Abstract
Fluoroquinolones are the highest priority, critically important antimicrobial agents. Resistance development can occur via different mechanisms, with plasmid-mediated quinolone resistance (PMQR) being prevalent in the livestock and food area. Especially, qnr genes, commonly located on mobile genetic elements, are major drivers for the spread of resistance determinants against fluoroquinolones. We investigated the prevalence and characteristics of qnr-positive Escherichia (E.) coli obtained from different monitoring programs in Germany in 2017. Furthermore, we aimed to evaluate commonalities of qnr-carrying plasmids in E. coli. We found qnr to be broadly spread over different livestock and food matrices, and to be present in various sequence types. The qnr-positive isolates were predominantly detected within selectively isolated ESBL (extended spectrum beta-lactamase)-producing E. coli, leading to a frequent association with other resistance genes, especially cephalosporin determinants. Furthermore, we found that qnr correlates with the presence of genes involved in resistance development against quaternary ammonium compounds (qac). The detection of additional point mutations in many isolates within the chromosomal QRDR region led to even higher MIC values against fluoroquinolones for the investigated E. coli. All of these attributes should be carefully taken into account in the risk assessment of qnr-carrying E. coli from livestock and food.
Keywords: E. coli, typing, genomes, plasmid, livestock, food, fluoroquinolones
1. Introduction
Antimicrobial resistance (AMR), especially against the highest priority, critically important substances (e.g., quinolones and fluoroquinolones) is a global threat for humans and animals. Food-producing animals are considered an important reservoir of AMR-carrying bacteria [1,2]. Therefore, annual monitoring programs in the EU are conducted to observe trends in the development and dynamics of resistances in specific target animals. Commensal Escherichia (E.) coli serves as an indicator bacterium among Enterobacteriaceae for estimating changes in the prevalence of resistance genes in food and livestock in European countries.
Quinolones and fluoroquinolones, further named (fluoro)quinolones, are antimicrobial agents, considered as clinically highly important substances [3], and are used for the treatment of animal infections and human diseases in Europe. In the last years, EFSA notified a steadily increasing trend in (fluoro)quinolone-resistant bacteria, isolated from food-producing animals. Therewith, a high proportion of Salmonella enterica and E. coli, mainly isolated from poultry, were classified as not susceptible against ciprofloxacin. Next to the livestock sector, the human sector also registered an increase in ciprofloxacin resistance from 1.7% (in 2016) to 4.6% (in 2018) in certain Enterobacteriaceae [4]. The prevailing trend in European countries indicates a spread of (fluoro)quinolone-resistant bacteria, which poses a threat to animal and human health.
Resistance development against (fluoro)quinolones can occur via various mechanisms ranging from alterations of chromosomal genes to the acquisition of specific transferable genes. Mutations in the chromosomal elements encoding the target enzyme DNA gyrase (gyrA, gyrB) and topoisomerase IV (parC, parE) can alter the susceptibility of the isolates considerably. Other resistance mechanisms are involved in an overexpression of quinolone efflux pumps, alteration of the membrane permeability, or enzymatic inactivation of specific (fluoro)quinolones. These mechanisms can be induced by plasmid-mediated quinolone resistances (PMQR) including pentapeptide-encoding qnr genes, efflux pump-encoding genes (e.g., qepA), and the aminoglycoside acetyltransferase-coding aac-(6′)-Ib-cr gene [5]. PMQRs represent an inevitable threat, as they play an important role in the dissemination of (fluoro)quinolone resistance genes through horizontal gene transfer and the development of new (fluoro)quinolone-resistant bacteria. Hence, they are assumed accountable for the increased resistance to (fluoro)quinolones [6,7]. Horizontal gene transfer (HGT) is an efficient mechanism for adaptation of bacteria to prevailing environmental conditions. The exchange of resistance genes is a common response of bacteria to overcome antimicrobial selection pressures. Among them, qnr represents a highly prevalent PMQR gene in livestock with a broad overall distribution [8]. It has frequently been reported that qnr genes in Enterobacteriaceae increase (fluoro)quinolone resistance by enhancing the degree of resistance at which they can be selected [9]. Furthermore, some reports have linked the presence of PMQRs with a successive development of chromosomal alterations in the genes gyrA, gyrB, or parC, known to be associated with increased (fluoro)quinolone resistance when mutated at specific positions [10,11]. In addition, qnr genes were often observed in combination with other mobile determinants involved in resistance development against other critically important antimicrobials (i.e., extended spectrum β-lactamases (ESBLs), carbapenemases, and colistin) [12,13,14]. The co-occurrence of genes associated with resistances against antimicrobial agents routinely used in human medicine is of great concern, as it limits the therapeutic options for treatment of infections [15]. To estimate the specific impact of qnr-carrying isolates for the emergence and dissemination of (fluoro)quinolone-resistant E. coli in livestock and food, a deeper understanding of the occurrence, genetic variability, and elements involved in their spread is needed.
This study was conducted to assess the prevalence and diversity of qnr-carrying isolates among (fluoro)quinolone-resistant, commensal E. coli gained during the annual German AMR monitoring in livestock and food in 2017. These isolates were characterised in detail for their resistance phenotype and genetic characteristics. Furthermore, the commonality of plasmids carrying qnr genes along with their diversity and transmissibility were determined. Finally, a potential association of qnr-genes with (fluoro)quinolone resistance enhancing point mutations among German livestock E. coli was evaluated.
2. Materials and Methods
2.1. Bacterial Isolates and Culture Conditions
In total, 2799 E. coli from different food and livestock sources [16], especially from faecal samples of deer and fattening pigs, from cecum contents of fattening pigs and veal calves, as well as samples from pork, veal, and game meat where analysed. Samples from faecal and cecum sources are analysed as the same source in this study. All isolates were recovered during the German AMR monitoring of commensal (ZoMo, unselective cultivation conditions) and ESBL and/or AmpC-producing E. coli from food and livestock (ESBL-monitoring, selective cultivation conditions using cefotaxime) in 2017. The isolates were investigated according to the European Commission Implementing Decision 2013/652/EU in the National Reference Laboratory for Antimicrobial Resistance (NRL-AR). The isolates constitute the positive findings of a representative collection of samples taken in all 16 German federal states. If not stated otherwise, all isolates were cultivated in lysogeny-broth-based media for 16–18 h at 37 °C for further characterisation.
2.2. Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MIC) were determined by using broth microdilution according to EUCAST recommendations on a standardised European antimicrobial test panel (EUVSEC/EUVSEC2; Sensititre™, TREK Diagnostic Systems, Altrincham, Cheshire, UK). The tested antimicrobials covered the substances and ranges fixed in the European Commission Implementing Decision No. 2013/652/EU [17]. The following antimicrobial agents were used in ranges as specified: ampicillin (1 to 64 mg/L), azithromycin (2 to 64 mg/L), cefepime (0.06 to 32 mg/L), ciprofloxacin (0.015 to 8 mg/L), colistin (1 to 16 mg/L), ertapenem (0.015 to 2 mg/L), cefoxitin (0.5 to 64 mg/L), gentamicin (0.5 to 32 mg/L), imipenem (0.12 to 16 mg/L), meropenem (0.03 to 16 mg/L), nalidixic acid (4 to 128 mg/L), cefotaxime (0.25 to 64 mg/L), ceftazidime (0.25 to 128 mg/L), temocillin (2 to 128 mg/L), tetracycline (2 to 64 mg/L), tigecycline (0.25 to 8 mg/L), trimethoprim (0.25 to 32 mg/L), chloramphenicol (8 to 128 mg/L), sulfamethoxazole (8 to 1024 mg/L), cefotaxime/clavulanic acid (0.06/4 to 64/4 mg/L), and ceftazidime/clavulanic acid (0.12/4 to 128/4 mg/L). For quality assessment, the E. coli strain ATCC 25,922 was included. MIC values were interpreted according to EUCAST epidemiological cut-off values (ECOFFs) [18].
2.3. Molecular Screening on qnr Genes
Genomic DNA from isolates exhibiting a non-wild-type phenotype for nalidixic acid (NAL ≥ 16 mg/L) and/or ciprofloxacin (CIP ≥ 0.06 mg/L) were subjected to boiling DNA preparation [19]. The DNA extracts were used for molecular screening of qnr genes. PCR amplification for detecting qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC was conducted using primers and conditions as previously described [20,21] (Table S1). Product amplification was performed in a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Feldkirchen, Germany).
2.4. Determination of Isolate-Specific Macrorestriction Patterns and Plasmid Profiles
For determination of the genetic relationship between isolates, macrorestriction profiles using pulsed-field gel electrophoresis (PFGE) according to the PulseNet laboratory protocol [22] were performed. For digestion, the restriction endonuclease XbaI (10 U/µL, Thermo Fischer Scientific, Darmstadt, Germany) was used. For plasmid profiling, bacteria were treated with S1 nuclease (180 U/µL, Thermo Fischer Scientific) and S1-PFGE was conducted as previously described [22]. Separation of DNA was conducted on a CHEF-DR III system (Bio-Rad Laboratories, Madrid, Spain). The Salmonella enterica (H9812) serovar Braenderup was used as a molecular weight standard for size determination. Detection of qnr gene-carrying fragments was performed by Southern blotting and DNA-DNA hybridisation of S1-PFGE agarose gels. Hybridisation was conducted using digoxigenin-labelled (Roche Diagnostics, Mannheim-Penzberg, Germany) PCR probes of qnr genes and were prepared as previously described [23] (Table S1).
2.5. Whole-Genome Sequencing (WGS) and Bioinformatics Analysis
Genomic DNA of the isolates was prepared using the PureLink Genomic DNA Mini Kit (Invitrogen-Thermo Fisher, Schwerte, Germany) according to the manufacturer’s recommendation. The sequencing library was generated with the Nextera DNA Flex Library Preparation Kit (Illumina®, San Diego, CA, USA) as previously described [24]. Short-read, paired-end, whole-genome sequencing was performed in 2 × 151 cycles with the Illumina® NextSeq™ 500/550 Mid Output Kit v2.5 (300 Cycles). After trimming of reads with aquamis (version 1.33) [25], unicycler (version 0.4.4) [26] was used for de novo assembly of raw reads. Quality assessment of genome assemblies was conducted using QUAST 5.0.2 [27]. Assembled contigs were analysed for virulence factors and resistance genes as well as for plasmid markers (i.e., replicon types) with AMRfinder (version 3.6.7) and its database [28] and abricate (version 0.9.8) [29] each, through bakcharak [30]. Cluster analyses and sequence alignments were conducted using PATRIC with the RASTtk-enabled Genome Annotation Service and default parameters [31] for the service “Codon Tree”. Visualisation was conducted in R with the packages ggplot2 (version 3.3.0) [32], ggtree (version 1.4.11), and treeio (version 3.10) [33].
The PointFinder tool [34] and the deposited database (updated—2 July 2019; access date—25 June 2020) were used to identify alterations in chromosomal genes that were confirmed to be associated with (fluoro)quinolone resistances for E. coli. In-silico-based multilocus sequence typing (MLST, according to the Achtmann scheme) was conducted using bakcharak [30] and the pubMLST database [35]. The screening for respective Inc groups was based on the PlasmidFinder database [36]. The detection of resistance genes was conducted with ResFinder 4.1 [37].
To determine the diversity of qnr-carrying plasmids, a reference database comprising all accessible qnr plasmid genomes of the NCBI RefSeq database (access date—17 April 2020) was developed. All available plasmids were checked for completeness through the keywords “complete sequence” or “complete plasmid”. Abricate was performed with the NCBI AMRfinder database to screen for qnr plasmids. The resulted database was used for the subsequent reference search with RefSNPer [38] as described elsewhere [39]. The trimmed reads of all individual isolates were mapped to each reference plasmid (using bowtie2 [40], version 2.3.5). Subsequently, the coverage breadth and depth of each reference plasmid as well as the number of single-nucleotide polymorphisms (SNPs) are computed with SAMtools version 1.10 and BEDTools version 2.29.0 [41], thus providing the qnr plasmids that most closely match each isolate.
2.6. Analysis and Statistics
Data analysis and visualisation was conducted using R (version 3.6.3). Choropleth figure was visualised with R (version 3.6.3) using the packages maptools (version 0.9-9), sp (version 1.4-0), rgeos (version 0.5-2), and rgdal (v1.4-8). The SpatialPolygonDataFrame of Germany was provided by the Bundesamt für Kartographie und Geodäsie database (https://gdz.bkg.bund.de/index.php/default/open-data/gebietseinheiten-1-2-500-000-ge2500.html, access date—24 March 2020). Dependencies between the presence of antimicrobial resistance genes were calculated in R (version 3.6.3). The occurrence of resistance genes was translated into binary data. Correlation between certain resistance determinants was predicted by Fisher’s exact test. A ρ-value of <0.05 was considered as a statistically significant correlation.
3. Results and Discussion
3.1. qnrS Is the Most Prevalent qnr Gene in E. coli from Livestock and Food
Antimicrobial resistance-testing (AST) revealed that 391 out of 2799 investigated isolates (14%) exhibited a non-wild-type phenotype (phenotypical resistance) against ciprofloxacin (CIP: MIC ≥ 0.06 mg/L) and/or nalidixic acid (NAL: MIC ≥ 16 mg/L) (Table S1). Of those, 80 isolates were recovered from the ESBL-monitoring and 23 from ZoMo. PCR screening revealed seven different qnr genes within the 103 qnr-positive E. coli (Table 1). Among them, qnrS (n = 95) was the most prevalent gene, followed by qnrB (n = 6), while qnrA and qnrVC occurred only once each. No qnrC- or qnrD-carrying isolates were detected. The rather low prevalence of (fluoro)quinolone-resistant E. coli of 14% in the investigated nonpoultry matrices is in good agreement with the data summarised by EFSA [42]. While CIP and NAL resistance were reported by several European countries at high levels in broiler and turkey, the EU medians in pigs and calves were rather low (6.2% and 4.2% for NAL and 7.4% and 8.4% for CIP per matrix, respectively) [43]. Regarding the detection of specific qnr genes, our result is in good agreement with previous reports in which qnrS and qnrB were the most frequently detected qnr genes in livestock sources [44,45]. In contrast to livestock, qnrA genes are often present in isolates from hospitalised patients [46].
Table 1.
Occurrence and frequencies of determined qnr genes within qnr-carrying, (fluoro)quinolone-resistant E. coli isolates.
| Gene *1 | Gene *2 | Occurrence | Frequency # |
|---|---|---|---|
| qnrA | qnrA1 | 1 | 1.0% |
| qnrB | qnrB1 | 3 | 5.8% |
| qnrB2 | 1 | ||
| qnrB19 | 2 | ||
| qnrS | qnrS1 | 92 | 92.2% |
| qnrS2 | 3 | ||
| qnrVC | qnrVC4 | 1 | 1.0% |
*1: determined with PCR, *2: determined with WGS, # frequency of genes as determined by PCR.
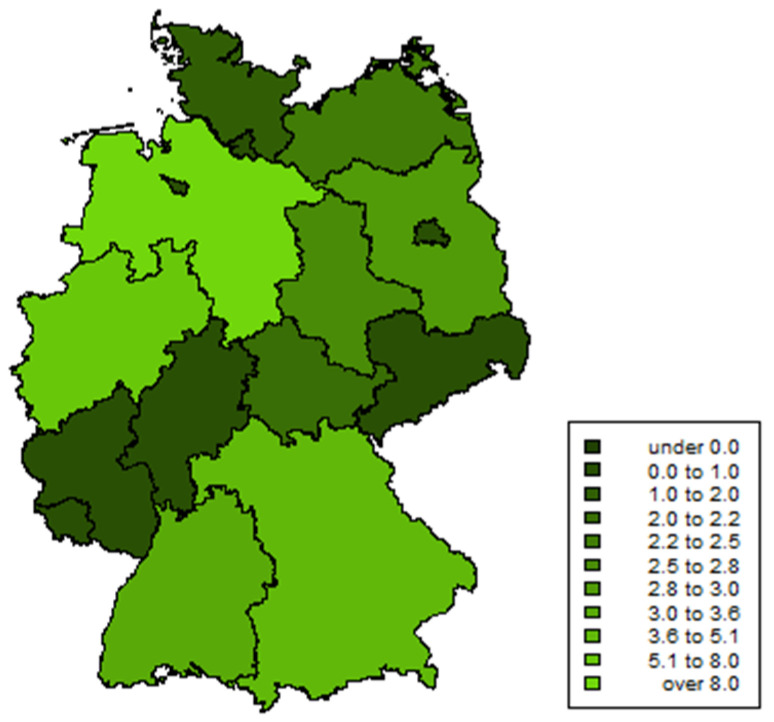
We detected qnr-positive E. coli in veal faeces (n = 56, 8.2% of all veal faeces samples from the ZoMo- and ESBL-monitoring in 2017), fattening pig faeces (n = 38, 3.6%), minced meat (n = 3, 4.7%), beef (n = 2, 5.1%), and pork (n = 2, 5.9%), as well as in deer faeces (n = 1, 0.2%) and deer meat (n = 1, 0.5%) (detailed data for each isolate are presented in Table S2). Overall, the highest relative proportion of (fluoro)quinolone-resistant and qnr-positive isolates during the monitoring program in 2017 was determined in Lower Saxony (9.3%), followed by North Rhine-Westphalia (7.9%). However, these two federal states have the highest livestock population and contributed most to the overall sample size. The prevalence for other federal states was below 5%. An overview on the regional prevalence of (fluoro)quinolone-resistant and qnr-positive isolates is given in Figure 1.
Figure 1.
Choropleth of the proportion of (fluoro)quinolone-resistant and qnr-positive E. coli recovered during the ZoMo- and ESBL-monitoring in Germany in 2017. Prevalence was calculated as the proportion of (fluoro)quinolone-resistant and qnr-positive E. coli isolates divided by all investigated samples per federal state.
The analysis of the XbaI-macrorestriction patterns revealed a high phylogenetic heterogeneity of E. coli carrying qnr genes. For further typing purposes, the isolates were subjected to WGS and bioinformatics analysis. Here, 45 different sequence types (STs) were determined by in silico analysis. ST10 (21%), ST2325 (6%), and ST58 (5%) represented the predominant types. Overall, a broad distribution of qnr-carrying isolates in different ST-types was observed. Of ST10, 12 isolates were gained from veal faeces, nine from pork faeces, and one from veal meat. Four isolates from veal faeces, one from fattening pig faeces, and one from veal meat were assigned to ST2325. ST58 was evenly distributed between isolates from faecal samples from veal and fattening pigs. Two isolates could not be assigned to previously described STs. The observed results respond well to prevailing reports, in which qnr genes were found to be prevalent in ST10 isolates of livestock and of human origin and support the hypothesis on their impact as possible distributors of plasmid-associated qnr genes between livestock and human [47]. To the best of our knowledge, ST2325 isolates have yet not been described to be associated with qnr genes.
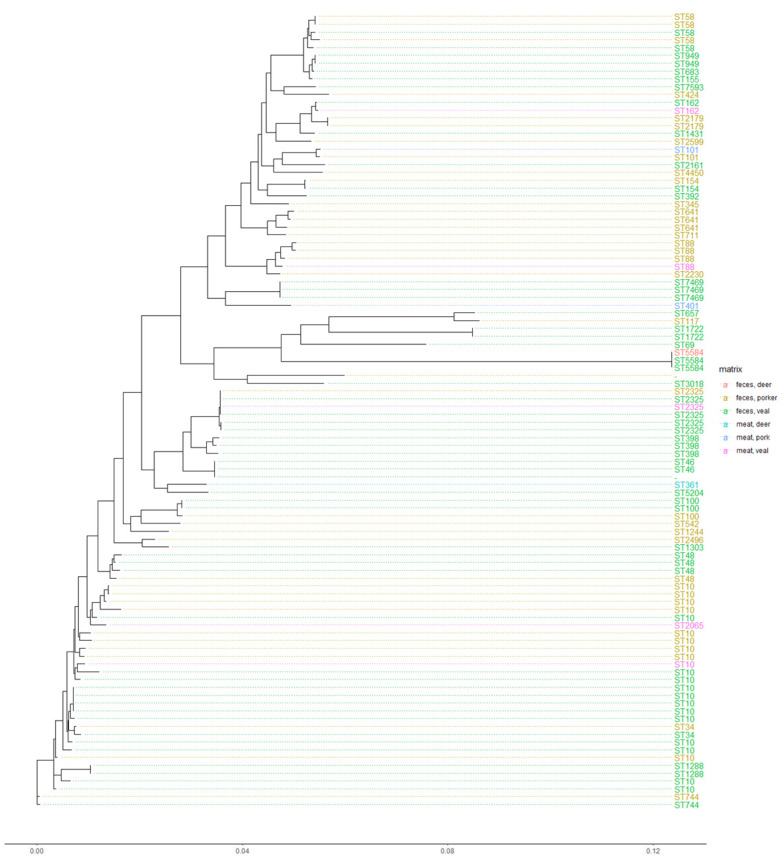
In Figure 2, the phylogenetic relationship of the isolates, based on WGS data of the individual isolates, is shown. As expected, the clusters correspond with the prevailing ST, but seemed not to be associated with a certain food/livestock matrix. In addition, diverse resistance profiles were observed in different clusters and STs, as well as over different matrices for the tested strains (Figure 2). Overall, we found a high diversity of E. coli carrying qnr. This widespread occurrence of qnr has been reported before [48]. Especially, E. coli of the clonal group ST10 are often associated with AMR plasmids [49] and often reported to carry qnr-positive plasmids [50]. Further, ST10 is characteristic for E. coli defined as ESBL [51]. As we analysed 80 E. coli isolates from the ESBL-monitoring, the observation of ST10 being related to ESBL E. coli was confirmed. Our findings support the current knowledge that resistance genes such as qnr can spread over different sources and are not restricted to certain E. coli sequence types.
Figure 2.
Phylogenetic relationship of (fluoro)quinolone-resistant and qnr-positive E. coli with metadata based on codon and protein differences, presented in a maximum-likelihood tree. The respective sequence type (ST) of the E. coli is shown as geom_tiplab and connected with dotted lines. The ST as well as the dots are coloured according to the matrix code of the recovered E. coli.
3.2. qnr-Carrying E. coli Isolates Exhibit Diverse Resistance Phenotypes Including Multidrug Resistances
In antimicrobial sensitivity testing, the qnr-carrying E. coli showed highly diverse resistance profiles. Most of them exhibited a phenotypic resistance for antimicrobial classes (Table S2) other than (fluoro)quinolones, which is linked to the fact that most isolates originated from ESBL-monitoring. Considering only the 23 ZoMo-isolates, the resistant phenotype ranged from resistance to only one to five different classes. When only ESBL-monitoring isolates were analysed, the numbers ranged from three to eight different classes.
Thus, for the ESBL-monitoring isolates, 90% exhibited resistance phenotype against more than three antimicrobial classes, 79% showed resistance phenotypes against five antimicrobial agents, and 34% exhibited increased MIC values against six to eight antimicrobial agents.
In general, besides ciprofloxacin (100% from ESBL-monitoring, 91% from ZoMo), resistant phenotypes for ampicillin (99% and 91%), cephalosporine (100% and 20%), and tetracycline (80% and 52%) were most common among the investigated E. coli. Table 2 shows the distribution of resistant phenotypes relative to the respective matrix. This distribution is presented graphically in the Supplementary material FS1. Overall, we only detected two isolates from the ZoMo, which were only resistant to (fluoro)quinolones. Thus, our data, in which (fluoro)quinolone resistance is frequently associated with multidrug resistance in E. coli, coincides well with the results of other studies [52]. As a result of a direct selection pressure, the treatment of livestock with a specific antimicrobial agent supports the maintenance of resistance genes directed against this antimicrobial agent [53]. As resistance genes can occur within a multidrug-resistant isolate or on a multiresistance plasmid, disseminating through selective forces, this could enhance the prevalence of other resistance genes on the same plasmid or within the same isolate. Our result supports that there is a potential risk of coselection, maintenance, transmission, and propagation of multidrug-resistant E. coli and their plasmids [15]. From our findings, one could assume that multidrug-resistant clones with (fluoro)quinolone resistance exist, especially in combination with ESBL genes, which might be of special concern.
Table 2.
Absolute number of the phenotypic resistance of qnr-carrying isolates and absolute number of isolates gained from the respective matrix. In brackets the absolute number of isolates from the ZoMo-/ESBL-Monitoring is indicated.
| Matrix | Matrix Occurrence | AMP | AZI | CHL | CIP | COL | FOT | GEN | MERO | NAL | SMX | TAZ | TET | TMP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| faeces, veal calves | 56 (52/4) |
54 (51/3) |
4 (4/0) |
19 (17/2) |
55 (52/3) |
1 (1/0) |
53 (52/1) |
6 (6/0) |
1 (1/0) |
8 (7/1) |
37 (36/1) |
52 (52/0) |
48 (44/4) |
40 (37/3) |
| faeces, deer | 1 (1/0) |
1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| faeces, pigs | 38 (24/14) |
38 (24/14) |
6 (5/1) |
11 (9/2) |
37 (24/13) |
0 | 27 (24/3) |
4 (4/0) |
0 | 10 (7/3) |
22 (18/4) |
27 (24/3) |
22 (17/5) |
20 (17/3) |
| meat, veal | 2 (2/0) | 2 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 2 |
| meat, deer | 1 (1/0) | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| meat, pork | 2 (0/2) | 1 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| minced meat | 3 (0/3) | 3 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
AMP—Ampicillin, AZI—Azithromycin, CHL—Chloramphenicol, CIP—Ciprofloxacin, COL—Colistin, FOT—Cefotaxime, GEN—Gentamicin, MERO—Meropenem, NAL—Nalidixic acid, SMX—Sulfamethoxazole, TAZ—Ceftazidime, TET—Tetracycline, TMP—Trimethoprim.
3.3. qnr-Carrying E. coli Isolates Are Associated with Highly Diverse Resistomes
Overall, the most abundant resistance genes of qnr-carrying isolates were blaEC (accession number: A0A244BQ89) (100% of all qnr-carrying strains), as well as different variants of the tet (96%) and blaCTX-M (74%) genes. In general, we found qnrS1 to be in frequent connection with tet(34) and tet(A). By analysing the nucleotide-sequence of the tet(34) gene of E. coli, a sequence coverage of only 76% to the reference (accession number: A7J11_00001) was detected. Besides this defective gene, tetracycline-resistant isolates usually carried other determinants like tet(A) or tet(B). Interestingly, none of the qnr-harbouring E. coli carried a PMQR (aac(6′)-Ib-cr, qepA, or oqxAB) other than qnr.
We further split the isolates by the ZoMo- and ESBL-monitoring source. For the ZoMo isolates, we found a significant correlation for the co-occurrence of qnrVC with blaOXA10, cmlA5, and the pesticide-resistance encoding gene qacF. For the other qnr gene variants, no significant correlation for co-occurrence was detected (Table S3). No statistically significant correlation was observed for qnrS1 and blaTEM-1 (p-value 0.059). However, the low number of ZoMo isolates hinders a thorough analysis for this potential correlation. When we analysed the ESBL-monitoring isolates, some co-occurrence of qnr and other resistance genes was detected (Table 3). Due to the selective isolation procedure, a high correlation of qnr and bla genes was observed. However, qnrA was rather associated with blaACC-1 and blaVIM-1 as well as qnrB with blaOXA-1, while qnrS correlated with blaCTX-M-65 and blaOXA-1. Other co-occurring resistance genes with qnr are presented in Table 3. Especially, qnrS1 was often detected in combination with multiple resistance genes as well as with the pesticide resistance encoding gene qacEΔ1.
Table 3.
p-value for co-occurrence of selected resistance genes and qnr genes in ESBL-monitoring isolates; p-values below 0.05 are highlighted in red and represent statistical significance. The value 1 reflects that the two genes were not detected.
| qnrA1 | qnrB1 | qnrB2 | qnrB19 | qnrS1 | qnrS2 | |
|---|---|---|---|---|---|---|
| aadA1 | 0.1625 | 0.00348101 | 1 | 1 | 0.00462503 | 0.41693038 |
| aph(3′)-lia | 1 | 1 | 1 | 1 | 0.1125 | 0.0375 |
| aph(3′)-XV | 0.0125 | 1 | 1 | 1 | 0.1125 | 1 |
| arr-3 | 1 | 1 | 1 | 1 | 0.011392405 | 0.00094937 |
| bla ACC-1 | 0.0125 | 1 | 1 | 1 | 0.1125 | 1 |
| bla CTX-M-65 | 1 | 1 | 1 | 1 | 0.060414269 | 0.00559883 |
| bla OXA-1 | 1 | 0.00012171 | 1 | 1 | 0.000000524 | 0.00925024 |
| bla VIM-1 | 0.0125 | 1 | 1 | 1 | 0.1125 | 1 |
| catA1 | 0.0625 | 0.00012171 | 1 | 1 | 0.000377371 | 1 |
| catB2 | 0.0125 | 1 | 1 | 1 | 0.1125 | 1 |
| catB3 | 1 | 1 | 1 | 0.0375 | 0.001022395 | 0.00282376 |
| dfrA25 | 1 | 1 | 0.0125 | 1 | 0.1125 | 1 |
| floR | 0.25 | 1 | 1 | 1 | 0.038946034 | 0.01387537 |
| mef(C) | 1 | 0.10966407 | 1 | 1 | 0.032132425 | 0.10966407 |
| mph(A) | 0.2125 | 1 | 1 | 1 | 0.090321713 | 0.00827653 |
| mph(G) | 1 | 0.10966407 | 1 | 1 | 0.032132425 | 0.10966407 |
| qacEΔ1 | 0.2 | 0.49289192 | 0.2 | 0.2 | 0.001541798 | 0.10029211 |
| sul1 | 0.175 | 0.44303798 | 0.175 | 0.175 | 0.006847169 | 0.44303798 |
This coexistence of multiple resistance genes can pose a higher risk, as their presence may contribute to a better adaption to different environmental conditions and enhance the persistence of the plasmid. It has been reported in previous publications that E. coli isolates from different livestock matrices carried both ESBL and PMQR genes, as they often coexist on the same plasmid. Mainly, a coexistence of qnr and blaCTX-M-15 as well as blaSHV was previously described [12]. In general, ESBL-producing E. coli are an emerging public-health threat and their rise will further reduce the available treatment options in human medicine. The co-occurrence of qnr and ESBL genes represent another risk as the bacteria exhibit resistances against antimicrobials of two important classes. Especially, the spread of plasmids bearing resistance determinants of both antimicrobial classes will further force the development of multidrug-resistant isolates. A correlation of qnr-positive ESBL E. coli was previously reported for human sources. The presence of genes conferring resistances against two critically important antimicrobial agents on the same plasmid or within one isolate can constitute an important issue for treatment failures when using the respective antimicrobial agents for therapeutic application in hospitalised patients. As Salah et al. mentioned, these plasmid-mediated resistances highly facilitate the spread and increase their frequency [54]. They also found that every qnr-positive strain investigated in their study was ESBL-producing. However, as we mainly screened ESBL-preselected E. coli, the observation of qnr in these ESBL-producers presumably relates on a conditional probability. Although the coexistence of multiple PMQR genes has been described as a frequent event [52], we did not detect PMQRs other than qnr.
As mentioned, a significant co-occurrence of qnrS1 and qacEΔ1 as well as of qnrVC and qacF was observed. These determinants confer resistance to quaternary ammonium compound disinfectants [55]. The awareness for this plasmid-associated antiseptic resistance gene is broadly present, as it enhances the tolerance to several disinfectants that might increase the ability of AMR-carrying isolates to persist in the environment [56]. Quaternary ammonium compounds are widely used as disinfectant in farm environments. It has been observed that qac genes are often associated with multidrug-resistant isolates [57]. Thus, they might support the evolution (i.e., adaptation to specific environmental conditions) of bacterial resistance to multiple antimicrobial agents. Here, the presence of the biocide resistance genes reveals another risk harboured by the qnr-carrying isolates, as it represents an additional determinant for resistances against biocides.
3.4. Virulence Genes Associated with qnr-Carrying E. coli
As this study was based on commensal E. coli, the number of virulence-associated genes in the isolates was expected to be low. In our samples, between 34 and 108 potential virulence factors (according to the Virulence Factor Database) per isolate were identified. Ninety-two isolates were found to carry fimH, a D-mannose-specific adhesin, type 1 fimbriae-encoding gene. Frequent detection of the potential surface virulence factors fimH is quite common among E. coli. However, FimH mediates adherence to cells and, therewith, helps the formation of bacterial biofilms [58]. It confers the possibility of colonisation and, when certain mutations occur, can represent a virulence factor [59,60]. Additionally, other surface virulence factor encoding genes such as afa (n = 2), focG (n = 1), paa (n = 1), pap (n = 1), and saf (n = 1) were found. Moreover, we detected one isolate with eae, an intimin-encoding gene. The protein, encoded by eae, plays a critical role on the intestinal colonisation and, therefore, STEC (shiga toxin-producing E. coli) or another aggregative E. coli pathogenesis [61]. Some isolates harboured toxin genes or toxin subunits such as astA (n = 16), cdtA (n = 3), cnf1 (n = 3), eltA (n = 1), and faeC (n = 1). The presence of astA has been detected in subgroups of enteroaggregative E. coli [62]. Further, two isolates possessed the hlyA gene, an important secretory virulence factor. Therewith, the presence of the virulence genes was unrelated to the different monitoring programs from where the E. coli was isolated. Thus, we showed that important virulence factors could sporadically occur in qnr-positive E. coli isolates. As many virulence factors are also located on mobile genetic elements, like plasmids, their potential spread with resistance determinants should be taken into account. Thus, through horizontal gene transfer, not only the resistance genes but additional virulence factors are spread. In these cases, antibiotic treatment failure, due to resistance, may give rise to potential impacts of certain virulence factors; therewith, representing an evolutionary pathway to pathogenicity [63]. However, most of the virulence factors detected in this study represent individual components of different complex systems. As the different virulence factors of E. coli are quite complex in their interaction, they may not have a high impact on the isolate’s pathogenicity on their own [64].
3.5. In-Silico-Based Prediction of Plasmids Types Carrying qnr
Plasmids play a major role in bacterial evolution and resistance gene transmission. Understanding factors influencing plasmid composition and evolution are essential for reliable assessments. Therefore, we detected the best-matching references to our qnr-carrying plasmids with the refSNPer tool. Therewith, reference plasmids that were covered by up to 100% and 90% were determined for 31 and 35 datasets, respectively, to their reference plasmids. Four WGS datasets showed no significant matches (best reference < 50% coverage) to any qnr-plasmid genome of the reference database and could be considered as new plasmids. The most frequently detected references were plasmid tig00003056 of E. coli strain AR_0162 (NZ_CP021681, n = 15), plasmid C of E. coli strain D9 (NZ_CP010155, n = 8), an unnamed plasmid of Shigella flexneri 1a strain0670 (NZ_CP020088, n = 8), and pKpvST101_6 of the Klebsiella pneumoniae strain KpvST101_OXA-48 (NZ_CP031373, n = 6).
Plasmids exhibiting nucleotide similarities of >80% to the best matched plasmid tig00003056 (NZ_CP021681) have been described in hospitalised patients in the USA (CP026200.1, CP044008.1), the UK (LT906492.1, LT882487.1), in Taiwan (CP046430.1), and in Pakistan (CP040574.1). Plasmid C (NZ_CP010155)-like genomes were found in China and Japan, isolated from wastewater (CP035315.1, CP045998.1, AP019678.1, MT219825.1, CP051432.1, CP046002.1) or dog faeces (NZ_CP010155). Both plasmids were mainly isolated from E. coli but also found in other Enterobacteriaceae. This geographical spread, as well as the different reservoirs, provide no further evidence on a common source/origin for qnr-carrying plasmids. In addition, NZ_CP020088-like plasmids seemed broadly distributed. They have been detected in Brazil (MK965545.1) and Norway (MH507589.1) in chicken and turkey meat, and in rook faeces in the Czech Republic (KF362122.2, MH121702.1). However, they also have been isolated from hospitalised patients in China (CP020088.1, KJ201886.1, CP012734.1, and CP020341.1). With NZ_CP020088-like plasmids being reported mainly in poultry origin, it supports our findings of this plasmid in the livestock reservoir. NZ_CP031373 was detected in the Netherlands (KX618696.1), Czech Republic (MH594478.1), and the UK (CP031373.2). Overall, the best matched plasmid-references to our identified qnr-carrying plasmids mainly originate from E. coli (n = 50) and Shigella flexneri (n = 20). However, similar plasmid-types were also identified in a broad range of other Enterobacteriaceae like Enterobacter, Salmonella, and Serratia. The broad distribution of these plasmids, closely related to our identified qnr-carrying plasmids, demonstrates the high ability of spread among Enterobacteriaceae. The diverse host adaption is clear evidence for a broad host spectrum of qnr-plasmids. Further, the distant locations of plasmid isolation demonstrate the putative exchange of global resistance transfer over plasmids, especially for qnr here.
As shown in Table 4 (and Table S4), the most abundant plasmid replicon types among our identified qnr reference plasmids were IncN (n = 12), IncY (n = 19), as well as a combination of IncX1 and IncX3 (n = 29). However, a total of 21 different plasmid-type combinations were identified. Therewith, IncN plasmids are reported as broad host range types, with the ability for conjugative transfer and the carriage of drug resistance genes. Close phylogenetic relationships from environmental and clinical samples are described for IncN plasmids [65]. Further, IncN plasmids are known for carrying a great variety of resistance genes against extended-spectrum β-lactams, sulphonamides, quinolones, aminoglycosides, tetracyclines, and streptomycin [66]. Here, we found the IncN reference plasmid to carry the blaTEM-1 resistance gene, encoding β-lactamase. IncN is widely found in Enterobacteriaceae and recognised as conjugative. Plasmids from the IncY type are known for frequently harbouring blaCTX-M-5 or blaSHV-2 resistance genes, associated with ESBL development [66]. Further, plasmids of these group have been shown to carry mcr-1 [67]. Here, we found the reference plasmid from the IncY group to harbour multiple resistance genes, including blaCTX-M-5 and blaTEM-1. Although plasmids belonging to the IncX group are known to be narrow host range plasmids, commonly found in Enterobacteriaceae, they often carry a wide spectrum of multidrug resistance enabling genes and were often found in the guts of animals [68]. Genes encoding carbapenemases as well as the colistin resistance genes mcr-1 and mcr-2 are frequently reported on IncX plasmids [69,70]. Dobiasova and Dolejska [71] found that IncX plasmids are widely distributed in E. coli in European animals and predominantly associated with (fluoro)quinolone resistance genes, particularly with qnrS. All of the prevalent plasmid incompatibility (Inc)-groups were often associated with multiple resistance genes besides qnr, which increases the risk of the dissemination of those plasmids. As shown in Table 4, the plasmids from the IncX group did carry multiple resistance genes next to qnrS. Thus, we detected blaTEM-1 and blaSHV located on the same plasmids. However, while we identified certain clusters of the mentioned plasmid Inc-groups, the qnr gene was rather broadly distributed over different plasmid types.
Table 4.
Inc group and resistance genes of the best matching reference plasmid to the most prevalent plasmids carrying qnr-genes detected in this study. Identified with RefSNPer.
| Plasmid Type and Resistance Genes on Matching Reference | Frequency |
|---|---|
| IncN | ∑ 12 |
| aac(3)-IId, qnrS1 | 1 |
| bla TEM-1 , qnrS1 | 5 |
| qnrB19 | 1 |
| qnrS1 | 5 |
| IncR, IncX1 | ∑ 1 |
| aadA2, blaTEM-1, dfrA12, floR, qnrS1, sul2, tet(A), tet(M) | 1 |
| IncX1 | ∑ 9 |
| aph(3′)-Ia, floR, qnrS2 | 8 |
| bla TEM-1 , qnrS1, tet(M) | 1 |
| IncX1, IncX3 | ∑ 14 |
| bla TEM-1 , qnrS1 | 14 |
| IncX3 | ∑ 6 |
| bla SHV , qnrS1 | 6 |
| IncY | ∑ 19 |
| aph(3″)-Ib, aph(6)-Id, bla CTX-M-15 , bla TEM-1 , qnrS1, sul2, tet(A) | 15 |
| aph(3″)-Ib, aph(6)-Id, bla CTX-M-15 , bla TEM-1 , dfrA14, qnrS1, sul2, tet(A) | 2 |
| aph(3″)-Ib, aph(6)-Id, bla CTX-M-15 , bla TEM-1 , qnrS1, sul2 | 2 |
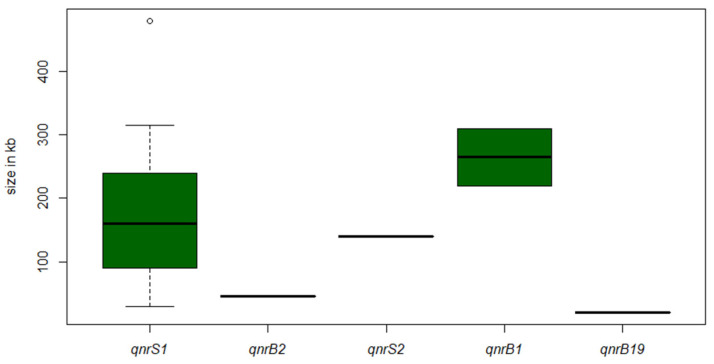
Further, we examined the size distribution of extrachromosomal DNA elements through S1-PFGE analysis and successive DNA–DNA hybridisation against the different qnr genes. Overall, we found a broad size diversity of plasmids carrying qnr (Figure 3).
Figure 3.
Boxplots of the size distribution of analysed plasmids harboring qnr, estimated through S1-PFGE. The white circle represents an outlier.
The qnrS1 gene was detected on plasmids ranging between 30 kb and 480 kb. Once again, this emphasises the ability of qnr to combine with different plasmid types of various sizes and, therewith, the adaptability to different niches.
Apart from qnr, the most frequently found resistance genes on the same reference plasmid were aph(3″)-Ib, aph(6)-Id, blaCTX-M-15, blaTEM-1, and sul2, independent of the monitoring program. The occurrence of this resistance gene combination was not exclusively for a certain Inc group, but rather broadly distributed. Hence, qnr genes are detectable on plasmids of various sizes and are seldom the only resistance gene located on the plasmid, but instead related to different resistance genes. The transfer of plasmids carrying qnr has been described as associated with the transfer of genes leading to multidrug resistance [72]. This suggests that provoking qnr resistance by overuse of an antimicrobial agent can support the expansion of multidrug-resistant isolates in livestock and food sources.
3.6. Frequent Detection of Point Mutations in the gyrA, parC and parE Genes of qnrS-Carrying E. coli
In our investigations, we detected isolates with phenotypes resistant against ciprofloxacin but lacking phenotype resistance against nalidixic acid (Table S2). Resistance to fluoroquinolones without resistance to quinolones is mainly associated with mutations in the chromosome within the gyrA and parC genes [58,59]. Further, the EFSA assumed that the resistance to ciprofloxacin without resistance against nalidixic acid indicates an increasing occurrence of plasmid-mediated quinolone resistance [2]. In general, besides PMQR genes, mutations in genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) are highly associated with increased (fluoro)quinolone resistance. Previous studies showed that a high level of resistance to (fluoro)quinolones is mainly associated with mutations in the gyrA and an additional mutation in the parC gene in E. coli [73,74]. In this study, 16 out of 103 qnr-carrying isolates were identified to exhibit a point mutation in the QRDR regions of gyrA (n = 9, two mutations; n = 5, one mutation) or parC (n = 2, two mutations; n = 10, one mutation) and parE (n = 4, one mutation) genes. Subsequently, these 16 isolates had high MIC values for nalidixic acid (mostly > 128 mg/L) and ciprofloxacin (mostly > 8 mg/L). Interestingly, we also detected three other isolates with a MIC for nalidixic acid > 128 mg/L, but no alteration in the chromosomal genes associated with (fluoro)quinolone resistance. However, all isolates but one had a point mutation in the parC region leading to the amino acid substation E62K. This strongly suggests an influence of this mutation for the (fluoro)quinolone resistance in E. coli. Vingopoulou et al. [75] described this ParC E62K substitution previously. They detected this new amino acid exchange in enrofloxacin-resistant E. coli isolated from dog otitis and faecal samples. As enrofloxacin is classified to the group of the (fluoro)quinolones, this would support the hypothesis of the E62KL exchange enhancing the (fluoro)quinolone resistance. In total, 87 of the analysed isolates were (fluoro)quinolone-resistant but carried only a qnr gene without any other PMQR genes associated with the development of the resistance. Possibly, the presence of qnr genes is sufficient to increase the MIC for NAL to a degree of resistance. One could also consider the development of yet unknown genes or point mutations in E. coli involved in this (fluoro)quinolone resistance. This clearly demonstrates the urgency of monitoring for PMQR to estimate resistance against (fluoro)quinolones and the possible capability of qnr to enhance (fluoro)quinolone resistance in E. coli.
4. Conclusions
In this study, we determined the prevalence of qnr-genes among (fluoro)quinolone-resistant E. coli from livestock and food in Germany and analysed the potential risks associated with the dissemination of the respective plasmids. While the prevalence of 3.7% of qnr-carrying isolates within (fluoro)quinolone-resistant E. coli was rather low, the risk linked with qnr-positive isolates is due to other reasons. Most of the qnr-positive E. coli also carried other resistance genes leading to a multidrug resistance phenotype of the isolate, especially when E. coli isolates from the ESBL-monitoring were analysed. Next to the occurrence of resistance genes, we detected genes leading to pesticide resistance or virulence genes within the same qnr-positive isolate. Further, we found that qnr-plasmids were widely distributed. Hence, the spread of the qnr-plasmid is not restricted to specific matrices or certain Inc groups. Further, we could confirm findings reporting that reported that the sole presence of qnr can lead to phenotypic (fluoro)quinolone resistance. We found isolates being resistant to nalidixic acid and ciprofloxacin that only carried a qnr gene without further PMQR or point mutations in the respective area of the chromosome. Since qnr is mostly identified on mobile genetic elements, this finding stresses the possible spread of this resistance determinant. The outgoing risk from qnr genes needs to be taken seriously, especially when evaluating (fluoro)quinolone resistance in E. coli isolated from livestock and food.
Acknowledgments
We gratefully thank Janina Malekzadah, Beatrice Baumann, and Katharina Thomas for their excellent laboratory assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9061308/s1. Supplemental Figure S1: Grouped bar chart of total number of resistant isolates per antimicrobial agent and matrix. Table S1: Sequences, annealing temperatures, and references of used primers for qnr detection. Table S2: Matrix and MIC values of investigated (fluoro)quinolone-resistant and qnr-positive E. coli isolates, as well as their resistance profile. Table S3: p-value of respective Fisher’s exact test; p-values < 0.05 represent statistical significance and are highlighted in red. The value 1 reflects that the two genes were not detected; columns with only 1 as value are not presented. Table (A) shows the correlation for genes detected in the ESBL-monitoring isolates. Table (B) shows the correlation for genes detected in the ZoMo isolates. Table S4: Inc-group and resistance genes of all the best-matching reference plasmids to the plasmids carrying qnr genes detected in this study. Identified with RefSNPer.
Author Contributions
K.J. and J.A.H. conceived the study and designed and conducted the experiments. J.A.H. supervised the project. J.A.H., M.G., A.K. and S.S. (Silvia Schmoger) performed and managed investigations regarding the monitoring programs. K.J. and C.D. performed the bioinformatics analyses. K.J., C.D. and J.A.H. analysed these bioinformatics data. K.J. provided the draft manuscript. J.A.H., B.M., D.M., A.K. and S.S. (Stefan Schwarz) decisively contributed to editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The research leading to these results has received funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 773830 (ARDIG). The study was further financially supported by a grant of the German Federal Institute for Risk Assessment (43-001, 1332-648).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aarestrup F.M. The livestock reservoir for antimicrobial resistance: A personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140085. doi: 10.1098/rstb.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 2008;14(Suppl. 1):117–123. doi: 10.1111/j.1469-0691.2007.01851.x. [DOI] [PubMed] [Google Scholar]
- 3.WHO . Antimicrobial Resistance Global Reporton Surveillance. WHO; Geneva, Switzerland: 2014. WHO Library Cataloguing-in-Publication Data. [Google Scholar]
- 4.EFSA The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020;18:e06007. doi: 10.2903/j.efsa.2020.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robicsek A., Jacoby G.A., Hooper D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 6.Cattoir V., Poirel L., Aubert C., Soussy C.J., Nordmann P. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas sp. Emerg. Infect. Dis. 2008;14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Wang T., Shao B., Shen J., Wang S., Wu Y. Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: Potential transfer to agricultural lands. Environ. Health Perspect. 2012;120:1144–1149. doi: 10.1289/ehp.1104776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Martínez J.M., Machuca J., Cano M.E., Calvo J., Martínez-Martínez L., Pascual A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Update. 2016;29:13–29. doi: 10.1016/j.drup.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P., Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 10.Martínez-Martínez L., Pascual A., Jacoby G.A. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 11.Wong M.H., Chan E.W., Liu L.Z., Chen S. PMQR genes oqxAB and aac(6’)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 2014;5:521. doi: 10.3389/fmicb.2014.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D., Wang H., Qi Y., Liang Y., Zhang J., Yu L. Characteristics of Klebsiella pneumoniae harboring qnrB32, aac(6’)-Ib-cr, gyrA and CTX-M-22 genes. Folia Histochem. Cytobiol. 2012;50:68–74. doi: 10.5603/FHC.2012.0009. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H.X., Tang D., Liu Y.H., Zhang X.H., Zeng Z.L., Xu L., Hawkey P.M. Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 2012;67:2350–2353. doi: 10.1093/jac/dks250. [DOI] [PubMed] [Google Scholar]
- 14.Wu H.H., Liu H.Y., Lin Y.C., Hsueh P.R., Lee Y.J. Correlation between levofloxacin consumption and the incidence of nosocomial infections due to fluoroquinolone-resistant Escherichia coli. J. Microbiol. Immunol. Infect. 2016;49:424–429. doi: 10.1016/j.jmii.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Dupouy V., Abdelli M., Moyano G., Arpaillange N., Bibbal D., Cadiergues M.C., Lopez-Pulin D., Sayah-Jeanne S., de Gunzburg J., Saint-Lu N., et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae from Dogs in France and Spain-Characterization of ESBL/pAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019;6:279. doi: 10.3389/fvets.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit . Zoonosen Monitoring Bericht 2017. Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL); Berlin, Germany: 2017. [(accessed on 14 May 2021)]. Available online: https://www.bvl.bund.de/SharedDocs/Downloads/01_Lebensmittel/04_Zoonosen_Monitoring_2017.pdf?_blob=publicationFile&v=4. [Google Scholar]
- 17.The European Commission Commission Implementing Decision of 12 November 2013 on the Monitoring and Reporting of Antimicrobial Resistance in Zoonotic and Commensal Bacteria (2013/652/EU) The European Commission; Official Journal of the European Union, EU; Brussels, Belgium: 2013. [Google Scholar]
- 18.Dortet L., Bonnin R.A., Pennisi I., Gauthier L., Jousset A.B., Dabos L., Furniss R.C.D., Mavridou D.A.I., Bogaerts P., Glupczynski Y., et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018;73:3359–3367. doi: 10.1093/jac/dky330. [DOI] [PubMed] [Google Scholar]
- 19.Holmes D.S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 20.Cattoir V., Poirel L., Rotimi V., Soussy C.J., Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007;60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 21.Kraychete G.B., Botelho L.A., Campana E.H., Picao R.C., Bonelli R.R. Updated Multiplex PCR for Detection of All Six Plasmid-Mediated qnr Gene Families. Antimicrob. Agents Chemother. 2016;60:7524–7526. doi: 10.1128/AAC.01447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PulsNet Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. [(accessed on 29 May 2021)];2017 Available online: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf.
- 23.Juraschek K., Borowiak M., Tausch S.H., Malorny B., Käsbohrer A., Otani S., Schwarz S., Meemken D., Deneke C., Hammerl J.A. Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli. Microorganisms. 2021;9:598. doi: 10.3390/microorganisms9030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borowiak M., Fischer J., Hammerl J.A., Hendriksen R.S., Szabo I., Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsenterica serovar Paratyphi, B. J. Antimicrob. Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 25.Deneke C., Brendebach H., Uelze L., Borowiak M., Malorny B., Tausch S.H. Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with AQUAMIS. Genes. 2021;12:644. doi: 10.3390/genes12050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.H., McDermott P.F., et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann T. Abricate. [(accessed on 29 May 2021)];2014 Available online: https://github.com/tseemann/abricate.
- 30.Deneke C. BakCharak. [(accessed on 9 January 2020)];2018 Available online: https://gitlab.com/bfr_bioinformatics/bakcharak.
- 31.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L., et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadley W. ggplot2: Elegant Graphics for Data Analysis. Springer; New York, NY, USA: 2016. [Google Scholar]
- 33.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T. GGTREE: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 34.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carattoli A., Zankari E., García-Fernández A., Voldby L.M., Kund O., Moller Aarestrup F., Hasman H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deneke C. refSNPer. [(accessed on 9 January 2020)];2019 Available online: https://gitlab.com/bfr_bioinformatics/refsnper/
- 39.Grützke J., Gwida M., Deneke C., Brendebach B., Projahn M., Schattschneider A., Hofreuter D., El-Ashker M., Malorny B., Al Dahouk S. Direct identification and molecular characterization of zoonotic hazards in raw milk by metagenomics using Brucella as a model pathogen. Microb. Genom. 2021;7:000552. doi: 10.1099/mgen.0.000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EFSA The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018;16:5182. doi: 10.2903/j.efsa.2018.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.EFSA The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019;17:e05598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kindle P., Zurfluh K., Nüesch-Inderbinen M., von Ah S., Sidler X., Stephan R., Kümmerlen D. Phenotypic and genotypic characteristics of Escherichia coli with non-susceptibility to quinolones isolated from environmental samples on pig farms. Porc. Health Manag. 2019;5:9. doi: 10.1186/s40813-019-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilani H., Ferjani S., Mansouri R., Boutiba-Benboubaker I., Salah Abbassi M. Occurrence of plasmid-mediated quinolone resistance determinants among Escherichia coli strains isolated from animals in Tunisia: Specific pathovars acquired qnr genes. J. Glob. Antimicrob. Resist. 2020;20:50–55. doi: 10.1016/j.jgar.2019.07.023. [DOI] [PubMed] [Google Scholar]
- 46.Potron A., Poirel L., Bernabeu S., Monnet X., Richard C., Nordmann P. Nosocomial spread of ESBL-positive Enterobacter cloacae co-expressing plasmid-mediated quinolone resistance qnr determinants in one hospital in France. J. Antimicrob. Chemother. 2009;64:653–654. doi: 10.1093/jac/dkp222. [DOI] [PubMed] [Google Scholar]
- 47.Kawamura K., Nagano N., Suzuki M., Wachino J.I., Kimura K., Arakawa Y. ESBL-producing Escherichia coli and Its Rapid Rise among Healthy People. Food Saf. 2017;5:122–150. doi: 10.14252/foodsafetyfscj.2017011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Tian G.B., Zhang R., Shen Y., Tyrrell J.M., Huang X., Zhou H., Lei L., Li H.Y., Doi Y., et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1 -positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017;17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 49.Madec J.Y., Haenni M. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid. 2018;99:72–81. doi: 10.1016/j.plasmid.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Segura W.D., Ramos H.P., de Faria Blanc Amorim R.E., da Silva Ribeiro Á.C., Pereira E.C., Cayo R., Gales A.C., Piantino Ferreira A.J., da Rocha Minarini L.A. In vitro and in vivo persistence of IncN plasmids carrying qnr genes in uropathogenic Escherichia coli isolates. J. Glob. Antimicrob. Resist. 2020;22:806–810. doi: 10.1016/j.jgar.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Falgenhauer L., Imirzalioglu C., Oppong K., Akenten C.W., Hogan B., Krumkamp R., Poppert S., Levermann V., Schwengers O., Sarpong N., et al. Detection and Characterization of ESBL-Producing Escherichia coli From Humans and Poultry in Ghana. Front. Microbiol. 2019;9:3358. doi: 10.3389/fmicb.2018.03358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu B., Qi Q., Zhang X., Cai Y., Yu G., Lv J., Gao L., Wei L., Chai T. Dissemination of Escherichia coli carrying plasmid-mediated quinolone resistance (PMQR) genes from swine farms to surroundings. Sci. Total Environ. 2019;665:33–40. doi: 10.1016/j.scitotenv.2019.01.272. [DOI] [PubMed] [Google Scholar]
- 53.Allen H.K., Stanton T.B. Altered Egos: Antibiotic Effects on Food Animal Microbiomes. Annu. Rev. Microbiol. 2014;68:297–315. doi: 10.1146/annurev-micro-091213-113052. [DOI] [PubMed] [Google Scholar]
- 54.Salah F.D., Soubeiga S.T., Ouattara A.K., Sadji A.Y., Metuor-Dabire A., Obiri-Yeboah D., Banla-Kere A., Karou S., Simpore J. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spin Lomé, Togo. Antimicrob. Resist. Infect. Control. 2019;8:104. doi: 10.1186/s13756-019-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan M.T., Chou C.C. Class 1 Integrons and the Antiseptic Resistance Gene (qacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health. 2015;12:6249–6260. doi: 10.3390/ijerph120606249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong T.Z., Zhang M., O’Donoghue M., Boost M. Presence of antiseptic resistance genes in porcine methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 2013;162:977–979. doi: 10.1016/j.vetmic.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 57.Zhang A., He X., Meng Y., Guo L., Long M., Yu H., Li B., Fan L., Liu S., Wang H., et al. Antibiotic and Disinfectant Resistance of Escherichia coli Isolated from Retail Meats in Sichuan, China. Microb. Drug Resist. 2016;22:80–87. doi: 10.1089/mdr.2015.0061. [DOI] [PubMed] [Google Scholar]
- 58.Raeispour M., Ranjbar R. Antibiotic resistance, virulence factors and genotyping of Uropathogenic Escherichia coli strains. Antimicrob. Resist. Infect. Control. 2018;7:118. doi: 10.1186/s13756-018-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahdar M., Rashki A., Miri H.R., Rashki Ghalehnoo M. Detection of pap, sfa, afa, foc, and fim Adhesin-Encoding Operons in Uropathogenic Escherichia coli Isolates Collected From Patients with Urinary Tract Infection. Jundishapur J. Microbiol. 2015;8:e22647. doi: 10.5812/jjm.22647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreux N., Denizot J., Martinez-Medina M., Mellmann A., Billig M., Kisiela D., Chattopadhyay S., Sokurenko E., Neut C., Gower-Rousseau C., et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang X., Sun H., Fan R., Fu S., Zhang J., Matussek A., Xiong Y., Bai X. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci. Rep. 2020;10:3275. doi: 10.1038/s41598-020-60225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eltai N.O., Al Thani A.A., Al Hadidi S.H., Al Ansari K., Yassine H.M. Antibiotic resistance and virulence patterns of pathogenic Escherichia coli strains associated with acute gastroenteritis among children in Qatar. BMC Microbiol. 2020;20:54. doi: 10.1186/s12866-020-01732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez-Etayo L., González D., Vitas A.I. The Aquatic Ecosystem, a Good Environment for the Horizontal Transfer of Antimicrobial Resistance and Virulence-Associated Factors Among Extended Spectrum β-lactamases Producing E. coli. Microorganisms. 2020;8:568. doi: 10.3390/microorganisms8040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaper J.B., Nataro J.P., Mobley H.L. Pathogenic Escherichia coli. Nat. Rev. Genet. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 65.Eikmeyer F., Hadiati A., Szczepanowski R., Wibber D., Schneiker-Bekel S., Rogers L.M., Brown C.J., Top E.M., Pühler A., Schlüter A. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid. 2012;68:13–24. doi: 10.1016/j.plasmid.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 66.Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H., Miao M., Yan J., Wang M., Tang Y.W., Kreiswirth B.N., Zhang X., Chen L., Du H. Expression characteristics of the plasmid-borne mcr-1 colistin resistance gene. Oncotarget. 2017;8:107596–107602. doi: 10.18632/oncotarget.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burmølle M., Norman A., Sørensen S.J., Hansen L.H. Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS ONE. 2012;7:e41259. doi: 10.1371/journal.pone.0041259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hasman H., Hammerum A.M., Hansen F., Hendriksen R.S., Olesen B., Agersø Y., Zankari E., Leekitcharoenphon P., Stegger M., Kaas R.S., et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eurosurveillance. 2015;20 doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 70.Falgenhauer L., Waezsada S.E., Gwozdzinski K., Ghosh H., Doijad S., Bunk B., Spröer C., Imirzalioglu C., Seifert H., Irrgang A., et al. Chromosomal Locations of mcr-1 and blaCTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg. Infect. Dis. 2016;22:1689–1691. doi: 10.3201/eid2209.160692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobiasova H., Dolejska M. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and β-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 2016;71:2118–2124. doi: 10.1093/jac/dkw144. [DOI] [PubMed] [Google Scholar]
- 72.Xia R., Ren Y., Xu H. Identification of Plasmid-Mediated Quinolone Resistance qnr Genes in Multidrug-Resistant Gram-Negative Bacteria from Hospital Wastewaters and Receiving Waters in the Jinan Area, China. Microb. Drug Resist. 2013;19:446–456. doi: 10.1089/mdr.2012.0210. [DOI] [PubMed] [Google Scholar]
- 73.Yamada M., Yoshida J., Hatou S., Yoshida T., Minagawa Y. Mutations in the quinolone resistance determining region in Staphylococcus epidermidis recovered from conjunctiva and their association with susceptibility to various fluoroquinolones. Br. J. Ophthalmol. 2008;92:848–851. doi: 10.1136/bjo.2007.129858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dasgupta N., Paul D., Chanda D.D., Chetri S., Chakravarty A., Bhattacharjee A. Observation of a New Pattern of Mutations in gyrA and parC within Escherichia coli Exhibiting Fluroquinolone Resistance. Indian J. Med. Microbiol. 2018;36:131–135. doi: 10.4103/ijmm.IJMM_17_181. [DOI] [PubMed] [Google Scholar]
- 75.Vingopoulou E.I., Siarkou V.I., Batzias G., Kaltsogianni F., Sianou E., Tzavaras I., Koutinas A., Saridomichelakis M.N., Sofianou D., Tzelepi E., et al. Emergence and maintenance of multidrug-resistant Escherichia coli of canine origin harbouring a blaCMY-2-IncI1/ST65 plasmid and topoisomerase mutations. J. Antimicrob. Chemother. 2014;69:2076–2080. doi: 10.1093/jac/dku090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material.