Abstract
G-quadruplexes (G4) are non-canonical secondary structures consisting in stacked tetrads of hydrogen-bonded guanines bases. An essential feature of G4 is their intrinsic polymorphic nature, which is characterized by the equilibrium between several conformations (also called topologies) and the presence of different types of loops with variable lengths. In cells, G4 functions rely on protein or enzymatic factors that recognize and promote or resolve these structures. In order to characterize new G4-dependent mechanisms, extensive researches aimed at identifying new G4 binding proteins. Using G-rich single-stranded oligonucleotides that adopt non-controlled G4 conformations, a large number of G4-binding proteins have been identified in vitro, but their specificity towards G4 topology remained unknown. Constrained G4 structures are biomolecular objects based on the use of a rigid cyclic peptide scaffold as a template for directing the intramolecular assembly of the anchored oligonucleotides into a single and stabilized G4 topology. Here, using various constrained RNA or DNA G4 as baits in human cell extracts, we establish the topology preference of several well-known G4-interacting factors. Moreover, we identify new G4-interacting proteins such as the NELF complex involved in the RNA-Pol II pausing mechanism, and we show that it impacts the clastogenic effect of the G4-ligand pyridostatin.
Subject terms: DNA, Protein-protein interaction networks, DNA-binding proteins, RNA-binding proteins
Introduction
In the last twenty years, G-quadruplex structures (G4) emerged as cis-acting factors impacting almost all DNA and RNA transactions. G4 in telomeric sequences were first shown to play essential roles in telomeres capping and telomeres replication by telomerase1. Now, G4 are associated with the firing of DNA replication origins2,3, transcription initiation and termination, mRNA processing, mRNA transport4–6, translation7 and mitochondrial maintenance8.
G4 are noncanonical secondary structures formed by stacked tetrads of Hoogsteen hydrogen-bonded guanines bases, which are stabilized through the coordination of physiologically relevant cations (Na+, K+). G4 can result from the intramolecular folding of a unique G-rich sequence or from the intermolecular assembly of different G-rich containing strands9. An essential feature of G4 is their intrinsic polymorphic nature: numerous in vitro studies have revealed their ability to adopt different conformations, also called topologies10. Indeed, depending on the length and the composition of the sequence, as well as the environmental conditions (including the nature and concentration of metal cations, and local molecular crowding), a G4-forming sequence can adopt different topologies, in which the strands are in parallel, antiparallel or hybrid conformations, with the co-existence of different types of loops (lateral, diagonal or propeller) of variable lengths9–11. In particular, this polymorphism is exacerbated for the human telomeric sequence and leads to intricate structural mixtures12.
In cells, the impact of G4 on cellular metabolism is mainly associated with protein or enzymatic factors that bind, stabilize or resolve these structures. The folding of G-rich sequences into a G4, on DNA and RNA molecules, is associated with the formation of DSBs, transcription and translation repression and the alteration of the RNA processing13–15. To handle these major threats, cells use a battery of DNA and RNA helicases to control G4 formation16,17. Notably, most of helicases resolving DNA G4 are associated, when mutated, with genetic disorders, progeria and cancer progression (WRN, BLM, FANCJ, RTEL), underlying the major impact of G4 structures on cell fitness18,19. In addition to helicases, the formation of G4 structures in cells is counteracted by proteins that bind single-stranded nucleic acids19,20 through their OB-fold, RRM or RGG interacting motifs21–23. Interestingly, RGG motif containing proteins also promote G4 stabilization24 and control mRNAs localization through interaction with G425. A major impact of G4 structures in cells is related to transcription4. Found enriched on promoters and transcriptional start sites (TSS)26,27, G4 structures have been shown to act predominantly as transcriptional repressors4,13,15, although some G4 have been also described as involved in transcription activation28,29. Furthermore, the presence of G4 motifs in the TSS proximal regions is associated with RNA-Pol II pausing sites and R-loops formation, two different factors promoting RNA-Pol II arrests and transcription-dependent DNA breaks30–34.
Given the increasing roles of G4 structures in cellular metabolism, extensive researches have been conducted in the last years in order to identify new G4-dependent mechanisms. Notably classical pull-down approaches identified hundreds of proteins associated to G-rich oligonucleotides forming G4 structures35–40. In solution, G-rich single-stranded molecules are in equilibrium between unfolded and folded states, and thus numerous identified G4 binding proteins are also able to recognize unfolded G-rich sequences20. In addition, G4 derived from single-stranded oligonucleotides can adopt different topologies9,11,41, that precludes to establish the specific contribution of each G4 topology to protein binding.
In this context, we have developed an approach to constrain the G4 into a single well-defined topology. The strategy is based on the use of a rigid cyclic peptide scaffold as a template for directing the intramolecular assembly of the anchored RNA or DNA oligonucleotides42–45. Moreover, such locked G4 display a thermal stability significantly higher than unconstrained G4 that strongly reduces the possibility to form unfolded single-stranded sequences. These constrained systems represent original tools, that we have used here for the identification and characterization of proteins interacting with a well-defined RNA or DNA G4 topology. In this study we identified through affinity purifications coupled to mass spectrometry (MS)-based quantitative proteomics a set of human proteins associated to locked G4 structures. Notably, this approach allowed us to identify NELF proteins as a new G4-interacting complex, leading us to investigate the impact of RNA-Pol II pausing mechanism into the response to G4 stabilization by G4 ligands.
Results
Identification and characterization of G4 associated proteins
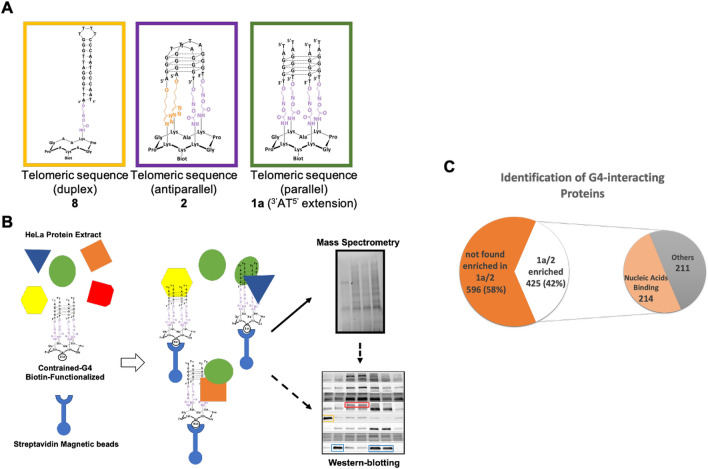
To identify human proteins interacting with G4 structures we performed classical pull-down assays followed by MS-based quantitative proteomic analysis. Various constrained G4 topologies based on the telomeric sequence (excepted for 5) were used (Supplementary Figure S1): systems 1, 6 and 7 depict a parallel topology and systems 2–5 have an antiparallel topology (Supplementary Figure S2)42–45. In our approach, biotin-functionalized G4-constrained molecules 1–7 and the biotin-functionalized duplex-DNA control 8, were incubated individually with a semi-total human protein extract prepared from HeLa cells46, before being trapped using streptavidin magnetic beads to isolate interacting proteins (Fig. 1B). In a first-round assay, and in order to validate our approach, western-blotting analyses were performed to test the interaction and the binding specificity of some well-established and depicted G4-binding proteins to constrained G4 constructions. From these analyses we observed that eIF4G, WRN, Nucleolin, Mre11, DHX36, hnRNP A1 and CNBP, all well-known G4-interacting proteins20, were enriched using constrained G4 structures compared to the duplex control 8. Conversely, the KU heterodimer, one of the most abundant human duplex-DNA-binding proteins47, was found enriched using duplex control 8 but was barely detectable on constrained G4 structures (Supplementary Figure S3A-B). Comparative analysis of the G4-interacting proteins enrichment on the six different constrained G4 structures 1a-3 and 5–7 shows a differential binding for these human proteins. Indeed, while eIF4G and Mre11 proteins are particularly enriched on constrained G4 2 and 3 (i.e. antiparallel topology with two lateral loops), DHX36 and hnRNP A1 proteins are abundant on constrained G4 1a, 6 and 7 (i.e. parallel topology without loops) (Supplementary Fig. 3A-B). Altogether, these first assays confirm that our pull-down strategy using constrained G4 structures allowed to both identify G4 binding proteins and to discriminate important aspects of their structural interaction with G4. Thus, our approach represents a powerful tool to find new proteins recognizing particular G4 conformations and prompted us to proceed to an extensive MS-based quantitative proteomic analysis of human proteins interacting with two particular constrained G4 structures, systems 1a (i.e. parallel without loops) and 2 (i.e. antiparallel with two lateral loops), compared to duplex control 8 (Fig. 1A-B).
Figure 1.
(A) Schematic representation of constrained DNA structures used in the pull-down assay. (B) Global strategy to identify constrained G4 interacting proteins from human cells. Biotin-functionalized G4-constrained molecules (1a and 2) and the biotin-functionalized duplex-DNA control 8 were individually mixed with a semi-total human protein extract from HeLa cells, then trapped by streptavidin magnetic beads to isolate interacting proteins. Protein identification was obtained from MS-based quantitative proteomic analysis and further characterized by western-blotting (arrow), or directly by western-blotting (dashed arrow). (C) Diagram showing the differential enrichment of human proteins on constrained G4 structures relative to control duplex DNA. G4 enriched proteins refer to proteins found enriched on 1a and/or 2 G4 constructions relative to the duplex control 8. 214 out of 425 proteins found enriched on constrained G4 have been shown to interact with nucleic acids. Differentially interacting proteins were sorted out using a fold change ≥ 2 and p-value < 0.05, allowing to reach a false discovery rate (FDR) inferior to 5% according to the Benjamini–Hochberg procedure.
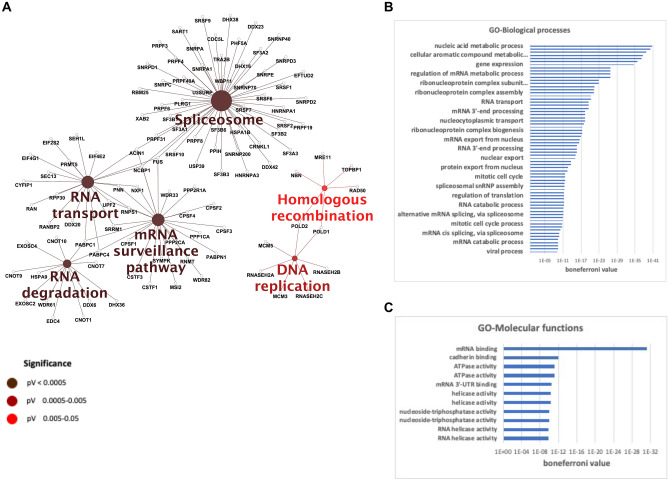
Based on three independent experiments, MS-based proteomic analyses identified a total of 1021 proteins interacting with constrained structures (see Materials and Methods for identification conditions) (Supplementary Table 1). This total corresponds to the sum of proteins interacting with 1a-2 and control 8 (duplex) (Fig. 1C and Supplementary Table 1). Using a label-free quantification and statistical filtering to compare the abundances of the proteins eluted from different constructions (see Materials and Methods for filter conditions), we identified 425 proteins enriched on constrained G4 structures 1a-2 compared to duplex control 8 (fold change ≥ 2 and p-value < 0.05, allowing to reach a false discovery rate (FDR) inferior to 5%) (Fig. 1C and Supplementary Table 2). These proteins belong to six significant enriched KEGG pathways cluster (p < 0.05): Spliceosome, RNA transport, RNA degradation, mRNA surveillance, DNA replication and Homologous Recombination (Fig. 2A). To go further, enriched GO Biological Processes and Molecular Functions terms were also determined. This analysis revealed that proteins enriched on contrained-G4 structures are mainly associated with DNA and RNA transactions (Fig. 2B-C), in agreement with current knowledge on genomic localization and biological function of G4. In line with these data, 214 out of 425 proteins enriched on G4 structures have been described as nucleic acid interacting factors, as indicated by terms from GO Molecular functions data analyses (Fig. 1C and Supplementary Table 3). Furthermore, the KEGG pathways clusters from these 214 nucleic acid binding proteins correspond to almost the same biological processes defined by the complete set of G4-interacting proteins (Supplementary Figure S4). An additional analysis of nucleic acid binding proteins enriched on constrained G4 structures defines five functional groups covering (i) ATP dependent DNA/RNA helicases activities, (ii) hnRNP proteins, (iii) proteins involved in the polyadenylation process, (iv) a large group of proteins involved in splicing and (v) proteins related to small nuclear ribonucleoprotein complexes (Table 1).
Figure 2.
Most significant pathways and processes covered by constrained G4 interacting proteins. (A) Enriched KEGG pathways. Gene Ontology terms, (B) GO-Biological processes and (C) GO-Molecular Functions for the 425 proteins found enriched on constrained G4 structures. A right-sided (Enrichment) test based on the hyper-geometric distribution was performed on the corresponding Entrez gene IDs for each gene list and the Bonferroni adjustment (p < 0.05).
Table 1.
Manually curated functional groups from nucleic acid binding proteins enriched on constrained G4 structures.
| Mapped IDs | Gene Name | Mapped IDs | Gene Name |
|---|---|---|---|
| DDX1 | ATP-dependent RNA helicase DDX1 | SF1 | Splicing factor 1 |
| DDX20 | Probable ATP-dependent RNA helicase DDX20 | SF3A1 | Splicing factor 3A subunit 1 |
| DDX23 | Probable ATP-dependent RNA helicase DDX23 | SF3A2 | Splicing factor 3A subunit 2 |
| DDX3X | ATP-dependent RNA helicase DDX3X | SF3A3 | Splicing factor 3A subunit 3 |
| DDX41 | Probable ATP-dependent RNA helicase DDX41 | SF3B1 | Splicing factor 3B subunit 1 |
| DDX42 | ATP-dependent RNA helicase DDX42 | SF3B2 | Splicing factor 3B subunit 2 |
| DDX52 | Probable ATP-dependent RNA helicase DDX52 | SF3B3 | Splicing factor 3B subunit 3 |
| DDX6 | Probable ATP-dependent RNA helicase DDX6 | SF3B6 | Splicing factor 3B subunit 6 |
| DHX16 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 | SRSF1 | Serine/arginine-rich splicing factor 1 |
| DHX29 | ATP-dependent RNA helicase DHX29 | SRSF10 | Serine/arginine-rich splicing factor 10 |
| DHX30 | Putative ATP-dependent RNA helicase DHX30 | SRSF11 | Serine/arginine-rich splicing factor 11 |
| DHX36 | ATP-dependent RNA helicase DHX36 | SRSF2 | Serine/arginine-rich splicing factor 2 |
| DHX38 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX38 | SRSF6 | Serine/arginine-rich splicing factor 6 |
| DHX40 | Probable ATP-dependent RNA helicase DHX40 | SRSF7 | Serine/arginine-rich splicing factor 7 |
| SRSF9 | Serine/arginine-rich splicing factor 9 | ||
| hnRNP A1 | Heterogeneous nuclear ribonucleoprotein A1 | ||
| hnRNP A2B1 | Heterogeneous nuclear ribonucleoproteins A2-B1 | SNRNP200 | U5 small nuclear ribonucleoprotein 200 kDa helicase |
| hnRNP A3 | Heterogeneous nuclear ribonucleoprotein A3 | SNRNP40 | U5 small nuclear ribonucleoprotein 40 kDa protein |
| hnRNP F | Heterogeneous nuclear ribonucleoprotein F | SNRNP70 | U1 small nuclear ribonucleoprotein 70 kDa |
| hnRNP H1 | Heterogeneous nuclear ribonucleoprotein H1 | SNRPA | U1 small nuclear ribonucleoprotein A |
| hnRNP H3 | Heterogeneous nuclear ribonucleoprotein H3 | SNRPA1 | U2 small nuclear ribonucleoprotein A |
| hnRNP L | Heterogeneous nuclear ribonucleoprotein L | SNRPC | U1 small nuclear ribonucleoprotein C |
| hnRNP R | Heterogeneous nuclear ribonucleoprotein R | SNRPD1 | Small nuclear ribonucleoprotein Sm D1 |
| SNRPD2,SNRPD1 | Small nuclear ribonucleoprotein Sm D2 | ||
| CPSF1 | Cleavage and polyadenylation specificity factor subunit 1 | SNRPD3 | Small nuclear ribonucleoprotein Sm D3 |
| CPSF2 | Cleavage and polyadenylation specificity factor subunit 2 | SNRPE | Small nuclear ribonucleoprotein E |
| CPSF3 | Cleavage and polyadenylation specificity factor subunit 3 | SNRPN | Small nuclear ribonucleoprotein-associated protein N;SNRPN |
| CPSF4 | Cleavage and polyadenylation specificity factor subunit 4 | ||
| CRNKL1 | Crooked neck-like protein 1 | ||
| CSTF1 | Cleavage stimulation factor subunit 1 | ||
| CSTF3 | Cleavage stimulation factor subunit 3 |
Next, using UniprotKB, gene Ontology (GO), G4IPDB48 databases and G4 search terms in PubMed, we determined that eighteen proteins found enriched on constrained G4 structures 1a-2 have been already implicated in G4 biology (Table 2).
Table 2.
Constrained G4 interacting factors found related to G4 on UniprotKB, gene Ontology (GO), PubMed abstract and G4IPDB48 data bases.
| Mapped IDs | Gene Name | PUBMED ID |
|---|---|---|
| ADAR | Double-stranded RNA-specific adenosine deaminase | 24813121 23381195 |
| CNBP | Cellular nucleic acid-binding protein | 23774591 28329689 24594223 26332732 31219592 |
| DDX1 | ATP-dependent RNA helicase | 29731414 |
| DDX42 | ATP-dependent RNA helicase | 31287417 |
| DHX36 | ATP-dependent RNA helicase | 29269411 28069994 25653156 25611385 24151078 22238380 21149580 18842585 16150737 |
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 | 30275516 |
| EWSR1 | RNA-binding protein | 21244633 21561087 22214309 |
| FUS | RNA-binding protein | 18776329 19749353 23521792 24251952 28575444 29434328 29800261 |
| hnRNP A1 | Heterogeneous nuclear ribonucleoprotein A1 | 9188487 19282454 20213319 24371143 24831962 26930004 28510424 29361764 30247678 31311954 |
| hnRNP A2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 15302914 17716999 |
| hnRNP A3 | Heterogeneous nuclear ribonucleoprotein A3 | 27623008 23381195 |
| hnRNP F | Heterogeneous nuclear ribonucleoprotein F | 29269483 |
| hnRNP H1 | Heterogeneous nuclear ribonucleoprotein H | 26930004 27623008 |
| MID1 | E3 ubiquitin-protein ligase Midline-1 | 21930711 |
| Mre11A (*) | Double-strand break repair protein MRE11 | 16116037 |
| RIF1 | Telomere-associated protein | 26436827 29348174 29357064 30510058 31197198 |
| SF3B3 | Splicing factor 3B subunit 3 | 23381195 |
| SRSF1 | Serine/arginine-rich splicing factor 1 | 24771345 |
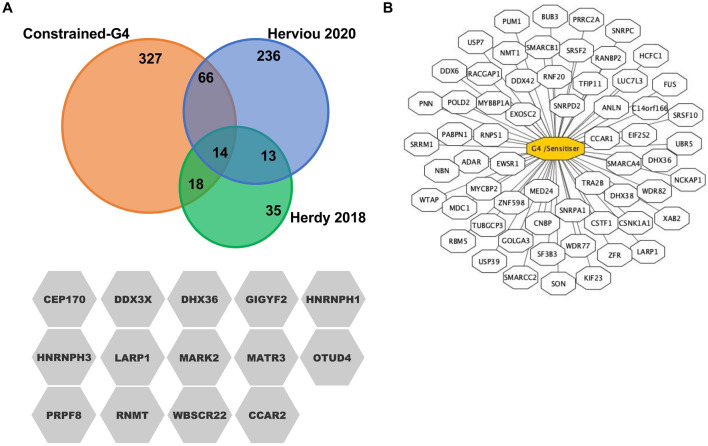
We compared our results with two recent studies that identified proteins associated with RNA G4 structures35,36 and found that 98 out of the 425 proteins identified in our study have been already shown to interact with G4 structures (Fig. 3A). Fourteen factors are in common in the three studies, including DHX36 and DDX3 proteins, two major G4 resolvases (Fig. 3A). Finally, in order to further investigate the association of constrained G4 interacting proteins identified here with G4 biological functions, we compared our list of 425 proteins with the list of 758 G4 sensitizers genes, the deficiency of which leads to increased sensitivity to G4 ligands, established by Zyner et al.49. From this analysis, we determined that at least 62 out of the 425 proteins enriched on constrained G4 structures 1a-2 were reported as G4 sensitiser proteins (Fig. 3B).
Figure 3.
Contrained-G4 interacting factors are associated with RNA-G4 binding activities and with the sensitisation to small molecules that stabilize G4 structures. (A) Venn diagram showing the overlap of our study (orange) with the RNA-G4 interacting proteins identified in Herdy35 (green) and Herviou36 (blue). 98 out of 425 proteins identified in our study were known to interact with RNA-G4 structures, with 14 indicated factors common to three studies. (B) Schematic representation of constrained G4 interacting proteins identified in our study that are associated to an increased sensitivity to G4 ligands, established by Zyner et al.49. From this analysis we determined that 62 out of 425 proteins were reported as G4 ligands sensitisers.
Different sets of proteins are enriched on particular G4 conformations
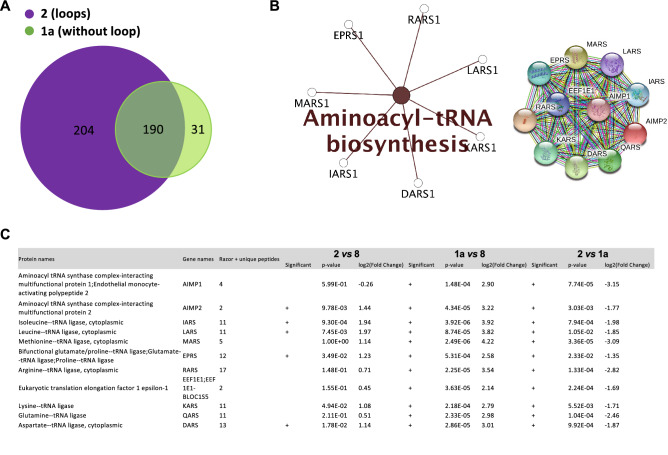
Statistical analysis of the abundances of proteins enriched on G4 structures 1a-2 shows differential interactions for some of them with the two constrained G4 conformations. Indeed, among the proteins with log2 (fold change 1a/2) ≥ 1 and p-values < 0.05, we established three groups of constrained G4 interacting proteins (Fig. 4A). The first and largest group is composed of 204 proteins found significantly enriched on construction 2 (i.e. antiparallel topology with two lateral loops) compared to construction 1a (i.e. parallel topology without loop). The second group comprises 190 proteins without significant enrichment on one particular conformation (−2 < fold change 1a/2 < 2). Finally, we found a third group with only 31 proteins significantly enriched on construction 1a relatively to structure 2 (Fig. 4A and Supplementary Tables 4.1, 4.2 and 4.3). KEGG analysis of the first group and Common proteins showed that both groups define similar significant enriched pathways than those covered by total constrained G4 interacting proteins (Spliceosome, mRNA surveillance, RNA degradation, DNA replication and repair) (Supplementary Figure S5). Surprisingly, the third group of proteins enriched on constrained G4 structure 1a defines a unique and highly enriched KEGG pathway cluster (p < 0.0005) consisting of factors involved in aminoacyl-tRNA biosynthesis50. Further analysis based on the STRING protein–protein interactions database51 unveiled that all of the components of the multi-tRNA synthetase complex (MSC) are enriched on constrained G4 1a (Fig. 4B-C). Indeed, in addition to eight cytoplasmic aminoacyl tRNA synthetase enzymes (methionyl MARS, glutaminyl DARS, lysyl KARS, arginyl RARS, isoleucyl IARS, leucyl LARS, aspartyl DARS and glutamyl-, prolyl EPRS-tRNA synthetase) composing this complex, our pull down assay also isolated the three non-enzyme components (AIMP1, AIMP2 and AIMP3, also known as EEF1E, proteins) of the MSC complex (Fig. 4B–C). Furthermore, western blotting analysis showed that AIMP1 interacts with constrained and unconstrained G4 structures in vitro (Fig. 5).
Figure 4.
Differential interaction of human proteins with constrained G4 structures adopting different topologies. (A) Diagram showing the differential enrichment of human proteins on 1a (green) and 2 (purple) constructions. Differential enrichment of proteins on structures 1a or 2 was determined through statistical analysis using the fold change ≥ 2 and p-value < 0.05, allowing to reach a false discovery rate (FDR) inferior to 5%. (B) KEGG pathway covered by the 31 proteins found enriched on constrained G4 structure 1a and functional interaction network analysis using STRING51 for the 31 proteins found enriched on construction 1a. A right-sided (Enrichment) test based on the hyper-geometric distribution was performed on the corresponding Entrez gene IDs for each gene list and the Bonferroni adjustment (p < 0.05). (C) MS-based quantitative proteomic analysis of the interaction of MSC-complex proteins with contrained-G4 structures (extracted from Supplementary Table 1). Differentially interacting proteins were sorted out using a fold change ≥ 2 and p-value < 0.05, allowing to reach a false discovery rate (FDR) inferior to 5% according to the Benjamini–Hochberg procedure.
Figure 5.
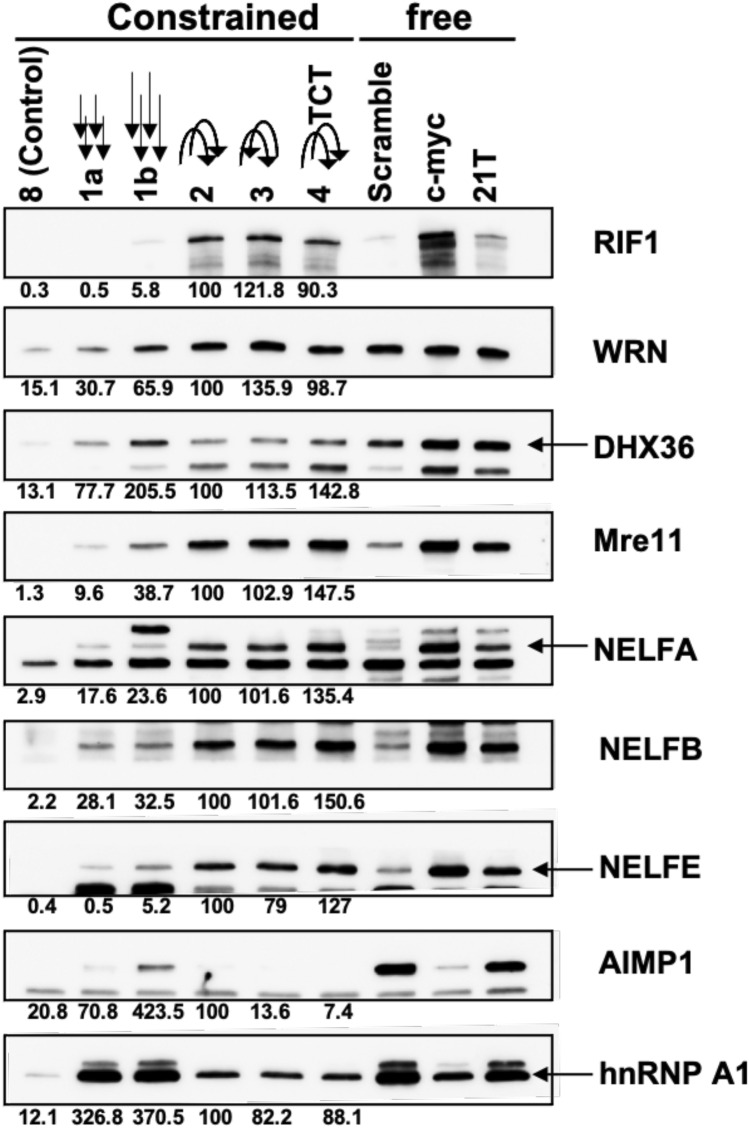
Impact of the orientation and nucleotide composition of connecting loops on the differential enrichment of proteins on G4 structures. Western-blotting analysis and quantification of the interaction of proteins found enriched on constrained G4 structures with modified molecules (1a, 1b, 2, 3, 4) and with unconstrained G4 (c-myc and 21 T). Arrows indicate the 5′-3′ strand orientation of single-stranded extensions or connecting loops present on different systems. The modification of the nucleotide composition of connecting loops in the system 4 is indicated by the sequence TCT. Construct 8 and scramble sequence were used as control for pull-down performed with constrained of free-G4 structures, respectively.
Impact of orientation and nucleotide composition of connecting loops on the differential association of proteins on G4 structures
Since most of the proteins characterized in this study are associated with construction 2 (i.e. antiparallel with two lateral loops), we explored the impact of the terminal single-stranded extension, loop sequence and loops orientation on the binding of proteins to constrained G4 molecules. First, western blotting analysis of the binding of nine selected factors, found enriched on constrained G4 structures relatively to duplex control 8, indicates that the relative orientation or the nucleotide loop composition has a not major impact on the binding of these proteins to constrained G4 structures (Fig. 5). Indeed, we found that protein signals obtained with systems 2 (i.e. with both loops oriented in the same 5′-3′ sense), 3 (i.e. with loops oriented in the 5′-3′ and 3′-5′ senses, respectively), and 4 in which the ATT sequence of the external loops was replaced by a TCT, are not significantly different (Fig. 5). Next, in order to explore the impact of the extension length, we constructed a new system 1b with a three-nucleotide extension consisting of the 5′ TTA 3′ sequence. Western-blotting analysis shows that the addition of a supplementary nucleotide on the terminal single-stranded extension of construction 1b significantly improves the binding of some factors (WRN, DHX36, Mre11, NELF-E and AIMP1). However, intensity signals observed on constrained G4 structures with connecting loops (i.e. systems 2–4) remain considerably stronger relatively to the signals obtained with both 1a and 1b constructions, except for AIMP1 (Fig. 5). These results indicate that longer single-stranded extensions improve the binding of proteins to type 1 constructions. Unconstrained G4 structures (free) formed by the telomeric and the c-myc promoter sequences were also used to further characterize their interaction (Fig. 5, right part). Most of the factors enriched on constrained G4 structures also present a significant interaction with non-constrained G4-forming sequences, relative to the scramble sequence, indicating a selective binding of these proteins to G4 structures. However, proteins showing a significant interaction with 1a and 1b systems (hnRNP A1, AIMP1, WRN and at lesser extent DHX36) also display a strong interaction with the scramble non-G4 forming oligonucleotide, confirming their ability to bind both G4 structures and single-stranded unfolded G-rich DNA.
Constrained G4 structures identify the NELF complex as a new G4 interacting factor
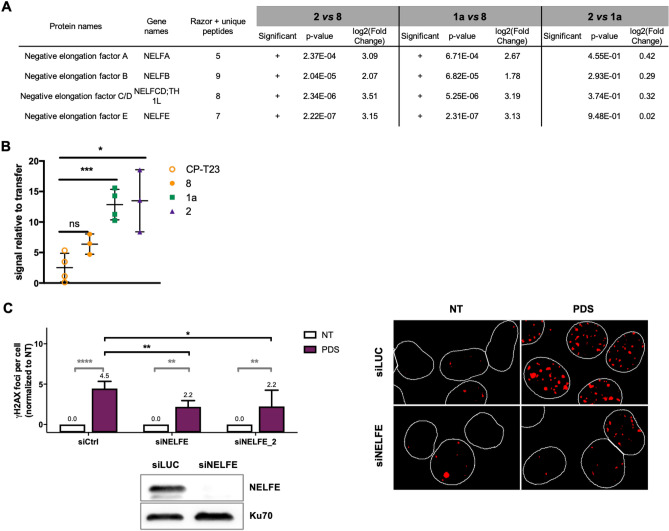
MS-based quantitative proteomic analyses identified NELF-A, NELF-B, NELF-E and NELF-C/D, all members of the NELF complex52, as enriched on constrained G4 structures relatively to the duplex control construction 8 (Fig. 6A, Supplementary Table 2). Selective interaction of NELF complex proteins with G4 structures was further confirmed through western blotting analysis performed using both constrained and unconstrained G4 structures (Fig. 5). In order to further characterize the interaction of the NELF complex with G4 structures, we performed the reverse experiment in which an immunoprecipitated NELF complex was used to investigate its interaction with constrained G4 structures. For that, the NELF complex was immunoprecipitated from a HeLa cell line overexpressing an ectopic Flag-tagged form of the NELFE protein. After elution by competition with a Flag peptide (Supplementary Figure S6), the NELF complex was use in pull-down experiments using constrained constructions and western blotting analyses were performed to quantify the relative signal of the bound NELFE protein. As shown in the Fig. 6B, the NELFE protein is significantly enriched on constrained G4 structures relatively to the cyclopeptide (CP-T23) and duplex control 8, indicating a selective binding of the NELFE protein to G4 structures. In addition, hybridization with an anti-NELFA antibody indicates that at least two proteins from the immunoprecipitated NELF complex are found enriched on constrained G4 structures relatively to the controls (CP-T23 and system 8) (data not shown).
Figure 6.
NELF complex interact with G4 structures and modulates the cellular response to G4 ligands. (A) MS-based quantitative proteomic analysis of the interaction of the NELF-complex proteins with contrained-G4 structures (extracted from Supplementary Table 1). Differentially interacting proteins were sorted out using a fold change ≥ 2 and p-value < 0.05, allowing to reach a false discovery rate (FDR) inferior to 5% according to the Benjamini–Hochberg procedure. (B) Quantification of the interaction of immunoprecipitated Flag NELF-E protein with constrained G4 structures (1a, 2) relative to cyclopeptide (CP-T23) and duplex control (8) constructions. Error bars represent SD from the means, n ≥ 3 independent experiments. p values were calculated using unpaired t-tests (without corrections for multiple comparisons). ns: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001. ns non-significant difference. (C) Quantification and representative images γH2AX foci fluorescence signal (red) detected HeLa cells transfected with control (Ctrl), or two NELF-E siRNAs (independent sequences) and treated with PDS (20 µM) for 4 h. Error bars represent SD from the means, n ≥ 3 independent experiments. p values were calculated using an unpaired multiple Student’s t test. ns: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001signals. Western-blotting analysis of NELF-E depletion in HeLa cells following siRNA treatment is shown.
The NELF complex facilitates DNA double-strand breaks induction by Pyridostatin
In human cells the NELF complex plays an essential role in the RNA-Pol II pausing mechanism53,54. RNA-Pol II pausing is a highly controlled mechanism regulating gene expression in eukaryotic cells53. During the pausing state, RNA-Pol II remains tightly associated with nascent RNA molecule in a promoter proximal region. Bioinformatic and pangenomic studies clearly establish that the formation of G4 structures, which are significantly enriched in promoter regions in human cells, correlates with the formation of R-loops and RNA-Pol II pausing sites, two processes associated with RNA-Pol II arrests and the induction of transcription dependent double-stranded DNA breaks (DSBs)30–32,55–60. Pyridostatin (PDS), one of the most selective and potent G4 stabilizers described so far, provokes a rapid induction of transcription dependent DSBs in human cells (61,62 and unpublished data). In order to investigate the impact of the NELF complex on PDS-induced DSBs, we quantified γH2AX signals (DSBs marker) through immunofluorescence studies in NELF proficient and deficient (siRNA-depleted NELF-E) HeLa cells. As shown in Fig. 6C, depletion of the NELFE protein, which also leads to the reduction of other NELF proteins in human cells52,63, provokes a significant reduction of PDS-induced DSBs signals relatively to the control cells. This result indicates that the NELF complex and RNA Pol II pausing favours DSB induction following G4 stabilization.
Discussion
The relationship between G4 structures/motifs and the progressive discovery of proteins that modulate the dynamic of G4 formation, such as helicases or other proteins involved in DNA and RNA transactions has expanded our knowledge to understand the ubiquitous function of G4 structures on cellular metabolism and cell fate5,6,13,15,17–20,64. In the present study, we identified through a MS-based quantitative proteomic analysis 425 human proteins significantly enriched on constrained G4 structures relative to a duplex DNA control, in which well-described G4 interacting factors were present. However, we found that some of well-known G4-interacting proteins, such as BLM and WRN, are not enriched on constrained G4 relatively to duplex control, thereby confirming the previously reported non-selective interaction of these proteins with G4s relative to duplex or hairpin DNAs65. Interestingly, numerous proteins interacting with constrained G4-DNA structures identified in this work are involved in RNA transactions: splicing, RNA degradation, transport and RNA surveillance pathways. Moreover, about one hundred proteins enriched from 425 total proteins identified here on constrained G4-DNA structures have been already identified in similar works using RNA G4 structures to trap G4-RNAs binding factors35–37. It is noteworthy that most RNA Binding Proteins (RBP) interact with RNAs through OB-fold, RGG, RRM and GARD motifs that have been also involved in the recognition of G4 structures21,22,66. Most G4 interacting proteins unfold G4 in order to counteract their impact on DNA and RNA transactions. Interestingly, 62 out of the 425 proteins identified through our approach have also been characterized as playing an important role in the resistance to cytotoxicity induced by G4-ligands49, and for many of them our work provides an evidence for a direct or indirect interaction with G4.
An important finding from our results concerns the differential enrichment of G4 interacting proteins on constrained G4 molecules mimicking parallel (systems 1, 6 and 7) versus anti-parallel conformations (systems 2–5). MS-based quantitative proteomic analysis identified 204 proteins significantly enriched on the G4 construction 2 relative to the G4 construction 1, and only 31 proteins with an inverse selectivity. Surprisingly, relative strand orientation and nucleotide composition of terminal connecting loops had no major impact on the binding of proteins to loops containing constructions (2–4), suggesting either a minor role of loops for the interaction with proteins recognizing antiparallel constrained G4, or a role related to their folding into a particular conformation independent on nucleotide composition or strand orientation. In agreement with the second hypothesis, proteins found enriched on system 2–4 relatively to system 1a-b, bind unconstrained G4 structures formed by the c-myc and telomeric sequences. In solution, both sequences have been shown to adopt different topologies and exhibit loops with different nucleotide compositions and strand orientations67,68. Altogether, these results suggest that while the folding of G-rich sequences into a G4 structure is required for the interaction with identify proteins, the presence of connecting loops is an important determinant of this interaction. In agreement with our data, although selective binding of proteins and small molecules to G4 structures are driven mainly through π–π interactions with tetrad faces, additional contacts with lateral grooves and connecting loops have been shown to stabilize both molecules and protein-G4 complexes22,69. Finally, although the addition of a supplementary nucleotide to construction 1a significantly increases the binding of system 2 enriched proteins on the system 1, we hypothesize that a part of the differential binding of human proteins to loops-containing systems 2–4 relative to loop-free constructions 1, 6 and 7 would be dependent on the particular conformations adopted by connecting loops.
No significant difference was found for 190 out of the 425 proteins interacting with systems 1a and 2, a result that would indicate that these proteins interact with common structural motifs such as tetrad faces or lateral groves. Although for systems 2–4 the interaction of proteins with tetrad faces could be hindered by lateral loops, future studies with constrained structures containing propeller loops will be needed to fully characterize this impact and the role of lateral groves.
An intriguing finding from our study is the characterization of the multi-tRNA synthetase complex (MSC) as a G4-interacting complex. The interaction of the MSC complex with G4 structures was confirmed by a recent publication that identified the MSC proteins in a molecular screen identifying RNA G4-interacting factors36. The MSC complex consists of eight Aminoacyl-tRNA Synthetases (ARSs) and three non-enzymatic ARS-interacting multi-functional proteins (AIMP1/p43, AIMP2/p38, and AIMP3/p18) that play an essential role in the protein synthesis by catalysing the activation of amino acids and linking them to their cognate transfer RNAs (tRNAs)50. AIMP1/p43 protein is a multifunction protein involved in various physiological and pathological processes. AIMP1 is the precursor of EMAP II, which was released after AIMP1 cleavage70. Surprisingly, the C-terminal segment of EMAP II are 50% identical to residues 205–364 of p42 protein from S. cerevisiae that was firstly identify as G4p1 a protein that shows a high and specific affinity for G4 nucleic acids71.
Finally, the interaction of NELF complex with G4 structures is a major finding of our study. To the best of our knowledge, this is the first time that a physical interaction between the NELF complex and G4 is reported. The NELF complex comprises the NELFA, NELFB, NELFC/D and NELFE proteins. Whether the interaction of NELF with G4 structures is direct or indirect needs to be elucidated. Nevertheless, the NELFE protein contains a RRM-domain (RNA Recognition Motif)52 that is present in different RNA binding proteins and interacts with G-rich tracts. These RRM motifs have also been involved in the interaction of several proteins with G4 structures, such as hnRNP A1, and in agreement with our findings, a NELF sub-complex formed by NELFA and NELFE proteins has been shown to selectively bind GC rich sequences72. Numerous bioinformatic and genomics analysis have established a strong correlation between G4 and RNA Pol II pausing occurring at the promoter proximal regions31,73. Transcriptional pausing consists in the arrest of RNA-Pol II mainly in a promoter proximal position and acts as a transcriptional checkpoint regulating gene expression53. It is induced by the association of two complexes, NELF and DSIF, with RNA-Pol II at the early stages of transcriptional elongation54. Although G4 motifs are found at pausing sites31, it is still unclear whether G4s act as signals for RNA-Pol II pausing. However, during transcription the impact of G4 on RNA Pol II progression could reflect their capacity to act as physical barriers, to promote the formation of other secondary structures such as R-loops or finally to drive the binding of protein complexes regulating RNA Pol II progression. In human cells, G4 motifs and RNA Pol II pausing have been also associated with transcription-dependent DNA breaks31,32,55, and the stabilisation of G4 structures by small molecules has been shown to provoke the formation of double-stranded DNA breaks that are, in part, dependent on RNA-Pol II transcription61. In this study, we show that the NELF complex regulates the formation of double-stranded DNA breaks induced by pyridostatin, a potent and selective G4 ligand. Altogether, our results suggest that G4 formation could facilitate the binding of the NELF complex to the chromatin to promote RNA Pol II pausing, and thus to act as a mediator of the response to G4 stabilization in human cells.
In this study, we show that biotin-functionalized constrained nucleic acids structures are powerful tools to identify proteins interacting with non-canonical secondary structures such as G4. We validated our approach by the identification of well-known G4 interacting factors and the functional characterization of new protein complexes related to G4 metabolism. Especially, the identification of NELF complex as a G4 interacting factor establishes a physical link between G4 structures and RNA Pol II pausing mechanism.
Future directions of our approach will concern the construction of constrained G4 structures mimicking intramolecular parallel G4 structures in order to refine the impact of propeller loops on protein binding. Finally, other constrained nucleic acid constructions may represent powerful tools to identify proteins interacting with other non-canonical secondary structures such as the i-motif or R-loops.
Material and methods
Constrained G4 and HP synthesis
The different constrained systems 1–8 were prepared according to the previously reported protocols (42–45). Systems 1a,b, and 6 with a parallel orientation of the G4-forming strands, were synthetized through a single oxime ligation reaction onto the cyclopeptide of the appropriate DNA or RNA sequences, respectively (45, 43). For system 7, two successive ligations (i.e. oxime and CuAAC reactions) were used to anchor on the cyclopeptide scaffold, DNA and RNA sequences, respectively (43). Systems 2–5 with an antiparallel orientation of the G4-forming strands, were prepared by anchoring on the cyclopeptide scaffold through the aforementioned ligations the appropriate bi-functionalized oligonucleotides from telomeric sequence (44) and HIV sequence (42). Controls 8 and CP-T23 were synthetized according to the oxime method by anchoring the appropriate sequences on the cyclopeptide scaffold (45). Biotinylated c-myc (5′GGA-GGG-TGG-GGA-GGG-TGG-GGA-A-TEG-biot) , 21T (5′ TTA-GGG-TTA-GGG-TTA-GGG-TTA-GGG-TT-TEG-biot) and scramble (5′ AAG-TGT-GTG-TGT-GTG-TGT-GTG-TGA-AG-TEG-biot) sequences were purchased from Eurogentec.
The CD spectra of the systems 1–7 which show typical CD signatures of parallel (positive peak at 263 nm and a negative peak around 240 nm for 1a, 1b, 6 and 7) an antiparallel (two characteristic positive peaks at 242 nm and 294 nm and a negative peak at 262 nm for 2, 3, 4 and 5) G-quadruplex are depicted in Supplementary Figure S2.
Cell culture
HeLa cells were grown in humidified atmosphere with 5% CO2 at 37 °C, in Dulbecco’s Modified Eagle Medium (Gibco). Culture medium was supplemented with 10% fœtal bovine serum (Eurobio), 100 U/mL penicillin (Gibco) and 100 µg/mL streptomycin (Gibco).
Plasmid constructions cell transfection and transduction
The pCMVsVg (envelope plasmid) were kindly provided by E. Gilson. pLPC-puro-N-Flag was a gift from Titia de Lange (Addgene plasmid # 12521; http://n2t.net/addgene:12521; RRID: Addgene_12521). The pLPC-puro-N-Flag-NELFE plasmid was obtained by amplification of the human NELFE cDNA (obtained from gene synthesis, GeneArt, LifeTechnlogies) using oligonucleotides: Fwd 5′-GACGATGACGATAAAGGATCCTTGGTGATACCCCCCGGACT-3′ and Rev- 5′CCCTCTAGATGCATGCTCGAGCTAGAAGCCATCCACAAGGTTTTCC-3′ and inserted in the pLPC-puro-N-Flag plasmid between XhoI and BamH1 restriction sites. Retroviral production was performed by transient transfection of HEK-GP2 293 cells (Clontech) with 0.8 µg pCMVsVg and 1.2 µg of pLPC-puro-N-Flag-NELFE plasmids using JetPrime reagent (polyplus). Retroviral particles were recovered from culture supernatant of HEK-GP2 293 cells 48 h and 72 h post transfection. For transduction, fifty thousand Hela cells were plated on six-well plates 24 h prior to transduction. Cell population was selected by puromycin resistance (1 µg/ml). The expression of Flag-NELF-E protein was verified by western blotting with Flag and NELF-E antibodies.
Pull-down from total protein extract with constrained G4s
First, proteins were bound to constrained G4s. Briefly, 1 mg of NHEJ protein extract (prepared as previously described46) was incubated with 10 μM of one of constrained G4s (Figure A) in 100 μL of binding buffer (20 mM Hepes (pH 7.5), 50 mM KCl, 0.01% NP40 and 0.5 mM EDTA) for 1.5 h at 4 °C under intermittent shaking (10 s at 1400 rpm every 2 min). During protein-G4 binding step, 1 mL of streptavidin-coupled magnetic beads (Promega, Z5481) per condition were washed three times during 10 min at 4 °C (under intermittent shaking) with binding buffer. Then, 100 μL of protein-G4 binding solution was put onto washed streptavidin-coupled magnetic beads for 30 min at 4 °C under intermittent shaking. After this step, supernatant was stored at − 80 °C as “unbound fraction” and beads were washed during 10 min three times at 4 °C (under intermittent shaking) with 0.1% NP40, 150 mM NaCl PBS. Finally, beads were incubated with 100 μL of 0.01% bromophenol blue, 15% glycerol, 2% SDS, 60 mM Tris–HCl (pH 8) for 10 min at 95 °C and supernatant were collected and stored at -80 °C before being used in mass spectrometry and western blotting assays.
MS-based quantitative proteomic analysis
Eluted proteins were stacked in a single band in the top of a SDS-PAGE gel (4–12% NuPAGE, Life Technologies) and stained with Coomassie blue R-250 before in-gel digestion using modified trypsin (Promega, sequencing grade) as previously described74. Resulting peptides were analyzed by online nanoliquid chromatography coupled to tandem MS (UltiMate 3000 and LTQ-Orbitrap Velos Pro, Thermo Scientific). Peptides were sampled on a 300 µm × 5 mm PepMap C18 precolumn and separated on a 75 µm × 250 mm C18 column (PepMap, Thermo Scientific) using a 120-min gradient. MS and MS/MS data were acquired using Xcalibur (Thermo Scientific).
Peptides and proteins were identified and quantified using MaxQuant (version 1.6.2.10,75) using the Uniprot database (Homo sapiens reference proteome, October 22nd 2018 version) and the frequently observed contaminant database embedded in MaxQuant. Trypsin was chosen as the enzyme and 2 missed cleavages were allowed. Peptide modifications allowed during the search were: carbamidomethylation (C, fixed), acetyl (Protein N-ter, variable) and oxidation (M, variable). Minimum peptide length was set to 7 amino acids. Minimum number of peptides and razor + unique peptides were set to 1. Maximum false discovery rates—calculated by employing a reverse database strategy—were set to 0.01 at peptide and protein levels. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE76 partner repository with the dataset identifier PXD 021003.
Statistical analysis were performed using ProStaR77. Proteins identified in the reverse and contaminant databases, proteins only identified by site, proteins identified with only 1 peptide and proteins exhibiting less than 3 intensity values in one condition were discarded from the list. After log2 transformation, intensity values were normalized by median centering before missing value imputation (slsa algorithm for partially observed values in the condition and DetQuantile algorithm set to first percentile for totally absent values in the condition); statistical testing was conducted using limma test. Differentially interacting proteins were sorted out using a log2 (fold change) cut-off of 1 and a p-value cut-off allowing to reach an FDR inferior to 5% according to the Benjamini–Hochberg procedure.
Immunoprecipitation of Flag-NELF-E and pull-down with constrained G4s
Four days after seeding, around 30.106 Flag-NELF-E expressing HeLa cells were harvested by scrapping in cold PBS. After centrifugation at 2000 g for 5 min at 4 °C, pelleted cells were lysed for 40 min at 4 °C with 1 mL of non-denaturing lysis buffer (50 mM Hepes (pH 7.5), 150 mM NaCl, 0.01% NP40) complemented with 1 mM DTT, 1X Halt Protease Inhibitor Cocktail (ThermoScientific), 23 U/mL benzonase. Cells were then centrifugated at 12,000 rpm at 4 °C for 10 min and protein concentrations in total extract were determined by measuring absorbance at 280 nm (Nanodrop). From total extract, 1 mg of proteins was used to performed incubation overnight at 4 °C with 5 μg of anti-Flag mouse antibody (Sigma) in 250 μL of complemented non-denaturing lysis buffer. Protein A/G-coupled magnetic beads (Pierce, 88802) were washed with complemented non-denaturing lysis buffer and 25 μL of beads were used to immunoprecipitated Flag antibody for 1 h at 4 °C. Supernatant was collected as “flag unbound fraction” and protein concentration was determined as described. Immunoprecipitated proteins were eluted overnight at 4 °C with 50 μg of Flag peptide (Sigma-Aldrich) in 100 μL complemented non-denaturing lysis buffer. Supernatant was collected as “flag eluted fraction” and protein concentration was determined as described. For protein-G4 binding step, 7 μg of flag eluted fraction were incubated with 1 μM of constrained G4s in 100 μL of binding buffer overnight at 4 °C, and pull-down was performed as described above. Finally, beads were incubated with 30 μL of 0.01% bromophenol blue, 15% glycerol, 2% SDS, 60 mM Tris (pH 8) for 10 min at 95 °C and supernatant were collected and stored at -80 °C before being used for western blotting assays.
Western blotting
For total pulled-down proteins and for Flag-eluted pulled-down proteins, respectively 45 μL and 10 μL of proteins were loaded for each condition and separated on gradient 4–12% polyacrylamide TGX Stain-Free pre-cast gels (Biorad) and transferred onto nitrocellulose membrane (Biorad). Before blocking (0.1% Tween-20, non-fat dry milk 5% and PBS), UV exposition of membrane was used to confirm homogeneous loading and to quantify transfer signal. The membrane was successively probed with primary antibodies and appropriate goat secondary antibodies coupled to horseradish peroxidase (described in table below). Chemidoc imager (Biorad) was used to perform UV and Clarity ECL (Biorad) detection. Digital data were processed and quantified using ImageLab (Biorad) or ImageJ softwares. Quantifications of antibody signal are relative to transfer signal.
| Target | Dilution | Species | Class | Reference | Manufacturer |
|---|---|---|---|---|---|
| KU70 | 0.2 µg/mL | Mouse | Monoclonal | MA5-13110 | Invitrogen |
| eIF4G | 1/1000 | Rabbit | Polyclonal | 2498 | Cell Signaling Technology |
| WRN | 1/2000 | Mouse | Monoclonal | W0393 | Sigma-Aldrich |
| Mre11 | 1/1000 | Mouse | Monoclonal | 611366 | BD bioscience |
| hnRNP A1 | 1/1000 | Rabbit | Polyclonal | GTX106208 | Gene Tex |
| DHX36 | 1/1000 | Rabbit | Polyclonal | HPA035399 | Sigma-Aldrich |
| CNBP | 8/1000 | Kindly provided by N. Calcaterra’s group | |||
| NCL23 | 1/1000 | Rabbit | Polyclonal | ab50279 | Abcam |
| NELFE | 1 μg/mL | Rabbit | Monoclonal | A301-913A | Bethyl laboratories |
| NELFB (COBRA) | 1 μg/mL | Rabbit | Monoclonal | A301-911A | Bethyl laboratories |
| Anti-rabbit | 1:10,000 | Goat | Polyclonal | 111-035-003 | Jackson Immunoresearch |
| Anti-mouse | 1:10,000 | Goat | Polyclonal | 115-035-003 | Jackson Immunoresearch |
RNA interferences
HeLa cells were seeded at 250.000 cells per well in a 6-wells plate. siRNA (Table) were transfected twice at 50 nM final concentration per well with Lipofectamine RNAiMax Reagent (Invitrogen) according to manufacturer’s instructions. Cells were treated and proteic extracts are realized 72 h after the first transfection.
| Target | Name | Sequence | Manufacturer |
|---|---|---|---|
| Luciferase | siLUC | 5′-CUUACGCUGAGUACUUCGATT-3′ | Eurofins |
| NELFE | siNELFE | 5′-AAGAUGGAGUCAGCAGAUCAG-3′ | Eurofins |
| NELFE | siNELFE_2 | 5′-GACCUUCUGGAGAAGAGCUTT-3′ | Eurofins |
Immunofluorescence
HeLa cells were seeded in 24-wells plate at 100.000 cells/well on glass coverslips (VWR, #631-0150). Twenty-four hours later, HeLa cells were treated with 20 μM pyridostatin (Sigma-Aldrich; CAS number 1085412-37-8) for 4 h, and then washed with PBS and fixed with 2% paraformaldehyde in PBS at room temperature for 10 min, washed with PBS and permeabilized for 15 min at room temperature with 10 mM Tris–HCl pH 7.5, 120 mM KCl, 20 mM NaCl, 0.1% Triton-X 100. Then, cells were washed with PBS and incubated for about 1 h at 37 °C in blocking buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2% BSA, 0.2% fish gelatin, 0.1% Triton-X 100) prior to incubation overnight at 4 °C with γH2AX (Phospho S139) antibody (Abcam, #81299) diluted at 0.7 µg/mL in blocking buffer. Cells were then washed with 0.1% Tween20-PBS and incubated with secondary goat anti-rabbit antibody coupled to AlexaFluor 488 (Thermo Fisher Scientific) diluted at 2 µg/mL in blocking buffer for 1 h at room temperature. At last, cells were washed with 0.1% Tween20-PBS and stained with 0.1 μg/mL DAPI for 20 min at room temperature, and coverslips were mounted with Vectashield mounting medium (Vector Laboratories). Nuclear γH2AX foci staining overlapping with DAPI staining were quantified with ImageJ software. Quantifications of nuclear γH2AX foci induced by pyridostatin are represented normalized to non-treated (NT) conditions.
KEGG pathway, gene ontology
The ClueGO v2.3.378,79 plugin for Cytoscape80 (v3.8) was used to determine networks of enriched KEGG pathways and Gene Ontology terms (Biological Process and Molecular Function). A right-sided (Enrichment) test based on the hyper-geometric distribution was performed on the corresponding Entrez gene IDs for each gene list and the Bonferroni adjustment (p < 0.05) was applied to correct for multiple hypothesis testing. The Kappa-statistics score threshold was set to 0.4 and GO term fusion was used to diminish redundancy of terms shared by similar proteins. Other parameters include: GO level intervals (3–8 genes) and Group Merge (50%).
Statistical analysis
All results provide from at least three independent experiments. Statistical analyses were performed with GraphPad Prism Software (version 8). For γH2AX quantifications analyses, multiple unpaired t-tests (without corrections for multiple comparisons) were performed between pairs of conditions. On all figures, significant differences between specified pairs of conditions are shown by asterisks (*: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.0005; ****: p-value < 0.0001). ns means non-significant difference.
Supplementary Information
Acknowledgements
This work was funded by grants from ANR (ANR-16-CE11-0006-01), and La Ligue Nationale Contre le Cancer (Equipe Labellisée 2018). Patrick Calsou is a researcher from INSERM. Proteomic experiments were partly supported by the ProFi grant (ANR-10-INBS-08-01). The Nanobio-ICMG platform (UAR 2607) is acknowledged for providing facilities for the synthesis and purification of oligonucleotides (R. Lartia) as well as for mass spectrometry analyses (A. Durand, L. Fort and R. Guéret).
Author contributions
D.G., J.F.R. and E.D. proposed the idea A.P., A.D., A.A., Y.C. and T.L. performed experiments. D.G., J.F.R., E.D., T.L., Y.C., S.B.and P.C., wrote the initial draft. All authors confirmed the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92806-8.
References
- 1.Riou JF, Gomez D, Lemarteleur T, Trentesaux C. G-quadruplex DNA: myth or reality? Bull. Cancer. 2003;90:305–313. [PubMed] [Google Scholar]
- 2.Prorok P, et al. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun. 2019;10:3274. doi: 10.1038/s41467-019-11104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valton AL, et al. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim N. The interplay between G-quadruplex and transcription. Curr. Med. Chem. 2019;26:2898–2917. doi: 10.2174/0929867325666171229132619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cammas A, Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharel P, Balaratnam S, Beals N, Basu S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA. 2020;11:e1568. doi: 10.1002/wrna.1568. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Perreault JP, Topisirovic I, Richard S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin) 2016;4:e1244031. doi: 10.1080/21690731.2016.1244031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falabella M, Fernandez RJ, Johnson FB, Kaufman BA. Potential roles for G-Quadruplexes in Mitochondria. Curr. Med. Chem. 2019;26:2918–2932. doi: 10.2174/0929867325666180228165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. RSC Biomolecular Sciences. Cambridge University Press; 2006. [Google Scholar]
- 10.Ma Y, Iida K, Nagasawa K. Topologies of G-quadruplex: Biological functions and regulation by ligands. Biochem. Biophys. Res. Commun. 2020;531(1):3–17. doi: 10.1016/j.bbrc.2019.12.103. [DOI] [PubMed] [Google Scholar]
- 11.Lightfoot HL, Hagen T, Tatum NJ, Hall J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019;593:2083–2102. doi: 10.1002/1873-3468.13547. [DOI] [PubMed] [Google Scholar]
- 12.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell. Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 14.Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21:459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer M, Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017;45:1173–1182. doi: 10.1042/BST20170097. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Brosh RM., Jr G-quadruplex nucleic acids and human disease. FEBS J. 2010;277:3470–3488. doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brazda V, Haronikova L, Liao JC, Fojta M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: A growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McRae EKS, Booy EP, Padilla-Meier GP, McKenna SA. On characterizing the interactions between proteins and guanine quadruplex structures of nucleic acids. J. Nucleic Acids. 2017;2017:9675348. doi: 10.1155/2017/9675348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahama K, et al. G-Quadruplex DNA- and RNA-specific-binding proteins engineered from the RGG domain of TLS/FUS. ACS Chem. Biol. 2015;10:2564–2569. doi: 10.1021/acschembio.5b00566. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiguro A, Kimura N, Watanabe Y, Watanabe S, Ishihama A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells. 2016;21:466–481. doi: 10.1111/gtc.12352. [DOI] [PubMed] [Google Scholar]
- 26.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 27.Hansel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 28.David AP, et al. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019;47:7901–7913. doi: 10.1093/nar/gkz527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Magis A, et al. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2019;116:816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy J, et al. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011;39:4975–4983. doi: 10.1093/nar/gkr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puget N, Miller KM, Legube G. Non-canonical DNA/RNA structures during transcription-coupled double-strand break repair: Roadblocks or bona fide repair intermediates? DNA Repair (Amst.) 2019;81:102661. doi: 10.1016/j.dnarep.2019.102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotsantis P, et al. RTEL1 regulates G4/R-loops to avert replication-transcription collisions. Cell Rep. 2020;33:108546. doi: 10.1016/j.celrep.2020.108546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miglietta G, Russo M, Capranico G. G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acids Res. 2020;48:11942–11957. doi: 10.1093/nar/gkaa944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herdy B, et al. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018;46:11592–11604. doi: 10.1093/nar/gky861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herviou P, et al. hnRNP H/F drive RNA G-quadruplex-mediated translation linked to genomic instability and therapy resistance in glioblastoma. Nat. Commun. 2020;11:2661. doi: 10.1038/s41467-020-16168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serikawa T, et al. Comprehensive identification of proteins binding to RNA G-quadruplex motifs in the 5' UTR of tumor-associated mRNAs. Biochimie. 2018;144:169–184. doi: 10.1016/j.biochi.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Vlasenok M, et al. Data set on G4 DNA interactions with human proteins. Data Brief. 2018;18:348–359. doi: 10.1016/j.dib.2018.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Hacht A, et al. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014;42:6630–6644. doi: 10.1093/nar/gku290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams P, Li L, Dong X, Wang Y. Identification of SLIRP as a G quadruplex-binding protein. J. Am. Chem. Soc. 2017;139:12426–12429. doi: 10.1021/jacs.7b07563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnat L, et al. Template-mediated stabilization of a DNA G-quadruplex formed in the HIV-1 promoter and comparative binding studies. Chemistry. 2017;23:5602–5613. doi: 10.1002/chem.201700417. [DOI] [PubMed] [Google Scholar]
- 43.Bonnat L, et al. Templated formation of discrete RNA and DNA:RNA hybrid G-quadruplexes and their interactions with targeting ligands. Chemistry. 2016;22:3139–3147. doi: 10.1002/chem.201504351. [DOI] [PubMed] [Google Scholar]
- 44.Bonnet R, Lavergne T, Gennaro B, Spinelli N, Defrancq E. Construction of anti-parallel G-quadruplexes through sequential templated click. Chem. Commun. (Camb.) 2015;51:4850–4853. doi: 10.1039/C4CC09744K. [DOI] [PubMed] [Google Scholar]
- 45.Murat P, et al. A novel conformationally constrained parallel g quadruplex. ChemBioChem. 2008;9:2588–2591. doi: 10.1002/cbic.200800457. [DOI] [PubMed] [Google Scholar]
- 46.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 47.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 48.Mishra SK, Tawani A, Mishra A, Kumar A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zyner KG, et al. Genetic interactions of G-quadruplexes in humans. Elife. 2019;8:e46793. doi: 10.7554/eLife.46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirande M. The Aminoacyl-tRNA synthetase complex. Subcell Biochem. 2017;83:505–522. doi: 10.1007/978-3-319-46503-6_18. [DOI] [PubMed] [Google Scholar]
- 51.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narita T, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Core L, Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 2017;68:745–757. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JJ, et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019;33:1751–1774. doi: 10.1101/gad.331231.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Aguilera A, Gomez-Gonzalez B. DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 2017;24:439–443. doi: 10.1038/nsmb.3395. [DOI] [PubMed] [Google Scholar]
- 59.Singh S, et al. Pausing sites of RNA polymerase II on actively transcribed genes are enriched with DNA double-stranded breaks. J. Biol. Chem. 2020;295(12):3990–4000. doi: 10.1074/jbc.RA119.011665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun J, et al. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J. Cell Biochem. 2008;103:1798–1807. doi: 10.1002/jcb.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zell J, Rota Sperti F, Britton S, Monchaud D. DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem. Biol. 2020;2:47–76. doi: 10.1039/D0CB00151A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamath-Loeb A, Loeb LA, Fry M. The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLoS ONE. 2012;7:e30189. doi: 10.1371/journal.pone.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 67.Ambrus A, et al. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 69.Haider SM, Neidle S, Parkinson GN. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011;93:1239–1251. doi: 10.1016/j.biochi.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 70.Zhou Z, Sun B, Huang S, Yu D, Zhang X. Roles of aminoacyl-tRNA synthetase-interacting multi-functional proteins in physiology and cancer. Cell Death Dis. 2020;11:579. doi: 10.1038/s41419-020-02794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frantz JD, Gilbert W. A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem. 1995;270:20692–20697. doi: 10.1074/jbc.270.35.20692. [DOI] [PubMed] [Google Scholar]
- 72.Vos SM, et al. Architecture and RNA binding of the human negative elongation factor. Elife. 2016;5:e14981. doi: 10.7554/eLife.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szlachta K, et al. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol. 2018;19:89. doi: 10.1186/s13059-018-1463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salvetti A, et al. Nuclear functions of nucleolin through global proteomics and interactomic approaches. J. Proteome Res. 2016;15:1659–1669. doi: 10.1021/acs.jproteome.6b00126. [DOI] [PubMed] [Google Scholar]
- 75.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 76.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wieczorek S, et al. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics. 2017;33:135–136. doi: 10.1093/bioinformatics/btw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.