Abstract
Deregulation of many homeobox genes has been observed in various cancers and has caused functional implications in the tumor progression. In this review, we will focus on the roles of the human muscle segment homeobox (MSX) transcription factor family in the process of tumorigenesis. The MSX transcription factors, through complex downstream regulation mechanisms, are promoters or inhibitors of diverse cancers by participating in cell proliferation, cell invasion, cell metastasis, cell apoptosis, cell differentiation, drug resistance of tumors, maintenance of tumor stemness, and tumor angiogenesis. Moreover, their upstream regulatory mechanisms in cancers may include: gene mutation and chromosome aberration; DNA methylation and chromatin modification; regulation by non-coding RNAs; regulation by other transcription factors and post-translational modification. These mechanisms may provide a better understanding of why MSX transcription factors are abnormally expressed in tumors. Notably, intermolecular interactions and post-translational modification can regulate the transcriptional activity of MSX transcription factors. It is also crucial to know what affects the transcriptional activity of MSX transcription factors in tumors for possible interventions in them in the future. This systematic summary of the regulatory patterns of the MSX transcription factor family may help to further understand the mechanisms involved in transcriptional regulation and also provide new therapeutic approaches for tumor progression.
Keywords: Muscle segment homeobox 1 (MSX1), muscle segment homeobox 2 (MSX2), transcription factor, cancer progression, transcriptional regulation
Introduction
The homeobox genes were first discovered in Drosophila and subsequently in mammals (1). Mammalian homeobox genes are named after their homologs in Drosophila and are grouped into several families such as distal-less homeobox (DLX), muscle segment homeobox (MSX), etc. (2). The human MSX family has two subtypes: MSX1 and MSX2, both MSX1 and MSX2 function as transcription factors and each of them has a homeodomain that plays an important role in the transcriptional regulation of target genes (3,4). Furthermore, the homeodomain is also a motif for protein-protein interactions which could affect their transcriptional activity (5-8). The expression of MSX1 or MSX2 is also often deregulated in many tumors (9,10) and the MSX family mediates various cellular processes in cancers such as proliferation (11), invasion (12), metastasis (13), apoptosis (14), differentiation (15), drug resistance (16), tumor stemness (17), tumor angiogenesis (8), etc. Furthermore, MSX transcription factors exhibit either promotive or inhibitive effects on cancers through complex downstream regulatory mechanisms (11,14,18,19).
In this review, we firstly elucidated on the biological characteristics of MSX family, upstream regulation on expression and regulation on transcriptional activity. Moreover, we observed the roles of the MSX family in various tumors. Finally, we presented our views on the clinical application values of the MSX family in cancer research, particularly in the monitoring of tumor prognosis, diagnosis and treatment.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-21-220).
Methods
We conducted a literature search of the PubMed database using the following keywords: “muscle segment homeobox 1 and cancer”, “muscle segment homeobox 2 and cancer”, “MSX1 and cancer”, “MSX2 and cancer”. We also reviewed articles on the concepts of “transcription factor”, “cancer progression”, “transcriptional regulation”. Original articles and review articles published in the English language between December 1986 to August 2020 were included.
Biological structures of the MSX family
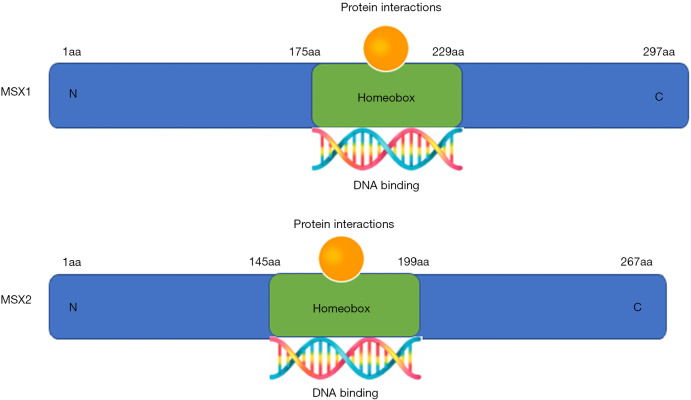
The MSX family which contains a homeodomain is highly conserved and present in all major classes of vertebrates, human MSX family includes two subtypes, MSX1 and MSX2 (20) (Figure 1). Both the MSX1 and MSX2 gene have 2 exons (21,22). The protein encoded by MSX1 consists of 297 amino acids and contains a homeodomain (aa175-229) (21). Similar to MSX1, the MSX2 protein is composed of 267 amino acids and also has a homeodomain (aa145-199) (22). Within the 60-amino-acid homeodomains of MSX1 and MSX2, human and murine sequences are the same, while only 2 amino acids are diverse in the MSX1 and MSX2 homeodomains (7,22,23). MSX2 and MSX1 share the same MSX binding-site and the mutation in MSX2 homeodomain (substitution of histidine for proline at residue 148) has no effect on the binding of MSX2 to the MSX binding-site (7). The three-dimensional structure of MSX1 shows that two atypical proline residues make the N-terminal arm of the homeodomain of MSX1 very stable and orderly. The N-terminal arm of MSX1 can track small groove of DNA, and the DNA bound by the homeodomain of MSX1 has a 28° bend compared to the normal 21° bend of the homeodomain of MSX2 (24). Since the N-terminal domain is unique, this allows MSX1 to have a more potent transcription capability (25). More studies are needed to elucidate on the transcriptional role of other regions other than homeodomain in the human MSX family (Figure S1) (26-30).
Figure 1.
Model of the protein structures of the muscle segment homeobox (MSX) family (MSX1 and MSX2): two-dimensional (2D) structures of MSX1 and MSX2. The human MSX family of transcription factors shares a homeodomain of the same length (green). The homeodomain is the DNA binding domain of MSX1 and MSX2, and also a motif for protein-protein interactions.
The regulatory mechanisms of expression
The expression of MSX1 or MSX2 in cancer tissues is often abnormal when compared to adjacent normal tissues (9,10). Since the mechanisms that regulate their expression levels in tumors are complex (Figure 2), five discovered important regulatory mechanisms are mainly discussed:
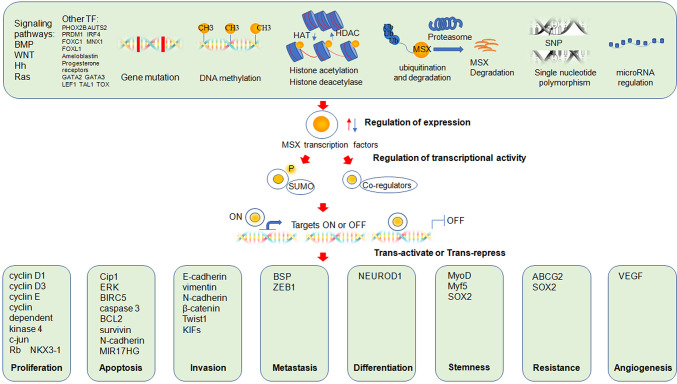
Figure 2.
Roles of MSX transcription factors in cancer and the known mechanism. Regulation of the expression of MSX transcription factors including downstream transcription factors of signaling pathways, individual transcription factors, gene mutation, DNA methylation, histone acetylation and deacetylation, ubiquitination and degradation, single nucleotide polymorphism or microRNA regulation may up- or down-regulate the expression of MSX transcription factors. Regulation of transcriptional activity: depending on the environment, MSX transcription factors function as transcriptional activators or repressors: (I) they may recruit co-regulators including Miz1, Dlx5, SHARP, MINT, RBM15, TBP, PAX3, YB1/p32, H1b, H1C to form transcriptional complexes and (II) post-translational modifications such as SUMOylation may affect transcription, resulting in different endings. By targeting different genes, MSX transcription factors are involved in: (I) cell proliferation (cyclin D1, cyclin D3, cyclin E, cyclin-dependent kinase 4, c-jun, Rb, NKX3-1); (II) cell apoptosis (Cip1, ERK, BIRC5, caspase 3, BCL2, survivin, N-cadherin, MIR17HG); (III) cell invasion (E-cadherin, vimentin, N-cadherin, β-catenin, Twist1, KIFs); (IV) cell metastasis (BSP, ZEB1); (V) cell differentiation (NEUROD1); (VI) tumor stemness (MyoD, Myf5, SOX2); (VII) drug resistance (ABCG2, SOX2); (VIII) angiogenesis (VEGF).
Gene mutation and chromosome aberration
In breast cancer, an 11 nucleotides deletion, the c.469+46_56del mutation in the intron of MSX1 gene was close to the splicing site, it interfered with the alternative splicing of pre-mRNA and affected the final product (31). Equally, van Nistelrooij et al. found that the down-regulated MSX1 expression caused by gene mutation may be associated with Barrett’s esophagus and esophageal adenocarcinoma (32). MSX1 was up-regulated in gastric cancer and the genes associated with single nucleotide polymorphism (SNP) sites in gastric cancer were screened through the database, where MSX1 is included (33). Chromosome deletions and rearrangements are common chromosome aberrations, Nagel et al. documented that in patients with natural killer (NK) cell leukemia, the transcription level of MSX1 was significantly down-regulated due to the deletion of chromosome 4p16 where MSX1 is located (34). Fluorescence in situ hybridization and gene chromatin immunoprecipitation (ChIP) analysis revealed that MSX1 in Hodgkin’s lymphoma cell lines was rearranged at site 4p16, yielding a lower expression of MSX1 (35).
DNA methylation and chromatin modification
Rauch et al. confirmed that in lung squamous cell carcinoma, the CpG island of MSX1 was hypermethylated and thus the expression of MSX1 was down-regulated (36). Furthermore, compared with adjacent normal tissues, MSX1 was found to be decreased and hypermethylated at the promoter region in colon adenocarcinoma (COAD) (9). Moreover, the methylation level of CpG island of MSX2 in gastric cancer tissues was found to be lower than in normal tissues and, therefore, MSX2 was upregulated in gastric cancer tissues (10,37). In endometrial cancer, the methylation status of MSX1 promoter was decreased, corresponding to its high expression (38). Histone acetyltransferase and histone deacetylase also regulate the expression of the MSX family. Nagel et al. reported that in mantle cell lymphoma, histone acetyltransferase plant homeodomain finger16 (PHF16) promoted MSX1 expression while histone deacetylase (HDAC) inhibited its expression (39). Moreover, Hamada et al. documented that histone acetyltransferases E1A-associated protein p300 and CREB-binding protein (CBP) were co-activators that promoted the expression of MSX2 in pancreatic cancer (40). Chromatin modification might therefore be an important mechanism by which MSX genes are deregulated in tumors.
Non-coding RNAs
MicroRNAs are small endogenous RNAs that cannot encode proteins, but they can inhibit or activate the translation and stabilization of target mRNAs (41,42). For example, in cultured human palate cells, microRNA-374a-5p down-regulated the expression of MSX1 (43). Liu et al. also reported that microRNA-203 up-regulated the expression of MSX2 in osteoblasts (44). Whether non-coding RNA can regulate MSX transcription factors in tumors is of great research value in the future.
Transcription factors
Several transcription factors including other homeobox genes exert different effects on the expression of MSX family. For example, Revet et al. reported that paired like homeobox2B (PHOX2B) down-regulated the expression of MSX1 in neuroblastoma (15). Additionally, Nagel et al. revealed that in NK leukemia, the activator of transcription and developmental regulator2 (AUTS2) and PR/SET domain1 (PRDM1) activated the expression of MSX1, while interferon regulatory factor4 (IRF4) acted as a suppressor of MSX1 expression (34). Forkhead box C1 (FOXC1) indicated the suppression of MSX1 expression in Hodgkin Lymphoma (45). Moreover, transcription factors FOXC1 and motor neuron pancreas homeobox1 (MNX1) were activators of MSX1 transcription in mantle cell lymphoma (39). In another study, MSX1 with SNP loci was shown to be regulated by forkhead box L1 (FOXL1) in gastric cancer (33). In odontogenic tumors, Sonoda et al. noted that ameloblastin suppressed the expression of MSX2 (46). A study on breast cancer revealed that progesterone receptors promoted the expression of MSX2 (47). In T-acute lymphoblastic leukemia (T-ALL), it has been established that GATA binding protein2 (GATA2) and FOXC1 mediated the activation of MSX1 transcription while GATA binding protein3 (GATA3), lymphoid enhancer binding factor1 (LEF1), TAL bHLH transcription factor1 (TAL1) and thymocyte selection associated high mobility group box (TOX) repressed MSX1 transcription (48).
Transcription factors regulate each other in the regulatory network of signaling pathways, the expression of MSX can also be regulated by corresponding transcription factors in specific signaling pathways such as growth factor signals in tumors (40). In colorectal carcinoma, WNT/β-catenin signaling activated MSX1 expression while the promoter of MSX1 was activated by β-catenin (18). Previous studies have suggested that MSX2 can also function as a downstream effector of WNT signaling in human ovarian endometrioid adenocarcinoma cells and human teratoma cells (49-51). The downstream factor of bone morphogenetic protein (BMP) signaling SMAD4 activated the expression of MSX1 in T-ALL (48). Additionally, in NK cell leukemia SMAD3 was activated by BMP signaling and SMAD3 inhibited MSX1 expression (34). Similarly, BMP4 activated the expression of MSX2 through SMAD4 or ERK1/2 or P38 in pancreatic cancer cells (40). The GLI family zinc finger 1 (GLI1), which is known to be involved in the hedgehog (Hh) signaling pathway, promoted the expression of MSX2 in gastric cancer (52). Several reports have confirmed that MSX2 acted as a regulatory target gene for Ras (12,53) even though its molecular mechanisms have not been established.
Post-translational modification
Song et al. revealed that protein inhibitor of activated STAT1 (PIAS1) enhanced the stability of MSX1 protein by inhibiting ubiquitination (54). Protein inhibitor of activated STAT Y (PIASy)-MSX1 interaction is also critical for MSX1 protein stabilization (8). Yuan et al. in their study on lung and breast cancer also showed that MSX2 was a substrate of F-box and WD repeat domain containing2 (FBXW2)-E3 ligase, whereas a combination of FBXW2 with MSX2 promoted the ubiquitination and degradation of MSX2 (55). O-linked-N-acetylglucosamine (O-GlcNAc) which is also a kind of post translational modification could down-regulate MSX1 expression (56). Therefore, the post-translational modification of the protein is also an important reason for the dysregulation of the expression of MSX transcription factors in tumors.
In addition to the five regulatory mechanisms we discussed above, retinoids (57) and MSX1 antisense RNA (58) also regulated the expression of MSX1 in development. Overall, the regulation of MSX gene expression is accomplished by abundant mechanisms.
The regulatory mechanisms of transcriptional activity
The regulatory mechanisms of transcriptional activity include protein-protein interactions and post-translational modification. Previous studies indicated that interactions between TATA-box binding protein (TBP) with MSX1 attenuated the inhibitory effect of MSX1 on the glycoprotein hormone α subunit (αGSU) gene expression in a DNA binding-independent manner (59). During tooth development, MSX1 formed a heterodimer with paired box 3 (PAX3), while the formation of non-transcriptional complex prevented MSX1 from binding to the DNA of its target gene (60). In addition, YB1/p32 synergistically enhanced the transcriptional activity of MSX1 through protein interactions (61). In particular, MSX1 in synergy with histone inhibited myogenic regulators (62). Furthermore, MSX1 and co-factor histone H1C can inhibited ZHX2 in Hodgkin’s lymphoma (35). PIASy can promote MSX1 to repress the reporter activity of vascular endothelial growth factor (VEGF) and this promotion further help MSX1 to mediate angiogenesis inhibition (8). In human embryo development research, MSX2 was shown to interact with two transcription factors, MSX-interacting-zinc finger1 (Miz1) (63) and distal-less homeobox5 (Dlx5) (64). Wu et al. documented that the affinity of MSX2-Miz1 complex to the MSX-DNA binding site on the target gene promoter was higher compared to that of MSX2 alone, thereby, enhancing the expression of the target gene (63). Newberry et al. found that MSX2-Dlx5 complex cannot bind the MSX binding site on the promoter of target gene, thus the expression of the target gene was inhibited (64). The SHARP and MSX2-interacting nuclear target protein (MINT) are involved in the transcriptional activity of MSX2 (65). A study on breast cell lines showed that RNA-binding motif protein 15 (RBM15) may regulate the transcriptional activity of MSX2 (14). Besides, post-translational modification small ubiquitin-related modification (SUMO) is considered to be an important regulatory mechanism for transcription (66). Song et al. showed that in 293T cells, PIAS1 promoted the SUMOylation of MSX1 and the SUMOylation inhibited the transcriptional activity of MSX1 (54). The regulatory mechanisms which can affect the transcriptional activity of MSX transcription factors in tumors need further study for the possibility of interventions in their transcriptional activity in practical applications.
The roles of MSX family in different types of tumors
The expression of MSX family in tumors
Expression levels of the MSX family vary in different tumors (Tables 1 and 2). In particular, the expression of MSX1 is lower in multiple tumors such as breast cancer (82,83), high-grade serous epithelial ovarian cancer (67), gastric cancer (37), cervical cancer (68,84), testicular germ cell tumors (TGCTs) (69), malignant NK cell leukemia (34), glioblastoma (70), neuroblastoma (15), Wilms tumor (73) and Hodgkin lymphoma (35). Moreover, MSX1 expression is upregulated in T-ALL (48), mantle cell lymphoma (MCL) (39), acute myeloid leukemia (AML) (39), pituitary adenomas (71), melanoma (72), myxoid liposarcoma (MLS) (17), endometrial cancer (38). Whereas in colorectal cancer, compared to the adjacent normal tissues, MSX1 was shown to be down-regulated in COAD (9). However, the expression of MSX1 was also found to be significantly high in some colorectal cancer tissues (18,85).
Table 1. The dysregulated expression of MSX1 in cancer.
| Type of cancer | Subtype of cancer | Expression | Reasons for dysregulation in cancer | Involved signaling pathways | References |
|---|---|---|---|---|---|
| Lung cancer | Lung squamous cell carcinoma | Down | Hypermethylation | – | (36) |
| Colorectal cancer | Colon adenocarcinoma | Down | Hypermethylation | – | (9) |
| Colorectal cancer | – | Up | – | WNT signaling | (18) |
| Ovarian cancer | High-grade serous epithelial ovarian cancer | Down | – | – | (67) |
| Breast cancer | – | – | Gene mutation | – | (31) |
| Esophageal cancer | Esophageal adenocarcinoma | Down | Gene mutation | – | (32) |
| Leukemia | T-cell acute lymphoblastic leukemia | Up | GATA2, FOXC1 | BMP signaling | (48) |
| Leukemia | Acute myeloid leukemia | Up | – | – | (39) |
| Leukemia | NK cell leukemia | Down | Chromosome loss, AUTS2, PRDM1, IRF4 | BMP signaling | (34) |
| Neuroblastoma | – | Down | PHOX2B | – | (15) |
| Lymphoma | Mantle cell lymphoma | Up | PHF16, FOXC1, MNX1 | – | (39) |
| Gastric cancer | – | Up | SNP | – | (33) |
| Cervical cancer | – | Down | – | – | (68) |
| Testicular cancer | Testicular germ cell tumors | Down | – | – | (69) |
| Glioblastoma | – | Down | – | – | (70) |
| Hodgkin’s lymphoma | – | Down | Chromosome rearrangement, FOXC1 | – | (35,45) |
| Pituitary cancer | Pituitary adenomas | Up | – | – | (71) |
| Melanoma | – | Up | – | – | (72) |
| Liposarcoma | Myxoid liposarcoma | Up | – | – | (17) |
| Wilms tumor | – | Down | – | – | (73) |
| Endometrial cancer | – | Up | Hypomethylation | WNT signaling | (38,74) |
MSX1, muscle segment homeobox 1; BMP signaling, bone morphogenetic protein signaling; FOXC1, forkhead box C1; GATA2, GATA binding protein 2; AUTS2, activator of transcription and developmental regulator 2; PRDM1, PR/SET domain 1; IRF4, interferon regulatory factor 4; PHOX2B, paired like homeobox 2B; PHF16, plant homeodomain finger 16; HDAC, histone deacetylase; SNP, single nucleotide polymorphism.
Table 2. The dysregulated expression of MSX2 in cancer.
| Type of cancer | Subtype of cancer | Expression | Reasons for dysregulation in cancer | Involved signaling pathways | References |
|---|---|---|---|---|---|
| Breast cancer | – | Up | Estrogen, progesterone | – | (75) |
| Breast cancer | – | Down | FBXW2, PR | (47,55) | |
| Pancreatic cancer | Pancreatic intraductal papillary mucinous tumor | Up | – | – | (76) |
| Pancreatic cancer | – | Up | P300, CBP | BMP signaling, Ras signaling | (40,53) |
| Gastric cancer | Diffuse-type gastric cancer | Up | Hypomethylation | Hh signaling | (10,52) |
| Odontogenic cancer | Ameloblastoma | Down | Ameloblastin | – | (46) |
| Melanoma | – | Down | – | – | (77) |
| Colorectal cancer | – | Up | – | – | (19) |
| Nasal cancer | Nasal inverted papilloma of the nose | Up | – | – | (78) |
| Prostate cancer | Prostate adenocarcinoma | Up | – | – | (79) |
| Lung cancer | – | Down | FBXW2 | – | (55) |
| Lung cancer | – | Up | – | – | (80) |
| Ovarian cancer | Ovarian endometrioid adenocarcinoma | Up | – | WNT signaling | (49) |
| Ovarian cancer | Ovarian yolk sac tumor | Up | – | – | (81) |
| Teratoma | – | Up | – | WNT signaling | (51) |
MSX2, muscle segment homeobox 2; FBXW2, F-box and WD repeat domain containing 2; PR, signaling progesterone receptor signaling; p300, E1A-associated protein p300; CBP, camp-response element-binding protein (CREB)-binding protein, BMP signaling, bone morphogenetic protein signaling; Hh, hedgehog.
The expression of MSX2 has also been shown to be elevated in most tumors such as breast cancer tissues (75), human pancreatic cancer (40,86), teratoma (51), ovarian cancer (49,81), gastric cancer (52), lung cancer (80), colorectal cancer (87), and inverted papilloma of the nose (78). In comparison, the expression of MSX2 in prostate cancer was higher than that in normal prostate tissues and its expression in metastatic cells was higher than that in non-metastatic cells (79). Conversely, its expression was lower in melanoma lung metastatic cells compared to melanoma cells that have not metastasized. However, the reasons for this occurrence have not been established (77). Finally, the expression of MSX2 was also reported to be low in lung and breast cancer (55). At the same time, the roles of MSX family also vary in different tumors. In some tumors, MSX family function as tumor promoters, but both MSX1 and MSX2 can exhibit inhibiting roles in other tumors.
MSX family in breast cancer
Overall, MSX1 has been reported to have an anticancer effect in breast cancer (82,83). In particular, MSX1 suppressed breast cancer cell growth via inducing the G1/S cell-cycle arrest and promoted apoptosis of breast cancer cells. Moreover, it suppressed the expression of β-catenin together with its downstream target genes so that inhibiting breast tumor cell migration and invasion (82). MSX1 can inhibit the expression of kinesin superfamily (KIFs) in breast cancer which leads to the occurrence and progression of breast cancer (83,88).
The dual role of MSX2 in breast cancer may be associated with subcellular localization and the downstream effectors it regulates (89). The MSX2-overexpressing cells presented a typical elongated and mesenchymal-like shape, accompanied by a reduced expression of the epithelial marker E-cadherin and a corresponding increase in the expression of mesenchymal markers, vimentin, and N-cadherin. This implies that MSX2 was significantly involved in the EMT process of breast cancer cells (90). Furthermore, in breast cancer cells, MSX2 was closely associated with the bone metastasis via improving the expression of bone sialoprotein (BSP) (91,92). Additionally, MSX2 promoted the apoptosis of breast cancer by upregulating the level of COP1-interactive protein1 (Cip1) and suppressing the level of the baculoviral IAP repeat containing5 (BIRC5). MSX2 also promoted apoptosis through the ERK pathway (14,93). Lastly, in breast cancer, MSX2 also acted the downstream of CHK1 to defend against Adriamycin (94). MSX2 might exhibit a central role in maintaining the characteristics of tumor stemness and drug resistance by inhibiting the expression of SRY-box transcription factor2 (SOX2) in breast cancer (95-97).
MSX family in pancreatic cancer
The role of MSX1 in pancreatic cancer has not yet been elucidated. However, MSX2 has been studied for its importance in pancreatic cancer, MSX2 promoted the invasion and metastasis of pancreatic cancer cells (40). The target gene of MSX2 may be Twist1, and further, it was speculated that MSX2 may promote EMT in pancreatic cancer through Twist1 (40). In another study, MSX2 was shown to inhibit the apoptosis of pancreatic cancer cells by down-regulating the expression of caspase 3 (12). Elsewhere, pancreatic cancer cells with highly expressed MSX2 were shown to be more likely to be resistant to gemcitabine and 5-fluorouracil (5-FU) via the downstream target gene adenosine triphosphate-binding cassette subfamily G member2 (ABCG2) which may be a molecular therapeutic target for overcoming the resistance of pancreatic cancer to chemotherapeutic drugs (16,98). The MSX2 expression level was used to predict the degree of malignancy and prognosis of pancreatic intraductal papillary mucinous tumor (IPMN), and thus providing a valuable clinical reference to predict the deterioration of IPMN (76). Moreover, MSX2 has been used to identify pancreatic cancer and pancreatitis (99). These results indicate that MSX2 has great potential in clinicopathological diagnosis and treatment.
MSX family in ovarian cancer
MSX1 has an inhibitory effect on ovarian cancer (11,67). For example, in high-grade serous epithelial ovarian cancer, MSX1 impeded the growth of ovarian cancer by promoting apoptosis of ovarian cancer cells. In addition, MSX1 was found to increase the sensitivity of cancer cells to cisplatin (67). A study on human ovarian cancer cell line, OVCAR3, found that the overexpression of MSX1 could significantly prolong the G1 phase of the cell cycle, while cyclin D1, D3, E, cyclin-dependent kinase 4, c-jun, and Rb were significantly inhibited, thus preventing cell proliferation (11). MSX1 has also been reported to inhibit tumor angiogenesis by repressing the VEGF promoter activity (8). On the contrary, MSX2 also exhibits a carcinogenic role in ovarian cancer. Specifically, MSX2 promoted the proliferation and metastasis of ovarian cancer cells from the basement membrane to the matrix as the downstream of WNT pathway, MSX2 also promoted neoplastic transformation of epithelial cells (49).
MSX family in colorectal cancer
Both MSX1 (18) and MSX2 (19) are involved in the progression of colorectal cancer. For instance, Horazna et al. found MSX1 was closely related to the occurrence of colorectal cancer (18). Another study revealed that the deficiency of MSX2 could result in the inhibition of cell proliferation and invasion, promotion of cell cycle arrest and apoptosis in colorectal cancer. MSX2 may regulate the malignant development of colorectal cancer through the AKT pathway (19,100,101). The exact mechanism through which MSX2 promotes colorectal cancer progression has not been established.
MSX family in gastric cancer
Yamashita et al. documented that MSX1 may be an anticancer gene in gastric cancer (37). The MSX1 has also been associated with the occurrence and development of gastric cancer. Therefore, MSX1 may be a suitable candidate biomarker for the diagnosis and prognosis of gastric cancer (33). Moreover, based on experimental studies, MSX2 promoted the growth of gastric cancer cells and could play an important role in promoting cancer. After interference with MSX2, diffuse gastric cancer cells showed a strong inhibition on cell growth. Therefore, an extended genome-wide search of the transcriptional downstream target genes of MSX2 may be of great significance for identifying the molecular targets of diffused gastric cancer (52).
MSX family in other cancers
In cervical cancer, MSX1 was found to act as a cancer suppressor by down-regulating Notch expression and up-regulating the expression of apoptosis-associated genes (68). Findings revealed that MSX1 could prevent p53 from exiting the nucleus via using its homeodomain to bind to p53 and this interaction induced the apoptosis of Hera cells (102). In NK cell leukemia, MSX1 inhibited oncogene miR-17-92a-1 cluster host gene (MIR17HG) which suppressed cell apoptosis (34,103). In endometrial cancer, MSX1 was a target gene of β-catenin and MSX1 induced G0/G1 arrest (74). In neuroblastoma, MSX1 exhibited an anticancer role by activating the Notch pathway that in turn induced growth inhibition and inhibiting NEUROD1 which is associated with neuronal differentiation in the neuroblastoma cell line (15,104,105). In another study, MSX1 was shown to be a negative regulator of cell migration and invasion by inhibiting the WNT/β-catenin pathway in glioblastoma (70). Besides, MSX1 plays a cancer-promoting role in some tumors. MSX1 was involved in MLS proliferation and stemness by regulating MyoD and Myf5 (17,106,107). In melanoma cells, MSX1 promoted tumor cell metastasis by upregulating zinc finger E-box binding homeobox1 (ZEB1) expression (72). Additionally, in T-ALL cells, as a carcinogenic factor, MSX1 promoted the proliferation, differentiation and metastasis of leukemia cells and activated the oncogenic NKL homeobox gene NK3 homeobox1 (NKX3-1) which may be involved in cell proliferation (48,108). In odontogenic tumors MSX2 may be involved in ameloblast differentiation (46). Additionally, MSX2 elevated tumor cell metastasis in inverted papilloma of the nose (78). MSX2 was closely correlated with the metastasis of prostate cancer (79). Through vitro experiments, differences in the expression of p21, BCL2, Survivin, and N-cadherin indicated that a higher expression of MSX2 induced the apoptosis of melanoma cells and reduces the invasiveness of melanoma cells (77).
Discussion
As mentioned above, both MSX1 and MSX2 are valuable biomarkers for the diagnosis and prediction of tumorigenesis in some tumors (33,76). In addition, based on clinical prognosis studies, we noted that in breast cancer (14) and melanoma (77), MSX2 in the cancer cell cytoplasm was more associated with clinical prognosis than MSX2 in the nucleus. However, the mechanisms involved require further evaluation. Specifically, the MSX family seems to be closely related to tumor resistance, MSX1 was shown to increase the sensitivity of ovarian cancer cells to cisplatin, which is beneficial to patients undergoing chemotherapy (67). Previous studies on pancreatic cancer revealed that the downstream gene, ABCG2, regulated by MSX2 is associated with chemotherapeutic resistance (16). In addition, understanding the epigenetic regulation of MSX family gene expression may elucidate on the mechanisms of tumorigenesis and, thus, implying that epigenetic modification therapies such as histone acetyltransferase inhibitors may be used as adjuncts (39). It’s imperative to develop new drugs for the treatment of tumors. Interestingly, MSX1 has shown great potential as a therapeutic target for breast cancer by inhibiting the expression of the breast cancer oncogene KIFs (83). Immunohistochemistry analysis revealed that MSX2 was more preferentially expressed in diffuse gastric cancer than in intestinal gastric cancer cells, while its effect on the growth of gastric cancer cells was independent. Lastly, blocking of MSX2 function could be an effective Hh-targeted cancer therapy, thus presenting a new strategy for the treatment of gastric cancer (52). Protein-protein interactions (35) could affect transcriptional activity of MSX transcription factors in cancers, we may find more partners to intervene in their transcriptional activity in practical applications. It has been reported the homeobox genes can control the level of glucose metabolism and respond to metabolic stress in tumor cells (109), studying the roles of MSX transcription factors in cell metabolism can also become our research direction, so it is worth further study. Homeodomain proteins can regulate the expression of tumor-derived factors to create a microenvironment that promotes the growth of cancer cells and may also enable cancer cells to escape immune destruction (110). This suggests that we can study whether the MSX family is involved in immunotherapy of tumors in the future.
Conclusions
Although the importance of the MSX family in tumor progression has been recognized by the scientific community in recent years, the study of its members is still in its “infancy stages” (11,40). Therefore, there is an urgent need to fully explore the structures and potential mechanisms of the partners they choose to interact with in tumors. In summary, generating novel information on the MSX family will elucidate on its transcriptional regulatory mechanisms and also provide new therapeutic strategies for tumors. In this article, we particularly focused on the impacts of the two members of human MSX family on various cancer types. Because of various downstream regulatory genes, the two members of the MSX family can promote or inhibit cancer progression in different tumors (11,14,18,19). The MSX family has great potential in the clinical diagnosis and treatment of cancer.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82072725 and No. 81872042 to XC, No. 81702442 to ZL, No. 81972332 to YC, No. 82002583 to MH, No. 82002591 to GF), the Natural Science Foundation of Jiangsu province (No. BK20170623 to ZL, No. BK20200273 to GF), the China Postdoctoral Science Foundation (No. 2020M670090ZX to ZL), the Postdoctoral Science Found of Jiangsu province (No. 2018K090B to ZL).
Ethical Statement: The authors are accountable for all aspects of the work and have ensured that questions related to the accuracy or integrity of all aspects of the work have been appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-21-220)
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-220). The authors have no conflicts of interest to declare.
References
- 1.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet 1986;20:147-73. 10.1146/annurev.ge.20.120186.001051 [DOI] [PubMed] [Google Scholar]
- 2.Haria D, Naora H. Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer. Cancer Hallm 2013;1:67-76. 10.1166/ch.2013.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Catron KM, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci U S A 1996;93:1764-9. 10.1073/pnas.93.5.1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newberry EP, Latifi T, Battaile JT, et al. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry 1997;36:10451-62. 10.1021/bi971008x [DOI] [PubMed] [Google Scholar]
- 5.Kimura M, Machida J, Yamaguchi S, et al. Novel nonsense mutation in MSX1 in familial nonsyndromic oligodontia: subcellular localization and role of homeodomain/MH4. Eur J Oral Sci 2014;122:15-20. 10.1111/eos.12105 [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Hu G, Wang H, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol 1997;17:2920-32. 10.1128/MCB.17.5.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL, Wang GL, Kundu R. DNA binding and transcriptional properties of wild-type and mutant forms of the homeodomain protein Msx2. Biochem Biophys Res Commun 1995;209:257-62. 10.1006/bbrc.1995.1497 [DOI] [PubMed] [Google Scholar]
- 8.Son MJ, Rho SB, Kim K, et al. Homeoprotein Msx1-PIASy Interaction Inhibits Angiogenesis. Cells 2020;9:1854. 10.3390/cells9081854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JY, Wang CL, Wang XM, et al. Comprehensive analysis of microRNA/mRNA signature in colon adenocarcinoma. Eur Rev Med Pharmacol Sci 2017;21:2114-29. [PubMed] [Google Scholar]
- 10.Cheng Y, Yan Z, Liu Y, et al. Analysis of DNA methylation patterns associated with the gastric cancer genome. Oncol Lett 2014;7:1021-6. 10.3892/ol.2014.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Park K, Kim S, et al. Msx1 gene overexpression induces G1 phase cell arrest in human ovarian cancer cell line OVCAR3. Biochem Biophys Res Commun 2001;281:1234-40. 10.1006/bbrc.2001.4474 [DOI] [PubMed] [Google Scholar]
- 12.Satoh K, Hamada S, Kimura K, et al. Up-regulation of MSX2 enhances the malignant phenotype and is associated with twist 1 expression in human pancreatic cancer cells. Am J Pathol 2008;172:926-39. 10.2353/ajpath.2008.070346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes GL, Javed A, Waller SM, et al. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res 2003;63:2631-7. [PubMed] [Google Scholar]
- 14.Lanigan F, Gremel G, Hughes R, et al. Homeobox transcription factor muscle segment homeobox 2 (Msx2) correlates with good prognosis in breast cancer patients and induces apoptosis in vitro. Breast Cancer Res 2010;12:R59. 10.1186/bcr2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Revet I, Huizenga G, Chan A, et al. The MSX1 homeobox transcription factor is a downstream target of PHOX2B and activates the Delta-Notch pathway in neuroblastoma. Exp Cell Res 2008;314:707-19. 10.1016/j.yexcr.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 16.Hamada S, Satoh K, Hirota M, et al. The homeobox gene MSX2 determines chemosensitivity of pancreatic cancer cells via the regulation of transporter gene ABCG2. J Cell Physiol 2012;227:729-38. 10.1002/jcp.22781 [DOI] [PubMed] [Google Scholar]
- 17.De Cecco L, Negri T, Brich S, et al. Identification of a gene expression driven progression pathway in myxoid liposarcoma. Oncotarget 2014;5:5965-77. 10.18632/oncotarget.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horazna M, Janeckova L, Svec J, et al. Msx1 loss suppresses formation of the ectopic crypts developed in the Apc-deficient small intestinal epithelium. Sci Rep 2019;9:1629. 10.1038/s41598-018-38310-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, An H, Yuan W, et al. Prognostic Relevance and Function of MSX2 in Colorectal Cancer. J Diabetes Res 2017;2017:3827037. 10.1155/2017/3827037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendall AJ, Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene 2000;247:17-31. 10.1016/S0378-1119(00)00081-0 [DOI] [PubMed] [Google Scholar]
- 21.Hewitt JE, Clark LN, Ivens A, et al. Structure and sequence of the human homeobox gene HOX7. Genomics 1991;11:670-8. 10.1016/0888-7543(91)90074-O [DOI] [PubMed] [Google Scholar]
- 22.Jabs EW, Müller U, Li X, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell 1993;75:443-50. 10.1016/0092-8674(93)90379-5 [DOI] [PubMed] [Google Scholar]
- 23.Bell JR, Noveen A, Liu YH, et al. Genomic structure, chromosomal location, and evolution of the mouse Hox 8 gene. Genomics 1993;16:123-31. 10.1006/geno.1993.1149 [DOI] [PubMed] [Google Scholar]
- 24.Hovde S, Abate-Shen C, Geiger JH. Crystal structure of the Msx-1 homeodomain/DNA complex. Biochemistry 2001;40:12013-21. 10.1021/bi0108148 [DOI] [PubMed] [Google Scholar]
- 25.Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res 2003;13:429-42. 10.1038/sj.cr.7290185 [DOI] [PubMed] [Google Scholar]
- 26.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296-303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bienert S, Waterhouse A, de Beer TA, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 2017;45:D313-9. 10.1093/nar/gkw1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 2009;30 Suppl 1:S162-73. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- 29.Studer G, Rempfer C, Waterhouse AM, et al. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020;36:2647. 10.1093/bioinformatics/btaa058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoni M, Kiefer F, Biasini M, et al. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep 2017;7:10480. 10.1038/s41598-017-09654-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sliwinski T, Synowiec E, Czarny P, et al. The c.469+46_56del mutation in the homeobox MSX1 gene--a novel risk factor in breast cancer? Cancer Epidemiol 2010;34:652-5. 10.1016/j.canep.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 32.van Nistelrooij AMJ, van Marion R, van Ijcken WFJ, et al. Germline variant in MSX1 identified in a Dutch family with clustering of Barrett's esophagus and esophageal adenocarcinoma. Fam Cancer 2018;17:435-40. 10.1007/s10689-017-0054-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T, Xu Y, Hou P. Identifying novel biomarkers of gastric cancer through integration analysis of single nucleotide polymorphisms and gene expression profile. Int J Biol Markers 2015;30:e321-6. 10.5301/jbm.5000145 [DOI] [PubMed] [Google Scholar]
- 34.Nagel S, Pommerenke C, Meyer C, et al. NKL homeobox gene MSX1 acts like a tumor suppressor in NK-cell leukemia. Oncotarget 2017;8:66815-32. 10.18632/oncotarget.18609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagel S, Schneider B, Meyer C, et al. Transcriptional deregulation of homeobox gene ZHX2 in Hodgkin lymphoma. Leuk Res 2012;36:646-55. 10.1016/j.leukres.2011.10.019 [DOI] [PubMed] [Google Scholar]
- 36.Rauch TA, Wang Z, Wu X, et al. DNA methylation biomarkers for lung cancer. Tumour Biol 2012;33:287-96. 10.1007/s13277-011-0282-2 [DOI] [PubMed] [Google Scholar]
- 37.Yamashita S, Tsujino Y, Moriguchi K, et al. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2'-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci 2006;97:64-71. 10.1111/j.1349-7006.2006.00136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Cui Y, Huang T, et al. Identification and Validation of MSX1 as a Key Candidate for Progestin Resistance in Endometrial Cancer. Onco Targets Ther 2020;13:11669-88. 10.2147/OTT.S271494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagel S, Ehrentraut S, Meyer C, et al. Oncogenic deregulation of NKL homeobox gene MSX1 in mantle cell lymphoma. Leuk Lymphoma 2014;55:1893-903. 10.3109/10428194.2013.864762 [DOI] [PubMed] [Google Scholar]
- 40.Hamada S, Satoh K, Hirota M, et al. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J Cell Physiol 2007;213:768-74. 10.1002/jcp.21148 [DOI] [PubMed] [Google Scholar]
- 41.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science 2011;331:550-3. 10.1126/science.1191138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu H, Zheng L, Pu J, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet 2015;24:2539-51. 10.1093/hmg/ddv018 [DOI] [PubMed] [Google Scholar]
- 43.Suzuki A, Li A, Gajera M, et al. MicroRNA-374a, -4680, and -133b suppress cell proliferation through the regulation of genes associated with human cleft palate in cultured human palate cells. BMC Med Genomics 2019;12:93. 10.1186/s12920-019-0546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Chen B, Li Y. microRNA-203 promotes proliferation, differentiation, and migration of osteoblasts by upregulation of Msh homeobox 2. J Cell Physiol 2019;234:17639-48. 10.1002/jcp.28387 [DOI] [PubMed] [Google Scholar]
- 45.Nagel S, Meyer C, Kaufmann M, et al. Deregulated FOX genes in Hodgkin lymphoma. Genes Chromosomes Cancer 2014;53:917-33. 10.1002/gcc.22204 [DOI] [PubMed] [Google Scholar]
- 46.Sonoda A, Iwamoto T, Nakamura T, et al. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem 2009;284:27176-84. 10.1074/jbc.M109.033464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knutson TP, Daniel AR, Fan D, et al. Phosphorylated and sumoylation-deficient progesterone receptors drive proliferative gene signatures during breast cancer progression. Breast Cancer Res 2012;14:R95. 10.1186/bcr3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagel S, Ehrentraut S, Meyer C, et al. Repressed BMP signaling reactivates NKL homeobox gene MSX1 in a T-ALL subset. Leuk Lymphoma 2015;56:480-91. 10.3109/10428194.2014.924119 [DOI] [PubMed] [Google Scholar]
- 49.Zhai Y, Iura A, Yeasmin S, et al. MSX2 is an oncogenic downstream target of activated WNT signaling in ovarian endometrioid adenocarcinoma. Oncogene 2011;30:4152-62. 10.1038/onc.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz DR, Wu R, Kardia SL, et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res 2003;63:2913-22. [PubMed] [Google Scholar]
- 51.Willert J, Epping M, Pollack JR, et al. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol 2002;2:8. 10.1186/1471-213X-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohta H, Aoyagi K, Fukaya M, et al. Cross talk between hedgehog and epithelial-mesenchymal transition pathways in gastric pit cells and in diffuse-type gastric cancers. Br J Cancer 2009;100:389-98. 10.1038/sj.bjc.6604846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi C, Akiyama N, Matsuzaki T, et al. Characterization of a human MSX-2 cDNA and its fragment isolated as a transformation suppressor gene against v-Ki-ras oncogene. Oncogene 1996;12:2137-46. [PubMed] [Google Scholar]
- 54.Song YJ, Lee H. PIAS1 negatively regulates ubiquitination of Msx1 homeoprotein independent of its SUMO ligase activity. Mol Cells 2011;32:221-6. 10.1007/s10059-011-1020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin Y, Xie CM, Li H, et al. The FBXW2-MSX2-SOX2 axis regulates stem cell property and drug resistance of cancer cells. Proc Natl Acad Sci U S A 2019;116:20528-38. 10.1073/pnas.1905973116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maury JJ, Chan KK, Zheng L, et al. Excess of O-linked N-acetylglucosamine modifies human pluripotent stem cell differentiation. Stem Cell Res 2013;11:926-37. 10.1016/j.scr.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 57.Shen R, Chen Y, Huang L, et al. Characterization of the human MSX-1 promoter and an enhancer responsible for retinoic acid induction. Cell Mol Biol Res 1994;40:297-312. [PubMed] [Google Scholar]
- 58.Blin-Wakkach C, Lezot F, Ghoul-Mazgar S, et al. Endogenous Msx1 antisense transcript: in vivo and in vitro evidences, structure, and potential involvement in skeleton development in mammals. Proc Natl Acad Sci U S A 2001;98:7336-41. 10.1073/pnas.131497098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park KS, Kim KK, Kim KE. Msx1 homeodomain transcription factor and TATA-binding protein interact to repress the expression of the glycoprotein hormone alpha subunit gene. Biochem Biophys Res Commun 2015;468:326-30. 10.1016/j.bbrc.2015.10.102 [DOI] [PubMed] [Google Scholar]
- 60.Ogawa T, Kapadia H, Wang B, et al. Studies on Pax9-Msx1 protein interactions. Arch Oral Biol 2005;50:141-5. 10.1016/j.archoralbio.2004.09.011 [DOI] [PubMed] [Google Scholar]
- 61.Song YJ, Lee H. YB1/p32, a nuclear Y-box binding protein 1, is a novel regulator of myoblast differentiation that interacts with Msx1 homeoprotein. Exp Cell Res 2010;316:517-29. 10.1016/j.yexcr.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 62.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 2004;304:1675-8. 10.1126/science.1098096 [DOI] [PubMed] [Google Scholar]
- 63.Wu L, Wu H, Ma L, et al. Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech Dev 1997;65:3-17. 10.1016/S0925-4773(97)00032-4 [DOI] [PubMed] [Google Scholar]
- 64.Newberry EP, Latifi T, Towler DA. Reciprocal regulation of osteocalcin transcription by the homeodomain proteins Msx2 and Dlx5. Biochemistry 1998;37:16360-8. 10.1021/bi981878u [DOI] [PubMed] [Google Scholar]
- 65.Newberry EP, Latifi T, Towler DA. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 1999;38:10678-90. 10.1021/bi990967j [DOI] [PubMed] [Google Scholar]
- 66.Zhao X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol Cell 2018;71:409-18. 10.1016/j.molcel.2018.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonito NA, Borley J, Wilhelm-Benartzi CS, et al. Epigenetic Regulation of the Homeobox Gene MSX1 Associates with Platinum-Resistant Disease in High-Grade Serous Epithelial Ovarian Cancer. Clin Cancer Res 2016;22:3097-104. 10.1158/1078-0432.CCR-15-1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yue Y, Zhou K, Li J, et al. MSX1 induces G0/G1 arrest and apoptosis by suppressing Notch signaling and is frequently methylated in cervical cancer. Onco Targets Ther 2018;11:4769-80. 10.2147/OTT.S165144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lind GE, Skotheim RI, Fraga MF, et al. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and SCGB3A1 (HIN-1). J Pathol 2006;210:441-9. 10.1002/path.2064 [DOI] [PubMed] [Google Scholar]
- 70.Tao H, Guo L, Chen L, et al. MSX1 inhibits cell migration and invasion through regulating the Wnt/beta-catenin pathway in glioblastoma. Tumour Biol 2016;37:1097-104. 10.1007/s13277-015-3892-2 [DOI] [PubMed] [Google Scholar]
- 71.Mizokami Y, Egashira N, Takekoshi S, et al. Expression of MSX1 in human normal pituitaries and pituitary adenomas. Endocr Pathol 2008;19:54-61. 10.1007/s12022-008-9021-7 [DOI] [PubMed] [Google Scholar]
- 72.Heppt MV, Wang JX, Hristova DM, et al. MSX1-Induced Neural Crest-Like Reprogramming Promotes Melanoma Progression. J Invest Dermatol 2018;138:141-9. 10.1016/j.jid.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chetcuti A, Aktas S, Mackie N, et al. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr Blood Cancer 2011;57:950-7. 10.1002/pbc.23003 [DOI] [PubMed] [Google Scholar]
- 74.Eppich S, Kuhn C, Schmoeckel E, et al. MSX1-A Potential Marker for Uterus-Preserving Therapy of Endometrial Carcinomas. Int J Mol Sci 2020;21:4529. 10.3390/ijms21124529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malewski T, Milewicz T, Krzysiek J, et al. Regulation of Msx2 gene expression by steroid hormones in human nonmalignant and malignant breast cancer explants cultured in vitro. Cancer Invest 2005;23:222-8. 10.1081/CNV-200055958 [DOI] [PubMed] [Google Scholar]
- 76.Satoh K, Hamada S, Kanno A, et al. Expression of MSX2 predicts malignancy of branch duct intraductal papillary mucinous neoplasm of the pancreas. J Gastroenterol 2010;45:763-70. 10.1007/s00535-010-0200-1 [DOI] [PubMed] [Google Scholar]
- 77.Gremel G, Ryan D, Rafferty M, et al. Functional and prognostic relevance of the homeobox protein MSX2 in malignant melanoma. Br J Cancer 2011;105:565-74. 10.1038/bjc.2011.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Depondt J, Shabana el H, Walker F, et al. Nasal inverted papilloma expresses the muscle segment homeobox gene Msx2: possible prognostic implications. Hum Pathol 2008;39:350-8. 10.1016/j.humpath.2007.06.017 [DOI] [PubMed] [Google Scholar]
- 79.Chua CW, Chiu YT, Yuen HF, et al. Differential expression of MSX2 in nodular hyperplasia, high-grade prostatic intraepithelial neoplasia and prostate adenocarcinoma. APMIS 2010;118:918-26. 10.1111/j.1600-0463.2010.02626.x [DOI] [PubMed] [Google Scholar]
- 80.Pacurari M, Qian Y, Porter DW, et al. Multi-walled carbon nanotube-induced gene expression in the mouse lung: association with lung pathology. Toxicol Appl Pharmacol 2011;255:18-31. 10.1016/j.taap.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki M, Tanaka M, Iwase T, et al. Over-expression of HOX-8, the human homologue of the mouse Hox-8 homeobox gene, in human tumors. Biochem Biophys Res Commun 1993;194:187-93. 10.1006/bbrc.1993.1802 [DOI] [PubMed] [Google Scholar]
- 82.Yue Y, Yuan Y, Li L, et al. Homeobox protein MSX1 inhibits the growth and metastasis of breast cancer cells and is frequently silenced by promoter methylation. Int J Mol Med 2018;41:2986-96. 10.3892/ijmm.2018.3468 [DOI] [PubMed] [Google Scholar]
- 83.Li TF, Zeng HJ, Shan Z, et al. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int 2020;20:123. 10.1186/s12935-020-01191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shim C, Zhang W, Rhee CH, et al. Profiling of differentially expressed genes in human primary cervical cancer by complementary DNA expression array. Clin Cancer Res 1998;4:3045-50. [PubMed] [Google Scholar]
- 85.Sun AJ, Gao HB, Liu G, et al. Identification of MSX1 and DCLK1 as mRNA Biomarkers for Colorectal Cancer Detection Through DNA Methylation Information. J Cell Physiol 2017;232:1879-84. 10.1002/jcp.25733 [DOI] [PubMed] [Google Scholar]
- 86.Duarte-Medrano G, Lopez-Mendez I, Ramirez-Luna MA, et al. Analysis of circulating blood and tissue biopsy PDX1 and MSX2 gene expression in patients with pancreatic cancer: A case-control experimental study. Medicine (Baltimore) 2019;98:e15954. 10.1097/MD.0000000000015954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu F, Ji F, Ji Y, et al. Dissecting the mechanism of colorectal tumorigenesis based on RNA-sequencing data. Exp Mol Pathol 2015;98:246-53. 10.1016/j.yexmp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 88.Lucanus AJ, Yip GW. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene 2018;37:833-8. 10.1038/onc.2017.406 [DOI] [PubMed] [Google Scholar]
- 89.Phippard DJ, Weber-Hall SJ, Sharpe PT, et al. Regulation of Msx-1, Msx-2, Bmp-2 and Bmp-4 during foetal and postnatal mammary gland development. Development 1996;122:2729-37. 10.1242/dev.122.9.2729 [DOI] [PubMed] [Google Scholar]
- 90.di Bari MG, Ginsburg E, Plant J, et al. Msx2 induces epithelial-mesenchymal transition in mouse mammary epithelial cells through upregulation of Cripto-1. J Cell Physiol 2009;219:659-66. 10.1002/jcp.21712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bellahcène A, Castronovo V. Expression of bone matrix proteins in human breast cancer: potential roles in microcalcification formation and in the genesis of bone metastases. Bull Cancer 1997;84:17-24. [PubMed] [Google Scholar]
- 92.Bellahcène A, Menard S, Bufalino R, et al. Expression of bone sialoprotein in primary human breast cancer is associated with poor survival. Int J Cancer 1996;69:350-3. [DOI] [PubMed] [Google Scholar]
- 93.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle 2009;8:1168-75. 10.4161/cc.8.8.8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu W, Huang M, Guo J, et al. The Role of CHK1 Varies with the Status of Oestrogen-receptor and Progesterone-receptor in the Targeted Therapy for Breast Cancer. Int J Biol Sci 2020;16:1388-402. 10.7150/ijbs.41627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu K, Lin B, Zhao M, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal 2013;25:1264-71. 10.1016/j.cellsig.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leis O, Eguiara A, Lopez-Arribillaga E, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012;31:1354-65. 10.1038/onc.2011.338 [DOI] [PubMed] [Google Scholar]
- 97.Piva M, Domenici G, Iriondo O, et al. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol Med 2014;6:66-79. 10.1002/emmm.201303411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoo BK, Chen D, Su ZZ, et al. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res 2010;70:3249-58. 10.1158/0008-5472.CAN-09-4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Satoh K, Hamada S, Kanno A, et al. Evaluation of MSX2 mRNA in brush cytology specimens distinguished pancreatic carcinoma from chronic pancreatitis. Cancer Sci 2011;102:157-61. 10.1111/j.1349-7006.2010.01759.x [DOI] [PubMed] [Google Scholar]
- 100.Yao YL, Shao J, Zhang C, et al. Proliferation of colorectal cancer is promoted by two signaling transduction expression patterns: ErbB2/ErbB3/AKT and MET/ErbB3/MAPK. PLoS One 2013;8:e78086. 10.1371/journal.pone.0078086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dufour G, Demers MJ, Gagné D, et al. Human intestinal epithelial cell survival and anoikis. Differentiation state-distinct regulation and roles of protein kinase B/Akt isoforms. J Biol Chem 2004;279:44113-22. 10.1074/jbc.M405323200 [DOI] [PubMed] [Google Scholar]
- 102.Park K, Kim K, Rho SB, et al. Homeobox Msx1 interacts with p53 tumor suppressor and inhibits tumor growth by inducing apoptosis. Cancer Res 2005;65:749-57. [PubMed] [Google Scholar]
- 103.Nagel S, Venturini L, Przybylski GK, et al. Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymphoma 2009;50:101-8. 10.1080/10428190802626632 [DOI] [PubMed] [Google Scholar]
- 104.Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol 2005;16:663-72. 10.1016/j.semcdb.2005.06.009 [DOI] [PubMed] [Google Scholar]
- 105.Taylor MK, Yeager K, Morrison SJ. Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development 2007;134:2435-47. 10.1242/dev.005520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang J, Kumar RM, Biggs VJ, et al. The Msx1 Homeoprotein Recruits Polycomb to the Nuclear Periphery during Development. Dev Cell 2011;21:575-88. 10.1016/j.devcel.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J, Abate-Shen C. Transcriptional repression by the Msx1 homeoprotein is associated with global redistribution of the H3K27me3 repressive mark to the nuclear periphery. Nucleus 2012;3:155-61. 10.4161/nucl.19477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kusy S, Gerby B, Goardon N, et al. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med 2010;207:2141-56. 10.1084/jem.20100745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Waeber G, Thompson N, Nicod P, et al. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol 1996;10:1327-34. [DOI] [PubMed] [Google Scholar]
- 110.Ko SY, Barengo N, Ladanyi A, et al. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest 2012;122:3603-17. 10.1172/JCI62229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as