Abstract
Background
Breast tumor stiffness, which can be objectively and noninvasively evaluated by ultrasound elastography (UE), has been useful for the differentiation of benign and malignant breast lesions and the prediction of clinical outcomes. Liquid biopsy analyses, including cell-free tumor DNA (ctDNA), exhibit great potential for personalized treatment. This study aimed to investigate the correlations between the UE and ctDNA for early breast cancer diagnosis.
Methods
Breast tumor stiffness in 10 patients were assessed by shear wave elastography (SWE), and the ctDNA of eight collected plasma specimens with different tumor stiffness were analyzed by whole-genome sequencing (WGS). Subsequently, the distribution of carcinoma-associated fibroblasts (CAFs) was investigated by detecting the expression levels of alpha-smooth muscle actin (α-SMA) in tissues of breast lesions. We validated the function of discoidin domain receptor 2 (DDR2) in breast tumor CAFs by knockout of fibroblast activation protein (FAP) with different tumor stiffness during cancer progression in vitro and vivo.
Results
The UE estimates of tumor stiffness positively correlated with CAF-rich (α-SMA+) tumors (P<0.05). Copy number profiles and percent genome alterations were remarkably different between benign and malignant breast lesions. Somatic genomic alterations or structural variants of DDR2, ANTXRL, TPSG1, and TPSB2 genes were identified in ctDNA of plasma from breast lesions with high SWE values and an increase in the CAF content obtained from clinical samples. Deletion of FAP in breast tumor CAFs by CRISPR/Cas9-mediated gene knockout and decreased tumor stiffness resulted in downregulated expression of DDR2 (P<0.05), which in turn led to decreasing the tumor stiffness and carcinogenesis process in vitro and in vivo.
Conclusions
These results have established proof of principle that WGS analysis of ctDNA could complement current UE approaches to assess tumor stiffness changes for the early diagnosis and prognostic assessment of breast cancer.
Keywords: Breast cancer, cell-free tumor DNA (ctDNA), copy number variation (CNV), tumor stiffness, discoidin domain receptor 2 (DDR2)
Introduction
Breast cancer is one of the most common cancers worldwide. Despite improvements in survival, breast cancer remains one of the leading causes of female cancer mortality. Clinical parameters and histopathological features of the tumor, including tumor size, tumor grade, metastasis of axillary lymph nodes, and specific gene amplification, are used to prognosticate clinical outcomes (1,2). Additionally, previous studies have revealed that tumor stiffness is associated with tumor progression (3); thus, tumor stiffness may help the diagnosis of breast lesions as well as the prediction of clinical outcome. Ultrasound elastography (UE) is an imaging technique that can visualize tissue elasticity (stiffness) in vivo (4,5) and provide additional information about breast lesions over conventional sonography and mammography (6-9). However, breast cancer is a heterogeneous disease in which the individual’s genetics, combined with the tumor microenvironment, molecular subtype, and histological type, contribute to disease development (10). The UE provides a means to measure tissue stiffness, which may function as a biomarker for the early diagnosis and therapies that target the tumor microenvironment.
Whole-genome sequencing (WGS) analysis of cell-free tumor DNA (ctDNA) provides a minimally invasive strategy for genomic profiling of tumor variations without needing a tumor biopsy, enabling the improvement of various aspects of breast cancer management, including early diagnosis and screening, therapy guidance, and disease surveillance. Furthermore, unlike traditional tissue biopsy, ctDNA provides a comprehensive reflection of the tumor heterogeneity, as it is derived from different tumor sites. Therefore, analyzing genome-scale variations between patients’ ctDNA and normal genomes provides opportunities for minimally invasive cancer diagnosis, prognosis, and tumor monitoring (11). As assay sensitivity improves with broader applications, ctDNA-based analyses will soon satisfy the need to make next-generation sequencing (NGS) technologies relevant to breast cancer treatment. The extensive and coordinated collaboration to systematically connect somatic mutations to clinical and pharmacologic data will be critical for this progress.
While most initial genetic variation studies have concentrated on individual nucleotide sequences, investigators have also found that large-scale changes occur in many locations throughout the genome (12). Compared to common single nucleotide variations or small insertions or deletions, copy number variations (CNVs) are relatively new in detecting genomic alterations. A CNV is also defined as a phenomenon in which sections of the genome are repeated, and the number of repeats in the genome varies between individuals. More importantly, CNV is a kind of variation that occurs at a much larger scale than variations in other categories.
Discoidin domain receptor 2 (DDR2) is a receptor tyrosine kinase that has been previously reported to overexpress on mesenchymal stem cells (MSCs) in many malignant tumors. In breast cancer, DDR2 mutation was associated with collagen synthesis, modification, and signaling, especially in the reorganization of collagen fibers at the stromal-tumor boundary, which was involved in breast cancer stiffness (13) and fibrotic conditions (14).
In the oncogenesis and development of malignant tumors, the stiffness of tumors increases gradually, and this is manifested in the increase in UE imaging shear wave elastography (SWE) values and the strain rate ratio. Whether CNV causes a change in the structural variation, such as large fragment insertion/deletion and gene rearrangement of tumor lesion tissue genes, remains unclear; whether this is reflected in the change in ctDNA when released into the blood, also remains unclear. This study aimed to investigate the possible correlations between the SWE values and copy number uniformities in genomes from breast cancer patients with malignant tumors and from individuals with benign tumors. A timely and accurate comparison of ctDNA CNV detection and UE testing was performed to evaluate the clinical significance in the early noninvasive diagnosis and personalized treatment of breast cancers.
Methods
Patient identification and clinicopathologic data
Study population
The Ethical Committee approved this study of the Affiliated Tumor Hospital of Xinjiang Medical University. The research was conducted on breast lesions and imaging data obtained from routine ultrasound (US) and UE by two expert radiologists before routine surgery. A total of 54 female patients who underwent surgery for breast masses without subsequent neoadjuvant chemotherapy from February 2015 to February 2017 were recruited for the study. Patients who were pregnant or lactating had any other cancer or refused gene detection or surgery was excluded. In total, 10 female patients who underwent WGS on ctDNA were enrolled (Figure S1). Patients’ age ranged from 40 to 65 years old, with an average age of 52 and a median age of 46.
Routine US and SWE
Both routine US and SWE examinations were performed using an ultrasound system (Logiq E9, GE Medical Systems, Wauwatosa, WI, USA) by two radiologists (>12 months of SWE experience). The routine US was performed using a linear transducer with a bandwidth of 9–15 MHz. Information on lesion size, margin, background echotexture, aspect ratio, calcification, and color Doppler were recorded. The images were evaluated according to the breast imaging-reporting and data system (BI-RADS) lexicon and assigned a BI-RADS score. Scores of 2 and 3 were considered benign, while scores of 4 and 5 were considered malignant (15).
After US examination, SWE was conducted with a 9-L linear transducer equipped with elastography software. Compressing the breast with the transducer can cause an increase in tissue stiffness and be therefore avoided when performing SWE. The tissue stiffness in the SWE image was displayed with a range from dark blue (lowest stiffness, 0 m/s) to red (highest stiffness, 7.1 m/s, in our study). A region of interest (ROI) with a diameter of 2 mm was utilized to cover the hardest region within or near the mass. The mean stiffness (Emean) average of three SWE images with the largest lesion diameters taken on two orthogonal planes were recorded and used for the analysis. According to previous research, an Emean <50 kPa was assigned to benign lesions and ≥50 kPa to malignant lesions (16). According to the SWE results, we allocated participants to three groups: group 1 (Emean <50 kPa), group 2 (50 kPa ≤ Emean <150 kPa), and group 3 (Emean ≥150 kPa).
Histopathologic diagnoses
A final diagnosis was determined by histopathology after surgical excision or US-guided core needle biopsy [BARD MAGNUM Reusable Core Biopsy Instrument with MN1620 (16 gauge) biopsy needles (Bard Peripheral Vascular, Inc., Tempe, AZ, USA)]. Histopathologic diagnoses of the specimens were obtained and used as reference standards. A specialized breast pathologist made all diagnoses with at least 25 years of experience who was blinded to the US results.
CNV detection by WGS for ctDNA
DNA extractions
Approximately 10 mL of whole blood was collected from the 10 participants before surgery. Samples were kept chilled and processed within 4 hours. The ctDNA was extracted from plasma using a QIAamp Circulating Nucleic Acid Kit (QIAGEN Hilden, Germany, Cat No. 55114) and stored at –70 °C. The quality of DNA samples was assessed using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s protocol.
Library preparation
A DNA library was constructed using the DNA Seq Library Preparation Kit-Illumina Compatible (Gnomegen, San Diego, CA, USA, K02422-L) started with 10 ng ctDNA. End repair, dA-tailing, adapter ligation, and polymerase chain reaction (PCR) amplification for 12 cycles were performed, following the manufacturer’s instructions with several purification steps to generate DNA libraries with different indexes for samples from each participant. Size selection was performed as the last step.
Following size selection, 1 µL of each library product was analyzed by capillary electrophoresis on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) to confirm that the expected fragment size had been obtained and to determine the concentration. A total of 10 purified and quantified libraries were mixed to form one library mixture, which was diluted to 26 pM in molecular biology grade water.
The libraries prepared from free-circulating DNA samples for WGS were sequenced, which was performed on a HiSeq X10 PE150 (Illumina, San Diego, CA, USA) with 45 M reads (6.8 Gb) for each participant sample.
Low-coverage WGS (LC-WGS) for ctDNA
To evaluate the tumor status by LC-WGS uniformity, we extracted DNA from the plasma of these 10 clinical samples and sequenced the DNA ~4× using 150 bp paired-end reads on a HiSeq X10 PE150 (Illumina), according to the standard manufacturing protocol. For the quality control (QC) of WGS data, ~86.24 million sequence reads were generated on average for each sample. A total of 76.24% of these reads were mapped to the reference genome (GRCh37, UCSC release hg19). Percentage of aligning rate were all greater than 90% (Table S1). After removing the PCR duplications and low-quality reads, ~40.72 M paired reads for each sample were obtained, resulting in 48.78% effective (unique, non-duplicated) reads for the following analysis.
Data processing and analysis
Raw paired-end reads of all samples were first examined with the sequencing QC. Reads that passed QC were aligned with the hg19/GRCh37 assembly of the human genome reference sequence (http://genome.ucsc.edu) with BWA (17) (Burrows-Wheeler aligner, version 0.7.12). Any PCR duplicate reads were removed using the “Mark Duplicates” function in Picard 1.10 (http://picard.sourceforge.net). Copy number analysis was performed using CNVnator. All chromosomes of the whole human genome reference were cut into sliding windows for every 10 thousand nucleotides and read counts (RCs) that were mapped onto windows were counted. The coefficient of variation (CV) was calculated as:
| [1] |
where , N is the number of 10 k bins of the corresponding chromosome, and xi is the corrected RC of the xih bin.
Comparing copy number uniformity between cancer patients and normal individuals
All calculated CVs from a chromosome in the normal individuals group were compared against those of the breast cancer participants for each chromosome. The Wilcoxon signed-rank test function embedded in R (version 3.1.3, https://cran.r-project.org) was applied when comparing two groups of CVs.
Cell line and culture
The mouse breast cancer cell line 4T1 was purchased from The Global Bioresource (https://www.atcc.org). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, St Louis, MO, USA). Cells were grown in a 5% CO2 incubator at 37 °C. To isolate primary mouse breast tumor cancer-associated fibroblasts (mCAFs), 4T1 xenograft breast tumors were dissected and minced. Minced pieces were transferred to ~20 mL of digestion media (DMEM, 1% FBS, 0.2% collagenase A (Roche, Basel, Switzerland), 0.2% trypsin (Gibco, Gaithersburg, MD, USA, 27250-018), 50 mg/mL gentamycin, and 5 mg/mL insulin) per tumor and rocked at 37 °C for 30–45 min. The digested tissue was then washed twice with serum-free media and treated with DNase for 5 min at room temperature. The tissue was resuspended in ice-cold serum-free media and serially centrifuged 4 times. Single-cell fractions were collected and plated for 25–30 min in DMEM with 10% FBS at 37 °C, 5% CO2, and 20% O2. After this treatment, the CAFs adhered to the plate, while other cells would not. The supernatant and nonadherent cells were removed, and CAFs were maintained in DMEM with 10% FBS at 37 °C, 5% CO2, and 20% O2. All spontaneously immortalized primary CAF cell lines were submitted to fluorescence-activated cell sorting (FACS) with alpha-smooth muscle actin (α-SMA) antibodies. All cell lines utilized were mycoplasma-free, as determined by quantitative PCR (qPCR) analyses every 6 months (18). For the coculture studies, mCAFs were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, 100 U penicillin, and 100 mg streptomycin. Then, the medium from mCAF subconfluent cultures was removed, and 4T1 cells were plated on top. Unattached cells were removed, and attached cells were washed with Krebs buffer and cultured in DMEM without serum. The medium was changed every 48 hours. Cells were passaged and liberated from wells with trypsin/ethylenediaminetetraacetic acid (EDTA) (0.25% trypsin/0.02% EDTA, Gibco).
FAP and DDR2 gene knock-out by CRISPR/Cas9 gene editing
The CRISPR/Cas9 system, based on an RNA-guided nuclease, has revolutionized how genome editing is performed (19). The CRISPR/Cas9 single guide sequences, which specifically target FAP (sgFAP) and DDR2 (sgDDR2), were designed from Vector Builder (www.vectorbuilder.com) and produced by Cyagen Biosciences (Suzhou, China). The guide sequences for FAP gRNA were 5'-GCCAATCTCATTTGACCAAC-3' and 5'-GTGGCTATTTCACTGATGGG-3'. The guide sequences for DDR2 gRNA were 5'-GGATCAGTCTGGATGGCTCC-3' and 5'-TGGACTCGCTGGGCAGGGAA-3'. The mCAF cell lines were transduced twice with sgRNA FAP lentiviruses. The 4T1 breast cancer cell lines were transduced twice with sgRNA DDR2 lentiviruses. The EGFP lentiviruses were used as a positive control, and transfection reagent was used as a negative control. Puromycin (Merck, Darmstadt, Germany) was added on day 4 by using the minimal toxic dose to select transduced cells. Then, after 3 days of screening and observing the well condition of cells, the second transduction was performed with Cas9 lentiviruses. After 3 days, hygromycin was added at the minimal toxic dose. The second screening was finished after all negative control cells had died. To minimize off-target situations, monoclonal screening was conducted. Single colonies were spread in 96-well plates and selected by a colony formation assay. The effect of FAP and DDR2 gene editing on cell proliferation was assessed by the Trypan Blue viable cell counting method over a 7-day time course. Lentiviral transduction efficiency was proven by reverse transcription-qPCR (RT-qPCR) and western blotting. Total protein from mCAFs and 4T1 were extracted using radioimmunoprecipitation assay (RIPA) buffer (Solabio, China), and protein expression were determined by western blotting, probing for FAP (Abcam, ab53066), DDR2 (Abcam, ab63337), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Yeason, China) with sensitive electrochemiluminescence (ECL) detection (Yeason, China).
Immunohistochemistry (IHC) analysis
Paraffin-embedded tissues collected from the 10 participants were cut into 4 µm sections and baked at 65 °C for 30 min. After being deparaffinized and rehydrated, the sections were submerged in EDTA (pH 8.0) and autoclaved for antigen retrieval; the sections were then treated with 3% H2O2, followed by incubating with 1% FBS. Mouse anti-human anti-α-SMA monoclonal primary antibody (1:50 dilution; Santa Cruz, CA, USA) was added, and the sections were incubated at 4 °C overnight. Horseradish peroxidase (HRP)-labeled secondary antibody (ZSGB-BIO, Beijing, China) was applied, and tissues were incubated at room temperature for 30 min; the sections were then incubated for 5 min incubation with 3,3'-diaminobenzidine (DAB) at room temperature for color development. Then, the sections were counterstained with hematoxylin and mounted using Permount medium (BIOS, Beijing, China). The sections were visualized and photographed under a light microscope. The proportion of positively stained cells was graded as follows: 0 (≤5% positively stained cells), 1 (>5–25% positively stained cells), 2 (>25–75% positively stained cells), and 3 (>75% positively stained cells). The staining intensity was determined on a scale of 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish-brown), and 3 (strong staining, brown). The staining index on behalf of the expression of α-SMA was calculated according to the following equation: staining index = (staining intensity × proportion of positively stained cells)/2. The sum of both scores was used to identify the expression grades: 0–1 indicated negative expression; 2–4 indicated weak expression; 5–8 indicated moderate expression; ≥9 indicated high expression.
Western blotting
Cultured cells were lysed with lysis buffer [1X RIPA buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium vanadate, 1 mM sodium fluoride, 10 µg/mL aprotinin, and 10 µg/mL leupeptin]. Lysates were sonicated twice for 30 s and centrifuged at 14,000 RPM for 10 min. Equal amounts of protein in lysates were run on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to polyvinylidene fluoride membranes (Millipore). After blocking with 5% milk in tris buffered saline with Tween 20 (TBST), membranes were incubated with primary antibodies overnight with gentle agitation, washed twice with TBS-0.5% Tween, and incubated with anti-mouse or anti-rabbit HRP secondary antibody for 1 hour at room temperature. Membranes were then washed 4 times with TBS-0.5% Tween and developed with ECL (Yeasen). The following specific antibodies were used: anti-FAP (1:1,000, Abcam), anti-DDR2 (1:1,000, Abcam), anti-α-SMA (1:1,000, Boster), and anti-GAPDH (1:6,000, Yeasen).
Orthotopic transplant model
The animal experiments were based on ethical considerations and integrity-based assumptions. Simultaneously, the experimental protocol was approved by the institutional animal ethics committee of Southern Medical University (Approval ID: No. 2019-005), which follows the guidelines for the care and use of laboratory animals published by the National Institutes of Health (No. 85-23, revised 1996) and the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA).
Female BALB/c mice (8-week-old) received breast transplants (mammary fat pad) of 106 4T1 breast tumor cells and DDR2-knockout 4T1 breast tumor cells. After 2 weeks, the primary tumor volume was histologically determined at autopsy. The tissue stiffness of breast transplants was measured by UE conducted with a 9-L linear transducer equipped with elastography software.
Bioinformatics and statistical analysis
All statistical analyses were performed in R (version 3.1.3). For each chromosome, all calculated CVs from that chromosome with an Emean <50 kPa (group 1) were compared against those with an Emean ≥150 kPa (group 3). The Wilcoxon signed-rank test function embedded in R (version 3.1.3) was applied when comparing two groups of CVs. Statistical analyses were performed with SPSS version 17 for Windows (SPSS Inc., Chicago, IL, USA). Student’s t-test assessed differences among elasticity scores for benign and malignant breast lesions. Relationships between the stiffness values and the histological features were compared using the independent t-test or one-way analysis of variance (ANOVA). Clinical and histological variables were compared among the tumor subtypes using the χ2 test or Fisher’s exact test. A P value <0.05 was considered statistically significant.
Results
Patient characteristics and UE-SWE diagnosis
Participant characteristics, including routine US results, SWE values, and pathology results, are detailed in Table 1 and Table S2.
Table 1. Patient characteristics and UE-SWE diagnosis.
| Clinicopathological parameters | Group 1, Emean <50 kPa (n=3) | Group 2, 50 kPa ≤ Emean <150 kPa (n=3) | Group 3, Emean ≥150 kPa (n=4) | Total (n=10) |
|---|---|---|---|---|
| Age (years) | ||||
| Mean | 49.00 | 52.00 | 50.00 | 51.70 |
| Median | 46.50 | 44.00 | 48.40 | 46.00 |
| Range | 40–54 | 42–65 | 41–56 | 40–65 |
| Size (mm) | ||||
| Mean | 21.30 | 21.20 | 23.00 | 22.05 |
| Median | 21.20 | 20.40 | 23.10 | 21.45 |
| Range | 19.50–23.30 | 17.60–25.50 | 15.70–30.10 | 15.70–30.10 |
| Aspect ratio, n (%) | ||||
| <1 | 3 (100.00) | 2 (66.67) | 2 (50.00) | 7 (70.00) |
| >1 | 0 (0.00) | 1 (33.33) | 2 (50.00) | 3 (30.00) |
| Margins, n (%) | ||||
| Well-defined | 3 (100.00) | 1 (33.33) | 0 (0.00) | 4 (40.00) |
| Ill-defined | 0 (0.00) | 2 (66.67) | 4 (100.00) | 6 (60.00) |
| BI-RADS, n (%) | ||||
| <4 | 3 (100.00) | 0 (0.00) | 0 (0.00) | 3 (30.00) |
| 4 | 0 (0.00) | 1 (33.33) | 1 (25.00) | 2 (20.00) |
| >4 | 0 (0.00) | 2 (66.67) | 3 (75.00) | 5 (50.00) |
| Pathologic results, n (%) | ||||
| Benign (breast FAs) | 3 (100.00) | 0 (0.00) | 0 (0.00) | 3 (30.00) |
| Malignant (breast invasive carcinoma) | 0 (0.00) | 3 (100.00) | 4 (100.00) | 7 (70.00) |
| Histological grade, n (%) | ||||
| I | 0 (0.00) | 1 (25.00) | ||
| II | 2 (66.67) | 1 (25.00) | ||
| III | 1 (33.33) | 2 (50.00) | ||
| ER positive, n (%) | 3 (100.00) | 4 (100.00) | ||
| PR positive, n (%) | 3 (100.00) | 4 (100.00) | ||
| HER2 positive, n (%) | 1 (33.33) | 2 (50.00) |
UE-SWE, ultrasound elastography shear wave elastography; BI-RADS, breast imaging-reporting and data system; FA, fibroadenoma; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
A total of 10 patients were enrolled in this study, including 3 participants with an Emean <50 kPa (group 1), 3 participants with 50 kPa ≤ Emean <150 kPa (group 2), and 4 participants with an Emean ≥150 kPa (group 3) with UE-SWE diagnosis. Among them, 7 participants (70%) were pathologically diagnosed with breast cancer (Emean ≥50 kPa). The size range was 15.70 to 30.10 mm, and the mean size was 22.10 mm. The other 3 participants (30%) with Emean <50 kPa had breast fibroadenoma (FA) (average Emean 33.86 kPa, range 30.46 to 38.78 kPa) (Figure 1). The sizes were 19.5–25.50 mm, and the mean size was 21.30 mm. The SWE values (Emean) of malignant lesions were significantly higher than that of benign lesions (168.49±49.05 vs. 33.86±4.36 kPa) (P<0.01) (Figure 2).
Figure 1.
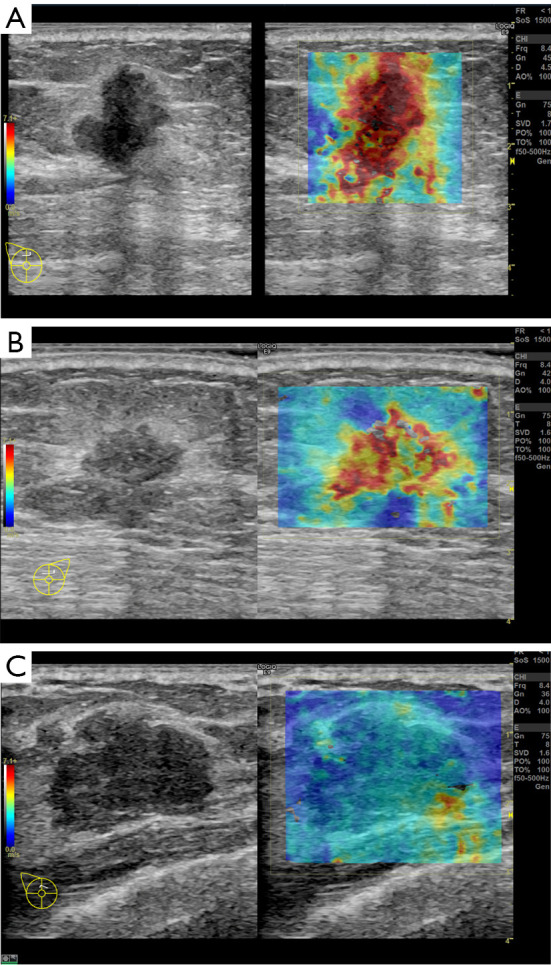
B-mode (left) and SWE (right) images of breast cancer (A,B) and breast FA (C) in split screen mode. (A) Grade II infiltrating ductal carcinoma. SWE depicts an almost red-colored mass, with an Emean of 149 kPa. (B) Grade II infiltrating ductal carcinoma. SWE depicts a yellow-to-red-colored mass with an Emean of 76 kPa. (C) Breast FA with an Emean of 19 kPa; SWE depicts a blue-to-yellow color for the lesion. SWE, shear wave elastography; FA, fibroadenoma.
Figure 2.
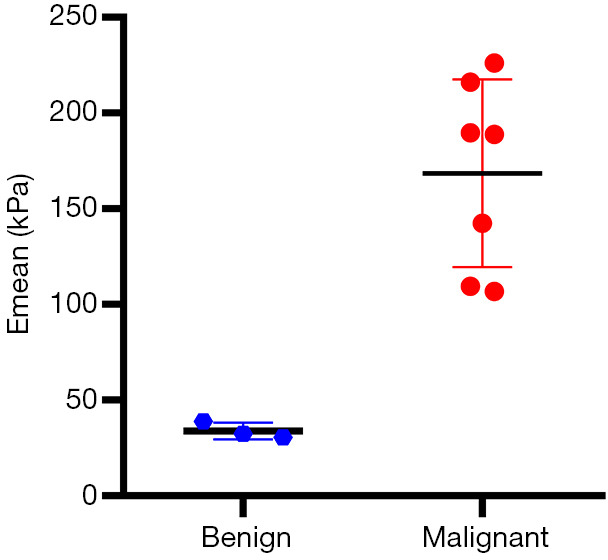
The SWE values (Emean) of malignant lesions was significantly higher than that of benign lesions (108.4±35.1 vs. 18.6±4.2 kPa) (P<0.01). SWE, shear wave elastography.
CAFs and α-SMA expression
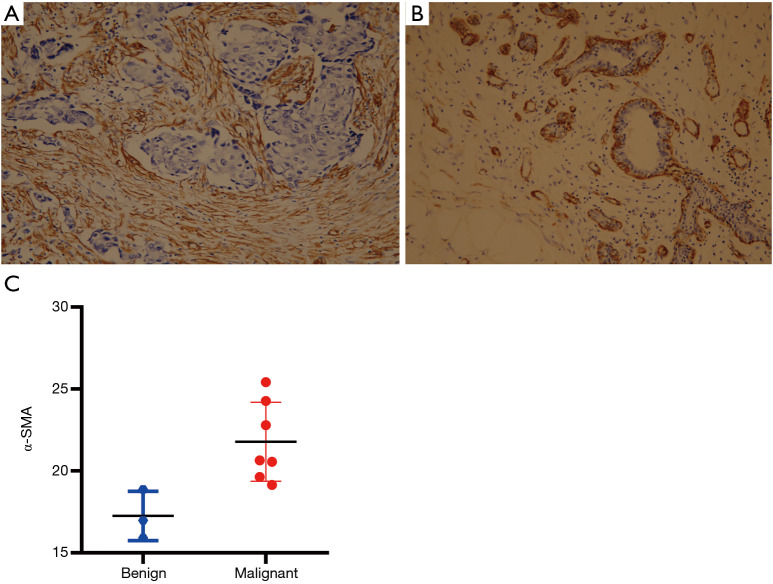
The expression of α-SMA reflects the number/density of CAFs at the tumor center, which underlies tumor stiffness and malignant behavior. Given the association between α-SMA and stiffness in breast tumors, the expression of α-SMA in breast lesions of the 10 participants was examined by IHC. The IHC analysis results showed that α-SMA was strongly expressed in breast cancer stromal cells in groups 2 and 3 (P<0.05) (Figure 3 and Table S2), indicating a decrease in the expression of α-SMA for benign tumors which possessed lower SWE values.
Figure 3.
α-SMA expression in malignant and benign breast lesions. (A) The expression of α-SMA in breast cancer stromal cells shows a strong positive signal. (B) The expression of α-SMA in stromal cells of benign breast lesions shows a negative signal. Magnification: 200×, staining method: PV9000. (C) α-SMA was strongly expressed in breast cancer stromal cells in groups 2 and 3 (P<0.05), indicating a decrease in the expression of α-SMA for benign tumors. α-SMA, alpha-smooth muscle actin.
Changes in CNV levels in chromosomes
DNA was extracted from the plasma of all 10 participants; in total, 7 patients had breast cancer, and 3 had benign lesions. Sequencing results from the 3 patients with benign lesions served as baselines to evaluate the changes in the CNV in patients with breast cancer.
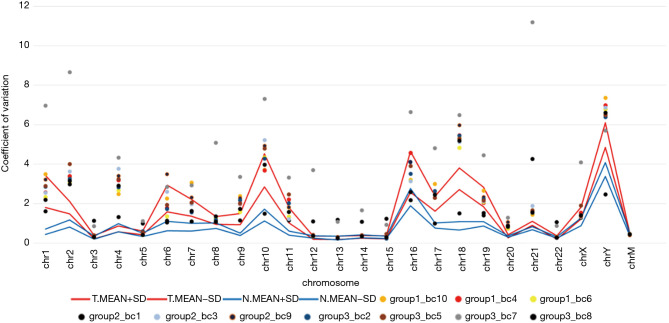
As described in the Materials and Methods, we calculated the CV of the unique RC for all 10 k bins on each chromosome from the LC-WGS dataset (20). These CVs should effectively reflect the stability of the genome. The copy number uniformity was compared between benign and malignant samples. As a result, the participants with breast cancer showed significantly higher CVs in most chromosomes (chr1, chr6, chr10, and so on) than those in the FA individuals (Figure 4).
Figure 4.
CV of RCs in 10 k windows distributed across all chromosomes. Dots are CVs of every sample (samples marked as group 1 are patients with benign tumors, and other samples are patients with malignant lesions), and lines are upper and lower indicators calculated as the mean ± SD of CVs within the benign tumor and cancer groups. CV, coefficient of variation; RC, read counts; SD, standard deviation.
However, we further analyzed the average CVs for three benign lesions and five malignant lesions (except group 2_bc1 and group 3_bc7 for low sequencing quality). The Wilcoxon signed-rank test (P=0.8237) indicated that there were no significant differences in the 10 k bin CVs between benign and malignant lesions. The CV of reading counts of every 10 k bin across samples and autosomes are presented in Table S3.
CNV analysis
Using the CNVnator platform, somatic CNVs within the genome of every individual were identified in this study cohort, comprising five breast cancer patients (groups 2 and 3) and three breast FA participants (group 1), except two samples (group 2_bc1 and group 3_bc7) that had low sequencing quality. As presented in Table 2, 395 CNV genes were identified within the 8 samples. All participants had CNVs in at least 150 genes (mean 200, range 152–279). Copy number amplifications were more commonly observed than deletions.
Table 2. CNVs within the genome of every individual.
| Sample | Total gene | Cancer-associated gene (n) | Cancer-associated gene | Coverage >2×, house keeping gene |
Coverage >4×, house keeping gene |
Coverage >8×, house keeping gene |
Coverage >8×, house keeping gene/total (%) |
|---|---|---|---|---|---|---|---|
| Score2_bc10 | 161 | 3 | HYDIN, PDE4DIP, NOTCH2 | 86 | 22 | 0 | 0.00 |
| Score2_bc4 | 163 | 3 | HYDIN, PDE4DIP, NOTCH2 | 82 | 13 | 0 | 0.00 |
| Score2_bc6 | 222 | 4 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL | 107 | 29 | 2 | 0.90 |
| Score4_bc3 | 152 | 6 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL, CFC1B, OR4F16 | 88 | 42 | 7 | 4.61 |
| Score4_bc9 | 192 | 5 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL, NPIPA7 | 84 | 27 | 0 | 0.00 |
| Score5_bc2 | 183 | 8 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL, CFC1B, OR4F16, TPSG1, TPSB2 | 78 | 15 | 3 | 1.64 |
| Score5_bc5 | 279 | 10 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL, CFC1B, NPIPA7, OR4F16, TPSG1, TPSB2, DDR2 | 109 | 34 | 13 | 4.66 |
| Score5_bc8 | 191 | 9 | HYDIN, PDE4DIP, NOTCH2, TCEB3CL, CFC1B, NPIPA7, TPSG1, TPSB2, ANTXRL | 97 | 27 | 3 | 1.57 |
| Total | 395 | 11 | – | – | – | – | – |
CNVs, copy number variations.
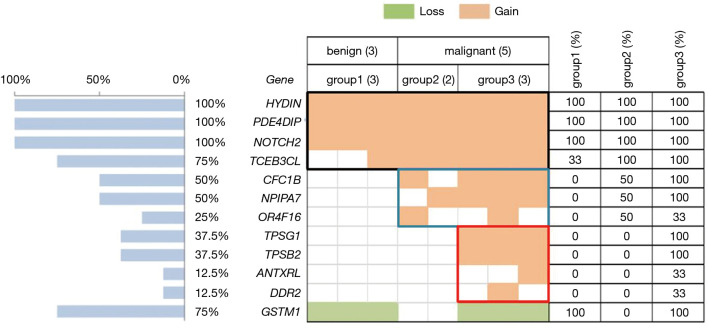
Copy number gains were identified in 11 tumor-related genes. In general, all of the samples had copy number gains in HYDIN (chr16), PDE4DIP, and Notch2. All CNVs in the PDE4DIP and Notch2 genes occurred in chromosome 1. The other genes with common copy number alterations were TCEB3CL (n=6, 75%), CFC1B (n=4, 50%), and NPIPA7 (n=4, 50%) (Figure 5).
Figure 5.
CNVs of breast tumors. Top, eight cases of breast lesions were divided into benign and malignant groups by pathology results and SWE values (Emean <50 kPa was assigned as benign and ≥50 kPa as malignant). The malignant cases are further labeled as either group 2 (50 kPa ≤ Emean <150 kPa) or group 3 (Emean ≥150 kPa). Center, 12 genes associated with cancer that we identified in our study. Colored rectangles indicate the CNV categories seen in the participants. Frame of black, blue, and red indicate copy number amplifications identified in all samples, the malignant group, and just group 3, respectively. Left, the percentage of cases with a CNV in each gene. Right panel, frequency of a CNV in each group. CNVs, copy number variations; SWE, shear wave elastography.
To identify genetic alterations in breast tumors contributing to different levels of tumor stiffness, we compared the CNVs in different stiffness groups. For the PDE4DIP and Notch2 genes, the comparison of the copy number amplification times between the benign (Emean <50 kPa) and malignant groups (Emean ≥50 kPa) were statistically indistinguishable using the t-test method (P=0.12 and P=0.94, respectively). In contrast, the copy number in the HYDIN gene had a higher fold amplification in the groups of breast cancer patients (Emean ≥50 kPa) (P=0.02).
In these 11 mutated genes, copy number amplifications of 7 tumor-related genes (CFC1B, NPIPA7, OR4F16, TPSG1, TPSB2, ANTXRL, and DDR2) were only detected in the groups of breast cancer patients. Additionally, in the malignant breast cancer group, CNVs of TPSG1, TPSB2, ANTXRL, and DDR2 occurred only in the group with a higher tumor stiffness (group 3) (Figure 5).
Bioinformatics analysis for DDR2
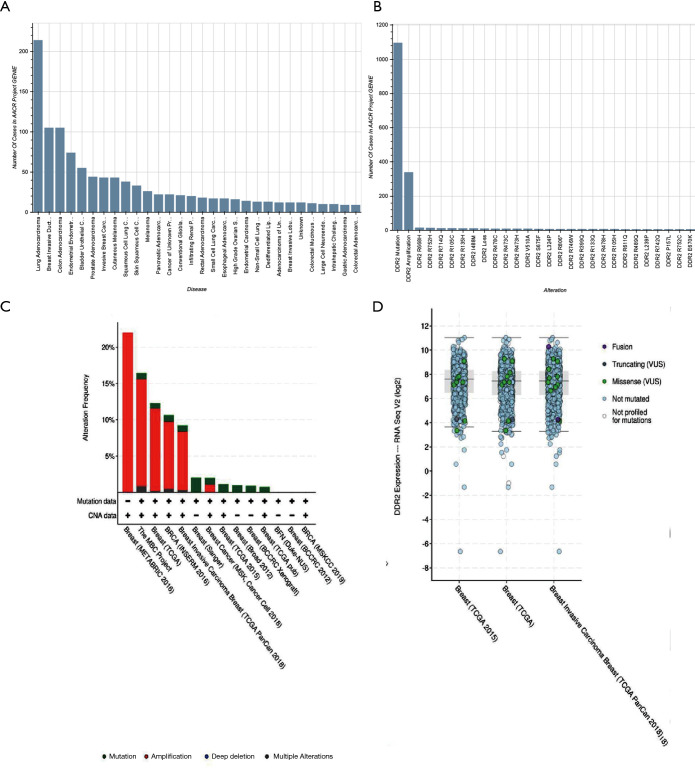
Using the online bioinformatics tool My Cancer Genome (https://www.mycancergenome.org/content/gene/DDR2/#ref-3), we performed bioinformatics analysis for DDR2 and found that similar to integrin receptors, DDR2 may play a role in modulating cellular interactions with the extracellular matrix (ECM). The DDR2 gene is mutated in 5.03% of malignant solid tumor patients. The most common alterations in DDR2 are DDR2 mutations (1.72%), DDR2 amplifications (0.52%), DDR2 I488M (0.02%), DDR2 R668H (0.02%), and DDR2 R105C (0.02%). There is an alteration of DDR2 in 2.19% of all cancers, with non-small-cell lung carcinoma, breast carcinoma, colorectal adenocarcinoma, melanoma, and uterine corpus neoplasm having the greatest prevalence of alterations (Figure 6A,B). Especially in breast cancer, amplification and mutation frequency of DDR2 was a common phenomenon in The Cancer Genome Atlas (TCGA) database (Figure 6C,D).
Figure 6.
Bioinformatics analysis for DDR2 by online bioinformatics tool My Cancer Genome. The most common alterations of DDR2 in pan-cancer (A,B). Bioinformatics analysis for DDR2 by online bioinformatics tool My Cancer Genome. The most common alterations of DDR2 in breast cancer (C,D). DDR2, discoidin domain receptor 2.
Validation of the function of the DDR2 gene in breast tumor mCAFs during cancer progression
The UE has been extensively evaluated as a noninvasive tool to assess tumor stiffness, and the measurement of tumor stiffness has high sensitivity and specificity for diagnosing breast tumors. Previous research has identified the relationship between UE (tumor stiffness) and CAF distribution with the expression features of α-SMA+ in patients with breast masses, whose data were used to evaluate its clinical significance in the early diagnosis of breast cancer.
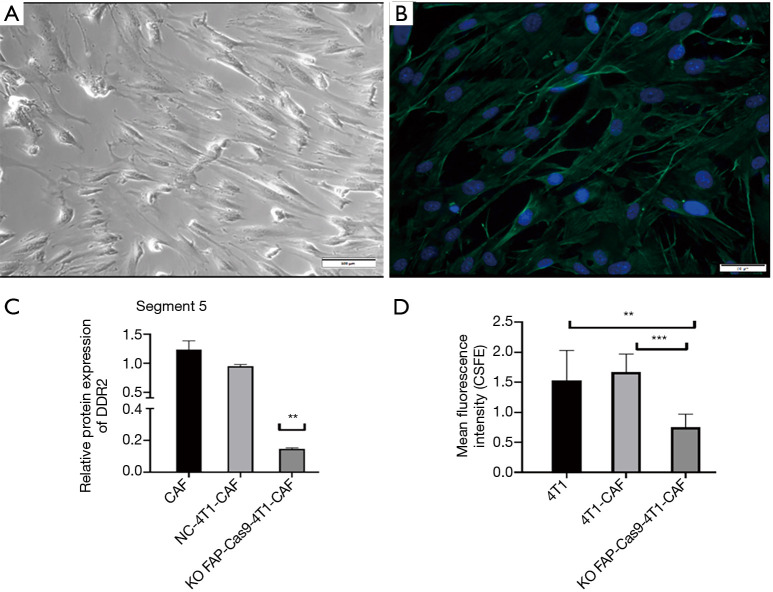
To confirm the biofunction of the DDR2 gene in breast tumor CAFs and to determine the correlation of DDR2 with tumor stiffness, we isolated primary mouse CAFs (mCAFs) from 4T1-GFP xenograft mice, which was verified by expression of α-SMA with immunofluorescence (Figure 7A,B). We then compared the expression of DDR2 in breast tumor CAFs by knocking out FAP via CRISPR/Cas9 technology. For the coculture studies, we found that the DDR2 gene and DDR2 protein expression were significantly upregulated in breast tumor CAFs during cancer progression; however, DDR2 was significantly downregulated in breast tumor CAFs with the FAP knockout via CRISPR/Cas9 (Figure 7C), which result in inhibition of cell proliferation (Figure 7D). These results indicated the action of DDR2 in CAFs and tumor cells within primary breast tumors in vivo.
Figure 7.
Primary mCAFs were isolated from 4T1-GFP xenograft mice and verified by expression of α-SMA with immunofluorescence (A,B) (A,B: magnification: 400×). The comparison of protein expression of DDR2 in breast tumor CAFs by knocking out FAP via CRISPR/Cas9 technology (C). Cell proliferation was inhibited in breast tumor CAFs with FAP knockout via CRISPR/Cas9 (D). **, P<0.05; ***, P<0.01. CAFs, carcinoma-associated fibroblasts; mCAF, mouse CAF; α-SMA, alpha-smooth muscle actin; DDR2, discoidin domain receptor 2; FAP, fibroblast activation protein.
Deletion of DDR2 in breast tumor CAFs resulted in decreased tumor stiffness measured by UE (1–5 score scale)
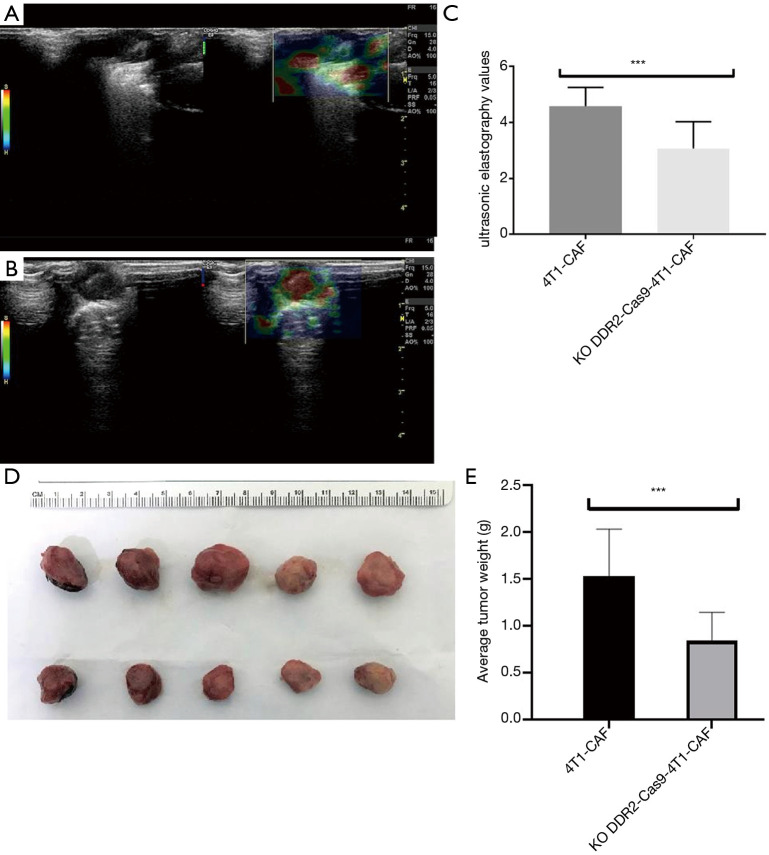
To determine if tumor stiffness regulation by DDR2 observed in the ex vivo culture of breast tumor CAFs was relevant in an in vivo setting, we deleted the DDR2 gene in breast tumor CAFs and assessed the stiffness properties of transplant tumors. We found that analyses of DDR2-knocked-out breast tumors, in which the DDR2 gene was deleted in the majority of CAFs, revealed that high UE scores were associated with hard stiffness (Figure 8A,B), and a significant difference was observed (P<0.05) (Figure 8C). These findings’ functional consequence was tumors with a diminished stiffness, with the most prominent changes in the stiffness in the breast tumor CAFs. Also, at the end of the experiment, the tumor volume and weight of the DDR2-knockout group were smaller than that of the 4T1-CAF group, and the difference was statistically significant (P<0.05) (Figure 8D,E).
Figure 8.
Deletion of DDR2 by CRISPR/Cas9 technology in breast tumor CAFs results in decreased tumor stiffness measured by UE (A,B,C) and inhibition of tumor growth (D,E). Representative B-mode (left) and UE (right) images in split-screen mode (A,B). ***, P<0.01. DDR2, discoidin domain receptor 2; CAFs, carcinoma-associated fibroblasts; UE, ultrasound elastography; US, ultrasound.
Discussion
Growing clinical diagnostic methods and biological features have been proposed to combine better predictive cancer characteristics and behaviors with improving the early diagnosis of cancer and determining a more individualized treatment strategy. US-based elastography has been extensively evaluated as a noninvasive tool to assess tumor stiffness, and tumor stiffness measurements have a high sensitivity and specificity for diagnosing breast tumors.
The tumor microenvironment, including the ECM and various stromal cells, such as CAFs and MSCs, has a great influence on tumor progression, metastasis, and drug resistance (21-23). Biophysical changes resulting from increased ECM remodeling increase tissue stiffness. Our previous study found that the expression level of α-SMA in breast cancer was positively correlated with the UE score (r=0.896, P<0.05) (24). Moreover, other researchers have also identified the relationship between UE and CAF distribution with the expression features of α-SMA+ in patients with breast masses, whose data were used to evaluate its clinical significance in the early diagnosis of breast cancer (25,26).
The technical means to detect CNVs rely largely on capturing coverage information across the genome. The coverage unevenness or uniformities of chromosomes could be an effective indicator of structural mutational levels triggered from ctDNA that originated from highly mutated tumors. The CNVs, as useful prognostic factors, were associated with subtypes, histological grade, and overall survival rate of breast cancer (27-29). In our study, CNVs were more common in the higher stiffness group, indicating that these patients have more genetic abnormalities. In this pilot study, we found that the frequency distribution of CNVs of cancer-associated genes TPSG1, TPSB2, ANTXRL, and DDR2 was only in group 3, participants who had increased tissue stiffness. That helps explain why patients with invasive breast cancer with a higher stiffness were associated with a higher histological grade and had significantly poorer survival compared to those with a lower level of stiffness (6,7,30). Tumors with higher stiffness may have further acquired other molecular mutations, leading to tumor evolution.
The action of DDR2 in CAFs, according to previous reports, controls ECM remodeling and thereby facilitates cell migration and tumor cell invasion in breast cancer (13). Binding of collagen to DDR2 results in the activation of downstream signaling pathways, perhaps the SRC and STAT signaling pathways. Like integrin receptors, DDR2 may play a role in modulating cellular interactions with the ECM (31). Combination treatment with an immunosuppressant and inhibitor of DDR2 may lead to tumor load reduction, suggesting DDR2 as a leading target for enhancing the response to immunosuppressive therapy (32). Here, we found that the DDR2 gene demonstrated a greater frequency of copy number gain in the ctDNA of breast cancer participants with higher tissue stiffness. Further studies should be directed towards investigating whether CNVs of DDR2 may cause alterations in the gene expression that contributes to cancer development.
During the development and progression of breast cancer, endogenous DDR2 expression is upregulated in CAFs and appears to be critical for their stiffness properties. Herein, we show that DDR2 plays an important role in tumor stiffness, both in breast tumor CAFs in culture and in tumor progression in vivo.
In our study, all participants in group 1 had FAs, the most common benign lesion of the breast. Previous studies have shown that there is a relationship between FAs and malignant phyllodes tumors, and these patients had an approximately two-fold increase in the relative risk of their FA developing invasive cancer after nearly 20 years (33,34). It is generally accepted that UE-SWE can distinguish between benign and malignant breast lesions. Thus, analyzing the relevance of UE-SWE and genetic alterations across FAs may provide insights into the molecular pathogenesis of fibroepithelial breast tumors and breast cancer. Unfortunately, the key genetic and molecular alterations associated with FA tumorigenesis remain unclear (35,36). Here, we found that CNVs of HYDIN, PDE4DIP, and Notch2 in ctDNA were also identified in all participants with FA, suggesting several similarities between FA and breast cancer genomic profiles.
There were some limitations in this study. Firstly, the present study’s ultimate small sample size possibly limited the power to detect more genomic alterations. However, in this research, we observed some recurrent mutations such as copy number alterations in the HYDIN and Notch2 genes. Another limitation is that we didn’t compare the CNV landscapes in ctDNA and matched tumor tissue, affecting research credibility. To our knowledge, some previous studies have shown the concordance of mutation patterns between ctDNA and tumor tissue (37).
Conclusions
In summary, we monitored the CNV landscape in ctDNA of breast tumor patients with different tumor stiffness by UE-SWE and identified several CNVs that occurred only in breast cancer samples. Furthermore, the DDR2 gene in CAFs was initially confirmed to be associated with the UE-SWE value and tumor stiffness. A combined analysis of UE-SWE and CNVs in ctDNA may be more beneficial for the early diagnosis and prognostic assessment of breast cancer.
Acknowledgments
Useful suggestions given by Prof. Yan Wang of QuestGenomics Ltd., Gnomegen LLC are also acknowledged.
Funding: This study was supported by the general programs from Shenzhen Science and Technology Innovation Committee (JCYJ20190814110207603, JCYJ20190814111801681); the National Natural Science Foundation of China (81972423); the project of free exploration from Shenzhen Science and Technology Innovation Committee (JCYJ20170307144246792, JCYJ20170307144103633); the clinical research start-up plan of Southern Medical University (LC2016YM018); the Incubation Programme of the National Natural Science Foundation of China from Shenzhen Hospital of Southern Medical University (PY2020ZY02); the Social Public Welfare Project of Science and Technology from Shenzhen Baoan District (2016CX301); the Grant of Shenzhen Key Laboratory of Viral Oncology (ZDSYS201707311140430); and the Grant of Shenzhen Sanming Medical Project (SM201702). The funding bodies did not influence the study design or the collection, analysis, or interpretation of data.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experimental protocol was approved by the institutional animal ethics committee of Southern Medical University (Approval ID: No. 2019-005), which follows the guidelines for the care and use of laboratory animals published by the National Institutes of Health (No. 85-23, revised 1996) and the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-443). All authors reported this study was supported by the general programs from Shenzhen Science and Technology Innovation Committee (JCYJ20190814110207603, JCYJ20190814111801681); the National Natural Science Foundation of China (81972423); the project of free exploration from Shenzhen Science and Technology Innovation Committee (JCYJ20170307144246792, JCYJ20170307144103633); the clinical research start-up plan of Southern Medical University (LC2016YM018); the Incubation Programme of the National Natural Science Foundation of China from Shenzhen Hospital of Southern Medical University (PY2020ZY02); the Social Public Welfare Project of Science and Technology from Shenzhen Baoan District (2016CX301); the Grant of Shenzhen Key Laboratory of Viral Oncology (ZDSYS201707311140430); and the Grant of Shenzhen Sanming Medical Project (SM201702).
References
- 1.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer 2009;9:108-22. 10.1038/nrc2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009;139:891-906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JY, Chang JK, Dominguez AA, Lee HP, Nam S, Chang J, Varma S, Qi LS, West RB, Chaudhuri O. YAP-independent mechanotransduction drives breast cancer progression. Nat Commun 2019;10:1848. 10.1038/s41467-019-09755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pediconi F, Galati F. Breast cancer screening programs: does one risk fit all? Quant Imaging Med Surg 2020;10:886-90. 10.21037/qims.2020.03.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng H, Qi X, Zhang T, Qi X, Yoshida EM, Guo X. Supersonic shear imaging for the diagnosis of liver fibrosis and portal hypertension in liver diseases: a meta-analysis. Expert Rev Gastroenterol Hepatol 2018;12:91-8. 10.1080/17474124.2018.1412257 [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Bai X, Chen Y, Jiang L, Hu B, Hu B, Yu L. Application of real-time elastography ultrasound in the diagnosis of axillary lymph node metastasis in breast cancer patients. Sci Rep 2018;8:10234-44. 10.1038/s41598-018-28474-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Zhang S, Zang L, Li J, Li J, Kang Y, Ren W. Combination of shear wave elastography and Ki-67 index as a novel predictive modality for the pathological response to neoadjuvant chemotherapy in patients with invasive breast cancer. Eur J Cancer 2016;69:86-101. 10.1016/j.ejca.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 8.Evans A, Sim YT, Pourreyron C, Thompson A, Jordan L, Fleming D, Purdie C, Macaskill J, Vinnicombe S, Pharoah P. Pre-operative stromal stiffness measured by shear wave elastography is independently associated with breast cancer-specific survival. Breast Cancer Res Treat 2018;171:383-9. 10.1007/s10549-018-4836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao Y, Ren GH, Yang W, Zheng WY, Wu YM, Li WJ, Li X, Li YJ, Guo X. Combination diagnosis with elastography strain ratio and molecular markers effectively improves the diagnosis rate of small breast cancer and lymph node metastasis. Quant Imaging Med Surg 2020;10:678-91. 10.21037/qims.2020.02.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Hu J, Hu G. Biomarker studies in early detection and prognosis of breast cancer. Adv Exp Med Biol 2017;1026:27-39. 10.1007/978-981-10-6020-5_2 [DOI] [PubMed] [Google Scholar]
- 11.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Killcoyne S, Del Sol A. Identification of large-scale genomic variation in cancer genomes using in silico reference models. Nucleic Acids Res 2016;44:e5. 10.1093/nar/gkv828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayer SV, Grither WR, Brenot A, Hwang PY, Barcus CE, Ernst M, Pence P, Walter C, Pathak A, Longmore GD. DDR2 controls breast tumor stiffness and metastasis by regulating integrin mediated mechanotransduction in CAFs. Elife 2019;8:e45508. 10.7554/eLife.45508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Jensen H, Johnston JJ, Di Maria E, Kloth K, Cristea I, Sapp JC, Darling TN, Huryn LA, Tranebjærg L, Cinotti E, Kubisch C, Rødahl E, Bruland O, Biesecker LG, Houge G, Bredrup C. Recurrent, activating variants in the receptor tyrosine kinase DDR2 cause Warburg-Cinotti syndrome. Am J Hum Genet 2018;103:976-83. 10.1016/j.ajhg.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Orsi C, Sickles EA, Mendelson EB, Morris EA. Breast Imaging Reporting and Data System: ACR BI-RADS breast imaging atlas. 5th ed. Reston: American College of Radiology, 2013. [Google Scholar]
- 16.Evans A, Whelehan P, Thomson K, McLean D, Brauer K, Purdie C, Jordan L, Baker L, Thompson A. Quantitative shear wave ultrasound elastography: initial experience in solid breast masses. Breast Cancer Res 2010;12:R104. 10.1186/bcr2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molla Kazemiha V, Bonakdar S, Amanzadeh A, Azari S, Memarnejadian A, Shahbazi S, Shokrgozar MA, Mahdian R. Real-time PCR assay is superior to other methods for the detection of mycoplasma contamination in the cell lines of the National Cell Bank of Iran. Cytotechnology 2016;68:1063-80. 10.1007/s10616-015-9862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, Wang L, Zeng L, Shao Y, Chen Y, Ma N, Lu W, Hu K, Han H, Yu Y, Huang Y, Liu M, Li D. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. Embo Mol Med 2016;8:477-88. 10.15252/emmm.201506039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang D, Peng Y, Lv W, Deng L, Zhang Y, Li H, Yang P, Zhang J, Song Z, Xu G, Cram DS, Wu L. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn 2014;16:519-26. 10.1016/j.jmoldx.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 21.Hua X, Yu L, Huang X, Liao Z, Xian Q. Expression and role of fibroblast activation protein-alpha in microinvasive breast carcinoma. Diagn Pathol 2011;6:111. 10.1186/1746-1596-6-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99-115. 10.1038/s41573-018-0004-1 [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Ke L, Lv X, Ling YH, Lu J, Liang H, Qiu W, Huang X, Liu G, Li W, Guo X, Xia W, Xiang Y. The prognostic significance of carcinoma-associated fibroblasts and tumor-associated macrophages in nasopharyngeal carcinoma. Cancer Manag Res 2018;10:1935-46. 10.2147/CMAR.S167071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Y, Guo L, Liu LS. A study on the relationship of differences on ultrasound elastography with histopathological alterations of α-SMA and CD34 in breast cancer between the Uygur and Han nationality. Journal of Xinjiang Medical University 2013;6:744-7. [Google Scholar]
- 25.Cui Y, Shen W, Zhang W, Wu L. Clinicopathological significance of stromal myofibroblasts in invasive ductal carcinoma of the breast. Tumour Biol 2004;25:290-5. 10.1159/000081394 [DOI] [PubMed] [Google Scholar]
- 26.Hao Y, Guo X, Ma B, Zhu L, Liu L. Relationship between ultrasound elastography and myofibroblast distribution in breast cancer and its clinical significance. Sci Rep 2016;6:19584. 10.1038/srep19584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 2006;45:1033-40. 10.1002/gcc.20366 [DOI] [PubMed] [Google Scholar]
- 28.Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, Choudhury AD, Gydush G, Reed SC, Rhoades J, Rotem D, Hughes ME, Dillon DA, Partridge AH, Wagle N, Krop IE, Getz G, Golub TR, Love JC, Winer EP, Tolaney SM, Lin NU, Adalsteinsson VA. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol 2018;36:543-53. 10.1200/JCO.2017.76.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Feizi N, Chi C, Hu P. Association analysis of somatic copy number alteration burden with breast cancer survival. Front Genet 2018;9:421. 10.3389/fgene.2018.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machida Y, Shimauchi A, Okuma H, Tozaki M, Isobe S, Fukuma E. Shear wave speed of the lesion in preoperative breast ultrasonography: association with disease-free survival of patients with primary operable invasive breast cancer. Acad Radiol 2018;25:1003-9. 10.1016/j.acra.2018.01.015 [DOI] [PubMed] [Google Scholar]
- 31.Toy KA, Valiathan RR, Núñez F, Kidwell KM, Gonzalez ME, Fridman R, Kleer CG. Tyrosine kinase discoidin domain receptors DDR1 and DDR2 are coordinately deregulated in triple-negative breast cancer. Breast Cancer Res Treat 2015;150:9-18. 10.1007/s10549-015-3285-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren T, Zhang J, Zhang J, Liu X, Yao L. Increased expression of discoidin domain receptor 2 (DDR2): a novel independent prognostic marker of worse outcome in breast cancer patients. Med Oncol 2013;30:397. 10.1007/s12032-012-0397-3 [DOI] [PubMed] [Google Scholar]
- 33.Dupont WD, Page DL, Parl FF, Vnencak-Jones CL, Plummer WD, Jr, Rados MS, Schuyler PA. Long-term risk of breast cancer in women with fibroadenoma. N Engl J Med 1994;331:10-5. 10.1056/NEJM199407073310103 [DOI] [PubMed] [Google Scholar]
- 34.Abe M, Miyata S, Nishimura S, Iijima K, Makita M, Akiyama F, Iwase T. Malignant transformation of breast fibroadenoma to malignant phyllodes tumor: long-term outcome of 36 malignant phyllodes tumors. Breast Cancer 2011;18:268-72. 10.1007/s12282-009-0185-x [DOI] [PubMed] [Google Scholar]
- 35.Tan J, Ong CK, Lim WK, Ng CC, Thike AA, Ng LM, et al. Genomic landscapes of breast fibroepithelial tumors. Nat Genet 2015;47:1341-5. 10.1038/ng.3409 [DOI] [PubMed] [Google Scholar]
- 36.Xie SN, Cai YJ, Ma B, Xu Y, Qian P, Zhou JD, Zhao FG, Chen J. The genomic mutation spectrums of breast fibroadenomas in Chinese population by whole exome sequencing analysis. Cancer Med 2019;8:2372-9. 10.1002/cam4.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Chang CW, Spoerke JM, Yoh KE, Kapoor V, Baudo C, Aimi J, Yu M, Liang-Chu MMY, Suttmann R, Huw LY, Gendreau S, Cummings C, Lackner MR. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin Cancer Res 2019;25:2254-63. 10.1158/1078-0432.CCR-18-1593 [DOI] [PubMed] [Google Scholar]