Abstract
Despite the growing knowledge of T cell responses in COVID-19 patients, there is a lack of detailed characterizations for T cell-antigen interactions and T cell functions. Here, with a predicted peptide library from SARS-CoV-2 S and N proteins, we found that specific CD8+ T cell responses were identified in over 75% of COVID-19 convalescent patients (15/20) and an epitope from the N protein, N361-369 (KTFPPTEPK), was the most dominant epitope from our selected peptide library. Importantly, we discovered 2 N361-369-specific T cell receptors (TCRs) with high functional avidity that were independent of the CD8 co-receptor. These TCRs exhibited complementary cross-reactivity to several presently reported N361-369 mutant variants, as to the wild-type epitope. Further, the natural functions of these TCRs in the cytotoxic immunity against SARS-CoV-2 were determined with dendritic cells (DCs) and the lung organoid model. We found that the N361-369 epitope could be normally processed and endogenously presented by these different types of antigen presenting cells, to elicit successful activation and effective cytotoxicity of CD8+ T cells ex vivo. Our study evidenced potential mechanisms of cellular immunity to SARS-CoV-2, and illuminated potential ways of viral clearance in COVID-19 patients. These results indicate that utilizing CD8-independent TCRs against SARS-CoV-2-associated antigens may provide functional superiority that is beneficial for the adoptive cell immunotherapies based on natural or genetically engineered T cells. Additionally, this information is highly relevant for the development of the next-generation vaccines with protections against continuously emerged SARS-CoV-2 mutant strains.
Keywords: CD8+T cell, HLA class I, Lung organoid, SARS-CoV-2, T cell epitope, TCR
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global pandemic of COVID-19.1,2 As of 30 January 2021, over 100 million positive cases of SARS-CoV-2 infection have been reported worldwide, with two million deaths (https://covid19.who.int/). As a newly emerged single stranded RNA virus of the highly contagious coronavirus subfamily, SARS-CoV-2 has a tendency to accumulate mutations over time,3,4 which might affect the transmission and virulence of the virus. The continuous emergence of new mutational variants of SARS-CoV-2 has brought additional challenges for both medical interventions and vaccine-based prophylaxis. Although growing evidences suggest that protective immunity can be acquired after COVID-19 vaccination,5, 6, 7, 8 it is relatively early to confirm whether they can provide long-lasting protective immunity at the present time. A better understanding of the immune response to SARS-CoV-2 infection, particularly the cellular immunity, is the key for advancing vaccine design.
SARS-CoV-2 contains four structural proteins, namely nucleoprotein (N), membrane protein (M), envelope protein (E) and spike glycoprotein (S).1 SARS-CoV-2 S protein mediates virus cell entry through binding to its host receptor, angiotensin-converting enzyme 2 (ACE2).1,9,10 It has been demonstrated that the S protein was capable of inducing not only humoral immunity but also cellular immunity,11, 12, 13, 14, 15, 16, 17, 18, 19 hence the S protein has always been the primary consideration for SARS-CoV-2 vaccine development.7,20,21 SARS-CoV-2 N protein has also been shown to elicit high levels of T cell responses in COVID-19 patients, highlighting the potential of both S and N proteins as predominant targets for cellular immunity in COVID-19.15,16,22,23 Recently, independent studies identified a number of SARS-CoV-2 T cell epitopes in COVID-19 convalescent patients, using predicted or established peptide libraries of the S and N proteins.16,19,22, 23, 24, 25, 26, 27, 28 However, there is still much to be learned from capturing both the breadth (i.e. numbers of recognized epitopes for T cells) and the depth (i.e. strength of T cell cytotoxicity) of T cell responses to SARS-CoV-2 infection.
The adaptive cellular immune responses rely on both CD4+ T cells to secreting cytokines for the functional support of CD8+ T cells and B cells, and CD8+ T cells to eliminate the viruses by killing infected cells. SARS-CoV-2-specific CD8+ T cells can be detected in convalescent patients.15,16,22,23,29, 30, 31, 32 However, their functions in the host's immune response to SARS-CoV-2, particularly in clearing infected cells, is poorly understood. It has been reported that the functional avidity of TCRs is a critical factor to be evaluated for determining the peptide-major histocompatibility complex (pMHC) recognition and posterior T-cell activation, which is essential for successful killing of virus-infected cells and effective viral clearance.33, 34, 35, 36 Therefore, it is important to identify SARS-CoV-2 specific TCRs with high functional affinity and the corresponding epitopes, to understand the mechanisms of T cell-mediated viral clearance in COVID-19 patients and to optimize the design of next-generation vaccines.
In this study, we evaluated SARS-CoV-2 specific CD8+ T cell responses in COVID-19 convalescent patients with the 3 eminent HLA-A alleles in the Asian population, HLA-A2, HLA-A24 and HLA-A11.37 A total of 15 epitopes derived from SARS-CoV-2 S and N proteins were identified, with most dominant epitope N361-369 (KTFPPTEPK) characterized in detail. We confirmed the functional avidity of N361-369-specific TCRs and their responses to the wild type and all clinically presented mutant variants of N361-369. Our study is, to our knowledge, the first to reveal that an immunodominant CD8+ T cell epitope derived from SARS-CoV-2 N protein can be endogenously processed and presented by DCs and lung organoids, leading to robust co-receptor-independent T cell activation and cytotoxicity, which are mediated by TCRs with high functional avidity. Such findings illustrated potential mechanisms of CD8+ T cell immunity against SARS-CoV-2 infection, which could be of great use to understand the process of viral control and to facilitate the next-generation vaccine development for COVID-19.
Materials and methods
Ethics statement
This project was approved by the ethics committee of Chongqing Medical University. Informed consents were obtained from all participants.
Cell culture
Lenti-X 293T cells was purchased from Takara Biomedical Technology, COS-7 and K-562 cells were purchased from American Type Culture Collection (ATCC). Lenti-X 293T and COS-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific, USA) supplemented with 10% Fetal Bovine Serum (FBS, Thermo Fisher Scientific, USA), 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco, USA). K-562 cells were cultured in RPMI-1640 (Thermo Fisher Scientific, USA) with 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. All cells were maintained at 37 °C in an incubator with 5% CO2.
PBMCs isolation
Blood samples of COVID-19 convalescent patients (Table S1) were obtained from Yongchuan Hospital of Chongqing Medical University and The Third Affiliated Hospital of Chongqing Medical University. Additionally, blood samples of healthy individuals were obtained from Chongqing Medical University (Table S2). Samples were collected and processed as described before.38 All blood was collected in ethylene diamine tetra acetic acid (EDTA) tubes and stored at room temperature prior to processing for PBMCs isolation and plasma collection. The whole blood was centrifuged for 15 min at 400×g to separate the cellular fraction and plasma. The plasma was then carefully separated from the cell pellets and stored at −20 °C. The PBMCs were isolated via density gradient centrifugation (Lymphoprep, STEMCELL Technologies, Canada). Isolated PBMC were cryopreserved and stored in liquid nitrogen until used in the assays.
Preparation and maturation of DCs
Monocytes were enriched from the PBMCs of healthy individuals by using the EasySep CD14 positive selection kit according to the manufacturer's instructions (STEMCELL Technologies, Canada). Monocytes were cultured in the DC medium for 6 days, which was made with RPMI-1640 medium supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine (Gibco, USA), 800 IU/ml GM-CSF (PeproTech, USA) and 800 IU/ml IL-4 (PeproTech, USA). On day 3, fresh DC media were added to the cultures. On day 6, cells were cryopreserved and stored in liquid nitrogen until used in the assays.
Lung organoid culture
All lung tissues used in this study were obtained after surgical resections from volunteers, and processed for organoid culture within 4 h. Normal lung tissue organoids were established as reported.39 Briefly, a piece of lung tissue was minced, washed with 10 ml Advanced DMEM/F12 (Thermo Fisher Scientific, USA) containing 1 × Glutamax (Thermo Fisher Scientific, USA), 10 mM HEPES (Thermo Fisher Scientific, USA) and antibiotics, termed as AdDF+++, and digested in 10 ml AO medium (Table S3) containing 1 μg/ml collagenase (Sigma–Aldrich, USA) on a shaker at 37 °C for 1 h. The digested lung tissue suspension was sheared using 5 ml plastic Pasteur pipettes before straining over a 100 μm filter. The retained tissue pieces were resuspended in 10 ml AdDF+++ following another shearing step. Strained suspensions were combined and centrifuged, and the pellet was resuspended in 10 ml AdDF+++ and washed once. Lung cell pellets were resuspended in 10 mg/ml cold Cultrex growth factor reduced basement membrane extract (BME), type 2 (Trevigen, USA). Drops of 40 μl BME-cell suspension were added into 24-well plates and solidified at 37 °C for 10–20 min. Then, 400 μl of AO medium was added to each well and culture in the 37 °C incubator with 5% CO2. The medium was changed every 4 days and the organoids were passaged every 14–18 days.
Peptide library
To define the SARS-CoV-2-specific CD8+ T cell epitopes of the S and N proteins derived from SARS-CoV-2 reference sequences (GenBank: MN908947). The NetMHCpan 4.0 algorithm40 was used to analyze the binding of 9-mer peptides derived from the full lengths of SARS-CoV-2 S and N proteins, with HLA-A∗02:01, HLA-A∗24:02 or HLA-A∗11:01. For each allele, binding peptides with high strength (Rank Threshold < 0.5) were selected, and a total of 72 predicted epitopes were generated. Additional 6 SARS-CoV epitopes were selected (Table S4).41,42 Individual peptide's HLA-A specificity was listed in Table S4. All 78 epitopes were synthesized (GenScript) and dissolved in the corresponding solvents for the subsequent experiments. Detailed information for peptide grouping was summarized in Table S5.
HLA-A typing for COVID-19 convalescent patients and healthy individuals
To screen the HLA-A∗11:01 phenotypes of the COVID-19 convalescent patients and health individuals, we used Dynabeads™ mRNA DIRECT™ Purification kit (Thermo Fisher Scientific, USA) to extract RNAs from the PBMC samples. Then, reverse transcription and PCR were performed to amplify HLA-A molecules, with available PCR primer information.43 The PCR products were ligated into the T vector (TransGen Biotech, China) and then transformed into the Trans5α competent cells (TransGen Biotech, China). Single bacterial clones of each sample were selected for PCR verification and sequencing. The sequencing results were analyzed in the IMGT/HLA (https://www.ebi.ac.uk/ipd/imgt/hla/) website.
The HLA-A2 and HLA-A24 typing of the COVID-19 convalescent patients and health individuals were analyzed using flow cytometry. For staining, 5 × 105 PBMCs were resuspended in PBS with 2% FBS (FACS buffer), and stained with the HLA-A2-PE (MBL, Clone BB7.2, K0186-5, Japan) or HLA-A24-PE (MBL, Clone 17A10, K0208-5, Japan) for 30 min at room temperature in the dark. Samples were washed twice with FACS buffer following staining. After the final wash, cells were resuspended in 200 μl FACS buffer. All samples were acquired on a FACSCelesta cytometer (BD Biosciences, USA). The data were analyzed using FlowJo software (TreeStar Inc, USA).
In vitro stimulation of IFN-γ-producing CD8+ T cells with selected peptides
PBMCs were plated in 24-well plates at 2 × 106 cells per well with 100 IU/ml IL-2 (PeproTech, USA) in the complete medium, which was RPMI-1640 supplemented with 10% FBS, 2 mM l-glutamine, 25 mM HEPES and 10 μg/ml gentamicin. The cells were subsequently stimulated with indicated pools of predicted SARS-CoV-2 peptides, at 5 μM for each peptide. Cells were cultured for 10 days to expand specific T cell populations to corresponding stimulations. Half of the media were refreshed every 3 days, or as required. At day 10, cells were harvested and tested for peptide-specific CD8+ T cells by the IFN-γ release ELISPOT assay. The remaining cells were cryopreserved and stored in liquid nitrogen until used in assays.
IFN-γ ELISPOT assays
IFN-γ ELISPOT assays were performed as reported and with minor modification.16 Briefly, ELISPOT plates (Millipore, USA) were coated with human IFN-γ antibody (Clone 1-D1K, 2 μg/ml, Mabtech, Sweden) overnight at 4 °C. T cells were rested in cytokine-free media overnight. Then 2 × 104 T cells were seeded per well in ELISPOT plates and stimulated for 24 h with pools of predicted SARS-CoV-2 peptides (5 μM each). Stimulation with an equimolar amount of DMSO was performed as the negative control, and Phorbol 12-myristate 13-acetate (PMA)/Ionomycin (P/I) (Sigma–Aldrich, USA) was used as the positive control. Subsequently, the plates were developed with human biotinylated IFN-γ detection antibody (Clone 7-B6-1, 1 μg/ml, Mabtech, Sweden), followed by incubation with Streptavidin-AP (1:1000, Mabtech, Sweden) and BCIP/NBT-plus substrate (Mabtech, Sweden). IFN-γ spots were quantified with the AID ELISPOT Reader (AID, Germany). To quantify positive peptide-specific responses, with 2 × mean spots of the unstimulated wells subtracted from the peptide-stimulated wells, and the results expressed as IFN-γ spots/2 × 104 PBMCs.
Flow cytometry analysis
For staining, 5 × 105 cells were re-suspended in FACS buffer, and stained with the anti-human antibody cocktail for 30 min at room temperature in the dark. CD3-BV510 (Clone SK7, Biolegend, USA), CD4-PerCP-Cy5.5 (Clone RPA-T4, Biolegend, USA), CD8-FITC (Clone HIT8a, Biolegend, USA) and CD137-BV421 (Clone 4B4-1, Biolegend, USA) were used for the activation induced T cell marker assay. The P64-tetramer-PE used for the identification of antigen-specific CD8+ T lymphocytes was generated using the PE QuickSwitch™ Quant HLA-A∗11:01 Tetramer Kit (MBL, tb-7305-k1, Japan). The IFN-γ Secretion Assay Detection Kit (MACS, 130-090-762, Germany) was used for labeling IFN-γ-secreting T cells, which were sorted into DNase and RNase free 96-well PCR plates (Bio-Rad, USA) at a single cell level by FACSAria III (BD Biosciences, USA).
Cytokine production was assessed by intracellular cytokine staining (ICS). Briefly, after target cells and effector cells were co-cultured in the well of a 96-well plate for 2 h, both Monensin (1:1000, Biolegend, USA) and Brefeldin A (1:1000, Biolegend, USA) were added to the culture. After 6 h stimulation, cells were washed with FACS buffer, and stained for cell surface markers (described above). Cells were then washed with FACS buffer twice following fixation and permeabilization. Next, cells were washed with Perm/Wash buffer (Biolegend, USA) twice and stained for 25 min at room temperature with anti-human antibody against IFN-γ (BV421, Clone 4S.B3). Cells were washed twice with Perm/Wash buffer and resuspended in FACS buffer, then data was acquired via the FACSCelesta cytometer and analyzed using the FlowJo software.
HLA binding assay
The HLA binding assay was performed using the PE QuickSwitch™ Quant HLA-A∗11:01 Tetramer kit following the manufacturer's instructions. Each lyophilized peptide was dissolved in 1 mM recommended solution. In each well of a 96-well plate, the following reagents were added sequentially and gently mixed via pipetting: 25 μl QuickSwitch™ Tetramer, 0.5 μl Peptide Exchange Factor and 0.5 μl of 1 mM peptide. Then, the plate was incubated for 4 h at room temperature while protected from light. The tetramer's exchange rates were measured by the BD FACSCelesta and calculated by QuickSwitch™ Calculator (https://www.mblintl.com/quickswitch-peptide- exchange -calculator/).
T cell receptor sequencing and analysis
For single T cell PCR, the TCRα and TCRβ chain genes were amplified as previously described,44 with minor modifications. Briefly, the reverse transcription (RT) reaction was performed in a two-step method following the PrimeScript™ II Reverse Transcriptase kit protocol (Takara, Japan). For the first step, single-cell sorted plates were thawed on ice and added with 5 μl mix containing dNTP mixture and the primers for TCRα and TCRβ, then incubated at 65 °C for 5 min and put on ice immediately. For the second step, the above plates were added with the RNase inhibitor and PrimeScript II Reverse Transcriptase (Takara, Japan) to a total volume of 10 μl. The RT program was 45 °C for 45 min and 70 °C for 15 min. The RT products were amplified by nested PCR following the PrimeSTAR® HS DNA Polymerase kit protocol (Takara, Japan), with primers for TCRα and TCRβ. Here, the 5 × PrimeSTAR® Buffer (Mg2+ plus) in the kit was replaced with the 2 × PrimeSTAR® GC Buffer (Mg2+ plus) (Takara, Japan). The first PCR program was as follows: 98 °C for 1 min, 98 °C for 10 s for 30 cycles, 52 °C for 5 s and 72 °C for 40 s. The resultant PCR mixtures were diluted 50-fold with water, and 2 μl of the diluted PCR mixtures were added to 18 μl of the nested PCR mixture as the template DNA. The nested PCR program was as follows: 98 °C for 1 min, 98 °C for 10 s for 35 cycles, 52 °C for 5 s and 72 °C for 30 s. The PCR products were then analyzed with the α-nest or β-nest primers by direct sequencing. The TCR repertoire was analyzed with the IMGT/V-Quest tool (http://www.imgt.org/).45
Enrichment of CD3+, CD4+ and CD8+ T cells
CD3+, CD4+ or CD8+ T cells were selected using the EasySep human CD3 positive selection kit II, EasySep human CD4+ T cell enrichment kit and EasySep human CD8+ T cell enrichment kit (STEMCELL Technologies, Canada), respectively, according to the manufacturer's instructions.
Construction of lentivirus vectors and transduction of CD3+ T cells
For the construction of TCR lentivirus vectors, TCRα or TCRβ chains were amplified from the corresponding nested PCR mixtures and cloned into the lentivirus vector pWPXL (Addgene Plasmid #12257). The constant regions were replaced by mouse counterparts with improved TCR pairing and TCR/CD3 stability that were convenient for the detection of TCR-T cells.46,47 The coding sequence of the S or N gene of SARS-CoV-2 and the human HLA-A∗11:01 gene were synthesized, and cloned into the pWPXL vector. Lentiviruses were generated by co-transfecting Lenti-X 293T cells with the lenti-vector and the packaging plasmids psPAX2 (Addgene Plasmid #12260) and pMD2.G (Addgene Plasmid #12259) at a ratio of 5:2:1, using the Xfect Transfection Reagent (Takara, Japan). The lentiviral supernatants were harvested in 48 h. Transduction of CD3+ T cells was conducted as delineated before,46 with minor modifications. CD3+ T cells were stimulated in T cell media with 1 μg/ml anti-CD3 antibody (Clone OKT3) and 1 μg/ml anti-CD28 antibody (Clone 15E8) for 2 days. Then, the transduction efficiency was assessed by flow cytometry using anti-mouse TCR-β chain constant region antibody (Clone H57-597).
TCR transduced T cell rapid expansion protocol
TCR transduced T cells were rapidly expanded with the method as described before.48 Briefly, in indicated experiments, sorted T cells were expanded using excess irradiated (50 Gy) allogeneic PBMC feeder cells (a pool from 3 to 5 different donors) at a ratio of 1–100 with 30 ng/ml anti-CD3 antibody (Clone OKT3) and 3000 IU/ml IL-2, in the 50/50 media consisted of a 1:1 mixture of the complete media and the AIM-V media (Gibco, USA). All cells were cultured at 37 °C with 5% CO2. Media were exchanged 3 days post-stimulation and then every other day. The cells were rapidly expanded in this way for two weeks before use.
Antigen presentation in human DCs
DCs were incubated in AIM-V for 1 h and then 5 μg/ml recombinant N or S proteins of SARS-CoV-2 were added. After 4 h, the media were replaced to DC media supplemented with DC maturation cytokines including 10 ng/ml lipopolysaccharide (LPS) (Sigma–Aldrich, USA) and 100 IU/ml IFN-γ (PeproTech, USA). After incubated overnight, DCs were washed and co-cultured with TCR-T cells for 24 h. The supernatants were used for measuring IFN-γ concentrations by ELISA.
TCR-T cytotoxicity assay
The ability of T cells to lyse target cells was measured using a Calcein-AM release assay. Target cells were labelled for 15 min at 37 °C with 10 μM Calcein-AM (Dojindo, Japan), and then co-cultured with T cells at indicated ratios for 24 h in the black-walled, 96-well flat-bottom plates (Corning, USA). The controls of these labelled target cells were separated into the spontaneous death group and the maximal killing group, the latter were treated with 0.1% Triton-X (Sigma–Aldrich, USA). After 24 h, 50 μl/well supernatant was removed, and the relative fluorescence units (RFUs) was measured by Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, USA). The percentage of lysed cells was calculated as follows: specific lysis = 100% × (Test RFUs – Spontaneous death RFUs)/(Maximal killing RFUs – Spontaneous death RFUs).
The ability of T cells to lyse organoids was also measured using LDH-Glo™ Cytotoxicity Assay Kit (Promega, USA) according to the manufacturer's instructions. The organoids were dissociated to single cells and counted, which was used to infer the number of organoids to allow co-cultured with T cells at a 4:1 of effector:target ratio. After co-cultured for 24 h in the 96-well flat-bottom plates, the relative luminescent units (RLUs) of the co-culture and the control groups were measured by the Varioskan LUX Multimode Microplate Reader. The percentage of lysed organoids was calculated as follows: specific lysis = 100% × (Test RLUs – Spontaneous death RLUs)/(Maximal killing RLUs – Spontaneous death RLUs).
Cytokine release ELISA
The antibody pairs used for cytokine detection are as follows: IFN-γ capture antibody (Clone MD-1, 2 μg/ml), IFN-γ detection antibody (Clone 4S.B3, 1 μg/ml, Biolegend, USA), IL-2 capture antibody (CloneMQ1-17H12, 4 μg/ml, Biolegend, USA), IL-2 detection antibody (Clone Poly5176, 1 μg/ml, Biolegend, USA), TNF-α capture antibody (CloneMAb1, 1 μg/ml, Biolegend, USA) and TNF-α detection antibody (Clone MAb11, 1 μg/ml, Biolegend, USA). The ELISA plate was coated with individual anti-cytokine capture antibodies for 12 h, then washed with phosphate-buffered saline with Tween (PBST) buffer for 3 times, followed by blocking with 3% BSA for at least 1 h at room temperature. The supernatants of T cell and target cell co-cultures were added into the plate at 50 μl per well. After incubation at room temperature for 1 h, the plate was washed 3 times with PBST. Biotinylated capture antibodies (50 μl/well) were added and the plate was incubated for 1 h at room temperature, then washed for 5 times. Streptavidin-ALP (1:1000) was added at 50 μl per well, with, and incubated for 1 h at room temperature in the dark. Then, the plate was washed 6 times and the pNPP solution (Mabtech, Sweden) was added with 50 μl per well. After 20 min incubation at room temperature in the dark, 50 μl stop solution was added per well. The plate was immediately analyzed by the Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, USA) for the OD405 value.
Statistical analysis
Statistical analyses of the data were performed using GraphPad Prism version 8.0 (GraphPad Software, Inc. La Jolla, CA, USA) software. Quantitative data in histograms and line charts were presented as mean ± standard deviation (mean ± SD). Statistical significance was determined using ANOVA for multiple comparisons. Student's t-tests were applied to compare two groups. P < 0.05 was considered statistically significant.
Results
Identification of SARS-CoV-2 specific CD8+ T cell epitopes
It has been reported that SARS-CoV-2 S and N proteins were the predominant T cell targets in COVID-19 patients.15 In order to predict SARS-CoV-2 specific CD8+ T cell epitopes, the NetMHCpan 4.0 algorithm was used to analyse the binding of 9-mer peptides derived from the full lengths of SARS-CoV-2 S and N proteins, with HLA-A∗02:01, HLA-A∗24:02 and HLA-A∗11:01, the 3 most frequent HLA class I A alleles in the Asian population. We synthesized a peptide library of 72 predicted SARS-CoV-2 epitopes, which overlapped 29 SARS-CoV epitopes, together with additional 6 confirmed SARS-CoV epitopes (see Table S4 in the supplemental material).41,42 Among these 78 peptides, 62 were from the S protein and 16 were from the N protein, with 3 different HLA-A restrictions (Table S5).
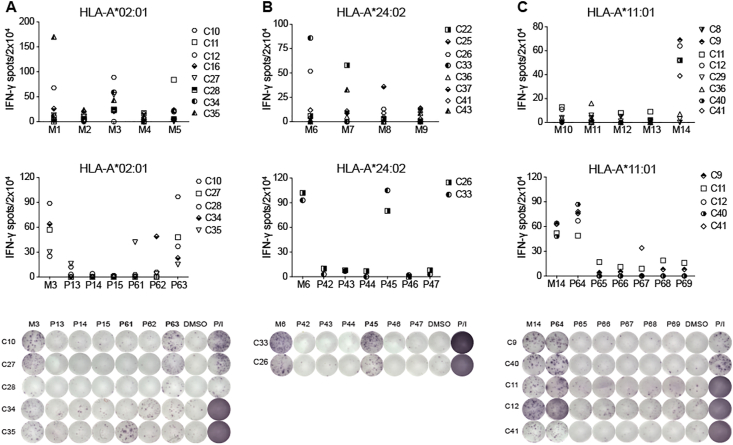
In parallel, we collected a total of 37 COVID-19 convalescent PBMC samples and subjected them for the HLA typing analysis. Among the 20 HLA-A2+ samples and the 12 HLA-A24+ samples revealed by flow cytometric assay, we selected 8 samples from each group. A matching 8 samples with HLA-A∗11:01+ genotype was identified by RT-PCR, which included 2 overlapping with the HLA-A2+ group and 2 with the HLA-A24+ group (Table S1). These PBMCs were stimulated for 10 days in the presence of individual peptide mixtures and IL-2, and the induction of IFN-γ secretion was determined by the enzyme-linked immunospot (ELISPOT) assay. We found that 75% COVID-19 convalescent PBMCs (15/20) demonstrated positive T cell responses when stimulated by corresponding peptide mixtures. Particularly, the HLA-A2+ group showed dominant responses to Mixture (M) 1/3/5, the HLA-A24+ group to M6/7 and the HLA-A∗11:01+ group to M14 (Fig. 1A–C and Fig. S1). Next, individual peptides from each positive mixture were applied to identify specific T cell epitopes, and 15 SARS-CoV-2 specific peptides were found to be recognized by the convalescent PBMCs (Fig. 1A–C, Figs. S2–3; Table 1 and Table S6). Among the 15 positive peptides, 5 peptides were derived from the N protein, and the rest were derived from the S protein (Table 1). Four dominant peptides were identified, with at least one peptide from each HLA group (Table 1). Two of these peptides, P63 (N338-346) and P77 (N219-227), were both HLA-A∗02:01-restricted, and they could induce IFN-γ release in 4/8 and 2/8 samples, respectively (Fig. 1A and Fig. S2; Table 1). The HLA-A∗24:02-restricted P45 (S448-456) was located at the receptor binding domain of the S protein and could be recognized by 2/8 samples tested (Fig. 1B; Table 1). Importantly, 5 out of 8 samples showed positive responses to the HLA-A∗11:01-restricted P64 (N361-369). Hence we termed P64, or N361-369, as the most dominant epitope in this study (Fig. 1C; Table 1). Of note, IFN-γ induction was not observed when these 4 dominant peptides were used to stimulate PBMCs of healthy individuals (Fig. S4; Table S2). Overall, these data demonstrated that 15 predicted peptides derived from the S and the N protein sequences could induce SARS-CoV-2-specific CD8+ T cell responses in about 75% of COVID-19 convalescent patient samples, and at least 4 peptides were identified to be dominant CD8+ T cell epitopes.
Figure 1.
CD8+ T cell responses of SARS-CoV-2 peptides. IFN-γ ELISPOT assay results for the PBMCs of COVID-19 convalescent patients with (A) HLA-A2+, (B) HLA-A24+ or (C) HLA-A∗11:01+ allele, stimulated with mixed peptides (top) and single reactive peptide (middle and bottom). A stimulation with an equimolar amount of DMSO was performed as negative control, and Phorbol 12-myristate 13-acetate (PMA)/Ionomycin (P/I) was performed as positive control. Results are expressed as number of peptide-specific IFN-γ spots/2 × 104 PBMCs of each patient, subtracted those from the corresponding DMSO groups. N = 8. For peptide grouping: 26 HLA-A∗02:01-restricted peptides were pooled into 5 mixtures (M1-M5), 22 HLA-A∗24:02-restricted peptides were pooled into 4 mixtures (M6-M9), and 30 HLA-A∗11:01-restricted peptides were pooled into 5 mixtures (M10-M14). Each mixture contained 3–6 peptides. Positive peptide mixture and single peptide with the most dominant reactivity were marked in bold. For the single dominant peptide, P63 was from M3, P45 was from M6 and P64 was from M14. C8–C41: COVID-19 convalescent patients. Data were representative of two independent experiments.
Table 1.
Identification of CD8+ T cell epitopes of SARS-CoV-2 in COVID-19 convalescent patients.
| Numb. | Epitope ID | Protein | Peptide Position | Epitope sequence | HLA restriction | Responded/tested donors |
|---|---|---|---|---|---|---|
| 1 | P64 | N | 361–369 | KTFPPTEPK | A∗11:01 | 5/8 |
| 2 | P63 | N | 338–346 | KLDDKDPNF | A∗02:01 | 4/8 |
| 3 | P77 | N | 219–227 | LALLLLDRL | A∗02:01 | 2/8 |
| 4 | P62 | N | 316–324 | GMSRIGMEV | A∗02:01 | 1/8 |
| 5 | P61 | N | 222–230 | LLLDRLNQL | A∗02:01 | 1/8 |
| 9 | P45 | S | 448–456 | NYNYLYRLF | A∗24:02 | 2/8 |
| 6 | P74 | S | 958–966 | ALNTLVKQL | A∗02:01 | 1/8 |
| 7 | P16 | S | 718–726 | FTISVTTEI | A∗02:01 | 1/8 |
| 8 | P4 | S | 983–991 | RLDKVEAEV | A∗02:01 | 1/8 |
| 10 | P52 | S | 1137–1145 | VYDPLQPEL | A∗24:02 | 1/8 |
| 11 | P49 | S | 169–177 | EYVSQPFLM | A∗24:02 | 1/8 |
| 12 | P53 | S | 312–320 | IYQTSNFRV | A∗24:02 | 1/8 |
| 13 | P54 | S | 193–201 | VFKNIDGYF | A∗24:02 | 1/8 |
| 14 | P55 | S | 1101–1109 | HWFVTQRNF | A∗24:02 | 1/8 |
| 15 | P57 | S | 268–276 | GYLQPRTFL | A∗24:02 | 1/8 |
Identification of specific CD8+ TCRs to SARS-CoV-2 dominant epitopes
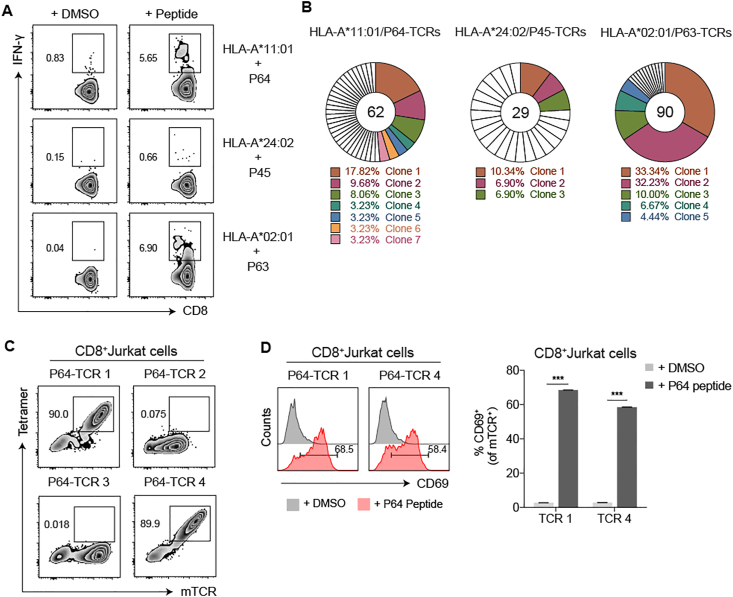
To identify the dominant epitope-specific TCRs, we selected P64, P45 and P63, the most dominant epitopes from the 3 HLA-A alleles. After stimulated with these epitopes, the populations of IFN-γ secreting T cells from the corresponding COVID-19 convalescent patient samples were sorted at single cell levels (Fig. 2A). The TCRs of sorted cells were amplified,44 and the sequences of the variable regions of α- and β-chains were analysed. In the P64-specific TCR repertoire, we found 7 dominant TCR clones (TCR 1–7), with the most dominant clone 1 (TCR 1) accounted for 17.82% (Fig. 2B). Similarly, 3 dominant P45 TCR clones and 5 dominant P63 TCR clones were obtained, with the most dominant clone accounted for 10.34% and 33.34% respectively (Fig. 2B). These results showed that COVID-19 convalescent patients with these three HLA-A restrictions could produce diverse T cell clones for the recognition of corresponding dominant epitopes.
Figure 2.
The isolation and validation of the SARS-CoV-2 specific TCRs of CD8+ T cells. (A) After stimulated by peptide mixtures and expanded for 10 days, single IFN-γ secreting T cell from three COVID-19 convalescent PBMC samples (C40: HLA-A∗11:01+, C33: HLA-A24+ and C27: HLA-A2+) were individually stimulated by P64, P45 and P63, from three corresponding HLA subtypes, and isolated by flow cytometric sorter. (B) The TCRs of sorted cells were amplified by RT-PCR and the TCR repertoire was analyzed with the IMGT/V-Questtool (http://www.imgt.org/) to obtain dominant TCR clones. Unique clones were marked in white. Different colors represented relative dominant clones (copy≧2). (C,D) Specificity verification for the top 4 P64-TCR clones in CD8+ Jurkat cells. P64-HLA-A∗11:01 tetramer (Tetramer) and anti-mouse TCR (mTCR) antibody were applied to evaluate the specific binding of P64-TCRs (C). CD69 was used to determine the specific activation of TCR-transduced CD8+ Jurkat cells, after co-cultured with HLA-A∗11:01 expressingCOS-7 cells pulsed with P64 (D). Grey represented DMSO-stimulated samples and pink represented P64-stimulated samples. Data were presented as mean ± SD, n = 3, ∗∗∗P < 0.001. Representative data of two independent experiments were shown.
Furthermore, the specificities of the P64 reactive TCRs were verified in CD8+ Jurkat cells, which were transduced with the top 4 dominant TCR clones (TCR 1–4). Using the P64-HLA-A∗11:01 tetramer, we confirmed that only TCR 1 and TCR 4 were P64-specific (Fig. 2C; Table S7). Also, with co-culture of the HLA-A∗11:01-expressing COS-7 cells loaded with P64, TCR 1- or TCR 4-transduced CD8+ Jurkat cells could be activated, indicated by elevated CD69 expression (Fig. 2D). Altogether, we successfully identified 2 different CD8+ TCRs targeting the SARS-CoV-2 dominant epitope P64 (N361-369) from the COVID-19 convalescent PBMCs.
Characterization of the functional avidity of the N361-369-specific TCRs
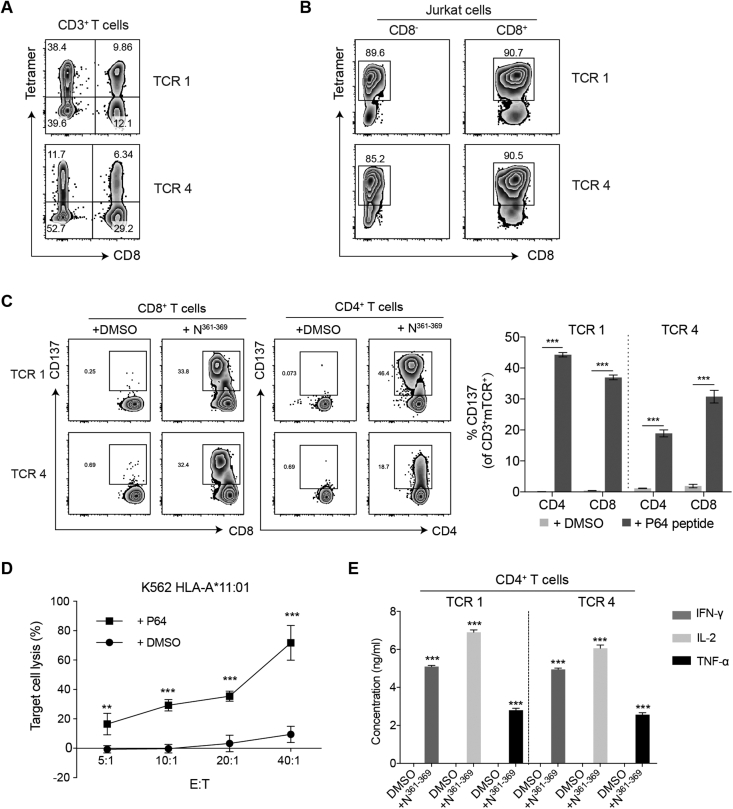
To further evaluate the functional activities of the N361-369-specific TCRs, TCR 1 and TCR 4 were individually transduced into the primary CD3+ T cells isolated from the PBMCs of healthy individuals. These cells were expanded for 12 days following the rapid expansion protocol (REP),48 before the evaluation of their transfection efficiency by N361-369-HLA-A∗11:01 tetramer. We found that 9.86% CD8+ T cells expressed TCR 1 and 6.34% expressed TCR 4 (Fig. 3A). Unexpectedly, we observed higher percentages of TCR-transduced CD4+ T cells, with 38.4% for TCR 1 and 11.7% for TCR 4 (Fig. 3A). These results indicated that TCR-transduced CD4+ T cells were able to recognize the N361-369-HLA complex with the absence of CD8 co-receptor. Also, the similar fluorescence intensities detected in the CD4+ T cells and in the CD8+ T cells implied that TCR 1 and TCR 4 might both have high avidity for the N361-369-HLA-A∗11:01 complex (Fig. 3A). To confirm the binding of these TCRs to N361-369-HLA-A∗11:01 was independent of CD8 co-receptor, TCR 1 and TCR 4 were separately transduced to CD8- and CD8+ Jurkat cells. Both TCR 1- and TCR 4-transduced CD8- Jurkat cells showed positive tetramer staining, and comparable fluorescent signal levels were found between CD8- and CD8+ Jurkat cells (Fig. 3B). These results were in line with those of the CD4+ and CD8+ T cells (Fig. 3A, B).
Figure 3.
Functional characterization of the N361-369-specific TCR 1 and TCR 4. (A) Flow cytometric analysis of the transfection efficiency of TCR 1 and TCR 4 in CD4+ and CD8+ T cells, by N361-369-HLA-A∗11:01 tetramer. (B) Binding assay of TCR-transduced CD8- and CD8+ Jurkat cells with N361-369-HLA-A∗11:01 tetramer. (C) Flow cytometric analysis of T cell activation evaluated by CD137 expression. TCR-transduced CD8+ T cells and CD4+ T cells were individually co-cultured with HLA-A∗11+ K-562 cells pulsed with N361-369, or an equimolar amount of DMSO, for 24 h. Expression of CD137 on CD8+ T cells (left), CD4+ T cells (middle) was evaluated and did further statistical analysis (right). (D) N361-369 loaded HLA-A∗11:01+ K-562 cells lysis by TCR 1-CD8+ T cells after 8 h at different E:T ratios. DMSO was used as negative control. Reported values are the mean of triplicates, with error bars indicating. E, Effector T cells. T, Target cells. (E) Detection of IFN-γ, IL-2 and TNF-α secretion from TCR 1 and TCR 4-transduced CD4+ T cells by ELISA, after co-cultured with HLA-A∗11:01+ K-562 cells loaded with N361-369 for 24 h. Data were presented as mean ± SD, n = 3, ∗∗P < 0.01, ∗∗∗P < 0.001. Representative data of two independent experiments were shown.
Next, we evaluated the functionality of TCR 1 and TCR 4 in CD8+ T cells and CD4+ T cells from expanded CD3+ T cells expressing TCR 1 or TCR 4. After co-cultured with HLA-A∗11:01+ K-562 cells pulsed with N361-369, remarkable levels of T cell activation were detected in TCR 1- and TCR 4-CD8+ T cells, indicated by the upregulation of T cell activation marker CD137 expression, compared to the DMSO controls (Fig. 3C). Similar results were obtained in TCR-transduced CD4+ T cells stimulated with N361-369 (Fig. 3C). Moreover, we evaluated the cytotoxicity of TCR 1- and TCR 4-transduced CD8+ T cells against HLA-A∗11:01+ K-562 cells, using Calcein-AM release assay. Significant levels of target cell lysis were detected in the presence of N361-369, relative to the DMSO controls (Fig. 3D). Increasing cytotoxic efficiency was observed at the effector to target (E:T) cell ratios of 20:1 or higher, and approximately 70% target cell lysis was achieved at E:T cell ratio of 40:1 (Fig. 3D). The functional activity of TCR 1- and TCR 4-transduced CD4+ T cells was analysed by the production of IFN-γ, IL-2 and TNF-α via cytokine secretion assay. We found that N361-369 could elicit significant induction of all three cytokines in these CD4+ T cells, comparing to an equimolar amount of DMSO (Fig. 3E). These results proved that the N361-369-specific TCR 1 and TCR 4 could mediate the cytotoxicity of CD8+ T cells and the effector cytokine secretion of CD4+ T cells.
Evaluation of the affinity and the cross-reactivity of the N361-369-specific TCRs
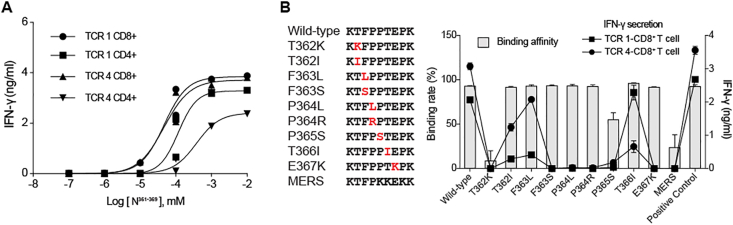
We addressed the functional avidity of TCR 1 and TCR 4 to the N361-369-HLA complex with the peptide concentrations required to induce a half-maximal activation (EC50) per IFN-γ production by the TCR-transduced T cells. The IFN-γ release assay revealed that the EC50 concentrations of TCR 1 and TCR 4 in CD8+ T cells were 46.83 nM and 47.17 nM, respectively, when stimulated by HLA-A∗11:01+ K-562 cells loaded with sequential ten-fold dilutions of the N361-369 peptide (Fig. 4A). Notably, TCR 1 and TCR 4 in CD4+ T cells also demonstrated high functional avidity, with EC50s of 118 nM and 380.9 nM, respectively, in a co-receptor independent manner (Fig. 4A). These data confirmed that TCR 1 and TCR 4 both had high affinity to the N361-369-HLA complex in CD8+ and CD4+ T cells.
Figure 4.
Evaluation of the affinity and the cross-reactivity of TCR 1 and TCR 4 to the N361-369 mutants. (A) EC50 analysis with IFN-γ secretion assessed by ELISA. HLA-A∗11+ K-562 cells loaded with titrating concentrations of the N361-369 peptide were co-cultured with the TCR-CD4+ T cells or the TCR-CD8+ T cells for 24 h. Individual EC50 of N361-369 was calculated using nonlinear regression analysis. (B) Nine N361-369 variants were aligned with the wild type peptide and the homologous MERS peptide (left). Individual mutations were marked in red. HLA-A∗11:01 binding affinity of each peptide was determined by the peptide exchange rates and shown by grey bars (n = 2). The reactivity of TCR 1- and TCR 4-CD8+ T cells to the N361-369 mutant peptides were tested by IFN-γ ELISA assay and shown in connecting lines (n = 3). Data were presented as mean ± SD. Representative data of two independent experiments were shown.
Given the clinical impact from continuous emergence of new SARS-CoV-2 mutations,3,4 the cross-reactivity of TCR 1 and TCR 4 to all reported N361-369 mutations were tested. We synthesized a total of 9 mutant peptides with clinically presented point mutations of residues between 361 and 369 amino acids of the N protein (Fig. 4B; Table S8).49 The HLA binding assay revealed that 7 different point mutations (T362I, F363L, F363S, P364L, P364R, T366I and E367K) did not affect the binding of N361-369 to HLA∗A11:01, whereas T362K markedly diminished the ability of the NT362K peptide to bind to HLA∗A11:01. These results, particularly the different reactivity of NT362I and NT362K, implied that the charge of amino acid side chain, at least, might influence the binding affinity of N361-369 to HLA∗A11:01. We also found that the N361-369 homologous peptide from MERS-CoV (NMERS), which exhibited 3 amino acids in difference to N361-369, bound to HLA∗A11:01 weakly (Fig. 4B). Meanwhile, IFN-γ production was evaluated for the reactivity of TCR 1- and TCR 4-CD8+ T cells, after co-culturing TCR-transduced cells with HLA-A∗11:01+ COS-7 cells loaded with each mutant peptide. We observed that N361-369-specific TCR-T cells responded differently to these mutant peptides. The T366I, F363L or T362I mutations of N361-369 could still induce high levels of IFN-γ production, but not other point mutations (Fig. 4B). Interestingly, we found that NT366I preferably led to an activation of the TCR 1-T cells similar to that of the wild type N361-369, and the TCR 4-T cells were mainly activated by NT362I or NF363L (Fig. 4B). These findings indicated that N361-369-specific TCRs derived from the COVID-19 convalescent patients had cross-reactivity to certain N361-369 variants. Altogether, the N361-369 is shown to be a T cell epitope with strong immunogenicity that can be recognized by a diversity of TCRs with high affinity. Such diverse TCRs may play critical protective roles in SASR-CoV-2 infection and decrease the mutation driven immune escape.
Confirmation of the endogenous presentation of the N361-369 epitope
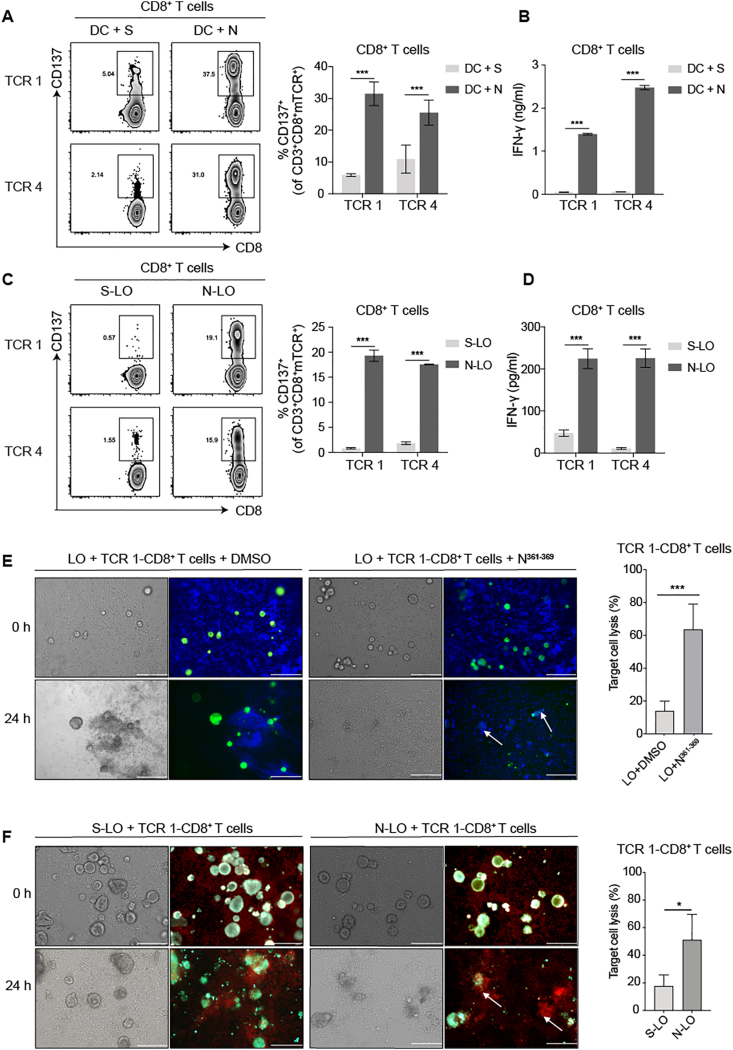
Furthermore, we studied whether the bioinformatically predicted N361-369 epitope could be naturally processed and presented from SARS-CoV-2 N protein. First, as dendritic cells (DCs) can ingest virus-infected cells or cellular fragments and cross-present antigens to activate CD8+ T cells, we tested whether the N361-369 specific-TCR-T cells could be activated by the N protein loaded mature DCs that were induced from the HLA-A∗11:01+ healthy donors’ monocytes. Substantial levels of T cell activation were detected in TCR 1- and TCR 4-CD8+ T cells, indicated by elevated CD137 expression, comparing to the S protein loaded groups (Fig. 5A). Also, we found significantly higher levels of IFN-γ secretion in the N protein loaded groups, relative to the S protein controls (Fig. 5B).
Figure 5.
The reactivity of TCR-transduced CD8+ T cells to the endogenously presented N361-369 epitope on APCs. The reactivity assay of TCR 1- and TCR 4-transduced CD8+ T cells following co-culturing with (A,B) HLA-A∗11:01+ DCs loaded with N or (C,D) HLA-A∗11:01+ lung organoids endogenously expressing N protein. (A,C) CD137 expression was evaluated by flow cytometry. (B,D) IFN-γ secretion was determined by ELISA. (E) Cytotoxicity assay of TCR 1-CD8+ T cells against HLA-A∗11:01+ lung organoids exogenously pulsed with N361-369 peptide (left). DMSO was used as negative control. T cells were stained with blue dye and lung organoids were stained in green. Target cell lysis was quantified using Calcein-AM release assay (right). (F) Cytotoxicity assay of TCR 1-CD8+ T cells against HLA-A∗11:01+ lung organoids endogenously expressing the N protein (left). Organoids expressing the S protein was applied as negative control. T cells were stained in red and lung organoids were stained in green. Target cell lysis was quantified by LDH release assay (right). Corresponding microphotographs of brightfield displayed all cells in view. White arrows show examples of organoids that are efficiently lysed. Bar: 200 μm. Data were presented as the mean ± SD, n = 3, ∗P < 0.05, ∗∗∗P < 0.001. Representative data of two independent experiments were shown.
Next, we verified whether N361-369 could be endogenously presented by lung epithelial cells that were infected by SASR-CoV-2. To this end, we established a series of lung tissue organoids, and HLA-A∗11:01+ lung organoids, referred as LO, were selected for the subsequent experiments (Figs. S5A–C). The expression of N protein in lung organoid (N-LO) was achieved by ectopically expressing the N protein via the lentiviral EGFP system, with the transfection efficiency confirmed by fluorescent detection (Fig. S5D). The lung organoids expressing S protein (S-LO) were used as control. After N361-369-specific TCR-T cells were co-cultured with N-LO or S-LO for 24 h, flow cytometric analysis demonstrated that only N-LO could induce an activation of these TCR-CD8+ T cells, indicated by enhanced CD137 expression and significantly elevated IFN-γ production (Fig. 5C, D). These results demonstrated that the N361-369 epitope could be naturally processed from the full length N protein, and presented on the surface of APCs by the intracellular HLA machinery, leading to the activation of N361-369 specific CD8+ T cells.
To evaluate the N361-369 activated CD8+ T cells could kill lung organoids, we co-cultured HLA-A∗11:01+ LO loaded with the N361-369 peptide with TCR 1-CD8+ T cells. After 24 h, we observed apparent organoid lysis, together with the recruitment of TCR 1-T cells, only with the presence of N361-369 (Fig. 5E). We found a significantly higher level of target cell lysis in the N361-369 loaded group, when these effects were quantified by Calcein-AM release assay (Fig. 5E). To determine whether the cytotoxic CD8+ T cells could eliminate infected lung organoids, we co-cultured HLA-A∗11:01+ N-LO or S-LO with TCR 1-CD8+ T cells. We found that lung organoids with endogenous expression of the N protein became more susceptible to the cytotoxicity of specific T cells, comparing to the S-LO group (Fig. 5F). Such difference of the cytotoxic effect was statistically significant, analysed by LDH release assay (Fig. 5F). Taken together, we identified a dominant T cell epitope, N361-369, that could activate specific CD8+ T cells, and the cytotoxicity of these T cells would result in the lysis of virus infected cells, executively managing the virus clearance in COVID-19 patients.
Discussion
Understanding the hallmarks of the cellular immunity against SARS-CoV-2 is critical for vaccine development. Here, we have identified immunodominant epitopes from SARS-CoV-2 S and N proteins that can induce specific CD8+ T cell responses, in the majority of COVID-19 convalescent patients with HLA-A∗02:01, HLA-A∗24:02 or HLA-A∗11:01. Importantly, we have discovered N361-369, a dominant epitope from the N protein, and 2 different CD8+ TCRs recognizing N361-369. These 2 TCRs displayed high functional avidity for the wild-type and the mutant variants of N361-369, which provided complementary protections for each other against SASR-CoV-2. We are the first to present evidence that N361-369 can be endogenously presented by HLA-A∗11:01 on APCs, which, in turn, can activate specific CD8+ T cells that may contribute to effective virus clearance in COVID-19 convalescent patients.
Both the S and N proteins of SARS-CoV-2 have been reported to effectively eliciting CD8+ T cell responses.15 Indeed, when stimulated by synthesized peptides corresponding to SARS-CoV-2 S and N, robust CD8+ T cell responses could be detected in nearly 75% of COVID-19 convalescent PBMCs (Fig. 1; Table S6). Strikingly, we found that 3 out of 4 dominant peptides were derived from the N protein, and accounted for approximately 50% of tested CD8+ T cell reactivity. These data suggested that the N protein might be a dominant target for CD8+ T cell recognition. Similar observations have been made.15,17 Of note, T cell responses to the 4 dominant CD8+ T cell epitopes identified in our study were not observed in uninfected individuals (Fig. S4), while cross-reactivities with other common coronaviruses in unexposed healthy donors had been reported. This difference might due to the refined sample size of healthy donors with corresponding HLA restrictions in our study. Contrast to the rather limited HLA-A phenotype information associated with available T cell epitopes from COVID-19 samples, we provided 15 SARS-CoV-2 epitopes for the 3 most prominent HLA class I A alleles in the Asian population (Table 1), offering more accurate information for vaccine designs. Of these dominant epitopes, the HLA-A∗11:01-restricted N361-369 (KTFPPTEPK) was found in 5/8 COVID-19 PBMCs (Fig. 1C; Table 1). Independently, this epitope has also been identified in multiple individuals in the British COVID-19 convalescent patients.22 These findings strongly suggested that N361-369 was highly immunogenic and capable to induce robust CD8+ T cell responses to SARS-CoV-2.
Presently, reports for SARS-CoV-2 inducing cellular immune responses were largely focused on the T cell-antigen reactivity. Detailed mechanisms regarding the cellular immunity in COVID-19 convalescent patients were needed for advancing the vaccine development and evaluation. In our study, we found 2 N361-369-specific TCRs with distinct functions. When ectopically transduced, they could elicit CD8+ T cell cytotoxicity upon target cells (Fig. 3D), and mediate the activation of T helper type 1 CD4+ T cells to secret cytokines in a CD8-independent manner (Fig. 3E). It has been reported that such independency could only be achieved when the TCR exhibited high-affinity binding to the cognate pMHC.33, 34, 35 In the affinity test of the N361-369-specific TCRs in the transduced CD8+ T or CD4+ T cells, we found that their EC50 concentrations ranged from 47.17 nM to 380.9 nM (Fig. 4A). These values are remarkably smaller than most of the reported thresholds of affinity for maximal T-cell activity to peptide epitopes, which were around 5–10 μM.50, 51, 52, 53 Hence, our data evidenced that the N361-369-specific TCRs had high affinity and functional avidity, both of which were key features to induce sufficient levels of T cell activation for antivirus responses. In addition, these results highlighted the possibility that the application of CD8-independent TCRs with natural or genetically engineered T cells might present desirable functional advancement, and the identification of our TCRs might contribute to the potential adoptive cell immunotherapies designed against SARS-CoV-2.
As a newly emerged single stranded RNA virus belonging to the highly contagious coronavirus subfamily, SARS-CoV-2 has a tendency to accumulate mutations over time.54,55 Here, another innovative piece of our finding was that the 2 distinct N361-369-specific TCRs could recognize multiple N361-369 mutations in complementary (Fig. 4B). Thus, our findings have revealed the high functional avidity of the 2 TCRs for N361-369, with compatible characteristics for clinical mutants of this epitope, providing practical tools to interpret potential mechanisms of CD8+ T cell responses against SARS-CoV-2, which are of clinical significance in the design and evaluation for the vaccine against the current SARS-CoV-2 mutants.
Viral antigens can be phagocytosed and cross-presented by DCs to prime CD8+ T cells, or directly processed and presented by infected cells to activate cellular immune responses. Having proven the strong immunogenicity of N361-369 in vitro, it is of physiological importance to confirm that the N protein can be processed and presented naturally. We proved that N361-369 could be presented by DC cells and elicited robust activation of specific CD8+ T cells (Fig. 5A, B). Similarly, marked CD8+ T cell cytotoxicity were observed with lung organoids either exogenously loaded or endogenously expressing the N361-369 epitope (Fig. 5C–F). The significance of CD8+ T cell response for viral clearance has been demonstrated in SARS-CoV infection.56,57 Therefore, our findings from DCs and the lung organoid model have, for the first time, revealed potential mechanisms of cellular immunity to eliminate SARS-CoV-2 infected lung cells.
At present, majority of COVID-19 vaccine candidates under development focus on SARS-CoV-2 S protein as the antigen, due to its location and functional significance in virus entry.5, 6, 7,20,58, 59, 60 Hereby, we have presented evidences of 4 dominant SARS-CoV-2 epitopes, most of which were derived from the N protein, highlighting the potential benefit of SARS-CoV-2 N in vaccine optimization. It should be mentioned that the sequence of the N361-369 epitope is identical to a fragment of the SARS-CoV N protein, as this region of the two coronaviruses share more than 90% homology.42 In fact, of the total 15 SARS-CoV-2 T cell epitopes presented in our study, 8 were independently confirmed with amino acid sequences identical to SARS-CoV (Table 1 and Table S4), indicating probable cross-protective effects of vaccines developed according to these CD8+ T cell epitopes.
A larger sample size with subgrouping of COVID-19 patients based on the intensities of their diseases may better represent epitope diversities and be advantageous to understand the correlations between various T cell immune responses and the disease severities. In addition to bioinformatics predicted peptide pools, the application of peptide libraries covering the entire SARS-CoV-2 protein sequences can add detailed resolution of the SARS-CoV-2-specific T cell responses and is currently under investigation. The magnitude and breadth of the observed cytotoxic effect of CD8+ T cells may be subjected for debate, and potential undesired pathological damage resulted from the T-cell immunity should be addressed in future studies.
In conclusion, this study has demonstrated potent and broad SARS-CoV-2-specific CD8+ T cell responses in the majority of COVID-19 convalescent patients. These confirmed immunodominant epitopes and the identified TCRs with functional superiority provide critical knowledge to understand the cellular immune response to SARS-CoV-2 and the mechanisms for eliminating infected cells, and revealed valuable candidates for next-generation vaccine designs.
Author contributions
A.J., and D.P. conceived and designed the study. Q.C., F.G., Y.W., T.L., S.L., J.H., and J.W. were responsible for collecting samples. C.H., M.S., X.H., Q.C., L.L., S.C., J.Z., J.Z., D.C., and Q.W. performed the experiments. C.H., L.L., and X.H. performed the single cell TCR cloning. M.S., and J.Z. established the lung organoids model. M.S., X.H., C.H., W.W., and K.T. performed data analysis and incorporation. C.H., A.J., and W.W. wrote the manuscript.
Conflict of interests
Patent has been filed for some of the epitopes presented here.
Funding
This study was supported by the Emergency Project from Chongqing Medical University and Chongqing Medical University fund, China (No. X4457) with the donation from Mr Yuling Feng.
Acknowledgements
We acknowledge the clinical laboratories of Yongchuan Hospital of Chongqing Medical University and The Third Affiliated Hospital of Chongqing Medical University for providing blood samples. We also thank all healthy individuals participated in this study. We appreciate the Department of Immunology of Toyama University for kindly providing technical advice.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.05.006.
Contributor Information
Da Pang, Email: pangda@ems.hrbmu.edu.cn.
Aishun Jin, Email: aishunjin@cqmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ. 2020;371:m4857. doi: 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 5.Chandrashekar A., Liu J., Martinot A.J., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbett K.S., Flynn B., Foulds K.E., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercado N.B., Zahn R., Wegmann F., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q., Bao L., Mao H., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y., Su B., Guo X., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R., Shan C., Duan X., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 13.Ju B., Zhang Q., Ge J., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 14.Chen X., Li R., Pan Z., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17(6):647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Bert N., Tan A.T., Kunasegaran K., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 17.Weiskopf D., Schmitz K.S., Raadsen M.P., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni L., Ye F., Cheng M.L., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shomuradova A.S., Vagida M.S., Sheetikov S.A., et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):1245–1257. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., Tostanoski L.H., Peter L., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbett K.S., Edwards D.K., Leist S.R., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Y., Mentzer A.J., Liu G., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti A.P., Kula T., Wang Y., et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–1107. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chour W., Xu A.M., Ng A.H.C., et al. medRxiv; 2020. Shared antigen-specific CD8+ T cell responses against the SARS-COV-2 spike protein in HLAA∗02:01 COVID-19 Participants. [Google Scholar]
- 25.Nelde A., Bilich T., Heitmann J.S., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22(1):74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 26.Poran A., Harjanto D., Malloy M., et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12(1):70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder T.M., Gittelman R.M., Klinger M., et al. medRxiv; 2020. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. [Google Scholar]
- 28.Ferretti A.P., Kula T., Wang Y.F., et al. medRxiv; 2020. COVID-19 patients form memory CD8+ T cells that recognize a small set of shared immunodominant epitopes in SARS-CoV-2. [Google Scholar]
- 29.Braun J., Loyal L., Frentsch M., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 30.Sekine T., Perez-Potti A., Rivera-Ballesteros O., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thieme C.J., Anft M., Paniskaki K., et al. Robust T cell response toward spike, membrane, and nucleocapsid SARS-CoV-2 proteins is not associated with recovery in critical COVID-19 patients. Cell Rep Med. 2020;1(6):100092. doi: 10.1016/j.xcrm.2020.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kared H., Redd A.D., Bloch E.M., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest. 2021;131(5):e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyons G.E., Moore T., Brasic N., Li M., Roszkowski J.J., Nishimura M.I. Influence of human CD8 on antigen recognition by T-cell receptor-transduced cells. Cancer Res. 2006;66(23):11455–11461. doi: 10.1158/0008-5472.CAN-06-2379. [DOI] [PubMed] [Google Scholar]
- 34.Tan M.P., Dolton G.M., Gerry A.B., et al. Human leucocyte antigen class I-redirected anti-tumour CD4(+) T cells require a higher T cell receptor binding affinity for optimal activity than CD8(+) T cells. Clin Exp Immunol. 2017;187(1):124–137. doi: 10.1111/cei.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campillo-Davo D., Flumens D., Lion E. The quest for the best: how TCR affinity, avidity, and functional avidity affect TCR-engineered T-cell antitumor responses. Cells. 2020;9(7):1720. doi: 10.3390/cells9071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chheda Z.S., Kohanbash G., Okada K., et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med. 2018;215(1):141–157. doi: 10.1084/jem.20171046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y., Li J., Mao W., et al. HLA common and well-documented alleles in China. HLA. 2018;92(4):199–205. doi: 10.1111/tan.13358. [DOI] [PubMed] [Google Scholar]
- 38.Weiskopf D., Angelo M.A., de Azeredo E.L., et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A. 2013;110(22):E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38(4):e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon P., Omokoko T.A., Breitkreuz A., et al. Functional TCR retrieval from single antigen-specific human T cells reveals multiple novel epitopes. Cancer Immunol Res. 2014;2(12):1230–1244. doi: 10.1158/2326-6066.CIR-14-0108. [DOI] [PubMed] [Google Scholar]
- 44.Hamana H., Shitaoka K., Kishi H., Ozawa T., Muraguchi A. A novel, rapid and efficient method of cloning functional antigen-specific T-cell receptors from single human and mouse T-cells. Biochem Biophys Res Commun. 2016;474(4):709–714. doi: 10.1016/j.bbrc.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin B.Y., Campbell T.E., Draper L.M., et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI insight. 2018;3(8):e99488. doi: 10.1172/jci.insight.99488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen C.J., Zhao Y., Zheng Z., Rosenberg S.A., Morgan R.A. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66(17):8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin J., Sabatino M., Somerville R., et al. Simplified method of the growth of human tumor infiltrating lymphocytes in gas-permeable flasks to numbers needed for patient treatment. J Immunother. 2012;35(3):283–292. doi: 10.1097/CJI.0b013e31824e801f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao W.M., Song S.H., Chen M.L., et al. The 2019 novel coronavirus resource. Yi Chuan. 2020;42(2):212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- 50.Tan M.P., Gerry A.B., Brewer J.E., et al. T cell receptor binding affinity governs the functional profile of cancer-specific CD8+ T cells. Clin Exp Immunol. 2015;180(2):255–270. doi: 10.1111/cei.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong S., Malecek K., Johnson L.A., et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc Natl Acad Sci U S A. 2013;110(17):6973–6978. doi: 10.1073/pnas.1221609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oren R., Hod-Marco M., Haus-Cohen M., et al. Functional comparison of engineered T cells carrying a native TCR versus TCR-like antibody-based chimeric antigen receptors indicates affinity/avidity thresholds. J Immunol. 2014;193(11):5733–5743. doi: 10.4049/jimmunol.1301769. [DOI] [PubMed] [Google Scholar]
- 53.Schmid D.A., Irving M.B., Posevitz V., et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol. 2010;184(9):4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 54.Villa T.G., Abril A.G., Sanchez S., de Miguel T., Sanchez-Perez A. Animal and human RNA viruses: genetic variability and ability to overcome vaccines. Arch Microbiol. 2021;203(2):443–464. doi: 10.1007/s00203-020-02040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8):e3000003. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88(19):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84(18):9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J., Wang W., Chen Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 59.Keech C., Albert G., Cho I., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu F.C., Li Y.H., Guan X.H., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.