Abstract
ABCC multidrug resistance-associated proteins (ABCCs/MRPs), a subfamily of ABC transporters, are involved in multiple physiological processes. Although these proteins have been characterized in some plants, limited efforts have been made to address their possible roles in Rehmannia glutinosa, a medicinal plant. Here, we scanned R. glutinosa transcriptome sequences and identified 18 RgABCC genes by in silico analysis. Sequence alignment revealed that the RgABCCs were closely phylogenetically related and highly conserved with other plant ABCCs/MRPs. Subcellular localization revealed that most of the RgABCCs were deposited in vacuoles and a few in plasma membranes. Tissue-specific expression of the RgABCCs indicated significant specific accumulation patterns, implicating their roles in the respective tissues. Differential temporal expression patterns of the RgABCCs exhibited their potential roles during root development. Various abiotic stress and hormone treatment experiments indicated that some RgABCCs could be transcriptionally regulated in roots. Furthermore, the transcription of several RgABCCs in roots was strongly activated by cadmium (Cd), suggesting possible roles under heavy metal stresses. Functional analysis of RgABCC1 heterologous expression revealed that it may increase the tolerance to Cd in yeast, implying its Cd transport activity. Our study provides a detailed inventory and molecular characterization of the RgABCCs and valuable information for exploring their functions in R. glutinosa.
Introduction
ATP-binding cassette (ABC) transporters are one of the largest known superfamilies of membrane transporters in all living organisms [1,2]. ABCC transporters, which belong to one subclass of the ABC transporter superfamily, are well known for their roles as multidrug resistance-associated proteins (MRPs), which are found in all eukaryotic organisms [3–7]. These proteins shuttle substrates as diverse as glutathione conjugates, xenobiotic compounds, intermediate metabolites and hormones across a variety of biological membranes [2,5,8,9]. ABCC transporters from several plants were shown to be responsible for a multitude of functions that included vacuolar sequestration of secondary metabolites [7,10] and heavy metals [2,9], phytohormone transport [5,11], development of plant tissues and response to various stresses [2,5,12].
Each ABCC protein contains at least one highly conserved ATPase domain as an energy source (~200 aa residues long), also referred to as a nucleotide binding domain (NBD) [13]. This ATPase domain comprises Walker A motif and Walker B motif on either end of an ABC signature motif [14]. Corresponding to their sequence similarity, ABCC transporter proteins are classified into two types (i.e., full- and half-molecule) according to their structure and forward orientation [15]. Typically, ABCC proteins, as full-molecule ABC transporters, usually contain two transmembrane domains (TMDs), and two NBDs represented as TMD1-NBD1-TMD2-NBD2 [2,13]. Few half-molecule ABCC transporters are composed of one TMD and one NBD and can be forward (TMD1-NBD1) [16,17]. To date, a number of ABCC transporters have been explored from approximately twenty plant species, such as Arabidopsis thaliana (AtABCC1-15) [18], Oryza sativa (OsABCC1-17) [19], Brassica napus (BnABCC1-48) [20], Vitis vinifera (VvABCC1-26) [21], Triticum aestivum (TaABCC1-18) [5], Populus trichocarpa (PtrABCC1-29) [22], and Fragaria vesca (FvABCC1-16) [2] (S1 Table).
Rehmannia glutinosa, a species of the Scrophulariaceae family, is a perennial herbaceous plant. Its tuberous roots contain a diverse range of pharmacologically active compounds, such as secondary metabolites, possessing various medicinal properties and economic value [23]. Although some ABCC/MRP transporters have been identified or functionally characterized in plants, few attempts have been made to characterize this subfamily from R. glutinosa. As the genome of R. glutinosa is still unknown, with the high-efficiency assembly of the transcriptome of this species and the development of bioinformatics tools, the investigation of its ABCC genes has become possible [24]. To address the importance of transporters in diverse physiological processes in R. glutinosa, the molecular structures, phylogeny and conservation of the putative R. glutinosa ABCC genes (RgABCC) were predicted by in silico analysis. We analysed RgABCC subcellular localization, investigated their spatio-temporal expression patterns in R. glutinosa and ascertained their responses to various abiotic stresses as well as plant hormones and Cd stresses. As yeast cadmium factor (YCF1), an ABCC/MRP transporter, was characterized for its possible role in the vacuolar transport of heavy metal sequestration and certain secondary metabolites [5,25], YCF defective (ΔYCF1) mutants have been utilized as an important resource to address the functional activity of ABCC/MRP orthologues across the kingdom [2,26,27]. Our study also analysed the transport activity of RgABCC1 by YCF1 functional complementation. This laid a basis for further revealing the molecular functions of this subfamily of transporters in R. glutinosa.
Materials and methods
In silico analysis
The R. glutinosa transcriptome data were archived at the NCBI SRA (accession numbers: PRJNA197434) and were assembled and annotated against public data by Li et al. [23]. Based on the transcriptome annotation, putative sequences encoding ABCC proteins of R. glutinosa were obtained, and their open-reading frames (ORFs) sequences were deduced using the ORFfinder tool [28]. The full-length RgABCC protein sequences were screened for the characteristic features of Walker A, B, and ABCC-MRP-like ATPase domains as defined in the NCBI conserved domains database (CDD) [29] and ScanProsite [30] tools. The RgABCC physicochemical properties were analysed by the ProtParam tool [31]. Topological analysis of the transmembrane helices (TMs) of the RgABCCs was conducted using the TMHMM-2.0 program [32], and their subcellular localizations were predicted using the Plant-mPLoc program [33]. The phylogeny of ABCCs was inferred using the neighbour-joining method implemented in the MEGA v7.0 package, applying 1000 bootstrap replicates and a Poisson correction [34]. The RgABCCs were named according to nomenclature guidelines and the inventory of A. thaliana ABCC sequences [14]. The domain topology and arrangement of the RgABCCs was predicted using the ScanProsite tool [30,35]. WebLogo3-generated sequence logos were analysed for the presence of representative ABCC domains [36]. Sequence alignment of the ABCC sequences was performed using ClustalX v2.0 software [37].
Plant material culture
The R. glutinosa cultivar “Wen 85–5” was cultured in pots in a greenhouse (under a constant temperature of 26°C with a 14-h light/10-h dark cycle and 65% humidity) at the College of Bioengineering, Henan University of Technology. To clone full-ORFs and test tissue-specific expression patterns of RgABCC genes, various tissues (including roots, stems, young leaves, functional leaves and old leaves) from five R. glutinosa plants were sampled at early root expansion stages (i.e., 80 days of cultivation), which is a key point in the transition from fibrous roots to tuberous roots and should be more vigorous in the plant’s gene expression according our previous researches [23,24,38]. To test RgABCC temporal expression during root development, roots from five independent plants were collected at the seedling (40 days of cultivation), root elongation (60 days of cultivation), root expansion (early, 80 days, middle, 100 days and late, 120 days of cultivation), and maturity (150 days of cultivation) stages [38]. All the samples were frozen in liquid nitrogen for RNA extraction.
For the various stress treatments, the potted R. glutinosa seedlings in the above greenhouse were treated at the seedling stage. For the heat treatment, the plants were exposed to 42°C for 24 h. The plants in the salinity and H2O2 stress treatments were watered with 100 mL of NaCl (150 mM) and H2O2 (10 mM) solution, respectively. The seedlings not subjected to stress treatment were used as the experimental control. After 24 h of incubation, roots from each plant were collected. For hormone treatments, the potted plants were sprayed with 20 mL of 0.1 mM abscisic acid (ABA), 2 mM ethylene (ETH), and 0.05% gibberellic acid (GA3), and the control plants were sprayed with distilled water. After 24 h of incubation, roots of each of these plants were collected. For Cd stress treatment, the seedlings were irrigated with 100 mL of 100 μM CdCl2 solution. The roots of these plants were collected at 0 (i.e., untreated samples as a control), 6, 12, 24, 36 and 48 h. All the samples were stored at −80°C prior to RNA extraction. Three replicates of five plants per pot were used for the above experiments.
Total RNA isolation and reverse transcription
Total RNA of each tissue sample was extracted using TRIzol™ reagent (Invitrogen, Carlsbad, USA), as recommended by the manufacturer. The RNA concentration was measured spectrophotometrically with a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA), and its integrity was evaluated through agarose gel electrophoresis. A 1-μg aliquot of total RNA from each sample was reverse-transcribed into cDNA using HiScript III Reverse Transcriptase (Vazyme, Nanjing, China) according to the manufacturer’s instructions.
Construction of RgABCC vectors
For the RgABCC destination vectors, primers for the amplification of the full ORF sequences of the RgABCCs extending from the gene’s upstream “ATG” start codon site to the downstream region, including the stop codon, were designed by using Oligo 7.0 software and are shown in S2 Table. The RgABCC cDNAs were amplified by polymerase chain reaction (PCR) using PrimeSTAR HS DNA Polymerase. The products were purified with the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit (Takara, Tokyo, Japan) and subcloned into the pMD-18 vector (Takara, Tokyo, Japan), which was then used to transform E. coli. The constructs were sequenced by the Sanger method (Sangon, Shanghai, China).
For subcellular localization, the ORF cDNAs of the RgABCCs were inserted into the pBI121 vector under the control of the CaMV35S promoter and fused with the N-terminus of the GFP gene to generate the CaMV35S:GFP-RgABCC constructs (S1 Fig). For heterologous expression, the cDNA of RgABCC1 was inserted into the pYES2 vector (Biovector Science Lab, China) under the control of the gal promoter to generate the pYES2-ABCC1 construct (S1 Fig).
Transient expression analysis
The CaMV35S:GFP-RgABCC constructs and the CaMV35S:GFP empty vector were transformed with Agrobacterium tumefaciens GV3101 strains using the freeze-thaw method [39]. For transient expression, the GV3101 strains, which were transformed into the vectors CaMV35S:GFP-RgABCCs and CaMV35S:GFP, were cultured for collection and then infiltrated into onion epidermal cells. The transfected epidermal regions were examined after 48 h of coculture with a fluorescence microscope (FV1000 MPE, Olympus) at an excitation wavelength of 488 nm to visualize GFP fluorescence.
Quantitative real-time PCR analysis
To determine gene expression, these RgABCC primers were designed with Beacon Designer 8.0 software (S3 Table). For quantitative real-time PCR (qRT-PCR) analysis, 2 μg total RNA was reverse-transcribed in a 20 μL reaction containing 5 U M-MLV reverse transcriptase (Takara, Tokyo, Japan) according to the manufacturer’s instructions. Each 25 μL reaction contained 0.2 μM of each primer, 12.5 μL SYBR Premix EX Taq (Takara, Tokyo, Japan) and 100 ng cDNA. The qPCR protocol was as follows: 95°C for 30 s, followed by 36 cycles of 95°C for 10 s, 58–60°C 30 s and 72°C for 30 s to determine the amplicon’s dissociation behaviour. Three biological replicates were included per sample, and three technical replicates were used for each biological replicate. The 2-ΔΔCT method [40] was applied to estimate relative transcript abundances, and the data were normalized to the RgActin gene (Genbank ID: EU526396.1) as an internal reference.
Functional complementation analysis
YCF1 in the wild-type BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) Saccharomyces cerevisiae strain was removed by the Cre-LoxP system method [41], and ΔYCF1 (MATa; ΔYCF1::KanMX2; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) mutant strains were generated. Using the lithium acetate/PEG transformation method [42], the ΔYCF1 mutant cells were transformed with pYES2-RgABCC1 (i.e., pYES2-RgABCC1) and pYES2 empty vectors (named pYES2-RgABCC1-ΔYCF1 and pYES2-ΔYCF1, respectively), and the wild-type BY4741 cells were transformed with pYES2 empty vectors (i.e., pYES2-WT). These cells were precultured in SD-Ura selective liquid medium containing 2% glucose at 30°C for 16 h. The transformed cells were grown overnight to an OD600 of 1.5. Aliquots of the cell suspensions were then serially diluted and spotted on solid medium with or without 60 μM CdCl2. Colonies were visualized after incubating the plates for 3 days at 30°C. In addition, the strains were grown overnight in liquid medium, and then the cultures were diluted in minimal medium to an OD600 of 0.1 in the presence of various concentrations of CdCl2 and incubated for an additional 24 h, after which growth was determined by measuring the OD600 [43].
Statistical analyses
All the data were subjected to one-way analysis of variance (ANOVA) using SPSS 22.0 software. Significant differences in the two comparison datasets were analysed by Student’s t-test (p<0.05 or 0.01). Multiple comparison tests were performed using the least significant difference (LSD) test at p < 0.05.
Results
Identification and characterization of RgABCC transporters
The R. glutinosa transcriptome was scanned to screen a set of 86 unigenes putatively encoding ABCC transporters (S4 Table). Using the CDD and ScanProsite tools, the full ORF of their translated amino acid sequences (Table 1) were confirmed, and 18 ABCCs from R. glutinosa were refined. Based on the phylogenetic relationship of ABCCs from R. glutinosa and A. thaliana (Fig 1A), we designated RgABCC1 through 18 (Table 1), which were submitted in NCBI Genbank (Accession numbers assigned MW355848 through MW355865). The size of these deduced proteins (Table 1) varied from 844 to 1,622 residues; their predicted molecular masses ranged from 94.97 to 182.32 kDa, their predicted pI values ranged from 5.70 to 8.66 and their TMs ranged from 4 to 17 (Table 1).
Table 1. Basic information about the RgABCCs in R. glutinosa.
| Name | Accession number | Length (nt|aa) | Moleculr weight (kDa) | pI | TMD | Topology | Subcelluar location |
|---|---|---|---|---|---|---|---|
| RgABCC1 | MW355848 | 4869|1622 | 182.32 | 6.75 | 14 | (TMD-NBD)2 | vacuole |
| RgABCC2 | MW355849 | 4869|1622 | 182.29 | 6.75 | 14 | (TMD-NBD)2 | vacuole |
| RgABCC3 | MW355850 | 2883|960 | 106.66 | 5.90 | 8 | (TMD-NBD)2 | plasma membrane |
| RgABCC4 | MW355851 | 4488|1495 | 167.64 | 7.49 | 17 | (TMD-NBD)2 | vacuole |
| RgABCC5 | MW355852 | 3114|1037 | 115.31 | 7.47 | 5 | (TMD-NBD)2 | plasma membrane |
| RgABCC6 | MW355853 | 4536|1511 | 169.34 | 6.18 | 15 | (TMD-NBD)2 | plasma membrane |
| RgABCC7 | MW355854 | 4527|1508 | 169.36 | 6.87 | 12 | (TMD-NBD)2 | vacuole |
| RgABCC8 | MW355855 | 4440|1479 | 165.64 | 7.34 | 12 | (TMD-NBD)2 | vacuole |
| RgABCC9 | MW355856 | 2913|970 | 107.78 | 5.70 | 4 | (TMD-NBD)2 | plasma membrane |
| RgABCC10 | MW355857 | 4440|1479 | 165.62 | 7.34 | 12 | (TMD-NBD)2 | plasma membrane |
| RgABCC11 | MW355858 | 3774|1257 | 140.97 | 8.66 | 7 | (TMD-NBD)2 | vacuole |
| RgABCC12 | MW355859 | 3774|1257 | 140.87 | 8.55 | 7 | (TMD-NBD)2 | vacuole |
| RgABCC13 | MW355860 | 4605|1534 | 170.43 | 8.48 | 14 | (TMD-NBD)2 | plasma membrane |
| RgABCC14 | MW355861 | 4488|1495 | 167.60 | 7.49 | 17 | (TMD-NBD)2 | vacuole |
| RgABCC15 | MW355862 | 4527|1508 | 169.33 | 6.89 | 12 | (TMD-NBD)2 | vacuole |
| RgABCC16 | MW355863 | 3774|1257 | 140.40 | 6.68 | 8 | (TMD-NBD)2 | vacuole |
| RgABCC17 | MW355864 | 2535|844 | 94.97 | 5.98 | 5 | TMD-NBD | vacuole |
| RgABCC18 | MW355865 | 2619|872 | 98.09 | 6.82 | 5 | TMD-NBD | vacuole |
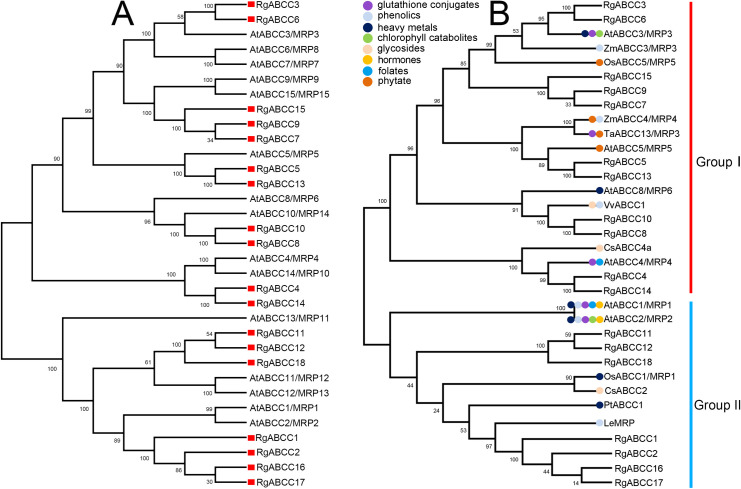
Fig 1. Phylogenetic alignment of plant ABCCs/MRPs.
(A) Phylogenetic tree of ABCCs/MRPs from R. glutinosa and A. thaliana; (B) Phylogenetic tree of the RgABCCs and functionally characterized ABCCs/MRPs from other plants (Note: Coloured dots indicate the class of transported substrates for the functionally characterized ABCCs/MRPs).
To assign their potential functional roles, a phylogenetic tree was constructed that compared the sequences of the RgABCCs with 16 functionally characterized ABCC/MRPs from other plants (Fig 1B and S5 Table). Phylogenetic analysis showed that these ABCCs/MRPs primarily formed a cluster with two separate clades (Group I and Group II). In Group I, RgABCC3 and RgABCC6 were clustered closely together with AtABCC3/MRP3, ZmABCC3/MRP3 and OsABCC3/MRP3; RgABCC7, RgABCC9 and RgABCC15 were clustered together with AtABCC9/MRP9, AtABCC15/MRP15, OsABCC6/MRP6 and OsABCC7/MRP7; RgABCC8 and RgABCC10 were closely related to AtABCC5/AtMRP5; in addition, RgABCC8 and RgABCC10 were closely related to VvABCC1, while RgABCC4 and RgABCC14 exhibited a close relationship to AtABCC4/MRP4 and CsABCC4a. In group II, seven RgABCCs (e.g., RgABCC1, RgABCC2, RgABCC11, RgABCC12, RgABCC16, RgABCC17 and RgABCC18) were found to be closely associated with AtABCC1/MRP1 and AtABCC2/MRP2, and four other known ABCCs (OsABCC1/MRP1, CsABCC2, PtABCC1 and LeMRP) and five RgABCCs (e.g., RgABCC1, RgABCC2, RgABCC16, RgABCC17 and RgABCC18) were found to be closely clustered with PtABCC1.
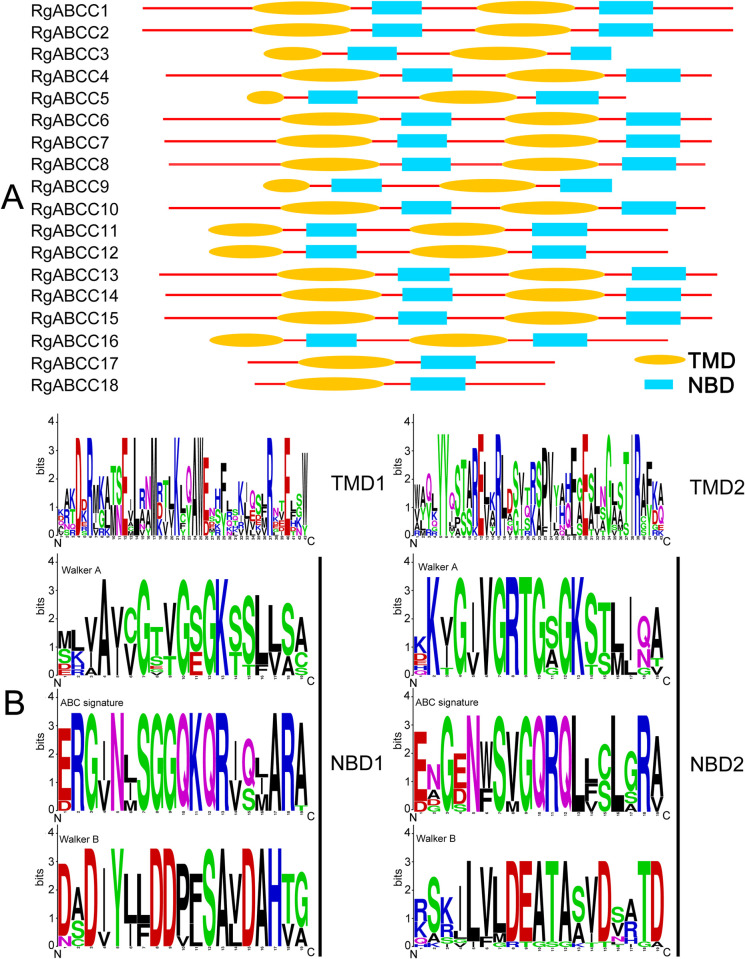
Conserved domain analysis showed that the specific signatures of the RgABCCs were present in 16 full-molecule (TMD-NBD-TMD-NBD) and 2 half-molecule (TMD-NBD) members (Fig 2A and Table 1). Sequence logos strongly suggested more conserved amino acids in the RgABCC domains (Fig 2B). All of the RgABCCs had a mainly conserved NBD1 represented by GTVGSGK amino acids of the Walker A motif, SGGQKQR of the ABC signature motif and IYLLD of the Walker B motif. Similarly, the NBD2 domains with the Walker A (GRTGSGK), the ABC signature (SVGQRQL) and the Walker B (ILVLD) motifs were also highly conserved and present among the full-molecule RgABCCs (Fig 2B). Based on homology analysis among the RgABCCs, RgABCC1 showed the highest identity (97.94%) with RgABCC2 (S6 Table). In contrast, the highest divergence was observed between RgABCC9 and RgABCC17, with 10.43% identity. To assess the conservation of the RgABCCs across various species, we collected 16 functionally characterized ABCC/MRPs from other plants (S7 Table). When a cross-species comparison was performed, the highest percentage identity of 81.18% was observed for RgABCC1 and PtrABCC1 of P. trichocarpa. Similar to A. thaliana, the highest percentage identity of 77.33% was observed for RgABCC1 or RgABCC2 with AtABCC2/MRP2.
Fig 2. Conserved domain analysis of the RgABCCs.
(A) Schematic diagram of the domain arrangement in different RgABCCs; (B) sequence logo of amino acids conserved in different domains (Note: The Y-bits represent conservation of amino acids at that position (height)).
Subcellular localization of the RgABCCs
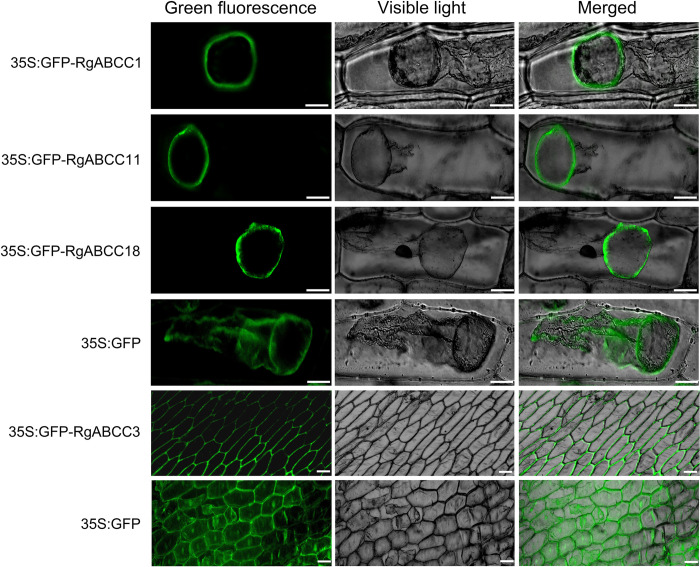
Based on in silico analysis, of these RgABCCs, 12 were predicted to localize to the vacuole and 6 to the plasma membrane (Table 1). Four of them (RgABCC1, RgABCC3, RgABCC11 and RgABCC18) were selected for experimental determination of their subcellular localization. The recombinant proteins from the CaMV35S:GFP-RgABCC constructs were transiently expressed in onion epidermis. Green fluorescence from the fusion proteins of CaMV35S:GFP-RgABCC constructs was mainly observed in the vacuoles (RgABCC1, RgABCC11 and RgABCC18) and plasma membranes (RgABCC3) (Fig 3), while the expression of the control CaMV35S:GFP was detected in the plasma membrane, cytoplasm, nucleus or other cell organelles (Fig 3). The results confirmed the predicted subcellular localizations of these RgABCCs.
Fig 3. Subcellular localization of RgABCCs.
The expression of CaMV35S:GFP-RgABCC fusion proteins in onion epidermal cells (Bars = 320 μm).
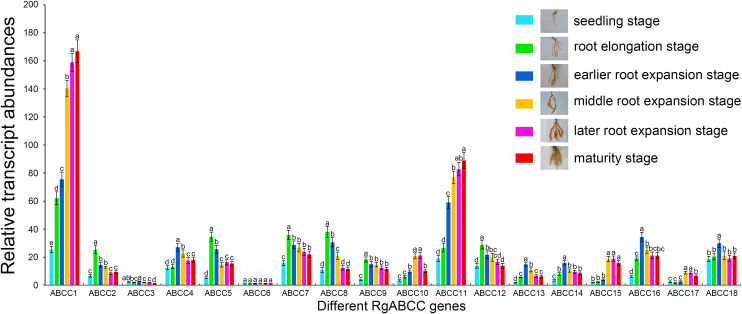
Spatio-temporal expression patterns of the RgABCCs in R. glutinosa
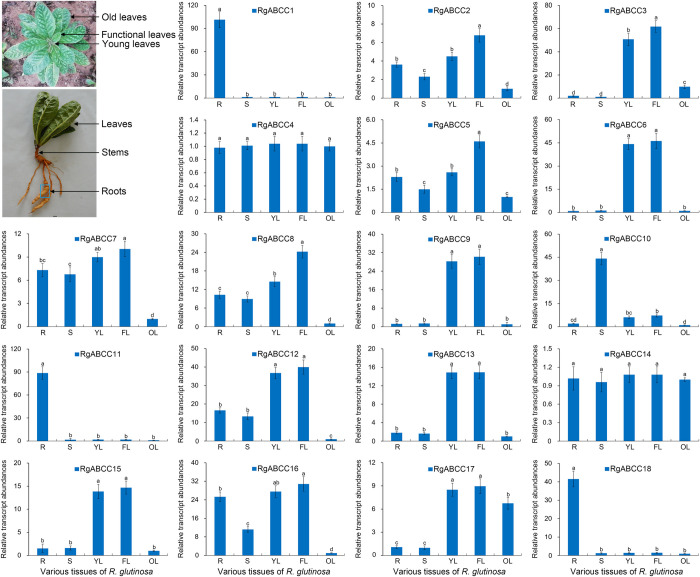
To gain insight into the expression patterns of the RgABCCs in various tissues of R. glutinosa, qRT-PCR analysis was performed (Fig 4 and S8 Table). Overall, among the RgABCCs, RgABCC4 and RgABCC14 were ubiquitously expressed in all tissues, exhibiting no significant difference in transcript abundances; the remaining 16 RgABCCs exhibited differential transcription responses in various tissues. For example, the transcript abundances of RgABCC12 in the roots, stems, young leaves and functional leaves were 16.56-, 13.34-, 36.72- and 39.92-fold higher, respectively, than those in the old leaves. Moreover, 15 of the remaining 16 RgABCCs (except RgABCC18) exhibited extremely low transcript abundances in the old leaves, i.e., the expression of 6 RgABCCs (i.e., RgABCC2, RgABCC5, RgABCC7, RgABCC8, RgABCC12 and RgABCC16) exhibited similar patterns in these tissues, which were the highest in functional leaves, followed by young leaves, roots and stems, with the lowest abundances in old leaves, whereas the 5 RgABCCs including RgABCC3, RgABCC6, RgABCC9, RgABCC13 and RgABCC15 exhibited the highest transcript abundances only in young leaves and functional leaves. For example, the abundances of RgABCC12 in the functional and young leaves, roots and stems were 16.56-, 13.34-, 36.72- and 39.92-fold those in the old leaves, whereas the abundances of RgABCC6 in the young leaves and functional leaves were 45.11- and 47.14-fold those in the roots. In addition, the transcript abundance of RgABCC17 was significantly higher in these leaves than in roots and stems (Fig 4 and S8 Table), and the abundance of RgABCC10 was the highest in the stems. Surprisingly, compared with those in other tissues, the transcript abundances in the roots were significantly highest for RgABCC1 (approximately 70-fold) followed by RgABCC11 (50-fold) and RgABCC18 (30-fold) (Fig 4). However, only RgABCC10 abundance was the highest in the stems compared with other tissues, exhibiting levels 21.85-, 7.27-, 6.07- and 44.13-fold in roots, stems, young leaves and functional leaves, respectively. The differential expression patterns of these RgABCCs suggested that they had diverse functions in R. glutinosa.
Fig 4. Transcript profiles of the RgABCCs in various tissues.
Note: “R, S, YL, FL and OL” represent “roots, stems, young, functional and old leaves, respectively; errors bars are standard deviation (SD), the same below.
Moreover, the expression patterns of the RgABCCs during root development of R. glutinosa were assessed by qRT-PCR analysis. As shown in Fig 5 and S9 Table, compared with the seedling stage, the transcript abundances of most RgABCCs (except RgABCC3 and RgABCC6) exhibited significantly differential increases at the other five stages. We found that the transcript abundances of RgABCC1 and RgABCC11 increased with the prolongation of cultivation days (especially at maturity); six RgABCCs (i.e., RgABCC2, RgABCC5, RgABCC7, RgABCC8, RgABCC9 and RgABCC12) showed the highest transcript abundances at the root elongation stage, five (RgABCC4, RgABCC13, RgABCC14, RgABCC16 and RgABCC18) exhibited the highest transcript abundances at the earlier root expansion stage, and three (RgABCC10, RgABCC15 and RgABCC17) showed the highest abundances at both the middle and later root expansion stages. For example, the transcript abundance of RgABCC8 at the root elongation stage was 3.49-fold that at the seedling stage, the abundance of RgABCC14 at the earlier root expansion stage was 4.73-fold that at the seedling stage, and the abundances of RgABCC15 at both the middle and later root expansion stages were 7.94- and 8.05-fold that at the root elongation stage, respectively. Furthermore, among the RgABCCs, the transcript abundances of RgABCC1 and RgABCC11 were prominently higher than those of other RgABCCs during root development, whereas those of RgABCC3 and RgABCC6 were much lower than those of other RgABCCs. For example, the transcript abundances of RgABCC1 and RgABCC11 were 152.86- and 79.58-fold that of RgABCC6 at the later root expansion stage, respectively. The results suggested that the transcription of the RgABCCs responded differently during the development of R. glutinosa roots.
Fig 5. Transcript profiles of the RgABCCs at various root development stages.
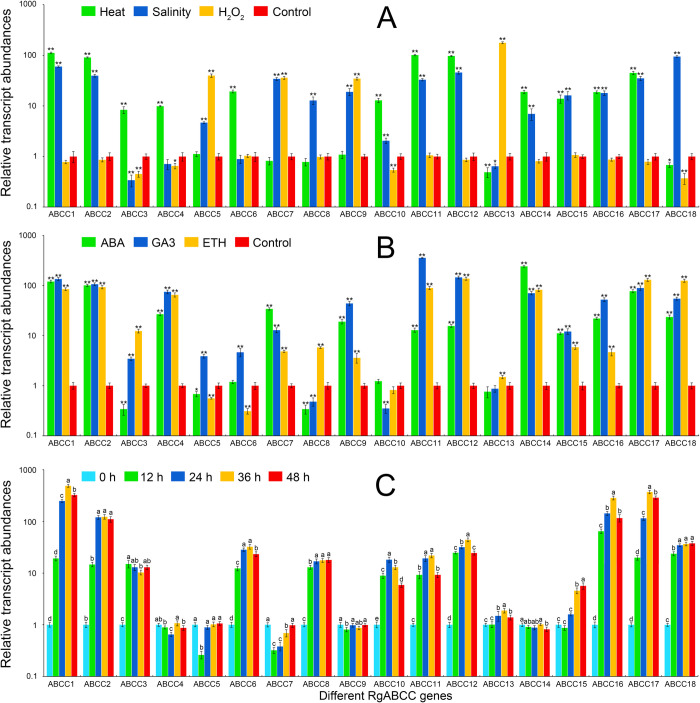
Transcript profiles of the RgABCCs under various stresses
To investigate the roles of the RgABCCs in the adaptation of plants to various abiotic stresses, qRT-PCR was performed to examine the responses of these genes to heat, salinity and H2O2 stresses in R. glutinosa roots. The transcript profiles of these genes exhibited differences under various stresses. As shown in Fig 6 and S10 Table, most RgABCC transcript profiles were changed under these stresses. Compared with the control, 14 of the RgABCCs in the roots, except for RgABCC7, RgABCC8, RgABCC13 and RgABCC18, were upregulated under heat stress; moreover, the abundance of RgABCC1 was the highest, peaking at 111.67-fold relative to the control. The increased transcript abundances of RgABCC5 and RgABCC9 were not obviously different relative to the control, whereas the abundances of RgABCC7, RgABCC8, RgABCC13 and RgABCC18 were decreased. Under salinity stress, 14 RgABCCs, except for RgABCC3, RgABCC4, RgABCC6 and RgABCC13, were upregulated. In particular, RgABCC18 had the highest abundance, peaking at 94.56-fold relative to the control, whereas RgABCC3 was downregulated 0.34-fold. However, under H2O2 stress, a few of the RgABCCs (i.e., RgABCC5, RgABCC7, RgABCC9 and RgABCC13) were highly upregulated, whereas other RgABCCs were downregulated or showed no response. For example, RgABCC13 had the highest abundance among the genes and peaked at 173.71-fold relative to the control, whereas RgABCC18 had the lowest abundance, 0.37-fold relative to the control. Obviously, the homologous RgABCCs exhibited different transcript profiles, implying a functional divergence of the homologous genes to these abiotic stress responses.
Fig 6. Transcript profiles of the RgABCCs under various conditions.
(A) three abiotic stresses; (B) three hormone treatments; (C) CdCl2 stress.
Moreover, the response of the RgABCCs to plant hormones (ABA, ETH and GA3) was also investigated in R. glutinosa roots (Fig 6 and S10 Table). When exposed to ABA conditions, 14 RgABCCs were upregulated, whereas only four RgABCCs (i.e., RgABCC3, RgABCC5, RgABCC8 and RgABCC13) were downregulated; in particular, RgABCC14 had the highest abundance and peaked at 240.93-fold relative to the control, whereas RgABCC8 was downregulated 0.34-fold. After GA3 treatment, 15 of the RgABCCs were upregulated (except for RgABCC8, RgABCC10 and RgABCC13); in particular, RgABCC11 had the highest abundance and peaked at 355.71-fold, whereas RgABCC10 was significantly downregulated 0.35-fold by GA3 induction (Fig 6). After ETH induction, 15 RgABCCs were upregulated, except for RgABCC5, RgABCC6 and RgABCC10; in particular, RgABCC12, RgABCC17, and RgABCC18 had higher abundances, peaking at 136.89-, 129.81- and 124.89-fold relative to the control.
To investigate the potential roles of the RgABCCs in the heavy metal stress response, these gene transcription patterns were assessed in R. glutinosa roots exposed to Cd stress at different time points. After Cd treatment, most of the RgABCCs exhibited different transcript abundances in response to Cd stress (Fig 6 and S10 Table). For example, RgABCC1 and RgABCC17 were significantly upregulated during the Cd treatment, peaking at 488.93- and 369.81-fold, respectively, at the 36-h point. RgABCC2 and RgABCC16 were also highly upregulated in response to the Cd treatment. In contrast, six RgABCCs, including RgABCC4, RgABCC5, RgABCC7, RgABCC9, RgABCC13 and RgABCC14, were mostly downregulated or showed no significant differences in the response to the Cd treatment.
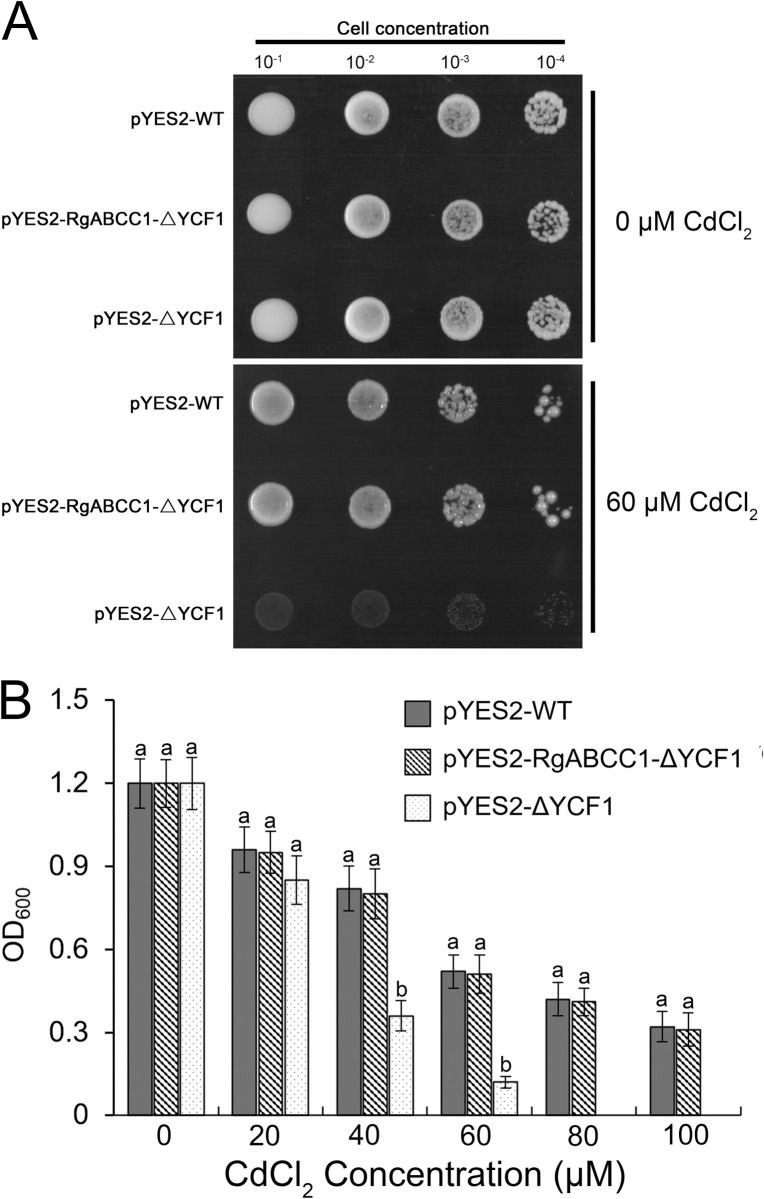
Functional complementation analysis of RgABCC1
To identify whether RgABCC1 was capable of Cd transport in cells, the ΔYCF1 mutant cells were transformed with constructs containing an empty vector pYES2 and pYES2-RgABCC1, which were named pYES2-RgABCC1-ΔYCF1 and pYES2-ΔYCF1, respectively; and the wild-type cells were transformed with an empty vector pYES2, which were named pYES2-WT. The yeast cells were cultured on SD-Ura solid medium lacking or containing 60 μM CdCl2 (Fig 7A). Under non-Cd condition, the growth phenotypes of the cells were not different; however, under Cd stress, the growth of the pYES2-RgABCC1-ΔYCF1 cells was similar to that of pYES2-WT, whereas the pYES2-ΔYCF1 cells grew much worse than those of pYES2-WT. Moreover, we measured the density phenotypes of the three cell types cultured in liquid medium lacking or containing CdCl2 (Fig 7B and S11 Table). In the medium lacking CdCl2, the growth of the PYES2-RgABCC1-ΔYCF1 and pYES2-ΔYCF1 cells was similar to that of pYES2-WT cells. In the presence of 20–100 μM CdCl2, the density of pYES2-ΔYCF1 cells decreased in a concentration-dependent manner. However, the densities of PYES2-RgABCC1-ΔYCF1 and pYES2-WT cells were not significantly different, at approximately 0.3 at OD600 in the presence of 100 μM CdCl2 (Fig 7B). These results indicated that the expression of RgABCC1 in ΔYCF1 mutants conferred tolerance to CdCl2.
Fig 7. Cd tolerance analysis for RgABCC1 heterologous expression in the ΔYCF1 yeast mutant.
(A) Drop assay of the strain expressing RgABCC1 on medium with 0 or 60 μM CdCl2; (B) Growth assay of the strain expressing RgABCC1 under CdCl2 stress at various concentrations.
Discussion
Identification and characterization of the RgABCCs
ABCC/MRPs, a subfamily of the larger ABC gene family, transport a wide range of molecules involved in plant growth and development, heavy metal detoxification, biotic and abiotic stresses, and the accumulation of endogenous active compounds and other physiological processes [1,14,44,45]. A number of ABCCs/MRPs have been well documented in plant species [2,5,10]. Our study identified a set of putative RgABCCs based on R. glutinosa transcriptome data. Phylogenetic analysis revealed that the RgABCCs had close relations to these functionally characterized homologous proteins from other plants (Fig 1B). To date, these functionally characterized ABCCs have been found to transport specific or multiple substrates [1,44–46]. For example, in the phylogenetic tree, OsABCC1/MRP1 and PtABCC1 of group II were prominently responsible for heavy metal transport [9,46], and two CsABCCs (i.e., CsABCC4a of Group I and CsABCC2 of Group II) were responsible for the transport of natural crocins [10], whereas AtABCC1/MRP1, AtABCC2/MRP2 and AtABCC3/MRP3 may be responsible for the transport of diverse substrates (such as heavy metals, glutathione conjugates and some secondary metabolites) [11,47–49]. Thus, we speculated that the RgABCCs, regardless of whether they were classified into group I or group II, could be responsible for the transport of specific or multiple substrates involved in various biological processes of R. glutinosa.
The structure of a typical ABCC protein is that of a full-molecule ABC transporter, which includes four core domains, i.e., two NBDs and two TMDs [14,50]. Among the RgABCCs, 16 possessed these typical core domains and were considered full-molecule ABCC transporters like most other plant ABCCs [2,5]. However, the functional unit of a few ABCC proteins showed half-molecule transport, and the configuration of the TMD-NBD has been reported in several plants [16,17,51]. Our study found that two RgABCCs (i.e., RgABCC17 and RgABCC18) showed half-molecule characteristics and had a topological pattern similar to that reported in the ABCC proteins of soybean and tomato [17,51]. Conservation analysis revealed that the RgABCCs were highly homologous to other plant ABCCs, specifically retaining the set of the architectural NBD domains, such as the Walker A motifs, the Walker B motifs and the ABC signatures [13,52,53]. Therefore, this implied that the potential functions of the RgABCCs are similar to those of the ABCC/MRPs of other plant species [17,51].
In plants, most ABCC/MRPs characterized to date are vacuole-localized proteins that mediate detoxification, sequestration and accumulation of endogenous or exogenous secondary metabolites and toxins [5,10,47], and a few of them have been reported to reside on the plasma membrane, controlling the plasma membrane anion channels of guard cells [12,13]. Here, subcellular localization prediction of the RgABCCs revealed that the majority of proteins were located in the vacuoles, while a few of proteins were located in the plasma membrane, in agreement with the homologous proteins in rice, A. thaliana and maize [5,7,12,13,16]. Transient expression analysis verified that three RgABCCs were localized to the vacuoles, while RgABCC3 was localized to the plasma membrane. Thus, subcellular localization analysis revealed that the RgABCCs might have potential transmembrane transport functions like other documented plant ABCCs [2,5,47,53]. R. glutinosa as a traditionally medicinal plant can produce extremely diverse specialized metabolites (such as phenolics, phenylethanoid glycosides and rehmanniosides) [23,24,38], which is mainly synthesized in the cytosol and must be transported to the vacuole by one or more vacuolar transporters [7,10,47]. As previous researches revealed that most plant ABCCs can transport numerous phenolics and glycosylated metabolites into the vacuoles [2,7,47,54], we thought most vacuole-localized RgABCCs might be related to the transport of diverse specialized metabolites in R. glutinosa.
Different spatio-temporal expression patterns of the RgABCCs reflected functional diversification
As preferential gene expression patterns suggest specificities for certain tissues/organs [2,5,54], investigation of the expression patterns in specific tissues/organs provides molecular clues for the roles of the RgABCCs and helps to explore their functions in R. glutinosa. Our data indicated that RgABCC1, RgABCC11 and RgABCC18 exhibited significantly higher transcript levels in the roots, implying potential transport activities in the root growth and development process, whereas RgABCC3 and RgABCC6 are preferentially expressed in the young and functional leaves, implying their functional roles in leaf development. Specifically, RgABCC10 had higher transcription in the stems than other RgABCCs, and its protein sequence exhibited a closer phylogenetic relationship to VvABCC1, an anthocyanidin glucoside transporter [54], implying a similar specific function in R. glutinosa stems. However, RgABCC4 and RgABCC14 transcripts were found to be equally abundant in various tissues, and both were present in the same subcluster of Group I from the phylogenetic tree as AtABCC4/MRP4 and CsABCC4a, which participate in the vacuolar transport of glutathione-conjugates, glycosides and folates [10,12,18], implying such potential transport activities for RgABCC4 and RgABCC14.
Due to the presence of multiple genes for the RgABCCs and their different transcript abundances during root development, we expected to uncover some vital functional members. Our data indicated that the transcript accumulation of RgABCC1 and RgABCC11, which were predicted to localize in the vacuoles, exhibited obviously higher expression during the entire root development process, and they formed a cluster (Group II) with several functional ABCC/MRPs, including AtABCC1/MRP1, AtABCC2/MRP2, CsABCC2 and LeMRP, by phylogenetic analysis. In A. thaliana, AtABCC1/MRP1 and AtABCC2//MRP2 are critical for vacuole transport and the accumulation of endogenous secondary metabolites, including anthocyanins, flavonoids and folates [7,55]. In Crocus sativus, CsABCC2 mediates the vacuolar accumulation of crocins [10]. In Lithospermum erythrorhizon, LeMRP was identified as being responsible for shikonin transport and accumulation during the hairy root development process [56]. As R. glutinosa roots are active parts for the accumulation of important medicinal secondary metabolites (e.g., phenolics and glycosides), higher expression of RgABCC1 and RgABCC11 in roots might be related to the transport and accumulation of some active ingredients, such as phenolics or glycosides, although further functional studies are required for verification. Moreover, RgABCC10, RgABCC15 and RgABCC17 exhibited preferential expression in the middle and later root expansion stages, implying their potential roles in root trait development. However, RgABCC3 and RgABCC6 exhibited extremely low transcript abundances during root development, whereas both genes were prominently expressed in young leaves and functional leaves, and RgABCC3 and RgABCC6 were closely related to AtABCC3/MRP3, which is involved in chlorophyll catabolite transport [47], and ZmABCC3, which is involved in anthocyanin transport [57]. Leaves are an important organ for the biosynthesis and accumulation of transitory nutrients and secondary metabolites that contribute to root expansion [58]. Thus, we speculated that the higher transcription of RgABCC3 and RgABCC6 in leaves might be involved in the transport of some nutrients and secondary metabolites to guarantee R. glutinosa tuberous root expansion. However, the RgABCCs exhibited different temporal expression patterns in R. glutinosa roots, demonstrating the functional specificities for these respective stages.
The RgABCCs involved in abiotic stress responses
Previous studies revealed that some ABCCs are involved in the molecular regulation of abiotic stresses [59–61]. In strawberry, most of the FvABCC subfamily expression was positively responsive to heat and salt treatment [2]. Our data indicated that the transcription of most RgABCCs was positively induced by heat and salt stresses, implying that the RgABCCs could be involved in the defence against heat and salt stresses. In particular, the transcription of RgABCC1 was prominently induced by heat treatment, implying that the high temperature stress could promote many secondary metabolisms of R. glutinosa, leading to produce more secondary metabolites and enhance the RgABCC transport activity; similarly, the transcription of RgABCC18 was highly induced by salinity treatment, implying its potential function in response to salinity stress. However, transcription was activated by H2O2 treatment (as an oxidant stress) in only four RgABCCs, especially RgABCC13, which was prominently activated by the stress. In wheat, TaABCC13 was significantly induced by H2O2 treatment [5], and RgABCC13 was closely clustered with TaABCC13, implying its importance in the antioxidant stress response in R. glutinosa.
Some ABCCs are involved in plant hormone transport and regulation [2,5,62]. Here, most RgABCCs were highly induced by ABA treatment; the result was the same for AtABCC13/MRP11 of A. thaliana [62] and FvABCCs of strawberry [2]. It was reported that in A. thaliana, AtABCC1/MRP1 and AtABCC2/MRP2 responded to ABA treatment and were critical for vacuolar sequestration of abscisic acid glucosyl ester [11]. The RgABCCs (RgABCC11, RgABCC12 and RgABCC18) were also highly induced by GA3 treatment; this finding is supported by previous transcription patterns observed for AtABCC13/MRP11 in A. thaliana [62] and wheat TaABCCs for this hormone [5]. The genes (RgABCC12, RgABCC17 and RgABCC19) were highly induced by ETH treatment. The results showed similar FvABCCs’ expression in strawberry [2]. Thus, the RgABCCs exhibited specific transcription patterns under these hormone treatments, implying their functional diversity in response to these stresses.
Many ABCCs in eukaryotes are involved in the transport of exogenous heavy metals [5,63]. The transcription of AtABCC6 gene was induced by Cd treatment during seedling development [64]. The transcription of four TaABCCs in wheat was activated under Cd exposure [5]. Additionally, two BnaABCCs were upregulated under Cd treatment [20]. Here, most RgABCCs were induced by Cd treatment, especially RgABCC1, which showed the highest expression. RgABCC1 clustered with AtABCC1 and AtABCC2, which are involved in detoxification and tolerance to Cd stress [48], implying that the genes could be specifically involved in Cd transport and Cd stress regulation.
RgABCC1 might participate in the tolerance to Cd stress
In yeast, YCF1 as an ABCC transporter is a crucial factor and is involved in Cd toxicity tolerance [25,65]. ΔYCF1 mutant has been classically utilized to confirm the functional role of ABCC transporters from some plants [13,25–27]. Some ABCC/MRPs are important for Cd and other heavy metal transports and have been found in several plants [5,20,30,33]. Here, the significant expression of RgABCC1 in R. glutinosa roots was induced by Cd stress, and its complemented expression in the yeast ΔYCF1 mutant resulted in a Cd tolerant phenotype, suggesting that RgABCC1 might be responsible for Cd transport (Fig 7). Some plant ABCC/MRPs can transport Cd into the vacuole, increasing the tolerance to Cd stress by sequestration [2,13,46,49]. Our data also revealed that RgABCC1 was localized in the vacuole, implying its involvement in the Cd transport into the vacuole and possibly enhancing Cd tolerance. It would be valuable to screen the remaining RgABCCs for the functional rescue of YCF1 sensitivity to Cd stress. Although the molecular function details of these RgABCCs have yet to be verified by further experimentation, our study provides molecular evidence that these transporters might play a general role in heavy metal transport in R. glutinosa.
Conclusion
Our study first identified a set of RgABCC subfamily genes, examined their spatial-temporal expression patterns, and revealed their differential transcription under various conditions, suggesting their importance in abiotic stress responses. Additionally, functional complementation analysis revealed that RgABCC1 may possess Cd transport activity. The insights provided herein serve as a better understanding of the RgABCC functions in R. glutinosa that could be used to decipher the transport of diverse specialized metabolites, promote growth and development, and enhance the tolerance to various abiotic stresses.
Supporting information
(CaMV35S:GFP-RgABCC1, a; CaMV35S:GFP-RgABCC3, b; CaMV35S:GFP-RgABCC11, c; CaMV35S:GFP-RgABCC18, d and PYES2-RgABCC1, e).
(DOCX)
(XLS)
The red fonts represent the sequences of these vector adapters.
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability
The gene sequences of RgABCCs are available in NCBI Genbank (Accession Nos. MW355848 - MW355865). The transcriptome analysis performed in this study are available in the NCBI Sequence Reads Archive (Accession No. PRJNA197434).
Funding Statement
This research was supported by grants from the National Natural Science Foundation of China (No. 81973417), the Science Foundation of Henan University of Technology (No. 2017RCJH05) and the Program for Innovative Research Team (in Science and Technology) of the University of Henan Province (No. 19IRTSTHN008).
References
- 1.Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol Plant. 2016; 9(3):338–55. 10.1016/j.molp.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 2.Shi M, Wang S, Zhang Y, Wang S, Zhao J, Feng H, et al. Genome-wide characterization and expression analysis of ATP-binding cassette (ABC) transporters in strawberry reveal the role of FvABCC11 in cadmium tolerance. Sci Hortic. 2020; 271:109464. 10.1016/j.scienta.2020.109464. [DOI] [Google Scholar]
- 3.Petrovic S, Pascolo L, Gallo R, Cupelli F, Ostrow JD, Goffffeau A, et al. The products of YCF1 and YLL015w (BPT1) cooperate for the ATP-dependent vacuolar transport of unconjugated bilirubin in Saccharomyces cerevisiae. Yeast. 2000; 16(6):561–71. [DOI] [PubMed] [Google Scholar]
- 4.Kolukisaoglu HU, Bovet L, Klein M, Eggmann T, Geisler M, Wanke D, et al. Family business: the multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta. 2002; 216(1):107–19. 10.1007/s00425-002-0890-6 [DOI] [PubMed] [Google Scholar]
- 5.Bhati KK, Sharma S, Aggarwal S, Kaur M, Shukla V, Kaur J, et al. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front Plant Sci. 2015; 6:488. 10.3389/fpls.2015.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P, Li Y, Zhao L, Hou Z, Yan M, Hu B, et al. Genome-wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in Pineapple (Ananas comosus (L.) Merr.) reveal the role of AcABCG38 in pollen development. Front Plant Sci. 2017; 8:2150. 10.3389/fpls.2017.02150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrens CE, Smith KE, Iancu CV, Choe J, Dean JV. Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2. Sci Rep. 2019; 9(1):437. 10.1038/s41598-018-37504-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panzeri D, Cassani E, Doria E, Tagliabue G, Forti L, Campion B, et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011; 191(1):70–83. 10.1111/j.1469-8137.2011.03666.x [DOI] [PubMed] [Google Scholar]
- 9.Song WY, Yamaki T, Yamaji N, Ko D, Jung K, Fujii-Kashino M, et al. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc Natl Acad Sci U S A. 2014; 111(44):15699–704. 10.1073/pnas.1414968111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demurtas OC, Francisco RB, Diretto G, Ferrante P, Frusciante S, Pietrella M, et al. ABCC transporters mediate the vacuolar accumulation of crocins in saffron stigmas. Plant Cell. 2019; 31(11):2789–2804. 10.1105/tpc.19.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burla B, Pfrunder S, Nagy R, Francisco RM, Lee Y, Martinoia E. Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 2013; 163(3):1446–58. 10.1104/pp.113.222547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein M, Geisler M, Suh SJ, Kolukisaoglu HU, Azevedo L, Plaza S, et al. Disruption of AtMRP4, a guard cell plasma membrane ABCC-type ABC transporter, leads to deregulation of stomatal opening and increased drought susceptibility. Plant J. 2004; 39(2):219–36. 10.1111/j.1365-313X.2004.02125.x [DOI] [PubMed] [Google Scholar]
- 13.Klein M, Burla B, Martinoia E. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 2006; 580(4):1112–22. 10.1016/j.febslet.2005.11.056 [DOI] [PubMed] [Google Scholar]
- 14.Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, et al. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008; 13(4):151–9. 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Ortiz C, Dutta SK, Natarajan P, Pena-Garcia Y, Abburi V, Saminathan T. Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp. PLoS One. 2019; 14(4):e0215901. 10.1371/journal.pone.0215901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang K, Li Y, Liu M, Meng Z, Yu Y. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene. 2013; 526(2):411–28. 10.1016/j.gene.2013.05.051 [DOI] [PubMed] [Google Scholar]
- 17.Ofori PA, Mizuno A, Suzuki M, Martinoia E, Reuscher S, Aoki K, et al. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS One. 2018; 13(7):e0200854. 10.1371/journal.pone.0200854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sánchez-Fernández R, Davies TGE, Coleman JOD, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001; 276(32):30231–44. 10.1074/jbc.M103104200 [DOI] [PubMed] [Google Scholar]
- 19.Jasinski M, Ducos E, Martinoia E, Boutry M. The ATP-binding cassette transporters: structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 2003; 131(3):1169–77. 10.1104/pp.102.014720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XD, Zhao KX, Yang ZM. Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene. 2018; 664:139–151. 10.1016/j.gene.2018.04.060 [DOI] [PubMed] [Google Scholar]
- 21.Çakır B, Kılıçkaya O. Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS One. 2013; 8(11):e78860. 10.1371/journal.pone.0078860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan C, Duan W, Lyu S, Li Y, Hou X. Genome-wide identification, evolution, and expression analysis of the ATP-binding cassette transporter gene family in Brassica rapa. Front Plant Sci. 2017; 8:349. 10.3389/fpls.2017.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Yang Y, Li X, Gu L, Wang F, Feng F, et al. Analysis of integrated multiple ’omics’ datasets reveals the mechanisms of initiation and determination in the formation of tuberous roots in Rehmannia glutinosa. J Exp Bot. 2015; 66(19):5837–51. 10.1093/jxb/erv288 [DOI] [PubMed] [Google Scholar]
- 24.Li M, Yang Y, Feng F, Zhang B, Chen S, Yang C, et al. Differential proteomic analysis of replanted Rehmannia glutinosa roots by iTRAQ reveals molecular mechanisms for formation of replant disease. BMC Plant Biol. 2017; 17(1):116. 10.1186/s12870-017-1060-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci U S A. 1997; 94(1):42–7. 10.1073/pnas.94.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang TL, Wu M. An ATP-binding cassette transporter related to yeast vacuolar ScYCF1 is important for Cd sequestration in Chlamydomonas reinhardtii. Plant Cell Environ. 2006; 29(10):1901–12. 10.1111/j.1365-3040.2006.01566.x [DOI] [PubMed] [Google Scholar]
- 27.Feng T, He X, Zhuo R, Qiao G, Han X, Qiu W, et al. Identification and functional characterization of ABCC transporters for Cd tolerance and accumulation in Sedum alfredii Hance. Sci Rep. 2020; 10(1):20928. 10.1038/s41598-020-78018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2002; 30(1):13–6. 10.1093/nar/30.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M, Derbyshire MK, Yamashita RA, Marchler-Bauer A. NCBI’s conserved domain database and tools for protein domain analysis. Curr Protoc Bioinformatics. 2020; 69(1):e90. 10.1002/cpbi.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigrist CJA, Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41(Database issue):D344–7. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. Methods Mol Biol. 1999; 112:531–52. 10.1385/1-59259-584-7:531 [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer EL, Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998; 6:175–82. [PubMed] [Google Scholar]
- 33.Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One. 2010; 5(6):e11335. 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: A reference implementation of a PROSITE scanning tool. Appl Bioinformatics. 2002; 1(2):107–8. [PubMed] [Google Scholar]
- 36.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004; 14(6):1188–90. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23(21):2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 38.Yang YH, Li MJ. Cloning, molecular characterization, and expression analysis of a nucleoporin gene (rgNUP98-96) from Rehmannia glutinosa. Genet Mol Res. 2015; 14(4):13022–32. 10.4238/2015.October.21.23 [DOI] [PubMed] [Google Scholar]
- 39.Wise AA, Liu Z, Binns AN. Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol Biol. 2006; 343:43–53. 10.1385/1-59745-130-4:43 [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)). Methods. 2001; 25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 41.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of LoxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic acids Res. 2002; 30(6):e23. 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SSDNA/PEG procedure. Yeast. 1995; 11(4):355–60. 10.1002/yea.320110408 [DOI] [PubMed] [Google Scholar]
- 43.Ghosh M, Shen J, Rosen BP. Pathways of As (III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999; 96(9):5001–6. 10.1073/pnas.96.9.5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinoia E, Grill E, Tommasini R, Kreuz K, Amrhein N. ATP-dependent glutathione-S-conjugate ‘export’ pump in the vacuolar membrane of plants. Nature. 1993; 364(6434):247–9. 10.1038/364247a0. [DOI] [Google Scholar]
- 45.Theodoulou FL, Kerr ID. ABC transporter research, going strong 40 years on. Biochem Soc Trans. 2015; 43(5):1033–40. 10.1042/BST20150139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun L, Ma Y, Wang H, Huang W, Wang X, Han L, et al. Overexpression of PtABCC1 contributes to mercury tolerance and accumulation in Arabidopsis and poplar. Biochem Biophys Res Commun. 2018; 497(4):997–1002. 10.1016/j.bbrc.2018.02.133 [DOI] [PubMed] [Google Scholar]
- 47.Tommasini R, Vogt E, Fromenteau M, Hörtensteiner S, Matile P, Amrhein N, et al. An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 1998; 13(6):773–80. 10.1046/j.1365-313x.1998.00076.x [DOI] [PubMed] [Google Scholar]
- 48.Park J, Song WY, Ko D, Eom Y, Hansen TH, Schiller M, et al. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012; 69(2):278–88. 10.1111/j.1365-313X.2011.04789.x [DOI] [PubMed] [Google Scholar]
- 49.Brunetti P, Zanella L, Paolis AD, Litta DD, Cecchetti V, Falasca G, et al. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot. 2015; 66(13):3815–29. 10.1093/jxb/erv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu Ü, et al. Multifunctionality of plant ABC transporters more than just detoxifiers. Planta. 2002; 214(3):345–55. 10.1007/s004250100661 [DOI] [PubMed] [Google Scholar]
- 51.Mishra AK, Choi J, Rabbee MF, Baek KH. In silico genome-wide analysis of the ATP-binding cassette transporter gene family in soybean (Glycine max L.) and their expression profiling. Biomed Res Int. 2019; 2019:8150523. 10.1155/2019/8150523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999; 293(2):381–99. 10.1006/jmbi.1999.2993 [DOI] [PubMed] [Google Scholar]
- 53.Wanke D, Kolukisaoglu HU. An update on the ABCC transporter family in plants: many genes, many proteins, but how many functions? Plant Biol (Stuttg). 2010; 12 suppl 1:15–25. 10.1111/j.1438-8677.2010.00380.x [DOI] [PubMed] [Google Scholar]
- 54.Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell. 2013; 25(5):1840–54. 10.1105/tpc.112.102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raichaudhuri A, Peng M, Naponelli V, Chen S, Sanchez-Fernandez R, Gu H, et al. Plant vacuolar ATP-binding cassette transporters that translocate folates and antifolates in vitro and contribute to antifolate tolerance in vivo. J Biol Chem. 2009; 284(13):8449–60. 10.1074/jbc.M808632200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y, Chu SJ, Luo YL, Fu JY, Tang CY, Lu GH, et al. Involvement of LeMRP, an ATP-binding cassette transporter, in shikonin transport and biosynthesis in Lithospermum erythrorhizon. Plant Biol (Stuttg). 2018; 20(2):365–73. 10.1111/plb.12666 [DOI] [PubMed] [Google Scholar]
- 57.Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell. 2004; 16(7):1812–26. 10.1105/tpc.022574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang YH, Li MJ, Chen XJ, Wang PF, Wang FQ, Lin WX, et al. De novo characterization of the Rehmannia glutinosa leaf transcriptome and analysis of gene expression associated with replanting disease. Mol Breeding. 2014; 34(3):905–15. 10.1007/s11032-014-0084-5. [DOI] [Google Scholar]
- 59.Ibraheem O, Botha CE, Bradley G. In silico analysis of cis-acting regulatory elements in 5’ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput Biol Chem. 2010; 34(5–6):268–83. 10.1016/j.compbiolchem.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 60.Hu W, Hu G, Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009; 176(4):583–90. 10.1016/j.plantsci.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 61.Nguyen VNT, Moon S, Jung KH. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J Plant Physiol. 2014; 171(14):1276–88. 10.1016/j.jplph.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 62.Guizani TE, Blanc N, Triki S, St-Pierre B, Ducos E. Expression pattern of AtABCC13/MRP11 reveals developmental, hormonal, and nutritional regulations. Biol Plantarum. 2014; 58(2):231–40. 10.1007/s10535-013-0387-0. [DOI] [Google Scholar]
- 63.Rea PA. MRP subfamily ABC transporters from plants and yeast. J Exp Bot. 1999; 50:895–913. 10.1093/jxb/50.Special_Issue.895. [DOI] [Google Scholar]
- 64.Gaillard S, Jacquet H, Vavasseur A, Leonhardt N, Forestier C. AtMRP6/AtABCC6, an ATP-binding cassette transporter gene expressed during early steps of seedling development and up-regulated by cadmium in Arabidopsis thaliana. BMC Plant Biol. 2008; 8(1):22. 10.1186/1471-2229-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song WY, Martinoia E, Lee J, Kim D, Kim DY, Vogt E, et al. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004; 135(2):1027–39. 10.1104/pp.103.037739 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(CaMV35S:GFP-RgABCC1, a; CaMV35S:GFP-RgABCC3, b; CaMV35S:GFP-RgABCC11, c; CaMV35S:GFP-RgABCC18, d and PYES2-RgABCC1, e).
(DOCX)
(XLS)
The red fonts represent the sequences of these vector adapters.
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
The gene sequences of RgABCCs are available in NCBI Genbank (Accession Nos. MW355848 - MW355865). The transcriptome analysis performed in this study are available in the NCBI Sequence Reads Archive (Accession No. PRJNA197434).