Abstract
Amyloid aggregation, formed by aberrant proteins, is a pathological hallmark for neurodegenerative diseases, including Alzheimer’s disease and Huntington’s disease. High-resolution holistic mapping of the fine structures from these aggregates should facilitate our understanding of their pathological roles. Here, we achieved label-free high-resolution imaging of the polyQ and the amyloid-beta (Aβ) aggregates in cells and tissues utilizing a sample-expansion stimulated Raman strategy. We further focused on characterizing the Aβ plaques in 5XFAD mouse brain tissues. 3D volumetric imaging enabled visualization of the whole plaques, resolving both the fine protein filaments and the surrounding components. Coupling our expanded label-free Raman imaging with machine learning, we obtained specific segmentation of aggregate cores, peripheral filaments together with cell nuclei and blood vessels by pre-trained convolutional neural network models. Combining with 2-channel fluorescence imaging, we achieved a 6-color holistic view on the same sample. This ability for precise and multiplex high-resolution imaging of the protein aggregates and their micro-environment without the requirement of labeling would open new biomedical applications.
Graphical Abstract
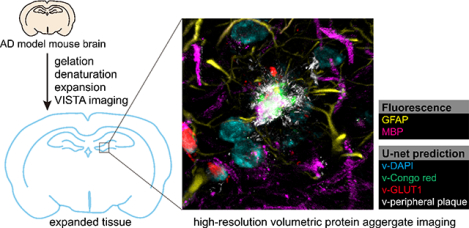
High spatial-resolution label-free imaging of in vivo protein aggregates by VISTA for studying the details of amyloid-β plaque morphologies. High multiplexity volumetric imaging was achieved by coupling VISTA imaging with U-net feature prediction.
Introduction
Protein aggregation is one major pathological hallmark in neurodegenerative diseases.(1–3) For example, in Alzheimer’s disease (AD), the amyloid plaques are made up of fibrillar aggregations containing misfolded Aβ peptides. There is, however, still much unknown about the mechanism of the structural formation and the neurotoxic effects of these aggregates in the neurodegenerative diseases.(4–6) For example, it remained controversial whether the oligomers of the misfolded proteins are more toxic than the densely-packed aggregates.(7–9) On the other hand, aggregates have also been shown to induce cellular malfunctions by physically disrupting functional proteins and organelles.(10) It is hence essential to obtain high-resolution holistic views of the protein aggregates together with their micro-environment to offer new structural-functional insights and to shed light on the pathologic understandings in neurodegeneration.
Extensive efforts have been devised to characterize the nanoscale structural details and to understand the functional implications of these protein aggregations.(11–17) For instance, with the combination of array tomography and stimulated emission depletion microscopy (AT-STED), ~0.1 μm resolution imaging on Aβ plagues has been shown.(12) In addition, single-molecule switching nanoscopy (SMSN) has demonstrated imaging of the fine features of the fibrillar arrangement in 30 μm thick brain sections.(11) However, super-resolution fluorescence imaging techniques (i.e. STORM, PALM, and STED) are still limited to a rather shallow penetration depth of within 100 μm and offer low multiplexity.(14,18) This limits the investigations of the native microenvironment around the plaques, which plays an irreplaceable role in AD progression.(19–23)
Recent reports have shown that AD pathology is also accompanied by neurofibrillary tangles and cerebral vasculature alterations in addition to the presence of Aβ plagues.(19–21) Moreover, reactive astrocytes and activated microglia are implicated in influencing the neuronal functions in AD.(22,23) Tissue clearing techniques, such as iDISCO, have been deployed for fluorescence imaging of the aggregates in deep tissues and to investigate the microenvironment of the plaques.(24–26) They indeed revealed remarkably complex morphology of the plagues and intricate interactions between the aggregates and their immediate environment, including blood vessels, microglia, and axons.(24–26) However, tissue labeling is generally prolonged and inhomogeneous, due to the slow penetration and inefficient targeting of the fluorescent probes.(27) In particular, the diffusion of large antibodies into the tissues often poses a rate-limiting step for the experiments.(27) Multiple rounds of screening are also often required to find an effective antibody with satisfying targeting efficiency and specificity.(24) Congo red, a smaller organic dye, presents better penetration, but has been shown to only stain positively for the cores of the plagues and could not reveal more detailed encompassing fibrillar structures.(28–30) It therefore remains highly demanded to have a novel imaging strategy that brings super-resolution imaging of the aggregates and their surroundings into deep tissues while addressing the issues arising from required fluorophore labeling inside tissues. Such high-resolution and high-throughput volumetric imaging should help offer a holistic view of various plaque morphology in their native environment together with their spatial interactions with the surrounding components.
Toward this goal, Raman spectro-microscopy provides specific chemical information in a label-free manner by probing chemical vibrations directly from the biomolecules.(31–34) Prominently, hyperspectral stimulated Raman scattering (hSRS) imaging has been applied to mapping the Aβ plaques in fresh tissues with high sensitivity and speed.(35) Utilizing the Amide I vibrational band, native and misfolded proteins were imaged and delineated. The spatial resolution of conventional stimulated Raman scattering (SRS) imaging is, nonetheless, limited to ~0.4 μm, which is much coarser than the size of the plague filaments and fibrils, which were suggested to be about 0.1 μm in diameter, in brain tissues.(11)
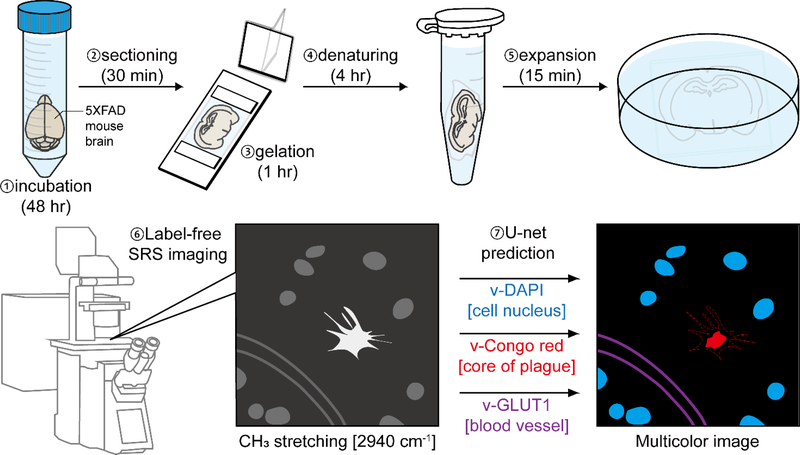
Recently, we developed a strategy for label-free super-resolved SRS imaging of endogenous protein-abundant structures in cells and tissues, by utilizing the Raman-compatible sample-polymer hybridization chemistry with subsequent isotropic sample expansion.(36) This new strategy, termed vibrational imaging of swelled tissues and analysis (VISTA), obtained an effective imaging resolution within 100 nm and offered a volumetric 3D view deep inside tissues.(36) Although sample expansion has gained rapid popularity for biological investigations,(37) so far no expansion imaging on protein aggregates has been reported. In this manuscript, we establish a VISTA-based pipeline for high-resolution, label-free, and specific interrogations of Aβ plagues, filaments with the nearby cell nuclei, and blood vessels throughout mouse tissues (Fig. 1). First, we established that VISTA could preserve the general structure of the protein aggregates and allow isotropic expansion of these dense structures for nanoscale morphological investigations. Second, with the integration of a pre-trained U-net convolutional neural network (CNN) from correlative VISTA and fluorescence imaging, high-resolution segmentation was also obtained for target-specific expanded imaging. Third, with tandem fluorescence microscopy, we achieved 6-color multiplex imaging for holistic visualization of the aggregate nano-morphology and their complex tissue surroundings of blood vessels, nuclei, myelin, astrocytes, and oligodendrocytes.
Fig. 1. Schematic illustration of proposed experimental workflow for high-resolution volumetric VISTA imaging of the amyloid-beta (Aβ) aggregates.
The brain tissues from 5XFAD mice are hybridized with the polymer gel and expanded following the VISTA protocol of incubation, gelation, denaturing, and expansion. The samples are subsequently examined by SRS imaging at 2940 cm−1 (i.e. VISTA imaging). The VISTA images are then segmented by the pre-trained U-net models with correlative VISTA images and fluorescence stains to predict different and distinct features, including cores of plagues (v-Congo red), cell nuclei (v-DAPI), and blood vessels (v-GLUT1).
Materials and methods
Stimulated Raman scattering (SRS) microscopy.
The pump and Stokes beams were produced by a picoEmerald laser system (Applied Physics & Electronics.) The tunable pump beam (770 nm – 990 nm, spectral bandwidth ~ 7 cm−1) and the Stokes beam (1031.2 nm, spectral bandwidth 10 cm−1 ) are produced at an 80 MHz repetition rate. The Stokes beam was modulated at 20 MHz by a built-in electro-optic modulator (EOM). The pump and Stokes beams are spatially and temporally coupled inside the picoEmerald laser system and introduced into an FV3000 microscope (Olympus). The beams were then focused by a 25x water objective (XLPLN25XWMP, 1.05 numerical aperture (NA), Olympus) onto the sample for imaging. Transmitted light was collected by an oil immersion condenser lens (1.4 NA, Olympus) and the Stokes beam was blocked out by a bandpass filter (893/209 BrightLine, 25 mm, Semrock) while the pump beam was guided to a 10 mm×10 mm Si photodiode (S3590–09, Hamamatsu). To increase the saturation threshold and to reduce the response time, the photodiode was reverse biased by 64 V. The output current was pre-filtered by a 19.2–23.6-MHz bandpass filter (BBP-21.4+, Mini-Circuits) and terminated with 50 Ω. The Raman loss signal of the pump beam was then demodulated by a lock-in amplifier (SR844, Stanford Research Systems or HF2LI, Zurich instrument) at the modulation frequency. The in-phase output is fed back into the Olympus IO interface box (FV30- ANALOG) of the microscope. Image acquisition speed is limited to 80 μs pixel dwell time with 30 μs time constant for the SR844 lock-in amplifier; and set to 20 μs pixel dwell time with 5 μs time constant for the HF2LI lock-in amplifier. The pump laser is tuned to 791.3 nm for imaging CH3 vibrational mode at 2940 cm-1. 16-bit grey scale images were acquired by Olympus Fluoview 3000 software. Volumetric images were acquired by a step size of 1 micron in the z-direction.
Reagents and materials.
Sodium acrylate (SA), acrylamide (AA), N,N′-methylenebisacrylamide (BIS, 2% in water), ammonium persulfate (APS), tetramethylethylenediamine (TEMED, Sigma-Aldrich), sodium dodecyl sulfate (SDS), Triton X-100, and deuterium oxide were obtained from Sigma-Aldrich. Tris(hydroxymethyl)aminomethane was obtained from Biosolve. Nuclease-free water and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Thermo Fisher. Paraformaldehyde (PFA, 16% in water) was purchased from Electron Microscopy Sciences.
Primary antibodies: anti-myelin basic protein in rat (ab7349) was purchased from Abcam; anti-glial fibrillary acidic protein in mouse (3670S) was purchased from Cell Signaling Technology; anti-glucose transporter 1 protein in rabbit (07–1401) was purchased from Sigma-Aldrich.
Secondary antibodies: goat anti-rat IgG, Alexa Fluor 568 (A-11077), goat anti-mouse IgG, Alexa Fluor 647 (A-21236); goat anti-rabbit IgG, Alexa Fluor 488 (A-11034) were purchased from Invitrogen.
Biological samples.
Cultured HeLa-CCL2 (ATCC) cells were cultured in DMEM (Gibco) with 10 percent fetal bovine serum (FBS, ThermoFisher) and 1 percent streptomycin penicillin (Sigma-Aldrich). Cells were seeded onto 14 mm glass-bottom microwell dishes (MatTek Corporation) or coverslips (12mm, #1.5, Fisher) for 24 h prior to transfection. Cells were first grown in regular DMEM complete medium until they reached 70–90% confluence. Transfection of 1-μg plasmids encoding mHtt-97Q-GFP was performed using Lipofectamine 3000 transfection reagent (ThermoFisher) according to the manufacturer’s manual. After 24–28 h of protein expression, the coverslips were harvest by fixing the cells in 30% AA in 4% PFA at 37 ˚C for 6–8 h. Further steps were carried out following the standard VISTA protocol.
9-months-old 5XFAD mouse (B6SJL-Tg (APPSwFlLon, PSEN1*M146L*L286V) 6799Vas/Mmjax) was purchased from the Mutant Mouse Resource & Research Centers and The Jackson Laboratory. All experimental procedures involving the use of live animals or their tissues were carried out in accordance with the NIH guidelines and approved by the Institutional Animal Care and Use Committee at the California Institute of Technology (Caltech). Rodent euthanasia via carbon dioxide narcosis was performed by filling the chamber with a flow of 100% CO2 in the order of 30–70% of the volume of the chamber per minute and by maintaining the flow for at least one minute after clinical death. Following confirmation of humane euthanasia, post-mortem tissue collection of the brain was performed by trained veterinary technical staff. The fresh mouse brain tissue was collected and fixed in 30% AA in 4% PFA under 4 °C for two days and 37 °C for one day before sectioning. It was cut into 150 μm sections by Leica VT 1200S vibratome and stored under 4 °C.
Sample gelation and expansion.
The gelation solution (7% SA, 20% AA, 0.1% BIS), initiator solution (10% APS) and accelerator solution (10% TEMED) were prepared by nuclease-free water and stored under −20 °C. The solutions were thawed on ice before the gelation process. Coverslips with the tissue sections or cells were placed between an extra layer of coverslips and immersed with 126 μL gelation solution, 7 μL of accelerator and initiator rapidly after the solution is mixed and vortexed, minimizing gelation prior to immersing the sample. The immersed sample was covered by a parafilm-wrapped coverslip, kept on a frozen heat block for 1 minute, and transferred to a 37 °C incubator for 1 hour. After gelation, the coverslip with sample-embedded gel was immersed in the denaturing buffer (200 mM SDS, 200 mM NaCl, and 50 mM Tris in nuclease-free water, pH 9) in a petri dish for 15 minutes at room temperature. After the sample-embedded gel detached from the coverslip, it was transferred into a 1.5 mL Eppendorf tube with 1 mL of denaturing buffer. The sample was denatured by heating to 70 °C for 3 hours and 95 °C for 1 hour in the tube. It was then transferred to H2O for initial expansion. The sample was kept in PBS under 4 °C until further needed.
Immunostaining.
The gel samples were transferred to a 1 mL PBST solution (1% Triton X-100 in PBS), and incubated with primary antibodies at 1:100–1:200 dilution under 37 °C for 24 hours, followed by washing with PBST three times at 37°C for 1–2 hours. The samples were then incubated with secondary antibodies at a 1:200 dilution with PBST at 37 °C for 16–18 hours, followed by washing with PBST three times at 37 °C for 1–2 hours before imaging.
Fluorescence imaging.
The fluorescence images of the processed samples with fluorescent labels were obtained with an 8 μs pixel dwell time by a 25×, 1.05 NA water-immersion objective with the Olympus Fluoview system. Single-photon confocal laser scanning imaging was performed with 488-, 561-, and 640-nm lasers (Coherent OBIS). The images were analyzed with Fiji or Imaris.
Feature prediction.
The machine learning prediction of the cellular features was based on a U-Net CNN convolutional neural network demonstrated in the work of Ounkomol et al.(38) Correlative fluorescence images and CH3 SRS images were collected on the same field of view with a z-direction step size of 1 μm. Before training, the SRS images were background subtracted by Fiji with rolling ball radius of 160 pixels, which is larger than the features in the images. The image sets were split randomly in a 1:3 ratio for testing and training sets. The images were subsampled into batches of 128 pixels x 128 pixels patches for training. The Adam optimizer optimized the mean squared error between the ground-truth (fluorescent) image and the predicted image with the learning rate set at 0.001. All models are trained for 50,000 epochs with a batch size of 32 images. All the training and predictions were run on a node equipped with Nvidia P100 GPU containing 16 GB of memory of High Performance Computing Center at Caltech.
Model accuracy.
The prediction accuracies of the models were quantified by the Pearson correlation coefficient (Pearson’s r) between the ground-truth images and the predicted images in the test sets. The Pearson’s r, is calculated as:
The Pearson’s r, for lectin and Congo red predictions were calculated for images that reached a minimum signal threshold, since the model could not predict random noise in the background.
Data and code availability.
All data supporting the findings of the present study are available in the article and its supplementary figures, or from the corresponding author upon request. The code for U-Net training, prediction, and evaluation in this paper is available at https://github.com/Li-En-Good/VISTA
Results and discussion
Characterizations of aggregate capture and expansion
We first characterized whether VISTA could be applied to imaging protein aggregates. Although we have recently successfully demonstrated the strategy of VISTA on visualizing protein-abundant structures in sample-hydrogel hybridized and expanded cells and tissues by SRS imaging at 2940 cm−1, there are two major concerns toward imaging of the aggregates that await validations. First, whether acrylamide monomer could efficiently penetrate into the dense aggregate structure (Fig. 1, step 1, incubation) to effectively capture and hybridize the proteins inside aggregates to the polymer gel network (Fig. 1, step 3, gelation). Second, whether the captured protein aggregates after the denaturing step (Fig. 1, step 4) could be expanded isotropically to achieve the desired resolution with well-preserved local structures (Fig. 1, step 5, expansion).
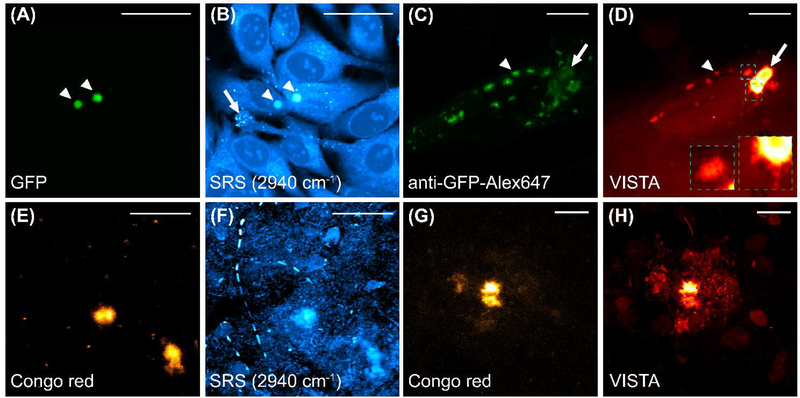
To establish VISTA on protein aggregates, we first tested whether the proteins inside the aggregates could be largely hybridized and retained onto the hydrogel network. We adopted two well-utilized model protein aggregate systems, the polyQ aggregates from the Huntington’s disease in HeLa cells (Fig. 2A–D) and the Aβ plaques from the AD in brain tissues from 5XFAD transgenic mice (Fig. 2E–H), which develop severe amyloid pathology.(39) We started on the cellular level, for which we transfected Hela cells with an mHtt-97Q-GFP plasmid to examine the sample-processing and expansion of the polyQ aggregates. Before sample treatment, fluorescence imaging of GFP confirmed the aggregation formation (Fig. 2A). Parallel SRS imaging at the 2940 cm−1 showed a clear contrast from the corresponding protein-rich aggregates (Fig. 2B). After sample treatment and expansion, the alexa647-anti-GFP antibody was used to capture the expanded polyQ aggregates (Fig. 2C) since GFP fluorescence is quenched during the protein denaturing procedure (Fig. 1, step 4). In parallel, VISTA imaging on the same field of view showed a clear contrast of both large and small aggregates (Fig. 2D, the representative ones are indicated by an arrow and an arrowhead, respectively), indicating an effective hybridization between the aggregates and the hydrogel. We note that the lower fluorescence signal in the large and much more dense aggregates (Fig. 2C, arrowed) is likely due to the decreased labeling efficiency from limited penetration of the large antibodies into the aggregates. We also note that the smaller bright spots in Fig. 2B (arrow indicated) are lipid droplets, which are not retained during the expansion process.
Fig. 2. SRS imaging at 2940 cm−1 for polyQ aggregates formed by the mHtt-97Q-GFP proteins (A-D) and Aβ plaques (E-H) with correlative fluorescence imaging of targeted labels before and after expansion.
(A-B) Correlative fluorescence imaging of GFP (A) and the SRS imaging at 2940 cm−1 of unprocessed live HeLa cells formed with polyQ aggregates (arrowheaded) after transfected with the mHtt-97Q-GFP plasmid. PolyQ aggregates are indicated by arrowheads and representative small lipids droplets are indicated by an arrow. (B) at the same field of view. (C-D) Correlative fluorescence imaging of Alexa647-anti-GFP antibody labeled polyQ aggregates (C) and SRS imaging at 2940 cm−1 (D, VISTA) on expanded HeLa cells. The arrowhead and arrow indicate a representative small and a large polyQ aggregate, respectively. Two zoomed-in views in colored dash boxes (D, insets, green, and cyan dash boxes designate the views from a large and a small aggregate respectively) show fine filaments at the outer shell of the aggregates. (E-F) Correlative fluorescence imaging of Congo red labeled Aβ aggregates (E) and the SRS imaging at 2940 cm−1 (F) of fixed and unprocessed 5XFAD mouse tissues at the same field of view. (G-H) Correlative fluorescence imaging of Congo red labeled Aβ aggregates (G) and the SRS imaging at 2940 cm−1 (H, VISTA) of processed and expanded 5XFAD mouse tissues. Merged images of correlative fluorescence and SRS sets are shown in Fig. S1. scale bars: 40 μm.
It is clear that the small fibril structures encompassing the aggregates could now be resolved after sample expansion, confirming the effectiveness of resolution increase in VISTA (Fig. 2D, zoomed-in insets of the green and cyan dash boxes). Interestingly, hollow-like structures were also observed in the large aggregate shown in Fig. 2D with adjusted image contrast (Fig. S2), indicating a lower protein concentration at the center of the large aggregates. We have also observed similar hollow structures in a number of unexpanded polyQ aggregates in live cells, but not with such clear contrast. Similarly, with the aid of correlative fluorescence imaging from Congo red staining (Fig. 2E, before treatment; Fig. 2G, after expansion), we established high specificity and sensitivity of VISTA imaging of the expanded Aβ plaques (Fig. 2F, SRS imaging at 2940 cm−1 before sample treatment; Fig. 2H, VISTA imaging after expansion). Both aggregation model systems confirmed that the aggregates could be clearly retained and expanded for sensitive visualization by VISTA.
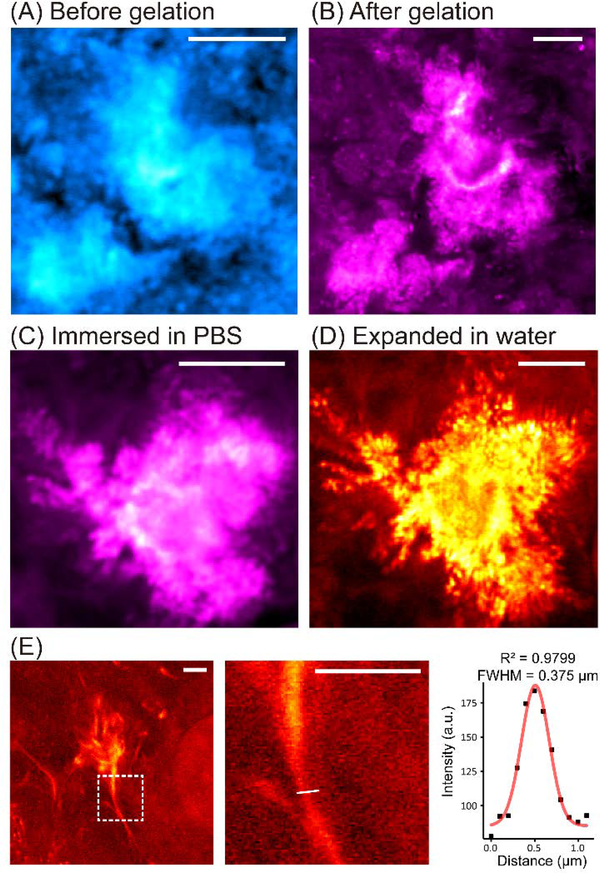
Since we aim to characterize the fine morphology of the Aβ plaques, we next focused on Aβ aggregates in 5XFAD mouse tissues to analyze the expansion ratios and whether potential distortion might be introduced into the dense aggregates during the expansion. We tracked the protein aggregates in the tissue samples during the sample processing and expansion. We first performed SRS imaging comparison for the same tissue samples before and after gelation (Fig. 3A, before; Fig. 3B, after). It is obvious that gelation did not cause any morphological change of the aggregates. In addition, we observed an enhancement in contrast due to the loss of signals in the vicinity of the aggregates after gelation (Fig. 3B). This should be due to the deprivation of previously reported surrounding lipid contents,(35,40) since the hybridization chemistry here only retained proteins. The removal of the lipids was also confirmed by a loss of lipid features in the hSRS spectra from the aggregates and their surrounding areas (Fig. S3A).
Fig. 3. Characterization of Aβ plaque expansion.
(A-B) SRS images (at 2940 cm−1) for the same plaques before (A) and after (B) gelation. No obvious changes of location and morphology are observed. (C-D) SRS images (at 2940 cm−1) for the same plaques after gelation and immersed in PBS (C) versus in water (D). No distortion is shown during expansion in water with much-enhanced resolution. (E) The intensity profile of a fibril structure (boxed, the zoomed-in view shown on the right, the white line indicates the cross section for intensity fitting) in the expanded Aβ plaques imaged by VISTA is fitted by a gaussian distribution with a FWHM of 375 nm. Scale bars: 10 μm in (A)-(D), 5 μm in (E).
We next compared SRS images from the same aggregate immersed in PBS (with only a slight expansion, Fig. 3C) and in water (expected with full expansion, i.e. VISTA, Fig. 3D). Such detailed comparisons from the same plaques designate that the expansion is isotropic in water and is effective in resolution enhancement since all the fibrillar structures that were blurred in regular resolution SRS imaging (Fig. 3A–C) became clear and resolvable in VISTA imaging (Fig. 3D). Images from the same aggregate-containing tissue before gelation and after water-expansion were also captured to confirm the isotropic expansion and the loss of lipids (Fig. S3B–D). The similarity of the contour comparison for tissues before and after expansion (Fig. 2C–D and Fig. S3B–D) showed a more than 90% similarity, confirming the minimum expansion distortion. We calculated the average expansion ratio to be about 3.4 times from the plaques, which is consistent with previous reports of ours and others on brain tissues.(36,41) This indicates our effective resolution of label-free VISTA imaging could reach about 95 nm, highly suited to investigate these fibrils with reported width around 100 nm.(11) Indeed, our analysis confirmed that VISTA could capture Aβ fibrils with neck width around 110 nm (Fig. 3E, obtained by dividing the fitted full width at half maximum (FWHM), 375nm, by the average expansion ratio, 3.4).
High-resolution Volumetric VISTA imaging of the protein aggregates
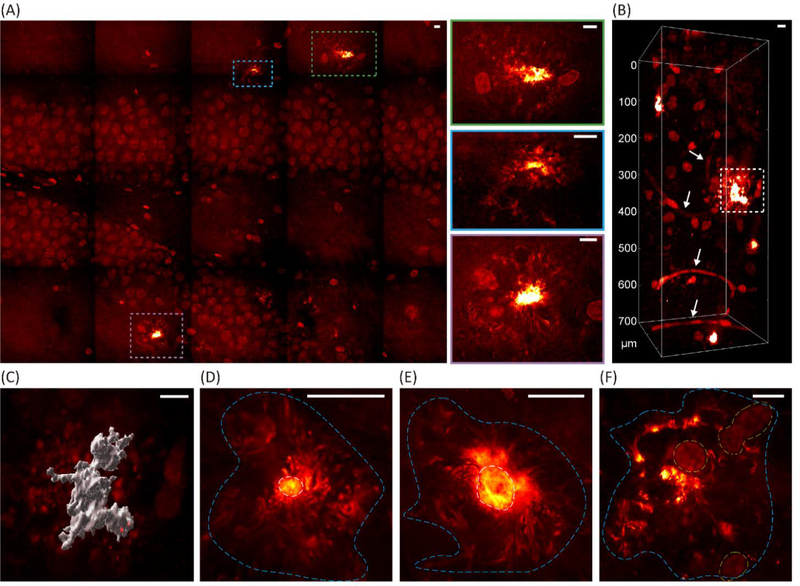
Confirmed that VISTA is effective for high-resolution label-free imaging of the aggregate structures with retained fidelity, we then set out to visualize and identify various morphologies of the Aβ plaques in their native environment in 3D. We investigated the plaques formed in the hippocampus, which has been implicated to show early critical symptoms during the course of AD.(21) For example, the hippocampal volumes were reported to reduce at an early stage of AD in human patients.(21) A large field-of-view mosaic VISTA imaging from the dentate gyrus easily captured the plaque distributions with their pronounced contrasts (Fig. 4A, plaques boxed with colored dash boxes). Zoomed-in views on the boxed regions are shown on the right with corresponding box colors (Fig. 4A). Despite the multi-component complexity in tissues, the protein aggregates showed a clear standout in the label-free image due to their higher local protein concentrations. In addition to aggregates, cell nuclei in the tissues were also distinctively captured by VISTA, which was confirmed by DAPI stain (Fig. S4A). It has been previously observed that the Aβ deposits are surrounded by a large number of microglia and astrocytes and could activate the prominent responses from these cells.(24,42,43) The ability to clearly visualize cell nuclei and aggregates at the same time without other signal interferences (i.e. lipids) could help us perform analysis of plaque-cell interactions.
Fig. 4. VISTA mapping for the locations, morphologies, and the surroundings of the Aβ plaques in the hippocampal tissues.
(A) A single-z mosaic VISTA mapping of an Aβ-plaque-containing hippocampal tissue section. Multiple observed plagues are highlighted in the colored dash boxes and shown in the zoomed-in views on the right side. (B) Deep imaging (700 μm) of the hippocampal tissues captures volume views of the plagues and the surrounding matrix. The arrows indicate blood vessels. (C) Surface rendering of the plaque indicated by the dashed box in (B). (D-F) Representative Aβ plaques with different morphological details. The areas with protein aggregates are circled by cyan dashed lines, core areas are circled by white dashed lines, and cell nuclei are circled by yellow dashed lines. Scale bars: 20 μm.
Indeed, volumetric imaging of the hippocampal tissues could nicely provide 3D views of the plaques deep into the tissues without any sacrifice on the resolution (Fig. 4B, 700 μm depth after expansion). It is also clear that the large plaques are surrounded by a high number of cells (boxed), while the smaller ones do not present such obvious enrichment of cell nuclei in the vicinity (Fig. 4B). The image depth should be further increased to a few mm after the upgrade of the condenser to a long working distance objective.(25,36) Additionally, we found that blood vessels are also a clear standout in the VISTA images (Fig. 4B, arrow indicated), the identity of which was confirmed by both lectin and anti-GLUT1 staining (Fig. S4B). Here, we showcased the capability of visualizing the intricate spatial relationship of the Aβ plaques and their surrounding components of cell nuclei and blood vessels. The change of blood flow, and hence the vascular alterations in morphology and diameters, are also an early sign of AD.(19,44) We envision this ability to perform high-resolution high-throughput investigations between these three components inside tissues could largely facilitate our understandings of the influence on the structural integrity with the deposition of the Aβ plaques.
To take a closer look at the plaque morphology, we generated the surface rendering of the aggregate boxed in Fig. 4B for more detailed visualization in 3D (Fig. 4C). Different layers of the same plaque could be also visualized by adjusting the rendering threshold (Fig. S5, the core of the plaque). Furthermore, we showed high-resolution label-free VISTA images for representative morphology of versatile plaques (Fig. 4D–F). While plaques with obvious core structures and diffusive fibrillar surroundings were most commonly observed (Fig. 4D), plagues with halo-like cores and ribbon-like surroundings were also captured by our VISTA imaging (Fig. 4E). Such observations agreed well with previous reports by super-resolution fluorescence imaging.(12) Moreover, we identified entirely diffusive plaques, which were Congo red-negative, with less well-defined cores together with their close proximity to cell nuclei (Fig. 4F). All these rich diffusive morphologies were absent in fluorescence images of Congo red stained plaques since it only stains the cores. We hence demonstrated that our label-free VISTA method offers superior imaging performance for close to 100 nm resolution imaging of Aβ plaques together with their key interacting components (e.g. cells and vessels) in a complex biological matrix.
High-resolution U-net segmentation of label-free VISTA images for multiplex imaging.
After identifying that our VISTA could allow clear imaging of aggregates, cell nuclei, and blood vessels with nice correlations to the corresponding post-expansion fluorescence stains and imaging (i.e. Congo red for the plaque cores (Fig. 2G–H), DAPI for cell nuclei (Fig. S4A), and lectin and GLUT1 antibody for the vessels (Fig. S4B)), we next investigated whether we could segment all these components into individual channels for specific and multiplex imaging. CNN has achieved tremendous success in computer vision problems.(45) The U-net architecture has been shown to be highly successful in segmenting, denoising, and predicting biomedical images.(38,46,47)
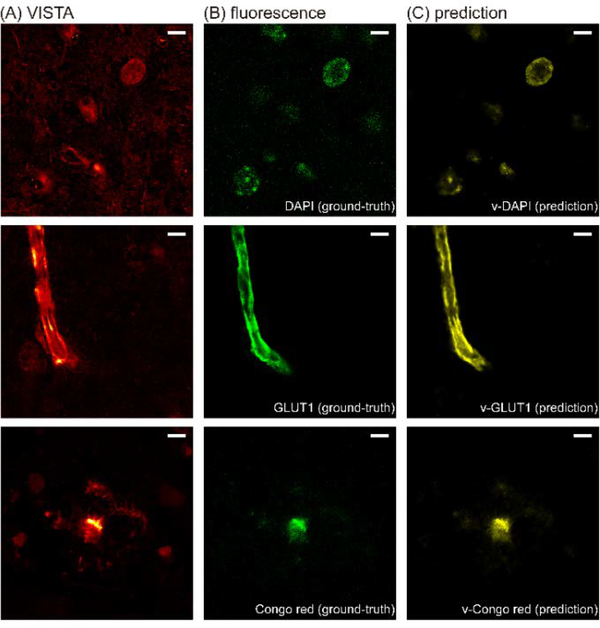
We adopted the reported U-net architecture(38) for modeling the features in our VISTA images by feeding in the corresponding fluorescence-VISTA image pairs (Fig. 5). The qualities and contrasts of the predicted images (Fig. 5C) from the input VISTA images (Fig. 5A) were comparable to that of the fluorescence ground-truth (Fig. 5B). The nice model performance was quantified by Pearson’s correlation coefficient (Pearson’s r) between the predicted images and the ground-truth images for the test sets (Fig. S6). We note that for calculating Pearson’s r for v-GLUT1 and v-Congo red sets, we set a threshold to ensure that the images we were comparing indeed contained signals because the random noise in the background could not be predicted by the U-net. Since the v-Congo red image only predicted the plaque cores, we then performed subtraction of the predicted channels from the original VISTA images to visualize the peripheral plaque filaments.
Fig. 5. VISTA images of the brain tissue with its corresponding ground-truth and prediction results.
(A) Label-free VISTA images. (B) The corresponding fluorescence ground-truth at the same field of view. (C) Prediction results by U-net.
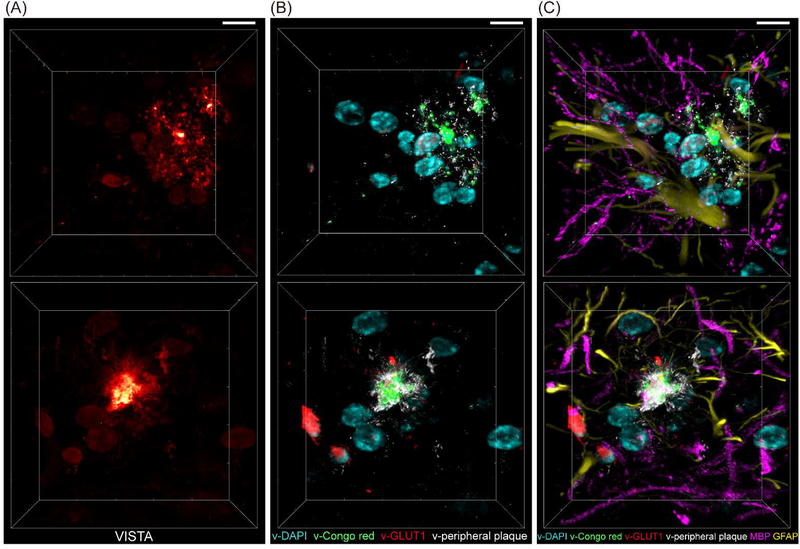
Through U-net segmentation of VISTA images in 3D (Fig. 6A), a total of four-channel volumetric predicted imaging could be obtained (Fig. 6B) for plaque cores (green), plaque filaments (white), cell nuclei (cyan), and blood vessels (red). With additional channels of tandem fluorescence imaging of anti-MBP (myelin basic protein) antibody and anti-GFAP (glial fibrillary acidic protein) antibody staining, we obtained 6-color high-resolution imaging from the same Aβ-plaque-bearing hippocampal tissues (Fig. 6C) that could allow us to perform high-resolution interrogations for the distribution of Aβ plaque cores (green) and periphery filaments (white) together with their immediate environment of blood vessels (red), cell nuclei (cyan), astrocytes (yellow), oligodendrocytes and myelin (magenta). For example, our further cross-channel analysis between the v-DAPI and GFAP channels revealed that a large portion (almost to a half) of the cells surrounding the Aβ plaques are astrocytes (Fig. S7&S8), which is consistent with the known effect of Aβ activated reactive astrocytes with astrocytic processes surrounding the plaque cores.(23,42) Such a multiplex imaging feat with minimum labeling requirements makes it extremely ideal for high-throughput and cost-effective large-scale biomedical investigations, for instance in human tissues, in the future.
Fig. 6. 3D multi-channel prediction of plaque-bearing hippocampus from two sets (top and bottom) of VISTA images.
(A) VISTA images. (B) Four-channel overlay for v-DAPI (cyan, nuclei), v-Congo red (green, for plaque core), v-GLUT1 (red, blood vessels), and v-Peripheral plaque (white) by U-net predicted high-resolution segmentation of the VISTA images in (A). (C) 6-color tandem fluorescence image of MBP (magenta, myelin basic protein) and GFAP (yellow, Glial fibrillary acidic protein) with 4-channel VISTA predictions. Scale bars: 30 μm.
In the context of the multiplex micro-environment imaging, we further observed plaques with different fine filament structures, which would not be detected by typical Congo red staining, nor readily resolved by regular-resolution SRS. We resolved aggregates with dotted filament structure and ribbon-like structure, each surrounded by astrocytes of different morphologies (Fig. 6, top row (dotted-like, v-peripheral plaque) vs bottom row (ribbon-like). Zoomed-in views are shown in Fig. S9). Differences in astrocyte morphologies may indicate that they belong to varied subtypes, or designate that they have different levels of activation.(48–50) While further biological investigations are still required, our results here show that it is very likely that Aβ plaques with varied nano-scale morphological structures may play distinct pathological roles and our multiplex VISTA imaging could help to subtype these plaques based on their fine filament structures, which are absent from the typical Congo red labeling and imaging.
Conclusions
In this paper, we presented a label-free VISTA method for high-resolution vibrational imaging of protein aggregates, in particular for Aβ plaques in the AD. The Aβ plaques were easily identified and imaged. We further imaged plaque filaments with a resolution of approximately 100 nm and demonstrated large-scale mosaic and deep tissue volumetric imaging capabilities. Plaque cores, peripheral filaments, and the surrounding vasculature and cell nuclei were observed without the need of any labels and were further specifically segmented by a CNN-based U-net method for 4-channel multiplex imaging. Finally, we demonstrated 6-channel imaging when tandem with fluorescence labels for further characterization of the proteins of interest, with observations for the correlation between varied fine plaque filament structures with distinct astrocyte morphologies. Our paper presents the first report for robust high-resolution imaging on the expanded aggregates with their interacting native environment in tissues. We envision this expanded view of the protein aggregates could provide a new perspective for understanding the aggregate structures in the complex tissue environment and would further our insights on the roles of protein aggregates in neurodegenerative diseases.
Supplementary Material
Acknowledgements
We thank Caltech Biological Imaging Facility for software support. We are grateful to Bryce Manifold (University of Washington) and the Caltech OLAR staff for technical support. Chenxi Qian acknowledges the support of the Natural Sciences and Engineering Research Council of Canada (NSERC Postdoctoral Fellowship). Lu Wei acknowledges the support of National Institutes of Health (DP2GM140919–01), Amgen (Amgen Early Innovation Award), and the start-up funds from California Institute of Technology.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest
Notes and references
- 1.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature Medicine. 2004. July;10(7):S10–7. [DOI] [PubMed] [Google Scholar]
- 2.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, et al. Formation of Neuronal Intranuclear Inclusions Underlies the Neurological Dysfunction in Mice Transgenic for the HD Mutation. Cell. 1997. August 8;90(3):537–48. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science. 2002. July 19;297(5580):353–6. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Komi Y. Layers of structure and function in protein aggregation. Nature Chemical Biology. 2015. June;11(6):373–7. [DOI] [PubMed] [Google Scholar]
- 5.Breydo L, Uversky VN. Structural, morphological, and functional diversity of amyloid oligomers. FEBS Lett. 2015. September 14;589(19 Pt A):2640–8. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Liu H, Dong P, Li D, Legant WR, Grimm JB, et al. Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. Kowalczykowski SC, editor. eLife. 2016. August 3;5:e17056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, et al. Aβ Oligomer-Induced Aberrations in Synapse Composition, Shape, and Density Provide a Molecular Basis for Loss of Connectivity in Alzheimer’s Disease. J Neurosci. 2007. January 24;27(4):796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang T, Li S, Xu H, Walsh DM, Selkoe DJ. Large Soluble Oligomers of Amyloid β-Protein from Alzheimer Brain Are Far Less Neuroactive Than the Smaller Oligomers to Which They Dissociate. J Neurosci. 2017. January 4;37(1):152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Checler F, Turner AJ. Journal of Neurochemistry special issue on Alzheimer’s disease: “amyloid cascade hypothesis−-20 years on”. Journal of Neurochemistry. 2012. January;120 Suppl 1:iii. [DOI] [PubMed] [Google Scholar]
- 10.Bäuerlein FJB, Fernández-Busnadiego R, Baumeister W. Investigating the Structure of Neurotoxic Protein Aggregates Inside Cells. Trends in Cell Biology. 2020. December 1;30(12):951–66. [DOI] [PubMed] [Google Scholar]
- 11.Mlodzianoski MJ, Cheng-Hathaway PJ, Bemiller SM, McCray TJ, Liu S, Miller DA, et al. Active PSF shaping and adaptive optics enable volumetric localization microscopy through brain sections. Nature Methods. 2018. August;15(8):583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Querol-Vilaseca M, Colom-Cadena M, Pegueroles J, Nuñez-Llaves R, Luque-Cabecerans J, Muñoz-Llahuna L, et al. Nanoscale structure of amyloid-β plaques in Alzheimer’s disease. Scientific Reports. 2019. March 26;9(1):5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bäuerlein FJB, Saha I, Mishra A, Kalemanov M, Martínez-Sánchez A, Klein R, et al. In Situ Architecture and Cellular Interactions of PolyQ Inclusions. Cell. 2017. September 21;171(1):179–187.e10. [DOI] [PubMed] [Google Scholar]
- 14.Takasaki KT, Ding JB, Sabatini BL. Live-Cell Superresolution Imaging by Pulsed STED Two-Photon Excitation Microscopy. Biophysical Journal. 2013. February 19;104(4):770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duim WC, Jiang Y, Shen K, Frydman J, Moerner WE. Super-resolution fluorescence of huntingtin reveals growth of globular species into short fibers and coexistence of distinct aggregates. ACS Chem Biol. 2014. December 19;9(12):2767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M, Williamson N, Mishra A, Michel CH, Kaminski CF, Tunnacliffe A, et al. Structural progression of amyloid-β Arctic mutant aggregation in cells revealed by multiparametric imaging. Journal of Biological Chemistry. 2019. February 1;294(5):1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski Schierle GS, van de Linde S, Erdelyi M, Esbjörner EK, Klein T, Rees E, et al. In Situ Measurements of the Formation and Morphology of Intracellular β-Amyloid Fibrils by Super-Resolution Fluorescence Imaging. J Am Chem Soc. 2011. August 24;133(33):12902–5. [DOI] [PubMed] [Google Scholar]
- 18.Möckl L, Moerner WE. Super-resolution Microscopy with Single Molecules in Biology and Beyond–Essentials, Current Trends, and Future Challenges. J Am Chem Soc. 2020. October 21;142(42):17828–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett RE, Robbins AB, Hu M, Cao X, Betensky RA, Clark T, et al. Tau induces blood vessel abnormalities and angiogenesis-related gene expression in P301L transgenic mice and human Alzheimer’s disease. PNAS. 2018. February 6;115(6):E1289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, et al. Caspase cleavage of tau: Linking amyloid and neurofibrillary tangles in Alzheimer’s disease. PNAS. 2003. August 19;100(17):10032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, et al. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer’s disease. Journal of Neurology, Neurosurgery & Psychiatry. 2001. October 1;71(4):441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartels T, Schepper SD, Hong S. Microglia modulate neurodegeneration in Alzheimer’s and Parkinson’s diseases. Science. 2020. October 2;370(6512):66–9. [DOI] [PubMed] [Google Scholar]
- 23.Habib N, McCabe C, Medina S, Varshavsky M, Kitsberg D, Dvir-Szternfeld R, et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nature Neuroscience. 2020. June;23(6):701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebmann T, Renier N, Bettayeb K, Greengard P, Tessier-Lavigne M, Flajolet M. Three-Dimensional Study of Alzheimer’s Disease Hallmarks Using the iDISCO Clearing Method. Cell Reports. 2016. July 26;16(4):1138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, et al. Sca l eS: an optical clearing palette for biological imaging. Nature Neuroscience. 2015. October;18(10):1518–29. [DOI] [PubMed] [Google Scholar]
- 26.Ando K, Laborde Q, Lazar A, Godefroy D, Youssef I, Amar M, et al. Inside Alzheimer brain with CLARITY: senile plaques, neurofibrillary tangles and axons in 3-D. Acta Neuropathol. 2014. September;128(3):457–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim S-Y, et al. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell. 2015. December 3;163(6):1500–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological Alterations in Alzheimer Disease. Cold Spring Harb Perspect Med. 2011. January 9;1(1):a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, Chen KS, Knox J, Inglis J, Bernard A, Martin SJ, et al. A learning deficit related to age and β-amyloid plaques in a mouse model of Alzheimer’s disease. Nature. 2000. December;408(6815):975–9. [DOI] [PubMed] [Google Scholar]
- 30.Urbanc B, Cruz L, Le R, Sanders J, Ashe KH, Duff K, et al. Neurotoxic effects of thioflavin S-positive amyloid deposits in transgenic mice and Alzheimer’s disease. PNAS. 2002. October 29;99(22):13990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurouski D, Duyne RPV, Lednev IK. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review. Analyst. 2015. July 13;140(15):4967–80. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, Hu F, Min W. Raman Imaging of Small Biomolecules. Annual Review of Biophysics. 2019;48(1):347–69. [DOI] [PubMed] [Google Scholar]
- 33.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science. 2010. December 3;330(6009):1368–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng J-X, Xie XS. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 2015. November 27;350(6264):aaa8870. [DOI] [PubMed] [Google Scholar]
- 35.Ji M, Arbel M, Zhang L, Freudiger CW, Hou SS, Lin D, et al. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Science Advances. 2018. November 1;4(11):eaat7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian C, Miao K, Lin L-E, Chen X, Du J, Wei L. Super-Resolution Label-free Volumetric Vibrational Imaging. bioRxiv. 2021. January 9;2021.01.08.425961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassie AT, Zhao Y, Boyden ES. Expansion microscopy: principles and uses in biological research. Nature Methods. 2019. January;16(1):33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ounkomol C, Seshamani S, Maleckar MM, Collman F, Johnson GR. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nature Methods. 2018. November;15(11):917–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006. October 4;26(40):10129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiskis J, Fink H, Nyberg L, Thyr J, Li J-Y, Enejder A. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy. Sci Rep. 2015. August 27;5:13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ku T, Swaney J, Park J-Y, Albanese A, Murray E, Cho JH, et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016. September;34(9):973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Nievas BG, Serrano-Pozo A. Deciphering the Astrocyte Reaction in Alzheimer’s Disease. Front Aging Neurosci. 2018;10:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeWitt DA, Perry G, Cohen M, Doller C, Silver J. Astrocytes Regulate Microglial Phagocytosis of Senile Plaque Cores of Alzheimer’s Disease. Experimental Neurology. 1998. February;149(2):329–40. [DOI] [PubMed] [Google Scholar]
- 44.Chui HC, Ramirez-Gomez L. Clinical and imaging features of mixed Alzheimer and vascular pathologies. Alz Res Therapy. 2015. February 27;7(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan S, Rahmani H, Shah SAA, Bennamoun M. A Guide to Convolutional Neural Networks for Computer Vision. Synthesis Lectures on Computer Vision. 2018. February 13;8(1):1–207. [Google Scholar]
- 46.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015. May 18 [cited 2020 Sep 25]; Available from: https://arxiv.org/abs/1505.04597v1
- 47.Manifold B, Thomas E, Francis AT, Hill AH, Fu D. Denoising of stimulated Raman scattering microscopy images via deep learning. Biomed Opt Express. 2019. August 1;10(8):3860–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017. January;541(7638):481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nature Reviews Neuroscience. 2015. June;16(6):358–72. [DOI] [PubMed] [Google Scholar]
- 50.Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, et al. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-β peptides. Nature Medicine. 2004. July;10(7):719–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of the present study are available in the article and its supplementary figures, or from the corresponding author upon request. The code for U-Net training, prediction, and evaluation in this paper is available at https://github.com/Li-En-Good/VISTA