Significance
Uncovering the genetic and cellular mechanisms of frontotemporal dementia (FTD) may lead to new therapeutic strategies. Mutations in the GRN gene are linked to FTD; the underlying cellular mechanisms are still unknown, but current research points to lysosomal dysfunction. Using the model organism Caenorhabditis elegans, we found a link between pgrn-1/GRN mutations, and sphingolipid metabolism and autophagy. We further identified small molecules that attenuated several pgrn-1/GRN phenotypes in vivo. These results offer sphingolipid metabolism as a potential mechanism contributing to FTD pathogenesis and suggest that efforts to restore sphingolipid homeostasis may be a beneficial therapeutic approach.
Keywords: frontotemporal dementia, progranulin, Caenorhabditis elegans
Abstract
In 2006, GRN mutations were first linked to frontotemporal dementia (FTD), the leading cause of non-Alzheimer dementias. While much research has been dedicated to understanding the genetic causes of the disease, our understanding of the mechanistic impacts of GRN deficiency has only recently begun to take shape. With no known cure or treatment available for GRN-related FTD, there is a growing need to rapidly advance genetic and/or small-molecule therapeutics for this disease. This issue is complicated by the fact that, while lysosomal dysfunction seems to be a key driver of pathology, the mechanisms linking a loss of GRN to a pathogenic state remain unclear. In our attempt to address these key issues, we have turned to the nematode, Caenorhabditis elegans, to model, study, and find potential therapies for GRN-deficient FTD. First, we show that the loss of the nematode GRN ortholog, pgrn-1, results in several behavioral and molecular defects, including lysosomal dysfunction and defects in autophagic flux. Our investigations implicate the sphingolipid metabolic pathway in the regulation of many of the in vivo defects associated with pgrn-1 loss. Finally, we utilized these nematodes as an in vivo tool for high-throughput drug screening and identified two small molecules with potential therapeutic applications against GRN/pgrn-1 deficiency. These compounds reverse the biochemical, cellular, and functional phenotypes of GRN deficiency. Together, our results open avenues for mechanistic and therapeutic research into the outcomes of GRN-related neurodegeneration, both genetic and molecular.
Frontotemporal dementia (FTD) is a devastating neurodegenerative disorder and the third most common cause of dementia (1). FTD is a leading cause of early-onset disorder, with patients usually diagnosed between 45 and 65 y of age (1, 2). Unlike other dementias, such as Alzheimer’s disease, FTD is a fast-progressing disease and affected patients usually die 2 to 5 y after clinical diagnosis. Advances in genetic screening techniques have identified many of the causative genes, revealing the complex heterogeneity of the underlying molecular mechanisms of the disease (1–3). Among them are autosomal-dominant heterozygous mutations in the GRN gene (4, 5) causing a severe reduction in the circulating levels of its product, progranulin (PGRN) (6). PGRN-deficient FTD is characterized by neuropathological inclusions of ubiquitinated TAR DNA-binding protein 43 (TDP-43) in the cytoplasm (4, 5, 7–10). The presence of these inclusions links FTD and amyotrophic lateral sclerosis (ALS) at the molecular level, as mutations in TDP-43 also cause ALS, and inclusions of the protein are also found in 97% of cases (11). Although more than a decade has passed since the identification of GRN’s involvement in FTD, there is no consensus on the molecular mechanisms linking low PGRN levels and disease pathology. Inversely, the overexpression of PGRN has been found to be protective in multiple models of neurodegeneration, such as ALS (7), Alzheimer’s (12), Parkinson’s (13), and Huntington’s diseases (14), suggesting that it possesses a broad neuroprotective role.
Many animal models of PGRN deficiency have been generated to develop a better understanding of the disease. A study in nematodes has linked PGRN loss to defects in apoptotic cell clearance and abnormal organismal stress response (15, 16). Furthermore, the various mouse models of Grn knockout all displayed exaggerated levels of microgliosis, lysosomal dysfunction, and lipofuscin deposits, but results have been inconsistent in regard to social and behavioral deficits (17–21). Other studies have suggested that the cleavage products of full-length PGRN, the granulin domains, actively promote PGRN pathology (8). Lipofuscin is an autofluorescent pigment that accumulates with age and is composed of lipid by-products derived from lysosomal degradation. Homozygous null mutations in the GRN gene result in neuronal ceroid lipofuscinosis (22–26), a lysosomal storage disorder, supporting a link between PGRN and lysosomes and that PGRN dosage affects the clinical phenotype. In GRN-related FTD, carrying a heterozygous mutation, there is good evidence for milder lysosomal disturbances (27–29), and this is supported by several studies showing a direct link between PGRN and lysosomal function (30–37).
Despite the growing evidence of the importance of PGRN in lysosome function, the mechanistic basis that links PGRN deficiency to lysosome dysfunction and the pathophysiology of FTD remains uncertain. We sought to identify these missing links by studying PGRN in vivo in the model organism Caenorhabditis elegans. This small nematode has been widely used for genetic, behavioral, and anatomical studies. Thanks to genetic conservation throughout evolution, many human genes have orthologs in nematodes (38), including GRN with its ortholog pgrn-1, which has led C. elegans to become a commonly used model to study human genetic disorders. Furthermore, its nervous system, consisting of only 300 neurons, provides a simple model to study complex human neurodegenerative disorders, and its biological similarities to mammalian and human biology make it a superb tool for in vivo drug screening (39–41).
In this study, we have used pgrn-1 mutant animals to model PGRN deficiency in vivo and to better elucidate the causes of the underlying pathology of the disease. We found that loss of pgrn-1 results in distinct aging phenotypes that recapitulate key features of FTD, such as lysosomal dysfunction and defects in autophagic clearance. As we explored possible explanations underlying the pathology of PGRN-deficient FTD-like phenotypes, the sphingolipid (SL) metabolic pathway emerged as a potential key regulator of PGRN’s lysosomal function. Using RNA-mediated interference (RNAi), we identified two genetic targets whose knockdown was capable of restoring many of the FTD-related phenotypes in our C. elegans models. In parallel, we performed a high-throughput drug screen of ∼3,850 compounds, many of which are already approved for human use and identified two capable of rescuing PGRN-deficient phenotypes in nematodes and cultured mammalian cells. Altogether, we identified genetic and chemical modifiers of PGRN deficiency and discovered the involvement of the SL pathway in PGRN pathology. These findings have the potential to open therapeutic options for FTD, and given the broad role of PGRN as a protective factor across the neurodegenerative disease spectrum, these results may extend to a range of neurological disorders.
Results
Loss of pgrn-1 Results in Distinct Phenotypes and Recapitulates Key Features of FTD.
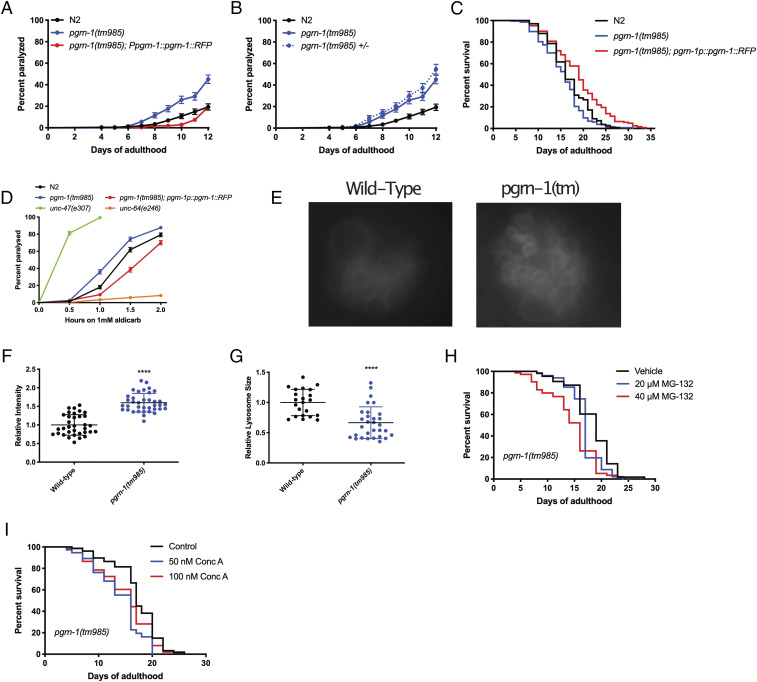
Characterization of pgrn-1(tm985) animals revealed that they exhibited an age-dependent paralysis phenotype, which could be rescued by the re-expression of full-length PGRN-1::red fluorescent protein (RFP) under the control of the endogenous pgrn-1 promoter (Fig. 1A). As the tm985 allele of pgrn-1 is a full deletion resulting in a null allele, we also tested the effect of a substitution mutation in the gene, which resulted in a missense change at the protein level. We selected the gk123284 allele, generated by the Million Mutation Project (42), which resulted in a G119E change in the second exon of pgrn-1. For simplicity, pgrn-1(tm985) mutants will be referred to as pgrn-1(tm), pgrn-1(gk123284) mutants will be referred to as pgrn-1(gk), and pgrn-1p::pgrn-1::RFP animals will be referred to as “PGRN-1 rescue.” Gene expression analysis revealed that pgrn-1(tm) animals had no residual expression of pgrn-1, while pgrn-1(gk) animals displayed a ∼50% reduction in pgrn-1 messenger RNA (mRNA) expression (SI Appendix, Fig. S1A). This second mutant, pgrn-1(gk), also displayed a paralysis phenotype (SI Appendix, Fig. S1B). Interestingly, in both cases, the mutant pgrn-1 alleles were dominant, as heterozygous animals displayed equal rates of paralysis as the homozygous mutants (Fig. 1B and SI Appendix, Fig. S1B). Overexpression of PGRN-1::RFP in pgrn-1(tm) animals displayed no paralysis phenotype compared with wild-type (WT) N2 animals, while its overexpression in a WT background resulted in a slight decrease in spontaneous age-related paralysis (SI Appendix, Fig. S1C). Analysis of the lifespan of these animals revealed that pgrn-1(tm) mutants showed a slight but statistically significant decrease in lifespan, whereas the rescue construct showed a slight increase in lifespan compared with WT animals (Fig. 1C), whereas pgrn-1(gk) mutants showed no lifespan effects (SI Appendix, Fig. S1D). We then subjected pgrn-1(tm) mutants to aldicarb, a potent acetylcholinesterase inhibitor commonly used to study neurotransmission in C. elegans (43). Hypersensitivity to aldicarb can be indicative of increased synaptic acetylcholine release, whereas the resistance to it can suggest the opposite. Here, we observed that both pgrn-1 mutant alleles displayed a heightened sensitivity to aldicarb, with the gk123284 point mutation being slightly less sensitive than the pgrn-1(tm) mutants (Fig. 1D and SI Appendix, Fig. S1E). However, this phenotype was not as severe as that of unc-47(e307) control animals, which have a mutation in their GABA transporter.
Fig. 1.
Loss of pgrn-1 results in distinct FTD-like phenotypes including lysosomal dysfunction. (A) pgrn-1(tm985) mutant animals display age-dependent paralysis which can be rescued by the overexpression of full-length PGRN-1::RFP (Mantel–Cox test: N2 versus pgrn-1(tm985), ****P < 0.0001; N2 versus pgrn-1(tm985); pgrn-1::rfp, ****P < 0.0001). (B) Heterozygous pgrn-1(tm985)/+ animals display the same levels of paralysis as homozygous mutant animals (Mantel–Cox test, n.s.). (C) Complete loss of pgrn-1 leads to a slight reduction in lifespan, whereas the re-expression of full-length PGRN-1 leads to an extension (Mantel–Cox test: N2 versus pgrn-1(tm985), *P < 0.05, N2 versus pgrn-1(tm985); pgrn-1::rfp, ***P < 0.001). (D) pgrn-1 mutant animals display hypersensitivity to aldicarb, while rescue animals expressing PGRN-1::RFP display resistance (Mantel–Cox test: N2 versus pgrn-1(tm985), ****P < 0.0001; N2 versus pgrn-1(tm985); pgrn-1::rfp, ****P < 0.0001). (E) Representative images of pgrn-1(tm985) coelomocyte lysosomes as visualized using an LMP-1::GFP reporter. (F and G) Loss of pgrn-1 leads to an increased fluorescence intensity of LMP-1::GFP lysosomes (Student’s t test, ****P < 0.0001) but smaller ones as evidenced by a reduction in size (Student’s t test, ****P < 0.0001). (H) Treatment pgrn-1 mutant animals with the proteasome inhibitor, MG-132, leads to a dose-dependent decrease in lifespan (Mantel–Cox tests: Vehicle versus 20 μM MG-132, *P < 0.05; Vehicle versus 40 μM MG-132, ****P < 0.0001). (I) Concanamycin A treatment results in a decrease in lifespan in pgrn-1(tm985) animals but is not dose dependent (Mantel–Cox tests: Vehicle versus 20 μM MG-132, ****P < 0.0001; Vehicle versus 40 μM MG-132, *P < 0.05).
In humans, PGRN is a secreted molecule, and previous studies have shown the same is true in C. elegans (16). As a result, we investigated whether the paralysis phenotypes we observed were due to PGRN-1 acting in a cell-autonomous or a non-cell autonomous manner. We performed the knockdown of pgrn-1 by RNAi in non-neuronal tissues and in neuronal tissues (44–46) and found that only the neuron-specific RNAi resulted in paralysis (SI Appendix, Fig. S1 F and G). This suggests that PGRN acts in a cell-autonomous manner, though it remains possible that there are feedback mechanisms from other tissues after the neuronal loss of PGRN. Furthermore, pgrn-1 mutants displayed an overactive food-seeking behavior when starved and crawl up the sides of Petri dishes, a phenotype which is also rescued by the pgrn-1–rescue construct (SI Appendix, Fig. S1H), suggesting neuronal deficits in their ability to properly recognize food sources (47).
One of the key features observed in FTD cases due to GRN mutations is lysosomal dysfunction suggesting a link between PGRN and lysosomal function. We therefore sought to investigate the presence of lysosomal dysfunction in our nematode model. Using a fluorescent reporter for lmp-1/LAMP1, LMP-1::green fluorescent protein (GFP), which localizes to lysosomal membranes, we observed significantly higher levels of GFP fluorescence in the coelomocytes of pgrn-1(tm) mutant animals than in WT animals (Fig. 1 E and F) at 5 d of adulthood and that they had correspondingly smaller-sized lysosomes (Fig. 1G). Other studies in human cells have shown that loss of PGRN results in increased lysosomal biogenesis (33), and therefore, it is possible the increase in LMP-1::GFP intensity could be analogous. To confirm the presence of lysosomal dysfunction, we hypothesized that these worms would be sensitive to impairment of proteasomal degradation (by MG132, a proteasome inhibitor) or through inhibition of lysosomal function (by Concanamycin A, a potent inhibitor of lysosomal vATPases and prevents acidification of the lysosomal lumen). Treatment of worms with MG132 reduced the lifespan of pgrn-1(tm) mutants in a dose-dependent manner (Fig. 1H) but had little effect on the survival of WT and pgrn-1–rescue animals (SI Appendix, Fig. S1 I and J. We observed that pgrn-1(tm) mutant animals displayed reduced lifespan when exposed to ConcA (Fig. 1I), but WT and pgrn-1–rescue animals were unaffected (SI Appendix, Fig. S1 K and L). Altogether, these results suggest that pgrn-1 mutant C. elegans recapitulate key features of FTD and are therefore a biologically relevant model to model this human disease.
Mutations in pgrn-1 Alter Autophagy in C. elegans.
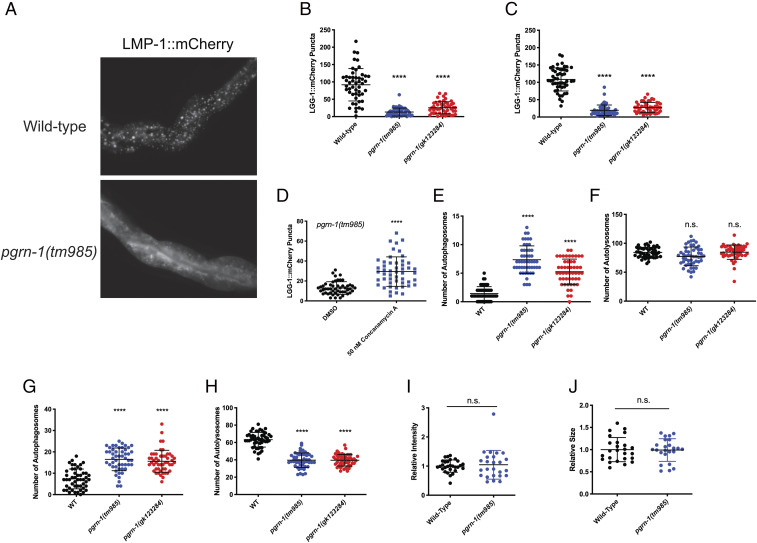
Given the presence of lysosomal defects in pgrn-1 mutants, we sought to better understand its consequences on the autophagic process. We utilized a fluorescent reporter of lgg-1/LC3 (LGG-1::mCherry), which is expressed in the intestinal cells of C. elegans. Under normal autophagy conditions, this transgenic reporter will form distinct red puncta in intestinal cells. However, this was not the case in pgrn-1 mutant animals: at days 5 and 10 of adulthood, LGG-1::mCherry remains diffuse in pgrn-1 mutants (Fig. 2A) and forms significantly fewer puncta than in WT animals (Fig. 2 B and C). There was no change in puncta formation at day 1 (SI Appendix, Fig. S2A). Normally, autophagy levels increase upon conditions of nutrient deprivation, such as starvation (48). However, in both pgrn-1 mutants, starvation did not increase levels of LGG-1::mCherry puncta (SI Appendix, Fig. S2B). However, treatment of pgrn-1 mutant worms with ConcA for 24 h led to increased numbers of LGG-1::mCherry puncta (Fig. 2D). Together, these results suggest that pgrn-1 mutations result in changes in autophagic degradation and may be independent of classical autophagy-inducing mechanisms.
Fig. 2.
Mutations in pgrn-1 result in changes in autophagic flux. (A) LGG-1::mCherry reporter shows the formation of distinct puncta in the intestines of WT animals, whereas it remains diffuse in pgrn-1 mutants. (B and C) Quantification of LGG-1::mCherry puncta at day 5 (B) and day 10 (C) adult animals shows a clear reduction in puncta in both pgrn-1 mutant strains (Student’s t test. Day 5: WT versus pgrn-1(tm985), ****P < 0.0001, WT versus pgrn-1(gk123284), ****P < 0.0001. Day 10: WT versus pgrn-1(tm985), ****P < 0.0001, WT versus pgrn-1(gk123284), ****P < 0.0001). (D) Treatment of day 5 pgrn-1(tm985); lgg-1::mCherry animals with 50 nM Concanamycin A results in an increase in the number of LGG-1::mCherry puncta (Student’s t test, ****P < 0.0001). (E and F) Visualization of neuronal autophagy using a dually tagged LGG-1 reporter, mCherry::lgg-1::gfp, shows that both pgrn-1 mutants display an increased number of autophagosomes (E) but no change in the number of autolysosomes (F) at day 5 of adulthood (Student’s t test. Autophagosomes count (E): WT versus pgrn-1(tm985), ****P < 0.0001, WT versus pgrn-1(gk123284), ****P < 0.0001. Autolysosome count (F): WT versus pgrn-1(tm985), n.s., WT versus pgrn-1(gk123284), n.s.). (G and H) Both pgrn-1 mutations also result in an increased number of autophagosomes at day 1 (G) but also a decreased number of autolysosomes (H) (Student’s t test. Autophagosome count (G): WT versus pgrn-1(tm985), ****P < 0.0001, WT versus pgrn-1(gk123284), ****P < 0.0001. Autolysosome count (H): WT versus pgrn-1(tm985), ****P < 0.0001, WT versus pgrn-1(gk123284), ****P < 0.0001). (I and J) At day 1, pgrn-1(tm985) animals had no significant change in LGG-1::GFP fluorescence intensity (I) or in the size of their lysosomes (J) (Student’s t test. Fluorescence intensity (I): WT versus pgrn-1(tm985), n.s.. Lysosome size (J): WT versus pgrn-1(tm985), n.s.).
To see if these changes were due to changes in autophagic flux, we used a tandem-tagged lgg-1/LC3 reporter, GFP::mCherry::LGG-1. This reporter can provide important information on the degradation process by measuring conversion of autophagosomes to autolysosomes (49). Since GFP emission is limited in low-pH environments, red and green puncta indicate an autophagosome, whereas red-only puncta indicate an autolysosome. Furthermore, this transgene is expressed only in C. elegans neurons, providing important information on the autophagic process specifically in these cells. We quantified autophagosomes by counting green puncta in the worms’ nervous system and observed that at day 5, there was an increase in autophagosomes in both pgrn-1 mutant animals compared with WT (Fig. 2E). There was, however, no significant change in the number of autolysosomes (Fig. 2F). At day 1, we still observed an increase in autophagosomes (Fig. 2G) but also saw a decrease in autolysosome numbers (Fig. 2H). This suggests that by day 5, pgrn-1 mutant worms exhibit defects in autophagosome–lysosome fusion which is not apparent at day 1. As seen in Fig. 1 E and F, mutant worms show changes in lysosome size and fluorescence intensity at day 5, but these defects were not seen at day 1 (Fig. 2 I and J). Given the increase in autophagosomes as early as day 1 of adulthood, without the appearance of lysosomal defects, these data point to autophagy defects appearing before lysosomal defects in pgrn-1–deficient nematodes.
Genetically Targeting the SL Biosynthetic Pathway Restores Defects in pgrn-1 Mutant Nematodes.
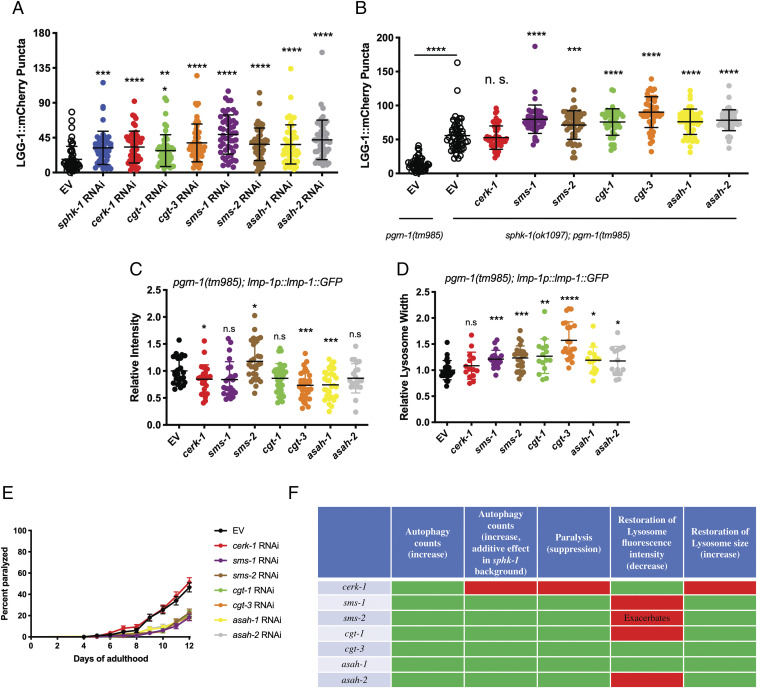
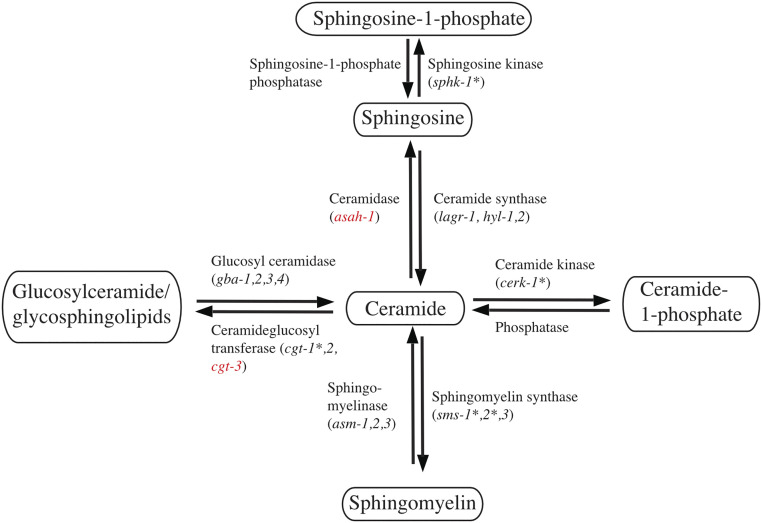
PGRN modulates the maturation and activity of at least two enzymes involved in SL metabolism, glucocerebrosidase (GBA) (50–53) and hexosaminidase A (54). In addition, it binds to prosaposin and regulates its trafficking and processing to saponins, a family of nonenzymatic, lysosomal proteins that promote SL catabolism (35, 53, 55, 56). Disruptions in SL metabolism may, therefore, contribute to the pathological phenotype of GRN-related disorders. To further explore this and to better understand the nature of the interaction between pgrn-1 and the SL pathway, we performed a small-scale RNAi screen of 17 genes involved in ceramide metabolism available from commercially available RNAi libraries. We scored the formation of intestinal LGG-1::mCherry puncta in pgrn-1(tm) animals in the presence of the RNAi after 5 d of treatment. Since nematode neurons are mostly resistant to RNAi treatment, we were limited to using this intestinal autophagy marker as a proxy for neuronal knockdown of the genes. We observed that a small subset of eight RNAis resulted in a partial increase of LGG-1 puncta (Fig. 3A). Interestingly, there was common thread linking all the genes whose RNAi knockdown increased puncta formation, as they all coded for enzymes that use ceramide as a substrate to convert it to a downstream product. Likewise, the RNAi knockdown of genes that catalyzed the reverse reactions and regenerate ceramide had no effect on puncta formation (SI Appendix, Fig. S3A). We also verified this genetically in neurons in the double GFP::mCherry::LGG-1 reporter by introducing a SL metabolic mutation, sphk-1(ok1097), in a pgrn-1(tm) background. We observed that the loss of sphk-1 restores autophagosome (SI Appendix, Fig. S3B) and autolysosome numbers (SI Appendix, Fig. S3C) in pgrn-1 mutants. We then wanted to see if targeting a combination of genes in the SL pathways could have an additive effect on restoring these phenotypes. Treating worms with two RNAis simultaneously is not a common methodological approach since it is difficult to control the efficacy of the knockdown of each individual RNAi or to control for the amount of each bacteria eaten by the animals. Therefore, we opted to generate a double mutant between pgrn-1(tm) and sphk-1, one of the genes that was identified in initial screen, and then treat these double mutants with individual RNAi. Thus, we constructed the genetic double mutant, pgrn-1(tm); sphk-1(ok1097), in conjunction with the intestinal LGG-1::mCherry reporter and observed that seven of the eight clones tested had an additive effect on the sphk-1 genetic mutant in restoring formation of LGG-1::mCherry puncta in a pgrn-1–null background (Fig. 3B).
Fig. 3.
Genetic manipulation of the SL biosynthetic pathway restores defects in pgrn-1 mutant animals. (A) RNAi-mediated knockdown of genes involved in SL biosynthesis, using ceramide as a reactant, partially restore LGG-1::mCherry puncta formation in the intestine of pgrn-1–null animals (one-way ANOVA, treated versus EV control, **P < 0.01, ***P < 0.001, ****P < 0.0001). (B) RNAi-mediated knockdown of genes involved in SL biosynthesis have an additive effect on LGG-1::mCherry puncta formation in a pgrn-1(tm985); sphk-1(ok1097) genetic background (one-way ANOVA, sphk-1(ok1097); pgrn-1(tm985) treated versus EV control, ***P < 0.001, ****P < 0.0001). (C and D) Knockdown of certain SL biosynthetic genes restore LMP-1::GFP lysosomal intensity and lysosomal width phenotypes in pgrn-1(tm985) mutant animals (one-way ANOVA, treated versus EV control, *P < 0.05, **P < 0.01, ***P < 0.001). (E) RNAi knockdown rescues paralysis phenotype of pgrn-1(tm985) mutant animals (Mantel–Cox test: EV versus cerk-1 RNAi, n.s.; EV versus sms-1 RNAi, ****P < 0.0001; EV versus sms-2 RNAi, ****P < 0.0001; EV versus cgt-1 RNAi, ****P < 0.0001; EV versus cgt-3 RNAi, ****P < 0.0001; EV versus asah-1 RNAi, ****P < 0.0001; EV versus asah-2 RNAi, ****P < 0.0001). (F) Summary table of RNAi knockdown effects on different phenotypes tested.
We further tested the eight genes of interest against other behavioral and molecular phenotypes characteristic of our pgrn-1(tm985) animals. Among these, we tested the ability of the RNAi clones to restore lysosomal defects seen in these animals. We observe that RNAi knockdown of several genes rescued the worms’ lysosomal defects by restoring size, LMP-1::GFP fluorescence intensity, or both (Fig. 3 C and D). We finally tested the ability of these RNAi clones to suppress the animals’ paralysis phenotype (Fig. 3E). The summary of the results of these RNAi knockdown assays is given in Fig. 3F. We observe that the knockdown of only two of the eight genes tested, cgt-3 and asah-1, are able to restore all the tested phenotypes in pgrn-1(tm) animals (Fig. 4).
Fig. 4.
Schematic representation of SL genes implicated in PGRN-related defects. SL genes whose knockdown is able to restore one or more PGRN-related phenotypes are denoted by an asterisks (*), while the ones able to restore all tested phenotypes are indicated in red.
High-Throughput, Unbiased Drug Screening in PGRN-Deficient Models.
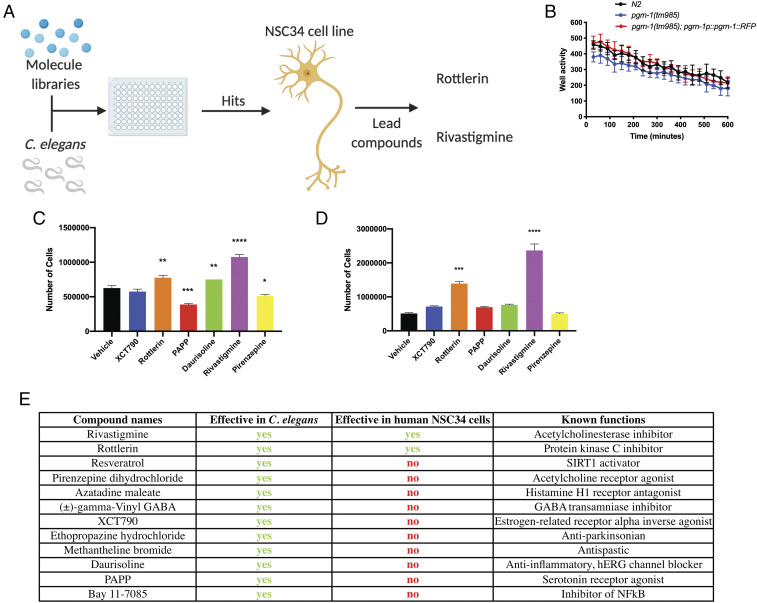
With the unmet need for new therapies to treat individuals with GRN-deficit mutations, we sought to apply our nematode model to close this gap and identify small molecules capable of compensating for the loss of PGRN. Using C. elegans for drug discovery has many advantages since they can be used for high-throughput in vivo drug screening, something that cannot be easily performed with larger animals such as mice. Also, although typical high-throughput drug screens are done using cell-based models, nematodes are able to rapidly assess the efficacy of compounds in the context of a whole organism, complete with multiple cell types and biological complexity. The success of this approach has been validated when our group has recently used C. elegans to identify and translate a compound into clinical trials for ALS (41).
Since pgrn-1(tm) nematodes are fully deficient in PGRN-1, our unbiased drug screening approach sought to chemically replace PGRN-1’s action. To do so, we screened ∼3,850 small-molecule compounds from the Prestwick, Sigma Aldrich Library of Pharmacologically Active Compounds (LOPAC), Microsource, and the BML Natural Products drug libraries in C. elegans, followed by validation in previously characterized GRN-deficient NSC34 cells (57) (Fig. 5A). We screened in C. elegans for their ability to restore the swimming defect in pgrn-1(tm) animals (Fig. 5B). Our primary screen (1 well/compound) resulted in 108 hits, and these were tested in triplicate during a secondary screen (3 wells/compound) to eliminate false positives, which further narrowed down our list to 34 compounds (SI Appendix, Table S1). In keeping with the unbiased nature of our screen, we selected the top 17 compounds, regardless of their known function, for further testing in pgrn-1 nematodes for their ability to restore lifespan (SI Appendix, Fig. S4 A–C) and suppress paralysis phenotypes (SI Appendix, Fig. S4 D–F) in pgrn-1(tm) mutants at a single dose of 20 µM, the same dose used in the high-throughput screen. In total, 12 compounds were considered hits from the unbiased, high-throughput screen.
Fig. 5.
High-throughput drug screening identifies small molecules able to ameliorate PGRN deficiency in nematodes and mammalian cell lines. (A) Schematic representation of the high-throughput drug screen for PGRN-compensating drugs. pgrn-1(tm) mutant nematodes were treated with ∼3,850 compounds in a phenotypic screen looking to restore motility. Hits were subsequently validated in Grn-deficient NSC34 cell lines to determine their effect in a mammalian model. Made in BioRender (https://biorender.com/). (B) pgrn-1(tm) nematodes have a swimming defect in liquid culture when compared with WT animals (two-way ANOVA, WT versus pgrn-1(tm), ****P < 0.0001) but PGRN-1 rescue animals did not (two-way ANOVA, WT versus pgrn-1p::pgrn-1::rfp, n.s.). (C and D) Survival of NSC34 cells treated with drug screen hits after 7 (C) and 14 d (D) of low serum growth (7 d, C: one-way ANOVA, drug treatment versus Vehicle; XCT790, n.s.; rottlerin, **P < 0.01; PAPP, ***P < 0.001; Daurisoline, **P < 0.01; Rivastigmine, ****P < 0.0001; Pirenzepine, *P < 0.05. 14 d, D: one-way ANOVA, drug treatment versus Vehicle; XCT790, n.s.; rottlerin, ***P < 0.001; PAPP, n.s.; Daurisoline, n.s.; Rivastigmine, ****P < 0.0001; Pirenzepine, n.s.). (E) Summary table of top 12 compounds and their effects on worm paralysis and NSC34 cells.
To validate the translational ability of the compounds and to see if they could also function in a mammalian system, we tested the 12 compounds in NSC34 cell line models of PGRN deficiency. These cells have been validated for relevant phenotypes after silencing of GRN expression by short hairpin RNA (57). The 12 drugs were tested for their ability to restore cell survival, and it was determined that 6 reduced cell survival and were excluded. Of the other six, five had the ability to promote cell growth after 7 d (Fig. 5C), but only two, rottlerin and rivastigmine, had a prolonged effect and significantly increased cell growth after 14 d (Fig. 5D). Rivastigmine is an acetylcholinesterase inhibitor (Fig. 5D) that is used for symptom management in Alzheimer’s disease and has previously been tested in a small clinical trial for FTD (58, 59). This compound was shown to be beneficial in managing behavioral symptoms but had no significant effect on cognitive impairment or deterioration. Our results test rivastigmine in a specific genetic model of PGRN deficiency, where it had not previously been explored. Rottlerin, on the other hand, is a natural product polyphenol isolated from the red kamala tree (Mallotus philippinensis). Rottlerin has been shown to have various cellular effects including activating autophagy, acting as an antitumor factor, as an antiproliferative compound, and as an uncoupler of mitochondrial oxidative phosphorylation (60–64). Studies have demonstrated that rottlerin is a protein kinase C delta (PKCδ) inhibitor (61, 62, 64), although the evidence in support of this activity is contradictory (63, 65). To date, there are no clinical data looking at rottlerin’s effect in human subjects, and although it has been shown to have a protective effect in Alzheimer’s (61) and Parkinson’s (66) disease models, its efficacy in the context of PGRN-deficient FTD has not been previously reported.
Small Molecules Restore PGRN-Deficient Phenotypes in C. elegans.
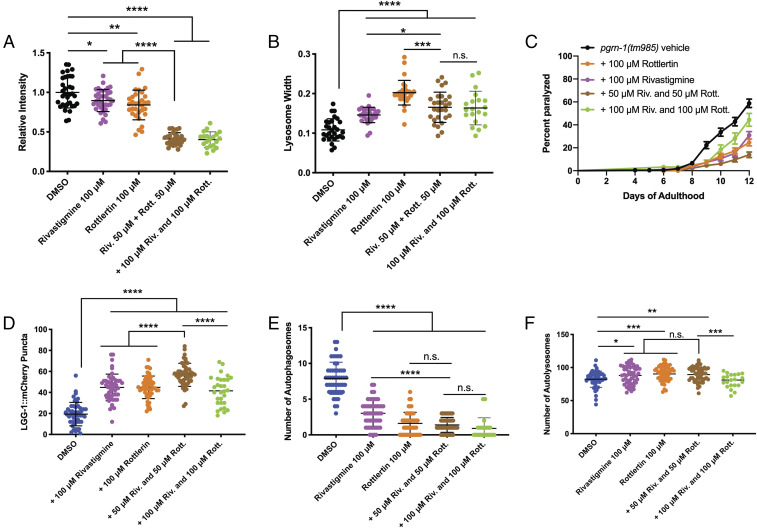
With our final two hits identified from the drug screen, we returned to our nematode models to further validate the compounds against other phenotypes in the PGRN-1–deficient nematodes. We first looked at the ability of the compounds to restore lysosomal phenotypes and observed that both rottlerin and rivastigmine were able to restore lysosomal fluorescence intensity and size phenotypes seen in the mutant animals (Fig. 6 A and B). We performed dose-dependent testing on both drugs and selected 100 µM as this was the concentration at which the drugs elicited a statistically significant effect on both phenotypes (SI Appendix, Fig. S5 A and B). Interestingly, the combination of both drugs at a half dose, 50 µM, had a stronger effect than either drug individually on lysosomal size (Fig. 6A), but for lysosomal width (Fig. 6B), the combination did not have this additive effect at both 50 and 100 µM. We then tested them against paralysis (Fig. 6C) and observed a significant effect from the individual drugs and a stronger effect from the combination of both (at 50 µM) compared with controls, though the combination at 100 µM was not. We next looked at the compounds’ influence on autophagy in pgrn-1(tm985) animals, beginning with the intestinal LGG-1::mCherry reporter, and observed that both drugs were able to increase LGG-1::mCherry puncta formation, and the combination at 50 µM had a stronger effect but at 100 µM did not (Fig. 6D). Finally, using the tandem-tagged autophagy reporter, GFP::mCherry::LGG-1, we also observed a restoration of autophagosomes (Fig. 6E) and autolysosomes (Fig. 6F). Here, however, the combination of both drug treatments at 50 µM had no additive effect on autolysosomes and only had a greater effect than rivastigmine alone on autophagosomes, likely because the rottlerin-only treatment had already reached a plateau. However, at 100 µM, the combination of both compounds had no effect on autolysosome formation. While the mechanisms of action (MOA) of rivastigmine is well known, rottlerin’s still remains complex. Though complete studies into both these compounds’ MOAs in the context of GRN-FTD should be performed, we tested another known acetylcholinesterase inhibitor, donepezil, and a PKC inhibitor, sotrastaurin, in pgrn-1(tm) worms and observed both compounds were able to reduce paralysis phenotypes (SI Appendix, Fig. S5C). Together, these data suggest that rottlerin and rivastigmine are promising compounds with therapeutic applications in PGRN-deficient FTD.
Fig. 6.
Rottlerin and Rivastigmine restore behavioral and molecular defects in vivo. (A and B) When used individually, rottlerin or rivastigmine treatment can restore lysosomal LMP-1::GFP fluorescence intensity (A) and lysosomal size (B) in day 5 pgrn-1(tm985) animals (A, Student’s t test: Vehicle versus 100 μM rivastigmine, *P < 0.05; Vehicle versus 100 μM rottlerin, **P < 0.01. (B) Student’s t test: Vehicle versus 100 μM rivastigmine, ****P < 0.0001; Vehicle versus 100 μM rottlerin, ****P < 0.0001). When used simultaneously, the combination has a greater effect than each drug individually on fluorescence intensity (A) but not on lysosomal width (B) (A: Student’s t test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott. ****P < 0.0001; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott. ****P < 0.0001. (B) Student’s t test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott. ***P < 0.001; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott. *P < 0.05). (C) Treatment of pgrn-1(tm985) animals with 100 μM rottlerin or rivastigmine, or the combination, results in a strong suppression of paralysis (Mantel–Cox test: Vehicle versus 100 μM rottlerin, ****P < 0.0001; Vehicle versus 100 μM rivastigmine, ****P < 0.0001; Vehicle versus 50 μM rivastigmine + 50 μM rottlerin ****P < 0.0001), and the simultaneous treatment had a stronger effect than the individual drugs (Mantel–Cox test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott. **P < 0.01; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott. ***P < 0.001). (D) Both compounds, individually and combined, are able to increase levels of LGG-1::mCherry punctae in pgrn-1 mutant animals’ intestines (Student’s t test: Vehicle versus 100 μM rivastigmine, ****P < 0.0001; Vehicle versus 100 μM rottlerin, ****P < 0.0001; Vehicle versus 50 μM riv. + 50 μM rott. ****P < 0.0001), and the combination had a stronger effect than the individual treatments (Student’s t test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott. ****P < 0.0001; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott. ****P < 0.0001). (E and F) Both compounds, individually and combined, are able to influence autophagosome (E) and autolysosome (F) levels of the dual mCherry::LGG-1::GFP reporter in pgrn-1 mutant animals’ neurons (E: Student’s t test: Vehicle versus 100 μM rivastigmine, ****P < 0.0001; Vehicle versus 100 μM rottlerin, ****P < 0.0001; Vehicle versus 50 μM riv. + 50 μM rott. ****P < 0.0001. (F) Student’s t test: Vehicle versus 100 μM rivastigmine, *P < 0.05; Vehicle versus 100 μM rottlerin, ***P < 0.001; Vehicle versus 50 μM riv. + 50 μM rott. **P < 0.01). The combination of both drugs had a stronger effect than rivastigmine alone on autophagosomes (E) but did not have an effect on autolysosome levels (E, Student’s t test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott., n.s.; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott., ****P < 0.0001. (F) Student’s t test: 100 μM rottlerin versus 50 μM riv. + 50 μM rott., n.s.; 100 μM rivastigmine versus 50 μM riv. + 50 μM rott., n.s.).
Discussion
Although it is known that a loss of PGRN leads to lysosomal dysfunction, the mechanisms that have been identified as causing this process are diverse. Previous studies have elucidated dysfunction related to lysosomal trafficking in PGRN-deficient cells, while others have investigated the effect of PGRN and its singular peptides on lysosomal protease activity (8). In this study, we aimed to use the small animal model, C. elegans, to better understand the biological consequences of PGRN deficiency and to identify genetic targets and small-molecule therapeutics capable of chemically compensating for it.
While others have previously used C. elegans to study the fundamental actions of PGRN in vivo in relation to its role to embryonic cell death and stress response (15, 16), we investigated whether PGRN-deficient nematodes had aging-related defects and if they recapitulated key features of FTD. Furthermore, while previous pgrn-1 studies have used the tm985 deletion mutant, we also employed the gk123284 allele from the Million Mutation Project. We selected this allele since the protein change, G119E, is in a highly conserved region of the protein next to a pair of characteristic cysteine residues of the granulin structural motif (67, 68), and it is likely that the replacement of a single hydrogen atom side chain (glycine) to a large, negatively charged side chain (glutamic acid) would destabilize the mutant protein enough to induce phenotypes. The reduced levels of pgrn-1 mRNA in pgrn-1(gk) mutants was unexpected since we expected changes to have been at the protein level. We observed, interestingly, that both the pgrn-1–null and pgrn-1–missense animals displayed age-dependent paralysis phenotypes, as do many other C. elegans models of age-dependent neurodegenerative diseases. These animals also displayed neuronal transmission defects, as are present in human FTD patients and in other dementias (69, 70). Of interest to modeling GRN-deficient FTD is the presence of lysosomal defects, which we now know is a characteristic feature of the disease. Morphologically, we observed that lysosomes in pgrn-1 mutant animals were smaller but greater in number than WT counterparts, which is consistent with previous studies (33). We also observed an increase in LMP-1/LAMP1 fluorescence signals, although it is unclear if this is due to an increase in lysosomal activity or if it is an artifact of the increase in lysosomal density in the nematode coelomocytes. We challenged these mutant worms with the proteasome inhibitor MG132 and the autophagy inhibitor Concanamycin A and observed a dose-dependent decrease in lifespan but no effect in WT or rescued animals. These data suggest that the morphological defects present in these animals’ lysosomes correlate with a defect in lysosomal function.
Although lysosomal defects are often studied in relation to PGRN deficiency, the role of autophagy in this process is less fully explored. We utilized an intestinal autophagy LGG-1/LC3 marker and observed that WT animals displayed LGG-1 puncta, whereas pgrn-1 mutants hardly displayed any, and instead, the florescent signals formed a diffuse pattern in the intestine. Autophagy can be induced through starvation, but this did not increase its level in pgrn-1 mutant animals. This suggested that the inhibition of autophagy in pgrn-1 mutant animals was independent of this classical autophagy-induction pathway. A limitation of the intestinal LGG-1 marker is that it does not necessarily represent what is happening in other tissues, nor does it offer much information on autophagic flux. We therefore turned to a dually tagged LGG-1 marker, tagged with both GFP and mCherry, which is expressed in nematode neurons. Using this tool, we were able to assess autolysosomes, autophagosomes, and the transition between the two in vivo. We observed that already at day 1 of adulthood, there are increases in the number of autophagosomes but no change in autolysosome number, whereas by day 5, there is also a decrease in the number of autolysosomes. Therefore, we can categorize these phenotypes into two categories: early phenotypes of autophagic flux and lysosomal acidification defects, which precede the later lysosomal morphology phenotypes. Our findings suggest an important contribution of PGRN in the regulation of autophagic flux and therefore cellular waste clearance. If this translates to mammalian brains, it may help explain the accumulation of TDP-43 aggregates seen in GRN-FTD.
While many links have been drawn between PGRN deficiency and metabolic dysfunctions, the role that SLs play in PGRN-related pathology has rarely been studied. These represented an attractive area of focus as there is significant overlap between the known functions of SLs and those of PGRN. Using a small-scale RNAi screen, we found that the knockdown of genes encoding proteins which utilize ceramide as substrate was able to partially restore LGG-1 puncta levels. We further narrowed down the list of genes by testing them in a sphk-1–null background for an additive effect on LGG-1 puncta formation, including for lysosomal intensity and size, as well as for an ability to suppress age-dependent paralysis. The RNAi knockdown of asah-1 (acid ceramidase) and cgt-3 (ceramide glucosyltransferase) were able to rescue all the tested phenotypes. Understanding the mechanism by which the RNAi knockdown of these two specific genes are able to restore pgrn-1 phenotypes was not in the scope of this study, but we have hypothesized potential causes of this effect. The most obvious explanation would be that a loss of PGRN results in a dysregulation of SL levels, which would have a number of downstream effects (71–77). As SLs are extensively involved in cell signaling, we could envisage a scenario where a dysregulation in SL levels could alter this signaling. Second, cgt-3 catalyzes the reverse reaction of GBA. This is intriguing given the known links between PGRN and GBA and their involvement in Gaucher disease pathology (51, 50). The connection with asah-1 and PGRN pathology is, however, less clear. Third, as SLs are also an important component of plasma membranes and are known to affect membrane dynamics (78–81), we could also propose a scenario where a loss of PGRN could lead to defects in membrane fusion. This could potentially explain the lysosomal morphology defects, as a change in membrane SL composition could alter the shape of the organelles, which could thereby affect autophagic flux by altering the dynamics of fusion between autophagosomes with lysosomes. Further metabolomic and biophysical studies in organisms other than C. elegans would be required to answer these questions and clarify the role of SLs in PGRN pathology. However, given that there is evidence for altered SL metabolism in GRN-related FTD (53, 52) and lysosomal storage disorders (51, 54, 50) and the demonstration here that manipulating SL metabolic pathways compensates for loss of PGRN, SL metabolism is clearly a target of interest for the development of potential FTD therapies.
One of the most important needs for neurodegenerative disease treatment is the development of new therapeutics that can rapidly be translated into a clinical setting. However, the traditional drug discovery process is slow and has a dismally high failure rate (82). C. elegans has recently emerged as an efficient tool for rapid drug discovery for neurodegenerative diseases. Since nematodes are multicellular organisms that are highly amenable to rapid genetic modification, they can be used to address some of the most important roadblocks in the drug discovery process. First, if a drug is found to be efficacious in a nematode, a living multicellular organism, we know the drug is reaching its biological target, which can give researchers an indication of its bioavailability. Second, we can use nematodes for efficient phenotype-based screens, obviating the reliance on known molecular targets which are difficult to determine in complex disorders such as FTD. Our approach to drug discovery is quite different from many of the current efforts, which seek to either increase the expression of PGRN protein or deliver the protein exogenously. We treated pgrn-1(tm) animals with ∼3,850 compounds from commercially available libraries to identify repurposed drugs that could serve to potentially treat GRN-deficient FTD. From this screen, we tested our drug hits in NSC34 cells to validate their effect in a mammalian cell system. From this, we identified two final hits, rottlerin and rivastigmine. Upon further investigation, the two drugs, either individually or in combination, were able to restore many of the molecular and behavioral defects seen in the pgrn-1(tm985) animals. While the identification of rivastigmine, a drug already tested clinically in FTD patients, was promising in that it helped to validate our approach, rottlerin was a surprising hit. A molecule that has never been tested in humans, rottlerin, also known as mallotoxin, is a natural compound isolated from the red kamala tree with obscure mechanisms of action. It has been shown to be protective in Alzheimer and Parkinson’s disease models. It was initially reported to be a PKCδ inhibitor, but many studies have called this function into question and have reported a larger list of activities: a broad inhibitor of PKCs, PKA, casein kinase II, MAPK-activated protein kinase 2, p38-regulated/activated kinase, protein kinase A, glycogen synthase kinase 3, an uncoupler of mitochondrial oxidative phosphorylation, and an activator of AMPK (60, 62, 63, 65, 83). The mechanism underlying the protective effect of rottlerin in PGRN-deficient nematodes and mammalian cells is, therefore, unclear; however, another study has shown that rottlerin can stimulate autophagy through activation of AMPK (83). As for rivastigmine, although it was already tested clinically with mixed results, many years have passed since the publication of this study. Over that time, our knowledge about PGRN and FTD have greatly advanced, and it may be of interest to retest this drug in a cohort of genetically confirmed GRN FTD patients. Since our results point toward the ability of rivastigmine and rottlerin to restore molecular defects associated with PGRN deficiency, the drug could be administered years before the onset of symptoms and before the occurrence of neurodegeneration in an attempt to slow or halt onset of the disease.
Together, our results point toward many potential therapeutic avenues for PGRN-deficient pathologies and neurodegenerative disorders in general. First, our work suggests that modulating the pathways involved in the biosynthesis of SLs may be a strategy for reversing certain aspects of the disease’s pathology. Second, we propose two small molecules, rottlerin and rivastigmine, that appear to restore defects associated with PGRN deficiency. Although therapeutic efforts are primarily focused on increasing PGRN levels or reducing the expression of TMEM106b (32), which in turn also increases levels of PGRN, our approaches appear to function in alternative pathways. Since our nematode models are deficient in pgrn-1 and its corresponding protein, our results suggest the ability to compensate for a lack of PGRN, chemically or genetically. While we acknowledge that further testing and validation of our genetic targets and small molecules will be required, we nonetheless anticipate that these findings will serve as a stepping-stone to the development of new therapeutics for PGRN-deficient pathologies.
Materials and Methods
C. elegans Strains and Maintenance.
All nematode strains were cultured and handled as per standard methods. All experiments were carried out at 20 °C and were repeated a minimum of three times. The following strains were used: Bristol N2 WT, XQ561 pgrn-1(tm985), XQ592 pgrn-1(gk123284), CF3778 pgrn-1(tm985); pgrn-1p::pgrn-1::rfp, RT258 pwIs50 [lmp-1::GFP + Cbr-unc-119(+)], TU3311 uIs60 [unc-119p::YFP + unc-119p::sid-1], VK1093 vkEx1093 [nhx-2p::mCherry::lgg-1], and MAH508 sqEx67 [rgef-1p::mCherry::GFP::lgg-1 + rol-6]. Genetic mutant worms were outcrossed five times to WT N2 worms before use, and pgrn-1(gk) animals were outcrossed seven times. Genotyping of deletion mutants was done by genomic PCR, whereas genotyping of point mutations was done by high-resolution melting (HRM) using HRM MeltDoctor reagents (Applied Biosystems) and analyzed on HRM software (Applied Biosystems). Verification by Sanger sequencing was performed by Genome Quebec (McGill University). Heterozygous animals were obtained by crossing homozygous mutants with WT N2 animals; the progeny from fertilized hermaphrodites were used and immediately frozen in lysis buffer after use for confirmation of their genotype by either PCR or HRM.
RNAi Experiments.
All C. elegans RNAi experiments were administered through feeding using standard protocols. For all assays, worms were grown on standard nematode growth media (NGM) plates, synchronized, and eggs were placed on NGM plates with either empty vector (EV) bacteria or bacterial clones expressing double-stranded RNA against the target gene. Worms were grown on RNAi plates, and the second generation was used for subsequent assays.
Gene expression Assays.
Synchronized, age-matched animals were collected in M9 buffer at day 1 of adulthood. Animals were washed with buffer four times to remove excess bacteria, and the supernatant was removed after the last wash step. Worms were then flash frozen at −80 °C in 500 µL TRIzol Reagent (Thermo Fisher Scientific). After thawing, worms were homogenized using a 27.5 G needle with a syringe and another 500 µL of TRIzol was added. Samples were let to incubate at room temperature for 5 min before adding 200 µL of chloroform and letting them sit for an additional 2 min. Samples were then centrifuged at 12,000 g for 15 min, allowing the phases to separate. The aqueous phase was collected, and extraction was completed using the RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized using the SuperScript VILO cDNA Synthesis Kit (Invitrogen), and gene expression to quantify pgrn-1 transcript levels was performed using TaqMan probes and standard TaqMan reagents (both probes and reagents were purchased from Applied Biosystems). act-5 was chosen as the housekeeping gene. Gene expression assays were run on a QuantStudio 7 Flex (Applied Biosystems) instrument, and data analysis was done using QuantStudio Real-Time PCR software.
Paralysis Assays.
For paralysis assays, 25 to 30 L4 larval animals were placed onto NGM plates and scored daily starting the following day, at day 1 of adulthood. Worms were counted as paralyzed if they failed to move their body when prodded with a platinum worm pick. Worms were considered dead if they failed to respond to heat stimuli or if exhibited no pharyngeal pumping; dead worms were censored from statistical analyses. For each paralysis assay, a minimum of 200 animals were scored per genotype and per condition. For paralysis assays with compound treatments, the compounds were mixed directly into the NGM to the appropriate concentration.
Lifespan Assays.
As with paralysis assays, 25 to 30 L4 animals were picked onto NGM plates and then scored from the first day of adulthood until death. Worms were considered dead if they failed to respond to mechanical or heat stimuli and if they showed no pharyngeal pumping. A minimum of 200 animals were scored per genotype or per condition. For assays done with compound treatments, the compounds were mixed directly into the NGM to the appropriate concentration.
Overactive Food-Seeking Behavior.
Worms were age matched, washed three times in M9 to remove excess bacteria, and placed on NGM plates without bacterial food. Worms were counted daily for the number of worms remaining on the plate, as well as the dead worms found stuck to the side of the plate. Animals that disappeared were censored from statistical analyses.
Aldicarb Assays.
Animals were picked onto NGM plates supplemented with 1mM aldicarb at day 1 of adulthood. Paralysis was scored every 30 min for 2 h. Animals were considered paralyzed if they failed to respond to gentle prodding by a platinum pick. A minimum of 200 animals were counted per genotype.
Microscopy Experiments.
All microscopy experiments were carried out by a Zeiss Axio Observer inverted microscope. For all microscopy experiments, animals were mounted onto 2% agarose pads and immobilized in 5 mM levamisole.
Intestinal autophagy counts.
Worms were synchronized on standard NGM plates until the L4 stage and were then maintained until the appropriate day. Worms were placed on empty NGM plates for about 30 min before experiments were carried out in order to get rid of excess intestinal bacteria. For RNAi experiments, animals were synchronized on standard NGM plates until the L4 stage and were then transferred onto RNAi plates until the appropriate day. For starvation assays, animals were maintained on standard NGM plates and were transferred to empty plates for 24 h before experiments were carried out. During data acquisition, fluorescent LGG-1::mCherry puncta were counted manually, and 50 animals were counted per genotype or condition.
Neuronal autophagy counts (dual GFP/mCherry LGG-1 reporter).
Synchronized worms were raised on standard NGM plates and maintained until the appropriate day and visualized by fluorescent microscopy. Total numbers of red and green puncta were then counted manually. Since green puncta were indicative of autophagosomes (AP) and red puncta were indicative both of AP and autolysosomes (AL), the number of AL was calculated by AL= (Total no. of red punctae) − (Total no. of green punctae).
Lysosome morphology tests.
For this assay, only lysosomes from the posterior coelomocytes were considered as there was minimal obstruction from other background tissues and intestinal fluorescence. Images were taken using the same camera settings across all replicates and were then analyzed in ImageJ. For lysosome intensity analyses, the background signal was subtracted from the lysosomal signal.
Liquid Culture Motility Testing.
Animals were synchronized to age match the populations and grown on standard NGM plates until day 1 of adulthood. Worms were collected and washed with M9 buffer to remove excess bacteria. They were then placed in standard 96-well plates in M9 buffer to a number of ∼50 to 70 worms/well. Motility was recorded unbiasedly using a WMicrotracker ONE instrument.
High-Throughput, Unbiased Drug Screening in C. elegans.
Small-molecule libraries from the Prestwick Chemical Library, Sigma Aldrich LOPAC Library, Microsource Drug Library, and the BML Natural Products Library from Enzo Life Sciences (together, totaling 3,942 molecules) were selected for screening. pgrn-1 mutant animals were grown on standard NGM plates, and the efficacy of the drugs was monitored using WMicrotracker ONE instruments. Drug treatment was acute, and all compounds were tested at 20 μM. Animals were only put in contact with the drugs at the start of the assay for an acute treatment. Each compound was tested once (1 drug/well). Molecules were considered as hits if they were able to increase motility of pgrn-1(tm985) animals based on a yes/no criteria. Hits were then retested in 3 wells/drug in a secondary screen to remove any false positives.
Drug Efficacy Testing in NSC34 Cells.
NSC34 cells expressing reduced levels of PGRN (shPGRN-NSC34) were plated in 6-well plates with 20,000 cells per well and cultured in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum (FBS). After 24 h, cells were replaced with 1% FBS. The experimental drugs were blinded with numbers. The next day, the cells were incubated with or without drugs in duplicates. After 7 and 14 d, cells were trypsinized and counted.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, funded by NIH Office of Research Infrastructure Programs (Grant P40 OD010440), and the National Bioresource Project for the nematode (Japan) for providing strains. We thank Dr. Aimee Kao for the pgrn-1–overexpressing strain.
Footnotes
Competing interest statement: J.J.D., A.B., and J.A.P. are named inventors on pending US provisional patent application no. 63/202,491 filed on June 14, 2021.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022115118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Bang J., Spina S., Miller B. L., Frontotemporal dementia. Lancet 386, 1672–1682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olney N. T., Spina S., Miller B. L., Frontotemporal dementia. Neurol. Clin. 35, 339–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrell J. R., et al., The frontotemporal dementia-motor neuron disease continuum. Lancet 388, 919–931 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Baker M., et al., Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Cruts M., et al., Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Meeter L. H., et al., Progranulin levels in plasma and cerebrospinal fluid in granulin mutation carriers. Dement. Geriatr. Cogn. Disord. Extra 6, 330–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beel S., et al., Progranulin reduces insoluble TDP-43 levels, slows down axonal degeneration and prolongs survival in mutant TDP-43 mice. Mol. Neurodegener. 13, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salazar D. A., et al., The progranulin cleavage products, granulins, exacerbate TDP-43 toxicity and increase TDP-43 levels. J. Neurosci. 35, 9315–9328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Swieten J. C., Heutink P., Mutations in progranulin (GRN) within the spectrum of clinical and pathological phenotypes of frontotemporal dementia. Lancet Neurol. 7, 965–974 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Wils H., et al., Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J. Pathol. 228, 67–76 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ling S. C., Polymenidou M., Cleveland D. W., Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami S. S., et al., Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 20, 1157–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Kampen J. M., Baranowski D., Kay D. G., Progranulin gene delivery protects dopaminergic neurons in a mouse model of Parkinson’s disease. PLoS One 9, e97032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tauffenberger A., Chitramuthu B. P., Bateman A., Bennett H. P., Parker J. A., Reduction of polyglutamine toxicity by TDP-43, FUS and progranulin in Huntington’s disease models. Hum. Mol. Genet. 22, 782–794 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Judy M. E., et al., A shift to organismal stress resistance in programmed cell death mutants. PLoS Genet. 9, e1003714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao A. W., et al., A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 108, 4441–4446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed Z., et al., Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 177, 311–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkau T. L., Leavitt B. R., Progranulin in neurodegenerative disease. Trends Neurosci. 37, 388–398 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Petkau T. L., et al., Synaptic dysfunction in progranulin-deficient mice. Neurobiol. Dis. 45, 711–722 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Yin F., et al., Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207, 117–128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin F., et al., Behavioral deficits and progressive neuropathology in progranulin-deficient mice: A mouse model of frontotemporal dementia. FASEB J. 24, 4639–4647 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almeida M. R., et al., Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging 41, 200.e1–200.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Canafoglia L., et al., Recurrent generalized seizures, visual loss, and palinopsia as phenotypic features of neuronal ceroid lipofuscinosis due to progranulin gene mutation. Epilepsia 55, e56–e59 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Faber I., Prota J. R., Martinez A. R., Lopes-Cendes I., França M. C. J., A new phenotype associated with homozygous GRN mutations: Complicated spastic paraplegia. Eur. J. Neurol. 24, e3–e4 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kamate M., Detroja M., Hattiholi V., Neuronal ceroid lipofuscinosis type-11 in an adolescent. Brain Dev. 41, 542–545 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Smith K. R., et al., Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 90, 1102–1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Götzl J. K., et al., Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 127, 845–860 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Valdez C., et al., Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum. Mol. Genet. 26, 4861–4872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward M. E., et al., Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci. Transl. Med. 9, eaah5642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao A. W., McKay A., Singh P. P., Brunet A., Huang E. J., Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18, 325–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evers B. M., et al., Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep. 20, 2565–2574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein Z. A., et al., Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95, 281–296.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka Y., et al., Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 26, 969–988 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zhou X., et al., Progranulin deficiency leads to reduced glucocerebrosidase activity. PLoS One 14, e0212382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., et al., Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol. 210, 991–1002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X., Sun L., Brady O. A., Murphy K. A., Hu F., Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol. Commun. 5, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paushter D. H., Du H., Feng T., Hu F., The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol. 136, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim W., Underwood R. S., Greenwald I., Shaye D. D., OrthoList 2: A new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics 210, 445–461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altun Z. F., Hall D. H., Herndon L. A., WormAtlas Hermaphrodite Handbook–Nervous System–General Description (WormAtlas, 2005). [Google Scholar]
- 40.Altun Z. F., Hall D. H., Herndon L. A., WormAtas Hermaphrodite Handbook–Introduction (WormAtlas, 2006). [Google Scholar]
- 41.Patten S. A., et al., Neuroleptics as therapeutic compounds stabilizing neuromuscular transmission in amyotrophic lateral sclerosis. JCI Insight 2, e97152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson O., et al., The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahoney T. R., Luo S., Nonet M. L., Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 1, 1772–1777 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M., Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7, 554–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conte D. Jr., MacNeil L. T., Walhout A. J. M., Mello C. C., RNA interference in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 109, 26.3.1–26.330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocheleau C. E., RNA interference: Systemic RNAi SIDes with endosomes. Curr. Biol. 22, R873–R875 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Avery L., You Y. J., C. elegans feeding. WormBook 1–23(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang L., et al., Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. U.S.A. 108, 4788–4793 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang J. T., Kumsta C., Hellman A. B., Adams L. M., Hansen M., Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife 6, e18459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jian J., et al., Progranulin recruits HSP70 to β-Glucocerebrosidase and is therapeutic against Gaucher disease. EBioMedicine 13, 212–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jian J., et al., Association between progranulin and Gaucher disease. EBioMedicine 11, 127–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arrant A. E., et al., Impaired β-glucocerebrosidase activity and processing in frontotemporal dementia due to progranulin mutations. Acta Neuropathol. Commun. 7, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdez C., Ysselstein D., Young T. J., Zheng J., Krainc D., Progranulin mutations result in impaired processing of prosaposin and reduced glucocerebrosidase activity. Hum. Mol. Genet. 29, 716–726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y., et al., Progranulin associates with hexosaminidase A and ameliorates GM2 ganglioside accumulation and lysosomal storage in Tay-Sachs disease. J. Mol. Med. (Berl.) 96, 1359–1373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholson A. M., et al., Prosaposin is a regulator of progranulin levels and oligomerization. Nat. Commun. 7, 11992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X., et al., Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat. Commun. 8, 15277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan C. L., et al., Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 10, 130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karakaya T., Fußer F., Prvulovic D., Hampel H., Treatment options for tauopathies. Curr. Treat. Options Neurol. 14, 126–136 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Moretti R., et al., Rivastigmine in frontotemporal dementia: An open-label study. Drugs Aging 21, 931–937 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Davies S. P., Reddy H., Caivano M., Cohen P., Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351, 95–105 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du Y., et al., Inhibition of PKCδ reduces amyloid-β levels and reverses Alzheimer disease phenotypes. J. Exp. Med. 215, 1665–1677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y. A., et al., Role of PKCbetaII and PKCdelta in blood-brain barrier permeability during aglycemic hypoxia. Neurosci. Lett. 468, 254–258 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Soltoff S. P., Rottlerin: An inappropriate and ineffective inhibitor of PKCdelta. Trends Pharmacol. Sci. 28, 453–458 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Zhang D., Anantharam V., Kanthasamy A., Kanthasamy A. G., Neuroprotective effect of protein kinase C delta inhibitor rottlerin in cell culture and animal models of Parkinson’s disease. J. Pharmacol. Exp. Ther. 322, 913–922 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Tapia J. A., Jensen R. T., García-Marín L. J., Rottlerin inhibits stimulated enzymatic secretion and several intracellular signaling transduction pathways in pancreatic acinar cells by a non-PKC-delta-dependent mechanism. Biochim. Biophys. Acta 1763, 25–38 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Fan Y., et al., Rottlerin protected dopaminergic cell line from cytotoxicity of 6-hydroxydopamine by inhibiting PKCdelta phosphorylation. Neurosci. Bull. 25, 187–195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hrabal R., Chen Z., James S., Bennett H. P., Ni F., The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat. Struct. Biol. 3, 747–752 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Tolkatchev D., et al., Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 17, 711–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huey E. D., Putnam K. T., Grafman J., A systematic review of neurotransmitter deficits and treatments in frontotemporal dementia. Neurology 66, 17–22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murley A. G., Rowe J. B., Neurotransmitter deficits from frontotemporal lobar degeneration. Brain 141, 1263–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolter T., Sandhoff K., Sphingolipids-their metabolic pathways and the pathobiochemistry of neurodegenerative diseases. Angew. Chem. Int. Ed. Engl. 38, 1532–1568 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Bryan L., Kordula T., Spiegel S., Milstien S., Regulation and functions of sphingosine kinases in the brain. Biochim. Biophys. Acta 1781, 459–466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan J. P., Sieburth D., Localized sphingolipid signaling at presynaptic terminals is regulated by calcium influx and promotes recruitment of priming factors. J. Neurosci. 32, 17909–17920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y., et al., The pleiotropic roles of sphingolipid signaling in autophagy. Cell Death Dis. 5, e1245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oaks J., Ogretmen B., Regulation of PP2A by sphingolipid metabolism and signaling. Front. Oncol. 4, 388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tidhar R., Futerman A. H., The complexity of sphingolipid biosynthesis in the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2511–2518 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Zheng W., et al., Ceramides and other bioactive sphingolipid backbones in health and disease: Lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta 1758, 1864–1884 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Bieberich E., Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 216, 114–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breslow D. K., Weissman J. S., Membranes in balance: Mechanisms of sphingolipid homeostasis. Mol. Cell 40, 267–279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goñi F. M., Alonso A., Effects of ceramide and other simple sphingolipids on membrane lateral structure. Biochim. Biophys. Acta 1788, 169–177 (2009). [DOI] [PubMed] [Google Scholar]
- 81.Kolter T., Sandhoff K., Principles of lysosomal membrane digestion: Stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21, 81–103 (2005). [DOI] [PubMed] [Google Scholar]
- 82.Cummings J., Reiber C., Kumar P., The price of progress: Funding and financing Alzheimer’s disease drug development. Alzheimers Dement. (N. Y.) 4, 330–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar D., Shankar S., Srivastava R. K., Rottlerin induces autophagy and apoptosis in prostate cancer stem cells via PI3K/Akt/mTOR signaling pathway. Cancer Lett. 343, 179–189 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.