Significance
The oceans are populated by an astronomical number of predominantly uncultured microbes, which altogether guarantee ecosystem function. Unicellular eukaryotic predators represent basal links in marine food webs and have so far been predominantly characterized as a functional group, despite having different ecologies and evolutionary histories. In order to better understand the ecoevolution of the ocean’s smallest predators, we have investigated four species belonging to an uncultured cosmopolitan family: marine stramenopiles (MAST)-4. Using state-of-the-art single-cell genomics and metaomics approaches, we found that members of this predatory family have different distributions in the surface ocean and different genes to degrade food, which likely represent niche adaptations. Our work highlights the importance of understanding the species-level ecology and genomics of tiny ocean predators.
Keywords: protists, MAST-4, biogeography, ecoevolution, phagocytosis
Abstract
Unicellular eukaryotic predators play a crucial role in the functioning of the ocean ecosystem by recycling nutrients and energy that are channeled to upper trophic levels. Traditionally, these evolutionarily diverse organisms have been combined into a single functional group (heterotrophic flagellates), overlooking their organismal differences. Here, we investigated four evolutionarily related species belonging to one cosmopolitan group of uncultured marine picoeukaryotic predators: marine stramenopiles (MAST)-4 (species A, B, C, and E). Co-occurrence and distribution analyses in the global surface ocean indicated contrasting patterns in MAST-4A and C, suggesting adaptation to different temperatures. We then investigated whether these spatial distribution patterns were mirrored by MAST-4 genomic content using single-cell genomics. Analyses of 69 single cells recovered 66 to 83% of the MAST-4A/B/C/E genomes, which displayed substantial interspecies divergence. MAST-4 genomes were similar in terms of broad gene functional categories, but they differed in enzymes of ecological relevance, such as glycoside hydrolases (GHs), which are part of the food degradation machinery in MAST-4. Interestingly, MAST-4 species featuring a similar GH composition (A and C) coexcluded each other in the surface global ocean, while species with a different set of GHs (B and C) appeared to be able to coexist, suggesting further niche diversification associated with prey digestion. We propose that differential niche adaptation to temperature and prey type has promoted adaptive evolutionary diversification in MAST-4. We show that minute ocean predators from the same phylogenetic group may have different biogeography and genomic content, which needs to be accounted for to better comprehend marine food webs.
Ocean microbes are fundamental for the functioning of the Earth’s ecosystems, playing prominent roles in the global cycling of carbon and nutrients (1). In particular, small phototrophic microbes are responsible for ∼50% of the primary production on the planet (2). In turn, heterotrophic microbes have a fundamental role in nutrient cycling and food-web dynamics (3). Heterotrophic flagellates, along with marine viruses, maintain prokaryotic and eukaryotic picoplankton at relatively stable abundances (4). At the same time, they transfer part of the organic matter they consume from lower to upper trophic levels, thus being a key component at the base of the ocean’s food web.
Among heterotrophic flagellates, marine stramenopiles (MASTs) play a prominent role in unicellular trophic interactions in the global ocean (5). MASTs are polyphyletic, including so far 18 subgroups (6). Except for a handful of strains, MASTs remain uncultured (7), which complicates the study of their cell physiology, ecology, and genomics. Studies using fluorescence in situ hybridization (8–10) and metabarcoding (5, 11) helped to determine MAST cell sizes (2 to 5 µm), vertical and horizontal distributions in the ocean, as well as metabolic activity. Further studies linked MAST’s cell morphology with environmental heterogeneity, for example, MAST-1B cell size varies with temperature (9). Other studies provided insight into the predatory behaviors of some MAST groups. For instance, MAST-4 prey on Synechococcus (5) and SAR11 (12), two of the most abundant microorganisms in the ocean (13, 14).
MAST-4 is a prominent clade within the MASTs, featuring small cells (2 to 3 µm), high relative abundance in comparison to other heterotrophic flagellates, and worldwide distribution (15). Due to these characteristics, MAST-4 can be considered as a model heterotrophic flagellate. MAST-4 is constituted by at least six recognized species: MAST-4A/B/C/D/E/F based on 18S ribosomal ribonucleic acid (rRNA) gene phylogenies (6). The biogeography of specific MAST-4 species has been partially elucidated: MAST-4 A, B, and C occur in temperate and warm waters (17 to 30 °C), whereas species E is typically found in colder waters (2 to 17 °C) (16, 17). This suggests that MAST-4 species have adapted to a different niche temperature. MAST-4 biogeography could also be controlled by bottom-up or top-down biotic factors, such as prey/food availability (e.g., bacteria, algae, and dissolved organic carbon) or predation, respectively. Several studies have pointed to a positive correlation between the abundances of prokaryotic and heterotrophic flagellates (4, 18–20). Yet, it is unclear to what extent such biotic relationships can generate biogeography in MAST-4.
Biogeographic patterns of MAST-4 species can provide insight into the drivers that have promoted their evolutionary diversification. Identifying species–specific gene functions, genes, or gene variants may point to differential adaptations conferring higher fitness in specific biotic or abiotic conditions. In a bacterivorous flagellate like MAST-4, a first approach for assessing species–specific adaptations is to analyze ecologically relevant genes (ERGs), which are those that could reflect associations with environmental heterogeneity or different ecological roles. Candidate ERGs include the enzymes present in the lysosome that are involved in the digestive processes that follow phagocytosis, allowing the degradation of a wide variety of substances such as proteins, carbohydrates, or nucleic acids among others (21). In heterotrophic flagellates, lysosomal enzymes are of particular relevance because different suites could potentially be associated with the degradation of different food items. Among them, glycoside hydrolases (GHs), commonly found in lysosomes, catalyze the hydrolysis of glycosidic bonds in complex sugars, allowing the cell to digest other organisms. For example, lysozyme (N-acetylmuramide glycanhydrolase) is a well-known enzyme under the GH category that catalyzes the breakdown of the peptidoglycan cell wall found in bacteria (22). Other studies have shown that each MAST lineage may have a different functional profile in terms of organic matter processing (16).
Genomes are key to obtaining ERGs from a species. Common genome sequencing protocols require thousands if not millions of cells; however, recovering this number of cells from uncultured protists such as MAST-4 is an almost impossible task. This issue is circumvented with single-cell genomics (SCG) (13, 23). The principles of this method consist in isolating single cells using, for example, flow cytometry, lysing the cells, and amplifying and sequencing their genomes producing single amplified genomes (SAGs). In previous work, SCG allowed the recovery of ∼20% of the genomes from individual MAST-4 cells, which increased to ∼80% genome recovery when genomes from different cells were coassembled (16, 24, 25). Here, we use the SAG collection produced by the Tara Oceans expedition (26), which generated ∼900 SAGs from eight stations in the Indian Ocean and the Mediterranean Sea. We compiled the largest collection to date of MAST-4 SAGs, totaling 69 SAGs (23 MAST-4A, 9 MAST-4B, 20 MAST-4C, and 17 MAST-4E). Using this dataset, together with other large metaomics datasets (metabarcoding, metagenomics, and metatranscriptomics) from the Tara Oceans and Malaspina 2010 expeditions (27), we address the following questions: How different are MAST-4 species at the genome level? Did MAST-4 species diverge via niche adaptation? If so, is such adaptation reflected in their genomes and potential ecological interactions? Can ERG composition and expression provide insights on MAST-4 niche diversification?
Results
MAST-4 Global Distributions and Associations.
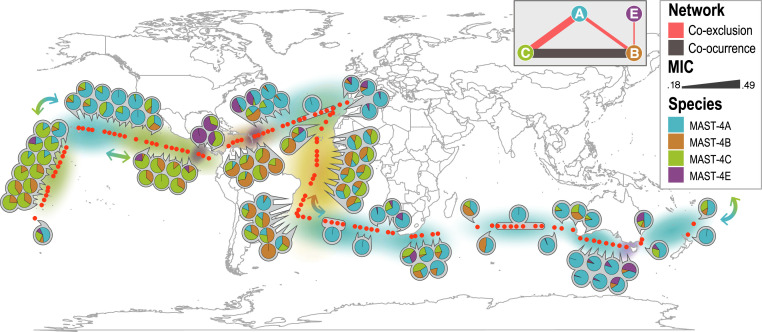
MAST-4A/B/C/E Operational Taxonomic Units (OTUs; “species” proxies) tended to display specific spatial distributions in the global ocean, in some cases markedly contrasting (Fig. 1). Specifically, species A and C were abundant and widespread across the global ocean, and even though both may appear in the same sample, they tended to exclude each other, as indicated by their association sign (Fig. 1). For example, in the Pacific Ocean when moving from equatorial waters to the north, there was a partial replacement between MAST-4C and A (see arrows in Fig. 1). Species B displayed a more restricted distribution and a lower abundance when compared to species C and A, being more prevalent in the tropical and subtropical Atlantic Ocean and in the tropical Pacific Ocean (Fig. 1). Our analyses indicated that species B co-occurred with species C, with both species coexcluding from species A (Fig. 1). Species E had a lower abundance than the other species in the tropical and subtropical global ocean, with a distribution being limited to a few locations, mostly coastal areas (Fig. 1). Species E had a weak negative association with MAST-4B (Fig. 1).
Fig. 1.
Distribution of MAST-4A/B/C/E species in the surface global ocean as inferred by OTUs based on the 18S rRNA gene (V4 region). Red dots show Malaspina stations while pie charts indicate the relative abundance of MAST-4 species at each station. (Top Right, Inset) The network shows the association patterns between each MAST-4 species as measured using MIC analyses. The width of the edges in the network shows association strength as indicated in the legend (MIC). Background color shows the most abundant MAST-4 species in the region. Arrows point to areas with an important switch of the abundant species; note that the most abundant species, A and C, alternate predominance in large oceanic regions.
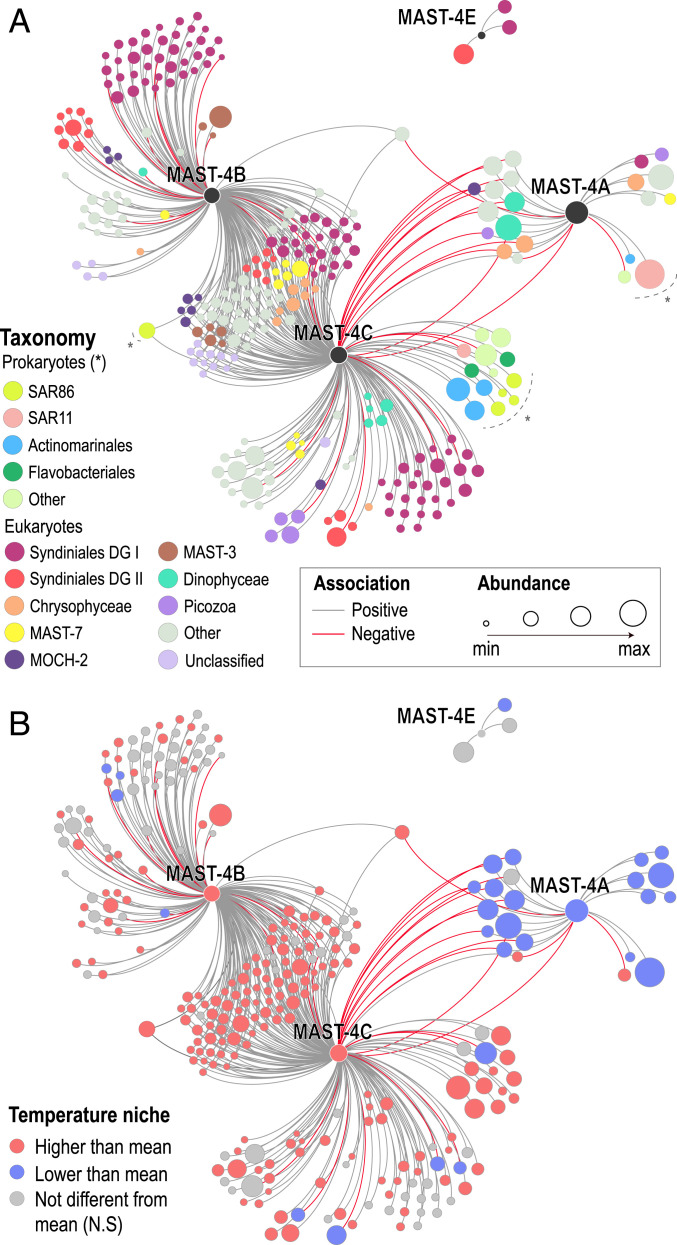
We have also investigated the association patterns between MAST-4A/B/C/E OTUs with other picoeukaryotes and prokaryotes. We found a total of 258 associations with other picoeukaryotic and 18 with prokaryotic OTUs that cannot be explained by the measured environmental factors (Fig. 2A). MAST-4C and MAST-4B displayed the largest number of associated OTUs, 191 and 174, respectively, while MAST-4A, despite being abundant and cosmopolitan, had only 23 associations. MAST-4E had only three associations to other taxa different from MAST-4 (Fig. 2A). Most associated taxa were related to a unique (59.3%), or two (38.9%) MAST-4 species (mostly species B and C) (Fig. 2A). The co-occurring species B and C displayed the largest number of shared associated taxa (total 98 taxa), which in most cases (97%) were positively associated (Fig. 2A). A lower number of associations (total of 13) was shared by the mutually excluding species A and C and, as expected, had opposite signs (50% positive and 50% negative; OTUs positively associated with MAST-4A were negatively associated to MAST-4C and vice versa) (Fig. 2A). A similar trend was observed between OTUs associated with species A and B (Fig. 2A).
Fig. 2.
Association network including MAST-4 species, associated prokaryotes, and other picoeukaryotes from the Malaspina expedition. Only OTUs with abundances >100 reads and occurrences >15% of the stations were considered in MIC analyses. A filtering strategy was applied to remove indirect (i.e., environmentally driven) and weak associations (Methods). Node size is proportional to the centered log-ratio transformed abundance sum (Methods). (A) Nodes are colored based on taxonomy. Legend: DG, Dino-Group. (B) Node color indicates whether specific OTUs displayed weighted mean temperatures significantly lower or higher than the unweighted mean temperature (24.5 °C), pointing to species with temperature distributions that differ from chance. Note that MAST-4A and both MAST-B/C tend to show co-occurrences with other OTUs that display coherent temperature preferences. N.S, not significant.
The most represented eukaryotic classes in the network included parasites (Syndiniales; 40.7% of the OTUs) and other MASTs (16.8%), including MAST-1/3/7/11/25 and other MAST-4 OTUs related to species B/C/E, which had different 18S-V4 sequences when compared to those from the SAGs. The most represented prokaryotic classes in the network included the heterotrophic species SAR86 (1.8%) and the small-sized marine Actinobacteria (Actinomarinales; 1.4%) (Fig. 2A). Other ecologically relevant classes that were present but displayed fewer OTUs were the eukaryote Picozoa (2.14%), which have similar physiological characteristics to MAST-4 (28, 29) or the prokaryotic SAR11 (0.71%), one of the most abundant bacteria in the ocean (14).
We analyzed the niche preference of individual MAST-4 OTUs as well as that of associated OTUs from other taxa in terms of temperature, salinity, NO2, NO3, PO4, SiO4, and fluorescence (Dataset S5). Adaptation to different temperature niches appeared as the main plausible driver explaining the coexclusion between species A and species B and C (Fig. 2B). The coexcluding species had different temperature preferences, with species B and C featuring a weighted mean temperature of 27.6 °C, while species A had a weighted mean temperature of 22.1 °C. Both values were significantly different from chance. In contrast, species E did not show any preference associated with temperature in our sample set covering the tropical and subtropical ocean. A fraction of the taxa positively linked to MAST-4 species showed temperature niche preferences that were coherent with those of species A, B, and C (Fig. 2B and Dataset S5). For example, taxa positively associated with species A displayed an average weighted mean temperature of 22 °C, while taxa positively associated with MAST-4B/C displayed an average weighted mean temperature of ∼26 °C. Both values differed when compared against the average unweighted mean temperature of the entire dataset: ∼24 °C. Note that detected associations reflecting only environmental preference were removed from the network, therefore remaining positive associations between microbes that prefer similar environmental conditions (e.g., temperature) indicate cases where the links between microbes could not be explained by their comparable environmental preferences. Overall, water temperature explained up to 35% of the variance in the distribution of MAST-4 species (ADONIS, P < 0.05).
Comparative Genomics of MAST-4 Species.
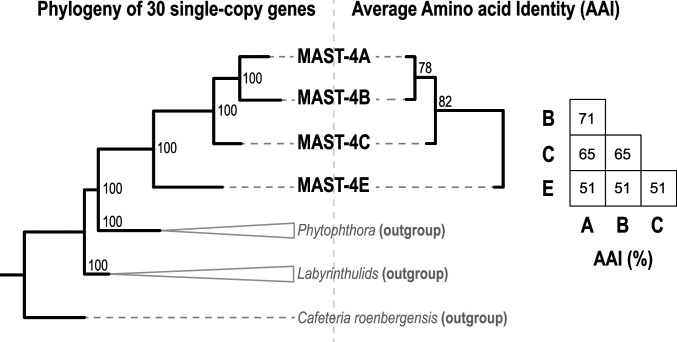
A total of 69 single-cell genomes from MAST-4A (n = 23), MAST-4B (n = 9), MAST-4C (n = 20), and MAST-4E (n = 17) were analyzed. All MAST-4E cells were isolated from the same Tara Oceans station (station 23) at the same depth (deep chlorophyll maximum—DCM) (Dataset S1). The other MAST-4 single cells were isolated from different Tara Oceans stations located in either the Indian Ocean or in the Adriatic Sea. These cells originated also from different depths, including surface or the DCM. Based on 18S rRNA gene similarity, genome tetranucleotide composition, and average nucleotide identity (ANI), cells of MAST-4A/B/C/E were independently coassembled (24). The two largest coassemblies were MAST-4A (47.4 megabases [Mb]) and MAST-4C (47.8 Mb), which contrasted in terms of size to MAST-4B (29 Mb) and MAST-4E (30.7 Mb). Accordingly, species A and C featured more predicted genes (15,508 and 16,260, respectively) than species B and E (10,019 and 9,042, respectively). MAST-4 multigene phylogenies based on 30 conserved single-copy predicted proteins (Dataset S3) as well as genome similarity based on Average Amino acid Identity (AAI) agreed with known phylogenetic relationships based on ribosomal RNA gene sequences (6) (Fig. 3). These results support our coassembly and gene prediction strategy, suggesting also a substantial amount of evolutionary divergence between MAST-4 species A/B/C/E.
Fig. 3.
Evolutionary divergence between the studied MAST-4. (Left) MAST-4 species phylogeny based on 30 single-copy protein genes from the BUSCO v3 eukaryota_odb9 database that were identified in the coassemblies (Methods and Dataset S3). (Right) Clustering of MAST-4 coassembled genomes and bootstrap support based on the AAI between predicted homologous genes. AAI values (%) between MAST-4 species are shown in the matrix.
All predicted MAST-4 genes were mapped against the Marine Atlas of Tara Oceans Unigenes (MATOU, a metatranscriptomics-based gene catalog of expressed eukaryotic genes clustered at 95% identity) (30) in order to do the following: 1) assess whether predicted MAST-4A/B/C/E genes have been previously recovered in global-ocean metaomics surveys, and 2) determine the presence of other environmental orthologs that could point to additional MAST-4 species that are prevalent in the ocean but were not considered in our work. We analyzed MATOU genes that had ≥75% nucleotide (N) similarity to MAST-4A/B/C/E genes. This threshold was used to recover environmental orthologs belonging to both MAST-4A/B/C/E as well as other MAST-4 species. The number of orthologs detected in MATOU for MAST-4A/B/C/E was variable, with species A showing orthologs for ∼25% of its genes, species B ∼20%, species C ∼33%, and species E ∼13% (Dataset S6). Not a single MATOU unigene had orthologs present in all the analyzed MAST-4 species, while 81.9% of the MAST-4 orthologs present in MATOU were associated with a single MAST-4A/B/C/E species (SI Appendix, Fig. S1). This suggests that other MAST-4 species different from MAST-4A/B/C/E are not abundant in the tropical, subtropical, and temperate open ocean and that the recovered orthologs mainly represent population/ecotype variation. Yet, the MAST-4 group seems to have a limited representation in MATOU (only orthologs for ≤one-third of MAST-4A/B/C/E genes were found), and more environmental genes should be sampled over different spatiotemporal scales than that of Tara Oceans in order to support our findings. In any case, MATOU results were coherent with our previous AAI results indicating a substantial genome differentiation among MAST-4A/B/C/E.
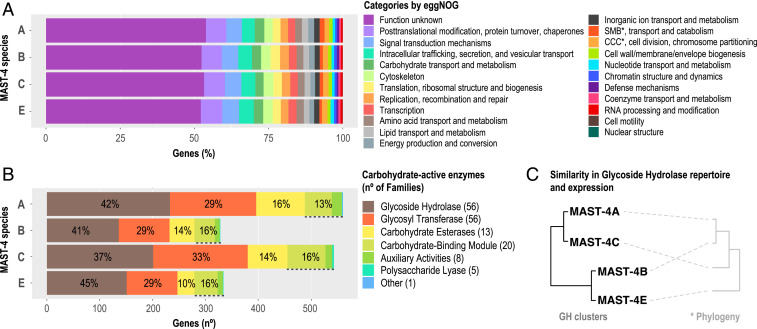
Predicted amino acid sequences were functionally annotated using the databases eggNOG (evolutionary genealogy of genes: Nonsupervised Orthologous Groups) and KEGG (Kyoto Encyclopedia of Genes and Genomes). eggNOG allowed the annotation of ∼75% of the genes from the four species, while ∼31% were annotated with KEGG. Considering that eggNOG includes environmental sequences, some with unknown functions, while KEGG is based on model or cultured organisms and annotated genes, these differences are not surprising. According to the broad eggNOG functional categories, MAST-4 species shared similar functional profiles (Fig. 4A). Yet, about half of the eggNOG hits had no function associated, as the reference sequences were environmental. Nevertheless, the existence of these hits further supports our coassembly and gene prediction approach. The most represented categories with known functions were “Posttranslational modification, protein turnover, chaperones” and “Signal transduction mechanisms,” which group important genes for the proper functioning of the cell, along with “Intracellular trafficking, secretion, and vesicular transport” and “Carbohydrate transport and metabolism,” which include pathways related to food ingestion and degradation (lysosomal reactions). Similarly, KEGG functional categories with the largest number of MAST-4 genes were “Global Metabolism,” “Signal Transduction,” and “Transport and Catabolism” (SI Appendix, Fig. S2). The first two comprise broad housekeeping functions and pathways, while the third covers vesicular processes such as endo- and phagocytosis. As expected, the potential for grazing is represented in all four MAST-4 genomes.
Fig. 4.
Functional profile of MAST-4 genes according to eggNOG and CAZy. Total MAST-4 genes analyzed were 15,508, 10,019, 16,260, and 9,042 for species A, B, C, and E, respectively. (A) eggNOG annotations indicated as percentage of genes falling into functional categories. SMB, Secondary Metabolites Biosynthesis; CCC, Cell Cycle Control. (B) Number of MAST-4 genes within CAZy categories and the corresponding percentage. The number of gene families considered within each CAZy category is indicated between parenthesis in the panel legend. (C) Clustering of MAST-4 species using Manhattan distances based on either their GH composition or the GH expression (in transcripts per million) results in the same clustering pattern. Note that MAST-4C and A are more similar in their GH content than E and B, which are more similar between themselves. *A schematic representation of the phylogeny of the studied MAST-4 is shown for comparison purposes (see Fig. 3 for more details).
The amino acid gene sequences were also annotated against the carbohydrate-active enzymes (CAZy) database, which targets functions affecting glycosidic bonds. A total of ∼3% of the total MAST-4 genes had a match against the CAZy database (Dataset S6), and the group with the largest number of genes in MAST-4 species was the GHs (Fig. 4B). We have analyzed the GH composition of MAST-4, given that different GH repertoires in species could be linked to different capacities to degrade prey bacteria or microalgae (16, 31). Most GH families were found in all MAST-4 species, but some were specific or missing in particular species (e.g., GH23 specific to MAST-4B or GH22 missing in MAST-4E) (Dataset S7). Clustering of MAST-4 species based on GH composition generated two groups, species A to C and B to E (Fig. 4C). Thus, MAST-4 genomes with contrasting geographic distributions (Fig. 1) and contrasting potential ecological interactions (Fig. 2A) were clustered together based on similar GH composition.
Global Expression of MAST-4 GHs.
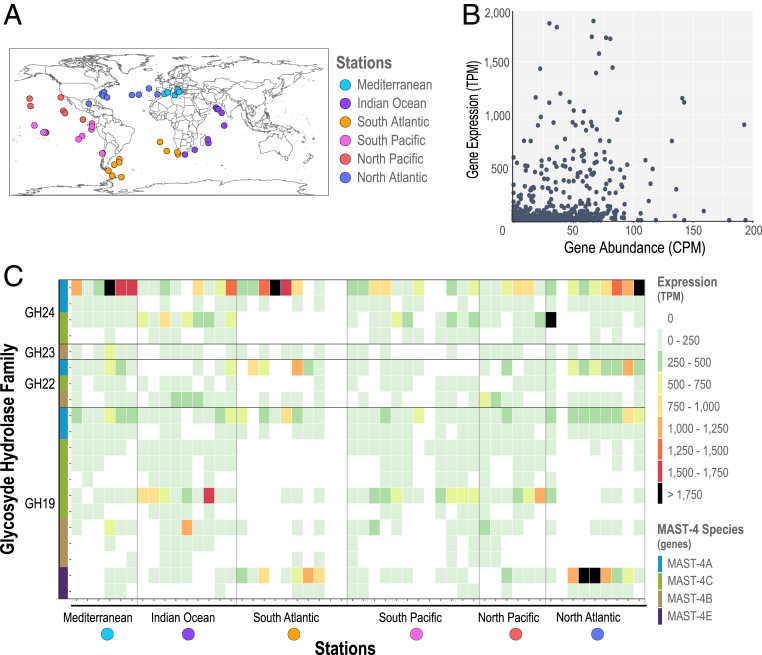
In MAST-4, GHs are most likely involved in the machinery to digest food after phagocytosis. We used metatranscriptomic and metagenomic data from the Tara Oceans expedition to assess the expression and abundance of MAST-4’s GH genes in the surface global ocean (Fig. 5A). We found that there was no obvious relationship between GH gene abundance and expression over the surface global ocean, indicating that differences in gene expression most likely represent up- or down-regulation of GH genes (Fig. 5B; see also Fig. 5C and SI Appendix, Fig. S3B). MAST-4’s GH gene expression was highly heterogeneous in the surface global ocean (Fig. 5C). The GH families with the highest expression were the lysozyme families GH22 and GH24, in charge of degrading the peptidoglycan in the bacterial cell wall (22, 32), as well as the chitinase family GH19, involved in the degradation of chitin (present in particulate detritus, crustaceans, and several other organisms in the ocean) (Fig. 5C). These GH genes tended also to display a higher expression mean than single-copy housekeeping genes within the same Tara Oceans stations (Dataset S8). Interestingly, the South Pacific displayed low or absent GH expression in all MAST-4 species, despite GH gene abundances that were similar to those found in other regions displaying higher expression (SI Appendix, Fig. S3B). We found also clear differences in expression between species: for example, while species’ A GHs were widely expressed in several regions, those GHs from species E were expressed only in specific samples, in particular in the North and South Atlantic. GH genes from species B and C were either not detected or had low expression in the South Atlantic samples, in contrast to specific GH genes from species A and E in the same region (Fig. 5C). In turn, specific GHs from species B and C had higher expression than A and E in the Indian Ocean.
Fig. 5.
Expression and abundance of GHs in MAST-4A/B/C/E in the upper global ocean. (A) Geographic location of the metagenomic and metatranscriptomic samples from Tara Oceans. (B) Gene abundance versus expression using normalized data for each gene and station. Note that the axes have different but proportional ranges of values. (C) Heatmap of the GH families in MAST-4 that had the highest expression. Samples are in the x-axis, grouped by ocean region and ordered following the expedition’s trajectory. Genes in the y-axis are organized by family, and each species is indicated with a color. GH22, GH23, and GH24 are families of lysozymes, and GH19 is a family of chitinases that can also act as lysozyme in some organisms.
Differences in abundance and expression were also found in GH genes belonging to the same family and within the same MAST-4 species. For example, species A had two genes belonging to the GH24 family; one gene (631 base pairs [bp]) was more expressed than the other (1,465 bp), despite gene abundances being similar across all samples (Fig. 5C and SI Appendix, Fig. S3B). These two genes shared 29.5% similarity at the amino acid level based on 73% coverage (153 amino acids) of the shorter gene. A similar pattern was observed in the two GH24 genes in MAST-4C: the shorter was more expressed than the longer (622 versus 1,198 bp). In fact, the short and long GH24 genes from species A and C are homologs, respectively: the short homologs have 79.4% identity (94% coverage) while the long homologs have 56.4% identity (87% coverage). In general, MAST-4 species with more than one gene belonging to the same GH family tended to express one particular variant over the others. One plausible explanation is that the underexpressed GHs are gene duplications. GH genes often undergo duplication and thus several copies can be present in the form of paralogs (33–35). After gene duplication, a redundant copy is generated and freed from selective pressure, allowing it to accumulate mutations (36) and potentially lead to new functions (37, 38).
Detecting Positive Selection Acting on MAST-4 Genes.
We analyzed whether there is evidence of positive selection leading to niche adaptation in the different MAST-4 species. For that, we analyzed nonsynonymous versus synonymous substitutions (dN/dS) in selected homologous genes in MAST-4A/B/C/E. Normally, the ratio dN/dS is used to test hypotheses related to the action of selection on protein-coding genes, where dN/dS >1 indicates that substitutions generating changes in amino acids are greater than substitutions that do not, suggesting the action of diversifying (i.e., positive) selection (39). A total of 692 alignments (homologous groups) were used for testing positive selection on both branch (whole sequence phylogeny) and codon analyses (gene site-specific) (40, 41) (Dataset S9; Methods). Overall, 60 gene alignments (8.7%) indicated positive selection in branch analyses, of which 57 alignments displayed selection in 1 branch and 3 in 2 branches (60 alignments, 63 total branches selected). MAST-4A and B appeared to be the most selected branches, 22 (34.9%) and 25 (39.7%) times, respectively, while MAST-4C and E had a low number of selected branches, 8 (12.7%) and 4 (6.3%) times, respectively. In codon analyses, 478 gene alignments (69.1%) displayed positive selection in one or more positions, ranging from 1 to 15 positively selected codons per alignment. In GH, a key part of the predatory machinery of the MAST-4, 1 alignment (0.14%) showed positive selection in branch analyses for family GH74 while 14 alignments (2%) displayed positive selection in codon analyses that included GH3, GH13, GH16, GH19, GH28, GH30, G74, GH78, GH79, and GH99 (Dataset S9). Of all of them, only GH19 belongs to one of the most expressed families according to the metatranscriptomic analyses. Overall, these analyses suggest that adaptive evolution promoted the diversification of MAST-4 into species A, B, C, and E or at least that it promoted the diversification of specific genes.
Discussion
Currents, waves, and wind promote the dispersal of plankton in the surface ocean. Given their typically large populations and small organismal sizes, microbial plankton species are expected to be widely distributed in the upper ocean. This is particularly relevant for the MAST-4 group, which features a moderate abundance [about 50 cells ml−1 in surface waters and ∼10% of the heterotrophic flagellates (42)] and minute size. Such characteristics in combination would guarantee dispersal and widespread distributions (43), decreasing the potential effects of dispersal limitation (44). These characteristics would also promote a coupling between environmental heterogeneity (selection) and species distributions (45). Thus, we expected that MAST-4 distributions would reflect, to a certain extent, the abiotic and biotic conditions in the ocean. This is coherent with previous findings indicating that 1) temperature is an important environmental variable driving MAST-4 distributions and 2) that dispersal limitation does not seem to affect the distributions of MAST-4 species (17). We expanded previous knowledge by determining the temperature distribution of species A, B, and C. Specifically, we show that species B and C occur in warmer temperatures (weighted mean = 27.6 °C), while species A is present in lower temperatures (weighted mean = 22.1 °C). In contrast, we did not find evidence that the distribution of species E was affected by temperature in the tropical and subtropical ocean. This is coherent with reports indicating that MAST-4E inhabits cold waters (17).
Even though temperature is a key variable structuring the global ocean microbiota, including MAST-4 (46–49), biotic variables could also affect the distributions of MAST-4 species. We found that the number of associations between MAST-4 OTUs and bacterial OTUs was low. Actually, most associations were not considered as they were either weak (low correlation) or they just represented similar or different environmental preference (mainly temperature) between MAST-4 and bacterial OTUs. Altogether, this suggests that MAST-4 abundance and occurrence is weakly coupled to bacterial distributions and abundance in the upper ocean, which agrees with previous studies where changes in the overall heterotrophic flagellate abundances were related to water temperature (42). We detected a substantial number of taxa that were positively associated with either MAST-4B/C or MAST-4A but not to both. Even though associated taxa tended to reflect the temperature preference of the species to which they were associated (B/C or A), their association to different MAST-4 cannot be simply explained by similar niche temperature, since we also detected associations to OTUs without a significant temperature preference. The vast majority of associations were between species A or B/C with other picoeukaryotes, such as Syndiniales’ Dino-Group I and II, which are known parasites (50), or MAST-3 and MAST-7, which are flagellates as well (6). These associations could either manifest a similar preference for an environmental variable different from temperature that covaries with MAST-4 distributions or reflect real ecological interactions, including parasitism. For instance, there is evidence of MAST-4A having a predator–prey relationship with Synechococcus (9) and possibly with SAR11 (12), which was not only reflected in our networks from the Malaspina expedition but also in previous studies from the Tara Oceans expedition (51). Results from Tara Oceans reported other taxa associated with MAST-4A that were corroborated by our results (MOCH-2, Chrysophyceae, MALVs, MAST-7). However, whether or not these associations reflect true ecological interactions needs to be proved with further experiments. Altogether, we did not find evidence that biotic interactions between MAST-4 and other microbes represent an important driver of MAST-4 biogeography.
Our results suggest that adaptation to different temperature niches and interspecific interactions between MAST-4 species (competition) are likely the main drivers determining MAST-4 biogeography. If so, differential adaptation should likely be reflected in the genomes of the MAST-4 species. Our analyses indicated that MAST-4 species differ in genome size: two bigger genomes (MAST-4A and C) with a partial genome size of ∼47 Mb and ∼80% completeness and two smaller genomes (MAST-4B and E) of ∼30 Mb and ∼70% completeness, which correspond to ∼59 and ∼42 Mb full estimated genomes, respectively. The observed differences in genome size need to be considered with care, as they may be reflecting incomplete genome assemblies. Nevertheless, our estimates of genome size were similar to those of Cafeteria roenbergensis (∼40 Mb) (52), a heterotrophic flagellate in the same cell-size range of MAST-4, and other Stramenopile genomes, for example the diatom Thalassiosira pseudonana (∼34.5 Mb) (53) or various Phytophthora species (Phytophthora plurivora, Phytophthora multivora, Phytophthora kernoviae, and Phytophthora agathidicida with 41, 40, 43, and 37 Mb, respectively) (54). This suggests that our partial genomes are likely large enough to be representative of the studied MAST-4 species. We found that differences in MAST-4 genome size were mirrored by the number of predicted genes in each species, which ranged between 9,042 and 16,260, even though larger genomes in eukaryotes do not always imply a greater number of genes (55). These differences in gene content between species may to some extent be linked to niche adaptation. Overall, none of the studied MAST-4 displayed any loss or gain of broad functional categories when compared to each other. In fact, they were similar in terms of the proportion of genes that belong to each functional trait, suggesting that MAST-4 metabolisms are broadly comparable, which agrees with other reported results in MAST-4 species A/C and E (16). Among the most represented functional categories in the MAST-4 genomes were those involved in phagocytosis and subsequent digestion. For instance, eggNOG’s “vesicular and carbon transport,” along with KEGG’s “transport and catabolism,” includes pathways for “Endocytosis,” “Phagosome,” “Lysosome,” “Peroxisome,” and “Autophagy (animal and yeast),” all related to vesicular forms of transport and prey digestion. Thus, MAST-4’s lifestyle as marine grazers (5, 56) is in agreement with their broad genomic functions associated with phagocytosis. Yet, homologs among species were very different at the DNA or amino acid level. In particular, when comparing MAST-4A/B/C/E gene predictions against the MATOU (30, 57), the vast majority of homologs were unique to one MAST-4 species. In fact, we did not find a single gene in MATOU with homologs in all MAST-4A/B/C/E species, which manifests the interspecific differences of MAST-4 in terms of genomic composition. The substantial differentiation between homologs was reflected by the AAI and phylogenomic results as well (SI Appendix, Fig. S1), which altogether indicate that MAST-4 experienced substantial evolutionary diversification.
MAST-4 is not exclusively bacterivorous and can feed on other small organisms, for example, Micromonas pusilla and Ostreococcus sp. (5), and perhaps complement its diet with noninfective viruses (58). A comparable diet has been observed in other heterotrophic flagellates (59). Such a variety of food items, which vary in quality and quantity, most likely require different metabolic machineries to digest them (16, 31), in particular different carbohydrate-active enzymes. For example, studies in Fungi have shown that the number and composition of CAZymes may determine the degradation capacity of different plant biomass sources (60). Here, we analyzed the GHs, one of the most efficient known catalysts of organic substances in living organisms (61) and likely important for MAST-4’s heterotrophic lifestyle. GHs genes accounted on average for 3% of the predicted genes in each MAST-4. Most of the GH gene families were found in the four species, but some were either exclusive of a single species or missing in others, which may be due to genome incompleteness. Similar patterns have been reported before, not only in a reduced number of MAST-4 species (16) but also in the fungal genus Saccharomyces (62), where the set of GH genes differs even in strains of the same species. Site (codon) analyses suggested positive selection in a few GH families in MAST-4 (e.g., within the GH19 gene family). Similarly, other GH families that are not lysozyme like, such as GH3, GH30, or GH74, appeared to have experienced positive selection as well, even though they were not as much expressed in the global ocean as the lysozyme. Altogether, this suggests the action of adaptive evolution in the machinery that MAST-4 uses to digest food and may reflect adaptations to the degradation of different compounds or prey.
The four MAST-4 species formed two groups based on GH composition (number of genes per family). One group consisted of species A and C and the other of species B and E. Interestingly, species A and C, with similar GH repertoires, showed spatial coexclusion in the upper global ocean, while species C and B, with different GH repertoires, were co-occurring (Fig. 1). These geographic distributions suggest that niche adaptation associated with different temperatures allowed MAST-4A and C to keep similar GH repertoires, while species adapted to similar temperatures that co-occur (C and B) were exposed to divergent selection diversifying their diets as a response to competition, which is reflected in their different GHs (31). We found that species A and B/C have different niche temperatures (A = 22.1 °C and B/C = 27.6 °C). Since temperature niche can be a phylogenetically conserved trait in specific microbes (63, 64), it would have been expected that the closely related MAST-4A and MAST-4B share a similar temperature preference. However, species A had a temperature preference 5 °C lower than that of B, suggesting that selection has promoted the adaptation of species A to lower temperatures perhaps to not compete with species C or that species C is a superior competitor and excludes species A from warmer waters. Furthermore, since MAST-4A, B, and C form a monophyletic group, they are expected to share a comparable GH repertoire. But instead, our analysis showed that the GH repertoire of B was closest to E, suggesting that evolution promoted the divergence of MAST-4B’s GH content.
The temperature distributions of the studied MAST-4 species, together with their different GH repertoires lead to two plausible evolutionary scenarios. MAST-4E, the deepest branching lineage, did not show a particular preference for either warm or cold waters in our data (Dataset S5), but other reports indicate it occurs in cold waters (17). Thus, during the MAST-4 diversification, species E would have either adapted to or remained in cold waters. Then, two evolutionary hypotheses emerge depending on whether the Last Common Ancestor (LCA) of MAST-4A/B/C originated in warm or cold waters: 1) The LCA of MAST-4A/B/C was adapted to warm waters and species C remained in warm waters. Then, the two most evolutionary derived species, A and B, diverged their niches as a result of competition with C; species A adapted to colder subtropical and temperate waters, while species B stayed in the tropics and avoided competition with C by changing its niche via diet modification, which is reflected in its GH composition; and 2) The LCA of MAST-4A/B/C inhabited cold (subtropical) waters and then C and B adapted independently to warmer tropical habitats with B modifying its niche to avoid competition with C by changing its GH repertoire and consequently its diet. Even though both evolutionary scenarios are possible, our dN/dS results using homologous proteins of the four MAST-4 species are more coherent with the first evolutionary scenario by indicating that MAST-4A and MAST-4B appear to have diverged the most, as they displayed the effects of significant positive selection in 75% of the total alignments with branch selection.
We also analyzed MAST-4’s GH distribution and expression in the surface global ocean, as this may shed light on whether species with similar GH composition express similar or different genes when they co-occur, possibly indicating prey preference depending on the presence/absence of competitors. We found that the different species displayed a large heterogeneity in their expression patterns. The tropical species that co-occurred the most, C and B, showed dissimilar expression patterns, with some genes being highly expressed only in one species, which is coherent with their difference in GH composition as well as with a scenario proposing different food preferences. Furthermore, species C and B showed differences in expression over specific ocean regions, suggesting that despite their co-occurrence, their GH activity is modulated differently. In turn, the coexcluding species A and C, which display the most similar GH composition, appeared to express different GHs over the upper global ocean, suggesting that they regulate GH expression perhaps as an adaptation to different preys or that GH expression is affected by the different temperatures in which these species occur. Overall, our evidence suggests that species A, B, and C regulate GH genes differently, perhaps as an adaptation to different diets or prey, even though some differences in GH expression only reflect the presence or absence of MAST-4 species in specific ocean regions.
Altogether, our results suggest that the evolutionary diversification of MAST-4 was promoted by divergent adaptive evolution toward different temperature and/or diet niches possibly as a response to competition and that biotic interactions with other species did not have a major influence in MAST-4 diversification. The previous possibly led to the emergence of the species associated with tropical (MAST-4B and C), subtropical-temperate (MAST-4A), and subpolar-polar (MAST-4E) waters. Furthermore, species B may have diverged in its diet as a response to competition with C, and as a result, it has a different GH composition from its closest evolutionary relatives, A and C. If future cultures of MAST-4 species are established, the previous scenarios could be tested by determining the temperature range of species growing in isolation or with interspecific competitors. Our work represents a significant contribution to understanding the evolution, diversity, biogeography, and function of the smallest predators in the ocean. This knowledge is fundamental to comprehending the base of marine food webs and the biotic and abiotic factors that may affect them, as well as the consequences in upper trophic levels.
Methods
Geographic Distribution of MAST-4 Species and Association Patterns.
The distribution of MAST-4 species as well as their association patterns were investigated using metabarcoding based on data from Logares et al. (49). This dataset includes surface water samples (3-m depth) from a total of 120 globally distributed stations located in the tropical and subtropical ocean that were sampled as part of the Malaspina 2010 expedition (27). Both the 18S [variant region 4 (V4) (65)] (66) and 16S [V4 to V5 region (67)] (68) rRNA genes were analyzed. OTUs were delineated as Amplicon Sequence Variants using Divisive Amplicon Denoising Algorithm 2 (DADA2) (69) and OTU tables were generated (see details in SI Appendix, SI Methods S1). OTUs were assigned taxonomy using the naïve Bayesian classifier method (70) together with the SILVA v132 database (71) as implemented in DADA2. Eukaryotic OTUs were also assigned taxonomy using Basic Local Alignment Search Tool (BLAST) searches against the Protist Ribosomal Reference database [version 4.11.1 (72)]. Streptophyta, Metazoa, nucleomorphs, chloroplasts, and mitochondria were removed from the OTU tables.
Associations between OTUs were inferred using Maximal Information Coefficient (MIC) as implemented in MICtools (73). Environmentally driven associations between OTUs were detected and removed using EnDED (74), with the methods Interaction Information and Data Processing Inequality. Furthermore, we removed associations between OTUs that were not present in ≥50% of the samples and featured a Jaccard index <0.25 or an MICe <0.4 (see details in SI Appendix, SI Methods S1). The distribution of OTUs across sea temperatures was explored using the niche.val function in the EcolUtils package (75). The abundance-weighted mean temperature was calculated for each OTU and used as an estimate of its temperature niche. We checked whether the obtained abundance-weighted mean temperature for each OTU was significantly different from chance (P < 0.05) using a null model with 1,000 randomizations.
Genome Reconstruction Using SAGs.
Plankton samples were collected during the circumglobal Tara Oceans expedition, and SAGs from different taxa were generated as previously described at the Single Cell Genomics Center (https://scgc.bigelow.org) (76) (see details in SI Appendix, SI Methods S2) (77). A total of 69 SAGs affiliating to MAST-4 species A/B/C/E were selected for downstream analyses. Each MAST-4 SAG was sequenced in one-eight of a lane using Illumina HiSeq2000 or HiSeq4000 at either the Oregon Health & Science University or the French National Sequencing Center (Genoscope). A total of 424.1 giga bases (Gb) of sequencing data were produced, averaging 6.1 (± 0.22) Gb per SAG. For each SAG, sampling location, depth, and date are reported in Dataset S1. Each SAG was de novo assembled using SPAdes (St. Petersburg genome assembler) 3.10 (78). Estimation of genome recovery was calculated with BUSCO v3 (Benchmarking Universal Single-Copy Orthologs) (79) using the Eukaryota_odb9 dataset (Dataset S2). In order to increase genome recovery, SAGs were also coassembled based on 18S rRNA gene similarity, ANI, and tetranucleotide frequencies (reference SI Appendix, SI Methods S2 for more details).
A total of 69 MAST-4 SAGs were coassembled: MAST-4A (23 SAGs), MAST-4B (9 SAGs), MAST-4C (20 SAGs), and MAST-4E (17 SAGs). Prior to coassembly, reads were digitally normalized using BBNorm (80). Normalized reads were coassembled with SPAdes 3.10. To extend contigs, coassemblies were rescaffolded with SSPACE (Scaffolding Pre-Assemblies After Contig Extension) v3 (81). Repetitive regions were masked, along with transfer ribonucleic acid (tRNA) sequences, using RepeatMasker (82) and tRNAscan-SE-1.3 (83). Quality and assembly statistics were computed with Quast (84) and are shown in Dataset S2. Coassembled SAGs were carefully checked for foreign DNA using emergent self-organizing maps (ESOM) (85) and EukRep (86) (Dataset S2 and SI Appendix, SI Methods S2). Coassembled genome completeness was estimated with BUSCO v3. For each coassembly, protein-coding genes were predicted de novo with AUGUSTUS 3.2.3 (87, 88) using the identified BUSCO v3 proteins as training set (89). Predicted genes were functionally annotated using 1) CAZy database from dbCAN v6 (90) and HMMER 3.1b2 (91), 2) KEGG [Release 2015-10-12; (92, 93)], and 3) eggNOG v4.5 (94), both using BLAST 2.2.28+. Gene sequences (Ns) were also mapped against the MATOU Version 1 (20171115) (30) using BLAST 2.2.28+ (see details in SI Appendix, SI Methods S2). MAST-4 genomes were clustered in terms of their composition of GHs with the hclust function in R based on “manhattan” distances.
Phylogenomics and Genome Differentiation.
We used two approaches to analyze the phylogenetic versus whole-genome differentiation among MAST-4 species. In the first approach, we randomly selected 30 conserved proteins (included in eukaryota_odb9, BUSCO v3) that were identified in all MAST-4 species (Dataset S3) as well as in other publicly available Stramenopile genomes: Phytophtora sojae (National Center for Biotechnology Information [NCBI]: txid67593), Phytophtora infestans (National Center for Biotechnology Information [NCBI]: txid403677), Schizochytrium aggregatum (Joint Genome Institute [JGI]: Schag1), Aurantiochytrium limacinum (JGI: Aurli1), and Cafeteria roenbergensis (52). Genes were aligned individually with Mafft (95) and concatenated with catfasta2phyml (96). Poorly aligned sequences and regions were removed using trimAl v1.4.rev22 (97). A phylogenetic tree was built with RAxML version 8.0.0 (98) (see details in SI Appendix, SI Methods S3). The second approach consisted of computing the AAI for each pair of MAST-4 using Enveomics based on the predicted genes (amino acids). Genomes were clustered by similarity using the pvclust (99) package in R with “maximum” as the distance method.
Abundance and Expression of Selected MAST-4 ERGs in the Ocean.
We investigated the distribution, abundance, and expression in the global ocean of selected ERGs, in this case, GH. For that, we mapped metagenomic (100) and metatranscriptomic (101) reads from Tara Oceans (a total of 52 surface water stations encompassing the 0.8 to 5 μm size fraction [total 104 samples]) against predicted genes from each MAST-4 species (Dataset S4). Metatranscriptomic reads were derived from sequencing polyA-enriched RNA (30, 102). The mapping was done with Burrows-Wheeler Aligner (103), and the abundance of genes and transcripts were expressed as counts per million or transcripts per million, respectively (see details in SI Appendix, SI Methods S4).
Calculation of dN/dS Ratios in Homologous Genes.
Homologous MAST-4 genes were identified using reciprocal protein BLAST (v. 2.2.28+). Gene sequences (amino acid) were aligned using Mafft 7.402 and then converted into a codon-based N alignment with Pal2nal (104). Alignments with one or more unknown Ns were discarded. For each homolog, a N-based phylogenetic tree was built using RAxML 8.2.12 (98). Positive selection was tested on each homolog with HyPhy 2.3.14 (105) using adaptive Branch-Site Random Effects Likelihood (aBSREL) (branch) (40) and Mixed Effects Model of Evolution (MEME) (site) (41) models considering the codon-based N alignment and the corresponding phylogenetic tree (see details in SI Appendix, SI Methods S5).
Supplementary Material
Acknowledgments
We thank all the scientists from the Malaspina 2010 expedition and the Tara Oceans 2009 to 2013 expedition, as well as the staff of the Single Cell Genomics Center (Bigelow Laboratory) for the generation of SAGs. Bioinformatics analyses were performed at the MARBITS platform of the Institut de Ciències del Mar (https://marbits.icm.csic.es/) and also on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. We thank Lidia Montiel and Pablo Sánchez for the assistance with bioinformatics. F.L. was supported by the Spanish National Program Formación de Personal Investigador 2016 (BES-2016-076317, Ministerio de Ciencia e Innovación, Spain). R.L. was supported by a Ramón y Cajal fellowship (RYC-2013-12554, Ministerio de Economía y Empresa, Spain). This work was supported by the projects INTERACTOMICS (Unveiling Core Ecological Interactions in Marine Microbial Communities Using Omics Approaches) (CTM2015-69936-P, MINECO, Spain, to R.L.), MicroEcoSystems (240904, Research Council of Norway, to R.L.), and MINIME (Microbial Evolution and Population Genomics in a Changing Ocean) (PID2019-105775RB-I00, Agencia Estatal de Investigación Spain, to R.L.). I.M.D. and A.L. were supported by the European Union’s Horizon 2020 research and innovation program under Marie Skłodowska-Curie Grant Agreement No. 675752 (SINGEK [Promoting Single Cell Genomics to Explore the Ecology and Evolution of Hidden Microeukaryotes]: http://www.singek.eu). We thank the Consejo Superior de Investigaciones Científicas (CSIC) Open Access Publication Support Initiative through the Unit of Information Resources for Research for helping to cover publication fees. We also thank the reviewers for providing valuable feedback that helped to improve our work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020955118/-/DCSupplemental.
Data Availability
DNA sequences and metadata from the Malaspina expedition are publicly available at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena; accession numbers PRJEB23913 (66) [18S rRNA genes] and PRJEB25224 (68) [16S rRNA genes]). DNA sequences from Tara Oceans are also stored at ENA with the accession numbers PRJEB6603 (76) for the SAGs, PRJEB6609 (101) for the metatranscriptomes, and PRJEB4352 (100) for the metagenomes (reference Datasets S1 and S4). Genome coassemblies, coding sequence predictions, and amino acid predictions have been deposited in FigShare (DOI: 10.6084/m9.figshare.13072322) (89). All other study data are included in the article and/or supporting information.
References
- 1.Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Li W. K. W., Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol. Oceanogr. 39, 169–175 (1994). [Google Scholar]
- 3.Worden A. Z., et al., Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Pernthaler J., Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3, 537–546 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Massana R., et al., Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 3, 588–596 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Massana R., del Campo J., Sieracki M. E., Audic S., Logares R., Exploring the uncultured microeukaryote majority in the oceans: Reevaluation of ribogroups within stramenopiles. ISME J. 8, 854–866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derelle R., López-García P., Timpano H., Moreira D., A phylogenomic framework to study the diversity and evolution of stramenopiles (=Heterokonts). Mol. Biol. Evol. 33, 2890–2898 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massana R., Terrado R., Forn I., Lovejoy C., Pedrós-Alió C., Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 8, 1515–1522 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Lin Y.-C., et al., Distribution patterns and phylogeny of marine stramenopiles in the north Pacific Ocean. Appl. Environ. Microbiol. 78, 3387–3399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piwosz K., Wiktor J. M., Niemi A., Tatarek A., Michel C., Mesoscale distribution and functional diversity of picoeukaryotes in the first-year sea ice of the Canadian Arctic. ISME J. 7, 1461–1471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piwosz K., Pernthaler J., Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the Southern Baltic Sea. Environ. Microbiol. 12, 364–377 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Garcia M., et al., Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J. 6, 703–707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krabberød A., Bjorbækmo M. F. M., Schalchian-Tabrizi K., Logares R., Exploring the oceanic microeukaryotic interactome with metaomics approaches. Aquat. Microb. Ecol. Aquat Microb Ecol 79, 1–12 (2017). [Google Scholar]
- 14.Giovannoni S. J., SAR11 bacteria: The most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 9, 231–255 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Martínez R., Rocap G., Logares R., Romac S., Massana R., Low evolutionary diversification in a widespread and abundant uncultured protist (MAST-4). Mol. Biol. Evol. 29, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Seeleuthner Y.et al.; Tara Oceans Coordinators , Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat. Commun. 9, 310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez-Martínez R., Rocap G., Salazar G., Massana R., Biogeography of the uncultured marine picoeukaryote MAST-4: Temperature-driven distribution patterns. ISME J. 7, 1531–1543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasol J. M., A framework for the assessment of top-down vs bottom-up control of heterotrophic nanoflagellate abundance. Mar. Ecol. Prog. Ser. 113, 291–300(1994). [Google Scholar]
- 19.Lee W. J., Patterson D. J., Abundance and biomass of heterotrophic flagellates, and factors controlling their abundance and distribution in sediments of Botany Bay. Microb. Ecol. 43, 467–481 (2002). [DOI] [PubMed] [Google Scholar]
- 20.de Meira B. R., et al., Abundance and size structure of planktonic protist communities in a Neotropical floodplain: Effects of top-down and bottom-up controls. Acta limnol. Bras. 29, e104 (2017). [Google Scholar]
- 21.Schulze H., Kolter T., Sandhoff K., Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta 1793, 674–683 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Manchenko G. P., Handbook of Detection of Enzymes on Electrophoretic Gels (ed. 2 CRC Press. Taylor & Francis Group, 2002). [Google Scholar]
- 23.Stepanauskas R., Single cell genomics: An individual look at microbes. Curr. Opin. Microbiol. 15, 613–620 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Mangot J. F., et al., Accessing the genomic information of unculturable oceanic picoeukaryotes by combining multiple single cells. Sci. Rep. 7, 41498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labarre A., et al., Comparative genomics reveals new functional insights in uncultured MAST species. ISME J. 15, 1767–1781 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieracki M. E., et al., Single cell genomics yields a wide diversity of small planktonic protists across major ocean ecosystems. Sci. Rep. 9, 6025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duarte C. M., Seafaring in the 21st century: The Malaspina 2010 circumnavigation expedition. Limnol. Oceanogr. Bull. 24, 11–14 (2015). [Google Scholar]
- 28.Moreira D., López-García P., The rise and fall of picobiliphytes: How assumed autotrophs turned out to be heterotrophs. BioEssays 36, 468–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuvelier M. L., et al., Widespread distribution of a unique marine protistan lineage. Environ. Microbiol. 10, 1621–1634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carradec Q.et al.; Tara Oceans Coordinators , A global ocean atlas of eukaryotic genes. Nat. Commun. 9, 373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berlemont R., Martiny A. C., Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 12, e1005300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madigan M. T., Martinko J. M., Stahl D. A., Clark D. P., Brock Biology of Microorganisms (ed. 13th, Benjamin Cummings, 2009). [Google Scholar]
- 33.Rabinovich M. L., Melnick M. S., Bolobova A. V., The structure and mechanism of action of cellulolytic enzymes. Biochemistry (Mosc.) 67, 850–871 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Naumoff D. G., GH97 is a new family of glycoside hydrolases, which is related to the α-galactosidase superfamily. BMC Genomics 6, 112 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumoff D. G., GH101 family of glycoside hydrolases: Subfamily structure and evolutionary connections with other families. J. Bioinformatics Comput. Biol. 8, 437–451 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Ohno S., Evolution by Gene Duplication (Popul, French Ed., 1971). [Google Scholar]
- 37.Walsh J. B., How often do duplicated genes evolve new functions? Genetics 139, 421–428 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner A., The fate of duplicated genes: Loss or new function? BioEssays 20, 785–788 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Yang Z., Bielawski J. P., Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith M. D., et al., Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 32, 1342–1353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murrell B., et al., Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangot J.-F., Forn I., Obiol A., Massana R., Constant abundances of ubiquitous uncultured protists in the open sea assessed by automated microscopy. Environ. Microbiol. 20, 3876–3889 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Finlay B. J., Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Vellend M., The Theory of Ecological Communities (MPB-57) (Princeton University Press, 2016). [Google Scholar]
- 45.Lindström E. S., Langenheder S., Local and regional factors influencing bacterial community assembly. Environ. Microbiol. Rep. 4, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Sunagawa S., et al., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Salazar G.et al.; Tara Oceans Coordinators , Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179, 1068–1083.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibarbalz F. M.et al.; Tara Oceans Coordinators , Global trends in marine plankton diversity across kingdoms of life. Cell 179, 1084–1097.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Logares R., et al., Disentangling the mechanisms shaping the surface ocean microbiota. Microbiome 8, 55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillou L., Alves-de-Souza C., Siano Dr R., González H., The ecological significance of small, eukaryotic parasites in marine ecosystems. Microbiol. Today 2010, 93–95 (2010). [Google Scholar]
- 51.Lima-Mendez G., et al., Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Hackl T., et al., Four high-quality draft genome assemblies of the marine heterotrophic nanoflagellate Cafeteria roenbergensis. Sci. Data 7, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armbrust E. V., et al., The genome of the diatom Thalassiosira Pseudonana: Ecology, evolution, and metabolism. Science 306, 79–86 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Vetukuri R. R., et al., Draft genome sequence for the tree pathogen Phytophthora plurivora. Genome Biol. Evol. 10, 2432–2442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Y., Lin S., Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PLoS One 4, e6978 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massana R., Guillou L., Terrado R., Forn I., Pedrós-Alió C., Growth of uncultured heterotrophic flagellates in unamended seawater incubations. Aquat. Microb. Ecol. 45, 171–180 (2006). [Google Scholar]
- 57.Villar E., et al., The Ocean gene atlas: Exploring the biogeography of plankton genes online. Nucleic Acids Res. 46, W289–W295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown J. M., et al., Single cell genomics reveals viruses consumed by marine protists. Front. Microbiol. 11, 524828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arndt H., et al., “Functional diversity of heterotrophic flagellates in aquatic ecosystems” in Flagellates Unity, Diversity Evolution, Leadbeater B. S. C., Green J. C., Eds. (CRC Press, 2000), pp. 1–29. [Google Scholar]
- 60.Sista Kameshwar A. K., Qin W., Comparative study of genome-wide plant biomass-degrading CAZymes in white rot, brown rot and soft rot fungi. Mycology 9, 93–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfenden R., Lu X., Young G., Spontaneous hydrolysis of glycosides. J. Am. Chem. Soc. 120, 6814–6815 (1998). [Google Scholar]
- 62.Turakainen H., Aho S., Korhola M., MEL gene polymorphism in the genus Saccharomyces. Appl. Environ. Microbiol. 59, 2622–2630 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martiny A. C., Treseder K., Pusch G., Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7, 830–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groussin M., Gouy M., Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol. Biol. Evol. 28, 2661–2674 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Stoeck T., et al., Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Malaspina-2010 expedition consortium, Data from “Picoplankton 18S rRNA genes from the tropical and sub-tropical global-ocean sampled during the Malaspina-2010 expedition .” European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB23913. Accessed 7 January 2019. [Google Scholar]
- 67.Parada A. E., Needham D. M., Fuhrman J. A., Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Malaspina-2010 expedition consortium, Data from “Picoplankton 16S rRNA genes from the tropical and sub-tropical global-ocean sampled during the Malaspina-2010 expedition .” European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB25224. Accessed 7 January 2019. [Google Scholar]
- 69.Callahan B. J., et al., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quast C., et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guillou L., et al., The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albanese D., Riccadonna S., Donati C., Franceschi P., A practical tool for maximal information coefficient analysis. Gigascience 7, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deutschmann I. M., et al., Disentangling environmental effects in microbial association networks. Research Square [Preprint]. 10.21203/rs.3.rs-57387/v1 (Accessed 20 August 2020). [DOI]
- 75.package R, Version 0.1. https://github.com/GuillemSalazar/EcolUtils. Accessed 10 August 2020.
- 76.Heywood J. L., Sieracki M. E., Bellows W., Poulton N. J., Stepanauskas R., Capturing diversity of marine heterotrophic protists: One cell at a time. ISME J. 5, 674–684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tara Oceans expedition consortium, Data from “Shotgun Sequencing of Single Cell Whole Genome Amplification from Tara Oceans samples corresponding to size fractions for protist .” European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB6603. Accessed 1 July 2017. [Google Scholar]
- 78.Bankevich A., et al., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waterhouse R. M., et al., BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. Accessed 28 January 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bushnell B., Rood J., Singer E., BBMerge–Accurate paired shotgun read merging via overlap. PLoS One 12, e0185056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Smit A., Hubley R., Green P., RepeatMasker Open-4.0. 2013-2015. http://www.repeatmasker.org. Accessed 28 January 2020.
- 83.Lowe T. M., Chan P. P., tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gurevich A., Saveliev V., Vyahhi N., Tesler G., QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ultsch A., Morchen F., ESOM-Maps: Tools for clustering, visualization, and classification with Emergent SOM. https://www.unimarburg.de/fb12/arbeitsgruppen/datenbionik/pdf/pubs/2005/ultsch05esom. Accessed 15 May 2016.
- 86.West P. T., Probst A. J., Grigoriev I. V., Thomas B. C., Banfield J. F., Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res. 28, 569–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stanke M., Morgenstern B., AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanke M., Diekhans M., Baertsch R., Haussler D., Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008). [DOI] [PubMed] [Google Scholar]
- 89.Latorre F., Data from “MAST-4 genomes: evolutionary diversification of tiny ocean predators.” Figshare. 10.6084/m9.figshare.13072322.v1. Deposited 9 October 2020. [DOI] [Google Scholar]
- 90.Yin Y., et al., dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445-51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eddy S. R., Bioinformatics Review Profile Hidden Markov Models (Bioinforma. Rev, 1998). [DOI] [PubMed] [Google Scholar]
- 92.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huerta-Cepas J., et al., eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.catfasta2phyml , Version 1.1.0. https://github.com/nylander/catfasta2phyml. Accessed 7 April 2020.
- 97.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki R., Shimodaira H., Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Tara Oceans expedition consortium, Data from “Shotgun sequencing of Tara Oceans DNA samples corresponding to size fractions for protists .” European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB4352. Accessed 1 November 2018. [Google Scholar]
- 101.Tara Oceans expedition consortium, Data from “Metatranscriptome sequencing from samples corresponding to size fractions for protists .” European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB6609. Accessed 1 November 2018. [Google Scholar]
- 102.Alberti A.et al.; Genoscope Technical Team; Tara Oceans Consortium Coordinators , Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data 4, 170093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pond S. L., Frost S. D. W., Muse S. V., HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences and metadata from the Malaspina expedition are publicly available at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena; accession numbers PRJEB23913 (66) [18S rRNA genes] and PRJEB25224 (68) [16S rRNA genes]). DNA sequences from Tara Oceans are also stored at ENA with the accession numbers PRJEB6603 (76) for the SAGs, PRJEB6609 (101) for the metatranscriptomes, and PRJEB4352 (100) for the metagenomes (reference Datasets S1 and S4). Genome coassemblies, coding sequence predictions, and amino acid predictions have been deposited in FigShare (DOI: 10.6084/m9.figshare.13072322) (89). All other study data are included in the article and/or supporting information.