Abstract
The fungal cyclohexadepsipeptides destruxins (DTXs), isaridins (ISDs), and isariins (ISRs) are nonribosomal peptides whose structures include a 19-membered ring composed of five amino acid residues and one α- or β-hydroxy acid residue. These cyclohexadepsipeptides contain unusual nonproteinogenic amino acid–building blocks and possess a range of antiviral, antibacterial, and other activities. The biosynthetic gene clusters for ISDs and ISRs have not been identified, and the biosynthesis of the nonproteinogenic (3S)-methyl-l-proline residue, which is found in DTXs, ISDs, and many other natural products, lacks full characterization. In an ongoing effort to identify compounds that can inhibit the Zika virus (ZIKV), we examined the extract of marine-derived fungus Beauveria felina SX-6-22 and discovered 30 DTXs, ISDs, and ISRs (1–30) including seven new compounds (1–7). The anti-ZIKV assays showed that 9–12 and 16–18 possess inhibitory activities against ZIKV RNA replication and NS5 (nonstructural protein 5) production in ZIKV-infected A549 cells. We sequenced the genome of B. felina SX-6-22 and identified three biosynthetic gene clusters detx, isd and isr, which are responsible for the biosynthesis of DTXs, ISDs, and ISRs, respectively. Comparative analyses of the three gene clusters clarified the biosynthetic relationships among these cyclohexadepsipeptides. Finally, we characterized the entire biosynthesis of nonproteinogenic building block (3S)-methyl-l-proline. The Δ1-pyrroline-5-carboxylate reductases (P5CRs), also used in the biosynthesis of l-proline, were demonstrated to catalyze the final reduction step in (3S)-methyl-l-proline formation, suggesting potential cross talk between primary and secondary metabolisms. These results provide opportunities for biosynthetic pathway engineering to generate new anti-ZIKV cyclohexadepsipeptides.
Keywords: anti-Zika virus activities, biosynthetic gene clusters, cyclohexadepsipeptides, destruxins, isaridins, isariins, (3S)-methylproline, nonproteinogenic amino acids
Abbreviations: α-d-HIC, α-d-hydroxyisocaproic acid; DTX, destruxin; ECL, enhanced chemiluminescence; GK, γ-glutamyl kinase; GPR, γ-glutamyl phosphate reductase; HBV, hepatitis B virus; ISD, isaridin; ISR, isariin; P5CR, pyrroline-5-carboxylate reductase; PCP, peptidyl carrier protein; TIC, total ion chromatography; UPLC, ultra-performance liquid chromatography; ZIKV, Zika virus
Destruxins (DTXs), isaridins (ISDs), and isariins (ISRs) are three types of structurally related cyclohexadepsipeptides with a 19-membered ring formed by five amino acid residues and one α-hydroxy or β-hydroxy acid residue (1, 2, 3). DTX structures are characterized by the presence of nonproteinogenic amino acid-building blocks including β-alanine, N-methyl-l-alanine, and N-methyl-l-valine (only a few DTXs without methylation) and the unusual (3S)-methyl-l-proline residues. The α-d-hydroxyisocaproic acid (α-d-HIC) residue in DTXs is usually modified by oxidation and decarboxylation. ISDs are structurally related to DTXs but contain different residues such as l-phenylalanine and N-methyl-l-phenylalanine at the corresponding positions in DTXs. In contrast to DTXs, the α-HIC residues in ISDs are unmodified without exception. ISRs differ from DTXs and ISDs with different amino acid building blocks and a β-hydroxy acid residue containing different length of alkyl side chain. Apart from the potent insecticidal activities (4, 5, 6), some of the cyclohexadepsipeptides also possess anti-hepatitis B virus (HBV) (7), V-ATPase inhibition (8), cytotoxicity (9, 10), anti-inflammatory, and antibacterial activities (11, 12). The significant biological activities and unusual chemical scaffolds of the cyclohexadepsipeptides attracted much attention for the total synthesis (8, 13). In biosynthetic aspects, only the biosynthetic gene cluster of DTXs from Metarhizium robertsii was reported in 2012 (14), whereas the biosynthetic gene clusters of ISDs and ISRs have not been identified despite the first discovery of ISD and ISR decades ago. Although the structural similarity between DTXs and ISDs and the fact that the three types of cyclohexadepsipeptides coexist in one strain (e.g., marine-derived fungi Beauveria felina AcSS8) are demonstrated (15), the biosynthetic relationships among DTXs, ISDs, and ISRs are still uncertain.
(3S)-Methyl-l-proline is an unusual building block incorporated in various DTXs and ISDs. The similar building blocks are also present in other natural products including paraherquamide E (16), bottromycins (17), scytalidamide B (18), and neoefrapeptins (19) (Fig. S1). However, the biosynthesis of (3S/R)-methyl-l-proline residues in nature lacks full characterization. In the biosynthetic investigation of the fungal alkaloids UCS1025A, (3S)-methyl-l-proline as the biosynthetic precursor of the pyrrolizidinone moiety is identified to be originated from l-isoleucine by the catalysis of an α-ketoglutarate (KG)-dependent nonheme iron dioxygenase UcsF and a reductase UcsG (20). However, the function of UcsG has not been experimentally characterized. In DTXs and ISDs, (3S)-methyl-l-proline residue and l-proline residue are alternatively present in the same position of the 19-membered ring, raising the question whether the (3S)-methyl-l-proline residue is generated by the similar biosynthetic steps as those in UCS1025A biosynthesis, or by the methylation of l-proline residue as proposed in the biosynthesis of bottromycins (21). These puzzles in the biosynthesis of DTXs and ISDs require thorough characterization.
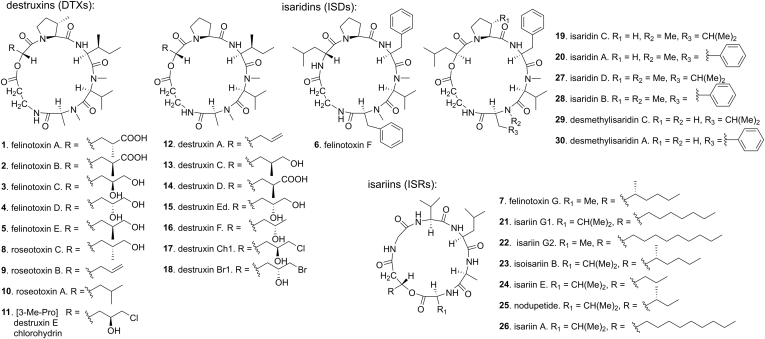
In the course of our continuous discovery of antiviral natural products from marine-derived microorganisms, we established a Zika-infected cell line model for screening anti-ZIKV compounds from the crude extracts of our in-house library of marine-derived fungi. Together with other crude extracts, the crude extract of sponge-associated fungus B. felina SX-6-22 (Fig. S2) was subjected for the screening, showing anti-ZIKV activity. A Global Natural Product Social Molecular Networking (GNPS) (22, 23, 24) analysis in association with the 1H NMR characterization revealed that the fungus produced an array of cyclodepsipeptides (Figs. S3 and S4). A large-scale fermentation of this fungus led to the discovery of 30 cyclohexadepsipeptides including seven new congeners (1–7). These analogues can be classified into three types: DTXs (1–5, 8–18), ISDs (6, 19, 20, 27–30; compound 6 was considered as a cyclohexapeptide-ISD analogue), and ISRs (7, 21–26) (Fig. 1). Their structures were determined on the basis of the NMR, MS, X-ray diffraction, and Marfey's analyses. A bioassay for anti-ZIKV activities in vitro revealed that a number of congeners exhibited considerable inhibition against the ZIKV RNA replication and expression of NS5 (nonstructural protein 5), which is the most conserved ZIKV protein essential for virus replication and an ideal drug target for potential therapeutics (25). Based on the genome sequencing, gene deletion, and chemotype analysis, three nonribosomal peptide synthetase (NRPS)-encoding gene clusters detx, isd, and isr in fungus B. felina SX-6-22 are identified to be responsible for the biosynthesis of DTXs, ISDs, and ISRs, respectively. In addition, the full biosynthesis of (3S)-methyl-l-proline is characterized by gene deletion and reconstitution of enzymatic reactions in vitro.
Figure 1.
Cyclohexadepsipeptides discovered from B. felina SX-6-22. Compounds 27–30 are detected and analyzed by LC-MS/MS.
Results
Discovery and structural elucidation of cyclohexadepsipeptides
The UPLC analysis in association with the NMR characterization of the crude extracts showed that the solid-state fermentation of fungal strain B. felina SX-6-22 produced a profile of peptides (Fig. S3). Further analysis using GNPS Molecular Networking tool and in-house marine natural product database afforded the molecular ion nodes that are classified into three peptide clusters (Fig. S4). Analysis of the MS/MS spectra extracted from raw data led to the identification of destruxin A (12) from cluster A; isaridin A (20), isaridins D (27) and B (28), desmethylisaridin C (29) and desmethylisaridin A (30) from cluster B; and isoisariin B (27) from cluster C (Fig. S5). The GNPS method also informed that a number of unidentified nodes that are speculated to be new metabolites. Extensive chromatographic separation after large-scale fermentation resulted in the isolation of 26 cyclohexadepsipeptides, including seven new congeners namely felinotoxins A-G (1–7) (Fig. 1).
Felinotoxin A (1) has a molecular formula of C31H51N5O9 as determined by the HRESIMS (m/z 638.3766 [M + H]+) and NMR data, showing nine degrees of unsaturation. The 1H NMR resonances for four D2O exchangeable protons and five α-protons in association with seven carbonyl and five α-carbon resonances in the 13C NMR spectrum were characteristic of a peptide. Diagnostic 1D and 2D NMR data (COSY, HMQC, HMBC, and NOESY) enabled to establish six building blocks, including a 2-hydroxy-4-methylpentanedioic acid (HMPDA), a 3-methylproline, an isoleucine, an N-methylvaline, an N-methylalanine, and a β-alanine residues. These data resemble those of destruxin D (14) (6) except for a proline residue in destruxin D to be replaced by a 3-methylproline residue in 1. The connection of the residues in 1 was fully established by the HMBC and NOE correlations, demonstrating a cyclohexadepsipeptide with the similar sequence as destruxin D (Supporting information). The absolute configuration of 1 was determined by the single-crystal X-ray diffraction analysis using Cu Kα radiation, of which the α-carbons of amino acid residues were identified as l-configuration, while S configuration for the β-carbon of 3-methylproline residue and R configuration for both α- and γ-carbons of the HMPDA residue were clarified (Fig. 2).
Figure 2.
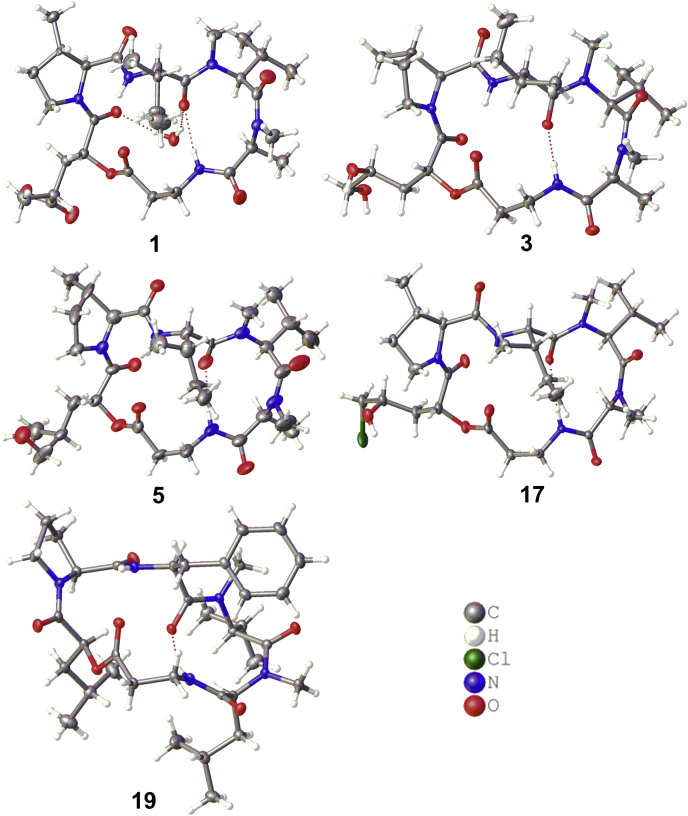
ORTEP view of the crystal structures of 1, 3, 5, 17, and 19. Hydrogen bonds are shown in red dashed lines.
A detailed analyses of the spectroscopic data (Tables S1–S7, Figs. S26–S90) in association with the Marfey’s analysis and X-ray diffraction established the structures of felinotoxins B-G (2–7), in addition to 19 known congeners (8–26). It is notable that the crystal structures of 1, 3, and 5 that belong to the DTX-type showed 2R configuration of the modified α-HIC residue (α-d-HIC), whereas the X-ray diffraction of isaridin C (19), an ISD-type congener whose stereochemistry was unidentified in the literature (1), revealed a 2S configuration of the modified α-HIC residue (α-l-HIC). Other ISD-type congeners such as isaridin G, desmethylisaridin G, and desmethylisaridin C1, whose structures are unambiguously assigned by the X-ray diffraction (3), also showed 2S configuration for the modified α-HIC residue. Interestingly, the crystal structures of 1, 3, 5, 17, and 19 show an intramolecular hydrogen bond between the carbonyl oxygen of l-isoleucine residue (or l-phenylalanine residue at the same position in 19) and the amide hydrogen of β-alanine residue (Fig. 2), suggesting a specifically conserved peptidyl conformation in both DTXs and ISDs. Although most of the known homologues have been reported for decades, it is surprising that some of them lack the configuration assignments or spectroscopic data. In this work, the NMR data of all known congeners have been assigned completely based on the 1D and 2D NMR data (Supporting information), while the absolute configurations of the amino acid residues as well as the α- and β-hydroxy acid moieties have been determined by Marfey’ analysis (Fig. S6), X-ray diffraction analysis, and NMR chemical shift characterization.
Anti-ZIKV bioassays
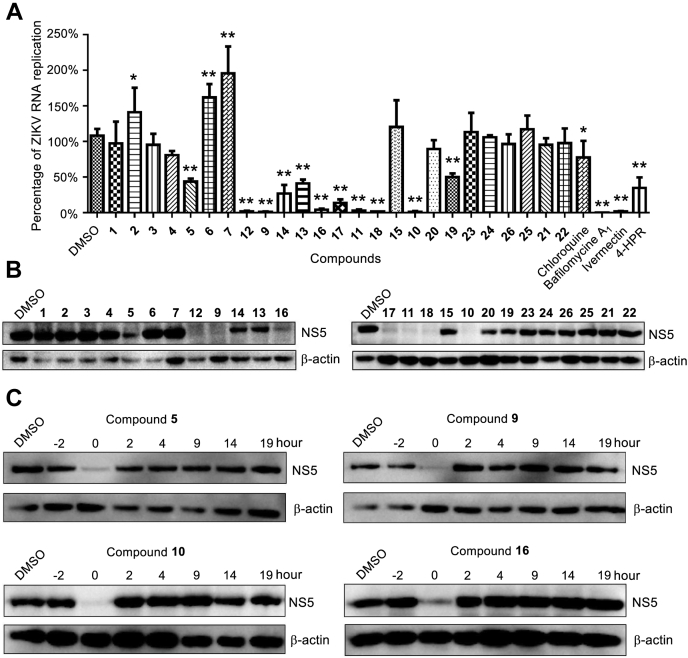
DTXs have been reported to suppress the secretion of hepatitis B virus surface antigen (HBsAg) in human hepatoma Hep3B cells (7). To investigate whether cyclohexadepsipeptides are active to inhibit RNA virus, we performed the anti-Zika virus (ZIKV) assays by testing their inhibition toward ZIKV total RNA replication and NS5 production levels in virus-infected A549 cells (25, 26). ZIKV is a member of enveloped positive-sense single-stranded RNA virus and is transmitted by mosquitos. Prior the bioassay, cyclohexadepsipeptides 1–26 were tested by the MTT method for their cytotoxic effects against the virus-uninfected A549 cells. All tested cyclohexadepsipeptides showed low cytotoxicity with the CC50 value higher than 100 μM. To test the anti-ZIKV activities of these low cytotoxic compounds, A549 cells were treated with each compound along with ZIKV (MR766) infection, and the ZIKV total RNA replication levels were determined by qRT-PCR at 24 h after infection, using ivermectin as the positive control and DMSO as the negative control. The qRT-PCR results showed that seven compounds (9–12 and 16–18) at 10 μM significantly inhibit the ZIKV total RNA replication (Fig. 3A). Furthermore, western blotting analysis of NS5 in the lysed A549 cells after infection for 24 h showed that these antiviral compounds also inhibit NS5 protein production effectively (Fig. 3B). It is noteworthy that the active congeners are mainly raised from DTX-type analogues, whereas ISDs and ISRs showed no or weak antiviral effects. In the tested DTXs, those bearing a halogen (Br or Cl) substitution such as 11, 17, and 18, or those containing an α-HIC or a 2-hydroxy-4-pentenoic acid moiety such as 9, 10, and 12 showed more potent activities than those (1–5 and 14–15) containing hydroxy or carboxylic groups on the α-HIC moieties.
Figure 3.
Anti-ZIKV activities of cyclohexadepsipeptides.A, the inhibitory activities of cyclohexadepsipeptides against ZIKV RNA replication in ZIKV-infected A549 cells. The ZIKV RNA replication levels in ZIKV-infected A549 cells treated with each compound were shown as the percentage of that in ZIKV-infected A549 cells treated with DMSO. The error bars were generated from three independent experiments for each compound, and the statistical significances were shown with ∗p < 0.05 and ∗∗p < 0.01. B, the inhibitory activities of cyclohexadepsipeptides against ZIKV NS5 production detected by western blotting, with β-actin as the negative control. C, the inhibition of NS5 production in ZIKV-infected A549 cells treated with 5, 9, 10, and 16 at different time points (not time periods), detected by western blotting.
We further selected 9, 10, and 16 to carry out a time-course detection for their inhibitory activities against NS5 production by treating the ZIKV-infected A549 cells with compounds at different time points, i.e. −2 to 0 (compound added 2 h before ZIKV infection), 0 to 2, 2 to 4, 4 to 9, 9 to 14, 14 to 19, and 19 to 24 h, respectively. Compound 5, which gave slightly less inhibition than 9, 10, and 16 in above assays, was also selected to test whether it can give different result. The NS5 production was analyzed after 24 h ZIKV infection by western blotting, showing that NS5 productions in all assays were only inhibited when compounds were added together with ZIKV infection (0–2 h), suggesting that the antiviral effects occur at early stage (virus entry or fusion) (Fig. 3C). Zika virus enters cells via clathrin-mediated pH-dependent endocytosis, while inhibition of endosome acidification can hamper the infection. To detect whether the antiviral compounds affect the endosome acidification, Lysotracker Red was used to probe acidic compartments and track acidic organelles in the cell (27). Compounds 5, 9, 10, and 16 markedly decreased cell-associated Lysotracker Red fluorescence (Fig. S17), indicating that the endosome acidification was restrained. These results allow to speculate that the antiviral compounds may prevent viral endosomal fusion at early stage of virus fusion.
Identification of the biosynthetic gene clusters of DTXs, ISDs, and ISRs
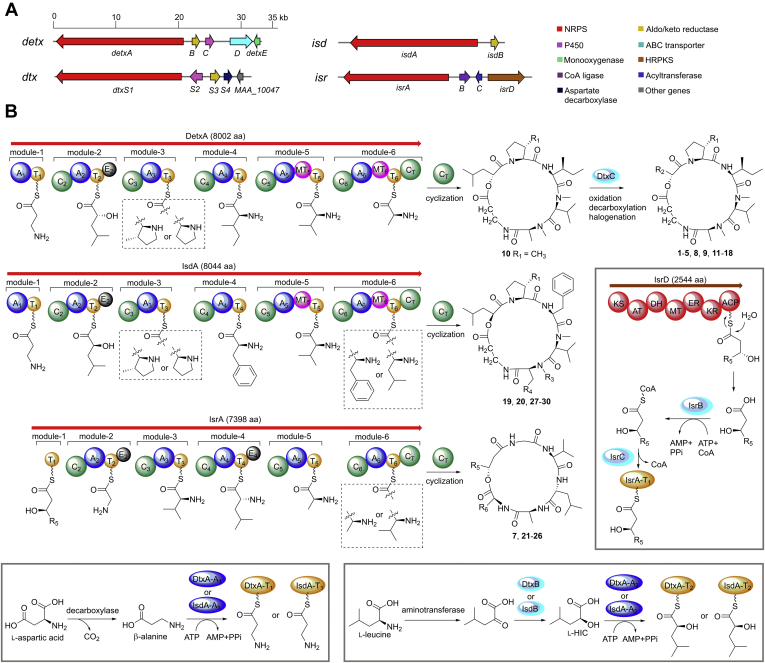
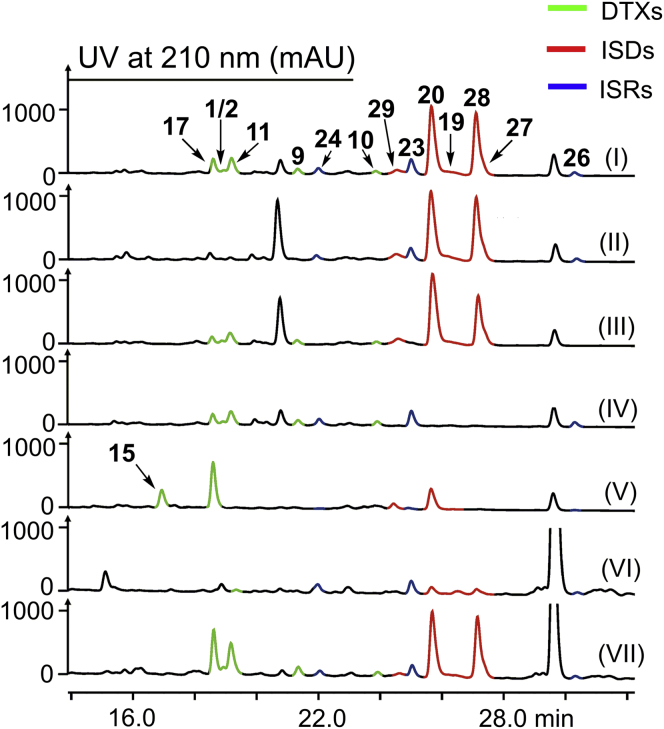
To investigate the biosynthesis of DTXs, ISDs, and ISRs, we sequenced the genomic DNA of B. felina SX-6-22. The bioinformatics analysis of the genome revealed 23 NRPS-encoding genes (Table S8), of which four genes (NRPS3, NRPS10, NRPS11, and NRPS14) encode NRPSs with six modules corresponding to the six amino acid residues in the cyclohexadepsipeptides. The amino acid sequence of NRPS3 shows high homology (70% identity) to that of NRPS11, suggesting their potential responsibilities for the biosynthesis of structurally similar DTXs and ISDs. A polyketide synthase (PKS)-encoding gene was located at the upstream of NRPS10 instead of NRPS14, suggesting that NRPS10 correlates to the biosynthesis of ISRs that contain polyketide chains. Thus, NRPS3 (designated detxA), NRPS11 (designated isdA), and NRPS10 (designated isrA) were proposed to encode the NRPSs for the biosynthesis of DTXs, ISDs, and ISRs, respectively (Fig. 4). To confirm the biogenetic functions of DetxA, IsdA, and IsrA, we carried out the gene deletion experiments using a CRISPR/Cas9-mediated homologous recombination system (Fig. S7). The regions encoding the peptidyl carrier protein (PCP or T domain) of module-3 and condensation domain (C domain) of module-4 within detxA, isdA, and isrA were deleted, respectively, generating detxA-inactivated mutant MM10024, isdA-inactivated mutant MM10025, and isrA-inactivated mutant MM10026. HPLC and total ion chromatography (TIC) of LC-MS analyses showed that the wild-type B. felina SX-6-22 produced all homologues including 1–30 (Fig. 5, lane I, Fig. S8). It is noted that some congeners from the wide-type strain are undetectable in HPLC due to the low yields, but TIC spectrum is highly sensitive to check all minor congeners. In the mutant MM10024, the productions of all DTXs including 1–5 and 8–18 were abolished (Fig. 5, lane II, Fig. S8), but this mutant still produced ISDs and ISRs. In the mutant MM10025, the productions of all ISDs including 6, 19, 20, and 27–30 (Fig. 5, lane IV, Fig. S8) were abolished, while the productions of all ISRs involving 7 and 21–26 were abolished in MM10026 (Fig. 5, lane III, Fig. S8). Thus, the roles of DetxA, IsdA, and IsrA to undertake the biosynthesis of DTXs, ISDs, and ISRs were unambiguously confirmed, respectively. By analyses of the accessory genes surrounding detxA, isdA, and isrA (Table S9), the gene clusters detx, isd, and isr for the biosynthesis of DTXs, ISDs, and ISRs, respectively, were postulated (Fig. 4B).
Figure 4.
Biosynthetic gene clusters and pathways of DTXs, ISDs, and ISRs from B. felina SX-6-22.A, biosynthetic gene clusters detx, isd, and isr compared to dtx from Metarhizium robertsii. B, proposed biosynthetic pathways of DTXs, ISDs, and ISRs. The “R5” is equal to “R” in Figure 1 for 7, 21–26.
Figure 5.
HPLC analysis of DTXs, ISDs, and ISRs from wild-type B. felina SX-6-22 and mutants. Lane I, wild-type B. felina SX-6-22; lane II, ΔdetxA mutant MM10024; lane III, ΔisrA mutant MM10026; lane IV, ΔisdA mutant MM10025; lane V, ΔdetxE mutant MM10027; lane VI, ΔSX-p5cr1 mutant MM10028; lane VII, ΔSX-p5cr2 mutant MM10029.
Comparative analysis of the detx, isd, and isrgene clusters
Although the biosynthetic gene cluster dtx of DTXs from M. robertsii ARSEF 23 has been reported (14), the biosynthetic gene clusters of ISDs and ISRs still remain unknown. The NRPS-encoding detxA, aldo-keto reductase-encoding detxB, P450-encoding detxC in the cluster detx show high homologies to dtxS1, dtxS3, and dtxS2 in cluster dtx, respectively (Fig. 4A). In comparison with DtxS1, DetxA possesses the same six-module NRPS assembly line with similar domain organization. The adenylation domains (A domains) A1–A6 in DetxA select β-alanine, α-HIC, l-proline/(3S)-methyl-l-proline, l-isoleucine, l-valine, and l-alanine, respectively (Fig. 4B). However, the predicted substrates of A1–A6 by NRPSsp are l-proline, l-tryptophan, l-proline, l-proline, l-leucine, and l-leucine, respectively (Table S10). This discrepancy is also observed previously as that the substrate prediction for fungal A domains is not as accurate as that for their bacterial counterparts (28). The presence of an epimerization domain (E domain) in module-2 and methyltransferase domains (MT domains) in both module-5 and -6 is well consistent with the modified α-d-HIC, N-methylvaline, and N-methylalanine residues in 1–5 and 8–18. The presence of (3S)-methyl-l-proline residues in 1–5/8–11 and corresponding l-proline residues in 12–18 may be attributed to the substrate promiscuity of A3 domain in module-3 (Fig. 4B). No homologue of dtxS4, which encodes an aspartate decarboxylase in the biosynthesis of the β-alanine that is selected by A1 domain in module-1, is found in detx cluster (Fig. 4A). The biosynthesis of HIC, which is the substrate of A2 domain in module-2, is proposed to be catalyzed by an aminotransferase and an aldo-keto reductase (DetxB/DtxS3) using l-leucine as a substrate (29). However, no aminotransferase-encoding gene is present in both detx and dtx gene clusters (Fig. 4A). The aminotransferase and aspartate decarboxylase are the enzymes commonly found in amino acid metabolism (30, 31), and these enzymes are proposed to participate in the generation of α-HIC and β-alanine residues in 1–5 and 8–18. Similar to the function of P450 DtxS2 in the biosynthesis of DTXs from M. robertsii ARSEF 23 (14), the P450 DetxC is proposed for the successive oxidation of HIC moiety in the biosynthesis of 1–5, 8, 9, and 11–18 (Fig. 4B). The coexistence of epimers 1/2 and 5/8 suggests the randomly selective oxidation of the α-HIC methyl groups by DetxC. DetxC-catalyzed decarboxylation affords a terminal vinyl group (as in 9 and 12), which is further catalyzed by DetxC to generate the epoxide group followed by hydroxylation and halogenation (enzymatic or nonenzymatic) generating structurally diverse congeners (3, 4, 11, 15–18) (Fig. 4 and Fig. S9). The detailed catalysis involved in these modification steps needs further experimental characterization.
The isd cluster contains the NRPS-encoding isdA and aldo-keto reductase-encoding isdB, which are homologous to detxA and detxB in detx, respectively. IsdA has the same domain organization as DetxA (Fig. 4B), and the predicted substrates (A1-A6) of IsdA by NRPSsp are the same as the predicted substrates of A1–A6 in DetxA (Table S10). Based on the structures of ISDs and DTXs, the A1–A3 and A5 domains in IsdA are depicted to select the same substrates as those in DetxA; both A4 and A6 domains in IsdA select phenylalanine (or leucine for A6) instead of l-isoleucine and l-alanine that are selected by A4 and A6 domains in DetxA. Indeed, the substrate selectivity-conferring codes of A1–A3 and A5 domains in IsdA are almost the same to those of A1–A3 and A5 domains in DetxA, whereas the substrate selectivity-conferring codes of A4 and A6 domains in IsdA differ from those of A4 and A6 domains in DetxA by five and four residues, respectively (Table S10). The differences between the substrate selectivity-conferring codes of A4 and A6 domains in IsdA and DetxA can be the references for more accurate substrate prediction for fungal A domains in future. The presence of l-proline residues in 6, 19, 20, 29, and 30 instead of (3S)-methyl-l-proline residues in 27 and 28 may be attributed to the substrate promiscuity of A3 in module-3, and the presence of l-phenylalanine residues in 6, 20, 28, and 30 instead of l-leucine residues in 19, 27, and 29 at corresponding positions may be attributed to the substrate promiscuity of A6 in module-6 (Fig. 4B). The presence of d-leucine instead of d-HIC residue in 6 suggests the substrate promiscuity of A2 domain in module-2. The α-l-HIC residues in ISDs are not oxidized by DetxC, suggesting that the l configuration of HIC or the different phenylalanine residues in ISDs preclude any possible substrate recognized by DetxC.
The isr gene cluster contains NRPS-encoding isrA, CoA ligase-encoding isrB, acyltransferase-encoding isrC, and PKS-encoding isrD. IsrA possesses six modules but lacks an A domain in module-1 (Fig. 4B). Based on the structures of ISRs, module-1 selects a high saturated polyketide as the starter unit. We proposed that the PKS IsrD generates the polyketide residue, which is activated by IsrB and transferred by IsrC to PCP1 in module-1 (Fig. 4B). Similar combinations of PKS, CoA ligase, and acyltransferase have been reported for their synergistical catalysis to transfer polyketide moieties to NRPS assembly lines in the biosynthesis of a variety of fungal lipopeptides, such as emericellamide in Aspergillus nidulans and scopularide in Scopulariopsis brevicaulis (32, 33, 34). The different length of the polyketide moieties in ISRs suggests the substrate promiscuities of IsrB and IsrC. The presence of E4 domain in module-4 and the absence of MT domains in module-5 and -6 are consistent with the three adjacent d-leucine, l-alanine, and l-valine residues in ISRs (Fig. 4B). The substrate selectivity-conferring codes and the predicted substrates of A2–A6 in IsrA differ largely from those of A2–A6 in DetxA and IsdA (Table S10), consistent with the distinct amino acid–building blocks of ISRs from those of DTXs and ISDs. The replacement of the l-alanine residue in 7 and 22 by the l-valine residue in 21, 23 to 26 at corresponding positions may be attributed to the substrate promiscuity of A6 domain in module-6. After the comparative analysis of detx, isd, and isr, all candidate genes for the biosynthesis of 1–30 have been proposed with the exception of those for the generation of (3S)-methyl-l-proline.
The biosynthesis of (3S)-methyl-l-proline
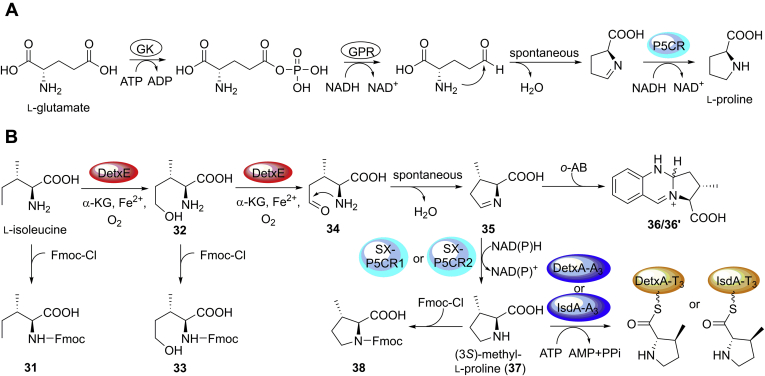
The generation of (3S)-methyl-l-proline in the biosynthesis of UCS1025A has been investigated (20). The α-ketoglutarate (KG)-dependent nonheme iron dioxygenase UcsF oxidizes l-isoleucine to (4S, 5S)-4-methyl-Δ1-pyrroline-5-carboxylate, which is depicted to be reduced by the reductase UcsG to generate (3S)-methyl-l-proline. However, the function of UcsG has not been characterized by gene inactivation or reconstitution in vitro. After analysis of detx and isd gene clusters and surrounding regions, we located an α-KG-dependent nonheme iron dioxygenase-encoding detxE that shows homology (identity of 42%) to UcsF in detx cluster, but no homologues of UcsG in the two clusters and surrounding regions were present (Table S9). Thus, we chose to explore whether detxE is involved in the biosynthesis of (3S)-methyl-l-proline residues in 1–5, 8–11, 27, and 28.
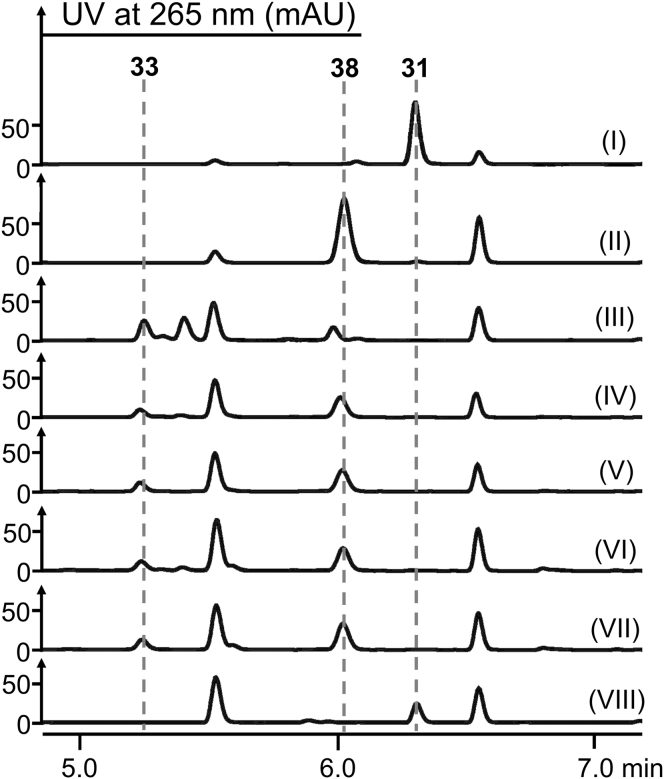
To explore the function of detxE, the detxE-deletion mutant MM10027 was generated by the CRISPR/Cas9-mediated homologous recombination system (Fig. S7). HPLC and LC-MS analyses showed that MM10027 abolished the production of the (3S)-methyl-l-proline-containing congeners including 1–5, 8–11, 27, and 28 but still produced 6, 12–18, 19, 20, 29, and 30 that contain the l-proline residue (Fig. 5, lane V, Fig. S10), confirming the involvement of detxE in the biosynthesis of (3S)-methyl-l-proline residues in both DTXs and ISDs. We further purified DetxE from Escherichia coli and reconstituted the catalysis of DetxE in vitro using l-isoleucine as the substrate. The reaction system including DetxE, l-isoleucine, α-KG, (NH4)Fe(SO4)2, and ascorbate was constructed, and possible intermediates or products were analyzed by 9-fluorenylmethyl chloroformate (Fmoc-Cl) and o-aminobenzaldehyde (o-AB) derivatization (35). The reaction of DetxE with l-isoleucine produced 5-hydroxy-l-isoleucine (32) and (4S, 5S)-4-methyl-Δ1-pyrroline-5-carboxylate (35) based on the detection of their Fmoc-derivative (33, Fig. 6, lane III) and o-AB-derivatives (36/36ʹ, Fig. S12), respectively, but no Fmoc-derivative of (3S)-methyl-l-proline was detected, suggesting that a reductase is required in the reduction of (4S, 5S)-4-methyl-Δ1-pyrroline-5-carboxylate (35) to (3S)-methyl-l-proline (37). The reaction of DetxE with d-isoleucine under the same reaction condition produced no oxidation product, whereas the reaction of DetxE with l-allo-isoleucine produced minor amount of oxidation product (Figs. S13 and S14), confirming l-isoleucine as the biosynthetic origin of (3S)-methyl-l-proline. Based on above result, the biosynthesis of (3S)-methyl-l-proline requires a reductase, whose encoding gene is probably out of the detx and isd gene clusters, to complete the last reduction step. In the biosynthesis of (4R)-methyl-l-proline in griselimycin, the α-KG-dependent nonheme iron dioxygenase GriE oxidizes l-leucine to (4R)-5-hydroxy-l-leucine, followed by the dehydrogenase GriF-catalyzed oxidation to generate (2S, 4R)-4-methylglutamate-5-semialdehyde, which is spontaneously cyclized to (3R, 5S)-3-methyl-Δ1-pyrroline-5-carboxylate, and the F420-dependent oxidoreductase GriH reduced (3R, 5S)-3-methyl-Δ1-pyrroline-5-carboxylate to generate (4R)-methyl-l-proline (Fig. S15) (36). The last reduction step catalyzed by GriH can also be achieved by the Δ1-pyrroline-5-carboxylate reductase (P5CR) ProC, which catalyzes the reduction step in l-proline biosynthesis in E. coli. In the most common biosynthetic pathway of l-proline in fungi and bacteria, l-glutamate is initially phosphorylated to γ-glutamyl phosphate by γ-glutamyl kinase (GK), followed by reduction to generate γ-glutamyl semialdehyde by γ-glutamyl phosphate reductase (GPR) and spontaneous cyclization to Δ1-pyrroline-5-carboxylate, and subsequent reduction to produce l-proline by P5CR (37, 38) (Fig. 7A). Given the structural similarities between the intermediates (4S, 5S)-4-methyl-Δ1-pyrroline-5-carboxylate (35) and Δ1-pyrroline-5-carboxylate, we propose that the P5CRs in l-proline biosynthesis catalyze the reduction step from (4S, 5S)-4-methyl-Δ1-pyrroline-5-carboxylate (35) to (3S)-methyl-l-proline (37). Only two P5CR-encoding genes, SX-p5cr1 and SX-p5cr2, were located by homologue searching of ProC in the genome of B. felina SX-6-22, and the SX-p5cr1-deletion mutant MM10028 and SX-p5cr2-deletion mutant MM10029 were constructed with the CRISPR/Cas9-mediated homologous recombination system. HPLC analysis showed that the productions of DTXs and ISDs, not ISRs, were significantly decreased in MM10028 compared with the wild-type B. felina SX-6-22 (Fig. 5, lane VI), and MM10029 showed the similar chemotype as the wild-type B. felina SX-6-22 (Fig. 5, lane VII). These results suggest that SX-p5cr1 instead of SX-p5cr2 is deeply involved in both l-proline and (3S)-methyl-l-proline biosynthesis in B. felina SX-6-22. The minor productions of DTXs and ISDs in MM10028 may be attributed to the complementation of other l-proline biosynthetic pathways (37, 38) or the catalysis of SX-P5CR2.
Figure 6.
HPLC analysis of enzymatic reactions in vitro. Lane I, Fmoc-Cl derivatization of l-isoleucine; lane II, Fmoc-Cl derivatization of (3S)-methyl-l-proline; lane III, Fmoc-Cl derivatization after the reaction catalyzed by DetxE with l-isoleucine; lane IV, Fmoc-Cl derivatization after the reaction catalyzed by DetxE and SX-P5CR1 with l-isoleucine and NADH; lane V, Fmoc-Cl derivatization after the reaction catalyzed by DetxE and SX-P5CR1 with l-isoleucine and NADPH; lane VI, Fmoc-Cl derivatization after the reaction catalyzed by DetxE and SX-P5CR2 with l-isoleucine and NADH; lane VII, Fmoc-Cl derivatization after the reaction catalyzed by DetxE and SX-P5CR2 with l-isoleucine and NADPH; lane VIII, Fmoc-Cl derivatization after the reaction catalyzed by boiled DetxE, SX-P5CR1 with l-isoleucine and NADH.
Figure 7.
Biosynthesis of (3S)-methyl-l-proline (37) in DTXs and ISDs.A, biosynthesis of l-proline by γ-glutamyl kinase (GK), γ-glutamyl phosphate reductase (GPR), and Δ1-pyrroline-5-carboxylate reductase (P5CR) using l-glutamate as the precursor. B, biosynthesis of (3S)-methyl-l-proline (37) by DetxE and SX-P5CR1 or SX-P5CR2 using l-isoleucine as the precursor. Fmoc-derivatives 31, 33, and 38 and o-AB-derivatives 36/36' (a pair of stereoisomers) are also shown.
To further confirm the involvement of P5CR in (3S)-methyl-l-proline biosynthesis, we purified SX-P5CR1 and SX-P5CR2 from E.coli and carried out the enzymatic reactions in vitro. When SX-P5CR1 was added into the reaction system including DetxE, l-isoleucine, α-KG, (NH4)Fe(SO4)2, ascorbate, and NADH or NADPH, (3S)-methyl-l-proline (37) was produced as evidenced by the detection of its Fmoc-derivative 38 (Fig. 6, lanes IV and V) in addition to the production of 5-hydroxy-l-isoleucine (Fmoc-derivative 33). Unexpectedly, the reaction with SX-P5CR2 that replaces SX-P5CR1 in the reaction system gave similar results (Fig. 6, lanes VI and VII, Fig. S16). Whether SX-p5cr2 is transcribed in a low level in B. felina SX-6-22 remains future exploration. No (3S)-methyl-l-proline and 5-hydroxy-l-isoleucine were produced when boiled DetxE was used in the reaction (Fig. 6, lane VIII). Thus, P5CRs in l-proline biosynthesis are demonstrated to catalyze the last reduction step in the biosynthesis of (3S)-methyl-l-proline in DTXs and ISDs (Fig. 7B), which is distinct from the biosynthesis of (4R)-methyl-l-proline (36) and proposed biosynthesis of (3S)-methyl-l-proline in UCS1025A (20). This is the first time to clarify the full biosynthesis of (3S)-methyl-l-proline by experimental characterization.
Discussion
Fungal strains have been shown as rich sources of DTXs, ISDs, and ISRs. DTXs are widely distributed in the fungal species of Metarhizium, Trichothecium, Aschersonia, and Beauveria genera (39, 40, 41), whereas ISDs and ISRs have been reported exclusively from B. felina (3, 42). B. felina AcSS8 is the only fungal strain reported to produce all the three types of cyclohexadepsipeptides, in which four DTXs, two ISDs, and two ISRs were characterized (15). In this study, the marine sponge-derived fungus B. felina SX-6-22 exhibits potent capability to produce cyclohexadepsipeptides with a large chemical diversity, of which a total of 16 DTXs (1–5, 8–18), seven ISDs (6, 19, 20, 27–30), and seven ISRs (7, 21–36) including seven new compounds (1–7) are characterized. A number of diastereoisomers (1/2, 3/4, 5/8, and 13/15) are purified by the chromatographic approach, suggesting that the oxidation on the α-HIC moiety of DTXs is stereochemically unspecific. The crystal structures of DTXs (1, 3, 5, and 17) and ISD (19) clarify the absolute configurations in these peptides with multiple stereogenic centers. The observation of a hydrogen bond between the carbonyl oxygen of l-isoleucine residue (or l-phenylalanine residue at the same position in 19) and the amide hydrogen of β-alanine residue in the crystal structures (Fig. 2) suggests a specifically conserved conformation of peptidyl skeleton in both DTXs and ISDs. Whether this conserved peptidyl skeleton conformation is related to the biological activities of DTXs and ISDs is worthy of further investigation. This work also extends the antiviral activities of DTXs to anti-ZIKV activities for the first time. Compounds 9–12 and 16–18 show the potent inhibitory effects against ZIKV total RNA level in the ZIKV (MR766 strain)-infected A549 cells with higher efficacy than the positive control ivermectin. These compounds also downregulate the production of NS5 that is one phenotype of blocking viral replication, and inhibit the virus entry or fusion by regulation of the endosome acidification.
Genome sequencing and gene deletion in association with the chemotype analysis reveal the NRPS-encoding gene clusters detx, isd, and isr responsible for the biosynthesis of DTXs, ISDs, and ISRs, respectively. These findings clarify that DTXs and ISDs are produced from two distinct NRPSs (DetxA and IsdA) instead of one NRPS with promiscuous A domains. The biosynthetic gene clusters (isd and isr) of ISDs and ISRs are reported in this study for the first time, although the first ISD and ISR were discovered in 2001 (11) and 1962 (43), respectively. DetxA, IsdA, and IsrA are all composed of six modules with similar domain organization, but unusual issues are raised in their domain catalysis. The A1, A2, A4–A6 in DetxA, A1, A5, and A6 in IsdA, A2–A5 in IsrA show strict substrate selectivity, whereas A3 (selects (3S)-methyl-l-proline or l-proline) in DetxA, A2 (selects α-HIC or l-leucine), A3 (selects (3S)-methyl-l-proline or l-proline), and A4 (selects l-phenylalanine or l-leucine) in IsdA, A6 (selects l-valine or l-alanine) in IsrA show substrate promiscuities. The C2 in DetxA and IsdA catalyzes the esterification rather than a typical amide bond formation between upstream β-alanine and downstream α-HIC; C2 in IsrA accepts upstream reduced polyketide substrates for the condensation; C3 in DetxA only accepts upstream α-HIC with d-configuration but C3 in IsdA only accepts upstream α-HIC with l-configuration; and CT domains in all three NRPSs accept different linear peptide intermediates to catalyze intramolecular cyclization and release the NRPS nascent products. Thus, DetxA, IsdA, and IsrA in this study provide excellent templates for comparative analyses regarding the catalytic mechanisms of NRPS domains, and for biosynthetic pathway engineering by module- or domain-swapping to rationally generate building block-hybrid cyclohexadepsipeptides. The nonproteinogenic (3S)-methyl-l-proline residue is present in many different natural products (Fig. S1), but the full biosynthesis of this unusual building block has not been characterized experimentally in previous research. In this study, we characterize the biosynthetic steps of (3S)-methyl-l-proline by gene deletion and reconstitution of enzymatic catalysis in vitro, revealing that P5CRs in l-proline biosynthesis catalyze the last reduction step using l-isoleucine as the precursor. This result suggests potential cross talk between primary and secondary metabolisms not only by sharing same biosynthetic building blocks but also by utilizing same biosynthetic machineries.
In summary, the discovery of 30 cyclohexadepsipeptides in B. felina SX-6-22, anti-ZIKV activity assays, and the biosynthetic investigation in this study enrich the structural diversity and biological activity of DTXs, and offer great opportunities for biosynthetic pathway engineering to generate new anti-ZIKV agents in future.
Experimental procedures
General procedures
Optical rotations were measured on an Autopol III automatic polarimeter (Rudolph Research Analytical). UV (ultraviolet) data were obtained from a NanoDrop One spectrophotometer (Thermo Scientific). IR (infrared) spectra were recorded by a Thermo Nicolet Nexus 470-FT-IR spectrometer (Thermo Scientific). NMR (nuclear magnetic resonance) spectra were recorded on Bruker Ultrashield Plus 400 MHz and 600 MHz spectrometers (Bruker Corporation) with TMS as an internal standard. HRESIMS (high-resolution electrospray ionization mass) data were measured on a Xevo G2 Q-TOF/YCA spectrometer (Waters). LC-MS was performed on a Thermo Q Exactive Orbitrap instrument (Thermo Fisher Scientific) with a C18 column (ACQUITY UPLC HSS T3 C18 1.8 μm, 2.1 × 100 mm). ECD (electronic circular dichroism) spectra were measured using a JASCO J-810/J-815 spectropolarimeter (Welltech Inc Company, CHN). The X-ray diffraction data were collected on a XtaLAB Synergy diffractometer system (1.54056 Å). UPLC (ultra-performance liquid chromatography) was performed on a Waters Acquity UPLC (Waters) with a C18 column (ACQUITY UPLC BEH C18 1.7 μm, 2.1 × 100 mm), and HPLC analysis was performed on a Waters 2695 with a C18 column (Hypersil BDS C18 5 μm, 4.6 × 250 mm). Semipreparative HPLC was performed on a Lab Alliance series III HPLC (Lab Alliance) with a semipreparative C18 column (Alltech Chrom, octadecylsilyl, 10 μm, 10 × 250 mm). Column chromatography was performed with solid phases including silica gel (200–300 mesh) or ODS (50 μm; YMC). B. felina strains were cultured in an SPX-150B-Z biochemical incubator (Boxun Industry and Commerce Co, Ltd), and E. coli strains were fermented in HYG-C shakers (Peiying Laboratory Equipment Co, Ltd). PCR and primer annealing were performed in a Veriti Certified Refurbished Thermal Cycler (Applied Biosystems).
Strains and culture conditions
Fungus B. felina SX-6-22 was isolated from the marine sponge Xestospongia testudinaria collected in the South China Sea by tissue inoculation and identified by analysis of the ITS (internal transcribed spacer) region of its rDNA. The ITS sequence data derived from the fungus are similar (99%) to the sequence of B. felina CBS 217.37 (accession No. MH855893). The fungus is preserved at the Marine Natural Products Research Laboratory of the Peking University.
Chromatographic analysis of secondary metabolites in B. felina SX-6-22
Sterile water (1 ml) was added to fresh mycelia (1 g) for homogenization, and the homogenate was diluted by 100 times for inoculation (50 μl/plate). The small-scale fermentation of B. felina SX-6-22 was carried out in PDA medium (potato dextrose agar, Becton/Dickinson and Company) by adding 3.3% sea salt at 25 °C for 15 days. After fermentation, mycelia and medium were extracted with appropriate amount of EtOAc for three times by ultrasound. Part of the wild-type fungal extract was dissolved in 70% MeOH for the HPLC and UPLC detection, and the other part was dissolved in DMSO-d6 for 1H NMR measurement. The UPLC analysis was performed using an elution program of 10% CH3CN in H2O to 100% CH3CN over 10 min, with a flowrate of 0.4 ml/min under the detection at 210 nm. The HPLC analysis was performed using an elution program of 5% CH3CN in H2O to 100% CH3CN over 60 min, with a flowrate of 1 ml/min under the detection at 210 nm.
Chemotype detection by molecular networking
Molecular networks were generated with GNPS from the targeted high-resolution CID spectra of all metabolites produced by the fungus detected in this study. MS/MS spectra were extracted by MZmine 2 using the following parameters: mass detection noise level 10; ADAP chromatogram builder group intensity threshold 1E3, min highest intensity 2E3, m/z tolerance 0.005 m/z or 20 ppm; chromatogram deconvolution wavelets, S/N threshold 3, min feature height 1000, peak duration range 0.05to 0.7, RT wavelet range 0.01 to 0.7, m/z range for MS/MS scan paring 0.05, RT range for MS/MS scan paring 0.2. Molecular networks parameters (GNPS) were set as follows: precursor ion mass tolerance 0.2 Da; fragment ion mass tolerance 0.02 Da; min pairs cosine score 0.4; minimum matched fragment ions 4; minimum peak intensity 1000. The molecular network resulting from GNPS was visualized in Cytoscape 2.8.2. The chemical assay of crude extract using GNPS database and in-house data enable to recognize a number of nodes within the three peptide clusters (Fig. S4), indicating that the crude extract contained three types of peptides. The MS2 of some nodes in the three clusters was the same to that of the reported compounds, leading to the identification of three peptide clusters corresponding to destruxins (marked in green), isaridins (marked in red), and isariins (marked in blue) (Fig. S5, A–C). These data promoted subsequent isolation of cyclodepsipeptides, and four inseparable compounds (27–30) were identified by MS2 (Fig. S5, D–G).
Large-scale fermentation and isolation
The large-scale fermentation was carried out in 100 flasks (500 ml), each containing rice (80 g/flask) and artificial seawater (50 ml/flask) (33 g sea salt dissolved in 1 l distilled H2O), which were sterilized before fungal inoculation. After cooling to room temperature, a small piece of PDA medium containing fungus was added to each flask and the fermentation was continued at 25 °C for 35 days. The fermented rice medium was exhaustively extracted with EtOAc. After filtration, extracts (40.5 g) were obtained under vacuum concentration and subjected to a silica-gel vacuum liquid chromatography (VLC) eluting with a solvent gradient (CH2Cl2-MeOH 50:1–1:1) to obtain ten fractions (A1–A10). According to the 1H NMR and HPLC MS/MS detection, fractions A9 and A10 mainly contain peptide components. A9 (3.1 g) was separated upon ODS column eluting with MeOH-H2O gradient (from 20% to 100%) to yield seven subfractions (B1–B7). B3 (1.1 g) was separated by ODS column eluting with MeOH-H2O (from 20% to 100%) to afford 12 (68.4 mg), 11 (186.0 mg), and the subfractions C1 to C6. C4 (190 mg) was separated by semipreparative HPLC (51% MeOH-H2O) to get 14 (168.9 mg), 13 (4.8 mg), 16 (4.7 mg). C5 (380 mg) was further purified by semipreparative HPLC (35% MeCN-H2O) to yield 17 (369.1 mg). C6 (65 mg) was separated by semipreparative HPLC (56% MeOH-H2O) to afford 1 (43.0 mg), 18 (3.6 mg), 5 (1.8 mg), 2 (3.8 mg), and 8 (2.1 mg). Compounds 20 (42.1 mg), 19 (3.9 mg), and 6 (3.9 mg) were isolated from B6 (58 mg) by the same protocol as for C6 using 73% MeCN-H2O as mobile phase. A silica gel flash column separation of B4 (12 mg) eluting with CH2Cl2-MeOH (30:1) to yield 9 (2.0 mg) and 10 (2.4 mg). A10 (235.5 mg) was separated by ODS eluting with MeOH/H2O (from 20% to 100%) and further purified by the semipreparative HPLC to obtain 23 (21.6 mg), 24 (10.4 mg), 21 (38.5 mg), 3 (10.6 mg), 4 (5.2 mg), 15 (10.2 mg), 7 (27.4 mg), 25 (8.9 mg), 26 (2.9 mg), and 22 (3.1 mg). All semipreparative HPLC purifications were carried out with the flowrate of 2 ml/min under the UV detection at 210 nm.
Marfey’s analysis
Compound 6 (0.5 mg) was hydrolyzed by heating in 6 N HCl (1 ml) at 110 °C for 24 h. After cooling to room temperature, the solvent was removed in vacuo. The residue was dissolved into the solution of 0.1 ml of H2O, 0.1 ml of 1 M NaHCO3, and 0.2 ml of 1% FDAA in acetone, and the solution was heated at 45 °C for 1 h. Then, 10 μl of 1 N HCl was added to quench the reaction. FDAA derivatives of standard l- and d-amino acids were prepared using the same procedures above. Each of the reaction mixtures was analyzed by UPLC at 340 nm. Analysis conditions: Acquity UPLC BEH C18 column (2.1 × 50 mm, 1.7 μM, 0.4 ml/min) with a linear gradient of MeCN in H2O containing 0.05% trifluoroacetic acid (0–10 min, 10:90–20:80; 10–15 min, 20:80–50:50; 15–17 min, 50:50–100:0). The retention times (Rt) of FDAA derivatives of standard amino acids were as follows: l-Pro (13.48 min), d-Pro (13.75 min), N-Me-l-Val (15.13 min), N-Me-d-Val (15.54 min), N-Me-l-Phe (15.40 min), N-Me-d-Phe (15.86 min), l-Phe (15.30 min), d-Phe (15.92 min), l-Leu (15.33 min), d-Leu (16.08 min), while the retention times of the amino acid FDAA derivatives of 6 were 13.48, 15.13, 15.40, 15.30, and 16.08 min, corresponding to l-Pro, N-Me- l-Val, N-Me-l-Phe, l-Phe, and d-Leu, respectively.
Crystallographic data
Crystallographic data for 1: C31H53N5O10, fw = 655.78, orthorhombic, space group P2(1)2(1)2(1), unit cell dimensions a = 11.6069(3) Å, b = 15.7016(4) Å, c = 19.6550(5) Å, α = β = γ = 90°, V = 3582.06(16) Å3, Z = 4, ρcalcg = 1216 cm3, μ = 0.751 mm−1, F (000) = 1416.0, crystal size 0.68 × 0.55 × 0.5 mm3. The 18,714 measurements yield 6579 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yield R1 = 0.0834, wR2 = 0.2318 [I ≥ 2σ (I)]. The Flack parameter was −0.07(10) in the final refinement for all 6579 reflections with 439 Friedel pairs. Crystallographic data have been deposited in the Cambridge Crystallographic Data Center with the deposition number CCDC 2031969. Crystallographic data for 3: C30H51N5O9, fw = 625.75, orthorhombic, space group P2(1)2(1)2(1), unit cell dimensions a = 8.86720(10) Å, b = 17.6481(3) Å, c = 20.5603(3) Å, α = β = γ = 90°, V = 3217.47(8) Å3, Z = 4, ρcalcg = 1292 cm3, μ = 0.787 mm−1, F (000) = 1352.0, crystal size 0.08 × 0.05 × 0.02 mm3. The 14,998 measurements yield 6012 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yield R1 = 0.0332, wR2 = 0.0772 [I ≥ 2σ (I)]. The Flack parameter was −0.00(9) in the final refinement for all 6012 reflections with 421 Friedel pairs. Crystallographic data have been deposited in the Cambridge Crystallographic Data Center with the deposition number CCDC 2031038. Crystallographic data for 5: C31H53N5O8, fw = 623.78, monoclinic, space group P2(1), unit cell dimensions a = 9.68172(18) Å, b = 11.2950(5) Å, c = 15.7305(3) Å, α = γ = 90°, β = 93.6814(17), V = 1716.67(8) Å3, Z = 2, ρcalcg = 1207 cm3, μ = 0.713 mm−1, F (000) = 676.0, crystal size 0.29 × 0.29 × 0.05 mm3. The 21,435 measurements yield 5886 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yield R1 = 0.0835, wR2 = 0.2310 [I ≥ 2σ (I)]. The Flack parameter was −0.08(16) in the final refinement for all 5886 reflections with 407 Friedel pairs. Crystallographic data have been deposited in the Cambridge Crystallographic Data Center with the deposition number CCDC 2031968. Crystallographic data for 17: C29H52ClN5O10, fw = 710.66, monoclinic, space group P2(1), unit cell dimensions a = 9.78542(7) Å, b = 10.92110(8) Å, c = 15.77201(10) Å, α = γ = 90°, β = 90.7265(6), V = 1685.38(2) Å3, Z = 2, ρcalcg = 1400 cm3, μ = 2.177 mm−1, F (000) = 752.0, crystal size 0.71 × 0.36 × 0.29 mm3. The 18,468 measurements yield 6094 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yield R1 = 0.0375, wR2 = 0.1069 [I ≥ 2σ (I)]. The Flack parameter was −0.039(8) in the final refinement for all 6094 reflections with 430 Friedel pairs. Crystallographic data have been deposited in the Cambridge Crystallographic Data Center with the deposition number CCDC 2051154. Crystallographic data for 19: C36H59N5O9, fw = 705.88, orthorhombic, space group P2(1)2(1)2(1), unit cell dimensions a = 10.26330(10) Å, b = 15.7135(2) Å, c = 23.8764(2) Å, α = β = γ = 90°, V = 3850.60(7) Å3, Z = 4, ρcalcg = 1.218 cm3, μ = 0.714 mm−1, F (000) = 1528.0, crystal size 0.22 × 0.09 × 0.05 mm3. The 21,902 measurements yield 7410 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yield R1 = 0.0353, wR2 = 0.0860 [I ≥ 2σ (I)]. The Flack parameter was 0.05(8) in the final refinement for all 7410 reflections with 465 Friedel pairs. Crystallographic data have been deposited in the Cambridge Crystallographic Data Center with the deposition number CCDC 2049507.
Genome sequencing and bioinformatics analysis
The wide-type B. felina SX-6-22 was cultured in PDA medium by adding 3.3% sea salt at 25 °C for 15 days for genomic DNAs preparation. Genomic DNAs of B. felina were extracted by EasyPure Genomic DNA Kit (TransGen Biotech). Shotgun sequencing was performed at GENEWIZ using Illumina Hiseq 2000 platform. The contigs were assembled by SSPACE 3.0 to yield 78 scaffolds, which were used for gene prediction by prodigal v2.6.3 and gene cluster analysis by anti-SMASH (antibiotics and secondary metabolite analysis shell). The functions of the coding sequences were annotated by BLAST on NCBI website. The domain organization and annotation of all NRPSs were further analyzed by anti-SMASH, BLAST, and InterProScan.
Mutants construction
The process for gene knockout is the same as that reported previously (44). Initial plasmid pYBC-01a was digested by BsaI (NEB) to yield linearized fragment with sticky ends for specific sgRNA fragment insertion. The paired sg-F/R primers of target genes (Table S11, e.g., detxA_sg-F/R, p5cr1_sg-F/R) were slowly annealed and phosphorylated to form a 72 bp short fragment containing a specific sgRNA sequence for different target genes, which were ligated into the above linearized plasmid by T4 ligase (NEB) respectively to yield pMM1029-pMM1034. Donor DNA (dDNA) fused with sequences flanking the knockout region for homologous recombination was constructed by three rounds of PCR as previous reports (44, 45).
Fresh mycelia (2 g) of B. felina SX-6-22 were homogenized by adding 1 ml of sterile H2O and then transferred into 50 ml of PDB medium (potato dextrose broth, Becton/Dickinson and Company) to germinate at 25 °C for 36 h, 220 rpm. Germinal mycelia were collected and rinsed with osmotic medium (1.2 M MgSO4, 10 mM sodium phosphate, pH 5.8) for three times, then transferred into 10 ml of sterile osmotic medium with 2 mg/ml catalase (TAKARA) and 3 mg/ml lysing enzyme (Sigma-Aldrich). After digestion at 25 °C for 12 h, 90 rpm, 6 ml of trapping buffer (0.6 M sorbitol, 0.1 M Tris HCl, pH 7.0) was added in the lysate gently. The mixture was centrifuged at 4000 rpm for 15 min at 4 °C, and the protoplast fraction was obtained from the interface and washed with 6 ml of STC buffer (1.2 M sorbitol, 10 mM CaCl2, 10 mM Tris HCI, pH 7.5) for twice. Protoplasts (100 μl) were gently mixed with dDNA (2 μg) and constructed plasmid (1 μg) for incubation on ice for 50 min, followed by the addition of 1.25 ml of PEG 4000 solution (60% PEG 4000, 50 mM CaCl2, 50 mM Tris-HCI, pH 7.5) and incubated at room temperature for 20 min. The solution was mixed with 3.75 ml of STC buffer and plated on the regeneration selection medium (SMM, 1.2 M d-sorbitol, 200 μg/ml hygromycin B), and the transformants were obtained after incubation at 25 °C for 7 days.
To confirm the constructed mutants, the transformants were inoculated in PDB medium with stationary incubation for about 5 days. Mycelia were collected, lyophilized, and resuspended in LETS buffer (10 mM Tris HCI, pH 8.0, 20 mM EDTA, 0.5% SDS, 0.1 M LiCl) for cell lysis. The cell lysate was extracted twice with phenol/chloroform/isoamylol. Genomic DNAs were precipitated with ethanol and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The genotypes of all mutants were verified by PCR. The crude extracts of all mutants were analyzed by HPLC using the same method as that for wild-type strain. The LC-MS analysis was carried out on a Thermo Q Exactive Orbitrap instrument using an elution program of 2% MeOH in H2O (0.05% formic acid) to 100% MeOH over 11 min at a flow rate of 0.3 ml/min.
Gene clone and protein purification
Total RNA of B. felina SX-6-22 grown on PDA (add 3.3% sea salt) plates for 15 days was extracted by TransZol Up Plus RNA Kit (TransGen Biotech) following user's manual, and cDNA was synthesized by EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). The coding sequences of DetxE, SX-P5CR1, and SX-P5CR2 were amplified from the cDNA of B. felina SX-6-22 and cloned into pMCSG7, pMCSG19, and pMCSG19 plasmids to generate pMM1035, pMM1036, and pMM1037, respectively, by LIC (ligation independent cloning) following previous published protocol (46, 47). Plasmids pMM1035-pMM1037 were transformed into E. coli BL21 (DE3) for His6 or MBP (maltose binding protein)-His6-tagged protein overproduction.
The cells were cultured at 37 °C, 220 rpm in 1 l of LB medium supplemented with 50 μg/ml ampicillin untill an OD600 of 0.4 to 0.5 was reached. The culture was cooled down to 16 °C and gene expression was induced by the addition of 500 μl IPTG (1 M stock). After incubation for 16 h, the cells were collected and resuspended by 50 ml of lysis buffer (15 mM imidazole, 50 mM Tris, 300 mM NaCl, 10% glycerol, pH 8.0) for sonication. After centrifugation at 14,000 rpm for 45 min, the supernatant was loaded onto a Ni-NTA agarose column (GE Healthcare Life Sciences). The Ni-NTA agarose was incubated with the supernatant at 4 °C for 2 h. Then the column was eluted with 30 ml of buffer A (50 mM Tris, 300 mM NaCl, 20 mM imidazole, pH 8.0), 10 ml of buffer B (50 mM Tris, 300 mM NaCl, 30 mM imidazole, pH 8.0), and 10 ml of buffer C (50 mM Tris, 300 mM NaCl, 200 mM imidazole, pH 8.0). The eluent buffer C containing recombinant proteins was concentrated with with a Vivaspin 6 centrifugal concentrator (Sartorius Stedim Biotech) and exchanged into storage buffer (50 mM Tris-HCl, 20% glycerol, pH 8.0) by PD-10 column (GE Healthcare). The DetxE, SX-P5CR1, and SX-P5CR2 proteins were individually analyzed by SDS-PAGE and concentrations were determined at 280 nm by NanoDrop (Thermo Scientific).
Reconstitution of the catalysis of DetxE, SX-P5CR1, and SX-P5CR2 in vitro
For reconstitution of the catalysis of DetxE, a 100 μl reaction system containing 2 mM of l-isoleucine (or other amino acids), 4 mM α-KG, 2 mM ascorbate, 0.5 mM (NH4)2Fe(SO4)2, and 100 μM of purified DetxE in 100 mM HEPES (pH 7.0) was constructed. The reaction was carried out at 25 °C for 3 h and quenched with 100 μl of MeOH. After centrifugation, the supernatant was used directly for LC-MS analysis using the same procedure as that for mutants analysis. For ortho-aminobenzaldehyde (o-AB) derivatization after reaction, 1 mg of o-AB was added to the 100 μl reaction system without MeOH quenching, and the mixture was incubated at room temperature for 1 h. Afterward, the insoluble o-AB was removed by filtration, and the filtrate was subjected to UPLC analysis (same procedure as that for crude extract analysis of wild-type strain). For 9-fluorenylmethyl chloroformate (Fmoc-Cl) derivatization, the reaction was quenched with 100 μl of MeOH. After centrifugation, 50 μl of 200 mM sodium borate (pH 8.0) followed by 20 μl of 10 mM Fmoc-Cl were added to the supernatant, and the mixture was incubated at room temperature for 5 min. Then 20 μl of 100 mM 1-adamantanamine was immediately added to the mixture. After centrifugation, the supernatant was analyzed by UPLC (same procedure as that for crude extract analysis of wild-type strain) and LC-MS (same procedure as that for mutants analysis).
For reconstitution of the catalysis of DetxE combined with SX-P5CR1 or SX-P5CR2, the same 100 μl of reaction system was constructed with the exception of the addition of SX-P5CR1 or SX-P5CR2 (final concentration for each: 50 μM) and NADH or NADPH (final concentration for each: 2 mM). The reactions, Fmoc-Cl derivatization, UPLC, and LC-MS analysis were carried out with the same procedures as those for the catalysis of DetxE. The standards of l-isoleucine, d-isoleucine, and l-allo-isoleucine were purchased from Aladdin, while standard of (2S,3S)-3-methylproline was purchased from Acros.
Anti-ZIKV assays
A549 cells were obtained from America type culture collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin at 37 °C in a 5% CO2 incubator. ZIKV African-lineage MR766 strain was obtained from ATCC. ZIKV NS5-specific monoclonal antibody (8B8) was from BioFront Technologies. Human β-actin monoclonal antibody and horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody were obtained from Bioeasy Biotechnology. RNA isolation kit and DNase I were from Magen. TransScript Probe One-Step qRT-PCR SuperMix was from TransGen Biotech. RIPA lysis buffer and enhanced BCA protein assay kit were from Beyotime. Enhanced chemiluminescence (ECL) substrate for western blot was purchased from Bidragon Immunotechnologies. LysoTracker Red was from Beyotime. The 96-well Glass Bottom plate (P96-1.5H-N) was from Cellvis.
A549 cells were seeded in a 24-well plate at a density of 2 × 105 per well and cultured in DMEM containing 10% FBS overnight. After being washed with 1 × phosphate-buffered saline (PBS) twice, A549 cells were treated with ZIKV [MR766 strain, MOI (multiplicity of infection) = 1] and different compounds (final concentration: 10 μM) were added at different time points. At 24 h post infection, the total RNA was isolated and quantified by qRT-PCR with chloroquine (final concentration: 10 μM), bafilomycin A1 (final concentration: 100 nM), ivermectin (final concentration: 10 μM), and 4-hydroxyphenylretinamide (final concentration: 10 μM) as the positive control and DMSO as the negative control, and western blotting was carried out with primary NS5 antibody (8B8) and β-actin antibody using β-actin as the internal control.
For qRT-PCR, total RNA from cultured cells was extracted using Rapure Total RNA kit (Magen), according to manufacturer's instructions. qRT-PCR was performed using probe one-step qRT-PCR Kit (Transgene). Primers and probes are ZIKV-F: CCCTCAAGTATAGCTGCAAGAG, ZIKV-R: AGAGTCAGGAAACGCATCAC, ZIKV-probe:/56-FAM/TTATGACTGCCACACCACCAGGAA/3IABkFQ/; β-actin-F: GGAAATCGTG CGTGACATTAAG, β-actin-R: AGCTCGTAGCTCTTCTCCA, β-actin-probe:/56-FAM/CTGGA CTTCGAGCAAGAGATGGCC/3IABkFQ/. Three independent experiments were carried out to generate the means ± standard deviations (error bars were shown in Fig. 3) for each compound, and the statistical significances were analyzed by one-way ANOVA: ∗p < 0.05 and ∗∗p < 0.01.
For western blotting, a total of 20 μg protein for each sample was separated by electrophoresis in SDS-PAGE, followed by transfer to a polyvinylidene difluoride (PVDF) membrane. Immunoblot analysis was carried out using primary mouse monoclonal antibodies specific to NS5 (8B8), HRP-conjugated anti-mouse secondary antibody, and an enhanced chemiluminescence (ECL) kit (Bidragon).
For endosome acidification detection, A549 cells were grown to 60% confluency in 96-well Glass Bottom plate and compounds (final concentration: 10 μM) were added to the culture medium. After 2 h or 4 h, cells were switched to the medium containing LysoTracker Red (final concentration 50 nM) for 30 min. The medium was changed to LysoTracker-free medium, and cell-associated Lysotracker Red fluorescence was observed by confocal microscopy with chloroquine (final concentration: 50 μM) and bafilomycin A1 (final concentration: 100 nM) as the positive control.
Data availability
Data supporting the findings of this study are available within the article and its Supplementary Information files.
Supporting information
This article contains supporting information (16, 17, 18, 19, 20, 36, 36).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (81991525-5, 81630089, 21861142006, 81872793), COMRA (DY135-B-05), and MOST of China (2018ZX09711001-001-008, 2019YFC0312502).
Author contributions
B. Y., Z. W., W. J., and H. J. Formal analysis; B. Y., Z. W., and W. J. Investigation; B. Y., Z. W., W. J., X. G., A. F., and H. J. Methodology; B. Y. Writing-original draft; D. L., D. Y., and A. F. Validation; M.M. and W. L. Conceptualization; M. M. and W. L. Supervision; M. M. and W. L. Funding acquisition; M. M. and W. L. Project administration; M. M. and W. L. Writing-review and editing.
Biographies

Bochuan Yuan is a graduate student at Peking University. He studies natural product biosynthesis and has characterized the biosynthesis of nonproteinogenic building block (3S)-methyl-l-proline, which is present in a series of cyclohexadepsipeptides from marine fungus Beauveria felina SX-6-22. Currently, he is an assistant professor in the Department of Pharmaceutical Sciences at Beijing Institute of Radiation Medicine, focusing on the discovery of antiradiation drug leads and radiation biosensor research.

Ziwei Wu is a graduate student at Zhejiang Ocean University. She was engaged in a collaboration project with Professor Wenhan Lin’s research group at Peking University. She is trained in natural product discovery by chromatographic methods and structural elucidation by spectroscopic analyses. She has discovered a series of cyclohexadepsipeptides with anti-Zika virus activities from a marine-derived fungus Beauveria felina SX-6-22.
Edited by Joseph Jez
Contributor Information
Ming Ma, Email: mma@bjmu.edu.cn.
Wenhan Lin, Email: whlin@bjmu.edu.cn.
Supporting information
References
- 1.Sabareesh V., Ranganayaki R.S., Raghothama S., Bopanna M.P., Balaram H., Srinivasan M.C., Balaram P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007;70:715–729. doi: 10.1021/np060532e. [DOI] [PubMed] [Google Scholar]
- 2.Liu B.L., Tzeng Y.M. Development and applications of destruxins: A review. Biotechnol. Adv. 2012;30:1242–1254. doi: 10.1016/j.biotechadv.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Du F.Y., Zhang P., Li X.M., Li C.S., Cui C.M., Wang B.G. Cyclohexadepsipeptides of the isaridin class from the marine-derived fungus Beauveria felina EN-135. J. Nat. Prod. 2014;77:1164–1169. doi: 10.1021/np4011037. [DOI] [PubMed] [Google Scholar]
- 4.Cavelier F., Verducci J., Andre F., Haraux F., Sigalat C., Traris M., Vey A. Selective apoptotic cell death effects of oral cancer cells treated with destruxin B. J. Pestic. Sci. 1998;52:81–89. [Google Scholar]
- 5.Langenfeld A., Blond A., Gueye S., Herson P., Nay B., Dupont J., Prado S. Insecticidal cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2011;74:825–830. doi: 10.1021/np100890n. [DOI] [PubMed] [Google Scholar]
- 6.Morais-Urano R.P., Chagas A.C., Berlinck R.G. Acaricidal action of destruxins produced by a marine-derived Beauveria felina on the bovine tick Rhipicephalus (Boophilus) microplus. Exp. Parasitol. 2012;132:362–366. doi: 10.1016/j.exppara.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Yeh S.F., Pan W., Ong G.T., Chiou A.J., Chuang C.C., Chiou S.H., Wu S.H. Study of structure–activity correlation in destruxins, a class of cyclodepsipeptides possessing suppressive effect on the generation of hepatitis B virus surface antigen in human hepatoma cells. Biochem. Biophys. Res. Commun. 1996;229:65–72. doi: 10.1006/bbrc.1996.1758. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida M., Takeuchi H., Ishida Y., Yashiroda Y., Yoshida M., Takagi M., Shin-ya K., Doi T. Synthesis, structure determination, and biological evaluation of destruxin E. Org. Lett. 2010;12:3792–3795. doi: 10.1021/ol101449x. [DOI] [PubMed] [Google Scholar]
- 9.Odier F., Vey A., Bureau J.P. In vitro effect of fungal cyclodepsipeptides on leukemic cells: Study of destruxins A, B and E. Biol. Cell. 1992;74:267–271. doi: 10.1016/0248-4900(92)90037-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y.M., Ju G.L., Xiao L., Zhang X.F., Du F.Y. Cyclodepsipeptides and sesquiterpenes from marine-derived fungus Trichothecium roseum and their biological functions. Mar. Drugs. 2018;16:519–531. doi: 10.3390/md16120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Che Y.S., Swenson D.C., Gloer J.B., Koster B., Malloch D. Pseudodestruxins A and B: New cyclic depsipeptides from the coprophilous fungus Nigrosabulum globosum. J. Nat. Prod. 2001;64:555–558. doi: 10.1021/np000547r. [DOI] [PubMed] [Google Scholar]
- 12.Chung Y.M., El-Shazly M., Chuang D.W., Hwang T.L., Asai T., Oshima Y., Ashour M.L., Wu Y.C., Chang F.R. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, induces the production of anti-inflammatory cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2013;76:1260–1266. doi: 10.1021/np400143j. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida M., Ishida Y., Adachi K., Murase H., Nakagawa H., Doi T. Solid-phase combinatorial synthesis and biological evaluation of destruxin E analogues. Chemistry. 2015;21:18417–18430. doi: 10.1002/chem.201502970. [DOI] [PubMed] [Google Scholar]
- 14.Wang B., Kang Q., Lu Y., Bai L., Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vita-Marques A.M., Lira S.P., Berlinck R.G.S., Seleghim M.H.R., Sponchiado S.R.P., Tauk-Tornisielo S.M., Barata M., Pessoa C., de Moraes M.O., Cavalcanti B.C., Nascimento G.G.F., de Souza A.O., Galetti F.C.S., Silva C.L., Silva M. A multi-screening approach for marine-derived fungal metabolites and the isolation of cyclodepsipeptides from Beauveria felina. Quim. Nova. 2008;31:1099–1103. [Google Scholar]
- 16.Stocking E.M., Sanz-Cervera J.F., Williams R.M., Unkefer C.J. Studies on the biosynthesis of paraherquamide A. Origin of the β-methylproline ring. J. Am. Chem. Soc. 1996;118:7008–7009. [Google Scholar]
- 17.Shimamura H., Gouda H., Nagai K., Hirose T., Ichioka M., Furuya Y., Kobayashi Y., Hirono S., Sunazuka T., Omura S. Structure determination and total synthesis of bottromycin A2: A potent antibiotic against MRSA and VRE. Angew. Chem. Int. Ed. Engl. 2009;48:914–917. doi: 10.1002/anie.200804138. [DOI] [PubMed] [Google Scholar]
- 18.Tan L.T., Cheng X.C., Jensen P.R., Fenical W. Scytalidamides A and B, new cytotoxic cyclic heptapeptides from a marine fungus of the genus Scytalidium. J. Org. Chem. 2003;68:8767–8773. doi: 10.1021/jo030191z. [DOI] [PubMed] [Google Scholar]
- 19.Fredenhagen A., Molleyres L.P., Bohlendorf B., Laue G. Structure determination of neoefrapeptins A to N: Peptides with insecticidal activity produced by the fungus Geotrichum candidum. J. Antibiot. (Tokyo) 2006;59:267–280. doi: 10.1038/ja.2006.38. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Tang M.C., Tang S., Gao S., Soliman S., Hang L., Xu W., Ye T., Watanabe K., Tang Y. Genome mining and assembly-line biosynthesis of the UCS1025A pyrrolizidinone family of fungal alkaloids. J. Am. Chem. Soc. 2018;140:2067–2071. doi: 10.1021/jacs.8b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crone W.J., Vior N.M., Santos-Aberturas J., Schmitz L.G., Leeper F.J., Truman A.W. Dissecting bottromycin biosynthesis using comparative untargeted metabolomics. Angew. Chem. Int. Ed. Engl. 2016;55:9639–9643. doi: 10.1002/anie.201604304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Elimat T., Figueroa M., Ehrmann B.M., Cech N.B., Pearce C.J., Oberlies N.H. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013;76:1709–1716. doi: 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivon F., Remy S., Grelier G., Apel C., Eydoux C., Guillemot J.C., Neyts J., Delang L., Touboul D., Roussi F., Litaudon M. Antiviral compounds from codiaeum peltatum targeted by a multi-informative molecular networks approach. J. Nat. Prod. 2019;82:330–340. doi: 10.1021/acs.jnatprod.8b00800. [DOI] [PubMed] [Google Scholar]
- 24.Hou X.M., Li Y.Y., Shi Y.W., Fang Y.W., Chao R., Gu Y.C., Wang C.Y., Shao C.L. Integrating molecular networking and 1H NMR to target the isolation of chrysogeamides from a library of marine-derived Penicillium fungi. J. Org. Chem. 2019;84:1228–1237. doi: 10.1021/acs.joc.8b02614. [DOI] [PubMed] [Google Scholar]
- 25.Lim S.P., Noble C.G., She C.C., Soh T.S., Sahili A.E., Chan G.K.Y., Lescar J., Arora R., Benson T., Nilar S., Manjunatha U., Wan K.F., Dong H., Xie X., Shi P. Potent allosteric dengue virus NS5 polymerase inhibitors: Mechanism of action and resistance profiling. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delvecchio R., Higa L.M., Pezzuto P., Valadão A.L., Garcez P.P., Monteiro F.L., Loiola E.C., Dias A.A., Silva F.J.M., Aliota M.T., Caine E.A., Osorio J.E., Bellio M., O’Connor D.H., Rehen S. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses. 2016;8 doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owczarek K., Chykunova Y., Jassoy C., Maksym B., Rajfur Z., Pyrc K. Zika virus: Mapping and reprogramming the entry. Cell Commun. Signal. 2019;17 doi: 10.1186/s12964-019-0349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller K.I., Qing C., Sze D.M., Neilan B.A. Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambellon E., Rijnen L., Lorquet F., Gitton C., van Hylckama V.J., Wouters J.A., Yvon M. The d-2-hydroxyacid dehydrogenase incorrectly annotated PanE is the sole reduction system for branched-chain 2-keto acids in Lactococcus lactis. J. Bacteriol. 2009;191:873–881. doi: 10.1128/JB.01114-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dusch N., Puhler A., Kalinowski J. Expression of the Corynebacterium glutamicum panD gene encoding l-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 1999;65:1530–1539. doi: 10.1128/aem.65.4.1530-1539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bezsudnova E.Y., Boyko K.M., Popov V.O. Properties of bacterial and archaeal branched-chain amino acid aminotransferases. Biochemistry. 2017;82:1572–1591. doi: 10.1134/S0006297917130028. [DOI] [PubMed] [Google Scholar]
- 32.Chiang Y.M., Szewczyk E., Nayak T., Davidson A.D., Sanchez J.F., Lo H.C., Ho W.Y., Simityan H., Kuo E., Praseuth A., Watanabe K., Oakley B.R., Wang C.C. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chooi Y.H., Tang Y. Adding the lipo to lipopeptides: Do more with less. Chem. Biol. 2010;17:791–793. doi: 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Lukassen M.B., Saei W., Sondergaard T.E., Tamminen A., Kumar A., Kempken F., Wiebe M.G., Sorensen J.L. Identification of the scopularide biosynthetic gene cluster in Scopulariopsis brevicaulis. Mar. Drugs. 2015;13:4331–4343. doi: 10.3390/md13074331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W., Cacho R.A., Chiou G., Garg N.K., Tang Y., Walsh C.T. EcdGHK are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold. J. Am. Chem. Soc. 2013;135:4457–4466. doi: 10.1021/ja312572v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukat P., Katsuyama Y., Wenzel S., Binz T., Konig C., Blankenfeldt W., Bronstrup M., Muller R. Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity. Chem. Sci. 2017;8:7521–7527. doi: 10.1039/c7sc02622f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner J.J., Fendt S.M., Becker D.F. The proline cycle as a potential cancer therapy target. Biochemistry. 2018;57:3433–3444. doi: 10.1021/acs.biochem.8b00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fichman Y., Gerdes S.Y., Kovacs H., Szabados L., Zilberstein A., Csonka L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. Camb. Philos. Soc. 2015;90:1065–1099. doi: 10.1111/brv.12146. [DOI] [PubMed] [Google Scholar]
- 39.Han P., Jin F., Dong X., Fan J., Qiu B., Ren S. Transcript and protein profiling analysis of the destruxin A induced response in larvae of Plutella xylostella. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozano-Tovar M.D., Garrido-Jurado I., Lafont F., Quesada-Moraga E. Insecticidal activity of a destruxin-containing extract of Metarhizium brunneum against Ceratitis capitate. J. Econ. Entomol. 2015;108:462–472. doi: 10.1093/jee/tov041. [DOI] [PubMed] [Google Scholar]
- 41.Kim H.M., Choi I.S., Lee S., Hwang I.M., Chun H.H., Wi S.G., Kim J.C., Shin T.Y., Kim J.C., Kim J.S., Kim J., Park H.W. Advanced strategy to produce insecticidal destruxins from lignocellulosic biomass Miscanthus. Biotechnol. Biofuels. 2019;12:188–198. doi: 10.1186/s13068-019-1530-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du F.Y., Li X.M., Zhang P., Li C.S., Wang B.G. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Mar. Drugs. 2014;12:2816–2826. doi: 10.3390/md12052816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vining L.C., Taber W.A. Isariin, a new depsipeptide from Isaria cretacea. Can. J. Chem. 1962;40:1579–1584. [Google Scholar]
- 44.Yuan B.C., Liu D., Guan X., Yan Y.C., Zhang J.P., Zhang Y.P., Yang D.H., Ma M., Lin W.H. Piperazine ring formation by a single-module NRPS and cleavage by an α-KG-dependent nonheme iron dioxygenase in brasiliamide biosynthesis. Appl. Microbiol. Biotechnol. 2020;104:6149–6159. doi: 10.1007/s00253-020-10678-w. [DOI] [PubMed] [Google Scholar]
- 45.Szewczyk E., Nayak T., Oakley C.E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S.A., Oakley B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 46.Donnelly M.I., Zhou M., Millard C.S., Clancy S., Stols L., Eschenfeldt W.H., Collart F.R., Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prieto C., Garcia-Estrada C., Lorenzana D., Martin J.F. NRPSsp: Non-ribosomal peptide synthase substrate predictor. Bioinformatics. 2012;28:426–427. doi: 10.1093/bioinformatics/btr659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the article and its Supplementary Information files.