Abstract
CNNM/CorB proteins are a broadly conserved family of integral membrane proteins with close to 90,000 protein sequences known. They are associated with Mg2+ transport but it is not known if they mediate transport themselves or regulate other transporters. Here, we determine the crystal structure of an archaeal CorB protein in two conformations (apo and Mg2+-ATP bound). The transmembrane DUF21 domain exists in an inward-facing conformation with a Mg2+ ion coordinated by a conserved π-helix. In the absence of Mg2+-ATP, the CBS-pair domain adopts an elongated dimeric configuration with previously unobserved domain-domain contacts. Hydrogen-deuterium exchange mass spectrometry, analytical ultracentrifugation, and molecular dynamics experiments support a role of the structural rearrangements in mediating Mg2+-ATP sensing. Lastly, we use an in vitro, liposome-based assay to demonstrate direct Mg2+ transport by CorB proteins. These structural and functional insights provide a framework for understanding function of CNNMs in Mg2+ transport and associated diseases.
Subject terms: Magnesium, Membrane proteins, Permeation and transport, X-ray crystallography
CNNM/CorB proteins are a conserved family of membrane proteins associated with Mg2+ transport. Here, structures of an archaeal CorB protein in apo state and with Mg2+-ATP bound and accompanying biophysical experiments suggest direct Mg2+ transport by CorB proteins.
Introduction
Magnesium (Mg2+), the most abundant divalent cation inside cells, is essential for a wide variety of biochemical processes, such as energy metabolism, maintenance of genomic stability, protein synthesis, and over 600 enzymatic reactions1. CNNMs (CBS-pair domain divalent cation transport mediators) are a conserved family of integral membrane proteins implicated in Mg2+ homeostasis and divalent cation transport2. CNNM2 and CNNM4 are found abundantly in the basolateral membrane of kidney and colon epithelial cells, where renal/intestinal (re)absorption of Mg2+ occurs3,4. In humans, mutations in CNNM proteins are linked to two genetic diseases: CNNM2 mutations cause hypomagnesemia3,5 while mutations in CNNM4 are associated with Jalili syndrome6. CNNM2-knockout mice are embryonic lethal7, while loss of CNNM4 leads to male infertility8 and susceptibility to cancer9,10. CNNMs are additionally implicated in hypertension7, non-alcoholic steatohepatitis11, and schizophrenia12.
CNNMs are also known as ancient conserved domain proteins (ACDPs) because their core domains are evolutionarily conserved in essentially all organisms, from animals, plants, fungi, and archaea to bacteria (Fig. 1a)13. The bacterial CorB protein was first identified in a screen for cobalt-resistant mutants in Salmonella typhimurium14. CorB was proposed to mediate Mg2+ efflux together with CorC and CorD, in which CorC is a soluble protein that shares high sequence similarity to the cytosolic domains of CorB. Subsequently, many other CorB orthologs have been implicated in Mg2+ transport. For example, the Staphylococcus aureus ortholog, MpfA, is thought to function as a Mg2+ exporter as deletion mutants are unable to grow in the presence of high concentrations of magnesium15,16. Disruption of the homologous gene (yhdP) in Bacillus subtilis leads to increased cellular Mg2+ content, again supporting a role in Mg2+ efflux17.
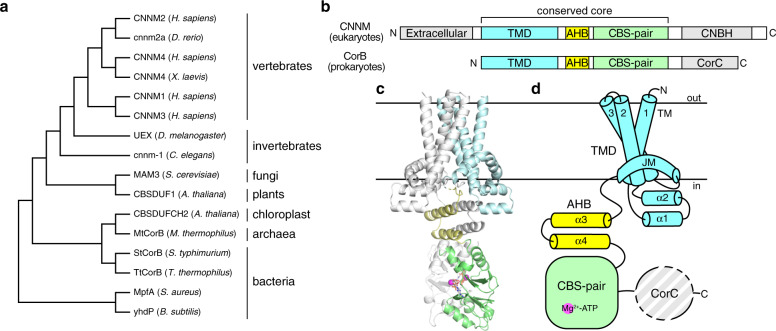
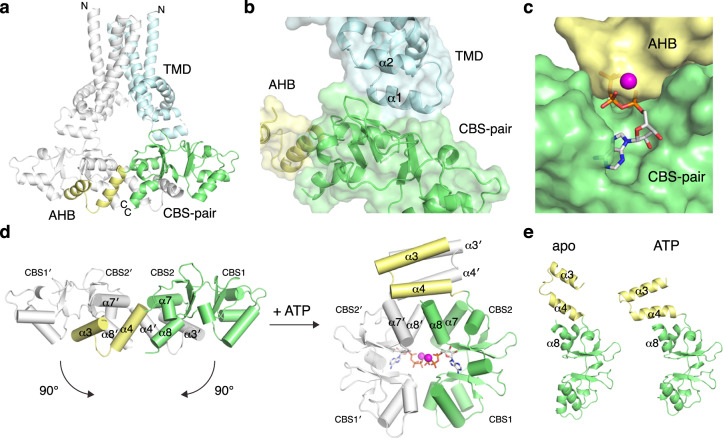
Fig. 1. Overall structure and domain organization of CNNM/CorB Mg2+ transporters.
a Phylogenetic analysis of representative CNNM/CorB orthologs generated using the neighbor-joining method. b Domain organization of eukaryotic CNNM and prokaryotic CorB. TMD transmembrane domain, AHB acidic helical bundle, CNBH cyclic nucleotide-binding homology domain, CorC cobalt resistance C domain. c Crystal structure of the Mg2+-ATP-bound MtCorB without the C-terminal CorC domain as a homodimer. One chain is colored by domains. d Topology of a MtCorB monomer showing the transmembrane and juxtamembrane helices of the TMD (residues 1–154, cyan), the two helices of the AHB (residues 166–199, yellow), the Mg2+-ATP-binding CBS-pair domain (residues 200–324, green), and the CorC domain (residues 325–426, grey).
Despite the clear association with Mg2+ transport, the field has not yet reached a consensus on the molecular mechanism of CNNM/CorB proteins18,19. Some papers report direct efflux of Mg2+ through Na+-coupled exchange4, others observed direct Mg2+ influx20,21, while still others propose an indirect mechanism through regulation of other Mg2+ channels (e.g., TRPM6/7)5. As all the studies to date have used measurements in cells3–5,15,21–23, which preclude differentiation of direct or indirect mechanisms, it remains unclear whether CNNM/CorB proteins are themselves Mg2+ transporters or regulators of other Mg2+ transporters/channels.
Structurally, CNNM/CorB proteins are defined by a conserved core consisting of a transmembrane domain (TMD) and a cytosolic cystathionine-β-synthase (CBS)-pair domain (Fig. 1b)24. The TMD constitutes the largest family of protein domains of unknown function (DUF21) in the Pfam database25. The CBS-pair domains are composed of tandem CBS motifs and found in a wide variety of proteins, including the bacterial Mg2+ channel MgtE, where they mediate dimerization and Mg2+-ATP binding26. The CBS-pair domains of CNNMs have been characterized structurally and shown to undergo conformational changes upon Mg2+-ATP binding27,28. Outside of the core, eukaryotic CNNM and prokaryotic CorB diverge. Eukaryotes have an extracellular domain and a cytosolic cyclic-nucleotide binding homology (CNBH) domain29. Prokaryotes lack the extracellular domain and have a distinct C-terminal CorC domain. Eukaryotic CNNMs contain an additional loop in the CBS-pair domain that allows binding of oncogenic phosphatase of regenerating liver (PRL) to regulate CNNM activities30–32. Only the soluble domains of CNNM/CorB have been characterized structurally until recently, during the preparation of this manuscript, the TMD fragment of a bacterial CorB ortholog (TpCorB) was reported23.
Here, we determine the crystal structure of an archaeal CorB ortholog in two distinct conformations (apo and Mg2+-ATP-bound). The TMD exists in an inward-facing conformation with a bound Mg2+ ion coordinated by a conserved π-helix. Major structural rearrangements in the cytosolic domains are observed upon Mg2+-ATP binding, suggesting a mechanism of Mg2+-ATP sensing. We employ a liposome-based transport assay to demonstrate the direct Mg2+ transport function of CorB proteins. Together, these structural and functional insights provide a framework for understanding the function of human CNNMs and associated diseases.
Results
Structure determination
To identify CNNM/CorB orthologs with suitable properties for structural studies, we performed small-scale screening of 20 prokaryotic CorB orthologs from diverse species (Supplementary Fig. 1). Each ortholog was expressed as a GFP-fusion protein and purified in six different detergents (DDM, LMNG, OGNG, LDAO, C12E9, and OG). Among them, two orthologs showed good expression and gel-filtration profiles: MtCorB from a thermophilic archaeon (Methanoculleus thermophilus) and TtCorB from a thermophilic, gram-negative bacterium (Tepidiphilus thermophilus). MtCorB and TtCorB share 26 and 25% sequence identities to the conserved core of human CNNM2, respectively (Supplementary Fig. 2). Attempts in crystallizing the full-length proteins yielded no suitable crystals. Thus, several deletion mutants were generated. MtCorB with a C-terminal CorC domain deletion (MtCorBΔC) purified in DDM yielded crystals in presence of Mg2+-ATP. However, upon structural elucidation, the unit cell only contained the soluble CBS-pair domain (Table 1).
Table 1.
Statistics of data collection and refinement.
| MtCorBCBS + Mg2+-ATP |
MtCorBΔCΔloop + Mg2+-ATP |
MtCorBΔC R235L | |
|---|---|---|---|
| Data collection | |||
| X-ray source | ALS 5.0.2 | CLS 08ID-1 | CLS 08B1-1 |
| Wavelength (Å) | 1.00003 | 0.97996 | 1.52154 |
| Space group | P4122 | P212121 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 52.10, 52.10, 112.28 | 61.05, 118.68, 177.31 | 139.17, 124.02, 86.26 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 92.8, 90.0 |
| Resolution (Å) | 50.00–2.05 (2.09–2.05)a | 50.00–3.25 (3.31–3.25)a | 50.00–3.80 (3.94–3.80)a |
| Redundancy | 22.0 (12.4) | 11.9 (9.1) | 6 (4.5) |
| Completeness (%) | 99.8 (97.3) | 99.2 (96.9) | 91.7 (78.7) |
| I/σI | 38.4 (1.4) | 22.0 (1.0) | 16.3 (0.8) |
| CC1/2 | 0.992 (0.721) | 0.993 (0.430) | 0.965 (0.510) |
| Refinement | |||
| Resolution (Å) | 38.19–2.05 | 49.31–3.25 | 46.27–3.80 |
| No. of reflections | 8835 | 14835 | 10821 |
| Rwork/Rfree | 0.206/0.252 | 0.226/0.268 | 0.243/0.285 |
| No. of atoms | |||
| Protein | 930 | 4786 | 4289 |
| Ligands | 37 | 299 | NA |
| Water | 56 | NA | NA |
| B-factors | |||
| Protein | 35.1 | 27.5 | 95.9 |
| Ligands | 27.3 | 42.7 | NA |
| Water | 37.3 | NA | NA |
| RMSDs | |||
| Bond lengths (Å) | 0.002 | 0.002 | 0.002 |
| Bond angles (°) | 0.54 | 0.50 | 0.44 |
| Ramachandran plots | |||
| Favored (%) | 97.5 | 96.7 | 95.5 |
| Allowed (%) | 2.5 | 3.3 | 4.5 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 |
| PDB code | 7MSU | 7M1T | 7M1U |
aHighest resolution shell is shown in parentheses.
After extensive construct and detergent optimizations, we ultimately obtained diffracting crystals of MtCorBΔC with an internal loop deletion (MtCorBΔCΔloop) purified in UDM in complex with Mg2+-ATP (Supplementary Fig. 1d). The crystals were obtained by vapor diffusion in presence of 0.1 M sodium citrate pH 5.5, 0.1 M Li2SO4, 0.1 M NaCl, 20 mM MgCl2, 34% PEG400 and 5 mM ATP. Initial phases were determined by molecular replacement using MtCNNMCBS structure solved earlier, which represented 40% of the crystallized construct (residues 199–322). Two α-helices (residues 166–198) preceding the CBS-pair domain were built-in manually, then PHENIX AutoBuild33 was used to place idealized helical fragments in the missing sections (residues 1–166). The resulting phases were of great quality that allowed tracing of most of the molecule, guided by strong densities of bulky residues (Supplementary Fig. 3a). The final structure, consisting of chain A (residues 5–322) and chain B (residues 4–154 and 160–322), is refined to 3.25 Å with an R-free of 0.27 (Table 1).
Architecture and domain organization
MtCorBΔC crystallizes as a homodimer with each protomer consisting of three distinct regions: a TMD, an acidic helical bundle (AHB), and a CBS-pair domain (Fig. 1c, d). The TMD and the AHB represent previously unknown structural regions that required de novo model building. Since we were able to visualize the linker connecting TMD and AHB, we were able to assign the two chains with confidence, with the right TMD connecting to the right CBS-pair domain. Our result shows a discrepancy with the hypothetical model generated by combining isolated TMD and CBS-pair domain structures of bacterial TpCorB, in which the right TMD was connected to the left CBS-pair domain23.
Although MtCorB is present as a dimer, it does not show C2 symmetry (Supplementary Fig. 3b). The individual TMD, AHB, and CBS-pair domains form symmetric dimers, but their arrangement is not. Overlaying the TMD of two polypeptide chains reveals asymmetry in the positions of the cytosolic domains, primarily due to differences in the linker between the TMD and AHB (Supplementary Fig. 3c). Together with previous observations that cytosolic domains of CNNMs form symmetric dimers27–30,32,34, these results suggest the interface between the transmembrane and cytosolic domains is dynamic, and the current asymmetric arrangement likely arises from crystal packing (Supplementary Fig. 3d).
TMD in an inward-facing conformation with a negatively charged cavity
The TMD is a dimer with each chain composed of three transmembrane helices (TM1–3), a pair of short helices exposed on the intracellular side, and a juxtamembrane (JM) helix that forms a belt-like structure (Fig. 2a). The homodimeric TMD exists in an inward-facing conformation with dimerization predominantly formed by hydrophobic contacts of TM2 and TM3 of each protomer. Electrostatic surface potential analysis reveals a large, negatively charged cavity that has a maximum diameter of approximately 10 Å and is lined with conserved polar residues (Fig. 2b, c and Supplementary Fig. 2). A well-defined electron density is observed in the cavity (Fig. 2d), and the density was modeled as an Mg2+ ion based on the TpCorB structure23. The Mg2+ is coordinated by hydroxyl groups of Ser21, Ser25, and Ser71; carboxyl group of Glu111; and the main-chain carbonyl groups of Ser21 and Gly110. Residues contributing to ion interactions are strongly conserved across species, suggesting these are conserved features of the CNNM/CorB proteins (Supplementary Fig. 2).
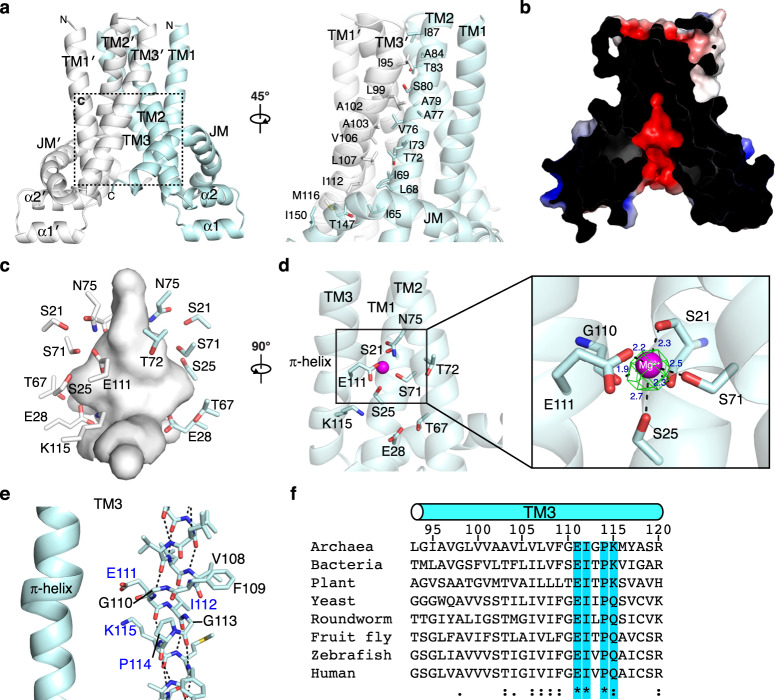
Fig. 2. TM domain in an inward-facing conformation with a negatively charged cavity.
a TM domain homodimerizes with interface formed by TM2 and TM3 of each protomer. b Electrostatic surface potential representation (±5 kT e−1) of MtCorB TM domain showing a cross-sectional view of the negatively charged cavity. c Close-up view of the polar residues forming the cavity. d A magnesium ion (Mg2+) bound in the cavity with Fo–Fc omits map contoured at 5.0σ. e π-helical turn preceding Pro114 in TM3. Highly conserved residues are highlighted in blue. f Conservation of residues in the π-helical turn from archaea to humans.
A second conserved feature is a π-helical turn in TM3, in which the helix is composed of an i + 5 instead of i + 4 configurations, resulting in a bulge in the helix (Fig. 2e and Supplementary Fig. 3a). The residues surrounding the helical turn are highly conserved with Glu111, Ile112, and Pro114 completely invariant from archaea to humans (Fig. 2f). Glu111 points toward the negative cavity and is involved in the coordination of the Mg2+ ion, whereas Pro114 acts as a helix-breaker that allows the shift in hydrogen bonding of the π-helical turn. The positively charged Lys115 after Pro114 is also highly conserved across species.
The archaeal MtCorB shares sequence identify of 37% with the bacterial TpCorB. The bacterial TMD also exists as a homodimer in an inward-facing conformation with similar TM rearrangements (Supplementary Fig. 4a). Although it was not pointed out in the bacterial study23, the bacterial structure also contains the acidic cavity and the conserved π-helical turn (Supplementary Fig. 4b, c). The polar residues in the acidic cavity and Mg2+-binding site are highly conserved with all five Mg2+-interacting residues being conserved except for Ser71, which is substituted by asparagine in bacteria.
A re-entrant JM helix encircles TM helices
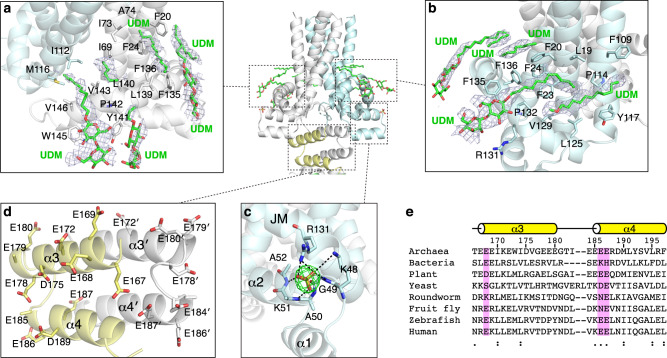
The re-entrant JM helix following TM3 wraps around TM1 and TM2 like a belt through several hydrophobic contacts (Fig. 3a, b). The curvature is imposed by a conserved Pro142 that makes a kink in the helix (Supplementary Fig. 2). The outer surface of the JM helix is amphipathic and rich in aromatic residues. Surrounding the JM helix, we observed several electron densities, which we modeled as UDM detergent molecules from the protein buffer. In total, we observed ten UDM molecules, which likely correspond to conserved lipid-binding sites important for assisting conformational changes during transport. Structural comparison to the bacterial JM helix shows that the bacterial one is longer and makes more extensive contacts with TM3 of the other protomer (Supplementary Fig. 4d). This discrepancy could be species-specific or caused by the absence of the following domains in the bacterial TMD structure23.
Fig. 3. Juxtamembrane helix and acidic helical bundle.
a, b UDM detergent molecules bound by the juxtamembrane helix. Simulated annealing 2Fo–Fc composite omit map contoured at 1.0σ. c A sulfate ion bound between JM and helix–turn–helix motif with Fo–Fc omit map contoured at 5.0σ. d Conservation of acidic residues in the AHB domain. e Sequence conservation of conserved glutamates in the AHB.
Underneath the JM helix resides the helix-turn-helix motif (α1 and α2) that connects TM1 and TM2 (Fig. 3c). The solvent-exposed portions contain several basic residues, which may be involved in binding phospholipid headgroups. Supporting this, a strong density was observed in this region and assigned as a sulfate molecule due to its presence in the crystallization buffer.
Acidic helical bundle
An unexpected feature of the MtCorB structure is the existence of the four-helical AHB domain following the TMD (Fig. 3d). The linker between the JM helix and AHB differs in the two protomers. It is extended and partially disordered in one and coiled in the other. The AHB itself is a symmetric dimer composed of two helices from each protomer and highly negatively charged. Thirteen of the 31 residues are acidic with three stretches of three or more consecutive glutamic acids, which likely represent cation binding sites. The negative charge of the AHB is well conserved across evolution with the exception of yeast and roundworms (Fig. 3e).
Structural basis of Mg2+-ATP binding by the CBS-pair domain
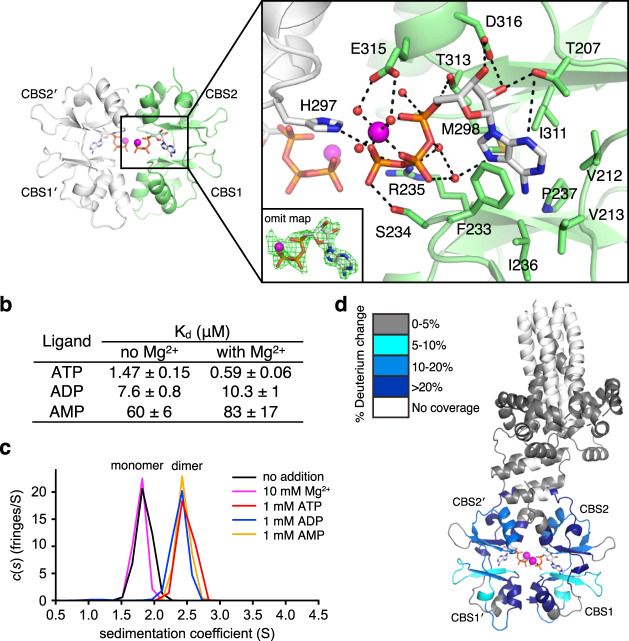
The isolated MtCorBCBS structure from the in situ cleavage of MtCorBΔC allowed us to visualize the detailed interactions of Mg2+-ATP binding at higher resolution (Fig. 4a and Table 1). As observed in the structure with the TMD, two molecules of Mg2+-ATP are bound to the central cavity of the CBS-pair dimer. The adenine bases are sandwiched between Phe233 and Ile311 in a hydrophobic pocket comprising Met298, Phe237, Ile236, Val212, and Val213. The ATP ribose forms hydrogen bonds with the side chains of Thr207, Asp316, and Thr313. The phosphate groups are stabilized with Arg235 and Ser234 with the bound Mg2+ coordinated in an octahedral arrangement with three phosphates and three water molecules.
Fig. 4. Mg2+-ATP binding to CBS-pair domain.
a Structural basis of Mg2+-ATP binding. Mg2+ ions and water molecules are shown in magenta and red, respectively. The Mg2+-ATP Fo–Fc omits map was contoured at 3.0σ. b Affinities of MtCorBΔC to adenosine nucleotides with and without 50 mM Mg2+ measured by ITC. c Dimerization of the MtCorB CBS-pair domain in the presence of 1 mM adenosine nucleotides as measured by SV-AUC experiments. d Conformational change in the MtCorBΔC in the presence and absence of Mg2+-ATP measured by HDX-MS. Regions that showed significant decreases in exchange (defined as >5%, 0.4 Da, and a two-tailed Student’s t test p < 0.01) in the presence of Mg2+-ATP are colored blue. Peptides in the CBS-pair domain dimerization interface show the most protection from deuterium exchange upon Mg2+-ATP binding.
Measurements of the affinity of adenosine nucleotides by isothermal titration calorimetry (ITC) showed MtCorBΔC has the highest affinity for ATP, followed by ADP and AMP (Fig. 4b and Supplementary Fig. 5). While the addition of Mg2+ has a relatively large effect on the binding of ATP to the isolated CBS-pair domain in human CNNMs27,35, the effect was more muted with MtCorB. In presence of Mg2+, the affinity for ATP was increased three-fold with MtCorBΔC and ten-fold with the isolated MtCorBCBS domain (Supplementary Fig. 5). These affinities are comparable to those measured from the isolated CBS-pair domains of TpCorB23.
We used analytical ultracentrifugation (AUC) to characterize the effect of ATP binding on MtCorB (Fig. 4c and Supplementary Fig. 6). As observed for human CNNMs27, dimerization of the CBS-pair domain was tightly coupled with Mg2+-ATP binding. While the MtCorBCBS in the absence of ATP sedimented as a monomer, the addition of adenine nucleotides triggered dimerization. The addition of Mg2+ alone had no effect on the sedimentation of MtCorBCBS.
Next, we turned to hydrogen–deuterium exchange mass spectrometry (HDX-MS) to detect conformational changes in full-length MtCorB upon ATP binding and deletion of the CorC domain. MtCorB and MtCorBΔC were first reconstituted into MSP1D1 nanodiscs (Supplementary Fig. 7a), and hydrogen exchange was monitored in the presence and absence of Mg2+-ATP. While peptide coverage of the hydrophobic TMD was limited, coverage of the cytosolic domains was excellent and revealed large conformational changes (Fig. 4d and Supplementary Fig. 7b, c). The addition of Mg2+-ATP significantly reduced the exchange in the CBS-pair domain dimerization interface and ATP-binding site. Similar changes were observed with both the full-length and truncated protein, which is consistent with the absence of a role of the C-terminal domain in Mg2+-ATP binding. This is different from human CNNMs where the C-terminal CNBH domain promotes CBS-pair domain dimerization and Mg2+-ATP binding27. Comparison of full-length and truncated MtCorB showed changes in the CBS2 motif, suggesting this is the primary site of interaction of the CorC domain.
Major structural rearrangements in MtCorB upon loss of Mg2+-ATP binding
Further insight into the conformation in the absence of bound ATP came from the crystal structure of a MtCorBΔC R235L mutant, homologous to a Jalili syndrome mutant (R407L) that is unable to bind ATP in human CNNM427. Crystals of MtCorBΔC R235L mutant purified in UDM were obtained by vapor diffusion in presence of 0.1 M sodium citrate pH 5.5, 50 mM Li2SO4, and 11% PEG4000, but they diffracted poorly to 6 Å. Subsequent additives screening improved the resolution to 3.8 Å with the addition of 10 mM MgSO4 (Table 1). The structure was determined by molecular replacement using individual TM and CBS-pair domains, while the AHB was rebuilt manually.
In the apo structure, the TMD remains in the same inward-facing conformation, but the cytosolic domains undergo major structural rearrangements (Fig. 5a). The CBS-pair domain has completely dissociated from the disc-like dimeric configuration to a more elongated, open conformation with different domain-domain contacts. To our knowledge, this dimeric configuration is not observed in any available CBS-pair domain structures26. In agreement with the AUC results, the loss of ATP binding disrupted the dimerization of the CBS-pair domain, which then makes contact with the helix-turn-helix motif from TMD (Fig. 5b). In addition, the dimeric AHB dissociates and binds to the CBS2 motif of the neighboring molecule, competing for the Mg2+-ATP binding site (Fig. 5c).
Fig. 5. Apo conformation of MtCorBΔC captured by R235L mutant.
a Overall structure of MtCorBΔC R235L mutant. One chain is colored by domains. b Novel domain–domain contact between TMD and CBS-pair domain. c AHB binds to CBS-pair domain and competes for Mg2+-ATP site. d Large conformational change in the AHB and CBS-pair domains upon Mg2+-ATP binding. e The apo and ATP-bound structures are similar with differences limited to the AHB and α8 helix.
The morphing of the two conformations allowed us to visualize the conversion of the apo to the Mg2+-ATP bound form (Supplementary Movie 1). The two conformations are interchangeable by a 90° rotation of both protomers (Fig. 5d). Despite the large dimeric rearrangement, the individual protomer in the two states is similar with differences limited to the AHB and α8 helix (Fig. 5e). Our result shows a discrepancy to the isolated CBS-pair domain structures of the bacterial TpCorB, in which minimal structural changes were observed upon nucleotide binding23. Perhaps, this was due to the absence of TMD and one of the AHB helices in the bacterial structures.
Liposome-based Mg2+ transport assays
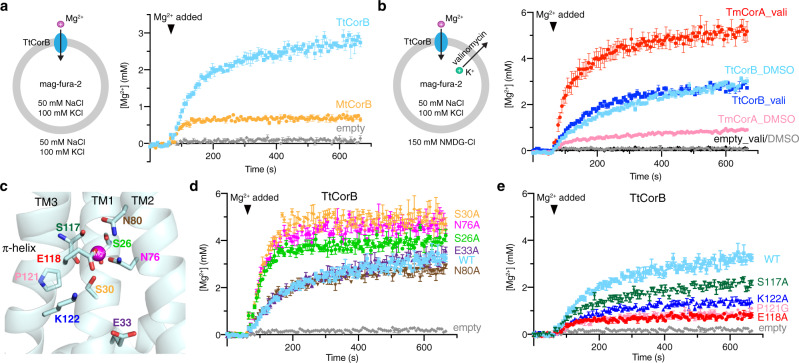
In order to address the most debated question in the field: whether CNNM/CorB proteins mediate direct or indirect Mg2+ transport, we carried out an in vitro, liposome-based Mg2+ transport assay using a modified version of assays used for CorA proteins36,37. We reconstituted CorB proteins into liposomes and encapsulated mag-fura-2, a ratiometric Mg2+ indicator (Fig. 6a). Upon addition of Mg2+, we observed a time-dependent change in the ratio of fluorescence from excitation at two wavelengths (330/369 nm), which could be used to calculate the Mg2+ concentration inside the liposomes. CorB liposomes showed a time-dependent increase in the fluorescence ratio while no fluorescence change was observed for liposomes devoid of protein. Assays with the bacterial CorB (TtCorB) showed more transport activity than the archaeal protein (MtCorB), possibly due to better compatibility with the lipid composition of the proteoliposomes. Thus, we used the bacterial protein for further functional studies. By varying the protein/lipid ratio and the concentration of Mg2+, we showed that the TtCorB-mediated Mg2+ transport is protein- and magnesium-dependent (Supplementary Fig. 8a, b).
Fig. 6. Liposome-based assay shows direct Mg2+ transport by CorB proteins.
a Liposomal assay showing direct transport of Mg2+ by MtCorB and TtCorB. Proteoliposomes (protein/lipid ratio of 1:30) or empty liposomes containing mag-fura-2 were equilibrated for 1 min before the addition of 5 mM MgCl2 to initiate Mg2+ uptake (arrowhead). The data points represent mean ± SEM (n = 3 independent measurements). b An inward negative membrane potential generated by valinomycin leads to enhanced Mg2+ transport by TmCorA (protein/lipid ratio of 1:270) but not TtCorB (protein/lipid ratio of 1:30), suggesting electroneutral transport of Mg2+ by TtCorB. The data points represent mean ± SEM (n = 3 independent measurements). c Homology model of TtCorB showing locations of conserved residues in the negatively charged cavity and Mg2+ binding site. d, e Liposomal Mg2+ transport assays of TtCorB mutants under the same experimental conditions as panel (a). Mutations in the Mg2+-binding site increased transport activity while mutations near the π-helical turn reduced transport. The data points represent mean ± SEM (n = 3 independent measurements).
Next, to test whether the TtCorB-mediated Mg2+ transport is electrogenic, we used valinomycin, a potassium-selective ionophore, to generate an inward negative membrane potential (Fig. 6b). We used TmCorA, a well-studied Mg2+ channel, as a positive control. As shown previously, TmCorA showed increased Mg2+ accumulation in presence of valinomycin36,37. In contrast, we did not observe an increase in Mg2+ transport for TtCorB. These results suggest TtCorB mediates Mg2+ transport through an electroneutral antiporter mechanism. We also examined the ion selectivity of TtCorB toward different divalent cations. Due to the differences in the affinity and response of mag-fura-2 to different ions, it is difficult to directly compare ions, but we could compare the ion selectivity of TtCorB with TmCorA. In our experimental conditions, TtCorB was more selective and showed less transport of Ca2+ and Zn2+ than TmCorA (Supplementary Fig. 8e). TtCorB and TmCorA showed similar transport activity with Mg2+.
Lastly, to probe the importance of the conserved residues lining the negatively charged cavity in TMD (Fig. 6c), we tested the effect of several cavity mutations on the Mg2+ transport activity. During purification, some mutants, especially ones involved in Mg2+ coordination, have abnormal gel-filtration profiles compared to wild-type proteins as shown by the earlier elution volumes (Supplementary Fig. 8c), suggesting the importance of these residues in maintaining the structural integrity of the protein. Despite that, all mutants showed high purity and incorporated well into liposomes (Supplementary Fig. 8d). Mg2+ transport assays were performed on all the mutants (Fig. 6d, e). Some mutants (E33A and N80A) have near-wildtype transport activity, while others, such as the Mg2+-binding site mutants (S26A, S30A, and N76A), have increased transport activity. On the other hand, mutations of the four conserved residues (S117A, E118A, P121G, and K122A) near the π-helical turn consistently reduced the Mg2+ transport activity. These results demonstrate the functional importance of the conserved residues in the Mg2+-binding site and π-helical turn.
Huang et al.23 had also tested the effect of several cavity mutations with an indirect, cell-based Mg2+ efflux assay through the expression of chimeric CNNM4/TpCorB proteins in HEK293 cells. In comparison, we have similar results for S117A and E118A equivalent mutants, which showed reduced Mg2+ efflux activity. In contrast, divergent results were obtained for S30A and E80A equivalent mutants. Both mutants had reduced efflux activity in the bacterial TpCorB, while our S30A mutant increased activity, and the N80A mutant has near-wildtype activity. Huang et al. did not measure transport activity with mutations equivalent to S26A, E33A, N76A, P121G, and K122A.
Homology modeling of human CNNM
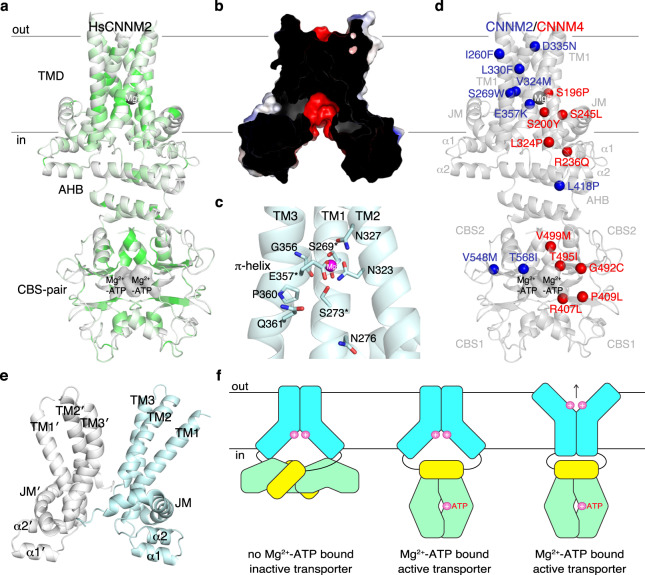
We used homology modeling to improve our understanding of mutations in human CNNM proteins (HsCNNMs) that cause diseases. Previous attempts to predict the structure of HsCNNM2 were confounded by the hydrophobicity of the HsCNNM2 JM helix38 (Supplementary Fig. 9a). Using the Mg2+-ATP bound structure of MtCorBΔC, we generated a homology model of HsCNNM2, which showed good geometry and allowed TM helices to be reliably positioned (Fig. 7a and Supplementary Fig. 9). The main differences lie in the length of linkers connecting the TM helices. The HsCNNM2 model reproduced the negatively charged cavity, π-helix in TM3, the Mg2+ ion binding site, and the presence of buried polar residues (e.g., Thr331, Asp335, and Ser348), supporting the importance of these features for CNNM function (Fig. 7b, c).
Fig. 7. Molecular modeling.
a Homology model of human CNNM2 (HsCNNM2) color-coded according to identity to CNNM/CorB proteins shown in Supplementary Fig. 2. Conservation is highest around the Mg2+ binding site, negatively charged cavity, and the CBS-pair dimerization interface. b Electrostatic surface potential analysis (±5 kT e−1) shows conservation of negatively charged cavity in HsCNNM2 in cross-sectional view. c Conservation of the polar residues in the Mg2+ binding site and π-helical turn of HsCNNM2. Asterisk denotes disease-causing mutations. d Mutations in CNNM2 and CNNM4 responsible for hypomagnesemia and Jalili syndrome cluster around the core of the TMD and nucleotide-binding site. For clarity, mutations are only shown on one chain. e Outward-facing conformation generated by MD simulations. f Proposed model of MtCorB transport and regulation. Loss of Mg2+-ATP binding, CBS-TMD contact inactivates TMD in the inward-facing conformation. Upon Mg2+-ATP binding, CBS-TMD contact is alleviated, thereby activating and allowing TMD to mediate Mg2+ transport.
We used the model to understand the molecular basis for human disease mutations (Fig. 7d). Nine missense mutations in HsCNNM2 are responsible for hypomagnesemia3,5,39,40. T568I in the CBS-pair domain prevents ATP binding and Mg2+ transport27,35. L418P in the dimerization interface of AHB likely disrupts the helical bundle. In the TMD, S269W, and L330F both likely disrupt TM helix packing, and E357K is a charge reversal mutation of the invariant glutamate that coordinates the Mg2+. D335N near the extracellular surface may be important in the outward-facing conformation. The high degree of sequence conservation (84% identity) between HsCNNM2 and HsCNNM4 also allowed us to use the model to interpret HsCNNM4 mutations that cause Jalili syndrome41–46. Of particular interest are mutations that occur in the TMD. S196P and S200Y both map to the negatively charged cavity. HsCNNM4 S196P affects the same residue as CNNM2 S269W, confirming the importance of this position for function. R236Q, S245L, and L324P are located at the inner surface of the membrane, in TM3, and the JM helix, respectively.
Molecular dynamics simulations
We used molecular dynamics simulations to study asymmetry and conformational changes in the MtCorBΔC structure. Unbiased MD simulations of the MtCorBΔC structure were first carried out (Supplementary Movie 2). As expected from the asymmetry in the structure, the interface between the TMD and cytosolic domains was flexible and large movements were observed. At the end of the simulation, the conformations of the two protomers were essentially switched while the individual domains all retained their symmetry. In addition, we also performed unbiased MD simulations of symmetric MtCorBΔC dimer (Supplementary Movie 3). In this case, the cytosolic domains appear more rigid than that of asymmetric dimer. We also observed an upward movement of the AHB towards the TMD, suggesting entrapment and stabilization by the helix-turn-helix motif of TMD.
The current structure resembles a transporter fold in the inward-facing conformation as supported by the hydration analysis of the structure (Supplementary Fig. 10a). In order to simulate the outward-facing conformation, we pulled an Mg2+ ion through the TMD dimerization interface while keeping a distance constraint with Glu111 of both protomers. This in turn causes the two helices to flip open, generating a possible outward-facing conformation (Fig. 7e). We then performed targeted MD to gain insight into the conformational changes from the inward to the outward-facing conformation (Supplementary Movie 4 and Supplementary Fig. 10b, c). The conserved residues Glu10, Ser80, and Thr83, which are buried in the membrane in the inward-facing conformation, become solvent-exposed and available for ion binding. The conservation of the π-helix and buried polar residues suggest that the mechanism of ion transport is conserved across all CNNM/CorB family members.
Discussion
Mg2+ is the most abundant divalent cation inside cells and essential for a wide variety of biochemical and enzymatic reactions1. Despite this, our structural knowledge of Mg2+ ion transporter is limited to three proteins: MgtE47, CorA48–50, and TRPM751. The structure of CorB proteins defines a different class of Mg2+ transporter, highly conserved and present in all organisms.
CNNM/CorB proteins had previously been suggested to be too small in ion transport due to the small number of predicted TM helices18. CorA is a pentamer with a total of 10 TM helices, MgtE is a dimer with 10, and TRPM7 has 24. Our structure shows that 6 TM helices suffice to form a large cavity (~1500 Å3) with ion binding sites deep within the membrane (Fig. 2). In fact, the current structure resembles a transporter fold in an inward-facing conformation with solvent accessible cavity toward the cytosol. This suggests that CNNM/CorB proteins might transport Mg2+ through a rocker-switch mechanism, in which the protein may exist in other conformational states, such as the occluded or outward-facing conformations. We have attempted to simulate the outward-facing conformation (Fig. 7e), but further studies are needed to validate and capture CNNM/CorB proteins in other conformational states.
The biggest controversy in the field surrounds the question: whether CNNMs/CorBs are Mg2+ transporters themselves or Mg2+ sensors that regulate other transporters? The dispute arises because all transport assays performed so far were done through overexpression of CNNMs/CorBs in cells3–5,15,21–23, which preclude the differentiation of direct or indirect Mg2+ transport. With our in vitro, liposome-based assay, we demonstrated direct Mg2+ transport of CorB proteins (Fig. 6). In addition, we showed that the transport likely occurs through an electroneutral antiporter mechanism, similar to that proposed in CNNMs4. Together with the structural insights, our results clearly demonstrate CNNM/CorB proteins are magnesium transporters themselves.
Throughout evolution, the CBS-pair domain of CNNM/CorB proteins remains associated with the TMD. The ATP-binding site is a disease mutational hotspot (Fig. 7d), and disruption of ATP-binding results in loss of Mg2+ efflux activity23,27,35. While the CBS-pair domain has no ATPase activity23,27, ATP-binding likely plays a regulatory role as was observed for the bacterial Mg2+ channel MgtE52. Using a disease-mimicking mutant, we were fortunate to capture the apo state of the transporter (Fig. 5). Structurally, the loss of Mg2+-ATP binding disrupts the dimerization of both CBS-pair and AHB domains. Dramatic changes in the dimeric rearrangement of CBS-pair domains upon nucleotide-binding have been well-documented in other CBS domain-containing proteins26 as well as human CNNMs27,28. However, the current dimeric configuration involving AHB contact has not been observed before and seems CNNM/CorB specific. The acidic residues in the AHB are reminiscent of the acidic patches in the cytosolic domains of CorA and MgtE that are involved in Mg2+ sensing and regulation of channel activity53,54. Disruption of the AHB dimer could play a regulatory role through the loss of Mg2+ binding sites. In addition, the CBS-TMD contact could result in an inactivation mechanism (Fig. 7f). For example, upon loss of ATP binding, the generation of the CBS-TMD contact would lock the TMD in the inward-facing conformation. This in turn inactivates the transporter as it is unable to change to other conformational states required for Mg2+ transport. Upon Mg2+-ATP binding, dimerization of the CBS-pair domain results in loss of CBS-TMD contact, and this, in turn, alleviates the inactivation of TMD, thereby allowing Mg2+ transport to occur.
The CNNM/CorB family of proteins is highly homologous in the conserved core from humans down to bacteria. Despite their common origin, eukaryotic CNNMs have evolved to be different. In animals, PRL phosphatases provide an additional level of regulation through binding the CNNM CBS-pair domain. PRLs are highly oncogenic and promote cancer metastasis through their suppression of CNNM Mg2+ efflux9,10. PRL binding itself is regulated by PRL phosphorylation in response to Mg2+ levels30,31. Although further studies are needed to fully understand the function of human CNNMs and their regulation by PRLs, the structural and functional insights from this study provide a framework for understanding CNNM function and the development of therapeutic approaches for regulating their activity.
Methods
Construction of phylogenetic tree
Amino acid sequences of various CNNM/CorB orthologs were aligned using MUSCLE55. The phylogenetic tree was generated using the neighbor-joining method and bootstrapping of 1000 replications in MEGAX (Version 10.1.8)56. The CNNM/CorB orthologs and their UniProt accession numbers are: CNNM2 (Homo sapiens; Q9H8M5), cnnm2a (Danio rerio; A2ATX7), CNNM4 (H. sapiens; Q6P4Q7), CNNM4 (Xenopus tropicalis; A0JPA0), CNNM1 (H. sapiens; Q9NRU3), CNNM3 (H. sapiens; Q8NE01), UEX (Drosophila melanogaster; A0A0B7P9G0), cnnm-1 (Caenorhabditis elegans; A3QM97), MAM3 (Saccharomyces cerevisiae; Q12296), CBSDUF1 (Arabidopsis thaliana; Q67XQ0), CBSDUFCH2 (Arabidopsis thaliana; Q84R21), MtCorB (M. thermophilus; A0A1G8XA46), StCorB (Salmonella typhimurium; Q8XFY3), TtCorB (T. thermophilus; A0A0K6IWT9), MpfA (S. aureus; A0A0H3JL60), and yhdP (B. subtilis; O07585).
Cloning of CorB orthologs
Codon-optimized cDNA of 20 CorB orthologs were synthesized (Bio Basic Inc., Markham, Canada) and sub-cloned into NcoI and XhoI sites of pCGFP-BC vector57 with a C-terminal GFP-His8-tag for small-scale expression and detergent screening. Promising orthologs were subcloned into NdeI and XhoI sites of pET29a vector (Millipore Sigma) with a C-terminal His6-tag for large-scale expression. Constructs for M. thermophilus CorB (UniProt entry A0A1G8XA46): MtCorB (residues 1–426), MtCorBΔC (residues 1–322), MtCorBΔCΔloop (residues 1–322 Δ259–262), and MtCorBΔC R235L. Construct for T. thermophilus CorB (UniProt entry A0A0K6IWT9): TtCorB (residues 1–445). For MtCorB constructs lacking TMD, MtCorBCBS+CorC (residues 199–417), MtCorBCBS (residues 199–322), and MtCorBCorC (residues 333–417) were subcloned into BamHI and XhoI sites of pGEX-6P-1 vector (GE Healthcare) with an N-terminal GST-tag. QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) was used for the generation of point mutations. A list of mutagenic primers is included in Supplementary Table 1.
Small-scale expression and detergent screening of CorB orthologs
CorB orthologs cloned in pCGFP-BC were transformed into Escherichia coli strain C41 (DE3). Cells were grown in Luria Broth (LB) at 37 °C to an optical density of 0.6 and induced with 1 mM IPTG for 4 h at 30 °C. The cell pellet was obtained by centrifuging at 5000g for 10 min. The pellet was re-suspended in lysis buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, pH 7.5) supplemented with cOmpleteTM protease inhibitor cocktail (Roche) and split into 6 fractions. Lysis was performed using a 24-probe sonicator. Each fraction was solubilized with a different detergent (DDM, n-dodecyl-β-D-maltopyranoside; LMNG, lauryl maltose neopentyl glycol; OGNG, octyl glucose neopentyl glycol; LDAO, lauryldimethylamine-N-oxide; C12E9, dodecyl nonaethylene glycol ether; OG, n-octyl-β-d-glucopyranoside) to a final concentration of 1%, then purified by IMAC in the same detergent (3× CMC). Elutions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and size-exclusion chromatography on SEPAX Zenic-C SEC-300 connected to fluorescence detector (Ex 480 nm/ Em 510 nm) in buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 3× CMC of detergent).
Expression and purification of MtCorB, TtCorB, and TmCorA
MtCorB and TtCorB constructs were transformed into E. coli strain C41 (DE3), while TmCorA50 was transformed into E. coli strain BL21 (DE3). MtCorB was grown in LB at 37 °C to an optical density of 0.6 and induced with 0.5 mM IPTG overnight at 18 °C. TtCorB and TmCorA were grown in LB at 37 °C to an optical density of 0.8 and induced with 0.5 mM IPTG for 4 h at 30 °C. The cell pellet was obtained by centrifuging at 5000g for 20 min. The pellet was re-suspended in lysis buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, pH 7.5) supplemented with cOmplete™ protease inhibitor cocktail (Roche), 10 µg/mL DNAse I, 1 mM CaCl2, and 0.1 mg/mL lysozyme. MtCorB cells were lysed by passing through Avestin Emulsiflex-C3 homogenizer (10,000–15,000 p.s.i.), while TtCorB and TmCorA cells were lysed by sonication. Cellular debris was removed by centrifugation at 27,000g for 10 min at 4 °C (this step is omitted for full-length MtCorB). Membranes were pelleted by ultracentrifugation at 150,000g for 1 h at 4 °C. The membrane fraction was collected, flash-frozen in liquid nitrogen, and stored at −80 °C for later use. The membrane fraction after thawing was solubilized in lysis buffer supplemented with 1% DDM (MtCorB & TmCorA) or UDM (TtCorB) for 1 h at 4 °C on a rotator, then ultracentrifuged at 150,000g for 30 min at 4 °C. The supernatant was loaded onto Qiagen Ni-NTA resin by batch binding, and incubated with gentle shaking for 1 h at 4 °C. The resin was then washed with wash buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, 30 mM imidazole, pH 7.5) containing 0.03% DDM or 0.05% UDM and eluted with elution buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, 300 mM imidazole, pH 7.5) containing 0.03% DDM or 0.05% UDM. The affinity-purified protein was further purified by size exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) in HPLC buffer (20 mM HEPES, 150 mM NaCl, pH 7.5) containing 0.03% DDM or 0.05% UDM. For TtCorB and TmCorA, the purification or HPLC buffers additionally contain 5 mM BME or 3 mM TCEP. The final purified proteins were concentrated using 50 kDa (MtCorBΔC) or 100 kDa (MtCorB, TtCorB, and TmCorA) cutoff concentrators (Amicon Ultra, Millipore), flash-frozen in liquid nitrogen, and stored at −80 °C for later use. The protein concentration is determined spectrophotometrically using Nanodrop, and purity is verified by SDS-PAGE.
Expression and purification of MtCorB constructs lacking TMD
Constructs were transformed into E. coli strain BL21 (DE3). Cells were grown in LB at 37 °C to an optical density of 0.8 and induced with 1 mM IPTG overnight at 20 °C. The cell pellet was obtained by centrifuging at 5000g for 20 min. The pellet was re-suspended in lysis buffer (50 mM HEPES, 500 mM NaCl, 5% glycerol, pH 7.5) supplemented with cOmplete™ protease inhibitor cocktail (Roche), 10 µg/mL DNAse I, 1 mM CaCl2, and 0.1 mg/mL lysozyme. Cells were lysed by sonication. Cellular debris was removed by centrifugation at 44,000g for 45 min at 4 °C. The supernatant was loaded onto Glutathione Sepharose resin (GE Healthcare), washed with lysis buffer, and eluted with lysis buffer containing 20 mM glutathione. The GST-tag was removed by overnight incubation with PreScission Protease, leaving an N-terminal Gly-Pro-Leu-Gly-Ser extension. The protein was further purified by size exclusion chromatography on a HiLoad 16/600 Superdex 75 pg column (GE Healthcare) in HPLC buffer (20 mM HEPES, 100 mM NaCl, pH 7.5). The protein was diluted to 5 µM, dialyzed overnight in dialysis buffer (20 mM HEPES, 100 mM NaCl, 5 mM EDTA, pH 7.5), and re-injected onto Superdex-75 to remove bound nucleotides. The final purified protein was concentrated to around 10 mg/mL (measured by NanoDrop), and the purity verified by SDS-PAGE.
Crystallization
Crystals of MtCorBCBS with Mg2+-ATP were obtained by equilibrating 0.4 µL of protein (20.8 mg/mL MtCorBΔC purified in 0.03% DDM; 5 mM ATP added prior to crystallization) and 0.4 µL of reservoir solution (0.1 M MOPS, pH 7.0; 9% PEG 8000; 20 mM MgCl2) in sitting-drop vapor diffusion system incubated at 22 °C. Rod-like crystals appeared after 2 weeks. The crystal was cryo-protected with reservoir solution supplemented with 5 mM ATP and 30% ethylene glycol, picked up in a nylon loop, and flash-cooled in an N2 cold stream.
Crystals of MtCorBΔCΔloop with Mg2+-ATP were obtained by equilibrating 1 µL of protein (19.5 mg/mL MtCorBΔCΔloop purified in 0.05% UDM; 5 mM ATP added prior to crystallization) and 1 µL of reservoir solution (0.1 M sodium citrate, pH 5.5; 0.1 M Li2SO4; 0.1 M NaCl; 20 mM MgCl2; 34% PEG400; 10 mM Na2HPO4) in hanging-drop vapor diffusion system incubated at 22 °C. Petal-like crystals appear after 1 week. The crystal was directly picked up in a nylon loop and flash-cooled in an N2 cold stream.
Crystals of MtCorBΔC R235L were obtained by equilibrating 1 µL of protein (18.4 mg/mL MtCorBΔC R235L purified in HPLC buffer containing 20 mM HEPES, 500 mM NaCl, 0.05% UDM, pH 7.5) and 1 µL of reservoir solution (0.1 M sodium citrate, pH 5.5; 50 mM Li2SO4; 11% PEG4000; 10 mM MgSO4) in hanging-drop vapor diffusion system incubated at 22 °C. Rod-like crystals appear after 1 day. The crystal was cryo-protected with reservoir solution supplemented with 25% glycerol and 0.6 M NaCl, picked up in a nylon loop, and flash-cooled in an N2 cold stream.
Data collection and structure determination
The MtCorBCBS dataset from a single crystal was collected using a Pilatus3 6 M detector at beamline 5.0.2 of Advanced Light Source (ALS). Data processing and scaling were performed with HKL-200058 with auto-corrections enabled. Initial phases were obtained by molecular replacement with Phaser59 in PHENIX60 using the CBS-pair domain structure of EcCorC (PDB: 5YZ2)61.
The MtCorBΔCΔloop dataset from a single crystal was collected using a Pilatus3 6 M detector at beamline 08ID-1 of the Canadian Macromolecular Crystallography Facility of the Canadian Light Source (CLS). The dataset showed anisotropic diffraction up to 3.25 Å. Data processing and scaling were performed with HKL-200058 with auto-corrections enabled, in which ellipsoid truncation was performed automatically. Resolution cut-off is based on CC1/2 = 0.362. Resolution limits after ellipsoid truncation were a* = 4.07 Å, b* = 3.71 Å, and c* = 3.25 Å. Initial phases for the CBS-pair domain were obtained by molecular replacement with Phaser59 in PHENIX60 using MtCorBCBS structure (determined in this study). Then AutoBuild33 was used to build in the missing domains (TMD and AHB).
The MtCorBΔC R235L dataset from a single crystal was collected using Pilatus3 6 M detector at beamline 08B1-1 of the CLS. Data processing and scaling were performed with HKL-200058 with auto-corrections enabled. Initial phases were obtained by molecular replacement with Phaser59 in PHENIX60 using the CBS-pair and TMD structures (determined in this study).
After molecular replacement, the models were subsequently improved through iterative cycles of manual building with Coot63 and refinement with phenix.refine64. TLS parameters were included at later stages of the refinement65. The final structures were validated with MolProbity66. Crystallographic data collection and structure refinement statistics are shown in Table 1. Structural images were prepared with PyMOL, Version 2.3.4 (Schrödinger LLC, New York). Electrostatic surface potentials were calculated using the APBS plugin within PyMOL67.
Isothermal titration calorimetry
ITC experiments were performed on a MicroCal VP-ITC titration calorimeter (Malvern Instruments Ltd.) at 20 °C with stirring at 310 rpm. TMD-containing constructs (15 µM final concentration) and ligands were prepared in HPLC buffer containing 0.05% UDM (±50 mM MgCl2). Cytosolic constructs (30 µM final concentration) were prepared in HPLC buffer (±10 or 50 mM MgCl2). The ligands were injected 19 times (5 µL for the first injection, 15 µL for subsequent injections), with 4 min intervals between injections. Results were analyzed using ORIGIN software (MicroCal) and fitted to a binding model with a single set of identical sites.
Analytical ultracentrifugation
Sedimentation velocity AUC experiments were performed at 20 °C using a Beckman Coulter XL-I Optima analytical ultracentrifuge and an An-60Ti rotor at 98,000g (35,000 rpm) for 18 h with scans performed every 60 s. A double-sector cell, equipped with a 12 mm Epon centerpiece and sapphire windows, was loaded with 380 and 400 µL of sample and HPLC buffer. MtCorBCBS (100 µM) with various ligands were monitored using interference optics. The data were analyzed with Sedfit v1501b68 using a continuous c(s) distribution. Numerical values for the solvent density, viscosity, and partial specific volume were determined using Sednterp69. Buffer density and viscosity were calculated to be 1.0039 g/cm3 and 0.01026 mPa·s, respectively (20 mM HEPES, 100 mM NaCl, pH 7.5). Partial specific volume for MtCorBCBS was calculated to be 0.7464 cm3/g. The frictional ratio (f/f0) value for MtCorBCBS was calculated using US-SOMO70 to be 1.26. Residual and c(s) distribution graphs were plotted using GUSSI71.
Production and purification of MSP1D1
pMSP1D1 was a gift from S. Sligar (Addgene plasmid 20061). MSP1D1 production was carried out according to published protocols72. In brief, E. coli BL21 (DE3) cells transformed with pMSP1D1 were grown in LB at 37 °C to an optical density of 0.8 and induced with 1 mM IPTG for 4 h at 30 °C. MSP1D1 was purified by nickel affinity chromatography according to standard conditions described in72. The polyhistidine tag was removed by overnight incubation with TEV protease and further purified by Superdex-75 size-exclusion column (GE Healthcare) in HPLC buffer (20 mM HEPES, 150 mM NaCl, pH 7.5).
Reconstitution into nanodisc
MtCorB and MSP1D1 were mixed with soybean polar extract (Avanti) solubilized in 40 mM DDM at a MtCorB:MSP1D1:lipid molar ratio of 2:10:550 in HPLC buffer. Detergent was removed by incubation with Bio-Beads (Bio-Rad SM-2 Resin) at 4 °C overnight with constant rotation. Bio-beads were removed via filtration and the reconstitution mixture was re-loaded onto Qiagen Ni-NTA resin to remove empty nanodiscs. The resin was washed with wash buffer (20 mM HEPES, 200 mM NaCl, 20 mM imidazole, pH 7.5) and eluted with wash buffer with 300 mM imidazole. The eluted protein was further purified by size exclusion chromatography on a Superdex 200 Increase 10/300 GL column (GE Healthcare) in HPLC buffer (20 mM HEPES, 150 mM NaCl, pH 7.5).
Hydrogen deuterium exchange mass spectrometry
HDX-MS reactions were performed in a similar manner as described previously73,74. In brief, HDX reactions for MtCorB and MtCorBΔC were conducted in a final reaction volume of 10 µL with a molar quantity of 20 pmol of MtCorB and MtCorBΔC. The reaction was started by the addition of 9.0 µL of D2O buffer (100 mM NaCl, 20 mM HEPES pH 7.5, 94% D2O (V/V)) to 1.0 µL of protein solution (final D2O concentration of 84.9%). The reaction proceeded for 3, 30, 300, or 3000 s at 20 °C, before being quenched with ice-cold acidic quench buffer, resulting in a final concentration of 0.6 M guanidine–HCl and 0.9% formic acid post quench. All conditions and timepoints were created and run in triplicate. Samples were flash-frozen immediately after quenching and stored at −80 °C until injected onto the ultra-performance liquid chromatography (UPLC) system for proteolytic cleavage, peptide separation, and injection onto a QTOF for mass analysis, described below.
Protein samples were rapidly thawed and injected onto a UPLC system kept in a Peltier has driven cold box at 2 °C (LEAP). The protein was run over two immobilized pepsin columns (Trajan; ProDx Protease column PDX.PP01-F32) and the peptides were collected onto a VanGuard Precolumn trap (Waters). The trap was eluted in line with an ACQUITY 1.7 μm particle, 100 × 1 mm2 C18 UPLC column (Waters), using a gradient of 5–36% B (Buffer A 0.1% formic acid, Buffer B 100% acetonitrile) over 16 min. MS experiments were performed on an Impact HD QTOF (Bruker) and peptide identification was done by running tandem MS (MS/MS) experiments run in data-dependent acquisition mode. The resulting MS/MS datasets were analyzed using PEAKS7 (PEAKS) and a false discovery rate was set at 1% using a database of purified proteins and known contaminants. HDExaminer Software (Sierra Analytics) was used to automatically calculate the level of deuterium incorporation into each peptide. All peptides were manually inspected for the correct charge state and presence of overlapping peptides. Deuteration levels were calculated using the centroid of the experimental isotope clusters. No correction was made for back exchange during analysis, so all deuterium incorporation values are relative. Differences in exchange in a peptide were considered significant if they met three of the following criteria: >5% change in exchange, >0.4 Da mass difference in exchange, a p value < 0.01 using a two-tailed Student’s t test, and change spanned by multiple peptides. The source data are provided as a Source Data file.
Proteoliposome reconstitution and Mg2+ transport assay
Proteoliposome were made following75 with modifications. A 3:1 mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (Avanti Polar Lipids) were dried into a thin film, followed by overnight incubation in a vacuum chamber. Dried lipids were solubilized to 5 mg/mL in buffer A (20 mM HEPES-KOH, 100 mM KCl, 50 mM NaCl, 0.5 mM EDTA-KOH, pH 7.5) supplemented with 35 mM CHAPS and incubated at room temperature for 2 h. UDM-purified MtCorB and TtCorB or DDM-purified TmCorA were mixed with 100 µL of solubilized lipids (0.5 mg) at a protein/lipid ratio of 1:30, 1:90, 1:270, or 1:810 (wt:wt) and incubated for 20 min at room temperature. For the no-protein liposome control, the same volume of HPLC buffer containing 0.05% UDM was added. Proteoliposomes were formed by adding the protein/lipid sample to a partially dehydrated Sephadex G-50 column (1.5 mL) equilibrated in buffer A. Membrane impermeable magnesium indicator, mag-fura-2 (Thermo Fisher; 1 mM stock in H2O) was added to a final concentration of 50 µM to the proteoliposomes. Dye encapsulation was performed through 1 freeze–thaw cycle (we observed reduced activity with an increased number of cycles) and subsequent extrusion through 0.8 µm polycarbonate filters. Extravesicular dye was removed by centrifuging the proteoliposomes through a partially dehydrated Sephadex G-50 column equilibrated in buffer A.
The transport assay was measured with a SpectraMax M5e fluorescence plate reader using SoftMax Pro 5 software. Totally, 10 µL of dye-encapsulated liposomes were diluted with 190 µL of reaction buffer (buffer A without EDTA) in a 96-well black bottom plate (Greiner) and baseline fluorescence was measured for 1 min at 25 °C. The uptake reaction was initiated by the addition of MgCl2, CaCl2, or Zn(CH3CO2)2, and recorded for 10 min. For mag-fura-2-encapsulated liposomes, fluorescence was measured using two excitation wavelengths (330 and 369 nm) and one emission wavelength (509 nm) every 6 s including 1 s shaking between reads. Triton X-100 and EDTA were added to 0.1% and 12.5 mM to obtain the maximum and minimum mag-fura-2 signals for conversion to [Mg2+]. All measurements were performed in triplicates. To test for electrogenic transport, the NaCl and KCl in the reaction buffer were replaced with 150 mM NMDG-Cl and 1 µM valinomycin (Sigma; 1 mM stock in DMSO) to generate an outward potassium gradient. For no valinomycin condition, an equal volume of DMSO was added.
Homology modeling of TtCorB and HsCNNM2
The homology model of TtCorB and HsCNNM2 were constructed with MODELLER Version 9.2576 using the MtCorBΔC structure as the template. The sequence alignment was performed using Clustal Omega77. The model was evaluated using PROCHECK78. Conservation coloring was done with ProtSkin (http://www.mcgnmr.mcgill.ca/ProtSkin/).
Molecular dynamics simulation
MtCorBΔC structure was used as the starting model for MD simulations. The transporter was embedded in a bilayer of 3POPC:1POPG lipids and solvated in 150 mM NaCl (neutral with 277 Na+ and 139 Cl− ions) using the web service CHARMM-GUI79,80. Most residues were assigned their standard protonation state at pH 7. The total number of atoms in the atomic model is on the order of 200,000. The all-atom CHARMM force field PARAM36 for protein81–83, lipids84, and ions85 was used. Explicit water was represented with the TIP3P model86. The model was refined using energy minimization for at least 2000 steps. All the simulations were performed under NPT (constant number of particle N, pressure P, and temperature T) conditions at 303 K and 1 atm, and periodic boundary conditions with electrostatic interactions were treated by the particle mesh Ewald method87 and a real-space cutoff of 12 Å. The simulations use a time step of 2 fs, with bond distances involving hydrogen atoms fixed using the SHAKE algorithm88. After minimization, positional restraints on all of the Cα atoms were gradually released after which a trajectory of 500 ns was generated using NAMD version 2.1189. The same process was applied to the model of symmetric MtCorBΔC dimer, in which the simulation ran for 124 ns.
Targeted molecular dynamics simulation
For preparing the target (open) model, an Mg2+ ion was pulled through the interface helices of MtCorBΔC while keeping a distance restraint with Glu111 of both protomers. This caused the two helices to open. All forces were then removed and the whole system was equilibrated for ~8 ns. For the targeted MD simulation, the MtCorBΔC structure was used as the starting model. During the simulation, forces were applied on the backbone of the transmembrane helices with a force constant of 200 kcal mol−1 A−2 for a period of 10 ns. This decreases the RMSD value linearly to reach the final target conformation.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Katharina Dürr and her lab members for assistance with the initial screening of CorB orthologs; Alexei Gorelik for advice on crystallographic data processing; Nazzareno D’Avanzo for assistance with liposome transport assay; Satchal Erramilli for the TmCorA plasmid and nanodisc protocol; Martin Schmeing and Camille Fortinez for crystallographic data collection at ALS; Canadian Light Source (CLS) staff: Michel Fodje, Shaun Labiuk, Bulat Gabidullin, and Kathryn Janzen. CLS is supported by the Canada Foundation for Innovation, the Natural Sciences and Engineering Research Council (NSERC), the National Research Council, the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. Y.S.C. is supported by a Canadian Institutes of Health Research Doctoral Research Award (GSD-167011). This work is supported by Natural Sciences and Engineering Research Council of Canada grant RGPIN-2020-07195 to K.G.
Source data
Author contributions
Y.S.C. designed experiments, cloned constructs, performed small-scale screenings, purified proteins, solved crystal structures, and performed liposome transport assays. G.K. assisted with crystal screening and performed ITC experiments. B.E.M. performed HDX-MS experiments. A.R. performed molecular dynamics simulations. R.F. performed AUC and ITC experiments of soluble domains. Y.S.C. and K.G. wrote the paper. K.G., J.E.B., and B.R. oversaw the research.
Data availability
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Atomic coordinates and structure factors for MtCorBCBS with Mg2+-ATP, MtCorBΔC with Mg2+-ATP, and MtCorBΔC R235L mutant have been deposited to the Protein Data Bank (www.rcsb.org) under accession numbers 7MSU, 7M1T, and 7M1U, respectively. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communciations thanks Luis Alfonso Martinez-Cruz and other, anonymous, reviewers for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-24282-7.
References
- 1.de Baaij JH, Hoenderop JG, Bindels RJ. Magnesium in man: implications for health and disease. Physiol. Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 2.Funato Y, Miki H. Molecular function and biological importance of CNNM family Mg2+ transporters. J. Biochem. 2019;165:219–225. doi: 10.1093/jb/mvy095. [DOI] [PubMed] [Google Scholar]
- 3.Stuiver M, et al. CNNM2, encoding a basolateral protein required for renal Mg2+ handling, is mutated in dominant hypomagnesemia. Am. J. Hum. Genet. 2011;88:333–343. doi: 10.1016/j.ajhg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamazaki D, et al. Basolateral Mg2+ extrusion via CNNM4 mediates transcellular Mg2+ transport across epithelia: a mouse model. PLoS Genet. 2013;9:e1003983. doi: 10.1371/journal.pgen.1003983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arjona FJ, et al. CNNM2 mutations cause impaired brain development and seizures in patients with hypomagnesemia. PLoS Genet. 2014;10:e1004267. doi: 10.1371/journal.pgen.1004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daneshmandpour Y, Darvish H, Pashazadeh F, Emamalizadeh B. Features, genetics and their correlation in Jalili syndrome: a systematic review. J. Med. Genet. 2019;56:358–369. doi: 10.1136/jmedgenet-2018-105716. [DOI] [PubMed] [Google Scholar]
- 7.Funato Y, Yamazaki D, Miki H. Renal function of cyclin M2 Mg2+ transporter maintains blood pressure. J. Hypertens. 2017;35:585–592. doi: 10.1097/HJH.0000000000001211. [DOI] [PubMed] [Google Scholar]
- 8.Yamazaki D, et al. The Mg2+ transporter CNNM4 regulates sperm Ca2+ homeostasis and is essential for reproduction. J. Cell Sci. 2016;129:1940–1949. doi: 10.1242/jcs.182220. [DOI] [PubMed] [Google Scholar]
- 9.Funato Y, et al. Membrane protein CNNM4-dependent Mg2+ efflux suppresses tumor progression. J. Clin. Investig. 2014;124:5398–5410. doi: 10.1172/JCI76614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazaki D, et al. Cnnm4 deficiency suppresses Ca(2+) signaling and promotes cell proliferation in the colon epithelia. Oncogene. 2019;38:3962–3969. doi: 10.1038/s41388-019-0682-0. [DOI] [PubMed] [Google Scholar]
- 11.Simon J. et al. Magnesium accumulation upon cyclin M4 silencing activates microsomal triglyceride transfer protein improving NASH. J. Hepatol.75, 34–45 (2021). [DOI] [PMC free article] [PubMed]
- 12.Thyme SB, et al. Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell. 2019;177:478–491. doi: 10.1016/j.cell.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CY, et al. Molecular cloning and characterization of a novel gene family of four ancient conserved domain proteins (ACDP) Gene. 2003;306:37–44. doi: 10.1016/S0378-1119(02)01210-6. [DOI] [PubMed] [Google Scholar]
- 14.Gibson MM, Bagga DA, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: the influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Mol. Microbiol. 1991;5:2753–2762. doi: 10.1111/j.1365-2958.1991.tb01984.x. [DOI] [PubMed] [Google Scholar]
- 15.Armitano J, Redder P, Guimaraes VA, Linder P. An essential factor for high Mg(2+) tolerance of Staphylococcus aureus. Front. Microbiol. 2016;7:1888. doi: 10.3389/fmicb.2016.01888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trachsel E, Redder P, Linder P, Armitano J. Genetic screens reveal novel major and minor players in magnesium homeostasis of Staphylococcus aureus. PLoS Genet. 2019;15:e1008336. doi: 10.1371/journal.pgen.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akanuma G, et al. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J. Bacteriol. 2014;196:3820–3830. doi: 10.1128/JB.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arjona FJ, de Baaij JHF. CrossTalk opposing view: CNNM proteins are not Na(+) /Mg(2+) exchangers but Mg(2+) transport regulators playing a central role in transepithelial Mg(2+) (re)absorption. J. Physiol. 2018;596:747–750. doi: 10.1113/JP275249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funato Y, Furutani K, Kurachi Y, Miki H. CrossTalk proposal: CNNM proteins are Na(+) /Mg(2+) exchangers playing a central role in transepithelial Mg(2+) (re)absorption. J. Physiol. 2018;596:743–746. doi: 10.1113/JP275248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy S, et al. The protein tyrosine phosphatase PRL-2 interacts with the magnesium transporter CNNM3 to promote oncogenesis. Oncogene. 2015;34:986–995. doi: 10.1038/onc.2014.33. [DOI] [PubMed] [Google Scholar]
- 21.Sponder G, Svidova S, Schweigel M, Vormann J, Kolisek M. Splice-variant 1 of the ancient domain protein 2 (ACDP2) complements the magnesium-deficient growth phenotype of Salmonella enterica sv. typhimurium strain MM281. Magnes. Res. 2010;23:105–114. doi: 10.1684/mrh.2010.0206. [DOI] [PubMed] [Google Scholar]
- 22.Goytain A, Quamme GA. Functional characterization of ACDP2 (ancient conserved domain protein), a divalent metal transporter. Physiol. Genomics. 2005;22:382–389. doi: 10.1152/physiolgenomics.00058.2005. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, et al. Structural basis for the Mg2+ recognition and regulation of the CorC Mg2+ transporter. Science Advances7, eabe6140 (2021). [DOI] [PMC free article] [PubMed]
- 24.Gimenez-Mascarell P, et al. Current Structural knowledge on the cnnm family of magnesium transport mediators. Int. J. Mol. Sci.20, 1135 (2019). [DOI] [PMC free article] [PubMed]
- 25.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ereno-Orbea J, Oyenarte I, Martinez-Cruz LA. CBS domains: Ligand binding sites and conformational variability. Arch. Biochem. Biophys. 2013;540:70–81. doi: 10.1016/j.abb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Chen YS, et al. Mg(2+)-ATP sensing in CNNM, a putative magnesium transporter. Structure. 2020;28:324–335. doi: 10.1016/j.str.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Corral-Rodriguez MA, et al. Nucleotide binding triggers a conformational change of the CBS module of the magnesium transporter CNNM2 from a twisted towards a flat structure. Biochem. J. 2014;464:23–34. doi: 10.1042/BJ20140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YS, et al. The cyclic nucleotide-binding homology domain of the integral membrane protein CNNM mediates dimerization and is required for Mg(2+) efflux activity. J. Biol. Chem. 2018;293:19998–20007. doi: 10.1074/jbc.RA118.005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulerez I, et al. Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis. EMBO Rep. 2016;17:1890–1900. doi: 10.15252/embr.201643393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozlov G, et al. PRL3 pseudophosphatase activity is necessary and sufficient to promote metastatic growth. J. Biol. Chem. 2020;295:11682–11692. doi: 10.1074/jbc.RA120.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gimenez-Mascarell P, et al. Structural basis of the oncogenic interaction of phosphatase PRL-1 with the magnesium transporter CNNM2. J. Biol. Chem. 2017;292:786–801. doi: 10.1074/jbc.M116.759944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimenez-Mascarell P, et al. Structural Insights into the intracellular region of the human magnesium transport mediator CNNM4. Int. J. Mol. Sci.20, 6279 (2019). [DOI] [PMC free article] [PubMed]
- 35.Hirata Y, Funato Y, Takano Y, Miki H. Mg2+-dependent interactions of ATP with the cystathionine-beta-synthase (CBS) domains of a magnesium transporter. J. Biol. Chem. 2014;289:14731–14739. doi: 10.1074/jbc.M114.551176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payandeh J, et al. Probing structure-function relationships and gating mechanisms in the CorA Mg2+ transport system. J. Biol. Chem. 2008;283:11721–11733. doi: 10.1074/jbc.M707889200. [DOI] [PubMed] [Google Scholar]
- 37.Stetsenko A, Guskov A. Cation permeability in CorA family of proteins. Sci. Rep. 2020;10:840. doi: 10.1038/s41598-020-57869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Baaij JH, et al. Membrane topology and intracellular processing of cyclin M2 (CNNM2) J. Biol. Chem. 2012;287:13644–13655. doi: 10.1074/jbc.M112.342204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Castano A, et al. Novel variant in the CNNM2 gene associated with dominant hypomagnesemia. PLoS ONE. 2020;15:e0239965. doi: 10.1371/journal.pone.0239965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franken GAC, et al. The phenotypic and genetic spectrum of patients with heterozygous mutations in cyclin M2 (CNNM2) Hum. Mutat. 2021;42:473–486. doi: 10.1002/humu.24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parry DA, et al. Mutations in CNNM4 cause Jalili syndrome, consisting of autosomal-recessive cone-rod dystrophy and amelogenesis imperfecta. Am. J. Hum. Genet. 2009;84:266–273. doi: 10.1016/j.ajhg.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polok B, et al. Mutations in CNNM4 cause recessive cone-rod dystrophy with amelogenesis imperfecta. Am. J. Hum. Genet. 2009;84:259–265. doi: 10.1016/j.ajhg.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirji N, et al. Jalili Syndrome: cross-sectional and longitudinal features of seven patients with cone-rod dystrophy and amelogenesis imperfecta. Am. J. Ophthalmol. 2018;188:123–130. doi: 10.1016/j.ajo.2018.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parveen A, et al. A novel pathogenic missense variant in CNNM4 underlying Jalili syndrome: Insights from molecular dynamics simulations. Mol. Genet. Genom. Med. 2019;7:e902. doi: 10.1002/mgg3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez Torres L., Schorderet D., Valmaggia C. & Todorova M. A novel mutation in CNNM 4 (G492C) associated with Jalili Syndrome. Acta Ophthalmol.93 (2015).
- 46.Prasad MK, et al. A targeted next-generation sequencing assay for the molecular diagnosis of genetic disorders with orodental involvement. J. Med. Genet. 2016;53:98–110. doi: 10.1136/jmedgenet-2015-103302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- 48.Payandeh J, Pai EF. A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 2006;25:3762–3773. doi: 10.1038/sj.emboj.7601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eshaghi S, et al. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 50.Lunin VV, et al. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan J, et al. Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl Acad. Sci. USA. 2018;115:E8201–E8210. doi: 10.1073/pnas.1810719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomita A, et al. ATP-dependent modulation of MgtE in Mg(2+) homeostasis. Nat. Commun. 2017;8:148. doi: 10.1038/s41467-017-00082-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payandeh J, Pfoh R, Pai EF. The structure and regulation of magnesium selective ion channels. Biochim. Biophys. Acta. 2013;1828:2778–2792. doi: 10.1016/j.bbamem.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Guskov A, et al. Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA. Proc. Natl Acad. Sci. USA. 2012;109:18459–18464. doi: 10.1073/pnas.1210076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng N, et al. The C2′- and C3′-endo equilibrium for AMP molecules bound in the cystathionine-beta-synthase domain. Biochem. Biophys. Res. Commun. 2018;497:646–651. doi: 10.1016/j.bbrc.2018.02.124. [DOI] [PubMed] [Google Scholar]
- 62.Karplus PA, Diederichs K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 2015;34:60–68. doi: 10.1016/j.sbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 66.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jurrus E, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laue, T. et al., eds. Analytical Ultracentrifugation in Biochemistry and Polymers Science. 90–125. (Royal Society of Chemistry, Cambridge, UK, 1992).
- 70.Brookes E, Demeler B, Rocco M. Developments in the US-SOMO bead modeling suite: new features in the direct residue-to-bead method, improved grid routines, and influence of accessible surface area screening. Macromol. Biosci. 2010;10:746–753. doi: 10.1002/mabi.200900474. [DOI] [PubMed] [Google Scholar]
- 71.Brautigam CA. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Ritchie TK, et al. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jenkins ML, et al. Structural determinants of Rab11 activation by the guanine nucleotide exchange factor SH3BP5. Nat. Commun. 2018;9:3772. doi: 10.1038/s41467-018-06196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucic I, et al. Conformational sampling of membranes by Akt controls its activation and inactivation. Proc. Natl Acad. Sci. USA. 2018;115:E3940–E3949. doi: 10.1073/pnas.1716109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nimigean CM. A radioactive uptake assay to measure ion transport across ion channel-containing liposomes. Nat. Protoc. 2006;1:1207–1212. doi: 10.1038/nprot.2006.166. [DOI] [PubMed] [Google Scholar]
- 76.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016;54:5 6 1–5 6 37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Madeira F, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 79.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 80.Wu EL, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 82.MacKerell AD, Jr., Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 83.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beglov D, Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys. 1994;100:9050–9063. doi: 10.1063/1.466711. [DOI] [Google Scholar]
- 86.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 87.Darden T, York D, Pedersen L. Particle mesh Ewald: an N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 88.Ryckaert J-P, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 89.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Atomic coordinates and structure factors for MtCorBCBS with Mg2+-ATP, MtCorBΔC with Mg2+-ATP, and MtCorBΔC R235L mutant have been deposited to the Protein Data Bank (www.rcsb.org) under accession numbers 7MSU, 7M1T, and 7M1U, respectively. Source data are provided with this paper.