Abstract
Hydrogels are engineered with biochemical and biophysical signals to recreate aspects of the native microenvironment and to control cellular functions such as differentiation and matrix deposition. This deposited matrix accumulates within the pericellular space and likely affects the interactions between encapsulated cells and the engineered hydrogel; however, there has been little work to study the spatiotemporal evolution of matrix at this interface. To address this, metabolic labeling is employed to visualize the temporal and spatial positioning of nascent proteins and proteoglycans deposited by chondrocytes. Within covalently crosslinked hyaluronic acid hydrogels, chondrocytes deposit nascent proteins and proteoglycans in the pericellular space within 1 d after encapsulation. The accumulation of this matrix, as measured by an increase in matrix thickness during culture, depends on the initial hydrogel crosslink density with decreased thicknesses for more crosslinked hydrogels. Encapsulated fluorescent beads are used to monitor the hydrogel location and indicate that the emerging nascent matrix physically displaces the hydrogel from the cell membrane with extended culture. These findings suggest that secreted matrix increasingly masks the presentation of engineered hydrogel cues and may have implications for the design of hydrogels in tissue engineering and regenerative medicine.
Keywords: chondrogenesis, extracellular matrix, hyaluronic acid hydrogels, tissue engineering
1. Introduction
The extracellular matrix (ECM) is fundamental to the structure and function of almost all tissues throughout the body. Within native tissues, resident cells constantly receive and respond to signals from their extracellular microenvironment, including biochemical and biophysical cues that are critical to tissue development, homeostasis and repair.[1] In articular cartilage, chondrocytes and progenitor cells reside in a protein and proteoglycan-rich ECM that is dominated by collagens and aggrecan at tissue maturity.[2] However, degenerative pathologies or acute injury often result in tissue degradation, which in turn impairs function of the affected joint.[3] The poor intrinsic healing capacity of articular cartilage motivates the development of tissue-engineered constructs in vitro to replace degenerated and injured cartilage tissue.[4]
Commonly, cartilage tissue engineering includes chondrocytes or mesenchymal stromal cells (MSCs) embedded into biomaterials that recreate certain aspects of the native cartilage ECM structure, organization and composition.[5] To engineer a tissue with high water content like articular cartilage, hydrogels, water-swollen polymer networks, are often used to recapitulate the chemical and biological features of cartilage ECM.[6] The choice of the hydrogel material and its biophysical properties modulate embedded cell behavior, including the formation and distribution of secreted matrix, and ultimately defines the properties of the engineered tissue construct. For example, hydrogels made of articular cartilage components, such as hyaluronic acid (HA), enhance the expression of cartilage-specific markers when compared to cells encapsulated within inert polymeric hydrogels including those fabricated from poly(ethylene glycol).[7] In addition to these biochemical signals, hydrogel density, degradation, and mechanical properties affect chondrogenesis, as well as matrix synthesis by encapsulated cells and the overall engineered tissue properties.[8] However, there has been little work done to date to investigate how the matrix produced by individual cells influences interactions with the engineered hydrogel locally and over time, likely due to limitations in visualizing and measuring secreted matrix dynamics.
Within hydrogels, matrix production is often reported using histological techniques and biochemical assays collected over time. However, such measures mask cell-to-cell heterogeneity, have limited resolution, and lack information regarding the spatial distribution of the secreted matrix. Recently, metabolic labeling was reported to visualize proteins that individual cells secrete and assemble within hydrogels[9] and tissues;[10] however, there are no studies regarding the imaging of proteins and proteoglycans around cells in combination with an understanding of the location of the engineered material to probe the spatiotemporal nature of this interface. Specifically, the secreted proteins and proteoglycans may influence cell–hydrogel interactions by either i) physically separating the cell from the hydrogel or ii) interpenetrating with the engineered hydrogel—an understanding of this behavior may influence future hydrogel design.
To probe this question, we used metabolic labeling to visualize newly produced proteins and proteoglycans that were secreted by chondrocytes embedded within covalently crosslinked HA hydrogels, and used encapsulated fluorescent beads to monitor the position of the hydrogel with respect to the nascent matrix during culture. The crosslink density of these hydrogels is readily tunable, allowing us to further probe the role of hydrogel crosslinking on secreted matrix properties, including its distribution and mechanics. Nondegradable hydrogels were selected to ensure stability and bead immobilization throughout culture. Understanding of these spatiotemporal changes at the chondrocyte–hydrogel interface will inform hydrogel design strategies for cartilage tissue engineering and repair. Further, although the work was completed in the context of encapsulated chondrocytes and cartilage formation, the implications of the study may extend into numerous applications where cells interface with engineered material systems.
2. Results and Discussion
2.1. Secreted Matrix Contains Proteins and Proteoglycans
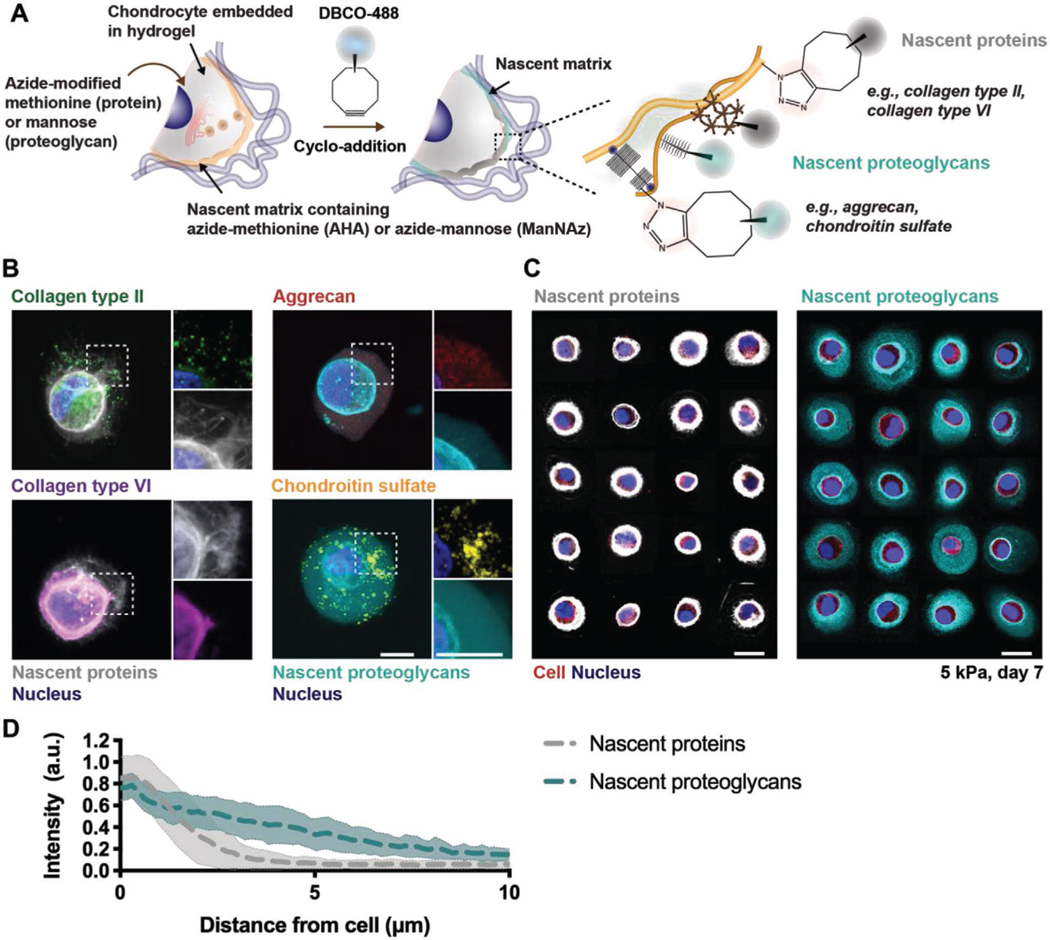
To visualize matrix secretion by cells encapsulated within hydrogels, we implemented a metabolic labeling approach that introduces azide-modified amino acids or sugar analogs to cultures for incorporation into matrix components as they are synthesized. Here, we separately used the methionine analog azidohomoalanine (AHA)[9a,b] or the sugar analog N-azidoacetyl-mannosamine-tetraacylated (ManNAz)[11] to label newly synthesized matrix. Subsequently, strain-promoted cyclo-addition with a fluorophore-modified cyclooctyne (DBCO-488) was performed to fluorescently label AHA-containing or ManNAz-containing matrix (Figure 1A). This metabolic labeling approach provided a global representation of all methionine- or mannose- containing matrix components synthesized by cells, as supported by the high structural analogy with specific proteins, such as collagens type II and type VI for the AHA labeling, or the proteoglycan aggrecan and associated chondroitin sulfate residues for the ManNAz labeling, that were also labeled through immunostaining (Figure 1B). This approach is very useful to provide a broad investigation of ECM components synthesized, which is important for our intended goal, rather than focusing only on specific molecules, which may miss others. Note that although we categorize these as nascent proteins (AHA) and nascent proteoglycans (ManNAz) due to our immunostaining results and the general abundance of these matrix components in cartilage tissue, other molecules (e.g., glycoproteins) will also be labeled and this should be considered when interpreting the results.
Figure 1.
Chondrocytes secrete extracellular matrix proteins and proteoglycans that are heterogeneous and can be visualized with metabolic labeling. A) Schematic of nascent protein and proteoglycan metabolic labeling. The azide-modified methionine analog (azidohomoalanine) or mannose analog (N-azidoacetylmannosamine tetraacylated) are added to the culture media and incorporated into nascent proteins (for example, collagen type II and collagen type VI) or proteoglycans (for example, aggrecan core protein and its associated chondroitin sulfate residues), respectively. The biorthogonal Cu(I)-free strain-promoted cyclo-addition between the azide and DBCO-modified fluorophore (DBCO-488) enables visualization of either the nascent proteins or proteoglycans. B) Representative images (magnifications on the right) of nascent proteins (white, co-stained with either collagen type II and VI) or proteoglycans (cyan, co-stained with aggrecan and chondroitin sulfate) deposited by bovine chondrocytes encapsulated in norbornene-modified hyaluronic acid (NorHA) hydrogels and cultured for 7 d in chondrogenic media (scale bars 10 μm). C) Representative images of nascent proteins (white, left) and proteoglycans (cyan, right) for a panel of chondrocytes (red) after 7 d of culture (scale bars 10 μm). D) Radial profiles of fluorescence intensities of nascent proteins (gray) and nascent proteoglycans (cyan) after 7 d of culture. Lines represent median intensity profiles, shaded areas represent standard deviation (n = 40 cells per group from two independent biological experiments).
To investigate the spatiotemporal accumulation of this nascent matrix, bovine chondrocytes were encapsulated within norbornene-modified hyaluronic acid (NorHA) hydrogels with an elastic modulus of 4.9 ± 0.4 kPa and cultured in chondrogenic media and the continuous presence of either AHA or ManNAz for 7 d. Individual cells exhibited a dense network of nascent proteins and proteoglycans surrounding the cell body (Figure 1C). Radial intensity profile plots across nascent matrix components revealed that nascent proteoglycans extend much further into the hydrogel (≥10 μm), whereas nascent protein fluorescence intensity diminished within 5 μm from the cell body (Figure 1D), suggesting that proteins and proteoglycans accumulate with different spatial distributions after being secreted by chondrocytes within hydrogels. This could be due to differences in the general size and assembly of these molecules, as many of the proteins are large fibrillar structures when compared to many of the proteoglycans. This is consistent with previous observations of greater diffusion of glycosaminoglycans when compared to proteins such as collagen.[8d,12]
Beyond spatial differences, the deposition of these ECM molecules is also cell type dependent.[9b,13] We explored similar studies to that performed with chondrocytes, but now with MSCs, which revealed less accumulation of nascent proteins and proteoglycans and distributions that were often asymmetric around the cell body (Figure S1, Supporting Information). When compared to chondrocytes, high cell-to-cell variability was observed, likely due to the heterogeneity of isolated MSC populations;[9b,14] thus, chondrocytes were used for subsequent studies. An important consideration when performing these labeling techniques is that proteoglycans such as aggrecan also possess proteins (i.e., aggrecan core protein). However, the concentration of methionine in aggrecan (<0.06%) is relatively low when compared to proteins most commonly found in cartilage (e.g., 1.5% methionine in collagen type II).[9b] Thus, the low number of methionine residues suggests that nascent protein labeling is unable to capture the detailed structure of proteoglycans. Taken together, these observations indicate that metabolic labeling through incorporation of azide-modified methionine and mannose can visualize structural differences in the accumulation of both nascent proteins and proteoglycans, allowing us to investigate the spatiotemporal accumulation of the pericellular matrix.
2.2. Secreted Matrix Accumulates over Time
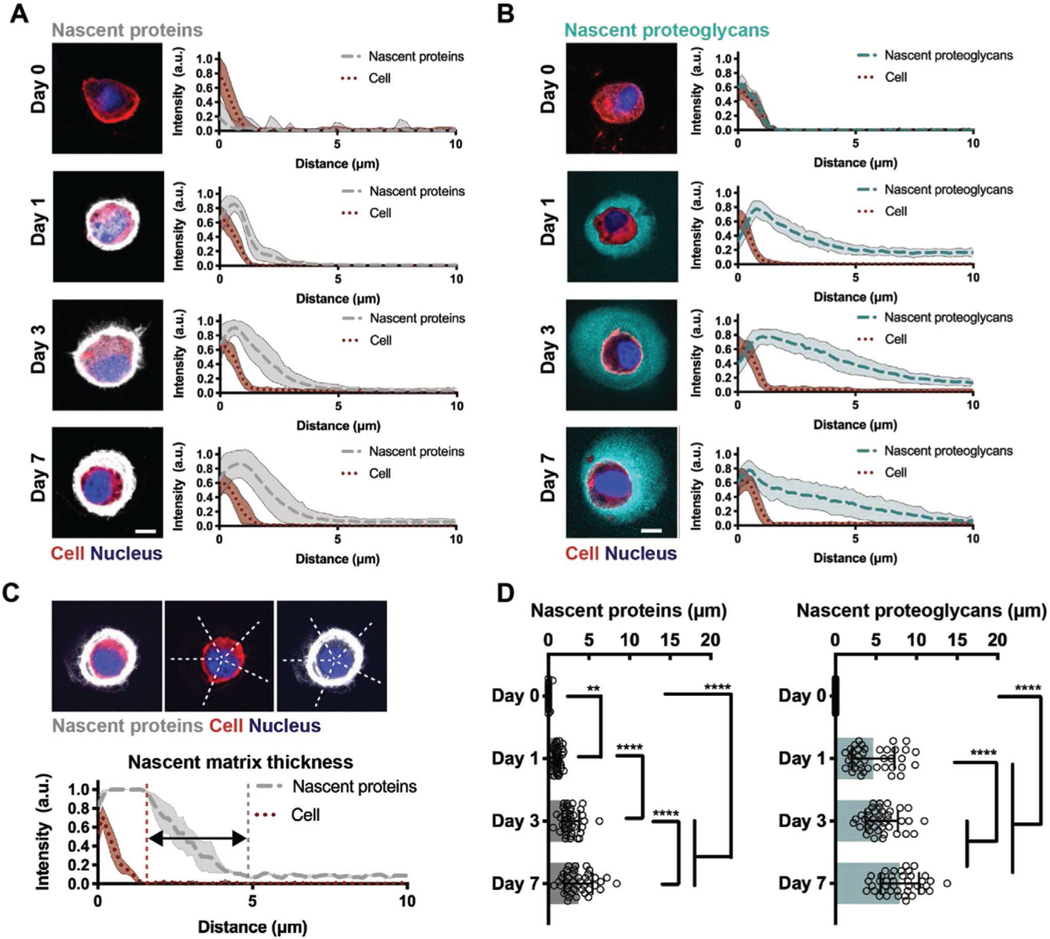
To assess the spatiotemporal changes that occur at the cell– hydrogel interface, we next monitored the time course of nascent matrix accumulation by encapsulated chondrocytes. No staining of nascent proteins was observed directly upon encapsulation (Day 0), but within 1 d of culture chondrocytes were surrounded by a nascent protein layer, consistent with the corresponding radial intensity profile plots of nascent protein fluorescence intensity relative to the cell boundary (Figure 2A). When culture in chondrogenic media was extended for 7 d, the thickness of this protein layer increased, indicating that nascent proteins accumulated in the pericellular space. Labeling of nascent proteoglycans revealed similar deposition and accumulation around the cell body; however, the nascent proteoglycans extended further into the hydrogel at each time point (Figure 2B), suggesting that the accumulation and distribution of nascent matrix are influenced by the size and/or assembly of its components.
Figure 2.
Nascent proteins and proteoglycans are deposited early and accumulate over time. A) Representative images of accumulated nascent proteins of encapsulated chondrocytes cultured in chondrogenic media for up to 7 d (left, scale bars 10 μm) and radial intensity profiles of nascent proteins (gray). Lines represent median intensity profiles; shaded areas represent standard deviation (n = 40 cells per group from two independent biological experiments). B) Representative images of accumulated nascent proteoglycans of encapsulated chondrocytes cultured in chondrogenic media for up to 7 d (left, scale bars 10 μm) and radial intensity profiles of nascent proteoglycans (cyan). Lines represent median intensity profiles, shaded areas represent standard deviation (n = 40 cells per group from two independent biological experiments). C) Representative cell and corresponding intensity profile plots illustrating the region used to quantify the nascent matrix thickness. D) Quantification of the average thickness of nascent proteins (left) and nascent proteoglycans (right) of encapsulated chondrocytes cultured for up to 7 d (n = 40 cells per group from two independent biological experiments, mean ± SD, **p ≤ 0.01, ****p ≤ 0.0001, two-way ANOVA with Bonferroni post hoc).
Intensity profile plots of nascent matrix components permitted quantification of the average thickness of nascent proteins and proteoglycans as the distance that the matrix extends past the cell body (Figure 2C). After one day, accumulated matrix thickness was higher for nascent proteoglycans (4.7 ± 2.2 μm) relative to proteins (1.1 ± 1.6 μm), and similar trends in nascent protein and proteoglycan accumulation were observed during the seven day culture period (Figure 2D). These findings highlight that, within hydrogels, nascent matrix deposition occurs rapidly (within 1 d) and again that accumulation is dependent on the type of its components, presumably due to differences in size and structure of chondrogenic matrix proteins and proteoglycans. Such differences in matrix distribution likely reflect intrinsic regulatory mechanisms during cartilage development and repair. For example, collagen type VI forms a distinct pericellular network while proteoglycans including aggrecan further distribute into the extracellular space, modulating biochemical and biomechanical signals to chondrocytes.[15] Thus, metabolic labeling provides a unique tool to visualize the organization and dynamics of both nascent proteins and proteoglycans.
2.3. Secreted Matrix Physically Displaces the Hydrogel from the Cell
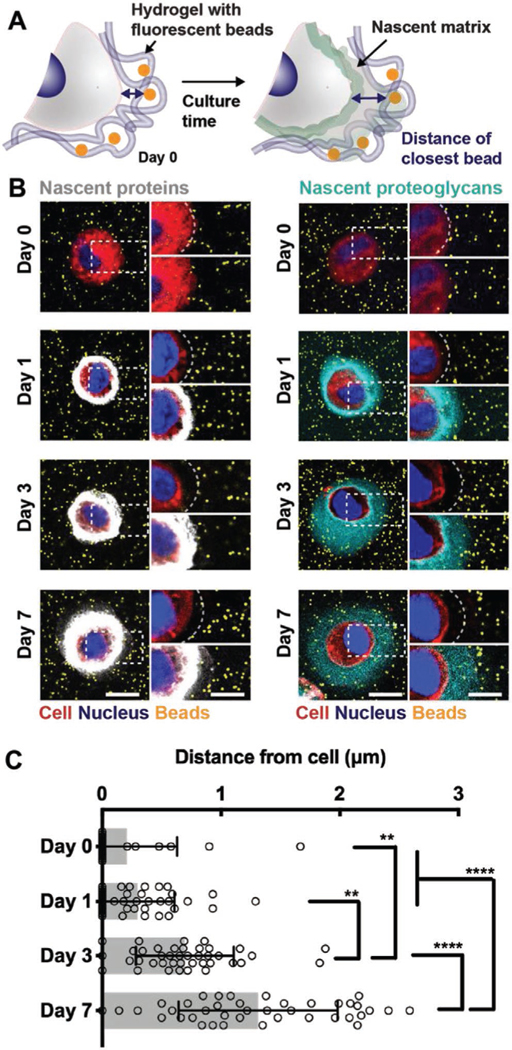
While previous studies indicate that chondrogenic matrix distribution is dependent on molecule diffusion,[8d,12] it is unclear how the nascent proteins and proteoglycans interact with the hydrogel that the cell is embedded within. We next sought to understand whether the nascent matrix alters this interface by pushing and thus physically displacing the cell from the engineered hydrogel or by integrating within the mesh of the hydrogel (Figure 3A). Fluorescent polystyrene beads that are bigger than the average hydrogel mesh size (nominal diameter of 0.2 μm) were physically immobilized in the hydrogel upon gelation. We confirmed their homogenous distribution and stability throughout the culture period of 7 d (Figure S2, Supporting Information) and showed that the addition of fluorescent beads did not affect the viability of encapsulated chondrocytes cultured for up to 7 d in media that was supplemented with either AHA or ManNAz (Figure S3, Supporting Information). Across all conditions and time points, the viability generally remained greater than 90%. The localization of immobilized beads was then evaluated relative to the accumulated nascent matrix during culture.
Figure 3.
Nascent proteins physically displace the hydrogel from the cell membrane while proteoglycans interdigitate further within the hydrogel. A) Schematic of hydrogel encapsulated beads to visualize the cell–hydrogel interface. Fluorescent beads (diameter 0.2 μm) are co-encapsulated with chondrocytes and nascent matrix labeling enables visualization and quantification of the cell–hydrogel interface. B) Representative images (magnifications on the right) of accumulated nascent proteins (white, left) and nascent proteoglycans (cyan, right) of chondrocytes co-encapsulated with fluorescent beads (orange) and cultured for up to 7 d (scale bars 10 μm). C) Quantification of the distance of the closest bead per cell for cultures up to 7 d and labeled for nascent proteins (see Figure S4 (Supporting Information) for methodology and Figure S5 (Supporting Information) for bead distance of chondrocytes when labeled for nascent proteoglycans, n = 40 cells per group from two independent biological experiments, mean ± SD, **p ≤ 0.05, ****p ≤ 0.0001, two-way ANOVA with Bonferroni post hoc).
Upon encapsulation (Day 0), we observed fluorescent beads adjacent to the cell membrane (Figure 3B), indicating direct initial contact between chondrocytes and the hydrogel. This separation between chondrocytes and the bead-containing hydrogel increased over the 7 d of culture in both nascent protein and proteoglycan-labeled groups, suggesting that the initial chondrocyte–hydrogel contact is lost during culture. Measuring the distance of the closest bead per cell enabled quantification of the chondrocyte-to-bead separation (Figure S4, Supporting Information). The gap between chondrocytes and immobilized beads increased during the 7 d culture period, reaching distances of 1.3 ± 1.0 μm (Figure 3C); however, this gap was smaller than the thickness of accumulated proteins (3.7 ± 1.9 μm) and proteoglycans (7.9 ± 3.2 μm) at day 7 (Figure 2D), indicating some distribution of these matrix molecules throughout the hydrogel. We also confirmed that the matrix labeling method itself did not affect the chondrocyte-to-bead separation, as the measured bead distances were similar for nascent proteoglycan-labeled cells (Figure S5, Supporting Information). Generally, these data suggest that nascent matrix alters the chondrocyte–hydrogel interface in two ways: i) physical displacement of the hydrogel through accumulation in the pericellular space and ii) interpenetration within the hydrogel through distribution further from the cell body.
2.4. Hydrogel Crosslink Density Influences the Chondrocyte-Hydrogel Interface
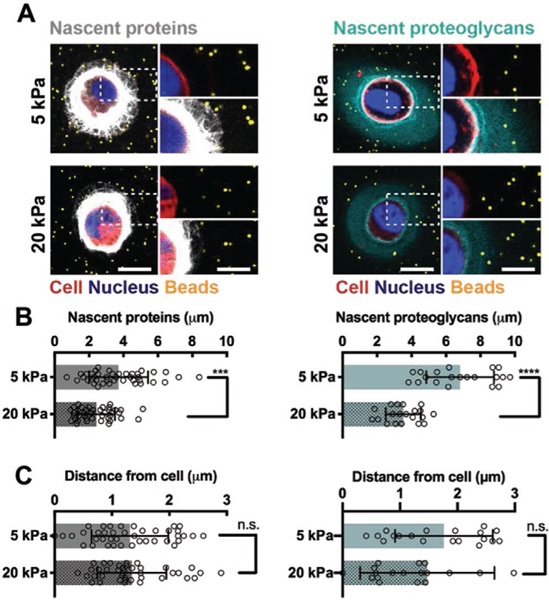
Given that nascent matrix accumulation alters the interactions between the cell and the hydrogel, we next sought to understand whether the properties of the engineered hydrogel impacts protein and proteoglycan deposition by culturing chondrocytes within hydrogels of varied crosslink density. The physical properties of the hydrogel may alter the cellular microenvironment, influencing the deposition and distribution of secreted matrix through changes in the diffusivity of the matrix (Figure S6, Supporting Information). To determine how higher crosslink density modulates nascent matrix accumulation, we encapsulated chondrocytes into hydrogels with higher crosslinking and higher elastic moduli (19.2 ± 1.3 kPa, defined as ‘20 kPa’) as measured in compression and compared these to the previously used 5 kPa hydrogels. Hydrogel moduli for both 5 and 20 kPa constructs were stable in chondrogenic media and enabled culture of viable chondrocytes for up to 7 d (Figure S7, Supporting Information).
After 1 day of culture, the thickness of accumulated matrix was reduced for chondrocytes in 20 kPa hydrogels compared to 5 kPa hydrogels (Figure S8, Supporting Information), and the same trends were maintained after 7 d of culture (Figure 4A). The increase in crosslink density also reduced the thickness of nascent matrix secreted by encapsulated MSCs (Figure S9, Supporting Information), and overall cell-to-cell variability in matrix accumulation decreased when compared to MSCs cultured within 5 kPa hydrogels (Figure S1, Supporting Information). This indicates that both chondrocytes and MSCs within hydrogels respond to hydrogel crosslink density, which controls the extent of matrix deposition and distribution, consistent with previous reports.[9b,16] Quantification of the nascent protein and proteoglycan thickness confirmed an approximately twofold decrease in thickness for chondrocytes encapsulated within 20 kPa hydrogels relative to those encapsulated within 5 kPa hydrogels (Figure 4B). Conversely, the bead distances from the cells were not significantly different between the 5 and 20 kPa hydrogels at day 7 (Figure 4C), indicating a similar separation of the hydrogel from the cell. Thus, this suggests that at higher crosslinking densities the nascent matrix is more restricted to the pericellular space and cannot extend into the hydrogel as it does in less crosslinked hydrogels, likely reflecting a potential change in local matrix structure and architecture. These observations are consistent with previous reports on increased matrix distribution in lower density hydrogels such as hyaluronic acid, agarose and poly(ethylene glycol) hydrogels;[8b,d,9b] however, it highlights that crosslink density differentially influences protein and proteoglycan distributions. Given this, monitoring newly secreted matrix components separately and over time may inform the design of hydrogels that enable matrix diffusion and connectivity, and as such enhance functional tissue maturation.[17]
Figure 4.
Hydrogel crosslink density influences the cell–hydrogel interface. A) Representative images (magnifications on the right) of accumulated nascent proteins (white, left) and nascent proteoglycans (cyan, right) of chondrocytes co-encapsulated with fluorescent beads (orange) and cultured in softer (5 kPa) and stiffer (20 kPa) hydrogels after 7 d of culture (scale bars 10 μm). B) Quantification of the average thickness of nascent proteins (left) and nascent proteoglycans (right) of encapsulated chondrocytes after 7 d of culture (n = 40 cells of two independent biological experiments (nascent proteins), n = 20 cells (nascent proteoglycans), mean ± SD, ***p ≤ 0.001, ****p ≤ 0.0001, two-tailed Student’s t-test). C) Quantification of the distance of the closest bead to chondrocytes after labeling for nascent proteins (left) or nascent proteoglycans (right) after 7 d of culture (n = 40 cells of two independent biological experiments (nascent proteins), n = 20 cells (nascent proteoglycans), mean ± SD with no significant difference by two-tailed Student’s t-test).
2.5. Hydrogel Crosslink Density Influences Micromechanical Properties at the Cell–Hydrogel Interface
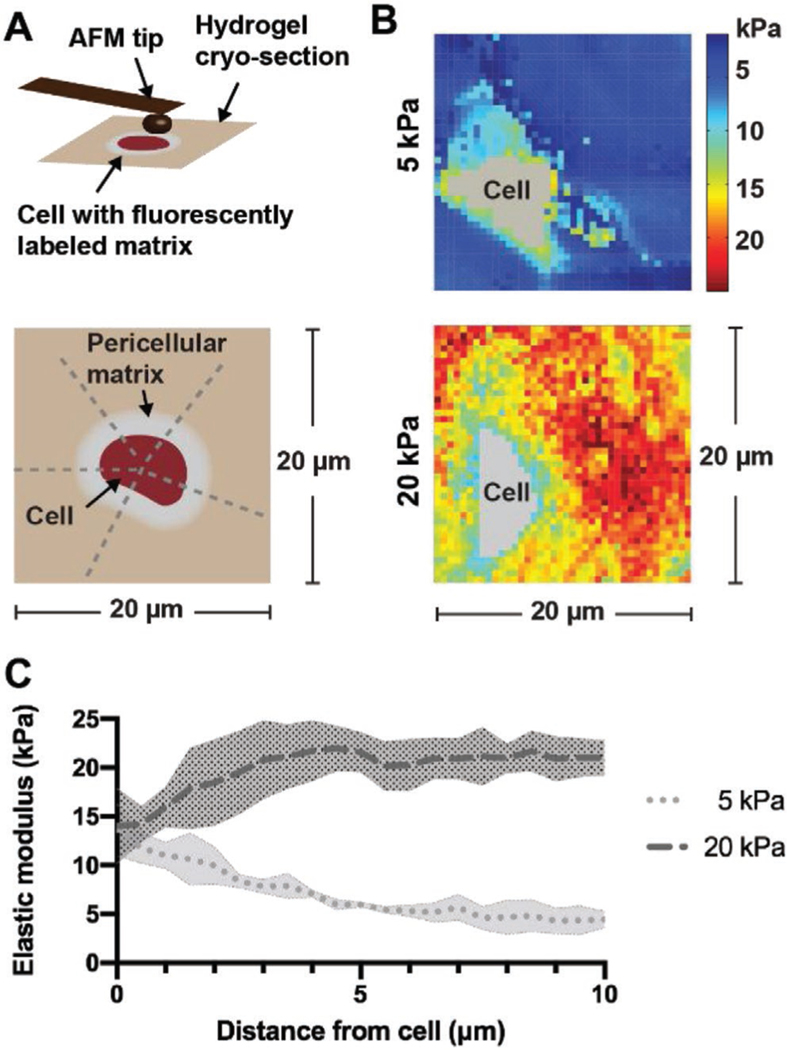
As the accumulation of matrix alters the biophysical interaction of the cell with the hydrogel,[18] we next asked how these nascent matrix-mediated changes in the cellular environment influence the local micromechanical properties. To begin to understand the mechanical landscape of nascent matrix deposited by cells in hydrogels, we used fluorescence-guided atomic force microscopy (AFM) upon metabolic labeling and cryosectioning.[19] This technique allowed us to identify the pericellular environment and measure the local elastic moduli around individual cells. Guided by the metabolically labeled matrix, we generated micromechanical maps of 20 μm × 20 μm dimension, and radial profile plots enabled quantification of the pericellular mechanics (Figure 5A).
Figure 5.
Hydrogel crosslink density alters pericellular mechanics at the cell–hydrogel interface. A) Schematic (top) illustrating fluorescence-guided atomic force microscopy (AFM) indentation of an 8 μm-thick unfixed cryosection and the regions (bottom) used to plot the elastic moduli of the pericellular matrix. B) Representative force maps (20 μm × 20 μm) of a region containing a chondrocyte within 5 and 20 kPa hydrogels after 7 d of culture in chondrogenic media. C) Radial profiles of the pericellular matrix elastic moduli measured after 7 d of culture. Lines represent median intensity profiles; shaded areas represent standard deviation (n = 4 cells per group from three independent biological experiments).
Micromechanical mapping confirmed that the elastic modulus was altered by varying the crosslink density as indicated by similar values measured under compression (5 kPa: 5.1 ± 2.4 kPa; 20 kPa: 19.2 ± 2.1 kPa). Using this technique, we observed distinct pericellular regions that extended into the hydrogel (5 kPa), whereas these regions were found more closely to the cell boundary in 20 kPa hydrogels (Figure 5B). These observations corresponded well with nascent matrix measurements in 5 and 20 kPa hydrogels after 7 d of culture (Figure 4B). Radial profile analysis across the pericellular environment showed decreased moduli as a function of distance from the cell boundary, which plateaued at 5 kPa, consistent with the elastic modulus of the initial hydrogel (Figure 5C). Conversely, in 20 kPa hydrogels the modulus was lower in the pericellular region and increased outward from the cell body. These findings confirm that nascent matrix distribution by cells encapsulated within hydrogels is influenced by the properties of the existing microenvironment and likely contributes to the mechanical properties of their pericellular environment.
Previous reports have used AFM indentation to explore changes in pericellular matrix mechanics of hydrogel-embedded cells by measuring heterogeneity as an indicator of differences in matrix stiffness.[20] However, the method established here allows characterization of individual cells and their local matrix environment, which could broadly be impactful toward determining how this mechanical environment is sensed by encapsulated cells. Specifically, we found that chondrocytes modulate their pericellular matrix mechanics, and that this process leads to pericellular matrix mechanics that are stiffer or softer than the initial hydrogel modulus. Previous work has studied the micromechanical environment of chondrocytes with regard to mechanical loading in which pericellular matrix structure and integrity regulated the mechanochemical changes that can be sensed by the cells.[21] This may also be important in the context of tissue engineering where in vivo physical cues, such as mechanical loading, are often recapitulated to enhance chondrogenic differentiation and matrix distribution.[22] Understanding how nascent proteins and proteoglycans contribute to the transduction of these extracellular signals may improve the design of biomaterials and mechanical loading regimes that enhance maturation of engineered cartilage constructs.
3. Conclusion
In this work, we assessed the influence of newly secreted matrix accumulation and engineered hydrogel properties on cell– hydrogel interactions of chondrocytes and MSCs undergoing chondrogenesis. Synthetic hydrogels are widely used as a system that recapitulates certain aspects of native tissue and supports chondrogenesis and maturation into functional tissue.[6] The aim of this study was to probe the interface between cells and hydrogels by using a nondegradable hydrogel system that enables quantitative measurements, but with the limitation that this culture model does not replicate the physiological or the pathological state of cartilaginous tissue. By labeling the nascent matrix and localizing the engineered hydrogel with immobilized fluorescent beads, we obtained a further understanding of the spatiotemporal changes at the chondrocyte–hydrogel interface. We found that within 1 day of culture secreted proteins and proteoglycans accumulated in the pericellular space and that this nascent matrix physically displaced the hydrogel from the cell membrane. With time, the newly secreted matrix distributed further throughout the hydrogel and altered the pericellular mechanics, and this was controlled by the crosslinking density of the hydrogel. These findings build on previous observations of matrix distribution within engineered hydrogels and demonstrate that these bulk changes impact the engineered pericellular environment of individual cells.
These observed differences and insights into this interface will be important when designing hydrogels to enhance chondrogenesis, including the modification with proteins or peptides that are engineered to directly interact with encapsulated cells.[23] Further, these tools may be useful for exploring how new approaches to modify hydrogels that sequester or interact with matrix components may influence matrix distribution and this interface, as well as the release of these components into the media. While this study sought to define the effect of matrix accumulation on physical interactions within relatively simple hydrogels, the inclusion of degradable crosslinkers or polymers[8c,24] and other parameters such as soluble factors can also alter matrix secretion and distribution.[25] Notably, nascent protein and/or proteoglycan labeling would also be able to identify matrix patterns in response to partial degradation in hydrogels crosslinked with varying ratios of degradable and nondegradable bonds[7a,26] or photodegradable linkages (e.g., ortho-nitrobenzyl (o-NB) linker groups).[27] In addition, juvenile bovine chondrocytes used in this study quickly deposited a dense pericellular matrix, but the ability of embedded cells, as indicated by encapsulated bovine MSCs, to synthesize and accumulate this matrix will be dependent on cell type, donor species and age, as well as in vitro passaging prior encapsulation.[28] Taken together, our findings implicate that the timing and accessibility of these engineered signals may be a critical parameter in the design of biomaterials for applications in tissue engineering and regenerative medicine.
4. Experimental Section
Hydrogel Synthesis:
NorHA was synthesized by first converting sodium hyaluronate (75 kDa, Lifecore, Chaska, MN) to HA tert-butyl ammonium salt (HA-TBA) using Dowex 50W proton exchange resin. HA-TBA was then reacted in the presence of (3 equivalent) 4-(dimethylamino)pyridine (1.5 equivalent) and ditert-butyl dicarbonate (Boc2O, 0.4 equivalent). The product was purified by dialyzing against deionized water for ≈2 weeks, adding sodium chloride (NaCl) (1 g NaCl per 100 mL of solution), and then precipitating with tenfold excess acetone at 4 °C, as previously described.[29] The precipitate was re-dissolved in deionized water, frozen at −80 °C, and then lyophilized. The macromer was characterized by 1H nuclear magnetic resonance (Bruker Advance 360 MHz, Bruker, Billerica, MA) and then stored at −20 °C (Figure S10, Supporting Information).
Hydrogel Formation:
Macromers were sterilized using a germicidal lamp in a laminar flow hood for 30 min. NorHA was dissolved in sterile phosphate buffered saline (PBS) containing 0.05 wt% 2-methyl1-[4-(hydroxyethoxy)phenyl]-2-methyl-1-propanone (Irgacure 2959, I2959, Ciba, Basel, Switzerland) and dithiothreitol (DTT, Millipore Sigma) for crosslinking. The degree of crosslinking was controlled via the ratio of thiols in DTT to norbornene groups.
Mechanical Characterization:
Hydrogels (5 mm diameter cylinders) underwent compression testing using a Dynamic Mechanical Analyzer Q800 (DMAQ800, TA Instruments, New Castle, DE). Samples were preloaded (0.01 N) and compressed via force ramp (0.5 N min−1) until they reached 70% of their initial thickness. The compressive moduli were calculated as the slope from 10% to 20% strain.
Fluorescence Recovery after Photobleaching (FRAP):
FRAP experiments were performed using a Leica SP5 II confocal microscope. Hydrogel films (≈660 μm thickness) containing 100 μM soluble fluorescein isothiocyanate-(FITC) dextran (average molecular weight 70 and 150 kDa) were placed between coverslips. A 40 μm diameter region was bleached with an Argon laser for 40 s; pre- and postbleaching images were recorded every 4 s. Data were analyzed using a custom MATLAB script that fit recovery using an exponential model
| (1) |
where f(t) is the normalized recovery profile, t is the time (s), τD is the characteristic diffusion time (s), and b is an approximation of the total fraction of the total fraction of FITC-dextran that is bleached.
Effective diffusivities were calculated according to
| (2) |
where Deff is the effective diffusivity (μm2 s−1) and ω represents the bleach spot radius (μm).[7b,30]
Cell Encapsulation and Culture:
Primary bovine chondrocytes and MSCs were isolated from juvenile bovine knees (Research 87, Boylston MA) as previously described.[31] Briefly, articular cartilage pieces were digested in collagenase for 20 h at 37 °C/5% CO2 and filtered (70 μm cell strainer). For MSC isolation, trabecular bone cubes were washed with Dulbecco’s modified Eagle’s medium (DMEM) (2 mg mL−1 heparin) to collect bone marrow, cells pelleted and cultured in high-glucose DMEM until colonies showed ≈80% confluency.[32] Chondrocytes or MSCs were then encapsulated at a density of 5 × 106 cells mL−1 in NorHA hydrogel films (≈660 μm thickness) that were cut into 5 mm × 5 mm constructs and cultured in 48-well plates. When stated, red-fluorescent polystyrene beads (nominal diameter ≈0.190 to 0.210 μm, emission/excitation 540/600 nm, Bangs Laboratories) were co-encapsulated with cells at ≈3 × 1010 beads mL−1 to visualize the hydrogels. Constructs were cultured in “AHA media,” a chondrogenic media consisting of glutamine-, methionine-, and cystine-free high-glucose DMEM, 0.1 × 10−6 m dexamethasone (Sigma-Aldrich), 4 × 10−3 m GlutaMAX supplement (Thermo Fisher), 0.201 × 10−3 m l-cystine (Sigma-Aldrich), 100 μg mL−1 sodium pyruvate (Cellgro), 1.25 mg mL−1 bovine serum albumin (BSA), 0.1% insulin-transferring-selenium (ITS) + premix, 50 μg mL−1 ascorbate 2-phosphate, 40 μg mL−1 l-proline, and 1% penicillin-streptomycin-amphotericin, further supplemented with 10 ng mL−1 TGF-β3, and 0.1 × 10−3 m AHA, or “ManNAz media” consisting of glutamine-, methionine-, and cystine-free high-glucose DMEM, 0.1 × 10−6 m dexamethasone, 4 × 10−3 m l-glutamine, 0.201 × 10−3 m l-cystine, 100 μg mL−1 sodium pyruvate, 1.25 mg mL−1 BSA, 0.1% ITS+ Premix, 50 μg mL−1 ascorbate 2-phosphate, 40 μg mL−1 l-proline, and 1% penicillin-streptomycin-amphotericin, further supplemented with 10 ng mL−1 TGFβ−3 and 0.05 × 10−3 m ManNAz. Constructs were cultured for up to 7 d in the designated media for either nascent protein or proteoglycan labeling.
Cell Viability:
For viability analysis, chondrocytes encapsulated in hydrogels were stained using a Live/Dead cell viability assay (Invitrogen) according to the manufacturer’s instructions. Viability was quantified from confocal stacks acquired using a Leica SP5 II confocal microscope and reported as the ratio of calcein-AM-stained cells to the total cell number.
Nascent Matrix Labeling and Quantification:
Constructs were harvested by first staining live cells using a fluorophore-conjugated cyclooctyne (DBCO-488) in PBS containing 1% BSA.[9a] Hydrogels were washed twice in PBS, followed by a 40 min incubation in 30 × 10−6 m DBCO-488 at 37 °C/5% CO2. After three washes with PBS, hydrogels were fixed in 10% formalin for 30 min at room temperature followed by three washes in PBS and storage at 4 °C. These fixed constructs were then stained with a plasma membrane stain (CellMask Deep Red, 1:1000 dilution, Invitrogen) and a nuclear stain (Hoechst 33 342, 5 μg mL−1, Thermo Fisher) for 40 min and were subsequently washed twice with PBS immediately prior to imaging. Constructs were imaged using a Leica SP5 II confocal microscope to acquire z-stacks of nuclei, cell membranes, beads, and labeled matrix. Bead encapsulation was validated by selecting random 50 μm × 50 μm squares at least 20 μm removed from the cells and matrix signals (n = 3 squares) in images acquired at 190 × 1.4 NA (0.15 μm per pixel) and quantifying mean bead densities at each timepoint (Figure S2, Supporting Information). Average local matrix thickness at the midsection of each cell was measured radially (n = 5 measurements per cell) as the distance that the matrix extended past the outer edge of the cell membrane. The bead-to-chondrocyte distance was quantified as the distance between the outer edge of the cell membrane to the closest proximal bead at the cell midsection. Radial intensity profiles were generated by collecting and averaging intensity profiles (n = 5 per cell) across the midsections of n = 20 cells per group, where each profile was normalized to its maximum intensity and truncated to only include the signal starting at the outer edge of the cell membrane (Figure 2C).
Immunostaining:
Before immunostaining, hydrogels were blocked in PBS containing 2% BSA. Primary antibodies were diluted in PBS containing 2% BSA and hydrogels were stained at 4 °C overnight. Antibodies and dilutions included anticollagen type II (1:100; DSHB II-II6B3), anticollagen type VI (1:250, Abcam ab6588), antichondroitin sulfate (1:200, Millipore MAB1581), and antiaggrecan (1:50, Abcam ab2778). After three PBS washes, the secondary antibody Alexa Fluor-594 IgG H&L (1:200; Abcam ab150080) was added for 2 h at room temperature. Hydrogels were washed three times, followed by DAPI staining (1:1.000; Invitrogen D1306, in PBS) for 20 min at room temperature.
Fluorescence-guided Nanoindentation:
To quantify the micromechanical properties of newly secreted matrix, chondrocytes were cultured for 7 d in either AHA or ManNAz containing chondrogenic media, dibenzocyclooctyne (DBCO)-labeled as described above, and snapfrozen. Hydrogels were then embedded in optimal cutting temperature medium upon cryoprotection with 30% sucrose and 8 μm thick, unfixed cryosections prepared using the Kawamoto’s film method.[33] Guided by the fluorescent labeling, AFM-nanomechanical mapping was performed using microspherical tips (R ≈ 2.25 μm, nominal k ≈ 0.03 N m−1, HQ:CSC38/tipless/Cr-Au, Cantilever B NanoAndMore) and an MFP-3D AFM (Asylum Research, Santa Barbara, CA) in PBS. For each map, a 40 × 40 indentation grid was acquired over a 20 μm × 20 μm ROI containing a well-defined cell. The effective indentation modulus, Eind, was calculated via the finite thickness-corrected Hertz model.[34]
Statistical Analysis:
Graphpad Prism 8.1.2 software was used for data analysis and graph plotting. Data were determined to be normally distributed. Analyses of significance between two experimental groups were performed using two-tailed Student’s t-tests and one-way ANOVA with Bonferroni post hoc testing for comparisons between more than two experimental groups. p-Values are indicated in the figures. All experiments were repeated as described in the figure legends.
Supplementary Material
Acknowledgements
C.L. and M.K. contributed equally to this work. The work was supported by the National Science Foundation (J.A.B.: DMR Award 1610525; M.K.: Graduate Research Fellowship; L.H.: CMMI Award 1751898), the National Institutes of Health (J.A.B., R.L.M.: R01 EB008722, M.K.: T32AR007132), and the Center for Engineering MechanoBiology through the National Science Foundation’s STC Program (CMMI: 15–48571). The authors would also like to thank Tianbi Duan for help with image acquisition and quantification. Corrections were made to the abstract, results section 2.1, and Figure 1 caption to clarify that other molecules such as glycoproteins would be labeled with this technique in addition to the proteoglycans on October 28, 2020 after initial online publication.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Claudia Loebel, Department of Bioengineering, University of Pennsylvania, 240 Skirkanich Hall, 210 S. 33rd Street, Philadelphia, PA 19104, USA Translational Musculoskeletal Research Center, Corporal Michael J Crescenz VA Medical Center, 900 Woodland Ave, Philadelphia, PA 19104, USA.
Dr. Mi Y. Kwon, Department of Bioengineering, University of Pennsylvania, 240 Skirkanich Hall, 210 S. 33rd Street, Philadelphia, PA 19104, USA
Chao Wang, School of Biomedical Engineering, Science and Health Systems Drexel University 3141 Chestnut Street, Bossone 718, Philadelphia, PA 19104, USA.
Lin Han, School of Biomedical Engineering, Science and Health Systems, Drexel University, 3141 Chestnut Street, Bossone 718, Philadelphia, PA 19104, USA
Robert L. Mauck, Department of Bioengineering, University of Pennsylvania, 240 Skirkanich Hall, 210 S. 33rd Street, Philadelphia, PA 19104, USA Translational Musculoskeletal Research Center, Corporal Michael J Crescenz VA Medical Center, 900 Woodland Ave, Philadelphia, PA 19104, USA; McKay Orthopaedic Research Laboratory, Department of Orthopaedic Surgery, Perelman School of Medicine, University of Pennsylvania, 308A Stemmler Hall, 3450 Hamilton Walk, Philadelphia, PA 19104, USA.
Jason A. Burdick, Department of Bioengineering, University of Pennsylvania, 240 Skirkanich Hall, 210 S. 33rd Street, Philadelphia, PA 19104, USA Translational Musculoskeletal Research Center, Corporal Michael J Crescenz VA Medical Center, 900 Woodland Ave, Philadelphia, PA 19104, USA; McKay Orthopaedic Research Laboratory, Department of Orthopaedic Surgery, Perelman School of Medicine, University of Pennsylvania, 308A Stemmler Hall, 3450 Hamilton Walk, Philadelphia, PA 19104, USA.
References
- [1].Frantz C, Stewart KM, Weaver VM, J. Cell Sci 2010, 123, 4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sophia Fox AJ, Bedi A, Rodeo SA, Sports Health: A Multidisciplinary Approach 2009, 1, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Buckwalter JA, Mankin HJ, Instr. Course Lect 1998, 47, 487; [PubMed] [Google Scholar]; b) Loebel C, Burdick JA, Cell Stem Cell 2018, 22, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Armiento AR, Stoddart MJ, Alini M, Eglin D, Acta Biomater. 2018, 65, 1. [DOI] [PubMed] [Google Scholar]
- [5].a) Tan AR, Hung CT, Stem Cells Transl. Med 2017, 6, 1295; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen FH, Rousche KT, Tuan RS, Nat. Clin. Pract. Rheumatol 2006, 2, 373. [DOI] [PubMed] [Google Scholar]
- [6].Vega SL, Kwon MY, Burdick JA, Eur. Cells Mater 2017, 33, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Chung C, Burdick JA, Tissue Eng., Part A 2009, 15, 243; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kwon MY, Wang C, Galarraga JH, Pure E, Han L, Burdick JA, Biomaterials 2019, 222, 119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Bryant SJ, Anseth KS, J. Biomed. Mater. Res 2002, 59, 63; [DOI] [PubMed] [Google Scholar]; b) Bian L, Hou C, Tous E, Rai R, Mauck RL, Burdick JA, Biomaterials 2013, 34, 413; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Feng Q, Zhu M, Wei K, Bian L, PLoS One 2014, 9, e99587; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Erickson IE, Huang AH, Sengupta S, Kestle S, Burdick JA, Mauck RL, Osteoarthritis Cartilage 2009, 17, 1639; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Buschmann MD, Gluzband YA, Grodzinsky AJ, Kimura JH, Hunziker EB, J. Orthop. Res 1992, 10, 745. [DOI] [PubMed] [Google Scholar]
- [9].a) Loebel C, Mauck RL, Burdick JA, Nat. Mater 2019, 18, 883; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McLeod CM, Mauck RL, Sci. Rep 2016, 6, 38852; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ferreira SA, Faull PA, Seymour AJ, Yu TTL, Loaiza S, Auner HW, Snijders AP, Gentleman E, Biomaterials 2018, 176, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Calve S, Witten AJ, Ocken AR, Kinzer-Ursem TL, Sci. Rep 2016, 6, 32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Laughlin ST, Bertozzi CR, Nat. Protoc 2007, 2, 2930; [DOI] [PubMed] [Google Scholar]; b) Yoon HI, Yhee JY, Na JH, Lee S, Lee H, Kang SW,Chang H, Ryu JH, Lee S, Kwon IC, Cho YW, Kim K, Bioconjugate Chem. 2016, 27, 927. [DOI] [PubMed] [Google Scholar]
- [12].Buxton AN, Zhu J, Marchant R, West JL, Yoo JU, Johnstone B, Tissue Eng. 2007, 13, 2549. [DOI] [PubMed] [Google Scholar]
- [13].Erickson IE, Huang AH, Chung C, Li RT, Burdick JA, Mauck RL, Tissue Eng., Part A 2009, 15, 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cote AJ, McLeod CM, Farrell MJ, McClanahan PD, Dunagin MC, Raj A, Mauck RL, Nat. Commun 2016, 7, 10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Roughley PJ, Mort JS, J. Exp. Orthop 2014, 1, 8; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cescon M, Gattazzo F, Chen P, Bonaldo P, J. Cell Sci 2015, 128, 3525. [DOI] [PubMed] [Google Scholar]
- [16].a) Lee HP, Gu L, Mooney DJ, Levenston ME, Chaudhuri O, Nat. Mater 2017, 16, 1243; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nicodemus GD, Skaalure SC, Bryant SJ, Acta Biomater. 2011, 7, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Sengers BG, Van Donkelaar CC, Oomens CW, Baaijens FP, Ann. Biomed. Eng 2004, 32, 1718; [DOI] [PubMed] [Google Scholar]; b) Erickson IE, Kestle SR, Zellars KH, Farrell MJ, Kim M, Burdick JA, Mauck RL, Acta Biomater. 2012, 8, 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guilak F, Alexopoulos LG, Upton ML, Youn I, Choi JB, Cao L, Setton LA, Haider MA, Ann. N. Y. Acad. Sci 2006, 1068, 498. [DOI] [PubMed] [Google Scholar]
- [19].a) Wilusz RE, DeFrate LE, Guilak F, Soc JR, Interface 2012, 9, 2997; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang C, Brisson BK, Terajima M, Li Q, Hoxha K. h., Han B, Goldberg AM, Sherry Liu X, Marcolongo MS, Enomoto-Iwamoto M, Yamauchi M, Volk SW, Han L, Matrix Biol. 2020, 85, 47; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xu X, Li Z, Leng Y, Neu CP, Calve S, Dev. Biol 2016, 418, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ferreira SA, Motwani MS, Faull PA, Seymour AJ, Yu TTL, Enayati M, Taheem DK, Salzlechner C, Haghighi T, Kania EM, Oommen OP, Ahmed T, Loaiza S, Parzych K, Dazzi F, Varghese OP, Festy F, Grigoriadis AE, Auner HW, Snijders AP, Bozec L, Gentleman E, Nat. Commun 2018, 9, 4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Haider MA, Schugart RC, Setton LA, Guilak F, Biomech. Model. Mechanobiol 2006, 5, 160; [DOI] [PubMed] [Google Scholar]; b) Hing WA, Sherwin AF, Ross JM, Poole CA, Osteoarthritis Cartilage 2002, 10, 297; [DOI] [PubMed] [Google Scholar]; c) Knight MM, Lee DA, Bader DL, Biochim. Biophys. Acta, Mol. Cell Res 1998, 1405, 67; [DOI] [PubMed] [Google Scholar]; d) McLeod MA, Wilusz RE, Guilak F, J. Biomech 2013, 46, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Fahy N, Alini M, Stoddart MJ, J. Orthop. Res 2018, 36, 52; [DOI] [PubMed] [Google Scholar]; b) Mauck RL, Soltz MA, Wang CC, Wong DD, Chao PH,Valhmu WB, Hung CT, Ateshian GA, J. Biomech. Eng 2000, 122, 252; [DOI] [PubMed] [Google Scholar]; c) Thorpe SD, Buckley CT, Vinardell T, O’Brien FJ, Campbell VA, Kelly DJ, Ann. Biomed. Eng 2010, 38, 2896; [DOI] [PubMed] [Google Scholar]; d) Steward AJ, Wagner DR, Kelly DJ, Eur. Cells Mater 2013, 25, 167; [DOI] [PubMed] [Google Scholar]; e) Vunjak-Novakovic G, Martin I, Obradovic B, Treppo S, Grodzinsky AJ, Langer R, Freed LE, J. Orthop. Res 1999, 17, 130. [DOI] [PubMed] [Google Scholar]
- [23].a) Bian L, Guvendiren M, Mauck RL, Burdick JA, Proc. Natl. Acad. Sci. USA 2013, 110, 10117; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mhanna R, Ozturk E, Vallmajo-Martin Q, Millan C, Muller M, Zenobi-Wong M, Tissue Eng., Part A 2014, 20, 1165; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Parmar PA, Chow LW, St-Pierre J-P, Horejs C-M, Peng YY, Werkmeister JA, Ramshaw JAM, Stevens MM, Biomaterials 2015, 54, 213; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Connelly JT, Garcia AJ, Levenston ME, Biomaterials 2007, 28, 1071. [DOI] [PubMed] [Google Scholar]
- [24].a) Bahney CS, Hsu CW, Yoo JU, West JL, Johnstone B, FASEB J. 2011, 25, 1486; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Anderson SB, Lin CC, Kuntzler DV, Anseth KS, Biomaterials 2011, 32, 3564; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Vega SL, Kwon MY, Song KH, Wang C, Mauck RL, Han L, Burdick JA, Nat. Commun 2018, 9, 614; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang T, Lai JH, Han LH, Tong X, Yang F, Tissue Eng., Part A 2014, 20, 2131. [DOI] [PubMed] [Google Scholar]
- [25].Kim M, Erickson IE, Choudhury M, Pleshko N, Mauck RL, J. Mech. Behav. Biomed. Mater 2012, 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bryant SJ, Anseth KS, J. Biomed. Mater. Res 2003, 64A, 70. [DOI] [PubMed] [Google Scholar]
- [27].Griffin DR, Kasko AM, J. Am. Chem. Soc 2012, 134, 13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].a) Somoza RA, Welter JF, Correa D, Caplan AI, Tissue Eng., Part B 2014, 20, 596; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lynch K, Pei M, Organogenesis 2014, 10, 289; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang Y-HK, Ogando CR, Wang See C, Chang T-Y, Barabino GA, Stem Cell Res. Ther 2018, 9, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gramlich WM, Kim IL, Burdick JA, Biomaterials 2013, 34, 9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].a) Kapitza HG, McGregor G, Jacobson KA, Proc. Natl. Acad. Sci. USA 1985, 82, 4122; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Loren N, Hagman J, Jonasson JK, Deschout H, Bernin D, Cella-Zanacchi F, Diaspro A, McNally JG, Ameloot M, Smisdom N, Nyden M, Hermansson AM, Rudemo M, Braeckmans K, Q. Rev. Biophys 2015, 48, 323. [DOI] [PubMed] [Google Scholar]
- [31].Mauck RL, Yuan X, Tuan RS, Osteoarthritis Cartilage 2006, 14, 179. [DOI] [PubMed] [Google Scholar]
- [32].Loebel C, Szczesny SE, Cosgrove BD, Alini M, Zenobi-Wong M, Mauck RL, Eglin D, Biomacromolecules 2017, 18, 855. [DOI] [PubMed] [Google Scholar]
- [33].Kawamoto T, Kawamoto K, Methods Mol. Biol 2014, 1130, 149. [DOI] [PubMed] [Google Scholar]
- [34].Dimitriadis EK, Horkay F, Maresca J, Kachar B, Chadwick RS, Biophys. J 2002, 82, 2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.