Abstract
Integral membrane proteins (IMPs) constitute ~30% of all proteins encoded by the genome of any organism and Escherichia coli remains the first-choice host for recombinant production of prokaryotic IMPs. However, the expression levels of prokaryotic IMPs delivered by this bacterium are often low and overproduced targets often accumulate in inclusion bodies. The targets are therefore often discarded to avoid an additional and inconvenient refolding step in the purification protocol. Here we compared expression of five prokaryotic (bacterial and archaeal) IMP families in E. coli and Saccharomyces cerevisiae. We demonstrate that our S. cerevisiae-based production platform is superior in expression of four investigated IMPs, overall being able to deliver high quantities of active target proteins. Surprisingly, in case of the family of zinc transporters (Zrt/Irt-like proteins, ZIPs), S. cerevisiae rescued protein expression that was undetectable in E. coli. We also demonstrate the effect of localization of the fusion tag on expression yield and sample quality in detergent micelles. Lastly, we present a road map to achieve the most efficient expression of prokaryotic IMPs in our yeast platform. Our findings demonstrate the great potential of S. cerevisiae as host for high-throughput recombinant overproduction of bacterial and archaeal IMPs for downstream biophysical characterization.
Keywords: Prokaryotic integral membrane proteins, S. cerevisiae expression Platform, Expression rescue, High-throughput recombinant protein production
Graphical abstract
Highlights
-
•
S. cerevisiae is superior to E. coli in expressing correctly folded and active IMPs.
-
•
S. cerevisiae completely rescues the expression of the family of zinc transporters.
-
•
Localization of the fusion tag affects expression yields and protein quality.
-
•
We provide a roadmap to efficient expression of prokaryotic IMPs in S. cerevisiae.
Abbreviations
- IMPs
Integral membrane proteins
- PDB
Protein Data Bank
- E. coli
Escherichia coli
- S. cerevisiae
Saccharomyces cerevisiae
- TEV
cleavage site for Tobacco Etch Virus protease
- ZIPs
zinc transporters (Zrt/Irt-like proteins)
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- RT
room temperature
- PMSF
phenylmethylsulfonyl fluoride
- L, P, C
leupeptine, pepstatin and chymostatin
- BCA
bicinchoninic acid
- CY-55
cyclohexyl-1-pentyl-β-D-maltoside
- DDM
n-dodecyl-β-D-maltopyranoside
- DM
n-decyl-β-D-maltopyranoside
- FC-12
n-dodecylphosphocholine
- FC-13
n-tridecylphosphocholine
- FC-16
n-hexadecylphosphocholine
- LDAO
lauryldimethylamine N-oxide
- FSEC
fluorescence-detection size-exclusion chromatography
- IMAC
immobilized metal affinity chromatography
- MECA
multi-electrode-cavity-array
- DPhPC
1,2-diphytanoyl-sn-glycero-3-phosphocholine
- IV
current-voltage
- TM
transmembrane
- TetR
tetracycline repressor protein
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PC
phosphatidylcholine
- CL
Cardiolipin
1. Introduction
Integral membrane proteins (IMPs) mediate efficient and selective transport of organic and inorganic molecules across cellular membranes, and many signaling processes rely on membrane-embedded receptors (Cournia et al., 2015). IMPs are critical components of each cell and constitute ~30% of the proteome across all kingdoms of life. In humans IMPs represent the largest class of drug targets, accounting for ~60% of currently approved pharmaceuticals (Gong et al., 2019).
Access to high-resolution structures of the membrane proteome is required to comprehend the membrane biology field (Cournia et al., 2015). While more than 120,000 structures are deposited in the Protein Data Bank (PDB; http://www.rcsb.org/) (Burley et al., 2019), currently just above 1% represents IMPs (http://www.blanco.biomol.uci.edu/mpstruc/). This bias reflects the main challenges associated with membrane protein structural biology, i.e., obtaining high expression levels of prime quality IMPs, followed by development of purification procedures that preserve protein fold and activity (Pandey et al., 2016). Overall, this laborious task typically involves screening for optimal expression and purification conditions for a large number of orthologue proteins to achieve optimal yields of pure IMPs.
Structural studies of prokaryotic IMPs have almost exclusively relied on recombinant production in Escherichia coli (E. coli) (Schlegel et al., 2014), which is partly attributed to fast and cheap cultivation protocols. Furthermore, E. coli has been extensively studied with a large toolbox of biochemical methods and this host remains one of the best characterized organisms for recombinant gene expression (Gileadi, 2017; Dilworth et al., 2018). Moreover, it is generally assumed that the expression host most closely related to the organism from which the desired protein originates is likely to deliver the protein sample in the highest amounts and the best quality (Bill et al., 2011). Hence, E. coli may intuitively provide the most suitable lipid environment for recombinant production of Gram-negative bacterial IMPs. In contrast, E. coli may not be optimal for IMPs from Gram-positive prokaryotes and in particular not for archaeal counterparts, as the latter are found in membranes with a lipid composition that differs significantly from those found in bacteria (Jain et al., 2014). Instead of fatty acids esterified to D-glycerol-3-phosphate, archaeal phospholipids are composed of repeating units of isoprene connected through an ether bond to L-glycerol-1-phosphate (Dowhan W et al., 2008).
In the present paper we challenge the dogma that E. coli offers the most optimal expression platform as we hypothesized that Saccharomyces cerevisiae (S. cerevisiae) may possess a number of advantageous features beneficial for expression of bacterial and archaeal IMPs. First, we avoid accumulation of overproduced IMPs in inclusion bodies, hereby circumventing the requirement for protein refolding, a laborious and inefficient recovery step, often impossible due to the particularly complicated nature of this class of proteins, which often results in targets being discarded at this initial stage (Schlegel et al., 2014; Lyons et al., 2016). Secondly, yeast possesses a more advanced machinery for IMPs biogenesis that may be superior to that found in E. coli (Freigassner et al., 2009).
To investigate the capacity for expression of IMPs in E. coli and S. cerevisiae, we selected prokaryotic targets belonging to five protein families of Gram-negative, Gram-positive or archaeal origin and maintaining different functions (Table 1). We performed an extensive comparison between the most commonly used E. coli systems (Studier and Moffatt, 1986) and our previously described S. cerevisiae platform (Pedersen et al., 1996). Specifically, we compared to what extent recombinant proteins could be produced in the two hosts, extracted from the cell membranes, and purified in a stable and active form in high yields. Based on the obtained results, we highlight the experimental conditions that have to be considered to achieve optimal expression of bacterial and archaeal IMPs in S. cerevisiae. To our knowledge, such a study comparing the capacity for production of bacterial and archaeal IMPs in a eukaryotic host with that in E. coli has not been conducted before. Our findings demonstrate the potential of S. cerevisiae for overcoming issues with poorly overproduced bacterial and archaeal IMPs for downstream studies.
Table 1.
Panel of selected integral membrane protein targets studied in this work. Summary of their origin, function, topological features and high-resolution structures (if available). Targets originating from archaeal species are shown in red, from Gram-negative bacteria are shown in black, while from Gram-positive bacteria in blue. UniProt IDs∗ originate from the UniProt database (https://www.uniprot.org/) (UniProt, 2019). ∗∗: The overall topology (given as number of transmembrane (TM) helices), length of N-/C-termini (aa) and their orientation (in: intracellular, out: outside) are based on available crystal structures in the Protein Data Bank (PDB; http://www.rcsb.org/) (Burley et al., 2019) or as indicated in the UniProt database. PDB IDs∗∗∗ originate from the above-mentioned PDB.
2. Materials and Methods
2.1. S. cerevisiae and E. coli strains
S. cerevisiae recombinant protein production was carried out in the PAP1500 strain (α ura3-52 trp1: GAL10-GAL4 lys2-801 leu2Δ1 hris3Δ200 pep4:HIS3 prb1Δ1.6R can1 GAL) (Pedersen et al., 1996). BL21(DE3) (Studier and Moffatt, 1986), BL21(DE3)-pLysS (Studier and Moffatt, 1986; Studier, 1991), C41 (Miroux and Walker, 1996) and C43 (Miroux and Walker, 1996) strains were used as hosts for recombinant protein production in E. coli.
2.2. Plasmid construction
For yeast-based expression, the pEMBLyex4 vector was used (Cesareni et al., 1987). The E. coli - S. cerevisiae shuttle plasmid, pPAP10286, carrying the pET52b(+) expression cassette (Supplementary Figure 1) (Dowhan W et al., 2008; Studier, 1991) was used for E. coli expression. Two sets of codon-optimized full-length cDNAs for each target were used, i.e., optimized for expression in E. coli and S. cerevisiae, respectively (GenScript, USA).
The cDNAs were amplified by PCR using primers listed in Supplementary Tables 1 and 2, and inserted into the pEMBLyex4 or pPAP10286 expression plasmid to encode proteins N-terminally fused to His10-GFP-TEV (cleavage site for Tobacco Etch Virus protease) or C-terminally fused to TEV-GFP-His10. All PCR reactions were performed using AccuPol DNA polymerase (Amplicon, Denmark). Both S. cerevisiae and E. coli expression plasmids were generated by homologous recombination in S. cerevisiae using the transformation protocol developed by Gietz and Schiestl (2007). PCR primers contained 5′-extensions designed for homologous recombination, hereby ensuring correct assembly of PCR fragments, and the SalI-, HindIII- and BamHI-digested plasmids (Supplementary Figure 1). Transformed yeast cells were selected on minimal medium containing 2% glucose, and supplemented with leucine (30 mg/L) and lysine (20 mg/L). Correct assembly of expression plasmids was verified by DNA sequencing (Eurofins MWG Operon, Germany). E. coli expression plasmids were subsequently transferred to the designated E. coli expression strain. Plasmids encoding histidine-tagged zinc transporters (Zrt/Irt-like proteins, ZIPs) for expression in E. coli were generated by insertion of NdeI/XhoI digested PCR products into similarly digested pET15b or pET52b(+) vectors (Merck, Germany) and did not contain the GFP-tag.
2.3. Expression screening and temperature optimization of protein production
E. coli pre-cultures were inoculated from frozen stocks, grown O/N at 30 °C in 50 mL of 2 x YT or LB medium supplemented with tetracycline and subsequently diluted to OD450 = 0.05. At OD450 = 0.6, half of the culture was transferred to 15 °C and the other half to 30 °C. After thermal equilibration, protein production was induced with a final concentration of 0.1, 0.5 or 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively.
Yeast pre-cultures were inoculated from frozen stocks in 5 mL of minimal medium containing leucine (30 mg/L) and lysine (20 mg/L), and grown at 30 °C until saturation (typically for 48 h). Subsequently, 200 μL of the culture was transferred to 5 mL of glucose minimal medium containing lysine, but lacking leucine, and grown for 24 h at room temperature (RT). Then 500 μL of the pre-culture was transferred to 50 mL of glucose minimal medium containing lysine, but lacking leucine, and grown for another 24 h at RT. The culture was used to inoculate 100 mL of expression media (YP medium containing 0.5% glucose and 3% glycerol) to OD450 = 0.05. At OD450 = 1.0, half of the culture was transferred to 15 °C and the other half to 30 °C. After thermal equilibration, recombinant protein production was induced by adding 10 mL of YP medium containing 20% galactose and 3% glycerol.
For both E. coli- and S. cerevisiae-based expression, OD450 was measured at different time points up to 120 h post induction. For each time point, whole cell GFP fluorescence was measured in 1 OD450 unit in a white micro plate in a Fluoroskan Ascent fluorometer (Thermo Scientific, USA) with excitation at 485 nm and emission at 520 nm.
2.4. Production of membrane proteins
Yeast cells were pre-cultured and grown at RT as described above. 2 L of YP supplemented with 0.5% glucose and 3% glycerol was inoculated to an OD450 of ~0.05. At OD450 = 1, the culture was transferred to 15 °C and galactose was added to a final concentration of 2%. Cells were harvested at their expression maximum as estimated by the expression screens.
Yeast cells from 2-L cultures were harvested at 1600 g for 10 min at 4 °C, at the protein accumulation optimum established by the expression screen. Cells were lysed by glass bead homogenization in ice-cold lysis buffer (1 M NaCl, 25 mM imidazole, 1 mM EGTA, 1 mM EDTA, 10% glycerol; pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 μg/mL leupeptine, pepstatin and chymostatin (L, P, C) in a 50-mL Falcon tube. Cell lysates were centrifuged at 3000 g for 10 min at 4 °C to remove cell debris. Crude membranes were pelleted from the supernatant by ultracentrifugation at 160,000 g for 90 min at 4 °C (using Sorvall T865 rotor; Thermo Scientific, USA). Crude membrane pellets were resuspended and homogenized in lysis buffer containing protease inhibitors and kept at −80 °C until use.
Protein concentration in crude membranes was determined by the bicinchoninic acid (BCA) assay according to specifications of the manufacturer (Sigma Aldrich, USA).
To normalize expression yields, GFP fluorescence in 25 μg of crude membranes was measured in a Fluoroskan Ascent microplate fluorometer (Thermo Scientific, USA) as described above. Fluorescence was converted to pmol of TEV-GFP-His10-tagged protein using a standard curve generated from the fluorescent signal of purified GFP mixed with crude membranes from PAP1500 yeast as previously established (Bomholt et al., 2013) and normalized to the molecular weight of the respective target protein to calculate its percentage of expression in isolated crude membrane fractions.
2.5. Small-scale crude membrane preparation and SDS-PAGE separation for comparing expression in S. cerevisiae and E. coli
Pairs of 1-L cell cultures were grown as described above at RT. At OD450 = 0.6, one of the cultures was transferred to 15 °C and the other one to 30 °C, and induced with IPTG (E. coli) or galactose (S. cerevisiae). Subsequently, 25 mL of cell culture samples were collected at 0, 24, 48, 72, 96, 108 and 120 h after induction.
Yeast cells from 25-mL cultures were harvested at 1000 g for 10 min at 4 °C and the cell pellet was resuspended in 400 μL of ice-cold lysis buffer (25 mM imidazole, 1 M NaCl, 1 mM EGTA, 1 mM EDTA, 10% glycerol; pH 7.5) containing 1 mM PMSF and 1 μg/mL L, P, C. Cells were opened with glass beads homogenization in a Bertin Precellys 24 homogenizer (4 cycles of 15 s; Bertin Instruments, France) with 2-min cooling breaks. Glass beads were subsequently washed several times with ice-cold lysis buffer and the homogenate was cleared from cell debris by centrifugation at 3000 g for 10 min at 4 °C. Crude membranes were isolated from the supernatant by ultracentrifugation at 210,000 g at 4 °C for 20 min (using a Beckman TLA 100 rotor; Beckman, USA). Crude membrane pellets were resuspended in 200 μL of lysis buffer supplemented with protease inhibitors and stored at −80 °C.
E. coli cells from 25-mL cultures were harvested at 6000 rpm for 20 min at 4 °C (using a Sorvall SLA-3000 rotor). Cell pellets were resuspended in 1 mL of ice-cold lysis buffer (25 mM imidazole, 1 M NaCl, 1 mM EGTA, 1 mM EDTA, 10% glycerol; pH 7.6) containing 1 mM PMSF and 1 μg/mL L, P, C, and cells were disrupted by sonication on ice for 15 min using pulsation with 5-sec intervals and amplitude 50% (using Sonopuls 3100 homogenizer; Bandelin, Germany). Cell debris was removed by centrifugation at 15,000 g for 10 min at 4 °C (using a Sorvall SS-34 rotor). Crude membranes were isolated from the supernatant by ultracentrifugation at 160,000 g for 20 min at 4 °C (using a Beckman TLA 100 rotor). Crude membranes were resuspended and homogenized in 500 μL of ice-cold lysis buffer supplemented with protease inhibitors and stored at −80 °C.
Crude membranes were analyzed by SDS-PAGE prepared according to Laemmli, U. K. (1970) (Laemmli, 1970) and stained with Coomassie blue (R-250). Prior to electrophoretic analysis, crude membrane samples were denatured with sample buffer containing 4% SDS and protease inhibitors at RT for 20 min before loading. Imaging of both in-gel GFP fluorescence and Coomassie blue stained gels was performed using the Image Quant LAS-4000 system (GE Healthcare, USA).
2.6. Detergent screens
Crude membranes were incubated in solubilization buffer (25 mM Tris-HCl, 10 mM imidazole, 0.5 M NaCl, 10% glycerol; pH 7.6) supplemented with protease inhibitors (1 mM PMSF and 1 μg/mL L, P, C) at a protein:detergent ratio (w/w) of 1:3. The screens included the following detergents: 5-cyclohexyl-1-pentyl-β-D-maltoside (CY-5), n-dodecyl-β-D-maltopyranoside (DDM), n-decyl-β-D-maltopyranoside (DM), n-dodecylphosphocholine (FC-12), n-tridecylphosphocholine (FC-13), n-hexadecylphosphocholine (FC-16) and lauryldimethylamine N-oxide (LDAO). Detergents were purchased from Affymetrix, UK (as Anagrade quality) or from Glycon Biochemicals, Germany. Solubilization was performed at slow rotation for 1 h at 4 °C. Unsolubilized material was removed by ultracentrifugation at 160,000 g for 20 min at 4 °C (using a Beckman TLA 100 rotor). GFP fluorescence in the resulting supernatant was monitored as described above and solubilization efficiency was estimated by normalization to the total GFP fluorescence in the sample prior to detergent treatment.
2.7. Fluorescence-detection size-exclusion chromatography
Solubilized crude membranes were analyzed by fluorescence-detection size-exclusion chromatography (FSEC) performed using a Superose 6 Increase 200 10/300 GL column (AqpZ) or a Superose 12 10/300 GL column (KcsA, NavAb, ClCec1) attached to an Äkta Purifier system (all from GE Healthcare, USA) coupled to a fluorescence detector (Shimadzu Prominence RF-20A; Shimadzu, Japan) to visualize the elution profile of the GFP-tagged proteins. All FSEC runs were conducted in 20 mM Tris-HCl, 0.15 M NaCl, 0.03% (w/v) DDM; pH 7.5.
2.8. Small-scale affinity purification
For small-scale purification of the target IMP-TEV-GFP-His10 fusions, the choice of detergent was based on the results from the respective solubilization screens and FSEC analysis. Crude membranes isolated from the 2-L yeast cultures were solubilized for 4 h at 4 °C by slow rotation in solubilization buffer (described below) containing the most optimal detergent in the final concentration of 1% (w/v). Unsolubilized material was removed by ultracentrifugation at 160,000 g at 4 °C for 1.5 h (using a Sorvall T-865 rotor). The resulting supernatant was subsequently diluted four times in the respective solubilization buffer and subjected to immobilized metal affinity chromatography (IMAC) using nickel chelated resin (see below). Following binding, the resin was washed and bound target IMPs were eluted with the below-mentioned buffers supplemented with imidazole and containing the primary detergent at a final concentration of 1.5 x CMC. The amount of recombinant protein in each fraction was determined by GFP fluorescence as described above. Essentially, three purification protocols were applied as described below.
-
1.
AqpZ. Solubilized protein was loaded onto 2 x 1-mL HisTrap FF columns (GE Healthcare) connected in series and eluted in IMAC buffer (25 mM Tris-HCl, 500 mM NaCl and 10% glycerol; pH 7.6) with a linear imidazole gradient (0–500 mM).
-
2.
ClCec1, NavAb and KcsA. Following O/N binding at 4 °C to 3 mL of high-affinity Ni-NTA resin (GenScript, USA), the material was packed into an Econo-column (Biorad, USA), flow-through was collected and the resin was washed with the IMAC buffer indicated above with a step imidazole gradient (i.e., 10, 30, 100, 250 and 500 mM, respectively) to remove impurities and elute bound target IMPs.
-
3.
TmZIP. FC-16-solubilized crude membranes containing His-tagged TmZIP were diluted in solubilization buffer (50 mM HEPES-NaOH, 500 mM NaCl, 20% glycerol, 1 mM PMSF and 1 μg/mL L, P, C; pH 7.5) to reduce detergent concentration to 0.003%. Diluted material was supplemented with imidazole to a final concertation of 50 mM, filtered using 0.45-μm cellulose acetate membrane filter (Sartorius, Germany) and subsequently loaded onto a 5-mL Ni-NTA HisTrap HP column (GE Healthcare, USA) connected to an Äkta Pure system (GE Healthcare, USA). The column was washed with 5 x CV of IMAC buffer (20 mM HEPES-NaOH, 250 mM NaCl, 20% glycerol, 0.003% FC-16; pH 7.5) and eluted with a linear imidazole gradient (0–500 mM).
2.9. Live cell bio-imaging
Localization of heterologously-expressed GFP-tagged membrane proteins was visualized by imaging GFP fluorescence in whole cells at 1000× magnification using a Nikon Eclipse E600 microscope (Nikon Instruments Inc., USA) coupled to an Magnafire camera model S99802 (Optronics, USA).
2.10. Electrophysiology
Single channel currents from DDM-solubilized, IMAC-pure KcsA and NavAb (1 mg/mL) were recorded using the Orbit Mini with EDR3 software (Nanion Technologies, Germany). Briefly, a four-electrode recording chip (MECA4) (50–100 μm; Ionera Technologies, Germany) was mounted in the Orbit Mini apparatus and recording buffer was applied to the chip. Planar lipid bilayers consisting of 10 mM 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC, Avanti Polar Lipids, USA) and 1 mM cholesterol dissolved in nonane were painted on the recording wells. 0.2 μL of KcsA or NavAb in recoding buffer (10 mM HEPES-NaOH, 100 mM KCl, 100 mM NaCl; pH 7.2) was added to the cis-side of the bilayer. Subsequently, alternating voltages were applied to facilitate insertion of the channels. Recordings were carried out at a sampling rate of 1.25 kHz and afterwards a low pass filter, Bessel (8-pole) with a 3-dB cutoff of 200 Hz, was applied. Mean current amplitudes were plotted as a function of tested voltages. Single channel conductances were determined as the slope factor of a linear regression to the current-voltage (IV) plot. Clampfit 10.7 (Molecular Devices, USA) and Prism 7 (GraphPad Software, USA) were used for data analysis.
3. Results
3.1. Target integral membrane proteins (IMPs)
The target proteins selected for the present paper belong to five different families and represent a wide variety of prokaryotic IMPs of bacterial (including Gram-negative and Gram-positive) and archaeal origin (Table 1). The five target protein families exert diverse biological functions, and possess different topologies with respect to the number of transmembrane (TM) helices and lengths and orientation of N-/C- termini. They furthermore differ in molecular weight and oligomeric state, as also reflected by available structural information (Table 1, Supplementary Figures 2 and 3). Due to our selection of prokaryotic IMPs included in this paper, the results may apply to the production of α-helical prokaryotic membrane proteins in general.
3.2. The E. coli and S. cerevisiae expression platforms
To compare the capacity of E. coli with S. cerevisiae for recombinant production of prokaryotic IMPs, we selected BL21(DE3), BL21(DE3)-pLys, C41 and C43 expression strains, and the pET vector system (Studier and Moffatt, 1986) for E. coli expression and our own PAP1500 platform for yeast expression. The BL21(DE3)/pET host-vector system is the most commonly used in recombinant protein production in E. coli, while the S. cerevisiae PAP1500 expression platform has previously been shown to successfully express many challenging classes of IMPs of eukaryotic origin (Pedersen et al., 1996; Bomholt et al., 2013; Gotfryd et al., 2018; Wang et al., 2019; Molbaek et al., 2015; Scharff-Poulsen and Pedersen, 2013; Bjorkskov et al., 2017).
To simplify construction and manipulation of E. coli expression plasmids, we used the pPAP10286 vector for engineering expression constructs with AqpZ, ClCec1, NavAb and KcsA. The pPAP10286 shuttle vector can replicate in both S. cerevisiae and E. coli (Supplementary Figure 1) and carries the E. coli expression cassette from pET52b(+). The ability of the plasmid to replicate in yeast enables fast, cheap and accurate assembly of prokaryotic expression constructs (Supplementary Figure 1), without introducing any “contaminating” sequences, such as restriction sites or site-specific recombination sequences. Expression of ZIP transporters was attempted using an unmodified pET15b or pET52b(+) vector to produce N- or C-terminally His-tagged targets, respectively.
3.3. E. coli expression host strain and temperature influence protein accumulation
To simplify optimization of protein production and purification, all target IMPs were expressed as TEV-GFP-His10 tag fusions. We used codon-optimized cDNAs, i.e., tuned for expression in an E. coli-based platform. GFP can be directly used for quantification and in vivo localization of properly folded fusion proteins (Drew et al., 2001) and the TEV protease cleavage site enables removal of the entire fusion tag following metal affinity chromatography.
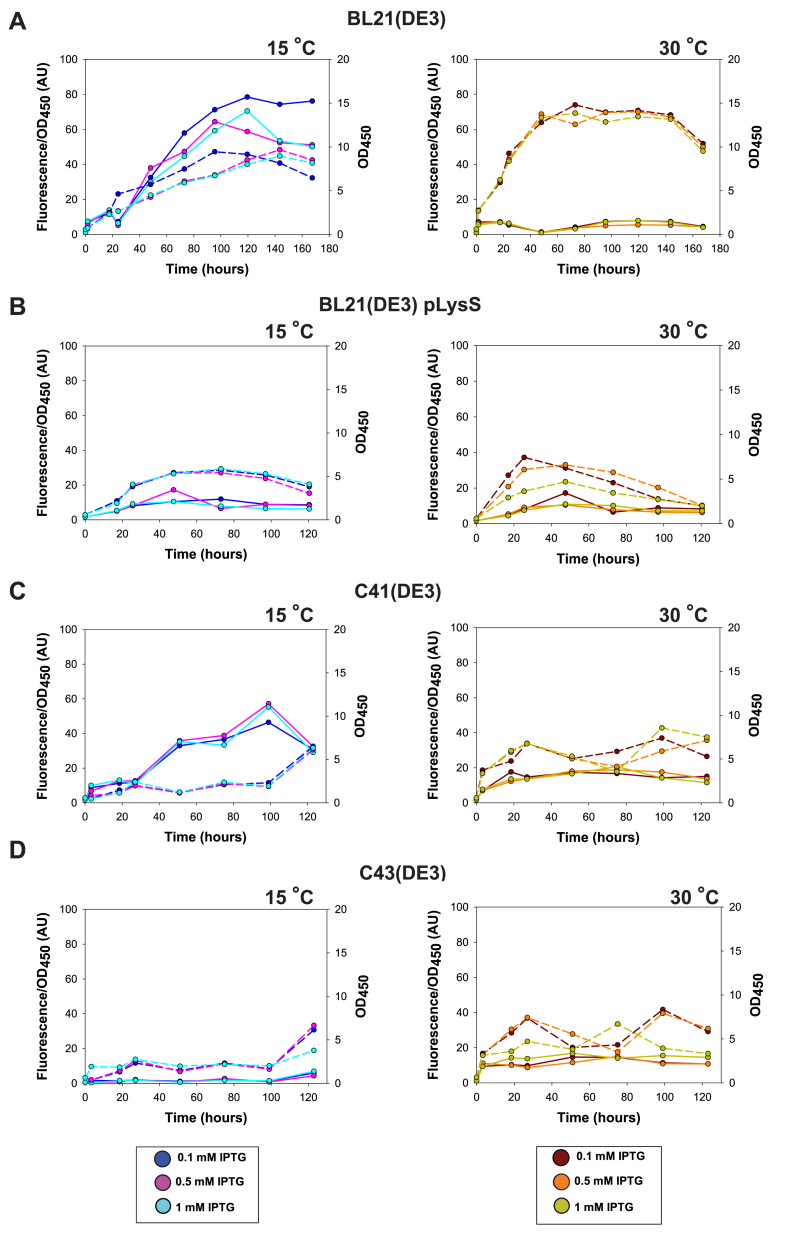
Expression of KcsA, NavAb and ClCec1 was screened in the four most used E. coli strains, i.e., BL21(DE3), BL21(DE3) pLysS, C41(DE3) and C43(DE3), at both 15 °C and 30 °C induction temperature, using a standard IPTG concentration of 0.5 mM and followed by assessment of GFP fluorescence accumulated in intact cells. The highest accumulation of ClCec1 was observed in BL21(DE3) at 30 °C and peaked 24 h after induction. NavAb accumulated to the largest extent in BL21(DE3) and C41(DE3) strains at 30 °C 8 h post induction, while the most pronounced KcsA expression was achieved in BL21(DE3) induced for 20 h at 30 °C (Fig. 1).
Fig. 1.
Effect of temperature on accumulation of ClCec1, KcsA and NabAb C-terminally fused to TEV-GFP-His10in four E. coli host strains.E. coli strains (A) BL21(DE3), (B) BL21(DE3) pLysS, (C) C41(DE3) and (D) C43(DE3) were grown O/N at 30 °C until OD450 = 1 and transferred to either 15 (blue lines) or 30 °C (red lines) prior to induction with 0.5 mM IPTG. Protein accumulation was measured by GFP fluorescence in the cell amount corresponding to 1 OD450 unit at each time-point after induction. Fluorescence measurements are shown as solid lines and OD450 measurements as dashed lines, respectively.
Applying AqpZ as a model protein, we subsequently investigated whether IPTG concentrations used for induction affect accumulation of recombinant protein in a specific E. coli host strain at a given temperature. Data in Fig. 2 show that accumulation of AqpZ in the BL21(DE3) strain was higher at 15 °C, while in the C41(DE3) and C43(DE3) strains the course of expression was elevated at 30 °C. The peak accumulation observed in C41(DE3) and C43(DE3) strains at 30 °C after 30 and 50 h, respectively, was comparable to that observed in BL21(DE3) strain at 15 °C after 120 h. However, the final OD450 was much higher for all strains grown at 30 °C. The effect of IPTG concentration tested for induction of AqpZ expression indeed appeared to be host strain- and temperature-specific, and should therefore be optimized independently for each of the strains used.
Fig. 2.
Effect of IPTG concentration and temperature on accumulation of AqpZ C-terminally fused to TEV-GFP-His10in four E. coli host strains.E. coli strains (A) BL21(DE3), (B) BL21(DE3) pLysS, (C) C41(DE3) and (D) C43(DE3) were grown at 30 °C until OD450 = 1, and transferred to either 15 (left panels) or 30 °C (right panels) prior to induction with 0.1, 0.5 or 1 mM IPTG. Protein accumulation was measured by GFP fluorescence in the cell amount corresponding to 1 OD450 unit at each time-point after induction. Fluorescence measurements are shown as solid lines and OD450 measurements as dashed lines, respectively. Color codes for the respective concentration of IPTG used for induction of the expression are indicated below each panel.
Expression originating from the pET52b(+)-based shuttle plasmid was tested by examining production of a soluble protein; the GFP-tagged tetracycline repressor protein (TetR; UniProt ID: P04483) from transposon Tn10 to ensure that the low levels of accumulation observed for the prokaryotic target IMPs in E. coli were not caused by using the pET52b(+)-derived yeast shuttle vector. As highlighted by the data presented in Supplementary Figure 4, the generally observed low accumulation of prokaryotic IMPs in E. coli, compared to the TetR, cannot be attributed to the use of the engineered pET52b(+)-pEMBLyex4 plasmid, but rather to the membrane embedded nature of the expressed targets.
3.4. Temperature-dependent accumulation of prokaryotic integral membrane proteins (IMPs) in S. cerevisiae
To determine how temperature affects the accumulation of recombinantly produced IMPs in S. cerevisiae, we investigated the time course of their accumulation at 15 °C and 30 °C. As seen in Fig. 3A–D, at 15 °C the accumulation increased over the entire production period and was much higher than that observed at 30 °C. In contrast, accumulation at 30 °C already peaked after ~24 h, followed by a rapid decline for all tested proteins.
Fig. 3.
Temperature-dependent accumulation of AqpZ, ClCec1, KcsA and NabAb C-terminally fused to TEV-GFP-His10in S. cerevisiae. Yeast cultures were grown at RT until OD450 = 1 before half of the culture was transferred to 15 and the other half to 30 °C. After thermal equilibration, recombinant protein production was induced by addition of galactose to the final concentration of 2%. Accumulation of (A) AqpZ, (B) ClCec1, (C) KcsA and (D) NavAb was measured by GFP fluorescence in the cell amount corresponding to 1 OD450 at each time-point after induction at 15 (blue lines) and 30 °C (red lines). (E) Live cell bio-imaging of yeast cells expressing the target proteins as C-terminal TEV-GFP-His10 fusions. Pairwise differential interference contrast and GFP fluorescence images are shown.
We used live cell bio-imaging to visualize the expressed GFP fusions in the yeast expression host. Fig. 3E illustrates that all target proteins accumulated in distinct intracellular compartments and not in the plasma membrane.
3.5. Prokaryotic integral membrane proteins (IMPs) accumulate to a high density in yeast membranes
The structures of the some of the IMP targets investigated in this work have already been determined using X-ray crystallography (Table 1) and the samples for structural characterization were produced using E. coli-based expression systems. Here we investigated whether the S. cerevisiae expression platform is capable of expressing the same targets, and subsequently compared both protein quantity and quality.
To achieve optimal protein production, yeast cells were induced at 15 °C and harvested at the established expression optimum for each target protein (Fig. 3A–D). The data in Supplementary Table 3 show that many target IMPs accumulated to a high density in yeast membranes relative to their yield in E. coli membranes. Because the molecular weight of the produced targets differs significantly, we also calculated the molar amounts accumulating per mg of crude membrane protein content. This revealed that the quantity of accumulated recombinant protein varied a lot between the different targets. From the estimated content of membrane-located recombinant protein, we also calculated the expected expression levels of the respective target IMPs (Supplementary Table 3). This would give rise to theoretical yields ranging from 0.9 to 149 mg of protein per liter shake flask culture. Such promising expression levels highlight the great potential for producing prokaryotic IMPs in large quantities needed for subsequent biophysical characterization.
3.6. Homogeneity of solubilized prokaryotic membrane proteins is detergent independent
Following the expression tests, we performed a screen to compare the ability of a selection of six non-ionic and zwitter-ionic detergents to extract the produced target IMPs from crude yeast membranes. The results shown in Fig. 4 (left panels) indicate that each target protein could be solubilized in all tested detergents. We identified AqpZ to be the most refractory to extraction, with an approximate solubilization efficiency of 20% in all applied surfactants (Fig. 4A). In contrast, ClCec1 generally solubilized well, with the highest extraction efficacies observed for FC-12, DDM and CY-5 (80–100%; Fig. 4B). KcsA could also be effectively extracted with 40–75% solubilization efficiency (Fig. 4C), with DDM being the most potent surfactant. Finally, for NavAb, solubilization was most pronounced in DM, LDAO and DDM (80–100%), whereas the other tested detergents resulted in solubilizations amounting to 50% in CY-5, and 30% in both FC-12 and FC-13 (Fig. 4D).
Fig. 4.
High-throughput solubilization screen and fluorescence-detection size-exclusion chromatography (FSEC) of AqpZ, ClCec1, KcsA and NabAb expressed in S. cerevisiae as C-terminal TEV-GFP-His10fusions. Crude yeast membranes overexpressing (A) AqpZ, (B) ClCec1, (C) KcsA and (D) NavAb were solubilized for 1 h at 4 °C in 5-cyclohexyl-1-pentyl-β-D-maltoside (CY-5), n-dodecyl-β-D-maltopyranoside (DDM), n-decyl-β-D-maltopyranoside (DM), n-dodecylphosphocholine (FC-12), n-tridecylphosphocholine (FC-13) or lauryldimethylamine N-oxide (LDAO) in a protein:detergent ratio (w/w) of 1:3. Normalized solubilization efficiencies (left panels) were estimated from GFP fluorescence in the supernatant after ultracentrifugation. Subsequently, detergent-solubilized material was separated by FSEC where GFP fluorescence was monitored during the elution (right panels). The colors of the chromatograms correspond to the respective colors of detergents shown in the bar diagrams (left panels). The void volume of the used columns is ~8 mL for Superose 6 Increase 200 10/300 GL and is ~9 mL for Superose 12 10/300 GL.
To maintain the activity of the extracted prokaryotic IMPs during purification, it is essential to apply a detergent that not only efficiently extracts the target from the membranes, but also preserves the sample in a homogeneous and non-aggregated state. As permitted by the GFP fusion tag, we used fluorescence-detection size-exclusion chromatography (FSEC) to assess the quality of detergent-solubilized samples. The results in Fig. 4 (right panels) show the FSEC profiles of each target protein in all six tested detergents, with color codes corresponding to those used in the detergent screen. In general, protein elution peaks were narrow and monodisperse, with only marginal amounts of aggregated protein eluting in the void volume, overall indicating a high protein quality. Only NavAb showed a double peak, revealing two populations of protein solubilized in LDAO, DM or DDM, and to some degree also in CY-5, FC-12 or FC-13.
3.7. IMAC yields pure fusion proteins from yeast expression
Based on the results from the detergent screen and FSEC analysis (Fig. 4), several detergents proved to be promising candidates for subsequent protein purification. Following solubilization in the respective detergents, i.e., AqpZ in LDAO, ClCec1 in DM, and both NavAb and KcsA in DDM, all tested IMP targets were purified by immobilized metal affinity chromatography (IMAC). Fig. 5 shows the IMAC elution profiles of the respective proteins. In all cases, the top fraction (containing also the highest amount of GFP) was separated by SDS-PAGE and the purity of the samples was visualized by in-gel GFP fluorescence and Coomassie blue staining.
Fig. 5.
Immobilized metal affinity chromatography (IMAC) purification of AqpZ, ClCec1, KcsA and NabAb expressed in S. cerevisiae as C-terminal TEV-GFP-His10fusions. Each fusion protein was extracted from the crude yeast membranes, solubilized in the respective detergent for 4 h at 4 °C and subjected to IMAC (see Materials and Methods section). IMAC profiles of (A) AqpZ, (B) ClCec1, (C) KcsA and (D) NavAb are shown. Protein elution was monitored by GFP fluorescence in the collected fractions. The dashed line indicates the used imidazole gradient. Inserts show the SDS-PAGE gels, where the top fraction from each elution was visualized by Coomassie blue staining (lane 1) and in-gel GFP fluorescence (lane 2).
AqpZ eluted as a single broad peak starting at 100 mM imidazole (Fig. 5A). SDS-PAGE separation of purified AqpZ revealed a highly pure protein band, with only minor bands of higher molecular weight visible by in-gel GFP fluorescence and Coomassie blue staining, indicating that AqpZ tetramers have not been fully dissociated and the sample also contains a population of higher oligomeric states.
ClCec1 eluted as one narrow and symmetrical peak at 500 mM imidazole (Fig. 5B). IMAC resulted in a large amount of pure protein, with only minor impurities visible in the Coomassie blue stained SDS-PAGE gel.
KcsA eluted mainly as a single peak with a shoulder at 100 mM imidazole (Fig. 5C). The IMAC profile revealed a minor amount of protein eluting at 50 mM imidazole and a second small elution peak at 250 mM imidazole, respectively. SDS-PAGE of the top fraction confirmed one band corresponding to KcsA, but a substantial amount of the fluorescent sample did not enter the gel. An impurity of higher molecular weight was only seen in the Coomassie blue stained SDS-PAGE gel.
The elution IMAC profile of NavAb resembled that of KcsA. However, the SDS-PAGE gel revealed more impurities only apparent in the Coomassie blue stained gel (Fig. 5D, lane 1). The SDS-PAGE gel also disclosed several bands of higher molecular weight visible by Coomassie blue and in-gel GFP fluorescence, hence not all of NavAb tetramers were fully dissociated after exposure to SDS (Fig. 5D).
The molecular weights of the purified fusion proteins, as determined by SDS-PAGE, correspond to the predicted molecular size of the monomeric proteins (i.e., Aqpz of 23 kDa, ClCec1 of 50 kDa, KcsA of 18 kDa and NavAb of 30 kDa, respectively), considering that correctly folded GFP contributes to the overall molecular weight of the fusion proteins only with 10–15 kDa (Geertsma et al., 2008).
3.8. The prokaryotic ion channels produced in S. cerevisiae are functional
The above-described data confirm that prokaryotic IMPs can be produced in yeast and purified to homogeneity. Both DDM-solubilized KcsA and NavAb channels were purified by IMAC and subsequently analyzed by FSEC to demonstrate that the detergent-solubilized samples maintained stability during purification (Supplementary Figure 5). While the IMAC and FSEC profiles indicate high protein quality, they do not provide information on the functionality of the purified channels. We therefore decided to investigate the activity of both KscA and NavAb.
After reconstitution into lipid bilayers, single channel current was recorded at a series of different voltages (Supplementary Figure 6A). A representative KcsA recording at −40 mV is shown in Fig. 6A. The reconstituted KcsA channel proved to be functional, and the corresponding recording displays channel openings and closings of at least three single channel molecules (O1–O3). Fig. 6B illustrates the mean single KscA channel current amplitude plotted as a function of voltage (I–V curve) and reveals a conductance of 77.3 ± 5.6 pS.
Fig. 6.
Single-channel recordings of immobilized metal affinity chromatography-pure KcsA and NavAb C-terminal TEV-GFP-His10fusions. Both channels were solubilized and purified in DDM prior to reconstitution into planar lipid bilayers on a 50- or 100-μm MECA 4 recording chip in the Orbit Mini system. (A) Representative single KcsA channel current recordings at −40 mV. At least three individual channels have been incorporated into the planar lipid bilayer. “C” indicates the closed state and “O1-3” indicates the open states, respectively. (B) The Current-Voltage relationship (I–V curve) for KcsA with a linear regression fitted to the data revealing a slope conductance of 77.3 ± 5.6 pS. (C) Representative single NavAb channel current recordings at +100 mV. (D) I–V curve for NavAb with a linear regression fitted to the data revealing a slope conductance of 57.6 ± 4.0 pS. Results are shown as mean current amplitudes ± SEM, where each data point was based on at least 3 single channel openings (n = 3–1300 events, from 3 independent experiments).
The reconstituted NavAb channel also displayed functionality and representative single-channel current recordings measured at ±120 mV reflect fast transitions between the open and the closed state (Fig. 6C), with an estimated single channel conductance of 57.6 ± 4.0 pS (Fig. 6D). The recordings also revealed that the activity was apparently voltage-dependent, with a larger open probability observed at positive voltages (Supplementary Figure 6B), suggesting that the NavAb channel shows a preference for opening at positive membrane potentials. It should be noted, however, that due to random insertion into the bilayer during reconstitution, the orientation of the channels cannot be predicted, thus the polarity of channel insertion is speculative.
3.9. N-terminal tagging improves accumulation of AqpZ in E. coli and S. cerevisiae
To test whether the position of the fusion tag affects the expression level and quality of recombinant prokaryotic IMPs produced in E. coli and S. cerevisiae, we used the previously described vectors (Supplementary Figure 1) to express AqpZ fused N-terminally to His10-GFP-TEV. Data in Fig. 7 show accumulation of this fusion protein in four BL21-derived E. coli host strains. It can be seen that BL21(DE3), C41(DE3) and C43(C43) strains were able to express the N-terminally tagged AqpZ to a larger degree at 15 °C than at 30 °C. By comparing Fig. 2, Fig. 7, it is evident that relocating the GFP-His10 tag from the C- to the N-terminus of AqpZ greatly increased accumulation of the protein. The induction curves also revealed that production levels were not significantly affected by the IPTG concentration used for induction, except for the expression in the BL21(DE3) strain, where accumulation was higher after induction with 0.1 mM IPTG, compared to 0.5 and 1.0 mM (Fig. 7A).
Fig. 7.
Effect of IPTG concentration and temperature on accumulation of AqpZ N-terminally fused to His10-GFP-TEV in four E. coli host strains.E. coli strains (A) BL21(DE3), (B) BL21(DE3) pLysS, (C) C41(DE3) and (D) C43(DE3) were grown at 30 °C until OD450 = 1, and transferred to either 15 (left panels) or 30 °C (right panels) prior to induction with 0.1, 0.5 or 1 mM IPTG. Protein accumulation was measured by GFP fluorescence in the cell amount corresponding to 1 OD450 unit at each time-point after induction. Fluorescence measurements are shown as solid lines and OD450 measurements as dashed lines, respectively. Color codes for the respective concentration of IPTG used for induction of the expression are indicated below each of the bottom panels.
Subsequently, we examined whether a similar trend can be observed in S. cerevisiae. Indeed, we observed that also in yeast N-terminally tagged AqpZ accumulated to a higher membrane density as compared to the C-terminal fusion (Fig. 8). Moreover, protein accumulation was also considerably higher at 15 °C than at 30 °C.
Fig. 8.
Temperature-dependent accumulation of AqpZ N-terminally fused to His10-GFP-TEV. Yeast cultures were grown at room temperature until OD450 = 1 before half of the culture was transferred to 15 and the other half to 30 °C. After thermal equilibration, recombinant protein production was induced by addition of galactose to the final concentration of 2%. (A) AqpZ accumulation was measured by GFP fluorescence in the cell amount corresponding to 1 OD450 at each time-point after induction at 15 (blue lines) and 30 °C (red lines). (B) Live cell bio-imaging of yeast cells expressing the protein as N-terminal His10-GFP-TEV fusion. Pairwise GFP fluorescence and differential interference contrast micrographs are shown. (C) A comparison of GFP fluorescence in crude cell membranes isolated from S. cerevisiae and E. coli grown at 15 and 30 °C, respectively (see Materials and Methods section). 50-mL culture samples were taken out at each indicated time-point, and crude membranes were isolated and separated by SDS-PAGE. In-gel GFP fluorescence on all of the four SDS-PAGE gels were imaged simultaneously. Arrows indicate the AqpZ monomer.
Whole cell fluorescence does not necessarily reflect accumulation of fusion proteins in the membranes, but may also potentially result from GFP that has been proteolytically liberated from the membrane protein. Hence, using in-gel GFP fluorescence we assessed membranes isolated from E. coli and S. cerevisiae expressing either the N- or C-terminally tagged AqpZ to investigate the combined effects of tag position, temperature, expression host and induction time on protein accumulation. To unbiasedly compare the accumulation of GFP-tagged AqpZ, all gels were imaged simultaneously (Fig. 8C). In accordance with the whole cell fluorescent measurements, the N-terminally tagged AqpZ displayed a higher accumulation in the membranes at 15 °C in both expression hosts. Additionally, the accumulation levels revealed that S. cerevisiae was superior for production of AqpZ (Fig. 8C). It is noteworthy, that only full-length proteins accumulated in the membranes, as the single band in the SDS-PAGE gel corresponds to the size of GFP-tagged AqpZ and no free GFP was observed in the SDS-PAGE gel.
3.10. N-terminal position of the fusion tag improves solubilization of AqpZ from E. coli and S. cerevisiae membranes
To investigate the influence of the GFP-His10 tag position on solubilization and quality of produced recombinant AqpZ, we performed a detergent screen and FSEC analysis of the His10-GFP-TEV-AqpZ fusion. Results from the screens presented in Fig. 9A revealed a significant increase in solubilization efficiency of the N-terminally tagged AqpZ compared to the C-terminal fusion (see Fig. 4 for comparison). Specifically, the N-terminally tagged AqpZ can be extracted with 80–100% efficiency, while only 20% of the C-terminal fusion could be solubilized. Although the FSEC profiles revealed a wider elution peak in LDAO, all obtained profiles were monodisperse and devoid of aggregates (Fig. 9B).
Fig. 9.
Solubilization screen and immobilized metal affinity chromatography (IMAC) purification of AqpZ expressed in S. cerevisiae as N-terminal His10-GFP-TEV fusion. (A) Crude yeast membranes overexpressing AqpZ were solubilized for 1 h at 4 °C in 5-cyclohexyl-1-pentyl-β-D-maltoside (CY-5), n-dodecyl-β-D-maltopyranoside (DDM), n-decyl-β-D-maltopyranoside (DM), n-dodecylphosphocholine (FC-12), n-tridecylphosphocholine (FC-13) or lauryldimethylamine N-oxide (LDAO) in a protein:detergent ratio (w/w) of 1:3. Normalized solubilization efficiencies were estimated from GFP fluorescence in the supernatant after ultracentrifugation. (B) Detergent-solubilized material was separated by fluorescence-detection size-exclusion chromatography (FSEC) where GFP fluorescence as described in Materials and Methods section. The colors of the chromatograms correspond to the respective colors of detergents shown in the bar diagrams (panel A). The void volume of the used Superose 6 Increase 200 10/300 GL column is ~8 mL. (C) IMAC profile of AqpZ solubilized in 1% (w/v) LDAO for 4 h at 4 °C. Protein elution was monitored by GFP fluorescence in the collected fractions. The dashed line indicates used imidazole gradient. Insert shows the SDS-PAGE gels, where the top fraction from the elution was visualized by Coomassie blue staining (lane 1) and in-gel GFP fluorescence (lane 2).
Subsequently, we attempted affinity purification of the LDAO-solubilized N-terminally tagged AqpZ produced in yeast, using identical IMAC protocol as applied for the C-terminal construct. The chromatogram in Fig. 9C revealed that the N-terminal AqpZ fusion eluted in a single narrow peak at ~60 mM imidazole, resulting in a pure protein sample as revealed by both Coomassie blue staining and in-gel GFP fluorescence.
3.11. Rescue of expression of recombinant prokaryotic ZIPs in S. cerevisiae
With the aim of producing prokaryotic ZIPs for structural studies, we constructed a library consisting of archaeal and Gram-negative bacterial members. We selected ten prokaryotic ZIPs representing the entire ZIP family, with the two types of predicted membrane topologies shown in Supplementary Figure 2. Utilizing our selection of BL21 (DE3) based E. coli host strains we attempted to produce the zinc transporters from pET vectors with both N- and C-terminal histidine tags. However, after numerous attempts none of the ten targets were detectable in either cytoplasmic or membrane fractions of E. coli even by immunoblotting (data not shown).
Based on the improved expression of AqpZ, ClCec1, KcsA and NavAb in S. cerevisiae, we hypothesized that yeast would also have the capability to express this particularly difficult family of IMPs. Initially, we performed small-scale expression screens, where accumulation of the protein in yeast was visualized by live cell bio-imaging 48 h after induction (Fig. 10A). The S. cerevisiae-based platform enabled successful expression of all selected ZIPs, in contrast to the lack of expression in E. coli. Again, based on GFP fluorescence detected in the yeast membranes and the percentage of crude membrane protein, as described previously for other target IMPs, the expected expression levels of accumulated ZIPs ranged from 0.8 to 12.9 mg of protein per liter shake flask culture (Supplementary Table 3).
Fig. 10.
Recombinant expression of bacterial zinc transporters (Zrt/Irt-like proteins, ZIPs) in S. cerevisiae. (A) Live cell bio-imaging of yeast cells expressing ten bacterial ZIPs as C-terminal TEV-GFP-His10 fusions. Yeast cultures were grown in 2-L scale and induced for protein expression for 48 h at 15 °C. Pairwise GFP fluorescence and differential interference contrast images are shown. (B) Crude yeast membranes overexpressing ten bacterial ZIPs were solubilized for 1 hour at 4 °C in n-dodecyl-β-D-maltopyranoside (DDM) or n-hexadecylphosphocholine (FC-16) in a protein:detergent ratio (w/w) of 1:3. Normalized solubilization efficiencies were estimated from GFP fluorescence in the supernatant after ultracentrifugation. (C) and (D) Immobilized metal affinity chromatography (IMAC) purification of C- and N-terminally GFP-tagged TmZIP, respectively. IMAC profiles after solubilization in 1% (w/v) FC-16 for 1 h at 4 °C are shown. Protein elution was monitored by absorbance at 280 nm. The dashed lines indicate the used imidazole gradient. Inserts show the SDS-PAGE gels, where the top fraction from each elution was visualized by Coomassie blue staining (lane 1) and in-gel GFP fluorescence (lane 2).
Following these expression tests, we performed a detergent screen using DDM and FC-16, a mild and a harsh surfactant, respectively. Fig. 10B shows that FC-16 was most efficient in protein extraction, with a solubilization efficacy higher than 70% for several ZIPs. DDM proved to be less effective, however, for PfZIP and TmZIP the achieved extraction efficacy was comparable with that observed for C-terminally tagged AqpZ (~30%). Based on the highest target content in the crude membrane fraction and the highest solubilization efficiency (Fig. 10B and C, and Supplementary Table 3), TmZIP emerged as the most promising target for downstream purification. Hence, we performed IMAC-based purification of both C- and N-terminally GFP-His10-tagged TmZIP variants from crude S. cerevisiae membranes solubilized in FC-16 (Fig. 10C and D, respectively). In case of both constructs, bound protein eluted at an imidazole concentration of ~300 mM and yielded high purity after a single purification step, as reflected by both Coomassie blue staining and in-gel GFP fluorescence. Although both TmZIP variants were purified mainly in their monomeric forms, small fraction of the GFP-fluorescent dimer (MW of ~110 kDa with electrophoretic mobility shifted to ~80 kDa) could also be detected. In addition, irrespective of tag location, the two IMAC-pure TmZIP fusions contained a significant population of non-fluorescent species (with electrophoretic mobility of ~50 kDa). Mass spectrometry and immunoblotting of this non-fluorescent fraction suggested that it originated from TmZIP misfolding or aggregation (data not shown), as malfolded GFP migrates with a different moiety than correctly folded GFP, hence using GFP as a folding reporter (Geertsma et al., 2008). However, this fraction could be completely removed during the reverse-IMAC step following TEV protease cleavage of the N-terminal fusion (Supplementary Figure 7), greatly improving the final purity of the produced TmZIP sample. It is also evident from Supplementary Figure 7 that the electrophoretic mobility of the GFP tagged TmZIP and TmZIP liberated after TEV protease digestion are higher that what would be expected from their molecular weight. This is commonly observed for membrane proteins due to altered binding of SDS (Rath et al., 2009). The approximate difference between the GFP tagged and non-tagged TmZIP is around 15 kDa in accordance with previous observations, that correctly folded GFP only contributes 10–15 kDa to the molecular weight of a corrected folded membrane protein (Geertsma et al., 2008).
4. Discussion
In the present paper we selected prokaryotic IMPs from bacteria and archaea representing five different protein families to systematically investigate the production levels in E. coli and in S. cerevisiae. Our results demonstrate that S. cerevisiae was a more robust expression host as all selected IMPs accumulated in the yeast membranes at a higher density than in E. coli. Strikingly, switching to the S. cerevisiae-based platform proved to be the only successful experimental strategy in overproducing ZIP transporters, providing complete rescue of the expression of all tested members of this IMP family. An important observation is that all tested targets accumulated to the highest density at the same expression conditions. Time consuming optimization of expression for each target was not required.
ZIP transporters remain rather understudied, largely due to difficulties in their production. Previously, ZIPs were selected as targets for a structural genomics study (Punta et al., 2009), where out of 52 homologous targets tested, only 4 displayed detectable expression levels (Lin et al., 2010). Furthermore, the generally accepted notion that members of this IMP family are refractory to overproduction in E. coli was also confirmed by another study (Ma et al., 2015). Although E. coli remains a widely used and robust expression system for expression of prokaryotic IMPs, problems can arise when attaching a poly-histidine tag to a terminus facing the periplasm, i.e., Nout or Cout. Reportedly, difficulties with membrane insertion and folding lead to severely impaired expression compared to tag placement on a cytoplasmic terminus (Rahman et al., 2007; Seppala et al., 2010), presumably due to violation of the positive inside rule (Baker et al., 2017). This problem is inevitable when working with proteins displaying Nout-Cout topologies as in case of ZIPs. Protein engineering studies have been successful in adding auxiliary TM helices and thus flipping the N- or C-terminus intracellularly to allow more efficient tagging and expression (Quick and Wright, 2002; Hsieh et al., 2010).
None of the 10 ZIPs investigated in this study was expressible in E. coli, including PaZIP, the only target predicted to possess Nin-Cout topology (Supplementary Figure 2F). However, it cannot be excluded that the location of the N-terminus in PaZIP is predicted incorrectly, as its sequence may not harbor an additional (false-positive) TM. Here, without employing any protein engineering, we switched to our S. cerevisiae platform and successfully rescued expression of all the attempted ZIP. This demonstrates clearly the strengths of the S. cerevisiae platform and likely its superior machinery for IMPs biogenesis, at least for the studied targets.
Microbial expression systems play a vital role in producing recombinant proteins for biotechnological and medical applications in addition to basic protein science. IMPs and their interactions are mainly of interest to the biomedical industry and to basic science as targets for medical treatments and structure-function studies, respectively (Yin and Flynn, 2016). The structures of prokaryotic IMPs deposited in the PDB (Burley et al., 2019) almost exclusively originate from samples produced in E. coli, reflecting the notion that this organism is an ideal host for production of proteins of prokaryotic origin. In contrast, S. cerevisiae has been used as a workhorse for production of eukaryotic IMPs for structural biology (Bill et al., 2011; Routledge et al., 2016).
E. coli has continuously been used for historical reasons. After it was found to express some recombinant IMPs well in a relatively easy to manipulate and cultivate manner, many laboratories pursued to use this organism as their standard expression host (Schlegel et al., 2014). Another reason given for choosing E. coli as the preferred expression host for expression of prokaryotic IMPs is that it offers low cost for culturing. However, the prices of E. coli and S. cerevisiae growth media are comparable, and both expression systems are applicable for large-scale cultivation in bioreactors.
Parallel expression screening of many targets requires fast and efficient generation, and, potentially, manipulation of recombinant expression plasmids. As exemplified in the present paper, yeast as expression host also offers an option of using homologous recombination to generate the expression plasmids directly in the S. cerevisiae production strain readily and without applying expensive enzymes.
Expressing IMPs in yeast has the indisputable advantage of avoiding accumulating the protein in inclusion bodies. Although IMPs can be expressed in very high quantities in inclusion bodies and the success rate for refolding β-barrel IMPs was shown to be high (Bannwarth and Schulz, 2003), it is often inefficient or impossible to refold α-helical IMPs, properly due to the complicated folding and hydrophobic nature of this class of proteins and therefore such efforts to refold are often avoided (Lyons et al., 2016; Kiefer, 2003; Krogh et al., 2001). Secondly, the eukaryotic machinery for membrane protein biogenesis in yeast may support correct folding of heterologous membrane proteins better than the simpler prokaryotic secretory pathway of E. coli (Pechmann et al., 2013; Bohnsack and Schleiff, 2010; Cross et al., 2009; Stephenson, 2005). In addition, the rate of protein synthesis and folding is much slower in S. cerevisiae than in E. coli, correlating with their generation time, which is approximately five times slower for yeast (Widmann and Christen, 2000). Consequently, the rates of protein synthesis in eukaryotic cells have been shown to be an order of magnitude slower as compared with prokaryotic hosts (Bill, 2014). Lowering the rate of recombinant protein production in E. coli strains C41 and C43 is the well characterized reason for their success. Both strains carry mutations in the promoter responsible for transcription of the T7 RNA polymerase gene, giving rise to slower recombinant protein synthesis rates than wild-type E. coli (Kwon et al., 2015).
The role of the lipid environment surrounding embedded proteins has been the focus of intense research and debate, as lipids have been proposed to be crucial for structure and function of IMPs (Martens et al., 2016). This may result from direct interactions between specific lipids and the protein or from the physico-chemical properties of the membrane itself, such as its fluidity, thickness, intrinsic curvature or lateral pressure (Renne and de Kroon, 2018). At the post transcriptional level, successful production of a prokaryotic IMP in S. cerevisiae at least requires that the protein can be targeted to the endoplasmic reticulum, membrane inserted through the Sec61 translocon and assembled into a stable and correctly folded form (Cymer et al., 2015). In this light, the S. cerevisiae host used here permitted expression of all included targets in their native higher oligomeric forms, which includes dimerization (ClCec1, ZIPs) and tetramerization (AqpZ, KcsA and NavAb).
Based on the previous paragraphs, it may seem counterintuitive to attempt producing prokaryotic IMPs in a eukaryotic host such as S. cerevisiae, as the lipid compositions of the Gram negative, Gram positive, archaeal and the yeast plasma membranes differ considerably. Bacterial and yeast membranes are both mainly made of phospholipids esterified to glycerol-3-phosphate, while the building blocks in archaeal membranes are isoprenes connected to glycerol-1-phosphate through an ether bond (Jain et al., 2014). As seen from Table 2, Table 3, major differences in the type and content of the phospholipids include the carbon length of the fatty acids and their degree of unsaturation. Specifically, yeast membranes compared with E. coli contain a higher percentage of C16 and C18 fatty acids, in particular their mono-unsaturated versions (Klose et al., 2012; Kanonenberg et al., 2019). The longer fatty acids found in yeast membranes result in an overall membrane thickness (9 nm) that is 3 nm thicker than the E. coli membrane and corresponds to a hydrophobic width of 30 Å and 20 Å, respectively (White and Wimley, 1999). The head group composition of the phospholipids also varies significantly between the two species, as phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) constitute the great majority in E. coli, while these are much less represented in yeast. Conversely, yeast membranes include substantially higher amounts of phosphatidylinositol (PI) and phosphatidylcholine (PC). Furthermore, cardiolipin (CL) is more enriched in the E. coli membranes in contrast to sphingolipids and sterols that are solely found in yeast (Klose et al., 2012; de Kroon et al., 2013). Presence of the latter component, e.g., in a form of cholesterol, has previously been shown to preferentially interact with IMPs to restrict their dynamics (Grouleff et al., 2015) and enhance stability (Fantini and Barrantes, 2013). Thus, concerning that protein dynamics and function are intimately entwined, this can further highlight the role of endogenous sterols of the yeast membranes in enhancing stability of overproduced prokaryotic IMPs that, identically to their eukaryotic counterparts, may also be sensitive to the presence of certain lipids. Indeed, despite the extensive differences in composition of the host membranes, our experiments demonstrate that the lipid environment in yeast is not prohibitive for accumulation of correctly folded prokaryotic IMPs. Moreover, only yeast was able to produce the prokaryotic ZIP transporters and deposit them in the membranes to a high concentration. Furthermore, also the other target proteins accumulated to a higher level in yeast even though they were well produced in E. coli. Importantly, our electrophysiological characterization of the KcsA and NavAb ion channels purified from S. cerevisiae showed that they display the same functional characteristics as those produced in E. coli (LeMasurier et al., 2001; Shaya et al., 2011; Callahan and Roux, 2018) and, to our knowledge, represent the first published single channel recordings of these proteins. Based on the portfolio of prokaryotic IMPs analyzed here, our yeast expression platform emerged to be more robust than the BL21(DE3)/pET expression system. Consequently, while our yeast system was able to produce all proteins to a high level at 15 °C, expression in E. coli differed significantly among the tested strains. As example, AqpZ showed the highest accumulation in C41 and C43 strains, while BL21 was superior at expressing ClCec1, KcsA and NavAb, and none of these expressed well in BL21(DE3) pLysS. Production of the ZIP transporters was even more challenging, as they all were refractory to production in all tested E. coli strains. Based on these results, a drawback of using the BL21(DE3)/pET expression system may implicate that it is necessary to screen a number of BL21-derived strains and expression conditions to achieve a satisfactory production level, as the optimal temperature for accumulation in the E. coli strains was observed to be IMP-dependent. These factors combined render the yeast-based expression system highly attractive and competitive.
Table 2.
Fatty acid composition of the E. coli inner membrane and the S. cerevisiae plasma membrane. CX:Y: fatty acid with X carbons and Y double bonds. Cyclo: fatty acids containing a cis-9, 10-methylene group. % of molar fractions (mol %) are shown. The table combined data from (Klose et al., 2012) and (Kanonenberg et al., 2019).
| C12:0 | C14:0 | C15:0 | C16:0 | C16:1 | C17:0cyclo | C17:0 | C18:0 | C18:1 | C19:0cyclo | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (mol %) | 0 | 9 | 5 | 32 | 16 | 19 | 7 | 5 | 5 | 6 |
| S. cerevisiae (mol %) | 15 | 50 | 5 | 25 |
Table 3.
Fatty acid head groups and non-fatty acid lipids found in the E. coli inner membrane and S. cerevisiae plasma membrane. PE: phosphatidylethanolamine. PG: phosphatidylglycine. PI: phosphatidylinositol. PC: phosphatidylcholine. PS: phosphatidylserine. CL: cardiolipin. Sterols: mainly ergosterol in yeast. Spingolipids: mainly inositolphosphorylceramide, mannosyl-inositol-phosphorylceramide and mannosyl-diinositol-phosphorylceramide. % of molar fractions (mol %) are shown (Klose et al., 2012; Kanonenberg et al., 2019).
| PE | PG | PI | PC | PS | CL | Sterols | Sphingolipids | |
|---|---|---|---|---|---|---|---|---|
| E. coli (mol %) | 70–75 | 20–25 | – | – | – | 5–10 | 0 | 0 |
| S. cerevisiae (mol %) | 17 | <1 | 17 | 11 | 0.5 | 0.25 | 12 | 12 |
The popularity of E. coli as host is undoubtedly due to the large number of successful functional studies and structures solved from recombinant membrane protein samples produced in this host, but it also has historical reasons. It was discovered often to produce recombinant proteins well, to be easy and cheap to manipulate and cultivate. Associated with its short generation time, laboratories continue to use this organism as their standard expression host for prokaryotic membrane proteins (Schlegel et al., 2014). However, the prices of E. coli and S. cerevisiae growth media are practically identical and protocols for large-scale cultivation in bioreactors are well established for both expression systems. The longer generation time in yeast does, however, inevitable increase production time relative to E. coli.
The present study focused on the production of prokaryotic IMPs in the membranes of the host organism since expressing membrane proteins in yeast has the advantage of avoiding expressing the protein in inclusion bodies. Membrane proteins can be expressed in very high quantities in inclusion bodies, but while the success rate for refolding β-barrel IMPs was shown to be high (Bannwarth and Schulz, 2003), it is often inefficient or impossible to refold α-helical IMPs properly due to the complicated folding and hydrophobic nature of this class of proteins (Lyons et al., 2016; Kiefer, 2003; Krogh et al., 2001). Secondly, yeast possesses eukaryotic machinery for membrane protein biogenesis that may be more compatible with correct folding of membrane proteins than the simpler prokaryotic secretory pathway of E. coli.
So what drawbacks may potentially emerge from using yeast as expression host for prokaryotic IMPs? Expression in yeast may potentially result in non-native posttranslational modifications including N- and O-glycosylation (Conde et al., 2004; Neubert et al., 2016) that may be detrimental for the downstream biophysical applications, e.g., crystallization (Mesters and Hilgenfeld, 2007). Here, we did not assess such modifications, but the very sharp bands observed for SDS-PAGE-resolved IMAC-pure samples do not implicate heavy protein glycosylation (e.g., Fig. 5, Fig. 9, Fig. 10).
Even though AqpZ expressed better in yeast than in E. coli and we were able to purify it as the C-terminally tagged fusion, re-localizing the tag to the N-terminus not only increased the expression level in both hosts, but also resulted in a remarkably improved solubilization in all six tested detergents. This agrees with the general strategy that target expression level and solubilization efficiency need to be tested with both C- and N-terminal tags to identify the most optimal fusion construct (Mohanty and Wiener, 2004).
Based on our data and previous experience with production of the prokaryotic IMPs, we present a roadmap generalizing strategies for enhanced production of recombinant prokaryotic IMPs in S. cerevisiae (Fig. 11). Starting with molecular biology, the following should be considered: codon optimization of the prokaryotic target proteins to the yeast host, selection of tag(s) and its/their position and choice of a suitable vector for a given expression strain. Based on these decisions, the expression plasmid can be assembled from PCR products directly in S. cerevisiae by homologous recombination. Here, we successfully used a poly-histidine tag in combination with GFP for rapid screening and quantification of expression levels. Correct construct design can then easily be verified “in vivo” by assessing live cell GFP fluorescence concomitantly with sequencing of the expression construct. The following step encompasses small-scale expression trials, where screening of the most optimal expression conditions is performed applying, e.g., assessment of GFP fluorescence in the cells and SDS-PAGE analysis to confirm integrity of accumulated targets. Subsequently, upon isolation of crude yeast membranes, detergent screen has to be conducted to identify the optimal surfactant that is able to efficiently extract the protein of interest in the most stable and native form. Here FSEC-based analysis proves to be useful, as it permits rapid determination of the monodispersity of expressed GFP fusions after solubilization, an important quality check in the whole strategy. Finally, affinity chromatography can be conducted, with the overall aim to obtain a protein sample of the highest possible purity and integrity that can be assessed by, e.g., SDS-PAGE analysis (in-gel GFP fluorescence, Coomassie blue staining, immunoblotting). Finally, if a sample of satisfactory quality is produced, it can be used for the subsequent characterization, e.g., activity measurements or structural determination that may require implementation of additional purification/reconstitution steps. However, prior to structure-function studies, an additional analysis may be required to further assess the quality of produced samples. Such validation may include, e.g., SEC-multi-angle light scattering (MALS), a technique useful in analyzing the homogeneity of proteins (Some et al., 2019). If any of the stages in the above-described expression strategy do not meet the success criteria, troubleshooting can be attempted.
Fig. 11.
Workflow of recombinant production of prokaryotic integral membrane proteins (IMPs) in yeast. The chart comprises four steps critical for a successful overproduction strategy. Each step includes end-points for verification and ideas for troubleshooting; a green arrow indicates that the criteria is met, a red arrow indicates the procedure was unsuccessful and needs to be optimized further before proceeding. Molecular biology: This step should include codon optimization of the cDNA encoding the prokaryotic IMP of interest, choosing the fusion tag (here a detachable GFP-His10 fusion is used as the reporter/affinity tag) and its position, application of a vector suitable for the selected yeast expression strain and engineering the final expression plasmid using PCR and homologous recombination. Correct construct assembly should be verified by DNA sequencing and the initial expression can be confirmed by live cell bio-imaging. Unsuccessful construction of the target should be followed by starting over with PCR and homologous recombination. Small-scale expression: In this step screening of optimal induction temperature, time course and concentration of inducer in chosen expression strains should be considered. Assessment can be performed by monitoring the course of GFP accumulation, live cell bio-imaging, SDS-PAGE-based analysis employing in-gel GFP fluorescence or immunoblotting. Unsuccessful protein accumulation should be followed by optimization of growth conditions (e.g., medium, additives or growth temperature), construct redesign (e.g., removal of large cytoplasmic termini, change of fusion tag position or introduction of a linker between target and tags). Detergent screen: Ability of detergents to efficiently solubilize the yeast membranes to isolate prokaryotic target IMP in a stable form should be verified. Extraction efficacy can be estimated directly in solubilized membranes (after removal of insoluble fraction by ultracentrifugation) using GFP fluorescence. This should be followed by testing the integrity of detergent-solubilized sample by fluorescence-detection size-exclusion chromatography (FSEC) to assess its level of aggregation and monodispersity. Additionally, sample quality (e.g., preservation of its native state) can be analyzed using non-reducing SDS-PAGE. Unsuccessful solubilization of target protein should be followed by change of solubilization conditions (pH, buffering system, reducing agent, ligands, additives, lipids) or by using alternative extraction methods (e.g., styrene maleic acid co-polymers (SMAs) or direct reconstitution into nanodiscs). Affinity chromatography: A purification protocol must be established for each individual IMP target, but overall it should aim at obtainment of pure sample eluting in one main peak. Sample purity should be assessed using SDS-PAGE (Coomassie blue staining, in-gel GFP fluorescence or immunoblotting) and, if necessary, the binding conditions should be revised (e.g., pH and buffering system, concentration of salt). Following successful affinity purification, activity and stability of the protein sample should be investigated. Unsuccessful purification should be followed by optimization of binding and elution conditions (pH, buffering system, concentration of salt, type of gradient) or changing the type of affinity resin used.
5. Future perspectives
Our comparative study demonstrates that S. cerevisiae is a promising starting organism for overproduction of prokaryotic IMPs, both for well characterized and difficult to express protein classes. Although there is not so much known on how specific lipids interact with IMPs to regulate their stability and activity, the natural diversity of membrane lipid composition might be one of the important factors to consider when attempting expression of prokaryotic targets in yeast and E. coli. Further improvements of our expression platform to excel at producing challenging targets could then include fine-tuning of the native membrane environment of the S. cerevisiae strain to suit a given type of IMP, both to improve expression levels and quality of the protein.
6. Conclusions
In summary, we showed that expression of KcsA, NavAb, AqpZ and ClCec1 was achievable in both E. coli and yeast, but switching to an S. cerevisiae-based platform improved production levels. We find it particularly important that we were able to express all of the selected prokaryotic ZIP transporters in S. cerevisiae that was not possible employing E. coli, demonstrating that the yeast-based system could be useful for overproduction of other challenging families of IMPs.
Overall, there are many strengths of the expression platform described herein, as reflected by robust recombinant protein overproduction of all targets. They could all be purified to high purity by a single affinity chromatography step and for two tested ion channels activity was preserved. Hence, this suggests that the powerful S. cerevisiae-based methodology presented here can be applied to a wider range of challenging prokaryotic IMP targets for subsequent biochemical and structural analysis.
CRediT authorship contribution statement
Sarah Spruce Preisler: Writing - original draft, generated the expression constructs, performed protein accumulation screens, performed protein purification, performed protein purification, designed the project, wrote the initial version of the manuscript and the final version in association, All authors contributed with comments on the final manuscript. Anders Drabaek Wiuf: Writing - original draft, generated the expression constructs, performed protein accumulation screens, performed protein purification, performed protein purification, All authors contributed with comments on the final manuscript. Marc Friis: Writing - original draft, performed protein purification. Lasse Kjaergaard: Writing - original draft, performed protein purification. Molly Hurd: Writing - original draft, performed activity measurements. Eva Ramos Becares: Writing - original draft, performed protein purification. Casper Normann Nurup: Writing - original draft, performed detergent screens and FSEC analysis. Frederik Bühring Bjoerkskov: Writing - original draft, performed detergent screens and FSEC analysis. Zsófia Szathmáry: Writing - original draft, performed protein accumulation screens. Pontus Emanuel Gourdon: Writing - original draft, designed the project. Kirstine Calloe: Writing - original draft, performed activity measurements. Dan A. Klaerke: Writing - original draft, performed activity measurements. Kamil Gotfryd: Writing - original draft, generated the expression constructs, performed protein accumulation screens, designed the project. Per Amstrup Pedersen: Writing - original draft, generated the expression constructs, performed protein accumulation screens, designed the project.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank David Soerensen for excellent technical assistance. We are also thankful to Per Maarten Hägglund for conducting mass spectrometric analysis of the TmZIP sample. The present work was supported by the Danish Innovation Foundation through the MEMENTO grant and Danish Council for Independent Research through research project 2 (#6108-00479A).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.02.001.
Contributor Information
Kamil Gotfryd, Email: kamil@sund.ku.dk.
Per Amstrup Pedersen, Email: PAPedersen@bio.ku.dk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Baker J.A. Charged residues next to transmembrane regions revisited: "Positive-inside rule" is complemented by the "negative inside depletion/outside enrichment rule. BMC Biol. 2017;15(1):66. doi: 10.1186/s12915-017-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth M., Schulz G.E. The expression of outer membrane proteins for crystallization. Biochim. Biophys. Acta. 2003;1610(1):37–45. doi: 10.1016/s0005-2736(02)00711-3. [DOI] [PubMed] [Google Scholar]
- Bill R.M. Playing catch-up with Escherichia coli: using yeast to increase success rates in recombinant protein production experiments. Front. Microbiol. 2014;5:85. doi: 10.3389/fmicb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill R.M. Overcoming barriers to membrane protein structure determination. Nat. Biotechnol. 2011;29(4):335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- Bjorkskov F.B. Purification and functional comparison of nine human Aquaporins produced in Saccharomyces cerevisiae for the purpose of biophysical characterization. Sci. Rep. 2017;7(1):16899. doi: 10.1038/s41598-017-17095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack M.T., Schleiff E. The evolution of protein targeting and translocation systems. Biochim. Biophys. Acta. 2010;1803(10):1115–1130. doi: 10.1016/j.bbamcr.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Bomholt J. Recombinant production of human Aquaporin-1 to an exceptional high membrane density in Saccharomyces cerevisiae. PloS One. 2013;8(2) doi: 10.1371/journal.pone.0056431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan K.M., Roux B. Molecular dynamics of ion conduction through the selectivity filter of the NaVAb sodium channel. J. Phys. Chem. B. 2018;122(44):10126–10142. doi: 10.1021/acs.jpcb.8b09678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G., Murray J.A.H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow J.K., editor. Genetic Engineering: Principles and Methods Volume 9. Springer US; Boston, MA: 1987. pp. 135–154. [DOI] [Google Scholar]
- Conde R. A search for hyperglycosylation signals in yeast glycoproteins. J. Biol. Chem. 2004;279(42):43789–43798. doi: 10.1074/jbc.M406678200. [DOI] [PubMed] [Google Scholar]
- Cournia Z. Membrane protein structure, function, and dynamics: a perspective from experiments and theory. J. Membr. Biol. 2015;248(4):611–640. doi: 10.1007/s00232-015-9802-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross B.C. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009;10(4):255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Cymer F., von Heijne G., White S.H. Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 2015;427(5):999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kroon A.I., Rijken P.J., De Smet C.H. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog. Lipid Res. 2013;52(4):374–394. doi: 10.1016/j.plipres.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Dilworth M.V. Microbial expression systems for membrane proteins. Methods. 2018;147:3–39. doi: 10.1016/j.ymeth.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Dowhan W B.M., Mileykovskaya E. Functional roles of lipids in membranes. In: Vance D.E., Vance J.E., editors. Biochemistry of Lipids, Lipoproteins and Membranes. fifth ed. Elsevier; Amsterdam: 2008. [Google Scholar]
- Drew D.E. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2001;507(2):220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- Fantini J., Barrantes F. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013;4(31) doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigassner M., Pichler H., Glieder A. Tuning microbial hosts for membrane protein production. Microb. Cell Factories. 2009;8:69. doi: 10.1186/1475-2859-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsma E.R. Quality control of overexpressed membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105(15):5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2(1):1–4. doi: 10.1038/nprot.2007.17. [DOI] [PubMed] [Google Scholar]
- Gileadi O. Recombinant protein expression in E. coli : a historical perspective. Methods Mol. Biol. 2017;1586:3–10. doi: 10.1007/978-1-4939-6887-9_1. [DOI] [PubMed] [Google Scholar]
- Gong J. Understanding membrane protein drug targets in computational perspective. Curr. Drug Targets. 2019;20(5):551–564. doi: 10.2174/1389450120666181204164721. [DOI] [PubMed] [Google Scholar]
- Gotfryd K. Human adipose glycerol flux is regulated by a pH gate in AQP10. Nat. Commun. 2018;9(1):4749. doi: 10.1038/s41467-018-07176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouleff J. The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim. Biophys. Acta. 2015;1848(9):1783–1795. doi: 10.1016/j.bbamem.2015.03.029. [DOI] [PubMed] [Google Scholar]
- Hsieh J.M. Bridging the gap: a GFP-based strategy for overexpression and purification of membrane proteins with intra and extracellular C-termini. Protein Sci. 2010;19(4):868–880. doi: 10.1002/pro.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Caforio A., Driessen A.J. Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 2014;5:641. doi: 10.3389/fmicb.2014.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanonenberg K. Shaping the lipid composition of bacterial membranes for membrane protein production. Microb. Cell Factories. 2019;18(1):131. doi: 10.1186/s12934-019-1182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer H. In vitro folding of alpha-helical membrane proteins. Biochim. Biophys. Acta. 2003;1610(1):57–62. doi: 10.1016/s0005-2736(02)00717-4. [DOI] [PubMed] [Google Scholar]
- Klose C. Flexibility of a eukaryotic lipidome-insights from yeast lipidomics. PloS One. 2012;7(4) doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kwon S.K. Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci. Rep. 2015;5:16076. doi: 10.1038/srep16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227(5259):680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeMasurier M., Heginbotham L., Miller C. KcsA: it’s a potassium channel. J. Gen. Physiol. 2001;118(3):303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. Selective electrodiffusion of zinc ions in a Zrt-, Irt-like protein, ZIPB. J. Biol. Chem. 2010;285(50):39013–39020. doi: 10.1074/jbc.M110.180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J.A. Expression strategies for structural studies of eukaryotic membrane proteins. Curr. Opin. Struct. Biol. 2016;38:137–144. doi: 10.1016/j.sbi.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Ma C. A versatile strategy for production of membrane proteins with diverse topologies: application to investigation of bacterial homologues of human divalent metal ion and nucleoside transporters. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0143010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 2016;23(8):744–751. doi: 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesters J.R., Hilgenfeld R. Protein glycosylation, sweet to crystal growth? Cryst. Growth Des. 2007;7(11):2251–2253. doi: 10.1021/cg7006843. [DOI] [Google Scholar]
- Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260(3):289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Mohanty A.K., Wiener M.C. Membrane protein expression and production: effects of polyhistidine tag length and position. Protein Expr. Purif. 2004;33(2):311–325. doi: 10.1016/j.pep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Molbaek K. High yield purification of full-length functional hERG K+ channels produced in Saccharomyces cerevisiae. Microb. Cell Factories. 2015;14:15. doi: 10.1186/s12934-015-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert P. Mapping the O-mannose glycoproteome in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2016;15(4):1323–1337. doi: 10.1074/mcp.M115.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem. Cell. Biol. 2016;94(6):507–527. doi: 10.1139/bcb-2015-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S., Willmund F., Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49(3):411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen P., Rasmussen J., Jorgensen P. Expression in high yield of pig alpha 1 beta 1 Na,K-ATPase and inactive mutants D369N and D807N in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271(5):2514–2522. doi: 10.1074/jbc.271.5.2514. [DOI] [PubMed] [Google Scholar]
- Punta M. Structural genomics target selection for the New York consortium on membrane protein structure. J. Struct. Funct. Genom. 2009;10(4):255–268. doi: 10.1007/s10969-009-9071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M., Wright E.M. Employing Escherichia coli to functionally express, purify, and characterize a human transporter. Proc. Natl. Acad. Sci. U.S.A. 2002;99(13):8597–8601. doi: 10.1073/pnas.132266599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. Topology-informed strategies for the overexpression and purification of membrane proteins. Mol. Membr. Biol. 2007;24(5–6):407–418. doi: 10.1080/09687860701243998. [DOI] [PubMed] [Google Scholar]
- Rath A. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106(6):1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne M.F., de Kroon A. The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett. 2018;592(8):1330–1345. doi: 10.1002/1873-3468.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge S.J. The synthesis of recombinant membrane proteins in yeast for structural studies. Methods. 2016;95:26–37. doi: 10.1016/j.ymeth.2015.09.027. [DOI] [PubMed] [Google Scholar]
- Scharff-Poulsen P., Pedersen P.A. Saccharomyces cerevisiae-based platform for rapid production and evaluation of eukaryotic nutrient transporters and transceptors for biochemical studies and crystallography. PloS One. 2013;8(10) doi: 10.1371/journal.pone.0076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel S. Bacterial-based membrane protein production. Biochim. Biophys. Acta. 2014;1843(8):1739–1749. doi: 10.1016/j.bbamcr.2013.10.023. [DOI] [PubMed] [Google Scholar]
- Seppala S. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328(5986):1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- Shaya D. Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc. Natl. Acad. Sci. U. S. A. 2011;108(30):12313–12318. doi: 10.1073/pnas.1106811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Some D. Characterization of proteins by size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) JoVE. 2019;148 doi: 10.3791/59615. [DOI] [PubMed] [Google Scholar]
- Stephenson K. Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway (review) Mol. Membr. Biol. 2005;22(1–2):17–28. doi: 10.1080/09687860500063308. [DOI] [PubMed] [Google Scholar]
- Studier F.W. Use of bacteriophage-T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219(1):37–44. doi: 10.1016/0022-2836(91)90855-Z. [DOI] [PubMed] [Google Scholar]
- Studier F.W., Moffatt B.A. Use of bacteriophage-T7 rna-polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Structure of the human ClC-1 chloride channel. PLoS Biol. 2019;17(4) doi: 10.1371/journal.pbio.3000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Widmann M., Christen P. Comparison of folding rates of homologous prokaryotic and eukaryotic proteins. J. Biol. Chem. 2000;275(25):18619–18622. doi: 10.1074/jbc.C000156200. [DOI] [PubMed] [Google Scholar]
- Yin H., Flynn A.D. Drugging membrane protein interactions. Annu. Rev. Biomed. Eng. 2016;18:51–76. doi: 10.1146/annurev-bioeng-092115-025322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.