Neural stem cells (NSCs) are mainly quiescent in the adult brain. Quiescent NSCs reenter the cell cycle and generate new neurons after they are activated. Recent studies suggest significant involvement of cellular proteostasis in quiescence of adult NSCs. We discuss regulation of quiescence by multiple signaling pathways and focus on the functional involvement of the lysosome, an organelle governing cellular degradation.
Keywords: adult neural stem cell, lysosome, proteostasis, quiescence, signaling
Abstract
Quiescence is a cellular strategy for maintaining somatic stem cells in a specific niche in a low metabolic state without senescence for a long period of time. During development, neural stem cells (NSCs) actively proliferate and self‐renew, and their progeny differentiate into both neurons and glial cells to form mature brain tissues. On the other hand, most NSCs in the adult brain are quiescent and arrested in G0/G1 phase of the cell cycle. Quiescence is essential in order to avoid the precocious exhaustion of NSCs, ensuring a sustainable source of available stem cells in the brain throughout the lifespan. After receiving activation signals, quiescent NSCs reenter the cell cycle and generate new neurons. This switching between quiescence and proliferation is tightly regulated by diverse signaling pathways. Recent studies suggest significant involvement of cellular proteostasis (homeostasis of the proteome) in the quiescent state of NSCs. Proteostasis is the result of integrated regulation of protein synthesis, folding, and degradation. In this review, we discuss regulation of quiescence by multiple signaling pathways, especially bone morphogenetic protein and Notch signaling, and focus on the functional involvement of the lysosome, an organelle governing cellular degradation, in quiescence of adult NSCs.
Abbreviations
- Ambra
activating molecule in Beclin‐1‐regulated autophagy
- Ascl
achaete‐scute homolog
- Atg
autophagy‐related gene
- BMP
bone morphogenetic protein
- BMPR
BMP receptor
- CSF
cerebrospinal fluid
- DG
dentate gyrus
- Dll1
delta‐like 1
- Dpp
decapentaplegic
- EGF
epidermal growth factor
- FGF
fibroblast growth factor
- FIP 200
FAK family kinase‐interacting protein 200
- FOXO
forkhead box O
- GABA
gamma amino butyric acid
- GFAP
glial fibrillary acidic protein
- GLAST
glutamate aspartate transporter
- Hes
hairy and enhancer of spirit
- HSC
hematopoietic stem cell
- Id
inhibitor of DNA binding
- IFN
interferon
- MiT‐TFE
microphthalmia‐transcription factor E
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- Ngn
neurogenin
- NICD
Notch intracellular domain
- NSC
neural stem cell
- OP‐1
osteogenic protein 1
- PTEN
phosphatase and tensin homolog
- RBPJ
recombination signal‐binding protein for immunoglobulin kappa J region
- ROS
reactive oxygen species
- SVZ
subventricular zone
- TFEB
transcriptional factor EB
- V‐ATPase
vacuolar ATPase
Introduction
Neural stem cells (NSCs) actively proliferate and give rise to all of the neurons and glial cells necessary to constitute the embryonic brain. Although NSCs decrease in number after production of mature brain tissues is complete, they persist and maintain multipotency in small areas of the adult brain. In the rodent brain, adult NSCs reside in the subventricular zone (SVZ) of the lateral ventricle and the hippocampal dentate gyrus (DG) (Fig. 1A,B) [1, 2, 3]. Adult NSCs are mainly quiescent; however, some adult NSCs, called active NSCs, proliferate and differentiate into mature neurons, which then integrate into the pre‐existing brain network [4, 5, 6, 7]. Quiescent NSCs become active NSCs after receiving activation signals, and the transition from quiescent to active is reversible rather than unidirectional [8, 9]. Quiescence of adult NSCs is maintained by extrinsic and intrinsic factors, and diverse signaling from local NSC niches is involved in this process (reviewed in Ref. [6]) (Fig. 1C,D). For example, bone morphogenetic protein (BMP) and Notch signaling, which are activated by ligands secreted or presented by neighbor cells, regulate adult NSC quiescence in both the SVZ and the DG. Here, we discuss how NSCs are regulated by these signaling pathways to maintain quiescence. The protein functions and stabilities of these signaling molecules influence the downstream outputs and their diverse responses. Recently, proteostasis (protein homeostasis [10]) was reported as a significant regulator of the maintenance of adult NSCs. Proteostasis is a consequence of integrated regulation of protein synthesis, proper folding, and protein degradation. We also discuss proteostasis in the context of quiescence, focusing especially on the function of lysosomes, an organelle involved in degradation of cellular components.
Fig. 1.
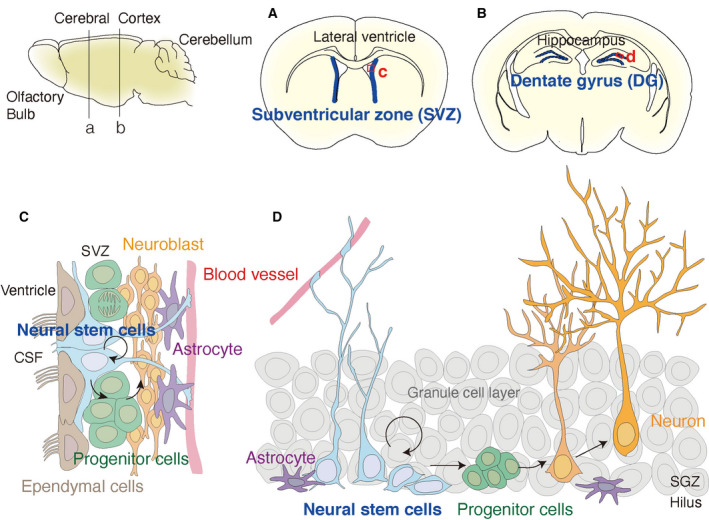
Two NSC niches, the SVZ and DG, in the adult mouse brain. Schematic representation of a lateral view of the whole adult mouse brain from the olfactory bulb (left) to cerebellum (right). Coronal planes dissected at lines (a) and (b) are shown in panels (A) and (B), respectively, on the right. The two NSC niches are labeled in blue lines, highlighting the SVZ near the lateral ventricle (A) and the DG in the hippocampus (B). Ventricles in the brain sections appear in white in panels (A) and (B). The detailed compositions of niches are represented in panels (C) and (D) as enlarged views marked by red squares (c in A) and (d in B), respectively. Panel (C) displays the SVZ niche, which is located near the lateral sides of the lateral ventricles. NSCs (blue) face the CSF in the lateral ventricle together with ependymal cells and elongate their projection to blood vessels (red). Panel (D) displays the DG niche, surrounded by the granule cell layers (gray) and hilus. NSCs are located in the subgranular zone next to the granule cell layer and elongate radial fibers, resulting in a radial glial morphology. NSCs in both niches self‐renew, differentiate into progenitor cells (green), and give rise to mature neurons (orange).
Signals that control quiescent NSCs
Bone morphogenetic protein signaling in adult NSCs has a long history of research. Previous studies revealed that BMP is a dominant inducer of quiescence in NSCs in vitro and in vivo [5, 8, 11]. This canonical BMP signaling upregulates the SMAD target factors Id1‐Id4 (Fig. 2A) as well as Hes1 and decreases cell proliferation [11, 12]. The SVZ exhibits enriched expression of BMP components including the BMP receptors BMPR Ia and BMPR II, BMP ligands, and the BMP inhibitor Noggin [13, 14]. Noggin is secreted from the ependymal cells, ciliated glial cells in the surface of lateral ventricle, in the adult brain [13]. In the DG, NSCs express the BMPR BMPR1a and BMPR2. These BMPR are activated by BMP2/4, members of the decapentaplegic (Dpp) subfamily of BMP ligands, which are secreted from dentate granule cells in the hippocampus [12]. Genetic deletion of Bmpr1a and Smad4, an effector of the canonical BMP pathway, using lentivirus‐mediated KO in Sox2+ cells in the DG immediately, induces NSC proliferation, followed by marked reduction in active NSCs and doublecortin (DCX)+ immature neurons in the DG within 3 weeks [12]. Blockade of BMP by Noggin recruits quiescent NSCs into the cell cycle in the DG [12, 15]. Thus, BMP signaling plays an essential role in the maintenance of quiescent NSCs. However, differential regulation of BMP ligand and receptor subtypes may cause adult NSCs to be differentially responsive to the BMP signal. For example, the cerebrospinal fluid (CSF) delivers many extrinsic factors to NSCs in the SVZ [13, 16]. One of these factors, BMP5, a member of the osteogenic protein 1 (OP‐1) subfamily of BMPs that is enriched in the CSF of young mice, was identified as a factor that enhances in vitro activation of quiescent NSCs from the SVZ in association with other growth factors [epidermal growth factor (EGF) and basic fibroblast growth factor] [17]. Thus, the Dpp subfamily and the OP‐1 subfamily of BMP ligands have opposing functions in regulating NSCs. Furthermore, posttranslational modification of BMPR1a receptor by palmitoylation modulates NSC function through receptor localization and signal transductions; different palmitoylated positions in BMPR1a differentially affect canonical and noncanonical BMP signaling, which lead to SMAD activation and extracellular signal‐regulated kinase activation, respectively [18]. In addition, age‐dependent different expression of BMP ligands, such as BMP2/4 and BMP6, in NSC niches, might affect the maintenance of quiescence in adult NSCs [19].
Fig. 2.
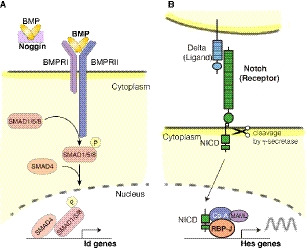
BMP and Notch signaling cascades. (A) Canonical pathway of BMP signaling. BMPR bind to BMP ligands, and transduce signals via SMAD molecules, which ultimately enhance Id gene expression. Noggin sequesters BMP and antagonizes BMP signaling. (B) Canonical pathway of Notch signaling. Notch receptors bind to ligands on neighboring cells, inducing gamma‐secretase‐mediated cleavage in the signal‐receiving cells (lower). Cleaved Notch receptor (NICD) translocates into the nucleus and activates the expression of Hes genes.
The Notch signaling pathway includes four Notch receptors (Notch1–Notch4), which are cleaved upon binding of Notch ligands (members of the delta or jagged families) expressed in neighboring differentiating cells (Fig. 2B). The intracellular domain of Notch (NICD) is transferred into the nucleus, where it binds to the transcriptional factor CBF‐1, suppressor of hairless, Lag‐2 [recombination signal‐binding protein for immunoglobulin kappa J region (RBPJ) in mice] to activate downstream genes such as Hes (reviewed in Ref. [20]). Notch1–Notch3 are expressed in the neurogenic niche of the adult mouse brain [21]. Significant involvement of Notch signaling in quiescence was demonstrated using Hes5‐GFP reporter mice, which express GFP under the control of Notch signaling. Both the SVZ [22] and DG [23] exhibit activated Notch signaling in heterogeneous cell populations, including active and quiescent NSCs. In canonical Notch signaling, RBPJ is the common downstream effector of all Notch receptors. Conditional knockout of RBPJ in the DG of GLAST‐Cre‐ERT2 Tg mice [24] and in the SVZ of Nestin‐Cre‐ERT2 Tg mice [25] or Hes5‐Cre‐ERT2 mice [26] results in enhanced generation of new neurons within a few weeks, mediated by NSC activation, followed by severe depletion of NSCs and the loss of neurogenesis within a few months. These reports suggest that Notch signaling regulates the maintenance of both active and quiescent NSCs in both the SVZ and DG. Importantly, previous studies showed that Notch1 and Notch2 play distinct roles in adult NSCs. Conditional KO of Notch1 decreased proliferation of neurogenic NSCs in the DG (using different Cre drivers: Nestin‐Cre‐ERT2 [27] and human GFAP‐Cre‐ERT2 [28]) and the SVZ (using Nestin‐Cre‐ERT2 [29] and Hes5‐Cre‐ERT2 Tg mice [26]). On the other hand, conditional KO of Notch2 induced abnormal activation of quiescent NSCs, resulting in exhaustion of the NSC pool in both the SVZ and DG of Hes5‐Cre‐ERT2 mice [26, 30]. These differential outputs were confirmed by the expression of active forms of the Notch receptors. Noch1ICD and Notch2ICD using a Cre‐inducible expression system in hGFAP‐Cre‐ERT2 Tg mice [28] and Hes5‐Cre‐ER T2 Tg mice [30] enhanced and decreased proliferation of NSCs, respectively. These results indicated that although Notch1 and Notch2 have distinct roles in adult NSCs, both Notch1 cKO and Notch2 cKO caused aging phenotypes by decreasing neurogenesis as well as reducing the NSC pool [26, 29, 30]. Another Notch receptor, Notch3, which is enriched in quiescent NSCs, affects NSC maintenance [31]. Depletion of Notch3 expression induced proliferation of adult NSCs in the SVZ [31], whereas ectopic Notch3 expression decreased proliferation of NSCs derived from the DG [32]. The downstream factors of these Notch receptors might shed light on the diverse functions of Notch signaling in adult NSCs. One example is Id4, originally identified as a target molecule of BMP signaling. Proteins in the inhibitor of DNA binding (Id) family, members of the HLH transcription factor family that lack a DNA‐binding motif, form heterodimers with other bHLH factors that sequester the partner protein in non‐DNA‐binding dimers (reviewed in Ref. [33]). Quiescent NSCs derived from the DG express high levels of Id4, and Id4 is a major effector of the Notch2 receptor and a quiescence‐inducing factor in NSCs [30, 34]. Id4 decreases the protein stability of achaete‐scute homolog (Ascl)1, a proneural bHLH factor expressed in active NSCs, by sequestering E protein, the binding partner for transcriptional activator function [34]. Another example of the differential output of Notch signaling is Hes1, a canonical Notch effector and bHLH transcriptional repressor that is expressed at a higher level in quiescent NSCs [35]. Oscillatory expression of Hes1 regulates the expression dynamics of the target genes, Dll1, Ngn2, and Ascl1, which contribute to the switching from proliferation to differentiation of embryonic NSCs [36, 37]. In adult NSCs, higher expression of Hes1 continuously inhibits Ascl1 expression and maintains NSCs in the quiescent state [35]. These reports suggest that the expression dynamics and protein stability of bHLH factors might contribute to differential outputs of Notch signaling.
The quiescent state of adult NSCs is also regulated by other signaling receptors, for example, activation of gamma‐aminobutyric acid (GABA) receptors by GABA secreted from NSC niche cells in the SVZ and DG [38, 39] and activation of integrin receptors by binding of Mfge8, a phagocytosis factor, which is secreted from quiescent NSCs of the DG [40]. Adult NSCs express GABAA receptors and tonically respond to GABA from niche cells, which are parvalbumin‐expressing (PV+) interneurons in the DG or neuroblasts in the SVZ that can dictate the NSC choice between quiescence and activation through nonsynaptic GABA signaling [39, 41]. On the other hand, quiescent NSCs secrete Mfeg8 and promote quiescence via suppression of the PTEN–Akt–mTOR1 pathway through binding to the integrin receptor [40]. Tropomyosin receptor kinase C (TrkC) receptor activation by binding of neurotrophin‐3 from the CSF or the nearby vasculature promotes NSC quiescence in the SVZ [42]. Together, these observations indicate the importance of combinations of multiple membrane receptors for maintaining NSC quiescence. The activation of these signaling pathways is ligand‐induced upon binding to membrane receptors, most of which induce the endocytic trafficking of receptors to lysosomes or recycling back to the cell surface, suggesting the important roles of lysosomes in NSC states. In the next part, we will discuss the involvement of lysosomes in quiescent NSCs.
The lysosome: a digestive organelle and a hub of nutrition signaling
Subsequent to ligand‐induced activation, membrane receptors are internalized in endosomes and ultimately degraded in lysosomes after endolysosomal trafficking or recycled into membrane surface (Fig. 3A). Lysosomes are membrane‐enclosed cytoplasmic organelles that degrade a variety of biological macromolecules, including proteins, lipids, carbohydrates, and nucleic acids (reviewed in Ref. [43, 44]). The lysosomal lumen is highly acidic; the low pH is maintained by a vacuolar ATPase (V‐ATPase) on the lysosomal membrane. Cells contain between 50 and 1000 lysosomes, in which more than 60 acidic hydrolases digest macromolecules delivered from the endolysosomal and autophagic pathways. Lysosomes were once thought of as static organelles involved in the waste disposal system. However, recent reports have shown that lysosomes act as regulatory hubs for cellular homeostasis by switching the metabolic state between catabolism and anabolism. In nutrient‐rich environments, multiple sensors of amino acid levels cause Rag GTPases to interact with the Ragulator–protein complex [45]. Then, the Rag–Ragulator complex recruits mTORC1 and other factors including transcriptional factor EB (TFEB), a master transcriptional regulator of lysosomal components, to the lysosomal membrane [46, 47]. TFEB is phosphorylated by mTORC1, resulting in inhibition of nuclear translocation of TFEB (Fig. 3B) [47]. TFEB, a member of the microphthalmia‐transcription factor E (MiT‐TFE) family of HLH leucine zipper transcription factors [48], activates lysosome‐related genes under the coordinated lysosomal expression and regulation (CLEAR) gene network [49]. Nutrient starvation promotes dephosphorylation of TFEB, leading to its nuclear translocation. In the nucleus, TFEB upregulates genes involved in lysosomal function and autophagy, resulting in the recycling and clearance of biomolecules inside cells. Thus, lysosomes control their functions to adapt to environmental cues. TFE3, another member of the MiT‐TFE family, is also involved in this process. Moreover, lysosomes regulate extracellular conditions by lysosomal exocytosis after fusion to the plasma membrane [50] and by the degradation of extracellular matrix protein for extracellular remodeling [51]. Lysosomes are also involved in a broad range of cellular functions such as lipid homeostasis and transfer to other organelles (reviewed in Ref. [52]), calcium signaling by lysosomal calcium channels (reviewed in Ref. [53]), and responses to stress such as proteostatic dysfunction (reviewed in Ref. [54]). Next, we will discuss the role of lysosomal functions in proteostatic regulation in quiescent stem cells.
Fig. 3.
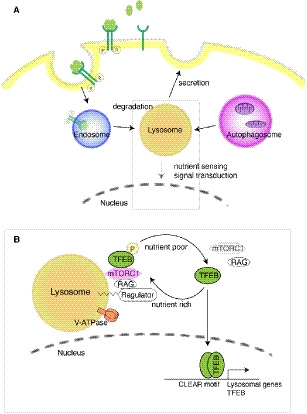
The lysosome functions as a degradative organelle and a signaling hub. (A) Lysosomes digest cargo from endosomes and autophagosomes in the acidic lumen. The endolysosomal pathway degrades biomolecules, including membrane receptors, in lysosomes following their internalization by endocytosis. Autophagy encloses cytoplasmic materials, including organelles, into autophagosomes, which fuse with lysosomes where their contents are digested. Lysosomes secrete their contents via lysosomal exocytosis. Lysosomes function as a signaling hub for nutrient sensing. The gray square is enlarged in panel B. (B) The lysosome is a hub where signaling molecules localize and transduce their signals. v‐ATPase maintains a low pH by pumping protons. Nutrient‐rich conditions activate Ragulator‐RAG, which recruits and activates mammalian target of rapamycin complex 1 (mTORC1) to lysosomes. Activated mTORC1 (pink) inhibits lysosomal biogenesis through inhibitory phosphorylation of TFEB. Low nutrient concentration results in inactivation of mTORC1 and activation of TFEB, thereby inducing expression of lysosomal genes.
Lysosomes in quiescent NSCs
Lysosomes are involved in the regulation of proteostasis (protein homeostasis). Proteostasis is maintained by the protein quality control machinery, which consists of protein synthesis on ribosome, proper protein folding assisted by molecular chaperones, and proteolysis [55]. Proteolysis is mainly mediated by the ubiquitin–proteasome and autophagy–lysosome pathways. Differential regulation of these two proteolytic pathways was recently reported in adult NSCs [56, 57] (Fig. 4). Transcriptome analyses of fluorescence‐activated cell sorting‐sorted active and quiescent NSCs derived from the SVZ revealed higher expression of lysosomal genes, including TFEB, and lower expression of both proteasomal and ribosomal genes in quiescent vs. active NSCs [56]. In regard to molecular chaperones, quiescent NSCs express higher levels of ER stress–related genes and lower levels of chaperonin TCP‐1 ring complex/chaperonin‐containing TCP‐1 subunits than active NSCs [56]. Quiescent NSCs exhibit much higher lysosomal proteolytic activity and lower proteasomal activity than active NSCs in vitro [57]. These results suggest that quiescent NSCs alter proteostasis, shifting the predominant sites of proteolysis from proteasomes to lysosomes in order to adapt to their environment. In in vitro cultures, BMP is a strong inducer of quiescence in proliferating NSCs [8]. BMP‐induced quiescent NSCs derived from the SVZ [56] and the DG [58] contain detergent‐insoluble aggregates, which might concentrate both proteasomes and their substrates for the rapid reactivation into active NSCs [58]. In this regard, these aggregates might be similar to proteasome storage granules observed in quiescent yeast cells [59]. BMP treatment of NSCs induces dephosphorylation and activation of TFEB, followed by nuclear localization and increased gene expression, thereby increasing lysosomal activity [57]. TFEB‐KO NSCs exhibit delayed entry into the quiescent state after BMP treatment with higher levels of activated membrane receptors [57]. Consistent with the in vitro results, conditional knockout of TFEB in adult NSCs (in GLAST‐Cre‐ERT2 mice) increases the number of active NSCs in the DG, concomitant with the accumulation of activated membrane receptors [57]. Ectopic expression of a constitutively active mutant of TFEB (caTFEB) or TFEB activation by mTORC1 inhibitors, rapamycin and Torin1, decreases NSC proliferation in vitro [57]. In vivo, ectopic caTFEB expression in NSCs of the DG by injection of lentivirus under the control of the Hes5 or GFAP promoter decreases the number of active NSCs in young adult mice [57]. These results suggest that lysosomal activation by TFEB leads to quiescence in young NSCs and maintains their quiescence in the DG. On the other hand, aged quiescent NSCs from the SVZ express less of the lysosomal protein Lamp‐1 than young quiescent NSCs, at levels similar to those in active NSCs [56]. Ectopic expression of caTFEB in vitro decreases the abundance of aggregates in primary NSC cultures from the SVZ in aged mice and promotes reactivation of quiescent NSCs in the presence of the growth factors EGF and FGF [56]. Rapamycin increases abundance of active NSCs in the SVZ of old mice by its dietary supplementation [56]. These results demonstrate that proteostatic regulation by lysosomes differs between young and aged NSCs (Fig. 4), and imply differential regulation of lysosomes in the SVZ and the DG. Differential outputs by TFEB activation in NSCs, including induction of quiescence in young NSCs of the DG [57] and enhancement of reactivation in aged NSCs of the SVZ [56], suggest an additional role of lysosomes in aged NSCs in alleviating severe damage caused by aging. Aged quiescent NSCs become more resistant to activation than young quiescent NSCs, a process in which inflammatory signals from niche cells are involved [60, 61]. Interferon (IFN)‐γ decreases NSC proliferation in vitro and inhibition of IFN response through deletion of IFN‐α and IFN‐γ leads to a similar fraction of active NSCs in young and old NSCs in the SVZ [61], implying an additional role for lysosomes in inflammatory responses in aged NSCs.
Fig. 4.
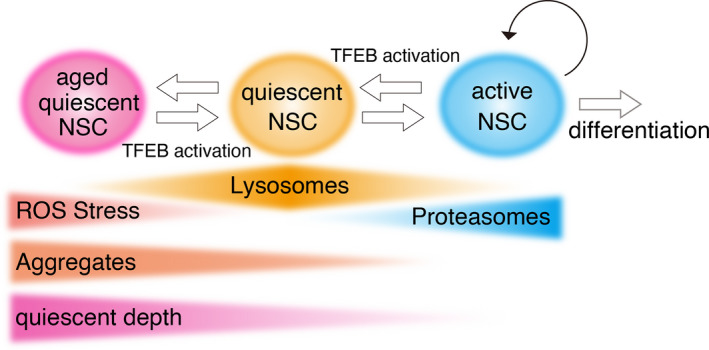
Lysosomal regulation of NSC quiescence. To maintain proteostasis, active NSCs have higher proteasomal activity and lower lysosomal activity, while quiescent NSCs have lower proteasomal activity and higher lysosomal activity. Quiescent NSCs contain more lysosomes, but lysosomal abundance decreases over the course of the aging process. On the other hand, the level of protein aggregates and ROS increases with age, in turn affecting the depth of quiescence. In active NSCs, TFEB activation induces quiescence, whereas in quiescent NSCs, it rejuvenates the cells and decreases the abundance of aggregates. Thus, lysosomes serve as a switch for maintaining NSC quiescence.
The autophagy–lysosomal pathway degrades cellular proteins and organelles. There are three types of autophagy in mammals: macroautophagy, microautophagy, and chaperone‐mediated autophagy [62]. Macroautophagy is a major lysosomal catabolic process induced by cellular stresses such as starvation and abnormal protein accumulation. Damaged cellular components and organelles are engulfed into autophagosomes and degraded after fusion to lysosomes. This process is called macroautophagy. Microautophagy and chaperone‐mediated autophagy directly incorporate cellular components and proteins into lysosomes without the use of autophagosomes. Several reports have shown that the autophagy–lysosomal pathway is involved in the adult NSC maintenance, progenitor cell differentiation, and neuronal maturation. Removal of the autophagy‐related gene (Atg) FIP200, an essential gene for autophagic induction, in NSCs of hGFAP‐Cre cKO mice, induced progressive loss of NSCs and defects in neurogenesis in the adult brain, concomitant with increases in mitochondria and reactive oxygen species (ROS) [63]. Deletion of Atg5, a gene important for autophagosome formation, in dividing neural progenitor cells, was analyzed using a retrovirus encoding Cre recombinase in the SGZ of adult mouse brain. Atg5 deletion decreases survival of NSCs and delays neuronal maturation [64]. Hypomorphic mutation of Atg16L1, another important gene for autophagosome formation, decreases autophagy and proliferation but increases Notch1ICD level in the SVZ of adult mouse brain [65]. Reduced expression of Beclin1 or activating molecule in Beclin‐1‐regulated autophagy (Ambra)1, both of which are involved in the initial step of autophagosome formation, decreases proliferation and increases apoptosis in the adult SVZ in heterozygote mice relative to wild‐type mice [66]. These reports suggest that autophagic flux exists in adult NSCs and maintains the survival and differentiation of these cells, as well as their progeny, in both the SVZ and DG. Recent reports demonstrated that upstream factors regulating autophagic genes are regulators of adult neurogenesis. For example, let‐7, an miRNA for cell‐cycle exit, affects migration of newly generated neurons and their morphology, both of which depend on autophagic activity [67]. Forkhead box O (FOXO) transcriptional factors are critical regulators for autophagic flux and proteostasis in the adult NSCs [68, 69, 70, 71]. Knockout of FoxO1, FoxO3, and FoxO4 in hGFAP‐Cre [68] and GLAST‐CreERT2 [70] mice induced an initial increase in proliferating NSCs and progenitor cells and a subsequent severe decline in the NSC pool associated with abnormal accumulation of autophagosomes. FoxO3 directly binds to numerous Atg, regulates proteostasis, and avoids protein aggregates via autophagic clearance in cultured neural stem and progenitor cells derived from the adult SVZ [71, 72].
Proteostatic regulation has also been reported in stem cell differentiation; NSC differentiation is associated with rewiring of chaperone networks [73], while embryonic stem cell differentiation decreases proteasomal activity [74]. Asymmetric cell division causes selective delivery of the cellular degradation machinery and damaged proteins, and contributes to cell fate decisions through proteostatic regulation in neural and hematopoietic stem cells (HSC) [58, 75, 76]. Proteomic approaches are essential for understanding such proteostatic regulations in detail. For example, recent proteomic studies revealed specific regulation of extracellular matrix in the SVZ niches in comparison with other brain regions; these differences were not identified by transcriptomic analysis. Detergent extraction methods for tissue sections enabled fractionation of proteins depending on their association strength with extracellular matrix. Several core matrix proteins are more detergent‐soluble in the neurogenic niches, suggesting the existence of mechanical regulation such as stiffness in the stem cell niche [77].
Lysosomes in other quiescent cells
Quiescence is a fundamental mechanism by which various types of cells, such as HSC and fibroblasts, maintain themselves in a low‐metabolism state for long periods of time [78]. Recent reports revealed the importance of lysosomes in the quiescent state. HSCs, the major source of multilineage hematopoietic cells, mostly remain quiescent in order to protect themselves from metabolic stresses, while their entry into the cell cycle is accompanied by an increase in mitochondrial activity (reviewed in Ref. [79]). Interestingly, deeply quiescent HSCs, which contain small punctate mitochondria with low mitochondrial activity, express elevated levels of lysosomal genes; however, this causes not the degradation of mitochondria, but rather their transient sequestration into enlarged lysosomes [80]. Autophagy, including mitophagy, involves many regulatory steps prior to fusion with lysosomes, but the mechanisms by which HSCs regulate lysosomal activity in the enlarged lysosomes remain unknown. Further investigation is required to elucidate the degradation of cargos such as membrane receptors [81]. In the adults, mitochondrial activity (as indicated by mitochondrial membrane potential) does not significantly differ between BMP4‐induced quiescent NSCs and active NSCs, whereas neuronally differentiated progenitor cells have higher levels of mitochondrial respiration than NSCs [82]. Lysosomes might affect the differentiation of neural progenitor cells by regulating mitochondrial activity. In addition, lysosomes play a role in cell division of HSCs [76]. As in NSCs, lysosomes are asymmetrically segregated in daughter cells during cell division of HSCs. This controls the fate of daughter cells, as cells receiving fewer lysosomes are prone to differentiate [76]. Lysosomes are co‐inherited with other factors, including autophagosomes, mitophagosomes, and Notch factors such as Numb and Notch1, which might act together.
Rat embryonic fibroblasts can be induced to undergo quiescence by serum starvation for 2 days [83]. In a transcriptome analysis aimed at identifying factors governing long‐term quiescence, lysosomal genes exponentially increased their expression 2 days after serum starvation and then increased continuously for 2 weeks [83]. Endosomal genes also exponentially increased their expression for 2 days but kept the same expression level at later time points. Lysosomal inhibition by chemicals increased the abundance of mitochondrial ROS and induced deeper quiescence, a state associated with lower responsiveness to serum stimulation. On the other hand, lysosomal activation by ectopic expression of Mitf, a member of the MiT/TFE family, in quiescent cells, reduced ROS levels and allowed more efficient reactivation by serum stimulation. These observations suggest that lysosomes maintain a quiescent state between shallow and deep quiescence that is associated with cellular metabolism and stresses, implying important links with cellular senescence and aging.
Conclusion
Here, we discussed recent findings related to adult NSC quiescence, ranging from signaling pathways to lysosomal regulation of quiescence. Recent reports have suggested that lysosomes are associated with quiescence in many types of cells. In a long‐lived quiescent state of Caenorhabditis elegans, which can survive for months without food, TFEB is a master regulator of reproductive quiescence that is required for entry into quiescence as well as survival and recovery [84]. Further investigations, using both proteomic approaches and protein analysis at the molecular level, are required to reveal the role of lysosomes in quiescence. Imaging in live cells with fluorescent probes for lysosomal activity could also be used to monitor lysosomal function and dynamics [51, 85, 86, 87]. Because the lysosome is a multifunctional organelle, the inhibition or activation of lysosomes alters multiple aspects of cellular functions, including metabolic changes and environmental stresses associated with aging. Lysosomes also regulate lipid metabolism in cells [52, 88, 89], suggesting that they are involved in lipid regulation in adult NSCs [90]. Taken together, these observations suggest that further studies of lysosomes will provide deep insights into stem cell quiescence.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
TK and RK wrote the manuscript.
Acknowledgements
This work was supported by a Grant‐in‐Aid for Scientific Research (B) (JSPS 20H03260) and SPIRITS 2020 of Kyoto University (to TK); a Grant‐in‐Aid for Scientific Research on Innovative Areas (16H06480) (to RK); and the Japan Agency for Medical Research and Development (AMED) under Grant Number JP18gm1110002h0002 (to RK).
Contributor Information
Taeko Kobayashi, Email: tkobayas@infront.kyoto-u.ac.jp.
Ryoichiro Kageyama, Email: rkageyam@infront.kyoto-u.ac.jp.
References
- 1. Altman J & Das GD (1965) Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neur 124, 319–336. [DOI] [PubMed] [Google Scholar]
- 2. Doetsch F, Caille I, Lim DA, Garcia‐Verdugo JM & Alvarez‐Buylla A (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716. [DOI] [PubMed] [Google Scholar]
- 3. Eriksson PS, Perfilieva E, Björk‐Eriksson T, Alborn M, Nordborg C, Peterson DA & Gage FH (1998) Neurogenesis in the adult human hippocampus. Nat Med 4, 1313–1317. [DOI] [PubMed] [Google Scholar]
- 4. Bond AM, Ming GL & Song H (2015) Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lim DA & Alvarez‐Buylla A (2016) The adult ventricular‐subventricular zone (V‐SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb Perspect Biol 8, a018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urban N, Blomfield IM & Guillemot F (2019) Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron 104, 834–848. [DOI] [PubMed] [Google Scholar]
- 7. Toda T, Parylak SL, Linker SB & Gage FH (2019) The role of adult hippocampal neurogenesis in brain health and disease. Mol Psychiatry 24, 67–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urban N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J et al. (2013) Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev 27, 1769–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urban N, van den Berg DL, Forget A, Andersen J, Demmers JA, Hunt C, Ayrault O & Guillemot F (2016) Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balch WE, Morimoto RI, Dillin A & Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319, 916–919. [DOI] [PubMed] [Google Scholar]
- 11. Joppe SE, Hamilton LK, Cochard LM, Levros LC, Aumont A, Barnabe‐Heider F & Fernandes KJ (2015) Bone morphogenetic protein dominantly suppresses epidermal growth factor‐induced proliferative expansion of adult forebrain neural precursors. Front Neurosci 9, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortiguela R, Marques‐Torrejon MA, Nakashima K et al. (2010) Signaling through BMPR‐IA regulates quiescence and long‐term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89. [DOI] [PubMed] [Google Scholar]
- 13. Lim DA, Tramontin AD, Trevejo JM, Herrera DG, García‐Verdugo JM & Alvarez‐Buylla A (2000) Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron 28, 713–726. [DOI] [PubMed] [Google Scholar]
- 14. Colak D, Mori T, Brill MS, Pfeifer A, Falk S, Deng C, Monteiro R, Mummery C, Sommer L & Gotz M (2008) Adult neurogenesis requires Smad4‐mediated bone morphogenic protein signaling in stem cells. J Neurosci 28, 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bonaguidi MA, Peng CY, McGuire T, Falciglia G, Gobeske KT, Czeisler C & Kessler JA (2008) Noggin expands neural stem cells in the adult hippocampus. J Neurosci 28, 9194–9204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peretto P, Dati C, De Marchis S, Kim HH, Ukhanova M, Fasolo A & Margolis FL (2004) Expression of the secreted factors noggin and bone morphogenetic proteins in the subependymal layer and olfactory bulb of the adult mouse brain. Neuroscience 128, 685–696. [DOI] [PubMed] [Google Scholar]
- 17. Silva‐Vargas V, Maldonado‐Soto AR, Mizrak D, Codega P & Doetsch F (2016) Age‐dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell 19, 643–652. [DOI] [PubMed] [Google Scholar]
- 18. Wegleiter T, Buthey K, Gonzalez‐Bohorquez D, Hruzova M, Bin Imtiaz MK, Abegg A, Mebert I, Molteni A, Kollegger D, Pelczar P et al. (2019) Palmitoylation of BMPR1a regulates neural stem cell fate. Proc Natl Acad Sci USA 116, 25688–25696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yousef H, Morgenthaler A, Schlesinger C, Bugaj L, Conboy IM & Schaffer DV (2015) Age‐associated increase in BMP signaling inhibits hippocampal neurogenesis. Stem Cells 33, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kageyama R, Ohtsuka T & Kobayashi T (2007) The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251. [DOI] [PubMed] [Google Scholar]
- 21. Irvin DK, Zurcher SD, Nguyen T, Weinmaster G & Kornblum HI (2001) Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the notch‐DSL signaling system during brain development. J Comp Neurol 436, 167–181. [PubMed] [Google Scholar]
- 22. Giachino C, Basak O, Lugert S, Knuckles P, Obernier K, Fiorelli R, Frank S, Raineteau O, Alvarez‐Buylla A & Taylor V (2014) Molecular diversity subdivides the adult forebrain neural stem cell population. Stem Cells 32, 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V & Giachino C (2010) Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445–456. [DOI] [PubMed] [Google Scholar]
- 24. Ehm O, Goritz C, Covic M, Schaffner I, Schwarz TJ, Karaca E, Kempkes B, Kremmer E, Pfrieger FW, Espinosa L et al. (2010) RBPJkappa‐dependent signaling is essential for long‐term maintenance of neural stem cells in the adult hippocampus. J Neurosci 30, 13794–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imayoshi I, Sakamoto M, Yamaguchi M, Mori K & Kageyama R (2010) Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci 30, 3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Engler A, Rolando C, Giachino C, Saotome I, Erni A, Brien C, Zhang R, Zimber‐Strobl U, Radtke F, Artavanis‐Tsakonas S et al. (2018) Notch2 signaling maintains NSC quiescence in the murine ventricular‐subventricular zone. Cell Rep 22, 992–1002. [DOI] [PubMed] [Google Scholar]
- 27. Ables JL, Decarolis NA, Johnson MA, Rivera PD, Gao Z, Cooper DC, Radtke F, Hsieh J & Eisch AJ (2010) Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J Neurosci 30, 10484–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breunig JJ, Silbereis J, Vaccarino FM, Sestan N & Rakic P (2007) Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA 104, 20558–20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Basak O, Giachino C, Fiorini E, Macdonald HR & Taylor V (2012) Neurogenic subventricular zone stem/progenitor cells are Notch1‐dependent in their active but not quiescent state. J Neurosci 32, 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang R, Boareto M, Engler A, Louvi A, Giachino C, Iber D & Taylor V (2019) Id4 downstream of Notch2 maintains neural stem cell quiescence in the adult hippocampus. Cell Rep 28, 1485–1498.e6. [DOI] [PubMed] [Google Scholar]
- 31. Kawai H, Kawaguchi D, Kuebrich BD, Kitamoto T, Yamaguchi M, Gotoh Y & Furutachi S (2017) Area‐specific regulation of quiescent neural stem cells by Notch3 in the adult mouse subependymal zone. J Neurosci 37, 11867–11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehret F, Vogler S, Pojar S, Elliott DA, Bradke F, Steiner B & Kempermann G (2015) Mouse model of CADASIL reveals novel insights into Notch3 function in adult hippocampal neurogenesis. Neurobiol Dis 75, 131–141. [DOI] [PubMed] [Google Scholar]
- 33. Wang LH & Baker NE (2015) E proteins and ID proteins: helix‐loop‐helix partners in development and disease. Dev Cell 35, 269–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blomfield IM, Rocamonde B, Masdeu MDM, Mulugeta E, Vaga S, van den Berg DL, Huillard E, Guillemot F & Urban N (2019) Id4 promotes the elimination of the pro‐activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. Elife 8, e48561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sueda R, Imayoshi I, Harima Y & Kageyama R (2019) High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev 33, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shimojo H, Ohtsuka T & Kageyama R (2008) Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. [DOI] [PubMed] [Google Scholar]
- 37. Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F & Kageyama R (2013) Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. [DOI] [PubMed] [Google Scholar]
- 38. Alfonso J, Le Magueresse C, Zuccotti A, Khodosevich K & Monyer H (2012) Diazepam binding inhibitor promotes progenitor proliferation in the postnatal SVZ by reducing GABA signaling. Cell Stem Cell 10, 76–87. [DOI] [PubMed] [Google Scholar]
- 39. Song J, Zhong C, Bonaguidi MA, Sun GJ, Hsu D, Gu Y, Meletis K, Huang ZJ, Ge S, Enikolopov G et al. (2012) Neuronal circuitry mechanism regulating adult quiescent neural stem‐cell fate decision. Nature 489, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Bond AM, Shade JE, Zhu Y, Davis CO, Wang X, Su Y, Yoon KJ, Phan AT, Chen WJ et al. (2018) Autocrine Mfge8 signaling prevents developmental exhaustion of the adult neural stem cell pool. Cell Stem Cell 23, 444–452.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu X, Wang Q, Haydar TF & Bordey A (2005) Nonsynaptic GABA signaling in postnatal subventricular zone controls proliferation of GFAP‐expressing progenitors. Nat Neurosci 8, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delgado AC, Ferron SR, Vicente D, Porlan E, Perez‐Villalba A, Trujillo CM, D'Ocon P & Farinas I (2014) Endothelial NT‐3 delivered by vasculature and CSF promotes quiescence of subependymal neural stem cells through nitric oxide induction. Neuron 83, 572–585. [DOI] [PubMed] [Google Scholar]
- 43. Ballabio A & Bonifacino JS (2020) Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21, 101–118. [DOI] [PubMed] [Google Scholar]
- 44. Holland LKK, Nielsen IO, Maeda K & Jaattela M (2020) SnapShot: lysosomal functions. Cell 181, 748.e1. [DOI] [PubMed] [Google Scholar]
- 45. Sancak Y, Bar‐Peled L, Zoncu R, Markhard AL, Nada S & Sabatini DM (2010) Ragulator‐Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pena‐Llopis S, Vega‐Rubin‐de‐Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR & Brugarolas J (2011) Regulation of TFEB and V‐ATPases by mTORC1. EMBO J 30, 3242–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC et al. (2012) A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA & Copeland NG (1998) The bHLH‐Zip transcription factor Tfeb is essential for placental vascularization. Development 125, 4607–4616. [DOI] [PubMed] [Google Scholar]
- 49. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- 50. Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M et al. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miao R, Li M, Zhang Q, Yang C & Wang X (2020) An ECM‐to‐nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev Cell 52, 21–37 e5. [DOI] [PubMed] [Google Scholar]
- 52. Thelen AM & Zoncu R (2017) Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol 27, 833–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li P, Gu M & Xu H (2019) Lysosomal ion channels as decoders of cellular signals. Trends Biochem Sci 44, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martini‐Stoica H, Xu Y, Ballabio A & Zheng H (2016) The autophagy‐lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci 39, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hartl FU, Bracher A & Hayer‐Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. [DOI] [PubMed] [Google Scholar]
- 56. Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, Dulken BW, Zhao X, Yeo RW, Ho TT et al. (2018) Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kobayashi T, Piao W, Takamura T, Kori H, Miyachi H, Kitano S, Iwamoto Y, Yamada M, Imayoshi I, Shioda S et al. (2019) Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat Commun 10, 5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morrow CS, Porter TJ, Xu N, Arndt ZP, Ako‐Asare K, Heo HJ, Thompson EAN & Moore DL (2020) Vimentin coordinates protein turnover at the aggresome during neural stem cell quiescence exit. Cell Stem Cell 26, 558–568.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Laporte D, Salin B, Daignan‐Fornier B & Sagot I (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kalamakis G, Brune D, Ravichandran S, Bolz J, Fan W, Ziebell F, Stiehl T, Catala‐Martinez F, Kupke J, Zhao S et al. (2019) Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell 176, 1407–1419.e14. [DOI] [PubMed] [Google Scholar]
- 61. Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV et al. (2019) Single‐cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen HM & Mizushima N (2014) At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci 39, 61–71. [DOI] [PubMed] [Google Scholar]
- 63. Wang C, Liang CC, Bian ZC, Zhu Y & Guan JL (2013) FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 16, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xi Y, Dhaliwal JS, Ceizar M, Vaculik M, Kumar KL & Lagace DC (2016) Knockout of Atg5 delays the maturation and reduces the survival of adult‐generated neurons in the hippocampus. Cell Death Dis 7, e2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM & Rubinsztein DC (2016) Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun 7, 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yazdankhah M, Farioli‐Vecchioli S, Tonchev AB, Stoykova A & Cecconi F (2014) The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis 5, e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Petri R, Pircs K, Jonsson ME, Akerblom M, Brattas PL, Klussendorf T & Jakobsson J (2017) let‐7 regulates radial migration of new‐born neurons through positive regulation of autophagy. EMBO J 36, 1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J et al. (2009) FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC et al. (2009) FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schaffner I, Minakaki G, Khan MA, Balta EA, Schlotzer‐Schrehardt U, Schwarz TJ, Beckervordersandforth R, Winner B, Webb AE, DePinho RA et al. (2018) FoxO function is essential for maintenance of autophagic flux and neuronal morphogenesis in adult neurogenesis. Neuron 99, 1188–1203.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Audesse AJ, Dhakal S, Hassell LA, Gardell Z, Nemtsova Y & Webb AE (2019) FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet 15, e1008097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Webb AE, Pollina EA, Vierbuchen T, Urban N, Ucar D, Leeman DS, Martynoga B, Sewak M, Rando TA, Guillemot F et al. (2013) FOXO3 shares common targets with ASCL1 genome‐wide and inhibits ASCL1‐dependent neurogenesis. Cell Rep 4, 477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vonk WIM, Rainbolt TK, Dolan PT, Webb AE, Brunet A & Frydman J (2020) Differentiation drives widespread rewiring of the neural stem cell chaperone network. Mol Cell 78, 329–345.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT et al. (2012) Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moore DL, Pilz GA, Araúzo‐Bravo MJ, Barral Y & Jessberger S (2015) A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334–1338. [DOI] [PubMed] [Google Scholar]
- 76. Loeffler D, Wehling A, Schneiter F, Zhang Y, Muller‐Botticher N, Hoppe PS, Hilsenbeck O, Kokkaliaris KD, Endele M & Schroeder T (2019) Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 573, 426–429. [DOI] [PubMed] [Google Scholar]
- 77. Kjell J, Fischer‐Sternjak J, Thompson AJ, Friess C, Sticco MJ, Salinas F, Cox J, Martinelli DC, Ninkovic J, Franze K et al. (2020) Defining the adult neural stem cell niche proteome identifies key regulators of adult neurogenesis. Cell Stem Cell 26. 277–293.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cheung TH & Rando TA (2013) Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Filippi MD & Ghaffari S (2019) Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood 133, 1943–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liang R, Arif T, Kalmykova S, Kasianov A, Lin M, Menon V, Qiu J, Bernitz JM, Moore K, Lin F et al. (2020) Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell 26, 359–376.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Luis TC, Lawson H & Kranc KR (2020) Divide and rule: mitochondrial fission regulates quiescence in hematopoietic stem cells. Cell Stem Cell 26, 299–301. [DOI] [PubMed] [Google Scholar]
- 82. Beckervordersandforth R, Ebert B, Schaffner I, Moss J, Fiebig C, Shin J, Moore DL, Ghosh L, Trinchero MF, Stockburger C et al. (2017) Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron 93, 560–573.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fujimaki K, Li R, Chen H, Della Croce K, Zhang HH, Xing J, Bai F & Yao G (2019) Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Proc Natl Acad Sci USA 116, 22624–22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gerisch B, Tharyan RG, Mak J, Denzel SI, Popkes‐van Oepen T, Henn N & Antebi A (2020) HLH‐30/TFEB is a master regulator of reproductive quiescence. Dev Cell 53, 316–329.e5. [DOI] [PubMed] [Google Scholar]
- 85. Ishii S, Matsuura A & Itakura E (2019) Identification of a factor controlling lysosomal homeostasis using a novel lysosomal trafficking probe. Sci Rep 9, 11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ba Q, Raghavan G, Kiselyov K & Yang G (2018) Whole‐cell scale dynamic organization of lysosomes revealed by spatial statistical analysis. Cell Rep 23, 3591–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang C, Taki M, Kajiwara K, Wang J & Yamaguchi S (2020) Phosphole‐oxide‐based fluorescent probe for super‐resolution stimulated emission depletion live imaging of the lysosome membrane. ACS Mater Lett 705–711. [Google Scholar]
- 88. Mansueto G, Armani A, Viscomi C, D'Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S et al. (2017) Transcription factor EB controls metabolic flexibility during exercise. Cell Metab 25, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Trivedi PC, Bartlett JJ, Mercer A, Slade L, Surette M, Ballabio A, Flibotte S, Hussein B, Rodrigues B, Kienesberger PC et al. (2020) Loss of function of transcription factor EB remodels lipid metabolism and cell death pathways in the cardiomyocyte. Biochim Biophys Acta Mol Basis Dis 1866, 165832. [DOI] [PubMed] [Google Scholar]
- 90. Knobloch M (2017) The role of lipid metabolism for neural stem cell regulation. Brain Plast 3, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


