroScan autoSCAN-4 system (NMDRM1 panel, Beckman Coulter). Selected antimicrobials (i.e., ceftazidime-avibactam, ceftolozane/tazobactam, and colistin) were evaluated by a broth microdilution Sensititre panel used for multidrug-resistant Gram-negative strains (DKMGN panel, Thermo Fisher Scientific). EUCAST criteria were used for determining susceptibility categories [22]. Finally, these isolates were analyzed by WGS-based typing and SNP-based phylogenetic reconstruction.
4.2. High Resolution Melting Assay
The selection of KPC-producing K. pneumoniae strains chosen for WGS investigation was made on the basis of the High Resolution Melting (HRM) assay results. The HRM was performed on the wzi hypervariable capsular gene as described by Perini et al. [23] using MeltingPlot software [24].
4.3. Whole-Genome Sequencing
A total of 35 strains (29 KPC-Ec and 6 KPC-Kp), representative of the epidemic event and of the post outbreak period, were processed for WGS analysis. In detail, inclusion criteria were: (i) isolates from ascertained infections (other than from screening rectal swabs) were chosen preferentially; (ii) KPC-Ec from all hospital wards involved in the epidemic event; (iii) KPC-Ec isolated in different periods of the outbreak (at the beginning, medium period, tail of the outbreak); (iv) In addition, 8 KPC-Ec from rectal swabs were included in order to evaluate the presence of the outbreak clone in the patients’ intestinal microbiota. Genomic DNA was extracted using a QIAamp DNA minikit (Qiagen) following the manufacturer’s instructions and sequenced using the Illumina Miseq platform with a 2 × 250 paired-end run after Nextera XT library preparation (Illumina Inc., San Diego, CA, USA).
4.4. CoreSNP Calling and Phylogenetic Analyses
For each of the 35 strains included in the study, the reads quality was assessed using FastqC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 29 June 2020), and the low quality terminal bases were trimmed using Trimmomatic software [25]. Reads were assembled using SPAdes software [26].
All the genome assemblies were submitted to the European Nucleotide Archive (ENA) with the project code PRJEB40388. All the ID codes are listed in Table S3.
The genome distance of Each KPC-Ec genome assembly was estimated using Mash software [27] against a collection of 3325 E. coli genomes retrieved from the PATRIC database [28] and the 50 most similar genomes were selected for subsequent analyses. All the selected genome assemblies (from this study and the PATRIC database) were aligned against the E. coli MG1655 reference genome using progressive Mauve and coreSNPs were called as described by Gona and colleagues [29]. Repeated regions in the reference genome assembly were detected using Blastn. Then coreSNPs localized within repeated regions were masked. From here, the obtained coreSNP alignment was called “Global coreSNP”.
Global coreSNP alignments were subjected to phylogenetic analysis using RAxML software with a 100 pseudo-bootstrap, after best model selection using ModelTest-NG [30]. The strains were then clustered on the basis of the Global phylogenetic tree and SNP distance. At first, we identified on the ML phylogenetic tree the largest highly supported (>75 bootstrap) monophyletic groups including study strains only.
4.5. Whole-Genome Sequencing-Based Typing
Resistance genes of the 29 KPC-Ec strains were identified using the ResFinder online tool [31] and SRST2 software [32] with the ARG-ANNOT dataset [33]. Virulence genes were detected by the VirulenceFinder online tool [34]. Plasmid incompatibility groups were detected using PlasmidFinder [35]. The Multi Locus Sequence Typing profiles of the 29 E. coli and six K. pneumoniae strains were determined in silico according to the Achtman and Pasteur schemes, respectively, using an in-house Perl script.
4.6. KPC-Harboring Contigs Comparison
For each identified cluster (see above) one representative strain was selected and the genome assembly was analyzed as follows. The contig harboring blaKPC gene was identified by Blastn search (E-value threshold: 0.00001). The extracted contigs were oriented on the basis of blaKPC gene orientation, then annotated using Prokka [36] and aligned with progressive Mauve [37]. The Tn4401 transposon was annotated using TETyper [38]. Lastly, the gene composition and synteny of extracted contigs were graphically represented using the R library genoPlotR [39].
4.7. Conjugation Assay
To assess the possible transferability of resistance determinants identified, a conjugation assay was performed using the E. coli J53 AzideR as the recipient strain at temperatures of both 25 °C and 37 °C, and with MER 0.5mg/L for the selection of transconjugants.
5. Conclusions
Although KPC-producing Escherichia coli (KPC-Ec) remains uncommon, and mainly reported as the cause of sporadic episodes of infection rather than outbreaks, the present work shows that the acquisition of blaKPC gene by a high-risk successful clone, as the ST131, can lead to even large and potentially difficult to manage epidemic events. The attention on the presence and circulation of carbapenemase-producing enterobacteria (CPE) should be always kept high, especially in healthcare settings. In this context, the application of WGS could be useful to better understand the evolution and dynamic of outbreaks sustained by CPE in order to promptly address infection control and contrast interventions.
Acknowledgments
Thanks to the Romeo and Enrica Invernizzi Foundation for its general support.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10060718/s1, Figure S1. Local prevalence of KPC-Kp and KPC-Ec in the period 2014–2016 in the hospital setting. Table S1. Escherichia coli (KPC-Ec) isolates selected for WGS analysis and co-isolation of Klebsiella pneumoniae (KPC-Kp) isolates from the same patients. Table S2. Klebsiella pneumoniae (KPC-Kp) isolates selected for WGS analysis and co-isolation of Escherichia coli (KPC-Ec) isolates from the same patients. Table S3. List of the ENA codes for the genome assemblies of the studied strains.
Author Contributions
A.P. conceived the study, performed microbiological experiments and sequencing, analyzed and interpreted the data and wrote the paper; L.P. conceived the study, performed microbiological experiments, analyzed and interpreted the data and drafted the paper; F.C. and M.P. performed bioinformatic analyses; E.M. performed microbiological experiments and provided the epidemiological data; V.M.M. performed microbiological experiments and the conjugation assay; R.M. conceived the study, supervised the activities and revised the manuscript; F.L. conceived the study, supervised the activities and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in the context of normal clinical routine. All information and metadata about patients had been anonymized. Samples were coded and analyses were performed with anonymized database.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome assemblies of the sequenced strains are available at the European Nucleotide Archive (ENA). The ID code for each genome submitted is reported in Supplementary Table S3.
Conflicts of Interest
All the authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brolund A., Lagerqvist N., Byfors S., Struelens M.J., Monnet D.L., Albiger B., Kohlenberg A., European Antimicrobial Resistance Genes Surveillance Network EURGen-Net Capacity Survey Group Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.9.1900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decraene V., Phan H.T.T., George R., Wyllie D.H., Akinremi O., Aiken Z., Cleary P., Dodgson A., Pankhurst L., Crook D.W., et al. A Large, Refractory Nosocomial Outbreak of Klebsiella pneumoniae Carbapenemase-Producing Escherichia coli demonstrates carbapenemase gene outbreaks involving sink sites require novel approaches to infection control. Antimicrob. Agents Chemother. 2018;62:e01689-18. doi: 10.1128/AAC.01689-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoesser N., Sheppard A.E., Peirano G., Anson L.W., Pankhurst L., Sebra R., Phan H.T.T., Kasarskis A., Mathers A.J., Peto T.E.A., et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci. Rep. 2017;7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoesser N., Sheppard A.E., Pankhurst L., De Maio N., Moore C.E., Sebra R., Turner P., Anson L.W., Kasarskis A., Batty E.M., et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grundmann H., Glasner C., Albiger B., Aanensen D.M., Tomlinson C.T., Andrasević A.T., Cantón R., Carmeli Y., Friedrich A.W., Giske C.G., et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): A prospective, multinational study. Lancet Infect Dis. 2017;17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 6.Petrella S., Ziental-Gelus N., Mayer C., Renard M., Jarlier V., Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 2008;52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urban C., Bradford P.A., Tuckman M., Segal-Maurer S., Wehbeh W., Grenner L., Colon-Urban R., Mariano N., Rahal J.J. Carbapenem-resistant Escherichia coli harboring Klebsiella pneumoniae carbapenemase beta-lactamases associated with long-term care facilities. Clin Infect Dis. 2008;46:e127–e130. doi: 10.1086/588048. [DOI] [PubMed] [Google Scholar]
- 8.Goren M.G., Navon-Venezia S., Chmelnitsky I., Carmeli Y. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv Medical Center, 2005 to 2008. Antimicrob. Agents Chemother. 2010;54:2687–2691. doi: 10.1128/AAC.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naas T., Cuzon G., Gaillot O., Courcol R., Nordmann P. When carbapenem-hydrolyzing beta-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob. Agents Chemother. 2011;55:4933–4934. doi: 10.1128/AAC.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris D., Boyle F., Ludden C., Condon I., Hale J., O’Connell N., Power L., Boo T.W., Dhanji H., Lavallee C., et al. Production of KPC-2 carbapenemase by an Escherichia coli clinical isolate belonging to the international ST131 clone. Antimicrob. Agents Chemother. 2011;55:4935–4936. doi: 10.1128/AAC.05127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giani T., Antonelli A., Caltagirone M., Mauri C., Nicchi J., Arena F., Nucleo E., Bracco S., Pantosti A., Luzzaro F., et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:30583. doi: 10.2807/1560-7917.ES.2017.22.31.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iacchini S., Sabbatucci M., Gagliotti C., Rossolini G.M., Moro M.L., Iannazzo S., D’Ancona F., Pezzotti P., Pantosti A. Bloodstream infections due to carbapenemase-producing Enterobacteriaceae in Italy: Results from nationwide surveillance, 2014 to 2017. Euro Surveill. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.5.1800159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piazza A., Caltagirone M., Bitar I., Nucleo E., Spalla M., Fogato E., D’Angelo R., Pagani L., Migliavacca R. Emergence of Escherichia coli Sequence Type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a Long-Term Care and Rehabilitation Facility (LTCRF) in Northern Italy. Adv. Exp. Med. Biol. 2016;901:77–89. doi: 10.1007/5584_2015_5017. [DOI] [PubMed] [Google Scholar]
- 14.Adler A., Miller-Roll T., Assous M.V., Geffen Y., Paikin S., Schwartz D., Weiner-Well Y., Hussein K., Cohen R., Carmeli Y. A multicenter study of the clonal structure and resistance mechanism of KPC-producing Escherichia coli isolates in Israel. Clin. Microbiol. Infect. 2015;21:230–235. doi: 10.1016/j.cmi.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Mavroidi A., Miriagou V., Malli E., Stefos A., Dalekos G.N., Tzouvelekis L.S., Petinaki E. Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents. 2012;39:247–250. doi: 10.1016/j.ijantimicag.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Landman D., Urban C., Bäcker M., Kelly P., Shah N., Babu E., Bratu S., Quale J. Susceptibility profiles, molecular epidemiology, and detection of KPC-producing Escherichia coli isolates from the New York City vicinity. J. Clin. Microbiol. 2010;48:4604–4607. doi: 10.1128/JCM.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung V., Loo V.G., Frenette C., Domingo M.C., Bourgault A.M., Mulvey M.R., Robson H.G. First Canadian outbreak of Enterobacteriaceae-expressing Klebsiella pneumoniae carbapenemase type 3. Can. J. Infect. Dis. Med. Microbiol. 2012;23:117–120. doi: 10.1155/2012/725151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Accogli M., Giani T., Monaco M., Giufrè M., García-Fernández A., Conte V., D’Ancona F., Pantosti A., Rossolini G.M., Cerquetti M. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J. Antimicrob. Chemother. 2014;69:2293–2296. doi: 10.1093/jac/dku132. [DOI] [PubMed] [Google Scholar]
- 19.Chavda K.D., Chen L., Jacobs M.R., Bonomo R.A., Kreiswirth B.N. Molecular Diversity and Plasmid Analysis of KPC-Producing Escherichia coli. Antimicrob. Agents Chemother. 2016;60:4073–4081. doi: 10.1128/AAC.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petty N.K., Ben Zakour N.L., Stanton-Cook M., Skippington E., Totsika M., Forde B.M., Phan M.D., Gomes Moriel D., Peters K.M., Davies M., et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA. 2014;111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben Zakour N.L., Alsheikh-Hussain A.S., Ashcroft M.M., Khanh Nhu N.T., Roberts L.W., Stanton-Cook M., Schembri M.A., Beatson S.A. Erratum for Ben Zakour et al., Sequential Acquisition of Virulence and Fluoroquinolone Resistance Has Shaped the Evolution of Escherichia coli ST131. MBio. 2016;7:e00958-16. doi: 10.1128/mBio.00958-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing . Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 6.0. EUCAST; Basel, Switzerland: 2016. [(accessed on 14 June 2021)]. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_6.0.pdf. [Google Scholar]
- 23.Perini M., Piazza A., Panelli S., Di Carlo D., Corbella M., Gona F., Vailati F., Marone P., Cirillo D.M., Farina C., et al. EasyPrimer: User-friendly tool for pan-PCR/HRM primers design. Development of an HRM protocol on wzi gene for fast Klebsiella pneumoniae typing. Sci. Rep. 2020;10:1307. doi: 10.1038/s41598-020-57742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perini M., Batisti Biffignandi G., Di Carlo D., Pasala A.R., Piazza A., Panelli S., Zuccotti G.V., Comandatore F. MeltingPlot, a user-friendly online tool for epidemiological investigation using High Resolution Melting data. BMC Bioinform. 2021;22:76. doi: 10.1186/s12859-021-04020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nurk S., Bankevich A., Antipov D., Gurevich A.A., Korobeynikov A., Lapidus A., Prjibelski A.D., Pyshkin A., Sirotkin A., Sirotkin Y., et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S., Phillippy A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L., et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gona F., Comandatore F., Battaglia S., Piazza A., Trovato A., Lorenzin G., Cichero P., Biancardi A., Nizzero P., Moro M., et al. Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb. Genom. 2020;6:e000347. doi: 10.1099/mgen.0.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inouye M., Dashnow H., Raven L.A., Schultz M.B., Pope B.J., Tomita T., Zobel J., Holt K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L., Rolain J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Micobiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Darling A.E., Mau B., Perna N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheppard A.E., Stoesser N., German-Mesner I., Vegesana K., Sarah Walker A., Crook D.W., Mathers A.J. TETyper: A bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb. Genom. 2018;4:e000232. doi: 10.1099/mgen.0.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy L., Kultima J.R., Andersson Siv G.E. genoPlotR: Comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assemblies of the sequenced strains are available at the European Nucleotide Archive (ENA). The ID code for each genome submitted is reported in Supplementary Table S3.