Abstract
Hong Kong utilized an elimination strategy with intermittent use of public health and social measures and increasingly stringent travel regulations to control SARS-CoV-2 transmission. By analyzing >1700 genome sequences representing 17% of confirmed cases from 23-January-2020 to 26-January-2021, we reveal the effects of fluctuating control measures on the evolution and epidemiology of SARS-CoV-2 lineages in Hong Kong. Despite numerous importations, only three introductions were responsible for 90% of locally-acquired cases, two of which circulated cryptically for weeks while less stringent measures were in place. We found that SARS-CoV-2 within-host diversity was most similar among transmission pairs and epidemiological clusters due to a strong transmission bottleneck through which similar genetic background generates similar within-host diversity.
One sentence summary:
Out of the 170 detected introductions of SARS-CoV-2 in Hong Kong during 2020, three introductions caused 90% of community cases.
Severe acute respiratory coronavirus 2 (SARS-CoV-2) emerged in late 2019 (1) and has caused over 170 million confirmed cases and over three million deaths worldwide (as of 1-July-2021) (2). Heterogeneity in disease severity (3-5) and high virus transmission rates (6, 7) necessitated extensive and diverse control strategies, which achieved varied degrees of success. Hong Kong (population 7.5 million) is among the few regions that have been relatively successful in prevention and control of community transmission using an elimination strategy (8), with intermittent public health and social measures combined with strict isolation of cases and quarantine of close contacts in designed facilities (9, 10). Increasingly stringent travel regulations have been implemented to limit importation of infections (Fig. 1). As of 1-July-2021, 11,855 laboratory confirmed cases have been detected resulting in 210 deaths. Using contact tracing data from January to April 2020 (waves one and two) we have shown that sustained community transmission in Hong Kong was largely driven by superspreading events, with 80% of community infection caused by 20% of cases (10). However, two large outbreaks have occurred in Hong Kong since this period (waves three and four). Here, we show that the three major surges in infections that occurred during waves two to four were the result of only three virus introductions (PANGO lineages (11) B.3, B.1.1.63, and B.1.36.27) out of a total of 170 captured through genome sequencing, resulting in 90.0% of the confirmed cases of SARS-CoV-2 observed in the community in Hong Kong. Using genomic data generated from travel-related (n=186) and community cases (n=1,713) during all four waves of the pandemic, comprising 51.4%, 21.1%, 23.6% and 13.7% of all cases in the corresponding period, respectively (figs. S1, S2 and S3, tables S1 and S2), we discuss the effects of intermittent use of public health and social measures on the evolution and epidemiology of SARS-CoV-2 in Hong Kong.
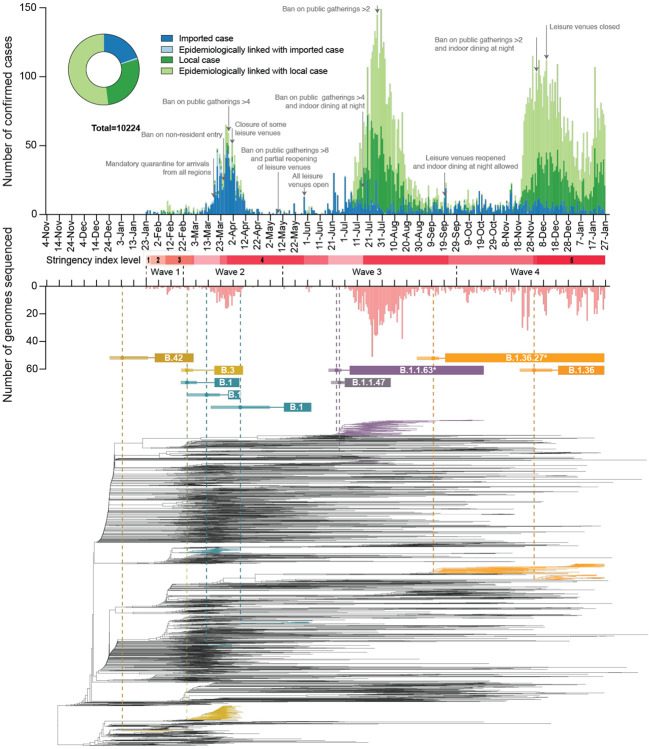
Fig. 1. Epidemiological summary and time-scaled phylogeny of SARS-CoV-2 in Hong Kong.
The number of genomes sequenced in this study is shown below in light red. Time-scaled phylogeny of SARS-CoV-2 genomes from Hong Kong, in which lineages HK-wave3 and HK-wave4A were subsampled to 100 and 65 sequences, showing B.1.1.63* and B.1.36.27*, respectively. *denotes the real clade contains more than one PANGO lineage (see Materials and Methods and table S4). Red shaded bars below the timeline delineate five stringency levels of control measures in Hong Kong based on the Oxford COVID-19 Government response tracker (level 1: <40; level 2 : 40–50; level 3: 50–60; level 4: 60–70; level 5: >70) (12).
Introductions, local spread, and delayed detection during waves one and two
During wave one (23-January-2020 to 22-February-2020), laboratory-confirmed cases did not exceed 10 per day (Fig. 1 and table S1) and were comprised of both travel-related (n=17/70, 24.3%) and community-acquired cases (n=53/70, 75.7%). The first recognised introduction was detected on 30-January-2020 among a family cluster where the index case had returned from Wuhan, China on 22-January, while the first community case without a known source (travel history or contact with a confirmed case) was also reported on 30-January. However, among two of the earliest locally circulating lineages, time-scaled phylogenetic analysis estimated a median time to most recent common ancestor (tMRCA) of 30-December-2019 (95% Highest Posterior Density Interval (HPD) 24-December-2019 – 1-January-2020) indicating direct ancestry to cases circulating prior to the earliest recognised introductions (Fig. 2A). Critically, these lineages had no recognised epidemiological or phylogenetic link to all other imported cases identified and sampled at the time, indicating that the source of these lineages initially entered Hong Kong undetected and triggered sustained community transmission during the first wave. Though the tMRCA indicates introduction could have occurred as early as 24-December-2019, which would represent one of earliest examples of transmission outside mainland China, it cannot be proven as the lineage diversity observed may have first accumulated in mainland China and subsequently been imported closer to the detection of the earliest cases.
Fig. 2. Descriptive and temporal dynamics of SARS-CoV-2 lineages in Hong Kong.
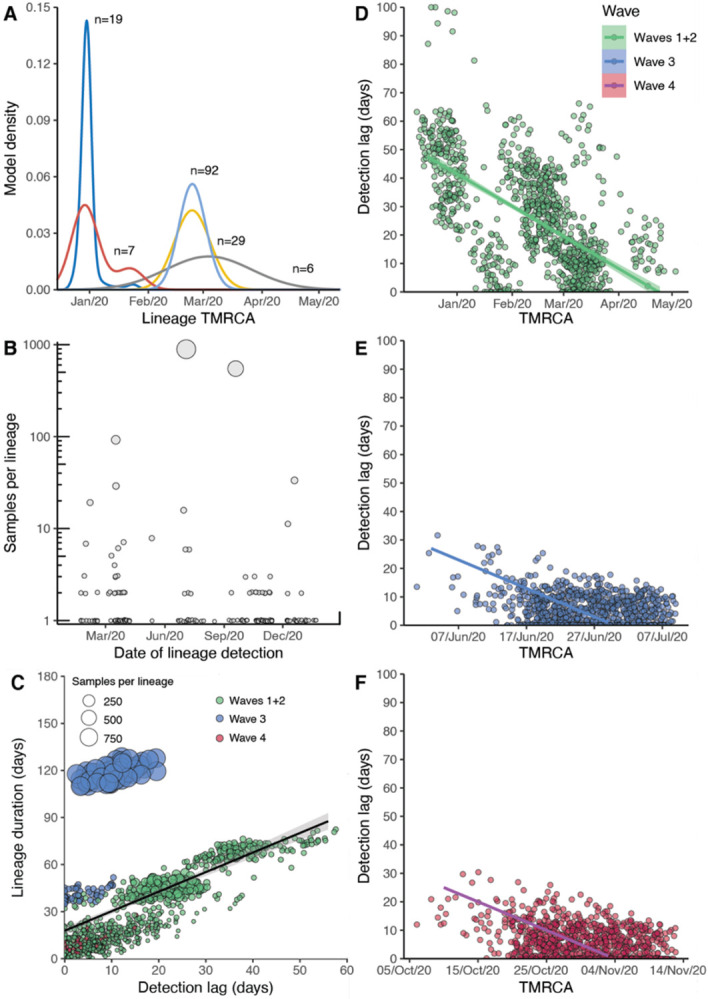
(A) Time to most recent common ancestor (tMRCA) among the five earliest locally circulating lineages of SARS-CoV-2 introduced into Hong Kong. (B) Number of SARS-CoV-2 genomic samples per lineage identified over time. Lineage size is ordered on a log10 scale, and plotted by the earliest confirmation date among the lineages. (C) Correlation between the detection lag of locally circulating lineages and the final lineage duration. Points represent a random sample of 1,000 lineages from a Bayesian posterior tree distribution (n=8,000). (D:E) Detection lag over time as a function of tMRCA across three epidemic periods (D) waves one and two, (E) wave three, (F) wave four. Overall, a significant reduction in detection lag was observed over time and across each epidemic wave. Points in each panel represent a random sample of 1,000 lineages from a Bayesian posterior tree distribution (n=8,000).
As SARS-CoV-2 was declared a global pandemic on 11-March-2020, Hong Kong experienced a substantial rise in travel-related cases (n=705/978, 72.1%, Fig. 1 and table S1) concomitant with large epidemics internationally (wave two). Introductions occurred mostly from outside of China (fig. S4), with a moderate increase observed in community cases (n=273/978, 27.9%). Again, the inferred common ancestry of community-linked lineages indicated circulation prior to or during early March 2020 (tMRCA), indicating a pattern of prolonged cryptic transmission that preceded increases in community spread (Fig. 2A). The largest of these local lineages comprised 92 genomes or 38.0% (n=92/242) of all sampled genomes during wave two and was classified as PANGO lineage B.3. This lineage was associated with a superspreading event from which contact tracing identified 106 community cases. Genomic analysis linked an additional 16 sporadic community cases, increasing the total inferred cluster size to 122 or 46.2% (n=122/326) of community cases during waves one and two.
Overall, during waves one and two, there were 38 lineages circulating in the community (out of a total of 61 unique imported clades). The median size for non-singleton local lineages was six cases, and the median duration of circulation was 10.5 days. Among local lineages without traced contact to an imported case, the median delay in lineage detection (time between lineage tMRCA and confirmation of the first detected case) was 25 days, though this was noticeably higher in January (median = 31 days), before significantly improving to eight days by early May 2020, likely related to early delays and subsequent improvements in case detection (Spearman’s test, rho (ρ) = −0.49, p < 0.001, Fig. 2D).
Travel measures and the suppression of overseas introductions
Following a peak of infections during 16–27 March 2020 (wave two; Fig. 1 and table S1), a rapid decline occurred as travel and community restrictions were expanded to include mandatory quarantine on arrival with strengthened testing, restrictions on non-resident entry, school closures, adjusted work arrangements, and bans on public gatherings (13). Travel restrictions began as early as 26-January-2020. First, all non-residents that visited Hubei province within two weeks were barred entry into Hong Kong. This was followed by a mandate for compulsory quarantine of passenger arrivals from regions affected with SARS-CoV-2, extending from mainland China to South Korea, Iran, Italy, and the Schengen region, and culminating in the barring of entry of non-residents during the peak of wave two in March. No community cases were reported from mid-April to 12-May-2020, and community measures were gradually relaxed to allow public gatherings of at most eight (from four) people with restricted opening of leisure venues (Fig. 1). Based on the Oxford COVID-19 Government response tracker (12), stringency of control measures reduced from level 4 to level 2 during this period (Fig. 1).
Reintroductions and local surge during waves three and four
The first large SARS-CoV-2 outbreak in Hong Kong occurred from July to September 2020 (wave three), resulting in >4,000 cases (table S1). With a predominant number of cases in the community (n=3,385/4,032, 84.0%), this third epidemic wave was preceded by a period of increased detection of travel-related cases during July 2020 (similar to waves one and two, Fig. 2B). While importations during wave two were predominantly from Europe, subsequent cases were mostly from Asia (fig. S4, table S3 and data S1). The number of laboratory-confirmed cases continued to rise, reaching a peak of >120 cases per day in late July (Fig. 1). Following the implementation of increasingly stringent public health and social distancing measures, the local epidemic subsided in September. However, beginning in early November a second resurgence (termed wave four) occurred, resulting in >6000 additional cases. Wave four peaked in December 2020 and slowly declined towards zero daily cases by April 2021.
Sequencing identified 170 virus lineages belonging to 71 PANGO lineages in Hong Kong within one year (Fig. 2 and data S2). However, 87.0% of those lineages were detected only in travel-related cases or one community case, and no variants of concern were detected in the community during waves three and four. Notably, a single introduction belonging to lineage B.1.1.63 resulted in over 90.0% of genomes sampled during wave three (92.4%, 881/953), forming a HK-wave3 clade (n=902) (also see fig. S1 and table S4). However, the earliest imported cases of the HK-wave3 clade were not sampled, indicating cryptic transmission prior to detection. In a similar trend, two lineages led to most of the fourth wave cases: over 74% genomes (552/704) formed one clade (HK-wave4A, B.1.36.27, table S4), and 4.7% (33/704) formed another (HK-wave4B, B.1.36).
By comparing the tMRCA of local non-singleton clusters in Hong Kong (n=7, 6.4% (7/109)) (Fig. 2 and data S3), we identified two introductions (HK-wave3 and HK-wave4A) that circulated in the community for 108 and 128 days, respectively. The delay in detection of local non-singleton lineages remained low during waves three and four (mean=2.9 days, 95% HPD 0 - 13 days) (Fig. 2E, F), with a detection delay of 11 and 10 days for the two large clades, HK-wave3 and HK-wave4A, respectively (data S3). A negative correlation between delay over time from wave one to wave four (Spearman’s test, rho (ρ) = −0.72, p < 0.001), indicates improvements in identifying cases during 2020 (Figs. 2D, E and F). Interestingly, the cryptic circulation of HK-wave3 occurred during periods of reduced stringency (level 2), however HK-wave4A introduction occurred during the late stages of wave three when stringency level 4 was enacted, and continued to circulate cryptically when stringency level was reduced to level 2 on presumed suppression of the second wave. Similarly, HK-wave4B introduction occurred during relatively stringent level 4 (Fig. 1), while a significant positive correlation between increasing lag in lineage detection and lineage duration was observed (Spearman’s test, rho (ρ) = 0.70, p < 0.001, Fig. 2C).
Contrasting patterns of epidemiology during waves three and four
To understand the effects of public health and social measures on community transmission, we characterized the two largest clades that circulated during waves three and four and identified contrasting pattens of genomic evolution and underlying transmission dynamics. HK-wave3 emerged during a period of reduced community measures (stringency level 2) and was controlled by increasing stringency to level 4. By contrast, HK-wave4A emerged under stringency level 4 and circulated cryptically during reduced level 3. Although the stringency level was rapidly raised to 5 on detection, HK-wave4A continued to circulate for several months.
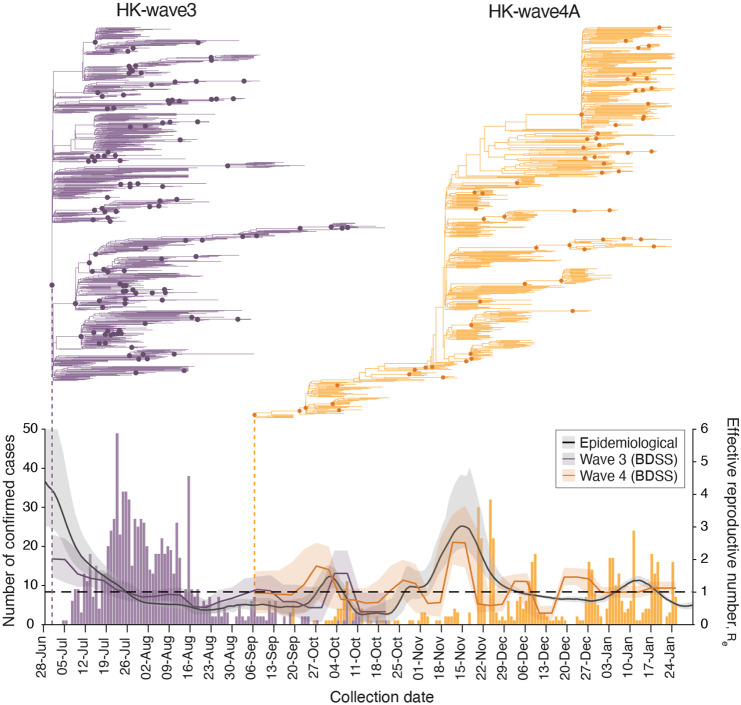
The effective reproductive number (Re) estimated using a birth-death skyline serial (BDSS) model (14) for 902 HK-wave3 genomes collected from all 18 districts in Hong Kong (5-July-2020 to 21-October-2020) showed that Re was significantly higher (~3) from the tMRCA in late June or early July (mean tMRCA = 1-July-2020, 95% HPD, 26-June-2020 to 4-July-2020) until recognition of the lineage on 5-July when stringency level was 2 (Fig. 3), highlighting a period of rapid virus expansion when leisure venues were open and public gatherings of up to 50 people were allowed. Notably, as the control stringency was increased on 15-July (from level 2 to 4), Re subsided to ~2 and subsequently decreased below ~1. An increasing number of cases continued to be reported among close contacts in residential care homes for the elderly or disabled, households, hospitals, dormitories and workplaces, whereas just 12.6% of the genomes were attributable to social interactions (table S5). Phylogenies reveal a rapid termination of transmission lineages among close contacts, leading to disappearance of all but one sub-lineage that continued to circulate with Re >1 until extinction in October 2020. These results indicate suppression with level 4 stringency during wave three, complimented by aggressive contact tracing, resulted in the elimination of the majority of wave three transmission chains despite the intermittent rise in cases among close contacts.
Fig. 3. Phylodynamics of waves three and four in Hong Kong.
Evolutionary relationships and effective reproduction number (Re(t)) of HK-wave3 (B.1.1.63) clade (purple) and HK-wave4A (B.1.36.27) clade (orange) estimated using tree-heights and sequenced incidence data. Purple and orange bars show the number of genomes by collection date. Black line shows the instantaneous effective reproduction number (Rt), estimated based on infection dates inferred from the reported dates of symptom onset (or dates of confirmation for asymptomatic cases).
HK-wave4A (mean tMRCA = 6-September-2020, 95% HPD 5–7 September-2020) continued to circulate for several months despite level 5 stringency (Fig. 3). In contrast to HK-wave3, a rapid expansion did not occur upon emergence. Instead, the HK-wave4A dynamics resembled the pattern of a propagated epidemic curve, with Re increasing with each subsequent peak during September and November, achieving a high of Re ~3 during mid to late November; Re fluctuated around 1 following this period. The structure of the phylogenetic tree suggests continual elimination of viral lineages with intermittent expansion coinciding with a superspreading event involving 28 dancing and singing venues in different districts of Hong Kong, resulting in a group of 732 cases epidemiologically linked to this superspreading event. Epidemiological data shows that up to 23.6% of genomes during this wave were related to social clusters (table S5) which is significantly higher than the HK-wave3 counterpart (p < 0.001, chi-square test). Human mobility levels inferred from Octopus, a smart card payment system used by >98% of the Hong Kong population aged 16 to 65 (15), showed adult and elderly mobility within Hong Kong resumed to pre-pandemic levels during the early stage of wave four in early to mid-November 2020 (~100% of the average level during 1–15 January 2020) (fig. S5). Taken together, our results suggest that community measures reduce Re, however prolonged community measures decrease the probability of lineage termination due to fatigue arising in the population (16, 17).
The instantaneous effective reproductive numbers (Rt) of local cases are consistent with Re of the two major transmission lineages during waves three and four (Fig. 3). During the short period of lineage co-circulation (Fig. 3), the relative reproductive number of HK-wave3 estimated using a relative fitness inference framework (15) was higher (mean = 1.28, credible interval [CrI] 0.88-1.93) when compared with HK-wave4A, though the difference in transmissibility was not significant (CI across 1.0). However, these estimates of comparative transmissibility may be affected by stochasticity, such as the occurrence of superspreading events as revealed through sequencing.
Within-host genetic variation determined by genetic background
By analyzing deep-sequence data from confirmed donor and recipient pairs using a beta-binomial statistical framework (18) we estimated the number of virions required to initiate infection was between one to three (Fig. 4A, Materials and Methods and table S8). This is consistent with data from transmission pairs estimated in UK and Austria (one to eight in UK and one to three in Austria (19, 20)) as well as between cats (two to five virions (21)), showing SARS-CoV-2 transmission bottleneck may be universally small. To understand the effects of strong transmission bottlenecks on within-host diversity, we estimated Jaccard distances between sample pairs and found that minor variants were not significantly different between established transmission pairs and epidemiologically clustered samples (Fig. 4B) but were significantly different between samples without epidemiological links. These results show that the SARS-CoV-2 within-host genetic variation is non-random and determined by genomic differences (i.e. consensus sequence), shedding new light on SARS-CoV-2 evolution.
Fig. 4. Transmission bottleneck and mutation profiles between cases.
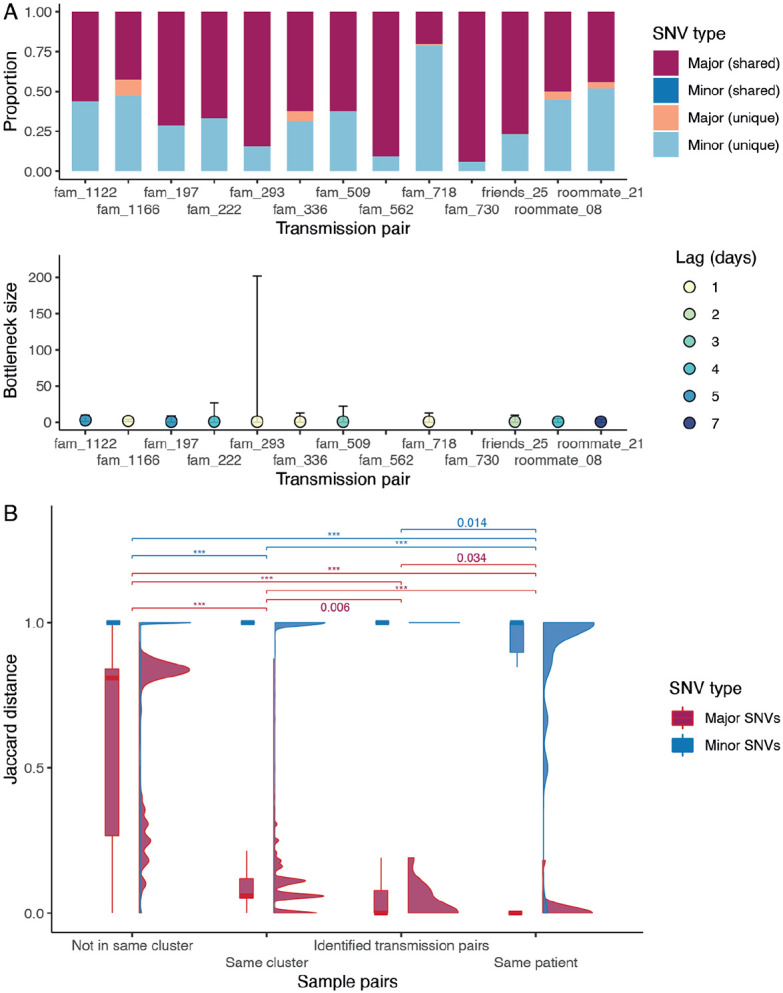
(A) Proportion of shared mutations and transmission bottleneck size estimations (with 95% confidence intervals) between samples from established transmission pairs. The estimates for transmission pairs fam_562 and fam_730 are not available due to a limited number of SNVs in the recipients’ samples. (B) Jaccard distance of the major and minor SNVs between different types of sample pairs. The distribution is shown in both boxplot and density plot. The between-group differences were tested by Wilcoxon tests, and p values < 0.05 are shown (p values < 0.001 are labelled as ***).
Conclusions
Sequencing allowed us to investigate the introduction and circulation pattens of SARS-CoV-2 transmission lineages under the elimination strategy in Hong Kong. Border control measures averted numerous introductions, and community outbreaks were typically associated with cryptic virus circulation during less stringent periods and expansion through superspreading events. Heightened control measures eliminated most transmission chains in the local community, but the proportion of social transmission during wave four was significantly higher than that of wave three, indicating control was less effective when prolonged. Although public health and social measures were promptly lifted when community cases could be traced and controlled, new waves continued as a result of new introductions rather than resurgence of previously circulating viruses. This shows that contact tracing was efficient, but averting outbreaks from new introductions requires heightened border control and enhanced community surveillance during periods of lower control level stringency.
Supplementary Material
Acknowledgments:
We gratefully acknowledge the staff from the originating laboratories responsible for obtaining the specimens and from the submitting laboratories where the genome data were generated and shared via GISAID. We thank Octopus Cards Limited for providing aggregate data of passenger numbers by public transportation means and aggregate data of transactions by retail categories for the research. We acknowledge the technical support provided by colleagues from the Centre for PanorOmic Sciences of the University of Hong Kong. We also acknowledge the Centre for Health Protection of the Department of Health for providing epidemiological data for the study. The computations were performed using research computing facilities offered by Information Technology Services, the University of Hong Kong. The funding bodies had no role in the design of the study, the collection, analysis, and interpretation of data, or writing of the manuscript.
Funding:
Health and Medical Research Fund, Food and Health Bureau of the Hong Kong SAR Government COVID190118 (LLMP)
Collaborative Research Fund of the Research Grants Council of the Hong Kong SAR Government C7123-20G (BJC)
National Institutes of Health grant U01AI151810 (LLMP)
National Institutes of Health grant HHSN272201400006C (JSMP, LLMP, BJC, DV)
National Institutes of Health grant 75N93021C00016 (JSMP, LLMP, DV)
Footnotes
Competing interests: BJC has consulted for Roche, Sanofi Pasteur, GSK, AstraZeneca and Moderna. The authors declare no other competing interests.
Data and materials availability:
Hong Kong SARS-CoV-2 genome sequences and associated metadata is deposited at gisaid.org. All anonymized data, sequence accession numbers, code, and analysis files are available in supplementary materials and in GitHub repository (https://github.com/hku-sph-covid-19-genomics-consortium/hk-sars-cov-2-genomic-epidemiology).
References
- 1.Wu F. et al. , A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verity R. et al. , Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 20, 669–677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell T. W. et al. , Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill 25, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J. T. et al. , Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 26, 506–510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizumoto K., Kagaya K., Zarebski A., Chowell G., Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 25, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti L. et al. , Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker M. G., Wilson N., Blakely T., Elimination could be the optimal response strategy for covid-19 and other emerging pandemic diseases. BMJ 371, m4907 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Cowling B. J. et al. , Impact assessment of non-pharmaceutical interventions against coronavirus disease 2019 and influenza in Hong Kong: an observational study. Lancet Public Health 5, e279–e288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adam D. C. et al. , Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat Med 26, 1714–1719 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Rambaut A. et al. , A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5, 1403–1407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale T. et al. , A global panel database of pandemic policies (Oxford COVID-19 Government Response Tracker). Nat Hum Behav, (2021). [DOI] [PubMed] [Google Scholar]
- 13.Leung G. M., Cowling B. J., Wu J. T., From a Sprint to a Marathon in Hong Kong. N Engl J Med 382, e45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadler T., Kuhnert D., Bonhoeffer S., Drummond A. J., Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc Natl Acad Sci U S A 110, 228–233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung K., Shum M. H., Leung G. M., Lam T. T., Wu J. T., Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhanwei D. et al. , Pandemic fatigue impedes mitigation of COVID-19 in Hong Kong. Research Square, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiuyan L. et al. , Community Psychological And Behavioural Responses To Coronavirus Disease 2019 Over One Year Of The Pandemic In 2020 In Hong Kong. Scientific Reports, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobel Leonard A., Weissman D. B., Greenbaum B., Ghedin E., Koelle K., Transmission Bottleneck Size Estimation from Pathogen Deep-Sequencing Data, with an Application to Human Influenza A Virus. J Virol 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lythgoe K. A. et al. , SARS-CoV-2 within-host diversity and transmission. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M. A., Koelle K., Reanalysis of deep-sequencing data from Austria points towards a small SARS-COV-2 transmission bottleneck on the order of one to three virions. bioRxiv, 2021.2002.2022.432096 (2021). [Google Scholar]
- 21.Braun K. M. et al. , Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog 17, e1009373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.