Abstract
With the development of nanomedicine, a mass of nanocarriers have been exploited and utilized for targeted drug delivery, including liposomes, polymers, nanoparticles, viruses, and stem cells. Due to huge surface bearing capacity and flexible genetic engineering property, filamentous bacteriophage and phage-mimetic nanoparticles are attracting more and more attentions. As a rod-like bio-nanofiber without tropism to mammalian cells, filamentous phage can be easily loaded with drugs and directly delivered to the lesion location. In particular, chemical drugs can be conjugated on phage surface by chemical modification, and gene drugs can also be inserted into the genome of phage by recombinant DNA technology. Meanwhile, specific peptides/proteins displayed on the phage surface are able to conjugate with nanoparticles which will endow them specific-targeting and huge drug-loading capacity. Additionally, phage peptides/proteins can directly self-assemble into phage-mimetic nanoparticles which may be applied for self-navigating drug delivery nanovehicles. In this review, we summarize the production of phage particles, the identification of targeting peptides, and the recent applications of filamentous bacteriophages as well as their protein/peptide for targeting drug delivery in vitro and in vivo. The improvement of our understanding of filamentous bacteriophage and phage-mimetic nanoparticles will supply new tools for biotechnological approaches.
Keywords: Filamentous phage, phage display, nanoparticles, targeting, drug delivery
1. Introduction
Since the presentation of “side chain theory of immunity” and “magic bullet concept” by Paul Ehrlich more than 100 years ago (Strebhardt & Ullrich, 2008; Bertrand et al., 2014; Vigevani & Valcárcel, 2014), a vast variety of novel drugs have been discovered. With the development of nanomedicine, more and more carriers have also been developed for targeted delivery of these new drugs to exert therapeutic effects, such as liposomes, dendrimers, polymers, micelles, virus-like particles, and even stem cells (Peer et al., 2007; Blanco et al., 2011; Wang et al., 2012; Cao et al., 2014). Normally, an ideal delivery vector should possess several special properties, such as good biocompatibility, proper hydrophilicity, targeting specificity, low toxicity, high uptake efficiency, and so on (Ma et al., 2012). However, all existing delivery systems have some inherent shortcomings more or less. For example, liposomes, which have been approved by US Food and Drug Administration (FDA) and widely used in clinics (Noble et al., 2014), are easily degraded in vivo and their large size (>100 nm) will hinder the penetration and diffusion (Longmire et al., 2011; Wen et al., 2013).
To date, safety and efficiency are two main evaluation criteria during drug delivery (Ryvolova et al., 2013), and how to improve their performance has been a hot topic of modern medical research. Although many therapeutic agents have been proposed for disease treatment, the therapeutic effect is still less than satisfactory. This phenomenon is mainly caused by the following several aspects: drugs are degraded before reaching the lesion sites; low target-specificity results in severe side effects; the quantity of drugs delivered into the cells is not sufficient for effective exertion and so on. Among all these factors, targeting is one of the key elements. Lately, as the excellent specificity, antibody has been proposed and applied for targeting delivery. However, because of “binding site barrier” and rapid clearance, antibody is not the best choice as a targeting motif in a targeted delivery system (Osdol et al., 1991). Subsequently, depending on the properties of peptide, such as small size, easy synthesis and typically non-immunogenicity (Hart et al., 1995; Bray, 2003; Ruoslahti, 2012; Bakhshinejad et al., 2014), it was widely used as targeting specific molecule from single target to complex multicomponent machinery (Dobbelstein & Moll, 2014). But the chemical synthesis of peptide is much more cost-effective than the production of antibodies. In case the peptides can be expressed and displayed directly on the surface of nanocarrier, such as filamentous phage, the cost of peptides will be negligible, and this is very meaningful for the development of novel drug delivery nanocarriers.
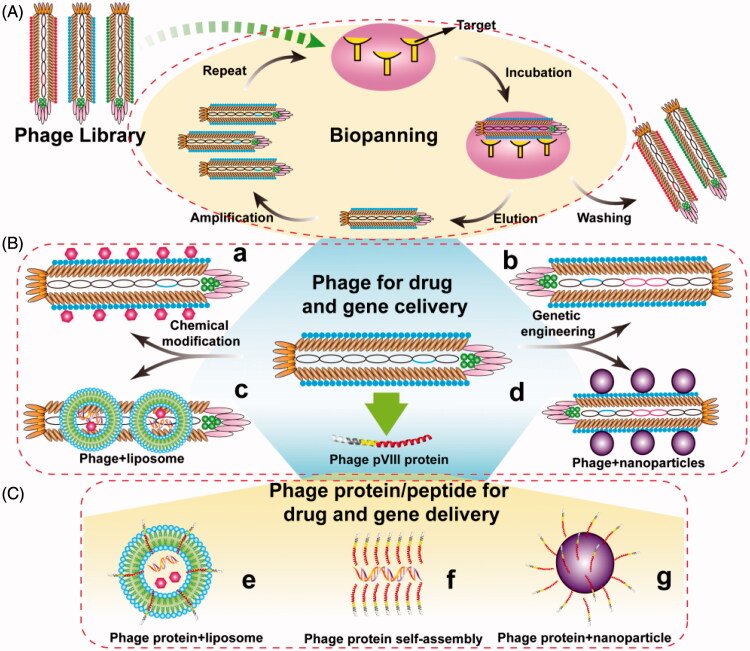
Bacteriophage (generally called phage), a kind of virus, was discovered by Frederick Twort and F´elix d’H´erelle in 1915 and 1917, respectively (Kaur et al., 2012). Compared with other viruses, one of the significant advantages of phage is their non-infectivity to mammalian cells. Besides, small genome, simple structure, and easy engineering are also the major properties of phage. So biologists, chemists, materials scientists, and medical scientists have paid more and more attentions to the phage. Since the technology of phage display was first elaborated by George P. Smith (1985) via inserting a fragment of EcoR I endonuclease on the pIII position of filamentous phage f1, different phage display systems have been exploited and applied, including phage vector-based display system and phagemid vector-based display system. Even each system has its own advantages and disadvantages, these two systems can allow small peptides to be displayed on the surface of phage in a manner of single or multivalent display. Based on phage display technology, a phage library has been constructed and used for selection of targeting phages or peptides through biopanning. The detailed process of biopanning is showed in Figure 1(A). The selected phages or peptides can be used to develop many new functional phages or phage-mimetic particles for a variety of applications (Figure 1), such as cell-targeting, tumor-homing and cell-penetrating, etc.
Figure 1.
General concept of using phage for drug and gene delivery. (A) Identification of target-recognizing peptide through bio-screening. A phage library is mixed with immobilized targets and incubated for a proper time. Unbound phages are then washed away with a washing buffer. Bound phages are eluted with an elution buffer, and amplified using medium containing preincubated E.coli bacteria and then acted as a new input library for next round bio-screening. After 3 ∼ 5 rounds, the selected phage clones are identified. (B) The paradigm of drug and gene delivery using phage particles. Phage can be chemically modified and/or genetically engineered to load drugs (a) and carry foreign genes (b), respectively. Phage can also be incorporated with other nanometer carriers for drug and gene delivery, such as liposomes (c) and nanoparticles (d). (C) The paradigm of drug and gene delivery using phage-borne proteins. Wild type or fused phage proteins can be inserted into liposomes (e) and polymer nanoparticles (g) to form phage-mimetic complexes, and even self-assembly into nanophage (f) to deliver drug and gene.
Until now, phages have been widely used in immunogenic vaccine delivery (Cruz et al., 1988; Henry et al., 2015), materials synthesis (Mao et al., 2003; Mao et al., 2004; Lee et al., 2009b; Qiu & Mao, 2010; Mao et al., 2012; Huang et al., 2015), cell growth, and differentiation (Merzlyak et al., 2009; Zhu et al., 2011; Qiu et al., 2013; Wang et al., 2013; Wang et al., 2014; Kim et al., 2017), molecular imaging (Deutscher, 2010; Carrico et al., 2012; Ghosh et al., 2012; Ma et al., 2017), and battery materials (Nam et al., 2006; Oh et al., 2014; Mohan & Weiss, 2016). One of the most important applications of phage is its use as the drug and gene delivery carrier (Figure 1(B,C)). For chemical drug delivery, filamentous phages can be loaded with a great deal of chemical drugs and have superior pharmacokinetic as well as delivery efficiency comparing with spherical nanoparticles (Lee et al., 2009a; Chauhan et al., 2011; Shukla et al., 2013). For gene delivery, foreign genes can be able to insert into the genome of phage by recombinant DNA technology, and can also be loaded by phage-mimetic nanoparticles through chemical or physical methods. Previous studies have identified that the single-strand genome of fd can be converted into double-stranded DNA in mammalian cells (Bakhshinejad et al., 2014). Most importantly, phage particles can be modified with targeting or internalizing peptides by phage display, which is a significant profile different from any other gene and drug delivery system. Moreover, with this unique characteristic, phage can also combine with other nanometer carriers to produce phage-mimetic nanoparticles and exploit plenty of new delivery systems (Figure 1(B)).
So far, there are a few review articles on phage display (Smith & Petrenko, 1997; Kehoe & Kay, 2005; Sergeeva et al., 2006; Hamzeh-Mivehroud et al., 2013), phage library (Gray & Brown, 2014), phage for imaging (Deutscher, 2010), phage self-assembly structures (Moona et al., 2015), nanomaterials for gene, and drug delivery (Biju, 2014). However, to our best knowledge, there is no review on the use of phage in gene and drug delivery. So in this review, we summarize the production of phage particles and the identification of targeting peptides using for drug delivery. Meanwhile, we also discuss the recent applications of phages and phage-mimetic nanoparticles for in vitro and in vivo drug delivery.
2. Mass production of phage nanofibers by infecting bacteria
Filamentous bacteriophage is a kind of virus that can only infects E.coli containing F pilus. The most studied filamentous bacteriophages are f1, fd, and M13, which contain a single-strand DNA (ssDNA) genome encapsulated by major coat proteins (pVIII) forming the backbone of filamentous bacteriophage, and minor coat proteins at two ends, one end composed of pIII and pVI, the other end composed of pVII and pIX.
Phages are divided into temperate and virulent phage on the basis of lifestyle. Virulent phage will initial the lysis of host cells. On the contrary, filamentous bacteriophages as temperate phages do not lyse host, moreover, it can protect the host from infection by other phages. The detailed process of filamentous phage infection is demonstrated as follows: At the beginning, phages absorb to the surface of host cells by interaction of pIII and F pilus, and the phage genome is injected into the cell with coat proteins staying outside. Subsequently, single-strand DNA (ssDNA) replicates into replicative form (RF) via a rolling circle mechanism, and new coat proteins are synthesized to assemble into phage particle by taking over host metabolism and molecular mechanism. Afterwards, pV binds to ssDNA and form a rod-like structure. Finally, with the help of pI, pVII, and pIX, ssDNA anchors to the inner membrane, and pVIII self-assembly on the surface of ssDNA. Then a whole phage is successfully assembled and released from the cell.
3. Identification of targeting using phage libraries
Since the first proposition of phage display by George P. Smith, this technology attracted more and more attentions. By genetic engineering of phage genome and assembly of phage proteins, foreign gene sequences which encode small peptides with specific targeting ability can be displayed on the surface of phage particles. The most commonly used coat proteins for phage display are pVIII and pIII (Wang & Yu, 2004; Kehoe & Kay, 2005). There are ∼2700 copies of major coat proteins pVIII with α-helical architecture (about 20°) arranged tightly along the phage particle. To avoid incorrect assembly, only short peptides (less than 10 amino acids) are allowed to display on every copy of pVIII. Actually, only ∼5 copies of pIII are responsible for infection and assembly termination. In addition, pVI (Hufton et al., 1999), pVII (Kwaśnikowski et al., 2005), and pIX (Gao et al., 2002b) can also applied for phage display.
Based on phage display technology, various phage display libraries have been constructed, such as random peptide library, phage antibody library and phage protein library, by using Kunkel mutagenesis, PCR reaction and ligation, codon sets reduction, and incorporation of unnatural amino acids. Phage library contains a reservoir of peptides that can be used for selecting versatile ligands in biomedical area, for example cell targeting drug carriers, directed location of gene delivery vectors, and targeting and tissue penetration of nanoparticles (Ruoslahti, 2012). Furthermore, the selecting peptides can also be used to overcome the obstacles in the process of drug delivery, including specific cell binding and internalization, endosome escape, and nuclei location (Han et al., 2016; Wang et al., 2016; Staquicini et al., 2017).
3.1. Cell-binding peptide
Originally, phage library was only used for selection of a given known protein or antibody solution (Devlin et al., 1990; Scott & Smith, 1990; Lam et al., 1991). But now, from biomolecules to inorganic nanoparticles, from known molecules to unknown targets, from in vitro to in vivo, phage library has already been applied to select all sorts of targets. One of its significant applications is to screen peptides that can bind with cell surface, named cell-binding peptides. On the surface of a cell, there are a large number of specific macromolecules, such as integrin (Hart et al., 1994), cadherin (Devemy & Blaschuk, 2009), HER2 (Houimel et al., 2001), EGFR (Li et al., 2005) and so on. The subtle differences of these molecules can be discriminated by small specific ligands which can be captured through bio-screening of phage libraries. For example, many targeting peptides bind to a specific antigen of cancer cells have been obtained using phage libraries and reviewed somewhere else (Sergeeva et al., 2006; Gray & Brown, 2014). Otherwise, the whole cells can also be immediately utilized for screening with phage library to obtain small peptides that bind with unknown specific targets of cells.
3.2. Cell-penetrating peptide
Many targeting molecules, named homing peptides (HPs), only help nanocarriers deliver their attached cargoes onto the surface of cell without penetrating it (Svensen et al., 2012). However, most gene and drug delivery systems need to penetrate into cells, or escape from endosomes and lysosomes, or even translocate into nucleus. So a new kind of peptide, named cell-penetrating peptide (CPP), has been discovered and verified. This peptide can be displayed on the surface of phages and phage-like particles which can help them internalize/penetrate into cells through endocytosis receptor-mediated endocytosis or receptor-independent endocytosis (Madani et al., 2011; Milletti, 2012). Since the trans activating transcription (Tat) protein of HIV-1 reported (Frankel & Pabo, 1988; Green & Loewenstein, 1988), more and more CPPs are discovered and used for translocation of therapeutic cargoes, including oligonucleotides, proteins and nanoparticles (Heitz et al., 2009; Milletti, 2012; Ruoslahti, 2012). Till now, lots of CPPs have been identified through bio-screening of phage libraries, such as HN-1 (Hong & Clayman, 2000), pep-7 (Gao et al., 2002a), 439a, 435 b (Kamada et al., 2007) and so on.
For internalizing and penetrating of phage and phage-like nanoparticles, many mechanisms of endocytosis have been proposed. In particular case, due to its own profiles of a delivery system, the special mechanism of endocytosis needs to be identified. In other words, a specific CPP only for a specific endocytic pathway should be selected and defined. For instance, through combination of phage display bio-screening and endocytic selection, two H1299 non-small cell lung cancer CPPs were identified and proved with two different mechanism of endocytosis (McGuire et al., 2014; Umlauf et al., 2014). This is the first report on the selection of endocytosis-specific peptide through phage library. While CPPs can also help therapeutic cargoes escape from endosomes and avoid degradating by lysosomes, which is crucial for gene delivery. Because, a majority of phage particles are sequestrated by endo-lysosomal degradative pathway (Stoneham et al., 2012). In addition, significant increase of gene delivery efficiency has been achieved through disruption of endosome and lysosome by virtue of lysosomotropic chemical agents and fusion or penetration of endosome membrane (Marsh & Helenius, 2006).
3.3. Nuclear location peptide
After escaping from endosome and lysosome, drugs will be released as proposed. But for gene delivery, nuclear envelope is the biggest barrier, which needs to be translocated. Luckily, nuclear localization signal (NLS) can be recognized by the nuclear transport proteins and help genes arrive at the nucleus. It has been reported that NLSs exist in the terminal proteins (TPs) of many bacteriophages, such as Ф29, Nf, PRD1, Bam35, Cp-1 and YS61 (Redrejo-Rodríguez et al., 2012; Redrejo-Rodríguez et al., 2013; Redrejo-Rodríguez & Salas, 2014). At present, no NLSs have been found in filamentous bacteriophage. And this may be the reason of low transduction efficiency (4%∼10%) in mammalian cells when transduced with filamentous bacteriophage (Poul & Marks, 1999; Larocca et al., 2001). So some new strategies have been proposed. For example, the inverted terminal repeats (ITRs) in adeno-associated virus were inserted into filamentous bacteriophages for improving expression efficiency of foreign genes (Hajitou et al., 2007).
In a word, drug and gene delivery is a sophisticated process, and most previous studies only endowed phage targeting ability to bind with cell surface or internalize into the cell. However, after entering into cells, phage particles need to escape from endosomes or lysosomes and arrive at nucleus. But the detailed mechanism is still unknown. Interestingly, phage library offers a reservoir of different peptides can exert different effects in drug and gene delivery through biopanning in vitro and in vivo (Krag et al., 2006).
4. Filamentous phage-mediated delivery
As demonstrated above, phage as a delivery system can be applied for treating different diseases, such as bacterial infection (Yacoby et al., 2006; Gravitz, 2012; Qadir, 2015; Bardy et al., 2016; Pires et al., 2016), tumor (Gandra et al., 2013a; Bakhshinejad et al., 2014; Bedi et al., 2014; Yata et al., 2014; Gross et al., 2016; Hou & Meng, 2017), Alzheimer’s disease (Frenkel & Solomon, 2002; Munke et al., 2017) and so on.
4.1. Targeted gene delivery by filamentous phage
With the enhancement of tolerance to antibiotics, phage therapy has demonstrated to be a new renaissance for antimicrobial therapy. As well known, lytic phage can infect bacteria and result in cell death by lysis, but at the same time, the released endotoxin will produce severe side effects (Slopek et al., 1982). Hence, non-lytic phage was engineered to deliver lethal genes and used for antibacterial therapy (Hagens & Bläsi, 2003; Westwater et al., 2003; Hagens et al., 2004). Beyond that, the genetically engineered M13 phage can also transfers genes into bacteria and renders them more sensitive to antibiotics (Lu & Collins, 2009; Edgar et al., 2012).
Originally, lambda phage was used to transducer mammalian cells for tumor therapy as early as 1975, but failed (Horst et al., 1975). Later, recombinant f1 phage containing urokinase type-plasminogen activator (u-PA) was used for transfection of simian COS-7 cells with the help of DEAE dextran (Yokoyama-Kobayashi & Kato, 1993) or lipopolyamine (Yokoyama-Kobayashi & Kato, 1994), leading to an increased efficiency of DNA transfection. Later, Andrew Baird (2011) proposed a detailed protocol about how to transfer mammalian cells using filamentous bacteriophage. However, all these phage vectors lack targeting capacity.
With the advent of “internalizing phages”, phage-mediated gene delivery is further developed. RGD peptide fused with phage proteins can mediate the internalization of DNA into cells (Hart et al., 1994). The first report about gene transfer of mammalian cells (COS-1 cells) by genetically targeted filamentous phage was published in 1999 by David Larroca et al. (1999), in which the phage was engineered with FGF2 and GFP as pIII fusion protein and report protein respectively. In addition, Poul and Marks (1999) further confirmed the feasibility of phage particles for gene transfer to SKBR3 breast tumor cells using the multivalently displayed anti-ErbB2 scFv as a target and GFP as a reporter, but the infection efficiency was very low. Soon after, David Larroca (2001) applied a multivalent phagemid vector for targeted delivery of GFP into PC-3 cells and improved the transduction up to 10%, which is still much lower than the traditional methods.
As well known, eukaryotic virus can provide superior gene delivery and transduction, despite their native tropism to mammalian cells. If the excellent properties can combine with phage particles, the gene transfer efficiency by target phage will definitely be improved. So a new system containing the engineered RGD-targeting filamentous phage and the cis-elements of adeno-associated virus (AAV) was introduced (Hajitou et al., 2006; Hajitou et al., 2007). This novel AAV/phage (AAVP) chimers were applied to enhance the delivery and expression of various genes, such as TNF-α for antivascular therapy using M21 cells (Tandle et al., 2009) and HSVtk suicide gene for cancer using SVEC4-10-transformed murine small vessel endothelial cells (Trepel et al., 2009). But the transduce efficiency is still low, ranging from 10% to 20% in different cancer cells (Trepel et al., 2009). With the purpose of advancing the efficiency of AAVP system, the stress-inducible Grp78 promoter was introduced instead of CMV promoter. Then after treating with histone deacetylation inhibitor and DNA methylation inhibitor, the efficiency of AAVP-mediated gene transfer was significantly improved (Kia et al., 2013). Later, a M13RGD8-AAVGFP hybrid phage was constructed using for gene delivery, and related analysis revealed that this hybrid phage could achieve selective delivery and induce GFP expression into MC3T3 cells (Yoo et al., 2016). Moreover, in order to improve anticancer safety therapy of AAVP system, this system was further modified by combining AAVP together with natural dietary genistein which had anticancer activity, and the results showed increased cell killing. (Tsafa et al., 2016).
In addition, the electrostatic repulsion between phage particles and cell surface as well as buffering capacity of phage may also affect the transduction efficacy. Therefore, a novel hybrid phage/polymer complex was developed and used for gene delivery through combining recombinant phage and cationic polymers (PDL and DEAE-DEX) (Yata et al., 2014). And the transduction efficiency of this novel hybrid phage/polymer was increased compared with no polymer modified phage. The researchers believed that this cationic polymer can help phage escape from endosomes. Hence it is feasible to deliver genes into mammalian cells using phage particles, and what we need to do is how to improve its transduction efficiency.
4.2. Targeted drug delivery by filamentous phage
Since 1996, monoclonal antibodies (mAbs) as therapeutics have already been reported and some of them have been approved by FDA (Reichert, 2008; Sievers & Senter, 2013). Except for targeting to the antigen on the surface of cells, mAbs or their fragments can also be displayed on a phage and act as new therapeutics for diagnosis and treatment of diseases. More importantly, phage possesses the ability to load antibodies and preserve their biological activities.
Apart from antibody, a substantial number of drugs are emerged as therapeutics for human diseases, such as antibiotics (chloramphenicol), anticancer drugs (doxorubicin), toxins, PDT agents (photosensitizer), radionuclides, cytokines, and so on. Due to higher toxicity to host and less sufficient of drug quantities, more and more drugs have been excluded from therapeutics. While, as a robust scaffold, filamentous phage can be applied to overcome these problems by chemical modification of chemical groups, including amino, carboxylic acid and phenol groups (Li et al., 2010), which can serve as linkers for drug decoration. Animal studies suggest that phage can carry drugs to its sidewall, penetrate the blood barrier, and then deliver the drug to brain (Carrera et al., 2004).
Previous studies have revealed that peptides can be conjugated to doxorubicin with NHS and EDC (Arap et al., 1998). Obviously, pVIII coat protein of phage with ∼2700 copies can also be decorated with drugs. In order to observe the visualization of phage infection, NHS chemistry was utilized for chemical conjugation of biotins on the phage particle to form biotinylated phages (BIO-phages) (Nakamura et al., 2001; Nakamura et al., 2002), which can be detected under confocal fluorescence microscopy by Biotin-Avidin-System (BAS). The first report on filamentous phage drug carrier is about the targeting eradication of bacteria, in which a large load of chloramphenicol (about 3000 molecules/phage) was linked to the lysines of phage on pVIII and delivered into the target cells by IgG-binding ZZ domain which was also displayed on pIII (Yacoby et al., 2006). In order to increase the loading capacity of filamentous phage, the phage coat carboxyl residues instead of the amine residues are used to conjugate chloramphenicol by EDC chemistry with over 40000 chloramphenicol molecules/phage (Yacoby et al., 2007), and thus leading to complete growth inhibition toward pathogens.
Besides antibacterial nanomedicines, the drug-carrying phage can also be applied for antitumor therapy. In the proof of concept study, the antibody-targeted phage is modified with cytotoxic drugs by a covalent bond or a cathepsin-B cleavable peptide, and then treated specific cancer cells. The results indicated that the potency of selective cell killing was significantly enhanced with a factor of >1000 over the corresponding free drugs (Bar et al., 2008).
Moreover, polymers which can load drugs and protect them from degradation are another good alternative for the conjugation of drugs with phage particles. For example, the FA-M13-PCL-P2VP nanoassemblies were developed, which composed of two main functional modules: one is M13 phage modified with folic acid constitute the shell acting as targeting moieties and drug carriers; the other is the PCL-P2VP copolymer loaded with doxorubicin constitute the core and used for drug protection and release. The results showed that the DOX-loaded particles also had a significantly higher tumor uptake and selectivity compared to free DOX (Suthiwangcharoen et al., 2011). Without release, some therapeutics just needed to be delivered to the target site, such as photosensitizer (PPa) and radionuclides. Through decorating with PPa on the surface via EDC and displaying with SKBR-3 cell-binding peptide (VSSTQDFP), Mao’s group prepared the p-PPPa complexes using for the selective killing of SKBR-3 cancer cells (Gandra et al., 2013a).
4.3. Phage-liposome complex for drug and gene delivery
Liposomes as artificial phospholipid vesicles have been developed as pharmaceutical carriers with biomedical profiles, such as good biocompatibility, little or no side effect, easy biodegradation, and large loading capacity (Torchilin, 2005; ElBayoumi & Torchilin, 2010). In order to optimize the characterization of liposomes, many new liposome formulations have been produced for longer half-life and targeting delivery. In fact, bacteriophages may be the good complementary to liposomes.
Recently, phage particles have been combined with liposomes to form new phage-liposome complexes. For instance, by multivalent electrostatic interactions, cationic liposomes are assembled on the surface of M13 phage displayed with negative charged peptides to form phage-liposome complexes (Ngweniform et al., 2009), which not only stabilize the liposome but also help ZnPC-loaded liposomes arrive specific cells. This provides a novel approach for delivering liposomes to desired targets. Through altering the ratio of liposome/phage, the structure evolution of phage-liposome complexes was studied, including “beads-on-rod” structure, nanoweb structure, short rings or spirals and phage matrix embedded with liposomes (Kalarical Janardhanan et al., 2010). Based on the targeting ability of engineered phage and loading capacity of liposomes for anticancer drugs, the nanoweb phage-liposome complexes are characterized and used as an efficient novel vehicle for drug (ZnNC) delivery. Compared with simple liposomes, nanoweb complexes can deliver more drugs to targeted cancer cells and result in increased death of cells.
5. Phage-mimetic nanoparticles-mediated delivery
5.1. Integrating targeting peptide/protein into liposome for targeted drug and gene delivery
With the excellent profiles of easy synthesis, good biocompatibility, flexible surface modification, low toxicity, and large drug/gene loading capacity, liposome as the first choice of gene and drug carrier has been used successfully in clinical trials (Noble et al., 2014). However, with the progress of liposome, quick clearance from circulation and nonspecific targeting has been the new stumbling blocks in liposome delivery. So some methods are developed and applied for liposome surface modification. Due to lower cost and facile chemical modifications, different length and density PEG was utilized to modify liposomes (Fang et al., 2005; Dos Santos et al., 2007; Maldiney et al., 2011). Although more attention has been paid to PEG coating and much progress was achieved, biomimetic coating was still adopted. One significant example is the short peptides that screened from phage libraries and used for targeting. By direct chemical conjugation, synthesized peptides were incorporated into liposomes for cell targeting in monomeric form (Pastorino et al., 2006; Stefanick et al., 2013; Noble et al., 2014) or multivalent form (Accardo et al., 2013; Gray et al., 2013; Avvakumova et al., 2014). In addition, peptides with different function were inserted into a same liposome to endow the complex more versatile. For example, rMSC-targeting peptide and a NLS peptide were encapsulated into a liposome protamine/DNA lipoplex (LPD), which improved its targeting capacity to rMSC and nuclei (Ma et al., 2013). These LPD nanoparticles were also used to deliver eye-specific genes to eyes for improving the vision of blind mice in vivo (Rajala et al., 2014).
However, the cost of synthesized peptides and the reproducibility of these systems are still major challenges for pharmaceutical applications. More importantly, the chemical modification of synthesized peptides makes the preparation process more complicated, even possibly alter the property and specificity of peptides (Emerich & Thanos, 2008). Therefore, a new alternative need to be recommended. Because of the “membranophilic” nature of phage major coat proteins, numerous studies have showed that the “wild-type” phage protein can be integrated into micelles and phospholipid bilayers (Stopar et al., 2003; Jayanna et al., 2009). So phage fusion pVIII coat proteins are directly inserted into liposomes and formed a new phage protein-liposome nanovehicles, which have been used in several drug and gene delivery (Bedi et al., 2011; Petrenko & Jayanna, 2014).
5.2. Transferring phage protein/peptide to nanoparticles for targeted drug and gene delivery
To date, a large number of nanoparticles are considered as potential drug and gene delivery carriers, such as polymeric nanoparticles (Nicolas et al., 2013), metal nanoparticles (Conde et al., 2012), dendrimers (Cheng et al., 2011), exosomes (Kooijmans et al., 2012) and so on. However, the deliver efficiency of these nanoparticles is too low due to nonspecific targeting ability. Therefore, many targeting peptides are conjugated on their surface to improve specific targeting (has been reviewed elsewhere (Pearce et al., 2012; Levine et al., 2013)) . Through transferring mesenchymal stem cells (MSCs)-targeting pVIII from phage to virus-mimetic magnetic silica nanoclusters (VMSNCs), the VMSNCs loaded with interesting genes can be targeting delivered to MSCs at a higher efficiency than commercially available vectors (Gandra et al., 2013b).
Beyond that, phage proteins themselves can assemble into nanoparticles for gene delivery. After inserting with specific targeting peptides, the phage will produce fusion pVIII (fpVIII) which has new targeting ability. Based on this, MCF-7 cells-targeting pVIII proteins self-assemble with polymeric PEG-PE to form phage-micelles, which have been used for targeting delivery of poorly soluble drugs (Wang et al., 2010; Vladimir, 2012). Similarly, Deepa Bedi et al. (2013) utilize fpVIII to encapsulate siRNA and form a kind of phage-mimetic nanoparticle, named “nanophage”, which can deliver siRNA to the target region and result in gene silencing.
6. In vivo applications of phage and phage-mimetic nanoparticles
Since the report of direct intralesional injection of bacteriophage in 6 patients with staphylococcal boils by Bruynhoge R and Maisin J in 1921, a large quantity of clinical applications of phage have been performed and reviewed elsewhere (Debattista, 2004; Fischetti et al., 2006; Kropinski, 2006; Skurnik & Strauch, 2006), most of them are mainly focus on antibacterial therapies. Subsequently, with the discovery of phage display technology by George P. Smith, many targeting peptides are explored from animals or patients in vivo using phage libraries. In 1996, the first in vivo selection of organ targeting peptide was selected (Pasqualini & Ruoslahti, 1996), and then a wide variety of organ and disease tissues were applied for targeting peptides exploration, including brain, kidney, lungs, liver and so on (Bábíčková et al., 2013). The first phage-mediated target gene transfer in vivo was reported in 2002 by Mechael A. Burg et al. (2002). Thereafter, by combining with camptothecin (CPT), EGF-targeted phagemid vector-mediated gene transfer efficiency was significantly improved up to 45% and its transduction in vivo was also assessed in PC-3 tumor xenografts of mice. Meanwhile, Frenkel and Solomon (2002) demonstrated that filamentous phage which delivered antibody to the brain could penetrate into the central nervous system in an intact form. Recently, it was reported that FDA-approved phase I clinical trial showed no safety concern (Chanishvili, 2012). In a word, all those above experiments show that no significant side effects are appeared in animals or humans in vivo after treated with phages.
During last several decades, a large quantity of administration methods have been utilized for phage delivery in vivo. One of the most common administration methods is intravenously delivery by directly injecting phage particles into blood. Phages are directly contacted with circulating blood first and then arrived different areas of the body. So this method is generally used for selection of peptides targeting vascular receptors of organs (Pasqualini & Ruoslahti, 1996; Arap et al., 2002; Yao et al., 2005; Jung et al., 2012) or tumor in vivo (Newton et al., 2006; Larimer & Deutscher, 2014). Because of the blood-brain barrier, intranasally delivered phage is exploited for targeting central nervous system (CNS) of brain (Frenkel & Solomon, 2002; Rakover et al., 2010; Lochhead & Thorne, 2012). Additionally, transdermal deliver (Prausnitz & Langer, 2008), intestinal delivery, oral administration, and intraperitoneal injection (Akita et al., 2006) are also used for in vivo delivery of phage and phage-mimetic nanoparticles. The closer administration to target organ or tissue, the stronger affinity will be obtained (Bábíčková et al., 2013).
Even a lack of reports on adverse effects, it does not mean that there is no any side effect of phage in animal and human therapy. After entering into animals or humans, bacteriophages can interact with host immune system including the phage immunogenicity and the immune-modulatory of phage (Górski et al., 2012). These interacts can induce specific immune response, and trigger innate or adaptive immune responses, such as phagocytosis, cytokines production and nonspecific antibodies production. All of these reactions may impact the effects of phage therapy in vivo. Obviously, as one kind of nanoscale biomaterial, phages need to be considered as nanoparticles for investigating their operation mechanism in vivo (Henein, 2013).
7. Conclusion
Filamentous bacteriophage has been exploited in the development of target drug delivery as virus-based delivery system. Phage enables target-selective delivery in several pathways: Firstly, the phage displayed with the target-specific peptides or antibodies can be used as nanocarriers of chemical drugs or gene drugs. Secondly, the phage displayed with target-specific peptides or antibodies can be conjugated with other vehicles (such as liposomes, inorganic nanoparticles) to form a novel delivery system. Third, the peptides or antibodies selected from a random phage library can be directly used as raw material to build new delivery systems by themselves or combining with other vehicles.
Though progress on filamentous phage-based delivery system has been made during last decades, the potential of this biomaterial needs further exploitation. Lately, the new concept of “self-navigating nanomedicines” based on filamentous bacteriophage was first proposed on the Tech Connect World Innovation Conference and Expo Techconnect Briefs 2017 (Petrenko & Gillespie, 2017), which may direct the future development of phage-based drug delivery system. As the research moves along, we believe that filamentous phage and phage-mimetic nanoparticles will play a crucial role for the development of precise and personal medicine.
Funding Statement
Zhigang Ju and Wei Sun would like to thank the grants from the National Natural Science Foundation of China (81703700 and 31760076), the Administration of Traditional Chinese Medicine of Guizhou Province (S20160829000), and the Guiyang University of Chinese Medicine (043-160002). This work was also supported by the Initial Fund Key Laboratories of Guizhou Province (grant no. 2011-4005), and the grants from the Science and Technology Department of Guizhou Province (LH [2016]7211) and Guizhou Normal University (0516006).
Disclosure statement
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- Accardo A, Ringhieri P, Tesauro D, et al. (2013). Liposomes derivatized with tetrabranched neurotensin peptides via click chemistry reactions. New J Chem 37:3528–34. [Google Scholar]
- Akita N, Maruta F, Seymour LW, et al. (2006). Identification of oligopeptides binding to peritoneal tumors of gastric cancer. Cancer Sci 97:1075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arap W, Kolonin MG, Trepel M, et al. (2002). Steps toward mapping the human vasculature by phage display. Nat Med 8:121–7. [DOI] [PubMed] [Google Scholar]
- Arap W, Pasqualini R, Ruoslahti E. (1998). Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 279:377–80. [DOI] [PubMed] [Google Scholar]
- Avvakumova S, Colombo M, Tortora P, et al. (2014). Biotechnological approaches toward nanoparticle biofunctionalization. Trends Biotechnol 32:11–20. [DOI] [PubMed] [Google Scholar]
- Bábíčková J, Tóthová Ľ, Boor P, et al. (2013). In vivo phage display-a discovery tool in molecular biomedicine. Biotechnol Adv 31:1247–59. [DOI] [PubMed] [Google Scholar]
- Baird A. (2011). Gene transfer into mammalian cells using targeted filamentous bacteriophage. Cold Spring Harb Protoc 2011:950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshinejad B, Karimi M, Sadeghizadeh M. (2014). Bacteriophages and medical oncology: targeted gene therapy of cancer. Med Oncol 31:1–11. [DOI] [PubMed] [Google Scholar]
- Bar H, Yacoby I, Benhar I. (2008). Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol 8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy P, Pantucek R, Benesik M, et al. (2016). Genetically modified bacteriophages in applied microbiology. J Appl Microbiol 121:618–33. [DOI] [PubMed] [Google Scholar]
- Bedi D, Gillespie JW, Petrenko VA, et al. (2013). Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Mol Pharm 10:551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi D, Gillespie JW, Petrenko VA. (2014). Selection of pancreatic cancer cell-binding landscape phages and their use in development of anticancer nanomedicines. Protein Eng Des Sel 27:235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi D, Musacchio T, Fagbohun OA, et al. (2011). Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes. Nanomedicine 7:315–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Wu J, Xu X, et al. (2014). Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev 66:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biju V. (2014). Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem Soc Rev 43:744–64. [DOI] [PubMed] [Google Scholar]
- Blanco E, Hsiao A, Mann AP, et al. (2011). Nanomedicine in cancer therapy: innovative trends and prospects. Cancer Sci 102:1247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray BL. (2003). Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat Rev Drug Discov 2:587–93. [DOI] [PubMed] [Google Scholar]
- Burg MA, Jensen-Pergakes K, Gonzalez AM, et al. (2002). Enhanced phagemid particle gene transfer in camptothecin-treated carcinoma cells. Cancer Res 62:977–81. [PubMed] [Google Scholar]
- Cao B, Yang M, Zhu Y, et al. (2014). Stem cells loaded with nanoparticles as a drug carrier for in vivo breast cancer therapy. Adv Mater Weinheim 26:4627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera MR, Kaufmann GF, Mee JM, et al. (2004). Treating cocaine addiction with viruses. Proc Natl Acad Sci USA 101:10416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico ZM, Farkas ME, Zhou Y, et al. (2012). N-Terminal labeling of filamentous phage to create cancer marker imaging agents. ACS Nano 6:6675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanishvili N. (2012). Phage therapy—history from Twort and d’Herelle through Soviet experience to current approaches. Bacteriophages 83:1. [DOI] [PubMed] [Google Scholar]
- Chauhan VP, Popović Z, Chen O, et al. (2011). Fluorescent nanorods and nanospheres for real-time in vivo probing of nanoparticle shape-dependent tumor penetration. Angew Chem Int Edit 123:11619–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhao L, Li Y, et al. (2011). Design of biocompatible dendrimers for cancer diagnosis and therapy: current status and future perspectives. Chem Soc Rev 40:2673–703. [DOI] [PubMed] [Google Scholar]
- Conde J, Doria G, Baptista P. (2012). Noble metal nanoparticles applications in cancer. J Drug Deliv 2012:751075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz V, Lal A, Mccutchan T. (1988). Immunogenicity and epitope mapping of foreign sequences via genetically engineered filamentous phage. J Biol Chem 263:5. [PubMed] [Google Scholar]
- Debattista J. (2004). Phage therapy: where East meets West. Expert Rev anti Infect Ther 2:815. [DOI] [PubMed] [Google Scholar]
- Deutscher SL. (2010). Phage display in molecular imaging and diagnosis of cancer. Chem Rev 110:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devemy E, Blaschuk OW. (2009). Identification of a novel dual E- and N-cadherin antagonist . Peptides 30:1539–47. [DOI] [PubMed] [Google Scholar]
- Devlin JJ, Panganiban LC, Devlin PE. (1990). Random peptide libraries: a source of specific protein binding molecules. Science 249:404–6. [DOI] [PubMed] [Google Scholar]
- Dobbelstein M, Moll U. (2014). Targeting tumour-supportive cellular machineries in anticancer drug development. Nat Rev Drug Discov 13:179–96. [DOI] [PubMed] [Google Scholar]
- Dos Santos N, Allen C, Doppen A-M, et al. (2007). Influence of poly(ethylene glycol) grafting density and polymer length on liposomes: relating plasma circulation lifetimes to protein binding. Biochim Biophys Acta 1768:1367–77. [DOI] [PubMed] [Google Scholar]
- Edgar R, Friedman N, Molshanski-Mor S, et al. (2012). Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Micro 78:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbayoumi TA, Torchilin VP. (2010). Current trends in liposome research. Methods Mol Biol 605:1. [DOI] [PubMed] [Google Scholar]
- Emerich DF, Thanos CG. (2008). Multifunctional peptide-based nanosystems for improving delivery and molecular imaging. Curr Opin Mol Ther 10:132–9. [PubMed] [Google Scholar]
- Fang C, Shi B, Pei Y-Y. (2005). Effect of MePEG molecular weight and particle size on in vitro release of tumor necrosis factor-alpha-loaded nanoparticles . Acta Pharmacol Sin 26:242–9. [DOI] [PubMed] [Google Scholar]
- Fischetti VA, Nelson D, Schuch R. (2006). Reinventing phage therapy: are the parts greater than the sum? Nat Biotechnol 24:1508–11. [DOI] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO. (1988). Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55:1189–93. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Solomon B. (2002). Filamentous phage as vector-mediated antibody delivery to the brain. Proc Natl Acad Sci USA 99:5675–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandra N, Abbineni G, Qu X. (2013a). Bacteriophage bionanowire as a carrier for both cancer-targeting peptides and photosensitizers and its use in selective cancer cell killing by photodynamic therapy. Small 9:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandra N, Wang DD, Zhu Y, et al. (2013b). Virus-mimetic cytoplasm-cleavable magnetic/silica nanoclusters for enhanced gene delivery to mesenchymal stem cells. Angew Chem Int Edit 125:11488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Mao S, Ditzel HJ, et al. (2002a). A cell-penetrating peptide from a novel pVII-pIX phage-displayed random peptide library . Bioorg Med Chem 10:4057–65. [DOI] [PubMed] [Google Scholar]
- Gao C, Mao S, Kaufmann G, et al. (2002b). A method for the generation of combinatorial antibody libraries using pIX phage display. Proc Natl Acad Sci USA 99:12612–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Kohli AG, Moser F, et al. (2012). Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth Biol 1:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Międzybrodzki R, Borysowski J, et al. (2012). Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res 83:41–71. [DOI] [PubMed] [Google Scholar]
- Gravitz L. (2012). Turning a new phage. Nat Med 18:1318–20. [DOI] [PubMed] [Google Scholar]
- Gray BP, Brown KC. (2014). Combinatorial peptide libraries: mining for cell-binding peptides. Chem Rev 114:1020–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray BP, Li S, Brown KC. (2013). From phage display to nanoparticle delivery: functionalizing liposomes with multivalent peptides improves targeting to a cancer biomarker. Bioconjugate Chem 24:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Loewenstein PM. (1988). Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 55:1179–88. [DOI] [PubMed] [Google Scholar]
- Gross AL, Gillespie JW, Petrenko VA. (2016). Promiscuous tumor targeting phage proteins. Protein Eng Des Sel 29:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst J, Kluge F, Beyreuther K, et al. (1975). Gene transfer to human cells: transducing phage lambda plac gene expression in GMI-gangliosidosis fibroblasts. Proc Natl Acad Sci USA 72:3531–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens S, Bläsi U. (2003). Genetically modified filamentous phage as bactericidal agents: a pilot study. Lett Appl Microbiol 37:318–23. [DOI] [PubMed] [Google Scholar]
- Hagens S, Habel A, Ahsen UV, et al. (2004). Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother 48:3817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajitou A, Rangel R, Trepel M, et al. (2007). Design and construction of targeted AAVP vectors for mammalian cell transduction. Nat Protoc 2:523–31. [DOI] [PubMed] [Google Scholar]
- Hajitou A, Trepel M, Lilley CE, et al. (2006). A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell 125:385–98. [DOI] [PubMed] [Google Scholar]
- Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, et al. (2013). Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today 18:1144–57. [DOI] [PubMed] [Google Scholar]
- Han L, Liu P, Petrenko VA, et al. (2016). A label-free electrochemical impedance cytosensor based on specific peptide-fused phage selected from landscape phage library. Sci Rep 6:22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart S, Harbottle R, Cooper R, et al. (1995). Gene delivery and expression mediated by an integrin-binding peptide. Gene Ther 2:552–4. [PubMed] [Google Scholar]
- Hart SL, Knight AM, Harbottle RP, et al. (1994). Cell binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. J Biol Chem 269:12468–74. [PubMed] [Google Scholar]
- Heitz F, Morris MC, Divita G. (2009). Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Brit J Pharmacol 157:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henein A. (2013). What are the limitations on the wider therapeutic use of phage? Bacteriophage 3:e24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KA, Arbabi-Ghahroudi M, Scott JK. (2015). Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold. Front Microbiol 6:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong FD, Clayman GL. (2000). Isolation of a peptide for targeted drug delivery into human head and neck solid tumors. Cancer Res 60:6551–6. [PubMed] [Google Scholar]
- Hou L, Meng X. (2017). Phage display technology and tumor targeted therapy. Cancer Res and Clin 29:214–16. [Google Scholar]
- Houimel M, Schneider P, Terskikh A, et al. (2001). Selection of peptides and synthesis of pentameric peptabody molecules reacting specifically with ErbB-2 receptor. Int J Cancer 92:748–55. [DOI] [PubMed] [Google Scholar]
- Huang J, Lin L, Sun D, et al. (2015). Bio-inspired synthesis of metal nanomaterials and applications. Chem Soc Rev 44:6330–74. [DOI] [PubMed] [Google Scholar]
- Hufton SE, Moerkerk PT, Meulemans EV, et al. (1999). Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands. J Immunol Methods 231:39–51. [DOI] [PubMed] [Google Scholar]
- Jayanna PK, Torchilin VP, Petrenko VA. (2009). Liposomes targeted by fusion phage proteins. Nanomedicine 5:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E, Lee NK, Kang SK, et al. (2012). Identification of tissue-specific targeting peptide. J Comput Aided Mol Des 26:1267–75. [DOI] [PubMed] [Google Scholar]
- Kalarical Janardhanan S, Narayan S, Abbineni G, et al. (2010). Architectonics of phage-liposome nanowebs as optimized photosensitizer vehicles for photodynamic cancer therapy. Mol Cancer Ther 9:2524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada H, Okamoto T, Kawamura M, et al. (2007). Creation of novel cell-penetrating peptides for intracellular drug delivery using systematic phage display technology originated from Tat transduction domain. Biol Pharm Bull 30:218–23. [DOI] [PubMed] [Google Scholar]
- Kaur T, Nafissi N, Wasfi O, et al. (2012). Immunocompatibility of bacteriophages as nanomedicines. J Nanotechnol 2012:1687–9503. [Google Scholar]
- Kehoe JW, Kay BK. (2005). Filamentous phage display in the new millennium. Chem Rev 105:4056–72. [DOI] [PubMed] [Google Scholar]
- Kia A, Yata T, Hajji N, et al. (2013). Inhibition of histone deacetylation and DNA methylation improves gene expression mediated by the adeno-associated virus/phage in cancer cells. Viruses 5:2561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kwon C, Jeon H. (2017). Genetically engineered phage induced selective H9c2 cardiomyocytes patterning in PDMS microgrooves. Materials (Basel) 10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijmans SA, Vader P, Van Dommelen SM, et al. (2012). Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine 7:1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krag DN, Shukla GS, Shen GP, et al. (2006). Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res 66:7724–33. [DOI] [PubMed] [Google Scholar]
- Kropinski AM. (2006). Phage therapy: everything old is new again. Can J Infect Dis Med Microbiol 17:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaśnikowski P, Kristensen P, Markiewicz WT. (2005). Multivalent display system on filamentous bacteriophage pVII minor coat protein. J Immunol Methods 307:135–43. [DOI] [PubMed] [Google Scholar]
- Lam KS, Salmon SE, Hersh EM, et al. (1991). A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354:3. [DOI] [PubMed] [Google Scholar]
- Larimer BM, Deutscher SL. (2014). Development of a peptide by phage display for SPECT imaging of resistance-susceptible breast cancer. Am J Nucl Med Mol Imaging 4:435. [PMC free article] [PubMed] [Google Scholar]
- Larroca D, Jensen-Pergakes K, Burg MA, et al. (2001). Receptor-targeted gene delivery using multivalent phagemid particles. Mol Ther 3:476–84. [DOI] [PubMed] [Google Scholar]
- Larroca D, Kassner PD, Witte A, et al. (1999). Gene transfer to mammalian cells using genetically targeted filamentous bacteriophage. FASEB J 13:727–34. [DOI] [PubMed] [Google Scholar]
- Lee SY, Ferrari M, Decuzzi P. (2009a). Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology 20:495101. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Yi H, Kim WJ, et al. (2009b). Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 324:5. [DOI] [PubMed] [Google Scholar]
- Levine RM, Scott CM, Kokkoli E. (2013). Peptide functionalized nanoparticles for nonviral gene delivery. Soft Matter 9:985–1004. [Google Scholar]
- Li K, Chen Y, Li S, et al. (2010). Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug Chem 21:1369–77. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao R, Wu X, et al. (2005). Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J 19:1978–85. [DOI] [PubMed] [Google Scholar]
- Lochhead JJ, Thorne RG. (2012). Intranasal delivery of biologics to the central nervous system. Adv Drug Deliv Rev 64:614–28. [DOI] [PubMed] [Google Scholar]
- Longmire MR, Ogawa M, Choyke PL, et al. (2011). Biologically optimized nanosized molecules and particles: more than just size. Bioconjug Chem 22:993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. (2009). Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci USA 106:4629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Wang DD, Lin Y, et al. (2013). Synergetic targeted delivery of sleeping-beauty transposon system to mesenchymal stem cells using LPD nanoparticles modified with a phage-displayed targeting peptide. Adv Funct Mater 23:1172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Nolte RJ, Cornelissen JJ. (2012). Virus-based nanocarriers for drug delivery. Adv Drug Deliv Rev 64:811–25. [DOI] [PubMed] [Google Scholar]
- Ma Z, Qin H, Chen H, et al. (2017). Phage display-derived oligopeptide-functionalized probes for in vivo specific photoacoustic imaging of osteosarcoma. Nano Medicine 13:111–21. [DOI] [PubMed] [Google Scholar]
- Madani F, Lindberg S, Langel U, et al. (2011). Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys 2011:414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldiney T, Richard C, Seguin J, et al. (2011). Effect of core diameter, surface coating, and PEG chain length on the biodistribution of persistent luminescence nanoparticles in mice. ACS Nano 5:854–62. [DOI] [PubMed] [Google Scholar]
- Mao C, Flynn CE, Hayhurst A, et al. (2003). Viral assembly of oriented quantum dot nanowires. Proc Natl Acad Sci USA 100:6946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Solis D, Reiss B, et al. (2004). Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 303:213. [DOI] [PubMed] [Google Scholar]
- Mao C, Wang F, Cao B. (2012). Controlling nanostructures of mesoporous silica fibers by supramolecular assembly of genetically modifiable bacteriophages. Angew Chem Int Edit 51:6411–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M, Helenius A. (2006). Virus entry: open sesame. Cell 124:729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcguire MJ, Gray BP, Li S, et al. (2014). Identification and characterization of a suite of tumor targeting peptides for non-small cell lung cancer. Sci Rep 4:4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak A, Indrakanti S, Lee S. (2009). Genetically engineered nanofiber-like viruses for tissue regenerating materials. Nano Lett 9:7. [DOI] [PubMed] [Google Scholar]
- Milletti F. (2012). Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov Today 17:850–60. [DOI] [PubMed] [Google Scholar]
- Mohan K, Weiss GA. (2016). Chemically modifying viruses for diverse applications. ACS Chem Biol 11:1167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moona JS, Kimb WG, Kimc C, et al. (2015). M13 bacteriophage-based self-assembly structures and their functional capabilities. Mini-Rev Org Chem 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munke A, Persson J, Weiffert T, et al. (2017). Phage display and kinetic selection of antibodies that specifically inhibit amyloid self-replication. Proc Natl Acad Sci USA 114:6444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Tsumoto K, Ishimura K, et al. (2001). A visualization method of filamentous phage infection and phage-derived proteins in escherichia coli using biotinylated phages. Biochem Bioph Res Co 289:252–6. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tsumoto K, Ishimura K, et al. (2002). The effect of an agglutogen on virus infection: biotinylated filamentous phages and avidin as a model. FEBS Lett 520:77–80. [DOI] [PubMed] [Google Scholar]
- Nam KT, Kim DW, Yoo PJ, et al. (2006). Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 312:885–8. [DOI] [PubMed] [Google Scholar]
- Newton JR, Kelly KA, Mahmood U, et al. (2006). In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia 8:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngweniform P, Abbineni G, Cao B, et al. (2009). Self-assembly of drug-loaded liposomes on genetically engineered target-recognizing M13 phage: a novel nanocarrier for targeted drug delivery. Small 5:1963–9. [DOI] [PubMed] [Google Scholar]
- Nicolas J, Mura S, Brambilla D, et al. (2013). Design, functionalization strategies and biomedical applications of targeted biodegradable/biocompatible polymer-based nanocarriers for drug delivery. Chem Soc Rev 42:1147–235. [DOI] [PubMed] [Google Scholar]
- Noble GT, Stefanick JF, Ashley JD, et al. (2014). Ligand-targeted liposome design: challenges and fundamental considerations. Trends Biotechnol 32:32–45. [DOI] [PubMed] [Google Scholar]
- Oh D, Qi J, Han B, et al. (2014). M13 virus-directed synthesis of nanostructured metal oxides for lithium-oxygen batteries. Nano Lett 14:4837–45. [DOI] [PubMed] [Google Scholar]
- Osdol W, Fujimori K, Weinstein J. (1991). An analysis of monoclonal antibody distribution in microscopic tumor nodules: consequences of a “binding site barrier”. Cancer Res 51:9. [PubMed] [Google Scholar]
- Pasqualini R, Ruoslahti E. (1996). Organ targeting in vivo using phage display peptide libraries. Nature 380:364–6. [DOI] [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Di Paolo D, et al. (2006). Targeting liposomal chemotherapy via both tumor cell-specific and tumor vasculature-specific ligands potentiates therapeutic efficacy . Cancer Res 66:10073–82. [DOI] [PubMed] [Google Scholar]
- Pearce TR, Shroff K, Kokkoli E. (2012). Peptide targeted lipid nanoparticles for anticancer drug delivery. Adv Mater Weinheim 24:3803–22. [DOI] [PubMed] [Google Scholar]
- Peer D, Karp JM, Hong S, et al. (2007). Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol 2:751–60. [DOI] [PubMed] [Google Scholar]
- Petrenko VA, Jayanna P. (2014). Phage protein-targeted cancer nanomedicines. FEBS Lett 588:341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko VA, Gillespie JW.. 2017. Self-navigating drug delivery nanovehicles driven by polyvalent multifunctional phages and their promiscuous proteins. Techconnect World Innovation Conference and Expo Techconnect Briefs, 14–17 May 2017. Washington, DC, Maryland: TechConnect.org, 134–137. [Google Scholar]
- Pires DP, Cleto S, Sillankorva S, et al. (2016). Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev 80:523–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poul MA, Marks JD. (1999). Targeted gene delivery to mammalian cells by filamentous bacteriophage. J Mol Biol 288:203–11. [DOI] [PubMed] [Google Scholar]
- Prausnitz MR, Langer R. (2008). Transdermal drug delivery. Nat Biotechnol 26:1261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir MI. (2015). Review: phage therapy: a modern tool to control bacterial infections. Pak J Pharm Sci 28:265–70. [PubMed] [Google Scholar]
- Qiu P, Qu X, Brackett DJ, et al. (2013). Silica-based branched hollow microfibers as a biomimetic extracellular matrix for promoting tumor cell growth in vitro and in vivo. Adv Mater Weinheim 25:2492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Mao C. (2010). Biomimetic branched hollow fibers templated by self-assembled fibrous polyvinylpyrrolidone structures in aqueous solution. ACS Nano 4:1573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala A, Wang Y, Zhu Y, et al. (2014). Nanoparticle-assisted targeted delivery of eye-specific genes to eyes significantly improves the vision of blind mice in vivo. Nano Lett 14:5257–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakover IS, Zabavnik N, Kopel R, et al. (2010). Antigen-specific therapy of EAE via intranasal delivery of filamentous phage displaying a myelin immunodominant epitope. J Neuroimmunol 225:68–76. [DOI] [PubMed] [Google Scholar]
- Redrejo-Rodríguez M, Muñoz-Espín D, Holguera I, et al. (2012). Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages. Proc Natl Acad Sci USA 109:18482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrejo-Rodríguez M, Muñoz-Espín D, Holguera I, et al. (2013). Nuclear localization signals in phage terminal proteins provide a novel gene delivery tool in mammalian cells. Commun Integr Biol 6:e22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrejo-Rodríguez M, Salas M. (2014). Multiple roles of genome-attached bacteriophage terminal proteins. Virology 468-470:322–9. [DOI] [PubMed] [Google Scholar]
- Reichert JM. (2008). Monoclonal antibodies as innovative therapeutics. Curr Pharm Biotechnol 9:423–30. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. (2012). Peptides as targeting elements and tissue penetration devices for nanoparticles. Adv Mater Weinheim 24:3747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvolova M, Drbohlavova J, Smerkova K, et al. (2013). Nanoparticles-based carriers for gene therapy and drug delivery. In: Mishra AK, ed. Nanomedicine for drug delivery and therapeutics. Hoboken (NJ): Wiley, 471–92. [Google Scholar]
- Scott JK, Smith GP. (1990). Searching for peptide ligands with an epitope library. Science 249:386–90. [DOI] [PubMed] [Google Scholar]
- Sergeeva A, Kolonin MG, Molldrem JJ, et al. (2006). Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev 58:1622–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Ablack AL, Wen AM, et al. (2013). Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Mol Pharm 10:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers EL, Senter PD. (2013). Antibody-drug conjugates in cancer therapy. Annu Rev Med 64:15–29. [DOI] [PubMed] [Google Scholar]
- Skurnik M, Strauch E. (2006). Phage therapy: facts and fiction. Int J Med Microbiol 296:5–14. [DOI] [PubMed] [Google Scholar]
- Slopek S, Durlakowa I, Weber-Dabrowska B, et al. (1982). Results of bacteriophage treatment of suppurative bacterial infections. I. General evaluation of the results. Arch Immunol Ther Ex 31:267–91. [PubMed] [Google Scholar]
- Smith GP. (1985). Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–17. [DOI] [PubMed] [Google Scholar]
- Smith GP, Petrenko VA. (1997). Phage display. Chem Rev 97:391–410. [DOI] [PubMed] [Google Scholar]
- Staquicini DI, Rangel R, Guzman-Rojas L, et al. (2017). Intracellular targeting of annexin A2 inhibits tumor cell adhesion, migration, and in vivo grafting. Sci Rep 7:4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanick JF, Ashley JD, Kiziltepe T, et al. (2013). A systematic analysis of peptide linker length and liposomal polyethylene glycol coating on cellular uptake of peptide-targeted liposomes. ACS Nano 7:2935–47. [DOI] [PubMed] [Google Scholar]
- Stoneham CA, Hollinshead M, Hajitou A. (2012). Clathrin-mediated endocytosis and subsequent endo-lysosomal trafficking of adeno-associated virus/phage. J Biol Chem 287:35849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopar D, Spruijt RB, Wolfs CJ, et al. (2003). Protein-lipid interactions of bacteriophage M13 major coat protein. Biochim Biophys Acta 1611:5–15. [DOI] [PubMed] [Google Scholar]
- Strebhardt K, Ullrich A. (2008). Paul Ehrlich's magic bullet concept: 100 years of progress. Nat Rev Cancer 8:473–80. [DOI] [PubMed] [Google Scholar]
- Suthiwangcharoen N, Li T, Li K, et al. (2011). M13 bacteriophage-polymer nanoassemblies as drug delivery vehicles. Nano Res 4:483–93. [Google Scholar]
- Svensen N, Walton JG, Bradley M. (2012). Peptides for cell-selective drug delivery. Trends Pharmacol Sci 33:186–92. [DOI] [PubMed] [Google Scholar]
- Tandle A, Hanna E, Lorang D, et al. (2009). Tumor vasculature-targeted delivery of tumor necrosis factor-alpha. Cancer 115:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchilin VP. (2005). Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov 4:145–60. [DOI] [PubMed] [Google Scholar]
- Trepel M, Stoneham CA, Eleftherohorinou H, et al. (2009). A heterotypic bystander effect for tumor cell killing after adeno-associated virus/phage-mediated, vascular-targeted suicide gene transfer. Mol Cancer Ther 8:2383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafa E, Al-Bahrani M, Bentayebi K, et al. (2016). The natural dietary genistein boosts bacteriophage-mediated cancer cell killing by improving phage-targeted tumor cell transduction. Oncotarget 7:52135–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf BJ, Mercedes JS, Chung CY, et al. (2014). Identification of a novel lysosomal trafficking peptide using phage display biopanning coupled with endocytic selection pressure. Bioconjugate Chem 25:1829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigevani L, Valcárcel J. (2014). Molecular biology. A splicing magic bullet. Science 345:624–5. [DOI] [PubMed] [Google Scholar]
- Vladimir P. (2012). Optimization of landscape phage fusion protein-modified polymeric Peg-Pe micelles for improved breast cancer cell targeting. J Nanomed Nanotechnol Suppl 4:008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AZ, Langer R, Farokhzad OC. (2012). Nanoparticle delivery of cancer drugs. Annu Rev Med 63:185–98. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang L, Li X, et al. (2013). Virus activated artificial ECM induces the osteoblastic differentiation of mesenchymal stem cells without osteogenic supplements. Sci Rep 3:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang M, Zhu Y, et al. (2014). Phage nanofibers induce vascularized osteogenesis in 3D printed bone scaffolds. Adv Mater Weinheim 26:4961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Yu M. (2004). Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr Drug Targets 5:1–15. [DOI] [PubMed] [Google Scholar]
- Wang L, Hu Y, Li W, et al. (2016). Identification of a peptide specifically targeting ovarian cancer by the screening of a phage display peptide library. Oncol Lett 11:4022–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Petrenko VA, Torchilin VP. (2010). Paclitaxel-loaded polymeric micelles modified with MCF-7 cell-specific phage protein: enhanced binding to target cancer cells and increased cytotoxicity. Mol Pharm 7:1007–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AM, Rambhia PH, French RH, et al. (2013). Design rules for nanomedical engineering: from physical virology to the applications of virus-based materials in medicine. J Biol Phys 39:301–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwater C, Kasman LM, Schofield DA, et al. (2003). Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother 47:1301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoby I, Bar H, Benhar I. (2007). Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother 51:2156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoby I, Shamis M, Bar H, et al. (2006). Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob Agents Chemother 50:2087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao VJ, Ozawa MG, Trepel M, et al. (2005). Targeting pancreatic islets with phage display assisted by laser pressure catapult microdissection. Am J Pathol 166:625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata T, Lee KY, Dharakul T, et al. (2014). Hybrid nanomaterial complexes for advanced phage-guided gene delivery. Mol Ther Nucleic Acids 3:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama-Kobayashi M, Kato S. (1993). Recombinant f1 phage particles can transfect monkey COS-7 cells by DEAE dextran method. Biochem Biophys Res Commun 192:935–9. [DOI] [PubMed] [Google Scholar]
- Yokoyama-Kobayashi M, Kato S. (1994). Recombinant f1 phage-mediated transfection of mammalian cells using lipopolyamine technique. Anal Biochem 223:130–4. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Jin HE, Choi DS, et al. (2016). M13 bacteriophage and adeno-associated virus hybrid for novel tissue engineering material with gene delivery functions. Adv Healthc Mater 5:88–93. [DOI] [PubMed] [Google Scholar]
- Zhu H, Cao B, Zhen Z, et al. (2011). Controlled growth and differentiation of MSCs on grooved films assembled from monodisperse biological nanofibers with genetically tunable surface chemistries. Biomaterials 32:4744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]