Significance
Osteogenesis imperfecta (OI), also known as brittle bone disease, commonly features low bone mass and frequent fractures. We and others have previously identified mutations that cause joint deformities in OI patients. However, mechanisms responsible for joint pathologies in OI are poorly understood. Here, we have established a mouse model of OI joint dysfunction that is caused by mutations in Fkbp10. Our mouse model recapitulates key features of FKBP10 OI patients, including reduced collagen cross-linking and joint deformities. In addition, we found that inflammation and altered matrix signaling is associated with joint abnormalities in Fkbp10 mutant animals. Overall, our work provides a valuable model that can be used to identify novel therapies to treat joint dysfunction in OI and OI-related diseases.
Keywords: osteogenesis imperfecta, Fkbp10, tendon, ligament, contracture
Abstract
Osteogenesis imperfecta (OI) is a genetic disorder that features wide-ranging defects in both skeletal and nonskeletal tissues. Previously, we and others reported that loss-of-function mutations in FK506 Binding Protein 10 (FKBP10) lead to skeletal deformities in conjunction with joint contractures. However, the pathogenic mechanisms underlying joint dysfunction in OI are poorly understood. In this study, we have generated a mouse model in which Fkbp10 is conditionally deleted in tendons and ligaments. Fkbp10 removal substantially reduced telopeptide lysyl hydroxylation of type I procollagen and collagen cross-linking in tendons. These biochemical alterations resulting from Fkbp10 ablation were associated with a site-specific induction of fibrosis, inflammation, and ectopic chondrogenesis followed by joint deformities in postnatal mice. We found that the ectopic chondrogenesis coincided with enhanced Gli1 expression, indicating dysregulated Hedgehog (Hh) signaling. Importantly, genetic inhibition of the Hh pathway attenuated ectopic chondrogenesis and joint deformities in Fkbp10 mutants. Furthermore, Hh inhibition restored alterations in gait parameters caused by Fkbp10 loss. Taken together, we identified a previously unappreciated role of Fkbp10 in tendons and ligaments and pathogenic mechanisms driving OI joint dysfunction.
Osteogenesis imperfecta (OI) is a heterogeneous genetic disorder that is commonly characterized by low bone mass, progressive musculoskeletal deformities, and increased susceptibility to fractures (1–3). In the past 15 y, 20 novel genetic causes of OI and OI-related diseases have been identified (4, 5). In most cases, OI is caused by dominant mutations in the two genes that encode type I procollagen (COL1A1 and COL1A2). However, we and others have previously identified recessively inherited mutations in genes that govern the posttranslational modification of type I procollagen (6–12). OI-causing mutations not only affect the skeleton but are also associated with defects in extraskeletal connective tissues such as tendons and ligaments, which adds to OI's phenotypic heterogeneity (2). Still, our understanding of the extraskeletal manifestations in OI and their underlying pathogenic mechanisms is limited. Antiresorptive therapies are the mainstays of OI treatment, with bisphosphonates as well as other newer anabolic therapies that primarily modify bone mass. However, there is an unmet need to investigate the impact of OI on tissues outside of bone, and to identify new therapies that target these extraskeletal manifestations.
FKBP10 belongs to a family of immunophilins that commonly share one or several repeats of the peptidylprolyl isomerase (PPIase) domain (13). FKBP10 is unique among the FKBP family, as it localizes to the endoplasmic reticulum where it is thought to function as a protein chaperone for type I procollagen and elastin (13, 14). Previous studies have shown that loss of function mutations in FKBP10 can lead to mild-to-severe OI including skeletal defects such as low bone mass and fractures. In addition, FKBP10 patients frequently display congenital contractures and kyphoscoliosis, also known as Bruck syndrome (9, 15–19). The impaired skeletal and joint phenotypes in FKBP10 OI patients negatively impact their mobility. Previous studies have shown that bisphosphonate treatment may have beneficial effects to reduce fractures and pain (4, 20). However, therapies to address joint abnormalities in FKBP10 OI patients are limited. Earlier studies have shown that loss of FKBP10 is associated with reduced telopeptide lysyl hydroxylation of type I procollagen and hence stable collagen cross-linking (21), which phenotypically overlap with PLOD2 mutations (22). Thus, it has been widely suggested that interactions between FKBP10 and PLOD2 during procollagen biogenesis is necessary for telopeptide lysyl hydroxylation. Indeed, studies showed that FKBP65 (encoded by Fkbp10) is required for the stability and activity of lysyl hydroxylase 2 (LH2, encoded by Plod2) in cell culture (23, 24). Still, the pathogenic mechanisms by which loss of FKBP10 leads to joint contractures remain unclear (25).
Tendons and ligaments are soft connective tissues that function to bridge muscle to bone and bone to bone, respectively, and provide stability during movement (26–28). As such, tendons and ligaments are designed to withstand high levels of biomechanical force (26). Tendon fibroblasts arise from mesenchymal progenitors that are marked by the expression of Scleraxis (Scx), a basic helix–loop–helix transcription factor that is essential for tendon development (29). In addition, the transcription factor Mohawk (Mkx) is necessary for the maintenance of tendon cell identity in postnatal stages (30, 31). Genetic ablation of either Scx or Mkx leads to defects in tendon development (30, 32, 33), which can lead to chondro-ossification of connective tissues (34). Similarly, aberrations in the extracellular matrix (ECM) of the tendon midsubstance have been shown to cause heterotopic ossification (HO) (35). The ECM serves as a reservoir for various growth factors and cytokines that are critical for the development and homeostasis of connective tissues (36, 37). Previous studies have shown that dysregulated matrix signaling may be an important mechanism that contributes to pathogenesis in defective connective tissues, including OI (35, 38–40). Furthermore, hyperactivation of Hedgehog (Hh) signaling in tendon stem/progenitors is sufficient to induce chondro-ossification in tendons and ligaments (41). However, whether altered cell signaling pathways contribute to disease pathogenesis in OI remains unclear.
Here we have generated a mouse model of FKBP10 OI joint dysfunction by conditional removal of Fkbp10 in tendons and ligaments. Whereas Fkbp10−/− mice display embryonic lethality (42), the deletion of Fkbp10 in Scx-Cre−expressing cells resulted in postnatal joint deformities following inflammation and ectopic chondrogenesis in tendons and ligaments. We also demonstrate that genetic inhibition of Hh signaling can attenuate joint deformities and gait defects resulting from Fkbp10 removal.
Results
Deletion of Fkbp10 in Tendons and Ligaments Causes Postnatal Joint Deformities and Heterotopic Ossification.
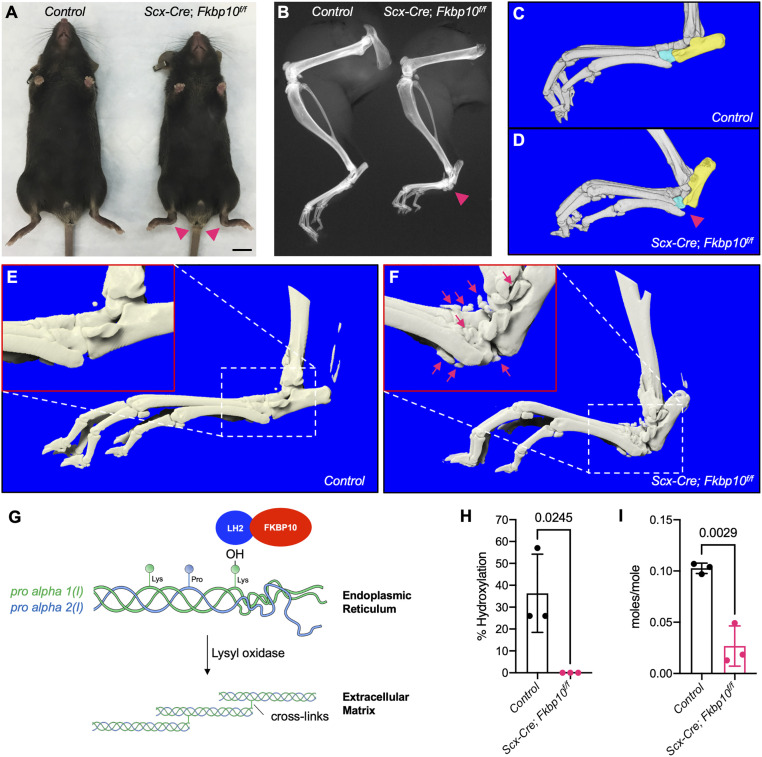
To determine the function of Fkbp10 in tendons and ligaments, we selectively deleted Fkbp10 using Scx-Cre (Scx-Cre;Fkbp10f/f, herein ScxCKO). ScxCKO mice were born at the expected Mendelian ratios and appeared indistinguishable from littermate controls at birth (Fkbp10f/+, Fkbp10f/f, or Scx-Cre;Fkbp10f/+ animals). At 2 mo of age, ScxCKO mice displayed a mild but consistent decrease in whole body length and weight (SI Appendix, Fig. S1). ScxCKO mice showed hind paw deformities by 2.5 mo of age which were evident by morphological assessment (Fig. 1A). Radiographs and three-dimensional (3D) reconstructed microcomputed tomography (microCT) images of these hind paw deformities revealed them to be associated with separation of the calcaneus and cuboid (Fig. 1 B–D). These joint deformities occurred in both hind paws of ScxCKO mice at 54.8 ± 2.7 d after birth (n = 7). Furthermore, ScxCKO mice showed HO surrounding the midfoot joints at between 11 and 13 mo of age (Fig. 1 E and F). Moreover, histology and Sclerostin (Sost) expression showed that HO occurred within the soft connective tissues in the midfoot joints (SI Appendix, Fig. S2). Taken together, these results suggest that loss of Fkbp10 in tendons and ligaments causes joint deformities and HO in postnatal mice.
Fig. 1.
Fkbp10 deletion causes joint deformities and HO. Representative (A) whole-body and (B) hindlimb radiography images at 2.5 mo of age (n = 8). (Scale bar, 1 cm.) (C and D) MicroCT 3D reconstruction of the foot at 4 mo of age. Aqua, cuboid; yellow, calcaneus (n = 3 per group). (E and F) Representative microCT 3D reconstruction of the foot at 11 mo to 13 mo of age (n = 4 per group). (G) Graphical summary of telopeptide lysyl hydroxylation of type I procollagen by the Fkbp10 and Lh2 complex and Lox-dependent collagen cross-linking. (H) C-telopeptide lysyl hydroxylation of type I procollagen and (I) collagen cross-linking (sum of HP and LP) in Achilles tendon at 2 mo of age (n = 3 per group). Red arrowheads, joint deformities; red box, high-magnification view of white boxed region; red arrows, HO. Data are presented as means ± SD. Actual P values from unpaired t test are shown. P < 0.05 were considered statistically significant.
Fkbp10 Is Required for Telopeptide Lysyl Hydroxylation of Type I Procollagen In Vivo.
Type I procollagen is subject to various posttranslational modification events including the conversion of telopeptide lysine to hydroxylysine, which subsequently serves as substrates for collagen cross-linking (Fig. 1G). Previous studies have shown that FKBP10 OI patients exhibit reduced telopeptide lysyl hydroxylation of type I procollagen and defective cross-linking in bone (19, 21). To determine whether telopeptide lysyl hydroxylation is affected in ScxCKO mice, we performed tandem mass spectrometry on Achilles tendons from 2-mo-old animals. C-telopeptide lysyl hydroxylation of type I procollagen and pyridinoline cross-linking was substantially reduced in the tendons of ScxCKO mice (Fig. 1 H and I), which corroborates earlier studies (21). The reduction of intermolecular cross-linking in tendons of Fkbp10 mutant mice prompted us to investigate whether collagen fibrillogenesis is affected. Electron microscopy imaging showed that the distribution of collagen fibril diameter was comparable between control and Fkbp10 mutants in 15-d-old animals (SI Appendix, Fig. S3 A–C). In contrast, Fkbp10 deletion increased the abundance of smaller collagen fibrils relative to controls in 30-d-old mice (SI Appendix, Fig. S3 D–F). Collectively, these results indicate that Fkbp10 is necessary for type I procollagen telopeptide lysyl hydroxylation, collagen cross-linking, and, hence, collagen fibrillogenesis in tendons of postnatal mice.
Fkbp10 Deletion Stimulates Site-Specific Ectopic Chondrogenesis.
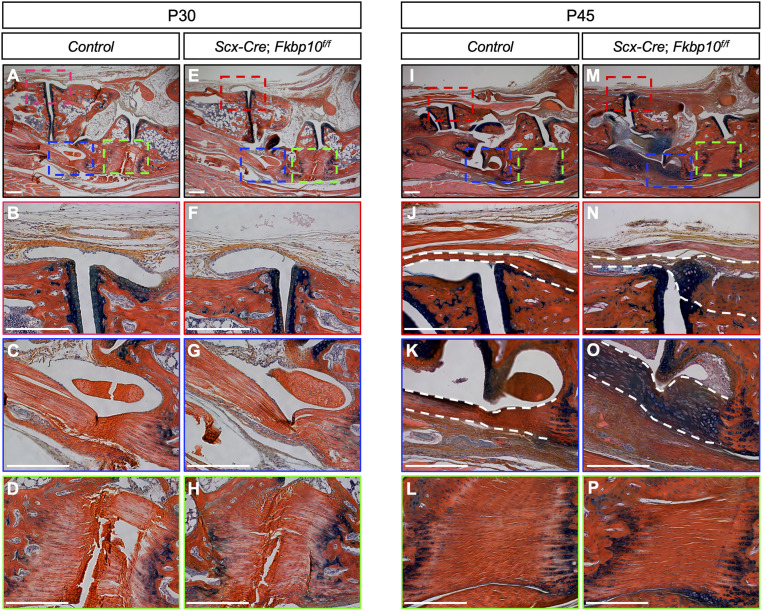
To investigate the cellular basis underlying the joint deformities in Fkbp10 mutants, we next performed histological analyses. Tendons and ligaments appeared comparable between ScxCKO and littermate controls at 1 mo of age (Fig. 2 A–H). However, at 1.5 mo of age, ScxCKO mice showed ectopic chondrogenesis in the midfoot joints between the metatarsal and tarsal bones, as evidenced by the increase in Alcian blue staining (Fig. 2 I–P). Specifically, ectopic chondrogenesis was observed in the tarsometatarsal ligaments (Fig. 2 J and N) and the long plantar ligaments (Fig. 2 K and O) that are located in the midfoot joints of ScxCKO mice, whereas the short plantar ligament connecting the calcaneus and cuboid showed normal histology at this stage (Fig. 2 L and P). In addition, we found that the majority of cells that contribute to aberrant chondrogenesis are Scx-Cre−expressing cells (SI Appendix, Fig. S4). Therefore, these results suggest that ectopic chondrogenesis in the midfoot joints of ScxCKO mice likely contributes to the onset of joint deformities. Furthermore, the short plantar ligament in ScxCKO animals is also affected by ectopic chondrogenesis at later stages (see Fig. 4 P and S), indicating progressive and wider-spread deformities.
Fig. 2.
Fkbp10 deletion triggers ectopic chondrogenesis. Hematoxylin, eosin/orange G, and Alcian blue staining of midfoot joints. (A and E) Low-magnification image of the midfoot joints in 30-d-old mice. High-magnification image of (B and F) metatarsal and cuneiform joint, (C and G) long plantar ligament, and (D and H) short plantar ligament. (I and M) Low-magnification image of the midfoot joints in 45-d-old mice. High-magnification image of (J and N) metatarsal and cuneiform joint, (K and O) long plantar ligament, and (L and P) short plantar ligament. Red, blue, and green boxes in A, E, I, and M indicate the location of high-magnification images in B, F, J, and N, in C, G, K, and O, and in D, H, L, and P, respectively. Dashed white lines demarcate regions affected by chondrogenic lesions. Representative images are shown (n = 4 per group). (Scale bar, 250 μm.)
Consistent with previous studies (43), we found stochastic expression of Scx-Cre in a subset of articular and growth plate chondrocytes in postnatal mice, using the R26mT/mG reporter mouse line (SI Appendix, Figs. S5A and S3B). To rule out any possibility that loss of Fkbp10 in chondrogenic cell populations contributes to the joint deformities observed in ScxCKO mice, we conditionally removed Fkbp10 using the Col2-Cre mouse line (hereafter Col2CKO). Col2CKO mice did not show joint deformities at 2.5 mo of age (SI Appendix, Fig. S5C). In addition, ectopic chondrogenesis was not observed in the midfoot joints of Col2CKO animals, up to 6 mo of age (SI Appendix, Fig. S5 D–K), despite efficient deletion of Fkbp10 protein in chondrocytes (SI Appendix, Fig. S6 A and B). Furthermore, the growth plate length of Col2CKO mice was comparable to that of littermate controls (SI Appendix, Fig. S6 C–E). Thus, Fkbp10 is dispensable for chondrogenesis, and cartilage-specific Fkbp10 deletion does not contribute to postnatal joint deformities.
RNA Sequencing Reveals Enhanced Fibrosis and Inflammatory Gene Expression in Fkbp10 Mutants.
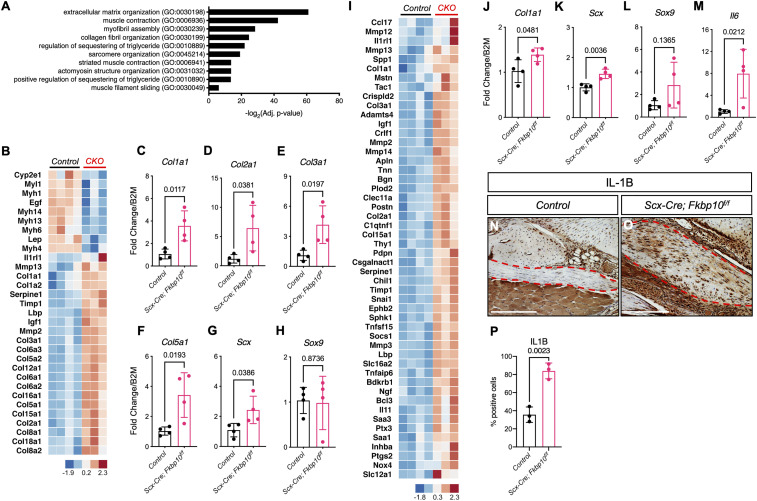
To examine the consequence of tendon- and ligament-specific removal of Fkbp10 on gene expression, we next performed RNA sequencing (RNA-seq) on midfoot joints isolated from ScxCKO mice at 2.5 mo of age. The data showed 517 up-regulated and 308 down-regulated genes with a threshold of a twofold change, and a false discovery rate-adjusted P value below 0.05 with differential gene expression analysis based on the negative binomial distribution (DESeq2) analysis. Gene ontology analysis showed a significant enrichment of differentially expressed genes involved in ECM organization (GO:0030198) (Fig. 3A). In addition, data analysis using the ingenuity pathway analysis (IPA) software (Qiagen) showed “Fibrosis” (Fig. 3B) as one of the top pathways affected. The qRT-PCR experiments confirmed the increase in Col1a1, Col2a1, Col3a1, and Col5a1 expression in Fkbp10 mutant mice at 2.5 mo of age (Fig. 3 C–F). We also found elevated Scx expression but similar Sox9 levels in the midfoot joints of Fkbp10 mutant mice (Fig. 3 G and H). Next, IPA analysis of potential upstream regulators predicted “activated tumor necrosis factor (TNF)” based on increased expression of fibrillar collagens, matrix metalloproteinases (MMPs), and inflammatory markers in ScxCKO mice (Fig. 3I). Interestingly, qRT-PCR experiments showed a mild but significant increase in Col1a1, Scx, and Il6 but not Sox9 expression in the midfoot joints of Fkbp10 mutant mice as early as 3 wk of age, in the absence of histological abnormalities. Furthermore, immunohistochemistry showed elevated levels of IL1B in the chondrogenic nodules of Fkbp10 mutant animals at 45 d of age (Fig. 3 N–P). Thus, these results suggest that Fkbp10 mutant mice display up-regulation of fibrosis and inflammatory genes in the midfoot joints that is likely due to localized tissue injury.
Fig. 3.
RNA-seq reveals fibrosis and inflammatory gene signatures in Fkbp10 mutants. (A) Gene ontology (biological process) analysis results of RNA-seq data. (B) Heatmap display of IPA showing “fibrosis” pathway genes. (C–H) The qRT-PCR confirmation of RNA-seq results for Col1a1, Col2a1, Col3a1, Col5a1, Scx, and Sox9 at 2.5 mo of age (n = 4 per group). (I) Heatmap display of IPA showing “activated TNF” pathway genes. (J–M) The qRT-PCR results of Col1a1, Scx, Sox9, and Il6 at 21 d of age (n = 4 per group). (N and O) Immunohistochemistry of IL-1B at 45-d-of-age. (P) Quantification of IL-1B immunohistochemistry results (n = 3 per group). Control, Cre-negative littermates; CKO, Scx-Cre;Fkbp10f/f. Bar indicates range of log2 expression values transformed to z scores. Data are presented as means ± SD. Actual P values from unpaired t test are shown. P < 0.05 were considered statistically significant. Dashed red lines demarcate regions affected by chondrogenic lesions. (Scale bar, 250 μm.)
Inhibition of Hh Signaling Attenuates Ectopic Chondrogenesis, Joint Deformities, and Gait Defects Caused by Fkbp10 Deletion.
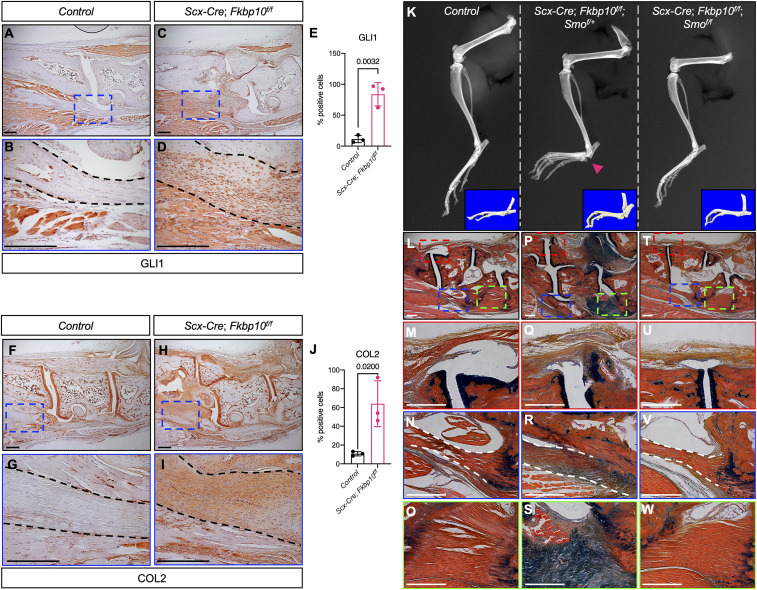
Previous studies have shown that connective tissue abnormalities due to either developmental defects or injury can cause elevated Hh signaling (34, 44). Furthermore, activated Hh signaling by overexpression of Smoothened (Smo) or deletion of Suppressor of fused (Sufu) in tendons or tendon progenitors, respectively, has been reported to stimulate chondro-ossification in the tendon midsubstance (41, 45). Thus, to test whether Hh signaling is altered in ScxCKO animals, we examined Gli1 expression by immunohistochemistry. Indeed, high levels of Gli1 were detected in the chondrogenic lesions of ScxCKO mice relative to comparable regions in control mice (Fig. 4 A–E). In addition, the Gli1 expression pattern largely overlapped with the ectopic expression of type II collagen in the midfoot joints of ScxCKO mice (Fig. 4 F–J). As Indian hedgehog (Ihh) is thought to be the main Hh ligand expressed in the skeleton of postnatal mice (45, 46), we also examined Ihh expression. Consistent with Gli1 expression, we found abundant levels of Ihh in regions of ectopic chondrogenesis in ScxCKO mice (SI Appendix, Fig. S7 A–D). Because Ihh can stimulate Bmp signaling, we determined pSmad1/5 levels. Interestingly, pSmad1/5 was detected in a subset of cells within the chondrogenic nodules of ScxCKO mice (SI Appendix, Fig. S7 E–H). These results indicate that Fkbp10 deficiency leads to localized dysregulation of the Hh pathway.
Fig. 4.
Genetic inhibition of Hh signaling delays joint deformities in Fkbp10 mutants. (A and C) Low- and (B and D) high-magnification images of GLI1 immunohistochemistry at 45 d of age. (E) Quantification of GLI1 immunohistochemistry (n = 3 per group). (F and H) Low- and (G and I) high-magnification images of COL2 at 45 d of age. (J) Quantification of COL2 immunohistochemistry (n = 3 per group). Blue box in A, C, F, and H indicates the location of high-magnification images in B, D, G, and I. Dashed black line demarcates chondrogenic lesions. Data are presented as means ± SD. Actual P values from unpaired t test are shown. P < 0.05 were considered statistically significant. (K) Hindlimb radiographs at 70 d of age in control (Left), Scx-Cre;Fkbp10f/f;Smof/+ (Center), and Scx-Cre;Fkbp10f/f;Smof/f (Right) mice. (Insets) The 3D microCT images of the foot. Red arrowhead, joint deformity. (L–W) Hematoxylin, eosin/orange G, and Alcian blue staining of midfoot joints. (L, P, and T) Low-magnification image of midfoot joints, and high-magnification image of (M, Q, and U) metatarsal and cuneiform joint, (N, R, and V) long plantar ligament, and (O, S, and W) short plantar ligament. Red, blue, and green boxes in L, P, and T indicate the location of high-magnification images in M, Q, and U, in N, R, and V, and in O, S, and W, respectively. Dashed white lines demarcate regions affected by chondrogenic lesions. Representative images are shown (n = 5 per group). (Scale bar, 250 μm.)
Next, to test whether inhibition of Hh signaling can prevent ectopic chondrogenesis in ScxCKO mice, in vivo, we genetically removed Smo in ScxCKO mice. ScxCKO mice with heterozygous removal of Smo showed joint deformities in the hind paw of 70-d-old animals (Fig. 4 K, Left and Center), and were phenotypically indistinguishable from ScxCKO mice. In contrast, homozygous removal of both Fkbp10 and Smo prevented joint deformities, and the radiographs and microCT images appeared similar to those of littermate controls (Fig. 4K, Right). Consistent with previous reports (47), Smo deletion had no effect on either cancellous or cortical bone formation in any of the groups examined (SI Appendix, Fig. S8). Histology showed ectopic chondrogenesis in the midfoot joints of Scx-Cre;Fkbp10f/f;Smof/+ mice compared to littermate controls (Fig. 4 L–S). Importantly, ectopic chondrogenesis in the midfoot joints was prevented in Scx-Cre;Fkbp10f/f;Smof/f mice (Fig. 4 T–W). At the molecular level, Sox9 expression was visibly increased within the chondrogenic lesions affecting the short plantar ligaments of Fkbp10 mutants, whereas comparable regions in either control or Scx-Cre;Fkbp10f/f;Smof/f mice showed very few Sox9-expressing cells (SI Appendix, Fig. S9 A–C). Similarly, IL-1B and P-p38 levels were enhanced in Fkbp10 mutants concomitant with increased cell death (SI Appendix, Fig. S9 D, E, G, H, J, and K), indicating tissue injury and inflammation. In contrast, inflammatory markers in Scx-Cre;Fkbp10f/f;Smof/f mice were comparable to those of littermate control, and cell death was restricted to the attachment sites (SI Appendix, Fig. S9 F, I, and L). Taken together, these results suggest that inhibition of Hh signaling may prevent joint deformities by attenuating ectopic chondrogenesis in Scx-Cre−expressing cells.
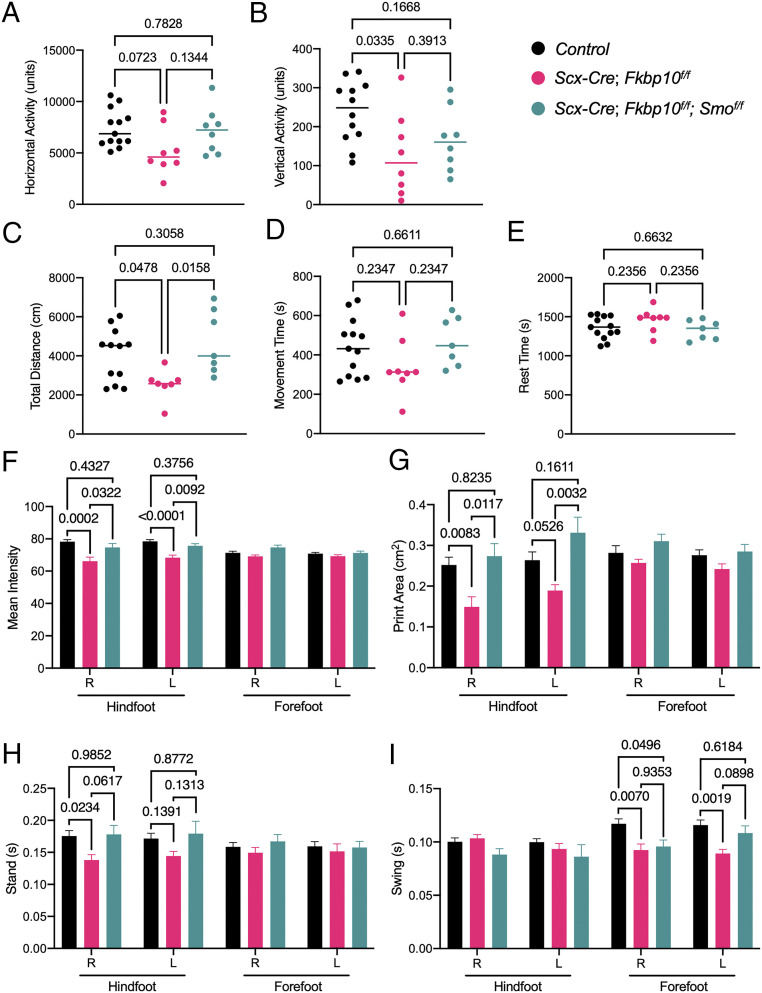
FKBP10 OI mutations negatively impact the mobility of patients with joint contractures. To determine whether the postnatal joint deformities in ScxCKO mice impact motor function, we next performed a series of behavior analyses. The open field assessment revealed that Fkbp10 deletion reduces horizontal and vertical activity as well as total distance traveled (Fig. 5 A–C), whereas movement and rest time are unaffected relative to littermate controls (Fig. 5 D and E). These results suggest that Fkbp10 removal leads to substantial defects in overall locomotor function. Inhibition of the Hh pathway rescued the decrease in total distance but not vertical activity in Fkbp10 mutant mice (Fig. 5 B and C). In addition, gait analysis using the CatwalkXT instrument showed that the hind paws of ScxCKO mice had decreased mean intensity and print area (Fig. 5 F and G), indicating a reduction in the weight placed on the hind paw during active movement. This reduction in hind paw intensity was accompanied by a decrease in stand (i.e., contact duration) for the hind paw (Fig. 5H) and a concomitant decrease in swing (i.e., noncontact duration) of the forepaw (Fig. 5I). The mean intensity, print area, and stand was unchanged in forepaws, which is consistent with the absence of joint deformities in the forefoot region (SI Appendix, Fig. S10). Importantly, alterations in mean intensity, print area, and stand time in Fkbp10 mutant mice were rescued to control levels in Fkbp10 and Smo double-conditional knockout mice (Fig. 5 F–H). Collectively, the data indicate that ScxCKO mice display a marked reduction in open field activity, similar to FKBP OI patients. Importantly, open field activity and gait abnormalities in Fkbp10 mutant mice were partially rescued through genetic inhibition of Hh signaling, indicating that dysregulated Hh may play a substantial role in FKBP10 OI joint dysfunction.
Fig. 5.
Hh pathway inhibition restores gait defects in Fkbp10 mutant mice. VersaMax open field assessment parameters showing (A) horizontal activity (units), (B) vertical activity (units), (C) total distance traveled (centimeters), (D) total movement time (seconds), and (E) total resting time (seconds) of animals at 70 d of age (n = 7 to 13 per group). Data are presented as means ± SD. Actual P values from Mann−Whitney test are shown. CatwalkXT assay parameters showing (F) mean intensity, (G) print area, (H) stand time, and (I) swing time of right (R) and left (L) hind paws or forepaws from mice at 70 d of age (n = 5 to 13 per group). Data are presented as means ± SEM. Actual P values from one-way ANOVA are shown. P < 0.05 were considered statistically significant.
Discussion
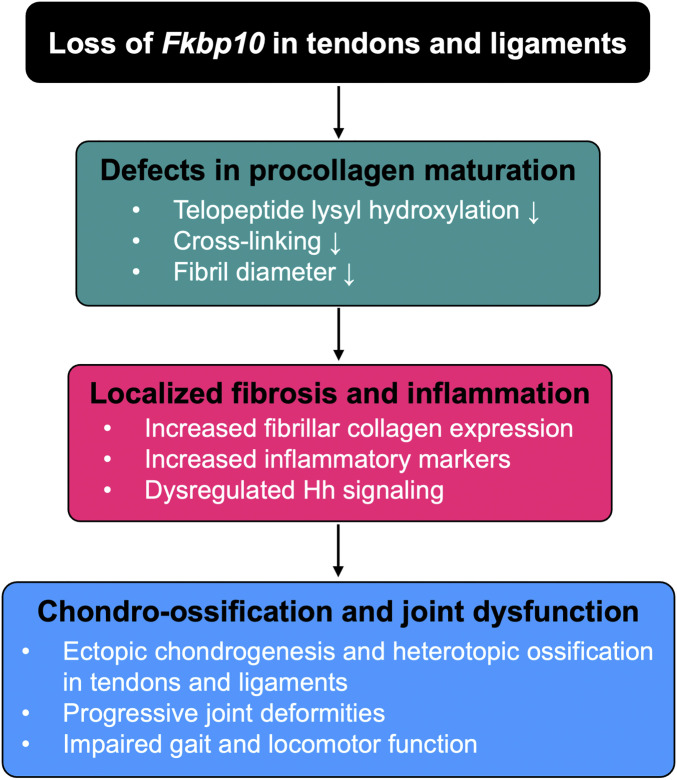
In this study, we demonstrate that the conditional removal of Fkbp10 in tendons and ligaments reduces telopeptide lysyl hydroxylation of type I procollagen and collagen cross-linking, which negatively affected tendon fibrillogenesis. The biochemical deficiencies and impaired fibril growth in Fkbp10 mutant mice were associated with site-specific ectopic chondrogenesis in connective tissues followed by joint deformities and HO. Transcriptomic analysis of midfoot joints by RNA-seq revealed enrichment of fibrosis and inflammation gene signatures in Fkbp10 mutants, indicating connective tissue injury. As previous studies have shown that tendon injuries are frequently associated with alterations in developmental signaling pathways, we investigated whether Hh signaling is dysregulated in Fkbp10 mutant animals. Indeed, we found abundant Gli1 expression in the ectopic chondrogenic nodules of Fkbp10 mutants. Furthermore, we discovered that inhibition of the Hh signaling pathway by genetic removal of Smo can attenuate ectopic chondrogenesis and prevent subsequent joint deformities. Importantly, Hh inhibition restored gait abnormalities in Fkbp10 mutants. Overall, we have investigated the stepwise progression of joint dysfunction (Fig. 6) and established the Fkbp10 mutant mice as a model of FKBP10 OI joint dysfunction. Thus, our tool may provide a valuable platform to discover new targets for therapeutic intervention in OI and OI-related joint diseases.
Fig. 6.
Graphical description of working model. Conditional ablation of Fkbp10 in tendons and ligaments reduces telopeptide lysyl hydroxylation of type I procollagen, cross-linking, and fibril diameter distribution in postnatal mice, suggesting defects in procollagen maturation. This is associated with increased fibrosis and inflammatory gene signatures as well as elevated Gli1 expression in the midfoot joints of Fkbp10 mutant mice. The biochemical and molecular changes due to Fkbp10 removal lead to localized chondro-ossification of tendons and ligaments followed by joint deformities and impaired gait and locomotor function.
The reduction in telopeptide lysyl hydroxylation of type I procollagen due to Fkbp10 deletion had little effect on tendons and ligaments during embryonic development. However, ectopic chondrogenesis was observed in the midfoot joints of Fkbp10 mutants (ScxCKO) postnatally, between 6 and 7 wk. Previous studies have shown that tendon fibril number, area, and diameter undergo rapid growth between birth and 6 wk of age (48). We found that ScxCKO mice showed decreased collagen fibril diameter distribution during this period of active tendon growth. Thus, Fkbp10 deletion likely affects tendon fibrillogenesis during early postnatal stages, which likely accounts for the late onset of joint deformities.
Ectopic chondrogenesis in ScxCKO mice site-specifically affected the lateral midfoot joints that articulate the calcaneus and cuboid, whereas the forefoot or medial midfoot joints were unaffected. The calcaneocuboid joint is a key component of the lateral longitudinal arch which is supported by both dorsal and plantar ligaments (49). In a study by Cambré et al. (50), the calcaneus−cuboid−metatarsal V joints displayed greater sensitivity to biomechanical loads relative to other joints of the foot, which may explain the localized induction of ectopic chondrogenesis in Fkbp10 mutant mice. Additional studies are warranted to examine whether the site-specific induction in chondrogenic nodule formation is influenced by differential responses to biomechanical loading.
Previous studies have shown that defects in tenocyte maturation or acute tendon injury can induce ectopic chondrogenesis and HO in tendons and ligaments, which is associated with dysregulated Hh signaling (34, 51). Furthermore, genetic activation of the Hh pathway in tendon progenitors is sufficient to stimulate HO (41). Consistent with these findings, we identified abundant Gli1 and Ihh expression in the chondrogenic lesions found in Fkbp10 mutants. The precise mechanism by which Hh signaling is dysregulated in Fkbp10 mutants remains unclear. One possibility is that abnormalities in the ECM enhance Hh ligand availability. Alternatively, defects in collagen fibrillogenesis may initiate an injury response and stimulate inflammation, which can lead to increased Hh signaling (52). Nonetheless, our data show that the simultaneous deletion of Fkbp10 and Smo with Scx-Cre can prevent ectopic chondrogenesis, joint deformities, and gait abnormalities. Taken together, these results suggest that Scx-Cre−expressing cells contribute to ectopic chondrogenesis in Fkbp10 mutants. Our data also show that using Hh inhibitors may have clinical implications for OI patients with joint defects. Previous studies have suggested the use of Hh antagonists as a potential therapy to treat HO (44). In addition, Food and Drug Administration-approved Hh inhibitors have been used clinically to treat basal cell carcinoma (53). While this remains an exciting possibility, the potential adverse effects of inhibiting Hh signaling must be taken into consideration, as they may impact the development and maintenance of joint tissues.
We have described a unique mouse model of OI joint dysfunction by conditional deletion of Fkbp10 in tendons and ligaments. Although the etiology of the joint contracture phenotypes in human OI patients remains to be further investigated, our animal model shows that joint deformities may be facilitated by ectopic chondrogenesis in connective tissues due to the loss of Fkbp10. Finally, we demonstrate that inhibition of the Hh signaling pathway may be a potential means of therapeutic intervention for FKBP10 OI patients with joint contractures. Future studies are warranted to determine whether targeting inflammatory pathways in addition to Hh signaling may be beneficial to prevent chondro-ossification in patients with FKBP10 OI as well as OI-related joint diseases.
Materials and Methods
Animals.
Scx-Cre (54), Col2-Cre (55), Fkbp10f/f (56), Smof/f (57), and R26tdT (JAX, 007909) and R26mT/mG (58) mice were previously described. Mice were housed in the Baylor College of Medicine vivarium in a pathogen-free environment with ad libitum access to food and water and were maintained under a 14-h light/10-h dark cycle. All studies were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee and Center for Comparative Medicine (protocol number AN-1506).
X-ray and MicroCT.
X-ray imaging was performed with the Kubtec XPERT80 (Kubtec Medical Imaging). MicroCT was performed using a SCANCO mCT-40 system (55-kVp and 145-mA X-ray source; Scanco Medical AG). The 3D reconstruction was completed with ORS Dragonfly. Bone parameters, including bone volume, total volume, trabecular number, trabecular thickness, and trabecular separation, were measured in the cancellous region of the distal femur by analyzing 75 slides, and cortical thickness was measured by analyzing 11 slides in the femur midshaft. Hindfoot joint deformities were determined by morphological assessment. Mice were examined daily, beginning at 1 mo of age, until deformities (hindfoot bending) were detected. Additionally, X-ray radiography imaging was used to confirm the separation of the calcaneus and cuboid.
Collagen Cross-Link and Mass Spectral Analyses.
Pyridinoline cross-links (hydroxylysylpyridinoline [HP] and lysylpyridinoline [LP]) were quantified by high performance liquid chromatography (HPLC) after hydrolyzing saline washed tendons in 6N HCl as described (59). Briefly, samples were dried and redissolved in 1% (vol/vol) n-heptafluorobutyric acid and run on an Agilent 1260 HPLC (Agilent Technologies) using a C18 RP-HPLC (Brownlee Aquapore RP-300 7u, 250 × 4.6 mm; PerkinElmer) running at 1 mL/min. Samples were eluted with a linear gradient of 17 to 21% acetonitrile in a running buffer of 0.01M n-heptafluorobutyric acid in water. CNBr peptides were run on 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) using the method of Laemmli. CB peptide bands were cut from SDS/PAGE gels and digested with trypsin in-gel (60). Peptides were analyzed by electrospray liquid chromatography–mass spectrometry (LC/MS) using an LTQ XL ion trap mass spectrometer (Thermo Scientific) equipped with inline LC using a C4 5-mm capillary column (300 μm × 150 mm; Higgins Analytical; RS-15M3-W045) eluted at 4.5 μL/min. The LC mobile phase consisted of buffer A (0.1% formic acid in Milli-Q water; Millipore) and buffer B (0.1% formic acid in 3:1 acetonitrile:n-propanol vol/vol). An electrospray ionization source introduced the LC sample stream into the mass spectrometer with a spray voltage of 4 kV. Proteome Discoverer search software (Thermo Scientific) was used for peptide identification using the National Center for Biotechnology Information protein database (https://www.ncbi.nlm.nih.gov/protein). Large collagenous peptides not found by Sequest had to be identified manually by calculating the possible tandem MS (MS/MS) ions and matching these to the actual MS/MS. Hydroxyproline and hydroxylysine calculations were done manually by scrolling or averaging the full scan over several minutes so that all of the posttranslational variations of a given peptide appeared together in the full scan.
Open Field and Gait Measurements.
Open field analysis was performed with the VersaMax Animal Activity Monitoring System (AccuScan Instruments). Briefly, mice were acclimated to the test room in their home cages for at least 30 min with the brightness and noise levels normalized to 50 lx and 60 dB, respectively. Mice were then transferred to individual chambers (dimensions: 40 cm × 40 cm × 30 cm) with clear plastic boundaries and were allowed to move freely for 30 min in the absence of the investigator. Movement parameters that measure horizontal, vertical, and regional movement were recorded by the VersaMax activity monitoring software. Chambers were cleaned with 30 to 50% ethanol to remove the scent of previously tested mice between each run.
Gait performance was measured using the CatwalkXT system (Noldus). Mice were acclimated to the test room in their home cages for at least 30 min. The walking platform was sanitized with 70% ethanol and wiped down until completely dry. Each mouse was moved to one end of the platform when the camera recording was active. Three full recordings of the mouse walking on the platform within the camera view in less than 10 s were required for a complete run. Data analyses were performed with the CatwalkXT software. Data are shown as mean ± SEM.
Histology and Immunohistochemistry.
Mice were killed, and foot and knee joints were dissected and fixed in 10% neutral buffered formalin for 24 h to 48 h at 4 °C with gentle shaking. Isolated tissues were briefly washed in phosphate-buffered saline (PBS), then decalcified using Immunocal (Statlab) for 72 h at 4 °C with gentle shaking followed by processing and paraffin embedding using standard protocols. Samples were sectioned at 7 μm and stained with hematoxylin and eosin or with hematoxylin, eosin/orange G, and Alcian blue. For cryosection, tissues were excised and fixed immediately in ice-cold 4% paraformaldehyde (pH 7.4 to 7.6) and incubated overnight at 4 °C with shaking. Fixed samples were washed in ice-cold PBS, then decalcified with Immunocal (Stat Lab) for 72 h at 4 °C with shaking. Samples were transferred to 15% sucrose/PBS solution and incubated for 1 h, followed by 30% sucrose/PBS. Embedding was performed using optimal cutting temperature (OCT) compound, then sectioned using a Leica cryostat equipped with Cryojane at 7-μm thickness.
Immunohistochemistry was performed using Fkbp10 (Protein Tech, 12172-1-AP), Gli1 (R&D, AF3455), Collagen Type II (Developmental Studies Hybridoma Bank, II-II6B3), Ihh (Abcam, ab39634), pSmad1/5 (Cell Signaling, 9516), P-p38 (Cell Signaling, 4511), Sost (R&D, BAF1589), and Sox9 (Millipore Sigma, AB5535) antibodies. Briefly, slides were baked at 55 °C to 60 °C for 2 h, then deparaffinized. Slides were then incubated in proteinase K solution (20 mg/mL) followed by 3% hydrogen peroxide in methanol for 10 min each, with PBS rinse in between. Blocking was performed using 5% normal donkey serum diluted in PBS for 1 h at room temperature. Primary antibodies were diluted in blocking solution at 1:100 and incubated overnight at 4 °C. Slides were washed three times with Tris-buffered saline, 0.1% Tween 20 (TBST) for 10 min each. Next, slides were incubated in biotinylated anti-Rabbit or anti-Goat antibodies diluted in blocking solution at 1:300 and incubated at room temperature for 1 h, followed by TBST wash three times for 10 min each at room temperature. Slides were incubated in streptavidin-horseradish peroxidase (HRP) diluted in blocking buffer at 1:300 dilution for 30 min. After TBST wash, slides were incubated in NovaRED or 3,3'Diaminobenzidine (DAB, DAKO) substrate for 2 min then cleared and mounted with a coverslip. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was performed using the In Situ Cell Death Detection Kit (Millipore Sigma, 12156792910) according to manufacturer’s instructions.
RNA-seq, qRT-PCR, and Data Analysis.
Midfoot joints (between the calcaneocuboid and tarsometatarsal joints) immediately distal to the calcaneus and cuboid were dissected and subsequently pulverized using Tissuelyser II (Qiagen). RNA was isolated with the Fibrous Connective Tissue Kit and on-column DNase I digestion (Qiagen). RNA-seq quality assessment, library preparation, and sequencing with Illumina HiSeq (150-bp paired-end reads) were performed by GENEWIZ. IPA (Qiagen) and GENIALIS software were used to perform canonical pathways, predicted upstream regulators, and gene ontology. Gene expression data were normalized per million transcripts. Differential gene expression was performed using the DESeq2 algorithm through the GENIALIS platform. The qRT-PCR was performed using the LightCycler 96 System (Roche) with gene-specific primers and FastStart SYBR Green I (Roche) following complementary DNA synthesis with iScript supermix (Bio-Rad) using 500 ng of RNA per group. Primer sequences are listed in Table 1.
Table 1.
The qRT-PCR primer sequence information
| Gene | Forward primer | Reverse primer |
| B2m | GGTCTTTCTGGTGCTTGTC | CGTATGTATCAGTCTCAGT |
| Col1a1 | TTGGGGCAAGACAGTCATCGAAT | TTGGGGTGGAGGGAGTTTACACGAA |
| Col2a1 | GCTCATCCAGGGCTCCAATGATGTAG | CGGGAGGTCTTCTGTGATCGGTA |
| Col3a1 | CCAGTGGCCATAATGGGGAA | ATCTCGACCTGGCTGACCAT |
| Col5a1 | TAAACAGAAGGGCTGTGTGATAAA | CTGCTGGGAAATATACAATGTACCGT |
| Scx | AAGACGGCGATTCGAAGTTAGAAG | TCTCTCTGTTCATAGGCCCTGCTCATAG |
| Sox9 | CCTTTTCTTTGTTGTTTTCGTTGTTG | AACACACGCACACATCCACATACAGT |
| Il6 | GGAAATCGTGGAAATGAGAAA | GAATTGGATGGTCTTGGTCCTTAG |
Statistical Analysis.
All data are presented as means ± SD with individual data points unless otherwise indicated. Normal distribution of data was examined using the Shapiro−Wilk test. Data with normal distribution were analyzed using unpaired t tests for comparing two groups, and one-way ANOVA for comparing more than two groups. For data whose residuals did not have a normal distribution, two groups were compared using the Mann−Whitney test. The representative images of at least three biological replicates per group are shown for all data regarding animal morphology, radiographs, microCT, histology, and immunohistochemistry. Prism 9.0.0 (GraphPad) was used for all statistical analysis. The P value was reported for relevant statistical analysis, with P < 0.05 considered as statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants P01 HD070394 (B.H.L, D.K., and D.E.), F32AR070612 (J.L.), and F31DE02248 (C.L.); the Baylor College of Medicine (BCM) Intellectual & Developmental Disabilities Research Center (Grants HD024064 and U54HD083092) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (https://www.nichd.nih.gov/Pages/index.aspx); the BCM Advanced Technology Cores with funding from the NIH (Grants AI036211, CA125123, and RR024574) (https://www.nih.gov/); the Rolanette and Berdon Lawrence Bone Disease Program of Texas (https://www.mdanderson.org/research/departments-labs-institutes/programs-centers/bone-disease-program-of-texas.html); and the BCM Center for Skeletal Medicine and Biology (https://www.bcm.edu/research/labs-and-centers/research-centers/center-for-skeletal-medicine-and-biology). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100690118/-/DCSupplemental.
Data Availability
All study data are included in the article, SI Appendix, and Dataset S1.
References
- 1.Lim J., Grafe I., Alexander S., Lee B., Genetic causes and mechanisms of osteogenesis imperfecta. Bone 102, 40–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marini J. C., et al., Osteogenesis imperfecta. Nat. Rev. Dis. Primers 3, 17052 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Rauch F., Glorieux F. H., Osteogenesis imperfecta. Lancet 363, 1377–1385 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Van Dijk F. S., Sillence D. O., Osteogenesis imperfecta: Clinical diagnosis, nomenclature and severity assessment. Am. J. Med. Genet. A. 164A, 1470–1481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etich J., et al., Osteogenesis imperfecta—Pathophysiology and therapeutic options. Mol. Cell Pediatr. 7, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asharani P. V., et al., Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am. J. Hum. Genet. 90, 661–674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Slot A. J., et al., Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 278, 40967–40972 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Morello R., et al., CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127, 291–304 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Alanay Y., et al., Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 86, 551–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabral W. A., et al., Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 39, 359–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dijk F. S., et al., PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet. 85, 521–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes A. M., et al., Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. N. Engl. J. Med. 362, 521–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis E. C., Broekelmann T. J., Ozawa Y., Mecham R. P., Identification of tropoelastin as a ligand for the 65-kD FK506-binding protein, FKBP65, in the secretory pathway. J. Cell Biol. 140, 295–303 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa Y., Vranka J., Wirz J., Nagata K., Bächinger H. P., The rough endoplasmic reticulum-resident FK506-binding protein FKBP65 is a molecular chaperone that interacts with collagens. J. Biol. Chem. 283, 31584–31590 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Barnes A. M., et al., Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum. Mutat. 33, 1589–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinlein O. K., Aichinger E., Trucks H., Sander T., Mutations in FKBP10 can cause a severe form of isolated Osteogenesis imperfecta. BMC Med. Genet. 12, 152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaheen R., et al., Mutations in FKBP10 cause both Bruck syndrome and isolated osteogenesis imperfecta in humans. Am. J. Med. Genet. A. 155A, 1448–1452 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Kelley B. P., et al., Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J. Bone Miner. Res. 26, 666–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarze U., et al., Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum. Mol. Genet. 22, 1–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X. J., et al., Novel mutations in FKBP10 in Chinese patients with osteogenesis imperfecta and their treatment with zoledronic acid. J. Hum. Genet. 62, 205–211 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bank R. A., et al., Defective collagen crosslinking in bone, but not in ligament or cartilage, in Bruck syndrome: Indications for a bone-specific telopeptide lysyl hydroxylase on chromosome 17. Proc. Natl. Acad. Sci. U.S.A. 96, 1054–1058 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puig-Hervás M. T., et al., Mutations in PLOD2 cause autosomal-recessive connective tissue disorders within the Bruck syndrome–Osteogenesis imperfecta phenotypic spectrum. Hum. Mutat. 33, 1444–1449 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Gjaltema R. A., van der Stoel M. M., Boersema M., Bank R. A., Disentangling mechanisms involved in collagen pyridinoline cross-linking: The immunophilin FKBP65 is critical for dimerization of lysyl hydroxylase 2. Proc. Natl. Acad. Sci. U.S.A. 113, 7142–7147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duran I., et al., HSP47 and FKBP65 cooperate in the synthesis of type I procollagen. Hum. Mol. Genet. 24, 1918–1928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umair M., et al., Homozygous sequence variants in the FKBP10 gene underlie osteogenesis imperfecta in consanguineous families. J. Hum. Genet. 61, 207–213 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Screen H. R., Berk D. E., Kadler K. E., Ramirez F., Young M. F., Tendon functional extracellular matrix. J. Orthop. Res. 33, 793–799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahara H., Inui M., Lotz M. K., Tendons and ligaments: Connecting developmental biology to musculoskeletal disease pathogenesis. J. Bone Miner. Res. 32, 1773–1782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nourissat G., Berenbaum F., Duprez D., Tendon injury: From biology to tendon repair. Nat. Rev. Rheumatol. 11, 223–233 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Schweitzer R., et al., Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855–3866 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Liu W., et al., The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol. 30, 4797–4807 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y., et al., The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 10538–10542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakabe T., et al., Transcription factor scleraxis vitally contributes to progenitor lineage direction in wound healing of adult tendon in mice. J. Biol. Chem. 293, 5766–5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murchison N. D., et al., Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697–2708 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Liu H., Xu J., Jiang R., Mkx-deficient mice exhibit hedgehog signaling-dependent ectopic ossification in the Achilles tendons. J. Bone Miner. Res. 34, 557–569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi Y., et al., Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219–1227 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Nelson C. M., Bissell M. J., Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 22, 287–309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickup M. W., Mouw J. K., Weaver V. M., The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15, 1243–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grafe I., et al., Excessive transforming growth factor-β signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 20, 670–675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neptune E. R., et al., Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 33, 407–411 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Sinha S., et al., Unsuspected osteochondroma-like outgrowths in the cranial base of Hereditary Multiple Exostoses patients and modeling and treatment with a BMP antagonist in mice. PLoS Genet. 13, e1006742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng H., et al., Tendon-derived cathepsin K-expressing progenitor cells activate Hedgehog signaling to drive heterotopic ossification. J. Clin. Invest. 130, 6354–6365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lietman C. D., et al., Connective tissue alterations in Fkbp10-/- mice. Hum. Mol. Genet. 23, 4822–4831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimoto Y., et al., Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep. 7, 45010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regard J. B., et al., Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat. Med. 19, 1505–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C. F., Breidenbach A., Aschbacher-Smith L., Butler D., Wylie C., A role for hedgehog signaling in the differentiation of the insertion site of the patellar tendon in the mouse. PLoS One 8, e65411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felsenthal N., et al., Development of migrating tendon-bone attachments involves replacement of progenitor populations. Development 145, dev165381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz A. G., Long F., Thomopoulos S., Enthesis fibrocartilage cells originate from a population of Hedgehog-responsive cells modulated by the loading environment. Development 142, 196–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalson N. S., et al., A structure-based extracellular matrix expansion mechanism of fibrous tissue growth. eLife 4, e05958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angin S., Simsek I. E., Comparative Kinesiology of the Human Body: Normal and Pathological Conditions (Elsevier, Waltham, MA, 2020). [Google Scholar]
- 50.Cambré I., et al., Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nat. Commun. 9, 4613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., et al., Inhibition of overactive TGF-β attenuates progression of heterotopic ossification in mice. Nat. Commun. 9, 551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhuo M., et al., Inflammation-induced JMJD2D promotes colitis recovery and colon tumorigenesis by activating Hedgehog signaling. Oncogene 39, 3336–3353 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Verkouteren B. J. A., et al., Eight years of experience with vismodegib for advanced and multiple basal cell carcinoma patients in the Netherlands: A retrospective cohort study. Br. J. Cancer 124, 1199–1206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blitz E., et al., Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell 17, 861–873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ovchinnikov D. A., Deng J. M., Ogunrinu G., Behringer R. R., Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26, 145–146 (2000). [PubMed] [Google Scholar]
- 56.Lietman C. D., et al., Fkbp10 deletion in osteoblasts leads to qualitative defects in bone. J. Bone Miner. Res. 32, 1354–1367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long F., Zhang X. M., Karp S., Yang Y., McMahon A. P., Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Eyre D., Collagen cross-linking amino acids. Methods Enzymol. 144, 115–139 (1987). [DOI] [PubMed] [Google Scholar]
- 60.Hanna S. L., Sherman N. E., Kinter M. T., Goldberg J. B., Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: An analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology (Reading) 146, 2495–2508 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article, SI Appendix, and Dataset S1.