Abstract
Aberrant alternative splicing is a hallmark of cancer, yet the underlying regulatory programs that control this process remain largely unknown. Here, we report a systematic effort to decipher the RNA structural code that shapes pathological splicing during breast cancer metastasis. We discovered a previously unknown structural splicing enhancer that is enriched near cassette exons with increased inclusion in highly metastatic cells. We show that the spliceosomal protein SNRPA1 interacts with hundreds of these enhancers to promote cassette exon inclusion. This interaction enhances metastatic lung colonization and cancer cell invasion, in part through SNRPA1-mediated regulation of PLEC alternative splicing, which can be counteracted by splicing modulating morpholinos. Together, our findings establish a non-canonical regulatory role for SNRPA1 as a pro-metastatic splicing enhancer in breast cancer.
One-sentence summary:
Through the systematic interrogation of alternative splicing programs we have identified an interaction between SNRPA1 and structured RNA elements near cassette exons that induces the expression of pro-metastatic mRNA isoforms in breast cancer.
Alternative splicing (AS) is a post-transcriptional regulatory mechanism critical for transcriptome and proteome diversity (1, 2). By increasing complexity at the protein level, AS can induce functional changes in the cell, including those important to cancer progression (3–8). Pathological changes in alternative splicing patterns are considered a hallmark of cancer, and specific transcript isoforms have been functionally implicated in tumorigenesis (9, 10). Similarly, dysregulation of AS has been described as a driver of breast cancer metastasis (11–16). While several trans-acting factors are involved in the control of individual AS events in breast cancer (11–13, 15, 17), the underlying regulatory pathways that drive aberrant RNA splicing programs and their major cis and trans regulatory factors are not well understood. Of special interest in this process is the role of regulatory information encoded by RNA secondary structure that governs splicing decisions (18–22). It is well-established that RNA structural elements play a critical role in post-transcriptional regulatory processes (23–27); however, bioinformatic strategies commonly used for the discovery of cis-regulatory elements fail to capture the contribution of RNA secondary structure to regulatory information.
Results
RNA structural elements impact alternative splicing
To capture alternative splicing events that are associated with breast cancer metastasis, we performed RNA-seq on a commonly used triple-negative model of breast cancer metastasis, MDA-MB-231 breast cancer cells (MDA-parental) and their lung metastatic derivative cell line, MDA-LM2 (28). We then used MISO (29), a probabilistic algorithm that quantifies splicing isoforms from RNA-seq data, to identify cassette exons that were differentially spliced between highly and poorly metastatic cells. The increased lung metastatic capacity of MDA-LM2 cells accompanies broad modulations in the AS landscape of the cell (Fig. 1A).
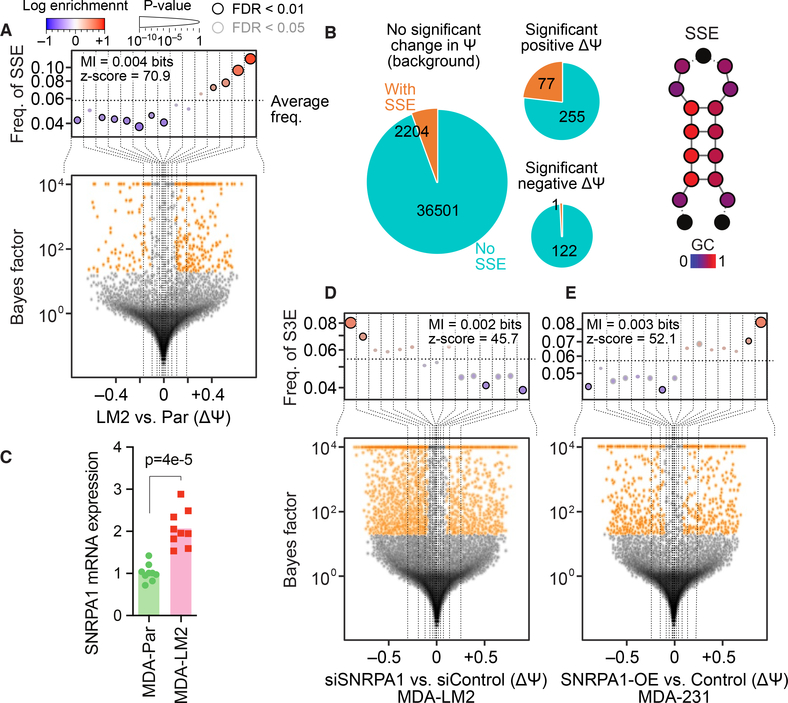
Fig. 1. Metastatic breast cancer-associated changes in cassette exon splicing are regulated by an RNA structural element and the RNA binding protein SNRPA1.
(A) Bottom: Volcano plot showing distribution of relative changes in alternative splicing (change in percent spliced in (ΔΨ)) in MDA-LM2 compared to MDA-parental breast cancer cells. Top: Enrichment of the SSE RNA structural element in exons (and flanking intronic sequences) as a function of ΔΨ between MDA-LM2 and MDA-parental cells. Cassette exons are binned according to ΔΨ (dotted vertical lines delineate the bins); the y-axis shows the frequency of the SSE that we identified in each bin (dotted horizontal line denotes the average SSE frequency across all cassette exons). Bins with significant enrichment (logistic regression, FDR < 0.05; red) or depletion (blue) of SSE elements are denoted with a black border. Also included are mutual information (MI) values and their associated Z scores. (B) Left: The distribution of instances of the SSE that we have identified across differentially spliced exons (in MDA-LM2 compared to MDA-parental cells). The area of each pie chart is proportional to the logarithm of all exon counts, and the slices represent exons that contain or lack an instance of this SSE. Right: Schematic of the SSE structure, with relative GC content indicated by color and dashed lines indicating the possible presence of additional nucleotides. (C) Bar graph showing relative SNRPA1 mRNA levels in MDA-parental and MDA-LM2 cells as measured by RT-qPCR. n = 9 biological replicates. P-value calculated using one-tailed Mann-Whitney U-test. (D) Enrichment of the SNRPA1-associated SSEs (S3Es) in exons (and flanking intronic sequences) with decreased ΔΨ in SNRPA1 knockdown MDA-LM2 cells compared to control cells. See (A) for description of volcano and enrichment plots. (E) Enrichment of SSEs in exons (and flanking intronic sequences) with increased ΔΨ in MDA-parental cells with SNRPA1 overexpression compared to control cells.
Previously, we developed Tool for Eliciting Informative Structural Elements in RNA (TEISER), a computational framework that uses both RNA structural and sequence information to identify cis-regulatory elements that are informative of transcriptomic changes (23). Here, we introduce pyTEISER (pythonic TEISER), which builds on the TEISER framework to better capture the inherent heterogeneity of RNA structural elements (e.g. (30)). In pyTEISER, structural elements are conceptualized as ensembles of closely-related context-free grammars, as measured by conditional information, that together define the underlying sequence and structural constraints. This approach allows for the integration of additional computational and experimental data to further constrain RNA secondary structures. These data include information from common in silico folding algorithms (such as RNAfold (31) or RNAstructure (32)) as well as RNA structural probing data (SHAPE or DMS-seq) (33). Furthermore, the modular programming of pyTEISER, along with an optimized context-free grammar library and predesigned tests, allows for a broader application of this approach to custom measurements of the RNA life-cycle, such as splicing and RNA processing.
Using pyTEISER, we discovered a previously unknown and highly specific RNA structural element, which is enriched (p < 3×10–53, logistic regression) in cassette exons, or the flanking 250 nucleotides of their neighboring introns, that are upregulated in highly metastatic cells (Figs. 1A and S1A–B). Its specific enrichment in differentially spliced exons suggests that this element functions as a “structural splicing enhancer” (SSE). We found 5670 instances of this SSE in or near 2282 exons that were measurable in the MDA-parental or MDA-LM2 cells (< 6% of all expressed cassette exons). Among the exons that show significantly higher inclusion in MDA-LM2 cells compared to MDA-parental, 23.2% had an instance of this SSE, as compared to only 5.8% of all other exons (Fig. 1B). These SSEs are highly GC rich (GC content 76.5%), with the terminal part of their stem consisting of G-C/U pairs (Fig. 1B). In silico predictions of SSE folding show that they should form stable secondary structures, often with a small bulge around the 5th position in the stem. We have visualized SSEs as a generic stem-loop structure with key positions colored on the basis of their GC content (Fig. 1B).
To assess the association between the SSE that we found and lung metastasis in more disease-relevant models of triple-negative breast cancer, we analyzed RNA-seq data from three established patient-derived xenograft (PDX) models of breast cancer; HCI-002 (poorly metastatic), and HCI-001, and HCI-010 (both highly metastatic) (34). The increased metastatic capacity of the HCI-001 and HCI-010 PDX models coincides with an increase in the inclusion of cassette exons that contain SSEs (Fig. S1C). In addition to these cell line and PDX models of breast cancer lung metastasis, we also analyzed RNA-seq data from triple-negative primary tumors and their matched lung metastases (35). Consistently, we found an upregulation of SSE-containing cassette exons in the paired metastases (Fig. S1D). Collectively, these findings in cell lines, PDXs, and clinical samples implicate SSEs in both the regulation of alternative splicing and in breast cancer progression.
Next, to determine if this structural element has an effect on AS patterns, we transfected breast cancer cells with an SSE mimetic RNA oligonucleotide consisting of three tandem SSE elements. These elements were of different primary sequence but were all predicted to form structures similar to the SSE that we have identified (Fig. S1E). We expected that this mimetic would act as a sponge for factors that bind endogenous SSEs, thereby inhibiting the function of these factors. In cells transfected with this mimetic compared to those transfected with a scrambled control oligonucleotide we observed a relative increase in the skipped exon variant of transcripts (decreased Ψ) containing an endogenous SSE in the cassette exon or the flanking intronic sequences (p < 2×10–18, logistic regression; Fig. S1F). This suggests that SSE elements can impact the alternative splicing of their host transcripts.
SNRPA1 interacts with the structural element SSE and controls alternative splicing
We next performed RNA co-precipitation followed by mass spectrometry to identify the protein(s) that interact with SSEs to mediate its effects. We incubated either the SSE mimetic RNA or a scrambled control RNA with whole cell lysate of MDA-LM2 highly metastatic breast cancer cells, and identified the proteins co-precipitating with these RNA baits with mass spectrometry. This analysis identified SNRPA1 as the RNA-binding protein with the greatest enrichment in the SSE compared to control RNA pull-down (Fig. S1G). SNRPA1 (small nuclear ribonucleoprotein polypeptide A’), is a component of the U2 snRNP, which recognizes the branchpoint site in the initial steps of pre-mRNA splicing (36–38). Hereafter we refer to the SSE that our study has uncovered as “SNRPA1-associated structural splicing enhancer” (S3E). Consistent with the splicing differences observed in MDA-parental compared to MDA-LM2 cells, we observed significantly higher levels of SNRPA1 mRNA and protein in MDA-LM2 compared to MDA-parental cells (Figs. 1C and S1H). This increase was specific to SNRPA1 and not other U2 snRNP components (Figs. S1I–J).
Next, to test if SNRPA1 affects the splicing of S3E-containing exons, we used siRNAs to knock down SNRPA1 in MDA-LM2 cells followed by RNA-seq. We observed a significant enrichment (p < 2×10–18, logistic regression) of S3Es in exons (and flanking intronic sequences) with decreased retention in SNRPA1 knockdown cells compared to control cells (Fig. 1D). Notably, a large number of the expressed exons in MDA-LM2 cells that contain an S3E had significantly decreased PSI upon SNRPA1 silencing (odds ratio = 2.4, p < 3×10–27, Fisher’s exact test). Conversely, upon SNRPA1 overexpression in MDA-parental cells, we found a significant enrichment of S3Es in exons (and flanking intronic sequences) with significantly increased retention (odds ratio = 3.1, p < 2×10–11, Fisher’s exact test), and an overall significant association between increased PSI and S3E occurrence (p < 3×10–16, logistic regression; Fig. 1E). Consistent with these results, exons with MDA-LM2-specific retention that exhibited increased skipping upon SNRPA1 knockdown showed significant enrichment of S3Es (odds ratio = 1.8, p < 3×10–35, Fisher’s exact test; Fig. S1K). In comparison, exons with MDA-LM2-specific retention that were not differentially spliced upon SNRPA1 knockdown did not show enrichment of S3Es (odds ratio = 1.08, p = 0.2). Similarly, S3E elements were enriched among exons that displayed both increased skipping upon SNRPA1 knockdown and increased retention upon SNRPA1 overexpression (odds ratio = 1.6, p < 10–23, Fisher’s exact test; Fig. S1K), but not among exons that were affected only by SNRPA1 knockdown or overexpression (odds ratio = 0.96, p = 0.5). Together, these observations suggested that SNRPA1 promotes the retention of S3E-containing exons in highly metastatic cells.
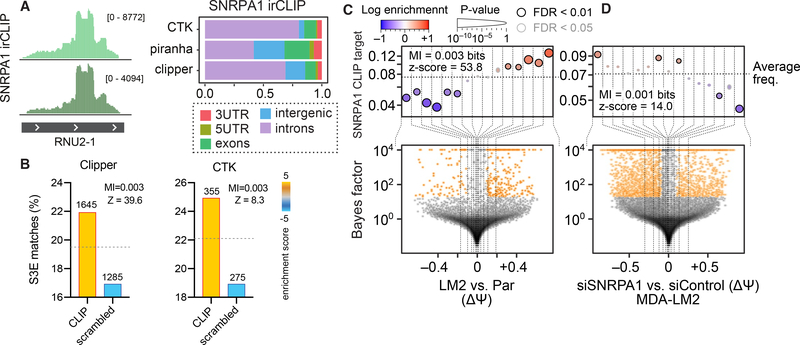
In order to test if this function could be the result of a direct interaction between SNRPA1 and S3E elements in vivo, we performed CLIP-seq (39) for SNRPA1 in MDA-LM2 cells. As expected, a robust interaction was observed between SNRPA1 and RNU2, the U2 small nuclear RNA that SNRPA1 binds in the spliceosome (40) (Fig. 2A). SNRPA1 binds stem loop IV in RNU2, which is structurally similar to S3Es (40). SNRPA1 bound hundreds of RNAs in addition to RNU2 (Fig. 2A). The SNRPA1 CLIP-seq data showed a significant enrichment (p < 10–13 (peaks called using Clipper) and p < 10–4 (CTK), hypergeometric test) of S3E elements among SNRPA1-bound sites, as well as an enrichment (p < 10–4 (CTK), hypergeometric test) of S3E elements in in silico folded SNRPA1-bound sites (Figs. 2B and S2A–B). Moreover, SNRPA1 binding sites were significantly enriched (p < 2×10–58, logistic regression) in and near cassette exons with increased retention in MDA-LM2 compared to MDA-parental cells (Fig. 2C), as well as in exons with decreased retention in SNRPA1 knockdown compared to control cells (p < 3×10–15, logistic regression; Fig. 2D). These observations are reproducible across multiple statistical methods used for quantification of differential splicing (Fig. S2C).
Fig. 2. SNRPA1 binds S3E elements in vivo.
(A) RNA-seq tracks showing reads from SNRPA1 irCLIP (39) mapped to RNU2–1. n = 2 biological replicates. The bar graph shows the relative distribution of the location of SNRPA1 binding sites identified in SNRPA1 irCLIP data using three peak finding methods. (B) Bar graphs showing enrichment of S3Es in SNRPA1 binding sites identified from CLIP data using Clipper (left) and CTK (right) peak finding algorithms. (C-D) Enrichment of SNRPA1 binding sites in exons (and flanking introns) with increased ΔΨ in MDA-LM2 compared to MDA-parental cells (C) and in exons (and flanking introns) with decreased ΔΨ in MDA-LM2 cells with SNRPA1 knockdown compared to control cells (D). See Fig. 1 for description of volcano and enrichment plots.
Notably, given the short distance requirements of UV-crosslinking (41), the non-RNU2 binding sites captured by SNRPA1 CLIP-seq are expected to originate from direct binding of SNRPA1 to these sites outside of the core spliceosome. In support of this, we detected very limited overlap between SNRPA1 binding sites and experimentally annotated human branchpoint sites (3 out of ~3,000 SNRPA1 binding sites) (42). We also performed CLIP-seq for SF3B1, another core U2 snRNP component that binds RNU2 (37, 38). Although, similar to SNRPA1, the majority of SF3B1 binding sites occur in introns (Fig. S2D), we observed only 1% overlap between SNRPA1 and SF3B1 binding sites as determined by CLIP-seq (Fig. S2E). Moreover, we found no substantial overlap of annotated alternatively spliced exons bound by SF3B1 or SNRPA1 (8% overlap; Fig. S2F). Furthermore, among the SF3B1-bound exons and flanking intronic sequences, we detected no significant change in alternative splicing events in SNRPA1-deficient or overexpressing cells compared to control cells (p = 0.35 and p = 0.88, respectively; logistic regression).
To support our hypothesis that SNRPA1 has a U2 snRNP-independent role in AS, we sought to determine the stoichiometry of U2 snRNP components. We performed mass spectrometry analysis of MDA-parental whole cell lysates supplemented with standard spike-ins, and found that SNRPA1 was the most abundant subunit of the U2 snRNP complex, and present in at least three-fold molar excess compared to other U2 snRNP components on average (Fig. S2G). Together, these results indicate that SNRPA1 likely acts as a regulator of alternative splicing extra-spliceosomally.
We also tested the specificity of SNRPA1 binding to exons with SNRPA1-dependent AS changes by performing RNA-seq on MDA-LM2 cells with knockdown of additional spliceosome components. For this, we tested SNRPB2 (U2 snRNP B"), which binds the U2 RNA hairpin IV along with SNRPA1 (40), as well as the more general spliceosome factor SNRPB (SmB/B'), a part of the Sm complex that is present in all spliceosomal snRNPs except U6 (43). We found no significant enrichment of SNRPA1 binding sites on exons adjacent to AS events induced by knockdown of either SNRPB2 or SNRPB (Fig. S2H), indicating that SNRPA1-mediated modulation of S3E-containing exons is not dependent on its spliceosomal role. Together, these data suggest that S3Es (i) regulate AS of a subset of transcripts in breast cancer cells, (ii) are bound by SNRPA1, and (iii) impact alternative splicing through direct interaction with SNRPA1.
SNRPA1 promotes invasion and in vivo metastatic colonization
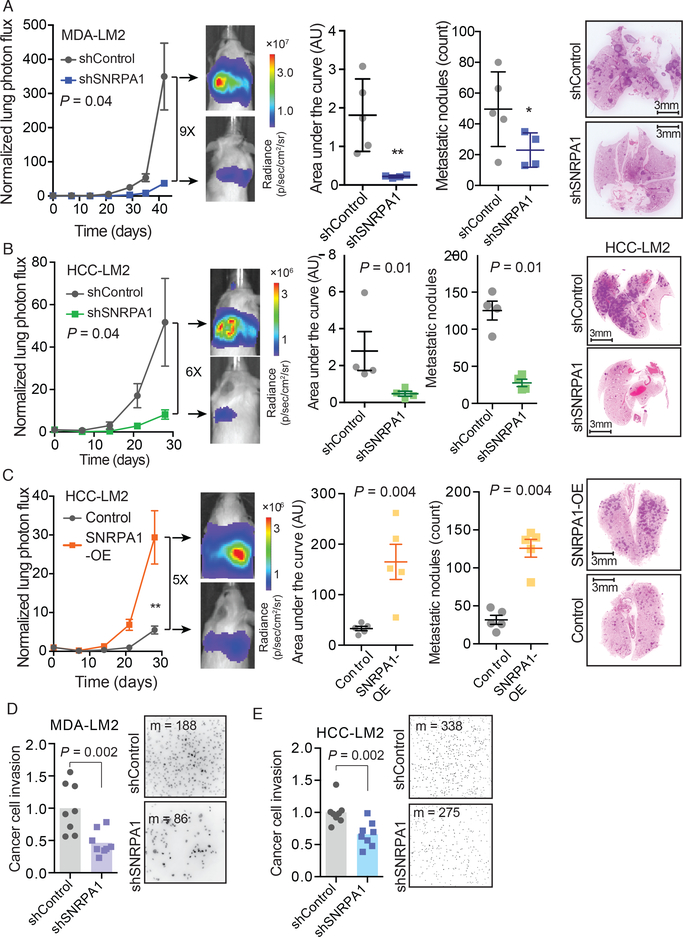
As SNRPA1 controls alternative splicing patterns in metastatic breast cancer cells, we tested if SNRPA1 impacts breast cancer progression with in vivo experimental metastasis assays. We injected SNRPA1-deficient MDA-LM2 cells (shRNA-mediated SNRPA1 knockdown) into the venous circulation of NOD-scid-gamma (NSG) mice followed by bioluminescence imaging to measure lung metastatic colonization over time. SNRPA1-depleted cells exhibited a 9-fold decrease (p = 0.04, two-way ANOVA) in metastatic capacity compared to cells expressing a control shRNA (Fig. 3A). Similarly, when a metastatic lung colonization assay was carried out using an independent breast cancer cell line, HCC1806-LM2, we found that SNRPA1 knockdown cells had reduced metastatic colonization capacity (6-fold, p = 0.04, two-way ANOVA) compared to control cells in this background (Fig. 3B).
Fig. 3. SNRPA1 promotes in vivo metastatic colonization and invasion.
(A) MDA-LM2 cells stably expressing an shRNA targeting SNRPA1 or a control shRNA were injected via tail vein into NSG mice. Bioluminescence was measured at the indicated times; area under the curve was measured at the final time point. Lungs were stained with H&E and nodules were counted. n = 4–5 mice per cohort. (B) HCC1806-LM2 cells stably expressing an shRNA targeting SNRPA1 or a control shRNA were injected via tail vein into NSG mice. Bioluminescence was measured at the indicated times; area under the curve was measured at the final time point. Lungs were stained with H&E and nodules were counted. n = 4 mice per cohort. (C) HCC1806-LM2 cells stably overexpressing SNRPA1 or mCherry (control) were injected via tail vein into NSG mice. Bioluminescence was measured at the indicated times; area under the curve was measured at the final time point. n = 5 mice per cohort. Two-way ANOVA was used to compare in vivo bioluminescence measurements in (A-C). One-tailed Mann-Whitney U-test was used to compare AU and metastatic nodule count measurements in (A-C). (D) MDA-LM2 cells stably expressing an shRNA targeting SNRPA1 or a control shRNA were subjected to transwell invasion assays. n = 8 biological replicates. (E) HCC1806-LM2 cells stably expressing an shRNA targeting SNRPA1 or a control shRNA were subjected to transwell invasion assays. n = 8 biological replicates. Median numbers of cells per view (m) are shown. P-values calculated using one-tailed Mann-Whitney U-test (D-E).
SNRPA1 knockdown did not result in a significant decrease in tumor growth of cells implanted in mammary fat-pads of mice, suggesting SNRPA1 does not significantly impact primary tumor growth (Fig. S3A). Additionally, we observed no significant differences in the in vitro proliferation rates of either of these cell lines upon SNRPA1 knockdown (Fig. S3B). Consistent with these results, when MDA-parental cells or HCC1806-LM2 cells with stable overexpression of SNRPA1 were injected into mice, they demonstrated significantly increased ability (6-fold, p < 0.05 and 5-fold, p < 0.01, respectively; two-way ANOVA) to colonize the lung compared to control cells (Figs. 3C and S3C). Importantly, silencing of SNRPB2 did not copy the phenotypic effect of SNRPA1 knockdown on in vivo metastatic colonization (Fig. S3D). These observations further support the existence of a non-canonical function for SNRPA1 as a pro-metastatic factor outside of its canonical role as a component of U2 snRNP.
We also performed transwell invasion assays with SNRPA1 knockdown and control cells in MDA-LM2 and HCC1806-LM2 cell lines and found that SNRPA1-depleted cells had significantly lower invasion capacity than control cells (relative fold change 0.46, p = 0.002 and 0.66, p = 0.002, respectively; one-tailed Mann-Whitney U-test; Figs. 3D–E). These results are consistent with a role of the SNRPA1 regulon in modulating cell mobility and invasion, potentially through modulating cellular interaction with the extracellular matrix (44). In line with these results, gene set overlap analysis of transcripts with decreased cassette exon inclusion in SNRPA1 knockdown cells revealed focal adhesion as the most significant gene ontology (GO) term (Fig. S3E). Focal adhesion is also the most significant GO term among the genes harboring exons with increased inclusion in SNRPA1 overexpressing cells, as well as those with exons with increased inclusion in MDA-LM2 compared to MDA-parental cells (Figs. S3E–F).
To assess if the S3E-SNRPA1 interaction-mediated alternative splicing program is more broadly associated with cell migratory potential, we analyzed alternative splicing events in human embryonic stem cell- (ESC) derived neural crest (NC) cells, a model for cell migration. We observed a significant enrichment (p < 8×10–6, logistic regression) of S3Es in the vicinity of exons showing higher inclusion in NC cells as compared to ESCs (Fig. S3G). This observation was specific to the migratory potential and not associated with increased cell proliferation capacity, as there was no enrichment (p = 0.13, logistic regression) in S3E elements in differentially spliced exons between ESC-derived non-neuronal ectoderm cells and ESCs, representing poorly and highly proliferative states, respectively (Fig. S3H). This finding is in agreement with our results that show modulating SNRPA1 expression does not impact cell proliferation in vitro or primary tumor growth in vivo. Collectively, these results suggest that the SNRPA1-mediated AS pathway may impact cancer cell invasion, mobility and metastasis via regulating focal adhesion activity.
SNRPA1 regulates alternative splicing of PLEC and ERRFI1
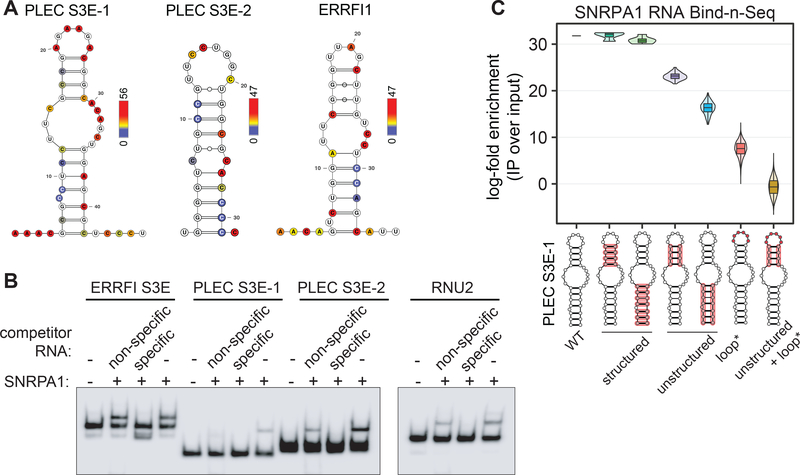
To further dissect the SNRPA1-S3E interaction of individual functional target candidates, we selected genes that contain an S3E, are bound by SNRPA1, and showed increased cassette exon inclusion in cells with higher levels of SNRPA1 (Fig. S4A). These criteria identified ERRFI1 and PLEC as the top two candidate genes, with ERRFI1 containing one and PLEC containing two SNRPA1 binding sites (PLEC S3E-1 and PLEC S3E-2). We verified the predicted secondary structure of the target S3E elements in ERRFI1 and PLEC by treating MDA-LM2 cells with dimethyl sulfate (DMS) and then analyzing the S3E-containing regions by targeted Mutational Profiling with sequencing (DMS-MaPseq) (33). The DMS probing results were generally consistent with the structures predicted by in silico folding (Fig. 4A), with increased DMS reactivity observed in the loop regions compared to stem regions of these structures (Fig. S4B). Importantly, the majority of mutations occurred at adenines and cytosines (Fig. S4C), and the mutation counts between biological replicates were highly correlated (Fig. S4D). Notably, this data demonstrates that pyTEISER is able to derive experimentally validated RNA structures.
Fig. 4. S3Es form stem loop structures in vivo and interact with SNRPA1.
(A) Predicted S3E structures in ERRFI1 and PLEC with nucleotides colored by their in vivo DMS reactivity; red indicates highest observed mutational frequency. n = 2 biological replicates. (B) RNA EMSA assessing the binding of recombinant SNRPA1 and the indicated S3Es. Non-specific and specific unlabeled competitor RNA was added at 100-fold molar excess. RNU2 is a canonical SNRPA1 binding RNA. (C) Violin plot representing the log-fold enrichment of SNRPA1-bound over input PLEC S3E-1 RNA variants from Bind-n-Seq, and grouped by S3E variant classes. Red highlights on S3E structures indicate mutated regions in each group.
RNA Electrophoretic Mobility-Shift Assays (EMSAs) indicate that SNRPA1 binds to the S3Es from PLEC and ERRFI1 in vitro, and that this interaction is not substantially outcompeted by the addition of an excess of a non-specific and unstructured RNA competitor, but is abrogated by addition of an excess of unlabeled specific competitor RNA (Fig. 4B). This demonstrates that SNRPA1 preferentially binds S3E-containing RNA and that additional factors are not required for this interaction in vitro. To further address the sequence and structural requirements for SNRPA1 binding, we generated a set of PLEC S3E-1 variants by introducing changes in the S3E primary sequence, structure or both. A Bind-n-Seq experiment (45) with this pool of in vitro transcribed S3E RNA variants and recombinant SNRPA1 revealed that changing the primary sequence but maintaining the structure of both stems of PLEC S3E-1 resulted in SNRPA1 binding indistinguishable from that of the wild-type S3E (Fig. 4C). In contrast, disrupting the structure of both PLEC S3E-1 stems reduced SNRPA1 binding. In addition to the expected effect of changing the S3E structure via stem mutations, we discovered that mutating the PLEC S3E-1 loop sequence also greatly reduced SNRPA1 binding.
We also sought to functionally assess the S3E structure and sequence requirements for SNRPA1-mediated alternative splicing in vivo. Modulating SNRPA1 abundance resulted in the differential inclusion of PLEC exon 31 and ERRFI1 exon 3. We obtained these results by three independent techniques: isoform-specific RT-qPCR, alternative exon-flanking RT-PCR, and isoform calling by RNA-seq using MISO (Figs. S5A–E). In all instances, increased SNRPA1 expression (in MDA-LM2 and SNRPA1 overexpressing cells) caused an increase in alternative exon inclusion, and conversely, SNRPA1 knockdown caused increased alternative exon skipping. We also used CRISPR/Cas9 to endogenously delete PLEC and ERRFI1 S3Es in MDA-LM2 cells, confirming the editing by PCR amplification of targeted loci. Mirroring the SNRPA1 knockdown results, ΔS3E cells showed increased alternative exon skipping (Fig. S5F), validating that S3Es are necessary for SNRPA1-mediated alternative splicing.
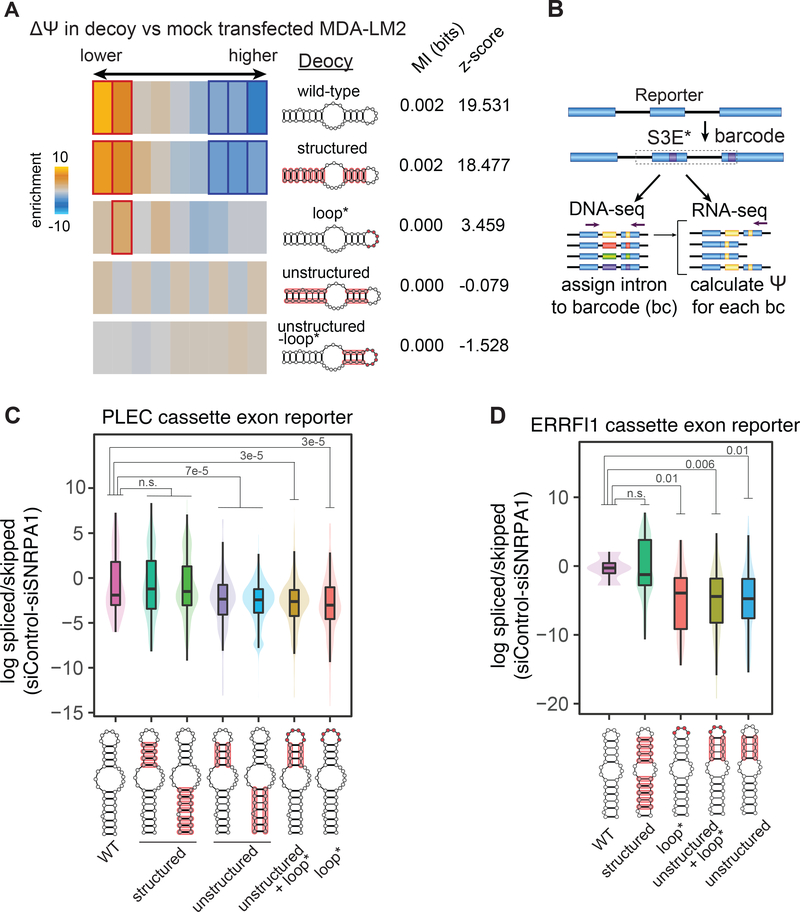
Next, we used in vitro transcribed PLEC S3E-1 variants (those employed in the Bind-n-Seq experiment) to repeat the mimetic transfection experiment described earlier. As shown in Fig. 5A, and in accordance with Fig. 4C, the S3E variants that were most preferentially bound by SNRPA1 in vitro also had the highest impact on S3E-containing transcript alternative splicing in MDA-LM2 cells. Consistent with this, the S3E variants that showed relatively low SNRPA1 binding in vitro did not significantly modulate the splicing landscape of the transfected cells. Finally, we constructed mini-gene reporters containing either a truncated PLEC exon 31 or a full-length ERRFI1 exon 3, along with their respective endogenous flanking intron and exon sequences. We replaced the wild-type S3E in each of these constructs with a library of S3E variants (specific to each gene’s S3E), designed to disrupt the S3E structure, sequence, or both. We also included unique exonic barcodes in the reporters, allowing us to track the effect of each individual S3E variant on reporter AS (Fig. 5B). The reporter libraries were stably expressed in MDA-LM2 cells, and we observed an increase in alternative exon skipping in both the PLEC and ERRFI1 reporter minigenes containing the S3E variants compared to those containing the wild-type S3E, as determined by RT-PCR (Fig. S5G). We also treated the library-expressing cells with control or SNRPA1-targeting siRNAs and determined the reporter PSI by RNA-seq. As shown in Figs. 5C–D, the results were consistent with the findings from our Bind-n-Seq and mimetic transfection experiments, confirming that the structure and not sequence of the S3E stems, in concert with the S3E loop sequence, are required for SNRPA1-mediated alternative splicing.
Fig. 5. Structure and sequence grammar of S3E-mediated alternative splicing.
(A) Heatmap showing the enrichment of S3E instances in exons grouped by their differential splicing upon transfection of PLEC S3E-1 variant RNA in MDA-LM2 cells, stratified by the variant groups. Experiment was performed in biological duplicates. Bolded rectangles represent statistically significant bins. Mutual information (MI) values and z-scores are shown. (B) Schematic of the mini-gene splicing reporter design, S3E variant library cloning and barcoding strategy. (C-D) Violin plots showing the log-fold change in minigene reporter exon inclusion versus exclusion ratio, in control siRNA versus siSNRPA1-transfected cells, stratified by the S3E variant groups. P-values, calculated using t-test (taking into account only barcodes expressed at >200 normalized count number), are shown. Red highlights on S3E structures indicate mutated regions in each group. n = 2 biological replicates.
ERRFI1 (ERBB receptor feedback inhibitor 1) is a scaffolding adaptor protein and acts as a negative regulator of EGFR signaling (46, 47). PLEC encodes the cytoskeletal linker protein Plectin, and mutations in PLEC underlie a group of genetic diseases primarily affecting skin and muscle (48). To assess the functional consequences of the alternative splicing of PLEC and ERRFI1, we used morpholino antisense oligonucleotides (MOs) to specifically target the junction between each SNRPA1-dependent cassette exon and its neighboring intron in these transcripts. These MOs are designed to induce increased skipping of the cassette exon but not substantial downregulation of total transcript levels, thereby mimicking SNRPA1 depletion.
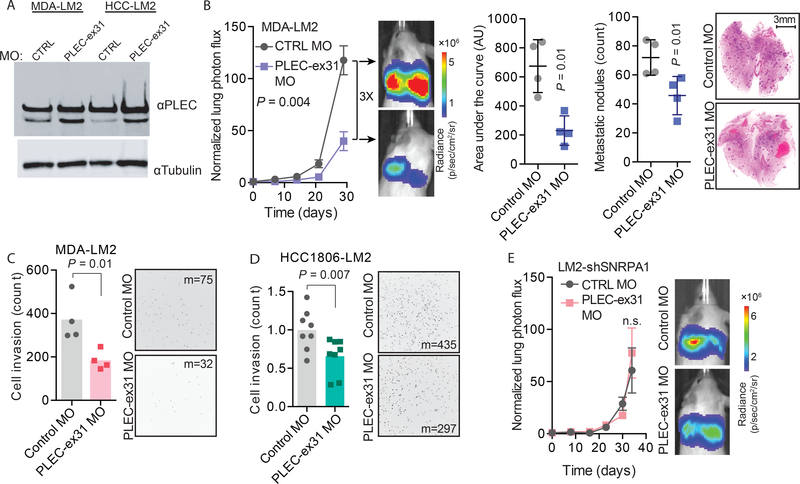
We tested the efficacy of these MOs by RT-qPCR and western blot, and observed the most robust isoform switching with the MO that targeted PLEC exon 31 (PLEC-ex31 MO) (Figs. 6A and S6A); therefore, we chose to further characterize the effects of PLEC isoform switching in metastasis. We performed experimental metastasis assays with MDA-LM2 cells transfected with either PLEC-ex31 MO or a non-targeting control MO and observed that cells with decreased PLEC exon 31 retention had 3-fold lower metastatic colonization capacity (Fig. 6B). Similarly, HCC1806-LM2 cells transfected with PLEC-ex31 MO exhibited significantly reduced metastatic colonization capacity (Fig. S6B).
Fig. 6. PLEC alternative splicing regulates breast cancer metastasis.
(A) Western blot for PLEC showing an increase in the Plectin Δexon 31:full length ratio upon transfection of PLEC-ex31 MO compared to a control MO in MDA-LM2 and HCC1806-LM2 cells. (B) MDA-LM2 cells transfected with a PLEC-ex31 MO or a control MO were injected via tail vein into NSG mice. Bioluminescence was measured at the indicated times; area under the curve was measured at the final time point. Lungs were stained with H&E and nodules were counted. n = 4 mice per cohort. (C) MDA-LM2 cells transfected with PLEC-ex31 MO or a control MO were subjected to transwell invasion assays. n = 4 biological replicates. (D) HCC1806-LM2 cells transfected with PLEC-ex31 MO or control MO were subjected to transwell invasion assays. n = 8 biological replicates. Median numbers of cells per view (m) are shown. (E) MDA-LM2 cells with shRNA-mediated SNRPA1 knockdown were transfected with a PLEC-ex31 MO or a control MO, and then injected via tail vein into NSG mice. Bioluminescence was measured at the indicated times. n = 4 mice per cohort. Two-way ANOVA was used to compare in vivo bioluminescence measurements in (B and E). One-tailed Mann-Whitney U-test was used to compare AU, metastatic nodule count, and cell invasion count measurements in (B-D).
We also tested the invasion capacity of MDA-LM2 cells and HCC1806-LM2 cells transfected with PLEC-ex31 MO or a control MO and found there was a significant reduction (relative fold change 0.49, p = 0.01 and 0.66, p = 0.007, respectively; one-tailed Mann-Whitney U-test) in the invasion capacity of the PLEC-ex31 MO transfected cells compared to control cells in both of these two cell lines (Figs. 6C–D). Transfection of PLEC-ex31 MO did not significantly affect the in vitro proliferation rates of either MDA-LM2 or HCC1806-LM2 cells compared to transfection of a control MO (Fig. S6C).
Finally, we performed an experimental metastasis assay with MDA-LM2 cells with stable SNRPA1 knockdown that were transfected with either PLEC-ex31 MO or a control MO. We observed no significant difference (relative fold change 1.3, p = 0.8, two-way ANOVA) in their metastatic colonization capacity (Fig. 6E), providing evidence that this effect is SNRPA1 dependent. Together, these observations are consistent with the longer isoform of PLEC acting as a promoter of breast cancer cell invasion and metastasis, and establishes that SNRPA1 can control the AS of PLEC exon 31.
SNRPA1 expression is associated with breast cancer progression
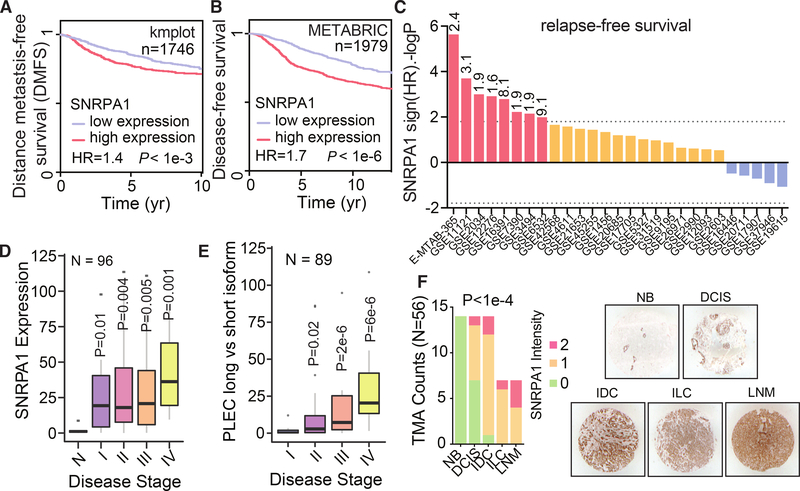
As our experimental results indicated that SNRPA1 plays a role in promoting breast cancer progression, we examined publicly available datasets from breast cancer patients and performed clinical association studies. Consistent with our experimental results, higher SNRPA1 levels are associated with decreased metastasis-free survival and relapse-free survival in multiple cohorts of breast cancer patients, both individually and in meta-analyses (Figs. 7A–C and S7A). Our analyses indicate that SNRPA1 expression may be upregulated in breast cancer possibly through genomic alterations, as SNRPA1 expression is significantly positively correlated (Pearson r = 0.4, p < 10–100) with SNRPA1 copy number in the TCGA breast cancer dataset and tends towards copy number gains (25 amplifications vs. 4 deep deletions; Fig. S7B). Furthermore, SNRPA1 levels are significantly increased across breast tumor stage and tumor grade (Figs. S7C–D), and there is a slight but significant difference in SNRPA1 expression across breast cancer subtypes (Fig. S7E).
Fig. 7. SNRPA1 expression is associated with poor clinical outcome in breast cancer.
(A) Kaplan-Meier survival curve showing correlation between tumor SNRPA1 levels and distant metastasis-free survival in a collection of breast cancer patient cohorts; n = 1746. (B) Kaplan-Meier survival curve showing correlation between tumor SNRPA1 levels and relapse-free survival in a collection of breast cancer patient cohorts; n = 1979. P-values calculated using log-rank test. (C) Shown are distribution of 10-year relapse-free survival P values (two-sided log rank test results reported as –logP for positive association and logP for negative) of the correlation of SNRPA1 expression and clinical outcome in the listed 27 breast cancer datasets. Red bars show associations that pass the statistical threshold (–log p > 1.8, FDR-corrected two-sided log-rank test), orange bars are trending positive, and blue bars are trending negative. The statistical threshold was adjusted as 27/number of datasets. For statistically significant datasets, the hazard ratio is also included at the top of each bar. (D) RT-qPCR was used to measure SNRPA1 mRNA levels across clinical samples composed of normal breast tissue and tissue from the indicated breast tumor stages; n = 96. (E) RT-qPCR was used to measure Ψ of PLEC exon 31 across clinical samples composed of normal breast tissue and tissue from the indicated breast tumor stages; n = 89. P-values calculated using one-tailed Mann-Whitney U-test. (F) SNRPA1 immunohistochemistry of tissue slices in an array of 56 sections across breast cancer progression (Cooperative Human Tissue Network, CHTN_BRCaProg2 TMA). Each section was scored from low (0) to high (2); scoring is displayed as a stacked bar graph stratified by tumor stage. P-value was calculated using a χ2 test. Also shown are representative images from each group. NB: non-neoplastic breast tissue; DCIS: ductal carcinoma in situ; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma; LNM: lymph node metastatic carcinoma.
Importantly, a multivariate survival analysis revealed that the association between increased SNRPA1 expression and poor survival persists even after controlling for covariates commonly used as prognostic factors, e.g., tumor stage and subtype (Fig. S7F). Interestingly, this same analysis showed a highly significant effect (hazard ratio = 1.38, p = 0.006, Cox proportional hazards model) of SNRPA1 expression levels on patient outcome compared to the expression levels of other core components of the spliceosome U2 complex (Fig. S7F).
To further validate these findings, we performed RT-qPCR for SNRPA1 in a panel of 96 clinical samples from different stages of the disease, and observed an upregulation of SNRPA1 expression as breast cancer stage increases (Fig. 7D). Together, these analyses and experiments support the clinical relevance of our findings in models of breast cancer progression. Notably, we used isoform specific primers to measure inclusion of PLEC exon 31 in the same panel of 96 clinical samples, and observed significantly higher PLEC exon 31 inclusion in late-stage breast cancer (log2 fold change 4.8 between stage IV and I, p = 6×10–6; one-tailed Mann-Whitney U-test; Fig. 7E). This is consistent with the Plectin ex31-containing isoform acting as a promoter of breast cancer metastasis in our in vivo mouse experiments. Finally, we performed immunohistochemistry for SNRPA1 on a tissue microarray with representation of all breast cancer stages; this showed a significant association between SNRPA1 expression and disease progression (3/7 of samples scoring higher SNRPA1 staining in metastatic breast cancer versus 0/14 in non-neoplastic breast tissue, p < 10–4, χ2 test; Fig. 7F). We note that this association is likely not limited to breast cancer, as expression of SNRPA1 and its splicing targets are associated with poor prognosis across multiple cancer types (Figure S8).
Discussion
Here, we have uncovered a non-canonical role for SNRPA1 in the regulation of alternative splicing that is mediated through its interaction with an RNA structural element. This RNA structural code consists of one of the first identified structural elements that plays a direct role in alternative splicing, and highlights the value of taking structural information into account when delineating the behavior of post-transcriptional regulatory networks. We also discovered that the S3E-binding protein SNRPA1 acts as a promoter of breast cancer metastasis in in vivo models, and found a significant association between SNRPA1 expression and clinical outcome in multiple cohorts of breast cancer patients.
Interestingly, although SNRPA1 is a core component of U2 snRNP, our data indicate that it can also act to regulate specific alternative splicing events in breast cancer through interactions with S3E elements. Our results suggest that SNRPA1 performs this function outside of its role as a core U2 snRNP factor, similar to what has been shown for U1 snRNP A, which has a spliceosome-independent function in controlling its own mRNA poly(A) site choice (49). Notably, SNRPA1 has also been reported to play a role in preventing the formation of R-loops, thereby promoting DNA repair (50), indicating that SNRPA1 functions in multiple capacities inside the cell. It would be interesting to examine the interplay between these multiple roles of SNRPA1 to explore any co-regulatory functions they may have in normal physiology and disease states.
Splicing dysregulation can be achieved via a variety of mechanisms; notably, mutations in core spliceosome factors and dysregulation of the activity of splicing regulators have both been well documented (8). Cancer-associated mutations in specific core splicing factors have been associated primarily with hematological malignancies, and the mechanisms that underlie this specificity have been traced to alterations in the splicing of specific transcripts (8).
It is interesting to note that many of the cancer-associated mutations in spliceosomal factors occur in components of U2 snRNP, of which SNRPA1 is a core element. Our results indicate that among the core U2 snRNP factors, SNPRA1 modulation has a strong clinical association with breast cancer progression and plays a functional role in metastasis. However, the function of SNRPA1 in promoting metastasis is outside of its role as a component of U2 snRNP and is mediated through direct interactions between SNRPA1 and its target RNAs via a structural cis-element. The crucial role of cis-regulatory elements in AS regulation is well documented (22), and mutations in these regions that affect the splicing of specific oncogenes and tumor suppressor genes have been reported (51, 52). However, to date, most studies have not taken global RNA structural information into account. We have used the abundant and underutilized structural information contained in cis-regulatory elements to identify this non-canonical pathway of splicing regulation mediated by SNRPA1.
Increasing the efficacy of therapeutic targeting of tumors is another challenge that can be addressed by the discovery of splicing regulatory networks involved in cancer pathogenesis. Identification of the aberrant alternative splicing events generated by pathologic activity of splicing factors such as SNRPA1 has the potential to uncover novel exon-exon junctions that encode tumor-specific antigens. In addition to possible new biological roles, such tumor-specific antigens are attractive targets for personalized cancer therapies (53). Antisense oligonucleotides (ASOs) are one type of molecule used to target pathological splicing variants, and have been recently implemented in the treatment of the genetic disorders spinal muscular atrophy and Duchenne muscular dystrophy (54). This same approach could be used to target transcripts that encode metastasis-promoting protein isoforms in cancer. Notably in this regard, SNRPA1 modulates the alternative splicing of exon 31 of PLEC, resulting in a switch in the ratio of expressed Plectin isoforms, which in turn impacts the metastatic capacity of breast cancer cells. Interestingly, the Δexon 31 Plectin isoform lacks the rod domain, a domain involved in Plectin oligomerization, but is not necessary for normal tissue function (55). This work suggests that the rod domain-containing Plectin isoform promotes breast cancer progression and may provide a viable target for splicomer therapies. Finally, the association of specific aberrant splicing patterns in tumors with patient outcome may lead to a better stratification of patients in the clinic.
MATERIALS AND METHODS
Cell culture
The MDA-MB-231 (MDA-parental, ATCC HTB-26) human breast cancer cell line, its highly metastatic derivative, MDA-LM2 (57), and 293LTV cells (Cell BioLabs LTV-100) were cultured in DMEM medium supplemented with 10% FBS, penicillin, streptomycin and amphotericin. The HCC1806-LM2 cell line (an in vivo selected highly lung metastatic derivative of the HCC1806 breast cancer line (ATCC CRL-2335)) was cultured in RPMI-1640 medium supplemented with 10% FBS, penicillin, streptomycin and amphotericin. Gene knockdowns were performed using stably expressed shRNA constructs or siRNA transfection. Gene overexpression was performed using stably expressed ORFs. Splicing inhibition was performed by morpholino transfection. Endogenous S3Es were deleted by transfecting gRNA-Cas9 RNPs (IDT), programmed with gRNAs flanking S3E regions.
Animal experiments
All animal studies were performed according to IACUC guidelines (IACUC approval number AN179718). Age-matched female NOD/SCID gamma mice (Jackson Labs, 005557) were used for metastatic lung colonization assays and orthotopic tumor growth assays. Metastasis was measured by bioluminescent imagining (IVIS) and histology performed by hematoxylin and eosin staining of lung tissue sections. Orthotopic tumor volume was assessed by caliper measurement.
Cell proliferation and invasion
Cell proliferation was measured by counting viable cells using trypan blue stain at day 1, 3 and 5 post-seeding. Cell invasion was assessed by seeding cells in Matrigel-coated transwell invasion chambers (Corning), and 24 hours post-seeding counting the number of cells that had invaded to the basal side of the transwell insert.
RT-qPCR
Total RNA was isolated using the Norgen Biotek total RNA isolation kit. SuperScript III (Invitrogen) was used to synthesize cDNA, and fast SYBR green master mix (Applied Biosystems) or Perfecta SYBR green supermix (QuantaBio) was used to perform qPCR. HPRT1 and 18S were used as endogenous controls.
Western Blotting
Cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail (Pierce). Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed using target-specific antibodies. Antibodies: beta-tubulin (Proteintech 66240–1-Ig), PLEC (Bethyl A304–506A).
High-throughput sequencing data generation
RNA-seq libraries were prepared with ScriptSeq v2 (Illumina), QuantSeq (Lexogen), or SMARTer Pico v2 (Takara) kits. Splicing isoform quantification was performed using MISO and rMATS packages. The bind-n-Seq experiment was performed as previously described (45), with minor modifications. Targeted DMS-MaPseq was performed as previously described (33), with minor modifications. SNRPA1 irCLIP-seq was performed as described (39), with minor modifications.
S3E pulldown and mass spectrometry
S3E-containing RNAs were incubated with MDA-LM2 cell nuclear lysate. After washing, the proteins bound to the S3E RNA bait were identified using mass spectrometry.
RNA EMSA
RNA probes containing the ERRFI1 and PLEC S3E-containing regions, along with RNU2, were in vitro transcribed 3’ biotinylated. These labeled RNAs were mixed with recombinant SNRPA1 protein (Abcam) and the resulting complexes were resolved by PAGE.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A. Goga, S. F. Tavazoie, and S. Tavazoie for reading earlier versions of this manuscript. We acknowledge the UCSF Center for Advanced Technology (CAT) and the Rockefeller Genomics Resource Center for high throughput sequencing and other genomic analyses. We thank S. F. Tavazoie for the gift of the HCC1806 and HCC1806-LM2 cell lines. We thank B. Hann and the Preclinical Therapeutics core as well as the Laboratory Animal Resource Center (LARC) at UCSF. We acknowledge support from our colleagues at the Helen Diller Family Comprehensive Cancer Center and the Breast Oncology Program.
Funding: This work was supported by grants from the NIH (R00CA194077 and R01CA240984) and ACS (130920-RSG-17-114-01-RMC) to H.G. and grants from CIHR (PJT-155966 and PJT-173317) and resource allocations from Compute Canada to H.S.N. This research was also supported by funding from the UCSF Helen Diller Family Comprehensive Cancer Center Breast Oncology Program (the content is solely the responsibility of the authors). L.F. was supported by NIH training grant T32CA108462-15. A.N. was supported by DoD PRCRP Horizon Award W81XWH-19-1-0594. S.Z. was supported by an HHMI medical research fellowship. H.S.N. holds a CIHR Canada Research Chair. M.D. and F.K.M. were supported by an MRC career development award to F.K.M. (MR/P009417/1). This research was also supported by funding from the UCSF Helen Diller Family Comprehensive Cancer Center Breast Oncology Program.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: All sequencing data have been deposited in the GEO database under the accession number GSE160957. The source code and documentation for pyTEISER has been deposited at Zenodo (56). Proteomics data has been deposited in the PRIDE data base under the accession number PXD024139.
REFERENCES AND NOTES
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ, Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. , Alternative isoform regulation in human tissue transcriptomes. Nature. 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anczuków O, Krainer AR, Splicing-factor alterations in cancers. RNA N. Y. N. 22, 1285–1301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Climente-González H, Porta-Pardo E, Godzik A, Eyras E, The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 20, 2215–2226 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Marzese DM, Manughian-Peter AO, Orozco JIJ, Hoon DSB, Alternative splicing and cancer metastasis: prognostic and therapeutic applications. Clin. Exp. Metastasis. 35, 393–402 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Siegfried Z, Karni R, The role of alternative splicing in cancer drug resistance. Curr. Opin. Genet. Dev. 48, 16–21 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Song X, Zeng Z, Wei H, Wang Z, Alternative splicing in cancers: From aberrant regulation to new therapeutics. Semin. Cell Dev. Biol. 75, 13–22 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Urbanski LM, Leclair N, Anczuków O, Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA. 9, e1476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Marabti E, Younis I, The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front. Mol. Biosci. 5 (2018), doi: 10.3389/fmolb.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, et al. , SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell. 60, 105–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Gao XD, Lee J-H, Huang H, Tan H, Ahn J, et al. , Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 28, 1191–1203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J, et al. , Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc. Natl. Acad. Sci. U. S. A. 115, E3426–E3435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman SN, Viner C, Rogan PK, Splicing mutation analysis reveals previously unrecognized pathways in lymph node-invasive breast cancer. Sci. Rep. 4, 7063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. , CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121, 1064–1074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bemmo A, Dias C, Rose AAN, Russo C, Siegel P, Majewski J, Exon-level transcriptome profiling in murine breast cancer reveals splicing changes specific to tumors with different metastatic abilities. PloS One. 5, e11981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alajati A, Sausgruber N, Aceto N, Duss S, Sarret S, Voshol H, et al. , Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res. 73, 5320–5327 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Harvey SE, Xu Y, Lin X, Gao XD, Qiu Y, Ahn J, et al. , Coregulation of alternative splicing by hnRNPM and ESRP1 during EMT. RNA N. Y. N. 24, 1326–1338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvinge H, Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett. 592, 2987–3006 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hiller M, Zhang Z, Backofen R, Stamm S, Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 3, e204 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kastner B, Will CL, Stark H, Lührmann R, Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol, a032417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y, Rio DC, Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus CJ, Graveley BR, RNA structure and the mechanisms of alternative splicing. Curr. Opin. Genet. Dev. 21, 373–379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, Salavati R, et al. , Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 485, 264–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodarzi H, Zhang S, Buss CG, Fish L, Tavazoie S, Tavazoie SF, Metastasis-suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature. 513, 256–260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, et al. , SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. 113, 10322–10327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, et al. , RNA–protein interaction detection in living cells. Nat. Methods. 15, 207–212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litterman AJ, Kageyama R, Le Tonqueze O, Zhao W, Gagnon JD, Goodarzi H, et al. , A massively parallel 3’ UTR reporter assay reveals relationships between nucleotide content, sequence conservation, and mRNA destabilization. Genome Res. 29, 896–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, et al. , Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest. 115, 44–55 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz Y, Wang ET, Airoldi EM, Burge CB, Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guenther U-P, Yandek LE, Niland CN, Campbell FE, Anderson D, Anderson VE, et al. , Hidden specificity in an apparently nonspecific RNA-binding protein. Nature. 502, 385–388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. , ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuter JS, Mathews DH, RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 11, 129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S, DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods. 14, 75–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. , Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 526, 131–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett AL, et al. , Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Invest. 128, 1371–1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams SG, Hall KB, Binding affinity and cooperativity control U2B”/snRNA/U2A’ RNP formation. Biochemistry. 53, 3727–3737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black DL, Chabot B, Steitz JA, U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 42, 737–750 (1985). [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Manley JL, Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 3, 1553–1561 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA, irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods. 13, 489–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price SR, Evans PR, Nagai K, Crystal structure of the spliceosomal U2B"-U2A’ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 394, 645–650 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Urdaneta EC, Beckmann BM, Fast and unbiased purification of RNA-protein complexes after UV cross-linking. Methods. 178, 72–82 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, et al. , Genomewide discovery of human splicing branchpoints. Genome Res. 25, 290–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fica SM, Nagai K, Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 24, 791–799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z, Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 5120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB, RNA Bind-n-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Mol. Cell. 54, 887–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anastasi S, Lamberti D, Alemà S, Segatto O, Regulation of the ErbB network by the MIG6 feedback loop in physiology, tumor suppression and responses to oncogene-targeted therapeutics. Semin. Cell Dev. Biol. 50, 115–124 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Segatto O, Anastasi S, Alemà S, Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J. Cell Sci. 124, 1785–1793 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Sutoh Yoneyama M, Hatakeyama S, Habuchi T, Inoue T, Nakamura T, Funyu T, et al. , Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur. J. Cell Biol. 93, 157–169 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Boelens W, The human U1 snRNP-Specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 72, 881–892 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Tanikawa M, Sanjiv K, Helleday T, Herr P, Mortusewicz O, The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis. 5, e280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calabrese C, Davidson NR, Demircioglu D, Fonseca NA, He Y, Kahles A, et al. , Genomic basis for RNA alterations in cancer. Nature. 578, 129–136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiraishi Y, Kataoka K, Chiba K, Okada A, Kogure Y, Tanaka H, et al. , A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28, 1111–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS, Alternative tumour-specific antigens. Nat. Rev. Cancer. 19, 465–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D, Mastaglia FL, Fletcher S, Wilton SD, Precision Medicine through Antisense Oligonucleotide-Mediated Exon Skipping. Trends Pharmacol. Sci. 39, 982–994 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Ketema M, Secades P, Kreft M, Nahidiazar L, Janssen H, Jalink K, et al. , The rod domain is not essential for the function of plectin in maintaining tissue integrity. Mol. Biol. Cell. 26, 2402–2417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.goodarzilab/pyteiser: v1.0.0 (Zenodo, 2021; https://zenodo.org/record/4533305). [Google Scholar]
- 57.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. , Genes that mediate breast cancer metastasis to lung. Nature. 436, 518–24 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. , A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124, 1283–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Fish L, Navickas A, Culbertson B, Xu Y, Nguyen HCB, Zhang S, et al. , Nuclear TARBP2 Drives Oncogenic Dysregulation of RNA Splicing and Decay. Mol. Cell. 75, 967–981.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butter F, Scheibe M, Mörl M, Mann M, Unbiased RNA-protein interaction screen by quantitative proteomics. Proc. Natl. Acad. Sci. U. S. A. 106, 10626–10631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. , Global quantification of mammalian gene expression control. Nature. 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Dermit M, Dodel M, Lee FCY, Azman MS, Schwenzer H, Jones JL, et al. , Subcellular mRNA Localization Regulates Ribosome Biogenesis in Migrating Cells. Dev. Cell. 55, 298–313.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB, Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 9 (2014), doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deigan KE, Li TW, Mathews DH, Weeks KM, Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. 106, 97–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.