Significance
During embryonic development, an early-stage progenitor cell could generate different types of descendant cells. Developmental biologists keep dreaming of making all the descendant cell types generated by each early-stage progenitor cell clear. Here, we focus on one germ layer, the endoderm. Early foregut endoderm generates foregut digestive organs, including the pancreas, liver, and digestive tract. The central point of this work is the establishment of a full-coverage, single-cell–resolution fate map of zebrafish early foregut endoderm through unbiased labeling of every single endodermal progenitor cell and tracing its descendant cells. Based on the maps, we reveal a link of the cell cycle state and pancreatic fate propensity, providing an example of how the maps could facilitate understanding cell fate determination.
Keywords: single-cell labeling and tracing, endoderm, fate map, pancreatic fate, cell cycle
Abstract
A progenitor cell could generate a certain type or multiple types of descendant cells during embryonic development. To make all the descendant cell types and developmental trajectories of every single progenitor cell clear remains an ultimate goal in developmental biology. Characterizations of descendant cells produced by each uncommitted progenitor for a full germ layer represent a big step toward the goal. Here, we focus on early foregut endoderm, which generates foregut digestive organs, including the pancreas, liver, foregut, and ductal system, through distinct lineages. Using unbiased single-cell labeling techniques, we label every individual zebrafish foregut endodermal progenitor cell out of 216 cells to visibly trace the distribution and number of their descendant cells. Hence, single-cell–resolution fate and proliferation maps of early foregut endoderm are established, in which progenitor regions of each foregut digestive organ are precisely demarcated. The maps indicate that the pancreatic endocrine progenitors are featured by a cell cycle state with a long G1 phase. Manipulating durations of the G1 phase modulates pancreatic progenitor populations. This study illustrates foregut endodermal progenitor cell fate at single-cell resolution, precisely demarcates different progenitor populations, and sheds light on mechanistic insights into pancreatic fate determination.
A progenitor cell could generate a certain type or multiple types of descendant cells during embryonic development. Characterizations of descendant cell loci and identities as well as developmental trajectories for every single progenitor cell remain one of the ultimate goals in developmental biology. Recent methodological innovations using DNA barcode labeling (1–5) combined with single-cell RNA sequencing (6–9) enable lineage tracing to be conducted on a large scale and at single-cell resolution. However, unbiased single-cell labeling and tracing with complete visibility and high spatial–temporal resolution in living embryos still remains technically challenging. At early postgastrulation, when molecular markers of organ progenitors are rarely available and cell fates are unspecified, understandings of endodermal cell fate determination are mostly contributed by studies on inductive signals and regulatory molecules (10–17). Previous pioneering studies have identified the regional source of pancreatic and liver progenitors in the early somite stage embryo (18–22). While providing a framework for characterizing the descendant cell types for foregut endoderm, these earlier fate maps lack the resolution at the single-cell level. In order to characterize the variety of descendant cells for each foregut endodermal progenitor cell and precisely demarcate progenitor populations of each foregut digestive organ, we establish unbiased, visible single endodermal cell labeling and descendant tracing techniques. Thus, single-cell–resolution fate and proliferation maps are generated, from which the pancreatic endocrine progenitors are found to be featured by a cell cycle state with a long G1 phase. This study obtains a single-cell–resolution, full-coverage fate map of early foregut endoderm on one hand, and on the other hand, it provides insight into endocrine pancreas development by identifying extended an G1 phase as a critical feature of its progenitors.
Results
Establishment of Unbiased Single-Cell Labeling Technique for Early Endoderm.
In this study, we referred to the zebrafish postgastrulational endoderm at the 4-somite stage as “early endoderm.” From the 4-somite stage to 48 h postfertilization (hpf), the early foregut endoderm rapidly developed into digestive organ primordia, including the pancreas, liver, foregut, and hepatopancreatic duct (HPD). To analyze the distribution of descendant cells for a single early endodermal cell, the Tg(sox17:hKikGR1)cq40 transgenic line was generated, using a humanized Kikume photoconvertible fluorescent protein hKikGR1 (23) driven by the promoter of SRY-box containing gene 17 (sox17) that marked all the early endodermal cells (24). Aided by the single-cell layer structure of zebrafish early endoderm, precise irradiation on a single early endodermal cell by the 405-nm laser could efficiently convert its fluorescence from green to red without affecting the fluorescence of surrounding cells (SI Appendix, Fig. S1A). Within the restricted time window from the 4-somite to 48 hpf, descendant cells of the photoconverted progenitor were identifiable according to their inherited red fluorescence (SI Appendix, Fig. S1A). These technical features made this approach feasible for single-cell labeling and tracing of the early endoderm.
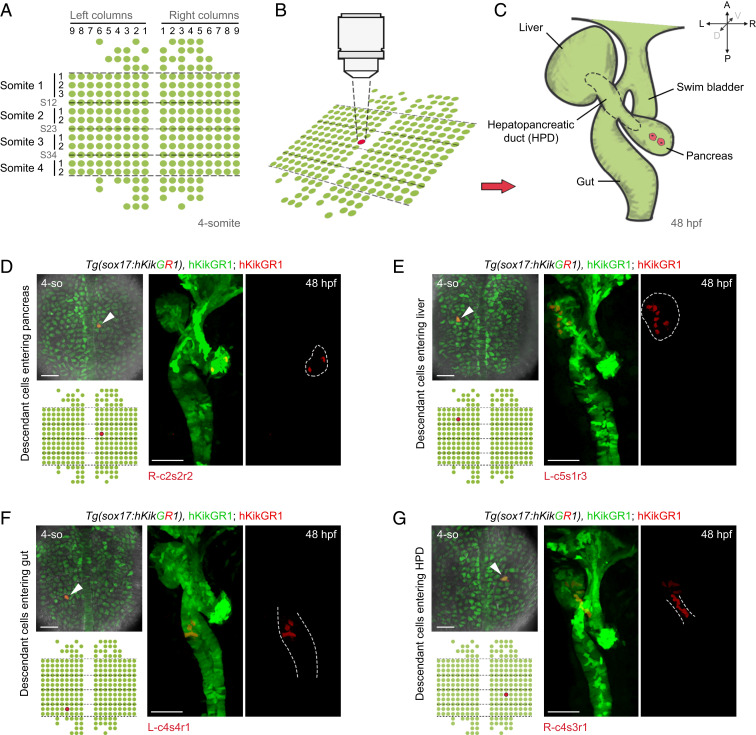
To roughly demarcate the progenitor region of foregut digestive organs within the early endoderm, photoconversions of multiple cells were performed. More than 99% of cells of foregut digestive organs at 48 hpf originated from the early endodermal cells beneath the somite-1 (s1) to somite-4 (s4) levels (SI Appendix, Fig. S1B). Although the number of these early endodermal cells varied between 216 and 10% and 216 + 5% in individual embryos, we used 216 cells as a unified standard number. These 216 progenitors plus a small portion of anterior and posterior cells, in total, 273 early endodermal cells as indicated in the standard map (Fig. 1A), composed the target region subjected to further single-cell–resolution labeling and descendant tracing. According to being left (L) or right (R) to the midline, column 1 (c1) to column 9 (c9) from medial to lateral, and row 1 (r1) to row 3 (r3) beneath the levels of a particular somite (s1 to s4) or the border of two adjacent somites (s12, s23, or s34), the position of each early endodermal cell was referred to as a unique code on a standard map (Fig. 1A), such as R-c2s2r2 (Fig. 1D, red) and L-c5s1r3 (Fig. 1E, red).
Fig. 1.
Establishment of single-cell labeling and descendant tracing techniques for endodermal progenitor cells. (A–C) Strategy of single-cell labeling and descendant tracing of early foregut endoderm. At the 4-somite stage, the 273 endodermal cells subjected to single-cell descendant tracing were aligned in a standard map. The dashed lines represent boundaries of somites (A). A single endodermal progenitor cell was labeled by green-to-red photoconversion (B). Then, localizations of its descendants at 48 hpf were detected based on the inheritance of fluorescent retention (C). (D–G) The cells at the positions R-c2s2r2 (D, n = 8), L-c5s1r3 (E, n = 8), L-c4s4r1 (F, n = 7), and R-c4s3r1 (G, n = 8), as indicated in the standard map, were individually labeled by photoconversions. Localizations of their descendant cells at 48 hpf were determined by red fluorescence according to C. Liver and pancreatic buds at 48 hpf stand to the left and right of the gut tube, respectively. (Scale bar, 50 μm.) 4-so, 4-somite stage.
The borderline of each digestive organ at 48 hpf was defined based on their anatomic details as previously reported (25, 26). The ductal segment connecting the gut to the pancreatic bud was referred to as pancreatic duct, while that connecting the gut to the liver bud was referred to as liver duct; together, they are referred to as the HPD (Fig. 1C, dashed outline). According to the single-cell labeling and descendant tracing strategy as described above, all the 273 endodermal progenitor cells in the standard map (Fig. 1A) were individually photoconverted (Fig. 1B) and traced to 48 hpf (Fig. 1C). The number and loci of descendant cells at 48 hpf were recorded. For example, the cell at the position R-c2s2r2 produced two pancreatic cells (Fig. 1D). More laterally, the cell at the position L-c5s1r3 produced seven liver cells (Fig. 1E). Posteriorly, the cell at L-c4s4r1 produced six gut cells (Fig. 1F). These examples represented progenitor cells that produced descendants with predominant commitment to a single digestive organ. However, many early cells might act as progenitors of multiple organs. For example, the cell at the position R-c4s3r1 produced five liver cells and six HPD cells (Fig. 1G), while the cell at R-c3s2r2 produced two pancreatic cells, one liver cell, five HPD cells, and seven gut cells (SI Appendix, Fig. S1C). Besides these foregut digestive organ progenitors, examples of the early endodermal cells predominantly contributing to the swim bladder (SI Appendix, Fig. S1D) and pharynx (SI Appendix, Fig. S1E) were also exhibited.
Generation of Single-Cell Fate and Proliferation Maps of Early Foregut Endoderm.
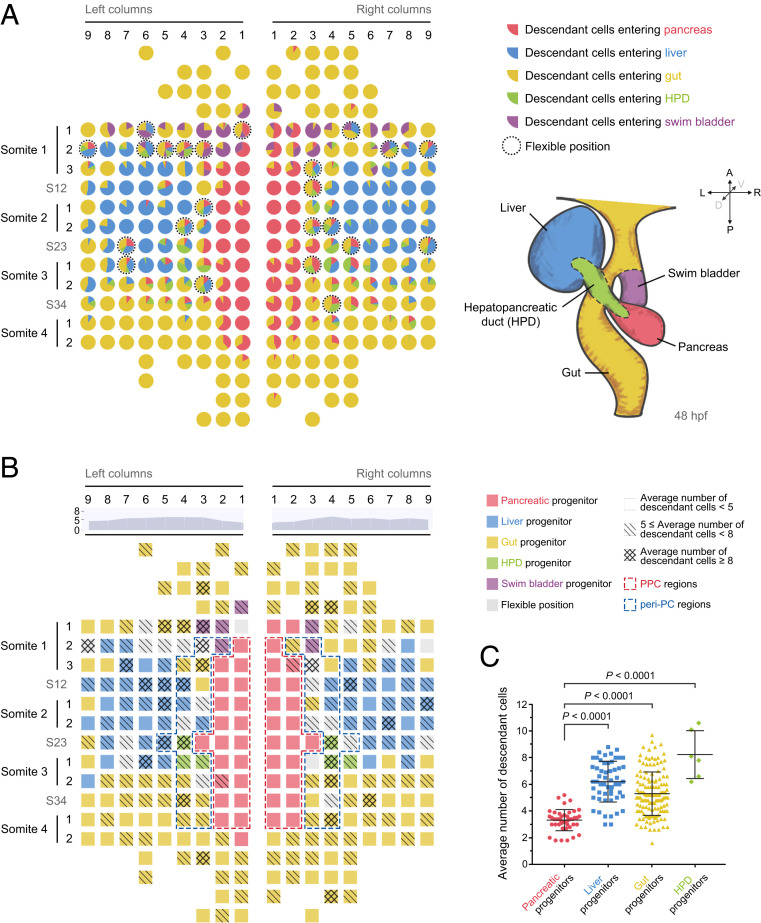
The single-cell labeling and descendant tracing procedures depicted above were carried out for each of the 273 early endodermal progenitor cells (Datasets S1 and S2). Each progenitor was traced for at least five independent repeats, and ratios of descendant cell distributions at 48 hpf were calculated (SI Appendix, Fig. S2 and Dataset S3) to generate a single-cell–resolution fate map (Fig. 2A).
Fig. 2.
Generation of single-cell–resolution fate and proliferation maps. (A) The single-cell–resolution fate map of early foregut endoderm. This map was established based on the descendant distribution ratios of every individual foregut progenitor cell. Each pie chart represents one early endodermal progenitor cell. Different colors represent different descendant cell fates. The size of a sector represents the ratio of descendant cells entering the corresponding lineage. The dotted circles indicate flexible positions. Borderlines of different digestive organs at 48 hpf are indicated on the lower right. red, pancreas; blue, liver; green, HPD; and purple, swim bladder. (B) The single-cell resolution map was established according to the number of descendant cells produced from 4-somite to 48 hpf. Different grids indicate different ranges of descendant numbers. The area chart indicates the average number of descendant cells within each column. The value of each column represents the average number of descendant cells produced by one progenitor in this column. Red and blue dashed frames represent the PPC and peri-PC regions, respectively. (C) The statistics show the average descendant cell number produced by pancreatic progenitors (n = 43), liver progenitors (n = 55), foregut progenitors (n = 139), and HPD progenitors (n = 6). Data are expressed as mean ± SD; P value is calculated using Student’s t test.
An early endodermal progenitor cell with more than half of its descendant cells localized in the pancreatic bud was defined as “pancreatic progenitor.” Similarly, liver and gut progenitors were defined. None of the early endodermal cells obtained more than half of its descendant cells in the HPD. Therefore, we defined a cell as an “HPD progenitor” only if the number of its HPD descendant cells was more than any other type of descendant cells. The early endodermal cells not included in the above four types of progenitors were termed as flexible positions. According to the fate map (Fig. 2A and SI Appendix, Fig. S2), pancreatic progenitors aligned within two columns on both sides of the dorsal midline between s1 and s4. The liver obtained two progenitor regions located bilaterally from c4 to c9 around the level of 2. The pancreas and liver progenitors were anteriorly and posteriorly enwrapped by the gut progenitors. The HPD progenitors localized at the junctional areas of pancreas, liver, and gut progenitor regions, spatially and functionally correlating with the future characters of HPD as connecting ducts. The majority of flexible positions were located at the borders of two or three progenitor populations (Fig. 2A, dotted circles).
To explore whether the pancreatic progenitors close to the dorsal midline mainly contribute to endocrine or exocrine pancreas, we performed single-cell labeling and tracing in combination with anti-Islet1 antibodies under the Tg(sox17:hKikGR1; ptf1a:EGFP) transgenic background (SI Appendix, Fig. S3 A and B). The Islet1 and Ptf1a label the endocrine and exocrine pancreas, respectively (19, 21). We found that none of the above defined pancreatic progenitors, such as L-c1s2r2 (SI Appendix, Fig. S3A), produced exocrine pancreatic descendant cells (SI Appendix, Fig. S3C and Dataset S4). We then briefly checked some early endodermal cells outside the pancreatic progenitor region that could generate pancreatic descendant cells. For some examples like L-c4s34 (SI Appendix, Fig. S3B), all of the pancreatic descendant cells were exocrine, while other examples such as R-c3s3r1 generated both exocrine and endocrine descendant cells (SI Appendix, Fig. S3C). According to the recorded data (SI Appendix, Fig. S3C and Dataset S4), we draw the conclusion that the “pancreatic progenitor” in the fate map (Fig. 2 A and B) is actually a pancreatic endocrine progenitor, which exclusively gives rise to pancreatic endocrine cells.
The single-cell labeling and descendant tracing strategy could identify not only the loci but also the number of descendant cells produced by each progenitor cell within the 36-h time window from the 4-somite stage to 48 hpf. Based on the number of descendant cells (Dataset S5), a single-cell–resolution proliferation map was generated (Fig. 2B). The proliferation map showed that the proliferative capacity of pancreatic progenitors turned out to be weaker than liver, gut, and HPD progenitors (Fig. 2 B and C). Each pancreatic progenitor produced, on average, 3.3 descendant cells, while the cells in the liver, gut, and HPD progenitor regions produced, on average, 6.2, 5.3, and 8.2 descendant cells, respectively (Fig. 2C). These data suggest a potential correlation between the low-proliferation cell cycle state and pancreatic endocrine fate propensity at as early as the 4-somite stage. Notably, although progenitors in the c1 and c2 produced a less-average number of descendant cells than other columns (Fig. 2B, area charts), the gut progenitors in c1 and c2 generated a number of descendant cells comparable to that of the gut progenitors in other columns. For example, gut progenitors in c1 and c6 produced, on average, 5.8 and 5.7 descendant cells, respectively. These results suggest that the low-proliferation cell cycle state is a feature of pancreatic progenitors but not associated with the location close to the dorsal midline.
The Cell Cycle State of Pancreatic Progenitors Is Featured by Long G1 Phase.
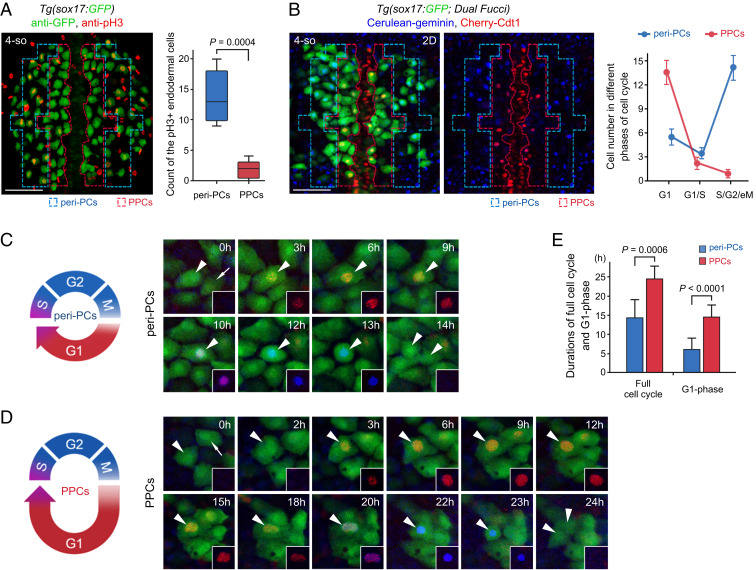
The link between the cell cycle state and cell fate has been reported in pluripotent stem cells, hematopoietic stem cells, and neurogenesis (27–33) but is not clear in the digestive organ progenitors. To study the link to pancreatic fate propensity, we first analyzed differences in the cell cycle state between pancreatic progenitor cells (PPCs, Fig. 2B, red dashed frames) and bilateral peri-pancreatic endodermal cells (peri-PCs, Fig. 2B, blue dashed frames). The peri-PCs included liver, gut, and HPD progenitors as well as flexible positions bilaterally adjacent to PPCs. The red-framed PPCs and blue-framed peri-PCs were with comparable the number of cells (Fig. 2B). The labeling of mitotic cells in late G2 through early anaphase using anti-phosphohistone H3 (pH3) (34) showed significantly fewer pH3+ cells in PPCs than peri-PCs (Fig. 3A). To further exhibit the cell cycle state of PPCs and peri-PCs, the fluorescence ubiquitin cell cycle indicator (Fucci) transgenic background, which produces a Cherry-tagged degron detectable at the G1 phase and a Cerulean-tagged degron detectable during S, G2, and early M phases (S/G2/eM phases) (30, 35), was applied. PPCs exhibited a higher proportion of cells in the G1 phase and a lower proportion of cells in the S/G2/eM phases than peri-PCs (Fig. 3B), indicative of a longer G1 phase (Fig. 3 C and D). Fucci live imaging on single cells of living embryos showed that one cell cycle duration (the time between two consecutive mitotic telophases) of peri-PCs was, on average, 14 h, while the G1 phase (from appearance to disappearance of Cherry epifluorescence) lasted ∼6 h (Fig. 3 C and E). By contrast, the average cell cycle and G1-phase durations of PPCs lasted 24 h and 15 h, respectively (Fig. 3 D and E), much longer than those of peri-PCs. These results demonstrate that the PPCs are featured by a cell cycle state with a long G1 phase in contrast to peri-PCs.
Fig. 3.
The cell cycle state of PPCs is featured by low proliferation and a long G1 phase. (A) Double immunostaining for pH3 (red) and GFP (green) in the Tg(sox17:GFP) embryos. The statistics show the number of cells double positive for pH3 and GFP in the peri-PC and PPC regions (n = 5). (B) The two-dimensional images of the Tg(sox17:GFP; Dual Fucci) transgenic line were captured covering the PPC and peri-PC regions at the 4-somite stage. The statistics show the average number of sox17+ cells in G1 phases (red), in G1/S transitions (red plus blue), and in S/G2/eM phases (blue). Note that five independent repeats represent five different embryos. (C–E) Diagrams and single-cell time-lapse live images of a full cell cycle of peri-PCs (C) and PPCs (D). The time windows between two consecutive mitotic telophases were exhibited. Arrowheads indicate the traced cells. The statistics show average durations of a full cell cycle and G1 phase of peri-PCs and PPCs (E, n = 10). (Scale bar, 50 μm.) 4-so, 4-somite stage. Data are expressed as mean ± SD; P value is calculated using Student’s t test.
Manipulations of the Cell Cycle Modulate Demarcation of Pancreatic Progenitors.
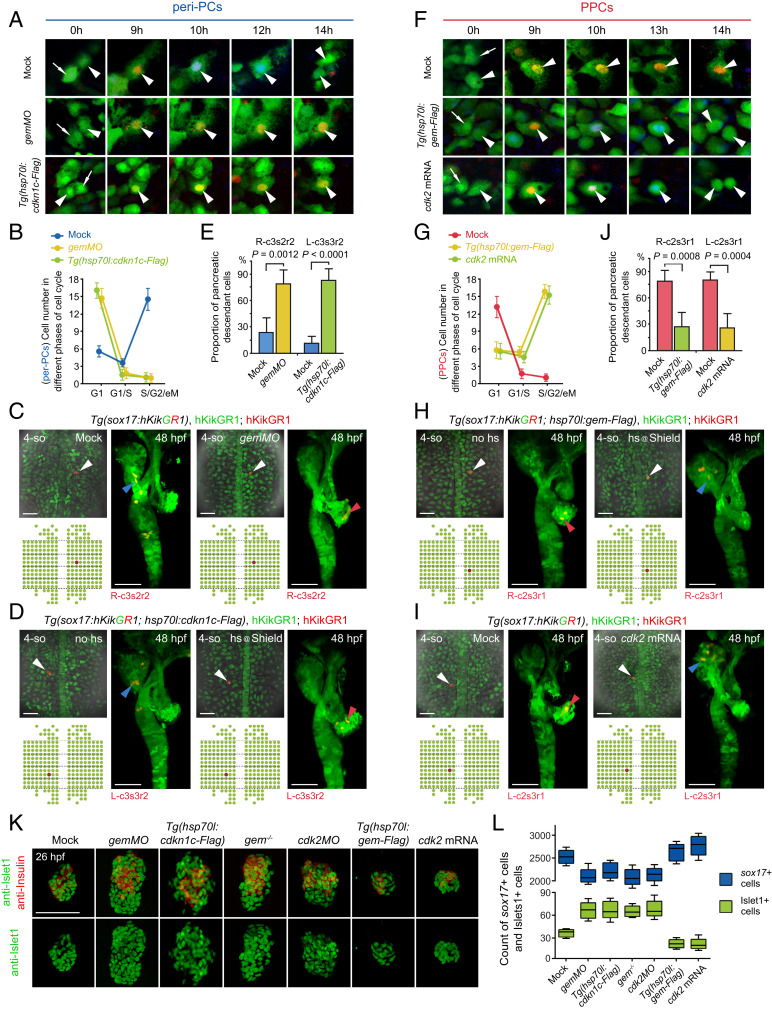
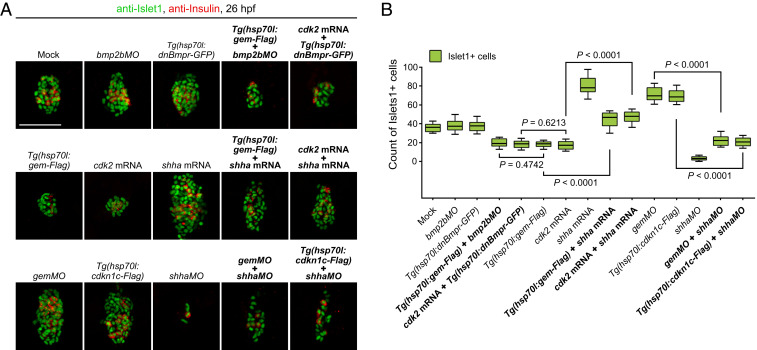
To explore whether the pancreatic fate propensity is coordinated with a long G1 phase, different molecules were applied to manipulate the cell cycle state. Geminin begins to accumulate at the G1 to S transition, which binds to Cdt1 to inhibit DNA rereplication (36–38). The application of antisense morpholino oligo against geminin (gemMO) (39) increased the overall fraction of G1-phase cells in the embryo (SI Appendix, Fig. S4A) and led to a reduced number of pH3+ cells in the peri-PC region (SI Appendix, Fig. S4 B and C), making it a potential tool to manipulate G1 duration. Further single-cell Fucci imaging in living embryos showed that gemMO extensively prolonged the cell cycle and G1 durations of peri-PCs (Fig. 4A), thus holding more peri-PCs in the G1 phase (Fig. 4B). Heat shock–inducible overexpression of Cdkn1c/p57Kip2, an inhibitor of the Cdk2/CyclinA-complex to block the G1 to S transition (40), achieved similar effects on the cell cycle state as gemMO did (Fig. 4 A and B). Consequently, single-cell labeling and descendant tracing showed that the proportion and number of pancreatic descendant cells of peri-PCs dramatically increased (Fig. 4 C–E). Therefore, these peri-PCs became pancreatic progenitors, thus enlarging pancreatic progenitor populations. The increases in the pancreatic progenitor populations were validated by the increased numbers of Islet1+ and Insulin+ pancreatic endocrine cells at 26 hpf, even though gemMO or Cdkn1c overexpression impaired cell division throughout the embryo and caused far fewer sox17+ cells than the control (Fig. 4 K and L). Additionally, this expansion of pancreatic endocrine cells could be phenocopied in a geminincq84 mutant generated using CRISPR/Cas9 (SI Appendix, Fig. S5 A–C) or by knocking down Cdk2 (Fig. 4 K and L), a key factor required for the G1 to S transition (41).
Fig. 4.
Manipulations of the cell cycle state modulate demarcation of pancreatic progenitors. (A–E) Extensive prolongation of the G1 phase of peri-PCs significantly increased their pancreatic fate propensity and enlarged pancreatic progenitor populations. Application of gemMO or heat shock–inducible overexpression of cdkn1c efficiently prolonged the G1 phase of peri-PCs as shown by single-cell live images (A, arrowheads), which was validated by the increased number of sox17+ cells in the G1 phase and reduced number of sox17+ cells in the S/G2/eM phases (B, n = 5). Consequently, examples of single-cell labeling and descendant tracing of peri-PCs showed increased descendant distributions to the pancreatic bud and gain of pancreatic progenitor identity (C and D), which were confirmed by the statistics (E, n = 5). (F–J) Shortening the G1 phase of PPCs significantly reduced their pancreatic fate propensity and pancreatic progenitor populations. Heat shock–inducible overexpression of geminin or general overexpression of cdk2 shortened the G1 phase of PPCs (F), which was validated by the reduced number of sox17+ cells in the G1 phase and increased number of sox17+ cells in the S/G2/eM phases (G, n = 5). As a result, examples of single-cell labeling and descendant tracing of PPCs showed decreased descendant distributions to the pancreatic bud and loss of pancreatic progenitor identity (H and I), which were confirmed by the statistics (J, n = 5). (K and L) After manipulations of the cell cycle state, antibody staining showed an increased and decreased number of Islet1+ and Insulin+ cells at 26 hpf (K), which were validated by statistics (L, n = 10). The corresponding alterations of the sox17+ cell number were counted by fluorescence activating cell sorter (FACS) and statistically analyzed (L, n = 5). (Scale bar, 50 μm.) 4-so, 4-somite stage; gem, geminin; hs, heat shock. Data are expressed as mean ± SD; P value is calculated using Student’s t test.
Vice versa, heat shock–inducible overexpression of geminin reduced the overall fraction of G1-phase cells in the embryo (SI Appendix, Fig. S4A) and led to an increased number of pH3+ cells in the PPC region (SI Appendix, Fig. S4 B and C). As shown by the single-cell live Fucci imaging, overexpression of geminin or Cdk2 reduced the durations of cell cycle and G1 phase of PPCs (Fig. 4F). Therefore, the number of PPCs in the G1 phase decreased and that in S/G2/eM phases increased (Fig. 4G). Consequently, the proportion and number of pancreatic descendant cells of PPCs significantly reduced, and they lost their pancreatic progenitor identities (Fig. 4 H–J), which meant a reduction of pancreatic progenitor populations and, in turn, led to a reduced number of Islet1+ and Insulin+ cells (Fig. 4 K and L).
At the 4-somite stage, the achaete-scute family bHLH transcription factor 1b (Ascl1b) is the only available digestive organ progenitor marker, which expresses in the pancreatic endocrine progenitors (42). Expression of ascl1b in the early endoderm was reduced by the overexpression of geminin or Cdk2 but became expanded by gemMO or Cdkn1c overexpression (SI Appendix, Fig. S6). Taken together, all the results above indicate the coordination of pancreatic endocrine fate with the cell cycle state of a long G1 phase, suggesting demarcation of pancreatic progenitor populations by the cell cycle state.
In zebrafish, sonic hedgehog (Shh) and bone morphogenetic protein (Bmp) signals positively and negatively regulate pancreatic fate, respectively (20, 21, 43, 44). Regarding the mechanistic insight into the link between cell cycle state and pancreatic fate propensity, we performed a pilot rescue experiment to test whether either of these two signals might be involved. The shh (sonic hedgehog signaling molecule) mRNA, but not bmpMO or the dominant-negative Bmpr, rescued the reduction of pancreatic endocrine cells caused by overexpression of geminin or Cdk2. Vice versa, the shhMO rescued the expansion of pancreatic endocrine cells caused by gemMO or Cdkn1c overexpression (Fig. 5 A and B). These pilot results indicate that Shh, but not Bmp, is potentially involved in the link between cell cycle and pancreatic fate. However, the underpinning mechanisms need further investigation.
Fig. 5.
The coordination of the cell cycle state and pancreatic fate propensity involves Shh signaling. (A) The shha mRNA but not bmpMO or the dominant-negative Bmpr rescued the reduction of pancreatic endocrine cells caused by overexpression of geminin or Cdk2. The shhaMO rescued the expansion of pancreatic endocrine cells caused by gemMO or Cdkn1c overexpression. (B) The results were confirmed by the statistics (n = 5). (Scale bar, 50 μm.) Data are expressed as mean ± SD; P value is calculated using Student’s t test.
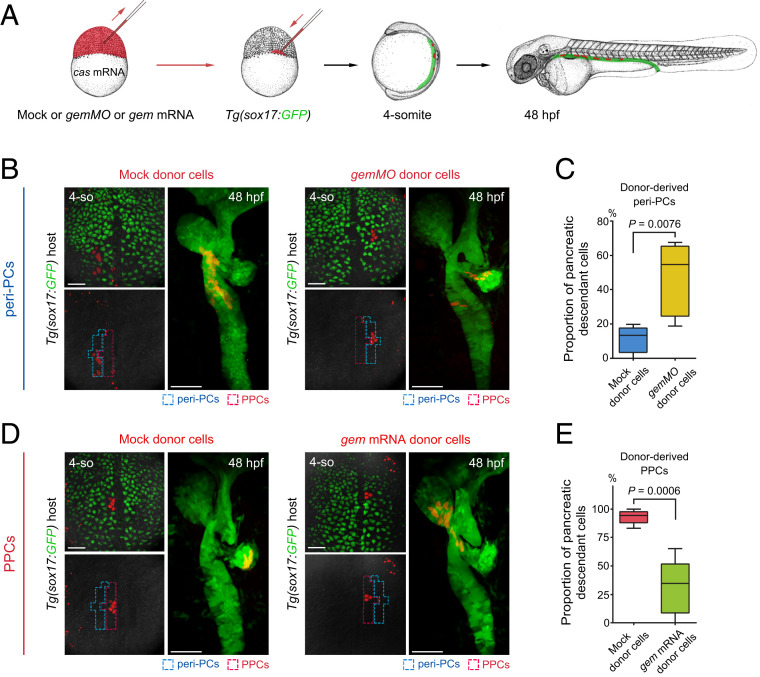
To achieve manipulations of the cell cycle state specifically in single or a few endodermal progenitor cells, cell transplantations were carried out to generate mosaic embryos. Because overexpression of casanova could drive nonendodermal cells into the endodermal lineage (14, 20), gemMO or geminin mRNA was coinjected with casanova mRNA and rhodamine dextran into donor embryos. The donor cells were transplanted into the Tg(sox17:GFP) host embryos. Then, host embryos with donor cells incorporated into peri-PCs or PPCs were collected at the 4-somite stage, and descendants of donor cells were traced to 48 hpf (Fig. 6A). In contrast to the mock donor cells, the gemMO donor cells that were incorporated into peri-PCs produced a significantly increased proportion and number of pancreatic descendant cells (Fig. 6 B and C), whereas the geminin mRNA donor cells that were incorporated into PPCs produced a significantly reduced proportion and number of pancreatic descendant cells (Fig. 6 D and E). These data demonstrate that manipulations of the cell cycle state specifically in PPCs or peri-PCs cell-autonomously modulate their pancreatic fate propensity.
Fig. 6.
Manipulations of the cell cycle state specifically in PPCs or peri-PCs cell-autonomously modulate their pancreatic fate propensity. (A) Schematic diagram of transplantation strategy. Donor embryos were injected with mock or gemMO or gem mRNA together with cas mRNA and rhodamine dextran (red). Transplantations into the Tg(sox17:GFP) host embryos were performed at the 1,000-cell stage. The host embryos with donor cells incorporated into peri-PCs or PPCs were identified and collected at the 4-somite stage and analyzed for descendant distributions at 48 hpf. (B and C) When donor cells were incorporated into peri-PCs, the gemMO donor cells produced more pancreatic descendant cells than mock donor cells (B), which were confirmed by the statistics (C, n = 5). (D and E) When donor cells were incorporated into PPCs, the gem mRNA donor cells produced fewer pancreatic descendant cells than mock donor cells (D), which were confirmed by the statistics (E, n = 5). (Scale bar, 50 μm.) 4-so, 4-somite stage; gem, geminin; cas, casanova. Data are expressed as mean ± SD; P value is calculated using Student’s t test.
Discussion
In summary, we delineate the distribution of descendant cells for every single early foregut endodermal cell, thus establishing a single-cell–resolution, full-coverage fate map and precisely demarcating progenitor regions of each digestive organ. In a long-term perspective, this map will promote studies on visible cell developmental behaviors, spatial trajectories, and mechanisms underlying cell fate determinations to achieve a single-cell level. We also provide an example here of how the maps and live imaging at the visible single-cell level facilitate an understanding of mechanisms underlying pancreatic fate determination and demarcation of pancreatic progenitors. According to the single-cell–resolution fate map, all the HPD progenitors are multilineage progenitors. These progenitors should receive multilineage developing signals in accordance with data suggesting the HPD system to be a reservoir of multipotent stem cells, possibly until adulthood (45).
Our pilot study shows that Shh is potentially involved in the link between cell cycle and pancreatic fate. Shh that promotes pancreatic fate is mainly produced by the notochord at the early somite stages (20, 46). Mutations in the Smoothened, a critical Hedgehog signal transducer, lead to loss of pancreatic endocrine cells (44). Shh is also known to induce pancreatic tumorigenesis through the transcriptional regulation of G1 cyclin (47). Besides extracellular inductive signals, intracellular factors have also been reported to couple cell cycle regulation to cell fate determinations. For instance, phosphorylation of Smad2/3 by Cdk4/6 prevents endoderm and allows neuroectoderm specification in human embryonic stem cells (30). Cdk1 can directly or indirectly regulate the expression of Oct4 and Nanog to drive lateral plate mesoderm formation (33). Neurog3 hyperphosphorylation driven by the actions of Cdk2 and Cdk4/6 in rapid cycling progenitors prevents efficient endocrine differentiation in mice (48, 49). A transcriptional coregulator Tis21 can induce neuronal differentiation by inhibiting the cyclin D1 activity in the mouse brain (28). Because the cell cycle state and pancreatic fate propensity are coordinated cell autonomously (Fig. 6), the mechanisms demonstrating how Shh is involved require further in-depth investigations, probably through a threshold of G1 duration in the early endodermal cell for the Shh signal to be received.
Diabetes is a key concern in global health. Thus, learning that an extended G1 phase is critical to endocrine pancreas development during the early somite stages is informative. The mechanisms underlying the demarcation of pancreatic progenitors by cell cycle might facilitate both in vitro and in vivo pancreatic cell differentiation, thus enhancing drug candidate development.
Materials and Methods
Zebrafish Strains and Generation of Transgenic Lines.
The zebrafish facility and studies were approved by the institutional review board. Zebrafish strains were maintained under standard laboratory conditions according to Institutional Animal Care and Use Committee (IACUC) protocols. All experimental protocols were approved. All results involved in the study were acquired according to ethical standards.
The Tg(sox17:hKikGR1)cq40 transgenic line was generated by insertion of sox17 promoter including 5.1 kb upstream of the zebrafish sox17 coding sequence (50) into a pBluescript-hKikGR1 vector. Constructs flanked by the I-SceI restriction sites were coinjected with I-SceI (NEB) into zebrafish embryos of the AB genetic background at the one-cell stage for transgenesis. The geminincq84 mutant was generated by CRISPR/Cas9 mutagenesis targeting exon 3 as previously described (51). The Tg(sox17:GFP)s870, Dual Fucciw141, Tg(hsp70l:cdkn1c-Flag)cq109, Tg(hsp70l:gem-Flag)cq110, Tg(ptf1a:EGFP)jh1, Tg(ins:GFP)zf5, and Tg(hsp70l:dnBmpr-GFP)w30 transgenic lines were established and/or used in this study. Primers for cloning sox17 promoter are Forward: GGAACAGTTTTGTACCTTTGTAAAAGTTG, Reverse: GCTAGCCCTCGACCA-AACACGCAC. Primers for cloning cdkn1c-Flag are Forward: ATGGCAAACGTG-GACGTATCAAG, Reverse: CTACTTGTCATCGTCGTCCTTGTAATCTCTAATAGTTTT-ACGTGGCGTTACT. Primers for cloning gem-Flag are Forward: ATGCTGTCCTGC-CAAGATCTGAGCT, Reverse: CTACTTGTCATCGTCGTCCTTGTAATCGGAACATTG-TTTGGGCACCCAG.
Endodermal Single-Cell Labeling and Descendant Tracing and Generation of Fate Map.
Single endodermal cell photoconversion was fulfilled by a 405-nm laser equipped on an LSM780 confocal microscope (Carl Zeiss). The Tg(sox17:hKikGR1) transgenic embryos at the 4-somite stage were mounted dorsally in 0.6% low–melting point agarose in the egg water. The irradiation region inside each cell was determined according to the shape of the cell. To achieve photoconversion and single-cell labeling, the irradiation region was focused in a single confocal plane and irradiated for 30 to 40 irradiation–detection cycles with 3 to 4 s for each cycle. The embryos were then incubated in the presence of 0.003% 1-phenyl-2-thiourea in the egg water to avoid pigmentation. At 48 hpf, the embryos were fixed with 4% formaldehyde, followed by mounting in 1.2% low–melting point agarose in 35-mm glass-bottom dishes. Z-stack images were captured using a 20× water immersion objective mounted on an LSM780 confocal microscope. Three-dimensional pictures were compiled using z-stack datasets and ZEN2010 software (Carl Zeiss). Descendant localizations and descendant number localized in each organ primordium were detected and counted based on fluorescent retention. The endodermal cells at each position in the standard map were labeled and traced as described above for at least five independent repeats. Then, according to the descendant number entering a certain organ primordium over the total descendant number produced by the labeled endodermal cell, ratios of descendant cells entering each organ primordium were calculated. Thus, a single-cell–resolution, full-coverage fate map was established for 273 early endodermal cells, including all the 216 foregut progenitors.
Expression Plasmids, mRNAs, and Morpholinos.
The full length of zebrafish geminin, cdk2 (cyclin-dependent kinase 2), casanova, and shha (sonic hedgehog signaling molecule a) cDNAs were amplified using PfuUltra High-Fidelity DNA polymerase (Stratagene) and subcloned into pCS2(+) vector for mRNA preparation. Linearized plasmids were used to synthesize mRNAs by the mMESSAGE mMACHINE Transcription Kit (Ambion) as previously described (16). geminin, cdk2, casanova, or shha mRNA were injected at the one-cell stage with 150 to 200 pg per embryo. The translation blocking morpholino antisense oligos against geminin (5′-CTTTGGTCTTCTGATGGA-ACTCATA-3′), cdk2 (5′-TTCTGAAAAGACTCCATGTCAAAAG-3′), bmp2b (5′-CGGTCTGCGTTCCCGTCGTCTCCTA-3′), shha (5′-CAGCACTCTCGTCAAAAGCCG-CATT-3′), or a general control morpholino (5′-CATTCGTCTACTGTTGGTCCTCAT-A-3′) were injected at the one-cell stage with 2 ng per embryo.
Fluorescent In Situ Hybridization, Antibody Staining, and Western Blot.
Zebrafish embryos were fixed overnight in 4% formaldehyde solution. A combination of fluorescent in situ hybridization and antibody staining were performed as previously described (52–54) using ascl1b antisense probes and anti-GFP antibodies (1:1,000, Abcam). Whole-mount antibody staining was performed using anti-pH3 (1:500, Millipore), anti-GFP (1:1,000, Abcam), anti-Islet1 (1:50, DSHB), and anti-Insulin (1:200, Abcam) primary antibodies and Alexa 488–conjugated donkey anti-mouse IgG (1:1,000, Invitrogen), Alexa 568–conjugated goat anti-rabbit IgG (1:1,000, Invitrogen), and Alexa 568–conjugated goat anti-guinea pig IgG (1:1,000, Invitrogen) secondary antibodies. Western blot was performed using anti-geminin (1:500, GenScript) and anti–α-Tubulin (1:1,000, Sigma) antibodies.
Fucci Time-Lapse Live Imaging.
The live Tg(sox17:GFP; Dual Fucci) transgenic embryos at the 4-somite stage were embedded in two layers of low–melting point agarose as previously described (55, 56). The inner layer is 0.2%, and the outer layer is 0.6%. The temperature of Thermo Plate (Tokai Hit) was set to 28.5 °C. Time-lapse scanning was performed with a 20× water immersion objective mounted on an LSM780 confocal microscope. Time-lapse images were acquired with a 5-min interval from the 4-somite to 28-somite stage. The imaging field needed to be adjusted if the tracked cell moved out. From the 28-somite stage to the end of tracking, the image acquisition interval was switched to 30 min.
Heat Shock Induction.
The Tg(hsp70l:cdkn1c-Flag), Tg(hsp70l:gem-Flag), or Tg(hsp70l:dnBmpr-GFP) embryos at the shield stage were heat shocked at 39 °C for 40 min and then returned to 28.5 °C for further development until fixation in ice-cold 4% formaldehyde solution at the time point of analysis.
Flow Cytometry and Cell Cycle Analysis.
The Tg(sox17:GFP) or Tg(ins:GFP) embryos at 26 hpf were deyolked and dissociated in 0.25% Trypsin-EDTA solution (Gibco) for 5 min. Cell suspensions were then filtered using 40-μm Falcon cell strainers. Flow cytometry was performed at room temperature using a MoFlo XDP cell sorter (Beckman Coulter). For cell cycle analysis, embryonic cells were incubated with 5 µg/mL Hoechst 33342 (Sigma) for 20 min at 37 °C. Data analyses were performed using Summit 5.2 software.
Transplantations.
Transplantations were performed as previously described (14, 20). Donor embryos were injected at the one-cell stage with 2 ng of gemMO or 200 pg of geminin mRNA, along with 200 pg of casanova mRNA and 2.5% rhodamine dextran (tetramethylrhodamine dextran, 70,000 MW, lysine fixable; Invitrogen). At the 1,000-cell stage, ∼20 cells from a donor embryo were transplanted into the blastoderm margin of Tg(sox17:GFP) host embryo. Transplanted host embryos were screened for donor incorporation into peri-PCs or PPCs at the 4-somite stage and then analyzed for the donor-derived descendant localizations.
Quantification and Statistical Analysis.
All statistical calculations were performed using IBM SPSS Statistics version 25. Variances for all groups of data are presented as ±SD. Different steps of the single-cell labeling and descendant tracing experiments were carried out by three different investigators according to a double-blind working model. All experiments comparing treatment samples were performed using randomly assigned siblings without investigator blinding. Sample sizes were chosen according to the estimation of effect sizes. After at least five repeats, data were analyzed for statistical significance using a comparison of means and unpaired two-tailed Student’s t tests. No data were excluded. A value of P < 0.05 was considered to be statistically significant. The exact sample size (n), P value for each experimental group, and statistical tests are indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank C.M. Bouldin, D. Kimelman, and H. Huang for fish lines and plasmids and H. Leng for technical assistance. This work was supported by the National Natural Science Foundation of China (Grant 31730060) and the Initiative Postdocs Supporting Program (Grant CQBX201807).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025793118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.McKenna A., et al., Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raj B., et al., Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spanjaard B., et al., Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalhor R., et al., Developmental barcoding of whole mouse via homing CRISPR. Science 361, eaat9804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan M. M., et al., Molecular recording of mammalian embryogenesis. Nature 570, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell J. A., et al., Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner D. E., et al., Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs J. A., et al., The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng G., et al., Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572, 528–532 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Ober E. A., Verkade H., Field H. A., Stainier D. Y., Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Calmont A., et al., An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev. Cell 11, 339–348 (2006). [DOI] [PubMed] [Google Scholar]
- 12.McLin V. A., Rankin S. A., Zorn A. M., Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207–2217 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Zaret K. S., Genetic programming of liver and pancreas progenitors: Lessons for stem-cell differentiation. Nat. Rev. Genet. 9, 329–340 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Dalgin G., et al., Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development 138, 4597–4608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C. R., et al., Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science 332, 963–966 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H., Ma J., Yang Y., Shi W., Luo L., EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Nicetto D., et al., H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363, 294–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tremblay K. D., Zaret K. S., Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev. Biol. 280, 87–99 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Ward A. B., Warga R. M., Prince V. E., Origin of the zebrafish endocrine and exocrine pancreas. Dev. Dyn. 236, 1558–1569 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Chung W. S., Stainier D. Y., Intra-endodermal interactions are required for pancreatic beta cell induction. Dev. Cell 14, 582–593 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung W. S., Shin C. H., Stainier D. Y., Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev. Cell 15, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorn A. M., Wells J. M., Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsutsui H., Karasawa S., Shimizu H., Nukina N., Miyawaki A., Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 6, 233–238 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reim G., Mizoguchi T., Stainier D. Y., Kikuchi Y., Brand M., The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev. Cell 6, 91–101 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Noël E. S., et al., Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev. Biol. 322, 237–250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delous M., et al., Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 8, e1002754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi T., Nowakowski R. S., Caviness V. S. Jr, The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 15, 6046–6057 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calegari F., Haubensak W., Haffner C., Huttner W. B., Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 25, 6533–6538 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange C., Huttner W. B., Calegari F., Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5, 320–331 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Pauklin S., Vallier L., The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton S., Linking the cell cycle to cell fate decisions. Trends Cell Biol. 25, 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurenti E., et al., CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 16, 302–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yiangou L., et al., Cell cycle regulators control mesoderm specification in human pluripotent stem cells. J. Biol. Chem. 294, 17903–17914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawicka A., Seiser C., Histone H3 phosphorylation–A versatile chromatin modification for different occasions. Biochimie 94, 2193–2201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouldin C. M., Kimelman D., Dual fucci: A new transgenic line for studying the cell cycle from embryos to adults. Zebrafish 11, 182–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGarry T. J., Kirschner M. W., Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Wohlschlegel J. A., et al., Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Luo L., Yang X., Takihara Y., Knoetgen H., Kessel M., The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature 427, 749–753 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Liu X., et al., NF-kappaB and Snail1a coordinate the cell cycle with gastrulation. J. Cell Biol. 184, 805–815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M. H., Reynisdóttir I., Massagué J., Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 9, 639–649 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Schnier J. B., Nishi K., Goodrich D. W., Bradbury E. M., G1 arrest and down-regulation of cyclin E/cyclin-dependent kinase 2 by the protein kinase inhibitor staurosporine are dependent on the retinoblastoma protein in the bladder carcinoma cell line 5637. Proc. Natl. Acad. Sci. U.S.A. 93, 5941–5946 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flasse L. C., et al., Ascl1b and Neurod1, instead of Neurog3, control pancreatic endocrine cell fate in zebrafish. BMC Biol. 11, 78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung W. S., Andersson O., Row R., Kimelman D., Stainier D. Y., Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 107, 1142–1147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tehrani Z., Lin S., Antagonistic interactions of hedgehog, Bmp and retinoic acid signals control zebrafish endocrine pancreas development. Development 138, 631–640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cardinale V., et al., The biliary tree–A reservoir of multipotent stem cells. Nat. Rev. Gastroenterol. Hepatol. 9, 231–240 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Roy S., Qiao T., Wolff C., Ingham P. W., Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr. Biol. 11, 1358–1363 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Morton J. P., et al., Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 5103–5108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azzarelli R., et al., Multi-site Neurogenin3 phosphorylation controls pancreatic endocrine differentiation. Dev. Cell 41, 274–286.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krentz N. A. J., et al., Phosphorylation of NEUROG3 links endocrine differentiation to the cell cycle in pancreatic progenitors. Dev. Cell 41, 129–142.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakaguchi T., Kikuchi Y., Kuroiwa A., Takeda H., Stainier D. Y., The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development 133, 4063–4072 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Hwang W. Y., et al., Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J., et al., Mammalian target of rapamycin complex 1 signaling is required for the dedifferentiation from biliary cell to bipotential progenitor cell in zebrafish liver regeneration. Hepatology 70, 2092–2106 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Chen J., et al., Cerebrovascular injuries induce lymphatic invasion into brain parenchyma to guide vascular regeneration in zebrafish. Dev. Cell 49, 697–710.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 54.He J., Mo D., Chen J., Luo L., Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nat. Protoc. 15, 3361–3379 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Upadhyay S., et al., A layered mounting method for extended time-lapse confocal microscopy of whole zebrafish embryos. J. Vis. Exp., 10.3791/60321 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C., et al., Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 44, 1162–1176 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.