Abstract
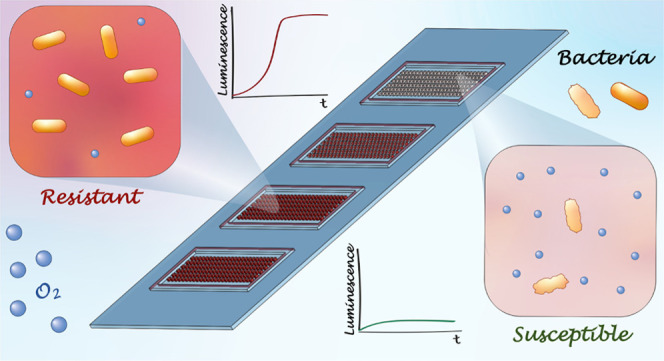
Rapid identification of a pathogen and the measurement of its antibiotic susceptibility are key elements in the diagnostic process of bacterial infections. Microfluidic technologies offer great control over handling and manipulation of low sample volumes with the possibility to study microbial cultures on the single-cell level. Downscaling the dimensions of cultivation systems directly results in a lower number of bacteria required for antibiotic susceptibility testing (AST) and thus in a reduction of the time to result. The developed platform presented in this work allows the reading of pathogen resistance profiles within 2–3 h based on the changes of dissolved oxygen levels during bacterial cultivation. The platform contains hundreds of individual growth chambers prefilled with a hydrogel containing oxygen-sensing nanoprobes and different concentrations of antibiotic compounds. The performance of the developed platform is tested using quality control Escherichia coli strains (ATCC 25922 and ATCC 35218) in response to clinically relevant antibiotics. The results are in agreement with values given in reference guidelines and independent measurements using a clinical AST protocol. Finally, the platform is successfully used for the AST of an E. coli clinical isolate obtained from a patient blood culture.
Keywords: antibiotic susceptibility testing, bacterial resistance, microfluidics, minimal inhibitory concentration, oxygen-sensing nanoprobes
Infectious diseases are a significant cause of morbidity and mortality worldwide.1 With the emergence of antibiotic resistance,2 bacterial infections are becoming an important threat to global health.3 Particularly in the case of bloodstream infections, which are often associated with severe sepsis or septic shock, early administration of effective antibiotics is extremely time-critical for successful treatment.4 Rapid antibiotic selection and proper dosage are therefore extremely important.
Standard growth-based assays for antibiotic susceptibility testing (AST) include manual methods, such as broth microdilution on a multiwell plate, and gradient methods on gel dishes or automated methods using commercial instruments (e.g., VITEK 2, bioMérieux SA). Regardless, these methods require between 104 and 108 CFU mL–1, leading to long preculturing times, which could result in several days until the AST can be performed and the results are obtained.5 Molecular AST techniques, mostly relying on the polymerase chain reaction, are rapid and sensitive but are only suitable for already well-characterized resistance genes. Moreover, they are relatively expensive and require well-trained personnel.6,7 Therefore, the development of new rapid and sensitive diagnostic tools is of great importance.8,9
Strategies for reducing the AST time include the miniaturization of test systems and improvements of their readout. Microfluidic devices typically operate with volumes of microliter to femtoliter and are therefore promising systems for AST.10,11 Loading the sample in small wells or the creation of water-in-oil emulsions on a microfluidic platform decreases the initial number of bacteria required to just one or few cells per compartment.12 This also significantly shortens the time required for preculturing and further analysis. Additionally, microfluidic techniques can be combined with molecular ASTs13,14 facilitating complementary bacterial identification or analysis.15
Droplet systems offer continuous compartmentalization of the bacterial samples in a very high throughput, enabling the analysis of thousands of samples16 and on the single-cell level.17 However, the pump systems, typically required to generate emulsions, are difficult to integrate with the point-of-care systems, while the alternative, formed with the use of gravitational forces, could be used, but it operates at a significantly lower throughput.18 In addition, droplet microfluidics requires inert oils and detergents to stabilize the droplets, yet shrinking of droplets and leakage of compounds (e.g., via micelle formation) is often observed.19
Alternatively, devices with integrated cultivation chambers or wells have been introduced.20−23 Here, the chamber number is fixed, given by the initial design of the device. The isolation, i.e., closing of the chamber, is achieved by valves24,25 or again, by sealing with non-water-miscible fluids (oils).15,26−28 The SlipChip technology represents another approach to isolate individual cultivation compartments. Such devices typically contain two parallel plates which can be moved to fill and close micro- to picoliter-volume wells.14,29 While the throughput is limited, chambers-based approaches have other significant advantages. The chambers can be supplied with compounds, medium, or antibiotics at a later time in the experiment. Hence, this provides the possibility of tracking bacterial growth in response to changes in cultivation conditions over time. Other designs use partially open chambers or channels without valves, designed to trap and immobilize bacterial cells for high-resolution single-cell imaging.30−33
Bacterial response to antibiotics is typically evaluated based on viability assays. There are several existing approaches that translate bacterial metabolic activity into a detectable optical signal. For example, adenosine triphosphate was determined in a bioluminescence assay,34 and pH-dependent color changes of a chromophore23,24 were successfully used for susceptibility testing. The conversion of resazurin to resorufin is a fluorescence assay widely employed in microfluidic AST systems.26−28 The assay is very sensitive but requires a high concentration of the reagent to generate sufficient signal during the long-term measurements. Considering that resorufin is also prone to photobleaching, the assay is more suitable for endpoint measurements rather than for dynamic changes of cell viability.35 Furthermore, it is necessary to evaluate the cross-reactivity of the tested antibiotics with the reagent.36
Many of the abovementioned systems represent excellent research tools, but practical drawbacks limit their use in clinical settings. In addition, the majority of microfluidic devices are made out of poly(dimethylsiloxane) (PDMS) and suffer from several limitations such as unstable surface properties, absorption of hydrophobic compounds into the polymer, evaporation of water through the polymer, and the limited possibility for mass production.19,37,38
In this study, we overcome these issues and introduce a microfluidic device made out of a cyclic olefin copolymer (COC) (Figure 1). The device incorporates hundreds of growth chambers and is easy to operate. The protocol only requires the pipetting of the sample and sealing of the device, making its production and assay protocol easily scalable and adaptable for low-cost mass production and suitable for potential clinical use. Moreover, using a gas-tight material such as COC enabled us to improve the readout to differentiate susceptibility and resistance of aerobic pathogens by monitoring their oxygen consumption. Thereby, the readout of the cell metabolic activity is based on oxygen-sensing nanoprobes embedded in the growth chambers. The cell metabolic activity and hence viability can be monitored via relative changes in the luminescence of the nanoprobes.
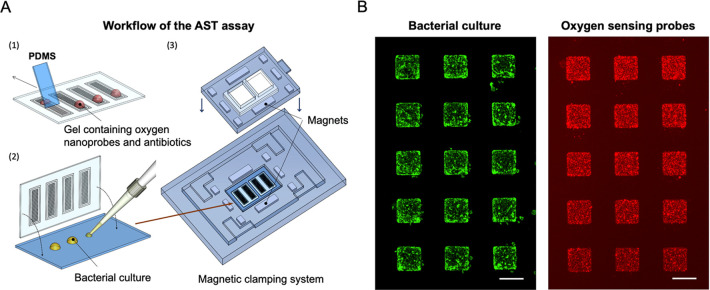
Figure 1.
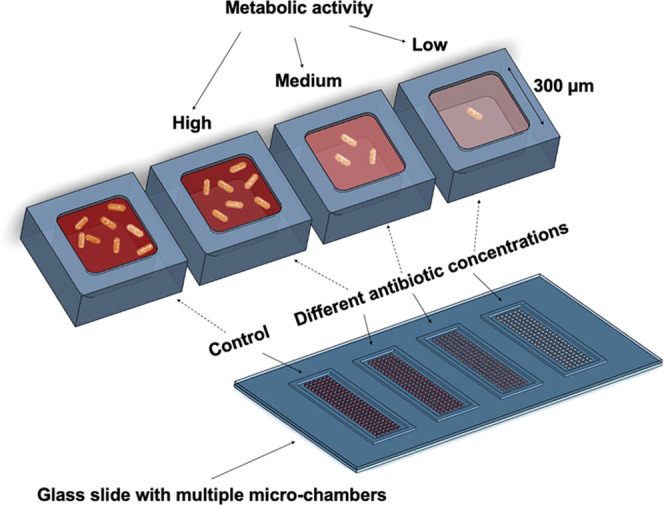
Conceptual sketch of the microfluidic system for antibiotic susceptibility testing. The platform consists of four sets of chamber arrays with hundreds of growth chambers (300 × 300 × 75 μm3) fabricated using the air-tight material, cyclic olefin copolymer. The chambers are filled with oxygen-sensing nanoprobes and different concentrations of the studied antibiotics embedded in the agarose gel. Changes in the metabolic activity of the bacteria result in changes in the luminescence signal of the nanoprobes, enabling the determination of susceptibility or resistance of the studied pathogens against the various antibiotic compounds.
In contrast to the above-described viability assays, the luminescence signal generated by oxygen-sensing nanoprobes is reversible. Therefore, it is not limited by the reagent concentration and provides an almost real-time indication of changes in the dissolved oxygen level in the surrounding environment.39 It was previously shown that these oxygen-sensing nanoprobes do not compromise cell viability and are compatible with oxygen measurements in shake flasks and microtitration plates.40,41
In the following, we demonstrate that the luminescence signal change generated using the oxygen-sensing nanoprobes is sensitive enough to assess bacterial viability in the miniaturized AST platform. The characterization of the platform is performed using two quality control strains of Escherichia coli, ATCC 25922 and ATCC 35218. Both strains constitutively produced a superfolder variant of the green fluorescent protein (sfGFP) for the simplified monitoring of bacterial growth and observation of morphological changes in response to antibiotic exposure. Finally, the platform was validated using a clinical isolate of an E. coli strain obtained from a positive blood culture of a septic patient.
Materials and Methods
Antibiotics for the AST Assay
Meropenem trihydrate, ciprofloxacin hydrochloride monohydrate (5 and 2.5 mg mL–1 stock solutions prepared in DI water), and amoxicillin (2 mg mL–1 stock solution prepared in phosphate buffer, pH 6) were purchased from Sigma-Aldrich. Solutions of antibiotics in media were prepared shortly before the experiments, from the aliquots of stock solutions (∼500 μL) stored at −20 °C. Each aliquot of the stock solutions was thawed only once, without refreezing.
Antibiotics for Plasmid Maintenance
Kanamycin sulfate was obtained from Sigma-Aldrich and stored in 1 mL aliquots at −20 °C. Ampicillin sodium salt was obtained from Glentham Life Sciences, dissolved in water, and stored in 1 mL aliquots at −20 °C.
Bacterial Strains
Escherichia coli, ATCC 25922 and ATCC 35218
These strains were modified to overexpress the gene for the superfolder green fluorescent protein (sfGFP). This provides the possibility to monitor changes in cell viability and morphology induced by the presence of antibiotic compounds. We transformed these strains by electroporation42 with the plasmid pSEVA271_sfgfp (lab collection) carrying the sfgfp gene, which was constructed based on plasmid pSEVA27143 (kanamycin resistance) adding a gene for sfGFP44 under control of a constitutive promoter (BioBrick part BBa_J23100).45
Escherichia coli, Clinical Isolate
The analyzed strain was obtained from the Division of Clinical Bacteriology and Mycology of the University Hospital Basel from a routine diagnostics. The isolate originated from a blood culture. Blood culture samples were incubated on a shaker (Virtuo, bioMérieux) in respective blood culture bottles (BactAlert FA/FN, bioMérieux) for a maximum of 6 days. The positive blood culture was subcultured and the species were identified using MALDI-TOF mass spectrometry (microflex, Bruker).
Fabrication of the COC Chamber Array Plate
The fabrication process began with transferring the microchamber design onto an initial master wafer. We used standard SU-8 photolithography processes according to the manufacturer’s guidelines. In short, SU-8 3050 was spin-coated at 2750 rpm for 30 s on a 4 in. silicon wafer substrate and subsequently soft-baked, resulting in a structure height of 75 μm. After exposure to UV light through a foil mask, the wafer was post-exposure-baked and developed in a developer bath. Finally, the wafer was hard-baked and silanized with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (PFOTS) to avoid unwanted adhesion.
The stamp for the thermal imprinting was prepared by transferring the design from the initial master wafer to a UV curable resist (Ormostamp, Micro Resist, Germany) deposited on a 4 in. glass wafer. The resulting stamp was silanized with PFOTS. The growth chambers were thermally imprinted into a 4 in. wafer format cyclic olefin copolymer (COC grade 8007) foil (Topas Advanced Polymers, Germany) with a thickness of 240 μm. The imprinting process was performed using a compact nanoimprinting tool (CNI V2.0, NILT, Denmark) by applying a pressure of 6 bar for 3 min at 130 °C. Finally, the COC wafers were diced into individual chips comprising four separate arrays each.
Bacterial Cultivation
Bacterial strains were stored as cryo-stocks containing 25% glycerol (−80 °C). Before the experiments, a small portion of the stock culture (10–20 μL) was transferred into 2 mL of MHB II (cation-adjusted) containing appropriate antibiotics for plasmid maintenance (ATCC 25922 [pSEVA271_sfgfp]: 50 μg mL–1 kanamycin sulfate and ATCC 35218 [pSEVA271_sfgfp]: 50 μg mL–1 kanamycin sulfate and 100 μg mL–1 ampicillin sodium salt). The liquid culture was grown at 37 °C using a shaking incubator (Minitron, Infors HT) with a shaking speed of 220 rpm. When an OD600 in the range of 0.2–0.4 was reached, the culture was diluted to an OD600 of 0.02 in a new cultivation medium.
The cultivation of the clinical isolate was performed under similar conditions, within a biosafety level 2 laboratory (no antibiotics were used for precultivation).
Clamping Device
The layout of the clamping device was designed to fit a standard microscopy holder for a 96-well plate. The system (Figure S4) is composed of two parts: the bottom part, which tightly fits a microscopy glass slide (24 × 40 mm2, #5; Thermo Scientific). Grooves in the bottom part guide the positioning of the top part precisely above the microfluidic device. Both parts contain eight openings to fit block magnets (four 20 × 5 × 2 mm3; four 10 × 3 × 2 mm3; eight 6 × 4 × 2 mm3, obtained from supermagnete.ch) that are used to hold both parts together. Within the top part, a PDMS slab with a poly(methyl methacrylate) support is incorporated. The PDMS slab allows for an even distribution of the force exerted by the magnets on the glass slide that is in contact with the COC chamber array plate. This maintains a proper sealing of the chambers during incubation and avoids any evaporation. Both parts were designed using CAD software (SOLIDWORKS 2019, Dassault systèmes) and printed using a 3D printer (Ultimaker 3, Ultimaker) with ABS plastic.
Preparation and Deposition of the Gel
The gel preparation process starts by heating a 1.5 mL centrifugation tube (Eppendorf) with a 100 μL aliquot of the ultralow gelling temperature agarose gel solution (6% in Mueller Hinton Broth II, cation-adjusted) to 85 °C in a Thermomixer (Eppendorf Thermomixer R Shaker, Eppendorf). Once the temperature was reached, 20 μL of the OXNANO oxygen-sensing nanoprobes (Pyroscience) were added to the agarose gel. A stock solution of the oxygen-sensing nanoprobes was prepared by dissolving the probe particles in DI water to a final concentration of 1 mg mL–1. Prior to the experiments, the stock solution was homogenized in an ultrasonic bath for at least 20–60 min.
The mixture of the gel and particles is heated using the thermomixer to 95 °C and held at the temperature for at least 5 min (to avoid potential bacterial contamination from the particle solution) and cooled down to 47 °C. The antibiotic solution in cultivation medium (2× the target concentration, see Table S1) was heated to 47 °C, and 120 μL of the solution was added to the gel and mixed using a vortex mixer. The mixture of the gel, particles, and antibiotics was kept at 47 °C and 20 μL of it was pipetted over each array and spread using a small piece of PDMS. Once the mixture cooled down in the chambers, the gel solidified and the arrays were directly used for the measurements.
All chemicals were prepared and handled in a sterile environment. Similarly, all of the tools were sterilized before use to avoid contamination.
Modifications in the Protocol
For the characterization of the system, we used bacterial strains (ATCC 25922 and ATCC 35218), constitutively, producing the fluorescent protein sfGFP. To maintain its production, the growth medium additionally contained kanamycin sulfate (during the preculture and AST). The preculture of the ATCC 35218 E. coli strain contained ampicillin sodium salt to maintain the resistance. However, ampicillin was not supplied during the AST.
Imaging of the Chamber Array Platform
The microchamber array was placed in the magnetic clamping system and mounted on an automated, fully motorized inverted wide-field microscope (Nikon Ti-E, controlled with NIKON NIS-Elements Advanced Research software) to perform time-lapse imaging. The environmental box of the microscope was preheated to 37 °C and maintained at this temperature during the 5-h on-chip cultivation. Individual chambers were imaged every 15 min for 3 h and every 30 min for the following 2 h using bright-field and luminescence microscopy. The chambers with entrapped air bubbles or visible damage of the gel were not imaged or evaluated. The imaging was performed using a 10× objective (Nikon, Plan Fluor NA:0.3, WD:16 mm) with a Lumencor Spectra X LED light source. The following optical configurations and settings were used: a cyan LED (30%), a 475/28 excitation filter, a 495 dichroic, a 525/50 emission filter, and an exposure time of 100 ms for the sfGFP fluorescence and a blue LED (30%), a 438/24 excitation filter, a 660 dichroic, a 785/60 emission filter, and an exposure time of 200 ms for the oxygen-sensing particles. Images were recorded with a Hamamatsu Orca Flash 4 camera.
Data Analysis
The luminescence intensities from the individual chambers were analyzed using FIJI image analysis software.46 Regions of interest (ROI) (315 × 315 μm2) were drawn around each chamber, and the mean gray value was obtained for each of the 18 time points. Considering that bacterial cells as well as nanoprobes were present also outside of the chambers, we did not perform background subtraction but instead subtracted the initial mean luminescence value obtained for the individual chambers at zero time point. The initial bacterial number per chamber (see Figure S3) was determined after the conversion of the images to RGB 8-bit, application of a threshold (selection of the cutoff pixel intensity value to distinguish between the luminescence signal and the background), and a watershed segmentation, using the particle analysis plugin from FIJI software.46
AST via Broth Microdilution
The AST results from the microchamber arrays were compared to the results obtained from a standard broth microdilution AST. This assay was performed following CLSI guidelines and as described previously47 but using a 384-well plate (a final volume of 40 μL per well) instead of a 96-well plate format. The inoculum was prepared as described for the microfluidic assay but using a final cell concentration of 5 × 105 CFU mL–1. The highest concentrations of antibiotics were 800 μg mL–1 (amoxicillin), 1.6 μg mL–1 (meropenem), and 0.8 μg mL–1 (ciprofloxacin) in the first well, which was then used for 10-step serial dilution (2 log). After incubation (37 °C, 18 h), the OD600 values of the plate were recorded using an Infinite 200 PRO plate reader (Tecan).
Demonstration of the Oxygen Sensing (Figure S1)
To show the dependence of the luminescence signal on the oxygen level, we prepared two DI water samples with different levels of dissolved oxygen. One sample was saturated with the nitrogen gas and was expected to have a lower oxygen level compared to the DI water sample, which was placed on a shaker (190 rpm, 24 °C). During the experiment, 330 μL of both samples together with 30 μL of the oxygen nanoprobe stock solution (1 mg mL–1) were pipetted in a standard 96-well plate (Thermo Fisher Scientific-Nunclon Delta Surface, 96 flat-bottom transparent polystyrene plates). The plates were immediately covered with a polyester sealing film (Starlab, Switzerland) and placed on the automated microscope for the luminescence readout. All other experimental details were as described above. The data collection started as the tape was removed, thus reinstating the oxygen level of both samples. We recorded the luminescence levels every 5 min, from eight wells per condition. Measurements were performed at 37 °C.
The measurements of the emission spectrum were performed on a microplate reader, Infinite M1000 PRO (Tecan), using Thermo Fisher Scientific-Nunclon Delta Surface, 96 flat-bottom transparent polystyrene plates. The sample with a reduced oxygen level was prepared again by saturation with nitrogen and immediate sealing after preparation. Three wells were loaded with 360 μL of samples containing 30 μL of the particle stock solution. The emission spectra of these samples were compared to the samples without pretreatment. The spectra for both samples between 500 and 850 nm were collected at an excitation wavelength of 438 nm in 5 nm steps. The chambers were sealed during the measurements. The spectra were recorded with three repetitions for each condition. All measurements were performed at 30 °C.
Results and Discussion
A conceptual sketch of the AST device is presented in Figure 1. The device comprises four sets of nanoliter-sized chamber arrays. These are prefilled with agarose gel containing oxygen-sensing nanoprobes and different concentrations of the selected, clinically relevant antibiotics. The bacterial sample is deposited on a standard glass slide and covered by the prefilled chamber array plate. The measurements are performed on an automated microscope, collecting luminescence signals from the individual chambers.
The dissolved oxygen acts as a quencher of the nanoprobe luminescence (Figure S1). The chambers containing metabolically active bacteria consume oxygen and thus provide a greater luminescence signal compared to the chambers containing bacterial cells with metabolism affected by the antibiotics. The AST workflow (Figure 2A) of the presented microfluidic platform starts by filling the individual chambers of the array with an agarose gel (2.5% w/v) containing oxygen-sensing nanoprobes and antibiotics diluted in the culture medium (MHB II, cation-adjusted). Using four arrays on the same device allows for four different conditions to be tested simultaneously. Each individual array consists of 144 chambers (width: 300 × 300 μm2, depth: 75 μm) and is surrounded by a trench to drain excess gel and bacterial culture once the device is assembled. Furthermore, this prevents cross-contamination between the arrays. A droplet (∼15 μL) of the liquid gel, maintained at 47 °C, is pipetted on the COC plate and spread over the array using a thin PDMS slab. We selected ultralow gelling temperature agarose to avoid high operating temperatures and risk of degrading the antibiotic compounds. Due to the low volume of the chambers (∼7 nL), the gel solidifies nearly immediately after its deposition. The gel matrix helps to reduce the evaporation of the medium during the experiments, simplifies manipulation with the filled COC plate, and also keeps the oxygen-sensing particles in fixed positions during the measurement, avoiding their sedimentation. Detailed figures of the COC chambers are found in Figure S2.
Figure 2.
(A) Experimental workflow. First, using a thin slab of PDMS, a small droplet of the liquid agarose (∼15 μL) containing oxygen-sensing probes and studied antibiotics is spread across the individual arrays. Next, 20 μL of a bacterial suspension (OD600 = 0.02) is deposited on a microscopy glass slide and overlaid with the microfluidic chamber arrays. Subsequently, the arrays are transferred into a magnetic clamping device where they are kept in contact during the AST measurement. Oxygen consumption during the bacterial growth is then monitored using the automated microscope via changes in the luminescence signal collected from the individual chambers. (B) (Left) Detailed micrograph of the array with the sfGFP-producing bacterial strain, growing predominantly in the chamber areas. (Right) The corresponding micrograph of the oxygen-sensing nanoprobes which are embedded in these chambers. Scale bars: 300 μm.
In the following step, the bacterial suspension (OD600 = 0.02) is pipetted on a microscopy slide (20 μL split into four 5 μL droplets) and overlaid with the COC plate containing chamber arrays. The mean cell number per array after the inoculation is between 25 and 30 CFU (Figure S3). The glass slide and chamber arrays are maintained in contact using a custom-made magnetic clamping system, which also serves as a holder for automated microscopy imaging (Figure S4). The bacterial cells entrapped in the chambers have sufficient medium and nutrients to grow for multiple generations.
Cells without access to the chambers have significantly lower nutrient supply. Therefore, after 5 h of cultivation, we observed only a negligible number of cells in the regions outside of the chambers (Figure 2B). Oxygen consumption during bacterial growth is monitored as an increase in the luminescence signal collected from the oxygen-sensing nanoprobes inside the individual chambers.
Similarly, as for the bacterial cells, there is a very low portion of the nanoprobes in the areas between the chambers. As shown in Figure 2B, the great majority of the probes are located within the chambers, allowing us to record real-time changes in the luminescence level during the bacterial cell culture in each chamber.
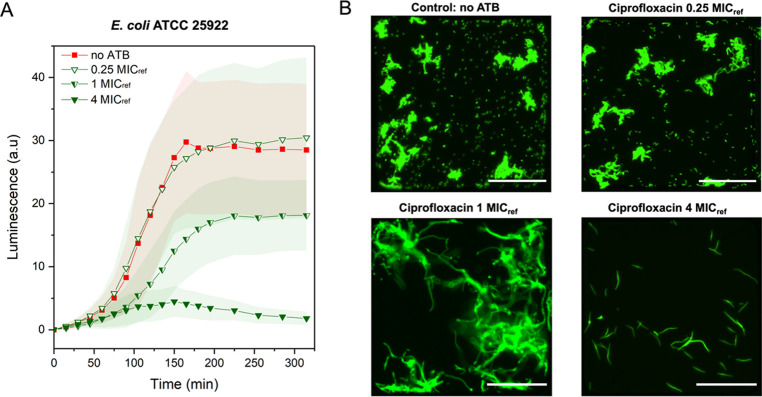
An example of the four chambers at the beginning of the culture and after 5 h of cultivation can be seen in Figure S5. By comparing respiration changes of the cultures containing different antibiotics and a control without antibiotic supplement, we are able to distinguish between resistant and susceptible bacterial strains. To characterize the system, we first performed AST using the fully susceptible quality control E. coli strain ATCC 25922. The strain was first tested against three different concentrations of ciprofloxacin (Figure 3A). The minimal inhibitory concentration (MIC) value for the E. coli ATCC 25922 against ciprofloxacin, listed in the EUCAST table,48 was used as a reference for the antibiotic concentration, MICref = 0.008 μg mL–1. Based on the luminescence signal originating from the oxygen-sensing probes, the oxygen consumption was reduced by approximately half at the MICref value, remained high at doses below the MICref (0.25 MICref), and dropped close to 0 when ciprofloxacin was added in excess (4 MICref), indicating a loss in bacterial viability.
Figure 3.
Characterization of the AST platform using the quality control strain. (A) Kinetic measurements of cell viability via oxygen-sensing nanoprobes immobilized in the agarose gel matrix. Comparison between control without antibiotic (no ATB) supplement E. coli ATCC 25922 culture (red, n = 15), and cultivation in the presence of ciprofloxacin at three different concentrations: 0.25 MICref, 1 MICref, and 4 MICref (green; numbers of analyzed chambers: n = 15, 24, 66, and 42, respectively). (B) Figures of the individual chambers show the bacterial morphology at the end of the measurement for each concentration of ciprofloxacin, MICref = 0.008 μg mL–1. Scale bars: 100 μm. Temperature: 37 °C; MHB II, cation-adjusted medium and an initial OD600 of 0.02 were maintained in all experiments. Error bars represent the standard deviations.
Moreover, we could clearly distinguish the signal difference in all tested conditions within the first 2.5 h of cultivation. The obtained results were further supported by the observed changes in morphology (Figure 3B). Within the chambers containing antibiotics supplied at sub-MICref doses, bacterial cells formed microcolonies surrounded by a high number of planktonic cells. Once the antibiotic concentration was supplied at the MICref concentration, bacteria continued to grow, but we did not observe any cell division, leading to elongated bacterial cells. An excess of ciprofloxacin (4 MICref) resulted in declined bacterial growth after about 2 h of cultivation. The results of the performed AST assay confirmed that the MIC value obtained for ciprofloxacin using the presented platform is in a similar range as given by EUCAST48 and a value obtained using an independent broth dilution AST (Table S1).
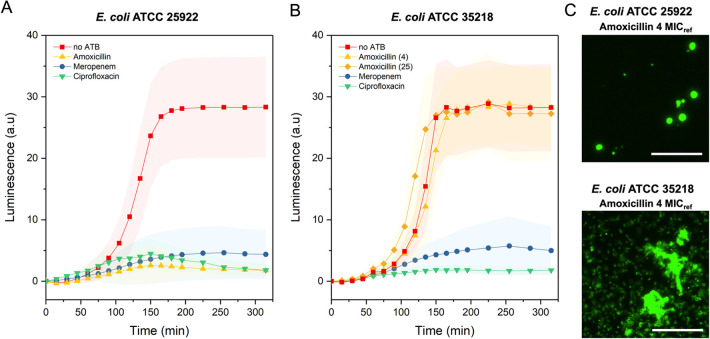
We further tested the strain susceptibility by exposure to excess meropenem and amoxicillin corresponding to 4 MICref concentration (for E. coli ATCC 25922), 0.016 and 4 μg mL–1, respectively (Figure 4A). In comparison to the bacterial culture without antibiotics, the presence of meropenem and amoxicillin in the chambers leads to a significant decrease in the measured luminescence signal, confirming the susceptibility of the tested strain to these antibiotics. Images showing the morphological changes for one selected chamber per each tested condition, including two examples of the luminescence signal from the oxygen-sensing nanoprobes, can be found in the Supporting Information (Figures S6–S12).
Figure 4.
Validation of the AST platform using quality control strains. (A) Comparison between control (no ATB) E. coli ATCC 25922 culture (red, n = 56) and cultivation in the presence of amoxicillin (yellow, n = 71), meropenem (blue, n = 76), and ciprofloxacin (green, n = 42) at a concentration 4 times greater than MICref (4 μg mL–1 for amoxicillin, 0.016 μg mL–1 for meropenem, and 0.008 μg mL–1 for ciprofloxacin). (B) Comparison between control (no ATB) E. coli ATCC 35218 culture (red, n = 74) and cultivation in the presence of meropenem (blue, n = 48), amoxicillin (yellow, (4), n = 43), and ciprofloxacin (green, n = 36) at a concentration 4 time greater than MICref (0.016 μg mL–1 for meropenem, 4 μg mL–1 for amoxicillin, and 0.008 μg mL–1 for ciprofloxacin) together with amoxicillin (yellow, (25), n = 55), supplied at a concentration 25 times greater than MICref. (C) Detailed micrographs from selected chambers after 225 min of cultivation showing the morphological difference between the susceptible (ATCC 25922) and resistant (ATCC 35218) E. coli strain in the presence of amoxicillin supplied at a concentration 4 times greater than MICref for amoxicillin. Scale bars: 50 μm. Temperature: 37 °C; MHB II, cation-adjusted medium and an initial OD600 of 0.02 were maintained in all experiments. Error bars represent the standard deviations. Here, n represents the number of analyzed chambers.
To further confirm the ability of the assay to correctly identify the bacterial resistance, we performed an AST assay using a TEM-1 β-lactamase-producing E. coli strain (ATCC 35218), which is resistant to β-lactam antibiotics such as amoxicillin. The results of the assay are presented in Figure 4B; the tested antibiotics as well as their reference MICs were identical to the previous AST with E. coli ATCC 25922.
Compared to the control culture without antibiotics, bacterial cultures supplied with meropenem and ciprofloxacin showed low values of the measured luminescence (oxygen consumption), confirming the susceptibility of the strain toward these antibiotics. Contrary, for the bacterial culture in the presence of amoxicillin, the respiration profile was in agreement with the culture without antibiotics. We observed a sharp increase in the luminescence shortly after the start of cultivation, which reached and maintained a maximum value at about 2.5 h.
A similar profile was observed when the concentration of amoxicillin was increased to 25 times greater than MICref. Based on these AST measurements, we could confirm that the E. coli ATCC 35218 strain is indeed resistant to amoxicillin. Additionally, the selected antibiotic concentration, corresponding to 4 times the MICref value, is sufficient to observe an adequate difference in respiration profiles necessary to determine antibiotic resistance or susceptibility.
These observations were further confirmed by the cell morphology (Figure 4C). Susceptible bacterial cells have a round shape, due to difficulties with the outer wall synthesis, which often result in cell lysis, while resistant cells maintain a typical rod shape morphology and growth in microcolonies or as planktonic cells. Images showing the morphology changes for one selected chamber per each tested antibiotic are in Figure S13.
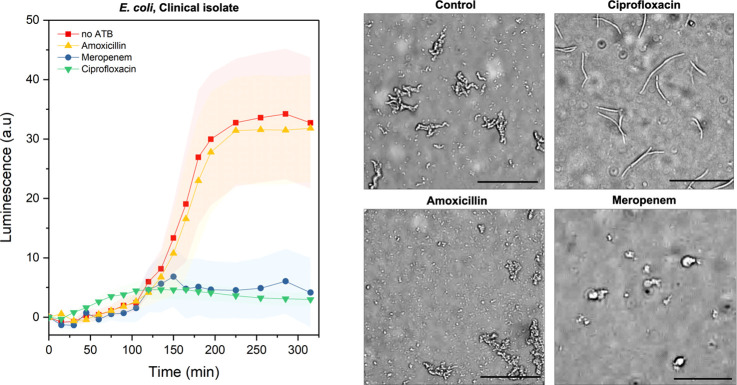
Further, we tested the presented microfluidic platform using a clinical isolate of E. coli. We performed the AST for this isolate against meropenem, ciprofloxacin, and amoxicillin at concentrations identical to the previous experiments. The resulting resistance profiles are presented in Figure 5. In comparison to the oxygen consumption for the culture supplied with an excess of different antibiotics and the control without the antibiotics, we could detect a reduced metabolic activity in the cultures containing meropenem and ciprofloxacin. We did not observe any decrease in bacterial respiration in the culture containing amoxicillin. In contrast, the luminescence profile is similar to that of the culture without the addition of the antibiotics, suggesting the resistance of this isolate against β-lactam antibiotics. Additionally, we were able to confirm the obtained results by observing the cell morphology. In the case of meropenem, we observed cell wall deformations; in the case of ciprofloxacin, we observed cell elongation. In the culture exposed to amoxicillin, we observed cell growth, division, and morphologies consistent with the control experiment, further suggesting the resistance of the studied clinical isolate against amoxicillin.
Figure 5.
Validation of the AST platform using a clinical isolate. Kinetic cell viability measurements via oxygen-sensing nanoprobes of the E. coli clinical isolate. Comparison between the standard bacterial culture without antibiotics (red, n = 29) and cultivation in the presence of amoxicillin (yellow, n = 48), meropenem (blue, n = 33), and ciprofloxacin (green, n = 47) at concentrations 4 times greater than reference MICref for E. coli. Corresponding bright-field micrographs show the bacterial morphology at the end of the assay, after 5 h of incubation. Inoculation was performed at an OD600 of 0.02 (20 μL). Temperature: 37 °C, MHB II, cation-adjusted. Error bars represent the standard deviations. Here, n represents the number of analyzed chambers. Scale bars: 50 μm.
Altogether, based on the results of the AST performed on the microfluidic platform, the tested strain was susceptible to ciprofloxacin and meropenem, while resistant to amoxicillin. These results were consistent with results we obtained by independent studies using the standard broth dilution technique (Table S1). The time-lapse images showing the morphology changes for one selected chamber per each tested condition can be found in the Supporting Information (Figures S14–S17).
Conclusions
In summary, we presented a simple to use AST platform based on the measurement of luminescence-mediated oxygen consumption. This was enabled through the use of air-tight microchambers made out of a thermoplastic COC. The response of the microbial cells was visible in real time, showing the clear difference between the susceptible and resistant bacterial strains.
We believe that the preparation and filling of the chambers with the gel, antibiotics, and nanoprobes can be further optimized to be compatible with mass production and long-term storage. Once the chambers are prepared, they serve as miniaturized agar plates, ready for inoculation. The bacterial cells are deposited in the chambers of the entire platform in a single step by simply pipetting the sample on a glass slide. This makes the AST simple to perform by medical professionals without any further training. The required cell number is in a similar range as required for the state-of-the-art technologies while we could obtain a significantly higher number of technical replicates in parallel. The characterization of the system was performed using the MIC values published in the clinical reference guidelines. For future clinical applications, the tested antibiotic concentrations would be selected based on the breakpoint concentrations.49 Susceptibility determination based on the breakpoint concentrations is an approach adopted by clinicians, directly providing the information regarding the pathogen resistance category (i.e., susceptible, intermediate, or resistant).
Automatization of the chamber filling system using pipetting robots or droplet spotting platforms would allow us to further increase the number of the tested conditions. We envision including more antibiotics or their dilutions to the system with an identical footprint. The platform requires approximately 3 (repetitions) × 30 CFU per condition; therefore, it is presumably possible to incorporate up to 48 different antibiotics at one platform and avoid long preculture times by filtering and hence up-concentrating the preculture. Additionally, the results can be obtained in about 2–3 h of cultivation, while many state-of-the-art technologies and the cultivation on agar plates require 16–24 h to determine resistance profiles.
Here, we used an automated microscope to visualize the cells and confirm the results of the nanoprobe-based signal during the system characterization, but the optical setup for the routine AST could be much more straightforward. The luminescence levels could be easily determined by implementing an optical fiber-based detector or coupling with a very simple optical setup. Recent advancements in the development of oxygen-sensing nanoparticles50 could further increase the sensitivity of the oxygen-based sensing platforms and allow us to simplify the optical setup for luminescence quantification.
Potentially, the system could be used with different types of gel matrices, providing a possibility to identify and cultivate fastidious organisms and slow-growing pathogens. Similarly, other viability reagents could be incorporated into the gel matrix to allow AST for the anaerobic pathogens as well. Furthermore, the clamping device can be opened after the initial assessment of cells and may allow further investigation of cells that were classified as “resistant”, hence opening the ways to investigate the underlying mechanisms of emergence of the resistance.
Acknowledgments
The authors thank Tania Roberts (D-BSSE, ETH Zurich) for her valuable help with setting up experiments with the clinical isolate. The authors thank the Clean Room Facility and the Single-Cell Facility (both at D-BSSE, ETH Zurich) for their valuable help and advice, particularly Erica Montani, Tom Lummen, and Mathias Markus Welsche.
Glossary
Abbreviations
- ATB
antibiotic
- AST
antibiotic susceptibility testing
- COC
cyclic olefin copolymer
- MHB II
Mueller Hinton Broth II, cation-adjusted
- MIC
minimal inhibitory concentration
- PDMS
poly(dimethylsiloxane)
- PFOTS
trichloro(1H,1H,2H,2H-perfluorooctyl)silane
- ROI
region of interest
- sfGFP
superfolder variant of the green fluorescent protein
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.1c00020.
Additional supporting figures illustrating the performance of the oxygen-sensing probes; microfluidic platform; and the clamping device as well as the cell morphology during the platform characterization and validation (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Financial support of the Swiss National Foundation SNF NRP72 (project 407240_167123) and the European Research Council (ERC Consolidator Grant No. 681587) is gratefully acknowledged. D.G.R. is grateful for financial support from the Banting Postdoctoral Fellowship program.
The authors declare no competing financial interest.
Notes
This manuscript has been deposited in preprint on bioRxiv.51
Supplementary Material
References
- Disease and Injury Incidence and Prevalence Collaborators; Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B.; Marshall B. Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nat. Med. 2004, 10, S122–S129. 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- Cassini A.; Högberg L. D.; Plachouras D.; Quattrocchi A.; Hoxha A.; Simonsen G. S.; Colomb-Cotinat M.; Kretzschmar M. E.; Devleesschauwer B.; Cecchini M.; Ouakrim D. A.; Oliveira T. C.; Struelens M. J.; Suetens C.; Monnet D. L.; Strauss R.; Mertens K.; Struyf T.; Catry B.; Latour K.; Ivanov I. N.; Dobreva E. G.; Tambic Andraševic A.; Soprek S.; Budimir A.; Paphitou N.; Žemlicková H.; Schytte Olsen S.; Wolff Sönksen U.; Märtin P.; Ivanova M.; Lyytikäinen O.; Jalava J.; Coignard B.; Eckmanns T.; Abu Sin M.; Haller S.; Daikos G. L.; Gikas A.; Tsiodras S.; Kontopidou F.; Tóth Á.; Hajdu Á.; Guólaugsson Ó.; Kristinsson K. G.; Murchan S.; Burns K.; Pezzotti P.; Gagliotti C.; Dumpis U.; Liuimiene A.; Perrin M.; Borg M. A.; de Greeff S. C.; Monen J. C.; Koek M. B.; Elstrøm P.; Zabicka D.; Deptula A.; Hryniewicz W.; Caniça M.; Nogueira P. J.; Fernandes P. A.; Manageiro V.; Popescu G. A.; Serban R. I.; Schréterová E.; Litvová S.; Štefkovicová M.; Kolman J.; Klavs I.; Korošec A.; Aracil B.; Asensio A.; Pérez-Vázquez M.; Billström H.; Larsson S.; Reilly J. S.; Johnson A.; Hopkins S. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. L.; Fitzgerald J. C.; Balamuth F.; Alpern E. R.; Lavelle J.; Chilutti M.; Grundmeier R.; Nadkarni V. M.; Thomas N. J. Delayed Antimicrobial Therapy Increases Mortality and Organ Dysfunction Duration in Pediatric Sepsis. Crit. Care Med. 2014, 42, 2409–2417. 10.1097/CCM.0000000000000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balouiri M.; Sadiki M.; Ibnsouda S. K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M.; Jupe J.; Mack H.; Coleman T. P.; Lawrence S. M.; Fraley S. I. Emerging Technologies for Molecular Diagnosis of Sepsis. Clin. Microbiol. Rev. 2018, 31, e00089-17 10.1128/CMR.00089-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini N.; Carletti S.; Ghidoli N.; Cichero P.; Burioni R.; Clementi M. The Era of Molecular and Other Non-Culture-Based Methods in Diagnosis of Sepsis. Clin. Microbiol. Rev. 2010, 23, 235–251. 10.1128/CMR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera B.; Anil Vishnu G. K.; Chatterjee S.; Sitaramgupta V. V. S. N.; Sreekumar N.; Nagabhushan A.; Rajendran N.; Prathik B. H.; Pandya H. J. Emerging Technologies for Antibiotic Susceptibility Testing. Biosens. Bioelectron. 2019, 142, 111552 10.1016/j.bios.2019.111552. [DOI] [PubMed] [Google Scholar]
- Maugeri G.; Lychko I.; Sobral R.; Roque A. C. A. Identification and Antibiotic-Susceptibility Profiling of Infectious Bacterial Agents: A Review of Current and Future Trends. Biotechnol. J. 2019, 14, 1700750 10.1002/biot.201700750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Banaei N.; Ren K. Microfluidics for Combating Antimicrobial Resistance. Trends Biotechnol. 2017, 35, 1129–1139. 10.1016/j.tibtech.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Khan Z. A.; Siddiqui M. F.; Park S. Progress in Antibiotic Susceptibility Tests: A Comparative Review with Special Emphasis on Microfluidic Methods. Biotechnol. Lett. 2019, 41, 221–230. 10.1007/s10529-018-02638-2. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Qin S.; Wu S.; Liang Y.; Li J. Microfluidic Systems for Rapid Antibiotic Susceptibility Tests (ASTs) at the Single-Cell Level. Chem. Sci. 2020, 11, 6352–6361. 10.1039/D0SC01353F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong T.; Backes N.; Kalwa U.; Legner C.; Phillips G. J.; Pandey S. Adhesive Tape Microfluidics with an Autofocusing Module That Incorporates CRISPR Interference: Applications to Long-Term Bacterial Antibiotic Studies. ACS Sens. 2019, 4, 2638–2645. 10.1021/acssensors.9b01031. [DOI] [PubMed] [Google Scholar]
- Schoepp N. G.; Schlappi T. S.; Curtis M. S.; Butkovich S. S.; Miller S.; Humphries R. M.; Ismagilov R. F. Rapid Pathogen-Specific Phenotypic Antibiotic Susceptibility Testing Using Digital LAMP Quantification in Clinical Samples. Sci. Transl. Med. 2017, 9, eaal3693 10.1126/scitranslmed.aal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athamanolap P.; Hsieh K.; O’Keefe C. M.; Zhang Y.; Yang S.; Wang T.-H. Nanoarray Digital Polymerase Chain Reaction with High-Resolution Melt for Enabling Broad Bacteria Identification and Pheno–Molecular Antimicrobial Susceptibility Test. Anal. Chem. 2019, 91, 12784–12792. 10.1021/acs.analchem.9b02344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W.; Sarkar S.; Lin Z. S.; McKenney S.; Konry T. Ultrafast Parallelized Microfluidic Platform for Antimicrobial Susceptibility Testing of Gram Positive and Negative Bacteria. Anal. Chem. 2019, 91, 6242–6249. 10.1021/acs.analchem.9b00939. [DOI] [PubMed] [Google Scholar]
- Postek W.; Gargulinski P.; Scheler O.; Kaminski T. S.; Garstecki P. Microfluidic Screening of Antibiotic Susceptibility at a Single-Cell Level Shows the Inoculum Effect of Cefotaxime on E. coli. Lab Chip 2018, 18, 3668–3677. 10.1039/C8LC00916C. [DOI] [PubMed] [Google Scholar]
- Kao Y.-T.; Kaminski T. S.; Postek W.; Guzowski J.; Makuch K.; Ruszczak A.; von Stetten F.; Zengerle R.; Garstecki P. Gravity-Driven Microfluidic Assay for Digital Enumeration of Bacteria and for Antibiotic Susceptibility Testing. Lab Chip 2020, 20, 54–63. 10.1039/C9LC00684B. [DOI] [PubMed] [Google Scholar]
- Payne E. M.; Holland-Moritz D. A.; Sun S.; Kennedy R. T. High-Throughput Screening by Droplet Microfluidics: Perspective into Key Challenges and Future Prospects. Lab Chip 2020, 20, 2247–2262. 10.1039/D0LC00347F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.; Chan C.-W.; Wang Y.; Yao X.; Mu X.; Lu X.; Zhou J.; Cai Z.; Ren K. Reliable and Reusable Whole Polypropylene Plastic Microfluidic Devices for a Rapid, Low-Cost Antimicrobial Susceptibility Test. Lab Chip 2019, 19, 2915–2924. 10.1039/C9LC00502A. [DOI] [PubMed] [Google Scholar]
- Lee J.; Park J.; Kim T. Dynamic Culture and Selective Extraction of Target Microbial Cells in Self-Assembled Particle Membrane-Integrated Microfluidic Bioreactor Array. Anal. Chem. 2019, 91, 6162–6171. 10.1021/acs.analchem.9b00762. [DOI] [PubMed] [Google Scholar]
- Huang H.-K.; Cheng H.-W.; Liao C.-C.; Lin S.-J.; Chen Y.-Z.; Wang J.-K.; Wang Y.-L.; Huang N.-T. Bacteria Encapsulation and Rapid Antibiotic Susceptibility Test Using a Microfluidic Microwell Device Integrating Surface-Enhanced Raman Scattering. Lab Chip 2020, 20, 2520–2528. 10.1039/D0LC00425A. [DOI] [PubMed] [Google Scholar]
- Cira N. J.; Ho J. Y.; Dueck M. E.; Weibel D. B. A Self-Loading Microfluidic Device for Determining the Minimum Inhibitory Concentration of Antibiotics. Lab Chip 2012, 12, 1052–1059. 10.1039/C2LC20887C. [DOI] [PubMed] [Google Scholar]
- Lee W.-B.; Chien C.-C.; You H.-L.; Kuo F.-C.; Lee M. S.; Lee G.-B. An Integrated Microfluidic System for Antimicrobial Susceptibility Testing with Antibiotic Combination. Lab Chip 2019, 19, 2699–2708. 10.1039/C9LC00585D. [DOI] [PubMed] [Google Scholar]
- Mohan R.; Sanpitakseree C.; Desai A. V.; Sevgen S. E.; Schroeder C. M.; Kenis P. J. A. A Microfluidic Approach to Study the Effect of Bacterial Interactions on Antimicrobial Susceptibility in Polymicrobial Cultures. RSC Adv. 2015, 5, 35211–35223. 10.1039/C5RA04092B. [DOI] [Google Scholar]
- Hsieh K.; Zec H. C.; Chen L.; Kaushik A. M.; MacH K. E.; Liao J. C.; Wang T. H. Simple and Precise Counting of Viable Bacteria by Resazurin-Amplified Picoarray Detection. Anal. Chem. 2018, 90, 9449–9456. 10.1021/acs.analchem.8b02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avesar J.; Rosenfeld D.; Truman-Rosentsvit M.; Ben-Arye T.; Geffen Y.; Bercovici M.; Levenberg S. Rapid Phenotypic Antimicrobial Susceptibility Testing Using Nanoliter Arrays. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E5787–E5795. 10.1073/pnas.1703736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi M.; Zaferani M.; Dogan B.; Zhang S.; Simpson K. W.; Abbaspourrad A. Nanoliter-Sized Microchamber/Microarray Microfluidic Platform for Antibiotic Susceptibility Testing. Anal. Chem. 2018, 90, 14137–14144. 10.1021/acs.analchem.8b03817. [DOI] [PubMed] [Google Scholar]
- Yi Q.; Cai D.; Xiao M.; Nie M.; Cui Q.; Cheng J.; Li C.; Feng J.; Urban G.; Xu Y.-C.; Lan Y.; Du W. Direct Antimicrobial Susceptibility Testing of Bloodstream Infection on SlipChip. Biosens. Bioelectron. 2019, 135, 200–207. 10.1016/j.bios.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Kaganovitch E.; Steurer X.; Dogan D.; Probst C.; Wiechert W.; Kohlheyer D. Microbial Single-Cell Analysis in Picoliter-Sized Batch Cultivation Chambers. New Biotechnol. 2018, 47, 50–59. 10.1016/j.nbt.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Mizoguchi M.; Matsumoto Y.; Saito R.; Sato T.; Moriya K. Direct Antibiotic Susceptibility Testing of Blood Cultures of Gram-Negative Bacilli Using the Drug Susceptibility Testing Microfluidic (DSTM) Device. J. Infect. Chemother. 2020, 26, 554–562. 10.1016/j.jiac.2020.01.014. [DOI] [PubMed] [Google Scholar]
- Li H.; Torab P.; Mach K. E.; Surrette C.; England M. R.; Craft D. W.; Thomas N. J.; Liao J. C.; Puleo C.; Wong P. K. Adaptable Microfluidic System for Single-Cell Pathogen Classification and Antimicrobial Susceptibility Testing. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 10270–10279. 10.1073/pnas.1819569116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltekin Ö.; Boucharin A.; Tano E.; Andersson D. I.; Elf J. Antibiotic Susceptibility Testing in Less than 30 Min Using Direct Single-Cell Imaging. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 9170–9175. 10.1073/pnas.1708558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T.; Zhao X. Rapid Identification and Susceptibility Testing of Uropathogenic Microbes via Immunosorbent ATP-Bioluminescence Assay on a Microfluidic Simulator for Antibiotic Therapy. Anal. Chem. 2015, 87, 2410–2418. 10.1021/ac504428t. [DOI] [PubMed] [Google Scholar]
- O’Brien J.; Wilson I.; Orton T.; Pognan F. Investigation of the Alamar Blue (Resazurin) Fluorescent Dye for the Assessment of Mammalian Cell Cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- Shenoy N.; Stenson M.; Lawson J.; Abeykoon J.; Patnaik M.; Wu X.; Witzig T. Drugs with Anti-Oxidant Properties Can Interfere with Cell Viability Measurements by Assays That Rely on the Reducing Property of Viable Cells. Lab. Invest. 2017, 97, 494–497. 10.1038/labinvest.2017.18. [DOI] [PubMed] [Google Scholar]
- Halldorsson S.; Lucumi E.; Gómez-Sjöberg R.; Fleming R. M. T. Advantages and Challenges of Microfluidic Cell Culture in Polydimethylsiloxane Devices. Biosens. Bioelectron. 2015, 63, 218–231. 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Berthier E.; Young E. W. K.; Beebe D. Engineers Are from PDMS-Land, Biologists Are from Polystyrenia. Lab Chip 2012, 12, 1224–1237. 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- Wang X. D.; Wolfbeis O. S. Fiber-Optic Chemical Sensors and Biosensors (2015-2019). Anal. Chem. 2020, 92, 397–430. 10.1021/acs.analchem.9b04708. [DOI] [PubMed] [Google Scholar]
- Flitsch D.; Ladner T.; Lukacs M.; Büchs J. Easy to Use and Reliable Technique for Online Dissolved Oxygen Tension Measurement in Shake Flasks Using Infrared Fluorescent Oxygen-Sensitive Nanoparticles. Microb. Cell Fact. 2016, 15, 45 10.1186/s12934-016-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner T.; Flitsch D.; Schlepütz T.; Büchs J. Online Monitoring of Dissolved Oxygen Tension in Microtiter Plates Based on Infrared Fluorescent Oxygen-Sensitive Nanoparticles. Microb. Cell Fact. 2015, 14, 161 10.1186/s12934-015-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R.; Sambrook J.. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press, 2012. [Google Scholar]
- Silva-Rocha R.; Martínez-García E.; Calles B.; Chavarría M.; Arce-Rodríguez A.; de las Heras A.; Páez-Espino A. D.; Durante-Rodríguez G.; Kim J.; Nikel P. I.; Platero R.; de Lorenzo V. The Standard European Vector Architecture (SEVA): A Coherent Platform for the Analysis and Deployment of Complex Prokaryotic Phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq J.-D.; Cabantous S.; Tran T.; Terwilliger T. C.; Waldo G. S. Engineering and Characterization of a Superfolder Green Fluorescent Protein. Nat. Biotechnol. 2006, 24, 79–88. 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Galagan J. E.; Calvo S. E.; Cuomo C.; Ma L.-J.; Wortman J. R.; Batzoglou S.; Lee S.-I.; Baştürkmen M.; Spevak C. C.; Clutterbuck J.; Kapitonov V.; Jurka J.; Scazzocchio C.; Farman M.; Butler J.; Purcell S.; Harris S.; Braus G. H.; Draht O.; Busch S.; D’Enfert C.; Bouchier C.; Goldman G. H.; Bell-Pedersen D.; Griffiths-Jones S.; Doonan J. H.; Yu J.; Vienken K.; Pain A.; Freitag M.; Selker E. U.; Archer D. B.; Peñalva M. Á.; Oakley B. R.; Momany M.; Tanaka T.; Kumagai T.; Asai K.; Machida M.; Nierman W. C.; Denning D. W.; Caddick M.; Hynes M.; Paoletti M.; Fischer R.; Miller B.; Dyer P.; Sachs M. S.; Osmani S. A.; Birren B. W. Sequencing of Aspergillus Nidulans and Comparative Analysis with A. Fumigatus and A. Oryzae. Nature 2005, 438, 1105–1115. 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I.; Hilpert K.; Hancock R. E. W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- European Committee for Antimicrobial Suceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Clin. Microbiol. Infect. 2000, 9, 509–515..
- Clinical and Laboratory Standards Institute (CLSI). M100Ed30 | Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; 2018.
- Ashokkumar P.; Adarsh N.; Klymchenko A. S. Ratiometric Nanoparticle Probe Based on FRET-Amplified Phosphorescence for Oxygen Sensing with Minimal Phototoxicity. Small 2020, 16, 2002494 10.1002/smll.202002494. [DOI] [PubMed] [Google Scholar]
- Jusková P.; Schmitt S.; Kling A.; Rackus D. G.; Held M.; Egli A.; Dittrich P. S.. Real-Time Respiration Changes as a Viability Indicator for Rapid Antibiotic Susceptibility Testing in a Microfluidic Chamber Array. bioRxiv. 10.1101/2021.01.02.425088 (accessed January 3, 2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.