Abstract
Objective
To evaluate the dynamics and longevity of the humoral immune response to SARS-CoV-2 infection and assess the performance of professional use of the UK-RTC AbC-19 Rapid Test lateral flow immunoassay (LFIA) for the target condition of SARS-CoV-2 spike protein IgG antibodies.
Design
Nationwide serological study.
Setting
Northern Ireland, UK, May 2020–February 2021.
Participants
Plasma samples were collected from a diverse cohort of individuals from the general public (n=279), Northern Ireland healthcare workers (n=195), pre-pandemic blood donations and research studies (n=223) and through a convalescent plasma programme (n=183). Plasma donors (n=101) were followed with sequential samples over 11 months post-symptom onset.
Main outcome measures
SARS-CoV-2 antibody levels in plasma samples using Roche Elecsys Anti-SARS-CoV-2 IgG/IgA/IgM, Abbott SARS-CoV-2 IgG and EuroImmun IgG SARS-CoV-2 ELISA immunoassays over time. UK-RTC AbC-19 LFIA sensitivity and specificity, estimated using a three-reference standard system to establish a characterised panel of 330 positive and 488 negative SARS-CoV-2 IgG samples.
Results
We detected persistence of SARS-CoV-2 IgG antibodies for up to 10 months post-infection, across a minimum of two laboratory immunoassays. On the known positive cohort, the UK-RTC AbC-19 LFIA showed a sensitivity of 97.58% (95.28% to 98.95%) and on known negatives, showed specificity of 99.59% (98.53 % to 99.95%).
Conclusions
Through comprehensive analysis of a cohort of pre-pandemic and pandemic individuals, we show detectable levels of IgG antibodies, lasting over 46 weeks when assessed by EuroImmun ELISA, providing insight to antibody levels at later time points post-infection. We show good laboratory validation performance metrics for the AbC-19 rapid test for SARS-CoV-2 spike protein IgG antibody detection in a laboratory-based setting.
Keywords: COVID-19, molecular diagnostics, diagnostic microbiology
Strengths and limitations of this study.
This paper describes a non-clinical laboratory evaluation and comparison of the ability of three different immunoassays to detect SARS-CoV-2 antibodies in the same samples, detecting different subtypes of antibodies against different targets of the viral antigenic repertoire that does not rely on PCR positivity as definition of expected test outcome, to provide a panel of known antibody positive and antibody negative serology for evaluation of newly developed immunoassays.
This study demonstrates AbC-19 lateral flow point-of-care detection of IgG antibodies to the full trimeric spike protein of SARS-CoV-2 virus, the antibodies made in response to the vaccines used globally, in a large cohort of subjects, more than 10 months post-infection, across a broad age range (18–78 years).
This study assesses correlation between approved laboratory-based assays and the AbC-19 lateral flow point-of-care lateral flow test for the detection of SARS-CoV-2 antibodies in characterised cohorts of known positive and negative plasma samples in an evaluation conducted according to Medicines and Healthcare products Regulatory Agency guidelines during a pandemic.
Longitudinal data detecting IgG antibodies more than 10 months from infection were collected as sequential samples over time through a convalescent plasma donation programme.
This study was conducted in a standardised setting with very experienced users on plasma characterised as positive or negative for the presence of antibodies using a reference standard, alongside one other assay which may introduce a possible spectrum bias and may not reflect the true performance metrics of the assay evaluated when translated to real-life settings, using finger prick blood samples and in which pretest probability would impact greatly on positive and negative predictive values.
Introduction
The WHO declared a pandemic in March 2020 due to SARS-CoV-2, identified in late 2019 in Wuhan, China, causing COVID-19 disease.1 2
A global race ensued to develop diagnostic assays, with the most common being viral RNA detection (reverse transcription-quantitative PCR (RT-qPCR) assays), to detect acute infection.3 RT-qPCR assays are labour and reagent intensive, limited by a short temporal window for positive diagnosis, and exhibit potential for false negative results.4 Evidence suggests sensitivity of RT-qPCR can be as low as 70%.5 False positive rates between 0.8% and 4.0% have been reported in the UK and are dependent on the cycle threshold values accepted as indicating infection, the number of SARS-CoV-2 genes analysed and the proportion of asymptomatic individuals tested.6 7 Lockdown measures and ‘flattening the curve’ strategies in the UK meant many infected individuals were instructed to self-isolate and were not offered a diagnostic RT-qPCR, with much of the testing limited to patients admitted to hospital, who perhaps reflect a more severely infected cohort. Consequently, a potentially large number of cases were unconfirmed or undetected.8
The ability to accurately detect SARS-CoV-2 specific antibodies, which develop after an immune response is evoked, is vital for building biobanks of convalescent sera for treatment, monitoring immune response to infection alongside surveillance studies and assessing responses to vaccination programmes.
Commercial serology immunoassays are mostly laboratory based and measure IgG antibody levels in plasma or serum. Alternatively, lateral flow immunoassays (LFIAs) require a finger prick blood sample and can be used at point-of-care (POC) or in the home; particularly important in the context of lockdown enforcement during the pandemic. A limited number of laboratory-based chemiluminescence immunoassays are approved for use in the UK including the Roche Elecsys Anti-SARS-CoV-2 IgG/IgA/IgM against the SARS-CoV-2 Nucleocapsid antigenic region (Roche Diagnostics, Basel, Switzerland) and the Abbott SARS-CoV-2 IgG assay against the same antigenic region (Abbott Diagnostics, Abbott Park, Illinois, USA).
The complexities of the humoral immune response to SARS-CoV-2 are a much-debated topic. In a US study, approximately 1 in 16 individuals lacked detectable IgG antibodies up to 90 days post-symptom onset, despite previous RT-qPCR-confirmed infection.9 Patients who remain asymptomatic may mount a humoral immune response which is short lived, with detectable levels of antibody falling rapidly.10 This, alongside the lack of RT-qPCR test availability across the UK, has hindered the development of a well-characterised gold standard serology test for IgG antibodies to SARS-CoV-2.
Herein, we describe the use of Roche and Abbott commercial immunoassays, as well as the EuroImmun Anti-SARS-CoV-2 ELISA-IgG against the S1 domain of the spike (antigenic) protein of SARS-CoV-2 (EuroImmun UK, London, UK) to characterise pre-pandemic and pandemic COVID-19 blood samples (n=880) from within Northern Ireland and report on longevity of IgG antibodies detected. Furthermore, we follow IgG antibody levels in convalescent plasma donors (n=101 individuals) for up to 11 months. Currently, there is no gold standard assay for comparison, therefore we aimed to establish a reference based on a positive COVID-19 antibody status. We present results of a laboratory evaluation of the UK-RTC AbC-19 with a target condition of antibodies against a cohort of 330 known IgG antibody positive samples according to this ‘positive by two’ system and 488 negative samples (223 pre-pandemic assumed negative and 265 known negative) for IgG to SARS-CoV-2.
Methods
Participant samples
The flow of participant samples is summarised in online supplemental figure 1. A small cohort (n=19) of anonymised plasma samples were obtained from a partner USA laboratory for initial protocol development only. All participants provided informed consent. An online recruitment strategy was employed, with the study advertised through internal Ulster University email, website and social media. A BBC Newsline feature providing the pandemic study email address also prompted interest from the general population. The first 800 respondents who expressed interest were provided with an online patient information sheet, consent form and health questionnaire and invited to register to attend a clinic. Participants were eligible for the study if they were over 18 years of age. Exclusion criteria included anyone with a blood disorder or contraindication to giving a blood sample, or anyone currently exhibiting symptoms of COVID-19. To enrich the cohort for samples potentially positive for SARS-CoV-2 IgG antibody, further participants were invited if they had previously tested PCR positive or had the distinctive symptom of loss of taste and smell. Blood sampling clinics were held at locations around Northern Ireland between May and July 2020 resulting in collection of 263 10 mL EDTA plasma samples from 263 separate study participants. Additional anonymised plasma samples were obtained from Southern Health and Social Care Trust (SHSCT) healthcare workers (n=195) and Northern Ireland Blood Transfusion Service (NIBTS, n=184) through convalescent plasma programmes. NIBTS convalescent plasma samples continued to be collected throughout 2020–early 2021, with a total of n=897 from n=676 individuals, including n=183 samples from the cross-sectional cohort. Individuals from this programme with a positive RT-qPCR result and EuroImmun starting value >6 were sequentially sampled over a period of up to 46 weeks resulting in a cohort of n=101 individuals, n=296 samples (including n=47 individuals from the cross-sectional cohort).
bmjopen-2020-048142supp001.pdf (3.1MB, pdf)
Pre-pandemic samples (prior to June 2019, n=136) were obtained from Ulster University Ethics Committee-approved studies with ongoing consent and from NIBTS (n=200, more than 3 years old). Plasma samples were used at no more than three freeze–thaw cycles for all analyses reported within this manuscript.
Clinical information
Basic demographic information and data with regard to probable or definite prior infection with SARS-CoV-2 were obtained from PANDEMIC Study participants through the secure online questionnaire requiring responses about positive RT-qPCR result and/or time from symptom onset. Anonymised participant samples from the USA, SHSCT and NIBTS were provided with age, gender and time since PCR positive, where a previous test had been carried out.
Laboratory-based immunoassays
Details of laboratory immunoassays are summarised in online supplemental methods and online supplemental table 1.
UK-RTC AbC-19 LFIA
All analyses were performed on UK-RTC AbC-19 Technical Transfer 3 devices at Ulster University according to manufacturer’s instructions (details in online supplemental table 1). Assays were performed as cohorts, with samples in batches of 10, with one researcher adding 2.5 µL of plasma to the assay and a second adding 100 µL of buffer immediately following sample addition. After 20 min, the strength of each resulting test line was scored from 0 to 10 according to a visual score card (scored by three researchers; online supplemental figure 2). A score ≥1 was positive. Details of samples used for analysis for detection of antibodies are available in online supplemental methods.
Statistical analysis
As per Daniel,11 a minimum sample size based on prevalence can be calculated using the following formula: , where n=sample size, Z=Z statistic for a chosen level of confidence, P=estimated prevalence and d=precision. Assuming a prevalence of SARS-CoV-2 of 10% and a precision of 5%, we estimate that the required sample size at 99% confidence (Z=2.58) to be 240 individuals. If the true prevalence is lower, 5%, the estimated required sample size given a precision of 2.5% is 506 individuals. A minimum sample size of 200 known positives and 200 known negatives is given within Medicines and Healthcare products Regulatory Agency guidelines for SARS-CoV-2 LFIA antibody immunoassays.12
Statistical analysis was conducted in in R V.4.0.2.13 To assess discordance between test results, data were first filtered to include individuals with an Abbott test result in the range ≥0.25–≤1.4, with a 2×2 contingency table produced that comprised all possible combinations of (concordant|discordant) test results (within|outside) of this range. A p value was derived via a Pearson χ2 test after 2000 p value simulations via the stats package.
AbC-19 LFIA performance analyses were performed using MedCalc online (MedCalc Software, Ostend, Belgium). Receiver operating characteristic curve analysis was performed via the pROC package. To compare test result (positive|negative) with age, a binary logistic regression model was produced with test result as outcome—a p value was then derived via χ2 analysis of variance. To compare time against test result (encoded continuously), a linear regression was performed. We calculated median per time period and then converted these to log (base 2) ratios against the positivity cut-off for each assay. All plots were generated via ggplot2 or custom functions using base R.14
Results
We analysed samples from a mixed cohort of individuals from the general public (n=279), Northern Ireland healthcare workers (n=195), pre-pandemic blood donations and research studies (n=223), and through a convalescent plasma programme (n=183). Antibody levels in plasma from these 880 individuals were assessed using the three SARS-CoV-2 immunoassays: EuroImmun IgG, Roche Elecsys IgG/IgM/IgA and Abbott Architect IgG (online supplemental table 1 and online supplemental figure 3). This included a cohort of 223 pre-pandemic plasma samples collected and stored during 2017 to end of May 2019 to determine assay specificity. Of the 657 participants whose samples were collected during the pandemic, 267 (40.64%) previously tested RT-qPCR positive with a range of 7–173 days since diagnosis. A total of 225 participants gave time since self-reported COVID-19 symptoms, with a range of 5–233 days from symptom onset, while 195 had no symptom or PCR data available. Samples collected in 2020 (n=657) ranged from 19 to 78 years of age with a median (IQR) of 43 years (±22), and n=454 were women and n=200 men (n=3, not disclosed). Pre-pandemic samples (n=223) ranged from 20 to 87 years of age with median (IQR) of 50 years (±20) and consisted of n=88 women and n=135 men.
Laboratory-based antibody immunoassays
A positive result for antibody on one or more of the three laboratory immunoassays was recorded for 385 of 657 (58.6%) participants who provided a sample during the pandemic. By EuroImmun ELISA, 346 were positive, 20 borderline and 291 were negative. The Roche assay detected 380 positive and 277 negative, while Abbott determined 310 positive and 347 negative (online supplemental table 2 and online supplemental figure 3). The median age across all age groups combined was lower for participants testing positive across each of the immunoassays (median (SD) for positive vs negative, respectively: EuroImmun, 41 (13.16) vs 48 (12.95); Roche, 42 (13.08) vs 48 (13.00); Abbott, 41 (13.18) vs 47 (13.09)) (online supplemental figure 4, p<0.0001). When segregated by age group, however, differences were less apparent in certain groups (online supplemental figure 5). Excluding the pre-pandemic cohort, this gap reduced but remained statistically significant: EuroImmun, 41 (13.18) vs 45 (12.49); Roche, 42 (13.15) vs 45 (12.49); Abbott, 41 (13.26) vs 44 (12.63) (p<0.01) (median (SD) for positive vs negative). Of note, out of 267 individuals with a previous positive RT-qPCR result for SARS-CoV-2 viral RNA, 14 (5.2%, online supplemental figure 3A) did not show detectable antibodies by any of the three immunoassays, with no association found with age, gender or time between test and blood draw (data not shown).
The three commercial laboratory immunoassays provide a ratio value that increases with IgG antibody titre. When correlation between these values is assessed, good overall agreement is observed between the three immunoassays (figure 1A–C and online supplemental figure 9). As highlighted by Rosadas et al,15 we also see significant disagreement in the Abbott 0.25–1.4 range when compared with EuroImmun and Roche (figure 1; Χ2 p values: EuroImmun vs Abbott, p<0.001; Roche vs Abbott, p<0.001).
Figure 1.
Two-way correlation scatter plots comparing (A) EuroImmun, (B) Abbott and (C) Roche immunoassays. Pearson χ2 test was used to assess correlations. The results for each test were log transformed to ensure results follow a normal distribution. Negative agreement shown as blue dots, red dots show positive agreement for the two immunoassays, while black dots show disagreement and grey dots as the EuroImmun borderline results. Vertical lines mark the Abbott test range 0.25–1.4. n=880. The graphs show positive correlations between all immunoassays evaluated, with the fewest disagreement of results between the log of Roche and the log of EuroImmun. Fit lines locally estimated scatterplot smoothing (LOESS), with 95% CI shaded.
Duration of humoral response to SARS-CoV-2
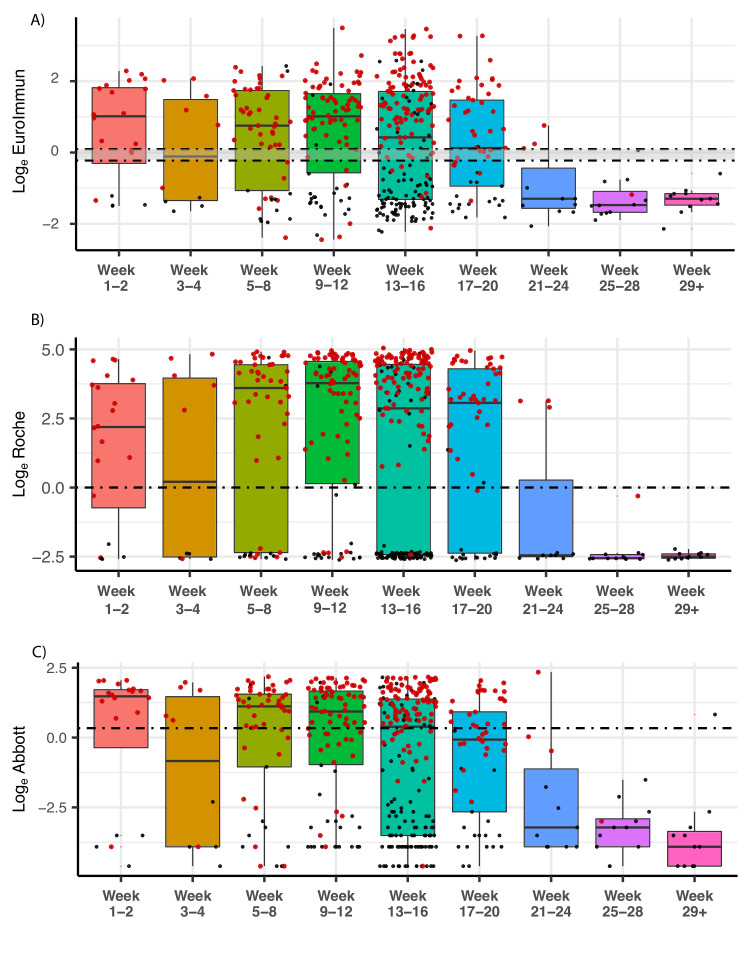
In a cross-sectional analysis of antibodies over time, we found IgG antibodies could still be detected in individuals (excluding pre-pandemic) across all three immunoassays used up to week 20 (day 140) (figure 2). We note a statistically significant decrease in signal with respect to time across each assay (p value (estimate slope)): EuroImmun, p=0.028 (−0.823); Roche, p=0.002 (−0.125); Abbott, p<0.0001 (−3.673). These remained statistically significant after adjustment for age. Antibody levels (expressed as a ratio of median result per time point divided by positivity cut-off; table 1) peaked at week 1–2 for EuroImmun (1.33) and Abbott (1.64), though reached highest levels at week 9–12 when measured by Roche (5.45). By week 21–24, median score for all tests had dropped below the positivity cut-off, though a small number of samples remained above the positive cut-off at these later time points (figure 2).
Figure 2.
SARS-CoV-2 antibody levels by (A) EuroImmun, (B) Roche and (C) Abbott, relative to weeks since first reported symptoms or positive PCR result (where data available, n=685). RT-PCR-positive individuals are denoted by red dots, while individuals with time since symptom data are denoted in black. Dashed lines delineate loge equivalent of positivity threshold (EuroImmun 1.1, Roche 1.0, Abbott 1.4) for each test, and the negativity threshold for EuroImmun (0.8; borderline result between the two lines). Black bars indicate median, within IQR boxes for EuroImmun/Roche/Abbott value. Red triangles indicate outliers, based on 1.5×IQR. RT-PCR, reverse transcription-PCR.
Table 1.
Antibody level ratios for assays over time
| Ratio median antibody level: assay positivity cut-off | ||||||||||
| Week | ||||||||||
| Pre-2020 | 1–2 | 3–4 | 5–8 | 9–12 | 13–16 | 17–20 | 21–24 | 25–28 | 29+ | |
| EuroImmun | −2.65 | 1.33 | 0.2 | 0.95 | 1.32 | 0.47 | 0.04 | −2.01 | −2.26 | −2.01 |
| Roche | −3.64 | 3.16 | 3.05 | 5.20 | 5.45 | 4.14 | 4.42 | −3.54 | −3.69 | −3.61 |
| Abbott | −5.54 | 1.64 | −0.51 | 1.12 | 0.86 | 0.08 | −0.59 | −5.13 | −5.13 | −6.13 |
| Sample number (n=) | 223 | 20 | 10 | 52 | 90 | 202 | 53 | 11 | 12 | 11 |
Antibody level ratios for assays over time show varying peak levels depending on test. Calculated by first establishing the median per time period, then calculating log2 ratio for each period versus each respective assay positivity cut-off.
Samples from the NIBTS convalescent plasma programme continued to be collected throughout 2020–early 2021. A total of n=897 samples from n=676 individuals were collected, 744 of 883 tested by EuroImmun were positive (>1.1, with values range of 0.051–34.361), 556 of 749 tested by Abbott were positive (>1.4, with values ranging from 0.01 to 8.85). Individuals with a positive RT-qPCR result and a EuroImmun result >6 were sequentially sampled (with median 3, range 2–9 samples per individual), and analysed by both EuroImmun (n=101 individuals) and Abbott immunoassays (n=75 individuals). Median age (IQR) for this cohort is 51 years (±21) with a range from 18 to 70 years and n=27 women, n=74 men. Longitudinal analysis shows persistence of detectable IgG antibodies until up to 302 days (43 weeks) by Abbott immunoassay (at which point this assay was discontinued at NIBTS) and 323 days (46 weeks) by EuroImmun ELISA, with a gradual decline over time (online supplemental figure 6). None of the individuals who were initially positive by EuroImmun SARS-CoV-2 S1 IgG assay dropped to below the EuroImmun positivity threshold (>1.1) over the course of the follow-up, while 26 who were initially positive by Abbott SARS-CoV-2 NP IgG fell below the Abbott threshold (>1.4).
UK-RTC AbC-19
Using the commercial immunoassays described, we established a well-characterised serology sample set of ‘known positive’ and ‘known negative’ for IgG antibodies to SARS-CoV-2 to evaluate performance metrics for the UK-RTC AbC-19 Rapid LFIA.
AbC-19 detects IgG antibodies against the spike protein antigen, so we therefore required all samples to be positive by the EuroImmun SARS-CoV-2 IgG ELISA, which likewise detects antibodies against the S1 domain.16 To develop this characterised cohort, samples were also required to be positive by a second immunoassay (Roche or Abbott). To analyse specificity of the AbC-19 LFIA for detection of SARS-CoV-2 IgG antibody, we assessed 350 plasma samples from participants classed as ‘known negative for SARS-CoV-2 IgG antibody’ on the AbC-19 LFIA. All samples were from individuals confirmed to be negative across all three laboratory assays (Roche, EuroImmun, Abbott). Using these positive n=304 and negative n=350 antibody cohorts, we determined a sensitivity for detecting SARS-CoV-2 IgG antibody of 97.70% (95% CI: 95.31% to 99.07%) and specificity of 100% (98.95% to 100.00%) for the AbC-19 LFIA (table 2).
Table 2.
UK-RTC AbC-19 LFIA performance metrics against known antibody positive and known antibody negative cohorts
| Total negative | True negative | False positive | Total positive | True positive | False negative | Sensitivity % (95 CI) |
Specificity % (95 CI) |
| Pre-pandemic (n=223) | |||||||
| 223 | 222 | 1 | n/a | n/a | n/a | n/a | 99.55 (97.53 to 99.99) |
| Initially reported cohorts (n=654) | |||||||
| 350 | 350 | 0 | 304 | 297 | 7 | 97.70 (95.31 to 99.07) |
100.00 (98.95 to 100.00) |
| Extended cohorts (n=818) | |||||||
| 488 | 486 | 2 | 330 | 322 | 8 | 97.58 (95.28 to 98.95) |
99.59 (98.53 to 99.95) |
LFIA, lateral flow immunoassay; n/a, not available.
Given a recent report of lower specificity in the AbC-19 LFIA17 and the possibility of introducing sample bias, we revised our inclusion criteria for the negative cohort. For the pre-pandemic cohort, we included samples from all 223 individuals, regardless of results on other laboratory immunoassays. When this assumed negative pre-pandemic cohort was used for laboratory evaluation for target condition of antibodies, we observed a specificity of 99.55% (97.53% to 99.99%, table 2). We expanded the negative cohort to include all samples that matched our criteria (samples collected during the pandemic to be negative by all three laboratory assays and all pre-pandemic samples regardless of other immunoassay results). The specificity observed on this extended negative cohort of 488 samples was 99.59% (98.53% to 99.95%, table 2). For sensitivity analysis on a positive cohort (samples positive by EuroImmun and one other test), we were able to analyse all samples previously untested due to limited testing capacity and tested a positive cohort of 330 samples giving a sensitivity of 97.58% (95.28% to 98.95%, table 2). When we sorted samples analysed in both negative (n=488) and positive cohorts (n=330) by RT-qPCR status and assessed AbC-19 LFIA sensitivity by including only those that were RT-qPCR positive (n=227), the test showed a sensitivity of 92.07% (87.76% to 95.23%; online supplemental table 3 and online supplemental figure 3B). However, of the n=18 RT-qPCR-positive individuals negative for IgG antibodies by AbC-19, n=12 showed no detectable antibodies by all three laboratory assays (EuroImmun, Roche or Abbott), suggesting that antibodies are not present in those samples (online supplemental figure 3C).
When used as intended by the public, the AbC-19 LFIA provides binary positive/negative results. However, when assessing LFIA in the laboratory, each test line was scored against a scorecard by three independent researchers (0 negative, 1–10 positive; online supplemental figure 2). When compared with quantitative outputs from the Abbott, EuroImmun and Roche assays, the AbC-19 LFIA shows good correlation (Abbott r=0.84 (p<0.001); EuroImmun r=0.86 (p<0.001); Roche r=0.82 (p<0.001); online supplemental figures 3 and 7–9).
Analytical specificity and sensitivity of AbC-19 LFIA
We observed no cross-reactivity across samples with known H5N1 influenza, respiratory syncytial virus, influenza A, influenza B, Bordetella pertussis, Haemophilus influenzae, seasonal coronavirus NL63 and 229E on the AbC-19 LFIA (n=34 samples, n=8 distinct respiratory viruses; online supplemental table 4). Against a panel of external reference SARS-CoV-2 serology samples, the AbC-19 LFIA detected antibodies with scores commensurate to the EuroImmun ELISA scores (online supplemental figure 10 and online supplemental table 5).
Discussion
Serological antibody immunoassays are an important tool in helping combat the SARS-CoV-2 pandemic. The duration of the humoral immune response is of particular importance, to inform an individual’s protection following both natural infection and vaccination. Using a large cohort of individuals across a wide age range (18–78 years), we assessed antibody levels across up to three laboratory immunoassays and perform a cross-sectional and longitudinal analysis over time. Our results show strong correlation between all three immunoassays, with shortcomings in the Abbott system output 0.25–1.4 range, as described previously, suggesting an overestimated positive cut-off (figure 1).15
Longitudinal studies on SARS-CoV-1 convalescent patients suggest that detectable IgG can still be present as long as 2 years after infection.18 There are conflicting reports of the longevity of the humoral response to SARS-CoV-2 infection which differ in the make-up of the cohort studied, the assays used and the length of time since symptom onset. The longevity of IgG antibodies to both spike and nucleocapsid protein more than 10 months after RT-qPCR-positive status (and beyond in a small number of samples (figure 2 and online supplemental figure 6) is consistent with that observed in other recent studies.19–21 In this study, samples were collected through a convalescent plasma programme (online supplemental figure 6), with individuals selected for sequential plasma donation based on an initial high EuroImmun assay score. In contrast to the time series analysis of healthcare workers recruited prospectively by Manisty et al,22 we observed no cases where EuroImmun ELISA-measured anti-spike antibody levels fell below threshold, while a large number of Abbott measured anti-nucleocapsid antibody levels dropped below the positivity threshold (34.7%, 26 of 75). However, this may be an overestimate given the shortcomings of the Abbott assay described above (figure 1). In a similar longitudinal study of 51 symptomatic participants, Dan et al estimated that half-life (t1/2) for IgG-spike (103 days) was longer than that for IgG-nucleocapsid (68 days), although with a considerable overlap of 95% CIs.23
In our more diverse cross-sectional cohort, we also note a statistically significant decline over time but levels remain detectable at 140 days (figure 2). We note that IgG levels reach their peak (Roche ratio 5.45 times threshold cut-off) as late as week 9–12 from first symptoms or a viral RNA RT-qPCR positive result, though this may be an artefact of lower number of participants at earlier time points (table 1). Robust antibody responses are produced in our cohorts across a wide age range (18–78 years old, figure 2 and online supplemental figure 6). We detect a slightly but significantly lower median age of participants testing positive (online supplemental figure 4); however, this is likely be due to cohort characteristics and not a true reflection of the population or indication of test performance.
A difficulty faced in validation of antibody diagnostic assays has been access to samples with known SARS-CoV-2 antibody status. As previously described, there is no clear gold standard reference against which to assess SARS-CoV-2 immunoassays. A positive RT-qPCR test has been used previously to indicate previous (COVID-19) SARS-CoV-2 infection, though this approach is limited by a high rate of false negatives and positives in RT-qPCR testing, failure in some cases to develop IgG antibodies (sero-silence or lack of antibody against the same antigenic component of the virus as the immunoassay used as a capture antigen) and the lack of RT-qPCR testing availability early in the pandemic.3 5 24 SARS-CoV-2 IgG antibodies were undetectable in 14 of 267 (5.2%) of previously RT-qPCR SARS-CoV-2 viral RNA-positive participants in this study. It is unclear if this is due to insufficient/absent antibody production in these individuals at the time the sample was taken, or due to a false positive PCR result which may occur in the UK at a rate between 0.8% and 4.0%.6 Self-assessment of symptoms for COVID-19 (disease) is a poor indicator of previous infection, even among healthcare workers.25 Additionally, the kinetics of a SARS-CoV-2 virus infection contributes to the loss of sensitivity of RT-qPCR to detect virus with time, contributing to false negative RT-qPCR test results for individuals who may be late to present for virus detection tests.5 26
To assess sensitivity and specificity of the AbC-19 LFIA for its ability to detect SARS-CoV-2 antibody in a laboratory evaluation, we developed a reference standard for SARS-CoV-2 antibodies, which does not rely on a single test as reference. A similar approach was used in a recent seroprevalence study in Iceland, whereby two positive antibody results were required to determine a participant sample as positive for SARS-CoV-2 antibody.24 Our evaluation of performance metrics for the UK-RTC AbC-19 LFIA to detect antibodies for SARS-CoV-2 gave 97.58% sensitivity and 99.59% specificity. In an evaluation of the AbC-19 tests, Mulchandani et al observed a specificity of 97.9% (97.2% to 98.4%) in a cohort of pre-pandemic samples and report a sensitivity of 92.5% (88.8% to 95.1%) for detecting previous infections (based on a previous RT-qPCR result) or 84.7% (80.6% to 88.1%) against the Roche Elecsys antibody test, which detects IgM/IgG/IgA SARS-CoV-2 antibodies to the nucleocapsid portion of SARS-CoV-2.25 In RT-qPCR-positive individuals from our cohorts, the AbC-19 test showed a similar sensitivity of 92.07% (87.76% to 95.23%, online supplemental figure 3, online supplemental table 3). However, we demonstrate the drawbacks of this approach given that in 12 of 18 AbC-19 false negatives, none of the four immunoassays used (EuroImmun, Roche, Abbott or AbC-19) detected antibodies, suggesting either a false RT-qPCR result, a failure to produce IgG antibodies or sero-reversion before sample collection in these individuals. Another recent evaluation of the AbC-19 LFIA by Moshe et al27 determined a sensitivity of 100% (98.1% to 100%) on laboratory sera, using a composite reference standard of antibody positive by either spike protein ELISA or hybrid double antigen binding assay (DABA) and specificity of 99.8% (98.9% to 100%) against pre-pandemic samples. However, when AbC-19 performance was analysed on matched finger prick and serum samples against the same antibody standard, a lower sensitivity was observed (finger prick 69% (53.8% to 81.3%), serum 92% (80% to 97.7%)).
In our study, strong correlation was observed in quantitative score between results on all immunoassays with the highest observed between EuroImmun and AbC-19 LFIA (online supplemental figures 8 and 9). This is to be expected, given both the AbC-19 LFIA and EuroImmun ELISA detect IgG antibodies against spike protein. Importantly, for the assessment of immunity to prior natural infection as well as to immunisation, IgG antibodies against SARS-CoV-2 spike protein detected by laboratory-based EuroImmun ELISA and AbC-19 LFIA are known to correlate with neutralising antibodies, which may confer future immunity.23 28 29 Previous evaluations of sensitivity and specificity reported by Public Health England (PHE) showed a EuroImmun sensitivity of 72% and specificity of 99%, Abbott with sensitivity of 92.7% and specificity of 100%, and Roche with sensitivity of 83.9% and specificity of 100%.30–32 The PHE analyses for each of these tests used previous infection (RT-qPCR-positive status) as a reference standard, the limitations of which are discussed above.
In the use of characterised ‘known positive’ and ‘known negative’ cohorts, one limitation of this study is its potential for spectrum bias, whereby our positive by two reference system may artificially raise the threshold for positive sample inclusion, possibly resulting in the overestimation of the sensitivity of any test evaluated.33 However, similar issues have been raised when using previous RT-qPCR result or definitive COVID-19 symptoms as inclusion criteria given these will likely skew a cohort towards more severe disease, especially given issues of RT-qPCR availability outside of hospital settings during the first wave.5 Importantly, our mixed origin of samples forming the cohort provides a positive cohort for assessing assay sensitivity that includes individuals from the general public, healthcare workers and from convalescent plasma programmes. In the absence of a clear gold standard test, our system relies on no single test (each with their individual shortcomings) and instead takes an average of three. Our analysis of specificity on only pre-pandemic individuals (n=223) shows similar specificity (99.55%) to the larger mixed ‘known negative cohort’ (n=488, sensitivity 99.59%). We also demonstrate a high level of analytical specificity of the AbC-19 test with no cross-reactivity against a panel of other respiratory viruses, including SARS-CoV-1 NL63 and 229E (online supplemental table 4).
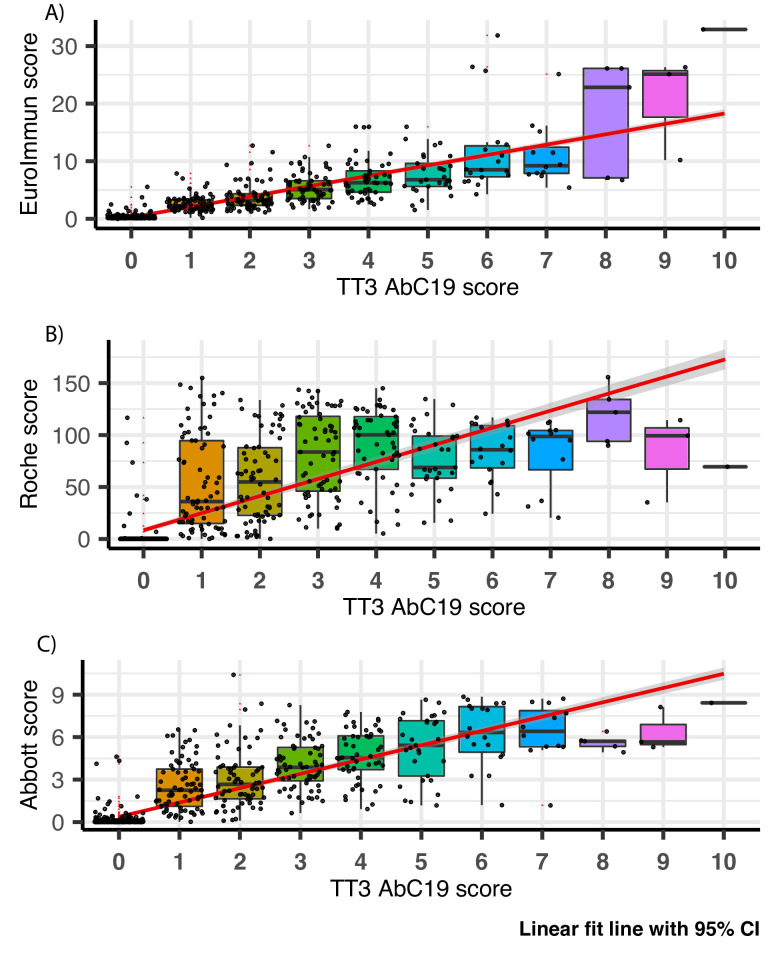
Our assessment of the AbC-19 LFIA in a laboratory setting, using characterised cohorts of known SARS-CoV-2 antibody positive and antibody negative plasma, shows good performance metrics for its ability to detect SARS-CoV-2 IgG antibodies following natural infection. We note our use of plasma from venous blood samples, as opposed to a finger prick blood sample as would be used in rapid testing scenarios.27 Additionally, when the AbC-19 LFIA was used on our cohort, a number of the positive results scored low (1 of 10 using the scorecard under laboratory conditions, figure 3), with a faint test band visible to a trained laboratory scientist but perhaps difficult to identify as positive by individuals performing a single test (online supplemental figure 10). This faint line may be reflective of the longer time from infection for the Northern Ireland cohort used. If the AbC-19 LFIA is to be used in clinical settings, it is important to determine if all users observe the same results as observed in this laboratory evaluation.
Figure 3.
AbC-19 extended cohort (n=818) correlation to (A) EuroImmun, (B) Roche and (C) Abbott scores. Box plots overlaid on scatter plot, comparing AbC-19 TT3 test scores with EuroImmun, Roche and Abbott quantitative antibody values. Red linear line of best fit with 95% CI shaded in grey. Black bars indicate median, within IQR boxes for EuroImmun/Roche/Abbott value. Red triangles indicate outliers, based on 1.5×IQR. TT3, Technical Transfer 3.
This assessment of the AbC-19 LFIA does not provide data on how this test will perform in a seroprevalence screening scenario, but instead provides metrics for the performance of the test, where presence of SARS-CoV-2 antibodies is of interest, as opposed to previous COVID-19 infection. An important potential use of the AbC-19 LFIA would be in monitoring the immune response to vaccination, with most vaccines using SARS-CoV-2 spike protein antigens.34
Conclusion
We present a comprehensive analysis of pre-pandemic and two large pandemic cohorts (more than 700 individuals) and in a longitudinal analysis, show that IgG antibodies to SARS-CoV-2 antigens are detectable more than 10 months from positive RT-qPCR test. We use antibody positive status as an alternative to RT-PCR-positive status as a standard for assessing SARS-CoV-2 antibody assays and show strong performance for the UK-RTC AbC-19 LFIA rapid POC test in detecting SARS-CoV-2 antibodies. User experience in future studies in the real world is important and may alter the performance characteristics. Also, the effect of operator training will have direct effects on test performance. We welcome further clinical evaluation of the AbC-19 LFIA in large cohorts of symptomatic and asymptomatic individuals alongside large studies assessing vaccination outcomes in individuals to fully validate its implementation across all intended use cases.
Supplementary Material
Acknowledgments
We are extremely grateful to all the people of Northern Ireland who took part in this study and gave blood during the pandemic. We are indebted to the phlebotomists: Geraldine Horrigan and Pamela Taylor who conducted the blood draws while ensuring the highest possible level of safety to the participants. We are also grateful to Kingsbridge Private Hospital Group for sponsorship and providing everything needed for blood collection including the clinical rooms. We acknowledge Dr Tony Byrne for use of his laboratory and Professor Gareth Davison for laboratory space and equipment during the pandemic within a locked down university.
Footnotes
Contributors: TM and JADM conceived the study. LJR, JSM and TM performed all laboratory analyses. LJR, SEM and KYN analysed data. KB performed all statistical analyses/interpretations and produced figures. NQ, FJ, GW and PS performed all Roche analyses and provided SHSCT cohort samples. MC, KM and SR performed all Abbott analyses and provided blood transfusion cohort samples. TM, RKP and MAN coordinated participant recruitment, consent and sampling. WPB and JADM developed online consent forms, questionnaires and databases. LJR, JSM, AMK, AA, GEW, DH, SS and CCS performed sample collection and processing. LJR and TM wrote the manuscript, with significant contributions from JSM, JADM, MAN and KB. All authors reviewed and approved the final manuscript.
Funding: Costs for assays and laboratory expenses only will be paid by UK-RTC as is normal practice (UU-UK-RTC-2020-001). The authors have not been paid or financially benefited from this study. The advisory roles within CIGA Healthcare were unpaid temporary roles. This manuscript and associated data within this paper have only been used to build confidence into the overall device design and performance assessment of the UK RTC AbC-19 devices and such work was never commissioned for any government contractual consideration.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request to the corresponding author.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All study participants provided informed consent. This study was approved by Ulster University Institutional Ethics Committee (REC/20/0043), South Birmingham REC (The PANDEMIC Study IRAS Project ID: 286041Ref 20/WM/0184) and adhered to the Declaration of Helsinki and Good Clinical Practice.
References
- 1.World Health Organisation . Rolling updates on coronavirus disease (COVID-19), 2020. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen
- 2.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet 2020;395:1101–2. 10.1016/S0140-6736(20)30788-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winichakoon P, Chaiwarith R, Liwsrisakun C, et al. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J Clin Microbiol 2020;58. 10.1128/JCM.00297-20. [Epub ahead of print: 23 04 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson J, Richter A, Deeks J. Testing for SARS-CoV-2 antibodies. BMJ 2020;370:m3325. 10.1136/bmj.m3325 [DOI] [PubMed] [Google Scholar]
- 6.Surkova E, Nikolayevskyy V, Drobniewski F. False-positive COVID-19 results: hidden problems and costs. Lancet Respir Med 2020;8:1167–8. 10.1016/S2213-2600(20)30453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omata M, Hirotsu Y, Sugiura H, et al. The dynamic change of antibody index against Covid-19 is a powerful diagnostic tool for the early phase of the infection and salvage PCR assay errors. J Microbiol Immunol Infect 2021. 10.1016/j.jmii.2020.12.009. [Epub ahead of print: 05 Jan 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black JRM, Bailey C, Przewrocka J, et al. COVID-19: the case for health-care worker screening to prevent Hospital transmission. Lancet 2020;395:1418–20. 10.1016/S0140-6736(20)30917-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen LR, Sami S, Vuong N, et al. Lack of antibodies to SARS-CoV-2 in a large cohort of previously infected persons. Clin Infect Dis 2020:ciaa1685. 10.1093/cid/ciaa1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long Q-X, Tang X-J, Shi Q-L, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020;26:1200–4. 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- 11.Daniel WW. Biostatistics : a foundation for analysis in the health sciences. 7th Edition. New York: John Wiley & Sons, Ltd, 1999: 720. [Google Scholar]
- 12.Medicines and Healthcare product Regulatory Agency . Target product profile: antibody tests to help determine if people have immunity to SARS-CoV-2, 2020. Available: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/881162/Target_Product_Profile_antibody_tests_to_help_determine_if_people_have_immunity_to_SARS-CoV-2_Version_2.pdf
- 13.The R Development Core Team . R: a language and environment for statistical computing. ISBN 3-900051-07-0 2017.
- 14.Wickham H. ggplot2 elegant graphics for data analysis. Berlin: Springer, 2016. [Google Scholar]
- 15.Rosadas C, Randell P, Khan M, et al. Testing for responses to the wrong SARS-CoV-2 antigen? Lancet 2020;396:e23. 10.1016/S0140-6736(20)31830-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UK-RTC and Abingdon Health . Charting the course to a post-COVID world. York, United Kingdom: Abingdon Health; 2020. https://www.abingdonhealth.com/guides/abc-19tm-rapid-test-white-paper/ [Accessed Nov 2020]. [Google Scholar]
- 17.Mulchandani R, Jones HE, Taylor-Phillips S, et al. Accuracy of UK Rapid Test Consortium (UK-RTC) "AbC-19 Rapid Test" for detection of previous SARS-CoV-2 infection in key workers: test accuracy study. BMJ 2020;371:m4262. 10.1136/bmj.m4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L-P, Wang N-C, Chang Y-H, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis 2007;13:1562–4. 10.3201/eid1310.070576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanshylla K, Di Cristanziano V, Kleipass F, et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe 2021. 10.1016/j.chom.2021.04.015. [Epub ahead of print: 03 May 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen MS, Hansen CB, Kristiansen MF. SARS-CoV-2 natural antibody response persists up to 12 months in a nationwide study from the Faroe Islands. medRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Yu D, Wu X. Twelve-month specific IgG response to SARS-CoV-2 receptor-binding domain among COVID-19 convalescent plasma donors in Wuhan. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manisty C, Treibel TA, Jensen M. Characterising heterogeneity and sero-reversion in antibody responses to mild SARS⍰CoV-2 infection: a cohort study using time series analysis and mechanistic modelling. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021;371:eabf4063. 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020;383:1724–34. 10.1056/NEJMoa2026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulchandani R, Taylor-Phillips S, Jones H. Self assessment overestimates historical COVID-19 disease relative to sensitive serological assays: cross sectional study in UK key workers. medRxiv 2020. [Google Scholar]
- 26.Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol 2020;5:eabe0367. 10.1126/sciimmunol.abe0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moshe M, Daunt A, Flower B, et al. SARS-CoV-2 lateral flow assays for possible use in national covid-19 seroprevalence surveys (react 2): diagnostic accuracy study. BMJ 2021;372:n423. 10.1136/bmj.n423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer AS, Jones FK, Nodoushani A, et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv 2020. 10.1101/2020.07.18.20155374. [Epub ahead of print: 20 Jul 2020]. [DOI] [Google Scholar]
- 29.Addetia A, Crawford KHD, Dingens A, et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol 2020;58. 10.1128/JCM.02107-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Public Health England . Evaluation of the Abbott SARS-CoV-2 IgG for the detection of anti-SARSCoV-2 antibodies, 2020. Available: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/890566/Evaluation_of_Abbott_SARS_CoV_2_IgG_PHE.pdf
- 31.Public Health England . Evaluation of the Euroimmun Anti-SARS-CoV-2 ELISA (IgG) serology assay for the detection of anti-SARS-CoV-2 antibodies, 2020. Available: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/893433/Evaluation_of_Euroimmun_SARS_CoV_2_ELISA_IgG__1_.pdf [DOI] [PMC free article] [PubMed]
- 32.Public Health England . Evaluation of Roche Elecsys AntiSARS-CoV-2 serology assay for the detection of anti-SARS-CoV-2 antibodies. London, United Kingdom: Public Health England; 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/891598/Evaluation_of_Roche_Elecsys_anti_SARS_CoV_2_PHE_200610_v8.1_FINAL.pdf [Accessed Nov 2020]. [Google Scholar]
- 33.Hall MK, Kea B, Wang R. Recognising bias in studies of diagnostic tests part 1: patient selection. Emerg Med J 2019;36:431–4. 10.1136/emermed-2019-208446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeyanathan M, Afkhami S, Smaill F, et al. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol 2020;20:615–32. 10.1038/s41577-020-00434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-048142supp001.pdf (3.1MB, pdf)
Data Availability Statement
Data are available on reasonable request to the corresponding author.