Abstract
The most prevalent and common sexually transmitted infection is caused by human papillomavirus (HPV) among sexually active women. Numerous genotypes of HPV are available, among which the major oncoproteins E6 and E7 lead to the progression of cervical cancer. The E7 oncoprotein interacts with cytoplasmic tumor suppressor protein PTPN14, which is the key regulator of cellular growth control pathways effecting the reduction of steady-state level. Disrupting the interaction between the tumor suppressor and the oncoprotein is vital to cease the development of cancer. Hence, the mechanism of interaction between E7 and tumor suppressor is explored through protein–protein and protein–ligand binding along with the conformational stability studies. The obtained results state that the LXCXE domain of HPV E7 of high and low risks binds with the tumor suppressor protein. Also, the small molecules bind in the interface of E7–PTPN14 that disrupts the interaction between the tumor suppressor and oncoprotein. These results were further supported by the dynamics simulation stating the stability over the bounded complex and the energy maintained during postdocking as well as postdynamics calculations. These observations possess an avenue in the drug discovery that leads to further validation and also proposes a potent drug candidate to treat cervical cancer caused by HPV.
Introduction
Human papillomaviruses causing sexually transmitted infection in women belong to the family of DNA viruses that are of 200 types and reported highly for the squamous epithelium. These viruses are differentiated into high and low-risk types based on the scientific diagnosis, which develops cancers and warts.1−3 The infection progressed by the fact that the high- and low-risk genotypes of HPV infect not only the cervix but also head, neck, vagina, penis, anus, vulva, and oropharynx, resulting in cervical cancer, skin cancer, mucosa verrucous hyperplasia, and orophayngeal cancer.4,5 The high-risk human papillomaviruses include HPV types 31, 35, 18, 16, 33, 39, 51, 56, and 59 from the genera alpha categorized to cause cancer, whereas the types 11, 6, 42, 44, 61, 54, 72, and 70 belong to the low-risk HPV types causing warts and cysts.2,6
The genome of the human papillomavirus is ∼8 kilobase pairs in size, which encodes a total of eight proteins, among which two are present in the late region and six are present in the early region. These proteins are responsible for regulatory functions like genome replication and transcription, cell signaling, cell cycle, alteration in the structure of infected cell, and variation in the immune system. It is clearly observed that these proteins are expressed during the infectious cycle possibly with the reduced expression at late times.7,8 The early proteins E6 and E7 termed to be oncogenic depict a major role in cell line transformation, transmembrane signaling, and regulation of the stability in chromosomes. Further, these E6 and E7 oncoproteins are responsible for association with tumor suppressors p53 and pRB, respectively, suggesting that these proteins involve in the inactivation of suppression activity of tumor, resulting in cancer development.9
It is reported that HPV 11 involves in the maintenance of genome through episomes, whereas the HPV types 16 and 18 are highly essential for the productive stage of the viral life cycle.1,10,11 The E7 oncoproteins encoded by HPV 16 and 18 bind to retinoblastoma with higher affinity than the oncoproteins encoded by the low-risk type HPV 11.12 Similar to HPV 16, low-risk HPV 11 is also observed to show less severity than the high-risk one, resulting in a reduction of the viral episomes partially. The contribution of HPV 11 in the viral cycle is not clearly understood, and also, the configuration of the early transcripts in the low-risk viruses results in the synthesis of lower-risk viruses in higher levels than high-risk viruses, which is also responsible for the similar effects on Rb. The LXCXE motif of the oncoprotein E7 recognizes the binding pocket of tumor suppressor proteins to bind. This binding is represented in the structure determination by the research group of Lee et al., in 1998. The HPV E7 oncoprotein is composed of three different conserved regions, namely, CR1, CR2, and CR3 consisting of an N-terminal end with the LXCXE motif and a C-terminal region of the protein, respectively. The CR3 region of the protein is structured compared to the CR1 and CR2 regions, which is reported to associate with the various host proteins like cyclin E, A, M2 pyruvate kinase, AP-1, p48, IRF-1, TBP, IGFBP-3, histone H1 kinase, and cyclin-dependent inhibitors like p21 and p27 other than Rb.13,14 These proteins are involved in the cellular transcription involving E2F1, E2F2, HDAC1, and HDAC2 contributing toward the immortalization of the host cell through the independent pathways of pRb.15
Recently, the protein tyrosine phosphatase PTPN14 tumor suppressor is identified to be the potential cellular target of the E7 proteins from various genotypes of HPV. This protein is also denoted PTPD2, PTP36, or PTP-Pez, which controls the protein tyrosine phosphorylation. This is found to be the major posttranslational modification related with the regulation of numerous biological progressions.16,17 PTPN14 inhibits the oncogenic activity of the Yes-associated protein and members of the Hippo pathway.18,19 White et al. states that E7 oncoprotein of HPV 16 targets PTPN14 for the degradation mediated in proteasome with the help of ubiquitin ligase UBR4 and gets degraded at the positive cancer cell lines of HPV.34 Szalmás et al. have reported that the oncoprotein E7 interacts with PTPN14, which is independent of the pRb involving the residues of HPV E7 CR3 terminal region.20 In this study, we emphasize on the interaction of PTPN14 with the high and low-risk HPV type 16 and 11 E7 oncoproteins with the reference of available HPV 18 E7–PTPN14. The CR3 terminal region of HPV E7, which is ordered, has been focused for interacting and helps in analyzing the difference in the virulence of E7 inhibition in PTPN14 tumor suppressor through computational approaches. We have performed protein–protein interactions to obtain the complex of high and low-risk type HPV with PTPN14, which was further used to analyze the intensity of E7 dominance. Further, conformational analysis depicts the changes observed in PTPN14 in contact with types 16 and 11 corresponding to high and low-risk HPV types. The compounds that have been already stated to inhibit the E7 oncoproteins of HPV in earlier work are docked at the interface of the protein–protein complex to block the interaction between both the macromolecules. Our study can provide better insights into the dominance of E7 in the development of cervical cancer with respect to HPV 16 E7 and warts/cysts with respect to HPV 11 E7, which helps to draw a conclusion in drug discovery approaches.
Results
Secondary Structure and Three-Dimensional Structure Prediction of HPV 11 E7
Since the three-dimensional structure of the HPV type 11 E7 is unavailable, we aimed to develop a model based on the obtained template of the structure through the BLAST search representing the similarity and identities. The secondary structure of type 11 E7 was anticipated with the assistance of PSIPRED server.21 The secondary structure of HPV 11 E7 predicted through the server is depicted in Figure S1, in which the confidence level of the prediction is represented in various shades of blue. The higher the darkness of the shade, the higher the confidence whereas less darkness represents the low confidence of the amino acid residues. In Figure S1, yellow color represents the strand, pink color represents the helix conformation, and the blue thin line represents the coil. Apart from these analyses, literature reports state that the E7 protein possesses intrinsic disorder in the N-terminal.22 Also, the sequence difference between HPV types 16 and 18 is presented in Figure S2 denoting a difference between the sequences.
To develop a three-dimensional structure, comparative modeling helps in developing suitable models for an extensive variety of targets. UniProt Knowledgebase was used for the retrieval of HPV 11 E7 sequence and implemented for the BLAST search. The blast results revealed that the template structure was observed to be 2EWL_A with the identity score of 49.02% and query coverage of 52%. The structure obtained as the template is the crystal structure of E7 protein from HPV type 45. The Modeller 9.20 has developed 10 rough models based on the template structure, and also the zinc ion which is available in the crystal structure is also included in the modeled structure due to its essential role in the conformation of the protein. The lowest DOPE score was set to be the minimal criteria to help in obtaining the best model representing the stability in thermodynamic aspect. The model has further carried out refinement and validation. The refinement of the modeled structure is carried out with the help of molecular dynamics (MD) simulation, and beforehand, the side chain of the modeled protein was set by the rotamers. It is clearly evident from the obtained model that HPV 11 E7 consists of the helix and antiparallel sheet with the CXXC sequence in two positions for forming the zinc finger motif. The modeled structure is depicted in Figure S3a, whereas the superimposed structure of query and template is represented in Figure S3c representing the root-mean-square deviation (RMSD) of 0.009 nm.
Assessment and Validation of the Modeled Structure
The validation and assessment of the modeled structure are very essential, and PDBsum Generate satisfies the need that identifies the reliability of the modeled structure in terms of φ and ψ torsional angles through the Ramachandran plot. This parameter is represented in Figure S3b denoting that 98% of the residues in the modeled regions fall in the most favored region and 2% residues fall in the additionally allowed region and only two glycine residues fall in the generously allowed/disallowed regions. This analysis helps in confirming that the obtained model is reliable in terms of backbone conformation. Along with this, the energy profile and Z-score value of the model are obtained with the ProSA program23 for calculating the interaction energy per residue with the distance-based pair potential, which is represented in Figure S3d. The analysis shows Z-score corresponding to −2.70 stating that the negative energy reflects the improved reliability of the model.
Conformational Stability of the Modeled HPV 11 E7 Oncoprotein
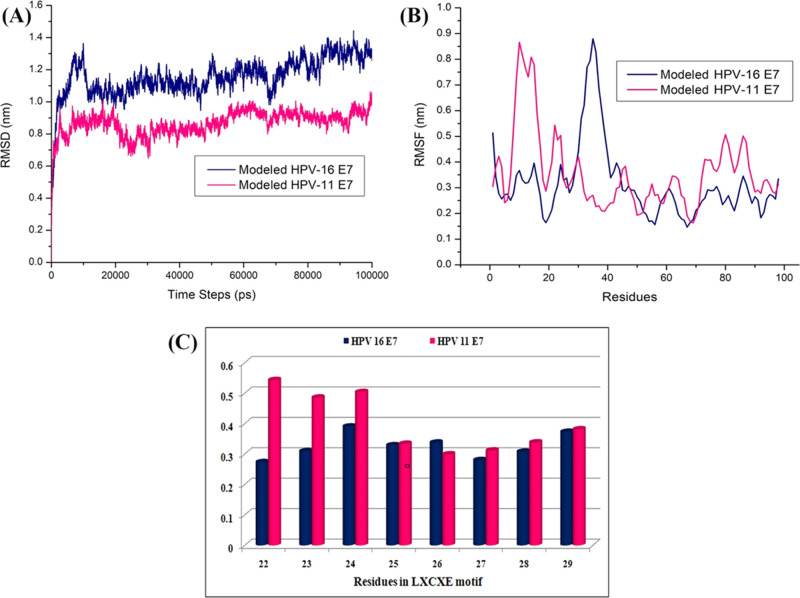
The modeled structure of HPV 11 E7 has been used further for stability analysis by subjecting it to the molecular dynamics simulation through GROMACS (Groningen Machine for chemical Simulations) for a period 100 ns in the explicit water model. This analysis has been compared with the stability check obtained for the modeled structure of HPV 16 E7 as mentioned in our earlier work.24 The stability of HPV type 11 E7 is found to be around 1.0–1.2 Å from its 20 ns period of simulation, whereas the deviations observed during the initial phase of time represent the time taken for equilibration. The root-mean-square deviations and fluctuations of both type 11 and 16 E7 are presented in Figure 1A,B, respectively. The blue and pink colors represented the modeled structure of HPV type 16 and 11 E7 oncoproteins, respectively. The molecular dynamics simulation of HPV 16 E7 states that initially, the protein gets deviated to get equilibrated till 0.9 nm around 0–20 ns, whereas after 20 ns, the equilibration of the protein reduced to 0.8 nm till 45 ns, and throughout the 100 ns simulation period, slight deviations are observed. But when the HPV 11 E7 is observed during the simulation period, it is clearly evident with the root-mean-square deviations that till 40 ns, the protein gets equilibrated for the initial stability around 1.0 nm and is maintained through the period of 100 ns simulation. When the simulation of HPV type 16 and 11 E7 are compared with each other in Figure 1A, it clearly shows that type 11 gets equilibrated easily and stabilized after 40 ns. Figure 1B represents the root-mean-square fluctuations (RMSF), in which blue color represents the type 16 E7 oncoprotein stating that the fluctuations are observed around the 0.2–0.5 nm throughout the simulation period. The fluctuations are observed during the 0.7 nm around the residue aspartic acid at position 41, which denotes the loop region. However, when the HPV type 11 is observed for fluctuations around the disordered region ranging from the residues 8 to 20 with 0.8 nm, the remaining region has represented fluctuations around 0.35–0.5 nm, stating that the fluctuations are not much compared with the 16 E7 oncoprotein throughout the simulation period of 100 ns. Figure 1C represents the residual fluctuation of the LXCXE domain of HPV types 16 and 11 with the blue and pink, respectively, and states that the fluctuation in Leu22, Tyr23, Cys24, and Tyr25 of type 16 is high, whereas that in the residue Tyr 26 and Glu27 of type 11 is high.
Figure 1.
(A) RMSD of the HPV modeled type 11 and 16 E7. (B) RMSF of the HPV modeled type 11 and 16 E7. (C) RMSF mean of the residues present in LXCXE motif.
LXCXE Domain of HPV E7 Binding to the PTP Domain of PTPN14
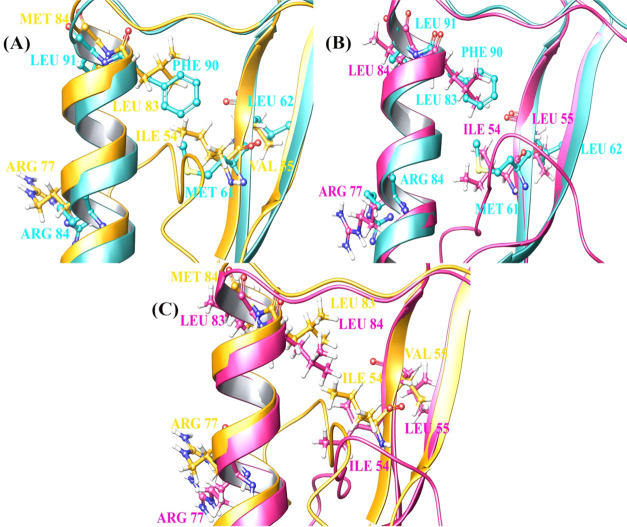
The LXCXE region of HPV E7 plays a major role in the binding toward tumor suppressor protein retinoblastoma. The modeled HPV 16 and 11 E7 were docked with PTPN14 and analyzed for strong interactions. The reported crystal structure of the PTPN14–HPV 18 E7 complex has been analyzed for the interactions and treated as a reference. The interaction reported reveals that residues of HPV E7 in positions 64, 91, 71, 62, 61, 87, 88, and 84 interact with the residues of PTPN14 in positions 1098, 1095, 1102, 1046, 1045, 1026, 1057, and 1031. Also, Yun et al. mentioned Phe90, Leu91, Met61, Leu62, and Arg84 as the hotspot residues of HPV E7 in interacting with PTPN14, which is present in the ordered region of the oncoprotein. From this, it is clearly evident that the LXCXE motif of HPV 18 E7 is not involved in the binding of tumor suppressor protein, whereas only the structured region stated to be ordered is involved. With this as a reference, and to identify the similar residues present in the ordered region of oncoprotein belonging to type 11 and 16 E7 proteins, alignment of E7 from three types has been performed. The alignment and superposition are represented in Figure 2. The active site obtained for the binding of the tumor suppressor with HPV E7 oncoprotein is the conserved region 2 encompassing the LXCXE motif, which is responsible for binding with the cellular targets. The conserved regions 2 and 3 of HPV are responsible for the degradation of tumor suppressor that ultimately leads to inhibit the cell cycle arrest. Figure 2A–C represents the alignment of HPV 18–16 E7 oncoprotein, HPV 18–11 E7 oncoprotein, and HPV 11–16 E7 oncoprotein, respectively. Cyan blue, yellow, and pink colors represent HPV 18 E7, HPV 16 E7, and HPV 11 E7 oncoproteins, respectively, and the residues which are represented as the hotspot of E7 for interaction are mentioned in Table 1.
Figure 2.
Superimposed structure of the hotspot residues in (A) HPV 18–16 E7, (B) HPV 18–11 E7, and (C) HPV 11–16 E7.
Table 1. Hotspot Residues Identified during the Alignment between the Types of E7.
| sl. no. | type of HPV | residues |
|---|---|---|
| 1 | HPV 18 E7 | Phe90, Leu91, Met61, Leu62, Arg84 |
| 2 | HPV 16 E7 | Leu83, Met84, Ile54, Val55, Arg77 |
| 3 | HPV 11 E7 | Leu83, Leu84, Ile54, Leu55, Arg77 |
Protein–Protein Interaction Study
The interaction between the tumor suppressor PTPN14 and HPV type 18 is determined by the research group of Yun and deposited in the Protein Data Bank with the ID: 6IWD. Yun et al. stated that E7 oncoprotein of HPV involves in the degradation of tumor suppressor proteins such as the retinoblastoma through the LXCXE motif present in the CR1 region of the C-terminal of E7.18 To define the binding mechanism of PTPN14 to the different types of HPV E7, multiple docking simulations of the high and low-risk types of HPV 11 and 16 were executed. Various conformations are obtained for the complex of PTPN14–E7 of both HPV type, and based on the pose energy, pose score, and cluster size, the best conformation is obtained for further analysis. The energy profile and the scores of the best conformation for the complexes are presented in Table 2. To validate the docking strategies performed, the complex HPV 18 E7–PTPN14 is used as a control by retrieving directly from the Protein Data Bank. Highly constituted results are obtained for the conformers docked with the help of Bioluminate, and these results are presented in Figures 3 and 4 for HPV type 16–PTPN14 complex and HPV type 11–PTPN14 complex, respectively. The E7 oncoprotein–PTPN14 complex involving types 11 and 16 was exhibited, whereas the PTPN14–HPV type 18 E7 was retrieved from the PDB. These results state that the PTP domain of PTPN14 was strongly bound with the residues HPV E7 with the help of hydrogen bonds and nonbonded contacts like van der Waals and electrostatic interactions. When the HPV 16 E7 is bound to PTPN14, numerous interactions are observed, but when the HPV 11 E7 is bound to PTPN14, less interactions are only observed, stating that the high risk possesses strong interactions in comparison to the low-risk HPV. To validate the residual interface and the stability of the complex, the obtained top hit complexes are subjected to molecular dynamics simulation.
Table 2. Energy Profile and Interaction Scores for the Protein–Protein Docking.
| sl. no. | complex description | cluster size | pose energy | pose score |
|---|---|---|---|---|
| 1 | HPV 16 E7–PTPN14 | 51 | –521.5870 | –276.321000 |
| 2 | HPV 11 E7–PTPN14 | 39 | –486.0251 | –202.843300 |
Figure 3.
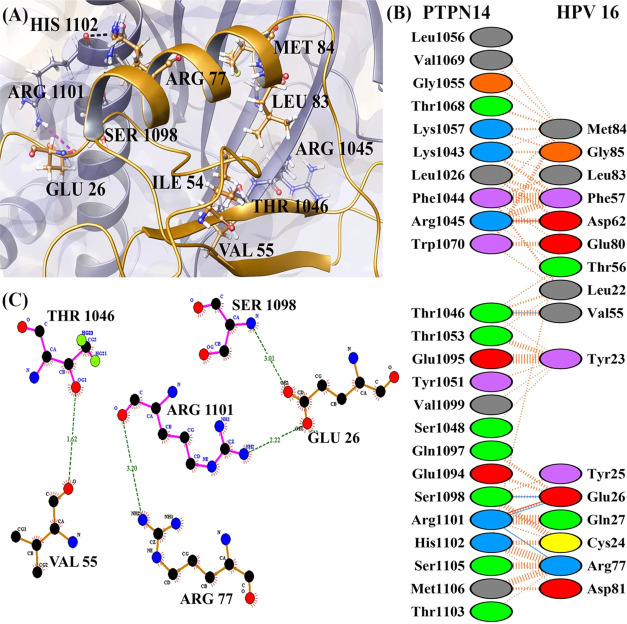
(A) Number of residues involved in interacting between PTPN14 and HPV 16 E7 oncoprotein. (B) Residue information about interaction between PTPN14 and HPV 16 E7 oncoprotein and (C) docked complex of PTPN14–HPV 16 E7.
Figure 4.
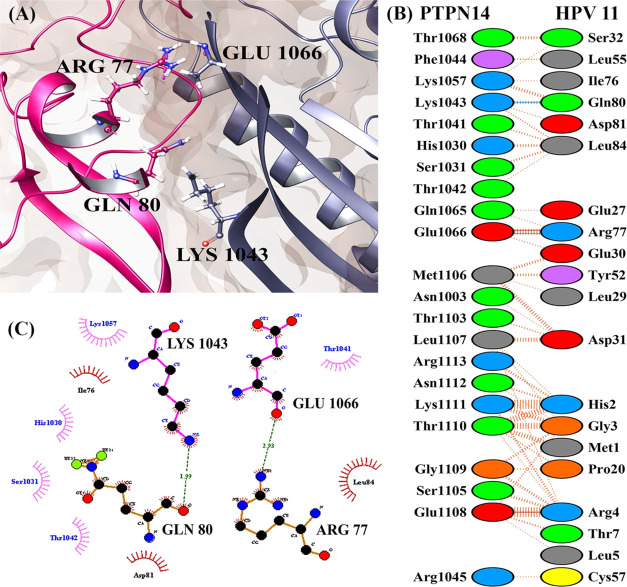
(A) Number of residues involved in interacting between PTPN14 and HPV 11 E7 oncoprotein. (B) Residue information about interaction between PTPN14 and HPV 11 E7 oncoprotein and (C) docked complex of PTPN14–HPV 11 E7.
Figures 3 and 4 present the interaction profiles of PTPN14–HPV 16 E7 and PTPN14–HPV 11 E7, respectively. The interactions in Figure 3 state that the residues Val55, Glu26, and Arg77 of the HPV 16 E7 oncoprotein interact with Thr1046, Ser1098, and Arg1101 of the PTPN14 tumor suppressor protein, respectively. When we analyze the PTPN14–HPV 18 E7 interaction, the residues among the structured region are involved and nonbonded contact is represented only in the residues in the N-terminal region. The lack of complete structure of HPV 18 E7, which contains the disordered region, is the differences from the interaction study by Yun et al.18 But when we analyze the interaction of PTPN14–HPV 16 E7, it clearly states that the N and C-terminal regions composed of CR1, CR2, and CR3 regions are actively involved. The residue Glu26 of the LXCXE domain is involved strongly in the hydrogen-bond formation along with the residues obtained from the literature report provided by Yun et al.18 Apart from these residues, the other four residues of the LXCXE domain like Leu22, Tyr23, Cys24, and Tyr25 also strongly interact with PTPN14 through nonbonded contacts, stating that these interactions play a major role in the suppression of the tumor suppressor activity. Likewise, the interaction of HPV 11 E7–PTPN14 depicted in Figure 4 represents that among the residues mentioned as hotspot in the literature, only Arg77 is involved in interacting with PTPN14. Only two hydrogen bonds were formed between the HPV 11 E7 and PTPN14 complex, denoting that there form lesser interactions. The LXCXE domain of the E7 oncoprotein type 11 E7 is not involved in the formation of hydrogen or nonbonded contacts representing the weakness of the interaction. This can be also the reason for the development of warts stating the mode of progression signifying the low-risk types. The interacting residues of all of the three oncoproteins and the PTPN14 are mentioned in Table 3.
Table 3. Interacting Residues of HPV E7 and PTPN14.
| sl. no. | complex | residues of PTPN14 involved in nonbonded contacts | residues of HPV E7 involved in nonbonded contacts | residues of PTPN14 forming hydrogen bonds | residues of HPV E7 forming hydrogen bonds |
|---|---|---|---|---|---|
| 1 | HPV 16 E7–PTPN14 | Leu1056, Val1069, Gly1055, Thr1068, Lys1057, Lys1043, Leu1026, Phe1044, Arg1045, Trp1070, Thr1053, Glu1095, Tyr1051, Val1099, Ser1048, Gln1097, Glu1094, His1102, Ser1105, Met1106, Thr1103 | Met84, Gly85, Leu83, Phe57, Asp62, Glu80, Thr56, Leu22, Tyr23, Tyr25, Gln27, Cys24, Asp81 | Thr1046, Ser1098, Arg1101, | Val55, Glu26, Arg77 |
| 2 | HPV 11 E7–PTPN14 | Thr1068, Phe1044, Lys1057, Thr1041, His1030, Ser1031, Thr1042, Gln1065, Met1106, Asn1003, Thr1103, Leu1107, Arg1113, Asn1112, Lys1111, Thr1110, Gly1109, Ser1105, Glu1108, Arg1045 | Ser32, Ile76, Leu55, Asp81, Leu84, Glu27, Glu30, Leu29, Tyr52, Asp31, Met1, His2, Gly3, Pro20, Arg4, Thr7, Leu5, Cys57 | Lys1043, Glu1066 | Arg77, Gln80 |
Molecular Dynamics Simulation of the PTPN14–HPV Complexes
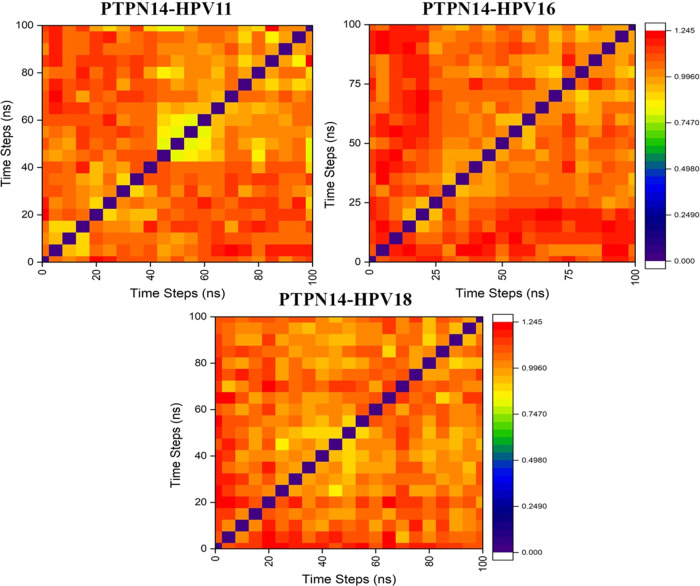
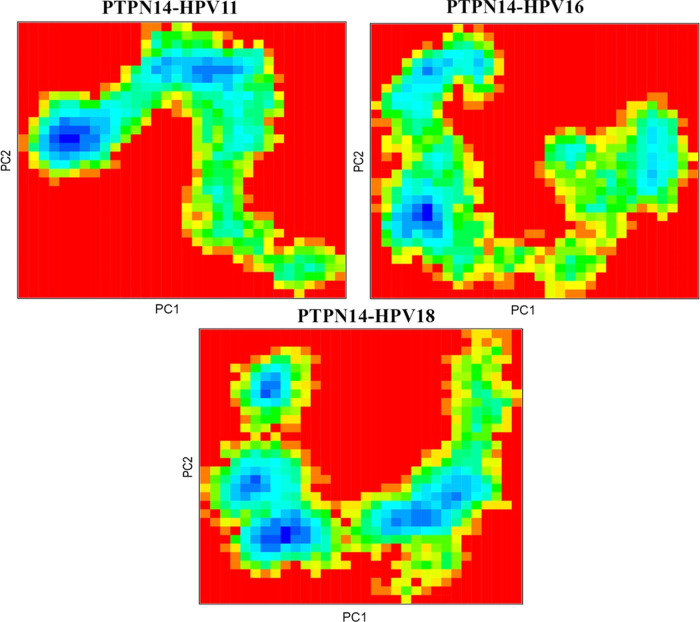
The protein–protein complexes obtained were employed for the molecular dynamics simulation, which reveals the behavior of the protein in dynamic environment over a period of 100 ns through GROMACS. The difference in the stability of the high-risk (HPV 16) and low-risk (HPV 11) genotypes with its binding toward PTPN14 is analyzed and observed that significant changes occurred. The changes in the structural features have been analyzed through the calculation of root-mean-square deviations and fluctuations over the backbone atoms. Deviations in the stability are observed in Figure S4a, stating that the deviations gradually increased after 20 ns from 0.2 to 0.6 Å and stabilized over the period of simulation till 0.3, 0.4, and 0.6 Å for HPV 18, HPV 16, and HPV 11, respectively. But around 38 ns, the deviations for the complexes with HPV 16 and 18 are observed but thereafter regained the stability till 80 ns. When the PTPN14–HPV 11 complex is observed, the deviations get fluctuated after 40 ns and stabilized over 80 ns. But when the simulation after 90 ns is observed for all of the three complexes, it is clearly evident that PTPN14–HPV 16 and PTPN14–HPV 18 have been involved in some deviations compared with PTPN14–HPV 11. It is also understood that the high-risk HPV 18 and HPV 16 when interacting with PTPN14 possess less RMSD of 0.2 and 0.4 Å, stating that it is strongly dominating the tumor suppressor protein. The RMSD of low-risk HPV is higher than that of high-risk HPV, with 0.7 Å stating less importance and deviation from binding. Also, we clearly examined the residual fluctuation over 100 ns period of simulation, which strongly represented the conformations of loop exhibits more fluctuation in all of the three HPV genotypes presented in Figure S4c, whereas the fluctuations of the PTPN14 when bound with all of the three genotypes are presented in Figure S4b. But when the tumor suppressor PTPN14 is observed, it is clearly evident that the residues between 1000 and 1050 possess strong fluctuations of about 0.55–0.7 Å. From the dynamics simulation, it is clearly understood that this analysis justifies that the interaction of high-risk HPV with PTPN14 is the major force driving the development of cervical cancer. The principle-component analysis presented in Figure S5 states that the subspace of the protein dynamics has been observed during the simulation period. The projections help in visualizing the stable states through the cluster. The clusters are clearly defined in all of the three complexes, stating that coverage of the motion of protein complex PTPN14–HPV 11 is wider compared with the other complexes. When the complexes are observed, it is evident that slight increase and decrease are achieved within the conformational space. Also, the conformational changes over the period of simulation are observed clearly at each 5 ns and obtained about 20 conformations and each superimposed with each other. These conformational changes at each 5 ns are represented through the heatmap analysis for the three complexes in Figure 5. The analysis of the conformational changes represents the correlation of root-mean-square deviations that can be visualized through the heatmap. The correlations are represented through different color codes ranging from 0.00 to 1.245 nm in Figure 5. These conformational analyses helped in gaining the average structure to be used for further docking strategies to identify the blockers to disrupt the interaction between oncoprotein and tumor suppressor protein. The Gibbs free energy landscape analysis has been calculated for the protein–protein complexes using the projections of the first and second eigenvectors, respectively, which is depicted in Figure 6. This analysis helps in the inspection of the direction of fluctuation observed in the systems for all Cα atoms of PTPN14–HPV 11 E7, PTPN14–HPV 16 E7, and PTPN14–HPV 18 complex structures from the MD trajectory. From the figure, it is clearly evident that the deeper blue color represents the lower energy, and it is observed that the main free energy is correlated well with the global energy. This denotes the stable conformation of these macromolecules.
Figure 5.
Heatmap analysis for the superpositioned poses obtained during each 5 ns interval for the 100 ns simulation of PTPN14–HPV 11 E7, PTPN14–HPV 16 E7, and PTPN14–HPV 18 E7.
Figure 6.
Free energy landscape analysis of the protein–protein docking complexes.
Ligand Binding Interface of High- and Low-Risk E7 with PTPN14
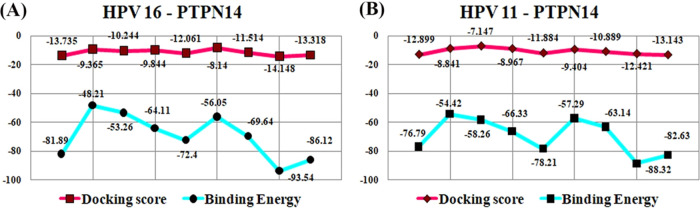
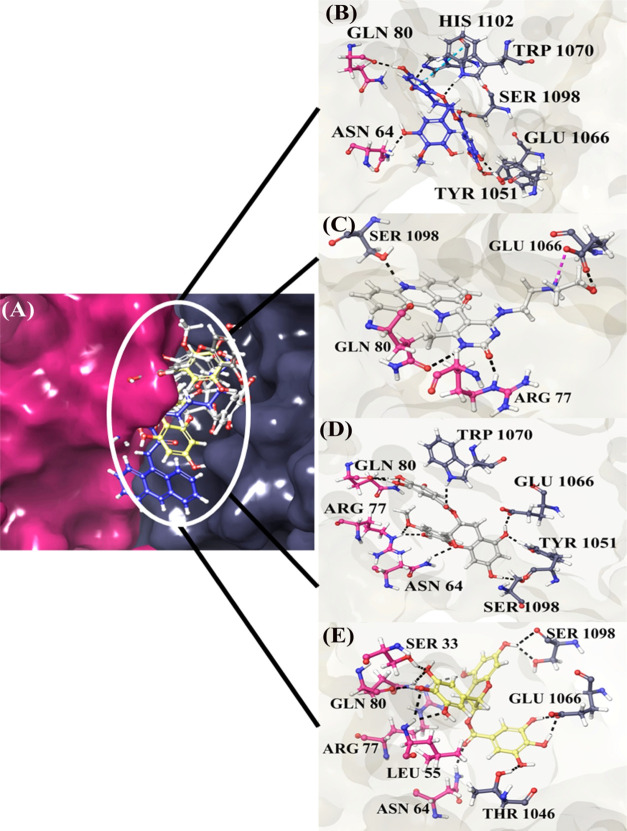
The protein–protein complex of PTPN14 with HPV 16 and HPV 11 E7 has been utilized for carrying out the docking studies to identify the inhibitors that can disturb the interaction of tumor suppressor and viral oncoprotein. In the protein–protein complex PTPN14–HPV 16 E7, interfacial residues are Thr1046, Ser1098, and Arg1101 in PTPN14 and Glu26, Arg77, and Val55 in E7 of HPV 16, whereas in the complex PTPN14–HPV 11 E7, the interfacial residues are Lys1043 and Glu1066 in PTPN14 and Gln80 and Arg77 of HPV 11. It is clearly observed that these residues possess strong interaction between each other and high binding affinity that helps in the identification of small-molecule inhibitors. The small molecules identified from our previous works listed in Table S1 are docked into the interface of the PTPN14–HPV E7 complex.25,26 The best conformation with the lowest docking score and high binding affinity was selected for each ligand. The obtained complex of PTPN14 with HPV E7 revealed that the interacting sites are composed of hydrophobic pockets as confirmed in our protein–protein docking results. The docking scores of both HPV 16 and 11 are presented in Table S2, whereas the post-docking binding free energy of the ligands of HPV 16 and HPV 11 with PTPN14 are provided in Tables S3 and S4, respectively. To gain better insights into the docking scores and binding energy, the scores were plotted in the graphical representation in Figure 7. The binding analysis states that all of the nine compounds involved in the blocking of interaction between the tumor suppressor and oncoprotein, but the top hit compounds are taken further for the conformation stability analysis. When the compounds ZINC49069570, 692320, ZINC49115270, and EGCG are analyzed for interaction in PTPN14–HPV 16 E7, there exhibits strong interaction with the residues Val55, Thr1046, Ser1098, and Glu1095, which is considered to be the hotspot residue responsible for the inhibition of tumor suppression. Also, when the PTPN14–HPV 11 E7 is observed, the residues Gln80, Arg77, Lys1043, and Glu1066 interact again with the compounds ZINC49069570, 692320, ZINC49115270, and EGCG, resulting in the blocking of the interaction between the tumor suppressor and the oncoprotein, which can result in the eradication of viral interaction. Further, Figures 8 and 9 present the top hit compound bounded at the interface of PTPN14–HPV 16 and PTPN14–HPV 11, respectively.
Figure 7.
Comparison of the binding energy and the docking scores for all of the nine compounds binding with PTPN14–HPV E7.
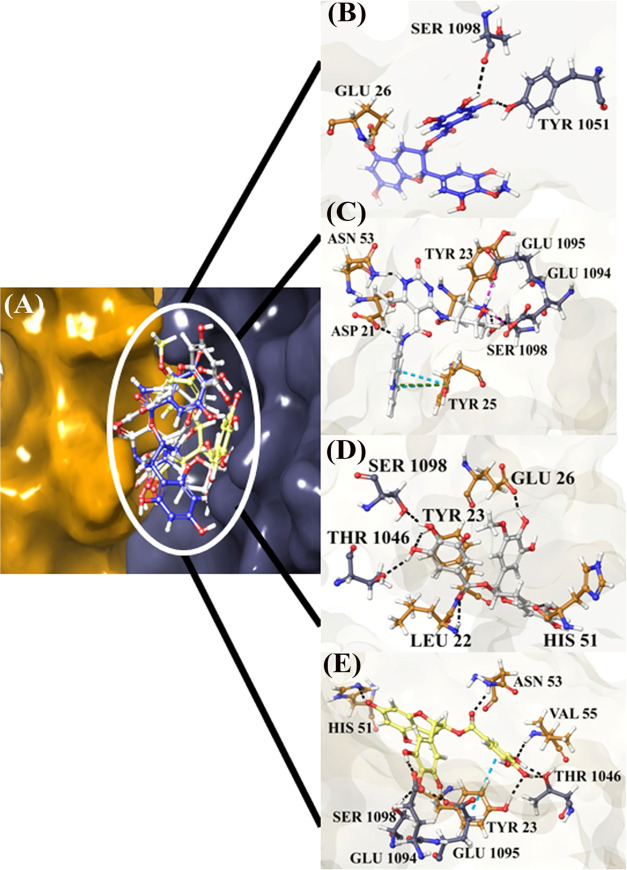
Figure 8.
PTPN14–HPV 16 E7 docked with the compounds for blocking the interaction between the tumor suppressor and oncoprotein. (A) Superimposed poses of top four compounds, (B) compound ZINC49069570, (C) compound 692320, (D) ZINC49115270, and (E) EGCG.
Figure 9.
PTPN14–HPV 11 E7 docked with the compounds for blocking the interaction between the tumor suppressor and oncoprotein. (A) Superimposed poses of top four compounds, (B) compound ZINC49069570, (C) compound 692320, (D) ZINC49115270, and (E) EGCG.
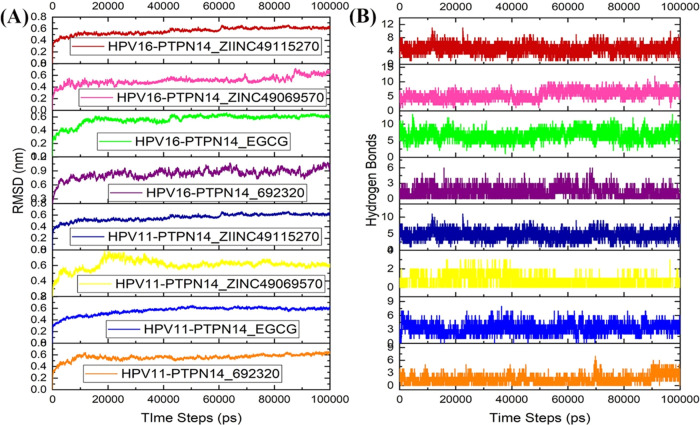
Conformational Stability Analysis on the Ligand Bound to the PTPN14–HPV Complex
The complex of PTPN14–HPV E7 of both the genotypes 16 and 11 docked with the small molecules EGCG, ZINC49069570, 692320, and ZINC49115270 has been subjected to conformational stability analysis for a period of 100 ns through dynamics simulation. It is clearly evident from Figure 10A that except the compounds 692320 and ZINC49069570 with HPV 16, all other complexes attained stability after 45 ns during the simulation period. The RMSD of the top hit compounds is observed to fall within the range of 0.4–0.6 nm except for 692320 HPV 16, which has the corresponding value of about 0.9 nm. It represents that the compound has been dominating the interaction of PTPN14 and HPV types 11 and 16 over the period of simulation. Figure 10B denotes that there is no much loss of interaction over the period of simulation, which denotes that the compounds block the interaction of the tumor suppressor and the oncoprotein that can help in the treatment of cervical cancer. The compounds ZINC49069570, 692320, ZINC49115270, and EGCG can be better inhibitors that can be carried forward for blocking the interaction with the human tumor suppressor and viral protein.
Figure 10.
(A) RMSD analysis of the best compounds with the PTPN14–HPV 11 and PTPN14–HPV 16 complexes. (B) Hydrogen-bond interaction of the best compounds with the PTPN14–HPV 11 and PTPN14–HPV 16 complexes.
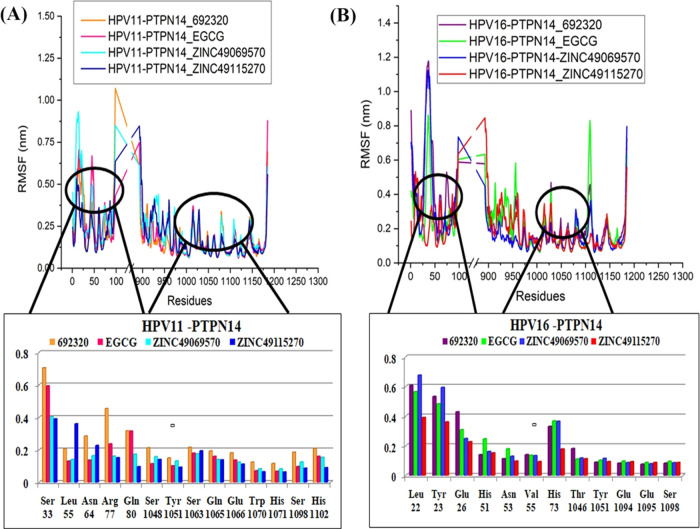
Observation of the RMSF analysis represented in Figure 11 states that there are some minimal fluctuations over the loop region, but these loop regions are not involved in the binding, and hence can be ignored. But the interacting residues of the tumor suppressor protein and virus with the compound are observed, stating that the fluctuation of the residues is obtained in both HPV 16 and HPV 11 during the course period of simulation. It is clearly evident that the residues of HPV possess higher fluctuation than the tumor suppressor, since these residues of HPV possess disordered region. The maximal fluctuation of the HPV 11–PTPN14 with the compound is around 1.0 nm, whereas the fluctuation of HPV 16–PTPN14 with the compound exhibits around 0.3 nm to the maximum of 1.2 nm. The interacting residues of both type E7 have been observed separately that helps in concluding with better insights.
Figure 11.
(A) RMSF analysis of the best compounds interacted with the PTPN14–HPV 11 complex and residual RMSF for the interacting residues. (B) RMSF analysis of the best compounds interacted with the PTPN14–HPV 16 complex and residual RMSF for the interacting residues.
Binding Free Energy Postdynamics Strategies
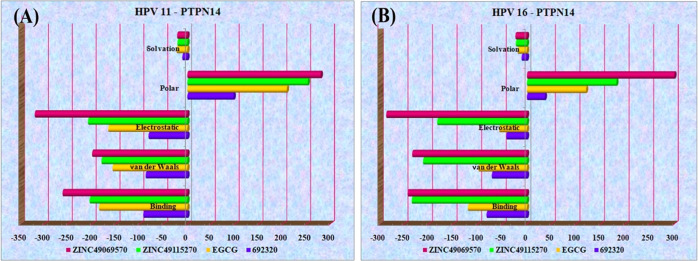
The postdynamics binding free energy helps in calculating the interaction energies that are involved in the analysis of the biomolecular complex, which estimates the free energy of binding and provides the breakdown of residual influence on binding. About 100 snapshots were selected over 100 ns simulation after every 1000 ps of constant intervals. The binding free energy over the molecular dynamics trajectories were obtained for the top four compounds that interact strongly. It is clearly apparent that the compounds ZINC49069570, 692320, ZINC49115270, and EGCG possess better binding energy with the HPV–PTPN14 complex. The regular components acquired over the binding free energy analysis of HPV type 16 and type 11 with the compound and PTPN14 are listed in Tables S5 and S6. The dominating strategy of the binding free energy post-dynamics involves van der Waals interactions, which represents the stronger interaction. The energies obtained were distributed in different representations over Figure 12A for HPV 11 and Figure 12B for HPV 16. The obtained results clearly state that the binding free energy obtained over the simulation studies is comparatively stronger, representing the strong binding of the ligand in the interface of HPV–PTPN14 complex, which in turn leads to the disruption of the interaction between oncoprotein and tumor suppressor.
Figure 12.
(A) Binding free energy over the simulation trajectory for HPV 11–PTPN14 with compounds. (B) Binding free energy over the simulation trajectory for HPV 16–PTPN14 with compounds.
Discussion
The most important cancer affecting women worldwide annually is cervical cancer, which is a widespread neoplastic disease that ranks second.27 This cancer is caused by unrelenting infection caused by the human papillomavirus in the cervix.28 Numerous genotypes like high-risk, low-risk, and intermediate-risk genotypes are available in human papillomavirus that has been classified based on the severity of malignancy and transformation.6,29 The infections are associated with a variety of clinical conditions ranging from innocuous lesions to cancer. The interconnection between the Human papillomavirus and cervical cancer was demonstrated first by the research group of German Virologist named zur Hausen around the 1980s.30 HPVs can contaminate the epithelial cells of the skin and the inner lining of the tissues that is categorized as the cutaneous or mucosal types. Further, the infectious cycle of the virus gets completed with the differentiation of the cells, which occurred through the assembly and release of the virus.25,31
Mostly, high-risk types of HPV cause cervical cancer, whereas the low-risk types cause genital warts or abrasions. Among the high-risk HPV types, the major types that are highly responsible for the development of cervical cancer are the HPV types 16 and 18. The low-risk type HPV 11 is responsible for the development of genital warts. These viruses encoded some proteins necessary for replication, cellular proliferation, virus assembly, and transformation.23,32 The viral oncoproteins E6 and E7 of high-risk HPV are the major carriers for the development of cervical cancer through its direct interference with the significant growth regulatory pathways. These oncoproteins are also represented to be the supreme target for impending therapeutic intrusion in tumor developed by human papillomaviruses.33 The E6 and E7 interact with p53 and pRb tumor suppressor protein, respectively, and inactivate them, which leads to the development of cervical cancer. Among both the oncoproteins, E7 oncoproteins interact with the varied number of cellular proteins whose involvement is in cell cycle control, regulation of cell proliferation, DNA damage, and gene expression.29 The E7 protein of HPV 16 interacts with ZER1, UBR4/p600, IF3, pRB, MDM2, and PTPN14. Vaccines Gardasil and Cervarix are available for uninfected adolescent people, but they are expensive. Also, the inadequate inhibitor to inhibit E7 oncoprotein is also the major striving force that provoked us to check the interactions between high and low-risk oncoproteins, which help in analyzing the transforming activity of the genotype.
Our study is completely focused on interpreting the interaction between E7 oncoproteins of HPV and tumor suppressor protein PTPN14 as well as identifying blockers to disrupt the interaction between the tumor suppressor and oncoproteins. E7 oncoprotein of HPV type 16 has been modeled through Modeller as mentioned in our previous work.24 Along with this, E7 of HPV type 11 is also modeled for the binding study of PTPN14 and E7 oncoprotein. The protein–protein interaction of PTPN14 and HPV 18 E7 determined by Yun et al. provided a strong base and reference for the study. It was mentioned that the residues Met61 and Met 62 in β1 followed by Arg84, Phe90, and Leu91 in α1 of HPV 18 E7 C-terminal domain bind hydrophobically with the Leu1026 residue in the loop of β3−β4, along with the residues Lys1043, Phe1044, Gly1055, Trp1070 from β5, β6, and β7 PTPN14.15 The HPV 16 and HPV 11 superimposed with the HPV 18 reveals that the hotspot residues in the C-terminal of HPV for all of the three oncoproteins are nonpolar amino residues that fall in the same position as HPV 18. Further, a protein–protein docking study of HPV types 11 and 16 with PTPN14 is carried out, which showed that hotspot residues observed from the research work of Yun are involved in the formation of hydrogen and hydrophobic contacts. When we observed the interaction between HPV 16 and PTPN14, it is clearly evident that Arg77 and Val55 as referred to the reported hotspot residues interact with Arg1101 and Thr1046 through the formation of hydrogen bonds. Along with these interactions, Glu26 interacts strongly with Ser1098 and Arg1101. At the same time, hydrophobic contacts are also observed between the important residues of PTPN14 and E7 oncoprotein of type 16, which can be a thriving force for the deprivation of the tumor suppressor function. When the interaction between HPV 11 and PTPN14 is analyzed, we found that only Arg77 and Gln80 of E7 interact with Glu1066 and Lys1043, stating that not much interactions are retrieved in this binding. This lack of interactions can also be the reason for the development of genital warts. Followed by the protein–protein interaction studies, the small molecule obtained from our previously published works has been used for blocking the interaction between the tumor suppressor and the viral oncoprotein. It is clearly evident from the results that the compounds interact with the residues that already establish binding with PTPN14, stating that the atoms of the compounds replace the binding between PTPN14 and HPV.
When the postdocking binding free energy is observed, we understood that the compounds ZINC49069570, 692320, ZINC49115270, and EGCG show better binding in comparison to the other five compounds. Further, molecular simulation studies have been carried out for the top four compounds exhibiting strong interaction over the disruption of interaction. These analyses reveal that the compounds ZINC49069570, 692320, ZINC49115270, and EGCG have strong affinity over blocking the interactions, whereas it is also depicted that no loss of hydrogen-bond interaction is observed over the course period of simulation. In the root-mean-square fluctuations of molecular dynamics, the fluctuation is not more than 0.7 nm but the fluctuations are observed mostly on the loop region for both the viral oncoprotein and the tumor suppressor. When the oncoprotein of type 16 is observed, it posses major interactions from the LXCXE domain with PTPN14 and the compounds interact with the residues present in LXCXE, stating that these compounds can act better for treating the infection caused by HPV type 16. When the oncoprotein of type 11 is observed, it clearly represents that the LXCXE is not involved in the interaction with the tumor suppressor as well as the compound, but the compounds interact with the interfacial residues of HPV 11 E7 and PTPN14, implicating that these compounds block the interactions. Also, the postdynamics binding free energy studies revealed that the compounds ZINC49069570, 692320, ZINC49115270, and EGCG can be potent due to their involvement over the dynamics trajectories in comparison to the compound 692320. Additionally, these compounds can be taken forward for further studies to bring potent small molecules for treating the infection caused by HPV and blocking the interaction between the tumor suppressor and oncoprotein of HPV.
Materials and Methods
Homology Modeling
The three-dimensional structure of the full-length E7 oncoprotein of type 11 human papillomavirus is not available in the Protein Data Bank (PDB), which provoked us to develop a 3D model.25,26 The protein sequences of type 11 E7 were retrieved from the database UniProtKB with ID P04020.34 The functions of the low-risk E6 and E7 oncoproteins in the maintenance of the genome of HPV 11 state that they may act in a way similar to the high-risk proteins. The protein sequences were blasted with the default parameters, which helps in finding the appropriate template for modeling. The crystal structure of the human papillomavirus type 45 E7 is obtained as the template protein with the best identity score of 49.02% and the lowest e-value of 1e–09 with the query coverage of 52% possessing PDB ID 2EWL.35 The academic version of modeler 9.20 was used for the generation of three-dimensional structure. Since the zinc ion is present in the crystal structure, it was added in the modeled structure without fail.27 A maximum of about 10 models are generated, and model with the lowest dope score was selected and subjected to quality check, Ramachandran plot, and ProSA profile for the validation of the obtained model.25 Further, the modeled structure is superimposed with the template structure without altering the coordinate systems of atomic position in the template.36,37
Preparation of Protein Structure
The three-dimensional crystal structure of the Protein Tyrosine Phospahatase Non-receptor 14 (PTPN14), a tumor suppressor protein, has been retrieved from the Protein Data Bank with the resolution of 1.80 Å possessing PDB ID 6IWD. This crystal structure represents the binding of the Conserved region 3 of the human papillomavirus type 18 in the PTP domain of the PTPN14.19 Also, the modeled structure of the HPV 16 E7 oncoprotein is used from the previously reported work.25,26 The obtained crystal structure along with the modeled E7 protein structure from type 16 and type 11 is not readily usable in the molecular modeling environment, and hence the protein is prepared with the protein preparation wizard present in Schrödinger, which is checked for the missing residues and loops.38 Preparation wizard focuses on two main components, namely, refinement and preparation. Once the chemical accuracy is ensured, hydrogen is added for neutralizing the side chain, which is either close to the binding site or the residues forming salt bridges.24,39 The force field OPLS-AA is implemented for use, which is represented to be more subtle for the refinement of steric clashes. The removal of waters, addition of hydrogen atoms, protonation, and tautomeric states of the histidine residues along with the flip assignment of the residues Asn, Gln, and His were carried out by Schrödinger. The nonhydrogen atoms were checked for the average RMSD to reach 0.3 Å with the help of minimization through the OPLS-AA force field.40,41
Protein–Protein Docking
The three-dimensional crystal structure of PTPN14 and modeled structures of HPV 11 and 16 E7 obtained from the average structure from molecular simulation have been carried out for the protein–protein docking studies with the help of Bioluminate module available in Schrödinger.34 The residues of PTPN14 vulnerable for the binding of HPV E7 are obtained from the literature and used for the binding of the oncoprotein of type 16. This docking strategy is an interface from the piper program. The PTPN14 was enforced for the preparation and the standard mode of docking has been implemented. Both the proteins are rotated into various number of orientations with respect to the PTPN14, which is considered to be the receptor and each orientation is translated for the retrieval of the best docking pose. The top rotations are clustered using the RMS distance between the matching atoms in each pair of the rotated structures. About 70 000 ligand orientations were generated, and this corresponds approximately to the sampling of every 5° in the space of Euler angles. The maximum number of 30 poses is obtained, and each pose behaves as the center of the cluster that results from the clustering of top results of the rigid docking of the ligand.42,43
Binding of Small Molecules to Block the Interaction of PTPN14 and E7
The possible binding patterns of potent inhibitors reported for the inhibition of E7 from the previously reported work have been carried out to examine the blocking mechanism of PTPN14–E7 complex. A total of nine molecules were used from the previously reported work of our group.24,39 New libraries have not been used in the study, since the binding mechanism of the identified compounds earlier is subjected to inhibit the E7 oncoprotein, but there emerges the next perspective to block the interaction between both the macromolecules (PTPN14–HPV E7). To use the compounds for both the purposes of inhibiting oncoprotein and blocking the interaction, the compounds in the earlier work have been utilized.24,39 This strategy helps to take these compounds for further study with experimental analysis. The active site of the protein–protein complex has been determined with the help of SiteMap module in Schrödinger, and it was observed that the residues identified during our previous works also fall within the active site for docking.44 The active-site-based grid generation was achieved for the docking of compounds with the extra precision docking protocol performed with the Grid-based Ligand Docking with energetics (Glide) module of Schrödinger.45,46 Different conformations were added and passed through the filters internally in which the ligand is centered initially and were allowed to rotate around three angles. Removal of different binding modes based on the docking scores of the entity and the geometrical filters is performed.39 Evaluation on the force field obtained with the grid and refinement of docking includes torsional and rigid body movements of the ligands through the OPLS-AA force field. To scale the ligands with the van der Waals force, 0.8 Å of the scaling factor and a 0.15 cutoff value of the partial charge are used, which are the default parameters. The conformational prediction and the orientations of the ligand within the active site of the receptor are executed by the process of docking. The results obtained through the induced fit docking strategies were used for further analysis.47
Conformational Stability Analysis
The conformational stability analyses of the protein–protein complex (PTPN14–E7 type 11 and PTPN14–E7 type 16) and the protein–ligand complexes were carried out for simulation with the help of GROMACS Version 2019, in which the force field GROMOS53a6 is incorporated. To carry out the protein–ligand simulation, PRODRG was executed for the development of ligand topology.48 These complexes were solvated with the simple point charge water model with the cubic box by applying proper periodic boundary condition. Appropriate numbers of sodium (Na+) and chlorine (Cl–) ions were added to neutralize the system before minimization of the complexes. The complexes obtained after neutralization were subjected to energy minimization with an indulgence of 1000 kJ/mol with the steepest descent integrator to eliminate unfavorable interactions. During the phases of equilibration, the conformational changes were restricted by applying the position restraints to all atoms.49 The energy-minimized systems were exposed to equilibration with NVT ensembles for a period of 100 ps, where the solvents and proteins with ions are treated for the temperature coupling with the help of V-rescale method. Further, the equilibrated structures are again treated with the constant pressure (NPT) at 1 bar atmospheric pressure.38 The Leapfrog algorithm is implemented for the integration of the Newton equation with the time step of 2 fs, whereas the coordinates were saved every 2 ps under constant pressure and temperature. Computation on the long-range electrostatic interactions with the particle mesh Ewald is used, whereas a cutoff distance of 1.2 nm is used for the short-range van der Waals and electrostatic interactions. The hydrogen atoms were retained in the system with the LINCS algorithm.50
Post-Docking and Postdynamics Binding Free Energy Calculations
The binding free energy of the complexes obtained after docking is calculated with the help of Prime MMGBSA approach, which helps in predicting the free energy of ligand binding to receptor. The Prime Molecular Mechanics Combined with the Generalized Born and Surface Area (MMGBSA) module available in Schrödinger is used for calculating the internal energy of the molecule and the implicit representation of the solvent continuum.39 Finally, the term of entropy is found negotiable in Prime, which uses the surface-generalized Born model engaging the Gaussian surface in place of the van der Waals surface for improved representation of the accessible surface area of the solvent.51
Further, the calculation on the binding free energy of the system obtained through the trajectories of the molecular dynamics simulation was subjected to MMPBSA calculation. This method is widely used for the binding free energy calculation from the snapshots of the molecular dynamics simulation trajectory. The molecular mechanics Poisson–Boltzmann surface area (MMPBSA) approach combines the energies based on the molecular mechanics with free energy based on the implicit solvent models. The binding free energy of the protein–ligand complex is estimated based on the difference between the free energy of the unbound component and the complex.
| 1 |
where ΔGbind is the total binding free energy, ΔEMM is the sum of gas-phase interaction energy between the receptor and ligand, ΔGsolv is the solvation energy involved in the transition of gas-phase energy to the solvation energy, and −TΔS is the change in conformational entropy associated with ligand binding
| 2 |
From eq 2, it is clearly evident that ΔEMM is the calculation of molecular mechanics on different components like ΔEcovalent (covalent energy), ΔEelec (electrostatic interaction), and ΔEvdW (van der Waals interaction energy)
| 3 |
Further, ΔGsolv is the sum of polar and nonpolar components, ΔGPB is the contribution of polar energy toward the solvation energy in the Poisson–Boltzmann implicit solvent model, and ΔGSA is the nonpolar energy toward the solvation energy in the solvent-accessible surface area.29,30 The residue that is involved significantly in the binding free energy provides an enhanced approach into the components participating in the interaction between protein and ligand.52
Principle-Component and Free Energy Landscape Analysis
Principle-component analysis is the commonly used computational technique that uses the trajectory of the molecular dynamics simulation and extorts the dominant modes present in the motion of the molecule.53−56 The dimensionality of the data will be reduced in the data obtained through the molecular dynamics simulation, which helps in recognizing the configurational space containing a few degrees of freedom where the harmonic motion occurs.57 The predicted motion of the protein correlates to the mode of vibration or the collective motions of group of atoms in the normal mode analysis.58 The significance of the principle-component analysis is that these data provide the appropriate way to analyze, visualize, and motion compare during the course period of simulation. The principle components of the protein were obtained through the diagnolization and solving of eigenvectors and eigenvalue for the covariance matrix. The eigenvectors and eigenvalues of the principle-component analysis represent the direction of protein motion and the magnitude of the motion, respectively. The matrix for illustrating the principle-component analysis was calculated using the simulation trajectories in the tool gmx covar that builds and diagnolizes the covariance matrix.59 Further, the gmx sham of the Gromacs modules is implemented to analyze the free energy landscape of the tumor suppressor and the oncoprotein complex.60
Acknowledgments
M.A. thanks University Grants Commission for providing RGNF fellowship (Award No. F1-17.1/2016-17/RGNF-2015-17-SC-TAM-18759). S.K.S. thanks Alagappa University, Karaikudi, Tamil Nadu, India, DBT, DST-PURSE, FIST, and Ministry of Human Resource Development RUSA—Phase 2.0 grant sanctioned vide Letter No. F.24-51/2014-U, Policy (TNMulti-Gen), Department of Education, Government of India, dated 09.10.2018, for providing the enhanced infrastructure facilities.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01619.
Secondary structure of HPV 11 E7 (Figure S1); multiple sequence alignment of the HPV types 16 and 18 representing the sequence difference in both the genotypes of HPV (Figure S2); structure of the modeled HPV 11 E7, superimposed structure of the template and the modeled structure, Ramachandran plot and ProSA score as an assessment for the modeled E7 oncoprotein (Figure S3); molecular dynamics simulation protein–protein complexes: RMSD, RMSF of PTPN14 residues, and RMSF of HPV E7 residues (Figure S4); principle-component analysis PTPN14 with HPV 11 and 18 E7, PTPN14 with HPV 16 and 11 E7, and PTPN14 with HPV 11 and 18 E7 (Figure S5); compounds obtained from the previously published works, with the references mentioned accordingly (Table S1); docking scores of the ligand with HPV types 11 and 16 bounded to PTPN14 (Table S2); scores of binding free energy post-docking (Tables S3 and S4); and postdynamics binding free energy scores of the complex between the ligand and PTPN14–HPV 16 and 11 type E7 (Tables S5 and S6) (PDF)
Author Contributions
M.A. and S.K.S. conceived the idea, designed the study, and wrote the manuscript. M.A. run the applications and obtained data. M.A. and S.K.S. contributed to the interpretation of the results and discussion
The authors declare no competing financial interest.
Notes
The commercial software package Schrödinger release 2018-4 was used for the protein–protein interaction studies and the binding of small molecules to block the protein–protein interaction. Also, the GROMACS version 2019.5, which is freely available, is installed in the Camarero DS400TR-54R system with Cent Operating system. This software is used for the molecular dynamics simulation studies. Further, the data used in the study are a continuation of further research work; hence, the data will be made available upon request.
Supplementary Material
References
- McLaughlin-Drubin M. E.; Munger K. The human papillomavirus E7 oncoprotein. Virology 2009, 384, 335–344. 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A.; Kundu R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020, 10, 3116 10.3389/fmicb.2019.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin M. E.; Huh K. W.; Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J. Virol. 2008, 82, 8695–8705. 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M.; Uxa S.; Stanko C.; Magin T. M.; Engeland K. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci. Rep. 2017, 7, 2603 10.1038/s41598-017-02831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R.; Song Y. J.; Sun S. Y.; Zhou Q.; Chen X. Z.; Zheng Q. L.; Cheng H. Influence of Human Papillomavirus E7 Oncoprotein on Maturation and Function of Plasmacytoid Dendritic Cells In Vitro. Virol. Sin. 2018, 33, 493–501. 10.1007/s12250-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd E. M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2002, 2, 342–350. 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- Doorbar J.; Foo C.; Coleman N.; Medcalf L.; Hartley O.; Prospero T.; Napthine S.; Sterling J.; Winter G.; Griffin H. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 1997, 238, 40–52. 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- Graham S. V. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010, 5, 1493–1506. 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim E. K.; Park J. S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res. Treat. 2005, 37, 319–324. 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.; Schmidt-Grimminger D. C.; Murant T.; Broker T. R.; Chow L. T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995, 9, 2335–2349. 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- Oh S. T.; Longworth M. S.; Laimins L. A. Roles of the E6 and E7 proteins in the life cycle of low-risk human papillomavirus type 11. J. Virol. 2004, 78, 2620–2626. 10.1128/JVI.78.5.2620-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. T.; Hubert W. G.; Ruesch M. N.; Laimins L. A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 8449–8454. 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. L.; Munger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin. Cancer Biol. 1996, 7, 327–337. 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- Lee J. O.; Russo A. A.; Pavletich N. P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 1998, 391, 859–865. 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Park J. S.; Kim E. J.; Kwon H. J.; Hwang E. S.; Namkoong S. E.; Um S. J. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 2000, 275, 6764–6769. 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- Barnard P.; McMillan N. A. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology 1999, 259, 305–313. 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- Yun H. Y.; Kim M. W.; Lee H. S.; Kim W.; Shin J. H.; Kim H.; Shin H. C.; Park H.; Oh B. H.; Kim W. K.; Bae K. H.; Lee S. C.; Lee E. W.; Ku B.; Kim S. J. Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus. PLoS Biol. 2019, 17, e3000367 10.1371/journal.pbio.3000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N. K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Szalmás A.; Tomaic V.; Basukala O.; Massimi P.; Mittal S.; Konya J.; Banks L. The PTPN14 Tumor Suppressor Is a Degradation Target of Human Papillomavirus E7. J. Virol. 2017, 91, e00057-17 10.1128/JVI.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Schüttelkopf A. W.; van Aalten D. M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta. Crystallogr. D: Biol. Crystallogr. 2004, 60, 1355–1363. 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Chaudhary N.; Aparoy P. Deciphering the mechanism behind the varied binding activities of COXIBs through Molecular Dynamic Simulations, MM-PBSA binding energy calculations and per-residue energy decomposition studies. J. Biomol. Struct. Dyn. 2017, 35, 868–882. 10.1080/07391102.2016.1165736. [DOI] [PubMed] [Google Scholar]
- Aarthy M.; Kumar D.; Giri R.; Singh S. K. E7 oncoprotein of human papillomavirus: Structural dynamics and inhibitor screening study. Gene 2018, 658, 159–177. 10.1016/j.gene.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Wadham C.; Gamble J. R.; Vadas M. A.; Khew-Goodall Y. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol. Biol. Cell 2003, 14, 2520–2529. 10.1091/mbc.e02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatterschide J.; Bohidar A. E.; Grace M.; Nulton T. J.; Kim H. W.; Windle B.; Morgan I. M.; Munger K.; White E. A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 7033–7042. 10.1073/pnas.1819534116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco E. L.; Schlecht N. F.; Saslow D. The epidemiology of cervical cancer. Cancer J. 2003, 9, 348–359. 10.1097/00130404-200309000-00004. [DOI] [PubMed] [Google Scholar]
- Shah M.; Anwar M. A.; Park S.; Jafri S. S.; Choi S. In silico mechanistic analysis of IRF3 inactivation and high-risk HPV E6 species-dependent drug response. Sci. Rep. 2015, 5, 13446 10.1038/srep13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G.; Granchi C.; Rizzolio F.; Tuccinardi T. Application of MM-PBSA Methods in Virtual Screening. Molecules 2020, 25, 1971 10.3390/molecules25081971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayanan V.; Singh S. K. Assessment of dual inhibition property of newly discovered inhibitors against PCAF and GCN5 through in silico screening, molecular dynamics simulation and DFT approach. J. Recept. Signal Transduction 2015, 35, 370–380. 10.3109/10799893.2014.956756. [DOI] [PubMed] [Google Scholar]
- Ahmad S.; Raza S.; Uddin R.; Azam S. S. Binding mode analysis, dynamic simulation and binding free energy calculations of the MurF ligase from Acinetobacter baumannii. J. Mol. Graphics Model. 2017, 77, 72–85. 10.1016/j.jmgm.2017.07.024. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Roman A.; Oldfield C. J.; Dunker A. K. Protein intrinsic disorder and human papillomaviruses: increased amount of disorder in E6 and E7 oncoproteins from high risk HPVs. J. Proteome Res. 2006, 5, 1829–1842. 10.1021/pr0602388. [DOI] [PubMed] [Google Scholar]
- Vijayalakshmi P.; Selvaraj C.; Singh S. K.; Nisha J.; Saipriya K.; Daisy P. Exploration of the binding of DNA binding ligands to Staphylococcal DNA through QM/MM docking and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2013, 31, 561–571. 10.1080/07391102.2012.706080. [DOI] [PubMed] [Google Scholar]
- White E. A.; Munger K.; Howley P. M.. High-Risk Human Papillomavirus E7 Proteins Target PTPN14 for Degradation mBio 2016, 7, e01530-16, 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D. W.; Minneci F.; Nugent T. C.; Bryson K.; Jones D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbyn M.; Weiderpass E.; Bruni L.; de Sanjose S.; Saraiya M.; Ferlay J.; Bray F. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Global Health 2020, 8, e191–e203. 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zurHausen H.; de Villiers E. M.; Gissmann L. Papillomavirus infections and human genital cancer. Gynecol. Oncol. 1981, 12, S124–S128. 10.1016/0090-8258(81)90067-6. [DOI] [PubMed] [Google Scholar]
- Daisy P.; Vijayalakshmi P.; Selvaraj C.; Singh S. K.; Saipriya K. Targeting multidrug resistant Mycobacterium tuberculosis HtrA2 with identical chemical entities of fluoroquinolones. Indian J. Pharm. Sci. 2012, 74, 217. 10.4103/0250-474X.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarthy M.; Panwar U.; Singh S. K. Structural dynamic studies on identification of EGCG analogues for the inhibition of Human Papillomavirus E7. Sci. Rep. 2020, 10, 8661 10.1038/s41598-020-65446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Mi W.; Xue Y.; Shi X.; Kutateladze T. G. The ZZ domain as a new epigenetic reader and a degradation signal sensor. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 1–10. 10.1080/10409238.2018.1564730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schaffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N.; Webb B.; Marti-Renom M. A.; Madhusudhan M. S.; Eramian D.; Shen M. Y.; Pieper U.; Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2006, 15 (1), 5–6. 10.1002/0471140864.ps0209s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A.; Katiyar S. P.; Singh S. K.; Dubey V. K.; Sundar D. A leishmaniasis study: structure-based screening and molecular dynamics mechanistic analysis for discovering potent inhibitors of spermidine synthase. Biochim. Biophys. Acta 2012, 1824, 1476–1483. 10.1016/j.bbapap.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Reddy K. K.; Singh S. K.; Tripathi S. K.; Selvaraj C.; Suryanarayanan V. Shape and pharmacophore-based virtual screening to identify potential cytochrome P450 sterol 14alpha-demethylase inhibitors. J. Recept. Signal Transduction 2013, 33, 234–243. 10.3109/10799893.2013.789912. [DOI] [PubMed] [Google Scholar]
- Pradiba D.; Aarthy M.; Shunmugapriya V.; Singh S. K.; Vasanthi M. Structural insights into the binding mode of flavonols with the active site of matrix metalloproteinase-9 through molecular docking and molecular dynamic simulations studies. J. Biomol. Struct. Dyn. 2018, 36, 3718–3739. 10.1080/07391102.2017.1397058. [DOI] [PubMed] [Google Scholar]
- Bandaru S.; Alvala M.; Nayarisseri A.; Sharda S.; Goud H.; Mundluru H. P.; Singh S. K. Molecular dynamic simulations reveal suboptimal binding of salbutamol in T164I variant of β2 adrenergic receptor. PLoS One 2017, 12, e0186666 10.1371/journal.pone.0186666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patidar K.; Deshmukh A.; Bandaru S.; Lakkaraju C.; Girdhar A.; Banerjee T.; Nayarisseri A.; Singh S. K.; et al. Virtual screening approaches in identification of bioactive compounds Akin to delphinidin as potential HER2 inhibitors for the treatment of breast cancer. Asian Pac. J. Cancer Prev. 2016, 17, 2291–2295. 10.7314/APJCP.2016.17.4.2291. [DOI] [PubMed] [Google Scholar]
- Maestro Version 2018; Schrödinger LLC: New York, NY, 2018.
- Beard H.; Cholleti A.; Pearlman D.; Sherman W.; Loving K. A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One 2013, 8, e82849 10.1371/journal.pone.0082849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Vijaya Prabhu S.; Suryanarayanan V.; Bhardwaj R.; Singh S. K.; Dubey V. K. Molecular docking and structure-based virtual screening studies of potential drug target, CAAX prenyl proteases, of Leishmania donovani. J. Biomol. Struct. Dyn. 2016, 34, 2367–2386. 10.1080/07391102.2015.1116411. [DOI] [PubMed] [Google Scholar]
- Tripathi S. K.; Selvaraj C.; Singh S. K.; Reddy K. K. Molecular docking, QPLD, and ADME prediction studies on HIV-1 integrase leads. Med. Chem. Res. 2012, 21, 4239–4251. 10.1007/s00044-011-9940-6. [DOI] [Google Scholar]
- Sharma N.; Murali A.; Singh S. K.; Giri R. Epigallocatechingallate, an active green tea compound inhibits the Zika virus entry into host cells via binding the envelope protein. Int. J. Biol. Macromol. 2017, 104, 1046–1054. 10.1016/j.ijbiomac.2017.06.105. [DOI] [PubMed] [Google Scholar]
- Stanley M. Pathology and epidemiology of HPV infection in females. Gynecol. Oncol. 2010, 117, S5–S10. 10.1016/j.ygyno.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Baseman J. G.; Koutsky L. A. The epidemiology of human papillomavirus infections. J. Clin. Virol. 2005, 32, 16–24. 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Boon S. S.; Banks L. High-risk human papillomavirus E6 oncoproteins interact with 14-3-3zeta in a PDZ binding motif-dependent manner. J. Virol. 2013, 87, 1586–1595. 10.1128/JVI.02074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger K.; Basile J. R.; Duensing S.; Eichten A.; Gonzalez S. L.; Grace M.; Zacny V. L. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 2001, 20, 7888–7898. 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- Haider S.; Parkinson G. N.; Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008, 95, 296–311. 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. C.; Jacobs D. J.. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. In Protein Dynamics; Humana Press: Totowa, NJ, 2014; pp 193–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosbie E. J.; Einstein M. H.; Franceschi S.; Kitchener H. C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. 10.1016/S0140-6736(13)60022-7. [DOI] [PubMed] [Google Scholar]
- Ali S.; Khan F. I.; Mohammad T.; Lan D.; Hassan M.; Wang Y. Identification and evaluation of inhibitors of lipase from Malassezia restricta using virtual high-throughput screening and molecular dynamics studies. Int. J. Mol. Sci. 2019, 20, 884 10.3390/ijms20040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.