Abstract
MicroRNAs (miRNAs) have been widely implicated in immune regulation, but evidence for the coordinated function of paralogous miRNA clusters remains scarce. Here, by using genetically modified mice with individual or combined cluster deficiencies, we found that three paralogous clusters of the miR-17~92 family of miRNAs collectively suppressed IL-12 production in macrophages. Accordingly, miR-17~92 family miRNAs deficiencies resulted in heightened production of IL-12 and thus enhanced the host defense against intracellular pathogen Listeria monocytogenes in vivo. Mechanistically, different members of the miR-17~92 family of miRNAs acted on a common target, PTEN, to inhibit IL-12 expression by modulating the PI3K-Akt-GSK3 pathway. In addition, the expression of miR-17~92 family miRNAs was collectively inhibited by the transcription factor RBP-J, and RBP-J-associated macrophage functional defects were genetically rescued by deleting three clusters of miR-17~92 family miRNAs on a RBP-J null background. Thus, our results illustrated key roles of three clusters of miR-17~92 family miRNAs in cooperatively controlling IL-12-mediated immune responses and identified miR-17~92 family miRNAs as functional targets of RBP-J in macrophages.
Keywords: miR-17~92 family miRNAs, microRNA, IL-12, RBP-J, Macrophages
Subject terms: miRNA in immune cells, Listeria, Monocytes and macrophages
Introduction
MicroRNAs (miRNAs) are a class of approximately 22-nucleotide-long noncoding RNAs that posttranscriptionally regulate gene expression in metazoan organisms by pairing with messenger RNAs to inhibit protein translation and/or promote mRNA degradation.1 miRNAs are generated from primary miRNA precursors (pri-miRNAs) that are cleaved by Drosha into ~70 nucleotide pre-miRNAs that are further processed by Dicer into mature miRNAs.2 Thus, the biogenesis of miRNAs can be controlled at multiple steps, including through the regulation of pri-miRNA transcription and miRNA processing.3 For approximately 25–40% of miRNAs, the pri-miRNAs are located in close proximity to several neighboring pri-miRNAs on chromatin to form miRNA clusters,4,5 which typically yield mature miRNAs with distinct seed regions that may either promote diverse effects or act in a coordinated manner to accomplish common functions. The miR-17~92 family of miRNAs consists of three paralogous miRNA clusters: miR-17~92, miR-106a~363, and miR-106b~25 (Fig. 1a, upper panel).6 Moreover, miR-17~92 family miRNAs comprise a total of 15 miRNA stem loops that represent 13 distinct mature miRNAs, which can be categorized into four different families according to the sequence alignment of their “seed” regions (a schematic depiction is presented in the lower panel of Fig. 1a).7,8 To date, miR-17~92 family miRNAs have reportedly played an important role in development and are regarded as oncogenes during tumorigenesis.7,8 It has been reported that, in the immune system, miR-17~92 family miRNAs are critical for lymphocyte homeostasis maintenance, T-follicular helper cell differentiation, B-cell development, invariant natural killer T (iNKT) cell ontogenesis, and monocyte differentiation and maturation.9–15 However, despite extensive investigation into individual miRNAs, relatively little is known about whether and how related miRNA clusters coordinate to regulate immune responses, and little is known about the specific functions of miR-17~92 family miRNAs in macrophage activation.
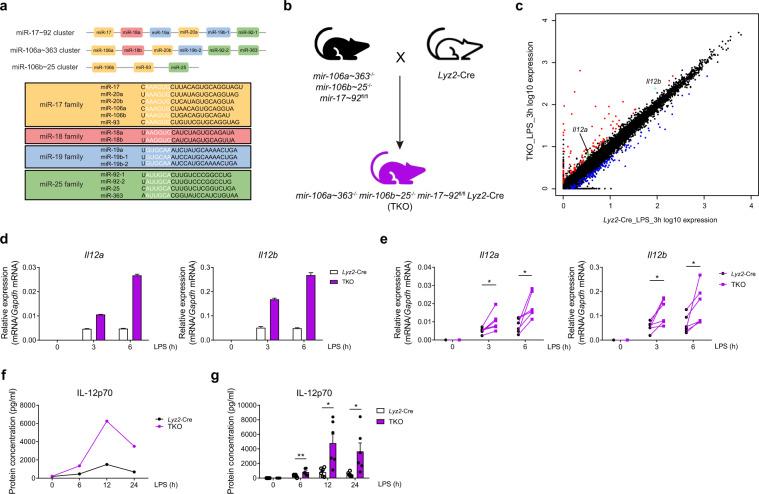
Fig. 1.
miR-17~92 family miRNAs inhibit IL-12 production in macrophages. a Schematic representation of the three miRNA clusters of miR-17–92 family miRNAs (upper panel) and their target sequences (lower panel). The mature miRNAs in different families are shown in different colors. Yellow: members of the miR-17 family; red: members of the miR-18 family; blue: members of the miR-19 family; green: members of the miR-25 family. The “seed region” of each mature miRNA is shown in white. b Outline of the mouse breeding strategy. mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox mice were bred with Lyz2-Cre mice to obtain mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre (TKO) mice. c RNA-seq analysis showing RNA expression in the TKO BMDMs versus the Lyz2-Cre cells after treatment with LPS for 3 h. RNAs upregulated in the TKO BMDMs are red, whereas the RNAs downregulated are blue. Gene Il12a is green, and gene Il12b is cyan. d–e qPCR analysis of Il12a and Il12b mRNA in the Lyz2-Cre and TKO BMDMs stimulated for the indicated periods with LPS. f–g ELISA results showing IL-12p70 in the supernatants from the Lyz2-Cre and TKO BMDMs stimulated for the indicated periods with LPS. Data are representative of six (d and f) or are pooled from six (e and g) independent experiments. Data are presented as the means ± SD; *P < 0.05, **P < 0.01 (paired Student’s t test)
Macrophages play an essential role in the maintenance of host homeostasis and in innate immune responses to various pathogens. Upon pathogen insult, the activation of macrophages by pathogen-associated molecular patterns, such as lipopolysaccharides (LPSs), leads to the production of a plethora of immune and inflammatory effector molecules, including those promoting subsequent adaptive responses such as interleukin 12 (IL-12). Bioactive IL-12, commonly designated IL-12p70, is a heterodimeric complex composed of the IL-12p40 subunit encoded by the Il12b gene and the IL-12p35 subunit encoded by the Il12a gene.16 IL-12 serves as a bridge between innate and adaptive immunity by promoting the differentiation of Th1 cells16 and thus is critically involved in host defense against intracellular pathogens such as Listeria monocytogenes.17 Nevertheless, despite the beneficial effects of IL-12 in host defense, overproduction of IL-12 has been observed in a number of human disease conditions and contributes to the pathogenesis of autoimmune and inflammatory disorders such as Crohn’s disease (CD) and rheumatoid arthritis (RA).18 Therefore, to ensure sufficiently robust responses while avoiding unnecessary inflammation, the production of IL-12 is tightly controlled by a variety of positive and negative regulatory mechanisms. As a secondary response gene, full-fledged induction of IL-12 genes is dependent on chromatin modification events involving the activation of the SWI/SNF complex19 and the stabilization of histone demethylase,20 which is accompanied by the recruitment of sequence-specific transcription factors in the NF-κB and IRF families.21 In addition, abundant inhibitory mechanisms are in place to curtail IL-12 expression at multiple levels, including signaling inhibition, epigenetic modification, and posttranscriptional regulation.16,22
In this study, using rigorous genetic approaches, we found that multiple miRNAs in the miR-17~92 family act in concert to inhibit IL-12 production by targeting key upstream signaling molecules, which has a significant biological impact on the host defense against intracellular bacteria. Moreover, we identified RBP-J as an inhibitor of miR-17~92 family miRNAs and miR-17~92 family miRNAs as alternative functional targets for RBP-J during the promotion of inflammatory macrophage polarization. These results revealed the critical roles of three paralogous clusters of miR-17~92 family miRNAs in cooperatively controlling IL-12-mediated immune responses.
Results
miR-17~92 family miRNAs inhibit IL-12 production in macrophages
To study the functions of miR-17~92 family miRNAs in macrophages, we first generated mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre triple knockout (TKO) mice by crossing mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox mice with Lyz2-Cre mice (Fig. 1b), and efficient deletion of all miR-17~92 family miRNAs was observed in the macrophages (Supplementary Fig. 1). Macrophage populations in homeostasis appeared grossly normal in the myeloid-specific TKO animals (Supplementary Fig. 2). To investigate whether miR-17~92 family miRNAs play a role in macrophage activation, we stimulated bone marrow-derived macrophages (BMDMs) obtained from the TKO and control Lyz2-Cre mice treated with LPS or untreated and performed high-throughput RNA sequencing. Interestingly, analysis of the resulting RNA-seq data revealed that the expression of genes (Il12a and Il12b) encoding the critical inflammatory cytokine IL-12 was upregulated in the TKO BMDMs upon LPS stimulation (Fig. 1c and Supplementary Fig. 3). Then, we experimentally confirmed that the expression of both Il12a and Il12b was significantly increased in the TKO BMDMs after LPS stimulation (Fig. 1d, e). Consistent with the mRNA expression results, we also detected the expressed protein level of IL-12p70 and found that IL-12p70 was significantly upregulated in the supernatants of the TKO BMDMs stimulated with LPS compared with the supernatants from the WT BMDMs (Fig. 1f, g). Thus, these results revealed that miR-17~92 family miRNAs inhibit IL-12 production in macrophages.
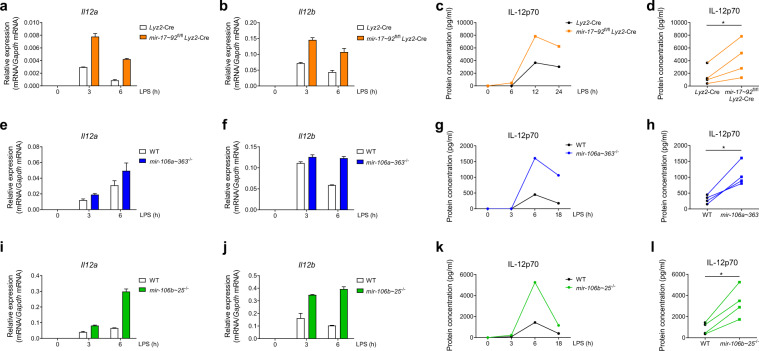
As miR-17~92 family miRNAs are composed of three paralogous clusters (Fig. 1a), we next distinguished the contribution of each miRNA cluster in the suppression of IL-12 production. As the mir-17~92−/− mice are not viable,6 we first generated mir-17~92flox/flox Lyz2-Cre (17/92 KO) mice in which the miR-17~92 cluster was specifically deleted in myeloid cells. We stimulated Lyz2-Cre and 17/92 KO BMDMs with LPS and examined the expression levels of Il12a and Il12b mRNA. We found that 17/92 KO BMDMs expressed higher levels of Il12a and Il12b than did cells from the Lyz2-Cre control mice (Fig. 2a, b). Concomitantly, the IL-12p70 protein concentration in the supernatants of the 17/92 KO BMDMs was enhanced compared with that of the Lyz2-Cre control BMDMs (Fig. 2c, d). Next, we determined the mRNA levels of Il12a and Il12b in mir-106a~363−/− (106a KO) and mir-106b~25−/− (106b KO) BMDMs upon LPS stimulation, and similar to the results obtained for the 17/92 KO BMDMs, we found that the expression levels of both Il12a and Il12b were increased in the 106a KO and 106b KO BMDMs compared with the WT cells (Fig. 2e, f, i, j). Consistent with these changes in mRNA levels, the protein level of IL-12p70 was also upregulated in the supernatants of the 106a KO and 106b KO BMDMs compared to the supernatants of the WT cells (Fig. 2g, h, k, l). Taken together, these results indicated that three paralogous clusters of miR-17~92 family miRNAs collectively restrain IL-12 production in macrophages.
Fig. 2.
Three clusters of miR-17~92 family miRNAs restrain Il-12 production in macrophages. a, b Results of the qPCR analysis of Il12a (a) and Il12b (b) mRNA in—Lyz2-Cre and mir-17~92flox/flox Lyz2-Cre BMDMs stimulated for the indicated periods with LPS. c ELISA results showing IL-12p70 in the supernatants from the Lyz2-Cre and mir-17~92flox/flox Lyz2-Cre BMDMs stimulated for the indicated periods with LPS. d Cumulative amounts of the IL-12p70 protein in the Lyz2-Cre and mir-17~92flox/flox Lyz2-Cre BMDMs stimulated with LPS for 12 h, as indicated in (c). e, f qPCR analysis of Il12a (e) and Il12b (f) mRNA in the wild-type (WT) and mir-106a–363−/− BMDMs stimulated for the indicated periods with LPS. g ELISA results showing IL-12p70 in the supernatants from the WT and mir-106a~363−/− BMDMs stimulated for the indicated periods with LPS. h Cumulative amounts of the IL-12p70 protein in the WT and mir-106a~363−/− BMDMs stimulated with LPS for 6 h, as indicated in (g). i, j Results from the qPCR analysis of Il12a (i) and Il12b (j) mRNA in the WT and mir-106b~25−/− BMDMs stimulated for the indicated periods with LPS. k ELISA results showing IL-12p70 in the supernatants from the WT and mir-106b~25−/− BMDMs stimulated for the indicated periods with LPS. l Cumulative amounts of the IL-12p70 protein in the WT and mir-106b~25−/− BMDMs stimulated with LPS for 6 h, as indicated in (k). Data are representative of 2–4 experiments (a–c, e–g, i–k) or are pooled from four (d, h, and l) independent experiments and are presented as the means ± SD; *P < 0.05 (paired Student’s t test)
miR-17~92 family miRNA deficiency protects the host from intracellular bacterial infection
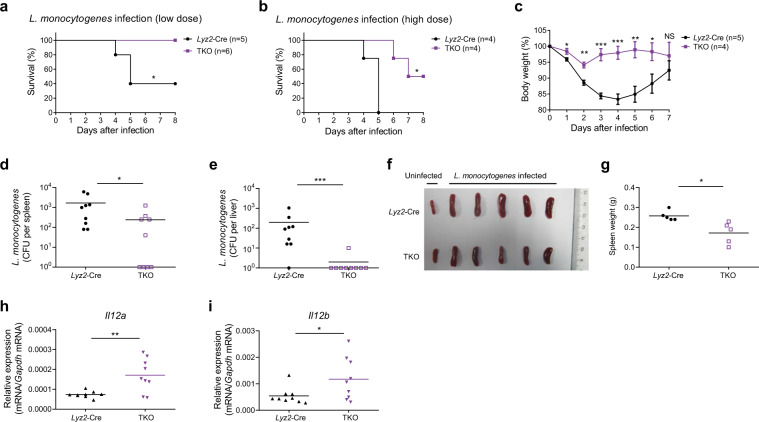
Having established that miR-17~92 family miRNAs suppress IL-12 production in vitro, we wished to determine the in vivo function of miR-17~92 family miRNAs and therefore subjected TKO mice to infection by L. monocytogenes, with a host defense that is dependent on IL-12-driven Th1 responses.17,23,24 Upon challenge with either a low dose or a high dose of L. monocytogenes, the TKO mice exhibited enhanced survival (Fig. 3a, b) and reduced body weight loss (Fig. 3c) compared with the survival and weight loss observed for the control animals. Moreover, the bacterial burden in the spleens and livers of the TKO mice was significantly lower than that in the Lyz2-Cre mice (Fig. 3d, e), and the spleens of the TKO mice were also smaller than were those of the Lyz2-Cre mice post infection (Fig. 3f, g), indicating more efficient bacterial clearance by the TKO mice. These enhanced immune responses in the TKO mice were likely not due to altered immune cell populations, as the splenocyte subsets were not apparently different in the Lyz2-Cre- and TKO-infected mice (Supplementary Fig. 4A–C). In line with the finding of heightened immunity, IFNγ production by CD4+ and CD8+ T cells in the TKO mice had a tendency to increase compared with that in Lyz2-Cre mice (Supplementary Fig. 5A, B). In addition, consistent with the in vitro results, both Il12a and Il12b were overexpressed in the peritoneal macrophages taken from the TKO mice post infection (Fig. 3h, i). In summary, these data suggest that miR-17~92 family miRNAs compromise the host defense against intracellular bacterial infections, possibly by curbing IL-12 expression.
Fig. 3.
miR-17~92 family miRNA deficiency enhances host resistance to Listeria monocytogenes infection. a, b Lyz2-Cre and mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre (TKO) mice were infected intravenously with a low dose (4 × 105) (a) or a high dose (6 × 105) (b) of Listeria monocytogenes, and animal survival was monitored daily. c Lyz2-Cre and TKO mice were infected intravenously with 2 × 105 Listeria monocytogenes, and the body weight of each mouse was measured daily. d–i Lyz2-Cre and TKO mice were infected intravenously with 5 × 104 Listeria monocytogenes, and the mice were sacrificed on day 6 post infection; bacterial load in the spleens (d) and livers (e) of the Lyz2-Cre and TKO mice was analyzed and reported in CFUs; photograph showing the spleens from uninfected and infected Lyz2-Cre and TKO mice (f) and the absolute weight of each spleen from the infected mice (g); results of the qPCR analysis of Il12a (h) and Il12b (i) mRNA in the peritoneal macrophages in the Lyz2-Cre and TKO mice. Each symbol represents an individual mouse; horizontal lines indicate the mean values. Data are pooled from two independent experiments (a, b, d, e, h, and i) or are representative of one independent experiment (c, f, and g). Data are presented as the means ± SEM (c); NS not significant (P > 0.05); *P < 0.05, **P < 0.01, and ***P < 0.001 (log-rank (Mantel–Cox) test (a, b), unpaired Student’s t test (c, g, h, and i), and Mann–Whitney U test (d, e))
miR-17~92 family miRNAs inhibit IL-12 production by alleviating the PTEN-mediated suppression of the Akt-GSK3 pathway
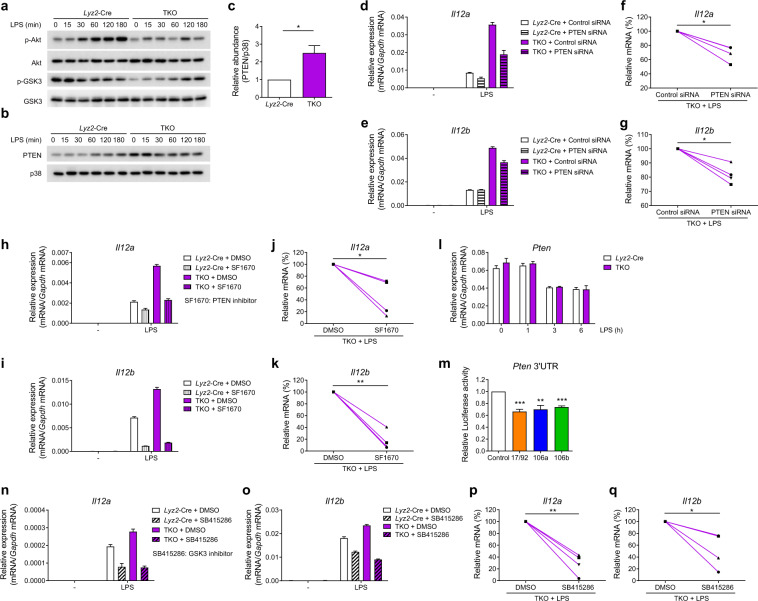
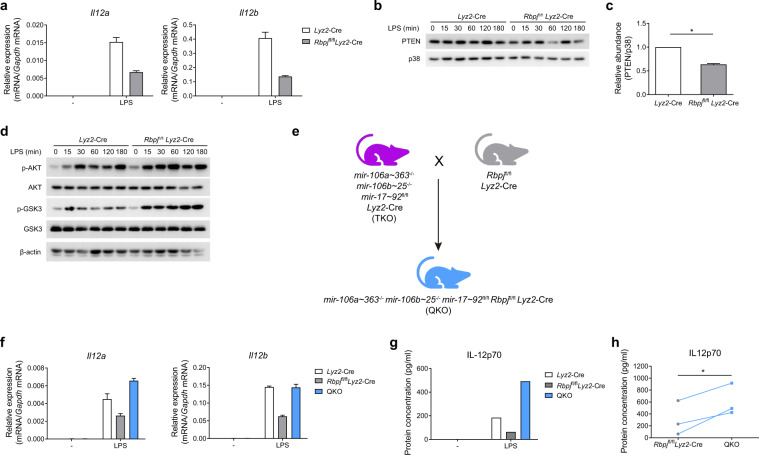
Next, we sought to investigate the mechanisms by which miR-17~92 family miRNAs suppress IL-12 production. First, we examined TLR4-induced canonical signaling pathways such as the NF-κB and MAPK pathways25 and found that their activation was not altered in the TKO BMDMs (Supplementary Fig. 6). Along with others, we previously described another pathway that regulates the TLR-induced production of cytokines, including IL-12: the PI3K-Akt-GSK3 axis.26,27 Therefore, in this study, we analyzed the activation of Akt and glycogen synthase kinase 3 (GSK3) and found that the phosphorylation of Akt and GSK3 was reduced in the TKO BMDMs (Fig. 4a). As phosphorylation of GSK3 is negatively correlated with its activity28,29 and because GSK3 has been shown to promote IL-12 expression,27 the observed decrease in GSK3 phosphorylation was consistent with the results above showing overproduction of IL-12 in the TKO BMDMs (Fig. 1d–g). To further investigate the mechanisms that regulate activation of the Akt pathway, we screened the expression of several well-characterized regulators of Akt and found that the protein level of phosphatase and tensin homolog (PTEN), a negative regulator of the PI3K-Akt pathway,30 was enhanced in the TKO BMDMs in the presence and absence of LPS stimulation (Fig. 4b). This increase in the basal protein expression of PTEN in resting macrophages prior to activation was consistently observed in multiple independent experiments (Fig. 4c). We next sought to determine whether the enhanced expression of PTEN in the TKO BMDMs contributed to the overproduction of IL-12 in these cells; therefore, we decreased PTEN expression using RNA interference (Supplementary Fig. 7A, B) and inhibited PTEN phosphatase activity by using a chemical inhibitor. Both knocking down the PTEN level (Fig. 4d–g) and inhibiting PTEN phosphatase activity (Fig. 4h–k) significantly reduced the expression of both Il12a and Il12b in the TKO BMDMs, indicating that the upregulated IL-12 expression in the TKO cells was PTEN dependent. Then, we further investigated the mechanisms of PTEN regulation and found that Pten mRNA levels were comparable in the Lyz2-Cre and TKO BMDMs (Fig. 4l), implying posttranscriptional regulation of PTEN by miR-17~92 family miRNAs. It has been reported that PTEN is a direct target of several miRNAs in the miR-17~92 cluster,6,9 and the results from our bioinformatics analysis using TargetScan 7.231 further suggested that PTEN was a predicted target of all the miRNAs in the miR-17~92 family. Next, we experimentally confirmed that PTEN was, indeed, the direct target of miR-17~92 family miRNAs by showing that overexpression of the miRNAs from each cluster of the miR-17~92 family reduced the activity level of a Renilla luciferase reporter containing the Pten 3′UTR (Fig. 4m). To further determine the role of GSK3 in IL-12 overexpression, we abrogated GSK3 activity by pharmacological inhibition and found inhibiting GSK3 led to significantly decreased Il12a and Il12b expression in the TKO BMDMs (Fig. 4n–q), indicating that GSK3 contributed to triple miRNA cluster-regulated IL-12 production. Overall, these data demonstrated that miR-17~92 family miRNAs inhibit IL-12 production by targeting PTEN to subsequently modulate the PI3K-Akt-GSK3 pathway.
Fig. 4.
miR-17~92 family miRNAs inhibit IL-12 production by targeting PTEN in macrophages. a Results from the immunoblotting analysis of phosphorylated and total Akt, phosphorylated and total GSK3 in whole-cell lysates of the Lyz2-Cre and mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre (TKO) BMDMs treated for the indicated periods with LPS. b Results from the immunoblotting analysis of PTEN and p38 (loading control) in the whole-cell lysates of the Lyz2-Cre and TKO BMDMs treated for the indicated periods with LPS. c Quantified PTEN protein abundance under unstimulated conditions in (b), according to the densitometry data from three independent experiments. d, e Results from the qPCR analysis of Il12a (d) and Il12b (e) mRNA in the Lyz2-Cre and TKO BMDMs transfected with control or PTEN siRNAs and stimulated with or without LPS for 3 h. f, g Cumulative amounts of Il12a (f) and Il12b (g) mRNA in the LPS-stimulated TKO BMDMs in (d) and (e), presented relative to amount of control siRNA in the transfected cells. h, i Results from the qPCR analysis of Il12a (h) and Il12b (i) mRNA in the Lyz2-Cre and TKO BMDMs stimulated with or without LPS for 3 h in the presence or absence of the PTEN inhibitor SF1670 (2 μM). j, k Cumulative amounts of Il12a (j) and Il12b (k) mRNA in the LPS-stimulated TKO BMDMs in (h) and (i), presented relative to the amounts in the DMSO-pretreated cells. l Results from the qPCR analysis of Pten mRNA in—Lyz2-Cre and TKO BMDMs stimulated for the indicated periods with LPS. m Results from the Luciferase reporter assays of Rluc gene expression containing the Pten 3′UTR in the 293T cells cotransfected with the luciferase reporter vector and negative control (Control), miR-17~92 cluster (17/92), miR-106a~363 cluster (106a), or miR-106b~25 cluster (106b) miRNA overexpression vector. The results are presented as the Rluc/Luc activity ratio and are normalized to the values in the control vector group. n, o Results from the qPCR analysis of Il12a (n) and Il12b (o) mRNA in the Lyz2-Cre and TKO BMDMs stimulated with LPS for 3 h in the presence or absence of the GSK3 inhibitor SB415286 (20 μM). p, q Cumulative amounts of Il12a (p) and Il12b (q) mRNA in the LPS-stimulated TKO BMDMs in (n) and (o), presented relative to amounts in the DMSO-pretreated cells. Data are representative of three to four independent experiments (a–c, e, h, i, l, n, and o) or are pooled from three to four independent experiments (c, f, g, j, k, m, p, and q). Data are presented as the means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001 (paired Student’s t test)
RBP-J suppresses the expression of miR-17~92 family miRNAs in macrophages
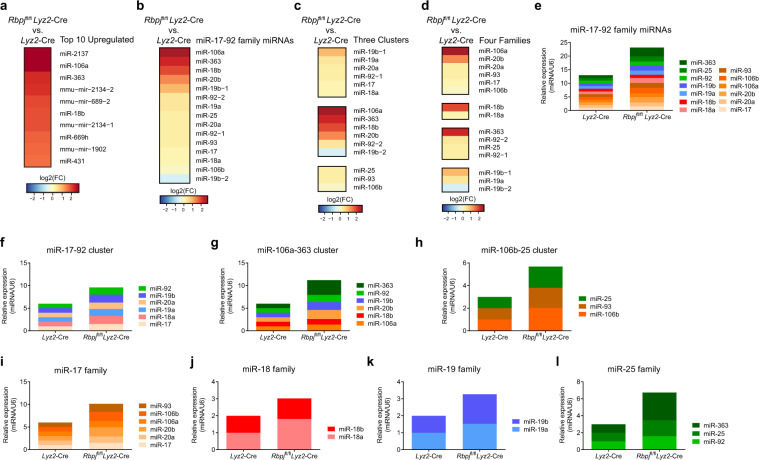
Having identified miR-17~92 family miRNAs as negative regulators of IL-12 production, we next wished to place miR-17~92 family miRNAs in the context of the immune regulatory network that controls IL-12 production and clarify the connections between miR-17~92 family miRNAs and other key regulators. In previous studies, we reported that RBP-J (recombinant recognition sequence binding protein at the Jκ site, also named CSL or CBF1), a master transcription regulator downstream of the Notch pathway,32–36 promoted IL-12 production by augmenting the eIF4E-mediated translation initiation of the interferon regulatory factor 8 (IRF8, also named ICSBP) protein.37 Here, we sought to determine whether RBP-J could regulate the expression of miR-17~92 family miRNAs. To identify RBP-J-regulated miRNAs in macrophages, we performed high-throughput small RNA sequencing using BMDMs obtained from Rbpjflox/flox Lyz2-Cre (RBP-J-KO) and from Lyz2-Cre mice. Interestingly, the analysis of small RNA-seq data showed that, among the top ten most upregulated miRNAs in RBP-J-KO BMDMs, three (miR-106a, miR-363, and miR-18b) belonged to the miR-17~92 family (Fig. 5a). By mining small RNA-seq expression profiles, we found that almost all 15 miRNAs of the miR-17~92 family were upregulated in RBP-J-KO BMDMs (Fig. 5b). Upon closer examination, the most upregulated miRNAs belonged mainly to the miR-106a~363 cluster (Fig. 5c), which was almost equally distributed among all four of the different families (Fig. 5d). Next, we validated the small RNA-seq results by quantitative polymerase chain reaction (qPCR) and found similar levels of upregulation for almost all miR-17~92 family miRNAs in the RBP-J-KO BMDMs (Fig. 5e–h). Consistent with the global expression profiling data, the quantitative measurements of individual miRNAs also showed that the miRNA expression levels in all four families were increased to a similar extent (Fig. 5i–l). Taken together, these results identified RBP-J as a suppressor of miR-17~92 family miRNAs.
Fig. 5.
RBP-J suppresses the expression of miR-17~92 family miRNAs in macrophages. a–d Heatmaps showing changes in the top ten upregulated miRNAs (a), all mature miRNAs (b), three miRNA clusters (c), and four miRNA families (d) of the miR-17~92 family in the Rbpjflox/flox Lyz2-Cre versus Lyz2-Cre BMDMs. e–l Results from the qPCR analysis of all mature miRNAs (e), miR-17~92 cluster (f), miR-106a~363 cluster (g), miR-106b~25 cluster (h), miR-17 family (i), miR-18 family (j), miR-19 family (k), or miR-25 family (l) of the miR-17~92 family in the Rbpjflox/flox Lyz2-Cre and Lyz2-Cre BMDMs. The results are normalized to the expression of the control small RNA U6 and are presented relative to those of the Lyz2-Cre BMDMs, which was set to 1. Data are representative of one to two independent experiments
RBP-J promotes IL-12 production partially by suppressing the expression of miR-17~92 family miRNAs
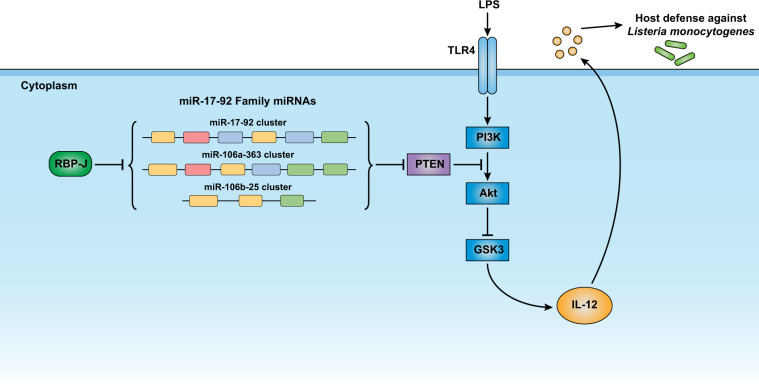
These results implied that miR-17~92 family miRNAs attenuated IL-12 production by targeting PTEN and that RBP-J suppressed the expression of miR-17~92 family miRNAs. Next, we sought to determine whether suppression of miR-17~92 family miRNAs contributed to the RBP-J-mediated enhancement of IL-12 expression (Fig. 6a). As miR-17~92 family miRNAs inhibit IL-12 production by regulating the PI3K-Akt-GSK3 pathway, we first analyzed the activation status of this pathway in the RBP-J-KO BMDMs. The results from the immunoblotting analysis showed significantly decreased PTEN protein (Fig. 6b, c) and subsequently enhanced phosphorylation of Akt and GSK3 in the RBP-J-KO BMDMs (Fig. 6d). These results indicate that RBP-J attenuates PI3K-Akt activation by sustaining PTEN protein expression, which possibly results from the alleviation of the repression that had been induced by the miR-17~92 family miRNA-mediated suppressive effects on PTEN. To genetically test the hypothesis that RBP-J modulates macrophage function by regulating miR-17~92 family miRNAs, we crossed Rbpjflox/flox Lyz2-Cre mice with TKO mice to obtain mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Rbpjflox/flox Lyz2-Cre myeloid-specific quadruple knockout (QKO) mice (Fig. 6e). Upon LPS stimulation, the IL-12 mRNA and protein levels were “rescued,” being upregulated in the QKO BMDMs compared to the RBP-J-KO BMDMs (Fig. 6f–h). These genetic data supported the notion that RBP-J promotes IL-12 expression, at least in part, via the suppression of miR-17~92 family miRNAs. In summary, our results support a model for the inhibition of IL-12 by miR-17~92 family miRNAs, which regulate the PTEN/PI3K-Akt-GSK3 pathway and are suppressed by RBP-J (Fig. 7).
Fig. 6.
RBP-J promotes IL-12 expression by inhibiting the expression of miR-17~92 family miRNAs. a Results from the qPCR analysis of Il12a and Il12b mRNA in the Lyz2-Cre and Rbpjflox/flox Lyz2-Cre BMDMs stimulated with or without LPS for 3 h. b Results from the immunoblotting analysis of PTEN and p38 (loading control) in the whole-cell lysates of the Lyz2-Cre and Rbpjflox/flox Lyz2-Cre BMDMs treated for the indicated periods with LPS. c Quantified PTEN protein abundance under unstimulated conditions in (b), according to the densitometry data from three independent experiments. d Results from the immunoblotting analysis of phosphorylated and total Akt, phosphorylated and total GSK3, and β-actin (loading control) in the whole-cell lysates of the Lyz2-Cre and Rbpjflox/flox Lyz2-Cre BMDMs treated for the indicated periods with LPS. e Outline of the mouse breeding strategy. mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre (TKO) mice were bred with Rbpjflox/flox Lyz2-Cre mice to obtain mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Rbpjflox/flox Lyz2-Cre (QKO) mice. f Results from the qPCR analysis of Il12a and Il12b mRNA in the Lyz2-Cre, Rbpjflox/flox Lyz2-Cre, and QKO BMDMs stimulated with or without LPS for 6 h. g ELISA measurements of IL-12p70 in supernatants from Lyz2-Cre, Rbpjflox/flox Lyz2-Cre, and QKO BMDMs stimulated with or without LPS for 6 h. h Cumulative amounts of the IL-12p70 protein in the LPS-stimulated Rbpjflox/flox Lyz2-Cre and QKO BMDMs, as indicated in (g). Data are representative of (a, b, d, f, and g) or are pooled from (c and h) three independent experiments. Data are presented as the means ± SD. *P < 0.05 (paired Student’s t test)
Fig. 7.
Model depicting the function, mechanisms and regulation of action of the miR-17–92 family miRNAs in macrophages. All three paralogous miRNA clusters of the miR-17–92 family target PTEN cooperate to enhance Akt activation, which results in the subsequent inhibition of GSK3. As a result of the GSK3-mediated promotion of IL-12 expression, miR-17–92 family miRNAs suppress IL-12 and enhance host susceptibility to intracellular pathogen infection. The expression of the three paralogous miRNA clusters of the miR-17–92 family is collectively inhibited by RBP-J, a master transcriptional factor of Notch signaling. Overall, miR-17–92 family miRNAs are regulated by RBP-J and act as critical negative regulators of IL-12 to prevent overactivation of macrophage inflammatory effector functions
Discussion
miR-17~92 family miRNAs have been widely studied in adaptive immune cells, including T cells and B cells.9–12,14,15 In innate immune cells, miR-17~92 family miRNAs reportedly regulate monocyte development.13,38 However, the functions of miR-17~92 family miRNAs in macrophage activation re main unclear. Here, we describe a regulatory circuit involving three RBP-J-suppressed paralogous clusters of miR-17~92 family miRNAs that cooperatively modulate the expression of the essential immune effector molecule IL-12. This study illustrates the previously uncharacterized functions of miR-17~92 family miRNAs in macrophage activation and in immune defense against intracellular bacteria. Furthermore, our results establish the connections between miR-17~92 family miRNAs and RBP-J in regulating IL-12 during macrophage activation.
Regarding their functions, miR-17~92 family miRNAs have predominantly been viewed as oncogenes.8,39 Amplification of these miRNAs has been detected in various cancer cells, in which they promote tumor angiogenesis by inducing cell proliferation and inhibiting apoptosis.7 In the immune system, miR-17~92 family miRNAs have been reported to be involved in the regulation of lymphocyte homeostasis,9 follicular helper T-cell differentiation,10 B-cell development,11,12 and iNKT cell ontogenesis.14 However, relatively little is known about the role of miR-17~92 family miRNAs in innate immune responses. To our knowledge, this study presents the first genetic evidence of the functions of miR-17~92 family miRNAs in macrophage activation, identifying a distinct biological role of miR-17~92 family miRNAs in macrophages that differs from their roles in other immune cells. Interestingly, the three clusters of miRNAs in this family act in a highly cooperative manner to collectively regulate the expression of a key immune effector molecule with significant in vivo functional outcomes in host antimicrobial defense.
In previous studies, various targets of miR-17~92 family miRNAs were reported, such as PTEN,9 Bim,6,9 E2F1,40,41 and PHLPP2.10 In this study, we identified PTEN as a common target of all three miRNA clusters in the miR-17~92 family and connected miR-17~92 family miRNAs targeting PTEN with the downstream PI3K-Akt-GSK3 pathway, which further regulates IL-12 production during macrophage activation. Given the multiple targets of miR-17~92 family miRNAs, we cannot totally exclude the contribution of other targets in the regulation of IL-12 production, and more work may be needed to fully clarify the roles of other factors in the future.
IL-12 is a critical proinflammatory cytokine secreted by macrophages and other immune cells that, on the one hand, promotes host immune defense and, on the other hand, is involved in the pathogenesis of autoimmune and inflammatory diseases. Thus, the production of IL-12 is strictly regulated. It is widely reported that IL-12 expression is regulated both positively and negatively by a variety of mechanisms, including epigenetic modification and signaling regulation, posttranscriptional regulation, and transcription factor regulation.16,20–22 We have previously reported that RBP-J controls the expression of the transcription factor IRF8 to promote downstream IL-12 production in macrophages.37 Here, we propose that RBP-J suppresses the expression of miR-17~92 family miRNAs to promote IL-12 production. Thus, we connect miR-17~92 family miRNAs with RBP-J in the regulation of the production of IL-12. However, whether miR-17~92 family miRNAs interact with other regulators of IL-12 and the mechanisms of this possible action are not yet clear, and further studies are needed to fully understand the regulatory network of IL-12. In summary, our findings highlight functions, as well as the regulatory effects, of miR-17~92 family miRNAs in macrophage activation and immune defense, implicating this family of miRNAs as being potential therapeutic targets in the treatment of immunity-related disorders.
Materials and methods
Mice
Experiments using mice were approved by the Institutional Animal Care and Use Committees at Tsinghua University. Mice with myeloid cell-specific deletion of Rbpj (Rbpjflox/floxLyz2-Cre) have been previously described.42 The mir-106a~363−/− (Jax stock 008461), mir-106b~25−/− (Jax stock 008460), and mir-17~92flox/flox (Jax stock 008458) mice were purchased from The Jackson Laboratory and were all on a C57BL/6J background. The mir-17~92flox/flox mice were crossed with the Lyz2-Cre mice to obtain mice with myeloid cell-specific deletion of the miR-17~92 cluster. The miR-17~92, miR-106a~363, and miR-106b~25 cluster triple KO mice were obtained by crossing mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox mice with Lyz2-Cre mice. The QKO mice were obtained by crossing mir-106a~363−/− mir-106b~25−/− mir-17~92flox/flox Lyz2-Cre mice with Rbpjflox/floxLyz2-Cre mice. The experiments were performed with mice that were 6–8 weeks of age using age- and gender-matched controls. Wild-type C57BL/6J mice were used as controls for the 106a KO and 106b KO mice, and Lyz2-Cre mice were used as controls for the 17/92 KO, TKO, RBP-J-KO, and QKO mice.
Cell culture and reagents
Murine BMDMs were obtained as previously described37 and maintained in DMEM supplemented with 10% FBS and 10% L929 cell supernatant, which is conditioned medium that provides macrophage colony-stimulating factor (M-CSF). Cell culture grade LPS was purchased from InvivoGen and was used at a concentration of 10 ng/ml unless otherwise specified. SF1670 and SB415286 were obtained from Selleck.
Reverse transcription and qPCR
RNA was extracted from whole-cell lysates with a total RNA purification kit (GeneMark) and was reverse transcribed to cDNA with a First Strand cDNA Synthesis Kit (TaKaRa). qPCR was performed in triplicate with an ABI StepOnePlus thermal cycler. The primary transcripts were measured with primers that amplify either exon–intron junctions or intronic sequences. The threshold cycle numbers were normalized to triplicate samples amplified with primers specific for glyceraldehyde-3-phosphate dehydrogenase (Gapdh). For the qPCR analysis of mature miRNA, cDNA was prepared from total RNA, which was isolated with TRIzol reagent (Invitrogen), with a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems). TaqMan MicroRNA assays were used according to the manufacturer’s recommendations (Applied Biosystems) for real-time PCR. The TaqMan U6 snRNA assay (Applied Biosystems) was used for normalization of the expression values. The primer sequences are listed in Supplementary Table 1.
RNA sequencing and analysis
Total RNA was isolated with a total RNA purification kit (GeneMark) from whole-cell lysates of Lyz2-Cre and TKO BMDMs treated with or without LPS for 3 h. RNA was isolated, prepared into a library, and sequenced with a BGISEQ-500 platform by BGI (BGI; Shenzhen, China). Total reads were cleaned and mapped to the mm10 reference genome and then normalized as fragments per kilobase of transcript per million mapped reads. Differentially expressed genes between Lyz2-Cre and TKO BMDMs were defined based on a false-discovery rate < 0.001 and an absolute value of log2 ratio (TKO/Lyz2-Cre) > 1.
Enzyme-linked immunosorbent assay
Cytokine secretion was quantified with an IL-12p70 ELISA kit from BD Biosciences according to the manufacturer’s instructions.
Listeria monocytogenes infection
Mice were infected intravenously with L. monocytogenes strain LM-OVA at a dose of 5 × 104 to 6 × 105 colony-forming units (CFU)/mouse, as previously described.43–45 Six days post infection, the bacterial burden in the spleens and livers was determined by counting the CFU of serially diluted homogenized spleens and livers cultured on brain heart infusion agar plates (BD Biosciences).
Immunoblot analysis
Whole-cell lysates were prepared as described previously.37 For the immunoblotting analysis, the lysates were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels that were transferred to a polyvinylidene fluoride membrane (Millipore) for probing with antibodies. The antibody against p38 (sc-535) was purchased from Santa Cruz Biotechnology. All the other antibodies were obtained from Cell Signaling Technology.
Luciferase reporter assay
The psiCHECK2 (Promega) reporter plasmid was cloned with 3′UTR fragments of Pten to generate Pten reporter plasmids. 293T cells were plated into 24-well plates at 1 × 105 cells per well 24 h before transfection with 10 ng reporter plasmid and 600 ng negative control, miR-17~92 cluster, miR-106a~363 cluster, or miR-106b~25 cluster miRNA overexpression vector using the Lipofectamine 2000 transfection reagent (Invitrogen). Luciferase assays were performed 24 h post transfection using a dual-luciferase reporter assay system (Promega) following the manufacturer’s protocol. The Renilla firefly luciferase (Rluc) activity was normalized to the firefly luciferase activity (Luc). The expression is presented as the Rluc/Luc ratio.
RNA-mediated interference
Small interfering RNA (siRNA) specifically targeting mouse Pten and nontargeting control siRNA were obtained from GenePharma. The siRNA was transfected into the mouse BMDMs through the use of TransIT TKO transfection reagent according to the manufacturer’s instructions (Mirus Bio). The cells were lysed 72 h post transfection for mRNA and protein extraction.
Small RNA-seq analysis
Total RNA was isolated, and the small RNA fractions were enriched with a mirVana miRNA isolation kit (Life Technologies) according to the manufacturer’s instructions. The miRNA libraries were constructed per the Illumina TruSeq small RNA library preparation kit. High-throughput sequencing was performed using an Illumina HiSeq 1500 instrument. The miRNA-seq reads were aligned to the mouse miRNA sequences in the miRBase database (release 21) using miRDeep2 software. Mature miRNA values were normalized by library size (corresponding to the counts per million (cpm) mapped miRNA reads). miRNAs with cpm values <5 in all conditions were eliminated from the analysis. RBP-J-regulated miRNAs were defined as those in which the cpm values of the miRNAs in the Rbpjflox/flox Lyz2-Cre BMDMs versus those in the Lyz2-Cre cells were greater than 1.2 (upregulated) or less than 0.6 (downregulated), with a P value less than 0.05.
Statistical analysis
P values were calculated with a two-tailed paired or unpaired Student’s t test, log-rank (Mantel–Cox) test, or Mann–Whitney U test and identified as not significant; P > 0.05; *P < 0.05; **P < 0.01; and ***P < 0.001. Statistical analyses were performed using GraphPad Prism 7.
Supplementary information
Acknowledgements
We thank B. Zhang (Tsinghua University) for advice and help with the small RNA-seq analysis. We thank Y. Zhang (University of Maryland) for help with the RNA-seq analysis. We thank C. Dong (Tsinghua University) for providing Listeria monocytogenes. We thank W. Guo (Zhejiang University) for providing mir-106a~363−/− and mir-106b~25−/− mice. This research was supported by the Ministry of Science and Technology of China National Key Research Projects (2015CB943201 to X.H. and 2015CB943200 to L.W.), National Natural Science Foundation of China grants (31821003, 31725010, 81571580, 91642115, and 81661130161 to X.H. and 31330027 to L.W.), funds from Tsinghua-Peking Center for Life Sciences (X.H., L.W., and X.Z.), funds from the Institute for Immunology at Tsinghua University (X.H. and L.W.), and funds from the National Institutes of Health (B.Z.).
Author contributions
X.Z. designed the research, performed the experiments, analyzed the data, and wrote the paper; S.S. performed small RNA-seq experiments; B.Z. contributed to the small RNA-seq experiments and provided advice on the experiments; X.W. provided advice and key reagents; L.W. provided miR-106a~363−/−, miR-106b~25−/− and mir-17~92flox/flox mice and contributed to paper preparation; X.H. conceptualized the project, designed the research, supervised the experiments, and wrote the paper.
Data availability
The small RNA-seq and RNA-seq data sets were deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession numbers GSE103220, GSE129613, and GSE133844.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41423-020-0363-5) contains supplementary material.
References
- 1.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 4.Altuvia Y, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014). [DOI] [PMC free article] [PubMed]
- 6.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17~92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT. miRiad roles for the miR-17~92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17~92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang SG, et al. MicroRNAs of the miR-17~92 family are critical regulators of TFH differentiation. Nat. Immunol. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai M, et al. Regulation of B-cell development and tolerance by different members of the miR-17~92 family microRNAs. Nat. Commun. 2016;7:12207. doi: 10.1038/ncomms12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benhamou D, et al. A c-Myc/miR17-92/Pten axis controls PI3K-mediated positive and negative selection in B cell development and reconstitutes CD19 deficiency. Cell Rep. 2016;16:419–431.. doi: 10.1016/j.celrep.2016.05.084. [DOI] [PubMed] [Google Scholar]
- 13.Fontana L, et al. MicroRNAs 17-5p–20a–106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 2007;9:775. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 14.Fedeli M, et al. miR-17∼92 family clusters control iNKT cell ontogenesis via modulation of TGF-β signaling. Proc. Natl Acad. Sci. 2016;113:E8286–E8295. doi: 10.1073/pnas.1612024114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labi V, Schoeler K, Melamed D. miR-17∼92 in lymphocyte development and lymphomagenesis. Cancer Lett. 2019;446:73–80. doi: 10.1016/j.canlet.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 17.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 2008;26:421–452.. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng MWL, et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 2015;21:719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin J, et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 2016;17:259–268. doi: 10.1038/ni.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goriely S, Neurath MF, Goldman M. How microorganisms tip the balance between interleukin-12 family members. Nat. Rev. 2008;8:81–86. doi: 10.1038/nri2225. [DOI] [PubMed] [Google Scholar]
- 22.Lyakh L, Trinchieri G, Provezza L, Carra G, Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunol. Rev. 2008;226:112–131.. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pamer EG. Immune responses to Listeria monocytogenes. Nat. Rev. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 24.Shi C, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Cao X. Cellular and molecular regulation of innate inflammatory responses. Cell. Mol. Immunol. 2016;13:711–721.. doi: 10.1038/cmi.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–574.. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 2005;6:777–784.. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen P, Frame S. The renaissance of GSK3. Nat. Rev. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 29.Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31:24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanigaki K, Honjo T. Two opposing roles of RBP-J in Notch signaling. Curr. Top. Dev. Biol. 2010;92:231–252.. doi: 10.1016/S0070-2153(10)92007-3. [DOI] [PubMed] [Google Scholar]
- 33.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 35.Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 2016;7:159–174. doi: 10.1007/s13238-016-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radtke F, Fasnacht N, MacDonald HR. Notch Signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Xu H, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351.. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Fuziwara C. S., Kimura E. T. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front. Med.2 64 (2015). [DOI] [PMC free article] [PubMed]
- 40.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 41.Novotny GW, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007;14:879. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 42.Hu X, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-γ pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung Y, et al. T cells and T cell tumors efficiently generate antigen-specific cytotoxic T cell immunity when modified with an NKT ligand. OncoImmunology. 2012;1:141–151.. doi: 10.4161/onci.1.2.18479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seregin SS, Chen GY, Laouar Y. Dissecting CD8+ NKT cell responses to Listeria infection reveals a component of innate resistance. J. Immunol. 2015;195:1112–1120. doi: 10.4049/jimmunol.1500084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji L, et al. Slc6a8-mediated creatine uptake and accumulation reprogram macrophage polarization via regulating cytokine responses. Immunity. 2019;51:272–284.e7. doi: 10.1016/j.immuni.2019.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The small RNA-seq and RNA-seq data sets were deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession numbers GSE103220, GSE129613, and GSE133844.