Abstract
Objective
Low back pain is a leading cause of disabilities worldwide, and intervertebral disc degeneration (IVDD)-related disorders have been recognised as one of the main contributors. Nevertheless, the underlying mechanism has not yet been fully understood. The aim of this study was to investigate the role of the miR-133a-5p/FBXO6 axis in the regulation of IVDD.
Methods
RT-qPCR, WB and IHC were performed to assess the expression of FBXO6 in human IVD tissues. Nucleus pulposus (NP) cells were treated with IL-1β to induce IVDD cellular model. Silence of FBXO6 was achieved using specific siRNAs. CCK-8 assay, flow cytometry, TUNEL assay, RT-qPCR and WB were used to evaluate the role and mechanism of FBXO6 in the process of IVDD. Online tools, GSE datasets and RT-qPCR were used to search the candidate miRNAs targeting FBXO6. The direct binding sites between FBXO6 and miR-133a-5p were further verified by a dual luciferase assay. RT-qPCR, WB and rescue experiments were conducted to identify the regulatory function of miR-133a-5p on the expression of aggrecan, collagen Ⅱ, MMP3, ADAMTS5, IL-6 and COX2. In addition, the role of the NF-κB pathway in regulating miR-133a-5p was studied using lentiviral shRNA, WB and RT-qPCR.
Results
Results showed that FBXO6 mainly expressed in the NP tissue of IVD and the expression of FBXO6 decreased with the process of IVDD as well as under IL-1β stimulation. The silence of FBXO6 led to the decreased expression of aggrecan and collagen Ⅱ and the increased expression of MMP3, ADAMTS5, IL-6 and COX2, which further induced the degeneration of NP cells. The bioinformatic analysis showed that miR-133a-5p was the candidate miRNA targeting FBXO6. miR-133a-5p was upregulated in IVDD tissues and significantly inhibited the expression of FBXO6. The inhibition of miR-133a-5p ameliorated the acceleration of IVDD induced by the silence of FBXO6 in vitro. Moreover, it was demonstrated that IL-1β regulated the expression of the miR-133a-5p/FBXO6 axis via the NF-κB pathway in NP cells.
Conclusion
miR-133a-5p was upregulated by IL-1β to aggravate intervertebral disc degeneration via sponging FBXO6. Inhibiting miR-133a-5p expression or rescuing FBXO6 expression may be promising strategies for the treatment of IVDD.
The translational potential of this article
This study suggests that the miR-133a-5p/FBXO6 axis could regulate NP cells proliferation, apoptosis, synthesis and degradation of extracellular matrix, which provides a promising therapeutic target and strategy for the treatment of IVDD.
Keywords: Intervertebral disc, miR-133a-5p, FBXO6, Nucleus pulposus cells, Interleukin (IL)-1β
1. Introduction
Low back pain (LBP) represents a leading cause of disabilities and occurs in approximately 80% of the worldwide population, causing enormous clinical and economical burdens on society [1,2]. LBP is strongly associated with intervertebral disc degeneration (IVDD), which bears responsibility for approximately 40% of all LBP cases [3]. It has been well documented that IVDD is characterised by elevated proinflammatory cytokines, enhanced aggrecan and collagen degradation, decreased cell density and extracellular matrix (ECM) [[4], [5], [6]]. Numerous studies have been conducted, but the underlying mechanism is still not fully understood.
FBXO6 (F-Box Protein 6) belongs to the F-box protein family, which has been found to play important roles in broad cellular processes, including apoptosis, autophagy and carcinogenesis [7,8]. For example, it was found that FBXO6 was able to inhibit endoplasmic reticulum stress-induced apoptosis [9] and to promote the growth and proliferation of gastric cancer cells [10]. The recent study [11] showed that FBXO6 expression was reduced in the articular cartilage of human and mouse osteoarthritis models and that the knockout of FBXO6 promoted the experimental osteoarthritis process, suggesting that FBXO6 participates in the regulation of chondrocytes. As NP cells share many similar characteristics with chondrocytes [12], we speculated that FBXO6 might participate in the process of IVDD and designed this study to investigate it.
Numerous studies have demonstrated that mRNAs are the target genes of mircoRNAs, which could inhibit mRNA translation or degrade mRNAs [13]. In addition, miRNAs are linked to a wide spectrum of diseases, including tumours [14], inflammation [15] and IVDD [16]. For example, Ji et al. found that miR-141 promoted IVDD development by targeting SIRT1 [16]. Similarly, in another study conducted by Feng et al., miR-29a was demonstrated to suppress the expression of MMP-2, inhibit the fibrosis process and reverse IVDD in animal models [17]. Nevertheless, the miRNA-related pathogenesis of IVDD is still largely unknown and warrants further investigation. miR-133a-5p has been proved to be associated with cancer progression, recurrence and distant metastasis [18,19]. Furthermore, a bioinformatic analysis showed that miR-133a-5p is the candidate miRNA targeting FBXO6. Thus, it is highly reasonable to speculate that miR-133a-5p could participate in the progression of IVDD.
Therefore, the aim of this study was to detect the roles of miR-133a-5p along with its targeting gene FBXO6 in the progression of IVDD. In addition, the pathway involved in the regulation of IL-1β on the miR-133a-3p/FBXO6 axis was also illustrated. This study may elucidate one of the molecular mechanisms in the degeneration of IVD and may provide a promising bio-therapeutic target for LBP.
2. Materials and methods
2.1. Ethics statement and clinical sample collection
All the experiments were performed according to protocol approved by the Ethics Committee of our institution (Protocol number: 2020–488). Written consents were obtained from all the patients before collection. Degenerative Nucleus pulposus (NP) tissues were collected from patients who had to receive discectomy because of lumbar disc herniation or lumbar stenosis in our hospital. No-degenerative NP tissues were collected from patients with thoracolumbar fracture or scoliosis who underwent spinal surgery by reason of spinal instability or neurological deficits. Magnetic resonance imaging (MRI) was performed on every patient ahead of surgery to assess IVD degeneration according to the classification system described by Pfirrmann et al. [20]. The specimens were classified into four groups: Normal or no degenerated (grade I/II), mildly degenerated (grade III), moderately degenerated (grade IV), and severely degenerated (grade V). Clinical features of the patients are listed in Table 1.
Table 1.
Clinical characteristics of patients.
| Number | Age(y) | Gender | Pfirrmann grade | Disc level |
|---|---|---|---|---|
| 1 | 11 | female | G1 | L1/2 |
| 2 | 12 | male | G1 | L1/2 |
| 3 | 32 | male | G2 | L2/3 |
| 4 | 19 | female | G3 | L5/S1 |
| 5 | 22 | male | G3 | L5/S1 |
| 6 | 32 | female | G3 | L4/5 |
| 7 | 67 | female | G4 | L4/5 |
| 8 | 68 | female | G4 | L5/S1 |
| 9 | 62 | male | G4 | L4/5 |
| 10 | 54 | female | G5 | L4/5 |
| 11 | 48 | female | G5 | L5/S1 |
| 12 | 66 | female | G5 | L4/5 |
2.2. Isolation of NP cells and IL-1β treatment
Human NP cells were isolated using the method described previously [21]. Briefly, Nucleus pulposus (NP) tissues were washed twice with phosphate buffered saline (PBS), cut into small pieces (1 mm3) and digested with 0.25% trypsin and 0.2% collagenase Ⅱ (Sigma, St. Louis, MO, USA) for 1–2 h at 37 °C. Then, the digestive solution was filtered using a cell strainer (100 μM) and centrifuged 1500 rpm and 5 min. After that, the NP cells were resuspended and cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) in a 37 °C, 5% CO2 environment. The medium was changed every two days. The second to fourth passage was used for subsequent experiments. To investigate the effect of proinflammatory cytokine on NP cell degeneration, cells were treated with IL-1β (Peprotech, New Jersey, USA) according to the experiment protocol [22]. For an in vitro model of IVDD, NP cells were treated with 10 ng/ml IL-1β for 24 h.
2.3. siRNA and cell transfection
The specific small interfering RNA (siRNA) of FBXO6 and its negative control was obtained from RIBOBIO Technology (Guangzhou, China). The target sequences of FBXO6 siRNAs are: siRNA-1: 5′-3′ GCACCTACCAACTCAAAGT, siRNA-2: 5′-3′TGACCATCCAACAGTGGAA, siRNA-3: 5′-3′ ACCGAGCTGTTGTCCAGAT. NP cells were seeded in 6-well plates and Lipofectamine™ RNAiMAX reagent (Thermo Fisher Scientific, IL, USA) was adopted for transfection according to the manufacturer's instructions.
2.4. RNA isolation and RT-qPCR
Following treatment, total RNA was extracted using Trizol (Thermo Fisher Scientific, Waltham, MA, USA). 2 μg DNA-free RNA was used to synthesise cDNA using SuperScript cDNA synthesis kits(YEASEN,Shanghai,China). RT-qPCR was achieved using a qPCR Master Mix (YEASEN,Shanghai,China) on a real-time PCR System following the manufacturer's instructions. Primers were designed and synthesised by Sangon Biotech(Shanghai,China). β-actin or U6 were used to normalise RNA expression. The primer sequences used are listed in Table 2.
Table 2.
Specific primers.
| Name | Forward | Reverse |
|---|---|---|
| FBXO6 | 5′-ATCCTACGAAATGTGCCTCAAG-3′ | 5′-CCAACACGAAGTAGTCAGCCG-3′ |
| IL-1β | 5′-AAGCTGATGGCCCTAAACAG -3′ | 5′-AGGTGCATCGTGCACATAAG -3′ |
| Aggrecan | 5′-ACTCTGGGTTTTCGTGACTCT-3′ | 5′-ACACTCAGCGAGTTGTCATGG-3′ |
| CollagenII | 5′-TGGACGCCATGAAGGTTTTCT-3′ | 5′-TGGGAGCCAGATTGTCATCTC-3′ |
| MMP3 | 5′-CTGGACTCCGACACTCTGGA-3′ | 5′-CAGGAAAGGTTCTGAAGTGACC-3′ |
| ADAMTS-5 | 5′-CCTGGTCCAAATGCACTTCAGC-3′ | 5′-TCGTAGGTCTGTCCTGGGAGTT-3′ |
| IL-6 | 5′-CCTGAACCTTCCAAAGATGGC-3′ | 5′-TTCACCAGGCAAGTCTCCTCA-3′ |
| COX-2 | 5′-TGAGCATCTACGGTTTGCTG-3′ | 5′-TGCTTGTCTGGAACAACTGC-3′ |
| Actin | 5′-CCTGGCACCCAGCACAAT-3′ | 5′-GGGCCGGACTCGTCATAC-3′ |
| P65 | 5′-ATGTGGAGATCATTGAGCAGC-3′ | 5′-CCTGGTCCTGTGTAGCCATT-3′ |
| IKKβ | 5′-GGAAGTACCTGAACCAGTTTGAG-3′ | 5′-GCAGGACGATGTTTTCTGGCT-3′ |
| miR-133a-5p | 5′-GGCGAGCTGGTAAAATGGAACCAAAT-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
| miR-4645–3p | 5′-CCTAGACAGTAGTTCTTGCCTGGTT-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACATA -3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
2.5. Protein extraction and western blotting
After treatment, cells were harvested in a mammalian protein extraction reagent buffer (Thermo Fisher Scientific, IL, USA) on ice. Total proteins were resolved on 8–12% SDS-PAGE gels and then transferred to PVDF membranes. The membranes were blocked with 5% non-fat dry milk in TBST and then incubated overnight at 4 °C with the first antibodies against IL-6 (Abcam, ab9324, 1:1000), COX2 (Abcam, ab15191, 1:1000), aggrecan (Abcam, ab3773, 1:200), collagen II (Abcam, ab188570, 1:2000), MMP3 (Abcam, ab53015, 1:1000), ATAMTS5 (Abcam, ab41037, 1:250), FBXO6 (Abcam, ab153853, 1:1000) and β-Tubulin (Servicebio, GB11017B, 1; 1000). After washing membranes three times using TBST, the membranes were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (YEASEN, Shanghai, China). After washing, immunolabeling was detected using the ECL reagent (Thermo Fisher Scientific, IL, USA).
2.6. Cell counting Kit-8 (CCK-8) assay
Cells were plated into 96-well plates at a density of 5 × 103 cells per well. Ten microliters of the CCK-8 reagent (YEASEN, Shanghai, China) were added and incubated for 1 h. The optical density (OD) was measured using a microplate reader (Tecan Sunrise, Austria) at 450 nm. The procedure was carried out thrice to obtain the mean values.
2.7. Flow cytometry analysis for cell apoptosis
NP cells were collected for the detection of apoptosis using an Annexin V-FITC Apoptosis Detection Kit (YEASEN, Shanghai, China) according to the manufacturer's instructions. After washing, NP cells were resuspended in a binding buffer at a concentration of 1 × 106 cells/ml. Five microliters of Annexin-V and ten microliters of PI were added to every 100 μL of the resuspended cell solution, which was then incubated for 15 min at room temperature in the dark and analysed by flow cytometry (Beckman Coulter, CA, USA).
2.8. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Staining
NP cells were fixed in 4% paraformaldehyde for 1 h at room temperature and then cultured with 0.5% TritonX-100 in phosphate-buffered saline (PBS) for 10 min. They were then washed 3 times with PBS and incubated following the instructions of an apoptosis detection kit (Servicebio, Wuhan, China) and stained with 4′,6-diamidino-2-phenylindole (DAPI). Apoptosis was detected under a light microscope (Leica DMI8, Wetzlar, Germany).
2.9. Transfections and dual luciferase assay
The 3′-UTR of FBXO6 mRNA containing predicted miR-133a-5p target sites or mutant binding sites was synthesized. The synthesized RNA was then PCR amplified and inserted into the pmirGLO dual luciferase expression vector (Promega, WI, USA). For the luciferase reporter assay, the inserted vectors for wildtype and mutant FBXO6 3′-UTR, miR-133a-5p mimic or inhibitor and their negative control were transfected into NP cells by a Lipofectamine 3000 regent (Thermo Fisher Scientific, IL, USA). Forty-eight hours after transfection, luciferase activity was detected by the Dual-Luciferase Reporter Assay System (YEASEN, Shanghai, China).
2.10. Lentiviral particle production and viral transduction
For gene silencing, HEK 293T cells were seeded in 10 cm plates, and then transfected with sh-control, sh-p65 or sh-IKKβ plasmids along with psPAX2 and pMD2.G (Invitrogen, CA, USA). Forty-eight hours later, lentiviruses containing the target gene shRNA were collected and used to transfect NP cells. The suppression efficiency of shRNA was tested by WB and RT–qPCR.
2.11. Bioinformatic analysis: MicroRNAs and FBXO6
In the present study, we used a bioinformatics approach to determine probable miRNAs that interact with the 3′UTR of individual FBXO6 genes. Three open-access online programmes, TargetScan (http://www.targetscan.org/vert_72/), miRDB (http://mirdb.org/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) along with IVDD GEO datasets (GSE116726, GSE63492) (https://www.ncbi.nlm.nih.gov/geo/) were used to identify putative miRNAs targeting FBXO6.
2.12. Statistical analysis
All quantitative data were presented as mean ± SD. Statistical analyses were performed using GraphPad Prism Software 6.0 (GraphPad Software Inc., San Diego, CA, USA). A one-way ANOVA and student's t-test or Wilcoxon rank sum test (if the data did not fit the gaussian distribution) were used to analyse the differences between groups. The comparisons of the CCK8 results were performed using a two-way ANOVA with repeated measures. The Pearson's correlation coefficient was applied to establish the relationship between the expression of FBXO6 and IL-1β or miR-133a-5p. Values of P < 0.05 were considered statistically significant.
3. Results
3.1. FBOX6 decreased with the development of IVDD and was suppressed by IL-1β in NP cells
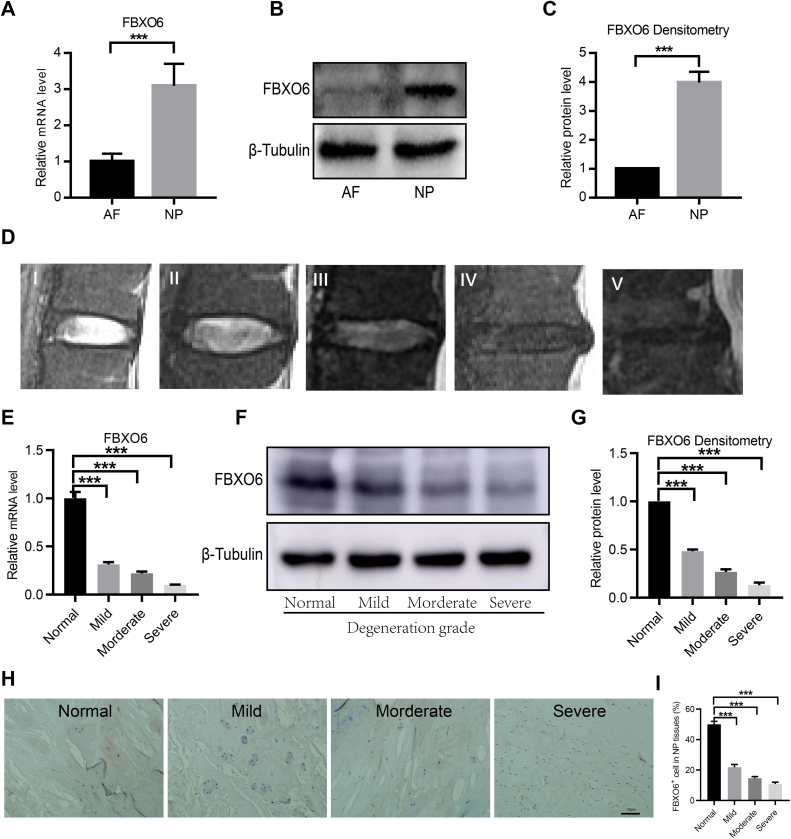
To determine the expression pattern of FBXO6 in nucleus pulposus (NP) and annulus fibrosus (AF) tissues, RT-qPCR and WB were used. RT-qPCR showed that the expression of FBXO6 mRNA was significantly higher in NP than that in AF tissues (Fig. 1A). WB also demonstrated that the protein level of FBXO6 was more prominent in NP tissues (Fig. 1B and C). Representative MRI images of IVD by Pfirrmann grade were shown in Fig. 1D. The expression of FBXO6 at different degenerative stages of IVD was evaluated by RT-qPCR, WB and IHC, which indicated that FBXO6 was highly expressed in non-degenerated IVD and decreased with the severity of IVDD by Pfirrmann grade (Fig. 1E–I).
Figure 1.
The expression pattern of FBXO6 in normal and degenerated IVD tissues (A) RT-qPCR analysis showed that FBXO6 mRNA expressed higher in NP than AF tissues (B/C) WB and densitometric analyses showed that the protein level of FBXO6 was higher in NP tissues (D) Specific MRI image of IVDD by Pfirrmann grades (E) The RT-qPCR analysis indicated that FBXO6 mRNA decreased with the severity of IVDD (F/G) The WB and densitometric analyses determined that the protein level of FBXO6 decreased with the progression of IVDD (H/I) IHC staining and quantitative analyses showed that the expression of FBXO6 in NP tissues decreased with the development of IVDD. ∗∗∗P < 0.001.
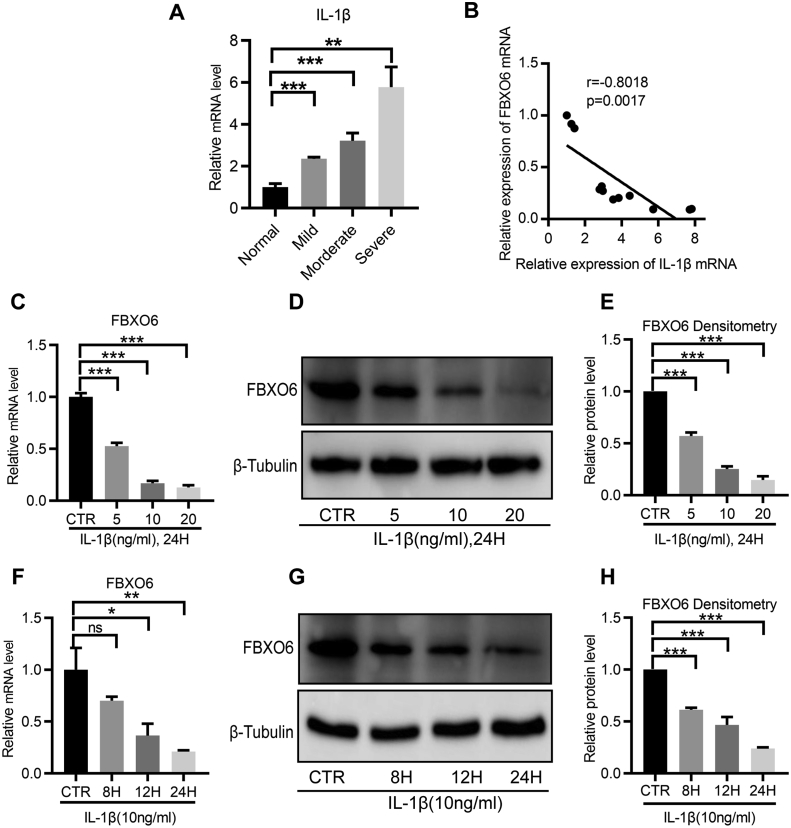
As the proinflammatory cytokine IL-1β plays a critical role in the pathogenesis of IVDD through suppressing synthesis of the normal IVD matrix and enhancing production of degradative enzymes [23,24], the expression of IL-1β in IVD tissues was detected by RT-qPCR. Consistent with previous studies, the results showed that IL-1β increased with the process of IVDD (Fig. 2A). More importantly, the statistical analysis showed that FBXO6 was negatively correlated with IL-1β (Fig. 2B), which suggested that IL-1β might participate in the regulation of FBXO6. Hence, IL-1β was used for further investigation. To evaluate the regulatory effect of IL-1β on FBXO6, NP cells were treated with IL-1β, which led to a dose and time dependent downregulation of FBXO6 at mRNA and protein levels (Fig. 2C–H).
Figure 2.
The expression of FBXO6 in NP cells was reduced by IL-1β (A) The expression level of IL-1β increased with the severity of IVDD (B) A negative correlation was found between the expression of FBXO6 and IL-1β (C–E) The RT-qPCR and WB showed that IL-1β reduced the FBXO6 expression in a dose-dependent manner (F–H) The RT-qPCR and WB analyses showed that IL-1β suppressed the expression of FBXO6 in a time-dependent manner. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. Silencing FBXO6 inhibited proliferation, enhanced apoptosis, suppressed ECM synthesis and accelerated ECM degradation
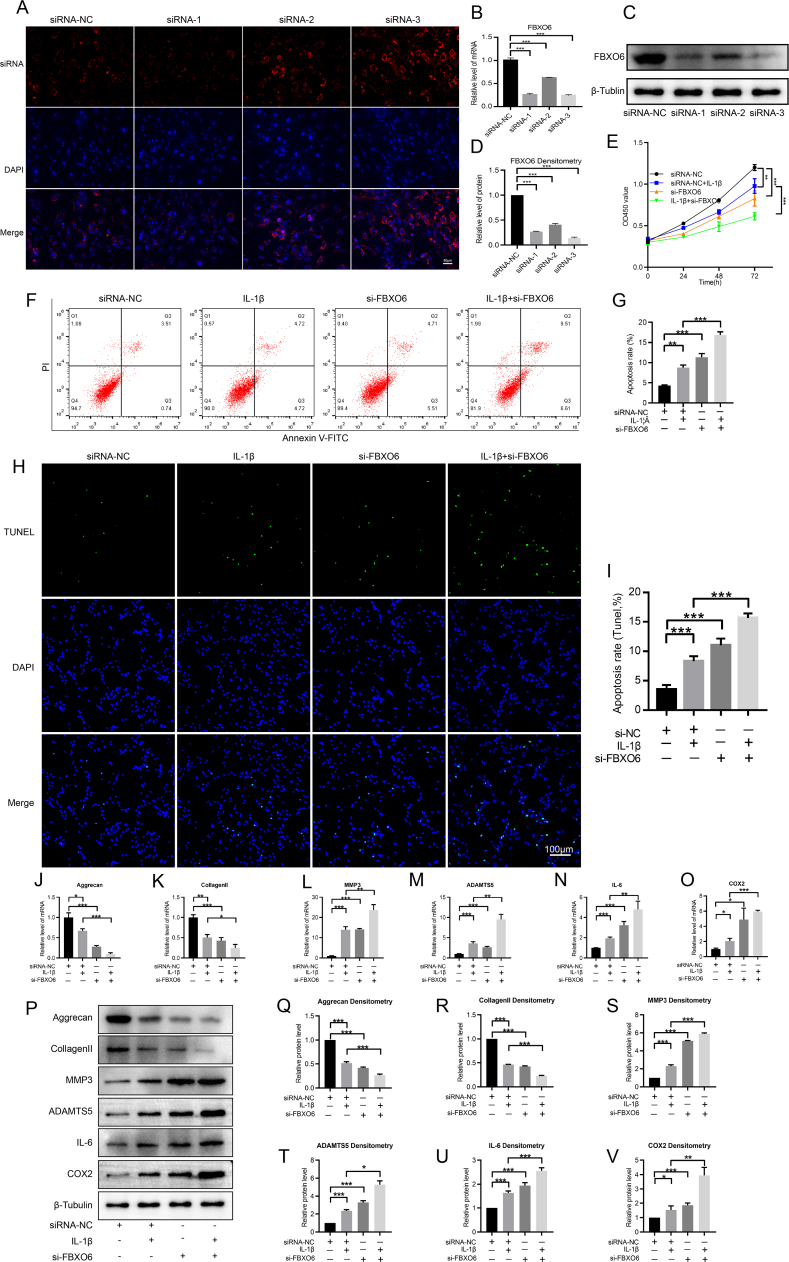
To evaluate the role of FBXO6 in the development of IVDD, FBXO6 siRNAs were developed, and the immunofluorescence detection of Cy3 showed high transfection efficiency in NP cells (Fig. 3A). RT-qPCR demonstrated that FBXO6 siRNA-3 had the highest transfection efficiency among the three siRNAs (Fig. 3B). Hence, it was used in the following experiments. First, CCK8 was used to evaluate the regulatory effect of FBXO6 on NP cells proliferation, and results showed that si-FBXO6 suppressed the proliferation of NP cells and enhanced the inhibitory effect of IL-1β on NP cell proliferation (Fig. 3C). Flow cytometry was then used to detect the apoptosis of NP cells, which showed that si-FBXO6 increased the apoptosis rate of IL-1β-stimulated NP cells (Fig. 3D and E). The TUNEL assay results also showed that the percentage of apoptosis induced by IL-1β was augmented by si-FBXO6 (Fig. 3F and G). Finally, the regulatory effect of FBXO6 on ECM synthesis and degradation in NP cells was evaluated. The RT-qPCR and WB showed that the expression of aggrecan and collagen Ⅱ decreased after IL-1β or si-FBXO6 treatment, while MMP3, ADAMTS5, IL-6 and COX2 increased after IL-1β or si-FBXO6 treatment. Moreover, si-FBXO6 enhanced the downregulations of aggrecan and collagen Ⅱ induced by IL-1β, as well as accelerated the upregulations of MMP3, ADAMTS5, IL-6 and COX2 induced by IL-1β (Fig. 3H-T).
Figure 3.
Regulatory effect of FBXO6 in NP Cells (A) High transfection efficiency was found in NP cells after transfection with siRNAs (B) RT-qPCR indicated that the expression of FBXO6 was significantly suppressed by the respective siRNA (C/D) WB and and densitometric analyses results showed that all the three siRNAs have good silence effect on the expression of FBXO6 and siRNA-3 has the highest silence efficiency (E) CCK8 test data determined that IL-1β treatment resulted in low proliferation of NP cells and si-FBXO6 was able to significantly aggravate this effect (F/G) Flow cytometry analysis showed that si-FBXO6 induced the apoptosis of NP cells, PI: propidium iodide, FITC: fluorescein isothiocyanate (H/I) TUNEL assay showed that si-FBXO6 induced the apoptosis of NP cells (J–O) RT-qPCR showed that aggrecan and collagen Ⅱ were decreased by IL-1β treatment and that si-FBXO6 aggravated the suppression of aggrecan and collagen Ⅱ by IL-1β; the expression of MMP3, ADAMTS5, IL-6 and COX2 were increased after IL-1β stimulation, and si-FBXO6 enhanced their downregulation induced by IL-1β (P–V) WB and densitometric analyses showed the regulatory effect of aggrecan, collagen II, MMP3, ADAMTS5, IL-6 and COX2 expressions by si-FBXO6 and IL-1β. The treatment condition of IL-1β was 10 ng/ml for 24 h; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.3. miR-133a-5p was the candidate miRNA that inhibited FBXO6 expression in IVDD
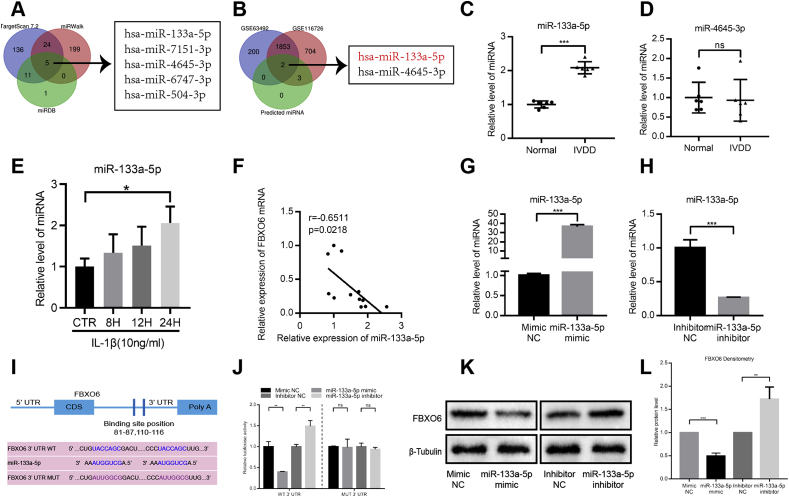
As miRNA plays a vital and broad role in controlling gene expression, we performed a comprehensive bioinformatic analysis using online tools (TargetScan, miRDB and miRWalk) along with GSE IVDD datasets(GSE116726, GSE63492)to search for potential microRNAs targeting the 3′ untranslated regions (3′ UTR) of FBXO6. The analysis showed that miR-133a-5p and miR-4645–3p were the potential miRNAs (Fig. 4A and B).
Figure 4.
miR-133a-5p was the inhibitory regulator of FBXO6 (A) TargetScan, miRDB and miRWalk were used to detect the related microRNAs targeting FBXO6 (B) GSE63492 and GSE116726 datasets were used to evaluate the related microRNAs targeting FBXO6 (C) Compared with normal IVD, miR-133a-5p expression level was upregulated in degenerated IVD specimens (D) RT-qPCR result showed that no significant difference of miR-4635–3p expression was found between normal and degenerated IVD (E) IL-1β induced the expression of miR-133a-5p (F) Negative correlation was found between the expression of FBXO6 and miR-133a-5p in IVD tissues (G/H) miR-133a-5p levels in NP cells after being transfected with the miR-133a-5p mimic or inhibitor were determined by the RT-qPCR (I) Schematic representation of miR-133a-5p's predicted binding sites in the 3′UTR of FBXO6 mRNA (J) The wild- or mutant-type FBXO6 3′UTR reporter plasmid was co-transfected with miR-133a-5p mimics or inhibitors into NP cells (K/L) FBXO6 expressions in NP cells after transfection with miR-133a-5p mimic or inhibitor. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To examine whether miR-133a-5p and miR-4645–3p were involved in the process of IVDD, we detected their expressions in normal and IVDD tissues by RT-qPCR. We found that the expression of miR-133a-5p increased in degenerated IVD tissues, and no significant difference was found in the expression of miR-4645–3p (Fig. 4C and D). Furthermore, IL-1β can increase the expression of miR-133a-5p in NP cells (Fig. 4E). The Pearson correlation analysis showed that miR-133a-5p was negatively correlated with FBXO6, suggesting that miR-133a-5p and FBXO6 might have some correlations during the progression of IVDD (Fig. 4F). Besides, miR-133a-5p can affect the process of proliferation and apoptosis in other cell types [18,19], so miR-133a-5p was selected for further study.
To assess whether FBXO6 could be regulated by miR-133a-5p, NP cells were transfected with miR-133a-5p mimic or inhibitor, respectively. RT-qPCR showed that the expression of miR-133a-5p increased in NP cells transfected with miR-133a-5p mimic, while decreased in NP cells transfected with miR-133a-5p inhibitor (Fig. 4G and H). To further confirm the functional interaction between miR-133a-5p and FBXO6, we predicated the binding sites between them through the miRDB website, and the result showed that there were two binding sites located at the 3′ UTR of FBXO6, 81–87 and 110–116, respectively (Fig. 4I). Furthermore, we established dual luciferase reporter vectors containing the wild or mutant-type miR-133a-5p seed sequences of FBXO6 3′UTR. The dual luciferase assay revealed that the miR-133a-5p mimic significantly repressed while miR-133a-5p inhibitor increased the luciferase activity of the wide-type FBXO6 reporter gene, but not for the mutant-type (Fig. 4J). The WB results demonstrated that the protein levels of FBXO6 decreased after miR-133a-5p mimic treatment, while increased following miR-133a-5p inhibitor treatment (Fig. 4K-L).
3.4. miR-133a-5p decreased ECM synthesis and increased ECM degradation in IVDD via directly targeting FBXO6
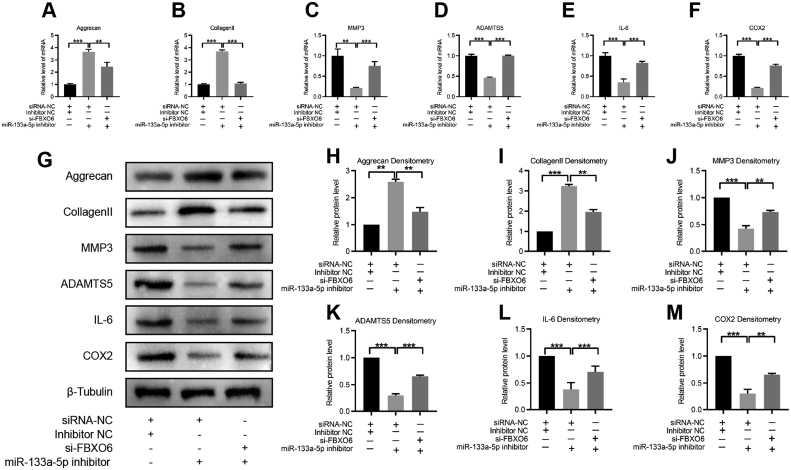
To investigate whether miR-133a-5p participated in IVDD by directly targeting FBXO6. miR-133a-5p inhibitor and si-FBXO6 were used to stimulate NP cells before the stimulation of IL-1β. Results showed that miR-133a-5p inhibition led to increased levels of aggrecan and collagen Ⅱ, which then were attenuated by si-FBXO6. Furthermore, the downregulations of MMP3, ADAMTS5, IL-6 and COX2 induced by miR-133a-5p inhibition could be partially rescued by the addition of si-FBXO6, suggesting that FBXO6 was essential for miR-133a-5p-induced ECM synthesis and degradation in NP cells (Fig. 5A-M). Taken together, the above results suggested that miR-133a-5p exerted its function in IVDD by directly targeting FBXO6.
Figure 5.
miR-133a-5p exerted its function in NP cells via sponging FBXO6 (A–F) RT-qPCR showed that the expressions of aggrecan and collagen Ⅱ were increased by the miR-133a-5p inhibitor, and si-FBXO6 alleviated the upregulation of aggrecan and collagen Ⅱ. The expressions of MMP3, ADAMTS5, IL-6 and COX2 were downregulated by the miR-133a-5p inhibitor and si-FBXO6 partly reversed their downregulations (G–M) WB and densitometric analyses showed the expressions of aggrecan, collagen Ⅱ, MMP3, ADAMTS5, IL-6 and COX2 by the miR-133a-5p inhibitor and si-FBXO6. The treatment condition of IL-1β was 10 ng/ml for 24 h; ∗∗P < 0.01, ∗∗∗P < 0.001.
3.5. Inflammatory environment regulated miR-133a-5p and FBXO6 via NF-κB signaling
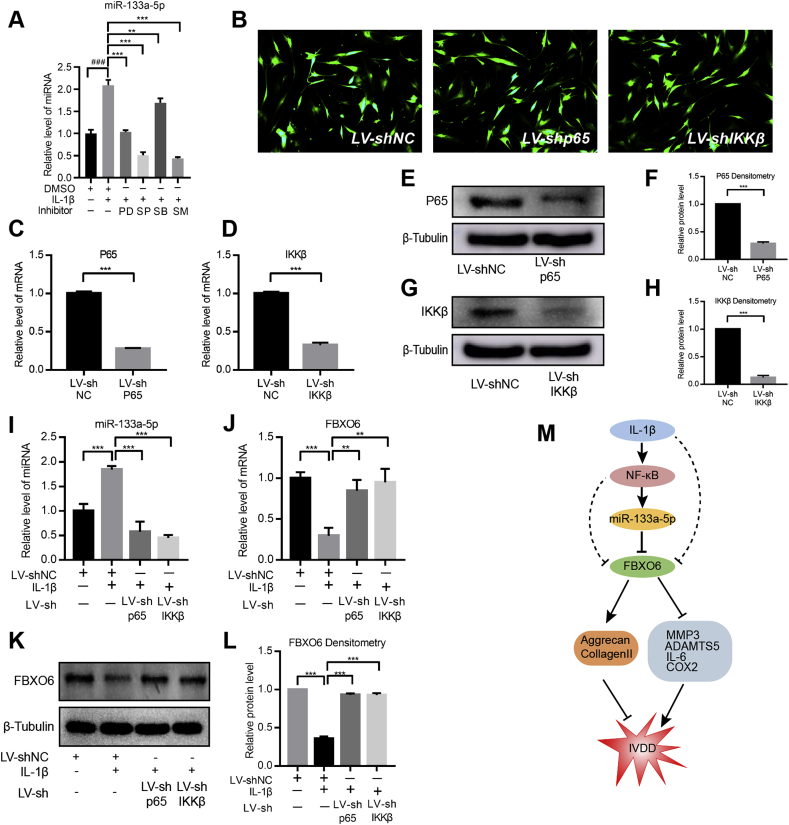
As it was illustrated that IL-1β regulated the expression of miR-133a-3p and FBXO6. Therefore, we explored the regulation mechanisms and signaling pathways. Previous studies have shown that IL-1β played its role by activating MAPK and NF-κB signaling, so we also elucidated the role of MAPK and NF-κB signaling on the regulation of miR-133a-5p and FBXO6. NP cells were treated with NF-kB inhibitor (SM7368, 20 μM), ERK1/2 inhibitor (PD98059, 20 μM, 1:1000 dilution), p38 inhibitor (SB203580, 20 μM, 1:1000 dilution) or c-Jun N-terminal kinase (JNK) inhibitor (SP600125, 20 μM, 1:1000 dilution) prior to the exposure of IL-1β. The RT-qPCR results showed that the MAPK and NF-κB inhibitors can partly abolish the promotion effect of IL-1β on miR-133a-5p expression, with the most significant change in the NF-κB inhibitor (Fig. 6A). Hence, the NF-κB pathway was selected for further analysis. Lentivirus p65-shRNA or IKKβ-shRNA were used to silence individual NF-κB signaling components. Results showed that there were robust GFP expression in the viral transduced cells, indicating a high level of transduction efficiency (Fig. 6B). Transduction with sh-p65 and sh-IKKβ led to significant decrease in the expression of p65 and IKKβ at mRNA and protein levels (Fig. 6C–H). The loss of function study showed that the suppression of NF-κB signaling components significantly abolished the regulatory effect of IL-1β on miR-133a-5p and FBXO6 expression (Fig. 6I-L). Taken together, these results suggested that the inflammatory environment in IVDD tissues activated NF-κB signaling pathway, then enhanced miR-133a-5p expression and finally suppressed the expression of FBXO6.
Figure 6.
The inflammatory environment regulated miR-133a-5p and FBXO6 via the NF-κB signaling pathway (A) The expression of miR-133a-5p following IL-1β treatment with or without ERK inhibitor (PD98059 (PD)) or JNK inhibitor (SP60025 (SP)) or p38 inhibitor (SB203580 (SB)) or NF-κB inhibitor (SM7368 (SM)) (B) Immunofluorescence detection of GFP in human NP cells transduced with either shp65 or shIkkβ showed a high transfection efficiency (C–D) RT-qPCR showed LV-shp65 and LV-shIkkβ significantly suppressed the expression of p65 and Ikkβ (E–H) A similar result was found by the WB and densitometric analyses (I) IL-1β-dependent induction in miR-133a-5p was significantly blocked by the suppression of the components of the NF-κB pathway (J–L) RT-qPCR and WB showed that the IL-1β-dependent decrease in FBXO6 was significantly relieved by the suppression of the components of the NF-κB pathway (M) Proposed illustration to point out the mechanism by which IL-1β upregulates miR-133a-5p expression to aggravate IVD degeneration via inhibiting FBXO6. The treatment condition of IL-1β was 10 ng/ml for 24 h; ‘###’ means P < 0.001 when compared with DMSO group; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
In this study, it was demonstrated that miR-133a-5p increased while FBXO6 decreased in the process of IVDD together with under the stimulation of IL-1β. Mechanically, miR-133a-5p aggravated IVD degeneration by targeting FBXO6, namely through the decreased expression of ECM (aggrecan, collagen Ⅱ), while increased expression matrix metalloproteinases (MMP3, ADAMTS5) and chemo-cytokine (IL-6, COX2) in NP cells. We also found that proinflammatory cytokine IL-1β regulated miR-133a-5p and FBXO6 expressions via the NF-κB signaling pathway.
FBXO6 plays important roles in broad biological processes, including circadian amplitude, apoptosis, autophagy, cell cycle, carcinogenesis and cancer metastasis [7,8,25,26]. The role of FBXO6 in different organs has tissue- and cell-specific characteristics. Du et al. [8] demonstrated that FBXO6 could enhance antiviral immunity by negatively regulating the homeostasis of IFN-I and ISG production via in vitro overexpression and knockout experiments. Another study elucidated that FBXO6 reduced the tumour growth and metastasis in colorectal cancer by inducing RIOK1 ubiquitination [27]. Importantly, Wang et al. [11] found that the knockout of FBXO6 in cartilage deteriorated the process of osteoarthritis both in human and mouse osteoarthritis models. In addition, a previous study indicated that NP cells had a chondrocyte-like phenotype, which shares common features with chondrocytes [28]. Therefore, we designed this study to evaluate the molecular role of FBXO6 on the pathogenesis of IVDD, generally for the regulation of NP cells proliferation, apoptosis, ECM synthesis and degradation. Relevant to cell proliferation, FBXO6 siRNA led to a decreased proliferation of NP cells. Moreover, flow cytometry and the TUNEL assay showed that the silence of FBXO6 resulted in increased NP cell apoptosis and enhanced the apoptosis of NP cells induced by IL-1β. For the regulatory effect of ECM by FBXO6, we treated NP cells with FBXO6 siRNA and detected the expressions of aggrecan, collagen Ⅱ, MMP3, ADAMTS5, IL-6 and COX2. Results showed suppression of FBXO6 by siRNA lead to increased MMP3, ADAMTS5, IL-6 and COX2 expression, while decreased expression of aggrecan and collagen Ⅱ, suggesting that FBXO6 can modulate ECM synthesis and degradation in NP cells.
As FBXO6 decreased in the process of IVDD, we determined to explore whether there was any upstream regulator that could inhibit the expression of FBXO6. Increasing evidence has shown that miRNAs can serve as inhibitory regulators of mRNA [29,30]. Therefore, we checked the candidate miRNAs sponging FBXO6 using online tools (TargetScan, miRDB and miRWalk) along with GSE IVDD datasets. Results showed that miR-133a-5p was predicted to be the sponge of FBXO6. Dual Luciferase Assay confirmed their binding sites and WB results showed the inhibitory effect of miR-133a-5p on the expression of FBXO6, which were consistent with previous studies. Furthermore, miRNAs have been proven to be involved in the pathology of IVDD, suggesting that miRNAs could be novel targets for treating patients with IVDD [[31], [32], [33], [34]]. For example, it was shown that miR-181a protected against IVDD by reducing inflammatory factors and promoting collagen Ⅱ expression [35]. Another study illustrated that the downregulation of miR-133a induced collagen Ⅱ loss by directly targeting MMP9 [36]. Similarly, in our study, we revealed that miR-133a-5p also participated in the regulation of IVDD. Loss of function studies demonstrated that miR-133a-5p inhibitor led to increased levels of collagen II and aggrecan and decreased levels of MMP3, ADAMTS5, IL-6 and COX2. Importantly, rescue experiments further confirmed that miR-133a-5p exerted its function in the regulation of IVDD via directly binding to FBXO6.
Previous studies have shown that IL-1β plays its role by activating MAPK and NF-κB signaling [37], so we assumed that NF-κB might participate in the induction of the miR-133a-5p/FBXO6 axis by IL-1β. Nuclear factor kappa B (NF-κB) is a family of transcription factors that plays a central role in the production of inflammatory mediators and catabolic gene expression [38,39]. NF-κB/Rel protein mainly includes the Rel A (p65), Rel B and c-Rel transcription factors, and phosphorylated NF-κB-p65 is another important factor that plays a vital role in promoting the activation of NF-κB [40]. IKK includes the three subunits, IKKα, IKKβ, and IKKγ, of which IKKβ plays a particularly important role in the activation of the classical NF-κB signaling pathway [41]. A previous study demonstrated that proinflammatory cytokine IL-1β can induce the phosphorylation of IKKβ and p65 NF-κB protein in NP cells [42]. Consistent with previous reports, our study also demonstrated that the stimulation of the miR-133a-5p/FBXO6 axis by IL-1β was alleviated following the silence of p65 and IKKβ, highlighting the importance of NF-κB in the regulation of the miR-133a-5p/FBXO6 axis under IL-1β stimulation.
It should be noted that there are several limitations of this study. First, only loss of function technologies, such as si-FBXO6 and the miR-133a-5p inhibitor, were used to demonstrate the gene function. In future studies, the overexpression of FBXO6 and miR-133a-5p mimic will be applied to further investigate their roles in IVDD; Second, only in vitro studies were designed, and therefore the potential therapeutic applications of the miR-133a-5p/FBXO6 axis in the treatment of IVDD still need to be elucidated by in vivo studies; Furthermore, the role of the miR-133a-5p/FBXO6 axis in IVDD was majorly investigated in NP and AF cells; however, the effect in the cartilage endplate cells was not determined and thus requires further investigations.
Collectively, our results demonstrated that miR-133a-5p regulated the process of IVDD and that it could be upregulated by IL-1β to aggravate IVDD via sponging FBXO6. This study clarified one of the mechanisms of inflammation-induced IVDD progression and the specific role of miR-133a-5p/FBXO6 axis in this regulatory network.
Declaration of competing interest
The authors have no conflicts of interest to disclose in relation to this article.
Acknowledgement
We thank the staffs of Department of Medical Genetics of Zhongshan Medicine School and Center for Genome Research, Sun Yat-Sen University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2021.05.004.
Contributor Information
Hua Wang, Email: wangh227@mail.sysu.edu.cn.
Zhao-Min Zheng, Email: zhzhaom@mail.sysu.edu.cn.
Author contributions
Hua Wang and He-Hai Pan designed the study. Xian-Fa Du and Hai-Tao Cui performed the experiments. Hao-Wen Cui, Shun-Lun Chen, Jian-Ru Wang, Ze-Min Li and Hui Liu collected the clinical samples. Xian-Fa Du, Hua Wang and Jun Long contributed to the data analysis and manuscript draft. Yong-Can Huang and Zhao-Min Zheng contributed to data analysis and corrected the manuscript. All authors read and approved the final manuscript.
Funding source
This work was supported by research grant from National Natural Science Foundation of China (Grant number: 81772386).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Buchbinder R., van Tulder M., Oberg B., Costa L.M., Woolf A., Schoene M. Low back pain: a call for action. Lancet. 2018;391(10137):2384–2388. doi: 10.1016/S0140-6736(18)30488-4. [DOI] [PubMed] [Google Scholar]
- 2.Abajobir A.A., Abate K.H., Abbafati C., Abbas K.M., Abd-Allah F., Abdulkader R.S. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vos T., Barber R.M., Bell B., Bertozzi-Villa A., Biryukov S., Bolliger I. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(9995):743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Liu H., Zheng Z.M., Zhang K.B., Wang T.P., Sribastav S.S. Role of death receptor, mitochondrial and endoplasmic reticulum pathways in different stages of degenerative human lumbar disc. Apoptosis. 2011;16(10):990–1003. doi: 10.1007/s10495-011-0644-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang S.Z., Rui Y.F., Lu J., Wang C. Cell and molecular biology of intervertebral disc degeneration: current understanding and implications for potential therapeutic strategies. Cell Prolif. 2014;47(5):381–390. doi: 10.1111/cpr.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyu F.J., Cui H., Pan H., Mc Cheung K., Cao X., Iatridis J.C. Painful intervertebral disc degeneration and inflammation: from laboratory evidence to clinical interventions. Bone Res. 2021;9(1):7. doi: 10.1038/s41413-020-00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaar J.R., Pagan J.K., Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14(6):369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du X., Meng F., Peng D., Wang Z., Ouyang W., Han Y. Noncanonical role of FBXO6 in regulating antiviral immunity. J Immunol. 2019;203(4):1012–1020. doi: 10.4049/jimmunol.1801557. [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Duan L.H., Luo P.C., Hu G., Yu X., Liu J. FBXO6-Mediated ubiquitination and degradation of Ero1L inhibits endoplasmic reticulum stress-induced apoptosis. Cell Physiol Biochem. 2016;39(6):2501–2508. doi: 10.1159/000452517. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Hou Y., Wang M., Wu B., Li N. A study on the functions of ubiquitin metabolic system related gene FBG2 in gastric cancer cell line. J Exp Clin Canc Res. 2009;28(1):78. doi: 10.1186/1756-9966-28-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G., Chen S., Xie Z., Shen S., Xu W., Chen W. TGFβ attenuates cartilage extracellular matrix degradation via enhancing FBXO6-mediated MMP14 ubiquitination. Ann Rheum Dis. 2020;79(8):1111–1120. doi: 10.1136/annrheumdis-2019-216911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv F., Leung V.Y., Huang S., Huang Y., Sun Y., Cheung K.M. In search of nucleus pulposus-specific molecular markers. Rheumatology. 2014;53(4):600–610. doi: 10.1093/rheumatology/ket303. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam S., Jeet V., Clements J.A., Gunter J.H., Batra J. Emergence of MicroRNAs as key players in cancer cell metabolism. Clin Chem. 2019;65(9):1090–1101. doi: 10.1373/clinchem.2018.299651. [DOI] [PubMed] [Google Scholar]
- 15.Pan Y., Hui X., Hoo R.L.C., Ye D., Chan C.Y.C., Feng T. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest. 2019;129(2):834–849. doi: 10.1172/JCI123069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji M.L., Jiang H., Zhang X.J., Shi P.L., Li C., Wu H. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9(1):5051. doi: 10.1038/s41467-018-07360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng G., Zha Z., Huang Y., Li J., Wang Y., Ke W. Sustained and bioresponsive two-stage delivery of therapeutic miRNA via polyplex micelle-loaded injectable hydrogels for inhibition of intervertebral disc fibrosis. Adv Healthc Mater. 2018;7(21) doi: 10.1002/adhm.201800623. [DOI] [PubMed] [Google Scholar]
- 18.Kojima S., Chiyomaru T., Kawakami K., Yoshino H., Enokida H., Nohata N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Canc. 2012;106(2):405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y., Pan J., Huang S., Peng X., Zou X., Luo Y. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J Exp Clin Canc Res. 2018;37(1):160. doi: 10.1186/s13046-018-0813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfirrmann C.W., Metzdorf A., Zanetti M., Hodler J., Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976. 2001;26(17):1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 21.Wang H., Tian Y., Wang J., Phillips K.L., Binch A.L., Dunn S. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 2013;288(23):16761–16774. doi: 10.1074/jbc.M112.446633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Che M., Xin J., Zheng Z., Li J., Zhang S. The role of IL-1β and TNF-α in intervertebral disc degeneration. Biomed Pharmacother. 2020;131:110660. doi: 10.1016/j.biopha.2020.110660. [DOI] [PubMed] [Google Scholar]
- 23.Risbud M.V., Shapiro I.M. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson Z.I., Schoepflin Z.R., Choi H., Shapiro I.M., Risbud M.V. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104–116. doi: 10.22203/ecm.v030a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan S., Cermak L., Pagan J.K., Rossi M., Martinengo C., di Celle P.F. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481(7379):90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santra M.K., Wajapeyee N., Green M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong X., Huang H., Qiu X., Ding Z., Feng X., Zhu Y. Targeting posttranslational modifications of RIOK1 inhibits the progression of colorectal and gastric cancers. eLife. 2018;7 doi: 10.7554/eLife.29511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Au T.Y.K., Lam T.K., Peng Y., Wynn S.L., Cheung K.M.C., Cheah K.S.E. Transformation of resident notochord-descendent nucleus pulposus cells in mouse injury-induced fibrotic intervertebral discs. Aging Cell. 2020;19(11) doi: 10.1111/acel.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Yu X., Shen J., Chan M.T., Wu W.K. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48(3):278–283. doi: 10.1111/cpr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y., Peng Y., He P., Zhang Q., Xu D. Urinary miRNAs as biomarkers for idiopathic osteonecrosis of femoral head: a multicentre study. J Orthop Translat. 2021;26:54–59. doi: 10.1016/j.jot.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X.Q., Tu W.Z., Guo J.B., Song G., Zhang J., Chen C.C. A bioinformatic analysis of MicroRNAs' role in human intervertebral disc degeneration. Pain Med. 2019;20(12):2459–2471. doi: 10.1093/pm/pnz015. [DOI] [PubMed] [Google Scholar]
- 32.Dong W., Liu J., Lv Y., Wang F., Liu T., Sun S. miR-640 aggravates intervertebral disc degeneration via NF-κB and WNT signalling pathway. Cell Prolif. 2019;52(5) doi: 10.1111/cpr.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R., Wen B., Sun D. miR-573 regulates cell proliferation and apoptosis by targeting Bax in nucleus pulposus cells. Cell Mol Biol Lett. 2019;24:2. doi: 10.1186/s11658-018-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Pan D., Liu S., Xing X., Zhou H., Zhang B. Identification of circ-FAM169A sponges miR-583 involved in the regulation of intervertebral disc degeneration. J Orthop Translat. 2021;26:121–131. doi: 10.1016/j.jot.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Shi X., Peng X., Li Y., Ma H., Li D. MicroRNA-181a exerts anti-inflammatory effects via inhibition of the ERK pathway in mice with intervertebral disc degeneration. J Cell Physiol. 2020;235(3):2676–2686. doi: 10.1002/jcp.29171. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y.Q., Zhang Z.H., Zheng Y.F., Feng S.Q. Dysregulated miR-133a mediates loss of type II collagen by directly targeting matrix metalloproteinase 9 (MMP9) in human intervertebral disc degeneration. Spine (Phila Pa 1976. 2016;41(12):E717–E724. doi: 10.1097/BRS.0000000000001375. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y.-L., Yan D.-Y., Wu C.-Y., Xuan J.-W., Jin C.-Q., Hu X.-L. Maslinic acid prevents IL-1β-induced inflammatory response in osteoarthritis via PI3K/AKT/NF-κB pathways. J Cell Physiol. 2021;236(3):1939–1949. doi: 10.1002/jcp.29977. [DOI] [PubMed] [Google Scholar]
- 38.Wuertz K., Vo N., Kletsas D., Boos N. Inflammatory and catabolic signalling in intervertebral discs: the roles of NF-κB and MAP kinases. Eur Cell Mater. 2012;23:103–119. doi: 10.22203/ecm.v023a08. [DOI] [PubMed] [Google Scholar]
- 39.Yongyun C., Jingwei Z., Zhiqing L., Wenxiang C., Huiwu L. Andrographolide stimulates osteoblastogenesis and bone formation by inhibiting nuclear factor kappa-Β signaling both in vivo and in vitro. J Orthop Translat. 2019;19:47–57. doi: 10.1016/j.jot.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu T.Y., Ju J.M., Mo L.H., Ma L., Hu W.H., You R.R. Anti-inflammation action of xanthones from Swertia chirayita by regulating COX-2/NF-κB/MAPKs/Akt signaling pathways in RAW 264.7 macrophage cells. Phytomedicine. 2019;55:214–221. doi: 10.1016/j.phymed.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Prescott J.A., Cook S.J. Targeting IKKβ in cancer: challeng es and opportunities for the therapeutic utilisation of IKKβ inhibitors. Cells. 2018;7(9) doi: 10.3390/cells7090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian Y., Yuan W., Fujita N., Wang J., Wang H., Shapiro I.M. Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-κB. Am J Pathol. 2013;182(6):2310–2321. doi: 10.1016/j.ajpath.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.