Abstract
Emerging infectious diseases can have devastating effects on host communities, causing population collapse and species extinctions. The timing of novel pathogen arrival into naïve species communities can have consequential effects that shape the trajectory of epidemics through populations. Pathogen introductions are often presumed to occur when hosts are highly mobile. However, spread patterns can be influenced by a multitude of other factors including host body condition and infectiousness.
White‐nose syndrome (WNS) is a seasonal emerging infectious disease of bats, which is caused by the fungal pathogen Pseudogymnoascus destructans. Within‐site transmission of P. destructans primarily occurs over winter; however, the influence of bat mobility and infectiousness on the seasonal timing of pathogen spread to new populations is unknown. We combined data on host population dynamics and pathogen transmission from 22 bat communities to investigate the timing of pathogen arrival and the consequences of varying pathogen arrival times on disease impacts.
We found that midwinter arrival of the fungus predominated spread patterns, suggesting that bats were most likely to spread P. destructans when they are highly infectious, but have reduced mobility. In communities where P. destructans was detected in early winter, one species suffered higher fungal burdens and experienced more severe declines than at sites where the pathogen was detected later in the winter, suggesting that the timing of pathogen introduction had consequential effects for some bat communities. We also found evidence of source–sink population dynamics over winter, suggesting some movement among sites occurs during hibernation, even though bats at northern latitudes were thought to be fairly immobile during this period. Winter emergence behaviour symptomatic of white‐nose syndrome may further exacerbate these winter bat movements to uninfected areas.
Our results suggest that low infectiousness during host migration may have reduced the rate of expansion of this deadly pathogen, and that elevated infectiousness during winter plays a key role in seasonal transmission. Furthermore, our results highlight the importance of both accurate estimation of the timing of pathogen spread and the consequences of varying arrival times to prevent and mitigate the effects of infectious diseases.
Keywords: Geomyces destructans, infectious disease, migration, pathogen seasonality, Pseudogymnoascus destructans, white‐nose syndrome
This paper examines when a virulent fungal disease of bat, white‐nose syndrome, spreads to new locations. The authors find that the fungus spreads to more new locations during the winter even though bats are mostly sedentary during this time.

1. INTRODUCTION
The seasonality of pathogen spread is important for understanding, predicting and controlling disease outbreaks. Pathogens infecting highly mobile hosts often have rapid rates of spread (Altizer et al., 2006, 2011; Conner & Miller, 2004; Dalziel et al., 2013). However, key trade‐offs exist between mobility and disease, which can affect the likelihood that hosts spread pathogens during periods of high mobility (Kiesecker et al., 1999; Norris & Evans, 2000; Shakhar & Shakhar, 2015; Wendland et al., 2010). For example, migration can affect host body condition and immune status which could increase host susceptibility to infection (Altizer et al., 2011). However, once a highly mobile host becomes infected, behavioural responses, such as sickness behaviour, are likely to decrease mobility (Bouwman & Hawley, 2010; Van Gils et al., 2007). Pathogen spread should therefore be most likely to occur when hosts are both highly infectious and mobile, but given trade‐offs caused by sickness behaviour, peak mobility and infectiousness may not temporally align (Shakhar & Shakhar, 2015). Understanding the relative importance of these factors in the seasonal timing of pathogen spread is critical because varying pathogen arrival times can have consequential effects on hosts that shape the trajectory of epidemics (Dalziel et al., 2018).
White‐nose syndrome (WNS) is an annual seasonal epidemic that occurs in bat communities during winter (Hoyt et al., 2020, 2021; Langwig, Frick, et al., 2015). The fungus that causes WNS, Pseudogymnoascus destructans, is psychrophilic and grows only at cool temperatures (1–20°C; Verant et al. 2012) which restricts fungal replication and infection into bats' epidermal tissue to periods when bats are in torpor (e.g. cool their body temperatures <20°C; Langwig et al., 2016; Langwig, Frick, et al., 2015; Meteyer et al., 2009). White‐nose syndrome causes a physiological disruption to bats that increases arousals during hibernation which results in the premature depletion of stored fat (Lorch et al., 2011; Reeder et al., 2012; Verant et al., 2014; Warnecke et al., 2012, 2013). The spread of P. destructans has resulted in dramatic declines in bat communities across North America, and threatens several species with extinction (Frick et al., 2010, 2015; Langwig et al., 2012, 2016).
Seasonal changes in host behaviour and physiology paired with seasonal differences in pathogen growth may influence the timing of pathogen spread to new communities. The seasonal timing of P. destructans spread among sites is unknown but may occur during autumn when bats are highly mobile during mating and move among caves and mines (Cope & Humphrey, 1977; Davis & Hitchcock, 1965; Glover & Altringham, 2008; Thomas et al., 1979), which are known reservoirs of the fungus (Hoyt et al. 2020). Autumn swarm behaviour is an important source of gene flow when individuals move large distances between otherwise disconnected populations, and contact among mating individuals could lead to pathogen transmission (Arnold, 2007; Glover & Altringham, 2008; Rossiter et al., 2012; Veith et al., 2004; Wilder et al., 2015). However, the spread of WNS has occurred relatively slowly compared with other disease systems with similarly mobile hosts. For example, WNS and West Nile virus were both introduced to New York, USA, but in 5 years West Nile virus had reached California, whereas WNS spanned less than half of eastern North America (Kilpatrick et al., 2006, 2007). In addition, P. destructans is less prevalent on bats during autumn, and infected individuals typically have reduced infection levels (i.e. 100‐ to 1,000‐fold lower than in midwinter; Langwig, Frick, et al., 2015, Figure 1), suggesting that spread may occur outside of the highly mobile autumn period. If infectiousness is more important than mobility in P. destructans spread among sites, then pathogen introduction to new communities could occur during winter when fungal loads on bats are highest (Langwig, Frick, et al., 2015). However, most species are thought to be highly philopatric to sites and relatively sedentary during winter, particularly at northern latitudes where temperatures fall well below freezing (Caceres & Barclay, 2000; Davis, 1970; Davis & Hitchcock, 1965; Fenton & Barclay, 1980; Fujita & Kunz, 1984). Therefore, further investigation is needed to disentangle the importance of host mobility and infectiousness in the spread of P. destructans.
FIGURE 1.
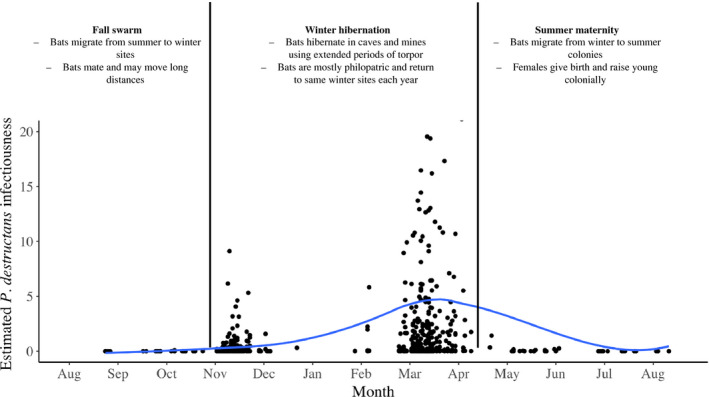
Mobility, migration and infectiousness of temperate bats infected with Pseudogymnoascus destructans. Data from Langwig, Frick, et al. (2015) and Hoyt et al. (2020) from Myotis lucifugus, Myotis septentrionalis, Eptesicus fuscus and Perimyotis subflavus. Each estimate of infectiousness (data point) is derived from the mean fungal loads of all positive individuals of a given species captured on a given date (‘average fungal loads’) multiplied by the fraction of individuals of that species positive for P. destructans (‘prevalence’), multiplied by the proportional abundance of the species in the sample. This estimate is commonly referred to as ‘propagule pressure’ in invasion ecology (Lockwood et al., 2005). For example, M. septentrionalis, which are rare (typically comprising <5% of captures), but have high fungal loads and high prevalence would receive a lower estimate of infectiousness than M. lucifugus, which is more common and has nearly equally high prevalence and loads. Blue line shows LOESS curve smoothing to data points to ease visualization. Temperate bats that use hibernation during winter are thought to have high mobility during fall swarm due to mating behaviour and migration between summer and winter grounds. These bats typically remain more sedentary during winter while they hibernate (Davis & Hitchcock, 1965; Thomas et al., 1979, 1990)
We investigated the timing of arrival of P. destructans into 22 hibernating bat communities in the Midwestern United States. We hypothesized that pathogen invasion would be more likely to occur during winter when bats are less mobile but highly infectious, resulting in the first detections of P. destructans at each site occurring in late, rather than early winter. Given the potential importance of seasonally varying arrival times on disease dynamics, we also assessed the influence of timing of P. destructans introductions on disease severity, hypothesizing that earlier introductions of P. destructans would result in higher infection prevalence, fungal loads and population impacts.
2. MATERIALS AND METHODS
2.1. Study sites and data collection
We studied patterns of P. destructans arrival at 22 sites in the Midwestern United States. Over a 5‐year period, we visited each site twice per winter and collected data on population dynamics and infection status of four hibernating bat species (Myotis lucifugus, Myotis septentrionalis, Eptesicus fuscus and Perimyotis subflavus). We used epidermal swab sampling to determine the presence and abundance of P. destructans on bats at two time points during hibernation [November (early hibernation) and March (late hibernation)] in each year. During each visit, we counted all bats present and identified bats to the species level. In addition, we installed HOBO U23 Pro v2 temperature (±0.2°C accuracy) and humidity (±3.5%–5%) loggers at one to four locations within a site to determine roost temperature and humidity.
2.2. Sample testing
We sampled bats using a standardized protocol (Langwig, Frick, et al., 2015) and stored swabs in RNAlater® for sample preservation until extraction. We tested samples for P. destructans DNA using real‐time PCR (Muller et al., 2013) and quantified fungal loads based on the cycle threshold (Ct) value to estimate a fungal load on each bat, with a cut‐off of 40 cycles. Quantification of serial dilutions of the DNA from 10 ng to 1,000 fg resulted in Ct scores ranging from 17.33 to 30.74 and a quantification relationship of Ct = −3.348 × log10 (P. destructans[ng]) + 22.049, r 2 = 0.986. We calculated prevalence as the proportion of bats of each species testing positive for P. destructans out of the number of individuals of that species sampled.
2.3. Statistical analysis
We used modified binomial power analyses to assess our ability to detect P. destructans arrival at each site where no positive samples were detected. We first calculated an expected early prevalence at each site where P. destructans was not detected in early hibernation based on a weighted mean prevalence. The weighted mean prevalence was calculated as the average prevalence of each species at sites where P. destructans was detected in early hibernation multiplied by the proportional abundance of a given species at each site (Table S1). We then calculated the probability of missing P. destructans at a site as the probability of getting all negatives in bats given the expected prevalence at a site multiplied by the probability of missing P. destructans in the unsampled bats (calculation shown in Appendix S1).
We investigated the effect of timing of P. destructans arrival on late winter prevalence by fitting a GLMM with a binomial distribution and a logit link. We included fixed effects for the timing of P. destructans detection (early or late hibernation) interacting with the effect of species, and included site as a random effect. We also examined the effect of timing of P. destructans detection on fungal loads using a linear mixed model with species and timing of detection as interacting fixed effects, and site as a random effect. Lastly, we assessed the effect of the timing of P. destructans introduction on log10 bat population growth rates, calculated as the annual change in late winter counts at each site, using linear mixed models with species interacting with timing of detection as fixed effects and site as a random effect. We explored the inclusion of autumn prevalence as a continuous variable (0 for all sites and species where P. destructans was not detected, prevalence in each species ranged between 0 and 0.25 for sites where P. destructans was detected during autumn) and the results were qualitatively similar to those shown, so we present the discretization (early or late winter P. destructans detection) for simplicity of visualization and interpretation.
Lastly, we investigated evidence of overwinter movements of bats among sites. At each site, we calculated our response variable, overwinter as the proportional change in the number of bats of each species at a site by dividing the late hibernation count by the early hibernation count. We then used a linear mixed model to assess the relationship between log10(overwinter ) and log10(early hibernation colony size) with an additive or interactive effect of species and site as a random effect.
3. RESULTS
During early hibernation, we sampled 706 total bats from 22 sites in the year that P. destructans was first detected (site M ± SE = 67.4 ± 1.79)(Table S1). We first detected P. destructans in early hibernation at 32% of sites (7/22), whereas in 68% of sites (15/22), the detection of P. destructans first occurred during our late hibernation visit (Figure 2). A total of 468 bats (site M ± SE = 31.1 ± 1.25) were sampled from sites that were negative for P. destructans during early hibernation and became positive later in the same winter. After accounting for the relatively high proportion of bats sampled in each site (an average of 38% of the total bats at each site) and variable estimated ‘true’ prevalence among sites (weighted mean prevalence of the species sampled from sites where P. destructans was detected in early hibernation: M = 0.07, range = 0.05, 0.10), the mean probability of missing P. destructans at each negative site was 0.063 (median = 0.039, range: 0.005–0.3266), suggesting fairly good power to detect P. destructans presence.
FIGURE 2.
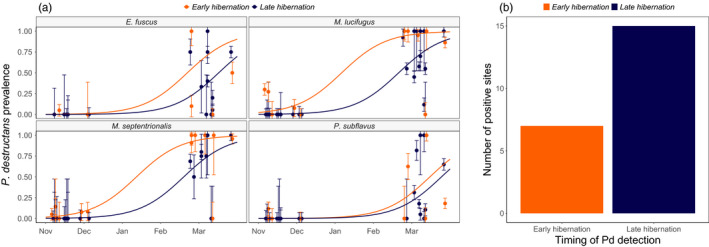
(a) Change in overwinter prevalence of Pseudogymnoascus destructans in the year that P. destructans was first detected at a site. Lines show fits of a GLMM with a binomial distribution with timing of first detection interacting with species and an additive effect of date with site as a random effect. (b) Twice as many sites had the first detection of P. destructans in late hibernation as in early hibernation
Across sites where P. destructans was first detected during early hibernation, we were more likely to detect P. destructans on M. lucifugus, M. septentrionalis and E. fuscus than on P. subflavus (P. subflavus intercept: −1.82 ± 0.66, M. lucifugus coeff: 1.79 ± 0.38 p < 0.0001, E. fuscus coeff: 1.40 ± 0.51, p = 0.006, M. septentrionalis coef: 1.44 ± 0.39, p = 0.0002, Appendix S1). In sites where P. destructans was first detected in late hibernation, we were also less likely to detect P. destructans on P. subflavus than on any other species (P. subflavus intercept: −1.69 ± 0.31, M. lucifugus coeff: 0.94 ± 0.22 p < 0.0001, E. fuscus coeff: 0.66 ± 0.30, p = 0.0264, M. septentrionalis coeff: 0.84 ± 0.26, p = 0.0014, Appendix S1).
The timing of P. destructans introduction influenced disease dynamics and population impacts for some, but not all, bat species (Figure 3). For M. septentrionalis, prevalence of P. destructans, fungal loads and population impacts during late winter were higher at sites where P. destructans was first detected in early hibernation (one‐tailed p‐values = 0.032, 0.005, 0.0004, respectively, Appendix S1). For M. lucifugus, prevalence was also higher in sites where P. destructans was first detected in early hibernation (one‐tailed p = 0.04, Appendix S1), although the effect on fungal loads and population impacts was less clear (Appendix S1). There was no clear effect of the timing of P. destructans introduction on prevalence, fungal loads or population impacts for either E. fuscus or P. subflavus, possibly because these species had lower prevalence and fungal loads than M. septentrionalis and M. lucifugus in the first year of detection (Figure 3; Appendix S1).
FIGURE 3.
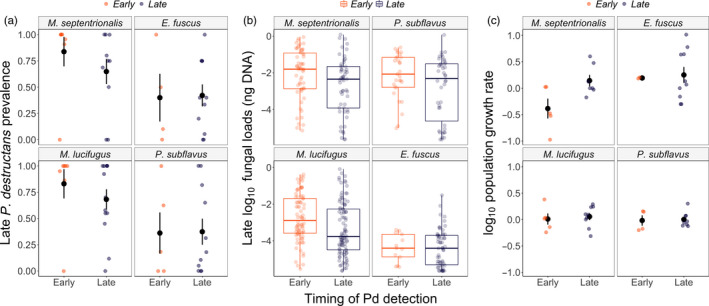
The timing of Pseudogymnoascus destructans introduction (early or late hibernation) influences late winter infection prevalence ±SE (a), late winter fungal loads (ng DNA) (b) and annual population impacts ±SE (from late winter in the year that P. destructans arrives to the subsequent winter) (c), in some species. In sites where P. destructans was first detected during late hibernation, Myotis septentrionalis had significantly higher infection prevalence, fungal loads and population impacts by the end of winter in the first year of WNS
We found that prior to the arrival of WNS, there were detectable changes in population counts between early and late hibernation (Figure 4; Appendix S1). Across species, as colony size decreased in early hibernation, immigration increased (early hibernation colony size coeff: −0.15 ± 0.06, t = −2.66, Appendix S1 5.1–5.4) such that smaller colonies had proportionally more immigrants than larger colonies, indicative of a general spreading out of bats across sites during the hibernation season. We found no support for an interactive model over an additive model (AIC = −9.63), suggesting no clear indication that the slope of this effect differed among species. Our findings were similar whether we used a log10 transformation of the response variable or modelled population growth rates using a linear mixed model with a Gamma error distribution (Appendix S1 5.4). The changes in overwinter counts occurred despite frigid minimum temperatures that were consistently below 5°C throughout this period (Figure S1), suggesting that some movement among hibernacula continues to occur during winter when bats are most infected with P. destructans.
FIGURE 4.
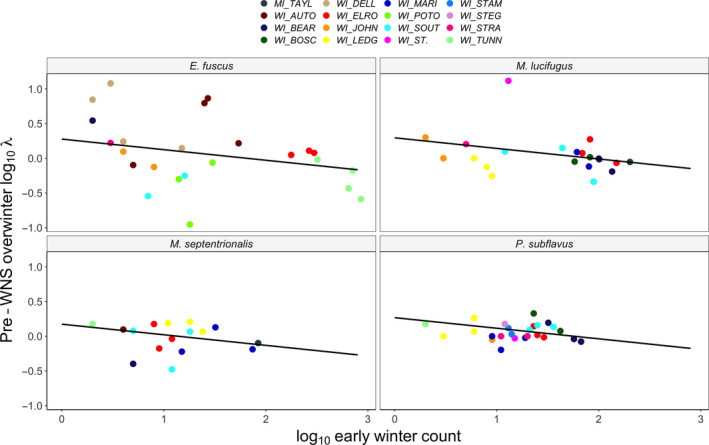
Pre‐WNS immigration rates show dynamic populations during winter. Panels show bat species and colours indicate different bat populations. Lines show the fitted relationship between log10 early winter counts of bats and the rate of change in the same population over winter. Model diagnostics are shown in Figure S2 and Appendix S1. Despite cold (<5°C) temperatures, there was detectable immigration and emigration among hibernation sites, consistent with bats moving from larger to smaller colonies over winter
Lastly, we explored the effect of multiple covariates on the probability that P. destructans arrived at a site outside of or during the hibernation season, including abiotic (vapour pressure deficit and temperature) and biotic (abundance of certain species, species richness). We found no clear effects that any of these variables modified the probability that P. destructans was first detected at a site during early hibernation (Figure S3; Appendix S1).
4. DISCUSSION
Our results indicate that host fungal burdens were important in determining the timing of pathogen spread. We found that P. destructans was more likely to be introduced to sites during winter hibernation than during autumn, suggesting that elevated infectiousness overwinter outweighed the increase in activity and long‐distance movements that occur during autumn. Bats were previously thought to be restricted in their movement among sites during hibernation due to extremely cold temperatures (<−5°C; Figure S1) and a lack of prey availability. However, we found evidence that limited movement may be occurring among hibernacula during the winter, and given increased fungal loads and prevalence on bats, provide a higher probability of successful invasion to new sites. The lower fungal loads and prevalence on bats during autumn when they are highly mobile have likely decreased the rate of geographic expansion of the fungus as the majority of spread occurs during the hibernation period. As P. destructans moves into less‐connected western bat populations (Lorch et al., 2016; Wilder et al., 2015), spread could be further dampened by larger distances among colonies (Weller et al., 2018; Wilder et al., 2015).
Although the absolute absence of a pathogen is difficult to confirm, large sample sizes paired with our finding that the timing of P. destructans introduction influenced disease dynamics, suggest that we did not simply fail to detect P. destructans using our qPCR at most sites during early winter. Our results also demonstrate that the timing of P. destructans introduction was more important for some species than others. Early introduction of P. destructans resulted in higher mortality in M. septentrionalis populations, but not M. lucifugus. Previous laboratory studies have shown that infection duration prior to death was 90–120 days for M. lucifugus (McGuire et al., 2016; Warnecke et al., 2012). The absence of clear mortality in M. lucifugus populations while M. septentrionalis suffered from higher impacts suggests that M. septentrionalis may have a shorter time from infection until death. Notably, infection in P. subflavus and E. fuscus were lower in the first year of WNS than Myotis spp., likely due to low environmental contamination (Hoyt et al., 2020; Langwig, Hoyt, et al., 2015) and fewer contacts over the hibernation period (Hoyt et al., 2018).
We found evidence of some overwinter movement among hibernacula prior to the arrival of WNS, with a general trend of bats spreading out from larger to smaller colony size sites. Other studies have found that some species (especially the larger E. fuscus) can move in very cold temperatures (Klüg‐Baerwald et al., 2016), although winter flight of Myotis spp. is thought to be a relatively rare occurrence (Davis & Hitchcock, 1965). We cannot exclude the possibility that bats in larger sites became more difficult to detect in late winter while bats in smaller sites became more apparent. However, sites were selected for inclusion in this study for their relative simplicity in searching and ability to census all bats present (e.g. they did not have sections that we were unable to access and many were square tunnels with no obvious hiding places). As a result, we think that failed detections are unlikely to fully explain this relationship. In addition, higher overwinter mortality is unlikely to be a potential explanation as there is no evidence to suggest elevated mortality in sites with larger population sizes prior to the arrival of WNS (Langwig et al., 2012). Interestingly, symptoms of WNS include mid‐hibernation emergence onto the winter landscape (Bernard & McCracken, 2017; Carr et al., 2014). This sickness behaviour may increase movements between hibernacula and the spread of P. destructans.
We found no clear effects of any covariate in explaining differences in timing of arrival. However, environmental conditions of hibernacula may be important determinants of P. destructans establishment into new communities outside of the hibernation season, as is suggested by the directionality and magnitude of a trend where warmer and wetter hibernacula tended to have a higher probability of autumn introduction. While bats with low fungal loads are likely moving among uninfected hibernacula during autumn, vapour pressure deficit and temperature may be important in determining whether P. destructans can successfully establish in a population (Lilley et al., 2018) and subsequent population persistence (Hopkins et al., 2021). Given the limited number of sites where P. destructans arrived outside of the hibernation season, it was difficult to ascertain the importance of a number of factors that have been implicated by other studies (Maher et al., 2012; Wilder et al., 2011, 2015). For example, species‐specific differences in sociality and transmission (Hoyt et al., 2018) may be important in determining whether a lightly infected bat could introduce P. destructans during fall, although we found no clear effects of species abundance or composition on the timing of first detection of P. destructans. Additional research is needed to determine which species and site‐specific differences may influence the timing of pathogen spread.
Importantly, many studies rely on disease stage (e.g. invading, epidemic or endemic) to draw inferences about geographic disease risk, population declines and community persistence (Hoyt et al., 2021). Given the high probability of P. destructans introduction to new sites during winter, early and midwinter surveys could miss the introduction of P. destructans, and thus falsely conclude pathogen absence during the first year of arrival. Our results suggest that the arrival time of P. destructans can substantially influence dynamics, and therefore could be responsible for unexplained variance in transmission, impacts and structure of remnant populations when sites are grouped by disease stage.
Our ability to predict the timing and patterns of pathogen spread are fundamental to the prevention and control of infectious disease outbreaks. The differential timing of initial pathogen arrival can have important effects on disease dynamics and lasting impacts to populations. This study suggests that host infectiousness is an important factor in determining successful pathogen spread and that incorrectly attributing pathogen spread to periods of high mobility would have masked the true underlying causes of among‐site variation in transmission and population impacts. Future studies examining the spatial and seasonal patterns of pathogen movement should consider the multitude of factors that might influence spread patterns.
AUTHORS' CONTRIBUTIONS
K.E.L. analysed the data and wrote the original draft; K.E.L., A.M.K. and J.R.H. revised the manuscript with input from all the authors; K.L.P. and J.T.F. tested the samples and K.E.L., J.P.W., H.M.K., J.A.R., J.E.D., W.H.S., A.M.K. and J.R.H. collected the data.
Supporting information
Supplementary Material
Appendix
ACKNOWLEDGEMENTS
The authors thank Steffany Yamada, MI DNR, Eric McMaster and the many landowners for site access. The research was funded by NSF grants DEB‐1115895, DEB‐1336290, DEB‐1911853 and the USFWS (F17AP00591). The authors have no competing interests to declare.
Langwig KE, White JP, Parise KL, et al. Mobility and infectiousness in the spatial spread of an emerging fungal pathogen. J Anim Ecol. 2021;90:1134–1141. 10.1111/1365-2656.13439
[The copyright line for this article was changed on 29 April 2021 after original online publication.]
A. Marm Kilpatrick and Joseph R. Hoyt contributed equally.
Handling Editor Isabella Cattadori
DATA AVAILABILITY STATEMENT
The datasets generated during this study are available in the VTechData repository https://doi.org/10.7294/WSDX‐AJ44 (Langwig, 2021).
REFERENCES
- Altizer, S. , Bartel, R. , & Han, B. A. (2011). Animal migration and infectious disease risk. Science, 331, 296–302. 10.1126/science.1194694 [DOI] [PubMed] [Google Scholar]
- Altizer, S. , Dobson, A. , Hosseini, P. , Hudson, P. , Pascual, M. , & Rohani, P. (2006). Seasonality and the dynamics of infectious diseases. Ecology Letters, 9, 467–484. 10.1111/j.1461-0248.2005.00879.x [DOI] [PubMed] [Google Scholar]
- Arnold, B. D. (2007). Population structure and sex‐biased dispersal in the forest dwelling vespertilionid bat, Myotis septentrionalis . The American Midland Naturalist, 157, 374–384. [Google Scholar]
- Bernard, R. F. , & McCracken, G. F. (2017). Winter behavior of bats and the progression of white‐nose syndrome in the southeastern United States. Ecology and Evolution, 7, 1487–1496. 10.1002/ece3.2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, K. M. , & Hawley, D. M. (2010). Sickness behaviour acting as an evolutionary trap? Male house finches preferentially feed near diseased conspecifics. Biology Letters, 6, 462–465. 10.1098/rsbl.2010.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, M. C. , & Barclay, R. M. R. (2000). Myotis septentrionalis . Mammalian Species, 634, 1–4. [Google Scholar]
- Carr, J. A. , Bernard, R. F. , & Stiver, W. H. (2014). Unusual bat behavior during winter in Great Smoky Mountains National Park. Southeastern Naturalist, 13, N18–N21. 10.1656/058.013.0211 [DOI] [Google Scholar]
- Conner, M. M. , & Miller, M. W. (2004). Movement patterns and spatial epidemiology of a prion disease in mule deer population units. Ecological Applications, 14, 1870–1881. 10.1890/03-5309 [DOI] [Google Scholar]
- Cope, J. B. , & Humphrey, S. R. (1977). Spring and autumn swarming behavior in Indiana bat, myotis‐sodalis. Journal of Mammalogy, 58, 93–95. [Google Scholar]
- Dalziel, B. D. , Kissler, S. , Gog, J. R. , Viboud, C. , Bjørnstad, O. N. , Metcalf, C. J. E. , & Grenfell, B. T. (2018). Urbanization and humidity shape the intensity of influenza epidemics in US cities. Science, 362, 75–79. 10.1126/science.aat6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel, B. D. , Pourbohloul, B. , & Ellner, S. P. (2013). Human mobility patterns predict divergent epidemic dynamics among cities. Proceedings of the Royal Society B: Biological Sciences, 280, 20130763. 10.1098/rspb.2013.0763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, W. (1970). Hibernation: Ecology and physiological ecology. Biology of Bats, 1, 265–300. [Google Scholar]
- Davis, W. H. , & Hitchcock, H. B. (1965). Biology and migration of the bat, Myotis lucifugus, in New England. Journal of Mammalogy, 46, 296–313. 10.2307/1377850 [DOI] [Google Scholar]
- Fenton, M. B. , & Barclay, R. M. R. (1980). Myotis lucifugus . Mammalian Species, 1–8. 10.2307/3503792 [DOI] [Google Scholar]
- Frick, W. F. , Pollock, J. F. , Hicks, A. C. , Langwig, K. E. , Reynolds, D. S. , Turner, G. G. , Butchkoski, C. M. , & Kunz, T. H. (2010). An emerging disease causes regional population collapse of a common North American bat species. Science, 329, 679–682. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- Frick, W. F. , Puechmaille, S. J. , Hoyt, J. R. , Nickel, B. A. , Langwig, K. E. , Foster, J. T. , Barlow, K. E. , Bartonička, T. , Feller, D. , Haarsma, A. J. , Herzog, C. , Horacek, I. , van der Kooij, J. , Mulkens, B. , Petrov, B. , Reynolds, R. , Rodrigues, L. , Stihler, C. W. , Turner, G. G. , & Kilpatrick, A. M. (2015). Disease alters macroecological patterns of North American bats. Global Ecology and Biogeography, 24, 741–749. 10.1111/geb.12290 [DOI] [Google Scholar]
- Fujita, M. S. , & Kunz, T. H. (1984). Pipistrellus subflavus . Mammalian Species, 1–6. 10.2307/3504021 [DOI] [Google Scholar]
- Glover, A. M. , & Altringham, J. D. (2008). Cave selection and use by swarming bat species. Biological Conservation, 141, 1493–1504. 10.1016/j.biocon.2008.03.012 [DOI] [Google Scholar]
- Hopkins, S. R. , Hoyt, J. R. , White, J. P. , Kaarakka, H. M. , Redell, J. A. , DePue, J. E. , Scullon, W. H. , Kilpatrick, A. M. , & Langwig, K. E. (2021). Continued preference for suboptimal habitat reduces bat survival with white‐nose syndrome. Nature Communications, 12, 1–9. 10.1038/s41467-020-20416-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, J. R. , Kilpatrick, A. M. , & Langwig, K. E. (2021). Ecology and impacts of white‐nose syndrome on bats. Nature Reviews Microbiology. 10.1038/s41579-020-00493-5 [DOI] [PubMed] [Google Scholar]
- Hoyt, J. R. , Langwig, K. E. , Sun, K. , Parise, K. L. , Li, A. , Wang, Y. , Huang, X. , Worledge, L. , Miller, H. , White, J. P. , Kaarakka, H. M. , Redell, J. A. , Görföl, T. , Boldogh, S. A. , Fukui, D. , Sakuyama, M. , Yachimori, S. , Sato, A. , Dalannast, M. , … Kilpatrick, A. M. (2020). Environmental reservoir dynamics predict global infection patterns and population impacts for the fungal disease white‐nose syndrome. Proceedings of the National Academy of Sciences of the United States of America, 117, 7255. 10.1073/pnas.1914794117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, J. R. , Langwig, K. E. , White, J. P. , Kaarakka, H. , Redell, J. , Kurta, A. , DePue, J. , Scullon, W. H. , Parise, K. L. , Foster, J. T. , Frick, W. F. , & Kilpatrick, A. M. (2018). Cryptic connections illuminate pathogen transmission within community networks. Nature. 10.1038/s41586-018-0720-z [DOI] [PubMed] [Google Scholar]
- Kiesecker, J. M. , Skelly, D. K. , Beard, K. H. , & Preisser, E. (1999). Behavioral reduction of infection risk. Proceedings of the National Academy of Sciences of the United States of America, 96, 9165–9168. 10.1073/pnas.96.16.9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, A. M. , Chmura, A. A. , Gibbons, D. W. , Fleischer, R. C. , Marra, P. P. , & Daszak, P. (2006). Predicting the global spread of H5N1 avian influenza. Proceedings of the National Academy of Sciences of the United States of America, 103, 19368–19373. 10.1073/pnas.0609227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, A. M. , LaDeau, S. L. , & Marra, P. P. (2007). Ecology of west nile virus transmission and its impact on birds in the western hemisphere. The Auk, 124, 1121–1136. 10.1093/auk/124.4.1121 [DOI] [Google Scholar]
- Klüg‐Baerwald, B. J. , Gower, L. E. , Lausen, C. , & Brigham, R. (2016). Environmental correlates and energetics of winter flight by bats in Southern Alberta, Canada. Canadian Journal of Zoology, 94, 829–836. 10.1139/cjz-2016-0055 [DOI] [Google Scholar]
- Langwig, K. E. (2021). Data from: Mobility and infectiousness in the spatial spread of an emerging pathogen. University Libraries, Virginia Tech. 10.7294/WSDX-AJ44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langwig, K. E. , Frick, W. F. , Bried, J. T. , Hicks, A. C. , Kunz, T. H. , & Marm Kilpatrick, A. (2012). Sociality, density‐dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white‐nose syndrome. Ecology Letters, 15, 1050–1057. 10.1111/j.1461-0248.2012.01829.x [DOI] [PubMed] [Google Scholar]
- Langwig, K. E. , Frick, W. F. , Hoyt, J. R. , Parise, K. L. , Drees, K. P. , Kunz, T. H. , Foster, J. T. , & Kilpatrick, A. M. (2016). Drivers of variation in species impacts for a multi‐host fungal disease of bats. Philosophical Transactions of the Royal Society B Biological Sciences, 371, 20150456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langwig, K. E. , Frick, W. F. , Reynolds, R. , Parise, K. L. , Drees, K. P. , Hoyt, J. R. , Cheng, T. L. , Kunz, T. H. , Foster, J. T. , & Kilpatrick, A. M. (2015). Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white‐nose syndrome. Proceedings of the Royal Society B: Biological Sciences, 282(1799). 10.1098/rspb.2014.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langwig, K. E. , Hoyt, J. R. , Parise, K. L. , Kath, J. , Kirk, D. , Frick, W. F. , Foster, J. T. , & Kilpatrick, A. M. (2015) Invasion dynamics of white‐nose syndrome white‐nose syndrome fungus midwestern United States, 2012–2014. Emerging Infectious Diseases, 21, 1023–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, T. M. , Anttila, J. , & Ruokolainen, L. (2018). Landscape structure and ecology influence the spread of a bat fungal disease. Functional Ecology, 32, 2483–2496. 10.1111/1365-2435.13183 [DOI] [Google Scholar]
- Lockwood, J. L. , Cassey, P. , & Blackburn, T. (2005). The role of propagule pressure in explaining species invasions. Trends in Ecology & Evolution, 20, 223–228. 10.1016/j.tree.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Lorch, J. M. , Meteyer, C. U. , Behr, M. J. , Boyles, J. G. , Cryan, P. M. , Hicks, A. C. , Ballmann, A. E. , Coleman, J. T. H. , Redell, D. N. , Reeder, D. M. , & Blehert, D. S. (2011). Experimental infection of bats with Geomyces destructans causes white‐nose syndrome. Nature, 480, 376–378. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- Lorch, J. M. , Palmer, J. M. , Lindner, D. L. , Ballmann, A. E. , George, K. G. , Griffin, K. , Knowles, S. , Huckabee, J. R. , Haman, K. H. , Anderson, C. D. , Becker, P. A. , Buchanan, J. B. , Foster, J. T. , & Blehert, D. S. (2016). First detection of bat white‐nose syndrome in Western North America. mSphere, 1(4). 10.1128/mSphere.00148-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher, S. P. , Kramer, A. M. , Pulliam, J. T. , Zokan, M. A. , Bowden, S. E. , Barton, H. D. , Magori, K. , & Drake, J. M. (2012). Spread of white‐nose syndrome on a network regulated by geography and climate. Nature Communications, 3. 10.1038/ncomms2301 [DOI] [PubMed] [Google Scholar]
- McGuire, L. P. , Turner, J. M. , Warnecke, L. , McGregor, G. , Bollinger, T. K. , Misra, V. , Foster, J. T. , Frick, W. F. , Kilpatrick, A. M. , & Willis, C. K. R. (2016). White‐nose syndrome disease severity and a comparison of diagnostic methods. EcoHealth, 13, 60–71. 10.1007/s10393-016-1107-y [DOI] [PubMed] [Google Scholar]
- Meteyer, C. U. , Buckles, E. L. , Blehert, D. S. , Hicks, A. C. , Green, D. E. , Shearn‐Bochsler, V. , Thomas, N. J. , Gargas, A. , & Behr, M. J. (2009). Histopathologic criteria to confirm white‐nose syndrome in bats. Journal of Veterinary Diagnostic Investigation, 21, 411–414. 10.1177/104063870902100401 [DOI] [PubMed] [Google Scholar]
- Muller, L. K. , Lorch, J. M. , Lindner, D. L. , O'Connor, M. , Gargas, A. , & Blehert, D. S. (2013). Bat white‐nose syndrome: A real‐time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans . Mycologia, 105, 253–259. [DOI] [PubMed] [Google Scholar]
- Norris, K. , & Evans, M. R. (2000). Ecological immunology: Life history trade‐offs and immune defense in birds. Behavioral Ecology, 11, 19–26. 10.1093/beheco/11.1.19 [DOI] [Google Scholar]
- Reeder, D. M. , Frank, C. L. , Turner, G. G. , Meteyer, C. U. , Kurta, A. , Britzke, E. R. , Vodzak, M. E. , Darling, S. R. , Stihler, C. W. , Hicks, A. C. , Jacob, R. , Grieneisen, L. E. , Brownlee, S. A. , Muller, L. K. , & Blehert, D. S. (2012). Frequent arousal from hibernation linked to severity of infection and mortality in bats with white‐nose syndrome. PLoS ONE, 7. 10.1371/journal.pone.0038920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter, S. J. , Zubaid, A. , Mohd‐adnan, A. , Struebig, M. J. , Kunz, T. H. , Gopal, S. , Petit, E. J. , & Kingston, T. (2012). Social organization and genetic structure: Insights from codistributed bat populations. Molecular Ecology, 21, 647–661. 10.1111/j.1365-294X.2011.05391.x [DOI] [PubMed] [Google Scholar]
- Shakhar, K. , & Shakhar, G. (2015). Why do we feel sick when infected—Can altruism play a role? PLoS Biology, 13, e1002276. 10.1371/journal.pbio.1002276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D. , Dorais, M. , & Bergeron, J. (1990). Winter energy budgets and cost of arousals for hibernating little brown bats, Myotis lucifugus . Journal of Mammalogy, 71, 475–479. 10.2307/1381967 [DOI] [Google Scholar]
- Thomas, D. W. , Fenton, M. B. , & Barclay, R. M. R. (1979). Social behavior of the Little brown bat, Myotis lucifugus. I. mating behavior. Behavioral Ecology and Sociobiology, 6, 129–136. 10.1007/BF00292559 [DOI] [Google Scholar]
- Van Gils, J. A. , Munster, V. J. , Radersma, R. , Liefhebber, D. , Fouchier, R. A. , & Klaassen, M. (2007). Hampered foraging and migratory performance in swans infected with low‐pathogenic avian influenza A virus. PLoS ONE, 2, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith, M. , Beer, N. , Kiefer, A. , Johannesen, J. , & Seitz, A. (2004). The role of swarming sites for maintaining gene flow in the brown long‐eared bat (Plecotus auritus). Heredity, 93, 342. 10.1038/sj.hdy.6800509 [DOI] [PubMed] [Google Scholar]
- Verant, M. L. , Boyles, J. G. , Waldrep, W. , Wibbelt, G. , & Blehert, D. S. (2012). Temperature‐dependent growth of Geomyces destructans, the fungus that causes bat white‐nose syndrome. PLoS ONE, 7. 10.1371/journal.pone.0046280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verant, M. L. , Carol, M. U. , Speakman, J. R. , Cryan, P. M. , Lorch, J. M. , & Blehert, D. S. (2014). White‐nose syndrome initiates a cascade of physiologic disturbances in the hibernating bat host. BMC Physiology, 14, 10. 10.1186/s12899-014-0010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke, L. , Turner, J. M. , Bollinger, T. K. , Lorch, J. M. , Misra, V. , Cryan, P. M. , Wibbelt, G. , Blehert, D. S. , & Willis, C. K. R. (2012). Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white‐nose syndrome. Proceedings of the National Academy of Sciences of the United States of America, 109, 6999–7003. 10.1073/pnas.1200374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke, L. , Turner, J. M. , Bollinger, T. K. , Misra, V. , Cryan, P. M. , Blehert, D. S. , Wibbelt, G. , & Willis, C. K. R. (2013). Pathophysiology of white‐nose syndrome in bats: A mechanistic model linking wing damage to mortality. Biology Letters, 9. 10.1098/rsbl.2013.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, T. J. , Rodhouse, T. J. , Neubaum, D. J. , Ormsbee, P. C. , Dixon, R. D. , Popp, D. L. , Williams, J. A. , Osborn, S. D. , Rogers, B. W. , Beard, L. O. , McIntire, A. M. , Hersey, K. A. , Tobin, A. , Bjornlie, N. L. , Foote, J. , Bachen, D. A. , Maxell, B. A. , Morrison, M. L. , Thomas, S. C. , … Navo, K. W. (2018). A review of bat hibernacula across the western United States: Implications for white‐nose syndrome surveillance and management. PLoS ONE, 13. 10.1371/journal.pone.0205647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, L. D. , Wooding, J. , White, C. L. , Demcovitz, D. , Littell, R. , Berish, J. D. , Ozgul, A. , Oli, M. K. , Klein, P. A. , Christman, M. C. , & Brown, M. B. (2010). Social behavior drives the dynamics of respiratory disease in threatened tortoises. Ecology, 91, 1257–1262. 10.1890/09-1414.1 [DOI] [PubMed] [Google Scholar]
- Wilder, A. P. , Frick, W. F. , Langwig, K. E. , & Kunz, T. H. (2011). Risk factors associated with mortality from white‐nose syndrome among hibernating bat colonies. Biology Letters, 7, 950–953. 10.1098/rsbl.2011.0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, A. P. , Kunz, T. H. , & Sorenson, M. D. (2015). Population genetic structure of a common host predicts the spread of white‐nose syndrome, an emerging infectious disease in bats. Molecular Ecology, 24, 5495–5506. 10.1111/mec.13396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Appendix
Data Availability Statement
The datasets generated during this study are available in the VTechData repository https://doi.org/10.7294/WSDX‐AJ44 (Langwig, 2021).


