Abstract
Fusidane-type antibiotics, represented by helvolic acid, fusidic acid and cephalosporin P1, are fungi-derived antimicrobials with little cross-resistance to commonly used antibiotics. Generation of new fusidane-type derivatives is therefore of great value, but this is hindered by available approaches. Here, we developed a stochastic combinational strategy by random assembly of all the post-tailoring genes derived from helvolic acid, fusidic acid, and cephalosporin P1 biosynthetic pathways in a strain that produces their common intermediate. Among a total of 27 gene combinations, 24 combinations produce expected products and afford 58 fusidane-type analogues, of which 54 are new compounds. Moreover, random gene combination can induce unexpected activity of some post-tailoring enzymes, leading to a further increase in chemical diversity. These newly generated derivatives provide new insights into the structure‒activity relationship of fusidane-type antibiotics. The stochastic combinational strategy established in this study proves to be a powerful approach for expanding structural diversity of natural products.
KEY WORDS: Fusidane-type antibiotics, Combinational biosynthesis, Triterpenoids, Fungi, Tailoring enzymes
Graphical abstract
Chemical diversification of fusidane-type antibiotics was accomplished using a stochastic combinational strategy through random assembly of all the post-tailoring genes derived from helvolic acid, fusidic acid and cephalosporin P1 pathways.
1. Introduction
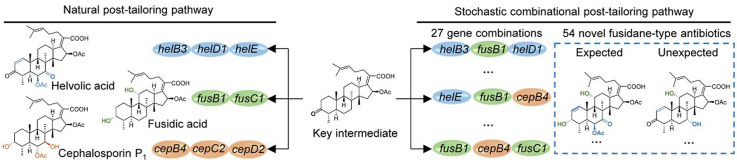
Fusidane-type antibiotics are a small group of fungi-derived triterpenoid antimicrobials, which are featured with a 4α-methyl group, 16β-acetoxyl group and 20-carboxylic acid in the protostadienol framework1. Helvolic acid (1)2, fusidic acid (2)3 and cephalosporin P1 (3)4 are the three most representative compounds of this class (Fig. 1A). They exhibit potent antibacterial activity against Gram-positive bacteria5, among which fusidic acid has been approved to clinically treat skin infections caused by Staphylococcus aureus6. Moreover, fusidic acid is the only clinically used drug that targets elongation factor G (EF-G) to inhibit bacterial protein synthesis7,8. This has made fusidane-type antibiotics have little cross-resistance to other commonly used antibiotics.
Figure 1.
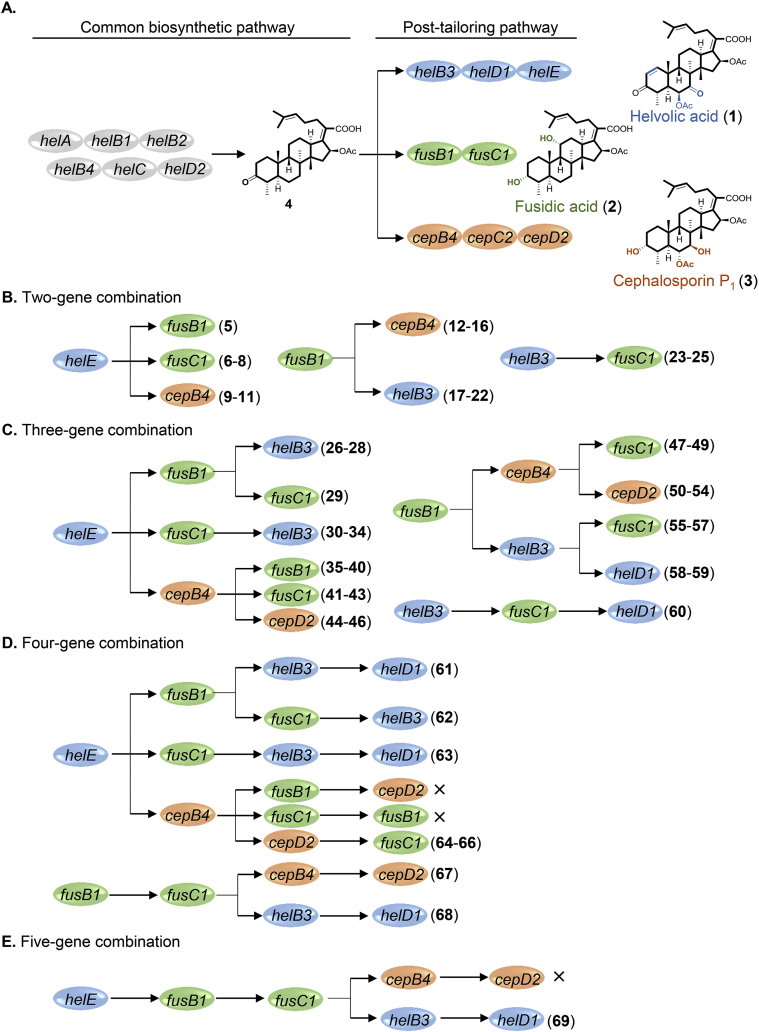
The stochastic combinational strategy for diversification of fusidane-type antibiotics. (A) Biosynthetic pathways of helvolic acid (1), fusidic acid (2) and cephalosporin P1 (3). (B) Two-gene combination. (C) Three-gene combination. (D) Four-gene combination. (E) Five-gene combination.
Because of the pharmaceutical prospects of fusidane-type antibiotics, the search for new analogues from nature has never been stopped. Recently, several new helvolic acid derivatives are discovered from Aspergillus species9, 10, 11. However, this process is highly haphazard and inefficient. On the other hand, chemical derivatization has been extensively employed for diversification of fusidane-type antibiotics5,12, 13, 14, 15, 16, 17. These studies are mainly focused on modification of the reactive groups including 16β-acetoxyl group, C17/C20 or C24/C25 double bond, 3-hydroxyl group and 20-carboxylic acid, however, analogues with greater antibacterial activity than that of fusidic acid have not yet been found. Combinational biosynthesis, designed to fuse the capabilities of combinational chemistry with the genetic power and enzymatic prowess of biosynthesis, serves as a promising complementary approach to chemical synthesis18. There are two major strategies in combinational biosynthesis: one is to alter the substrate specificity by enzyme engineering, which is usually applied in multi-modular polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) through swapping of the entire domains, modules and subunits of different enzymes; the other is to reprogram the natural biosynthetic pathway by mixing genes from different species19. Despite of the great success of combinational biosynthesis in generating natural product analogues, the compatibility between the combined enzymes derived from different origins is a key constraint, and we are still far from programing any desired small molecule at will. To bypass the restriction of the compatibility between enzymes, a strategy using enzymes derived from similar or related biosynthetic pathways have been developed and successfully applied for antitumor indolocarbazole compounds and decalin-containing diterpenoid pyrones20,21. However, the systematic study on the compatibility of the enzymes from related biosynthetic pathways is still lacked.
Recently, we have exhaustively characterized the complete biosynthetic pathway for fusidic acid, helvolic acid and cephalosporin P122, 23, 24. We have shown that these three compounds share an early-stage biosynthetic pathway involving six conserved genes (helA, helB1, helC, helB2, helD2 and helB4) for construction of a common intermediate 16β-acetyloxy-29-norprotosta-17(20)Z,24-dien-3-one-21-oic acid (4), which then diverges to corresponding final products under the action of the rest genes in individual gene clusters (Fig. 1A). Notably, these post-tailoring enzymes exhibit broad substrate promiscuity, allowing for recruiting combinational biosynthetic approach to generate new fusidane-type antibiotics.
Here, we adopt a stochastic combinational strategy by introducing all the possible post-tailoring gene combinations into a strain harboring the six conserved genes. We observe that among a total of 27 gene combinations, 24 combinations produce the expected products. Moreover, random gene combination can trigger novel enzymatic activity of some post-tailoring enzymes and generate unexpected products, leading to a further increase in chemical diversity. Our study has demonstrated that the stochastic combinational biosynthesis based on random combination of post-tailoring genes from pathways sharing a common intermediate is an efficient approach to expand chemical diversity of natural products.
2. Results
2.1. Stochastic combination of post-tailoring genes from hel, fus and cep gene clusters
Despite of the wide application of combinational biosynthesis in diversification of natural products, systematic studies by a stochastic strategy involving all the possible gene combinations have not yet been carried out. To test the potential of compatibility between the post-tailoring enzymes from different pathways sharing a common intermediate and to maximize the chemical diversity of fusidane-type antibiotics, we adopted a stochastic combinational strategy by introducing all the possible combinations of post-tailoring genes from helvolic acid, fusidic acid and cephalosporin P1 biosynthetic pathways into the six-gene expression strain (Fig. 1A). Among the eight post-tailoring genes from hel, fus and cep clusters22, 23, 24 (Supporting Information Table S1), fusC1 and cepC2 encode the short chain dehydrogenases/reductases (SDRs) with the same function, thus either fusC1 or cepC2 is used for gene combination. Since both helB3 and cepB4 encode P450 enzymes that catalyze dual oxidation at C6 and C7, they are avoided being simultaneously used. In addition, the acyltransferases HelD1 and CepD2 have to be used after HelB3 and CepB4, respectively. Due to the above restrictions, a total of 27 gene combinations have been designed, including 6 two-gene combinations, 11 three-gene combinations, 8 four-gene combinations and 2 five-gene combinations (Fig. 1B‒E).
2.2. Diversification of fusidane-type antibiotics via random combinational biosynthesis
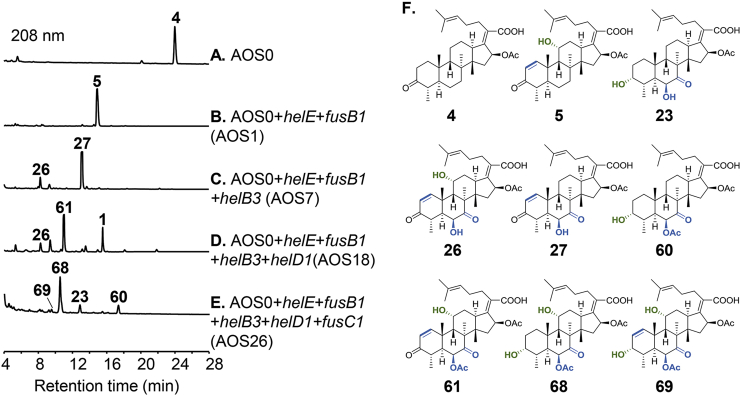
We have previously established the Aspergillus oryzae NSAR1 transformant harboring the six conserved genes helA, helB1, helC, helB2, helD2 and helB4, which produces the common intermediate 4 of fusidane-type antibiotics24. This six-gene transformant, termed as AOS0 (Fig. 2A), was used as the parent strain for construction of all the gene combinations. We first carried out the two-gene combinations. helE is derived from the helvolic acid pathway and responsible for C1/C2 double bond formation, and fusB1 from the fusidic acid pathway accounts for C11α-hydroxylation. When these two genes were combined and introduced into AOS0, a new major peak (5) was observed in the resulting transformant AOS1 (Fig. 2B). Isolation and full NMR analysis proved this compound to be the expected C11α-hydroxylated and C1/C2 dehydrogenated product of 4. Moreover, when we stepwise introduced helB3, helD1 and fusC1 into AOS1 to carry out three-, four- and five-gene combinations, all the resulting transformants AOS7, AOS18 and AOS26 generated the expected products 26, 61 and 69, respectively (Fig. 2C‒F). Additionally, compounds 1, 23, 27, 60 and 68 arising from the action of partial combinational genes were also observed. Using the above strategy, we have completed the construction of 27 gene combinations in AOS0 (Supporting Information Fig. S1 and Table S2). As shown in Figs. S1, 24 of the total 27 gene combinations gave their target compounds, leading to isolation of 58 fusidane-type analogues, among which 54 are new compounds (Supporting Information Fig. S2). All these new compounds were isolated and structurally determined by full NMR analysis and calculation of 13C chemical shifts (Supporting methods, Supporting Information Figs. S3‒S65 and Tables S3‒S69). Though there are still three combinations that did not produce the target products (Fig. 1D and E), the stochastic combination of post-tailoring genes from pathways with a common intermediate seems to be an effective approach to generate diversified natural products.
Figure 2.
HPLC analysis of metabolites from representative examples of two-gene, three gene, four-gene and five-gene combination transformants. (A) A. oryzae harboring helA, helB1, helB2, helC, helB4 and helD2 (AOS0). (B) Two-gene combination transformant AOS1 with addition of helE and fusB1 into AOS0. (C) Three-gene combination transformant AOS7 with addition of helE, fusB1 and helB3 into AOS0. (D) Four-gene combination transformant AOS18 with addition of helE, fusB1, helB3 and helD1 into AOS0. (E) Five-gene combination transformant AOS26 with addition of helE, fusB1, helB3, helD1 and fusC1 into AOS0. (F) Structures of isolated fusidane-type antibiotics from A. oryzae transformants.
2.3. Induction of novel enzymatic activity by random gene combination
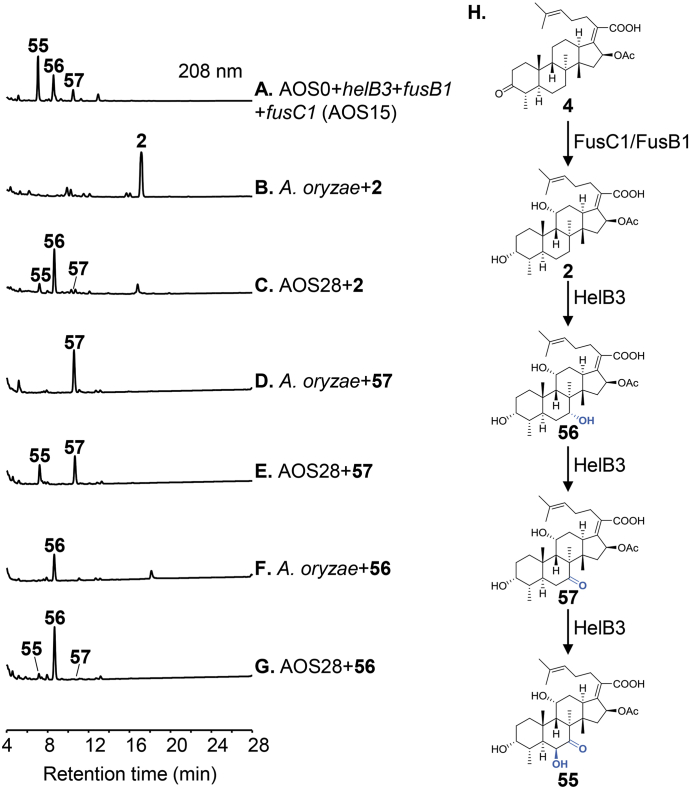
Production of 54 new fusidane analogues from 24 gene combinations suggests that many combinations produce more than one new compound. Careful characterization of these combinations reveals that some enzymes in these combinations show novel enzymatic activity. helB3 encodes a P450 enzyme that catalyzes the dual oxidation to afford the 6β-OH and 7-keto in the helvolic acid biosynthesis. However, when this gene is combined with fusB1 and fusC1 derived from the fusidic acid biosynthesis (AOS15), two monooxygenated products 56 and 57 were produced in addition to the predicted dually oxidized product 55 (Fig. 3A and H). To confirm this, we constructed the helB3 expression strain AOS28 and performed a feeding experiment using compound 2. We observed an efficient transformation of 2 to 55, 56 and 57 by AOS28, but not by the untransformed A. oryzae (Fig. 3B and C). Isolation of 56 and 57 allowed us to obtain further insights into the oxidation sequence of the multifunctional HelB3 by feeding AOS28 with 56 and 57, respectively (Fig. 3D‒G). These results showed that the multi-step oxidation catalyzed by HelB3 begins with formation of 7α-OH followed by oxidation to 7-keto, and finally terminates with attachment of the 6β-OH group (Fig. 3H).
Figure 3.
HPLC analysis for verification of the oxidation sequence of the multifunctional P450 enzyme HelB3. (A) Three-gene combination transformant AOS15 with addition of helB3, fusB1 and fusC1 into AOS0. (B) A. oryzae incubated with 2. (C) AOS28 harboring helB3 incubated with 2. (D) A. oryzae incubated with 57. (E) AOS28 incubated with 57. (F) A. oryzae incubated with 56. (G) AOS28 incubated with 56. (H) HelB3-mediated dual oxidation at C6 and C7.
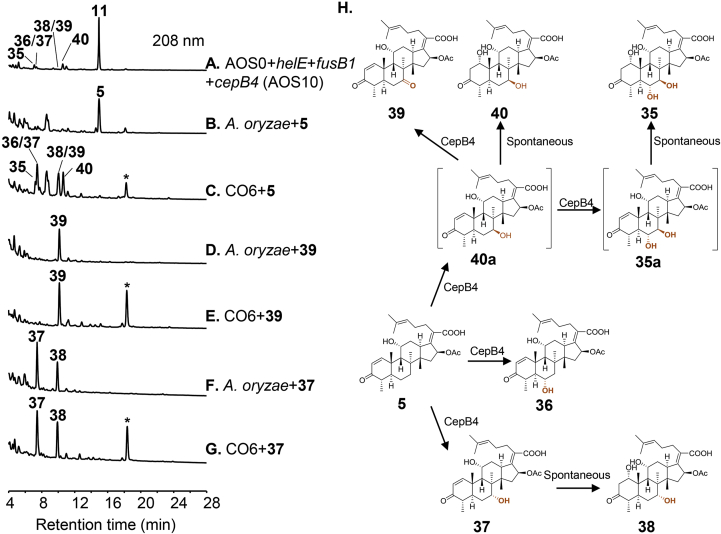
Aside from HelB3, novel activity of the P450 enzyme CepB4 and the acetyltransferase CepD2 were also observed during random combinational biosynthesis. We have previously shown that CepB4 is a multifunctional P450 enzyme that catalyzes the dual hydroxylation at C6 and C7 of 4 to afford 6α-OH and 7β-OH. This process proceeds with C7β-hydroxylation followed by C6α-hydroxylation22. In addition, a shunt pathway starting with the C6α-hydroxylation followed by formation of 6-keto is also observed22. However, when CepB4 is combined with HelE and FusB1 (AOS10), in addition to exerting the original function to produce the predicted compounds 35, 36 and 40, CepB4 also produced three additional compounds 37, 38 and 39 featured with 7α-OH or 7-keto (Fig. 4A and H). This result suggests that CepB4 is capable of catalyzing C7α-hydroxylation. As 37 and 39 contain 11α-OH and the double bond at C1 and C2, we presumed that the presence of these groups in the substrate might affect its positioning in the active site of CepB4, which, therefore, expands the catalytic function of CepB4. In order to test the hypothesis, we performed feeding experiments using the cepB4-harboring strain CO622. When fed with 5, CO6 indeed yielded 37, 38 and 39 (Fig. 4B and C). Additionally, 35, 36 and 40 were also observed in the feeding experiment. To rule out the possibility that 37 is formed by reduction of 39, feeding CO6 and A. oryzae with 39 was carried out and formation of 37 was not detected in both cases (Fig. 4D and E). On the other hand, feeding CO6 with 37 did not give 39, suggesting 39 is likely to be derived from the C7β-hydroxylated product 40a (Fig. 4F‒H). In addition, we observed an efficient conversion of 37 to 38 in both CO6 and control strain (Fig. 4F and G), suggesting this might be a non-enzymatic process. Compounds 35, 38 and 40 bear 1α-OH instead of C1/C2 double bond (Fig. 4H). We reasoned that 7-OH and 11-OH might weaken the stability of products, and the α,β-unsaturated carbonyl in the products were more readily to undergo 1,4-addition reaction to form β-hydroxy ketone25. This was confirmed with appearance of 38 when 37 was incubated in Tris-HCl buffer of pH 8.0, but not pH 5.0 or 7.0 (Supporting Information Fig. S66). And we also found that the pH value of culture broth can reach 8.0 when cultured for 3 days, despite that the initial pH of fermentation medium is 5.5.
Figure 4.
HPLC analysis for functional study of the P450 enzyme CepB4. (A) Three-gene combination transformant AOS10 with addition of helE, fusB1 and cepB4 into AOS0. (B) A. oryzae incubated with 5. (C) CO6 harboring cepB4 incubated with 5. (D) A. oryzae incubated with 39. (E) CO6 incubated with 39. (F) A. oryzae incubated with 37. (G) CO6 incubated with 37. (H) CepB4-mediated oxidation of C6 and C7. *The peak is annotated as the impurity peak on the basis of MS analysis (Supporting Information Fig. S76).
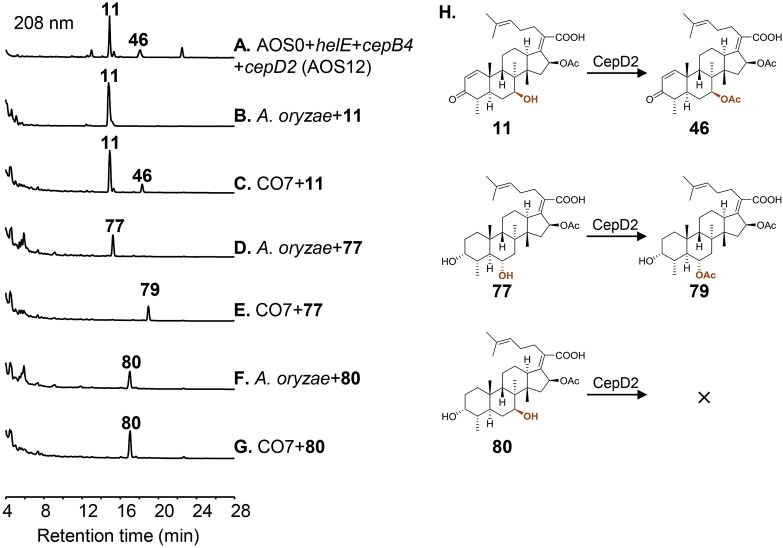
Another unexpected reaction is the acetylation of 7β-OH catalyzed by CepD2. In cephalosporin P1 biosynthesis, we have revealed that CepD2 specifically transfers acetyl group to 6α-OH, but not to 7β-OH, and the acetyl group at 6α-OH can spontaneously shift to 7β-OH under alkaline condition22. However, when CepD2 was combined with CepB4 and HelE, the resulting strain AOS12 generated a product 46 only bearing 7β-acetoxyl group (Fig. 5A and H). To determine whether acetylation of 7β-OH is catalyzed by CepD2 or the endogenous enzyme from A. oryzae NSAR1, the cepD2 harboring strain CO722 and A. oryzae NSAR1 were employed to perform feeding experiments. HPLC‒MS analysis demonstrated that CepD2 can convert the mono-hydroxylated product 11 at 7β-position to 46, while A. oryzae NSAR1 cannot (Fig. 5B and C). Combined with the results that CepD2 acetylates the 6α-hydroxylated product 77 (Fig. 5D and E), but not the 7β-hydroxylated form 80 (Fig. 5F and G), we proposed that the function of CepD2 is expanded with the presence of 3-keto or/and the C1/C2 double bond in the substrate.
Figure 5.
HPLC analysis for functional study of the acetyltransferase CepD2. (A) Three-gene combination transformant AOS12 with addition of helE, cepB4 and cepD2 into AOS0. (B) A. oryzae incubated with 11. (C) CO7 harboring cepD2 incubated with 11. (D) A. oryzae incubated with 77. (E) CO7 incubated with 77. (F) A. oryzae incubated with 80. (G) CO7 incubated with 80. (H) CepD2-mediated acetylation of 6α-OH and 7β-OH.
2.4. Structure‒activity relationship analysis of fusidane-type antibiotics
By systematic combinational biosynthesis, we obtained 58 fusidane-type antibiotics (Fig. S2). Combining with previously isolated 22 analogues during biosynthesis of helvolic acid, fusidic acid and cephalosporin P1 (Supporting Information Fig. S67), we therefore constructed a library consisting of 80 fusidane-type antibiotics, providing an opportunity to systematically analyze the structure‒activity relationship of fusidane-type antibiotics. We first tested the anti-S. aureus 209P activity of all these compounds by determining the minimum inhibitory concentration (MIC). As shown in Table 1, most compounds exhibited potent inhibitory activity. Although more active compounds than fusidic acid were not found, we obtained several new insights into the structure‒activity relationship. Previous study has shown that 3α-OH has better antibacterial activity than 3-keto, and 3-keto is better than 3β-OH. We confirmed that most compounds follow this rule (Supporting Information Figs. S68 and S69A), however, for compounds derived from helvolic acid pathway that possess 6β-OH/7-keto or C1/C2 double bond or both, reduction of 3-keto to 3α-OH has a negative effect on their antibacterial activity (Fig. S69B). We also observed that C1/C2 double bond generally has a negative effect on the antibacterial activity (Supporting Information Fig. S70A), but it can enhance the activity of compounds from helvolic acid pathway that possess 3-keto or 6β-OH/7-keto or both (Fig. S70B). Similarly, C6 or/and C7 oxidation generally decreases the activity (Supporting Information Figs. S71 and S72A), but 6β-OH/7-keto increase the activity of the compounds possessing 3-keto or C1/C2 double bond or both (Fig. S72B). These results clearly indicate that there exists a synergistic effect of the C1/C2 double bond, 6β-OH/7-keto and 3-keto on anti-S. aureus 209P activity for compounds derived from helvolic acid pathway. This synergistic effect will be destroyed when modified with 3-keto reduction or C11α-hydroxylation from cephalosporin P1 or fusidic acid pathway.
Table 1.
Inhibitory effects of fusidane-type antibiotics on S. aureus 209P and M. luteus.
| Compd. | MIC (μg/mL) |
Compd. | MIC (μg/mL) |
Compd. | MIC (μg/mL) |
|||
|---|---|---|---|---|---|---|---|---|
| S. aureus 209P | M. luteus | S. aureus 209P | M. luteus | S. aureus 209P | M. luteus | |||
| 1 | 4 | 64 | 29 | 1 | 4 | 57 | 128 | 32 |
| 2 | 0.125 | 2 | 30 | 128 | 32 | 58 | 128 | >128 |
| 3 | 0.5 | 8 | 31 | 8 | 8 | 59 | >128 | >128 |
| 4 | 16 | 32 | 32 | 8 | 8 | 60 | 64 | 8 |
| 5 | 16 | 4 | 33 | 32 | 4 | 61 | 128 | 64 |
| 6 | 4 | 32 | 34 | 32 | 16 | 62 | 128 | >128 |
| 7 | 4 | 32 | 35 | >128 | 128 | 63 | 128 | 64 |
| 8 | 2 | 64 | 36 | >128 | 32 | 64 | 16 | 64 |
| 9 | 32 | 64 | 37 | >128 | 128 | 65 | 4 | 8 |
| 10 | 128 | >128 | 38 | >128 | 64 | 66 | 32 | >128 |
| 11 | 16 | 8 | 39 | >128 | 64 | 67 | 2 | 32 |
| 12 | >128 | >128 | 40 | >128 | >128 | 68 | 64 | 32 |
| 13 | >128 | >128 | 41 | 32 | 128 | 69 | 128 | >128 |
| 14 | 64 | 128 | 42 | 128 | 64 | 70 | 32 | 16 |
| 15 | 32 | 32 | 43 | 16 | 8 | 71 | >128 | 128 |
| 16 | 32 | 64 | 44 | 16 | 32 | 72 | >128 | 128 |
| 17 | >128 | >128 | 45 | 64 | 64 | 73 | 1 | 16 |
| 18 | 128 | >128 | 46 | 16 | >128 | 74 | 16 | >128 |
| 19 | >128 | >128 | 47 | 64 | >128 | 75 | 16 | 32 |
| 20 | 32 | 128 | 48 | 8 | 64 | 76 | 128 | 128 |
| 21 | 8 | 128 | 49 | 32 | 64 | 77 | 4 | 8 |
| 22 | 128 | 64 | 50 | 16 | >128 | 78 | 16 | 64 |
| 23 | 16 | 4 | 51 | 8 | >128 | 79 | 2 | 32 |
| 24 | 2 | 2 | 52 | 8 | 32 | 80 | 16 | 16 |
| 25 | 16 | 4 | 53 | 16 | 128 | Tobramycin | 0.03 | – |
| 26 | 128 | >128 | 54 | 8 | 32 | Ampicillin | – | 0.125 |
| 27 | 1 | 16 | 55 | 32 | >128 | |||
| 28 | 8 | 8 | 56 | 2 | >128 | |||
‒Not applicable.
C11α-hydroxylation is the key modification that confers the potent antibacterial activity to fusidic acid (Supporting Information Fig. S73A), however, in most cases, we observed that it has a negative effect when C6 or/and C7 oxidation occurs (S73B). In addition, we have previously shown that acetylation of 6-OH has an opposite effect on the antibacterial activity of helvolic acid and cephalosporin P122,24. In helvolic acid, 6β-OH acetylation decreases the activity whereas acetylation of 6α-OH in cephalosporin P1 increases the activity. Here we further confirmed the opposite effects of 6β-OH and 6α-OH acetylation in a series of compounds including C6/C7 dually oxidized or monooxygenated products (Supporting Information Fig. S74). These results suggest that the configuration of 6-OH is a crucial factor that affects the antibacterial activity of acetylated products. Moreover, we also found that 1α-OH has a negative effect on antibacterial activity (Supporting Information Fig. S75).
In addition, we also evaluated the antibacterial activity of these compounds against Gram-positive bacterium Micrococcus luteus (Table 1). We found that the structure‒activity relationship towards M. luteus is slightly different from that against S. aureus. Among them, compound 24 exhibits potent anti-M. luteus activity with a MIC value of 2.0 μg/mL, which is equivalent to that of fusidic acid. This suggests that we can use different activity screening systems to find more active compounds.
3. Discussion
With uncovering of numerous biosynthetic gene clusters and advances in synthetic biology, combinational biosynthesis has received much more attention and becomes a widely used approach for diversification of natural products. Since this approach is mainly based on combination of enzymes, modules and domains from different sources, the compatibility among the combined components is the key constraint, and we are still unable to create any desired molecule at will thus far. In this study, we developed a stochastic combinational strategy based on random combination of the post-tailoring genes, which are derived from related biosynthetic pathways sharing a common intermediate. These genes are highly compatible with each other as they directly or indirectly act on their common intermediate. Among a total of 27 gene combinations, 24 combinations afford the expected products though some have low yields. Our study thus demonstrated that the stochastic combinational strategy established in this work would be a powerful approach for chemical diversification of natural products.
The most important finding of this study is that random gene combination does not only maximize the chemical diversity of target compounds, but also induces novel enzymatic activity of the combined enzymes. The multifunctional P450 CepB4 has been previously shown to catalyze 6α- and 7β-dual hydroxylation in cephalosporin P1 biosynthesis22. However, in the present study, when the substrate is equipped with the C1/C2 double bond and 11α-hydroxyl group, we found that CepB4 can simultaneously give 7α- and 7β-monohydroxylated products (Fig. 4H). We inferred that the ferryl-oxo intermediate [FeIV=O, porphyrin π cation radical] locates at an equal distance from 7α-H and 7β-H, and can activate both C–H bonds at C7 to yield a pair of epimers. In addition, the catalytic capability of CepD2 is expanded with functionalization of the substrate as well. Our previous work has confirmed that CepD2 can only region-specifically attach an acetyl group to 6α-OH22. However, when the substrate is processed by HelE, CepD2 is able to achieve acetylation of 7β-OH (Fig. 5H). Similar phenomena that catalytic capacities of enzymes vary with manipulation of substrates are also observed. The multifunctional P450 enzyme TcP450-1 shows poor reaction specificity, and can catalyze C-11/C-19 hydroxylation of cortexolone. However, when 17α-OH in cortexolone is acetylated, the specificity of TcP450-1 mediated C-19 hydroxylation is dramatically increased26. All these results indicate that manipulation of substrates by random gene combination is a feasible option to induce the new function of enzymes for further enriching chemical diversity.
There have been a few reports on the backbone modifications of fusidane-type antibiotics and their resulting effects on the antibacterial activity1,9,10,22, 23, 24. In this work, we isolated a number of fusidane-type antibiotics through backbone modifications, and obtained comprehensive insights into the structure‒activity relationship of anti-S. aureus 209P activity. These results have provided clues to the evolution-guided optimization of helvolic acid, fusidic acid and cephalosporin P1 biosynthesis. Generally, 3α-OH is a key factor for the activity of fusidane-type antibiotics, however, the enzyme to convert 3-keto to 3α-OH is omitted in helvolic acid pathway. Instead, it evolves by introducing the C1/C2 double bond and 6β-OH/7-keto, which exclusively have the synergistic effect on the anti-S. aureus 209P activity with 3-keto, to enhance the antibacterial property of helvolic acid. On the other hand, formation of 3α-OH followed by C11α-hydroxylation gives the most potent fusidane-type antibiotic fusidic acid. However, it is puzzling that C6α-/C7β-hydroxylation is evolved after 3α-OH formation in cephalosporin P1 biosynthesis, which is detrimental to the anti-S. aureus 209P activity. It is worth noting that the structure‒activity relationship of fusidane-type antibiotics against M. luteus is slightly different, which might be attributed to the difference of cell permeability or EF‒G crystal structure between M. luteus and S. aureus 209P. Although no more potent derivatives than fusidic acid are obtained, the stochastic combinational strategy has enabled us to easily introduce reactive functional groups into the common intermediate in the same time, which provides a library of fusidane-type antibiotics for chemical modification so as to improve antimicrobial properties.
In conclusion, based on the biosynthetic mechanisms of helvolic acid, fusidic acid, and cephalosporin P1, we constructed 27 A. oryzae NSAR1 transformants through the stochastic combinational strategy, and a total of 58 compounds were isolated, including 54 new compounds. Unexpectedly, we found that catalytic functions of CepB4 and CepD2 can be expanded with modification of substrates. With large number of fusidane-type antibiotics in hand, systematic anti-bacterial activity evaluation was performed, and provided new insights into the structure‒activity relationship. Our work demonstrates the powerful ability of the stochastic combinational strategy for structural diversification, and provides a series of fusidane-type antibiotics for activity screening and chemical modifications.
4. Experimental
4.1. General materials and experimental procedures
Acetonitrile (CH3CN) was purchased from Oceanpak Alexative Chemical Co., Ltd. (Gothenburg, Sweden). Methanol (MeOH) was purchased from Yuwang Industrial Co., Ltd. (Yucheng, China). Ethyl acetate (EtOAc) was purchased from Fine Chemical Co., Ltd. (Tianjin, China). Formic acid was purchased from Kemiou Chemical Reagent Co., Ltd. (Tianjin, China).
Primer synthesis and DNA sequencing were performed by TSINGKE Biotech Co., Ltd. (Guangzhou, China). Plasmid extraction kits and DNA purification kits were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Both KOD-FX DNA polymerase and KOD-Plus DNA polymerase (TOYOBO, Osaka, Japan) were used for PCR, and ClonExpress® MultiS One Step Cloning Kit (Vazyme, Nanjing, China) was used to construct recombinant plasmids. Yatalase™ Fungal Cell Lytic Enzyme was purchased from TaKaRa Co., Ltd. (Dalian, China).
UV spectra, IR spectra and optical rotations were recorded on the JASCO V-550 UV/Vis spectrometer, JASCO FT/IR-480 plus spectrometer and JASCO P1020 digital polarimeter from JASCO International Co., Ltd. (Tokyo, Japan), respectively. HRESIMS was performed on the quadrupole orthogonal time-of-flight (Q-TOF) tandem mass spectrometer (Waters, Manchester, U.K.). NMR data were obtained with Bruker AV 400/600 spectrometers (Bruker BioSpin Group, Faellanden, Switzerland) using the following solvent signals (CDCl3: δH 7.26/δC 77.0; CD3OD: δH 3.30/δC 49.0; pyridine-d5: δH 7.21/δC 123.5; acetone-d6: δH 2.05/δC 29.8) as internal standards.
HPLC‒MS analysis of metabolites was performed on an Ultimate 3000 HPLC system (Dionex, Germering, Germany) and an amaZon SL ion trap mass spectrometer (Bruker Daltonics Inc., Billerica, Bostone, USA) equipped with an electrospray ionization source using a Cosmosil 5C18-MS-II column (250 mm × 4.6 mm, 5 μm, Nacalai Tesque, Inc., Kyoto, Japan). The mobile phases for HPLC‒MS analysis were H2O containing 0.1% formic acid (A) and CH3CN containing 0.1% formic acid (B), and the gradient elution was 50%–100% B (0–30 min), 100%–100% B (30–40 min), 100%–50% B (40–42 min), and 50%–50% B (42–50 min) with the flow rate of 1 mL/min. Medium pressure liquid chromatography (MPLC) was performed with a UV detector, a dual pump gradient system, and a Dr. Flash II fraction collector system (Lisui E-Tech Co., Ltd., Shanghai, China). Semi-preparative HPLC was carried out on an Ultimate 3000 HPLC system (Dionex) equipped with a UV detector, using an YMC-Pack ODS-A column (250 mm × 10 mm, 5 μm, YMC Co., Ltd., Tokyo, Japan).
4.2. Strains and media
The quadruple auxotrophic A. oryzae NSAR1 (niaD‒, sC‒, ΔargB, adeA‒)27 was used as the host for heterologous gene expression. The mycelia of A. oryzae transformants were inoculated into 10 mL DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, 0.05% MgSO4·7H2O) and cultured at 28 °C and 220 rpm for 1–2 days as seed broth. Then the seed broth was transferred to the 100 mL modified Czapek-Dox (CD) medium (0.3% NaNO3, 0.2% KCl, 0.05% MgSO4·7H2O, 0.1% KH2PO4, 0.002% FeSO4·7H2O, 1% polypeptone, 2% starch, pH 5.5) and grown at 28 °C and 220 rpm for 5 days to induce the expression of exogenous genes under the amyB promoter.
Escherichia coli DH5α (TaKaRa) was used for molecular cloning. The E. coli cells carrying target plasmids were grown in Luria‒Bertani (LB) medium supplemented with 100 mg/L ampicillin at 37 °C and 200 rpm.
4.3. Construction of fungal expression plasmids
The gene expression cassette containing the amyB promoter, the target gene, and the amyB terminator was amplified from the pTAex3-based plasmids, which have been established in our previous work, and then inserted into the HindIII-linearlized pPTRI or pBarI. All the primers are listed in Supporting Information Table S70, and all the plasmids are listed in Supporting Information Table S71.
4.4. Transformation of A. oryzae NSAR1
The transformation of A. oryzae NSAR1 was conducted via the PEG-mediated strategy. 100 μL spore suspension of the parent strain was inoculated in 10 mL DPY medium and cultured at 28 °C and 220 rpm for 2 days. Subsequently, the seed broth was transferred to 100 mL DPY medium and continues to grow for 24 h. The mycelia were harvested by filtration, and then digested by 1% Yatalase in 0.6 mol/L (NH4)2SO4, 50 mmol/L maleic acid, pH 5.5 at 30 °C for 3 h. The resulting protoplasts were collected by centrifugation, and washed once with Solution 2 (1.2 mol/L sorbitol, 50 mmol/L CaCl2·2H2O, 35 mmol/L NaCl, 10 mmol/L Tris-HCl, pH 7.5), and then the concentration was adjusted to 1 × 107 cells/mL. 200 μL protoplast suspension and about 10 μL plasmids (10 μg) were incubated on ice for 30 min followed by addition of 1.3 mL Solution 3 (60% PEG4000, 50 mmol/L CaCl2·2H2O, 10 mmol/L Tris-HCl, pH 7.5) in three times, and then the mixture is placed at room temperature for 20 min. After that, 5 mL Solution 2 was added, and the mixture was subjected to centrifugation at 1500 rpm for 10 min. The precipitates were suspended in 200 μL Solution 2 and spread on the lower selective medium (0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, 2% glucose and 1.2 mol/L sorbitol as well as 0.2 μg/mL pyrithiamine hydrobromide and/or 50 μL/mL glufosinate-ammonium, pH 5.5) with 1.5% agar, and then covered with the selective upper medium containing 0.8% agar. The transformants could be obtained after incubation at 30 °C for 4–6 days. All the transformants used in the work are listed in Supporting Information Table S72.
4.5. HPLC analysis and isolation of metabolites
To analyze the metabolites from different transformant strains, 10 mL seed broth was inoculated into 100 mL modified CD medium in a 500 mL flask and cultured at 28 °C and 220 rpm for 5 days (CRYSTAL IS-RDS4 incubator shaker, Addison Texas, USA). Then the fermentation broth was collected by filtration and extracted with EtOAc. The resulting crude extract was subjected to HPLC‒MS analysis. For isolation and purification of metabolites, extract was subjected to MPLC and semi-preparative HPLC (Supporting Methods).
4.6. Feeding experiments
The A. oryzae NSAR1 transformant strain (CO6, CO7 or AOS28) was inoculated into 10 mL DPY medium and cultured for 1–2 days. The seed broth was then transferred into 100 mL modified CD medium, followed by addition of 1.0 mg substrate in 50 μL DMSO after growing for 12 h. Subsequently, the transformant strain was cultured for additional 4 days, and the fermentation broth was extracted twice with EtOAc and concentrated under reduced pressure. The crude extract was dissolved in methanol for HPLC‒MS analysis.
4.7. Stability testing of compound 37 under different pH conditions
To 1 mL of 1 mol/L Tris-HCl buffer (pH 5.0, 7.0 and 8.0) was added 1.0 mg of 37 in 20 μL DMSO, respectively. The mixture was incubated at 28 °C and 220 rpm for 4 days, and then extracted three times with EtOAc. The concentrated crude extract was dissolved in methanol for HPLC‒MS analysis.
4.8. Antibacterial assay
In vitro inhibitory effect against Gram-positive strains S. aureus 209P and M. luteus were evaluated by 2-fold dilution assay. After growing on beef extract agar medium at 37 °C for 1 day, S. aureus 209P and M. luteus were collected with normal saline, and then adjusted to a concentration of 5 × 106 cells/mL with beef extract medium (0.3% beef extract, 0.2% yeast extract, 1% tryptone and 0.5% NaCl, pH 7.5) and MH liquid medium (purchased from Huankai Microbial Sci. & Tech., Co., Ltd., GuangZhou, China), respectively. 200 μL of seed broth was transferred into the first test well of each line in the 96-well, and 100 μL was added in the other wells in the same line. All compounds were dissolved in DMSO and adjusted to 50 g/L. 0.5 μL sample solution was added into the first well with the initial concentration of 128 μg/mL, then 100 μL was transferred to the second well, and so on. The concentration of the compound ranged from 128 to 0.0625 μg/mL. Tobramycin and ampicillin were used as the positive control, respectively, and DMSO was used as the negative control. The 96-well microtiter plates were placed at 37 °C for 24 h. The MIC was defined as the minimal concentration at which no turbidity could be observed.
Acknowledgments
We thank Prof. K. Gomi (Tohoku University) and Prof. K. Kitamoto (The University of Tokyo) for providing the A. oryzae NSAR1 heterologous expression system. This work was financially supported by grants from National Key Research and Development Program of China (2018YFA0903200 and 2018YFA0903201), the National Natural Science Foundation of China (31870032, 81925037, 31761143016, 31670036 and 31800021), the 111 Project of Ministry of Education of the People's Republic of China (B13038), Chang Jiang Scholars Program (Young Scholar) from the Ministry of Education of China (Hao Gao, 2017), National High-level Personnel of Special Support Program (2017RA2259, China), the Guangdong Natural Science Funds for Distinguished Young Scholar (2019B151502014, China), Guangdong Science and Technology Planning Project (2020A0505100041, China), Guangdong Special Support Program (2016TX03R280, China), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Y036, China), K. C. Wong Education Foundation (Hao Gao, 2016, China), and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (JSPS KAKENHI Grant Number JP16H06443, JP20KK0173, and JP20H00490).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2020.12.007.
Contributor Information
Dan Hu, Email: thudan@jnu.edu.cn.
Hao Gao, Email: tghao@jnu.edu.cn.
Ikuro Abe, Email: abei@mol.f.u-tokyo.ac.jp.
Xinsheng Yao, Email: tyaoxs@jnu.edu.cn.
Author contributions
Dan Hu, Hao Gao, Ikuro Abe and Xinshen Yao designed the research. Xiaojun Song and Jianming Lv performed the experiments. Huiyun Huang and Guodong Chen performed quantum chemical calculation of NMR shifts. Xiaojun Song, Jianming Lv, Zhiqin Cao, Huiyun Huang, Guodong Chen, Takayoshi Awakawa, Dan Hu, Hao Gao, Ikuro Abe and Xinshen Yao analyzed the data, and Xiaojun Song, Jianming Lv, Dan Hu, Hao Gao, Ikuro Abe and Xinsheng Yao wrote the paper.
Conflicts of interest
The authors declare that they have no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhao M., Godecke T., Gunn J., Duan J.A., Che C.T. Protostane and fusidane triterpenes: A mini-review. Molecules. 2013;18:4054–4080. doi: 10.3390/molecules18044054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chain E., Florey H., Jennings M., Williams T. Helvolic acid, an antibiotic produced by Aspergillus fumigatus, mut. helvola Yuill. Br J Exp Pathol. 1943;24:108–119. [Google Scholar]
- 3.Godtfredsen W., Jahnsen S., Lorck H., Roholt K., Tybring L. Fusidic acid: A new antibiotic. Nature. 1962;193:987. doi: 10.1038/193987a0. [DOI] [PubMed] [Google Scholar]
- 4.Burton H., Abraham E. Isolation of antibiotics from a species of Cephalosporium. Cephalosporins P1, P2, P3, P4 and P5. Biochem J. 1951;50:168–174. doi: 10.1042/bj0500168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Von Daehne W., Godtfredsen W.O., Rasmussen P.R. Structure‒activity relationships in fusidic acid-type antibiotics. Adv Appl Microbiol. 1979;25:95–146. doi: 10.1016/s0065-2164(08)70148-5. [DOI] [PubMed] [Google Scholar]
- 6.Jones R.N., Mendes R.E., Sader H.S., Castanheira M. In vitro antimicrobial findings for fusidic acid tested against contemporary (2008–2009) Gram-positive organisms collected in the United States. Clin Infect Dis. 2011;52:S477–S486. doi: 10.1093/cid/cir163. [DOI] [PubMed] [Google Scholar]
- 7.Bodley J.W., Zieve F.J., Lin L., Zieve S.T. Formation of the ribosome‒G factor‒GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun. 1969;37:437–443. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 8.hou J., Lancaster L., Donohue J.P., Noller H.F. Crystal structures of EF-G–ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong F.D., Huang X.L., Ma Q.Y., Xie Q.Y., Wang P., Chen P.W. Helvolic acid derivatives with antibacterial activities against Streptococcus agalactiae from the marine-derived fungus Aspergillus fumigatus HNMF0047. J Nat Prod. 2018;81:1869–1876. doi: 10.1021/acs.jnatprod.8b00382. [DOI] [PubMed] [Google Scholar]
- 10.Zhang P.L., Wang G., Xu F.Q., Liu J.S., Wang J.T., Zhang R. Aspergilolide, a steroid lactone produced by an endophytic fungus Aspergillus sp. MBL1612 isolated from Paeonia ostii. Nat Prod Res. 2019;33:2133–2138. doi: 10.1080/14786419.2018.1488706. [DOI] [PubMed] [Google Scholar]
- 11.Zaman K.H.A.U., Hu Z., Wu X., Hou S., Saito J., Kondratyuk T.P. NF-κB inhibitory and antibacterial helvolic and fumagillin derivatives from Aspergillus terreus. J Nat Prod. 2020;83:730–737. doi: 10.1021/acs.jnatprod.9b01190. [DOI] [PubMed] [Google Scholar]
- 12.Prehn K., Tybring L., Forchhammer J. Fusidic acid and derivatives. Inhibition of protein synthesis in cell-free systems from Escherichia coli and Bacillus stearothermophilus. Acta Pathol Microbiol Scand. 1967;71:135–140. [Google Scholar]
- 13.Godtfredsen W., Von Daehne W., Tybring L., Vangedal S. Fusidic acid derivatives. I. Relationship between structure and antibacterial activity. J Med Chem. 1966;9:15–22. doi: 10.1021/jm00319a004. [DOI] [PubMed] [Google Scholar]
- 14.Dziwornu G.A., Kamunya S., Chibale K., Ntsabo T., Chibale K., Ntsabo T. Novel antimycobacterial C-21 amide derivatives of the antibiotic fusidic acid: Synthesis, pharmacological evaluation and rationalization of media-dependent activity using molecular docking studies in the binding site of human serum albumin. Medchemcomm. 2019;10:961–969. doi: 10.1039/c9md00161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P.P., He H., Hong W.D., Wu T.R., Huang G.Y., Zhong Y.Y. The biological evaluation of fusidic acid and its hydrogenation derivative as antimicrobial and anti-inflammatory agents. Infect Drug Resist. 2018;11:1945–1957. doi: 10.2147/IDR.S176390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvold T., Sørensen M.D., Björkling F., Henriksen A.S., Rastrup-Andersen N. Synthesis and conformational analysis of fusidic acid side chain derivatives in relation to antibacterial activity. J Med Chem. 2001;44:3125–3131. doi: 10.1021/jm010899a. [DOI] [PubMed] [Google Scholar]
- 17.Riber D., Venkataramana M., Sanyal S., Duvold T. Synthesis and biological evaluation of photoaffinity labeled fusidic acid analogues. J Med Chem. 2006;49:1503–1505. doi: 10.1021/jm050583t. [DOI] [PubMed] [Google Scholar]
- 18.Kim E., Moore B.S., Yoon Y.J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11:649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun H.H., Liu Z.H., Zhao H.M., Ang E.L. Recent advances in combinatorial biosynthesis for drug discovery. Drug Des Dev Ther. 2015;9:823–833. doi: 10.2147/DDDT.S63023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez C., Zhu L., Braña A.F., Salas A.P., Rohr J., Méndez C. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc Natl Acad Sci U S A. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukada K., Shinki S., Kaneko A., Murakami K., Irie K., Murai M. Synthetic biology based construction of biological activity-related library of fungal decalin-containing diterpenoid pyrones. Nat Commun. 2020;11:1830. doi: 10.1038/s41467-020-15664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Z.Q., Lv J.M., Liu Q., Qin S.Y., Chen G.D., Dai P. Biosynthetic study of cephalosporin P1 reveals a multifunctional P450 enzyme and a site-selective acetyltransferase. ACS Chem Biol. 2020;15:44–51. doi: 10.1021/acschembio.9b00863. [DOI] [PubMed] [Google Scholar]
- 23.Cao Z.Q., Li S.Y., Lv J.M., Gao H., Chen G.D., Awakawa T. Biosynthesis of clinically used antibiotic fusidic acid and identification of two short-chain dehydrogenase/reductases with converse stereoselectivity. Acta Pharm Sin B. 2018;9:433–442. doi: 10.1016/j.apsb.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv J.M., Hu D., Gao H., Kushiro T., Awakawa T., Chen G.D. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat Commun. 2017;8:1644. doi: 10.1038/s41467-017-01813-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen J.L., Hashtroudi H. Base-catalyzed hydration of α,β-unsaturated ketones. J Org Chem. 1976;41:3299–3302. [Google Scholar]
- 26.Wang J., Zhang Y., Liu H., Shang Y., Zhou L., Wei P. A biocatalytic hydroxylation-enabled unified approach to C19-hydroxylated steroids. Nat Commun. 2019;10:3378. doi: 10.1038/s41467-019-11344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin F.J., Maruyama J., Juvvadi P.R., Arioka M., Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett. 2004;239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.