Abstract
Introduction
Antimicrobial peptides are potential therapeutics as anti-bacteria, anti-viruses, anti-fungi, or anticancers. However, they suffer from a short half-life and drug resistance which limit their long-term clinical usage.
Methods
Herein, we captured the encapsulation of antimicrobial peptide HA-FD-13 into boron nitride nanotube (BNNT) (20,20) and its release due to subsequent insertion of BNNT (14,14) with molecular dynamics simulation.
Results
The peptide-BNNT (20,20) van der Waals (vdW) interaction energy decreased to −270 kcal·mol−1 at the end of the simulation (15 ns). However, during the period of 0.2–1.8 ns, when half of the peptide was inside the nanotube, the encapsulation was paused due to an energy barrier in the vicinity of BNNT and subsequently the external intervention, such that the self-adjustment of the peptide allowed full insertion. The free energy of the encapsulation process was −200.12 kcal·mol−1, suggesting that the insertion procedure occurred spontaneously.
Discussion
Once the BNNT (14,14) entered into the BNNT (20,20), the peptide was completely released after 83.8 ps. This revealed that the vdW interaction between the BNNT (14,14) and BNNT (20,20) was stronger than between BNNT (20,20) and the peptide; therefore, the BNNT (14,14) could act as a piston pushing the peptide outside the BNNT (20,20). Moreover, the sudden drop in the vdW energy between nanotubes to the value of the −1300 Kcal·mol−1 confirmed the self-insertion of the BNNT (14,14) into the BNNT (20,20) and correspondingly the release of the peptide.
Keywords: drug delivery, boron nitride nanotubes, molecular dynamics simulation, drug release, antimicrobial peptide
Introduction
Peptides are oligomers of short-chain amino acid sequences interconnected by amide or peptide bonds formed through dehydration condensation reaction.1,2 They are generally classified into four main groups based on their structures, including α-helical, β-sheet, the extended structures, and loops.3 The desired peptides for a given target having tailored chemical structures and activities can be obtained through appropriate genetics and chemical engineering.4 Depending on their microstructure and functional groups,5 diverse peptides have been developed for various applications in biomedicine,6 bioengineering, and nanotechnology.7,8 Because of their highly selective character and being relatively safe, peptides have been widely considered in pharmaceutical research.9
Bioactive peptides are a class of peptides that largely contribute to biological processes and have considerable physiological health benefits.10 They may act as hormones or drugs given their activities, such as anti-oxidative, mineral binding, anti-hypertensive, antimicrobial, anti-thrombotic, and immunomodulatory.11–13 Antimicrobial peptides have emerged as anti-infective drugs with the ability to protect living organisms against the invasion of pathogens.14 The antimicrobial activities of these peptides depend on their features, such as their charge, hydrophobicity, chain conformation, and polar angle.15–17 However, most antimicrobial peptides possess an amphipathic structure; they are cationic at physiological pH and accumulate onto and subsequently cause the disintegration or permeabilization of the bacterial membrane.18,19 These antimicrobial peptides edge over conventional antibiotics because of their broad range of antimicrobial activities (anti-bacteria, anti-viruses, anti-fungi, and anti-cancers) and their lower drug resistance.20
Besides the advantages mentioned above for antimicrobial peptides, some drawbacks such as short half-life, toxicity, hemolytic activity, drug resistance, and the blood‐brain barrier would limit their long-term usage in clinical applications.21–23 For example, the antimicrobial peptide indolicidin rich with tryptophan would lead to hemolytic anemia, despite providing a broad antimicrobial activity window.24 As another example, gram-negative bacterial pathogens (CAMPS) would resist cationic antimicrobial peptides through surface remodeling (polysaccharide modification, phospholipid modification, and capsule production), forming biofilms, efflux pumps, proteolytic degradation, binding and sequestering CAMPs, and modulation of CAMP expression.25 In this sense, nano-based drug delivery systems have been introduced and practiced to overcome barriers and thereby support peptide-based therapy by preventing antimicrobial drug resistance,26,27 sustain and target drug release,28,29 facilitating the penetration of the drug into the cells, bacteria, and tissues,30 and drug protection from oxidization, destructive reactions, and environmental tension.31
Boron nitride nanotubes (BNNT)-based drug delivery systems have attracted considerable attention as nanocarriers due to their distinct features such as their large interior volume adequate for drug encapsulation,32,33 their ability to surface functionalization for targeted delivery,28,29 non-toxicity, and biocompatibility.34,35 Several theoretical reports have addressed the performance of BNNT as a drug carrier. Table 1 summarizes the research performed on BNNT playing the role of drug carrier. For instance, Mirhaji et al studied the effect of water/ethanol ratio on the encapsulation process of the anticancer drug of docetaxel inside the Armchair BNNT (13, 13) utilizing molecular dynamics (MD) simulation.36 The Lennard-Jones energies calculated for the systems of BNNT-docetaxel attained at water/ethanol ratio of 60%, and 75% were 0 and −239 kcal·mol−1, respectively. Moreover, van der Waals (vdW) interaction between docetaxel and the BNNT took minimum at these ratios. Likewise, Khatti and Hashemianzadeh captured the penetration rate of the platinum drug into the BNNT and the functionalized BNNT with 18 hydroxyl groups on one edge.37 The hydroxyl groups facilitated the encapsulation of platinum into BNNT cavity through both hydrogen bonding and the van der Waals interaction between the drug and functionalized BNNT. Similar behavior is reported for the encapsulation of gemcitabine inside BNNT (18, 0) by Roosta et al.38 MD simulation proved spontaneous insertion of the drug into the cavities of the BNNT, featured by a negative vdW interaction energy value of −46.1 kcal·mol−1. They also quantified the release performance of encapsulated gemcitabine using fullerene (C60), which resulted in a total potential energy difference of −140.1 kcal mol−1.
Table 1.
The Summary of the Researches Performed on Evaluation of BNNT as Drug Carrier in Water at 300K and 1atm
| Author | Simulated System | Method | Calculated Energy |
|---|---|---|---|
| Mortazavifar et al39 | Hydroxyurea (HU) drug into BNNT (6,6) | MD DFT |
Adsorption energy=−34.36 kJ·mol−1 Interaction energy=−130.73 kJ·mol−1 Deformation energy=96.37 kJ·mol−1 vdW energy between HU and BNNT=−20 kJ·mol−1 |
| Roosta et al40 | Cisplatin drug into BNNT | DFT Monte carlo |
vdW energy =12.84 kcal·mol−1 Solvation free energy of drug-BNNT= −4.128 kcal·mol−1 Electrostatic energy =−2803.992 Kcal·mol−1 |
| Hasanzade and Raissi41 | 6-Thioguanine on BNNT with length and diameter of about 13.64 and 5.14Å | DFT | Adsorption energy= –47.31 Kj·mol−1 Deformation energy of BNNT= 179.41 kJ·mol−1 Solvation energy= –142.53 kJ·mol−1 Deformation energy of drug= 88.18 Kj·mol−1 |
| Zarghami et al33 | Protein SmtA into BNNT (28,28) | MD | vdW energy between protein and BNNT=−189.63 kcal·mol−1 Free energy of the protein=−160 kcal·mol−1Potential energy of the protein=−1900 kcal·mol−1 |
| Zarghami et al42 | The peptide cRW3 into BNNT (12,12) | MD | vdW energy between protein and BNNT= −142.7 kcal·mol−1 Free energy of the protein=−90 kcal·mol−1 Potential energy of the peptide=−50 kcal·mol−1 |
| Iranmanesh-Zarandy and Dehestani43 | Paracetamol Drug on BNNT (10,10),length=2.2nm | MD | Free energy= −1085.91 kJ·mol−1 vdW energy between drug and BNNT= −239.219 kJ·mol−1 Lennard-Jones energy=−2000 kJ·mol−1 Bonded energy= −973.56 kJ·mol−1 Binding free energy= −67.06 kJ·mol−11 |
| Sedghamiz et al44 | Mechlorethamine drug into BNNT (10,0) | MD | Total energy=−7500 kcal·mol−1 vdW energy = 1700 kcal·mol−1 Electrostatic energy= −10,550 kcal·mol−1 |
| Roosta et al38 | Gemcitabine in BNNT (18,0) | MD | Total potential energy=- 948.3 kcal·mol−1 Internal energy= −905.7 kcal·mol−1 Non-bond energy= −42.5 kcal·mol−1 VdW energy between drug and BNNT =- 42.2 kcal·mol−1 Electrostatic energy between drug and BNNT = −0.4 kcal·mol−1 |
| El Khalifi et al45 | Carboplatin drug in BNNT (10,10) | MD | The changes of Interaction enrgy between BNNT and water during the insertion of the drug into the BNNT= 650 kcal·mol−1 |
| Mirhaji et al36 | Docetaxel drug in BNNT (13,13) | MD | Potential energy=−700,000 kcal·mol−1 Lennard-Jones energy=−150 kcal·mol−1 |
| Khatti and Hashemianzadeh37 | Platinum drug in BNNT(40,0), length=14 Å | MD | vdW energy =−43 kcal·mol−1 Electrostatic energy=0.018 kcal·mol−1 |
| Mehrjouei et al46 | Cisplatin in BNNT (12,12) | MD | vdW energy= - kcal·mol−1 Electrostatic energy=−55 kcal·mol−1 Total energy=−62 kcal·mol−1 |
| Saikia et al47 | The protein Pyrazinamide on BNNT (5,5) | MD DFT |
Adsorption energy = −0.854eV |
Theoretical investigations on the BNNT-drug systems proved the presence of non-bonded interaction energies such as vdW and electrostatic interactions between the drug and BNNT supporting the encapsulation and release processes. The MD simulation, in this regard, is a promising tool that has been widely applied to investigate the performance of complex nano-sized systems bridging diverse areas of cell biology and materials science.48,49 Besides the great ability that the MD offers to evaluate the thermal and mechanical features of nanostructures.50–55 The 13-amino acid peptide (named HA-FD-13) with the ID code of 2l24 (chosen from protein data bank (https://www.rcsb.org/structure/2L24)) is an α-Helical antimicrobial peptide (α-AMP) derived from the fusion domain (FD) of the hemagglutinin (HA) of the influenza virus. This antimicrobial peptide has the positive charge needed for penetration into the anionic microbial membranes.56 However, like other kinds of antibiotics, drug resistance arising from long-term and overdose drug usage would limit its repeated usage. Therefore, the AMP-nanocarrier complexes are used to overcome this obstacle. In the present work, the MD simulation was applied to model and pattern the encapsulation process of α-AMP inside the BNNT (20,20) by calculating the vdW interactions between the BNNT and the peptide. Computational analyses were based on the potential of mean force (PMF) of the encapsulated peptide. Then, the release behavior of the encapsulated peptide in the presence of the BNNT (14,14), a releasing agent which acts as piston pushing out the peptide from BNNT (20,20), was evaluated by computing the distances between the center of mass (CoM) of the peptide and nanotubes and vdW interaction energy between the BNNT (14,14) and BNNT (20,20).
Simulation Method
In the present study, MD simulation of the encapsulation and release processes of the peptide HA-FD-13 into and from the BNNT was performed through the Large-Scale Atomic/Molecular Simulator (LAMMPS) software57 by applying CHARMM27 force field.58 Tersoff potential was used to consider the interaction between boron and nitrogen atoms of nanotubes.59 The partial charge of the B and N atoms are 1 and −1, respectively. Here the BNNT was neutralized, and the total charge was approximately zero. The temperature and the pressure of the systems were adjusted to about 300K and 101.3 KPa through applying NPT Ensemble. The inner and outer cutoff distances for the Lennard-Jones potential and Coulombic potential were considered 8 and 12 Ȧ, respectively. Lennard-Jones’ parameters for cross vdW interactions between non-bonded atoms were estimated using Lorentz–Berthelot combination rule.60 The visualization was obtained using visual molecular dynamics (VMD).61 The α-Helical antimicrobial peptide (α-AMP) named HA-FD-13 (containing amino acids ILE-PHE-GLY-ALA-ILE-ALA-GLY-PHE-ILE-LYS-ASN-ILE-TRP-NH2) with the ID code of 2l24 was chosen from the protein data bank. This antimicrobial peptide has a positive charge so that it can penetrate the anionic microbial membranes. The simulation steps of this research are described as following:
1) At the first step, the insertion process of the peptide HA-FD-13 into the BNNT and subsequently the stability of encapsulated peptide HA-FD-13 in BNNT was studied. Considering the size of HA-FD-13, the BNNT with chirality of armchair (20,20) and the length and diameter of 40 Ȧ and 27.14 Ȧ, respectively, have been selected as a nanocarrier according to the size of the antimicrobial peptide, which means that the minimum possible size of the BNNT was chosen so that the peptide would be encapsulated in the cavity of the nanotube beside the capability of the BNNT to penetrate cells. At the beginning of this simulation, the peptide HA-FD-13 was situated at the initial distance of 2 Ȧ from the nanotube. The axial direction of the nanotube was parallel to the z-axis of the simulation box. The complex of BNNT/HA-FD-13 was immersed in the simulation box consisting of TIP3P water molecules and counter-ions to neutralize the simulated solution with periodic boundary conditions. To assess the peptide’s encapsulation process, in the first stage, the system’s minimization was done while the nanotube and the peptide were fixed. Second, the system was relaxed in the NPT Ensemble at 300K. Next, the MD runs were performed in the NVT ensemble for 15 ns with the time step of 1 fs while the nanotube and nanotube and the peptide were fixed and unfixed, respectively. The vdW interaction between the peptide HA-FD-13 and the BNNT was calculated according to the below equality as:61
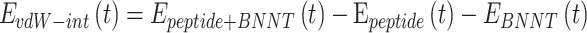 |
(1) |
where 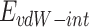 refers to vdW energy between HA-FD-13 and the nanotube,
refers to vdW energy between HA-FD-13 and the nanotube, 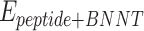 is vdW interaction of HA-FD-13 combined with the BNNT.
is vdW interaction of HA-FD-13 combined with the BNNT. 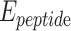 and
and 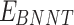 stand for vdW energies of HA-FD-13 and the BNNT, respectively.
stand for vdW energies of HA-FD-13 and the BNNT, respectively.
The root-mean-square deviation (RMSD) of the peptide was calculated to verify the changes in its conformation during the encapsulation process. The RMSD (in length units) was obtained from below Eq:62
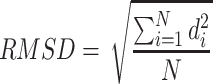 |
(2) |
where di is the distance between atom i in the two structures, and N is the total number of equivalent atoms.
To evaluate the stability of the encapsulated peptide inside BNNT, an external force was loaded on the encapsulated HA-FD-13 along the z-axis of the nanotube to pull it out from the BNNT in the direction opposite to the penetrating process. The spring constant k and pulling velocity were chosen equal to 15 kcal mol−1 Ȧ−2 and 0.005 Ȧ ps−1, respectively.63 The pulling process was simulated ten times to compute the potential of mean force profile (PMF) using Jarzynski’s equality as below:58
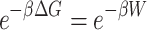 |
(3) |
where ΔG and W correspond to the free energy discrepancy between two states and the performed work on the system, respectively. β is equal to (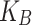 T)−1 where
T)−1 where 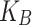 stands for the Boltzmann constant.
stands for the Boltzmann constant.
2) At the second step, the release behavior of the encapsulated peptide from the BNNT (20,20) using the BNNT (14,14), having a diameter of 18.52 Å was verified. The BNNT (14,14) was placed at the initial distance of 2 Ȧ from the BNNT (20,20), which acted as a piston pushing out the peptide from BNNT (20,20). The axial directions of nanotubes were parallel to the z-axis of the simulation box. The minimization of the system was done in the NVT ensemble at 300 K while nanotubes were fixed. Then, the MD run was performed in the NPT ensemble for 0.15 ns with the time step of 1 fs. The vdW interaction between the BNNT (20,20) and the BNNT (14,14) was computed as below:61
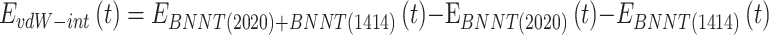 |
(4) |
Where 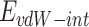 refers to vdW energy between BNNT (20,20) and BNNT (14,14),
refers to vdW energy between BNNT (20,20) and BNNT (14,14), 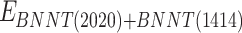 is vdW interaction of BNNT (20,20) combined with BNNT (14,14).
is vdW interaction of BNNT (20,20) combined with BNNT (14,14). 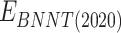 and
and 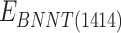 stand for vdW energies of BNNT (20,20) and the BNNT (14,14), respectively.
stand for vdW energies of BNNT (20,20) and the BNNT (14,14), respectively.
Results and Discussion
Localization of the Peptide HA-FD-13 Within the BNNT-Peptide Complex
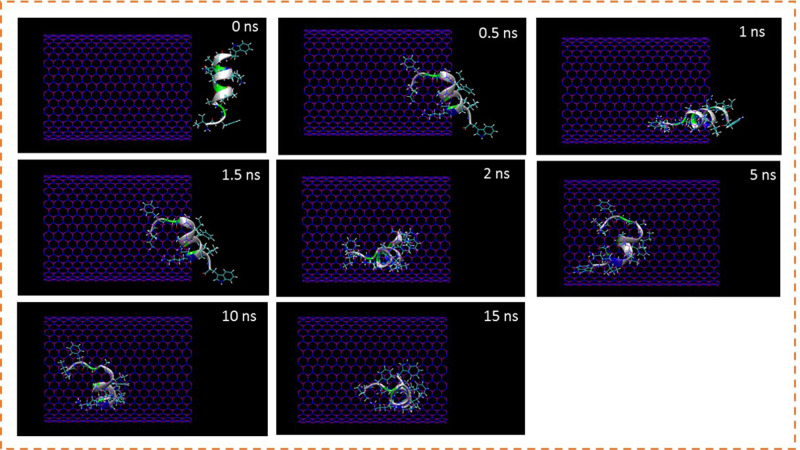
The insertion process of the peptide HA-FD-13 inside the cavity of BNNT (20,20) was verified through the MD simulation. The snapshots of this process were captured using VMD software to show the peptide positions at various times in the simulation box (Figure 1). As shown in Figure 1, the peptide HA-FD-13 was successfully encapsulated inside the BNNT; however, it experienced a slower insertion process during the period of 0.2 ns 1.8 ns of simulation time in which half of the peptide was inside the BNNT. This occurred due to the ability of the peptide to self-adjust to pass the maximum energy barrier in the vicinity of the nanotube. Moreover, the peptide remained stably encapsulated through the π-π interaction as a kind of vdW interaction between the conjugated aromatic rings of the peptide and the interior walls of the nanotube up to the end of the simulation time (15 ns).
Figure 1.
Representative snapshots of insertion of the peptide HA-FD-13 into an armchair (20,20) BNNT at various times. For clarity, molecules of water have not been shown.
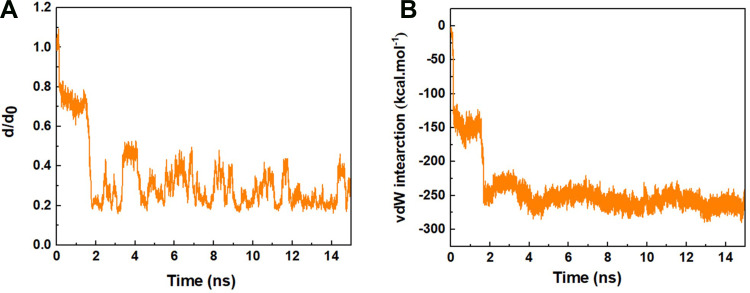
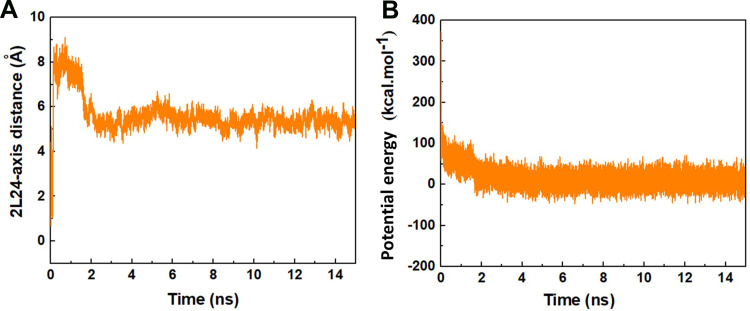
Figure 2A represents the normalized center of mass (CoM) distance between the peptide HA-FD-13 and the BNNT (20,20), d/d0, as a function of simulation time. The sudden two-step decrease of the d/d0 value from 1 to 0.2 at the beginning of the simulation up to 1.8 ns showed that the peptide was immediately under the influence of the vdW interaction between the peptide and the BNNT, which subsequently caused the peptide adsorption toward the cavity of the nanotube. The negligible change in the value of the d/d0 during the period between the 0.2 ns and 1.8 ns corresponded to the self-adjusting of the peptide residues to pass the energy barrier illustrated in Figure 1. A similar decreasing trend in the value d/d0 at the beginning of the simulation was reported in work performed by Kang et al.63 They observed that the protein SmtA experienced the conformational variations to be self-adjusted before entering the carbon nanotube (CNT). After the complete encapsulation of the peptide HA-FD-13 inside the BNNT at 2 ns, the normalized CoM distance between the peptide and the BNNT fluctuated around the specified range of 0.2–0.5 up to the end of the simulation at 15 ns which revealed the stability of the confined peptide inside the nanotube.
Figure 2.
(A) d/d0 (normalized CoM distance where d0 is the initial CoM distance) between the peptide HA-FD-13 and BNNT as a function of simulation time, (B) vdW interaction between the peptide HA-FD-13 and the BNNT (20,20) as a function of simulation time.
In accordance with the neutralized nature of the BNNT (20,20), it was concluded that the value of electrostatic interactions such as dipole-dipole and hydrogen bonding interactions between the BNNT and the peptide was obtained zero. Hence, the vdW interaction energy was considered as adsorbance energy of the peptide HA-FD-13 inside the BNNT. As can be observed in Figure 2B, the vdW interaction energy between the peptide and the BNNT dwindled dramatically in two steps: first, it was decreased from the value of 0 kcal·mol−1 to the approximate value of −150 kcal·mol−1 at initial 0.2 ns of the simulation, after the plateau in vdW energy value for the duration of 1.6 ns, for the second decrease, the vdW interaction energy dropped to the value of −270 kcal·mol−1. The observed plateau in the curve of vdW energy was in excellent agreement with the changes in the value of d/d0 where half of the peptide was inside the BNNT, and it did not move towards the cavity of the nanotube, and subsequently, the peptide-BNNT vdW interaction energy was not significantly varied for the duration of 1.6 ns. In other words, the presence of barrier energy arising from the higher accumulation of hydrogen bonding networks in the vicinity of the BNNT led to the 1.6 ns pause in the encapsulation process. A similar energy barrier in the vicinity of the BNNT is observed in our previous work in which the encapsulation of the peptide cRW3 into the BNNT is investigated.42 After completing the peptide encapsulation, the vdW energy between the peptide and the BNNT fluctuated smoothly in the minimum specified range due to the higher π-π interaction between the conjugated aromatic rings of the peptide and the interior walls of the BNNT. The decreasing trend observed for the encapsulation process of the drug inside BNNT has been similarly reported by Mehrjouei et al.46 They reported that the vdW energy between the Cisplatin and BNNT decreased and reached the value of −2282 kJ·mol−1 at the end of the simulation.
Calculation of the Free Energy by MD Simulation
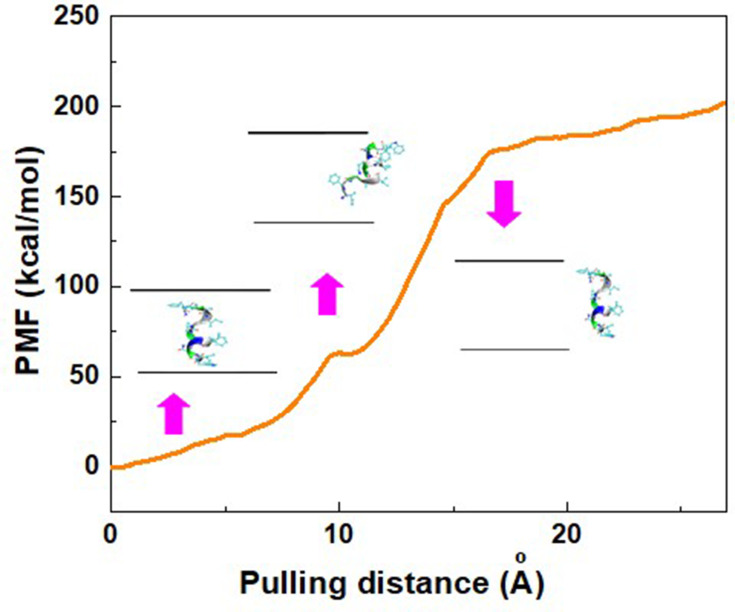
After the completion of the simulation, when the peptide HA-FD-13 was completely inserted in the cavity of BNNT (20,20) at 15 ns, the PMF profile of the confined peptide was computed by pulling it out using the MD simulation at the speed of 0.005 Ȧ ps−1, which was chosen in accordance with the speed of the encapsulation process. This simulation was repeated five times to obtain the average value of work (W) at each pulling distance, depicted as a PMF curve in Figure 3. Moreover, the peptide positions along the z-axis of the BNNT are illustrated in this Figure As shown, the free energy of this simulated system increased during the pulling distance and reached the maximum value of 200.12 kcal·mol−1 at 24 Å. This increasing trend of the PMF showed that the encapsulation process of the peptide into the BNNT was a spontaneous phenomenon with a free energy of −200.12 kcal·mol−1. A similar spontaneous process was reported in work performed by Veclani and Melchior.64 They calculated the free energy value of −9.5 Kcal·mol−1 for the adsorption of the ciprofloxacin on the surface of CNTs. As another proof, the protein SmtA has been inserted into BNNT (28,28) with the free energy of −160 kJ·mol−1 in previous work.33
Figure 3.
Potential of mean force (PMF) for the system of BNNT-peptide computed from five pullings through the MD simulation. The images represent that the positions of the peptide HA-FD-13 correspond to the z-coordinate along the BNNT at some key position. Pink arrows connect the exact position of the peptide with corresponding pulling distance.
As another observation, the PMF did not experience the significant changes and remained plateau at the pulling distance higher than 24 Å when the peptide was out of the BNNT. It can be explained that, at a pulling distance higher than 24 Å, when the distance between the CoM of the peptide and the entrance of the BNNT was higher than the cutoff distance (12 Å), the effect of the vdW interaction between the BNNT disappeared so that the displacement of the peptide to the higher distances did not affect the value of the PMF and free energy. The revealed small peak at the pulling distance of 10 Å, when half of the peptide was in the BNNT, was due to the energy barrier present in the vicinity of the BNNT.
According to the definition of the free energy of the hole system containing water molecules, BNNT (20,20) and drug Isatin (ΔG = ΔH - TΔS where ΔH and ΔS are the variations of total enthalpy and total entropy, respectively), it was observed that the value of -TΔS equaled to ΔG-ΔH=−200.12-(−1818.28) = 1618.16 kcal·mol−1. Therefore, it can be concluded that the encapsulation process was an enthalpy-driven phenomenon due to the greater contribution of the enthalpy change to the free energy of the whole system while the decrement of the entropy value during the encapsulation process.65
The Time-Dependent Conformation of the Peptide HA-FD-13
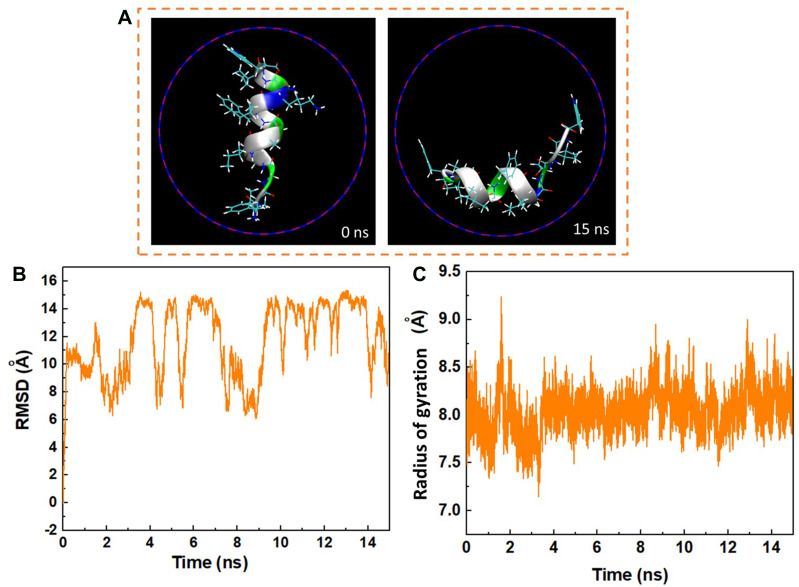
The variations in the conformation of the peptide HA-FD-13 at 0 and 15 ns of the MD simulation are depicted in Figure 4A. The left image of Figure 4 is related to the immersed peptide in an aqueous solution in its natural conformation at 0 ns. The right image corresponds to the state in which the peptide conformation was adjusted to the interior wall of the BNNT at 15 ns. Similar rearrangement and alteration in the conformation of the confinement drug doxorubicin inside the CNTs with various chirality and diameters were reported by Zhang et al.66
Figure 4.
(A) Axial views of the peptide HA-FD-13 at 0 ns and 15 ns in the MD simulation. For the sake of clarity, molecules of water have not been shown. (B) Root mean square deviation (RMSD) of the peptide HA-FD-13 as a function of simulation time. (C) Radius gyration of the peptide HA-FD-13 as a function of simulation time in the BNNT/HA-FD-13 complex.
The root mean square deviation (RMSD) relating to the changes in the conformation of peptide HA-FD-13 in BNNT (20,20) as a function of simulation time is demonstrated in Figure 4B. As observed in this figure, the RMSD varied significantly by 0.2 ns, revealing that the peptide conformation variations are influenced by the high peptide-BNNT vdW interaction. Then, for the duration of 1.6 ns, the RMSD did not significantly vary, which shows that not only did the peptide not experience the intense conformational changes, but also it did not move towards the BNNT. After this duration, when the peptide was inserted inside the BNNT, the value of RMSD fluctuated in the specified range so that the aromatic rings of the peptide can be placed parallel to the interior wall of the BNNT, causing the strong π–π stacking interaction. A similar structural variation during the encapsulation of the ssDNA oligomer inside the CNT was observed by Shen et al.67 They observed that the value of RMSD of the biomolecule slightly fluctuated, resulting in the creation of the π–π stacking interaction after complete encapsulation. In another work,68 they reported that the variations in RMSD for the encapsulated biomolecule occurred due to the different affinities of hydrophobic and the hydrophilic residues to the solvent and the nanotube, respectively. In two works done by Raffaini and Ganazzoli,69,70 they reported that the conformation of the encapsulated oligopeptide adjusted to the inner wall of the single wall CNT.
In addition to RMSD values, the alteration of the gyration radius of the peptide HA-FD-13 during the encapsulation process provides beneficial details about the peptide’s structural changes. Figure 4C shows the alteration of the gyration radius of the peptide during the simulation time. As observed, the peptide’s gyration radius varied continuously in the range of 7–9 Å due to the strong peptide-BNNT vdW interaction as the simulation started.
The alterations in the distance between the CoM of the peptide HA-FD-13 and the central axis of the BNNT as a function of simulation time are represented in Figure 5A. As seen, the peptide in the simulation box was situated in a position so that the CoM of the peptide was adjusted on the central axis of the BNNT at the beginning of the simulation. After initiation of the simulation, the peptide moved toward one side of the BNNT resulted in the immediate enhancement of the peptide-axis distance to the value of 9 Å. This sudden movement was attributed to the strong vdW interaction between the BNNT wall and the peptide. However, this conformation was not permanent and altered through the peptide’s adsorption to the other side wall of the nanotube as a sudden drop of the peptide-axis distance. After the complete self-insertion of the peptide in BNNT, the distance between the CoM of the peptide and the central axis of the nanotube slightly fluctuated, which showed the stability of the peptide in nanotube due to peptide-BNNT vdW interaction.
Figure 5.
(A) The distance between the CoM of the peptide HA-FD-13 and the central axis of BNNT as a function of simulation time. (B) The potential energy of the peptide HA-FD-13 in the BNNT-peptide complex as a function of simulation time.
Figure 5B shows the alteration in the potential energy of the peptide HA-FD-13 during the procedure of encapsulation into the BNNT. It was seen that the potential energy of the peptide decreased, which was in favor of the self-insertion of the peptide. Moreover, the continuous fluctuation of the potential energy in the range from −50 Kcal·mol−1 to 50 Kcal·mol−1for encapsulated peptide was due to and continuous conformational changes of the peptide in the cavity of the nanotube under the effect of vdW.63
The Release of the Encapsulated Peptide HA-FD-13 Using the BNNT (14,14)
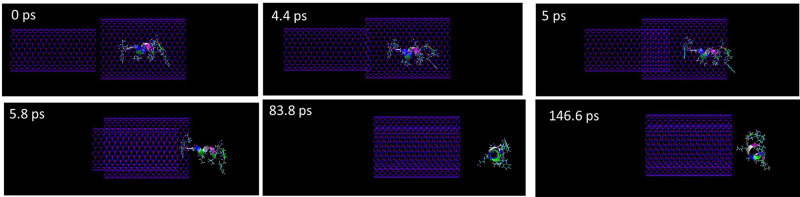
Besides the stability of the encapsulated peptide, the release of the encapsulation process is another key parameter to evaluate the nano-based drug delivery system. Therefore, the release of the encapsulated peptide HA-FD-13 from the BNNT (20,20) using the BNNT (14,14) was simulated. Figure 6 demonstrates the snapshots of the release process at various simulation times. The BNNT (14,14) was situated at a distance of 2 Å from the BNNT (20,20) to adjust their z-axis. It can be observed that, as the BNNT (14,14) started to enter into the BNNT (20,20), the peptide was released so that the release process was completed at 83.8 ps. This observation confirmed that the vdW interaction between the BNNT (14,14) and BNNT (20,20) was stronger than between BNNT (20,20) and the peptide so that the BNNT (14,14) could act as a piston pushing the peptide outside the BNNT (20,20). Moreover, when the encapsulated peptide was released its structure changed to the initial conformation (α-Helix) in the absence of CNT. This means that once this peptide was released, it could serve as an antibiotic.
Figure 6.
Representative snapshots of the release of the encapsulated peptide HA-FD-13 using an armchair (14,14) BNNT at various times. For clarity, molecules of water have not been shown.
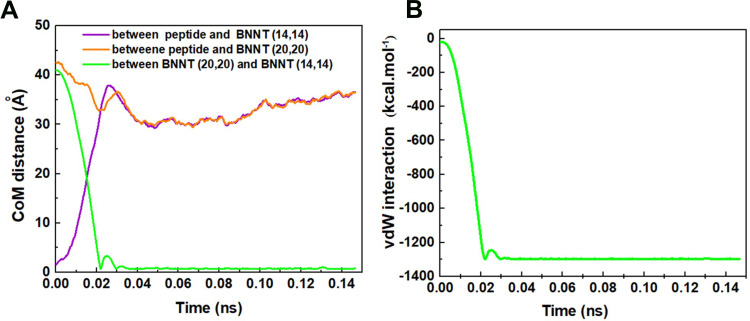
The changes of CoM distance between the peptide HA-FD-13 and BNNT (20,20), peptide HA-FD-13 and BNNT (14,14), BNNT (20,20), and BNNT (14,14) as a function of simulation time are depicted in Figure 7A. Expectedly, by entering the BNNT (14,14) into the BNNT (20,20), their CoM was adjusted. The curves of the CoM distances between the peptide and both nanotubes overlapped. Moreover, the CoM distance between the BNNT (20,20) and BNNT (14,14) decreased dramatically to the value of zero. The changes of vdW energies between the BNNT (20,20) and (14,14) during the simulation are illustrated in Figure 7B. The sudden drop of the vdW energy between nanotubes to the value of the −1300 Kcal·mol−1 is in excellent agreement with the self-insertion of the BNNT (14,14) into the BNNT (20,20) and subsequently release of the peptide.
Figure 7.
(A) the changes of CoM distance between the peptide HA-FD-13 and BNNT (20,20), peptide HA-FD-13 and BNNT (14,14), BNNT (20,20) and BNNT (14,14), (B) vdW interaction energy between the BNNT (14,14) and BNNT (20,20) as a simulation time.
Conclusion
In summary, the encapsulation process of the antimicrobial peptide HA-FD-13 with the ID code of 2l24 inside the BNNT (20,20) was studied, followed by the release behavior of the encapsulated peptide using the BNNT (14,14) and by employing the molecular dynamics simulation. It was observed that the peptide-BNNT (20,20) van der Waals (vdW) interaction energy and the normalized center of mass (CoM) distance between the peptide and the BNNT (d/d0) dwindled to the values of the −270 kcal·mol−1 and 0.3, respectively, at the end of the simulation (15 ns). However, during the period of 0.2–1.8 ns, when half of the peptide was inside the nanotube, the encapsulation process was paused due to the energy barrier in the vicinity of the BNNT and subsequently the self-adjustment of the peptide. The obtained negative value of −200.12 kcal·mol−1 for free energy confirmed that the insertion procedure occurred spontaneously. The changes of the root mean square deviation (RMSD) showed that for the duration of 1.6 ns, not only did the peptide not experience the intense conformational changes, it also did not move toward the BNNT. After the peptide’s complete encapsulation, the value of RMSD fluctuated in the specified range so that the aromatic rings of the peptide situated parallel to the interior wall of the BNNT causing the strong π–π stacking interaction. The decreasing trend of the peptide’s potential energy was in favor of the self-insertion of the peptide inside the BNNT. In terms of the peptide release, it was observed that, as the BNNT (14,14) started to enter into the BNNT (20,20), the peptide was released so that the release process was completed at 83.8 ps. Moreover, the sudden drop of the vdW energy between nanotubes to the value of the −1300 Kcal·mol−1 is in great agreement with the self-insertion of the BNNT (14,14) into the BNNT (20,20) and subsequently release of the peptide.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bodanszky M. Peptide Chemistry. Berlin, Germany: Springer-Verlag; 1993. [Google Scholar]
- 2.Mondal S, Das S, Nandi AK. A review on recent advances in polymer and peptide hydrogels. Soft Matter. 2020;16:1404–1454. [DOI] [PubMed] [Google Scholar]
- 3.Powers J-PS, Hancock RE. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24(11):1681–1691. doi: 10.1016/j.peptides.2003.08.023 [DOI] [PubMed] [Google Scholar]
- 4.Tamerler C, Sarikaya M. Genetically designed peptide-based molecular materials. Acs Nano. 2009;3(7):1606–1615. doi: 10.1021/nn900720g [DOI] [PubMed] [Google Scholar]
- 5.Knorr R, Trzeciak A, Bannwarth W, Gillessen D. New coupling reagents in peptide chemistry. Tetrahedron Lett. 1989;30(15):1927–1930. doi: 10.1016/S0040-4039(00)99616-3 [DOI] [Google Scholar]
- 6.Kuzmicheva G, Belyavskaya V. Peptide phage display in biotechnology and biomedicine. Biochem (Mosc) Suppl Ser B Biomed Chem. 2017;11(1):1–15. doi: 10.1134/S1990750817010061 [DOI] [PubMed] [Google Scholar]
- 7.De La Rica R, Matsui H. Applications of peptide and protein-based materials in bionanotechnology. Chem Soc Rev. 2010;39(9):3499–3509. doi: 10.1039/b917574c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzmicheva G, Belyavskaya V. Peptide phage display in biotechnology and biomedicine. Biomed Khim. 2016;62(5):481–495. doi: 10.18097/PBMC20166205481 [DOI] [PubMed] [Google Scholar]
- 9.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 10.Honigberg MC, Chang L-S, McGuire DK, Plutzky J, Aroda VR, Vaduganathan M. Use of glucagon-like peptide-1 receptor agonists in patients with type 2 diabetes and cardiovascular disease: a review. JAMA Cardiol. 2020;5(10):1182–1190. doi: 10.1001/jamacardio.2020.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahidi F, Zhong Y. Bioactive peptides. J AOAC Int. 2008;91(4):914–931. doi: 10.1093/jaoac/91.4.914 [DOI] [PubMed] [Google Scholar]
- 12.Daliri EB-M, Oh DH, Lee BH. Bioactive peptides. Foods. 2017;6(5):32. doi: 10.3390/foods6050032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P, Cui Q, Zhang Y, et al. A review of pedal peptide/orcokinin-type neuropeptides. Curr Protein Pept Sci. 2021;22(1):41–49. doi: 10.2174/1389203721666201109112758 [DOI] [PubMed] [Google Scholar]
- 14.Liu W-P, Chen Y-H, Ming X, Kong Y. Design and synthesis of a novel cationic peptide with potent and broad-spectrum antimicrobial activity. Biomed Res Int. 2015;2015:578764. doi: 10.1155/2015/578764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay S, Prasad AB, Mehta CH, Nayak UY. Antimicrobial peptide polymers: no escape to ESKAPE pathogens—a review. World J Microbiol Biotechnol. 2020;36(9):1–14. doi: 10.1007/s11274-020-02907-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitchner MA, Santiago-Ortiz LE, Necelis MR, et al. Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim Biophys Acta. 2019;1861(10):182984. doi: 10.1016/j.bbamem.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gough M, Hancock R, Kelly NM. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect Immun. 1996;64(12):4922–4927. doi: 10.1128/IAI.64.12.4922-4927.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei J, Sun L, Huang S, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11(7):3919. [PMC free article] [PubMed] [Google Scholar]
- 21.Moravej H, Moravej Z, Yazdanparast M, et al. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist. 2018;24(6):747–767. doi: 10.1089/mdr.2017.0392 [DOI] [PubMed] [Google Scholar]
- 22.Pardridge WM. Recent developments in peptide drug delivery to the brain. Pharmacol Toxicol. 1992;71(1):3–10. doi: 10.1111/j.1600-0773.1992.tb00512.x [DOI] [PubMed] [Google Scholar]
- 23.Pizzolato-Cezar LR, Okuda-Shinagawa NM, Machini MT. Combinatory therapy antimicrobial peptide-antibiotic to minimize the ongoing rise of resistance. Front Microbiol. 2019;10:1703. doi: 10.3389/fmicb.2019.01703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbalakshmi C, Bikshapathy E, Sitaram N, Nagaraj R. Antibacterial and hemolytic activities of single tryptophan analogs of indolicidin. Biochem Biophys Res Commun. 2000;274(3):714–716. doi: 10.1006/bbrc.2000.3214 [DOI] [PubMed] [Google Scholar]
- 25.Band VI, Weiss DS. Mechanisms of antimicrobial peptide resistance in gram-negative bacteria. Antibiotics. 2015;4(1):18–41. doi: 10.3390/antibiotics4010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AW. Biofilms and antibiotic therapy: is there a role for combating bacterial resistance by the use of novel drug delivery systems? Adv Drug Deliv Rev. 2005;57(10):1539–1550. doi: 10.1016/j.addr.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 27.Zarrintaj P, Ramsey JD, Samadi A, et al. Poloxamer: a versatile tri-block copolymer for biomedical applications. Acta Biomater. 2020. [DOI] [PubMed] [Google Scholar]
- 28.Yoosefian M, Etminan N. Density functional theory (DFT) study of a new novel bionanosensor hybrid; tryptophan/Pd doped single walled carbon nanotube. Physica E Low Dimens Syst Nanostruct. 2016;81:116–121. doi: 10.1016/j.physe.2016.03.009 [DOI] [Google Scholar]
- 29.Skandani AA, Al-Haik M. Reciprocal effects of the chirality and the surface functionalization on the drug delivery permissibility of carbon nanotubes. Soft Matter. 2013;9(48):11645–11649. doi: 10.1039/C3SM52126E [DOI] [PubMed] [Google Scholar]
- 30.Ramos MADS, Da Silva PB, Spósito L, et al. Nanotechnology-based drug delivery systems for control of microbial biofilms: a review. Int J Nanomedicine. 2018;13:1179. doi: 10.2147/IJN.S146195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernkop-Schnürch A, Bratengeyer I, Valenta C. Development and in vitro evaluation of a drug delivery system protecting from trypsinic degradation. Int J Pharm. 1997;157(1):17–25. doi: 10.1016/S0378-5173(97)00198-1 [DOI] [Google Scholar]
- 32.Arsawang U, Saengsawang O, Rungrotmongkol T, et al. How do carbon nanotubes serve as carriers for gemcitabine transport in a drug delivery system? J Mol Graph Model. 2011;29(5):591–596. doi: 10.1016/j.jmgm.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 33.Zarghami MD, Bagheri B, Nasiriasayesh A, et al. Insight into the self-insertion of a protein inside the boron nitride nanotube. ACS Omega. 2020;5(49):32051. doi: 10.1021/acsomega.0c05080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Wu P, Rousseas M, et al. Boron nitride nanotubes are noncytotoxic and can be functionalized for interaction with proteins and cells. J Am Chem Soc. 2009;131(3):890–891. doi: 10.1021/ja807334b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen ML, Zettl A. The physics of boron nitride nanotubes. Phys Today. 2010;63(11):34–38. doi: 10.1063/1.3518210 [DOI] [Google Scholar]
- 36.Mirhaji E, Afshar M, Rezvani S, Yoosefian M. Boron nitride nanotubes as a nanotransporter for anticancer docetaxel drug in water/ethanol solution. J Mol Liq. 2018;271:151–156. doi: 10.1016/j.molliq.2018.08.142 [DOI] [Google Scholar]
- 37.Khatti Z, Hashemianzadeh SM. Boron nitride nanotube as a delivery system for platinum drugs: drug encapsulation and diffusion coefficient prediction. Eur J Pharm Sci. 2016;88:291–297. doi: 10.1016/j.ejps.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 38.Roosta S, Nikkhah SJ, Sabzali M, Hashemianzadeh SM. Molecular dynamics simulation study of boron-nitride nanotubes as a drug carrier: from encapsulation to releasing. RSC Adv. 2016;6(11):9344–9351. doi: 10.1039/C5RA22945F [DOI] [Google Scholar]
- 39.Mortazavifar A, Raissi H, Shahabi M. Comparative prediction of binding affinity of hydroxyurea anti-cancer to boron nitride and carbon nanotubes as smart targeted drug delivery vehicles. J Biomol Struct Dyn. 2019;37(18):4852–4862. doi: 10.1080/07391102.2019.1567385 [DOI] [PubMed] [Google Scholar]
- 40.Roosta S, Hashemianzadeh SM, Ketabi S. Encapsulation of cisplatin as an anticancer drug into boron-nitride and carbon nanotubes: molecular simulation and free energy calculation. Mater Sci Eng C. 2016;67:98–103. doi: 10.1016/j.msec.2016.04.100 [DOI] [PubMed] [Google Scholar]
- 41.Hasanzade Z, Raissi H. Carbon and boron nanotubes as a template material for adsorption of 6-Thioguanine chemotherapeutic: a molecular dynamics and density functional approach. J Biomol Struct Dyn. 2020;38(3):697–707. doi: 10.1080/07391102.2019.1585951 [DOI] [PubMed] [Google Scholar]
- 42.Zarghami MD, Bagheri B, Yousefi F, et al. Boron nitride nanotube as an antimicrobial peptide carrier: a theoretical insight. Int J Nanomedicine. 2021;16:1837. doi: 10.2147/IJN.S298699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iranmanesh‐Zarandy Z, Dehestani M. Molecular dynamics simulation of paracetamol drug adsorption on boron nitride nanotube: effects of temperature, nanotube length, diameter, and chirality. ChemistrySelect. 2019;4(27):7866–7873. doi: 10.1002/slct.201900644 [DOI] [Google Scholar]
- 44.Sedghamiz E, Jamalizadeh E, Hosseini SMA, Sedghamiz T, Zahedi E. Molecular dynamics simulation of boron nitride nanotube as a drug carrier. Arab J Sci Eng. 2014;39(9):6737–6742. doi: 10.1007/s13369-014-1228-y [DOI] [Google Scholar]
- 45.El Khalifi M, Bentin J, Duverger E, Gharbi T, Boulahdour H, Picaud F. Encapsulation capacity and natural payload delivery of an anticancer drug from boron nitride nanotube. Phys Chem Chem Phys. 2016;18(36):24994–25001. doi: 10.1039/C6CP01387B [DOI] [PubMed] [Google Scholar]
- 46.Mehrjouei E, Akbarzadeh H, Shamkhali AN, Abbaspour M, Salemi S, Abdi P. Delivery of cisplatin anticancer drug from carbon, boron nitride, and silicon carbide nanotubes forced by Ag-nanowire: a comprehensive molecular dynamics study. Mol Pharm. 2017;14(7):2273–2284. doi: 10.1021/acs.molpharmaceut.7b00106 [DOI] [PubMed] [Google Scholar]
- 47.Saikia N, Jha AN, Deka RC. Interaction of pyrazinamide drug functionalized carbon and boron nitride nanotubes with pncA protein: a molecular dynamics and density functional approach. RSC Adv. 2013;3(35):15102–15107. doi: 10.1039/c3ra42534g [DOI] [Google Scholar]
- 48.Katiyar RS, Jha PK. Molecular simulations in drug delivery: opportunities and challenges. Wiley Interdiscip Rev Comput Mol Sci. 2018;8:e1358. [Google Scholar]
- 49.Singh A, Vanga SK, Orsat V, Raghavan V. Application of molecular dynamic simulation to study food proteins: a review. Crit Rev Food Sci Nutr. 2018;58(16):2779–2789. doi: 10.1080/10408398.2017.1341864 [DOI] [PubMed] [Google Scholar]
- 50.Bagheri B, Dehaghani MZ, Karami Z, et al. Correlation between surface topological defects and fracture mechanism of γ-graphyne-like boron nitride nanosheets. Comput Mater Sci. 2020;110152. [Google Scholar]
- 51.Dehaghani MZ, Mashhadzadeh AH, Salmankhani A, et al. Fracture toughness and crack propagation behavior of nanoscale beryllium oxide graphene-like structures: a molecular dynamics simulation analysis. Eng Fract Mech. 2020;235:107194. doi: 10.1016/j.engfracmech.2020.107194 [DOI] [Google Scholar]
- 52.Zarghami dehaghani M, Salmankhani A, Hamed mashhadzadeh A, Habibzadeh S, Abida O, Reza Saeb M. Fracture mechanics of polycrystalline beryllium oxide nanosheets: a theoretical basis. Eng Fract Mech. 2021;244:107552. doi: 10.1016/j.engfracmech.2021.107552 [DOI] [Google Scholar]
- 53.Bagheri B, Dehaghani MZ, Safa ME, et al. Fracture fingerprint of polycrystalline C3N nanosheets: theoretical basis. J Mol Graph Model. 2021;106:107899. doi: 10.1016/j.jmgm.2021.107899 [DOI] [PubMed] [Google Scholar]
- 54.Salmankhani A, Karami Z, Mashhadzadeh AH, et al. A theoretical scenario for the mechanical failure of boron carbide nanotubes. Comput Mater Sci. 2021;186:110022. doi: 10.1016/j.commatsci.2020.110022 [DOI] [Google Scholar]
- 55.Albooyeh A, Dadrasi A, Mashhadzadeh AH. Effect of point defects and low-density carbon-doped on mechanical properties of BNNTs: a molecular dynamics study. Mater Chem Phys. 2020;239:122107. doi: 10.1016/j.matchemphys.2019.122107 [DOI] [Google Scholar]
- 56.Zhu S, Aumelas A, Gao B. Convergent evolution-guided design of antimicrobial peptides derived from influenza A virus hemagglutinin. J Med Chem. 2011;54(4):1091–1095. doi: 10.1021/jm1010463 [DOI] [PubMed] [Google Scholar]
- 57.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J Comput Phys. 1995;117(1):1–19. doi: 10.1006/jcph.1995.1039 [DOI] [Google Scholar]
- 58.Park S, Schulten K. Calculating potentials of mean force from steered molecular dynamics simulations. J Chem Phys. 2004;120(13):5946–5961. doi: 10.1063/1.1651473 [DOI] [PubMed] [Google Scholar]
- 59.Los J, Kroes J, Albe K, Gordillo R, Katsnelson M, Fasolino A. Extended tersoff potential for boron nitride: energetics and elastic properties of pristine and defective h-BN. Phys Rev B. 2017;96(18):184108. doi: 10.1103/PhysRevB.96.184108 [DOI] [Google Scholar]
- 60.Hirschfelder JO, Curtiss CF, Bird RB, Mayer MG. Molecular Theory of Gases and Liquids. New York: Wiley; 1964. [Google Scholar]
- 61.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 62.Sargsyan K, Grauffel C, Lim C. How molecular size impacts RMSD applications in molecular dynamics simulations. J Chem Theory Comput. 2017;13:1518–1524. [DOI] [PubMed] [Google Scholar]
- 63.Kang Y, Liu Y-C, Wang Q, Shen J-W, Wu T, Guan W-J. On the spontaneous encapsulation of proteins in carbon nanotubes. Biomaterials. 2009;30(14):2807–2815. doi: 10.1016/j.biomaterials.2009.01.024 [DOI] [PubMed] [Google Scholar]
- 64.Veclani D, Melchior A. Adsorption of ciprofloxacin on carbon nanotubes: insights from molecular dynamics simulations. J Mol Liq. 2020;298:111977. doi: 10.1016/j.molliq.2019.111977 [DOI] [Google Scholar]
- 65.Zhang D, Gullingsrud J, McCammon JA. Potentials of mean force for acetylcholine unbinding from the alpha7 nicotinic acetylcholine receptor ligand-binding domain. J Am Chem Soc. 2006;128(9):3019–3026. doi: 10.1021/ja057292u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L, Peng G, Li J, et al. Molecular dynamics study on the configuration and arrangement of doxorubicin in carbon nanotubes. J Mol Liq. 2018;262:295–301. doi: 10.1016/j.molliq.2018.04.097 [DOI] [Google Scholar]
- 67.Shen J-W, Tang T, Wei X-H, et al. On the loading mechanism of ssDNA into carbon nanotubes. RSC Adv. 2015;5(70):56896–56903. doi: 10.1039/C5RA01941A [DOI] [Google Scholar]
- 68.Shen J-W, Wu T, Wang Q, Kang Y. Induced stepwise conformational change of human serum albumin on carbon nanotube surfaces. Biomaterials. 2008;29(28):3847–3855. doi: 10.1016/j.biomaterials.2008.06.013 [DOI] [PubMed] [Google Scholar]
- 69.Raffaini G, Ganazzoli F. Surface topography effects in protein adsorption on nanostructured carbon allotropes. Langmuir. 2013;29(15):4883–4893. doi: 10.1021/la3050779 [DOI] [PubMed] [Google Scholar]
- 70.Raffaini G, Ganazzoli F. Separation of chiral nanotubes with an opposite handedness by chiral oligopeptide adsorption: a molecular dynamics study. J Chromatogr A. 2015;1425:221–230. doi: 10.1016/j.chroma.2015.11.045 [DOI] [PubMed] [Google Scholar]