Abstract
Aims/hypothesis
The incidence of type 1 diabetes is increasing more rapidly than can be explained by genetic drift. Viruses may play an important role in the disease, as they seem to activate the 2′-5′-linked oligoadenylate (2′-5′A) pathway of the innate antiviral immune system. Our aim was to investigate this possibility.
Methods
Innate antiviral immune pathways were searched for type 1 diabetes-associated polymorphisms using genome-wide association study data. SNPs within ±250kb flanking regions of the transcription start site of 64 genes were examined. These pathways were also investigated for type 1 diabetes-associated RNA expression profiles using laser-dissected islets from two to five tissue sections per donor from the Diabetes Virus Detection (DiViD) study and the network of Pancreatic Organ Donors (nPOD).
Results
We found 27 novel SNPs in genes nominally associated with type 1 diabetes. Three of those SNPs were located upstream of the 2′-5′A pathway, namely SNP rs4767000 (p = 1.03 × 10−9, OR 1.123), rs1034687 (p = 2.16 × 10−7, OR 0.869) and rs739744 (p = 1.03 × 10−9, OR 1.123). We also identified a large group of dysregulated islet genes in relation to type 1 diabetes, of which two were novel. The most aberrant genes were a group of IFN-stimulated genes. Of those, the following distinct pathways were targeted by the dysregulation (compared with the non-diabetic control group): OAS1 increased by 111% (p < 1.00 × 10−4, 95% CI −0.43, −0.15); MX1 increased by 142% (p < 1.00 × 10−4, 95% CI −0.52, −0.22); and ISG15 increased by 197% (p = 2.00 × 10−4, 95% CI −0.68, −0.18).
Conclusions/interpretation
We identified a genetic predisposition in the 2′-5′A pathway that potentially contributes to dysregulation of the innate antiviral immune system in type 1 diabetes. This study describes a potential role for the 2′-5′A pathway and other components of the innate antiviral immune system in beta cell autoimmunity.
Graphical abstract
Supplementary Information
The online version contains peer-reviewed but unedited supplementary material available at 10.1007/s00125-021-05469-5.
Keywords: 2′-5′A pathway, Interferon α, 2′-5′ Oligoadenylate synthetase, Ribonuclease L, RNase L, Toll-like receptor 7, Type 1 diabetes, Type 1 interferon, Type 2 diabetes, Virus
Introduction
Type 1 diabetes is characterised by autoimmune T cell mediated destruction of pancreatic beta cells. Environmental factors may contribute to damage of beta cells and, particularly, viruses have been suspected to be implicated [1]. During a viral infection, the expression of type 1 IFN among the islet cells seems specific to beta cells [2]. Supporting the beta cell susceptibility hypothesis, it has been shown that treating alpha and beta TC3 cell lines with IFN-α and/or polyinosinic:polycytidylic acid [poly (I:C)], mimicking a viral infection, results in a six times higher 2′-5′ oligoadenylate synthetase (OAS) response in beta cells compared with alpha cells [3]. Why beta cells are more vulnerable than alpha cells to viral infection may be due to nitric oxide toxicity, antigen mimicry and/or possibly dysregulation of IFN-stimulated genes (ISGs) such as those in the 2′-5′-linked oligoadenylate (2′-5′A) pathway. The cellular tolerance to infection is known to be regulated by the innate antiviral immune response and varies among tissues and cell types [4]. This is especially problematic for beta cells due to their high metabolic activity resulting in vulnerability (e.g. during a viral infection).
In individuals with type 1 diabetes, the expression of IFN-α is increased in the islets of Langerhans and peripheral blood. Inflammation mediated by IFN signalling can potentially drive programmed cell death in beta cells and initiate autoreactivity against beta cell antigens [5]. Type 1 IFNs display differential tissue expression and binding affinity to their receptor complexes, which give rise to various outcomes including inflammation and antiviral, antiproliferative and immunomodulatory activity. Thus, type 1 IFNs and ISGs may be the link between genetics, the immune system, and environmental factors that have been shown to contribute to autoimmunity in type 1 diabetes [6].
Stimulation of pathogen-recognition receptors (PRRs) by pathogen-associated molecular patterns (PAMPs) induce the production of proinflammatory cytokines and type I IFN, which leads to transcription of ISGs. Some of the well-described ISGs include ISG15, MX1, MX2, OAS1, OAS2, OAS3, OASL and RNASEL. OAS/RNase L, IFN-stimulated protein of 15 kDa (ISG15), and Mx dynamin-like GTPase pathways show varying responsiveness to type 1 IFNs [7]. The 2′-5′A pathway, consisting of OAS proteins and the RNase L enzyme, is an RNA degradation pathway, which is induced by type 1 IFNs and activated by double-stranded RNA (dsRNA), an intermediate product of viral protein synthesis that is present in the cytosol of the cell. The activation of OAS by dsRNA results in polymerisation of ATP into 2′-5′A molecules. These 2′-5′A molecules activate latent RNase L, resulting in cleavage of single-stranded RNA (ssRNA) from viruses. However, RNase L can also inhibit protein synthesis of any other virus by cleaving host mRNA/ribosomal RNA, inhibiting translation. RNA cleaved by RNase L can then activate other cytoplasmic PRRs, resulting in further induction of type 1 IFN gene expression [8] and damage to the beta cell from within.
This study uses data from the genome-wide association study (GWAS) and islets transcriptomics from the Diabetes Virus Detection (DiViD) and the network of Pancreatic Organ Donors (nPOD) studies to investigate genetic variations and expression patterns in genes associated with innate antiviral immune pathways in relation to type 1 diabetes.
Methods
Tissue collection
Pancreas biopsies (n=5) were collected in the DiViD study as described previously [9]. Individuals with new-onset type 1 diabetes, 25–35 years of age, underwent a surgical minimal pancreatic tail resection by laparoscopy 3–9 weeks after diagnosis. The DiViD study was approved by the Norwegian Government’s Regional Ethics Committee (reference 2009/1907). Tissue from donors who were autoantibody-positive (n=12), had longstanding type 1 diabetes (n=20), had longstanding type 2 diabetes (n=8), or who were non-diabetic (control group; n=18) was used for RNA analysis and was acquired from nPOD with approval from the University of Tennessee Health Science Center local Institutional Review Board (reference 10-00848-XM).
RNA analysis
RNA analysis was conducted on snap-frozen tissue obtained from the DiViD [9] and nPOD [10] tissue collections. Optimal cutting temperature embedded tissue slides were subjected to laser capture of islets conducted as previously described [11]. In brief, all islets in two to five tissue sections from each donor were captured, pooled, and RNA extracted using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, Grand Island, NY, USA). Quality and quantity of RNA was determined on a Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA, USA); RNA integrity numbers are shown in electronic supplementary material (ESM) Table 1. Samples with sufficient quantity and quality of RNA were then subjected to gene expression analysis using Affymetrix expression arrays (GeneChip Human Gene 2.0 ST; Thermo Fisher) and global scaling as normalising method as previously described [12].
GWAS analysis
GWAS analysis included 64 genes that were researched for genetic associations with type 1 diabetes. Immunochip SNPs for type 1 diabetes were retrieved from Onengut-Gumuscu et al [13]. A cut-off p value <0.01 was used to retrieve nominally significant SNPs. SNPs within ±250 kb flanking regions of the transcription start site of the examined genes were identified. We used Encyclopedia of DNA Elements (ENCODE) regulatory features from the University of California Santa Cruz genome browser [14] (http://genome.ucsc.edu/) and RegulomeDB [15] to identify potential regulatory SNPs that were likely to affect the expression of the associated genes. We also integrated data from multiple expression quantitative trait locus (eQTL) studies [16] to identify SNPs associated with changes in expression (cis-eQTL) of the associated gene. The cis-eQTL effects were calculated using linear regression models in the selected tissues. Validated eQTLs from Westra et al [17] and GTEx2015_v6. GTEx2015_v6 eQTLs were computed using a ±1 Mb cis window around the transcription start site. Significance was determined using a Q value threshold. At least 70 samples per tissue are necessary to achieve the statistical power needed for this type of analysis. Predicted eQTL was calculated for pancreas and whole blood. Genotype–tissue expression predictions calculation were performed in tissues with at least ten samples. No Q value filtering was performed.
Statistical analysis
Statistical analysis for RNA expression was performed using GraphPad Prism 8.0.2 (GraphPad, La Jolla, Ca, USA) and data are shown as mean ± SEM. Outliers were detected in each group using the ROUT method and a total of 21 datapoints across all genes and groups were identified and removed. All groups were tested for normal distribution by the D’Agostino–Pearson and Anderson Darling test. For comparison between groups, one-way ANOVA was used with Dunnett’s multiple comparison test and a 95% CI. A p value of <0.05 was considered significant (shown as *p < 0.05, **p < 0.01 and ***p < 0.001 in figures).
Results
Selection of genes for analysis were based on their direct and/or indirect association upstream and downstream of the 2′-5′A pathway by examining the literature. We first conducted the GWAS study with 64 genes and, based on the resulting data, we selected 41 genes out of the original 64 for analysis of the RNA levels in islets from the nPOD and DiViD study. The individuals included in the GWAS study were all under the age of 17 years at diagnosis. In the nPOD study, the age at diagnosis was 3–26 years. The DiViD study participants were aged 24–35 years at diagnosis.
SNPs in genes of the innate antiviral immune system show association to type 1 diabetes
GWAS identified 22 genes with 167 (158 unique) associated SNPs having p values <0.01 and ORs for the minor allele ranging from 0.5724 to 1.1841 (see ESM Table 2). Of these SNPs, 27 were nominally associated with type 1 diabetes. There were 15 genes that had at least one SNP with an eQTL p value <0.05 in either whole blood (validated eQLT) or pancreas (predicted eQTL). By comparing those 15 genes to the genes analysed for RNA expression in islets, seven genes (denoted by superscript ‘a’ in Tables 1 and 2) overlapped and were identified with significant type 1 diabetes SNPs with an eQTL signal.
Table 1.
Total number of SNPs related to the innate antiviral immune system and associated with type 1 diabetes
| Gene | Total no. of T1D SNPs (±250 kb) | No. of SNPs with eQTL signal | SNP most strongly associated with T1D | T1D SNP p value | OR (minor/major allele) |
|---|---|---|---|---|---|
| ADARa,b | 9 | 7 | rs4845625 | 1.32 × 10−3 | 1.06/0.94 |
| CASP3 | 2 | 1 | rs17075783b | 4.46 × 10−3 | 0.91/1.10 |
| CYBA | 4 | 2 | rs4782429b | 4.21 × 10−3 | 0.92/1.09 |
| EIF2AK2 | 4 | 4 | rs2247010b | 3.73 × 10−3 | 0.94/1.06 |
| EIF3H | 1 | 1 | rs1446534b | 1.69 × 10−4 | 0.92/1.08 |
| GSPT1 | 2 | 1 | rs350234b | 1.64 × 10−2 | 1.05/0.95 |
| IFIH1a | 90 | 50 | rs2111485 | 3.81 ×10−18 | 0.85/1.18 |
| IFNGa | 4 | 3 | rs11614178 | 1.55 × 10−3 | 0.96/1.05 |
| IL1R1 | 8 | 6 | rs4850992b | 5.58 × 10−3 | 0.75/1.34 |
| IRF7a | 2 | 1 | rs17758b | 1.01 × 10−2 | 0.87/1.15 |
| MX2a,b | 2 | 1 | rs7278439b | 1.24 × 10−3 | 0.95/1.05 |
| NOD2 | 4 | 4 | rs2302759b | 4.11 × 10−3 | 0.94/1.07 |
| OAS1a | 4 | 3 | rs4767000b | 1.03 × 10−9 | 1.12/0.89 |
| OAS3a | 3 | 2 | rs4767000b | 1.03 × 10−9 | 1.12/0.89 |
| TNFRSF1A | 15 | 3 | rs10849451b | 1.59 × 10−3 | 1.06/0.94 |
aGenes that also showed dysregulation on RNA expression level
bNewly discovered in association with type 1 diabetes
T1D, type 1 diabetes
Table 2.
SNPs with an OR above 1.10 that were associated with type 1 diabetes
| Gene | SNP details | Validated eQTL p value in whole blood | Predicted eQTL p value in pancreas | Predicted eQTL p value in whole blood | |||
|---|---|---|---|---|---|---|---|
| SNP | p value (without Q value filtering) | OR (minor allele) | OR (major allele) | ||||
| IFIH1a | rs77088072b | 9.94 × 10−3 | 0.793 | 1.261 | 1.40 × 10−3 | 4.30 × 10−3 | |
| rs984971 | 2.40 × 10−16 | 0.849 | 1.178 | 2.50 × 10−2 | |||
| rs2068330 | 3.64 × 10−16 | 0.850 | 1.177 | 2.30 × 10−2 | |||
| rs3788964 | 3.99 × 10−9 | 0.858 | 1.165 | 2.80 × 10−2 | |||
| rs1549020b | 2.56 × 10−12 | 0.863 | 1.158 | 4.80 × 10−3 | |||
| rs3747517 | 4.98 × 10−11 | 0.871 | 1.148 | 3.50 × 10−2 | 2.20 × 10−2 | ||
| rs13422767 | 5.33 × 10−8 | 0.873 | 1.146 | 1.50 × 10−2 | |||
| rs13023380 | 2.56 × 10−12 | 0.876 | 1.142 | 4.60 × 10−2 | |||
| rs7608315 | 9.56 × 10−11 | 0.884 | 1.131 | 2.50 × 10−2 | |||
| rs13022749b | 1.60 × 10−8 | 0.893 | 1.120 | 5.10 × 10−3 | |||
| IFNAR1a | rs2834136b | 1.36 × 10−2 | 0.854 | 1.171 | |||
| IFNGa | rs7132610b | 3.18 × 10−3 | 1.125 | 0.889 | |||
| IL1R1 | rs79184888b | 5.83 × 10−3 | 0.753 | 1.328 | |||
| rs7565504b | 6.70 × 10−3 | 0.754 | 1.326 | ||||
| rs6731416b | 6.70 × 10−3 | 0.754 | 1.326 | ||||
| rs140672656b | 6.33 × 10−3 | 0.784 | 1.275 | ||||
| IRF7a | rs118067545b | 8.48 × 10−3 | 0.836 | 1.196 | |||
| rs3993797b | 1.64 × 10−2 | 0.887 | 1.128 | ||||
| OAS1a | rs1034687b | 2.16 × 10−7 | 0.869 | 1.150 | 2.19 × 10−11 | ||
| rs1015249b | 1.25 × 10−7 | 0.895 | 1.117 | 2.7 × 10−13 | |||
| rs739744b | 1.03 × 10−9 | 1.123 | 0.890 | 1.11 × 10−21 | |||
| OAS2a | rs1034687b | 2.16 × 10−7 | 0.869 | 1.150 | |||
| rs4767000b | 1.03 × 10−9 | 1.123 | 0.890 | ||||
| rs739744b | 1.03 × 10−9 | 1.123 | 0.890 | ||||
| OAS3a | rs1034687b | 2.16 × 10−7 | 0.869 | 1.150 | 1.30 × 10−2 | ||
| rs739744b | 1.03 × 10−9 | 1.123 | 0.890 | 1.32 × 10−4 | |||
| SP1 | rs17125267b | 1.03 × 10−2 | 0.838 | 1.194 | |||
| rs11170466 | 1.47 × 10−5 | 1.184 | 0.845 | ||||
The SNPs in this table were selected based on their eQTL p value in the specific tissues and their OR being above 1.10 for either the minor or the major allele
aGenes that also showed dysregulation on RNA expression level
bNewly discovered in association with type 1 diabetes
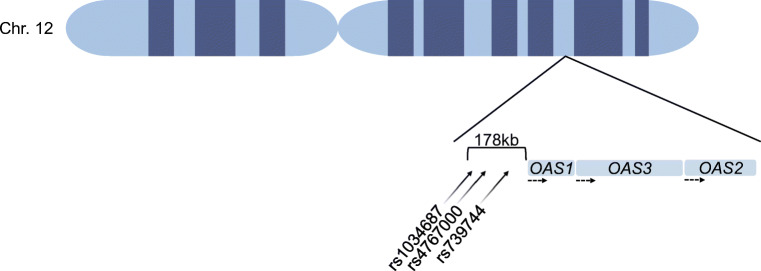
Four SNPs with a strong type 1 diabetes association and an eQTL signal were seen in ADAR (encoding RNA-specific adenosine deaminase), IL1R1 (encoding IL-1 receptor type 1), IRF7 (encoding IFN regulator 7) and the OAS gene family (Table 1). In ADAR, SNP rs4845625 (eQTL p = 1.32 × 10−3) had an OR of 1.06 for the minor allele and was located 135 kb downstream of the gene. In IL1R1, the novel SNP rs4850992 (eQTL p = 5.58 × 10−3) had an OR of 0.75 for the minor allele and was located 85 kb upstream of the gene. In IRF7, the novel SNP rs17758 (eQTL p = 1.01 × 10−2) had an OR of 0.87 for the minor allele and was located 30 kb upstream of the gene. SNP rs4767000 (eQTL p = 1.03 × 10−9) was located upstream of OAS1 and OAS3 with an OR of 1.12 for the minor allele. In OAS2, SNP rs4767000 (p value = 1.03 × 10−9, Table 2) also showed a significant p value with an OR of 1.12 for the minor allele but it did not have an eQTL signal. Three SNPs were located upstream of the OAS genes (Fig. 1). These SNPs were associated with and novel to type 1 diabetes as well as related to OAS1, OAS2 and OAS3. In addition to the aforementioned rs4767000, the others were rs1034687 (p = 2.16 × 10−7, OR 0.869) and rs739744 (p = 1.03 × 10−9, OR 1.123).
Fig. 1.
Overview of the OAS family of genes on chromosome 12 and the location of SNPs rs1034687, rs4767000 and rs739744
As shown in Table 2, the gene encoding IFN induced with helicase domain 1 (IFIH1) had four SNPs with significant predicted eQTLs in the pancreas: rs77088072 (p = 9.94 × 10−3, OR 0.79); rs3788964 (p = 3.99 × 10−9, OR 0.86); rs3747517 (p = 4.98 × 10−11, OR 0.87); and rs13422767 (p = 5.33 × 10−8, OR 0.87). The SNP rs3788964 was located 35 kb upstream of the transcription start site in IFIH1, rs13422767 was located 24 kb downstream, and rs77088072 (a novel SNP in relation to type 1 diabetes) and rs3747517 were located inside the IFIH1 gene. The SNP rs2111485 (Table 1) was located 14 kb downstream of IFIH1 and was the SNP with the strongest eQTL signal associated with type 1 diabetes out of 90 SNPs found in this gene.
For IL1R1, rs4850992 (p = 5.58 × 10−3, OR 0.75) was located 87 kb upstream of the gene (Table 1). The SNP rs7565504 (p = 6.70 × 10−3, OR 0.75) was located 155 kb upstream of IL1R1, and the last SNP of interest in this gene was rs6731416 (p = 6.70 × 10−3, OR 0.75) located 160 kb upstream from transcription start site (Table 2). All three SNPs were to the best of our knowledge novel in relation to type 1 diabetes.
Genes of the innate antiviral immune system show dysregulated RNA expression associated with type 1 diabetes
The heatmap revealed a strong dysregulation of most of the genes in individuals with new-onset type 1 diabetes from the DiViD study (Fig. 2). We selected 41 out of the 64 innate antiviral immune system genes analysed in the GWAS study for expression analysis in DiViD and nPOD islet tissue. Out of those 41 genes, two were novel in association with type 1 diabetes. Two genes showed lower RNA expression levels in autoantibody-positive individuals (p value <0.05). Thirteen genes showed either higher or lower expression levels in individuals with new-onset type 1 diabetes. Six genes showed higher expression levels in individuals with longstanding type 1 diabetes. Two genes showed higher expression levels and one gene was downregulated in longstanding type 1 diabetes. In general, the most aberrant genes were a group of IFGs.
Fig. 2.
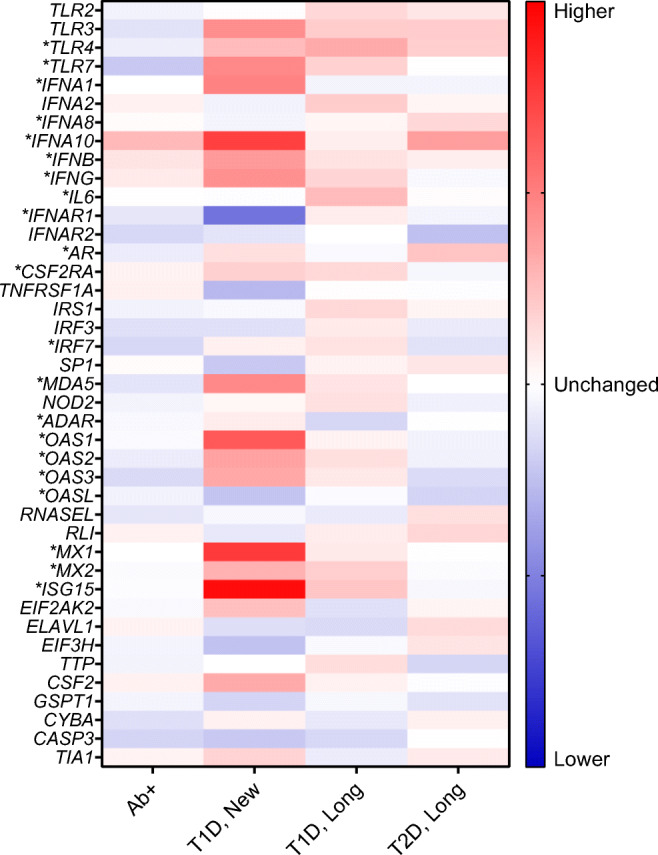
Heatmap of RNA expression in the analysed genes. Genes were normalised to the non-diabetic control group by calculating the ratio of the means in each group. Blue indicates lower RNA expression compared with the control group and red indicates higher RNA expression level. A total of 21 genes showed significantly (*) dysregulated RNA expression levels in either autoantibody-positive (Ab+), new-onset type 1 diabetes (T1D, New), longstanding type 1 diabetes (T1D, Long) or longstanding type 2 diabetes (T2D, Long) compared with controls
PAMP recognition and IFN induction are increased in new-onset type 1 diabetes
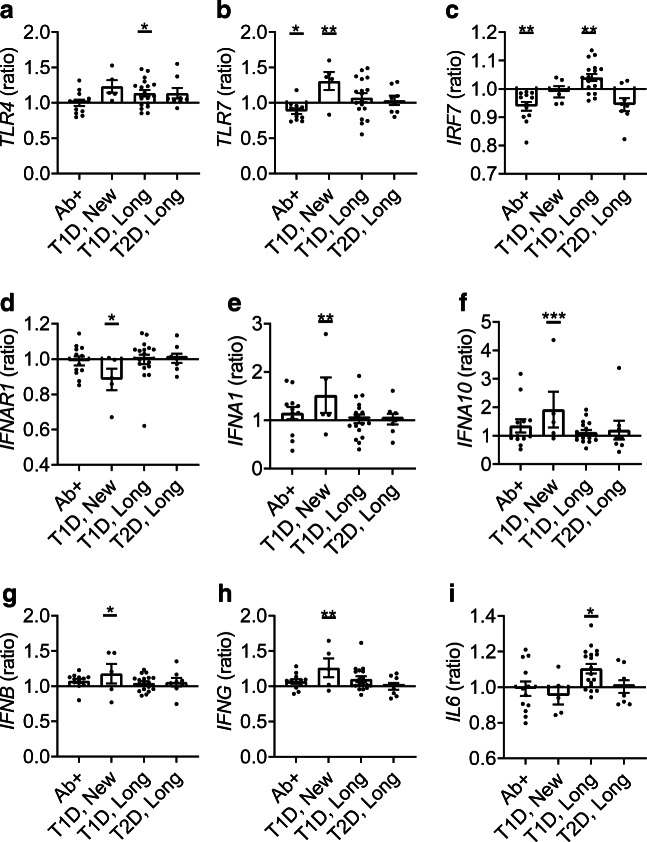
Expression of the nine genes involved in the initial innate antiviral immune response is shown in Fig. 3. The expression of the gene encoding toll-like receptor 7 (TLR7) was significantly increased by 70% in individuals with new-onset type 1 diabetes compared with the non-diabetic control group (p = 9.60 × 10−3, 95% CI −0.42, −0.05, Fig. 3b). IFNA1 expression was increased by 75% (p = 1.30 × 10−3, 95% CI −0.44, −0.09, Fig. 3e) and IFNA10 expression was increased by 136% (p < 1.00 × 10−4, 95% CI −0.50, −0.18, Fig. 3f) in individuals with new-onset type 1 diabetes. Expression of IFNB (p = 4.00 × 10−2, 95% CI −0.30, −0.006, Fig. 3g) and IFNG (p = 2.30 × 10−3, 95% CI −0.32, −0.06, Fig. 3h) was increased by 58% and 65%, respectively, in new-onset type 1 diabetes. Interestingly, in the same group, expression of the gene encoding IFN-α and -β receptor subunit 1 (IFNAR1) was decreased by 47% (p = 1.80 × 10−2, 95% CI −0.04, −0.05, Fig. 3d).
Fig. 3.
Genes involved in PAMP recognition and IFN induction. The RNA expression levels of TLR4 (a), TLR7 (b), IRF7 (c), IFNAR1 (d), IFNA1 (e), IFNA10 (f), IFNB (g), IFNG (h) and IL6 (i) were transformed, and a mean was calculated. The ratio between the mean expression level in each group (autoantibody-positive [Ab+], new-onset type 1 diabetes [T1D, New], longstanding type 1 diabetes [T1D, Long], longstanding type 2 diabetes [T2D, Long]) and the non-diabetic control group was graphed. *p < 0.05, **p < 0.01 and ***p < 0.001 vs control group
In individuals with longstanding type 1 diabetes, gene expression level was increased by 47% for TLR4 (p = 2.90 × 10−2, 95% CI −0.29, −0.01, Fig. 3a) and 36% for IL6 (p = 1.30 × 10−2, 95% CI −0.18, −0.02, Fig. 3i). IRF7 expression was decreased by 17% in autoantibody-positive individuals (p = 6.10 × 10−3, 95% CI 0.02, 0.14, Fig. 3c); the expression of this gene was increased in longstanding type 1 diabetes by 14% (p = 4.00 × 10−2, 95% CI −0.11, −0.002, Fig. 3c).
Multiple ISGs are highly elevated in new-onset type 1 diabetes
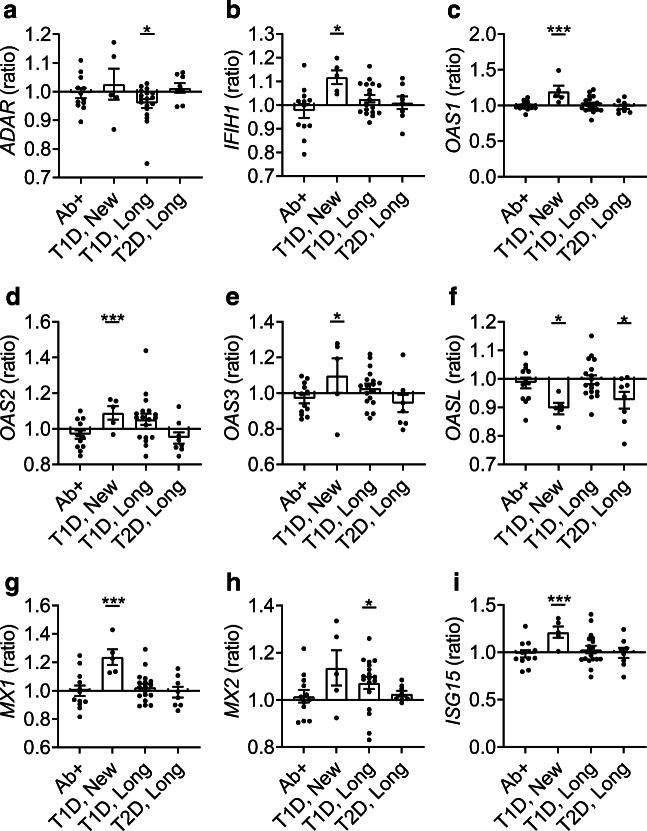
In Fig. 4, the nine genes shown are upregulated or downregulated by the genes (shown in Fig. 3) involved in the initial response of the innate antiviral immune pathway. In individuals with new-onset type 1 diabetes, the expression of OAS1 was increased by 111% (p < 1.00 × 10−4, 95% CI −0.43, −0.15, Fig. 4c), the expression of MX1 was increased by 142% (p < 1.00 × 10−4, 95% CI −0.52, −0.22, Fig. 4g) and the expression of ISG15 was increased by 197% (p = 2.00 × 10−4, 95% CI −0.68, −0.18, Fig. 4i) compared with non-diabetic control individuals. OASL expression was decreased by 23% (p = 2.40 × 10−2, 95% CI −0.06, −0.08) in individuals with new-onset type 1 diabetes (Fig. 4f). MX2 expression was increased by 24% in individuals with longstanding type 1 diabetes (p = 3.80 × 10−2, 95% CI −0.18, −0.004, Fig. 4h). Moreover, ADAR expression was decreased by 17% in individuals with longstanding type 1 diabetes (p = 4.60 × 10−2, 95% CI 0.001, 0.2, Fig. 4a).
Fig. 4.
Cytokine genes and ISGs that were upregulated by IFN induction. The RNA expression levels of ADAR (a), IFIH1 (b), OAS1 (c), OAS2 (d), OAS3 (e), OASL (f), MX1 (g), MX2 (h) and ISG15 (i) were transformed, and a mean was calculated. The ratio between the mean expression level in each group (autoantibody-positive [Ab+], new-onset type 1 diabetes [T1D, New], longstanding type 1 diabetes [T1D, Long], longstanding type 2 diabetes [T2D, Long]) and the non-diabetic control group was graphed. *p < 0.05 and ***p < 0.001 vs control group
Discussion
Our study is the first to investigate all major innate antiviral immune response pathways by combining analysis of GWAS data and analysis of RNA expression levels in islets from the DiViD and nPOD study populations. The GWAS study identified 168 type 1 diabetes-associated SNPs (159 unique SNPs), of which 27 were novel in relation to type 1 diabetes and are predicted to affect the expression of the associated genes. The islet RNA expression analysis identified two genes (ADAR and MX2) that were novel in having a direct relation to type 1 diabetes, out of 19 dysregulated genes. The highest degree of dysregulation (13 genes) was seen in individuals with new-onset type 1 diabetes compared with non-diabetic individuals. In autoantibody-positive individuals two genes were dysregulated in those with longstanding type 1 diabetes, six genes were dysregulated and in those with type 2 diabetes, three genes were dysregulated at the mRNA level. We found multiple ISGs, such as the whole OAS family of genes, that were aberrant in individuals with type 1 diabetes and these genes had several SNPs associated with the disease. Another interesting result was found in genes upstream of the 2′-5′A pathway, such as those encoding type 1 IFN and toll-like receptors.
The gene with the highest number of SNPs in our study was IFIH1, which is also referred to as MDA5 (encoding melanoma differentiation-associated 5 [MDA5] protein). This gene, a cytosol PRR that recognises dsRNA and prevents early replication of viruses, had 50 SNPs predicted to affect its expression. Several SNPs in IFIH1, which could be linked to risk of and protection against type 1 diabetes, have previously been identified and may modify the structure and function of IFIH1 [18].
The toll-like receptor genes and IRF7 are important in the induction of type 1 IFN and upregulation of ISGs, especially the 2′-5′A pathway, and for further immune response through PRRs such as MDA5 [19]. Tight regulation of IRF7 expression and activity is imperative for appropriate type 1 IFN production [20]. A total of three SNPs were found in IRF7 and they could interfere with IRF7 expression or its ability to bind a promoter region.
IFNAR1 was downregulated in individuals with new-onset type 1 diabetes and it had an SNP associated with type 1 diabetes. Moreover, several IFNA isoforms, as well as IFNB, were upregulated. IFN-α and IFN-β share the IFN-α and -β receptor subunit 1 as a common cell surface receptor. Interestingly, the receptor for the two cytokines, IFN-α and IFN-β, being downregulated but the cytokines themselves being upregulated could result in inflammation and destruction of beta cells in the islets of individuals with type 1 diabetes.
Beta cells are especially vulnerable to infection, potentially through dysregulation of the innate antiviral immune system leading to inflammation and destruction [2]. Dysregulation of IFN-α or the 2′-5′A pathway caused by either a genetic predisposition or environmental factors has been associated with type 1 diabetes in different ways. Moreover, studies mimicking a viral response by poly (I:C) or type 1 IFN have shown that RNase L increases the expression of proinflammatory genes in the pancreas [21] and this might also contribute to the pathogenesis of type 1 diabetes.
The OAS genes were highly overexpressed in islets of individuals with new-onset type 1 diabetes and had several eQTL SNPs. In these individuals, OAS1, OAS2 and OAS3 were all upregulated whereas the OASL gene, which has an OAS-like domain but lacks the synthetase activity, was downregulated. We found three novel SNPs within the promoter region of OAS1, OAS2 and OAS3 that could potentially affect the expression of the OAS family of genes. High levels of OAS1, OAS2 and OAS3 indicate an antiviral response or a higher basal protein level in the cell. Whether OAS1, OAS2 and OAS3 are able to properly activate RNase L [22] or other downstream proteins such as retinoic acid-inducible I or MDA5 is crucial for antiviral responses. Therefore, dysregulation of OAS1, OAS2, OAS3 and OASL could indicate lack of the antiviral response and/or a higher degree of virus replication in the cell [23]. In addition, other studies have shown that the 2′-5′A pathway could potentially regulate expression of ISG15 [24], activate MDA5 and even influence IFN expression [8]. The OAS/RNase L activation has also been associated with programmed cell death even without the presence of a pathogen [25].
We found that MX1 was upregulated in new-onset type 1 diabetes and that MX2 was upregulated in longstanding type 1 diabetes as well as having two eQTL SNPs. Both MX1 and MX2, members of a gene family encoding dynamin-like large GTPases, suppress a wide but different variety of virus. Despite the similarities in sequence and structure, MX1 and MX2 have different mechanisms of action against viruses as well as which type of viruses they eliminate [26]. To the best of our knowledge, there is no previous evidence of a direct association between MX1 and MX2 and type 1 diabetes. Hence, this finding is novel and could be central for understanding virus-induced type 1 diabetes.
The majority of ISGs were upregulated in individuals with new-onset type 1 diabetes; however, in longstanding type 1 diabetes ADAR was downregulated and MX2 was upregulated (both ISGs). The protein encoded by ADAR has a dual role as it either modifies or regulates the innate antiviral immune response. It can prevent an unwanted MDA5 and type 1 IFN feedback loop by preventing MDA5 and OAS activation through A-to-I editing of dsRNA. However, if ADAR is not regulated correctly, the A-to-I editing activity could either prevent or create an overactivation of an antiviral immune response.
Regarding type 2 diabetes, we only found three dysregulated genes at the mRNA level when compared with healthy control individuals, indicating that viral infection is not likely to contribute to this disease. OASL was downregulated and has shown effects on C-reactive protein, γ glutamyltransferase and LDL-cholesterol, which are all related to cardiovascular events that are frequent in individuals with type 2 diabetes as well as inflammation [27].
Lundberg et al investigated the RNA expression of several innate antiviral immune system genes in laser-dissected islet tissue from the DiViD study, of which 20 samples were also analysed in our study. However, nPOD islet tissue and GWAS data were not included in this study, and a smaller control group (n=5) was used [28]. As in our study, Lundberg et al [28] found the following genes to be significantly elevated: IFIH1; MX1; IL6; ISG15; and OAS1. This verifies an involvement of the innate antiviral immune response in new-onset type 1 diabetes.
A limitation of this study is the age difference between the individuals included in the GWAS study and the RNA expression study. It is well known that in type 1 diabetes there are different endotypes, which can be defined according to age [29, 30]. Thus, an endotype can be defined as those individuals diagnosed before the age of 7 years, those diagnosed after the age of 13 years, and those diagnosed between 7 and 12 years of age that can belong to either of the two endotypes. The pathogenesis of an endotype is defined by the type of infiltrating immune cells causing insulitis and by how much proinsulin and insulin are colocalised in beta cells [29]. In the current study, a direct association between the GWAS, the nPOD and DiViD cohorts should only be stated with caution, as the participants are from different endotypes. It is known that genetic variants in type 1 diabetes, typically located near genes and acting both in pancreatic beta cells and immune cells, are more prevalent in the endotypes diagnosed before the age of 7 years [30]. We were not able to correct for age, sex, BMI, HLA and/or autoantibody status due to small sample sizes; however, all this information can be found in ESM Table 1. This information can be combined with the information shown in ESM Figs 1 and 2, wherein each data point from the nPOD and DiViD analysis has an identifier correlating with information shown in ESM Table 1.
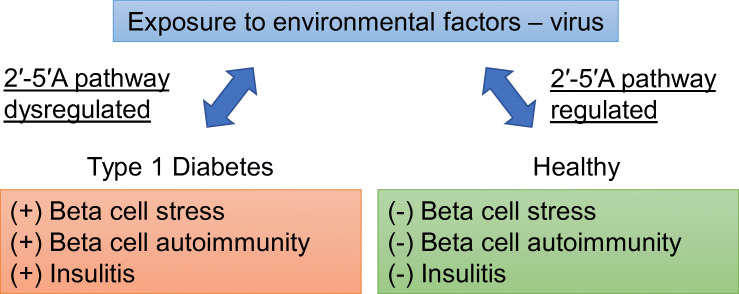
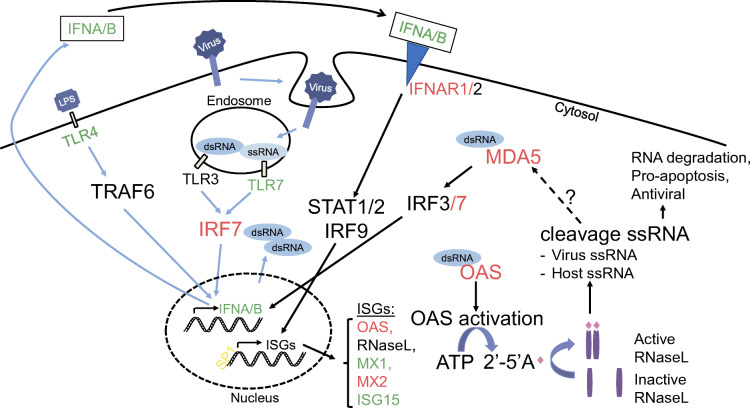
In conclusion, innate antiviral immune sensors capable of detecting and clearing ssRNA, were downregulated in autoantibody-positive individuals and upregulated in individuals with new-onset type 1 diabetes. These genes included TLR7, OAS, MX1, multiple type 1 IFN genes and IFIH1 (Fig. 5). In conjunction, we found polymorphisms in OAS, MX1 and IFIH1 that indicate predisposition to type 1 diabetes. The DiViD study previously found footprints of virus in islets from individuals with type 1 diabetes [31]. Thus, we hypothesise that environmental factors (e.g. virus) in predisposed and/or autoantibody-positive individuals stimulate dysregulated 2′-5′ pathway through toll-like receptor 7 and type 1 IFN in the innate immune system, causing inflammation in the islets and progression to diabetes. Identification and avoidance of diabetogenic viruses that can stimulate the toll-like receptor 7 pathway in early disease development may prevent or delay type 1 diabetes. It is our hope that further research on the role of toll-like receptors, the 2′-5′A pathway and other ISGs will lead to better prognostic markers and new therapeutic drugs.
Fig. 5.
The activation and the feedback loop of the innate antiviral immune system during a viral infection. The RNA dysregulated genes/proteins where SNPs were also found in the GWAS study are highlighted in red. Genes/proteins that were only dysregulated at the RNA level are highlighted in green. SP1 is highlighted in yellow because we only found SNPs in this gene associated with type 1 diabetes. The figure illustrates the multitude of genes involved in the potential overactivation in response to an infection in type 1 diabetes. LPS, lipopolysaccharide
Supplementary Information
(PDF 297 kb)
Acknowledgements
The authors thank N. Lenchik from the Department of Medicine, University of Tennessee, Memphis, TN, USA for technical support in collecting gene expression data.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
KP, MHJ and KB contributed to the conception and design of the study. KP, LK, SK, ICG, FP and KDJ acquired the data. KP analysed and interpreted data and drafted the manuscript. All authors revised the article critically and approved the final version. KP is the guarantor of the work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Abbreviations
- 2′-5′A
2′-5′-Linked oligoadenylate
- DiViD
Diabetes Virus Detection
- dsRNA
Double-stranded RNA
- eQTL
Expression quantitative trait locus
- GWAS
Genome-wide association study
- ISG
IFN-stimulated gene
- MDA5
Melanoma differentiation-associated 5
- nPOD
Network of Pancreatic Organ Donors
- OAS
2′-5′ Oligoadenylate synthetase
- PAMP
Pathogen-associated molecular pattern
- Poly (I:C)
Polyinosinic:polycytidylic acid
- PRR
Pathogen-recognition receptor
- ssRNA
Single-stranded RNA
Funding
The DiViD study was funded by the South-Eastern Norway Regional Health Authority, the Novo Nordisk Foundation, and through the PEVNET (Persistent Virus Infection in Diabetes Network) Study Group funded by the European Union’s Seventh Framework Program (FP7/2007-2013) under grant agreement no. 261441 PEVNET. Additional grant support was from the National Institutes of Health (UC4 DK104155) and the JDRF (47-2013-520). nPOD, a collaborative type 1 diabetes research project, is sponsored by the JDRF (grant no. 25-2013-268, 25-2012-380 and 25-2007-874). Lastly, the study was supported by Bagger-Sørensen Foundation.
Data availability
All data generated and analysed during this study are included in this published article and its ESM. Raw data are published online (https://www.dropbox.com/s/egshju5x6d4j0or/GWAS%20study.xlsx?dl=0;https://www.dropbox.com/s/1r7lrxba1fb0atk/nPOD%20and%20DiViD%20studies.csv?dl=0).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Roth R, Lynch K, Hyoty H, et al. The association between stressful life events and respiratory infections during the first 4 years of life: The Environmental Determinants of Diabetes in the Young study. Stress Health. 2019;35(3):289–303. doi: 10.1002/smi.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnevie-Nielsen V, Buschard K, Dyrberg T. Differential responsiveness to interferon-alpha in beta-cells and non-beta cells. Diabetes. 1996;45(6):818–821. doi: 10.2337/diab.45.6.818. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Zheng DJ, Field LL, Bonnevie-Nielsen V. Murine pancreatic beta TC3 cells show greater 2', 5'-oligoadenylate synthetase (2'5'AS) antiviral enzyme activity and apoptosis following IFN-alpha or poly(I:C) treatment than pancreatic alpha TC3 cells. Exp Diabetes Res. 2009;2009:631026. doi: 10.1155/2009/631026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marroqui L, Lopes M, dos Santos RS, et al. Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic alpha and beta cells. Elife. 2015;4:e06990. doi: 10.7554/eLife.06990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues KB, Dufort MJ, Llibre A, et al. Innate immune stimulation of whole blood reveals IFN-1 hyper-responsiveness in type 1 diabetes. Diabetologia. 2020;63(8):1576–1587. doi: 10.1007/s00125-020-05179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnevie-Nielsen V, Larsen ML, Frifelt JJ, Michelsen B, Lernmark A. Association of IDDM and attenuated response of 2',5'-oligoadenylate synthetase to yellow fever vaccine. Diabetes. 1989;38(12):1636–1642. doi: 10.2337/diab.38.12.1636. [DOI] [PubMed] [Google Scholar]
- 7.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448(7155):816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogvold L, Edwin B, Buanes T, et al. Pancreatic biopsy by minimal tail resection in live adult patients at the onset of type 1 diabetes: experiences from the DiViD study. Diabetologia. 2014;57(4):841–843. doi: 10.1007/s00125-013-3155-y. [DOI] [PubMed] [Google Scholar]
- 10.Campbell-Thompson M, Wasserfall C, Kaddis J, et al. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes Metab Res Rev. 2012;28(7):608–617. doi: 10.1002/dmrr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson SJ, Rodriguez-Calvo T, Gerling IC, et al. Islet cell hyperexpression of HLA class I antigens: a defining feature in type 1 diabetes. Diabetologia. 2016;59(11):2448–2458. doi: 10.1007/s00125-016-4067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Kakoola DN, Lenchik NI, Desiderio DM, Marshall DR, Gerling IC. Molecular phenotyping of immune cells from young NOD mice reveals abnormal metabolic pathways in the early induction phase of autoimmune diabetes. PLoS One. 2012;7(10):e46941. doi: 10.1371/journal.pone.0046941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onengut-Gumuscu S, Chen WM, Burren O, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47(4):381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348(6235):648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed]
- 17.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284(20):13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Domizio J, Blum A, Gallagher-Gambarelli M, Molens JP, Chaperot L, Plumas J. TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood. 2009;114(9):1794–1802. doi: 10.1182/blood-2009-04-216770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Zheng Y, Hou C, et al. DNA methylation impairs TLR9 induced Foxp3 expression by attenuating IRF-7 binding activity in fulminant type 1 diabetes. J Autoimmun. 2013;41:50–59. doi: 10.1016/j.jaut.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Zeng C, Yi X, Zipris D, et al. RNase L contributes to experimentally induced type 1 diabetes onset in mice. J Endocrinol. 2014;223(3):277–287. doi: 10.1530/JOE-14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Banerjee S, Wang Y, et al. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci U S A. 2016;113(8):2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Zhang Y, Ghosh A, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40(6):936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li XL, Blackford JA, Judge CS, et al. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J Biol Chem. 2000;275(12):8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- 25.Birdwell LD, Zalinger ZB, Li Y, et al. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J Virol. 2016;90(6):3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fribourgh JL, Nguyen HC, Matreyek KA, et al. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe. 2014;16(5):627–638. doi: 10.1016/j.chom.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middelberg RP, Ferreira MA, Henders AK, et al. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundberg M, Krogvold L, Kuric E, Dahl-Jorgensen K, Skog O. Expression of interferon-stimulated genes in insulitic pancreatic islets of patients recently diagnosed with type 1 diabetes. Diabetes. 2016;65(10):3104–3110. doi: 10.2337/db16-0616. [DOI] [PubMed] [Google Scholar]
- 29.Leete P, Oram RA, McDonald TJ, et al. Studies of insulin and proinsulin in pancreas and serum support the existence of aetiopathological endotypes of type 1 diabetes associated with age at diagnosis. Diabetologia. 2020;63(6):1258–1267. doi: 10.1007/s00125-020-05115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inshaw JRJ, Cutler AJ, Crouch DJM, Wicker LS, Todd JA. Genetic variants predisposing most strongly to type 1 diabetes diagnosed under age 7 years lie near candidate genes that function in the immune system and in pancreatic beta-cells. Diabetes Care. 2020;43(1):169–177. doi: 10.2337/dc19-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogvold L, Edwin B, Buanes T, et al. Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64(5):1682–1687. doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 297 kb)
Data Availability Statement
All data generated and analysed during this study are included in this published article and its ESM. Raw data are published online (https://www.dropbox.com/s/egshju5x6d4j0or/GWAS%20study.xlsx?dl=0;https://www.dropbox.com/s/1r7lrxba1fb0atk/nPOD%20and%20DiViD%20studies.csv?dl=0).