Abstract
Objective
To find the genetic etiology of premature ovarian insufficiency (POI) in a patient with primary amenorrhea and hypergonadotropic hypogonadism.
Design
Case report.
Setting
University hospital.
Patient(s)
A Belgian woman aged 32 years with POI at the age of 17, her parents, and her sister whose POI was diagnosed at age 29.
Intervention(s)
Analysis of a panel of 31 genes implicated in POI (POIGP) using next-generation sequencing (NGS), Sanger sequencing, and in vitro functional study.
Main Outcome Measure(s)
Gene variants, family mutational segregation, and in vitro functional impact of the mutant proteins.
Result(s)
The analysis of the gene panel using NGS identified the presence of two novel follicle-stimulating hormone receptor (FSHR) missense mutations at a compound heterozygous state in the affected patient: c.646 G>A, p.Gly216Arg, and c.1313C>T, p.Thr438Ile. Sanger sequencing showed the presence of each mutation at heterozygous state in the patient’s parents and at heterozygous compound state in the affected sister. Both substituted amino acids (Gly216 and Thr438) were conserved in FSHR of several vertebrate species as well as in other glycoproteins receptors (TSHR and LHCGHR), suggesting a potentially important role in glycoprotein receptor function. An in vitro functional study showed similar results for both variants with more than 90% reduction of their cell surface expression and a 55% reduction of their FSH-induced cyclic adenosine 3′:5′ monophosphate (cAMP) production compared with the wild-type FSHR.
Conclusion(s)
The analysis of a gene panel of 31 genes implicated in POI allowed us to identify two novel partially inactivating mutations of FSHR that are likely responsible for the POI phenotype of the proband and of her affected sister.
Key Words: Gene panel, genetics, FSH receptor, premature ovarian insufficiency, next-generation sequencing
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/posts/xfre-d-20-00030
Premature ovarian insufficiency (POI) is a heterogeneous syndrome affecting around 1% of women at the age of 40 and 1 in 1,000 women at age 30 (1, 2, 3). According to the European Society of Human Reproduction and Embryology (ESHRE) guidelines on POI, the diagnosis is made based on the presence of menstrual disturbances before the age of 40 years, consisting of primary amenorrhea (PA), secondary amenorrhea, or oligomenorrhea for at least 4 months, associated to hypergonadotropic hypogonadism with FSH levels >25 IU/L on two occasions more than 4 weeks apart (3). The etiology of POI may be iatrogenic, autoimmune or genetic but remains however undetermined in the large majority of cases (2, 4, 5). Established genetic causes of POI involve chromosomal abnormalities, mainly X chromosome numerical and/or structural defects, FMR1 premutations and other much rarer gene mutations involved in the follicular development and folliculogenesis process (2, 6). Elucidating the genetic etiologies is crucial to gain more insight in the physiopathology of POI and to provide an appropriate genetic counseling and management to affected patients and their relatives.
In the last decades, different genomic approaches have contributed to uncovering several genes implicated in the development of POI. Sanger sequencing of candidate genes, largely used in the past, appeared to be time consuming and hardly efficient when used as a first line for the search of genetic etiologies of nonsyndromic POI (6, 7, 8). More recently, next-generation sequencing (NGS) techniques, including whole exome sequencing (WES) or gene panels (GP) allowing massive parallel sequencing of targeted causal and candidate genes of POI, have turned out to be powerful genetic tools and have contributed to a large expansion of the identification of novel causal or candidate gene variants in affected patients (2, 9, 10, 11, 12, 13, 14, 15, 16, 17). These genes have been shown to be implicated directly or indirectly in gonadal and follicular development through very different mechanisms involving mitochondrial and immune function, metabolism, apoptosis, DNA replication and repair, mRNA processing, cell cycle progression, meiosis, and hormonal signaling (2, 5). We present here two novel FSH receptor (FSHR) inactivating mutations identified in two sisters with hypergonadotropic hypogonadism by using a POI gene panel of 31 genes.
Materials and methods
Patients
The proband is a Belgian patient who presented PA with normal breast development, whose POI was diagnosed when she was 17 years old. She came to our fertility clinic for a consultation regarding assisted reproduction with oocyte donation when she was 32 years old. Before consulting our center, she had a diagnostic laparoscopy showing a normal pelvic state with small ovaries. After unsuccessful attempts at ovulation induction with clomiphene citrate, she underwent controlled ovarian stimulation (COS) for in vitro fertilization (IVF) with increasing doses of human menopausal gonadotropins (hMG; Menopur) up to 300 IU per day. The cycle was canceled after 13 days of ovarian stimulation because no ovarian response was obtained (the maximal estradiol level was of 72 ng/L, and no follicular growth ≥10 mm was observed).
The family history revealed that POI had been diagnosed after hormone contraception arrest in the proband’s sister at the age of 29 years. Her sister had been taking contraceptive pills since the age of 18 years because she had experienced very irregular menstruation throughout the 2 years after spontaneous menarche (at the age of 16). The sister became pregnant via oocyte donation; ovarian stimulation with clomiphene citrate and COS for IVF with increasing doses of hMG (Menopur) for 18 days up to 300 IU per day had yielded no ovarian response. The patient’s mother was menopaused at 50 years old. There was no consanguinity in the family, and no other history of delayed puberty, abnormal sexual differentiation, infertility, or POI.
The study was approved by the ethics committee of Erasme Hospital (study registration number: P2016/196/CCB B406201628264). The patient and her relatives gave their written informed consent to be tested for a genetic etiology of POI.
POI evaluation
At the time of the study, the proband was 163 cm tall with a body mass index of 29 kg/m2. No dysmorphic features were noted. A transvaginal pelvic ultrasound showed a normal uterus and bilateral small ovaries with the presence of 4 + 5 small antral follicles. Biological analysis showed FSH levels of 74 IU/L and antimüllerian hormone (AMH) levels of 1.4 μg/L (AMH Gen II Elisa; Beckman Coulter). The patient’s conventional and molecular (array-based comparative genomic hybridization) karyotype were normal, and no fragile X premutation was found. Autoimmunity was assessed by the measurement of antiadrenal, antiovarian, and anti–thyroid peroxidase antibodies which turned out to be in the normal range.
The proband’s sister was 162 cm tall. Hormone tests that had been performed for secondary amenorrhea evaluation at the age of 29 showed FSH levels of 76 IU/L and AMH levels of 5.8 μg/L (AMH Gen II Elisa; Beckman Coulter).
Massive parallel sequencing targeted for POI (gene panel)
The POI gene panel (POIGP) was developed in 2016 for diagnostic purposes in the assessment of genetic etiologies of noniatrogenic POI with normal karyotype and no FMR1 premutation. This panel encompasses the coding regions and 11 base pairs (bp) flanking intronic sequences of 31 POI candidate and causal genes (Supplemental Table 1, available online), and it is reimbursed by the Belgian health insurance for this indication. It was performed at the Brussels Interuniversity Genomics High Throughput Core (http://www.brightcore.be/).
We mechanically fragmented 1 μg of genomic DNA to an average length of 220 bp with a Covaris M2200 machine. The DNA libraries were prepared using the kappa hyper Prep Kit according to the manufacturer’s instructions and captured together with Roche Nimblegen SeqCap EZ Choice XL enrichment probes. The captured fragments were then amplified on an Illumina cBOT machine and sequenced in 2 times 125-bp paired-end mode on an Illumina Hiseq 1500 sequencer.
After demultiplexing, the quality of reads was determined with FastQC (version 0.010.1; Babraham Bioinformatics). Reads were aligned to the human reference genome hg19 (ucsc.hg19.fasta) with BWA-MEM (version 0.7.10; Burrows-Wheeler Aligner). Aligned reads were sorted and quality controlled with SAMtools (version 0.1.9-44428cd). Duplicate reads were marked with Picard (version 1.97; Broad Institute). The reads were further optimized by GATK (version 2.7; Broad Institute) and underwent quality control with Picard (version 1.97). The coverage in the sequenced regions was determined with SAMtools (version 19-44428cd) and the in-house developed software Rscrip (Rv2.15.1). Variant calling was performed with GATK (version 2.7). Variants were annotated and analyzed with Highlander (version 14.9; Helaers and Vikkula). Variants of unknown significance (CL3), possibly pathogenic (CL4) and pathogenic (CL5) were further evaluated through different in silico predictions tools integrated in Highlander (FATHMM, LRT, Mutation Assessor, Mutation Taster, PolyPhen-2, SIFT) (18).
Sanger sequencing
Sanger sequencing was performed to validate the NGS results in the affected sisters and their parents. We amplified DNA using a standard polymerase chain reaction (PCR). The PCR products were purified with BigDye XTerminator Purification Kit (Applied Biosystems/Thermo Fischer Scientific) and analyzed on a 3130XL Genetic Analyzer (Applied Biosystems).
Functional characterization of FSHR mutations
Plasmid construction
We introduced FSHR gene variants into complementary DNA (cDNA) of human wild-type FSHR inserted in an expression vector (pSVL; Pharmacia) by the QuickChange site mutagenesis method (Stratagene) as described elsewhere (19). The mutagenic primers for FSHR point mutations were designed for each mutation as follows: for c.1313C>T (T438I), forward primer 5ʹ-GGCTGTGATGCTGCAGGCTTTTTCACTGTCTTTGCCAG-3ʹ and reverse primer 5ʹ-AAGCCTGCAGCATCACAGCCTGCCCCAATTTGCCAGTC-3ʹ; for c.646 G>A (G216R), forward primer 5ʹ-GAAGAATTGCCTAATGATGTTTTCCACAGAGCCTCTGG-3ʹ and reverse primer 5ʹ-AACATCATTAGGCAATTCTTCTAGATTATTATTATCGC-3ʹ. The success of mutagenesis was confirmed with Sanger sequencing.
FSHR membrane localization assay
We seeded COS-7 cells (ATCC CRL-1651) at a density of 250,000 in 3 × 3 cm culture wells and incubated them for 3 days in a 5% CO2 atmosphere at 37°C with 3 mL of culture medium containing Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum 10%, sodium pyruvate solution 1%, penicillin + streptomycin 1% + Fungizone 1%. Plasmids encoding human wild-type FSHR, empty vector (pSVL), G216R, and T438I mutations were transfected (12 μg/well) into COS-7 cells using Lipofectamine 2000 (27 μL/well) at day 2. The mutants were transfected separately and cotransfected at similar proportions (6 + 6 μg/well), mimicking the compound heterozygous state of the two affected patients. The effectiveness of transfection was assessed by the detection of fluorescence from green fluorescent protein 24 hours after transfection at day 3. Triplicates were used for each assay.
The assessment of the COS-7 cells’ surface expression of FSHR wild type, mutant and empty vector (pSVL) was performed at day 4 by FACScan flow cytofluorometer (Becton Dickinson) as described elsewhere (20, 21, 22). Briefly, the COS-7 cells were detached from each well with 1 mL of phosphate-buffered saline (PBS) containing 5 mM ethylenediaminetetraacetic acid (EDTA) and transferred into Falcon 2052 tubes. Each tube was supplemented by 2 mL of PBS/0.1% bovine serum albumin (BSA), then the cells were centrifuged at 560 × g, at 4°C for 3 minutes, and the supernatant was removed by inversion. The cells were then incubated for 30 minutes at room temperature with 100 μL PBS/0.1% BSA containing mouse monoclonal antibodies (5B2) 10X diluted, which recognize an epitope located in the extracellular domain (ECD) of the FSHR. The cells were washed again with 2 mL PBS/0.1% BSA and centrifuged as previously, then incubated for 30 minutes on ice in the dark with 100 μL PBS/BSA containing fluorescein-conjugated γ-chain-specific goat anti-mouse IgG 100X diluted (Sigma-Aldrich) and propidium iodide (0.3 μg/mL). The cells were washed and centrifuged once again and resuspended in 250 μL PBS/0.1% BSA. The fluorescence of 10,000 cells was then assayed by FACScan flow cytofluorometer. The results are expressed by the mean of fluorescence in arbitrary units (AU).
Cyclic AMP production assay
The cAMP production quantification was performed as described by Costagliola et al. (21) and was adapted for our experiment. The culture medium was removed on day 4 (48 hours after transfection) and replaced by 1 mL of Krebs-Ringer-HEPES buffer (KRH) for 30 minutes at 37°C. Transfected COS-7 cells were then incubated for 30 minutes in 1 mL of fresh KRH followed by 1 hour in 1 mL of fresh KRH supplemented with 25 μM phosphodiesterase inhibitor (Rolipram; Laboratory Logeais) with and without two different concentrations (1 and 0.01 IU/mL) of recombinant FSH (Ovaleap 900 IU/1.5 mL) (Theramex). The choice of the FSH concentration was based on previously published experiments (19) showing that in a similar setting cAMP production by FSHR reached its maximal level at 1 IU/mL (saturating concentration) and approximately half of it at 0.01 IU/mL (nonsaturating concentration).
For COS-7 cells cotransfected with both mutant FSHRs, the experiments were only performed with the saturating concentration of FSH (1 IU/mL). After 1 hour of incubation, the medium was discarded and replaced with 1 mL of 0.1 M HCl to release cAMP produced from cells in the supernatant. The cell extracts were dried in a vacuum concentrator, resuspended in water, and diluted appropriately for cAMP measurement. The results are expressed as picomoles of cAMP per milliliter. All experiments were performed in triplicate.
Statistical analysis
Statistical analyses were performed by STATA 15 software (Stata Corporation). The results of triplicate experiments are expressed as mean ± standard deviation. Analysis of variance (ANOVA) was used to compare the means between different groups (FSHRwt, pSVL, T438I, G216R, and T438+G216R) in each of the three experiments (FACS, AMPc production at 0.1 IU/mL, and 1 IU/mL of FSH). Bonferroni correction was used as post hoc test of ANOVA to adjust the P value between two means (23). P<.05 was considered statistically significant.
Results
NGS and Sanger sequencing
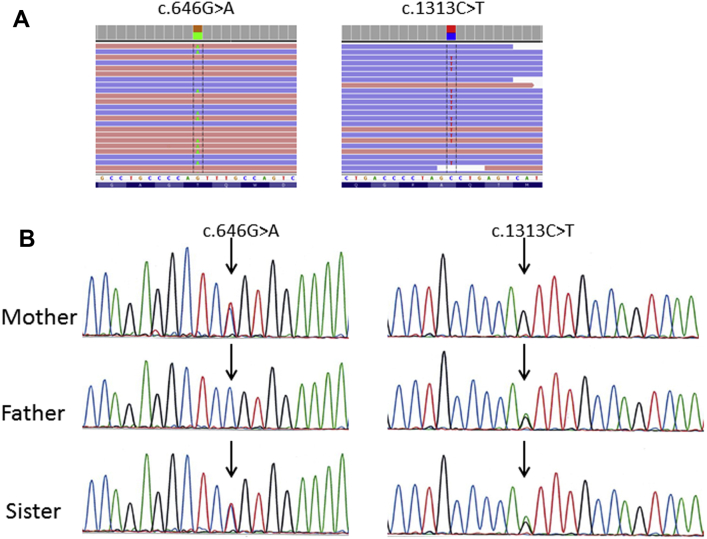
The analysis of the gene panel showed the presence of two novel nonsynonymous FSHR gene mutations in the proband. The first mutation is a substitution of guanine by alanine at position 646 in exon 8, predicted to result in a glycine to arginine substitution at the residue 216 of the ECD of the FSHR (c.646G>A, p.Gly216Arg). The second mutation is a substitution of cytosine by thymine at position 1313 in exon 10, predicted to induce a threonine to isoleucine substitution at the residue 438 located at the first extracellular loop (ECL1) of the transmembrane domain (TMD) (c.1313C>T, p.Thr438Ile) (Fig. 1A). The frequency of c.646G>A variant was 0.0008% in the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org). The c.1313C>T variant was not reported in ExAC database, nor was found in the Genome Aggregation database (gnomAD; http://gnomad.broadinstitute.org). Neither variant was found in our in-house database (including more than 2,000 individuals), nor were they reported in the human gene mutation database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php). Different in silico predictive algorithms predicted the two mutations to be damaging or probably damaging (Supplemental Table 2, available online).
Figure 1.
Next generation sequencing (NGS) of the proband, and Sanger sequencing of the proband’s parents and sister. (A) NGS performed in the affected proband (POI) identified two nonsynonymous mutations in the FSHR gene: c.646G>A (exon 8), p.Gly216Arg (ECD) and c.1313C>T (exon 10), p.Thr438Ile (ECL1). (B) Sanger sequencing showed paternal inheritance of c.1313C>T variant and maternal inheritance of c.646G>A. Both parents were unaffected heterozygous carriers; the affected sister carried both mutations at the heterozygous compound state.
The UCSC Genome Browser on human Feb.2009 (GRCh37/hg19) assembly showed the conservation of both residues (Gly216 and Thr438) in the FSHR of several vertebrate species. The assessment of the sequencing alignment of the three glycoprotein receptors using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that both amino acid residues were conserved in thyroid-stimulating hormone receptor (TSHR) and luteinizing hormone/ choriogonadotropin receptor (LHCGR). In addition, the Thr438 residue belongs to a sequence of seven amino acids present in the three glycoprotein hormone receptors and is also well conserved in several vertebrate species, supporting its potential functional importance. Sanger sequencing confirmed the c.1313C>T variant inheritance from the father and the c.646 G>A variant from the mother. The sister carried both mutations at the compound heterozygous state (Fig. 1B).
In vitro functional study
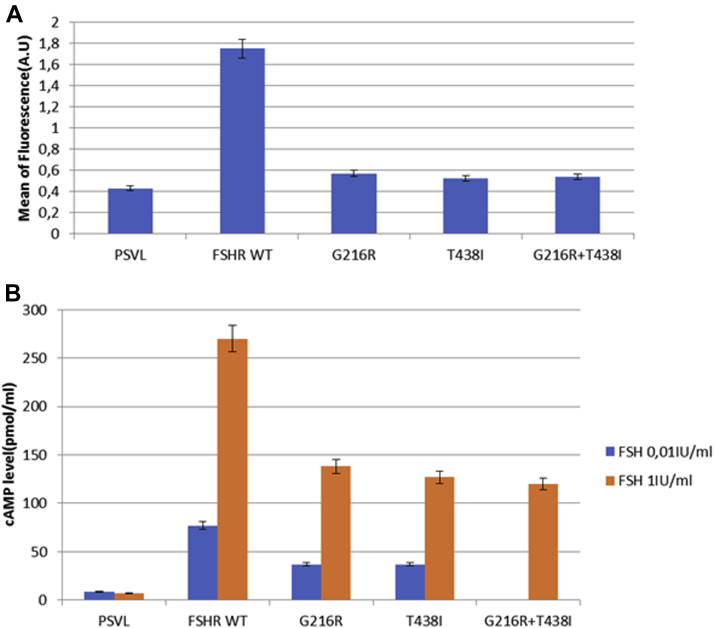
Flow cytometry analysis showed a drastic reduction of cell surface expression of mutated FSHRs compared with the wild type. This reduction was of 89.4%, 93%, and 92 % for Gly216Arg, Thr438Ile, and Gly216Arg + Thr438Ile, respectively (Fig. 2A). The production of cAMP at baseline was similar among the cells transfected by plasmids containing the empty vector (pSVL), FSHR WT, Gly216Arg variant, Thr438Ile variant, and Gly216Arg + Thr438Ile variants. Recombinant FSH induced a dose-dependent stimulation of cAMP production in COS-7 cells transfected with expression vectors encoding the wild-type or the mutated receptors.
Figure 2.
Functional testing of the mutated FSHR. Functional testing of the FSHR variants was performed using Cos-7 cells transiently transfected with cDNA encoding the wild-type FSHR (FSHR WT), the Gly216Arg variant, the Thr438Ile variant, the Gly216RArg+Thr438Ile variants (mimicking the compound heterozygous state in the affected patients), or the empty plasmid (PSVL). Data of two representative experiments are presented (mean ± standard deviation). Each experiment was performed two times in triplicate. (A) Cell surface expression of the FSHR variants quantified by flow cytometry (A.U.: arbitrary units of fluorescence). The cell surface expression was statistically significantly reduced for both variants compared to FSHR WT (P<.0001). Cells cotransfected with both variants (Gly216Arg + Thr438Ile) showed results similar to those obtained in cells transfected by either Thr438Ile or Gly216Arg. (B) FSH-induced cAMP production measured in the cell culture medium after cell incubation with two different FSH concentrations: 0.01 IU/mL (nonsaturating concentration) and 1 IU/mL (saturating concentration). A statistically significant decrease in cAMP production was observed for both variants compared with FSHR WT at both FSH concentrations (P<.0001 and P<.001 for FSH 0.01 IU/mL and 1 IU/mL, respectively). The cAMP produced by cells cotransfected with both variants showed results similar to those obtained in cells transfected by either Thr438Ile or Gly216Arg at saturating FSH concentration.
At nonsaturating concentrations of FSH (0.01 IU/mL), the cAMP production was 58% lower in COS-7 cells transfected with G216R and T438I variants compared with cells transfected with the wild-type receptor. At saturating concentrations of FSH (1 IU/mL), the cAMP production was, respectively, 50% and 54% lower in COS-7 cells transfected with G216R and T438I variants compared with cells transfected with the wild-type receptor (Fig. 2B). The cAMP production by COS-7 cells cotransfected by both variants (Gly216Arg + Thr438Ile) was evaluated after incubation with saturating FSH concentration (1 IU/mL) and was comparable with the cAMP produced by COS-7 cells transfected by each of the variants at the same concentration of FSH, showing 55% reduction compared with cells transfected with the wild-type receptor (Fig. 2B).
Discussion
We report here two novel mutations of the FSHR gene in two sisters presenting with hypergonadotropic hypogonadism. Both sisters were compound heterozygotes, having inherited a different mutated allele from each parent.
Both FSH and its receptor (FSHR) play crucial roles in female fertility, especially for follicular growth beyond the primary stage in humans and preantral stage in rodents (24, 25, 26). A glycoprotein hormone receptor, FSHR belongs to the rhodopsin-like G protein-coupled receptors (GPCRs). Glycoprotein hormone receptors include two other receptors LHCGR and TSHR and are characterized by a complex structure including a seven-helical transmembrane as well as a hinge region containing sulfated tyrosine residues important for ligand recognition and signal transduction (22, 27).
The FSHR biallelic inactivating mutations are one of the well-established, very rare causes of POI. The first homozygous FSHR inactivating mutation (c.566C>T, p.Ala189Val) was identified by linkage studies in six Finnish families of affected women presenting hypergonadotropic hypogonadism, primary amenorrhea, delayed puberty, and hypoplastic ovaries (28). Histologic examination of the ovaries showed primordial and primary follicles without any further follicular development, indicating that the first steps of folliculogenesis were independent of FSH action (29) in accordance with the block in folliculogenesis observed in mice with targeted disruption of the FSHR gene (25).
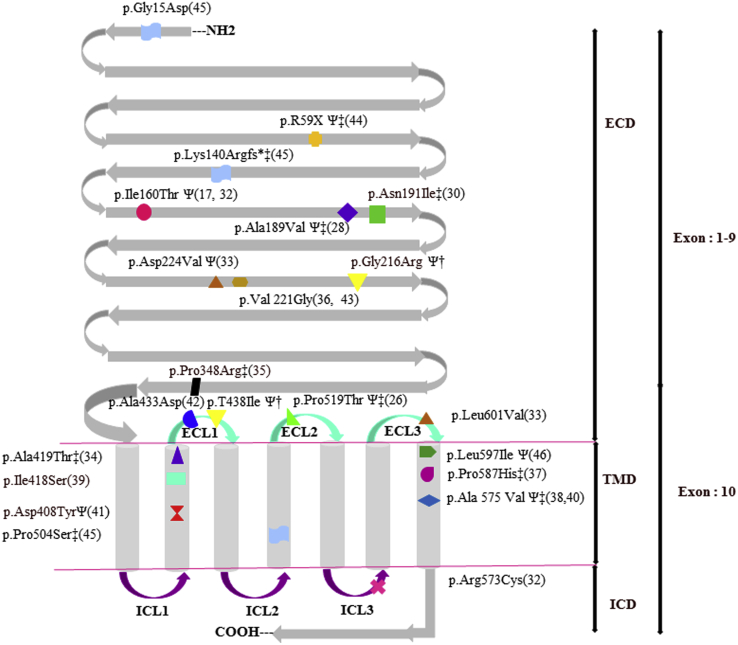
Twenty inactivating mutations have been reported so far, located in the ECD or TMD of the receptor associated with different clinical phenotypes (Fig. 3) (17, 26, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46). Of note, six of these mutations were recently identified by WES (39, 41, 42, 45). Patients’ phenotypes have been related to the level of inactivation of the receptor. Most patients carrying FSHR inactivating mutations associated with complete loss of cAMP production have displayed PA with delayed pubertal development and streak ovaries (26, 28, 35, 37, 38, 45, 46). Conversely, in the presence of partially inactivating FSHR mutations, the patients’ phenotype can vary from PA with partial or normal breast development to secondary amenorrhea and normal-sized ovaries containing preantral and even small antral follicles (32, 33, 41, 46).
Figure 3.
Inactivating mutations in FSHR. The gene encoding FSHR receptor contains 10 exons that encode a protein of 695 amino acids (aa). Exons 1 to 9 encode the extracellular domain (ECD); exon 10 encodes a C-terminal part of the ECD, the transmembrane domain (TMD), and the intracellular domain (ICD). The ECD contains 349 aa + signal peptide of 17 aa; the TMD contains 264 aa including seven transmembrane α-helices, three intracellular loop (ICLs), and three extracellular loop (ECLs); the ICD contains 65aa (24, 48). Twenty previously inactivating mutations of FSHR as well as the two mutations identified in our two patients (†) are presented in this figure. ‡Mutations inducing a total loss of FSHR function. ΨMutations inducing an alteration of FSHR cell surface expression based on functional studies.
In our study, both sisters had a normal development of secondary sex characteristics, and small antral follicles were identified by transvaginal ultrasound in the ovaries of the proband concordant with the partial inactivation of the two FSHR variants. Both sisters had AMH levels within the normal range, in accordance with previously reported AMH levels in a series of patients homozygous for the Ala189Val inactivating mutation of FSHR (47). In contrast to most other forms of POI characterized by a follicular depletion and undetectable levels of AMH, women with POI related to inactivating mutations of FSHR are rather considered as FSH resistant; they have early growing follicles, but their development is arrested at the small antral stage (29, 47). In those patients, AMH secreted by the granulosa cells of the small growing follicles remains detectable.
Both sisters failed to respond to 300 IU of hMG during COS for IVF. The daily dose of 300 IU FSH is generally recognized as the maximal efficient daily dose of FSH during COS. In addition, increased doses of exogenous FSH up to 600 IU/day have not been proven to be efficient in patients with other FSHR partial and complete inactivating mutations (17, 26, 32, 33). The absence of ovarian response to COS contrasts with the residual in vitro production of 50% cAMP by the mutant FSHRs. Although in vivo and in vitro situations cannot be directly compared, it should be emphasized that the in vitro saturating concentration of FSH (1 IU/mL) very likely exceeds the in vivo FSH concentration delivered to the follicular granulosa cells with a daily dose of 300 IU of exogenous hMG. The choice of the in vitro model used could also have an impact on the in vitro assessment of the functional characteristics of the FSHRs. Other FSHR mutations with comparable residual activities in vitro were associated with even more severe clinical phenotypes (delayed puberty with primary amenorrhea and incomplete breast development) (41). Moreover, additional impaired downstream signaling pathways such as ERK1/2 could also be involved (46). Eventually, the severity of FSHR inactivation could also be modulated by other genetic factors inherent to the genetic characteristics of each patient.
The proband received a POI diagnosis at age 17 because of PA. Her sister’s menarche occurred at age 16 with very irregular cycles up to the age of 18; she took oral contraceptives onward, with the diagnosis of POI made at contraception arrest at age 29. It is difficult to draw definitive conclusions about phenotypical differences between the two sisters because the proband started hormone treatment at age 17 while in PA, and her sister presented early signs of ovarian dysfunction with a late onset of menarche and hormonal contraception begun at 18 years after few episodes of bleeding.
Performing gene panel analysis allowed us to exclude simultaneously the potential contribution of 31 genes implicated in POI because no variant was identified in any of the tested genes (Supplemental Table 1). Although it might be informative to perform WES to further investigate the potential implication of additional genes that might modify the timing and progress of POI, the identification of two novel partially inactivating mutations of FSHR at a compound heterozygous state in the two affected patients provides a very likely causative explanation for the development of POI.
The two FSHR variants were located respectively in the ECD (Gly216Arg) and the first ECL (ECL1) of the TMD (Thr438Ile) of the receptor. Both implicated residues were preserved in several vertebrate species as well as in LHCGR and TSHR. For both mutations, flow cytometry showed a reduction of approximately 90% in the fluorescence intensity detected on the cell surface, suggesting an important reduction in cell surface expression of the mutated receptor compared with the wild type. Six previously identified inactivating mutations of FSHR inducing amino acid changes in the ECD were associated with an alteration in the cell surface expression of the receptor (17, 28, 32, 33, 44, 46). The Thr438Ile mutation is located in the first ECL of the TMD. Another FSHR mutation in ECL1 [c.1298C>A (Exon 10); p.Ala433Asp] has been recently identified by WES in a Brazilian patient with POI but was not tested in vitro (42).
It is interesting that both Thr438 and Ala433 residues are included in the same stretch of seven amino acids (Ala433, Ile434, Asp435, Trp436, Gln437, Thr438, and Gly439) and are well conserved in the three glycoprotein receptors as well as in several vertebrate species. In our experiments the reduction of cell surface expression of each of the two mutated receptors was associated with an approximately 55% reduction in FSH-induced cAMP production. The coexpression of both mutated FSHR did not show any change in FSH-induced cAMP production at saturating FSH concentration compared with the one observed for each mutated FSHR, suggesting that no synergistic effect occurred between the two FSHR variants for this FSH concentration. The setting of the present study does not permit evaluating the implications of other potential molecular mechanisms related to the partial inactivation of the mutant FSHRs such as receptor internalization or desensitization, which would be interesting to further explore in future studies.
In conclusion, we identified two novel mutations of FSHR present at compound heterozygous state in two sisters with hypergonadotropic hypogonadism. The functional characterization of both mutations showed reduced cell surface expression and FSH-induced cAMP production of the mutant receptors compared with the wild-type FSHR, demonstrating their causative role in the phenotype of the affected sisters. These findings enlarge the mutational spectrum of FSHR and expand its contribution to the development of POI.
Acknowledgments
The authors thank the patient and her family for their participation; Claude Massart, Jacqueline Van Sande, and Jean-Yves Springael for technical assistance and support for in vitro functional study; Judith Racapée for statistical analysis; and Xavier Peyrassol for assistance with the artwork.
Footnotes
A.S. has nothing to disclose. J.D. has nothing to disclose. V.J. has nothing to disclose. M.M. has nothing to disclose. D.D. has nothing to disclose. A.G. has nothing to disclose. M.B. has nothing to disclose. S.V. has nothing to disclose. S.C. has nothing to disclose. A.D. has received a research grant from Ferring Pharmaceuticals.
Supported by a grant from Fonds Erasme for medical research, Brussels, Belgium.
J.D. present address: Institut de Pathologie et de Génétique (IPG), Gosselies, Belgium.
Supplementary data
References
- 1.Luborsky J.L., Meyer P., Sowers M.F., Gold E.B., Santoro N. Premature menopause in a multi-ethnic population study of the menopause transition. Hum Reprod. 2003;18:199–206. doi: 10.1093/humrep/deg005. [DOI] [PubMed] [Google Scholar]
- 2.Tucker E.J., Grover S.R., Bachelot A., Touraine P., Sinclair A.H. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37:609–635. doi: 10.1210/er.2016-1047. [DOI] [PubMed] [Google Scholar]
- 3.Webber L., Davies M., Anderson R., Bartlett J., Braat D., Cartwright B. ESHRE guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31:926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- 4.Jiao X., Zhang H., Ke H., Zhang J., Cheng L., Liu Y. Premature ovarian insufficiency: phenotypic characterization within different etiologies. J Clin Endocrinol Metab. 2017;102:2281–2290. doi: 10.1210/jc.2016-3960. [DOI] [PubMed] [Google Scholar]
- 5.Huhtaniemi I., Hovatta O., Marca A La, Livera G., Monniaux D., Persani L. advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol Metab. 2018;29:400–419. doi: 10.1016/j.tem.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Qin Y., Jiao X., Simpson J.L., Chen Z.J. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiotiu D., Mercadal B.A., Imbert R., Verbist J., Demeestere I., De Leener A. Variants of the BMP15 gene in a cohort of patients with premature ovarian failure. Hum Reprod. 2010;25:1581–1587. doi: 10.1093/humrep/deq073. [DOI] [PubMed] [Google Scholar]
- 8.Alvaro Mercadal B., Imbert R., Demeestere I., Gervy C., De Leener A., Englert Y. AMH mutations with reduced in vitro bioactivity are related to premature ovarian insufficiency. Hum Reprod. 2015;30:1196–1202. doi: 10.1093/humrep/dev042. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca D.J., Patiño L.C., Suárez Y.C., De Jesús Rodríguez A., Mateus H.E., Jiménez K.M. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril. 2015;104:154–162.e2. doi: 10.1016/j.fertnstert.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Bouilly J., Beau I., Barraud S., Bernard V., Azibi K., Fagart J. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab. 2016;101:4541–4550. doi: 10.1210/jc.2016-2152. [DOI] [PubMed] [Google Scholar]
- 11.Hyon C., Mansour-Hendili L., Chantot-Bastaraud S., Donadille B., Kerlan V., Dodé C. Deletion of CPEB1 gene: a rare but recurrent cause of premature ovarian insufficiency. J Clin Endocrinol Metab. 2016;101:2099–2104. doi: 10.1210/jc.2016-1291. [DOI] [PubMed] [Google Scholar]
- 12.Patiño L.C., Beau I., Carlosama C., Buitrago J.C., González R., Suárez C.F. New mutations in non-syndromic primary ovarian insufficiency patients identified via whole-exome sequencing. Hum Reprod. 2017;32:1512–1520. doi: 10.1093/humrep/dex089. [DOI] [PubMed] [Google Scholar]
- 13.França M.M., Funari M.F.A., Nishi M.Y., Narcizo A.M., Domenice S., Costa E.M.F. Identification of the first homozygous 1-bp deletion in GDF9 gene leading to primary ovarian insufficiency by using targeted massively parallel sequencing. Clin Genet. 2018;93:408–411. doi: 10.1111/cge.13156. [DOI] [PubMed] [Google Scholar]
- 14.Tucker E.J., Grover S.R., Robevska G., Bergen J Van Den, Hanna C., Sinclair A.H. Identification of variants in pleiotropic genes causing “ isolated ” premature ovarian insufficiency: implications for medical practice. Eur J Hum Genet. 2018;26:1319–1328. doi: 10.1038/s41431-018-0140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao X., Ke H., Qin Y., Chen Z. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol. 2018;29:795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Laissue P. The molecular complexity of primary ovarian insufficiency etiology and the use of massively parallel sequencing. Mol Cell Endocrinol. 2018;460:170–180. doi: 10.1016/j.mce.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Flageole C., Toufaily C., Bernard D.J., Ates S., Blais V., Chénier S. Successful in vitro maturation of oocytes in a woman with gonadotropin-resistant ovary syndrome associated with a novel combination of FSH receptor gene variants: a case report. J Assist Reprod Genet. 2019;36:425–432. doi: 10.1007/s10815-018-1394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits G., Olatunbosun O., Delbaere A., Pierson R., Vassart G., Costagliola S. Ovarian hyperstimulation syndrome due to a mutation in the follicle-stimulating hormone receptor. N Engl J Med. 2003;349:760–766. doi: 10.1056/NEJMoa030064. [DOI] [PubMed] [Google Scholar]
- 20.Costagliola S., Rodien P., Ludgate M., Alerts E. Genetic immunization against the human thyrotropin receptor causes thyroiditis and allows production of monoclonal antibodies recognizing the native receptor. J Immunol. 1998;160:1458–1465. [PubMed] [Google Scholar]
- 21.Costagliola S., Panneels V., Bonomi M., Koch J., Many M.C., Smits G. Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. EMBO J. 2002;21:504–513. doi: 10.1093/emboj/21.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smits G., Campillo M., Govaerts C., Janssens V., Richter C., Vassart G. Glycoprotein hormone receptors: determinants in leucine-rich repeats responsible for ligand specicity. EMBO J. 2003;22:2692–2703. doi: 10.1093/emboj/cdg260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong R.A. When to use the Bonferroni correction. Ophthalmic Physiol Opt. 2014;34:502–508. doi: 10.1111/opo.12131. [DOI] [PubMed] [Google Scholar]
- 24.Simoni M., Gromoll J., Nieschlag E. The follicle-stimulating hormone receptor : biochemistry, molecular biology, physiology, and pathopysiology. Endocr Rev. 1997;18:739–773. doi: 10.1210/edrv.18.6.0320. [DOI] [PubMed] [Google Scholar]
- 25.Dierich A., Sairam M.R., Monaco L., Fimia G.M.G.,A., LeMeur M.S.-C.P. Impairing follicle-stimulating hormone ( FSH) signaling in vivo : targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci USA. 1998;95:13612–13617. doi: 10.1073/pnas.95.23.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meduri G., Touraine P., Beau I., Lahuna O., Desroches A., Vacher-Lavenu M.C. Delayed puberty and primary amenorrhea associated with a novel mutation of the human follicle-stimulating hormone receptor: clinical, histological, and molecular studies. J Clin Endocrinol Metab. 2003;88:3491–3498. doi: 10.1210/jc.2003-030217. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X., Liu H., Chen X., Chen P., Fischer D., Sriraman V. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci USA. 2012;109:12491–12496. doi: 10.1073/pnas.1206643109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aittomaki K., Lucena J.L., Pakarinen P., Sistonen P., Tapanainen J., Gromoll J. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82:959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 29.Aittomäki K., Herva R., Stenman U.H., Juntunen K., Ylöstalo P., Hovatta O. Clinical features of primary ovarian failure caused by a point mutation in the follicle-stimulating hormone receptor gene. J Clin Endocrinol Metab. 1996;81:3722–3726. doi: 10.1210/jcem.81.10.8855829. [DOI] [PubMed] [Google Scholar]
- 30.Gromoll J., Simoni M., Nordhoff V., Behre H.M., De Geyter C., Nieschlag E. Functional and clinical consequences of mutations in the FSH receptor. Mol Cell Endocrinol. 1996;125:177–182. doi: 10.1016/s0303-7207(96)03949-4. [DOI] [PubMed] [Google Scholar]
- 31.Kotlar T.J., Young R.H., Albanese C., Crowley W.F., Scully R.E., Jameson J.L. A mutation in the follicle-stimulating hormone receptor occurs frequently in human ovarian sex cord tumors. J Clin Endocrinol Metab. 1997;82:1020–1026. doi: 10.1210/jcem.82.4.3870. [DOI] [PubMed] [Google Scholar]
- 32.Beau I., Touraine P., Meduri G., Gougeon A., Desroches A., Matuchansky C. A novel phenotype related to partial loss of function mutations of the follicle stimulating hormone receptor. J Clin Invest. 1998;102:1352–1359. doi: 10.1172/JCI3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Touraine P., Beau I., Gougeon A., Meduri G., Desroches A., Pichard C. New natural inactivating mutations of the follicle-stimulating hormone receptor: correlations between receptor function and phenotype. Mol Endocrinol. 1999;13:1844–1854. doi: 10.1210/mend.13.11.0370. [DOI] [PubMed] [Google Scholar]
- 34.Doherty E., Pakarinen P., Tiitinen A., Kiilavuori A., Huhtaniemi I., Forrest S. A novel mutation in the FSH receptor inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab. 2002;87:1151–1155. doi: 10.1210/jcem.87.3.8319. [DOI] [PubMed] [Google Scholar]
- 35.Allen L.A., Achermann J.C., Pakarinen P., Kotlar T.J., Huhtaniemi I.T., Jameson J.L. A novel loss of function mutation in exon 10 of the FSH receptor gene causing hypergonadotrophic hypogonadism: clinical and molecular characteristics. Hum Reprod. 2003;18:251–256. doi: 10.1093/humrep/deg046. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura Y., Maekawa R., Yamagata Y., Tamura I., Sugino N. A novel mutation in exon8 of the follicle-stimulating hormone receptor in a woman with primary amenorrhea. Gynecol Endocrinol. 2008;24:708–712. doi: 10.1080/09513590802454927. [DOI] [PubMed] [Google Scholar]
- 37.Kuechler A., Hauffa B.P., Köninger A., Kleinau G., Albrecht B., Horsthemke B. An unbalanced translocation unmasks a recessive mutation in the follicle-stimulating hormone receptor (FSHR) gene and causes FSH resistance. Eur J Hum Genet. 2010;18:656–661. doi: 10.1038/ejhg.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Achrekar S.K., Modi D.N., Meherji P.K., Patel Z.M., Mahale S.D. Follicle stimulating hormone receptor gene variants in women with primary and secondary amenorrhea. J Assist Reprod Genet. 2010;27:317–326. doi: 10.1007/s10815-010-9404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katari S., Wood-Trageser M.A., Jiang H., Kalynchuk E., Muzumdar R., Yatsenko S.A. Novel inactivating mutation of the FSH receptor in two siblings of Indian origin with premature ovarian failure. J Clin Endocrinol Metab. 2015;100:2154–2157. doi: 10.1210/jc.2015-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai S.S., Achrekar S.K., Sahasrabuddhe K.A., Meharji P.K., Desai S.K., Mangoli V.S. Functional characterization of two naturally occurring mutations (Val514Ala and Ala575Val) in follicle-stimulating hormone receptor. J Clin Endocrinol Metab. 2015;100:E638–E645. doi: 10.1210/jc.2014-3662. [DOI] [PubMed] [Google Scholar]
- 41.Bramble M.S., Goldstein E.H., Lipson A., Ngun T., Eskin A., Gosschalk J.E. A novel follicle-stimulating hormone receptor mutation causing primary ovarian failure: a fertility application of whole exome sequencing. Hum Reprod. 2016;31:905–914. doi: 10.1093/humrep/dew025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.França M.M., Lerario A.M., Funari M.F.A., Nishi M.Y., Narcizo A.M., de Mello M.P. A novel homozygous missense FSHR variant associated with hypergonadotropic hypogonadism in two siblings from a Brazilian family. Sex Dev. 2017;11:137–142. doi: 10.1159/000477193. [DOI] [PubMed] [Google Scholar]
- 43.Banerjee A.A., Achrekar S.K., Joseph S., Pathak B.R., Mahale S.D. Functional characterization of two naturally occurring mutations V221G and T449N in the follicle stimulating hormone receptor. Mol Cell Endocrinol. 2017;440:69–79. doi: 10.1016/j.mce.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Liu H., Xu X., Han T., Yan L., Cheng L., Qin Y. A novel homozygous mutation in the FSHR gene is causative for primary ovarian insufficiency. Fertil Steril. 2017;108:1050–1055. doi: 10.1016/j.fertnstert.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 45.He W., Du J., Yang X., Li W., Tang W., Dai C. Novel inactivating mutations in the FSH receptor cause premature ovarian insufficiency with resistant ovary syndrome. Reprod Biomed Online. 2019;38:397–406. doi: 10.1016/j.rbmo.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Liu H., Guo T., Gong Z., Yu Y., Zhang Y., Zhao S., Qin Y. Novel FSHR mutations in han chinese women with sporadic premature ovarian insufficiency. Mol Cell Endocrinol. 2019;492 doi: 10.1016/j.mce.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Kallio S., Aittomki K., Piltonen T., Veijola R., Liakka A., Vaskivuo T.E. Anti-mullerian hormone as a predictor of follicular reserve in ovarian insufficiency: special emphasis on FSH-resistant ovaries. Hum Reprod. 2012;27:854–860. doi: 10.1093/humrep/der473. [DOI] [PubMed] [Google Scholar]
- 48.Desai S.S., Roy B.S., Mahale S.D. Mutations and polymorphisms in FSH receptor: functional implications in human reproduction. Reproduction. 2013;146:235–248. doi: 10.1530/REP-13-0351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.