Summary
Acyl-CoA/protein interactions are essential for life. Despite this importance, their global scope and selectivity remains undefined. Here we describe CATNIP (CoA/AcetylTraNsferase Interaction Profiling), a chemoproteomic platform for the high-throughput analysis of acyl-CoA/protein interactions in endogenous proteomes. First, we apply CATNIP to identify acetyl-CoA-binding proteins through unbiased clustering of competitive dose-response data. Next, we use this method to profile the selectivity of acyl-CoA/protein interactions, leading to the identification of specific acyl-CoA engagement signatures. Finally, we apply systems-level analyses to assess the features of protein networks that may interact with acyl-CoAs, and use a strategy for high-confidence proteomic annotation of acetyl-CoA binding proteins to identify a site of non-enzymatic acylation in the NAT10 acetyltransferase domain that is likely driven by acyl-CoA binding. Overall our studies illustrate how chemoproteomics and systems biology can be integrated to understand the roles of acyl-CoA metabolism in biology and disease.
Keywords: Chemical proteomics, activity-based protein profiling, acetyl-CoA, acetylation, epigenetics, metabolism, pharmacology, systems biology, NAT10, malonylation
In Brief
Levy et al. developed CATNIP, a competitive chemoproteomic approach for analyzing acyl-CoA/protein interactions. Systems analysis of CATNIP data revealed diverse acyl-CoA/protein interaction signatures across the human proteome. Comparing CATNIP and published lysine acylation datasets enabled enzymatic and non-enzymatic regulatory functions of acyl-CoAs to be annotated.
Introduction
Acyl-CoAs are essential for life. These metabolites serve as fundamental cellular building blocks in the biosynthesis of lipids, intermediates in energy production via the TCA cycle, and essential precursors for reversible protein acetylation. Each of these functions are physically dependent on acyl-CoA/protein interactions, which can regulate protein activity via a variety of mechanisms (Fig. 1). For example, the interaction of acyl-CoAs with lysine acetyltransferase (KAT) active sites allows them to serve as enzyme cofactors or, alternatively, competitive inhibitors (Dyda et al., 2000; Montgomery et al., 2015). Binding of acyl-CoAs to the allosteric site of pantothenate kinase (PanK) enzymes can exert positive or negative effects on CoA biosynthesis (Hong et al., 2007). Acyl-CoAs can also non-enzymatically modify proteins, a covalent interaction that often causes enzyme inhibition (Kulkarni et al., 2017; Wagner et al., 2017). These examples illustrate the ability of acyl-CoA signaling to influence biology and disease. However, the global scope and selectivity of these metabolite-governed regulatory networks remains unknown.
Figure 1.
Diverse consequences of acyl-CoA interactions on protein activity and signaling. Metabolic acyl-CoAs can interact with proteins as cofactors, inhibitors, allosteric modulators, or covalent modifiers.
A central challenge of studying acyl-CoA/protein interactions is their pharmacological nature (Kulkarni et al., 2019). These transient binding events are invisible to traditional next-generation sequencing and proteomic methods. To address this, our group recently reported a competitive chemical proteomic (“chemoproteomic”) approach to detect and analyze acyl-CoA/protein binding (Montgomery et al., 2016; Montgomery et al., 2014). This method applies a resin-immobilized CoA analogue (Lys-CoA) as an affinity matrix to capture CoA-utilizing enzymes directly from biological samples. Pre-incubating proteomes with acyl-CoA metabolites competes capture and allows their relative binding affinities to enzymes of interest to be assessed. In our initial application of this platform we studied the susceptibility of KATs to metabolic feedback inhibition by CoA, evaluating competition by quantitative immunoblot (Montgomery et al., 2016). The signal amplification afforded by immunodetection enables low abundance KATs to be readily quantified, but limits broad profiling or discovery applications. We reasoned such applications could be enabled by integrating CoA-based affinity reagents with i) multidimensional chromatographic separation, to efficiently sample rare KAT enzymes, ii) quantitative LC-MS/MS proteomics, for unbiased identification of CoA-interacting proteins, and iii) systems analysis of the acyl-CoA-binding proteins identified, for data-driven analysis of putative interaction networks. We term this approach CATNIP (CoA/AcetylTraNsferase Interaction Profiling). Here we describe the development and application of CATNIP to globally analyze acyl-CoA/protein interactions in endogenous human proteomes. First, we demonstrate the ability of CATNIP to identify acetyl-CoA-binding proteins through unbiased clustering of competitive dose-response data. Next, we apply this method to profile diverse protein-CoA metabolite interactions, enabling the identification of biological processes susceptible to altered acetyl-CoA levels. Finally, we utilize systems-level analyses to assess the features of protein networks that may interact with acyl-CoAs and develop a strategy for high-confidence annotation of direct acetyl-CoA binding proteins. Overall our studies illustrate the power of integrating chemoproteomics and systems biology analysis, and provide a resource for understanding the signaling roles of acyl-CoAs in biology and disease.
Results
Validation of CATNIP for the global study of acyl-CoA/protein interactions
In order to deeply sample acyl-CoA/protein interactions on a proteome-wide scale, we first integrated CoA-based protein capture methods with LC-MS/MS (Fig. 2a). In this workflow, whole cell extracts are first incubated with Lys-CoA Sepharose. This affinity matrix enables active site-dependent enrichment of many different classes of CoA-binding proteins, (Montgomery et al., 2016) making it ideal for broad profiling studies. Next, enriched proteins are subjected to tryptic digest and analyzed using MudPIT (multidimensional protein identification technology), a proteomics platform that combines strong cation exchange and C18 reverse phase chromatography to pre-fractionate tryptic peptides, followed by ionization and data-dependent MS/MS (Washburn et al., 2001b). The separation afforded by this approach significantly decreases sample complexity, allowing the identification of rare, low abundance peptides from complex proteomic mixtures. To facilitate the identification of acyl-CoA/protein interactions, competition experiments are performed in which proteomes are pre-incubated with a CoA metabolite prior to capture (Leung et al., 2003). Decreased enrichment in competition samples compared to controls (as assessed by quantitative spectral counting) signals a metabolite-protein interaction. These interacting proteins can then be further classified into pharmacological or biological networks using either conventional metrics (fold-change, gene ontology, etc) or systems-based analysis tools.
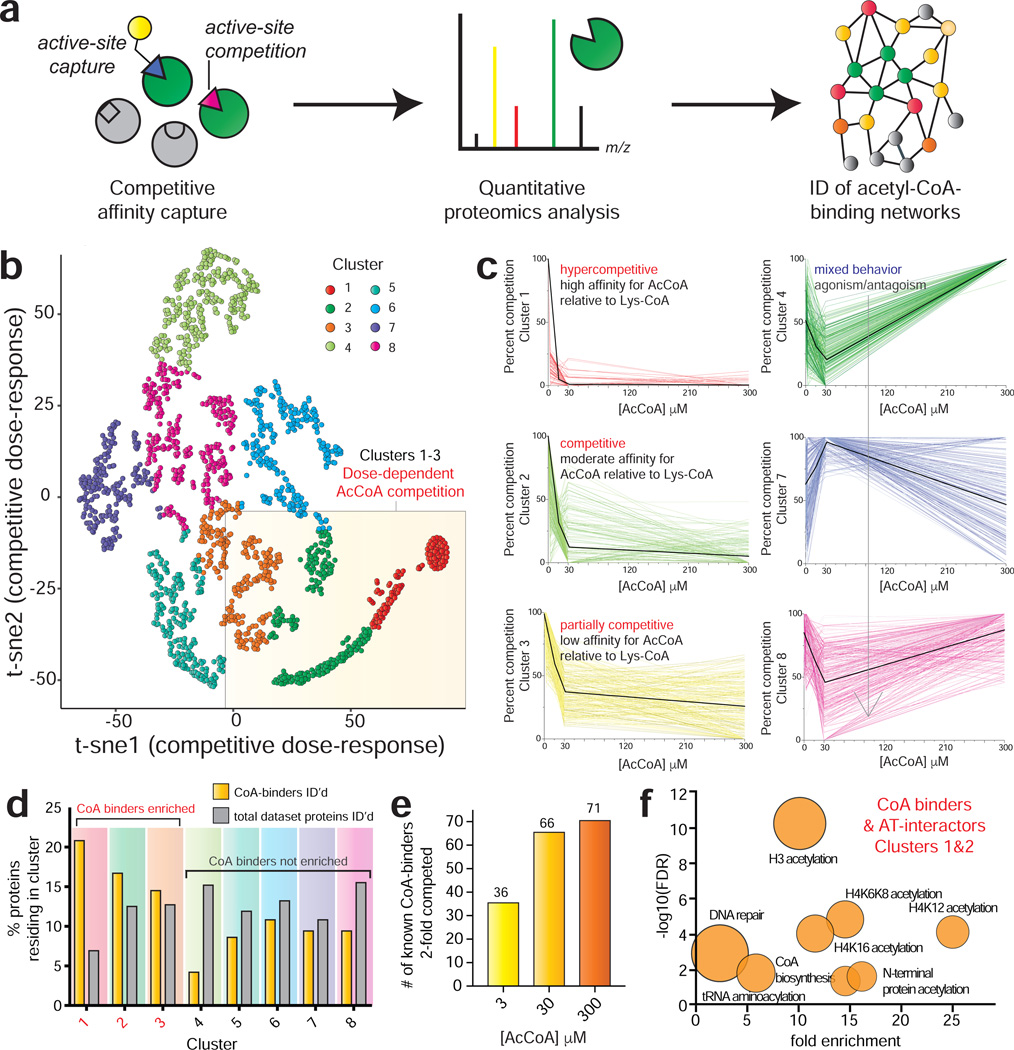
Figure 2.
Profiling acetyl-CoA/protein interactions using CATNIP. (a) Schematic for chemoproteomic analyses of acyl-CoA/protein interactions. (b) Normalized dNSAF values across 4 acetyl-CoA concentrations (0, 3, 30, 300μM) were t-SNE transformed and plotted in two dimensions for all proteins competed in CATNIP experiments (n=3 LC-MS/MS experiments). (c) Dose-response profiles of acetyl-CoA CATNIP clusters. Colored lines indicate the capture profiles of individual proteins at each concentration of acetyl-CoA competitor. Black lines indicate the mean capture profile for all proteins in a given cluster. (d) Clusters 1–3 are enriched in Uniprot-annotated CoA-binding proteins (“CoA binders”) as well as members of acetyltransferase complexes (”AT interactors”). (e) Analysis of annotated CoA binders exhibiting 2-fold competition at each concentration. Fold-change in d and e were calculated by QPROT. (f) Gene ontology analysis of CoA binders and AT interactors lying in CATNIP clusters 1 and 2. Fold enrichment of functional terms are plotted versus statistical significance (−log10[FDR]). Circle size reflects the number of proteins matching a given term. Functional enrichment was performed with the tool DAVID (https://david.ncifcrf.gov).
As an initial model, we explored the utility of CATNIP to globally profile acetyl-CoA/protein interactions in unfractionated HeLa cell proteomes. Proteomes were pre-incubated with acetyl-CoA or vehicle (buffer) control, followed by enrichment using Lys-CoA Sepharose. These experiments assessed competition at 3, 30, and 300 μM acetyl-CoA, which spans the physiological concentration range of acetyl-CoA in the cytosol and mitochondria. Protein capture in each condition was quantified using distributed normalized spectral abundance factor (dNSAF), a label-free metric that normalizes spectral counts relative to overall protein length (Fig. S1a-c, Table S1) (Zhang et al., 2015). Each condition was analyzed in triplicate, constituting 12 experiments, >144 hours of instrument time, and over 1.1 million non-redundant peptide spectra collected. We limited our analysis to high-confidence protein identifications (≥4 spectral counts in vehicle [0 μM] sample). The capture of Uniprot annotated CoA-binding proteins or members of AT complexes (termed ‘AT interactors’) did not correlate with overall protein abundance or gene expression (Nagaraj et al., 2011), consistent with the ability of chemoproteomic methods to sample functional activity metrics (e.g. unique pharmacology, active-site folding/conformation, integration into complexes, posttranslational modification) rather than raw quantity (Fig. S1d-i) (Moellering and Cravatt, 2012).
To analyze acetyl-CoA binding in a systematic manner throughout the proteome, we first grouped proteins into subsets based on their dose-dependent competition profiles. Chemoproteomic capture data from 0, 3, 30, and 300 μM acetyl-CoA competition was transformed, plotted in two dimensions, and subjected to t-SNE clustering (van der Maaten and Hinton, 2008). Eight protein clusters were identified, each of which exhibited a distinct dose-dependent competition signature (Fig. 2b-c, Fig. S2a-b, Table S1). The capture of proteins within clusters 1–3 were antagonized by acetyl-CoA in a dose-dependent fashion. Proteins in cluster 1 displayed hypersensitivity to acetyl-CoA competition, while proteins in clusters 2 and 3 exhibited moderate and partial competition, respectively. The remaining clusters exhibited more complicated capture profiles (clusters 4–8, Fig. 2c, Fig. S2b). To determine which of these competition signatures were most characteristic of acetyl-CoA binding, we first analyzed each cluster for the presence of known CoA-binding proteins and AT interactors. Cluster 1, composed of proteins whose capture is hypercompetitive to pre-incubation with acetyl-CoA, contains only 7% of the total proteins identified in this experiment. However, 25% of proteins in this cluster are annotated CoA-binding proteins and AT interactors, a disproportionate enrichment (Fig. 2d, Fig. S2c). Clusters 2 and 3 were also enriched in annotated CoA binders and AT interactors, while all other subsets were not (Fig. 2d). Consistent with this specific enrichment, assessments of background using a capped pulldown resin found 88% of background proteins identified lie in clusters 4–8 (Table S1). Examining our entire dataset, we found the total number of CoA-binding proteins and AT interactors competed 2-fold by acetyl-CoA) almost doubled going from 3 to 30 μM, but was only modestly increased by higher concentrations of competitor (Fig. 2e). Proteins in clusters 1 and 2 exhibit almost complete loss of capture at 30 μM acetyl-CoA (Fig. 2c). This suggests the occupancy of most acetyl CoA-binding sites accessible to our method are saturated at this intermediate concentration (~30 μM), in line with literature measurements of binding affinity and Michaelis constants (Scheer et al., 2011). Clusters 1 and 2 include proteins that bind to acetyl-CoA directly (CREBBP, NAA10), allosterically (PANK1), and indirectly via protein-protein interactions (NAA25, JADE1; Table S1). This indicates that proteins with disparate modes of acetyl-CoA interaction can display similar dose-dependent competition signatures. Gene ontology analysis of annotated CoA binders in clusters 1 and 2, whose enrichment was hypercompetitive to acetyl-CoA pre-incubation, revealed an enrichment in terms related to histone and N-terminal acetyltransferases as well as CoA biosynthetic enzymes (Fig. 2f, Fig. S2d-e). The strong enrichment of KATs is consistent with their propensity to interact with the bisubstrate Lys-CoA capture agent (Lau et al., 2000). A similar analysis of proteins in cluster 3, which exhibits partial competition by acetyl-CoA, identified a disproportionate number of mitochondrial CoA-binding enzymes (Fig. S2f). This decreased sensitivity to acyl-CoA competition may reflect evolutionary adaptation to subcellular metabolite concentrations, as acetyl-CoA is found at millimolar concentrations in mitochondria (Chen et al., 2016). Overall, these studies validate the ability of CATNIP to detect bona fide acetyl-CoA/protein interaction signatures and establish key parameters for studying the pharmacology of CoA metabolites.
Applying CATNIP to define the unique pharmacological signatures of acetyltransferase enzymes
In addition to acetyl-CoA (1), cells produce a physiochemically diverse range of CoA metabolites whose concentrations directly reflect the metabolic state of the cell. Many of these species make regulatory interactions with proteins, including the long chain fatty acyl (LCFA) palmitoyl-CoA, a classic feedback inhibitor of acetyl-CoA carboxylase, (Greenspan and Lowenstein, 1968) short chain fatty acyl (SCFA) butyryl-CoA, which can potently inhibit KATs or be used as a substrate, (Carrer et al., 2017; Montgomery et al., 2015) and negatively charged succinyl-CoA, which can covalently inhibit many mitochondrial enzymes (Kulkarni et al., 2017; Wagner et al., 2017). However, despite their physiological relevance, few studies have interrogated the comparative pharmacology of acyl-CoA/enzyme interactions. We hypothesized that the ability of CATNIP to report on the binding affinity of ligands relative to Lys-CoA could address this gap and enable the generation of pharmacological fingerprints of acyl-CoA-protein interactions across the proteome. To explore this hypothesis, we performed competitive chemoproteomic capture experiments in the presence of additional metabolites including: i) CoA (2), an acetyltransferase feedback inhibitor, ii) butyryl-CoA (3), a short chain fatty acyl-CoA, iii) crotonyl-CoA (4), a SCFA-CoA containing a latent acrylamide electrophile, iv) acetic-CoA (5), a stable analogue of malonyl-CoA, and v) palmitoyl-CoA (6), a LCFA-CoA we have previously shown can potently inhibit KATs in vitro (Fig. 3a, Table S2). For these experiments, CoA metabolites were equilibrated with proteomes (1 h) prior to Lys-CoA capture. A dosage of 30 μM was selected to enable a comparison of each ligand’s competition profile to that of acetyl-CoA, which showed substantial interaction with proteins in clusters 1–3 at this concentration. For palmitoyl-CoA a lower concentration (3 μM) was used, in order to ensure solubility and accurately reflect the limited free (non-protein/membrane bound) LCFA-CoA present in cells.
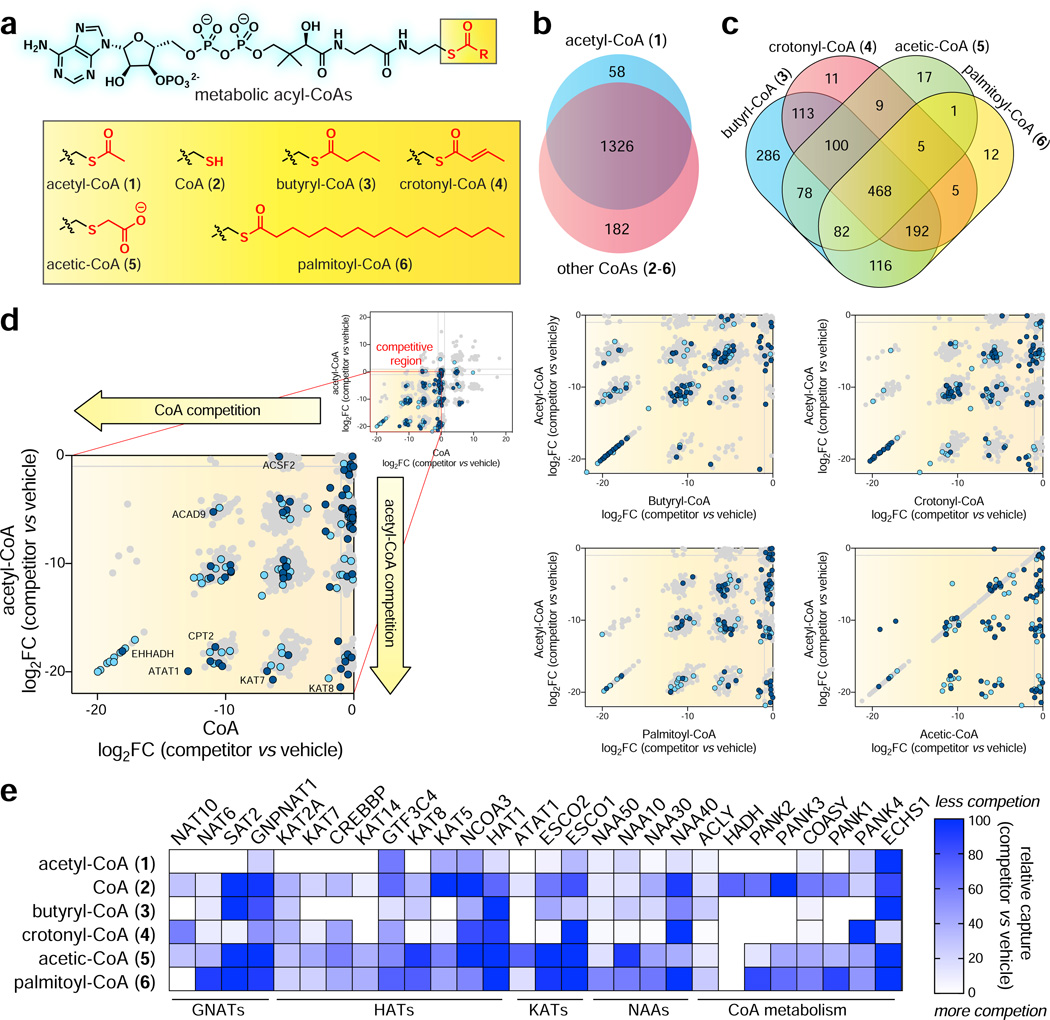
Figure 3.
Applying CATNIP to profile the comparative pharmacology of CoA metabolites. (a) CoA metabolites (1-6) analyzed in this study. (b) Venn diagram depicting overlap between proteins whose capture was competed more than four-fold by acetyl-CoA (1) or all other CoAs (2-6). (c) Venn diagram depicting overlap between proteins whose capture was two-fold competed by acyl-CoAs 3-6. (d) Comparison of acyl-CoA (x-axis) and acetyl-CoA (y-axis and acyl-CoA competition. Uniprot-annotated CoA binders and AT interactors are highlighted in dark blue and light blue, respectively. Log2FC values for b-d were calculated using QPROT. (e) Comparative CATNIP analysis highlights distinct signatures of metabolite interaction amongst families of CoA binders. Relative capture was calculated by comparing average dNSAF values. White = more competition by CoA metabolite, blue = less competition by CoA metabolite. All graphs are from n=3 control/competitor datasets.
As an initial rough measure of acyl-CoA selectivity, we performed a global analysis of proteins displaying robust interaction (QPROT log2FC ≤ −2) with competitors. Evaluating 1757 proteins quantified in Lys-CoA enrichments, we found 1566 (89%) were ≥4-fold competed by at least one CoA/acyl-CoA metabolite (1-6, Fig. 3b, Table S2). 1170 proteins (66%) displayed significant competition by one or more ligands using a more stringent statistical cutoff (QPROT FDRdown ≤0.1). In general, the majority of proteins found to interact with acetyl-CoA (1) also displayed competition by 2-6, suggestive of ligand-binding promiscuity amongst CoA-binding proteins. Examining physiochemically distinct ligands 3-6, a handful of selective interactions were observed for each acyl-CoA (Fig. 3c). Notably, targets of butyryl-CoA (3) and 4-6 showed substantial overlap. This may reflect butyryl-CoA’s metabolic stability in lysates, or ability to make high affinity interactions with many proteins at the concentration applied. To compare the magnitude of protein-ligand interactions, we plotted the competition (log2 fold change, competitor v. control) of each individual ligand relative to acetyl-CoA (Fig. 3d). Most proteins interacted more strongly with acetyl-CoA (1) than other ligands, with the exception of butyryl-CoA (3). This is consistent with the fact that acetyl-CoA (1) and butyryl-CoA (3) exhibited the greatest number of unique interaction partners in our comparative analysis. This promiscuous binding extended to KATs (Fig. S3). Notable exceptions were HADHB, interacted only with butyryl-CoA, and ECHS1, which interacted only with crotonyl-CoA (Table S2). HADHB encodes the thiolase subunit of the mitochondrial trifunctional protein, which is involved in the oxidation of fatty acids >8 carbons long (Middleton, 1994). Its ability to specifically interact with butyryl-CoA, but not acetyl-CoA, could represent a mechanism allowing cells to sense blockade of the terminal steps of SCFA-CoA catabolism, triggering feedback inhibition of fatty acid oxidation. ECHS1 encodes an acyl-CoA dehydrogenase, and was the only protein found to be specifically competed by crotonyl-CoA. This is consistent with the substrate specificity of this enzyme, which shows rapid turnover of crotonyl-CoA relative to longer chain enoyl-CoA thioesters (Yamada et al., 2015). To facilitate a more granular analysis, we grouped CoA-binding proteins by biological function or fold and compared their quantitative metabolite-binding signatures upon interaction with 1-6. Histone, lysine, and GNAT acetyltransferases displayed a diversity of ligand binding signatures (Fig. 3e). For example, the capture of enzymes such as CREBBP and NAT10 was strongly competed by multiple metabolites, while others (KAT8 and HAT1) displayed selectivity for the enzyme cofactor acetyl-CoA (Fig. 3e). Selectivity did not correlate with enzyme function/fold, capture abundance, or acetyl-CoA interaction cluster (Fig. 3e, Table S3), suggesting this metabolite interaction fingerprint represents a unique and intrinsic feature of individual enzymes. The promiscuous ligand binding of CREBBP is notable, as this KAT and its homologue EP300 have been found to utilize several acyl-CoAs as alternative cofactors (Chen et al., 2007; Sabari et al., 2018). The PANK family of proteins catalyze the phosphorylation of pantothentate (vitamin B5) to phosphopantothenate, which constitutes a key step in CoA biosynthesis. Previous biochemical studies have found PANK1 to be allosterically inhibited by acetyl-CoA but not CoA, while PANK2 is strongly inhibited by both ligands (Rock et al., 2002; Zhang et al., 2006). We found acetyl-CoA interacted strongly with each enzyme but did not observe substantial disparity between CoA binding to the two enzyme isoforms. This may reflect differential binding of metabolites to these enzymes in the complex proteomic milieu compared to biochemical assays or, alternatively, a limitation of our single concentration measurements. Overall, these studies validate the ability of chemoproteomics to study acyl-CoA/protein interactions and provide an initial snapshot of their proteome-wide selectivity.
Evaluating the dynamic activity of acetyltransferases in response to metabolic perturbation
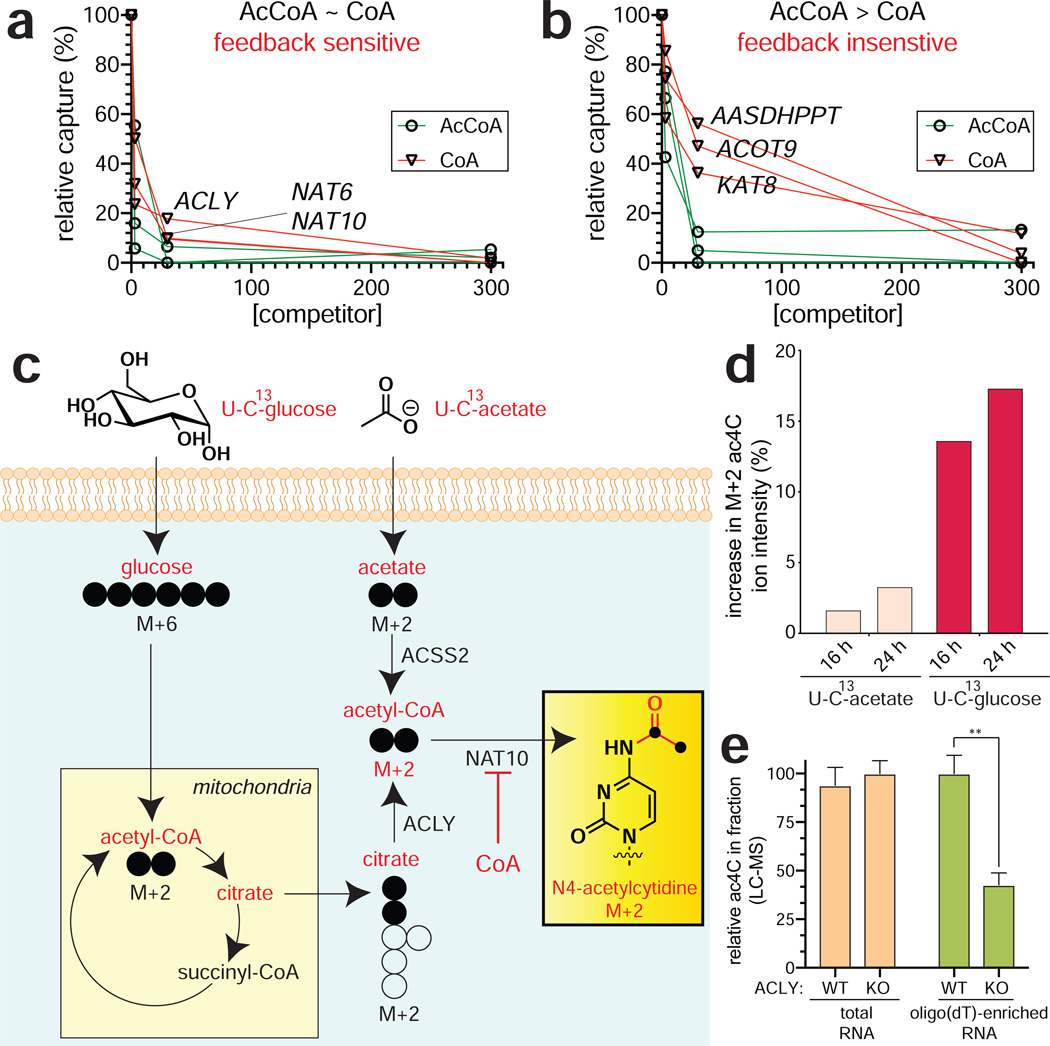
Coenzyme A (2) is one of the most abundant metabolites in cells. In addition to functioning as an obligate precursor for acyl-CoAs, it also serves as a potent feedback inhibitor AT enzymes. Previously, we used quantitative immunoblotting of Lys-CoA capture experiments to probe the sensitivity of eight ATs to product inhibition by testing their relative binding to acetyl-CoA (cofactor) and CoA (inhibitor) (Montgomery et al., 2016). This inspired us to apply CATNIP to extend this comparison proteome-wide. Capture experiments were performed in the presence of escalating doses of CoA (3, 30, 300 μM), transformed, and clustered using an identical pipeline as in our acetyl-CoA binding experiments above (Table S4). Two clusters (3 and 8) exhibited readily interpretable dose-dependent competition profiles, with several additional clusters (1, 2, and 5) displaying hypersensitivity at low concentrations (3 μM) of CoA (Fig. S4a-b). Dose-dependent cluster 3 contained KAT2A, CREBBP, and PANK2, all of whom have been shown to be biologically or biochemically susceptible to metabolic feedback inhibition by CoA (Hong et al., 2007; Marino et al., 2014; Tanner et al., 2000). Further examination of this cluster revealed three proteins that were most sensitive to CoA, exhibiting >50% loss of capture in the presence of 3 μM ligand and >80% loss of capture in the presence of 30 μM ligand: ACLY, NAT6, and NAT10 (Fig. 4a). The unusually strong CoA interaction profile of these three proteins was distinct from that of other proteins in the cluster and within the KAT superfamily, most of which are competed much more efficiently by acetyl-CoA (Fig. 4b, Fig. S4c). The binding of ACLY to both CoA and acetyl-CoA is consistent with the reversible activity of the enzyme, which has been previously observed in biochemical assays (Inoue et al., 1968). NAT6 (NAA80) is a recently de-orphanized enzyme which has been determined to acetylate the N-terminus of actin, whose metabolic sensitivity has not been explored (Drazic et al., 2018). NAT10 is an RNA acetyltransferase that has been found to catalyze acetylation of cytidine in ribosomal, transfer, and messenger RNA, forming the minor nucleobase N4-acetylcytidine (ac4C) (Arango et al., 2018; Ito et al., 2014; Sharma et al., 2015). The identification of NAT10-CoA interactions by CATNIP is consistent with our previous studies, which found NAT10 binds acetyl-CoA and CoA with similar affinities and may be susceptible to metabolic feedback inhibition (Montgomery et al., 2016). Of note, no related N4-acylations of cytidine (e.g. butyrylation) have been identified in RNA, suggesting NAT10 may also be subject to inihibition by SCFA- and LCFA-CoAs.
Figure 4.
Applying CATNIP to profile AT feedback inhibition. (a) Exemplary dose-response profiles of proteins that interact strongly with CoA. (b) Exemplary dose-response profiles of proteins more weakly with CoA. (c) Scheme for isotopic tracing of metabolic source of the acetate group in ac4C. Heavy (U-13C) glucose or acetate were applied in separate metabolic labeling experiments. Incorporation into ac4C was assessed by mass isotopomer analysis of digested total RNA. (d) The major source of ac4C’s N4-acetyl group is glucose. (e) Disruption of ACLY reduces levels of ac4C in poly(A)-enriched, but not total RNA. Values represent ≥ 3 replicates, analyzed by two-tailed student’s t-test (ns = not significant, *P<0.05, **P<0.01, ***P<0.001).
To gain further insight into the metabolic regulation of NAT10, we determined the source of the acetate group post-transcriptionally introduced into ac4C in proliferating cancer cell lines (Fig. 4c). Treatment of cells with isotopically-labeled acetyl-CoA precursors, followed by RNA digest and LC-MS/MS analysis revealed the majority of NAT10-dependent cytidine acetylation is derived from glucose (Fig. 4d). Since the production of glucose-derived acetyl-CoA in human cells is highly dependent on ACLY activity (Wellen et al., 2009), we next examined how stable knockout of ACLY impacted ac4C levels in RNA. Analysis of wild-type and ACLY knockout human glioblastoma cells (Zhao et al., 2016) revealed similar levels of ac4C in total RNA (Fig. 4e). However, LC-MS analysis of poly(A)-enriched RNA fractions from these cell lines indicated an ACLY-dependent decrease in ac4C. Of note, the relative abundance of ac4C is ~8-fold lower in oligo(dT)-enriched RNA than total RNA, indicating additional work will be needed to understand the physiological-relevance of this finding. ACLY-dependent deposition is also observed for another acetyl-CoA derived RNA nucleobase, 5-methoxycarbonylmethyl-2-thiouridine (mcm5S2U), whose production is catalyzed by the AT enzyme Elp3 (Fig. S4d-e) (Lin et al., 2019). The observation that ac4C and mcm5s2U are sensitive to the metabolic state of the cell is consistent with the findings of Balasubramanian and coworkers, who reported that starvation conditions reduced the levels of these two modifications transfer RNA (van Delft et al., 2017). The observation that ACLY perturbation influences the acetylation of oligo(dT)-enriched RNA, but not total RNA, suggests inhibitory CoA/acyl-CoAs may interact distinctly with different functional forms of NAT10. These studies illustrate the ability of CATNIP to link acetylation events to the metabolic state of the cell.
Unbiased CATNIP analysis reveals annotation and mechanistic features of acyl-CoA-binding
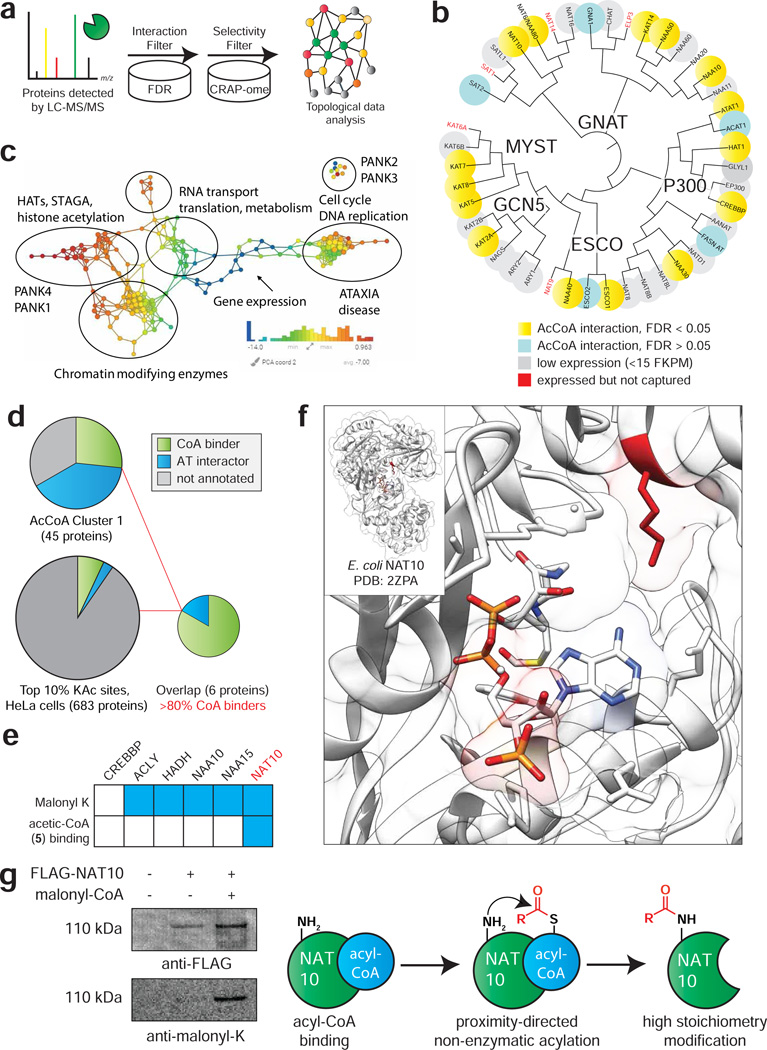
The annotation of the cellular acyl-CoA binding proteome has never been directly assessed using experimental methods. Therefore, we next set out to develop a workflow for analysis of CATNIP data that could enable the de novo identification of known acyl-CoA dependent enzymes and identify uncharacterized proteins that share these properties. To differentiate acyl-CoA interacting proteins from background, our initial criteria were: 1) significant competition (QPROT log2FC ≤ −2 and FDR ≤ 0.05) of enriched proteins by at least one CoA ligand, and 2) absence of enrichment in the ‘CRAPome’ common contaminant database (Fig. 5a) (Mellacheruvu et al., 2013). Of 1757 proteins detected in Lys-CoA Sepharose capture experiments, 650 (37.0%) passed these cut-offs (Table S2), including the majority of annotated ATs that were enriched by Lys-CoA (Fig. 5b). Acetyltransferases not identified were mostly found to be poorly expressed in HeLa cells by RNA Seq (Fig. 5b) (Nagaraj et al., 2011) and did not display obvious structural similarities in GNAT consensus elements (Fig. S5a) (Dyda et al., 2000). To examine whether unique patterns of acyl-CoA binding in this filtered dataset are associated with distinct biological processes, we further analyzed proteins using Topological Data Analysis (TDA) (Lum et al., 2013). TDA functions as a geometric approach that can be used to identify shared properties of complex multidimensional datasets that may not be apparent by other methods, and has previously been used to detect biologically-relevant modules in protein complexes from immunoprecipitation LC-MS/MS data (Sardiu et al., 2015). Therefore, we applied TDA to analyze the multidimensional CoA metabolite competition profiles for each protein in our filtered subset, and then annotated the TDA clusters with enriched pathways identified by gene ontology analysis using DAVID (https://david.ncifcrf.gov) and ConsensusPathDB (http://cpdb.molgen.mpg.de/). To further increase stringency we required competition with ≥3 CoA metabolites (248 proteins; Table S2). Applying this analysis revealed that histone acetyltransferases, which bind to CoAs directly, form a distinct cluster relative to PANK2 and PANK3, which are allosterically regulated by CoA metabolites. This analysis also identified many proteins involved in RNA metabolism and cell cycle whose association with CoA metabolites has not been previously characterized (Fig. 5c). This suggests that multiple proteins involved in these processes may directly or indirectly bind to acyl-CoAs, and potentially be subject to differential regulation by levels of CoA metabolism. Moreover, these studies demonstrate the utility of TDA for clustering and visual representation of ligand-protein interaction networks.
Figure 5.
Applying CATNIP to assess the annotation of the CoA-binding proteome. (a) Schematic for differentiating CATNIP-enriched proteins from background based on significant competition and absence from common contaminant databases. (b) Proteins from diverse AT families display statistically significant CoA/acyl-CoA competition. (c) Topological network analysis of proteins exhibiting significant interaction with ≥ 3 CoA metabolites. Protein nodes are colored based on the metric PCA2. Color bar: red = high values; blue = low values. Node size is proportional to the number of proteins in the node. (d) Combining CATNIP competition (QPROT Log2FC ≤ −2 and FDRdown<0.05) and acetylation stoichiometry filters greatly enriches CoA-binders and AT-interactors relative to either measure alone. (e) Comparing annotated sites of lysine malonylation (top) and competition by malonyl-CoA mimic 5 (bottom) for CoA-binders detected by de novo CATNIP analysis. (f) A conserved site of lysine malonylation lies in close proximity to the acetyl-CoA binding site of bacterial NAT10. (g) Overexpressed FLAG-NAT10 is malonylated in the presence of malonyl-CoA, consistent with a non-enzymatic acylation mechanism.
Next, we sought to examine the annotation of the human CoA-binding proteome. Specifically, we wished to incorporate additional criteria allowing us to differentiate proteins that directly bind to CoA metabolites, such as ATs, from proteins that are indirectly captured via protein-protein interactions, such as non-catalytic members of AT complexes. Such an approach would potentially provide a pipeline for novel AT discovery, as well as insights into how well the CoA-binding proteome is currently characterized. To accomplish this, we first classified proteins in our statistically significant filtered subset based on their dose-dependent acetyl-CoA competition profiles determined above, whose clustering we found could enrich known ATs and CoA-binding proteins (Fig. 2b, d). Approximately 7% (42/650) of proteins showing statistically significant competition by any single CoA metabolite also resided in cluster 1, whose capture is hypersensitive to competition by acetyl-CoA (Fig. 5d, Table S5). This included 12 direct CoA-binders, 14 AT-interactors, and 16 proteins whose interaction with CoA metabolites had not been previously characterized. Amongst this protein subset, terms related to histone acetyltransferases and CoA biosynthesis were clearly differentiated as the most highly enriched biological process (Fig. S5d, Table S5). To differentiate between direct and indirect CoA interactions, we first explored the use of high salt washes to disrupt proteins captured through protein-protein interactions. Salt sensitivity data was obtained for 20 of the 42 proteins in our stringent subset, nine of whose capture was insensitive to ionic strength. Amongst these were 6 proteins whose interaction with CoA had not been characterized. Notably, most of these proteins were found to contain either adenine cofactor (SAM, NADP+) or RNA binding sites, consistent with potential for capture by Lys-CoA Sepharose (Table S5). As an orthogonal measure, we further investigated these 42 proteins for sites of high stoichiometry acetylation. We hypothesized this criteria may provide identify functional binding events, since acyl-CoA interaction can underlie both enzymatic and non-enzymatic autoacylation. Using a recently published dataset, (Hansen et al., 2019) we identified 6 out of 42 proteins that contain a modified lysine lying near the top 10% of all acetylation stoichiometries measured in HeLa cells (>0.17% stoichiometry, Fig. 5d). Five of these proteins were Uniprot annotated CoA binders (ACLY, CREBBP, HADH, NAT10, NAA10), while one was a member of an AT complex (NAA15). This analysis suggests that a multi-pronged approach assessing i) statistically significant competition, ii) dose-response clustering, iii) acetylation stoichiometry, and iv) stringent washing can meaningfully annotate the CoA-binding proteome, with the major caveat that additional stringency will also lead to filtering of many ‘true’ positives.
Finally, we asked whether CATNIP could help identify acyl-CoA binding events that drive non-enzymatic acylation. Our previous studies have found lysine malonylation and succinylation serve as markers of non-enzymatic acylation, due to the high reactivity of their acyl-CoA precursors (Kulkarni et al., 2017). This led us to hypothesize that if an enzyme 1) binds a malonyl-CoA analogue and 2) possesses overlapping malonylation and high stoichiometry acetylation sites, then acyl-CoA binding may be responsible for driving non-enzymatic acylation. Examining the six proteins above, only one (NAT10) exhibited statistically significant competition by the malonyl-CoA surrogate acetic-CoA. In line with this, while 5/6 of these proteins were found to harbor sites of lysine malonylation, (Colak et al., 2015) only in the case of NAT10 were the high stoichiometry acetylation and malonylation sites found on the same residue (K426). This lysine lies within NAT10’s GNAT domain and is highly conserved from eukaryotes to bacteria (Fig. S5f). Analyzing the position of K426 using the structure of a NAT10 orthologue shows it lies proximal to the acetyl-CoA binding site (Chimnaronk et al., 2009), potentially priming it for non-enzymatic acetylation (Fig. 5f). Consistent with this, we find FLAG-NAT10 overexpressed in HEK-293 cells is readily malonylated upon incubation with malonyl-CoA (Fig. 5f; note that we used malonyl- and not acetyl-CoA to decouple enzymatic and non-enzymatic mechanisms, as no known ATs use malonyl-CoA as a cofactor). Such a non-enzymatic mechanism would reconcile the paradoxical finding that NAT10 undergoes functional lysine acetylation in its active site (Cai et al., 2017), but its only biochemically validated substrates are RNA cytidine residues. Further work will be needed to evaluate what impact K426 malonylation has on NAT10 activity, as well as the global extent to which malonylation is dependent on specific malonyl-CoA/protein interactions versus solvent accessibility of lysine residues. These studies demonstrate how CATNIP may be interfaced with PTM analyses to identify functionally-relevant acyl-CoA/protein interactions.
Discussion
Chemoproteomics has recently emerged as a powerful method for the interrogation of metabolite signaling. Here we describe the development and application of CATNIP, a systems chemoproteomic approach for the high-throughput analysis of acyl-CoA/protein interactions. We first validate the ability of CATNIP to identify protein subsets enriched in CoA-binding, and then apply this method to probe the selectivity of acyl-CoA/protein interactions, visualize acyl-CoA interactive biological networks, and characterize the interplay between direct acyl-CoA binding and covalent lysine acylation. These studies highlight the ability of CATNIP to identify biological processes conditionally regulated by acetyl-CoA and provide a novel hypothesis generation tool for the functional interrogation of metabolite-protein interactions in biology and disease.
To explore the utility of CATNIP for discovery applications, we developed an unbiased workflow to applying chemoproteomic data for the de novo annotation of acetyl-CoA binding proteins. Critical to this endeavor was the integration of CATNIP and acetylation stoichiometry datasets, (Hansen et al., 2019) which allowed the identification of a protein subset highly enriched in CoA-binders and AT interactors that was obscure to either method alone (Fig. S4d). An interesting finding was the absence of any ‘unexpected interactors,’ i.e. unannotated proteins with CATNIP profiles indicative of CoA-binding, within this highly curated subset. This suggests the current CoA-binding proteome is well-annotated, with the caveat that this conclusion is entirely dependent on the unique workflow applied here, and therefore does not preclude the discovery of novel acyl-CoA-binding proteins by new experimental methods (e.g. structurally distinct capture probes) or computational analyses. With regards to the latter, it is important to note that many authentic acyl-CoA-binding proteins sampled by CATNIP do not exhibit high stoichiometry acetylation sites (e.g. ATAT1) or fall outside of dose-dependent cluster 1 (e.g. KAT2A). Our studies demonstrate how acetylation stoichiometry may serve as a useful guide to high-confidence annotation of acyl-CoA binding, while simultaneously raising the possibility of mining additional CoA binders and AT interactors from CATNIP data.
Acyl-CoA/protein interactions can play many potential functional roles (Fig. 1). Inspired by recent chemoproteomic studies showing that inositol polyphosphate binding can trigger non-enzymatic protein pyrophosphorylation, (Wu et al., 2016) we wondered whether acyl-CoA binding may similarly be a major driver of non-enzymatic lysine acylation. Examining the lysine malonylation, a putative non-enzymatic PTM derived from the electrophilic metabolite malonyl-CoA, (Kulkarni et al., 2017) we identified NAT10 as a unique case in which these PTMs could be correlated with proximity to an acyl-CoA binding site. However, this approach is far from predictive and, even in our curated dataset of high confidence acyl-CoA-binding proteins, found many sites of malonylation mapping far from the annotated active site (Fig. 5e, Fig. S5f) (Colak et al., 2015). Although additional studies are needed, our data suggests for many non-enzymatic acylations factors independent of acyl-CoA binding affinity such as lysine nucleophilicity, surface accessibility, and exposure to high local concentrations of electrophilic CoAs may be important determinants for covalent modification.
Finally, it is important to note some limitations of our current method, and steps that may be taken to optimize it for future applications. To facilitate the development of CATNIP, our initial study employed ion trap mass spectrometers for protein identification. Transitioning CATNIP to higher resolution instruments will be important for improving the throughput and quantitative applications of our method. An important characteristic of CATNIP is that it reports on relative, rather than absolute, binding affinities due to differences in the inherent binding affinity of individual proteins to the Lys-CoA capture matrix. This means CATNIP is best suited to gauging the comparative pharmacology of individual acyl-CoA binding proteins (i.e. for a series of ligands, which ones interact strongly with protein of interest), rather than rank order comparisons of absolute ligand-protein binding affinity across the proteome. Such biases are an intrinsic feature of chemoproteomic methods and extend even to label-free approaches such as LiP-MS and CETSA, (Piazza et al., 2018; Sridharan et al., 2019). Future studies of acyl-CoA/protein interactions may benefit from the integration of multiple approaches. Clustering analysis indicated that many CoA binders and AT interactors display similar competition profiles, implying CATNIP as currently constituted is not able to discriminate between direct and indirect interactors. In addition to using acetylation stoichiometry as an orthogonal measure for the de novo assignment of direct acyl-CoA binding, it may be possible to conclusively distinguish indirect binding based on more intricate studies of ionic competition (i.e. high salt) or by complementing matrix-based pulldown with covalent capture using clickable photoaffinity probes (Montgomery et al., 2014). Alternatively this may be solved by optimized computational analysis, in which the proteins identified from multiple competitive ligands are compared using topological scoring (TopS) (Sardiu et al., 2019) to determine enrichment of proteins and direct interactions from a range of concentrations or ligand types. Although we focused here on studying the interactions of proteins with endogenous acyl-CoA metabolites, recently multiple classes of drug-like KAT inhibitors have been reported, (Baell et al., 2018; Lasko et al., 2017) and we anticipate our method will be immediately useful for understanding the pharmacological specificity and potency of these small molecule chemical probes. Such studies are underway and will be reported in due course.
STAR METHODS
LEAD CONTACT
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jordan L. Meier (jordan.meier@nih.gov)
MATERIALS AVAILABILITY STATEMENT
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HeLa S3 (female) and HEK-293T (female) cells were obtained from ATCC. LN229 (female; ACLY WT and ACLY KO) cells were a kind gift of K. Wellen (University of Pennsylvania). HeLa S3 and HEK-293T were cultured at 37 °C under 5% CO 2 atmosphere in a growth medium of DMEM supplemented with 10% FBS and 2 mM glutamine, while LN229 cells were cultured at 37 °C under 5% CO 2 atmosphere in a growth medium of RPMI supplemented with 10% FBS and 2 mM glutamine. All cell lines were routinely tested for mycoplasma contamination.
METHOD DETAILS
General synthetic methods
Amine-functionalized Lys-CoA-Ahx was synthesized as described previously (Montgomery et al., 2016). Acetic-CoA was synthesized from 2-bromoacetic acid and CoA in a single-step as described previously (Montgomery et al., 2014). Prior to utilization all acyl-CoAs were analyzed for purity by LC-MS and re-purified via HPLC if necessary. CoAs were quantified using the molar extinction coefficient (ε) for Coenzyme A of 15, 000 M−1cm−1 at λmax of 259 nm. Analytical analyses of Lys-CoA and all acyl-CoAs were performed using a Shimadzu 2020 LC-MS system.
Preparation of Lys-CoA Sepharose and capped resins
Lys-CoA Sepharose (1) was prepared using NHS-Activated Sepharose 4 Fast Flow resin essentially according to the manufacturer’s protocol (GE Healthcare Life Sciences, Instructions 71–5000-14 AD) (Montgomery et al., 2016). Briefly, amine-functionalized Lys-CoA-Ahx was prepared as a 3.4 mM solution in PBS. Resin was washed with cold 1 mM HCl prior to coupling, before addition of the ligand solution at a ratio of 2:1 resin:ligand volume. The pH was adjusted to ~7–8 by addition of 20x PBS, and the mixture was then rotated at 4°C overnight. The resin was pellet ed at 1400 rcf for 3 minutes, and the supernatant was discarded prior to addition of 3 resin volumes of 0.1 M Tris-HCl [pH 8.5], and the mixture was rotated for 3 hr at room temperature. Resin was washed 3x each with alternating solutions of 0.1 M Tris-HCl [pH 8.5] and 0.1M Sodium Acetate, 0.5 M NaCl [pH 4.5] (6 washes total). Capped resins were prepared analogously but substituting 1,6-hexanediamine for Lys-CoA-Ahx. Resins were stored as a 33% solutions in aqueous 20% EtOH at 4°C.
Preparation of HeLa cell lysates
HeLa cells used to prepare proteomic extracts were grown by Cell Culture Company (formerly National Cell Culture Center, Minneapolis, MN). Proteomes were prepared by re-suspending cell pellets in ice-cold PBS containing protease inhibitor cocktail (EDTA-free, Cell Signaling Technology # 5871S). Samples were then lysed by sonication using a 100 W QSonica XL2000 sonicator (3 × 1 s pulse, amplitude 1, 60 s resting on ice between pulses). Lysates were pelleted by centrifugation (20,817 r.c.f. x 30 minutes, 4 °C) and quantified using the Qubit 4.0 Fluorometer and Qubit Protein Assay Kit. Quantified proteomes were diluted and stored at −80 °C in between uses.
Procedure for CATNIP affinity capture, competition and LC-MS/MS studies
Affinity capture using Lys-CoA Sepharose was carried out essentially as previously reported (Montgomery et al., 2016; Montgomery et al., 2015). Briefly, 33 μl of capture resin was washed once with 1 ml of PBS, prior to addition of 500 μl of clarified lysates (1.5 mg/ml, pretreated with vehicle or competitor ligand for 30 min on ice). This mixture was rotated for 1 hr at room temperature, pelleted at 1400 rcf, and supernatant discarded. Sepharose capture resins were subjected to a series of mild washes using ice cold wash buffer (50 mM Tris-HCl [pH 7.5], 5% glycerol [omitted in LC-MS/MS experiments], 1.5 mM MgCl2, 150 mM NaCl, 3 × 500 μl). Salt sensitivity experiments were performed in a paired fashion as a separate biological replicate and used washes containing either 150 or 500 mM NaCl. Following the final wash, enriched resin was collected on top of centrifugal filters (VWR, 82031–256). Capped resin capture experiments were performed completely analogously. For LC-MS/MS analysis of captured proteins, enriched resin was transferred from centrifugal filters to fresh 1.7-ml tubes using 400 μl of tryptic digest buffer (50 mM Tris-HCl [pH 8.0], 1 M urea). Digests were initiated by addition of 0.4 μl of 1 M CaCl2 and 4 μl of trypsin (0.25 mg/ml) and allowed to proceed overnight at 37°C with shaking. After extraction, tryptic peptide samples were acidified to a final concentration of 5% formic acid, lyophilized, and frozen at −80°C until LC-MS/MS analysis.
MudPIT LC-MS/MS analysis of and database searching of Lys-CoA enriched proteomes
Lyophilized peptide samples from Lys-CoA Sepharose enriched HeLa proteomes were analyzed independently in triplicate by Multidimensional Protein Identification Technology (MudPIT), as described previously (Florens and Washburn, 2006; Washburn et al., 2001a). Briefly, dried peptides were resuspended in 100μL of Buffer A (5% acetonitrile (ACN), 0.1% formic acid (FA)) prior to pressure-loading onto 100 μm fused silica microcapillary columns packed first with 9 cm of reverse phase (RP) material (Aqua; Phenomenex), followed by 3 cm of 5-μm Strong Cation Exchange material (Luna; Phenomenex), followed by 1 cm of 5-μm C18 RP. The loaded microcapillary columns were placed in-line with a 1260 Quartenary HPLC (Agilent). The application of a 2.5 kV distal voltage electrosprayed the eluting peptides directly into LTQ linear ion trap mass spectrometers (Thermo Scientific) equipped with a custom-made nano-LC electrospray ionization source. Full MS spectra were recorded on the eluting peptides over a 400 to 1600 m/z range followed by fragmentation in the ion trap (at 35% collision energy) on the first to fifth most intense ions selected from the full MS spectrum. Dynamic exclusion was enabled for 120 sec (Zhang et al., 2009). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the XCalibur data system (Thermo Scientific). RAW files were extracted into .ms2 file format (McDonald et al., 2004) using RawDistiller v. 1.0, in-house developed software (Zhang et al., 2011). RawDistiller D(g, 6) settings were used to abstract MS1 scan profiles by Gaussian fitting and to implement dynamic offline lock mass using six background polydimethylcyclosiloxane ions as internal calibrants (Zhang et al., 2011). MS/MS spectra were first searched using ProLuCID(Xu et al., 2015) with a mass tolerance of 500 ppm for peptide and fragment ions. Trypsin specificity was imposed on both ends of candidate peptides during the search against a protein database combining 36,628 human proteins (NCBI 2016–06-10 release), as well as 193 usual contaminants such as human keratins, IgGs and proteolytic enzymes. To estimate false discovery rates (FDR), each protein sequence was randomized (keeping the same amino acid composition and length) and the resulting “shuffled” sequences were added to the database, for a total search space of 73,642 amino acid sequences. A mass of 15.9949 Da was differentially added to methionine residues.
DTASelect v.1.9 (Tabb et al., 2002) was used to select and sort peptide/spectrum matches (PSMs) passing the following criteria set: PSMs were only retained if they had a DeltCn of at least 0.08; minimum XCorr values of 1.9 for singly-, 2.7 for doubly-, and 2.9 for triply-charged spectra; peptides had to be at least 7 amino acids long. Results from each sample were merged and compared using CONTRAST (Tabb et al., 2002). Combining all replicate injections, proteins had to be detected by at least 2 peptides and/or 2 spectral counts. Proteins that were subsets of others were removed using the parsimony option in DTASelect on the proteins detected after merging all runs. Proteins that were identified by the same set of peptides (including at least one peptide unique to such protein group to distinguish between isoforms) were grouped together, and one accession number was arbitrarily considered as representative of each protein group.
NSAF7 (Zhang et al., 2010) was used to create the final reports on all detected peptides and non-redundant proteins identified across the different runs. Spectral and protein level FDRs were, on average, 0.31±0.10% and 1.0±0.35%, respectively. QPROT (Choi, et al, 2015) was used to calculate a log fold change and false discovery rate for the dosed samples compared to the vehicle control.
Partitioning clustering
To group proteins based on their abundance profile across the four treatment conditions (i.e. 0, 3μM, 30μM and 300μM), first each individual protein was normalized in each condition to the highest value across the four conditions (i.e. the highest value equals to 100%). To spatially map the proteins in the dataset, a t-distributed stochastic neighbor embedding (t-SNE), a nonlinear visualization of the data was applied. Then, k-means clustering was applied to this transformed matrix using the Hartigan-Wong algorithm and a maximum number of iterations set at 50000. To determine the best partition, the numbers of clusters, k, were continuously increased from 3 to 20. The result showed that the optimal number of clusters was obtained when k=8, after carefully inspecting all the clusters and their silhouette and Hartigan indexes. All computations were run using R environment using k-means function for the partition and daisy function to compute all the pairwise dissimilarities (Euclidean distances) between observations in the dataset for the silhouette.
Dose response curves
Normalized dNSAF values for each protein were plotted as a function of ligand concentration in Origin Pro 2018 for each cluster. The curves were averaged in Origin and the average was displayed on the graph.
Topological data analysis
The input data for TDA were represented in a matrix, with each column corresponding to a CoA ligand and each row corresponding to a protein. Values were distributed spectral counts values for each protein. A network of nodes with edges between them was then created using the TDA approach based on Ayasdi platform (AYASDI Inc., Menlo Park CA as described previously (Sardiu et al., 2015). Two types of parameters are needed to generate a topological analysis: First is a measurement of similarity, called metric, which measures the distance between two points in space (i.e. between rows in the data). Second are lenses, which are real valued functions on the data points. Here, Variance Norm Euclidean was used as a distance metric with 2 filter functions: Neighborhood lens 1 and Neighborhood lens2. Resolution 30 and gain 3 were used to generate Fig. 5c.
Pathway analysis.
Proteins that were changing in at least one of the CoA ligands with a Z-score less than −2 and FDR less or equal to 0.05 were considered for the analysis. Using this criteria, 671 proteins were identified and used for the pathway analysis. As expected, HATs acetylate histone was one of the top 30 enriched pathways (p-value of 4.55e-12) in the ConsensusPathDB (http://cpdb.molgen.mpg.de/) database.
Bioinformatic analyses of CATNIP data and correlation with literature datasets
A list of annotated CoA-binders was defined by searching the Uniprot database using query terms related to this function including “CoA binding,” “CoA,” “Coenzyme A,” “Acetyltransferase” “HAT,” “NAT,” “NAA,” “GNAT.” A similar analysis was performed to annotate AT interactors, using query terms including “HAT complex,” “KAT complex,” “NAA complex,” “NAT complex,” and “acetyltransferase complex.” Results were then manually curated with irrelevant proteins and duplicates removed, resulting in the term list provided in Table S1. Correlation of CATNIP enrichment to HeLa cell gene expression and protein abundance (Fig. S1 d-i) was performed using literature RNA-Seq and deep proteomic datasets (Nagaraj et al., 2011). Venn diagrams comparing overlap between proteins competed 2-fold by acetyl-CoA and all other ligands (Fig. 3b), or metabolic acyl-CoAs 3-6 (Fig. 3c) were generated by identifying a list of proteins showing a (−log2FC) value ≥1 for each ligand and then assessing overlap using an online Venn diagram tool accessible at http://bioinformatics.psb.ugent.be/webtools/Venn/. Protein subsets were interrogated for enrichment of molecular functions and pathways using the online informatics tools DAVID (david.ncifcrf.gov) and ConsensusPathDB (http://cpdb.molgen.mpg.de/CPDB/rlFrame). For analysis of acetylation stoichiometry, filtered protein subsets were cross-referenced with a list of peptide hits falling in the top 10% of all HeLa cell lysine acetylation stoichiometries measured in a recently published analysis (Hansen et al., 2019). For analysis of lysine malonylation, filtered protein subsets were cross-referenced with a list of malonylated peptides derived from a recently published analysis. Figures of E. coli NAT10 orthologue complexed with acetyl-CoA was generated using Chimera.
Isotopic tracing experiments to determine metabolic source of N4-acetylcytidine (ac4C)
HeLa S3 cells were cultured at 37 °C under 5% CO 2 atmosphere in a growth medium of DMEM supplemented with 10% FBS and 2 mM glutamine. HeLa S3 cells were plated in 10 cm dishes (3 ×106 cells in 10 ml RPMI media/dish) and allowed to adhere for 24 h. After this, media was removed, cells were washed once with PBS (10 ml), and switched to either i) heavy glucose media (glucose-free DMEM containing 2 mM glutamine, 25 mM U-13C6-glucose, 0.2 mM acetate), ii) heavy acetate media (glucose-free DMEM containing 2 mM glutamine, 25 mM glucose, 0.2 mM U-13C2-acetate) or iii) regular glucose media (glucose-free DMEM containing 2 mM glutamine, 25 mM glucose, 0.2 mM acetate). Cells were incubated with the tracer for 16 h or 24 h at 37 °C and total RNA was harvested using TRIzol reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. Digestion of total RNA (220 μg) was performed as previously described (Sinclair et al., 2017). Briefly, RNA was incubated with 1U/10 μg RNA of nuclease P1 (Sigma-Aldrich) in 100 mM ammonium acetate [pH 5.5] for 16 hr at 37 °C. Five microliter of 1 M ammoniu m bicarbonate [pH 8.3] and 0.5U/10 μg RNA of Bacterial Alkaline Phosphatase (ThermoFisher Scientific) were added for 2 hr at 37 °C. Following digestion, sample volumes were adjusted to 150 μL with RNase-free water and spin filtered to remove enzymatic constituents (Amicon Ultra 3K, #UFC500396). Filtrate and washes (200 μL x 3, RNase-free water) were collected and lyophilized. Lyophilized samples were reconstituted in 250 μL H2O containing internal standards (D3-ac4C, 500 nM; 15N3-C, 5 μM, Cambridge Isotopes). Individual samples (15 μL for ac4C analyses, 5 μL for major bases) were then analyzed via injection onto a C18 reverse phase column coupled to an Agilent 6410 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode (Agilent Technologies). Quantification was performed based on nucleoside-to-base ion transitions using standard curves of pure nucleosides and stable isotope labelled internal standards.
LN229 ac4C analysis and MS
LN229 wild-type (WT) and ACLY knockout (ACLY KO) cell lines (kind gift of K. Wellen laboratory, University of Pennsylvania) were cultured at 37 °C under 5% CO 2 atmosphere in a growth medium of RPMI supplemented with 10% FBS and 2 mM glutamine as previously described (Lee et al., 2018). For assessment of ac4C levels, total RNA was isolated from LN229 cells using TRIzol reagent (ThermoFisher Scientific). Enrichment of polyadenylated RNA [poly(A) RNA] for UHPLC-MS, was carried using two rounds of selection with Oligo-(dT)25 Dynabeads (ThermoFisher Scientific) according to the manufacturer’s instructions. 300 ng of total or poly(A)-enriched RNA was used to evaluate the levels of ac4C and mcm5s2U by LC-MS/MS using a similar method as described (Arango et al., 2018). Briefly, prior to UHPLC-MS analysis, 300 ng of each oligonucleotide was treated with 0.5 pg/μl of internal standard (IS), isotopically labeled guanosine, [13C][15N]-G. The enzymatic digestion was carried out using Nucleoside Digestion Mix (New England BioLabs) according to the manufacturer’s instructions. Finally, the digested samples were lyophilized and reconstituted in 100 μl of RNAse-free water, 0.01% formic acid prior to UHPLC-MS/MS analysis. The UHPLC-MS analysis was accomplished on a Waters XEVO TQ-S™ (Waters Corporation, USA) triple quadruple mass spectrometer equipped with an electrospray source (ESI) source maintained at 150 °C and a capillary voltage of 1 kV. Nitrogen wa s used as the nebulizer gas, which was maintained at 7 bars pressure, flow rate of 500 l/h and at temperature of 500°C. UHPLC-MS/MS analysis was performed in ESI positive-ion mode using multiple-reaction monitoring (MRM) from ion transitions previously determined for ac4C and mcm5s2U (Basanta-Sanchez et al., 2016). A Waters ACQUITY UPLC™ HSS T3 guard column, 2.1× 5 mm, 1.8 μm, attached to a HSS T3 column, 2,1×50 nm, 1.7 μm were used for the separation. Mobile phases included RNAse-free water (18 MΩcm−1) containing 0.01% formic acid (Buffer A) and 50:50 acetonitrile in Buffer A (Buffer B). The digested nucleotides were eluted at a flow rate of 0.5 ml/min with a gradient as follows: 0–2 min, 0–10%B; 2–3 min, 10–15% B; 3–4 min, 15–100% B; 4–4.5 min, 100 %B. The total run time was 7 min. The column oven temperature was kept at 35oC and sample injection volume was 10 ul. Three injections were performed for each sample. Data acquisition and analysis were performed with MassLynx V4.1 and TargetLynx. Calibration curves were plotted using linear regression with a weight factor of 1/x.
Ectopic overexpression, non-enzymatic malonylation, and immunoprecipitation of NAT10
HEK-293T cells were plated in 10 cm dishes (2.5 ×106 cells/dish in 10 ml DMEM media and allowed to adhere and grow for 24 h. 3xFLAG-tagged NAT10 was overexpressed using FuGENE® 6 transfection reagent (Promega #E2691) according to the manufacturer’s instructions. Overexpression was carried out by incubating the cells for 24 hr at 37 °C under 5% CO2 atmosphere, after which time the cells were harvested, and lysed in potassium phosphate buffer, pH 8, sonicated using a 100 W QSonica XL2000 sonicator (3 × 1 s pulse, amplitude 1, 60 s resting on ice between pulses), and quantified using the Qubit Broad Sensitivity Protein Kit (Thermo Fisher # Q33211). Lysates were incubated with 0 or 0.25 mM malonyl-CoA for six hours at 37°C. Anti-FLAG pulldo wn was performed using FLAG immunoprecipitation kit (Sigma-Aldrich, FLAGIPT1–1KT) according to the manufacturer’s instruction. 450 μg of lysate was incubated with the anti-FLAG resin overnight at 4 °C. Eluted protein was ran on SDS-PAGE and immunoblotted against anti-FLAG-tag and anti-Malonyl-Lysine antibodies. For immunoblotting, SDS–PAGE gels were transferred to nitrocellulose membranes (Novex, Life Technologies # LC2001) by electroblotting at 30 V for 1 h using a XCell II Blot Module (Novex). Membranes were blocked using StartingBlock (PBS) Blocking Buffer (Thermo Scientific) for 30 min and incubated overnight at 4 °C in primary antibody. The membranes were washed with TBST buffer and incubated with secondary HRP-conjugated antibody (Cell Signaling #7074) for 1 h at room temperature. The membranes were again washed with TBST and treated with chemiluminescence reagents (Western Blot Detection System, Cell Signaling) for 1 min, and imaged for chemiluminescent signal using an ImageQuant Las4010 Digital Imaging System (GE Healthcare).
QUANTIFICATION AND STATISTICAL ANALYSIS
The number of replicates, fold-change, mean +/− standard deviation (SD), and p-values are reported in the figures and legends. Experiments were performed with three biological replicates from at least two independent experiments when possible. LC-MS experiments were performed using an n=3, with fold-enrichments and -log10FDR calculated by analyzing dNSAF values using the statistical framework QPROT as described in Methods. Statistical analysis of non-proteomic data (i.e. t-tests) were performed with Graphpad Prism software.
DATA AND CODE AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via Pride (Deutsch, et. al, 2017; Perez-Riverol, Y., et. al, 2019) partner repository with the dataset identifier PXD013157 and 10.6019/PXD013157. Original data underlying this manuscript may also be accessed after publication from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1355. Review access can be obtained using the following username and password:
Username: reviewer59307@ebi.ac.uk
Password: 3npaE9w9
Supplementary Material
Table S1, related to Figure 2. Proteins identified after treatment with 0, 3, 30, 300 uM acetyl-CoA and Lys-CoA Sepharose capture that were used for k-means clustering and identification of false positive identifications.
Table S2, related to Figure 3. Proteins identified from multiple competitive ligands interacted with the proteome at 30uM or 3uM for palmityoyl-CoA for 1 hour before Lys-CoA Sepharose pull down. Proteins that were competed significantly by one ligand are shown where significance thresholds are defined as log2(fold change) ≥2 and false discovery rate of ≤0.05.
Table S3, related to Figure 3. Selectivity of various protein-ligand interactions analyzed by protein fold, family, and interaction clusters.
Table S4, related to Figure 4. Proteins identified after interacting with 0, 3, 30, or 300uM CoA for 1 hour prior to Lys-CoA Sepharose pulldown. Identified proteins were clustered using k-means clustering after t-SNE transformation.
Table S5, related to Figure 5. Identification of proteins insensitive to high salt washes (0.5M NaCl) by Lys-CoA sepharose pulldowns.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal MultiMab Malonyl-Lysine antibody | Cell Signaling Technologies | Cat# 14942 |
| Anti-FLAG (DYKDDDDK) Tag Antibody (HRP-linked) | Cell Signaling Technologies | Cat# 2044 |
| Anti-rabbit IgG antibody (HRP-linked) | Cell Signaling Technologies | Cat # 7074 |
| Bacterial and Virus Strains | ||
| n/a | ||
| Biological Samples | ||
| Cell pellets from HeLa S3 epithelial suspension cell line | Cell Culture Company | Cat # HA48 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| NHS-Activated Sepharose 4 Fast Flow resin | GE Healthcare | Cat# 71-5000-14 AD |
| Amine-functionalized Lys-CoA-Ahx | Montgomery et al,, 2016 | n/a |
| U-13C6-glucose | Cambridge Isotope Laboratories | Cat# CLM-1396 |
| U-13C2-acetate | Cambridge Isotope Laboratories | Cat# CLM-440 |
| acetyl-CoA | Sigma-Aldrich | Cat# A2056 |
| coenzyme A | Sigma-Aldrich | Cat# |
| butyryl-CoA | Sigma-Aldrich | Cat# B1508 |
| crotonyl-CoA | Sigma-Aldrich | Cat# 28007 |
| palmitoyl-CoA | Sigma-Aldrich | Cat# P9716 |
| acetic-CoA | This study | n/a |
| Critical Commercial Assays | ||
| Anti-FLAG IP kit | Sigma-Aldrich | Cat# FLAGIPT1-1KT |
| TRIzol reagent | ThermoFisher Scientific | Cat#15596026 |
| oligo-(dT)25 Dynabeads | ThermoFisher Scientific | Cat# 61005 |
| Qubit Broad Sensitivity Protein Assay Kit | ThermoFisher Scientific | Cat# Q33211 |
| Deposited Data | ||
| Proteomic datasets for CATNIP (Lys-CoA) pulldowns and competitions | ProteomeXchange Consortium; PRIDE | PXD013157 |
| Experimental Models: Cell Lines | ||
| HeLa S3 cells | ATCC | CCL-2.2 |
| HEK-293T cells | ATCC | CRL-3216 |
| LN229 ACLY knockout cells | Zhao et al. 2016 | n/a |
| LN229 ACLY wild-type cells | Zhao et al. 2016 | n/a |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| DTASelect v.1.9/ CONTRAST for protein detection | Tabb et al.,2002 | |
| NSAF7 | Zhang, et al., 2010 | |
| QSPEC/QPROT for the statistical analysis of protein differentiation | Choi, et al., 2015 | http://http//sourceforge.net/p/qprot/ |
| tSNE and k-means algorithms for proteins separation and clustering | Van der Maaten, et al., 2008 | R source |
| TDA software for the topological analysis | Lum et al., 2013. | https://platform.ayasdi.com/workbench (AYASDI Inc., Menlo Park CA) |
| DAVID gene ontology for functional enrichment | DAVID tool | http://david.abcc.ncifcrf.gov/ |
| ConsensusPathDB | http://cpdb.molgen.mpg.de/CPDB/rlFrame | |
| Other | ||
Highlights.
Global profiling of acetyl-CoA binding proteins with chemoproteomic probes
t-SNE clustering allows enrichment of direct and indirect acetyl-CoA binders
Identification of specific and promiscuous acyl-CoA interaction signatures
Development of a de novo acetyltransferase annotation workflow
Significance.
Acyl-CoAs are essential for life. These central metabolites interact with proteins to regulate critical biological processes ranging from energy production to gene expression. However, despite their importance, the scope and selectivity of these interactions remains unknown. Here we report the development of CATNIP (CoA/AcetylTraNsferase Interaction Profiling), an approach that combines chemoproteomic profiling with systems-level analysis for the proteome-wide interrogation of acyl-CoA/protein interactions. We validated the ability of this approach to identify CoA-utilizing enzymes based on their dose-dependent metabolite competition profiles and detect unique interaction signatures of enzymes with acyl-CoAs responsible for substrate utilization or metabolic inhibition. Applying CATNIP to profile the susceptibility of acetyltransferases led to the characterization of two RNA modifications sensitive to disruption of the acetyl-CoA biosynthetic enzyme ACLY. Finally, we demonstrated an unbiased workflow for analysis of CATNIP binding data that can be used to detect candidate acyl-CoA regulated protein networks, identify proteins that directly bind acetyl-CoA, and define proteins whose functional interaction with acyl-CoAs may lead to non-enzymatic acylation. Our studies illustrate the utility of integrating chemical biology and systems biology to rapidly characterize protein-metabolite interactions and provide a powerful first-in-class resource for studying the signaling functions of acyl-CoAs. Moreover, our experimental demonstration that acyl-CoAs show selectivity in their interactions with the AT superfamily suggests manipulating them may re-direct the activity of specific enzyme subsets and provides a rationale for the development of approaches to modulate acyl-CoA metabolism in cells and living organisms.
Acknowledgements
The authors thank K. Wellen (University of Pennsylvania) and N. Snyder (Drexel University) for helpful discussions. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (ZIA BC011488-04), the Stowers Institute for Medical Research, and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number RO1GM112639 to MPW. In addition, this project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Interests
The authors declare no contributing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. (2018). Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 175, 1872–1886 e1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB, Leaver DJ, Hermans SJ, Kelly GL, Brennan MS, Downer NL, Nguyen N, Wichmann J, McRae HM, Yang Y, et al. (2018). Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257. [DOI] [PubMed] [Google Scholar]
- Basanta-Sanchez M, Temple S, Ansari SA, D’Amico A, and Agris PF (2016). Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic acids research 44, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Liu X, Zhang C, Xing B, and Du X. (2017). Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun 483, 624–629. [DOI] [PubMed] [Google Scholar]
- Carrer A, Parris JL, Trefely S, Henry RA, Montgomery DC, Torres A, Viola JM, Kuo YM, Blair IA, Meier JL, et al. (2017). Impact of a High-fat Diet on Tissue Acyl-CoA and Histone Acetylation Levels. J Biol Chem 292, 3312–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Wang T, Birsoy K, and Sabatini DM (2016). Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell 166, 1324–1337 e1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, and Zhao Y. (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics 6, 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, and Tanaka I. (2009). RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28, 1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Kim S, Fermin D, Tsou C, Nesvizhskii AI (2015). QPROT: statistical method for testing differential expression using protein-level intensity data in label-free quantitative proteomics. J Proteomics 129, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H, Cheng Z, Park J, Wan X, Liu X, et al. (2015). Proteomic and Biochemical Studies of Lysine Malonylation Suggest Its Malonic Aciduria-associated Regulatory Role in Mitochondrial Function and Fatty Acid Oxidation. Mol Cell Proteomics 14, 3056–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, Boczkowska M, Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et al. (2018). NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci U S A 115, 4399–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F, Klein DC, and Hickman AB (2000). GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct 29, 81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, and Washburn MP (2006). Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol 328, 159–175. [DOI] [PubMed] [Google Scholar]
- Greenspan MD, and Lowenstein JM (1968). Effects of magnesium ions, adenosine triphosphate, palmitoylcarnitine, and palmitoyl coenzyme A on acetyl coenzyme A carboxylase. J Biol Chem 243, 6273–6280. [PubMed] [Google Scholar]
- Hansen BK, Gupta R, Baldus L, Lyon D, Narita T, Lammers M, Choudhary C, and Weinert BT (2019). Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat Commun 10, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong BS, Senisterra G, Rabeh WM, Vedadi M, Leonardi R, Zhang YM, Rock CO, Jackowski S, and Park HW (2007). Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J Biol Chem 282, 27984–27993. [DOI] [PubMed] [Google Scholar]
- Inoue H, Suzuki F, Tanioka H, and Takeda Y. (1968). Studies on ATP citrate lyase of rat liver. 3. The reaction mechanism. J Biochem 63, 89–100. [DOI] [PubMed] [Google Scholar]
- Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, and Suzuki T. (2014). Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem 289, 35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RA, Montgomery DC, and Meier JL (2019). Epigenetic regulation by endogenous metabolite pharmacology. Curr Opin Chem Biol 51, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, et al. (2017). Discovering Targets of Non-enzymatic Acylation by Thioester Reactivity Profiling. Cell Chem Biol 24, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. (2017). Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, and Cole PA (2000). HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell 5, 589–595. [DOI] [PubMed] [Google Scholar]
- Lee JV, Berry CT, Kim K, Sen P, Kim T, Carrer A, Trefely S, Zhao S, Fernandez S, Barney LE, et al. (2018). Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca(2+)-NFAT signaling. Genes Dev 32, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Hardouin C, Boger DL, and Cravatt BF (2003). Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol 21, 687–691. [DOI] [PubMed] [Google Scholar]
- Lin TY, Abbassi NEH, Zakrzewski K, Chramiec-Glabik A, Jemiola-Rzeminska M, Rozycki J, and Glatt S. (2019). The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat Commun 10, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Singh G, Lehman A, Ishkanov T, Vejdemo-Johansson M, Alagappan M, Carlsson J, and Carlsson G. (2013). Extracting insights from the shape of complex data using topology. Sci Rep 3, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. (2014). Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 53, 710–725. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCross MJ, Venable J, Graumann J, Johnson JR, Cociorva D, and Yates JR (2004). MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom 18, 2162–2168. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton B. (1994). The mitochondrial long-chain trifunctional enzyme: 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and 3-oxoacyl-CoA thiolase. Biochem Soc Trans 22, 427–431. [DOI] [PubMed] [Google Scholar]
- Moellering RE, and Cravatt BF (2012). How chemoproteomics can enable drug discovery and development. Chem Biol 19, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC, Garlick JM, Kulkarni RA, Kennedy S, Allali-Hassani A, Kuo YM, Andrews AJ, Wu H, Vedadi M, and Meier JL (2016). Global Profiling of Acetyltransferase Feedback Regulation. J Am Chem Soc 138, 6388–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC, Sorum AW, Guasch L, Nicklaus MC, and Meier JL (2015). Metabolic Regulation of Histone Acetyltransferases by Endogenous Acyl-CoA Cofactors. Chem Biol 22, 1030–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC, Sorum AW, and Meier JL (2014). Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation. J Am Chem Soc 136, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, and Mann M. (2011). Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I, Kochanowski K, Cappelletti V, Fuhrer T, Noor E, Sauer U, and Picotti P. (2018). A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell 172, 358–372 e323. [DOI] [PubMed] [Google Scholar]
- Rock CO, Karim MA, Zhang YM, and Jackowski S. (2002). The murine pantothenate kinase (Pank1) gene encodes two differentially regulated pantothenate kinase isozymes. Gene 291, 35–43. [DOI] [PubMed] [Google Scholar]
- Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, et al. (2018). Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol Cell 69, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Gilmore JM, Groppe BD, Dutta A, Florens L, and Washburn MP (2019). Topological scoring of protein interaction networks. Nat Commun 10, 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Gilmore JM, Groppe BD, Herman D, Ramisetty SR, Cai Y, Jin J, Conaway RC, Conaway JW, Florens L, and Washburn MP (2015). Conserved abundance and topological features in chromatin-remodeling protein interaction networks. EMBO Rep 16, 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, and Schomburg D. (2011). BRENDA, the enzyme information system in 2011. Nucleic acids research 39, D670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, and Lafontaine DL (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic acids research 43, 2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair WR, Arango D, Shrimp JH, Zengeya TT, Thomas JM, Montgomery DC, Fox SD, Andresson T, Oberdoerffer S, and Meier JL (2017). Profiling Cytidine Acetylation with Specific Affinity and Reactivity. ACS Chem Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan S, Kurzawa N, Werner T, Gunthner I, Helm D, Huber W, Bantscheff M, and Savitski MM (2019). Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat Commun 10, 1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteom Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Langer MR, and Denu JM (2000). Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39, 11961–11969. [DOI] [PubMed] [Google Scholar]
- van Delft P, Akay A, Huber SM, Bueschl C, Rudolph KLM, Di Domenico T, Schuhmacher R, Miska EA, and Balasubramanian S. (2017). The Profile and Dynamics of RNA Modifications in Animals. Chembiochem 18, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. (2008). Visualizing dta using t-SNE. J Mach Learn Res 9, 2579–2605. [Google Scholar]
- Wagner GR, Bhatt DP, O’Connell TM, Thompson JW, Dubois LG, Backos DS, Yang H, Mitchell GA, Ilkayeva OR, Stevens RD, et al. (2017). A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell Metab 25, 823–837 e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, and Yates JR (2001a). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, and Yates JR 3rd (2001b). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, and Thompson CB (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Chong LS, Perlman DH, Resnick AC, and Fiedler D. (2016). Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc Natl Acad Sci U S A 113, E6757–E6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. (2015). ProLUCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Aiba K, Kitaura Y, Kondo Y, Nomura N, Nakamura Y, Fukushi D, Murayama K, Shimomura Y, Pitt J, et al. (2015). Clinical, biochemical and metabolic characterisation of a mild form of human short-chain enoyl-CoA hydratase deficiency: significance of increased N-acetyl-S-(2-carboxypropyl)cysteine excretion. J Med Genet 52, 691–698. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, and Florens L. (2009). Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal Chem 81, 6317–6326. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, and Florens L. (2010). Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem 2010, 2272–2281. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, and Florens L. (2011). Improving proteomics mass accuracy by dynamic offline lock mass. Anal Chem 83, 9344–9351. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, and Florens L. (2015). Improving label-free quantitative proteomics strategies by distributing shared peptides and stabilizing variance. Anal Chem 87, 4749–4756. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO, and Jackowski S. (2006). Biochemical properties of human pantothenate kinase 2 isoforms and mutations linked to pantothenate kinase-associated neurodegeneration. J Biol Chem 281, 107–114. [DOI] [PubMed] [Google Scholar]
- Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, Carrer A, Sengupta A, Campbell SL, Kuo YM, et al. (2016). ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep 17, 1037–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 2. Proteins identified after treatment with 0, 3, 30, 300 uM acetyl-CoA and Lys-CoA Sepharose capture that were used for k-means clustering and identification of false positive identifications.
Table S2, related to Figure 3. Proteins identified from multiple competitive ligands interacted with the proteome at 30uM or 3uM for palmityoyl-CoA for 1 hour before Lys-CoA Sepharose pull down. Proteins that were competed significantly by one ligand are shown where significance thresholds are defined as log2(fold change) ≥2 and false discovery rate of ≤0.05.
Table S3, related to Figure 3. Selectivity of various protein-ligand interactions analyzed by protein fold, family, and interaction clusters.
Table S4, related to Figure 4. Proteins identified after interacting with 0, 3, 30, or 300uM CoA for 1 hour prior to Lys-CoA Sepharose pulldown. Identified proteins were clustered using k-means clustering after t-SNE transformation.
Table S5, related to Figure 5. Identification of proteins insensitive to high salt washes (0.5M NaCl) by Lys-CoA sepharose pulldowns.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via Pride (Deutsch, et. al, 2017; Perez-Riverol, Y., et. al, 2019) partner repository with the dataset identifier PXD013157 and 10.6019/PXD013157. Original data underlying this manuscript may also be accessed after publication from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1355. Review access can be obtained using the following username and password:
Username: reviewer59307@ebi.ac.uk
Password: 3npaE9w9