Abstract
Coumarins belong to a group of secondary metabolites well known for their high biological activities including antibacterial and antifungal properties. Recently, an important role of coumarins in plant resistance to pathogens and their release into the rhizosphere upon pathogen infection was discovered. It is also well documented that coumarins play a crucial role in the Arabidopsis thaliana growth under Fe-limited conditions. However, the mechanisms underlying interplay between plant resistance, accumulation of coumarins and Fe status, remain largely unknown. In this work, we investigated the effect of both mentioned factors on the disease severity using the model system of Arabidopsis/Dickeya spp. molecular interactions. We evaluated the disease symptoms in Arabidopsis plants, wild-type Col-0 and its mutants defective in coumarin accumulation, grown in hydroponic cultures with contrasting Fe regimes and in soil mixes. Under all tested conditions, Arabidopsis plants inoculated with Dickeya solani IFB0099 strain developed more severe disease symptoms compared to lines inoculated with Dickeya dadantii 3937. We also showed that the expression of genes encoding plant stress markers were strongly affected by D. solani IFB0099 infection. Interestingly, the response of plants to D. dadantii 3937 infection was genotype-dependent in Fe-deficient hydroponic solution.
Keywords: abiotic stress, biotic stress, fraxetin, iron deficiency, scopoletin, pathogen, plant–environment interactions, mineral nutrition
1. Introduction
The secretion of phenolic compounds from roots into the rhizosphere has long been recognised as a component of the acidification-reduction strategy to acquire iron (Fe), occurring in all plant species except grasses [1]. However, the molecular mechanisms underlying these processes remained elusive until recently, when several research groups including our team, independently demonstrated the important role of plant secondary metabolites called coumarins for the growth of a model plant Arabidopsis thaliana (hereafter Arabidopsis) under Fe-limited conditions [2,3,4,5,6,7,8,9]. It was proven that coumarins are involved in Fe chelation and that secretion of coumarins by Arabidopsis roots is induced under Fe-deficiency. The biological roles of novel enzymes involved in coumarin biosynthesis, which in parallel maintain Fe homeostasis in plants, were elucidated. A key enzyme for the biosynthesis of Arabidopsis major coumarin called scopoletin and its derivatives is Feruloyl-CoA 6′-Hydroxylase1 (F6′H1) that belongs to a large enzyme family of the 2-oxoglutarate and Fe(II)-dependent dioxygenases [3,5,10]. Our group elucidated the biological role of another member of this family, encoded by a strongly Fe-responsive gene (At3g12900), as a scopoletin 8-hydroxylase (S8H) involved in the last step of fraxetin biosynthesis [7]. Fraxetin is a coumarin derived from the scopoletin pathway, containing two adjacent hydroxyl groups in the ortho-position that can efficiently solubilise Fe from the hydroxide precipitates [3,8]. We proved S8H to be involved in coumarin biosynthesis as part of the Fe acquisition machinery [7].
Fe is a crucial micronutrient for every kind of living organism. It plays an essential role in metabolic processes such as DNA synthesis, respiration, photosynthesis, and it is a cofactor of many enzymes. The role of Fe homeostasis in resistance to infections was also shown across all kingdoms of life—different types of pathogens are likely to compete with their hosts for the acquisition of Fe [11]. Mechanisms of Fe homeostasis in plants, pathogens, and beneficial microorganisms play key roles in plant-microbe interactions [12]. Moreover, one-third of the world’s agricultural area is composed of calcareous soils, in which high pH leads to the precipitation of Fe that is finally not available and generate severe plant growth perturbation. Therefore, Fe deficiency is a widespread agricultural problem that reduces plant growth and crop yields, particularly in alkaline soils [13].
In addition to the important role in maintaining Fe homeostasis, coumarins can affect plant growth and fitness directly through their high biological activities including antibacterial and antifungal properties. Scopoletin that is one of the major Arabidopsis coumarins accumulating in roots [5,7,14], was shown to possess antimicrobial activity against various phytopathogens like Ralstonia solanacearum [15], Alternaria alternata [16], Botrytis cinerea [17], Fusarium oxysporum f. sp. batatas [18], Sclerotinia sclerotiorum [19], Aspergillus flavus, and Aspergillus niger [20], Ceratocystis fimbriata f. sp. platani [21] and acting against human pathogens [22] including multidrug-resistant Pseudomonas aeruginosa strains [23], Salmonella typhi [24] and clinical isolates of Staphylococcus aureus [25].
Recently, an important role for coumarins in microbiome modulation was demonstrated. It was shown that plant-derived coumarins shape the composition of Arabidopsis root microbial communities (rhizobiome) in Fe-starved plants, and possibly protect plants from pathogenic fungi [26]. Coumarins were shown to influence a reduced synthetic community (SynCom) of Arabidopsis root-isolated bacteria in synthetic media [27,28] and were proved to alter the rhizobiome and improve plant growth in Fe-limiting soil [29]. Even if, a role of root-exuded coumarins in structuring the rhizobiome was uncovered and their release into the rhizosphere upon pathogen infection was confirmed, the precise mechanisms underlying the above-described processes are only beginning to be discovered [26,28,30,31].
In the literature, there are examples of pathogens causing more severe disease symptoms on plants grown under high-Fe conditions when compared to Fe-deficient plants. One of the examples could be plant pathogenic bacteria Dickeya dadantii 3937, for which sufficient Fe uptake is essential to manifest full virulence on plants [32]. This bacterial pathogen causes soft rot and blackleg disease devastating potato and numerous other crops [33,34,35]. Bacteria from the Dickeya genus produce compounds called siderophores that form complexes with Fe and make it available to the microorganism. Taking into account that plants also produce Fe-chelating compounds, siderophore production is a part of the competitive relationships between plants and microorganisms that can promote infection [32]. It was demonstrated that Fe nutrition strongly affects the disease caused by D. dadantii 3937 also in a model plant Arabidopsis [36,37,38]. In Fe-starved Arabidopsis plants, authors observed a reduction in the expression of major bacterial virulence genes and finally a lower progression in disease symptoms on inoculated plants. The results obtained from a study of Arabidopsis response to D. dadantii 3937 infection highlight the major importance of the competition between plant and bacterial cells for Fe uptake during infection [36,38].
The above results reinforced the important role of coumarins in plant responses to disturbed Fe availability as well as their involvement in plant resistance and their release into the rhizosphere upon pathogen infection. The physiological functions of coumarins are strictly related to plant adaptation to various biotic and abiotic environmental stresses. Here, we investigated the Arabidopsis/Dickeya spp. pathosystem to better understand the relation between coumarins, plant Fe status, siderophores production and plant resistance to selected pathogenic bacteria.
2. Results
We used as a model, Arabidopsis wild-type plants (WT Col-0) and its mutants (s8h and f6′h1) defective in enzymes involved in coumarin biosynthesis (S8H and F6′H1, respectively) and pdr9 mutant defective in coumarin transport to the rhizosphere (PDR9: Pleiotropic drug resistance 9 [2]). As plant pathogens, we included two Dickeya spp. strains: (1) a reference strain D. dadantii 3937 of medium virulence, (2) and Dickeya solani IFB0099 isolated from infected potato plant in Poland [39,40,41]. Interestingly, the selected strains differed in their ability to chelate Fe ions [40]. The ability to chelate Fe ions by D. dadantii 3937 strain was shown to be twice as high as the ability to chelate Fe ions by D. solani IFB0099 strain on CAS-agar medium [40].
2.1. Differential Susceptibility of Arabidopsis Plants Grown in Fe-Deficient Hydroponics to Tested Dickeya spp. Strains
To strictly control the growth conditions and maintain the nutrient composition of media, we conducted the hydroponic cultures (as described by [7] and [42]). Arabidopsis plants were cultivated in controlled conditions either under optimal (40 μM Fe2+) or Fe-deficient conditions (0 μM Fe2+) that induces coumarins accumulation [42] and subsequently were inoculated with D. dadantii 3937 and D. solani IFB0099 spp. It was reported previously that Arabidopsis roots release more phenolic-related compounds at later stages of life [43], therefore plants were inoculated at the flowering stage. We inoculated the Arabidopsis WT Col-0 and mutant lines defective in coumarin biosynthesis (f6′h1, s8h) with both Dickeya spp. strains, and evaluated disease progression according to the visual symptoms scoring with disease severity scale 0–5 (DSS) which allowed us to quantify the susceptibility of Arabidopsis plants to both bacterial strains (Figure 1).
Figure 1.

Disease severity scale (DSS) on Arabidopsis thaliana wild-type (WT) Col-0 leaves inoculated with D. dadantii 3937 and/or D. solani IFB0099. Example images representing stages of DSS were taken 48 h after inoculation. The DSS was assigned to 0–5 scale and was defined as: (0) for no signs of symptoms of the disease and the wound has healed (observed for the mock control); (1) the necrotic tissue was observed in the inoculation site only; (2) the necrotic tissue observed in the inoculation site and max. 3 mm wide around it; (3) the maceration visible around the inoculation site spreading further with possible chlorosis of the leaf; (4) visible maceration of the whole leaf, possible chlorosis of other leaves, no maceration of the limb; (5) visible maceration of the whole limb and the leaf.
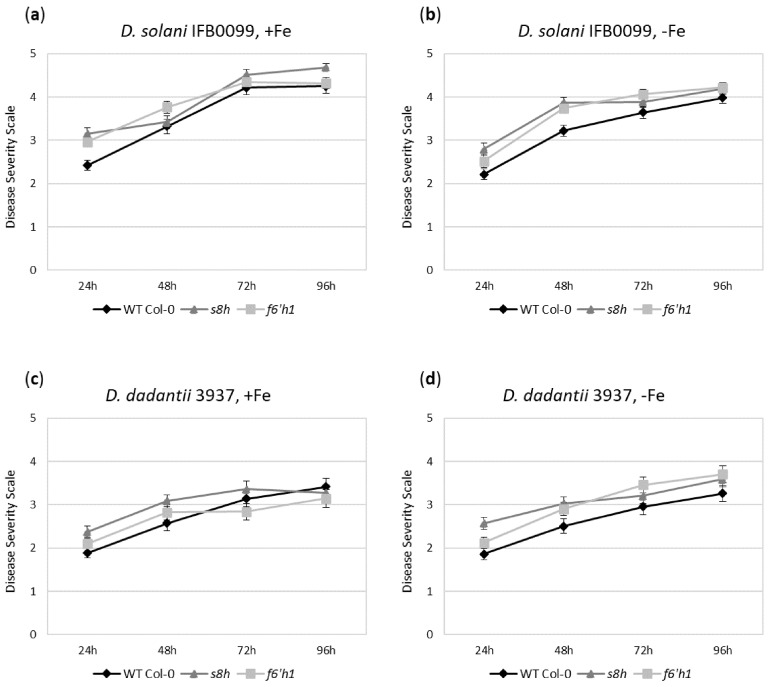
We observed that independently of genetic characteristic, all Arabidopsis genotypes inoculated with D. solani IFB0099 strain developed more severe disease symptoms (DSS up to 4.5) (Figure 2a,b) when compared to D. dadantii 3937 (DSS up to 3.5) (Figure 2c,d). It is worth emphasizing that in case of all plant genotypes inoculated with D. solani IFB0099 strain, the symptoms of infection were more pronounced in conditions with optimal Fe availability (Figure 2a,b). Both mutants, in particular the s8h line, showed slightly higher disease symptoms after inoculation with D. solani IFB0099 when compared to WT Col-0 plants. Interestingly, when WT Col-0 plants were inoculated with D. dadantii 3937 strain, they developed the most severe disease symptoms at 96 h after inoculation when grown in optimal Fe solution (Figure 2c), while the opposite trend was detected for both Arabidopsis mutants with impaired biosynthesis of coumarins. During the D. dadantii 3937 infection, both Arabidopsis mutant lines showed a tendency to exhibit more severe disease symptoms in Fe-deficient hydroponics (Figure 2d) and not in the Fe-sufficient solution as WT Col-0 plants. This was particularly striking at 72 and 96 h after inoculation for the f6′h1 mutants (Figure 2c,d), lacking the functional F6′H1 gene, which does not synthesise scopoletin and its derivatives.
Figure 2.
Disease progression caused by Dicekya solani IFB0099 (a,b) and Dicekya dadantii 3937 (c,d) strains on Arabidopsis thaliana wild-type (WT) Col-0 plants, s8h and f6′h1 mutant lines grown in optimal (+Fe; 40 µM Fe2+) (a,c) or Fe-deficient (−Fe, 0 µM Fe2+) (b,d) hydroponic cultures by visual symptom scoring (Disease Severity Scale, DSS). The values represent the mean values of DSS originating from two independent experiments, in each experiment numerous individuals (n = 5–9) per plant genotypes (three leaves per plant) were inoculated for each time point. It is worth noting that the results averaged the DSS values obtained for two independent mutant lines for each tested gene. The mock-inoculated plants (with 0.85% NaCl) did not show the symptoms of the disease progression throughout the experiment. Error bars represent ± standard error (SE).
2.2. Inoculation of Arabidopsis WT Col-0 Grown in Fe-Deficient Hydroponics with Dickeya spp. Cause Decrease in the Expression of S8H and F6′H1 Genes Involved in Coumarin Biosynthesis
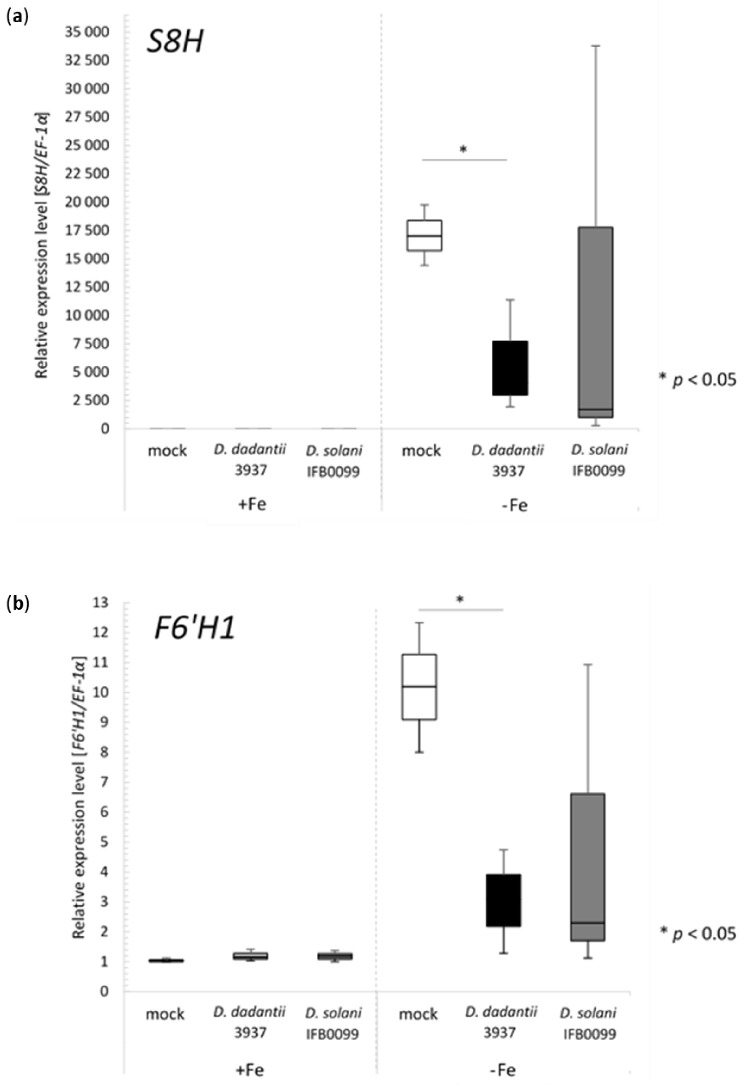
Next, to get insight into the expression levels of genes encoding key enzymes (S8H, F6′H1) involved in the biosynthesis of coumarins that are accumulated mostly in the underground part of a plant, we performed the qPCR analysis with cDNA reverse transcribed using RNA isolated from the WT Col-0 roots grown under different Fe-regimes and inoculated with D. dadantii 3937 or D. solani IFB0099 strains. As expected, we observed up-regulation of both genes (S8H and F6′H1) in the Fe-deficient condition (Figure 3a,b). In particular, the S8H gene, which is known to be one of the most strongly Fe-responsive genes, was induced several hundred times in all treatments tested in Fe-deficient conditions compared to Fe-sufficient condition (Figure 3a).
Figure 3.
The relative expression levels of S8H (a) and F6′H1 (b) genes were analysed in the roots of Arabidopsis thaliana wild-type (WT) Col-0 grown in hydroponics under optimal (40 μM Fe2+) or Fe-deficient conditions (0 μM Fe2+) and inoculated with Dickeya dadantii 3937 and Dickeya solani IFB0099 strains. Control plants were mock-inoculated with a 0.85% NaCl solution. As a reference, the EF-1α (ang. elongation factor-1α, At5g60390) gene was selected [44]. The pairwise t-test was used. Error bars, ±SD, from three biological replicates. * p < 0.05.
Interestingly, the expression of the S8H gene was significantly higher in the roots exposed only to abiotic stress (mock-inoculated WT Col-0 plants grown under Fe deficiency) when compared with those exposed to abiotic and biotic stress (Dickeya spp.- inoculated ones grown under Fe deficiency). It should be noted that the expression of S8H was relatively lower when combined environmental stress, composed of Fe-deficiency and bacterial infection, was applied (Figure 3a: right panel). This relative decrease was particularly significant when D. dadantii 3937 was used to inoculate WT Col-0 plants (p < 0.05). A similar trend was observed for the F6′H1 gene, for which a relative reduction in the expression levels was observed when the same two stress factors, Fe-deficiency and bacterial inoculation, were simultaneously applied. The F6′H1 expression was approximately 2-fold lower in WT Col-0 plants subjected to biotic stress when compared to plant exposed only to abiotic stress under Fe-deficient condition (Figure 3b). In this case, the inoculation with D. dadantii 3937 also had a significant effect on the relative reduction of F6′H1 expression in the WT Col-0 genetic background (p < 0.05). That is an interesting observation considering that this strain expressed a greater ability than D. solani IFB0099 to chelate Fe ions, as shown previously by the CAS-agar plate assay [40].
2.3. Fe-Chelation in CAS Agar Plate Assay
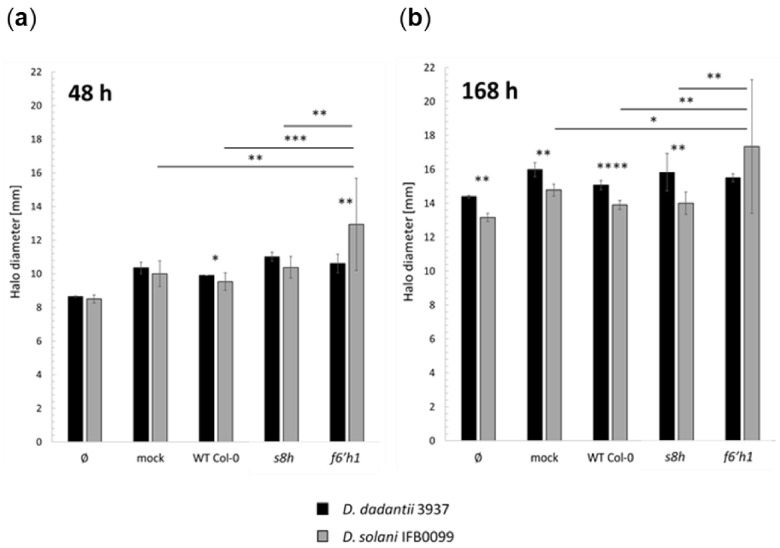
To test the potential of plant-produced compounds to affect the bacterial siderophore production, we observed the growth and halo production of D. dadantii 3937 and D. solani IFB0099 strains on CAS-agar plates supplemented with homogenates prepared from the leaves of s8h and f6′h1 mutants (Figure 4a,b) that were grown in Fe-depleted hydroponic solution (0 µM Fe2+).
Figure 4.
Siderophore production by Dickeya dadantii 3937 and Dickeya solani IFB0099 after incubation for (a) 48 and (b) 168 h on CAS-agar plates supplemented with leaf homogenates of Arabidopsis thaliana wild-type (WT) Col-0 and s8h or f6′h1 mutant plants. The halo diameters were measured [mm] on control CAS-agar medium with no supplements (Ø), mock (phosphate buffer) and CAS-agar medium supplemented with leaf homogenates prepared from the Col-0 WT plants or f6′h1, s8h mutants. The experiment was performed twice, with 12 replicates (a set of pooled 3 plants was used for the biological replicates). The results are presented as average halo diameter. Error bars represent ± standard deviation (SD). Statistical significance is marked with asterisks * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 based on student’s T-test for independent samples.
Here, we selected leaves to prepare homogenates, as organs in which inoculation is conducted. No supplements or homogenates were added as a negative control (Ø), as a mock the phosphate buffer was used. It seems that both leaf homogenates and phosphate buffer itself can induce the production of bacterial siderophores. As could be expected, in most cases D. dadantii 3937 strain that was previously characterised by a higher ability to chelate Fe ions [40], produced larger halos compared to D. solani IFB0099 (Figure 4a,b). But the most interestingly, we observed the opposite effect when homogenates originating from the leaves of f6′h1 mutant were used as a supplement. We observed a significant increase in D. solani IFB0099 halos’ diameter on agar plates supplemented with the f6′h1 mutant leaf homogenates (Figure 4a,b).
2.4. Differential Susceptibility of Soil-Grown Arabidopsis Plants to Tested Dickeya spp. Strains
To shed light on the relationship between Fe homeostasis, coumarin accumulation and plant immunity in more physiological conditions, we grew a set of Arabidopsis mutants defective in coumarin accumulation (f6′h1, s8h) in the non-sterile soil environment. Here, we included in the experiment the pdr9 mutant that is defective in coumarin transport to the rhizosphere [2]. Since soil mixture composition can alter Arabidopsis susceptibility to plant pathogens as was shown for the Pseudomonas syringae infection [45], we decided to estimate the disease symptoms caused by D. dadantii 3937 and D. solani IFB0099 on Arabidopsis plants grown in two different soil mixes (#1 and #2) derived from the commercial products that differ mainly in the level of salinity that can affect the availability of nutrients including Fe, and the content of some macro- and micronutrients like chlorides, phosphorus, potassium or calcium (Table 1).
Table 1.
Chemical analysis of used soil mixes: (a) pH, salinity, macro- and (b) micronutrients.
| (a) | ||||||||
|
Soil Mix
no. |
pH in H2O | NaCl g/dm3 Soil | NO3 | Cl | P | K | Ca | Mg |
| mg/dm3 Soil | ||||||||
| #1 | 6.8 | 2.65 | 224 | 13.6 | 34.6 | 70.1 | 2960 | >400 (548) 1 |
| #2 | 6.9 | 1.59 | 220 | 10.5 | 44.5 | 91.8 | 2509 | >400 (498) 1 |
| (b) | ||||||||
|
Soil Mix
no. |
Cu | Zn | Mn | Fe | B | |||
| mg/dm3 Soil | ||||||||
| #1 | 1.1 | 1.1 | 1.3 | 51.4 | 0.4 | |||
| #2 | 1.0 | 1.3 | 2.3 | 49.2 | 0.4 | |||
1 Results above upper limit of the method range for Mg = 400 mg/dm3.
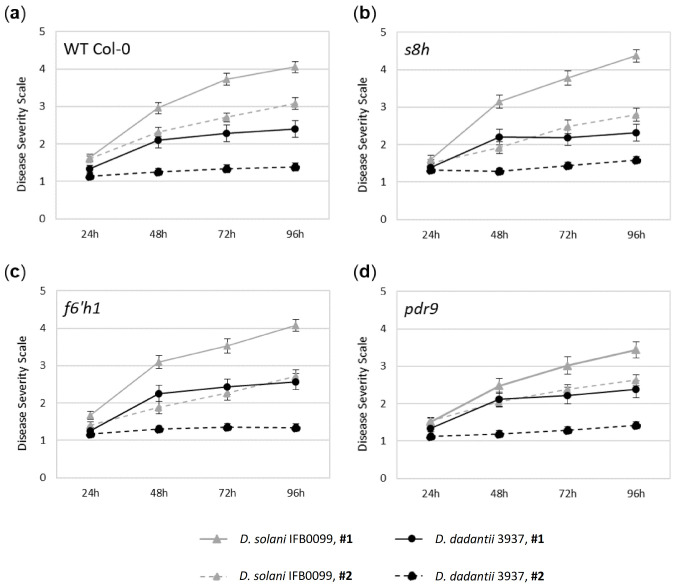
Arabidopsis plants of all genotypes (WT Col-0, f6′h1, s8h and pdr9) had better performance on soil mix #1, on which the plant rosettes were much bigger. In case of soil mix #2, the spontaneous plant wilt and die was also observed. This is an interesting observation since the conducted chemical analysis did not show any dramatic differences in the compositions of both soil mixes (Table 1). However, regardless of the soil in which the plants were grown, the Arabidopsis of all tested genotypes inoculated with D. solani IFB0099 strain developed more severe infection symptoms (DSS up to 4.5), compared to those challenged with D. dadantii 3937 reference strain (DSS up to 2.5) (Figure 5). However, most interestingly, we detected a variation in the disease symptoms between plant genotypes inoculated with the D. solani IFB0099 strain. In soil mix #1 characterised by a slightly higher salinity, the pdr9 mutants inoculated with D. solani IFB0099 showed the mildest infection symptoms among all plant genotypes with the severity score up to 3.5 (Figure 5d) compared with the DSS up to 4.0 for WT Col-0 and f6′h1 plants (Figure 5a,c) and 4.5 for s8h mutant line (Figure 5b). In soil mix #2, a slightly lower DSS (lower than 3) was observed for all mutant lines inoculated with D. solani IFB0099 (Figure 5b–d) compared to WT Col-0 plants (DSS up to 3) (Figure 5a). We did not observe such a variation in the infection symptoms on plants inoculated with the D. dadantii 3937 strain.
Figure 5.
Disease progression caused by Dickeya dadantii 3937 and Dickeya solani IFB0099 strains on Arabidopsis thaliana (a) wild-type (WT) Col-0 plants, (b) s8h, (c) f6′h1 and (d) pdr9 mutant lines by visual symptom scoring (Disease Severity Scale, DSS). Plants were grown in two types of soil mixes (#1 and #2, see Table 1). The values represent the mean values of DSS originating from two independent experiments, in each experiment numerous individuals (n = 6–8) per plant genotypes (three leaves per plant) were inoculated for each time point. The mock-inoculated plants (with 0.85 % NaCl) did not show the symptoms of the disease progression throughout the experiment. Error bars represent ± standard error (SE).
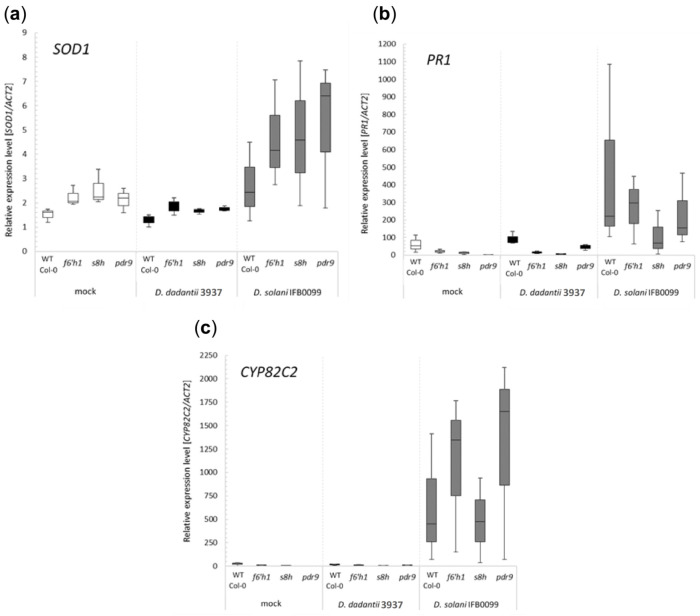
2.5. Expression of Selected Plant Stress-Response Genes Is Strongly Induced in Arabidopsis Mutants Defective in Coumarin Accumulation Inoculated with D. solani IFB0099
Next, to measure the expression levels of selected genes being the plant stress markers, we analysed by qPCR the expression of genes related to oxidative stress (SOD1, AT1G08830), plant defence (pathogenesis-related PR1, At2g14610) and modulation of jasmonate-induced root growth inhibition (CYP82C2, At4g31970). We used RNA isolated from the leaves of Arabidopsis WT Col-0 and three mutant lines (f6′h1, s8h, pdr9) grown in soil mix #1, in which the disease symptoms after D. solani IFB0099 or D. dadantii 3937 strains infection were more prominent compared to soil mix #2 (Figure 5).
We observed a strong increase in expression of selected plant stress markers in plants inoculated with D. solani IFB0099 strain, which indicate that inoculation of Arabidopsis plants with this pathogen particularly induce the plant defence systems (Figure 6a–c). Interestingly, the expression levels of two out of three tested genes encoding plant stress markers (SOD1, PR1) were differentially induced among mutants and WT Col-0 plants, those inoculated with Dickeya spp. strains and mock-inoculated ones (Figure 6a,b). The level of the SOD1 gene was visibly higher in all mutant lines compared to WT Col-0 plants (Figure 6a). While in the case of the PR1 gene, we observed the opposite effect, where its expression was higher in WT Col-0 plants compared to mutants with disturbed coumarin biosynthesis (f6′h1, s8h) or transport (pdr9) (Figure 6b). The transcript levels of CYP82C2 were specifically induced by D. solani IFB0099 infection in all tested plant genotypes (Figure 6c).
Figure 6.
Relative expression levels of plant stress marker genes (a) SOD1 (AT1G08830), (b) PR1 (At2g14610) and (c) CYP82C2 (At4g31970) analysed in the leaves of Arabidopsis thaliana wild-type (WT) Col-0 and mutant plants (f6′h1, s8h, pdr9) grown in soil mix #1 and inoculated with Dickeya dadantii 3937 and Dickeya solani IFB0099. 0.85% NaCl treated plants were used as a negative control. As a reference, the ACT2 (At3g18780) gene was used [44]. Error bars, ±SD, from three biological replicates.
3. Discussion
Microorganisms that urgently need Fe for their growth, replication, metabolism and the infectious disease process, have evolved numerous strategies for Fe acquisition such as siderophore production. At the same time, plants are constantly subjected to various environmental stresses, including Fe-deficiency or pathogen attack during which Fe itself plays an important role. During a microbial infection, there is a competition between host and pathogen for the necessary nutritional resources. Numerous studies have shown that Fe ions play a key role in such competitive relationships [12,46,47,48,49]. It was also shown recently by several groups, including our research team, that the secretion of coumarins is essential for Fe acquisition under Fe-deficient conditions in a model plant Arabidopsis thaliana [7]. Plants and microorganisms have evolved a set of active strategies for Fe uptake from the soil that are based on acidification, chelation and reduction processes. Root exudation is one of such important processes determining the interaction of plants with the soil environment and microbiome. Coumarins that are secreted to the rhizosphere by roots are involved in several processes determining plant interactions with the soil environment, both with biotic and abiotic factors.
In this study, we evaluated the disease symptoms caused by Dickeya spp. strains in Arabidopsis lines differing in coumarin accumulation that were grown under various growth conditions and Fe availability. The use of selected Arabidopsis mutants and bacterial strains of different origin enabled us to compare [1] the variation in disease symptoms among plant genotypes under numerous environmental scenarios, [2] and the expression of stress-related genes in plant genetic backgrounds with disturbed production and distribution of coumarins.
The presented analyses provided interesting insights into the differences in responses of the following plant genotypes: control plants (WT Col-0), coumarin-reach plants (WT Col-0 growing in Fe deficient environment), coumarin-deficient plants (f6′h1), fraxetin-deficient plants (s8h) and coumarin-hyperaccumulating plants (pdr9) to D. dadantii 3937 and D. solani IFB0099 strains. The developed model system of Arabidopsis/Dickeya spp. was applied to investigate the effect of two abiotic factors (Fe availability and coumarin content) on the disease severity. These studies are in line with the latest Top 10 Questions, selected by the International Congress on Molecular Plant-Microbe Interactions (IC-MPMI) community that met in Glasgow in 2019, covering the need to understand how the abiotic environment influence specific plant-microbe interactions [50]. It should be also highlighted that most of the previously published data describe the interaction between Arabidopsis plants and D. dadantii 3937. According to our best knowledge D. solani, which is an important plant pathogenic bacterium causing a loss in potato yield in Europe [34,51,52] was not tested before with a model plant Arabidopsis. Moreover, recent results show that D. solani strains cause severe disease symptoms in temperate climates, and are more aggressive than other blackleg-causing bacteria from genus Dickeya and Pectobacterium spp. [33,35,53,54].
To shed light on the strong relationship between Fe homeostasis (abiotic factor), coumarin accumulation and plant susceptibility to plant pathogenic bacteria, we grew WT Col-0 plants and two Arabidopsis mutants defective in coumarin accumulation (s8h and f6′h1) in the hydroponic cultures with strictly controlled Fe content and inoculated them with Dickeya spp. strains (biotic stress factor). We observed that all tested Arabidopsis genotypes (WT Col-0, s8h and f6′h1) inoculated with D. solani IFB0099 strain developed more severe disease symptoms than plants inoculated with D. dadantii 3937. The disease symptoms associated with D. solani IFB0099 infection were much more pronounced in Fe-sufficient hydroponics. A similar effect was observed for D. dadantii 3937-inoculated wild-type plants (WT Col-0). This is in line with the literature data showing that D. dadantii belongs to the pathogens causing more severe disease symptoms on plants grown under high-Fe conditions when compared to Fe-deficient environmental condition [32]. The most noticeable was a detection of the opposite effect for Arabidopsis mutants with impaired biosynthesis of coumarins. During D. dadantii 3937 infection of s8h and f6′h1 mutant plants, the more severe disease symptoms were observed in Fe-deficient hydroponics, and not in the Fe-sufficient cultures, particularly at 72 h after inoculation. Both of these mutants (s8h and f6′h1) are defective in enzymes involved in the biosynthesis of coumarins, which are secondary metabolites important for Fe uptake in plants [4]. Consequently, the Dickeya spp. cells, which compete for Fe with the plant cells, can uptake Fe with higher efficiency and accumulate more of it than those infecting WT plants. It was shown before by other groups [32,37] that Fe uptake is important for bacteria ability to macerate plant tissue and the production of virulence factors. As a result of this, the Dickeya spp. strains infecting Arabidopsis mutants, which are defective in Fe acquisition, cause more severe disease symptoms in these plants. It is worth noticing that qPCR analysis of the corresponding genes (S8H and F6′H1) in the WT Col-0 genetic background, proved that their expression levels were relatively lower when a combined environmental stress, composed of Fe-deficiency and bacterial infection, was applied. Taking into account that D. dadantii 3937 has a higher ability to chelate Fe ions and the expression of several bacterial genes involved in siderophore-mediated Fe uptake is controlled by the Fe availability [32], it can be concluded that (1) coumarins produced by plants influence more strongly pathogens for which siderophores production play a particularly important role in the pathogenesis process, (2) disorders of coumarin biosynthesis are more important for the disease symptoms under Fe-deficiency conditions.
Next, we explored the potential of coumarins and other factors possibly present in the selected plant homogenates to affect the bacterial siderophore production by measuring the halos’ diameter produced by the Dickeya spp. strains on CAS-agar plates. We tested leaf homogenates prepared from a set of Arabidopsis plants (WT Col-0, s8h and f6′h1). In siderophore production assay, we observed greater halos produced by D. dadantii 3937 than that produced by D. solani IFB0099 for all tested Arabidopsis genotypes, except for the f6′h1 mutants. It seems like coumarins and other possible factors present in leaf homogenates and phosphate buffer can induce, directly or indirectly, the production of bacterial siderophores. The existence of not-characterised yet interplay between coumarins and the bacterial siderophore production needs further investigation. It is important to continue and develop research on the role of coumarins as novel elements of chemical communication and to test if, in coumarin-deficient plants, the induction of other compensatory pathway occurs.
To better understand the responses of coumarin-deficient plants to combined environmental stimuli, we grew a set of Arabidopsis mutants defective in coumarin accumulation (f6′h1, s8h) and coumarin transport to the rhizosphere (pdr9) in more physiological conditions. The inoculation was conducted on plants grown in two soil mixes with some differences in chemical compositions (Table 1). It has to be highlighted that plants grew significantly better in soil mix #1, in which all rosettes were much larger. We can speculate that the lack of any of the elements of soil mix #2 is limiting plant growth. However, considering that the soil mix #2 consist of half of the peat moss, which is a natural product of organic origin, we can suspect the significant differences in the microbiomes of the tested soil mixes. These interesting questions should be clarified in the future. Importantly, regardless of the soil mix in which plants were grown, the Arabidopsis of all tested genotypes inoculated with the D. solani IFB0099 strain developed more severe infection symptoms compared to D. dadantii 3937 reference strain. For both bacterial strains, the symptoms of infection were more pronounced on plants with a better growth on soil mix #1 compared to plants grown in soil mix #2. Interestingly, we detected a variation in the disease symptoms between plant genotypes inoculated with D. solani IFB0099 strain, particularly on plants grown in the soil mix #1. The pdr9 mutant plants that hyperaccumulate coumarins in their tissues, showed the mildest infection symptoms among all plant genotypes when inoculated with D. solani IFB0099 strain. While both Arabidopsis mutants (f6′h1 and s8h) defective in coumarin biosynthetic genes, showed stronger symptoms of infection. The explanation of this phenomenon can be that coumarins are known for their antimicrobial activity, however, the observed genotype-specific mode of action needs further investigations. We did not observe such a clear variation in the infection symptoms on plants inoculated with D. dadantii 3937 strain that is characterised by a higher ability to chelate Fe ions (the CAS-plate assays presented in Figure 4 and [40]).
In this work, we also analysed the expression profiles of three plant genes (PR1, SOD1, CYP82C2) which products are involved in the plant tissue response to a wide range of stresses including biotic and abiotic factors. During D. solani IFB0099 infection of Arabidopsis, the expression of PR1, which is considered to be one of the markers for salicylic acid (SA)-dependent systemic acquired resistance (SAR) [55,56], was strongly induced in the leaves of all infected genotypes. This increase in the PR1 gene expression was most pronounced in the WT Col-0 genetic background. A similar PR1 expression profile was observed in D. dadantii 3937- and mock-inoculated plants, but the levels of PR1 expression were much lower in these experimental setups. In parallel, we observed in D. solani IFB0099-inoculated plants an induction in the expression of the SOD1 gene, which encodes a cytosolic copper/zinc superoxide dismutase CSD1 that can be regulated by biotic and abiotic stresses and detoxify superoxide radicals [57]. This indicates that plants infected with D. solani IFB0099 induce the defensive mechanism by increasing the production of critical antioxidant enzymes protecting organisms from reactive oxygen species. Interestingly, plants with impaired biosynthesis or coumarin accumulation have a higher expression of SOD1, although the observed differences are not statistically significant. Furthermore, the expression of CYP82C2 in D. solani IFB0099-inoculated Arabidopsis WT Col-0 and its mutants, was also clearly upregulated in comparison to D. dadantii 3937- and mock-inoculated plants. Since CYP82C2 was shown to be involved in several aspects of jasmonic acid (JA) responses [58], it seems likely that infection with D. solani IFB0099 pathogen induces activation of the JA-dependent response.
The above results and data previously published obtained from a study of Arabidopsis response to D. dadantii 3937 infection highlight the major importance of the competition between plant and bacterial cells for Fe uptake during infection [36,38]. It was demonstrated that Fe nutrition strongly affects the disease caused by soft rot-causing plant pathogenic bacteria with a large plant host range including Arabidopsis. Plants have evolved various strategies to acquire Fe from their environment and mechanisms tightly regulating Fe uptake, transport and storage [11,13,59] including the production of Fe-mobilizing phenolic compounds like coumarins [1,2,3,4,5,7,8,60,61,62]. The production of exudates, which is dependent on the external environment, at the same time is genetically regulated in plants. It was shown by Micallef et al. [63] that natural populations of Arabidopsis originating from various geographical localization, called accessions, release a unique set of compounds into their rhizosphere. The authors detected that the rhizobacterial community composition and the relative abundance of particular ribotypes were also accession-dependent. They hypothesised that the observed natural variation in root exudation could partly explain the genotypic influence on bacterial communities in the rhizosphere [63]. Many studies of plant-microbe interactions revealed that plants are not only able to shape their rhizosphere microbiome, but also highlight this root-associated microbial community to be referred to as the second genome of the plant, which is crucial for plant health [64]. Our research group detected previously the existence of natural variation in the accumulation of antimicrobial coumarins, namely scopoletin and scopolin, among Arabidopsis accessions [14]. Lately, we also detected a significant variation in the content of other simple coumarins like umbelliferone and esculetin together with their glycosides: skimmin and esculin, respectively [65]. It was also shown recently that a natural variation exists in Arabidopsis tolerance to Dickeya spp. [66]. The significantly different susceptibility groups were uncovered within a small set of eight Arabidopsis accessions following inoculation with D. dadantii 3937, which suggested that tolerance associated loci might be present in this model plant. Even though Dickeya spp. are causative agents of severe diseases in a wide range of plant species and major economic losses, little data concerning potential resistance genes are available [67,68,69]. These data strongly suggest that Arabidopsis with its extensive genetic natural variation and a set of powerful genetic tools including web-accessible collections of mutants, provides an excellent model to study the interplay between secondary metabolites production, exudate profiles, Fe homeostasis and interaction with beneficial and plant pathogenic microbes.
4. Materials and Methods
4.1. Plant Material
The Arabidopsis thaliana accession Columbia was used as the wild type (Arabidopsis WT Col-0) together with a set of T-DNA insertional mutant lines in the Col-0 background: [1] s8h-1 (SM_3.27151); s8h-2 (SM_3.23443); [2] f6′h1-1 (SALK_132418); f6′h1-2 (SALK_050137C) and [3] pdr9-1 (SALK_050885). Seeds of all lines are available at the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/, accessed on 15 April 2021).
4.2. Bacterial Strains, Media, Growth Conditions
The strains used in this study, Dickeya dadantii 3937 IFB0459 and Dickeya solani IFB0099, are available at the collection of bacterial pathogens located at the IFB UG & MUG, in Poland. For the plant inoculation, the bacteria were grown overnight in lysogeny broth (LB) [70] liquid medium at 28 °C with agitation at 120 rpm. Then the bacterial cultures were centrifuged in Eppendorf tubes (5 min, 6500 rpm), washed in sterile 0.85% NaCl and centrifuged again. The bacterial suspension (at least 15 mL) was prepared in sterile 0.85% NaCl and adjusted to 1 MacFarland Unit (Densitometer DEN-1/DEN-1B, Buch & Holm Herlev, Denmark), approximately 108 cfu/mL.
4.3. Hydroponic Cultures
After a few days’ stratification at 4 °C, Arabidopsis WT Col-0 plants were grown in a controlled environment (16 h light at 22 °C/~100 μmol m−2 s−1 and 8 h dark at 20 °C) in a modified 1 × Heeg solution [71], as described in details in [42] with the following modifications. Approximately 3-weeks-old plants in tube lids, filled with the solidified Heeg medium, were transferred from tip boxes with control solution (40 µM Fe2+) to the modified 50 mL Falcon centrifuge tubes filled with optimal (40 μM Fe2+) or Fe-deficient (0 μM Fe2+) medium. The roots were passing through the 1-cm diameter hole drilled in Falcons’ lid to support the seedling holder as proposed by Conn et al. [72]. Hydroponic solutions were replenished by the addition of a fresh medium.
4.4. Soil Cultures
Arabidopsis seeds were first stratified in Petri dishes on water-saturated Whatman paper followed by a cold treatment for 4 d at 4 °C and then planted into two different soil mixes derived from commercial products. Soil mix #1 consists of commercial soil 1 and vermiculite (3–6 mm in diameter) in a proportion of 3:1, respectively. Soil mix #2 consists of commercial de-acidified peat moss mix with commercial soil 2 and vermiculite in a proportion 2:1:1, respectively. Chemical analysis were conducted by the Regional Agro-Chemical Station in Gdansk, Poland (OSCh-R, http://www.oschrgdansk.pl/, accessed on 15 April 2021). Prior to sowing seeds, both soil mixes were soaked with general-purpose fertiliser (Substral, The Scotts Miracle-Gro Company, Marysville, OH USA). Arabidopsis plants were grown for five weeks under a photoperiod of 16 h light (120 μmol m−2 s−1) at 22 °C and 8 h dark at 20 °C, before being inoculated.
4.5. Siderophore Production in the Presence of Plant Extracts
Homogenates from leaves of Arabidopsis thaliana were prepared from WT Col-0 plants, s8h and f6′h1 mutants. Briefly, the pooled leaves (~300 ± 40 mg) stored at −80 °C were thawed and homogenised in Bioreba bags (BIOREBA AG, Reinach, Switzerland) with 3 mL of 50 mM phosphate buffer pH 7.0 with the use of hand homogeniser (BIOREBA AG, Reinach, Switzerland). Then, the homogenate was centrifuged twice in Eppendorf tubes (8500 rpm, 2 min). The supernatant was filtered with a syringe 0.22 µm filter into sterile Eppendorf tubes (in total 3.5 mL) and immediately used on plates. Siderophore production of bacterial strains was determined on chrome azurol S–agar (CAS-agar) plates [73] supplemented with 100 µL of each homogenate with sterile spreader 15 min before bacteria inoculation. The overnight cultures of bacterial strains were centrifuged and resuspended in sterile 0.85% NaCl and adjusted to 0.5 MacFarland (Densitometer DEN-1/DEN-1B, Buch & Holm). 10 µL of each bacterial suspension was put on the CAS-agar plates and incubated at 28 °C for up to 168 h. We measured the halo diameters developed on CAS-agar plates supplemented with plant homogenates every 24 h.
4.6. Plant Inoculation with Bacterial Strains
Approximately 5-weeks-old plants, grown in soil or hydroponically, were inoculated with the bacterial suspensions of either D. dadanti 3937 or D. solani IFB0099 with the use of laboratory pincers. The pincers were sterilised before use and approximately 1 cm of the pincer tip was dipped into the bacterial suspension (the final inoculum of 108 cfu/mL) and immediately the plant leaf was pinched with the pincers. At least 8 leaves were inoculated with each bacterial strain and mock. We pinched the middle parts of the selected representative leaves (3 leaves per plant). The negative control (mock) were plants inoculated only with sterile 0.85% NaCl. For the mock-inoculated plants no symptoms development was observed throughout the experiment. The number of bacteria inoculated into the plant leaf with pincers was about 2 × 107 cfu/leaf and it was stable throughout experiments (data not shown). After inoculation, trays with plants were placed in the boxes filled with one litre of water, which were covered with transparent lids, to enable 100% humidity. Next, boxes were placed for 96 h in phytotron at 28 °C (16 h light at 28 °C/~100 μmol m−2 s−1 and 8 h dark at 26 °C), which is an optimal temperature for the development of disease symptoms by bacteria from the Dickeya spp. The whole rosettes were collected for each genotype grown in soil mix #1, frozen in liquid nitrogen and stored at −80 °C until further analysis. The roots of Col-0 plants grown hydroponically were gently removed from the agar droplets with tweezers and then rinsed in a beaker with distilled water. After drying on a paper towel, roots were frozen in liquid nitrogen and placed at −80 °C.
4.7. Quantification of Arabidopsis Plants Susceptibility to Dickeya spp. Strains by Visual Symptom Scoring (Disease Severity Scale, DSS)
The Arabidopsis plants were scored for soft rot/ blackleg symptoms development on leaves daily at 0 h, 24 h, 48 h, 72 h and 96 h post-inoculation. Developed symptoms of the disease were assigned to 0–5 scale (Figure 1), for which “0” meant no signs of symptoms of the disease and the wound has healed (it was observed for the negative control leaves); “1” the necrotic tissue was observed in the inoculation site only; “2” the necrotic tissue observed in the inoculation site and max. 3 mm wide around it; “3” the maceration visible around the inoculation site spreading further with possible chlorosis of the leaf; “4” visible maceration of the whole leaf, possible chlorosis of other leaves, no maceration of the limb; “5” visible maceration of the whole limb and the leaf.
4.8. RNA Extraction and Expression Analysis by qRT-PCR
Total RNA was extracted from plant material harvested 48 h after inoculation. The plant tissue was homogenised in liquid nitrogen using sterile mortars cleaned with isopropanol and baked for 4 h at 180 °C. A commercially available E.Z.N.A.® Plant RNA Kit (Omega Bio-tek, Inc., Norcross, GA USA) was used following the instructions of the manufacturer and including an additional step to remove the genomic DNA contamination from the mixture with RNase-Free DNase I Set (Omega Bio-tek, Inc., Norcross, GA USA). 500 ng RNA for RNA isolated from leaves or 200 ng RNA for RNA isolated from roots was used for reverse transcription by Maxima First Strand reverse transcriptase cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific, Waltham, MA USA). qPCR was performed using LightCycler® 480 Real-Time PCR System (Hoffmann-La Roche, Basel, Switzerland) and SYBR® Green Master Mix (Thermo Fisher Scientific, Waltham, MA USA), using the gene-specific primers shown in Table 2. Primers’ specificities were confirmed by the analysis of the melting curves. Relative transcript levels (RLT) of the plant genes in leaf tissues were normalised to the transcript level of the house-keeping ACTIN2 gene (At3g18780). As a reference for the root tissues, the EF-1α (ang. elongation factor-1α, At5g60390) gene was selected [44].
Table 2.
Primer sequences for plant genes used in qPCR reactions.
| Name | Sequence (5′-3′) | Description |
|---|---|---|
| 3g18780For | CTTGCACCAAGCAGCATGAA | Primer for ACT2 gene 1 |
| 3g18780Rev | CCGATCCAGACACTGTACTTCCTT | Primer for ACT2 gene 1 |
| AT2G14610_F | TTCTTCCCTCGAAAGCTCAAGA | Primer for PR1 gene |
| AT2G14610_R | GTGCCTGGTTGTGAACCCTTA | Primer for PR1 gene |
| AT4G31970_F | GATGGTGAGAATGGTGGCCG | Primer for CYP82C2 gene |
| AT4G31970_R | GCCTCTTCGGCATCTTCAGG | Primer for CYP82C2 gene |
| AT1G08830_F | TCAACCCCGATGGTAAAACAC | Primer for SOD1 gene |
| AT1G08830_R | TCACCAGCATGTCGATTAGCA | Primer for SOD1 gene |
| At5g60390_F | TGAGCACGCTCTTCTTGCTTTCA | Primer for EF-1α gene 1 |
| At5g60390_R | GGTGGTGGCATCCATCTTGTTACA | Primer for EF-1α gene 1 |
| S8HqPCR_F | GCCGAGACACTTGGCTTCTT | Primer for S8H gene |
| S8HqPCR_R | CAGCAGCTCCACCGAAACA | Primer for S8H gene |
| F6H1qPCRf | TGATGAGGACAGAGTCGCTGAA | Primer for F6′H1 gene |
| F6H1qPCRr | CACTTGAAAGAACCCCCATTTC | Primer for F6′H1 gene |
1 Reference [44].
5. Conclusions
We investigated here the possible interactions between plant resistance, coumarin content and Fe status by using the plant pathogenic Dickeya spp. strains and a set of selected Arabidopsis mutants defective in coumarin biosynthesis (f6′h1, s8h) and their transportation (pdr9). We studied the effect of disturbed coumarin accumulation and Fe deficiency on the disease severity using a model system of Arabidopsis/Dickeya spp. interactions. Arabidopsis plants grown in hydroponic cultures with different Fe regimes and two soil mixes were inoculated with Dickeya spp. strains or treated with NaCl as a control. Under all conditions tested, Arabidopsis plants inoculated with D. solani IFB0099 developed more severe disease symptoms compared to plants inoculated with D. dadantii 3937 strain. While the response of plants to D. dadantii 3937 infection was genotype-dependent in Fe-deficient hydroponic solution. Subsequently, we showed that the expression of genes encoding plant stress markers was also strongly induced by D. solani IFB0099 infection. Interestingly, the inoculation of WT Col-0 plants grown in Fe-deficient hydroponics with both Dickeya spp. strains cause a decrease in the expression of S8H and F6′H1 genes involved in coumarin biosynthesis.
Dickeya spp. was chosen as a plant pathogenic bacteria causing soft rot disease that can infect a broad spectrum of plants, whereas plant genotypes were selected due to their disturbed coumarin accumulation in roots and exudate profiles, which may have an impact on specific microbial consortia selection in the rhizosphere and influence plant response to pathogen attack. This may play a particularly important role for plant development and growth under Fe deficiency. The molecular mechanisms underlying these fascinating interactions are not yet well understood. We believe that Arabidopsis/Dickeya spp./pathosystem, together with a set of various Arabidopsis mutants defective in coumarin biosynthesis and its significant natural genetic variation, will be in future beneficial in uncovering a role of root-exuded coumarins in structuring the rhizobiome and plant resistance to pathogens.
Author Contributions
Conceptualization, M.P., E.L., and A.I.; methodology, M.P., J.S., and A.I.; validation, I.P., M.P., and J.S.; formal analysis, I.P.; investigation, I.P., M.P., J.S., and D.S.; writing—original draft preparation, M.P. and A.I.; writing—review and editing, E.L. and A.I.; supervision, E.L. and A.I.; project administration, A.I.; funding acquisition, A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the NARODOWE CENTRUM NAUKI (Polish National Science Centre) grant number 2014/15/B/NZ2/01073 (OPUS to AI) and grant number 2019/35/O/NZ1/02751 (PRELUDIUM BIS to EL).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clemens S., Weber M. The Essential Role of Coumarin Secretion for Fe Acquisition from Alkaline Soil. Plant Signal. Behav. 2016;11:e1114197. doi: 10.1080/15592324.2015.1114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fourcroy P., Sisó-Terraza P., Sudre D., Savirón M., Reyt G., Gaymard F., Abadía A., Abadia J., Alvarez-Fernández A., Briat J.-F. Involvement of the ABCG37 Transporter in Secretion of Scopoletin and Derivatives by Arabidopsis Roots in Response to Iron Deficiency. New Phytol. 2014;201:155–167. doi: 10.1111/nph.12471. [DOI] [PubMed] [Google Scholar]
- 3.Rajniak J., Giehl R.F.H., Chang E., Murgia I., von Wirén N., Sattely E.S. Biosynthesis of Redox-Active Metabolites in Response to Iron Deficiency in Plants. Nat. Chem. Biol. 2018;14:442–450. doi: 10.1038/s41589-018-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Celma J., Lin W.-D., Fu G.-M., Abadía J., López-Millán A.-F., Schmidt W. Mutually Exclusive Alterations in Secondary Metabolism Are Critical for the Uptake of Insoluble Iron Compounds by Arabidopsis and Medicago Truncatula. Plant Physiol. 2013;162:1473–1485. doi: 10.1104/pp.113.220426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid N.B., Giehl R.F.H., Döll S., Mock H.-P., Strehmel N., Scheel D., Kong X., Hider R.C., von Wirén N. Feruloyl-CoA 6′-Hydroxylase1-Dependent Coumarins Mediate Iron Acquisition from Alkaline Substrates in Arabidopsis. Plant Physiol. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sisó-Terraza P., Luis-Villarroya A., Fourcroy P., Briat J.-F., Abadía A., Gaymard F., Abadía J., Álvarez-Fernández A. Accumulation and Secretion of Coumarinolignans and Other Coumarins in Arabidopsis Thaliana Roots in Response to Iron Deficiency at High PH. Front. Plant Sci. 2016;7:1711. doi: 10.3389/fpls.2016.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siwinska J., Siatkowska K., Olry A., Grosjean J., Hehn A., Bourgaud F., Meharg A.A., Carey M., Lojkowska E., Ihnatowicz A. Scopoletin 8-Hydroxylase: A Novel Enzyme Involved in Coumarin Biosynthesis and Iron-Deficiency Responses in Arabidopsis. J. Exp. Bot. 2018;69:1735–1748. doi: 10.1093/jxb/ery005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai H.-H., Rodríguez-Celma J., Lan P., Wu Y.-C., Vélez-Bermúdez I.C., Schmidt W. Scopoletin 8-Hydroxylase-Mediated Fraxetin Production Is Crucial for Iron Mobilization. Plant Physiol. 2018;177:194–207. doi: 10.1104/pp.18.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanholme R., Sundin L., Seetso K.C., Kim H., Liu X., Li J., De Meester B., Hoengenaert L., Goeminne G., Morreel K., et al. COSY Catalyses Trans-Cis Isomerization and Lactonization in the Biosynthesis of Coumarins. Nat. Plants. 2019;5:1066–1075. doi: 10.1038/s41477-019-0510-0. [DOI] [PubMed] [Google Scholar]
- 10.Kai K., Mizutani M., Kawamura N., Yamamoto R., Tamai M., Yamaguchi H., Sakata K., Shimizu B. Scopoletin Is Biosynthesized via Ortho-Hydroxylation of Feruloyl CoA by a 2-Oxoglutarate-Dependent Dioxygenase in Arabidopsis Thaliana. Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- 11.Curie C., Briat J.-F. Iron Transport and Signaling in Plants. Annu. Rev. Plant Biol. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- 12.Verbon E.H., Trapet P.L., Stringlis I.A., Kruijs S., Bakker P.A.H.M., Pieterse C.M.J. Iron and Immunity. Annu. Rev. Phytopathol. 2017;55:355–375. doi: 10.1146/annurev-phyto-080516-035537. [DOI] [PubMed] [Google Scholar]
- 13.Grillet L., Schmidt W. Iron Acquisition Strategies in Land Plants: Not so Different after All. New Phytol. 2019;224:11–18. doi: 10.1111/nph.16005. [DOI] [PubMed] [Google Scholar]
- 14.Siwinska J., Kadzinski L., Banasiuk R., Gwizdek-Wisniewska A., Olry A., Banecki B., Lojkowska E., Ihnatowicz A. Identification of QTLs Affecting Scopolin and Scopoletin Biosynthesis in Arabidopsis Thaliana. BMC Plant Biol. 2014;14:280. doi: 10.1186/s12870-014-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L., Ding W., Xu Y., Wu D., Li S., Chen J., Guo B. New Insights into the Antibacterial Activity of Hydroxycoumarins against Ralstonia Solanacearum. Molecules. 2016;21:468. doi: 10.3390/molecules21040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun H., Wang L., Zhang B., Ma J., Hettenhausen C., Cao G., Sun G., Wu J., Wu J. Scopoletin Is a Phytoalexin against Alternaria Alternata in Wild Tobacco Dependent on Jasmonate Signalling. J. Exp. Bot. 2014;65:4305–4315. doi: 10.1093/jxb/eru203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Oirdi M., Trapani A., Bouarab K. The Nature of Tobacco Resistance against Botrytis Cinerea Depends on the Infection Structures of the Pathogen. Environ. Microbiol. 2010;12:239–253. doi: 10.1111/j.1462-2920.2009.02063.x. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu B., Miyagawa H., Ueno T., Sakata K., Watanabe K., Ogawa K. Morning Glory Systemically Accumulates Scopoletin and Scopolin after Interaction with Fusarium Oxysporum. Z. Naturforsch. C J. Biosci. 2005;60:83–90. doi: 10.1515/znc-2005-1-216. [DOI] [PubMed] [Google Scholar]
- 19.Prats E., Bazzalo M.E., León A., Jorrín J.V. Fungitoxic Effect of Scopolin and Related Coumarins on Sclerotinia Sclerotiorum. A Way to Overcome Sunflower Head Rot. Euphytica. 2006;147:451–460. doi: 10.1007/s10681-005-9045-8. [DOI] [Google Scholar]
- 20.Ndam Y.N., Nyegue M.A., Mounjouenpou P., Kansci G., Kenfack M.J., Eugène E.E. LC-MS Quantification of Scopoletin in Cassava(Manihot Esculenta Crantz) Varieties, Local Derived Foods, and Activity on Some Food Spoilage Fungi. J. Food Process. Preserv. 2020;44:e14387. doi: 10.1111/jfpp.14387. [DOI] [Google Scholar]
- 21.Modafar C.E., Clérivet A., Vigouroux A., Macheix J.J. Accumulation of Phytoalexins in Leaves of Plane Tree(Platanus Spp.) Expressing Susceptibility or Resistance ToCeratocystis Fimbriata f. Sp.Platani. Eur. J. Plant Pathol. 1995;101:503–509. doi: 10.1007/BF01874474. [DOI] [Google Scholar]
- 22.Gnonlonfin G.J.B., Sanni A., Brimer L. Review Scopoletin—A Coumarin Phytoalexin with Medicinal Properties. Crit. Rev. Plant Sci. 2012;31:47–56. doi: 10.1080/07352689.2011.616039. [DOI] [Google Scholar]
- 23.Napiroon T., Bacher M., Balslev H., Tawaitakham K., Santimaleeworagun W., Vajrodaya S. Scopoletin from Lasianthus Lucidus Blume(Rubiaceae): A Potential Antimicrobial against Multidrug-Resistant Pseudomonas Aeruginosa. J. Appl. Pharm. Sci. 2018;8:1–6. doi: 10.7324/JAPS.2018.8901. [DOI] [Google Scholar]
- 24.Acharya D., Bogati B., Risal P. Scopoletin reduces intracellular survival of Salmonella typhi within U937 human macrophage cell line in vitro. Afr. J. Microbiol. Res. 2013;1:47–51. [Google Scholar]
- 25.Mogana R., Adhikari A., Tzar M.N., Ramliza R., Wiart C. Antibacterial Activities of the Extracts, Fractions and Isolated Compounds from Canarium Patentinervium Miq. against Bacterial Clinical Isolates. BMC Complement. Med. Ther. 2020;20:55. doi: 10.1186/s12906-020-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stringlis I.A., Yu K., Feussner K., de Jonge R., Van Bentum S., Van Verk M.C., Berendsen R.L., Bakker P.A.H.M., Feussner I., Pieterse C.M.J. MYB72-Dependent Coumarin Exudation Shapes Root Microbiome Assembly to Promote Plant Health. Proc. Natl. Acad. Sci. USA. 2018;115:E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai R.-R., Wu X.-M., Xu J.-Y. Current Natural Products with Antihypertensive Activity. Chin. J. Nat. Med. 2015;13:721–729. doi: 10.1016/S1875-5364(15)30072-8. [DOI] [PubMed] [Google Scholar]
- 28.Voges M.J.E.E.E., Bai Y., Schulze-Lefert P., Sattely E.S. Plant-Derived Coumarins Shape the Composition of an Arabidopsis Synthetic Root Microbiome. Proc. Natl. Acad. Sci. USA. 2019;116:12558–12565. doi: 10.1073/pnas.1820691116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbort C.J., Hashimoto M., Inoue H., Niu Y., Guan R., Rombolà A.D., Kopriva S., Voges M.J.E.E.E., Sattely E.S., Garrido-Oter R., et al. Root-Secreted Coumarins and the Microbiota Interact to Improve Iron Nutrition in Arabidopsis. Cell Host Microbe. 2020;28:825–837.e6. doi: 10.1016/j.chom.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stassen M.J.J., Hsu S.-H., Pieterse C.M.J., Stringlis I.A. Coumarin Communication along the Microbiome-Root-Shoot Axis. Trends Plant Sci. 2021;26:169–183. doi: 10.1016/j.tplants.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Stringlis I.A., de Jonge R., Pieterse C.M.J. The Age of Coumarins in Plant-Microbe Interactions. Plant Cell Physiol. 2019;60:1405–1419. doi: 10.1093/pcp/pcz076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Expert D. WITHHOLDING AND EXCHANGING IRON: Interactions between Erwinia Spp. and Their Plant Hosts. Annu. Rev. Phytopathol. 1999;37:307–334. doi: 10.1146/annurev.phyto.37.1.307. [DOI] [PubMed] [Google Scholar]
- 33.Motyka A., Zoledowska S., Sledz W., Lojkowska E. Molecular Methods as Tools to Control Plant Diseases Caused by Dickeya and Pectobacterium Spp: A Minireview. New Biotechnol. 2017;39 Pt B:181–189. doi: 10.1016/j.nbt.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Potrykus M., Golanowska M., Sledz W., Zoledowska S., Motyka A., Kolodziejska A., Butrymowicz J., Lojkowska E. Biodiversity of Dickeya Spp. Isolated from Potato Plants and Water Sources in Temperate Climate. Plant Dis. 2016;100:408–417. doi: 10.1094/PDIS-04-15-0439-RE. [DOI] [PubMed] [Google Scholar]
- 35.Toth I.K., van der Wolf J.M., Saddler G., Lojkowska E., Hélias V., Pirhonen M., Tsror (Lahkim) L., Elphinstone J.G. Dickeya Species: An Emerging Problem for Potato Production in Europe. Plant Pathol. 2011;60:385–399. doi: 10.1111/j.1365-3059.2011.02427.x. [DOI] [Google Scholar]
- 36.Dellagi A., Rigault M., Segond D., Roux C., Kraepiel Y., Cellier F., Briat J.-F., Gaymard F., Expert D. Siderophore-Mediated Upregulation of Arabidopsis Ferritin Expression in Response to Erwinia Chrysanthemi Infection. Plant J. 2005;43:262–272. doi: 10.1111/j.1365-313X.2005.02451.x. [DOI] [PubMed] [Google Scholar]
- 37.Kieu N.P., Aznar A., Segond D., Rigault M., Simond-Côte E., Kunz C., Soulie M.-C., Expert D., Dellagi A. Iron Deficiency Affects Plant Defence Responses and Confers Resistance to Dickeya Dadantii and Botrytis Cinerea. Mol. Plant Pathol. 2012;13:816–827. doi: 10.1111/j.1364-3703.2012.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segond D., Dellagi A., Lanquar V., Rigault M., Patrit O., Thomine S., Expert D. NRAMP Genes Function in Arabidopsis Thaliana Resistance to Erwinia Chrysanthemi Infection. Plant J. 2009;58:195–207. doi: 10.1111/j.1365-313X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- 39.Golanowska M., Potrykus M., Motyka-Pomagruk A., Kabza M., Bacci G., Galardini M., Bazzicalupo M., Makalowska I., Smalla K., Mengoni A., et al. Comparison of Highly and Weakly Virulent Dickeya Solani Strains, With a View on the Pangenome and Panregulon of This Species. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potrykus M., Golanowska M., Hugouvieux-Cotte-Pattat N., Lojkowska E. Regulators Involved in Dickeya Solani Virulence, Genetic Conservation, and Functional Variability. Mol. Plant Microbe Interact. 2014;27:700–711. doi: 10.1094/MPMI-09-13-0270-R. [DOI] [PubMed] [Google Scholar]
- 41.Sławiak M., Łojkowska E., Wolf J.M.V.D. First Report of Bacterial Soft Rot on Potato Caused by Dickeya Sp. (Syn. Erwinia Chrysanthemi) in Poland. Plant Pathol. 2009;58:794. doi: 10.1111/j.1365-3059.2009.02028.x. [DOI] [Google Scholar]
- 42.Ihnatowicz A., Siwinska J., Meharg A.A., Carey M., Koornneef M., Reymond M. Conserved Histidine of Metal Transporter AtNRAMP1 Is Crucial for Optimal Plant Growth under Manganese Deficiency at Chilling Temperatures. New Phytol. 2014;202:1173–1183. doi: 10.1111/nph.12737. [DOI] [PubMed] [Google Scholar]
- 43.Chaparro J.M., Badri D.V., Bakker M.G., Sugiyama A., Manter D.K., Vivanco J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE. 2013;8:e55731. doi: 10.1371/annotation/51142aed-2d94-4195-8a8a-9cb24b3c733b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassan J.A., de la Torre-Roche R., White J.C., Lewis J.D. Soil Mixture Composition Alters Arabidopsis Susceptibility to Pseudomonas Syringae Infection. Plant Direct. 2018;2:e00044. doi: 10.1002/pld3.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalhais L.C., Dennis P.G., Fan B., Fedoseyenko D., Kierul K., Becker A., von Wiren N., Borriss R. Linking Plant Nutritional Status to Plant-Microbe Interactions. PLoS ONE. 2013;8:e68555. doi: 10.1371/journal.pone.0068555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemanceau P., Expert D., Gaymard F., Bakker P.A.H.M., Briat J.-F. Advances in Botanical Research. Volume 51. Academic Press; Cambridge, MA, USA: 2009. Chapter 12 Role of Iron in Plant–Microbe Interactions; pp. 491–549. [DOI] [Google Scholar]
- 48.Marschner P. Plant-Microbe Interactions in the Rhizosphere and Nutrient Cycling. In: Marschner P., Rengel Z., editors. Nutrient Cycling in Terrestrial Ecosystems. Soil Biology. Volume 10. Springer; Berlin/Heidelberg, Germany: 2007. pp. 159–182. [DOI] [Google Scholar]
- 49.Masclaux C., Expert D. Signalling Potential of Iron in Plant—Microbe Interactions: The Pathogenic Switch of Iron Transport in Erwinia Chrysanthemi. Plant J. 1995;7:121–128. doi: 10.1046/j.1365-313X.1995.07010121.x. [DOI] [Google Scholar]
- 50.Harris J.M., Balint-Kurti P., Bede J.C., Day B., Gold S., Goss E.M., Grenville-Briggs L.J., Jones K.M., Wang A., Wang Y., et al. What Are the Top 10 Unanswered Questions in Molecular Plant-Microbe Interactions? MPMI. 2020;33:1354–1365. doi: 10.1094/MPMI-08-20-0229-CR. [DOI] [PubMed] [Google Scholar]
- 51.Motyka-Pomagruk A., Zoledowska S., Sledz W., Lojkowska E. The Occurrence of Bacteria from Different Species of Pectobacteriaceae on Seed Potato Plantations in Poland. Eur. J. Plant Pathol. 2021;159:309–325. doi: 10.1007/s10658-020-02163-x. [DOI] [Google Scholar]
- 52.Van der Wolf J.M., Nijhuis E.H., Kowalewska M.J., Saddler G.S., Parkinson N., Elphinstone J.G., Pritchard L., Toth I.K., Lojkowska E., Potrykus M., et al. Dickeya Solani Sp. Nov., a Pectinolytic Plant-Pathogenic Bacterium Isolated from Potato(Solanum Tuberosum) Pt 3Int. J. Syst. Evol. Microbiol. 2014;64:768–774. doi: 10.1099/ijs.0.052944-0. [DOI] [PubMed] [Google Scholar]
- 53.Degefu Y., Potrykus M., Golanowska M., Virtanen E., Lojkowska E. A New Clade of Dickeya Spp. Plays a Major Role in Potato Blackleg Outbreaks in North Finland. Ann. Appl. Biol. 2013;162:231–241. doi: 10.1111/aab.12020. [DOI] [Google Scholar]
- 54.Golanowska M., Łojkowska E. A Review on Dickeya Solani, a New Pathogenic Bacterium Causing Loss in Potato Yield in Europe. BioTechnologia. 2016;97:109–127. doi: 10.5114/bta.2016.60781. [DOI] [Google Scholar]
- 55.Hamamouch N., Li C., Seo P.J., Park C.-M., Davis E.L. Expression of Arabidopsis Pathogenesis-Related Genes during Nematode Infection. Mol. Plant Pathol. 2011;12:355–364. doi: 10.1111/j.1364-3703.2010.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Loon L.C., Bakker P.A., Pieterse C.M. Systemic Resistance Induced by Rhizosphere Bacteria. Annu. Rev. Phytopathol. 1998;36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 57.Kliebenstein D.J., Monde R.A., Last R.L. Superoxide Dismutase in Arabidopsis: An Eclectic Enzyme Family with Disparate Regulation and Protein Localization. Plant Physiol. 1998;118:637–650. doi: 10.1104/pp.118.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F., Jiang H., Ye S., Chen W.-P., Liang W., Xu Y., Sun B., Sun J., Wang Q., Cohen J.D., et al. The Arabidopsis P450 Protein CYP82C2 Modulates Jasmonate-Induced Root Growth Inhibition, Defense Gene Expression and Indole Glucosinolate Biosynthesis. Cell Res. 2010;20:539–552. doi: 10.1038/cr.2010.36. [DOI] [PubMed] [Google Scholar]
- 59.Kobayashi T., Nishizawa N.K. Iron Uptake, Translocation, and Regulation in Higher Plants. Annu. Rev. Plant Biol. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt H., Günther C., Weber M., Spörlein C., Loscher S., Böttcher C., Schobert R., Clemens S. Metabolome Analysis of Arabidopsis Thaliana Roots Identifies a Key Metabolic Pathway for Iron Acquisition. PLoS ONE. 2014;9:e102444. doi: 10.1371/journal.pone.0102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai H.-H., Schmidt W. One Way. Or Another? Iron Uptake in Plants. New Phytol. 2017;214:500–505. doi: 10.1111/nph.14477. [DOI] [PubMed] [Google Scholar]
- 62.Zamioudis C., Hanson J., Pieterse C.M.J. β-Glucosidase BGLU42 Is a MYB72-Dependent Key Regulator of Rhizobacteria-Induced Systemic Resistance and Modulates Iron Deficiency Responses in Arabidopsis Roots. New Phytol. 2014;204:368–379. doi: 10.1111/nph.12980. [DOI] [PubMed] [Google Scholar]
- 63.Micallef S.A., Shiaris M.P., Colón-Carmona A. Influence of Arabidopsis Thaliana Accessions on Rhizobacterial Communities and Natural Variation in Root Exudates. J. Exp. Bot. 2009;60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Perkowska I., Siwinska J., Olry A., Grosjean J., Hehn A., Bourgaud F., Lojkowska E., Ihnatowicz A. Identification and Quantification of Coumarins by UHPLC-MS in Arabidopsis Thaliana Natural Populations. Molecules. 2021;26:1804. doi: 10.3390/molecules26061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rigault M., Buellet A., Masclaux-Daubresse C., Fagard M., Chardon F., Dellagi A. Quantitative Methods to Assess Differential Susceptibility of Arabidopsis Thaliana Natural Accessions to Dickeya Dadantii. Front. Plant Sci. 2017;8:394. doi: 10.3389/fpls.2017.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Austin S., Lojkowska E., Ehlenfeldt K., Kelman A., Helgeson J.P. Fertile Interspecific Somatic Hybrids of Solanum: A Novel Source of Resistance to Erwinia Soft Rot. Phytopathology. 1988;78:1216–1220. doi: 10.1094/Phyto-78-1216. [DOI] [Google Scholar]
- 68.Pajerowska K.M., Parker J.E., Gebhardt C. Potato Homologs of Arabidopsis Thaliana Genes Functional in Defense Signaling--Identification, Genetic Mapping, and Molecular Cloning. Mol. Plant Microbe Interact. 2005;18:1107–1119. doi: 10.1094/MPMI-18-1107. [DOI] [PubMed] [Google Scholar]
- 69.Zimnoch-Guzowska E., Marczewski W., Lebecka R., Flis B., Schäfer-Pregl R., Salamini F., Gebhardt C. QTL Analysis of New Sources of Resistance to Erwinia Carotovora Ssp. Atroseptica in Potato Done by AFLP, RFLP, and Resistance-Gene-Like Markers. Crop Sci. 2000;40:1156–1167. doi: 10.2135/cropsci2000.4041156x. [DOI] [Google Scholar]
- 70.Lennox E.S., Luria S.E., Benzer S. On the Mechanism of Photoreactivation of Ultraviolet-Inactivated Bacteriophage. Biochim. Biophys. Acta. 1954;15:471–474. doi: 10.1016/0006-3002(54)90003-7. [DOI] [PubMed] [Google Scholar]
- 71.Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. Analysis of the Arabidopsis O-Acetylserine(Thiol)Lyase Gene Family Demonstrates Compartment-Specific Differences in the Regulation of Cysteine Synthesis. Plant Cell. 2008;20:168–185. doi: 10.1105/tpc.107.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conn S.J., Hocking B., Dayod M., Xu B., Athman A., Henderson S., Aukett L., Conn V., Shearer M.K., Fuentes S., et al. Protocol: Optimising Hydroponic Growth Systems for Nutritional and Physiological Analysis of Arabidopsis Thaliana and Other Plants. Plant Methods. 2013;9:4. doi: 10.1186/1746-4811-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwyn B., Neilands J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available within the article.