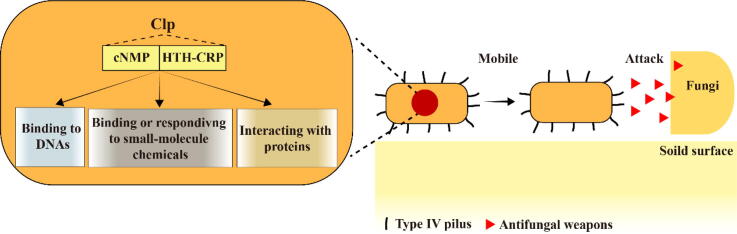
Graphical abstract
Lysobacter enzymogenes, a robust bacterial warrior uses a “mobile-attack” antifungal strategy comprised of twitching motility powered by the type IV pilus and antifungal weapons. This strategy is controlled by a “busy” transcription factor Clp via multiple previously uncharacterized mechanisms, including binding with DNA in unique modes, interacting directly or responding to diverse small-molecule chemicals, as well as associating specifically with structurally-distinct proteins.
Keywords: Transcription factor, Clp, Lysobacter, DNA binding, Ligand binding, Protein interactions
Abstract
Cyclic AMP receptor protein (CRP) is a well-characterized group of global transcription factors in bacteria. They are known to regulate numerous cellular processes by binding DNA and/or cAMP (a ligand called bacterial second messenger) to control target gene expression. Gram-negative Lysobacter enzymogenes is a soilborne, plant-beneficial bacterium without flagella that can fight against filamentous fungi and oomycete. Driven by the type IV pilus (T4P) system, this bacterium moves to nearby pathogens and uses a “mobile-attack” antifungal strategy to kill them via heat-stable antifungal factor (HSAF) and abundant lyases. This strategy is controlled by a unique “busy” transcription factor Clp, which is a CRP-like protein that is inactivated by binding of c-di-GMP, another ubiquitous second messenger of bacteria. In this review, we summarize the current progress in how Clp initiates a “mobile-attack” strategy through a series of previously uncharacterized mechanisms, including binding to DNA in a unique pattern, directly interacting with or responding to various small molecules, and interacting specifically with proteins adopting distinct structure. Together, these characteristics highlight the multifunctional roles of Clp in L. enzymogenes, a powerful bacterial warrior against fungal pathogens.
1. Introduction
Cyclic AMP receptor (CRP)-family proteins function as global regulatory transcription factors in bacteria [1], among which Escherichia coli CRP and Pseudomonas aeruginosa Vfr (virulence factor regulator) are arguably the two most characterized representatives of this family of proteins [1], [2]. Both CRP and Vfr contain a conserved N-terminal cAMP binding domain and a C-terminal helix-turn-helix (HTH) DNA binding domain that carries the consensus binding sequence of TGTGA-N6-TCACA [3]. Upon binding cAMP, a well-known nucleotide second messenger, the DNA binding domain of E. coli CRP is activated, resulting in the expression changes of genes associated with carbon metabolism [4]. P. aeruginosa Vfr also forms a complex with cAMP to jointly regulate the transcription of several gene subsets related to flagella-driven motility, type IV pili-driven twitching motility, virulence-associated type III secretion system (T3SS), biofilm formation, and bacterial pathogenicity [5], [6], [7]. In contrast, the Vfr homolog in the biocontrol P. fluorescens participates in fungal antagonism by down-regulating the production of 2, 4-diacetylphloroglucinol, pyrrolnitrin, and pyoluteorin, the three major antifungal compounds [8]. Other than interacting with cAMP, Vfr also senses intracellular redox signals (i.e. glutathione) to upregulate the expression of T3SS genes in P. aeruginosa and promote host infection [9]. Moreover, Vfr can establish concrete communication with quorum sensing (QS) pathway that is a cell density-dependent feature involved in bacterial infection [10]. Vfr positively regulates the las QS through transcriptional control of lasR encoding the las QS regulator by binding to its promoter region [10]. However, the transcription control of lasR by Vfr is cAMP independent [5]. Therefore, Vfr seems to control virulence gene expression via both cAMP-dependent and cAMP-independent pathways, which enables P. aeruginosa to precisely regulate its virulence program in response to specific host or environmental signals [5].
Clp is a CRP-like protein that contains domains similar to CRP [11]. While Clp shares the same domain organization as CRP and Vfr, it does not interact with cAMP, but specifically interact with cyclic di-GMP (c-di-GMP), as documented in Xanthomonas axonopodis pv. citri and Xanthomonas campestris pv. campestris, the two model strains in plant pathology [12], [13], [14], [15]. C-di-GMP is a ubiquitous second messenger mainly produced by Gram-negative bacteria [16] to control a wide range of cellular processes by binding to many target proteins or riboswitches [17], [18], [19]. The biosynthesis and degradation of c-di-GMP are achieved by the families of diguanylate cyclase (DGC) and phosphodiesterase (PDE) enzymes, respectively. While DGCs form a dimer and synthesize c-di-GMP by condensing two GTP molecules via the GGDEF catalytic domain [20], [21], PDEs directly hydrolyze c-di-GMP to GMP via the HD-GYP catalytic domain or cleave c-di-GMP to pGpG and further to two molecules of GMP using the EAL catalytic domain [22], [23], [24]. Previous biochemical and modeling studies uncovered the c-di-GMP binding site of Clp that is located between the cNMP binding domain and the HTH DNA binding domain [12], [15]. Although the binding ligand is different, Clp also exhibits different binding affinity to promoter DNA after binding c-di-GMP to control the transcription of target genes associated with virulence and biofilm formation in X. campestris [11], [25]. Like Vfr in P. aeruginosa, Clp in X. campestris is associated with the QS mechanism by binding a molecule of cis-unsaturated fatty acid, also known as the diffusible signal factor (DSF). The genes in the rpf (regulation of pathogenic factors) signaling system encode enzymes for DSF biosynthesis and proteins composed of RpfF, RpfC and RpfG, which act as sensor and response regulators [26], [27]. Among them, RpfF encodes enoyl coenzyme A (enoyl-CoA) hydratase responsible for DSF synthesis [25], [26]. Importantly, the RpfC-RpfG pair forms a two-component system responsible for DSF sensing and signal transduction, with RpfC acting as a histidine kinase anchored in the inner membrane, while RpfG as a cytoplasmic response regulator carrying a HD-GYP domain responsible for c-di-GMP degradation [28]. At high cell density, DSF is accumulated extracellularly and is bound by RpfC via its sensor domain located in the periplasm [29]. This results in the autophosphorylation of RpfC and subsequent phosphorylation of RpfG via phosphate group transfer [29]. The PDE activity of RpfG is consequently activated to degrade c-di-GMP, shifting the ratio of c-di-GMP bound and c-di-GMP free Clp to the latter, and leading to c-di-GMP specific signaling output in X. campestris [11], [24].
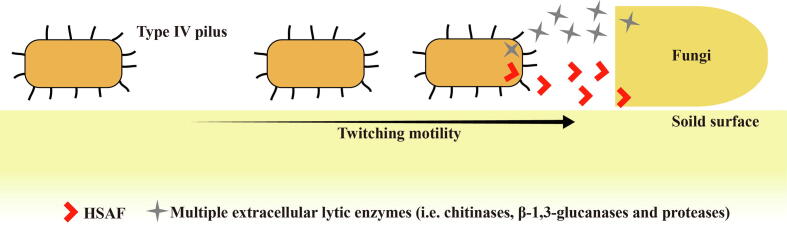
Species of the genus gammaproteobacterial Lysobacter are ubiquitous in the environment. Their natural habitat includes agricultural soil, water, and plant surfaces [30], [31]. This genus includes more than forty species and is receiving increasing attention in crop protection because several of its members (represented by L. enzymogenes, L. antibioticus, L. brunescens, L. gummosus, and L. capacisi) are effective biocontrol agents against crop fungal and/or bacterial diseases [32], [33], [34], [35], [36], [37]. The biocontrol species of Lysobacter kill pathogens through secreted antibiotics and lyases [30], [38], with L. enzymogenes being the most studied species in this genus, due to the available mature genetic manipulation technology [39], [40]. L. enzymogenes kills filamentous fungi and oomycetes through the various antifungal weapons we will discuss, thereby distinguishing itself as a bacterial warrior. However, due to the absence of flagellar gene FliC, this bacterium lacks the surface-attached flagellum evolutionally [41], [42], but produces another type of surface-attached thin appendages called type IV pilus (T4P) [43]. The extension and retraction of T4P on solid surface promotes the twitching movement of L. enzymogenes to draw near ecologically-relevant pathogens [44]. Upon contacting with fungi or oomycetes, L. enzymogenes kill them by secreting multiple antifungal factors, including abundant lyases to degrade fungal cell walls [45] and/or kill them via a broad-spectrum antifungal antibiotic HSAF (heat-stable antifungal factor) to disrupt the polar growth of fungi [46], [47], [48]. Therefore, L. enzymogenes appears to employ twitching motility and antimicrobial factors (HSAF and lyase) to establish a “mobile-attack” strategy against filamentous fungi and oomycetes (Fig. 1).
Fig. 1.
Schematic diagram of the “mobile-attack” strategy designed by L. enzymogenes. L. enzymogenes moves to nearby fungi by twitching motility driven by the type IV pilus. While in motion, it not only produces a variety of extracellular lyases to degrade the fungal cell walls but also secretes an antifungal antibiotic HSAF to inhibit fungal growth.
In this review, we summarize the latest developments in how Clp controls the “mobile-attack” strategy, emphasizing that Clp is a “busy” worker, involved in numerous cellular tasks to enable efficient fungal killing by L. enzymogenes.
2. Clp is “busy” in regulating numerous gene expression and binding to multiple DNA fragments
In an earlier study, Kobayashi and colleagues reported that Clp is essential for the biocontrol activity of L. enzymogenes towards bipolaris leaf-spot of tall fescue and pythium damping-off of sugarbeet [45]. Phenotypic analyses reveal that Clp is indispensable to produce diverse lytic enzymes, namely, chitinase, ß-1, 3-glucanase and protease, all of which can degrade the corresponding components of fungal cell walls [45]. Therefore, controlling the production of lyase is a reasonable strategy to prove the Clp-dependent biocontrol activity against the above-mentioned fungi or oomycete. In a later study, Kobayashi and Yuen further found that L. enzymogenes can establish a similar pathogenic relationship with Magnaporthe poae by colonizing its fungal hyphae that is controlled by Clp [49]. Recently, Zhao and colleagues found that L. enzymogenes cells can colonize and invade the hyphae of Pythium aphanidermatum and that this parasitic trait of L. enzymogenes is also governed by Clp [50].
How does L. enzymogenes achieve fungal antagonism via Clp? We showed previously that Clp can activate the expression of the HSAF biosynthesis operon and is required for producing HSAF in L. enzymogenes [51], [52]. Also, disruption of the key HSAF biosynthesis operon gene in L. enzymogenes leads to the diminution of HSAF-dependent antifungal activity and its effective invasion of the P. aphanidermatum hyphae [50]. Moreover, Clp is also essential for the T4P-driven twitching motility required for L. enzymogenes to move to nearby fungi to form biofilms, resulting in its colonization in plants or fungal hyphae [43], [50]. In summary, we believe that the “mobile-attack” anti-fungal strategy adopted by L. enzymogenes is controlled by Clp.
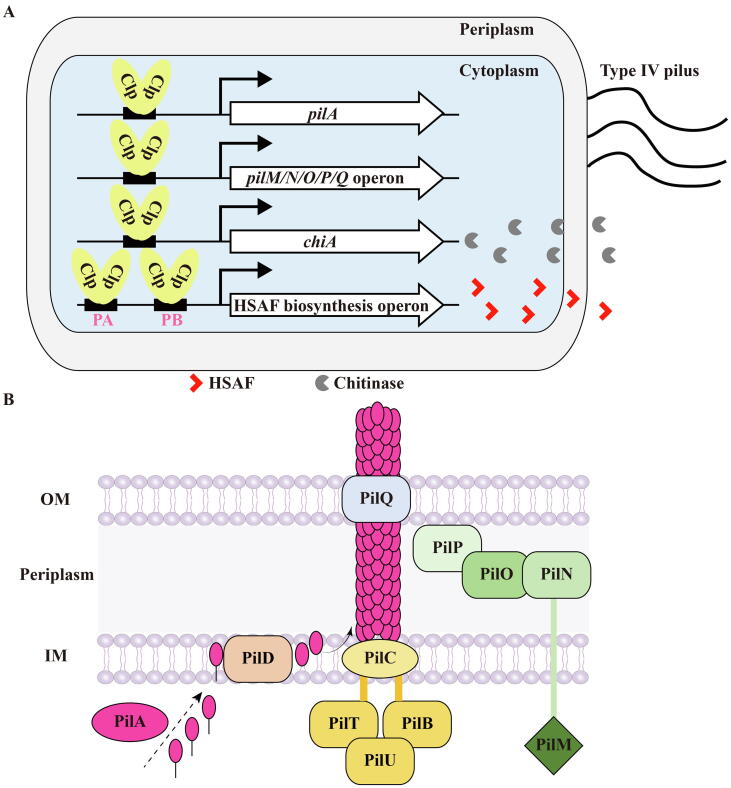
There is strong evidence to suggest that the “mobile-attack” fungal strategy is fulfilled by the DNA binding and gene activating activity of Clp. For example, an earlier transcriptome study revealed that among the approximately 4000 genes of L. enzymogenes OH11, more than 700 genes are directly or indirectly controlled by Clp at the transcription level, demonstrating Clp is “busy” in controlling gene expression [51]. Among them, Clp directly binds to the promoter sequence upstream of the pilA and pilMNOPQ operon genes that encode key T4P structural components (Fig. 2A&B) [53]. Such direct DNA binding enables Clp to activate transcription of these T4P genes and facilitate the T4P formation and twitching motility of L. enzymogenes OH11 (Fig. 2A&B) [53]. We also identified two Clp binding sites (designated PA and PB) upstream of the HSAF biosynthesis operon promoter and showed that both are involved in the expression activation of the HSAF biosynthesis operon by Clp (Fig. 2A) [54]. Clp can also directly bind to the promoter of chiA to up-regulate the transcription of chiA [55], thereby producing a large amount of extracellular chitinase to digest chitin, a key component of fungal cell walls (Fig. 2A). Taken together, Clp is “busy” by directly or indirectly binding to multiple promoter DNAs of target genes to execute the “mobile- attack” antifungal strategy to regulate the expression of numerous genes.
Fig. 2.
Clp carries out the “motile attack” strategy by binding with different DNA fragments in L. enzymogenes. (A) A single gene or gene operon responsible for pilus assembly, chitinase production, and HSAF generation is controlled by Clp via binding to their cognate promoter regions. (B) A cartoon showing the components necessary for the synthesis and function of T4P pilus. The positions of PilA, PilM, PilN, PilO, PilP, PilQ are roughly shown, and their expression regulations by Clp are described in panel A. PilD, a peptidase used to cleave off the leader peptide of prepilins; PilC, an integral membrane protein functioning as a platform protein; PilB and PilT, ATPases responsible for pilus extension and retraction, respectively. PilQ, an outer membrane secretin protein involved in the export of pilus subunits. Abbreviations: OM, outer membrane; IM, inner membrane.
3. Clp is “busy” in binding or responding to various small-molecule chemicals
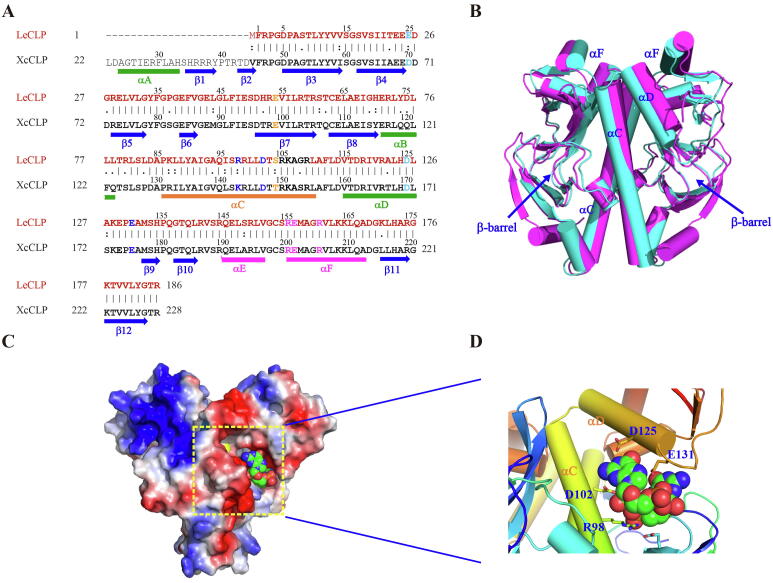
Like X. campestris Clp (XcCLP), L. enzymogenes Clp (LeCLP) also binds to c-di-GMP [54]. To compare the XcCLP and LeCLP proteins, we first used the pairwise sequence alignment scheme in the ClustalW2 program (EMBL-EBI, https://www.ebi.ac.uk/msd-srv/ssm/) to align their primary sequences. It can be seen from Fig. 3A that the two sequences are very comparable, with an identity of 73.7%, a similarity of 79.98%, and a root mean square value of 1.623 Å. In addition, except that LeCLP lacks the N-terminal αA helix and β1 and β2 strands and is shorter than 24 amino acid residues, no gap was found to exist between these two sequences. We then used the Phyre2 structure modeling program (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id = index) to predict the monomer structure of LeCLP, and further used the Symmetry Dock program (http://bioinfo3d.cs.tau.ac.il/SymmDock/) to obtain the dimer form. It can be clear seen from Fig. 3B that the two CLP structures are very similar with only some minor local structural change (Fig. 3B).
Fig. 3.
Structural comparison between the X. campestris Clp (XcCLP) and L. enzymogenes Clp (LeCLP). (A) Sequence alignment of XcCLP and LeCLP. The sequence of XcCLP is represented by black letters, and the sequence of LeCLP is represented in brown letters. The secondary structural elements of XcCLP are shown schematically below the alignment. The α-helix appears as a green cylinder, except for those involved in the DNA-binding helix-turn-helix domains that are magenta and that responsible for CLP dimerization is orange; and the β-strand appears as a blue arrow. Identical residues are connected by vertical lines, similar residues by two dots, and different residues by single dot. Residues that may be involved in DNA binding are colored in magenta, while those that form salt bridges with c-di-GMP from a model study are highlighted in blue. The residues that are close to the b-barrel and inhibit c-di-GMP binding are colored orange. (B) The superimposition between XcCLP (magenta) and LeCLP (cyan) structures shown in cylinder. ß-barrel regions are indicated by the blue arrow. (C) XcCLP dimer is drawn in electrostatic surface (positive, blue, and negative, red), and c-di-GMP molecule drawn in van der Waals (nitrogen atoms in blue, oxygen atoms in red, and carbon atoms in green). The docked region is framed in yellow and enlarged in the lower-right corner. (D) The interactions between LeCLP and c-di-GMP from one of the docked complex models drawn in cartoon representation. Residues potentially participating in these interactions are drawn as sticks and labeled with blue letters. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, we used the PatchDock program to obtain forty LeCLP-c-di-GMP complex structures. The stacked structure coordinates of c-di-GMP (PDB: 2RDE) was used for docking research. No structure with c-di-GMP locating in the cAMP binding pocket in the β-barrel region of the XcCLP could be observed; it was found that each ligand is found to be sandwiched between the αC dimerization helix and αD helix in the HTH DNA binding domain. We used one of these complex structures for a more detailed study, and presented it in the electrostatic representation shown in Fig. 3C. The positively charged area (blue region) of the DNA-binding domain is clearly observed in the figure. As shown in Fig. 3D, c-di-GMP is located in a wedge between the dimeric αC helix and αD of the DNA binding domain. Some amino acids (shown in blue letters in Fig. 3A) were found to surround the c-di-GMP ligand. The D125 in the model LeCLP-c-di-GMP complex (D170 in XeCLP) was found to be important since a D170A substitution resulted in a ten-fold loss in affinity [12]. But it is important to note that, so far, most modelling programs can only carry out a lock-and-key approach, but cannot carry out induced-fit modeling. In addition, the c-di-GMP can adopt a wide range of different conformations [18], and we only selected the fully stacked c-di-GMP conformation for docking studies. Therefore, the modelled LeCLP-c-di-GMP complex structure can only be considered as an approximation; the real complex structure can only be obtained by X-ray crystallization or NMR methods.
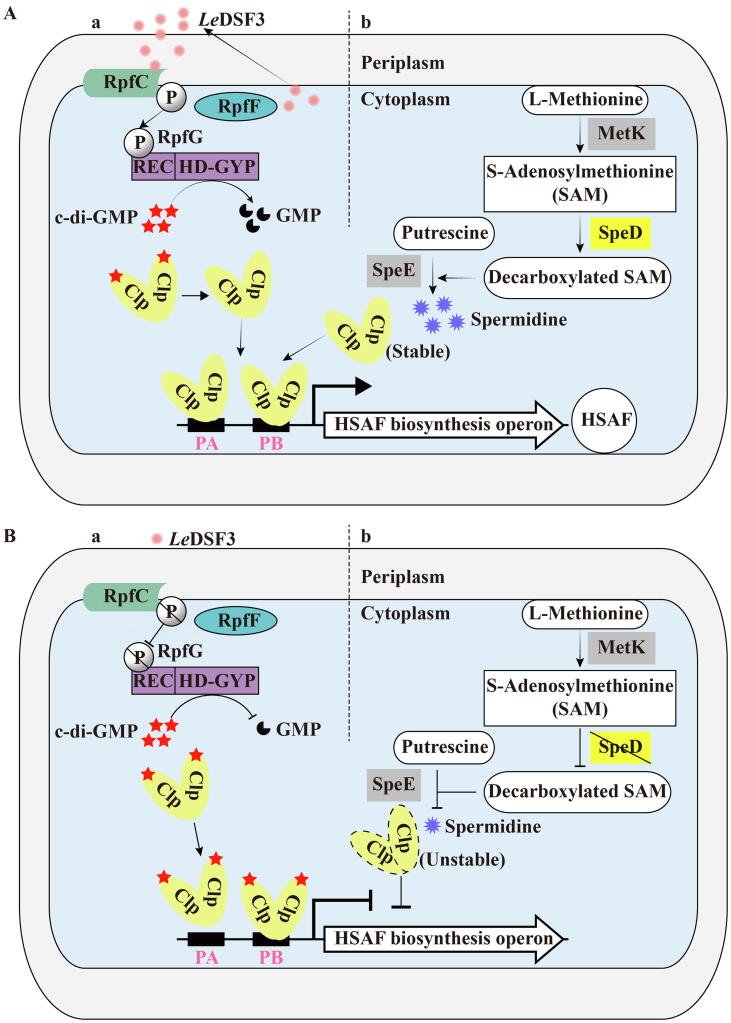
When combined with c-di-GMP, the capability of L. enzymogenes Clp to bind DNA is impaired, especially at the lower-affinity site (PA). This results in decreased transcription of the operon, therefore reducing HSAF synthesis (Fig. 4) [54]. This biochemical finding is consistent with our observation that elevated levels of c-di-GMP inhibit HSAF biosynthesis [56]. According to our earlier report, Clp can also respond to two small molecules to fine-tune HSAF production.
Fig. 4.
Clp controls the expression of the HSAF biosynthesis operon by binding to c-di-GMP and responding to fatty-acid signals of LeDSF3 and spermidine. (A) Clp-dependent activation of HSAF biosynthesis operon expression at high cell density (high LeDSF3 level, panel a) or in the presence of cellular spermidine generated by a pathway described in the panel b. (B) Clp inhibits HSAF production by binding to c-di-GMP at low cell density (low LeDSF3 level, panel a) or decreasing the level of cellular spermidine by inactivating SpeD as described in the panel b. The details are described in context.
A well characterized small molecule responding to Clp is the diffusible fatty-acid signal, called LeDSF3, which is significantly involved in the regulation of HSAF biosynthesis [48], [57]. The synthesis of LeDSF3 is carried out by an X. campestris RpfF homolog as described previously [48]. Although produced inside the cell, LeDSF3 can diffuse to the extracellular environment [57]. Similar to what we have previously described for X. campestris, at high cell density, extracellular LeDSF3 will accumulate and act as an intra-bacterial signal to activate the expression of HSAF biosynthesis operon through the defined “RpfC-RpfG-Clp” pathway. In this, RpfC is a hybrid histidine kinase comprising a sensory input domain, a histidine kinase (HisKA) domain, a response regulator receiver (REC) domain, and a histidine phosphotransfer (HPT) domain [58]. The sensory input domain of RpfC comprises five transmembrane helices with periplasmic and cytoplasmic loops, which are responsible for sensing LeDSF3 signaling [58]. After the sensory input domain senses the signal, the HisKA domain of RpfC is auto-phosphorylated, followed by the transfer of a phosphoryl group to the REC domain of the RpfG regulator [58]. Phosphorylation of RpfG then exhibits enhanced PDE activity, enabling it to degrade c-di-GMP more efficiently. The reduced c-di-GMP levels thus deprive Clp of c-di-GMP and activate the expression of HSAF biosynthesis operon (Fig. 4Aa & Ba) [54], [57].
Another small molecule responsive to Clp is spermidine, which is a common polyamine compound in cells. Polyamines consist of an aliphatic hydrocarbon chain with one or more amine groups, and are ubiquitous in almost all living organisms to carry out key physiological functions [59]. Intriguingly, we found that full scale HSAF production and HSAF-mediated fungal inhibition also require spermidine [60].
However, according to our previous study, spermidine cannot directly interact with Clp [60]. Therefore, what is the relationship between regulation of HSAF biosynthesis by spermidine and Clp? As documented before, spermidine biosynthesis requires SpeD that encodes an S-adenosylmethionine decarboxylase [61], speD is a neighbor gene of clp in L. enzymogenes genome and this genome arrangement also seems to occur in many other bacteria [60]. Further, the mutation of speD leads to a decrease in the production of spermidine and HSAF, while supplementation of physiological range spermidine in culture can rescue both defects shown in this mutant [60]. Moreover, although Clp level was significantly reduced in the speD mutant, this defect could also rescued by adding spermidine to mutant cultures [60]. In summary, SpeD seems to be the main factor for maintaining spermidine levels in the body, which indirectly stabilizes the transcription factor Clp to promote HSAF production and fungal antagonism (Fig. 4Ab & Bb) [60]. However, how spermidine specifically stabilizes Clp levels need further investigation.
Overall, as described above, we propose that Clp is also very “busy” in sensing or responding to various small molecules, which leads to the full production of HSAF and the effective killing of fungi by L. enzymogenes.
4. Clp is “busy” in physically interacting with diverse proteins
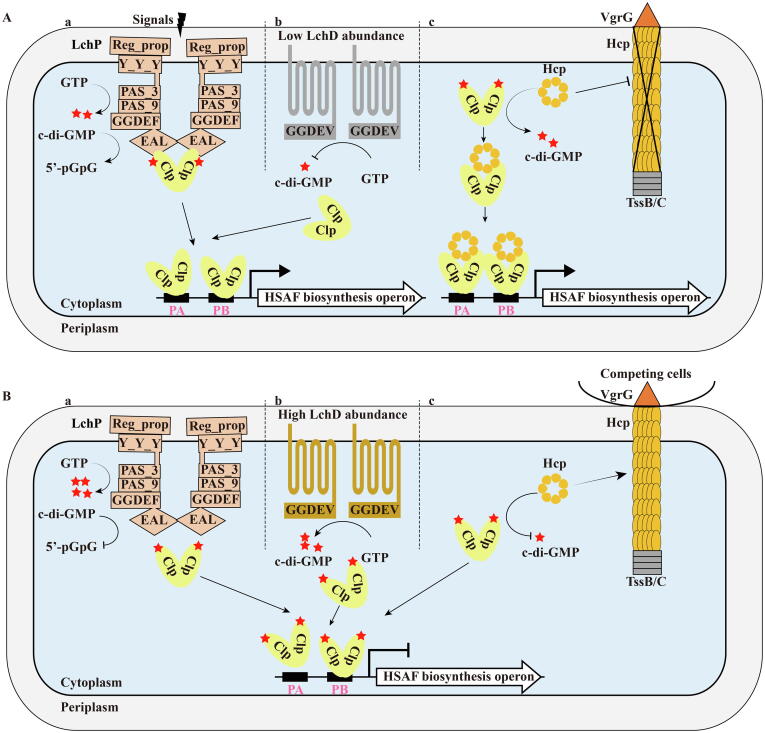
Besides binding to multiple DNA regions and responding to several small molecules, Clp was also found to be “busy” interacting with many proteins involved in HSAF biosynthesis regulation. The first Clp binding protein is LchP, which is an inner member-anchored protein containing both cytoplasmic GGDEF and EAL domains responsible for the synthesis and degradation of c-di-GMP, respectively [54]. Interestingly, LchP mainly acts as a functional PDE to regulate HSAF biosynthesis in natural L. enzymogenes cells [54]. It physically combines with Clp to form a stable LchP-Clp complex, thereby enhancing the PDE activity of LchP [54], allowing the accumulation of Clp without bound c-di-GMP and leads to elevated expression of the HSAF biosynthesis operon. On the other hand, when the intracellular concentration of c-di-GMP increases, the LchP-Clp interaction is impaired, resulting in the disintegration of the complex that inhibits the LchP PDE activity, thereby forming a negative feedback. The increasing c-di-GMP level then inhibits HSAF biosynthesis via Clp, as described above (Fig. 5Aa&Ba) [54]. Therefore, the LchP-Clp interaction is crucial and allows Clp to act as a molecular switch to regulate the enzymatic activity of LchP.
Fig. 5.
Clp regulates HSAF biosynthesis via binding to a variety of proteins. (A) Activation of HSAF biosynthesis under nearby fungi or nutrient-poor conditions. Clp physically interacts with the c-di-GMP-degrading enzyme LchP to stimulate its PDE activity to reduce c-di-GMP level. LchD is a type of DGC, but it is less active under conditions producing abundant HSAF, resulting in a decrease in c-di-GMP level (b). The reduction of c-di-GMP shifts the ratio of c-di-GMP-bound Clp and c-di-GMP-free Clp towards the latter, causing Clp to activate expression of HSAF biosynthesis operon (a, b). Type VI secretion system is not assembled under HSAF production conditions, and its structural component, the inner tube Hcp, binds and protects Clp from c-di-GMP inhibition, thereby promoting HSAF biosynthesis operon expression (c). (B) HSAF biosynthesis is inactivated. In the absence of fungi or in rich nutrients, cellular c-di-GMP will be elevated to disassemble the Clp-LchP complex. This step inhibits the PDE activity of LchP and further increases the level of c-di-GMP (a). Similarly, LchD level will accumulate and becomes more efficient when combined with Clp, which will promote the DGC activity of LchD, leading to elevated c-di-GMP synthesis (b). Increased level of c-di-GMP binds to Clp to inhibit its DNA binding affinity to the promoter region of the HSAF biosynthesis operon, resulting in a decrease in HSAF production (a, b). Under nutrient-rich conditions, HSAF production is blocked, and Hcp is mainly used to assemble T6SS. Under this case, Hcp cannot protect Clp from c-di-GMP inhibition (c). The details are described in context.
The second Clp binding protein is LchD. It is also an inner-membrane anchored protein, but unlike LchP, it is DGC possessing an active GGDEF domain involved in c-di-GMP synthesis and HSAF production [62]. LchD also physically interacts with Clp [62], but this combination will stimulate the DGC activity of LchD to obtain a higher level of c-di-GMP. Elevated level of c-di-GMP not only helps the separation of Clp from the LchD-Clp complex, but it is also more effective in binding Clp [62]. The resulting Clp-c-di-GMP complex cannot activate the expression of the HSAF biosynthesis operon, resulting in a decrease in HSAF production as described above (Fig. 5Ab & Bb) [54].
The third Clp-binding protein is Hcp, which is a common structural component of the inner tube of the type VI secretion system (T6SS). T6SS is a form of a contact-dependent (“short-range”) killing weapon, often used by proteobacteria [63], [64], [65], [66]. When a cell is in contact with another cell, the developed T6SS will pierce the cell wall and cell membrane of the prey (target) cell, and transmits a toxin-laden shell (known as the inner tube) to it [66], [67]. L. enzymogenes also possesses a complete T6SS gene cluster that can work together to assemble a functional T6SS, as evidenced by the secretion of Hcp when L. enzymogenes is cultured in a nutrient-rich medium (LB) that likely mimics an environment rich in other microorganisms [68]. However, under the same nutritional conditions, the production of “long-range” weapon is hindered [68]. In contrast, in the HSAF-producing medium (1/10 TSB), which is likely to be present in filamentous oomycetes and fungi, T6SS is not assembled and Hcp secretion is not detected [68]. The unique feature of the Hcp-Clp binding under the conditions of HSAF production and T6SS inactivity is that this combination can jointly regulate HSAF production to promote HSAF-based antifungal activity [68]. In terms of mechanism, Hcp-Clp binding seems to protect Clp from binding to c-di-GMP, thereby preventing c-di-GMP from inactivating Clp. Therefore, the increased in Clp concentration without bound c-di-GMP leads to higher HSAF operon expression and HSAF production, as described above (Fig. 5Ac&Bc) [54]. Thus, when T6SS, a typical “short-range” weapon, is not used, the accumulation of Hcp, one of its structural components, can be used as a co-activator of Clp to enhance the production of HSAF, dubbed a “long-range” weapon. In general, the “busy” roles of Clp in binding to a variety of proteins to participate in crucial biological functions in Gram-negative bacteria are unprecedented in the CRP family transcription factors.
5. Summary and outlook
In the past decades, a large number of genetic, biochemical, and structural studies have clearly demonstrated that the CRP-family transcription factors, namely CRP and Clp, control diverse bacterial gene transcription by combining with various DNA fragments and ligands. This review provides new mechanisms for this family of transcription factors by focusing on the L. enzymogenes Clp, which is involved in DNA binding, protein–protein interaction and sensing/responding to small molecules, and emphasizes its “busy” roles in the bacterial warrior, L. enzymogenes. However, there are still several unresolved issues. For example, does Clp regulate the transcription of HSAF operon by directly stabilizing polymerase binding like CRP family transcription factors? Why is L. enzymogenes Clp evolved to bind c-di-GMP but not cAMP? What is the structural basis of the combination of Clp-c-di-GMP? Why can’t Clp physically interact with RpfG, but dynamically perceive c-di-GMP through RpfG? How can Clp interact specifically with a variety of structurally distinct protein partners such as LchP, LchD and Hcp? How does the Hcp-Clp complex prevent Clp from binding to c-di-GMP? Further biochemical and structural approaches to answer these questions will not only promote our understating of the molecular evolution and roles of the global CRP family transcription factors in bacteria, but also help develop strategies to inactivate this family of proteins to control infection of pathogenic bacteria or enhance antibiotic production in the beneficial bacterium of L. enzymogene.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Prof. Mark Gomelsky (University of Wyoming, USA) and Prof. Daolong Dou (Nanjing Agricultural University, China) for critical suggestions on the review organization. This study was supported by the National Natural Science Foundation of China [32072470 and 31872016 to G.Q.; 32001955 to L. L.; 32070139 to D. S.]; the Natural Science Foundation of Jiangsu Province [BK20190026 and BK20181325 to G.Q.]; Fundamental Research Funds for the Central Universities [KJJQ202001, KYRC2021003, KYZ202106, KYT201805 and KYTZ201403 to G.Q.; KJQN202111 to L. L.]; Innovation Team Program for Jiangsu Universities [2017 to G.Q.]. The funders had no role in the study design.
Contributor Information
Kangwen Xu, Email: 2018202015@njau.edu.cn.
Long Lin, Email: linlong@njau.edu.cn.
Danyu Shen, Email: shendanyu@njau.edu.cn.
Shan-Ho Chou, Email: shchou@nchu.edu.tw.
Guoliang Qian, Email: glqian@njau.edu.cn.
References
- 1.Zheng D., Constantinidou C., Hobman J.L., Minchin S.D. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serate J., Roberts G.P., Berg O., Youn H. Ligand responses of Vfr, the virulence factor regulator from Pseudomonas aeruginosa. J Bacteriol. 2011;193(18):4859–4868. doi: 10.1128/JB.00352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunasekera A., Ebright Y.W., Ebright R.H. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992;267(21):14713–14720. [PubMed] [Google Scholar]
- 4.Gosset G., Zhang Z., Nayyar S., Cuevas W.A., Saier M.H. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol. 2004;186(11):3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs E.L., Brutinel E.D., Jones A.K., Fulcher N.B., Urbanowski M.L., Yahr T.L. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J Bacteriol. 2010;192(14):3553–3564. doi: 10.1128/JB.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatson S.A., Whitchurch C.B., Sargent J.L., Levesque R.C., Mattick J.S. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J Bacteriol. 2002;184(13):3605–3613. doi: 10.1128/JB.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta N., Ferrell E.P., Kanack K.J., West S.E., Ramphal R. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol. 2002;184:5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q., Ji Y., Xiao Q.i., Chng S., Tong Y., Chen X. Role of Vfr in the regulation of antifungal compound production by Pseudomonas fluorescens FD6. Microbiol Res. 2016;188-189:106–112. doi: 10.1016/j.micres.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Zhang C., Du X., Zhou Y., Kong W., Lau G.W. Glutathione activates type III secretion system through Vfr in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2019;9 doi: 10.3389/fcimb.2019.0016410.3389/fcimb.2019.00164.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albus A.M., Pesci E.C., Runyen-Janecky L.J., West S.E., Iglewski B.H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179(12):3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He YW, Ng AY, Xu M, Lin K, Wang LH, et al. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol Microbiol 2007;64:281–92. [DOI] [PubMed]
- 12.Chin K.-H., Lee Y.-C., Tu Z.-L., Chen C.-H., Tseng Y.-H., Yang J.-M. The cAMP receptor-like protein Clp is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396(3):646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 2012;13:614–29. [DOI] [PMC free article] [PubMed]
- 14.Leduc J.L., Roberts G.P. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191(22):7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao F., He Y.-W., Wu D.-H., Swarup S., Zhang L.-H. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192(4):1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325(6101):279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 17.Römling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou S.-H., Galperin M.Y. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol. 2016;198(1):32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R, Sliusarenko O, Kazmierczak BI. Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog 2017;13:e1006594. [DOI] [PMC free article] [PubMed]
- 20.Paul R., Weiser S., Amiot N.C., Chan C., Schirmer T. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryjenkov D.A., Tarutina M., Moskvin O.V., Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187(5):1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt A.J., Ryjenkov D.A., Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187(14):4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christen M., Christen B., Folcher M., Schauerte A., Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280(35):30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 24.Bellini D., Caly D.L., McCarthy Y., Bumann M., An S.Q. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol. 2014;91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He YW, Xu M, Lin K, Ng YJ, Wen CM, et al. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol Microbiol 2006;59:610–22. [DOI] [PubMed]
- 26.Barber C.E., Tang J.L., Feng J.X., Pan M.Q., Wilson T.J.G., Slater H. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24(3):555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- 27.He Y.-W., Zhang L.-H. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev. 2008;32(5):842–857. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 28.Slater H., Alvarez-Morales A., Barber C.E., Daniels M.J., Dow J.M. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol. 2000;38:986–1003. doi: 10.1046/j.1365-2958.2000.02196.x. [DOI] [PubMed] [Google Scholar]
- 29.Cai Z., Yuan Z.H., Zhang H., Pan Y., Wu Y. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog. 2017;13:e1006304. doi: 10.1371/journal.ppat.1006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen P., Cook F.D. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Evol Microbiol. 1978;28(3):367–393. [Google Scholar]
- 31.Puopolo G., Tomada S., Pertot I. The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J Appl Microbiol. 2018;124(1):15–27. doi: 10.1111/jam.13607. [DOI] [PubMed] [Google Scholar]
- 32.Panthee S., Hamamoto H., Paudel A., Sekimizu K. Lysobacter species: a potential source of novel antibiotics. Arch Microbiol. 2016;198(9):839–845. doi: 10.1007/s00203-016-1278-5. [DOI] [PubMed] [Google Scholar]
- 33.de Bruijn I., Cheng X.u., de Jager V., Expósito R.G., Watrous J., Patel N. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genom. 2015;16(1) doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y., Qian G., Ye Y., Wright S., Chen H., Shen Y. Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org Lett. 2016;18(10):2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- 35.Ling J., Zhu R., Laborda P., Jiang T., Jia Y., Zhao Y. LbDSF, the Lysobacter brunescens quorum-sensing system diffusible signaling factor, regulates anti- Xanthomonas XSAC biosynthesis, colony morphology, and surface motility. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.0123010.3389/fmicb.2019.01230.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puopolo G., Cimmino A., Palmieri M.C., Giovannini O., Evidente A. Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J Appl Microbiol. 2014;117:1168–1180. doi: 10.1111/jam.12611. [DOI] [PubMed] [Google Scholar]
- 37.Laborda P., Ling J., Chen X., Liu F. ACC deaminase from Lysobacter gummosus OH17 can promote root growth in Oryza sativa nipponbare plants. J Agric Food Chem. 2018;66(14):3675–3682. doi: 10.1021/acs.jafc.8b00063. [DOI] [PubMed] [Google Scholar]
- 38.Xie Y., Wright S., Shen Y., Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012;29(11):1277. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian G.-L., Hu B.-S., Jiang Y.-H., Liu F.-Q. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agr Sci China. 2009;8(1):68–75. [Google Scholar]
- 40.Qian G., Wang Y., Qian D., Fan J., Hu B., Liu F. Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J Microbiol Biotechnol. 2012;28(2):549–557. doi: 10.1007/s11274-011-0846-8. [DOI] [PubMed] [Google Scholar]
- 41.Fulano A.M., Shen D., Kinoshita M., Chou S.-H., Qian G. The homologous components of flagellar type III protein apparatus have acquired a novel function to control twitching motility in a non-flagellated biocontrol bacterium. Biomolecules. 2020;10(5):733. doi: 10.3390/biom10050733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fulano A.M., Shen D., Zhang E.-H., Shen X.i., Chou S.-H., Minamino T. Functional divergence of flagellar type III secretion system: a case study in a non-flagellated, predatory bacterium. Comput Struct Biotechnol J. 2020;18:3368–3376. doi: 10.1016/j.csbj.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia J., Chen J., Chen Y., Qian G., Liu F. Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol. 2018;102(2):833–846. doi: 10.1007/s00253-017-8619-4. [DOI] [PubMed] [Google Scholar]
- 44.Lin L., Zhou M., Shen D., Han S., Fulano A.M., Chou S.-H. A non-flagellated biocontrol bacterium employs a PilZ-PilB complex to provoke twitching motility associated with its predation behavior. Phytopathol Res. 2020;2(1) doi: 10.1186/s42483-020-00054-x. [DOI] [Google Scholar]
- 45.Kobayashi D.Y., Reedy R.M., Palumbo J.D., Zhou J.-M., Yuen G.Y. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl Environ Microbiol. 2005;71(1):261–269. doi: 10.1128/AEM.71.1.261-269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S., Du L., Yuen G., Harris S.D. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2006;17(3):1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu F., Zaleta-Rivera K., Zhu X., Huffman J., Millet J.C., Harris S.D. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51(1):64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian G., Wang Y., Liu Y., Xu F., He Y.-W., Du L. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microb. 2013;79(21):6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi D.Y., Yuen G.Y. The role of clp-regulated factors in antagonism against Magnaporthe poae and biological control of summer patch disease of Kentucky bluegrass by Lysobacter enzymogenes C3. Can J Microbiol. 2005;51(8):719–723. doi: 10.1139/w05-056. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y., Qian G., Chen Y., Du L., Liu F. Transcriptional and antagonistic responses of biocontrol strain Lysobacter enzymogenes OH11 to the plant pathogenic oomycete Pythium aphanidermatum. Front Microbiol. 2017;8:1025. doi: 10.3389/fmicb.2017.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Zhao Y., Zhang J., Zhao Y., Shen Y., Su Z. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol. 2014;98(21):9009–9020. doi: 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P., Chen H., Qian G., Liu F. LetR is a TetR family transcription factor from Lysobacter controlling antifungal antibiotic biosynthesis. Appl Microbiol Biotechnol. 2017;101(8):3273–3282. doi: 10.1007/s00253-017-8117-8. [DOI] [PubMed] [Google Scholar]
- 53.Chen J., Shen D., Odhiambo B.O., Xu D., Han S., Chou S.-H. Two direct gene targets contribute to Clp-dependent regulation of type IV pilus-mediated twitching motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol. 2018;102(17):7509–7519. doi: 10.1007/s00253-018-9196-x. [DOI] [PubMed] [Google Scholar]
- 54.Xu GG, Han S, Huo CM, Chin KH, Chou SH, et al. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res 2018;46:9276–88. [DOI] [PMC free article] [PubMed]
- 55.Xu H., Chen H., Shen Y., Du L., Chou S.-H., Liu H. Direct regulation of extracellular chitinase production by the transcription factor Le Clp in Lysobacter enzymogenes OH11. Phytopathology. 2016;106(9):971–977. doi: 10.1094/PHYTO-01-16-0001-R. [DOI] [PubMed] [Google Scholar]
- 56.Ren X., Ren S., Xu G., Dou W., Chou S.-H., Chen Y.u. Knockout of diguanylate cyclase genes in Lysobacter enzymogenes to improve production of antifungal factor and increase its application in seed coating. Curr Microbiol. 2020;77(6):1006–1015. doi: 10.1007/s00284-020-01902-x. [DOI] [PubMed] [Google Scholar]
- 57.Han Y., Wang Y., Tombosa S., Wright S., Huffman J., Yuen G. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol. 2015;99(2):801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan R.P., Dow J.M. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19(3):145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Gevrekci A. The roles of polyamines in microorganisms. World J Microbiol Biotechnol. 2017;33:204. doi: 10.1007/s11274-017-2370-y. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y., Zhang T., Ning Y., Shen D., Yang N., Li Y. Spermidine plays a significant role in stabilizing a master transcription factor Clp to promote antifungal activity in Lysobacter enzymogenes. Appl Microbiol Biotechnol. 2019;103(4):1811–1822. doi: 10.1007/s00253-018-09596-9. [DOI] [PubMed] [Google Scholar]
- 61.Hobley L., Li B., Wood J.L., Kim S.H., Naidoo J., Ferreira A.S. Spermidine promotes Bacillus subtilis biofilm formation by activating expression of the matrix regulator slrR. J Biol Chem. 2017;292(29):12041–12053. doi: 10.1074/jbc.M117.789644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu K, Shen D, Yang N, Chou SH, Gomelsky M, et al. (2021) Coordinated control of the type IV pili and c-di-GMP-dependent antifungal antibiotic production in Lysobacter by the response regulator PilR. Mol Plant Pathol. In press. [DOI] [PMC free article] [PubMed]
- 63.Mougous J.D., Cuff M.E., Raunser S., Shen A., Zhou M. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hachani A., Wood T.E., Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 65.Galán J.E., Waksman G. Protein-injection machines in bacteria. Cell. 2018;172(6):1306–1318. doi: 10.1016/j.cell.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernandez R.E., Gallegos-Monterrosa R., Coulthurst S.J. Type VI secretion system effector proteins: effective weapons for bacterial competitiveness. Cell Microbiol. 2020;22:e13241. doi: 10.1111/cmi.13241. [DOI] [PubMed] [Google Scholar]
- 67.Liang X., Kamal F., Pei T.-T., Xu P., Mekalanos J.J., Dong T.G. An onboard checking mechanism ensures effector delivery of the type VI secretion system in Vibrio cholerae. Proc Natl Acad Sci U S A. 2019;116(46):23292–23298. doi: 10.1073/pnas.1914202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang M., Ren S., Shen D., Yang N., Wang B. An intrinsic mechanism for coordinated production of the contact-dependent and contact-independent weapon systems in a soil bacterium. PLoS Pathog. 2020;16:e1008967. doi: 10.1371/journal.ppat.1008967. [DOI] [PMC free article] [PubMed] [Google Scholar]