Graphical abstract
Keywords: Hearing impairment, Genetic heterogeneity, Next generation sequencing, Diagnosis, Pathogenic variant
Abstract
Introduction
Hearing impairment (HI) is characterized by complex genetic heterogeneity. The evolution of next generation sequencing, including targeted enrichment panels, has revolutionized HI diagnosis.
Objectives
In this study, we investigated genetic causes in 22 individuals with non-GJB2 HI.
Methods
We customized a HaloplexHS kit to include 30 genes known to be associated with autosomal recessive nonsyndromic HI (ARNSHI) and Usher syndrome in North Africa.
Results
In accordance with the ACMG/AMP guidelines, we report 11 pathogenic variants; as follows; five novel variants including three missense (ESRRB-Tyr295Cys, MYO15A-Phe2089Leu and MYO7A-Tyr560Cys) and two nonsense (USH1C-Gln122Ter and CIB2-Arg104Ter) mutations; two previously reported mutations (OTOF-Glu57Ter and PNPT1-Glu475Gly), but first time identified among Tunisian families; and four other identified mutations namely WHRN-Gly808AspfsX11, SLC22A4-Cys113Tyr and two MYO7A compound heterozygous splice site variants that were previously described in Tunisia. Pathogenic variants in WHRN and CIB2 genes, in patients with convincing phenotype ruling out retinitis pigmentosa, provide strong evidence supporting their association with ARNSHI. Moreover, we shed lights on the pathogenic implication of mutations in PNPT1 gene in auditory function providing new evidence for its association with ARNSHI. Lack of segregation of a previously identified causal mutation OTOA-Val603Phe further supports its classification as variant of unknown significance. Our study reports absence of otoacoustic emission in subjects using bilateral hearing aids for several years indicating the importance of screening genetic alteration in OTOF gene for proper management of those patients.
Conclusion
In conclusion, our findings do not only expand the spectrum of HI mutations in Tunisian patients, but also improve our knowledge about clinical relevance of HI causing genes and variants.
Introduction
According to the World Health Organization 2019 report, 466 million people around the globe (over 5% of the world population) have disabling hearing impairment (HI) of whom 34 million are children. It is expected that this number will double by 2050 (https://www.who.int/en/news-room/fact-sheets/detail/deafness-and-hearing-loss). HI is characterized by complex heterogeneity in both genetic and clinical aspects with over 121 implicated genes in non-syndromic HI (Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. https://hereditaryhearingloss.org. Last assessed on 4/20/2020) which complicates genetic screening process [1]. It is imperative to understand the molecular basis of HI as genetic factors are responsible for over 50% of HI cases [2] meanwhile environmental and age related HI account for the remaining percentage [3], [4], [5]. Isolated and syndromic HI account for 70% and 30% of prelingual HI cases respectively [6]. Autosomal recessive non-syndromic HI (ARNSHI) accounts for nearly 80% of hereditary HI cases [7]. ARNSHI is the most common genetic pattern for HI specifically in countries with a high rate of consanguinity as in North African Mediterranean countries. To date, 76 genes have been associated with ARNSHI (Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. https://hereditaryhearingloss.org. Last assessed on 4/20/2020). On the other side, Usher syndrome (USH) is the most common form of syndromic HI with an estimated prevalence varying from 3 to 6.2 per 100,000 [8]. USH is inherited in an autosomal recessive pattern and is characterized by combined HI and retinitis pigmentosa (RP). USH has three main classifications namely USH1, USH2 and USH3 that differ clinically in terms of auditory and retinal phenotypes and also genetically with at least 10 associated genes. Genetic variants in those genes could also cause nonsyndromic hearing impairment (NSHI).
Identification of HI causal mutations is crucial for early diagnosis, clinical follow-up and genetic counseling. For a long time, molecular diagnostic tests for ARNSHI and USH in North Africa have been carried out using mainly routine screening techniques such as RFLP, microarray and Sanger sequencing for candidate genes [8], [9], [10]. Addressing HI complex heterogeneity for ARNSHI and USH necessitates sequencing multiplex cascade of all coding regions in large number of genes for the identification of causative mutations. Next-generation sequencing (NGS) methods provide the best alternative approach as they could analyze large number of gene positions in a single experiment offering faster and more cost-effective diagnosis. Nowadays and since many genes associated with hereditary HI have already been identified, the more appropriate choice for comprehensive genetic diagnosis of HI would be targeted exome customized panels combined with NGS sequencing [11]. The effectiveness of this technology for diagnosis of HI has been successfully evaluated with the identification of novel genes and variants associated with HI and therefore HI panels became widely used [8], [11], [12], [13], [14]. Given the large number of novel genes and variants of HI identified using NGS methods, a major interpretation challenge is imposed for precise genetic diagnosis. The American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published recommendations and guidelines for the interpretation of sequence variants [15]. Although these guidelines were intended to be used universally for all Mendelian disorders, certain criteria require gene- or disease-specific knowledge. For this reason, the ClinGen Hearing Loss Clinical Domain Working Group was established as expert panel to evaluate gene disease associations and to standardize variant interpretation in hereditary HI and related syndromes [16].
In the current study, in order to improve our knowledge about clinical relevance of HI causing genes and variants, we performed targeted enrichment with focused gene panels and high-throughput sequencing for 22 patients. The custom designed HaloplexHS (high sensitivity) panel for hereditary diseases provided an efficient and effective tool for molecular diagnosis for known HI genes in Tunisian families.
Material and methods
Patients
22 unrelated patients with HI were enrolled in the present study. We recruited additional family members for co-segregation analysis. Based on family pedigree analysis, HI inheritance pattern was recessively transmitted. Audiological tests through air and bone conduction pure tone audiometry as well as ocular fundus examination (OFE) were performed for all the 22 patients. Moreover, the otoacoustic emissions test (OAEs) which detects response of the outer hair cells (OHCs) to environmental sound was also carried out for one of our patients. In order to confirm USH cases, the Ganzfeld electroretinogram (ERG) and the optical coherence tomography (OCT) ophthalmological investigations were performed.
Peripheral blood samples, from all affected and unaffected subjects, were obtained in EDTA- tubes and genomic DNA was extracted using standard phenol–chloroform technique. All patients were firstly screened for GJB2 mutations through Sanger sequencing using the following primers; Forward: 5′-TCTTTTCCAGAGCAAACCGC-3′ and Reverse: 5′-CTGGGCAATGCGTTAAACTGG-3′.
Ethics statement
Informed consent from all participants or their legal representatives was obtained in accordance with the guidelines of the Regional Committee of the Protection of Persons, Sfax, Tunisia (CPP SUD N°28/2019).
Custom HaloPlexHS panel design, library preparation and next generation sequencing
We customized HaloPlex targeted sequencing approach in order to develop an efficient platform for the detection of mutations in HI genes. We limited our design to the most common causative genes previously reported in ARNSHI and Usher syndrome in North-African region [9]. Probes were designed to capture all coding exons and additional 10 bp flanking the intronic sequence using Agilent’s SureDesign software (www.agilent.com/genomics/suredesign). Table 1 lists names and theoretical coverage for the 30 genes responsible for HI that were included in this panel.
Table 1.
The coverage of target HI genes included in custom HaloPlex-HS panel.
| Target ID | Regions | Coverage | Target ID | Regions | Coverage |
|---|---|---|---|---|---|
| CDH23 | 68 | 99.83% | OTOF | 47 | 99.88% |
| CIB2 | 6 | 100.00% | PCDH15 | 39 | 99.47% |
| COL11A2 | 66 | 99.89% | PJVK (DFNB59) | 6 | 100.00% |
| DCDC2 | 10 | 99.88% | PNPT1 | 28 | 99.62% |
| EPS8 | 20 | 100.00% | PTPRQ | 42 | 99.24% |
| EPS8L2 | 20 | 100.00% | SLC22A4 | 10 | 100.00% |
| ESPN | 13 | 99.86% | TBC1D24 | 7 | 100.00% |
| ESRRB | 8 | 100.00% | TECTA | 23 | 99.96% |
| GIPC3 | 6 | 100.00% | TMC1 | 20 | 100.00% |
| GPR98 (ADGRV1) | 90 | 99.73% | TMPRSS3 | 12 | 99.82% |
| LHFPL5 | 3 | 99.72% | TPRN | 4 | 100.00% |
| LRTOMT | 10 | 99.59% | USH1C | 28 | 99.58% |
| MYO15A | 64 | 99.87% | USH1G | 3 | 100.00% |
| MYO7A | 48 | 100.00% | USH2A | 71 | 99.87% |
| OTOA | 30 | 100.00% | WHRN (DFNB31) | 12 | 99.63% |
Sequence capture was performed using the HaloPlexHS Target Enrichment System (Agilent Technologies, Inc. Santa Clara, CA, Version C1) according to manufacturer's standard protocol for Illumina Sequencing. Precise DNA concentrations of the 22 samples were verified with fluorometry-based Qubit dsDNA HS Assay kit. The gDNA library was created by digesting 50 ng gDNA with 16 different restriction enzymes. An Enrichment Control DNA (ECD) sample was used. The HaloPlex probe capture library was then added and hybridized to the targeted fragments. During this step, each sample was uniquely indexed. The Illumina sequencing motifs incorporated in the targeted fragments include molecular barcode sequences. The target DNA-HaloPlex probe hybrids were circularized and then captured using streptavidin beads. Elution of captured DNA libraries was achieved through addition of NaOH. Only circular DNA targets were amplified and purified using AMPure XP beads. At the end of this step, regions of interest were ready for sequencing.
High throughput sequencing was carried out using Illumina MiSeq instrument (Illumina Inc., San Diego, CA) which is designed for paired-end sequencing by synthesis chemistry. Samples were pooled in equimolar amounts. Each amplicon in our prepared library already contained Illumina adaptor sequences required for multiplexed sequencing. The instrument was set to generate FASTQ files only, without adapter trimming. HaloPlex HS index sequences were manually added through editing the sample sheet.
Data analysis and variant interpretation
FASTQ files were analyzed using Agilent’s SureCall workflow-based application version 4.0.1.46 which carries out alignment, annotation and variants categorization in HaloPlex enriched NGS sample data from Illumina sequencing. Prior to alignment, sequence reads were trimmed to remove low quality bases towards the ends, Illumina adaptor sequences and to mask enzyme footprints. Burrows-Wheeler Aligner algorithm (BWA MEM) for Illumina data was used to align raw sequencing reads to the human reference genome (build hg19/GRCh37) while the SNPPET SNP was used for variant calling. We used filters in SureCall to reduce number of false positive single nucleotide polymorphisms (SNPs), multiple nucleotide polymorphisms (MNPs) and indels. We selected minimum variant call quality threshold of 100 and allele frequency (AF) of 0.1. Variants should be also observed in both direct and reverse strands. Annotated variants were then classified according to their impact’s severity (nonsense, frame-shift indels, splice sites, missense and inframe indels).
Evaluating and understanding the clinical significance of the obtained variants are crucial steps towards proper diagnosis with the greatest clinical sensitivity. In order to achieve this goal, the Hearing Loss Variant Curation Expert Panel was created within the Clinical Genome Resource. This panel adapted the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines [15] for the proper interpretation of variants in HI genes [16]. According to those guidelines, variants were classified based on typical types of variant evidence (e.g. population data, computational data, functional data, segregation data, etc.) into five categories namely “Pathogenic or Likely Pathogenic”, “Benign or Likely Benign” and “Uncertain significance”. The AF in the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/) and the genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/) were used to evaluate variant’s frequency. The pathogenicity of novel missense variants was analyzed with prediction algorithms such as SIFT (http://sift.bii.a-star.edu.sg/) and Provean (http://provean.jcvi.org).
Segregation validation by Sanger sequencing
Segregation analysis for detected variants was performed in cases where DNA samples from relatives were available. Variants selected and suspected to be pathogenic were amplified at optimal condition for each primer pair and then validated by Sanger sequencing on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems by Life Technologies, Thermo Fisher Scientific, Inc) using primers listed in Table S1.
Molecular modelling and functional analysis for the novel missense mutations
To analyze the structural and functional impact of the three novel missense mutations ESRRB-Tyr295Cys, MYO15A-Phe2089Leu and MYO7A-Tyr560Cys, the sequence of the wild-type ESRRB (UniProtKB - O95718-3), MYO15A (UniProtKB - Q9UKN7) and MYO7A (UniProtKB Q13402-1) were submitted to the Phyre2 server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id = index). The computer assisted molecular modeling and Spdb viewer simulations were performed on the mutant version to allow local regional changes for full-length ESRRB (4 3 3), MYO15A (3530) and MYO7A (2215) amino acids. Energy minimization and analysis were conducted using the Yasara virtual reality workstation (http://www.yasara.org). Visualization and generation of high quality 3D graphics were performed using PyMOL program.
Results
Patients clinical data
According to audiological tests, all the 22 patients exhibited bilateral severe (70–89 dB) to profound (>90 dB) sensorineural HI with presence of HI family history. The OAEs test performed showed preserved OAEs indicating a normal outer hair cell function. For the ophthalmological examinations, the OFE test revealed normal eye fundi in almost all patients except for two patients (Family.10_10 and Family.11_11) who showed typical retinal degeneration confirming USH cases. ERG examination has as well confirmed significant bilateral retinopathy in patient (Family.10_10). For patient (Family.3_P3) no signs in favor of retinitis pigmentosa were shown in the ERG examination. OCT scans of our 3-years-old patient (Family.12_P12) showed alterations of the retinal pigment epithelium (RPE) confirming that this patient suffers from USH condition.
Targeted NGS data quality and variants detection
Using the custom HaloPlexHS panel diagnostic tool, we managed to screen for 30 HI genes in 22 patients. The raw data quality was checked using MiSeq Sequencing Analysis Viewer software. Approximately 6 Gbp sequences were obtained from the Haloplex capture assay. We obtained cluster densities from 800,000 to 900,000 clusters/mm2. More than 89% of generated bases were of Q30 value.
Based on the recessive inherited model and the filtration criteria, detailed in methods section, we were able to detect a set of 11 potential pathogenic variants (Table 2): five novel mutations (ESRRB-Tyr295Cys, MYO15A-Phe2089Leu, MYO7A-Tyr560Cys, USH1C-Gln122Ter and CIB2-Arg104Ter), two previously reported mutations but first time to be identified in Tunisian families (OTOF-Glu57Ter [17], [18], [19] and PNPT1-Glu475Gly [20]), and four other variants previously reported in Tunisia (WHRN-Gly808AspfsX11 [21], SLC22A4-Cys113Tyr [22] and two splice site variants found in a compound heterozygous state in MYO7A gene [18], [22], [23], [24], [25], [26], [27], [28], [29], [30]).
-
-
Patients with non-syndromic HI
Table 2.
Potential pathogenic mutations identified in Tunisian families.
| ID | Variant | Transcript | cDNA Position | Effect | Previous reports of the mutation | AF In EXAC (Global) | dbsnp | AF In gnomAD (Global) | SIFT | Provean | Clinvar |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | CIB2-Arg104Ter | NM_006383 | c.310C > T | Stop_gained | Novel | 8.244e-06 | rs1054728914 | 3.98e-6 | D | – | NR |
| P2 | ESRRB-Tyr295Cys | NM_004452 | c.884A > G | Non_synonymous | Novel | 2.537e-05 | rs780275423 | 9.02e-6 | T | De | NR |
| P3 | WHRN-Gly808AspfsX11 | NM_001083885 | c.2423delG | Frame_shift Premature stop codon | Tunisia [17] | – | – | – | – | – | NR |
| P4 | OTOF-Glu57Ter | NM_194322 | c.169G > T | Stop_gained | Saudi Arabia [18], [20] Libya [21] | – | rs397515591 | 4e-6 | – | – | P |
| P5 | MYO15A-Phe2089Leu | NM_016239.3 | c.6265 T > C | Non_synonymous | Novel | – | – | – | D | De | NR |
| P6 | SLC22A4-Cys113Tyr | NM_003059.2 | c.338G > A | Non_synonymous | Tunisia [32] | 0.00005 | rs768484124 | 0.000121 | D | De | NR |
| P7 | PNPT1-Glu475Gly | NM_033109.5 | c.1424A > G | Non_synonymous | Morocco [20] | – | rs397514599 | – | D | De | P |
| P8 | OTOA-Val603Phe | NM_144672.3 | c.1807G > T | Non_synonymous | Algeria [19] | 4.947e-05 | rs775686301 | 3.98e-6 | D | N | VUS |
| P9 | USH1C-Gln122Ter | NM_001297764 | c.360C > T | Stop_gained | Novel | – | – | – | – | – | NR |
| P10 | MYO7A-Tyr560Cys | NM_000260.4 | c.1679A > G | Non_synonymous | Novel | – | – | – | D | De | NR |
| P11 | MYO7A - SS | NM_000260.4 | c.470 + 1G > A | Splice donor variant | Tunisia [30], [9], [23] Algeria [22] Saudi Arabia [52] | – | rs797044510 | 6.37e-5 | – | – | P |
| c.2283-1G > T | Splice acceptor variant | Algeria [29], Morocco [28], France [27] Tunisia [24] | – | rs397516295 | – | – | – | P |
D: Damaging, T: Tolerated, De: Deleterious, N: Neutral.
NR: Not Reported, P: Pathogenic, VUS: Variant with Uncertain significance.
AF: Allele frequency.
*Novel mutations globally.
Three novel variants have been identified to cause non-syndromic HI.
The homozygous truncating mutation c.310C > T in CIB2 leads to the replacement of Arginine amino acid (AA) in position 104 respectively into stop codon. CIB2-Arg104Ter has been reported in dbSNP database (rs1054728914). This variation was also found in Exome Aggregation Consortium (ExAC) and Genome Aggregation Database (gnomAD) with AF of 0.000008242 and 0.000004, respectively (Family.1_P1).
The homozygous c.884A > G change in ESRRB gene is a mutation that results in codon change of tAt/tGt and AA change of Tyr295Cys. The ESRRB-Tyr295Cys is a very rare SNP (dbSNP ID rs780275423) with an AF of 0.00002537 and 9.02e-6 in ExAC and gnomAD databases respectively (Family.2_P2).
The c.6265 T > C in the MYO15A gene leads to the replacement of the phenylalanine aa in position 2089 with leucine aa p.(Phe2089Leu) and this was not annotated in neither dbSNP nor in gnomAD databases (Family.5_P5).
Furthermore, we have also identified two previously identified variants but first time found in Tunisian families. The homozygous variant c.169G > T, detected in gene OTOF (Family.4_P4), has the dbSNP ID rs397515591. The codon change Gag/Tag results in an AA change of OTOF-Glu57Ter with a high impact and stop gained for transcript NM_194322. This variant was reported in Saudi siblings and Libyan families [17], [18], [19] and is classified as likely pathogenic by the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/).
The homozygous PNPT1 (Family.7_P7) missense mutation (c.1424A > G) was previously found in a consanguineous Moroccan family with three siblings affected by severe HI. This alteration leads to an exchange, at the protein level, of a negatively charged glutamic acid at position 475 with glycine. The c.1424A > G variant was reported as pathogenic in ClinVar, annotated in dbSNP (ID rs397514599) and not reported in the gnomAD database.
Two other identified pathogenic variants were previously reported in consanguineous Tunisian families, the homozygous frameshift mutation c.2423delG in the WHRN (Family.3_P3) gene and the c.338G > A variant in SLC22A4 gene (Family.6_P6) [21], [31]. The homozygous frameshift mutation c.2423delG in the WHRN gene, reported by Tlili et al. 2005, changes the reading frame at position 808 with introduction of a novel stop codon p.(Gly808AspfsX11).
The c.338G > A variant causes missense change in the protein from cysteine to Tyrosine aa p.(Cys113Tyr) [32]. This SLC22A4-Cys113Tyr has the dbSNP ID rs768484124 and presents an AF of 0.00002185 in gnomAD databases. A well-established functional study supported the pathogenicity and the damaging effect of this SLC22A4 alteration [31].
-
-
Patients with USH
Two novel variants have been identified to cause syndromic HI, USH. The novel homozygous truncating mutation c.360C > T in USH1C (Family.10_P10) leads to the replacement of Glutamine AAs in position 122 into stop codon.
For family.11_P11, NGS allowed the identification of a novel missense mutation in MYO7A gene at exon 14 (c.1679A > G). This variation causes the change of tyrosine to cysteine (Tyr560Cys). This variant was not found neither in gnomAD nor in dbSNP data bases.
Two previously identified mutations in Tunisia have been detected in family.12_P12 in a compound heterozygous state. The known variant, c.470 + 1G > A, was detected in both dbSNP database, with ID rs797044510 and gnomAD, with AF of 6.37e-5. This substitution was predicted to alter the splice donor site of intron 5 resulting in exon 5 skipping in the mature transcript. This variant is predicted as pathogenic in ClinVar. The second splice acceptor variant, c.2283-1G > T, was annotated in dbSNP (ID rs397516295) but not found in gnomAD.
Segregation analysis and pathogenicity assessment
All variants reported in this study were either absent or having low MAF ≤ 0.00007 in the SNP database, the 1000 Human Genome Database, ExAC and gnomAD databases. Absence of those genetic variants from controls in various databases represents a moderate evidence for pathogenicity (PM2). For loss of function (LOF) variants we followed the detailed guidelines for interpreting the pathogenic criterion PVS1 [32]. The co-segregation study, for genes definitively known to cause HI, in multiple affected family members supports deleterious effect for genetic variants (PP1 strong or moderate, depending on the number of affected members). Affected members were found to be homozygous for 11 identified variants. Both parents were carriers for those variants. The unaffected members were either wild type or carrier (Figure S1). According to our results, these 11 variants met the pathogenic or likely pathogenic classification criteria (Table 3).
Table 3.
Variants segregation and pathogenicity assessment according to the ACMG guidelines.
| Patient ID | Mutation | Segregation | Pathogenic criteria |
Benin criteria | ACMG Classification |
|||
|---|---|---|---|---|---|---|---|---|
| Very Strong (PVS1) | Strong (PS1-PS4) |
Moderate (PM1-PM6) |
Supporting (PP1-PP5) |
Strong (BS1-BS4) |
||||
| P1 | CIB2-Arg104Ter | Yes | PVS1 | – | PM2-PP1* | PP3 | – | Pathogenic |
| P2 | ESRRB-Tyr295Cys | Yes | – | – | PM2-PP1* | PP3 | – | Likely Pathogenic |
| P3 | WHRN-Gly808AspfsX11 | Yes | PVS1 | PP1** | PM2 | – | – | Pathogenic |
| P4 | OTOF-Glu57Ter | Yes | PVS1 | PP1** PP5*** |
PM2 | PP3 | – | Pathogenic |
| P5 | MYO15A-Phe2089Leu | yes | – | PP1** | PM1-PM2 | PP2-PP3 | – | Pathogenic |
| P6 | SLC22A4-Cys113Tyr | yes | – | PS3-PP1** | PP3 | – | Pathogenic | |
| P7 | PNPT1-Glu475Gly | yes | – | PP1** | PM2 | PP3-PP5 | – | Pathogenic |
| P8 | OTOA-Val603Phe | No | – | – | PM2 | – | BS4 | Uncertain significance |
| P9 | TMC1- Met195Arg | No | – | – | PM2 | PP3 | BS4 | Uncertain significance |
| P10 | USH1C- Gln122Ter | Yes | PVS1 | PP1** | PM2 | – | – | Pathogenic |
| P11 | MYO7A-Tyr560Cys | yes | – | PP1** | PM1-PM2 | PP2-PP3 | – | Pathogenic |
| P12 | MYO7A - SS | Yes | PVS1 | PP5*** | PM2 | PP1-PP3 | – | Pathogenic |
| MYO7A - SS | yes | PVS1 | PP5*** | PM2 | PP1-PP3 | – | Pathogenic | |
* PP1 Moderate: CIB2 and ESRRB (2 affected members).
** PP1 Strong: WHRN, OTOF and USH1C (3 affected members).
***PP5 using strength “Strong” because ClinVar classifies this variant as Pathogenic.
SS : Splice Site Variant.
We have as well detected a previously identified missense variant OTOA-Val603Phe (Family.8_P8) that found in an Algerian family with severe-to-profound HI [33], as well as a non-previously reported variation TMC1-Met195Arg (P9); both of them did not segregate with the disease since they were absent or found in heterozygous state in other patients of the studied families (Table 3).
Discussion
Tunisia is among North African countries characterized by a high consanguinity rate which increased autosomal recessive HI frequency in Tunisian population [34]. In this study, 22 patients were screened for genetic variants in 30 known autosomal recessive HI causative genes using the HaloPlexHS custom-designed kit. All patients’ data as well as all identified variants are summarized in Table 4. Custom target-enrichment and high throughput parallel sequencing technologies have been widely used for molecular diagnosis of both syndromic and isolated HI [8], [12], [13], [35]. Our study is the first of its kind to use a custom HaloPlexHS kit for the detection of various HI causing variants. With its ultra-high sensitivity, Agilent’s HaloPlexHS customized target enrichment panel enables the detection of genetic variants including rare ones. A comparative study on custom targeted NGS panel [36] revealed that HaloPlex platform has the lowest workflow complexity and the highest alignment rate to target regions (over 90% of raw sequencing files were aligned to the human genome) in addition to the highest on-target specificity (over 99% of its mapped reads were aligned to targeted regions). Moreover, this targeted-capture NGS kit enables deeper sequencing coverage with the greatest average normalized coverage. The aim of our study was not only to assess the feasibility of this tool to identify inherited mutations, but also to extend our knowledge about the clinical relevance of HI genes and mutations. In fact, the spectrum of HI-causing genes shows huge genetic and phenotypic diversities. One example that best describes this diversity is the presence of a group of genes in which mutations can cause either NSHI or USH. It is therefore essential to provide proper diagnosis prior to offering optimal patient care. ClinGen gene curation process is publicly available and is designed to help in evaluating gene-disease strength relationship (https://www.clinicalgenome.org/).
Table 4.
Summary of all recruited patients’ information.
| Patient ID | Pedigree ID | Consanguineous Relations | Pathogenic Variant | Age | Gender | Affected Members | Segregation Analysis | Ophthalmological investigation Tests / Figures / Outcomes |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients with non-syndromic HearingImpairment | ||||||||||
| P.1 | Family 1 | First Cousins | CIB2-Arg104Ter | 24 | Male | 2 | + | OFE | Data not shown | No signs in favor of RP |
| P.2 | Family 2 | First Cousins | ESRRB-Tyr295Cys | 22 | Male | 2 | + | OFE | Data not shown | No signs in favor of RP |
| P.3 | Family 3 | First Cousins | WHRN-Gly808AspfsX11 | 34 | Male | 3 | + | OFE ERG |
Data not shown Fig. 3 |
No signs in favor of RP |
| P.4 | Family 4 | First Cousins | OTOF-Glu57Ter | 27 | Male | 3 | + | OFE | Data not shown | No signs in favor of RP |
| P.5 | Family 5 | Third cousins | MYO15A-Phe2089Leu | 19 | Female | 4 | + | OFE | Data not shown | No signs in favor of RP |
| P.6 | Family 6 | Third cousins | SLC22A4-Cys113Tyr | 33 | Male | 3 | + | OFE | Data not shown | No signs in favor of RP |
| P.7 | Family 7 | > Third cousins | PNPT1-Glu475Gly | 28 | Male | 5 | + | OFE | Data not shown | No signs in favor of RP |
| P.8 | Family 8 | First Cousins | OTOA-Val603Phe | 30 | Female | 5 | – | OFE | Data not shown | No signs in favor of RP |
| P.9 | Data Not Shown | First Cousins | TMC1-Met195Arg | 22 | Female | 5 | – | OFE | Data not shown | No signs in favor of RP |
| Patients with Usher Syndrome | ||||||||||
| P.10 | Family 10 | > Third Cousins | USH1C-Gln122Ter | 32 | Female | 3 | + | OFE ERG |
Data not shown Fig. 1 |
Significant bilateral retinopathy |
| P.11 | Family 11 | First Cousins | MYO7A-Tyr560Cys | 40 | Male | 7 | + | OFE | Data not shown | Typical retinal degeneration |
| P.12 | Family 12 | – |
MYO7A c.470 + 1G > A MYO7A c.2283-1G > T |
3 | Female | 1 | + | OFE OCT |
Fig. 2 | Normal eyes fundi Alterations of the RPE and of the boundary line between IS and OS in the perimacular region |
RP: Retinitis Pigmentosa / OCT: Optical Coherence Tomography.
RPE: Retinal Pigment Epithelium / ERG: Ganzfeld Electro-Retinogram.
OFE: Ocular fundus examination / IS / OS: Inner and Outer Segments.
Expanding the spectrum of HI mutations in Tunisian patients
In this study, we successfully identified and confirmed the segregation of five novel pathogenic mutations and two previously reported pathogenic variants but first time identified in Tunisian population.
The Arg104Ter mutation in CIB2 gene was found in homozygous state in our patient (Family.1_P1) as well as a second affected member, absent in unaffected members, while both parents were carriers for this particular variant (Figure S1). According to ACMG this variant is predicted as pathogenic since it represents a nonsense variant that affects the CIB2 gene, which is a known mechanism of HI. Furthermore, this variation has a low MAF in ExAC and gnomAD databases. To the best of our knowledge, this novel mutation also represents the second CIB2-mutation identified in Tunisian family.
The novel homozygous missense variation (c.884A > G) in ESRRB gene; which is a well-known gene associated with HI, leads to the change of tyrosine AA at position 295 into cysteine (Family.2_P2). The pathogenicity of p.(Tyr295Cys) mutation is supported by the homozygous state in two affected members and the heterozygous state in their parents (Figure S1) in addition to various prediction programs (SIFT and Provean) used to validate this finding. This variant has low MAF in the SNP, ExAC and gnomAD databases. According to the adapted ACMG/AMP standards and guidelines concerning HI, we classified this novel variation as likely pathogenic (Table 3). Mutations in ESRRB seem to be a rare cause for ARNSHI in Tunisian population. To the best of our knowledge, we believe that this novel missense variation represents the second ESRRB mutation reported in Tunisia since 2011 [37]. Mutations in ESRRB gene were also reported in population of Pakistan, Turkey and Czech Republic [38], [39], [40]. According to UniProt, the ESRRB-Tyr295Cys variant is located in an essential region for ESRRB transcriptional activity and interaction with other proteins such as the Nuclear Receptor Coactivator 3 protein (NCOA3). Our molecular modeling analysis showed that ESRRB-Tyr295Cys is located in a loop region, known as patternless regions, which connect two regular secondary structures. The substitution of the aromatic amino acid Tyr, located on the ESRRB solvent exposed areas within the protein's surface, with Cys amino acid may cause loss of hydrogen bonds in the protein core and/or disturbing correct folding (Figure.S2.A). Therefore, this variation would interrupt the interaction between ESRRB and RNA polymerase II complexes, blocking the NCOA3 co-recruitment to ESRRB, KLF4, NANOG and SOX2 enhancer regions that should trigger ESRRB-dependent gene activation involved in self-renewal and pluripotency.
We have as well identified a novel variant Phe2089Leu (Family.5_P5) in MYO15A gene, the third most critical HI causing gene. In fact, MYO15A has been associated with HI for the first time as early as 1998 [41]. Over the 192 recessive variations previously reported in this particular gene in HI patients from all over the world, 82 variants were reported to be pathogenic, 73 reported to be likely pathogenic and 17 variants of unknown significance VUS [42], [43]. In Tunisian population, the regional prevalence of MYO15A mutations in GJB2 negative HL patients is around 2.3%. The MYO15A-Phe2089Leu variant was not found in any database. According to UniProt protein this variation is located in the MYO15_HUMAN Tail which represent a crucial domain for normal hearing function by maintaining normal structure and function of vestibular hair cells [44] and also encompass over 64 previously identified pathogenic variants [43]. Based on our analysis, the Phe2089 is located in a beta sheet and its substitution with Leu in this position may disturb the secondary rigid structure (Figure.S2.B). Our segregation analysis confirmed the pathogenicity of this novel variant in MYO15A gene (Figure S1).
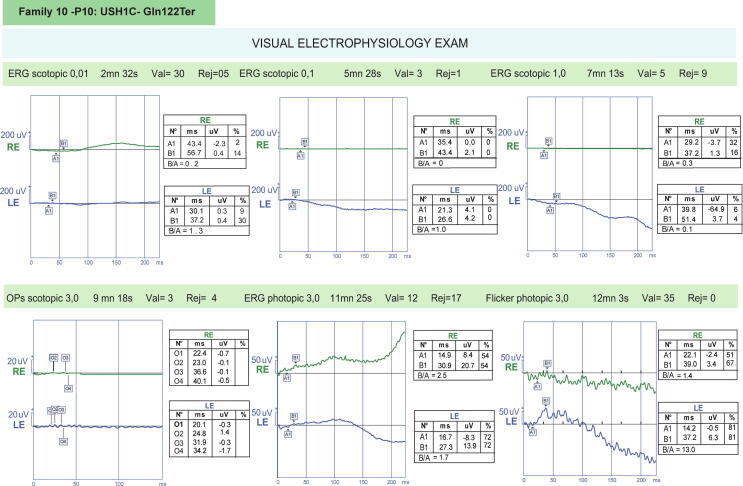
The c.360C > T variant was reported in this study affecting two transcripts of USH1C gene (namely NM_005709 and NM_153676) in a Tunisian patient (Family.10_P10). This mutation was detected in 3 siblings (Figure S1). It was previously discussed that the shorter USH1C transcript (NM_005709) is expressed in both retina and inner ear whereas the longer transcript (NM_153676) which includes the PDZ domain, a region thought to cause NSHI, is exclusively expressed in inner ear [45]. Variants in common exons of both transcripts lead to USH type 1C whereas variants in unique exons of the longer transcript lead to NSHI [46], [47]. Based on ClinGen gene curation process, mutations in this gene were definitely classified as USH type 1 causing mutations. Detailed ophthalmologic examinations in two affected members in this family were performed to determine the phenotype associated with the Gln122Ter mutation. The ERG recorded in both patients showed almost absence of responses in scotopic and in photopic with absence of 30 Hz flicker in both eyes. These traces confirm the evidence of a significant bilateral retinopathy in both affected members (Data of Family.10_P10 is shown in Fig. 1).
Fig. 1.
ERG recordings in Family.10_P10 patient with retinitis pigmentosa. Severely reduced scotopic and photopic ERG flash responses with attenuated 30 Hz flicker in both eyes.
Herein, we also identified and established the segregation of a novel homozygous pathogenic variant in five patients with USH1B (Figure S1). The c.1679A > G (Family.11_P11) is localized in the motor domain of the MYO7A gene resulting in a substitution of Tyr560, a highly conserved residue, to Cys (MYO7A-Tyr560Cys). Clinical exploration in this family (an audiometric examination, an ophthalmological examination and a caloric test) confirmed that all 7 patients suffered from congenital profound deafness, retinitis pigmentosa as well as a vestibular dysfunction (data not shown). In fact, according to genetics home reference (https://ghr.nlm.nih.gov/gene/MYO7A#resources), over 200 pathogenic variants have been previously detected in Usher syndrome type I patients, precisely USH1B, accounting for more than 50% of USH1 cases [48]. Among those variants, 124 pathogenic variants have been located in the MYO7A motor domain which supports the pathogenicity of our newly discovered variant (PM1-ACMG according to Varsome). Over 20 mutations have been previously identified in North African population, particularly in the Tunisian population, where molecular studies showed that mutations in MYO7A gene represents the major cause for USH1 [23]. Our data further confirms the heterogeneity in MYO7A HI-causing mutation among the Tunisian population. The MYO7A-Tyr560Cys variant is located in the Myosin motor domain which represents a hot spot region with 84 pathogenic non-VUS variants with only 9 benign variants (Pathogenicity rate approaching 90.3%). This mutation is located in a non-structured region. Tyrosine560 is a highly conserved amino acid which establishes two hydrogen bonds with both His553 and Asp455 (Figure.S2.C). The former His aa belongs to the same strand while the latter Asp aa lies upstream of the relay in the HP helix. These two residues, as well as the two hydrogen bonds, are conserved in all myosin motor domains. Replacement with cysteine will disrupt those hydrogen bonds and would probably affect the Myosin head motor domain stability. Moreover, this substitution is likely to prevent phosphorylation as this highly conserved Tyr residue is located in the surface and it is therefore totally exposed to the solvent.
In addition to those five novel pathogenic variants, we detected two previously reported mutations but identified for the first time in Tunisian families. The PNPT1-Glu475Gly variant identified in (Family.7_P7) segregated in 5 siblings born from consanguineous parents with ARNSHI (Figure S1). This variant which leads to substitution of the residue Glu475 located at the functional domain of the protein, precisely in the second RNase-PH domain, was identified in a consanguineous Moroccan family experiencing severe HI [20]. This missense variant was not annotated in both dbSNP and gnomAD databases. It was also absent in over 400 control individuals analyzed in the published study from Germany, Morocco and Turkey [20]. It was proven that congenital severe HI in the Moroccan family caused by this hypo-functional variation, would be explained by the disruption of the mitochondrial-RNA-import [20]. Even though the genetic data reported the functional importance of this residue (Glu475) suggesting the implication of this missense variation in HI, the PNPT1-ARNSHI relationship was classified as limited, when reviewed by the ClinGen Hearing Loss Working Group. According to the ACMG criteria, we classified this variant as likely pathogenic. ClinVar and UniProt classified this variant as pathogenic. Our findings provide further solid evidence for the PNPT1 gene implication in the auditory function and thus its association with ARNSHI.
The second previously reported homozygous mutation in OTOF gene (Family.4_P4) resulted in p.(Glu57Ter) change. To our knowledge this is the first OTOF mutation identified in a Tunisian family (Figure S1). This variant is labeled as likely pathogenic by the ClinVar database and has an AF of 4e-6 in gnomAD database. This mutation has been already reported in two Saudi siblings [18] as well as three other families from various regions in Saudi Arabia with severe to profound HI being described [17]. It was also reported in a single Libyan family with ARNSHI and other HI probands [19]. All families reported in Almontashiri et al. (2018) study had tribal origin, so they argued that pathogenic variants identified only once in this cohort may either have drifted further with tribal migration, along the same trade and tribal migration routes, or into neighboring Middle Eastern countries (Syria, Lebanon, Jordan, Palestine, Libya, and Tunisia to the north).
Correlation between OTOF mutations and hearing aids
More than 90 pathogenic variants were reported in OTOF gene [33] with over 50% of subjects having OTOF mutations causing nonsyndromic auditory neuropathy (AN) commonly with normal OAEs [19], [49], [50]. One of our patient (27-year-old) had preserved OAEs indicating a normal outer hair cell function and suggesting that AN would be the most likely diagnosis in this case. However, concerning her sister (18-year-old) who has been using bilateral hearing aids for eleven years, OAEs were absent. OAEs disappearance might be caused by damage from hearing aid use. Since patient care differs between AN and non-AN groups, finding its genetic cause is important. Genetic screening for mutations in OTOF are needed when no neurological syndrome is present with preserved OAEs for an effective genetic counseling and medical guidance [51], [52].
Importance of genetic testing for early USH-diagnosis
In this study, NGS enabled the identification of two compound heterozygous mutations previously reported in the MYO7A gene, the c.470 + 1G > A, a splice donor site variant, and the c.2283-1G > T a splice acceptor site variant in a three-years-old girl (Family.12_P12). Their parents were carriers for the c.470 + 1G > A (mother) and the c.2283-1G > T (father) pathogenic alterations (Figure S1). The splicing defect c.470 + 1G > A was first reported in 1997 in Tunisia [30]. This mutation was later detected in other Tunisian families [9], [23], in Algerian population [22] as well as in a Saudi family [53]. The second splice site variant, c.2283-1G > T was first identified in a family from Algeria [29], a patient from Morocco [28], France [27] and then using whole exome sequencing, Riahi et al. identified this splice acceptor site mutation for the first time in Tunisian families with USH1B [24]. Pathogenicity of this variant was confirmed by different bioinformatics tools and functional analysis [26].
A recent study of patients suffering from USH1B caused by the homozygous splice site mutation (c.470 + 1G > A) indicated that the age of onset of visual symptoms was between five and six years old [53]. Therefore, it is important to note that children with USH1 are most likely clinically misdiagnosed and considered as patients with NSHI since visual defect is only detectable in later years [54] such was the case of our three years-old patient. Based on our genetic diagnosis result, we asked the parents for an extensive ophthalmological examination. OFE of this patient was normal in both eyes, whereas optical coherence tomography (OCT) showed alterations of the retinal pigment epithelium (RPE) and of the boundary line between inner and outer segments (IS and OS) in the perimacular region (Fig. 2). A recent study of the retinal findings in a group of patients with MYO7A-associated USH1 showed that the most common qualitative retinal abnormality was the external layer damage in macular area, which represents an early and closely symptom associated with USH1 [55]. Our findings would assist in offering adapted educational orientation for this patient.
Fig. 2.
Ophthalmologic examinations of a 3-years-old patient with compound heterozygous SS-MYO7A (Family.12_P12). (A) Fundus photographs of both the left eye (LE) and the right eye (RE) are normal. (B) Optical coherence tomography (OCT) scans showing alterations of the retinal pigment epithelium (RPE) and of the boundary line between inner and outer segments (IS and OS) in the perimacular region. Lesions are indicated with arrows.
Further evidence supporting the association of WHRN and CIB2 genes with ARNSHI
WHRN gene was previously described to be associated with autosomal recessive Usher syndrome type 2 [56], [57], [58] and the role of this gene in USH has been scored as having a “definitive” clinical validity with highly supportive evidence (https://www.clinicalgenome.org/). Mutations in WHRN were also described as ARNSHI causing variants [21], [56], [59], [60]. Variants affecting the C-terminal region of the WHRN protein [21], [61] have been reported to segregate with HI. However, based on ClinGen Hearing Loss Working Group updates, there is currently moderate evidence to support the WHRN-ARNSHI gene-disease relationship. In our study, high throughput sequencing in one consanguineous Tunisian family with ARNSHI revealed a 1-bp deletion (c.2423delG, NM_015404.1) in exon 16 of the WHRN gene leading to frameshift with a premature stop codon Gly808AspfsX11. Subsequent genotyping of this deletion in related family members confirmed its co-segregation with the disease. The c.2423delG was found in homozygous state in three more affected members and in heterozygous state in both unaffected parent and siblings (Figure S1). This mutation has been previously reported by Tlili et al. (2005) in a different Tunisian family with ARNSHI. To determine the phenotype associated with the c.2423delG mutation, we performed detailed ophthalmologic examinations in affected members. Ocular fundus examination was normal and no night blindness was reported either. Ganzfeld-ERG recorded particularly in two patients (34 and 44 years of age) showed normal response flash visual-evoked potential in both eyes. Plot in photopic and scotopic with ample flicker 30 Hz and normal latency indicate that there are no signs in favor of retinitis pigmentosa in both patients. Ophthalmological examination’s results of our patient (Family.3_P3) are shown in Fig. 3. Our findings present strong evidence supporting the WHRN-ARNSHI relationship.
Fig. 3.
Plot in photopic and scotopic with ample flicker 30 Hz and normal latency. There are no signs in favor of retinitis pigmentosa in Family.3_P3.
CIB2 gene was first associated with autosomal recessive Usher Syndrome Type 1 [62]. Then in 2018, a multi-ethnic cohort study on patients with CIB2-related-ARNSHI called into question any involvement of CIB2 in the physiopathology of USH [63]. It was shown that the p.(Glu64Asp) variant in the Usher family previously described by Riazuddin et al., (2012) is only two AAs away from the p.(Arg66Trp) variant that has been linked to ARNSHI in three families [63]. Booth et al., (2018) argued that it would be expected to see retinal and vestibular phenotypes in families segregating those variants if CIB2 played a role in USH, however, all patients of this study showed normal ocular phenotype with no vestibular dysfunction. According to the ClinGen Hearing Loss Working Group, CIB2 gene was associated only with ARNSHI excluding the CIB2-autosomal recessive USH Type 1 relationship. Up till now, a total of 9 CIB2 variants (missense, in-frame indels, nonsense, frameshift) were reported in patients with ARNSHI [60], [62], [64], [65], [66]. Here, we report an additional novel nonsense mutation Arg104Ter in CIB2 gene in a patient with ARNSHI. Figure-logical evaluations of all affected individuals revealed absence of RP. Our results provide additional evidence and insight for CIB2 gene-ARNSHI association.
Further evidence supporting the uncertain clinical significance of a variant previously reported as causal mutation in OTOA gene
We identified the p.Val603Phe variant in OTOA gene which was previously reported in an Algerian family [33]. In Ammar-Khodja et al. 2015 study, two related patients carrying a homozygous missense mutation (c.1807G > T: p.Val603Phe) in OTOA gene were reported. This missense mutation was predicted to be pathogenic by PolyPhen-2, SIFT and Mutation Taster programs, and also predicted to affect the acceptor site of intron 18 by NNSPLICE prediction program [33]. Furthermore, the clinical significance of this variant in ClinVar is uncertain. According to ClinGen gene curation process, cautions should be used when evaluating missense variants in this gene as it has a pseudogene. Segregation analysis of this previously reported HI variant proved that it was absent in one affected member and found in heterozygous state in two other affected members in the same family. It is important to note that genetic heterogeneity of HI is common even within the same family. However, even though a second variant in OTOA or a different gene could explain the deafness in this family, we believe that lack of segregation of this OTOA variant, further support its classification as variant of unknown significance According to the ACMG/AMP guidelines (Table 3).
In conclusion, our findings do not only exhibit the usefulness of the targeted NGS approach in the identification and diagnosis of genetic mutations affecting HI in Tunisian families (11 identified pathogenic variants, 5 of which are novel and two newly identified in Tunisian population), but would also have global impact with introduction of new evidence which improve our knowledge concerning clinical relevance of HI causing genes and variants.
Finally, we believe that our findings would be useful for the curation activities of the ClinGen Hearing Loss working group especially in re-evaluating HI gene-disease validity as well as variants pathogenicity.
Ethics approval
This study was approved by the Regional Committee of the Protection of Persons, Sfax, Tunisia. Ethical committee number for the study: CPP SUD N°28/2019.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Acknowledgments
Acknowledgments
The authors would like to thank Dr. Mhamed Grati for his helpful comments.
The authors would like to thank Dr. Dorra Driss for her valuable contribution in the molecular modelling and functional analysis for the novel missense mutations. We sincerely thank the SEED project for the assistance during the manuscript preparation and the journal's choice. The SEED project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 856592.
The authors would like as well to thank Dr. Leila Largueche for her collaboration in the ophthalmological examination of patient 12.
This work was financially supported by the Ministry of Higher Education and Research of Tunisia (LR15CBS07, PAQ-Post PFE and VRR projects).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.01.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Pedigrees and Sequence chromatograms showing segregation’s analysis of eleven detected variants in known deafness genes in patients with nonsyndromic hearing impairment and Usher Syndrome. −/−: homozygous mutant; +/−: heterozygous; +/+: homozygous wild type.
References
- 1.K. Lebeko, J. Bosch, J. J. N. Noubiap, C. Dandara, et A. Wonkam, « Genetics of hearing loss in Africans: use of next generation sequencing is the best way forward », Pan Afr. Med. J., vol. 20, avr. 2015, doi: 10.11604/pamj.2015.20.383.5230. [DOI] [PMC free article] [PubMed]
- 2.H. Mahboubi, S. Dwabe, M. Fradkin, V. Kimonis, et H. R. Djalilian, « Genetics of hearing loss: where are we standing now? », Eur. Arch. Oto-Rhino-Laryngol. Off. J. Eur. Fed. Oto-Rhino-Laryngol. Soc. EUFOS Affil. Ger. Soc. Oto-Rhino-Laryngol. - Head Neck Surg., vol. 269, no 7, p. 1733‑1745, juill. 2012, doi: 10.1007/s00405-011-1910-6. [DOI] [PubMed]
- 3.G. A. Gates et J. H. Mills, « Presbycusis », Lancet Lond. Engl., vol. 366, no 9491, p. 1111‑1120, sept. 2005, doi: 10.1016/S0140-6736(05)67423-5. [DOI] [PubMed]
- 4.Bouzid A. Down-expression of P2RX2, KCNQ5, ERBB3 and SOCS3 through DNA hypermethylation in elderly women with presbycusis. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2018;23(4):347–356. doi: 10.1080/1354750X.2018.1427795. [DOI] [PubMed] [Google Scholar]
- 5.Bouzid A. CDH23 Methylation Status and Presbycusis Risk in Elderly Women. Front Aging Neurosci. 2018;10:241. doi: 10.3389/fnagi.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. J. Smith, J. F. Bale, et K. R. White, « Sensorineural hearing loss in children », The Lancet, vol. 365, no 9462, p. 879‑890, mars 2005, doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed]
- 7.L. Zelante et al., « Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans », Hum. Mol. Genet., vol. 6, no 9, p. 1605‑1609, sept. 1997. [DOI] [PubMed]
- 8.Aparisi M.J. Targeted next generation sequencing for molecular diagnosis of Usher syndrome. Orphanet J Rare Dis. 2014;9 doi: 10.1186/s13023-014-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakchouk I. NADf chip, a two-color microarray for simultaneous screening of multigene mutations associated with hearing impairment in North African Mediterranean countries. J. Mol. Diagn. JMD. 2015;17(2):155–161. doi: 10.1016/j.jmoldx.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 10.A. A. Gibriel, M. H. Abou-Elew, et S. Masmoudi, « Analysis of p.Gly12Valfs*2, p.Trp24* and p.Trp77Arg mutations in GJB2 and p.Arg81Gln variant in LRTOMT among non syndromic hearing loss Egyptian patients: implications for genetic diagnosis », Mol. Biol. Rep., vol. 46, no 2, p. 2139‑2145, avr. 2019, doi: 10.1007/s11033-019-04667-0. [DOI] [PubMed]
- 11.Zazo Seco C. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur J Hum Genet. 2017;25(3):308–314. doi: 10.1038/ejhg.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. E. Shearer et R. J. H. Smith, « Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care », Otolaryngol. Neck Surg., vol. 153, no 2, p. 175‑182, août 2015, doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed]
- 13.Shang H. Targeted Next-Generation Sequencing of a Deafness Gene Panel (MiamiOtoGenes) Analysis in Families Unsuitable for Linkage Analysis. Biomed Res Int. 2018;2018:3103986. doi: 10.1155/2018/3103986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y. Soares de Lima et al., « Syndromic hearing loss molecular diagnosis: Application of massive parallel sequencing », Hear. Res., vol. 370, p. 181‑188, déc. 2018, doi: 10.1016/j.heares.2018.10.008. [DOI] [PubMed]
- 15.S. Richards et al., « Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology », Genet. Med. Off. J. Am. Coll. Med. Genet., vol. 17, no 5, p. 405‑424, mai 2015, doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed]
- 16.A. M. Oza et al., « Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss », Hum. Mutat., vol. 39, no 11, p. 1593‑1613, nov. 2018, doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed]
- 17.Almontashiri N.A.M. Recurrent variants in OTOF are significant contributors to prelingual nonsydromic hearing loss in Saudi patients. Genet. Med. Off. J. Am. Coll. Med. Genet. 2018;20(5):536–544. doi: 10.1038/gim.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Dallol et al., « Utilization of amplicon-based targeted sequencing panel for the massively parallel sequencing of sporadic hearing impairment patients from Saudi Arabia », BMC Med. Genet., vol. 17, no Suppl 1, p. 67, oct. 2016, doi: 10.1186/s12881-016-0329-8. [DOI] [PMC free article] [PubMed]
- 19.Rodríguez-Ballesteros M. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008;29(6):823–831. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- 20.S. von Ameln et al., « A Mutation in PNPT1, Encoding Mitochondrial-RNA-Import Protein PNPase, Causes Hereditary Hearing Loss », Am. J. Hum. Genet., vol. 91, no 5, p. 919‑927, nov. 2012, doi: 10.1016/j.ajhg.2012.09.002. [DOI] [PMC free article] [PubMed]
- 21.Tlili A. Identification of a novel frameshift mutation in the DFNB31/WHRN gene in a Tunisian consanguineous family with hereditary non-syndromic recessive hearing loss. Hum Mutat. 2005;25(5) doi: 10.1002/humu.9333. [DOI] [PubMed] [Google Scholar]
- 22.Abdi S. Diversity of the Genes Implicated in Algerian Patients Affected by Usher Syndrome. PLoS ONE. 2016;11(9) doi: 10.1371/journal.pone.0161893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.I. Ben-Rebeh et al., « Genetic analysis of Tunisian families with Usher syndrome type 1: toward improving early molecular diagnosis », Mol. Vis., vol. 22, p. 827‑835, juill. 2016. [PMC free article] [PubMed]
- 24.Riahi Z. Whole Exome Sequencing Identifies Mutations in Usher Syndrome Genes in Profoundly Deaf Tunisian Patients. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnet C. Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J. Rare Dis. 2011;6:21. doi: 10.1186/1750-1172-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. Jaijo et al., « Functional analysis of splicing mutations in MYO7A and USH2A genes », Clin. Genet., vol. 79, no 3, p. 282‑288, mars 2011, doi: 10.1111/j.1399-0004.2010.01454.x. [DOI] [PubMed]
- 27.Roux A.-F. Four-year follow-up of diagnostic service in USH1 patients. Invest Ophthalmol Vis Sci. 2011;52(7):4063–4071. doi: 10.1167/iovs.10-6869. [DOI] [PubMed] [Google Scholar]
- 28.Jaijo T. MYO7A mutation screening in Usher syndrome type I patients from diverse origins. J Med Genet. 2007;44(3) doi: 10.1136/jmg.2006.045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A.-F. Roux et al., « Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90% », J. Med. Genet., vol. 43, no 9, p. 763‑768, sept. 2006, doi: 10.1136/jmg.2006.041954. [DOI] [PMC free article] [PubMed]
- 30.A. Adato et al., « Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins », Am. J. Hum. Genet., vol. 61, no 4, p. 813‑821, oct. 1997, doi: 10.1086/514899. [DOI] [PMC free article] [PubMed]
- 31.M. Ben Said et al., « A mutation in SLC22A4 encoding an organic cation transporter expressed in the cochlea strial endothelium causes human recessive non-syndromic hearing loss DFNB60 », Hum. Genet., vol. 135, no 5, p. 513‑524, mai 2016, doi: 10.1007/s00439-016-1657-7. [DOI] [PMC free article] [PubMed]
- 32.Tayoun A.N.A. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.F. Ammar-Khodja et al., « Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing », Mol. Genet. Genomic Med., vol. 3, no 3, p. 189‑196, mai 2015, doi: 10.1002/mgg3.131. [DOI] [PMC free article] [PubMed]
- 34.S. Ben Arab, S. Masmoudi, N. Beltaief, S. Hachicha, et H. Ayadi, « Consanguinity and endogamy in Northern Tunisia and its impact on non-syndromic deafness », Genet. Epidemiol., vol. 27, no 1, p. 74‑79, juill. 2004, doi: 10.1002/gepi.10321. [DOI] [PubMed]
- 35.T. Atik et al., « Comprehensive Analysis of Deafness Genes in Families with Autosomal Recessive Nonsyndromic Hearing Loss », PLoS ONE, vol. 10, no 11, 2015, doi: 10.1371/journal.pone.0142154. [DOI] [PMC free article] [PubMed]
- 36.E. Samorodnitsky et al., « Comparison of Custom Capture for Targeted Next-Generation DNA Sequencing », J. Mol. Diagn. JMD, vol. 17, no 1, p. 64‑75, janv. 2015, doi: 10.1016/j.jmoldx.2014.09.009. [DOI] [PMC free article] [PubMed]
- 37.M. Ben Saïd et al., « A novel missense mutation in the ESRRB gene causes DFNB35 hearing loss in a Tunisian family », Eur. J. Med. Genet., vol. 54, no 6, p. e535‑e541, nov. 2011, doi: 10.1016/j.ejmg.2011.06.008. [DOI] [PubMed]
- 38.M. Ansar et al., « A novel autosomal recessive non-syndromic deafness locus (DFNB35) maps to 14q24.1–14q24.3 in large consanguineous kindred from Pakistan », Eur. J. Hum. Genet., vol. 11, no 1, p. 77, janv. 2003, doi: 10.1038/sj.ejhg.5200905. [DOI] [PMC free article] [PubMed]
- 39.R. W. J. Collin et al., « Mutations of ESRRB Encoding Estrogen-Related Receptor Beta Cause Autosomal-Recessive Nonsyndromic Hearing Impairment DFNB35 », Am. J. Hum. Genet., vol. 82, no 1, p. 125‑138, janv. 2008, doi: 10.1016/j.ajhg.2007.09.008. [DOI] [PMC free article] [PubMed]
- 40.D. Šafka Brožková, J. Laštůvková, E. Machalová, J. Lisoňová, M. Trková, et P. Seeman, « DFNB35 due to a novel mutation in the ESRRB gene in a Czech consanguineous family », Int. J. Pediatr. Otorhinolaryngol., vol. 76, no 11, p. 1681‑1684, nov. 2012, doi: 10.1016/j.ijporl.2012.08.006. [DOI] [PubMed]
- 41.Wang A. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280(5368):1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- 42.M. Farjami et al., « The worldwide frequency of MYO15A gene mutations in patients with non-syndromic hearing loss: A meta-analysis », Iran. J. Basic Med. Sci., vol. 23, no 7, p. 841‑848, juill. 2020, doi: 10.22038/IJBMS.2020.35977.8563. [DOI] [PMC free article] [PubMed]
- 43.A. U. Rehman et al., « Mutational Spectrum of MYO15A and the Molecular Mechanisms of DFNB3 Human Deafness », Hum. Mutat., vol. 37, no 10, p. 991, oct. 2016, doi: 10.1002/humu.23042. [DOI] [PMC free article] [PubMed]
- 44.D. W. Anderson et al., « The motor and tail regions of myosin XV are critical for normal structure and function of auditory and vestibular hair cells », Hum. Mol. Genet., vol. 9, no 12, p. 1729‑1738, juill. 2000, doi: 10.1093/hmg/9.12.1729. [DOI] [PubMed]
- 45.E. Verpy et al., « A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C », Nat. Genet., vol. 26, no 1, p. 51‑55, sept. 2000, doi: 10.1038/79171. [DOI] [PubMed]
- 46.X. M. Ouyang et al., « Mutations in the alternatively spliced exons of USH1C cause non-syndromic recessive deafness », Hum. Genet., vol. 111, no 1, p. 26‑30, juill. 2002, doi: 10.1007/s00439-002-0736-0. [DOI] [PubMed]
- 47.H. Duzkale et al., « A systematic approach to assessing the clinical significance of genetic variants », Clin. Genet., vol. 84, no 5, p. 453‑463, nov. 2013, doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed]
- 48.L. M. Astuto et al., « Genetic Heterogeneity of Usher Syndrome: Analysis of 151 Families with Usher Type I », Am. J. Hum. Genet., vol. 67, no 6, p. 1569, déc. 2000, doi: 10.1086/316889. [DOI] [PMC free article] [PubMed]
- 49.R. Varga et al., « OTOF mutations revealed by genetic analysis of hearing loss families including a potential temperature sensitive auditory neuropathy allele », J. Med. Genet., vol. 43, no 7, p. 576‑581, juill. 2006, doi: 10.1136/jmg.2005.038612. [DOI] [PMC free article] [PubMed]
- 50.R. Varga et al., « Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene », J. Med. Genet., vol. 40, no 1, p. 45‑50, janv. 2003, doi: 10.1136/jmg.40.1.45. [DOI] [PMC free article] [PubMed]
- 51.I. Rouillon et al., « Results of cochlear implantation in two children with mutations in the OTOF gene », Int. J. Pediatr. Otorhinolaryngol., vol. 70, no 4, p. 689‑696, avr. 2006, doi: 10.1016/j.ijporl.2005.09.006. [DOI] [PubMed]
- 52.M. Tekin, D. Akcayoz, et A. Incesulu, « A novel missense mutation in a C2 domain of OTOF results in autosomal recessive auditory neuropathy », Am. J. Med. Genet. A., vol. 138A, no 1, p. 6‑10, sept. 2005, doi: 10.1002/ajmg.a.30907. [DOI] [PubMed]
- 53.E. Abdelkader, L. Enani, P. Schatz, et L. Safieh, « Severe retinal degeneration at an early age in Usher syndrome type 1B associated with homozygous splice site mutations in MYO7A gene », Saudi J. Ophthalmol., vol. 32, no 2, p. 119‑125, avr. 2018, doi: 10.1016/j.sjopt.2017.10.004. [DOI] [PMC free article] [PubMed]
- 54.H. Yoshimura, M. Miyagawa, K. Kumakawa, S. Nishio, et S. Usami, « Frequency of Usher syndrome type 1 in deaf children by massively parallel DNA sequencing », J. Hum. Genet., vol. 61, no 5, Art. no 5, mai 2016, doi: 10.1038/jhg.2015.168. [DOI] [PMC free article] [PubMed]
- 55.« Retinal Physician - Retinal Gene Therapies in Clinical Trials », Retinal Physician. https://www.retinalphysician.com/issues/2019/march-2019/retinal-gene-therapies-in-clinical-trials (consulté le sept. 23, 2020).
- 56.Mburu P. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34(4):421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- 57.I. Ebermann et al., « A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss », Hum. Genet., vol. 121, no 2, p. 203‑211, avr. 2007, doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed]
- 58.Besnard T. Non-USH2A mutations in USH2 patients. Hum Mutat. 2012;33(3):504–510. doi: 10.1002/humu.22004. [DOI] [PubMed] [Google Scholar]
- 59.Mustapha M. DFNB31, a recessive form of sensorineural hearing loss, maps to chromosome 9q32-34. Eur J Hum Genet. 2002;10(3):210. doi: 10.1038/sj.ejhg.5200780. [DOI] [PubMed] [Google Scholar]
- 60.Yan D. Spectrum of DNA variants for nonsyndromic deafness in a large cohort from multiple continents. Hum Genet. 2016;135(8):953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olsson K.S. Common founder effects of hereditary hemochromatosis, Wilsońs disease, the long QT syndrome and autosomal recessive deafness caused by two novel mutations in the WHRN and TMC1 genes. Hereditas. 2017;154 doi: 10.1186/s41065-017-0052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Riazuddin et al., « Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48 », Nat. Genet., vol. 44, no 11, p. 1265‑1271, nov. 2012, doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed]
- 63.K. T. Booth et al., « Variants in CIB2 cause DFNB48 and not USH1J », Clin. Genet., vol. 93, no 4, p. 812‑821, avr. 2018, doi: 10.1111/cge.13170. [DOI] [PMC free article] [PubMed]
- 64.K. Patel et al., « Correction: A Novel C-Terminal CIB2 (Calcium and Integrin Binding Protein 2) Mutation Associated with Non-Syndromic Hearing Loss in a Hispanic Family », PLoS ONE, vol. 10, no 10, oct. 2015, doi: 10.1371/journal.pone.0141259. [DOI] [PMC free article] [PubMed]
- 65.C. Z. Seco et al., « Novel and recurrent CIB2 variants, associated with nonsyndromic deafness, do not affect calcium buffering and localization in hair cells », Eur. J. Hum. Genet. EJHG, vol. 24, no 4, p. 542‑549, avr. 2016, doi: 10.1038/ejhg.2015.157. [DOI] [PMC free article] [PubMed]
- 66.G. Bademci et al., « Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort », Genet. Med. Off. J. Am. Coll. Med. Genet., vol. 18, no 4, p. 364‑371, avr. 2016, doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pedigrees and Sequence chromatograms showing segregation’s analysis of eleven detected variants in known deafness genes in patients with nonsyndromic hearing impairment and Usher Syndrome. −/−: homozygous mutant; +/−: heterozygous; +/+: homozygous wild type.