Summary
Floral pigmentation patterning is important for pollinator attraction as well as aesthetic appeal. Patterning of anthocyanin accumulation is frequently associated with variation in activity of the Myb, bHLH and WDR transcription factor complex (MBW) that regulates anthocyanin biosynthesis.
Investigation of two classic mutants in Antirrhinum majus, mutabilis and incolorata I, showed they affect a gene encoding a bHLH protein belonging to subclade bHLH‐2. The previously characterised gene, Delila, which encodes a bHLH‐1 protein, has a bicoloured mutant phenotype, with residual lobe‐specific pigmentation conferred by Incolorata I.
Both Incolorata I and Delila induce expression of the anthocyanin biosynthetic gene DFR. Rosea 1 (Myb) and WDR1 proteins compete for interaction with Delila, but interact positively to promote Incolorata I activity. Delila positively regulates Incolorata I and WDR1 expression. Hierarchical regulation can explain the bicoloured patterning of delila mutants, through effects on both regulatory gene expression and the activity of promoters of biosynthetic genes like DFR that mediate MBW regulation.
bHLH‐1 and bHLH‐2 proteins contribute to establishing patterns of pigment distribution in A. majus flowers in two ways: through functional redundancy in regulating anthocyanin biosynthetic gene expression, and through differences between the proteins in their ability to regulate genes encoding transcription factors.
Keywords: anthocyanin, Antirrhinum majus, floral pigment patterning, MBW complex, transcriptional regulation
Introduction
The diversity of coloration and patterning of animal‐pollinated flowers is truly remarkable. Complex patterns serve as nectar guides, attracting pollinators and directing them towards pollen and nectar rewards. Most of our understanding of pigmentation patterning comes from the study of regulation of anthocyanin production, which provides red/purple/blue colours to plants. Patterns of anthocyanin production across petals arise primarily through transcriptional regulation of the anthocyanin biosynthetic genes (Schwinn et al., 2006), which are regulated directly by a transcriptional activation complex comprised of R2R3‐Myb, bHLH and WDR proteins; the MBW complex (Baudry et al., 2004; Koes et al., 2005; Ramsey & Glover, 2005; Gonzalez et al., 2008). The R2R3‐Myb proteins are particularly important for pattern formation because they are encoded by gene families with members that are expressed differentially in response to a variety of developmental and environmental cues (Schwinn et al., 2006; Hoballah et al., 2007; Albert et al., 2011; Yuan et al., 2014; Bombarely et al., 2016; Esfeld et al., 2018). The bHLH transcription factors are also encoded by small gene families and they too have the potential to contribute to pigment patterning (Dellaporta et al., 1988; Chandler et al., 1989; Ludwig et al., 1989; Goodrich et al., 1992; Hellens et al., 2010).
MBW complexes also regulate proanthocyanidin biosynthesis (Baudry et al., 2004; Bogs et al., 2007; Terrier et al., 2009; Liu et al., 2014), vacuolar hyperacidification (Quattrocchio et al., 2006; Butelli et al., 2019), and trichome and root hair development in Brassica species (Payne et al., 2000; Zhao et al., 2008) with each process controlled by a specific subgroup of Myb proteins (Zhang et al., 2003; Xu et al., 2014; Zhang & Hulskamp, 2019). Most plants have at least two bHLH transcription factors that operate within MBW complexes, belonging to two distinct clades within subgroup IIIf of plant bHLH proteins (Heim et al., 2003; Feller et al., 2011). These are described as bHLH‐1 (represented by ZmR/ZmLc, AtGL3, AtEGL3, AtMYC1, PhJAF13, AmDel) and bHLH‐2 (represented by ZmIn, AtTT8, PhAN1, CsNoemi) (Albert et al., 2014). In many groups of plants the activity of bHLH‐2 genes is essential for anthocyanin biosynthesis (Spelt et al., 2000, 2002; Park et al., 2004; Hellens et al., 2010; Butelli et al., 2019; Strazzer et al., 2019), In other plants, including Arabidopsis (Gonzalez et al., 2008; Feyissa et al., 2009) and Antirrhinum (Goodrich et al., 1992), the bHLH‐1 proteins act also in controlling anthocyanin biosynthesis directly (Zhang et al., 2003; Zhang & Hulskamp, 2019). bHLH‐2 proteins appear to be essential for controlling proanthocyanidin biosynthesis in Arabidopsis (Nesi et al., 2000), petunia (Spelt et al., 2002), pea (Hellens et al., 2010), morning glory (Park et al., 2007), medicago (Li et al., 2016) and citrus (Butelli et al., 2019), and vacuolar acidification in petunia (Spelt et al., 2002) and citrus (Butelli et al., 2019; Strazzer et al., 2019). It is not yet clear whether bHLH‐1 proteins that regulate anthocyanin biosynthesis also contribute to regulating these other pathways. The existence of genes encoding both clades of bHLH proteins in monocots and dicots suggests that there has been selection to maintain two types of bHLH protein which may work by distinct mechanisms or have distinct functions (Pesch et al., 2015; Zhang et al., 2019).
Antirrhinum majus – a classic model for understanding floral pigmentation
In bicoloured flowers, colour patterning differs between individual petals or zones within petals and is common in zygomorphic flowers such as legumes (banner, wings, keel), iris (standards, falls), violas, Aquilegia (blades, spurs) and orchids (tepals, labellum). However, suitable models with bicoloured floral patterns that are also amenable to genetic analyses have not been analysed extensively. Wild‐type Antirrhinum majus has bilaterally symmetrical flowers consisting of self‐coloured fused petals in the corolla tube which separate into two hind petals, two lateral petals and a single ventral petal comprising the corolla lobes. However, mutants of the bHLH‐1 gene Delila have bicoloured flowers (Goodrich et al., 1992), providing a model genetic system for investigating bicolour patterning. Three R2R3Myb proteins, Rosea 1 (Ros 1), Rosea 2 and Venosa, of which Ros 1 has the strongest activity, control flower pigmentation in A. majus (Schwinn et al., 2006). Here we report the identification and functional characterisation of Incolorata I, a gene encoding a bHLH‐2 protein in A. majus, and relate this to bicoloured patterning of its flowers.
Materials and Methods
Plant material
The del mutant, line JI:8, is closely related to the wild‐type line, JI:7. The incolorata I mutant, line Jl:569, was obtained from Professor Linnert, Frei University of Berlin in 1984 and outcrossed to JI:7. Segregation in the F2 allowed the selection of ivory inc I : del homozygotes. The mutabilis mutant was identified from several individuals segregating in an F2 population from a cross between a stock line carrying decipiens and JI:7 (Stubbe, 1966). The origins of delrec (JI:602; JI22) and del23 (JI:23) have been described by Goodrich et al. (1992) and Martin et al. () and lines JI:32, JI:33, JI:56, JI:520, and JI:522 have been described by Fincham & Harrison, (1967), Coen et al., (1986) and Almeida et al., (1989).
Construction of a cDNA library from RNA from lobes of del flowers
RNA was extracted from red petal lobes from a del line (Jl:8). cDNA was synthesised from polyA+ RNA. EcoRl linkers were ligated to the cDNA ends and cloned into the EcoRl site of λgt10. The resultant library contained c. 6 × 106 PFU
Screening of the cDNA library
Approximately 5 × 105 PFU were screened using a 32P‐labelled fragment from a cDNA clone of An1 (Spelt et al., 2000) amplified from petals of line V26. Three positive plaques were identified. A 2.4 kb KpnI fragment from the largest cDNA insert was subcloned into the KpnI site of pBluescript (Stratagene) and named pJAM1494.
RNA preparation for RNA gel blots
RNA was extracted from petals and RNA gel blots were run as described by Martin et al. (1985). Probes for biosynthetic genes were prepared as described in Martin et al. (1991) and Schwinn et al. (2006).
Isolation of genomic clones
A library of genomic DNA from JI:522 was prepared in λEMBL4 (Martin et al., 1991) and screened with pJAM1494. DNA inserts from positive plaques were subcloned as EcoRI fragments into pBluescript. DNA was also isolated from inc I1 : del and inc I2 : del lines digested with EcoRI, size‐fractionated and cloned into λNM1149. Inserts were screened with pJAM1494 and the positive EcoRI inserts were subcloned into pGEM‐T (Promega) and sequenced.
Amplification of inc I, delila and Delila‐like alleles from genomic DNA was performed with iProof polymerase (Bio‐Rad) and gene‐specific primers (Supporting Information Table S1). Sequences have been submitted to GenBank with the following accession numbers: del23, genomic DNA, MW027119: inc I1, genomic DNA, MW027120; Incolorata I, cDNA, MW027121; WDR1, cDNA, MW027122.
Phylogenetic tree
Deduced amino acid sequences were aligned using Muscle (Edgar, 2004) to generate a maximum likelihood phylogenetic tree using PhyML (Guindon et al., 2010) with 1000 bootstrap replicates using Geneious (10.0.9) software. Amino acid sequence alignments are shown in Dataset S1.
Biolistic transformation of inc I petals
Complementation assays of inc I1 : del petals were performed by biolistic transformation of petals (Albert et al., 2015). GFP‐ER was used as an internal control for identifying transformed cells (Haselhoff et al., 1997).
Isolation of AmWDR1
WDR1 was isolated by 3′ Rapid Amplification of cDNA Ends (RACE) PCR (Frohman et al., 1988) from line JI:75 using degenerate oligonucleotides K112 and K113 for first and second amplification rounds, respectively. The 5′ end of the WDR1 gene was isolated using a Universal Genome Walker kit (Clontech) and gene‐specific primers K127 and K128 for the first and second amplification rounds, respectively. The full‐length WDR1 cDNA was amplified from RNA from petals using primers K133 and K134 and cloned into pJAM1502. For all primers see Table S1.
RNA isolation and qRT‐PCR
Whole petals of wild‐type (JI:522), roseadorsea and incolorata I flowers, or dissected wild‐type (JI:522) and delila (JI:8) petals (tubes and lobes) were sampled. Total RNA was extracted using Plant RNeasy Isolation kits (Qiagen). For qRT‐PCR analysis, cDNA was prepared from 1 µg total RNA using IScript™ gDNA clear cDNA synthesis kits (Bio‐Rad), and diluted 20‐fold for analysis. qRT‐PCR using SsoAdvanced™ Universal SYBR® Green Supermix (Bio‐Rad), gene‐specific primers (Table S1) were normalised to the geometric mean of Cyclophilin and EF1α (Albert et al., 2014). Means ± SEM of three biological replicates were calculated. Two‐tailed Student’s t‐tests between wild‐type and the corresponding mutant samples were performed to determine significant differences. Data were transformed (log10) to account for unequal variance.
Full‐length transcripts of Delila and Delila‐like were amplified with gene‐specific primers (Table S1) and 2GRobust polymerase (Kapa Biosciences), from cDNA synthesised using Superscript II™ reverse transcriptase (Life technologies) and oligo (dT)12–18.
Transient dual luciferase assays in Nicotiana benthamiana
The ability of different transcription factors to activate the promoter of the Pallida gene encoding dihydroflavonol‐4‐reductase (DFR) was assessed by combining transient expression via agroinfiltration of N. benthamiana leaves with a quantitative dual luciferase reporter system.
Effector plasmids encoding transcription factors were obtained by cloning coding sequences first into the pDONR207 entry clone (Thermo Fisher Scientific/Invitrogen) and subsequently transferred to the destination vectors pJAM1502 (Ros, Inc1, WDR1) or pMDC32 (Del) using Gateway cloning technology (Thermo Fisher Scientific/Invitrogen). Both vectors are Gateway‐compatible binary plasmids that allow strong, constitutive expression of the genes of interest driven by double CaMV 35S promoters (Curtis & Grossniklaus, 2003; Luo et al., 2007).
The reporter constructs p2532, p2534 and p2536 were obtained by PCR amplification of the DFR promoter from different A. majus accessions followed by cloning into the pGreen II 0800‐LUC vector as KpnI–NcoI fragments to control the expression of the firefly‐derived luciferase reporter gene. This vector also contains a Renilla luciferase gene under the control of a CaMV 35S promoter as an internal control to normalise the values of the experimental reporter gene for variations caused by transfection efficiency (Hellens et al., 2005). The reporter constructs containing deletions in the DFR promoter (p2551, p2557, p2565 and p2571) were obtained using the Q5 Site‐Directed Mutagenesis Kit (New England Biolabs) using plasmid p2532 as a template and primers listed in Table S1.
Effector and reporter plasmids were transformed into Agrobacterium tumefaciens strain GV3101. Reporter plasmids were co‐transformed with the helper plasmid pSoup (Hellens et al., 2005). Liquid cultures were grown overnight with selection (kanamycin 50 mg l−1, rifampicin 25 mg l−1) and harvested by centrifugation. Cells were washed and resuspended in 10 mM MgCl2, 10 mM MES pH 5.6, 200 μM acetosyringone to A600 = 0.2. Agrobacterium suspensions were infiltrated into the abaxial surface of expanded leaves of 3‐wk‐old N. benthamiana plants. For each combination, five injected areas were treated as biological replicates. Agroinfiltrated leaves were harvested after 3 d and luciferase activity was measured immediately using the Dual‐Glo Luciferase Assay System kit (Promega). Leaf discs of 4 mm in diameter were collected in 1.5 ml white transparent tubes containing 100 μl of PBS. A volume of 75 μl of luciferase assay reagent was added and firefly luminescence was measured on a Glomax 20/20 single tube luminometer (Promega) after 10 min. Renilla luminescence was measured on the same instrument 10 min after the addition to the same sample of 75 μl of Stop & Glo reagent. Results were expressed as the ratio of firefly to Renilla luciferase activity (Luc/Ren).
Results
Classic mutants incolorata I and mutabilis are allelic and affect anthocyanin biosynthesis
The classic delila mutant of Antirrhinum majus has bicoloured flowers with red lobes, pigmented with anthocyanin, and an ivory‐coloured tube (Wheldale, 1907; Baur, 1910). Del encodes a bHLH transcription factor that regulates anthocyanin pigmentation in both tubes and lobes (Goodrich et al., 1992). However, knockout mutants of del have fully pigmented lobes and consequently the bicolour pattern in del mutants has been predicted to arise from the activity of a second, unknown protein, active in the petal lobes (Goodrich et al., 1992; Schwinn et al., 2006). Two mutants affect lobe pigmentation: incolorata I (inc I) and mutabilis (mut) (Stubbe, 1966; Linnert, 1972). The inc I mutant was originally identified by its ivory flowers and complete absence of anthocyanins in vegetative tissues (Fig. 1) (Stubbe, 1966; Linnert, 1972). The inc I mutant line was obtained from the IPK Gatersleben collection of Antirrhinum germplasm in 1984 and outcrossed to JI:7 (a full red, self‐coloured line (Inc I/Inc I : Del/Del)). Segregation in the F2 showed 9/56 plants segregating for del in addition to full red (43/56: Inc I/Inc I and Inc I/inc I) and ivory inc I : del homozygotes (4/56). This suggested that the original inc I mutant line was homozygous for del, which was confirmed by crossing a reselected inc I line to JI:8 (del/del). All F1 plants lacked tube pigmentation (del/del) but had full red lobes, confirming that inc I was a recessive mutation, visible only in combination with homozygous del alleles (Fig. 1).
Fig. 1.
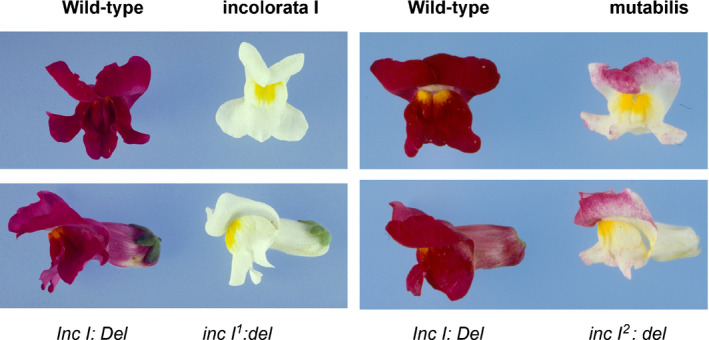
Phenotypes of incolorata I and mutabilis mutants compared with wild‐type full red flowers (JI:7) of Antirrhinum majus. Phenotypes are written above the images and their genotypes are shown below the images. The del knockout mutant has a colourless tube but fully pigmented lobes.
The mutabilis mutation was described originally by a series of alleles in the IPK Gatersleben collection, linked (2 cM) to Del (Stubbe, 1966). While these original mutants have been lost from the collection, we identified an individual with the mutabilis phenotype from an F2 population from a cross between a stock line carrying decipiens (Stubbe, 1966) and wild‐type (JI:7). The rediscovered mutabilis mutant has pale pink blushing/speckling on the petal lobes (Fig. 1), but completely lacks tube pigmentation because it is homozygous for del. The speckling of the lobes is much more evident in plants grown in the field than in those grown in the glasshouse (Fig. S1) with strong pigmentation on the abaxial epidermis of the lobes except where tissue has been shaded before unfolding, suggesting that this allele is sensitive to light. An F2 population from a cross between the mutabilis mutant (mut:del) and a full red wild‐type line (JI:7 Mut:Del), segregated for flower colour patterning. Phenotypes fell into three categories: ivory tube with pale, speckled lobes (mut:del parental phenotype; 56 plants), full red tube and lobes (wild‐type parental phenotype; 200 plants) and ivory tube with red lobes (del phenotype; 8 plants). Recombinants with a red tube and pale, speckled lobes were not observed, indicating that Del activity is epistatic to Mut in regulating pigmentation. Further attempts to replace del with Del alleles in the mut mutant line by outcrossing proved unsuccessful, confirming that the mut phenotype, like that of inc I, is observed only in the presence of nonfunctional del alleles.
Crosses between inc I and mut did not complement the pigmentation phenotype, showing the mutations to be allelic (Fig. S2). The gene was named Incolorata I, and the incolorata I mutant allele was named inc I1 (Stubbe, 1966; Linnert, 1972). The mutabilis mutant carries a weak allele of inc I, designated inc I2.
The redundancy of Inc I with Del in regulating anthocyanin biosynthesis suggested that the genes encode similar proteins. Del encodes a bHLH transcription factor (Goodrich et al., 1992), belonging to the R/JAF13 bHLH‐1 clade of subgroup IIIf (Heim et al., 2003). In other species, a second class of bHLH proteins (TT8/AN1 bHLH‐2 clade) regulates anthocyanin and proanthocyanidin biosynthesis (Nesi et al., 2000; Spelt et al., 2002). We screened cDNA libraries generated from lobe tissue with a del background with a cDNA probe of the AN1 gene from petunia line V26. Multiple cDNA clones corresponding to a new bHLH sequence were identified: the longest cDNA insert was subcloned as a 2.4 kb KpnI insert in pBluescript (pJAM1494). Sequencing confirmed this contained a full‐length cDNA. The wild‐type Incolorata I gene was identified in a 24 kb genomic DNA insertion in λEMBL4 from line JI:522. Fragments of the gene were also cloned from inc I1 genomic DNA and sequenced. A 4 bp duplication (CATG) was identified within exon three of the inc I1 mutant line, resulting in a frame‐shift, confirming this gene to be Incolorata I. We were unable to identify any sequence variants in genomic DNA that could account for the weak phenotype of the inc I2 allele but RNA gel blots from wild‐type and inc I2 mutant flowers (Fig. S3) revealed an absence of Inc I transcript in the mutant, suggesting that the inc I2 mutant phenotype results from considerably reduced levels of Inc I expression.
Previous genetic data suggested that Mut/Inc I and Del were closely linked, 2 cM apart (Stubbe, 1966). From our crosses of inc I2 : del × Inc I : Del a total of 264 F2 plants had 64 del/del individuals of which 56 had speckled/ivory flowers (inc I2 homozygotes) and 8 (del/del) individuals had red lobes and so carried at least one Inc l allele. Assuming all the latter to be heterozygous Inc I : inc I2, we calculated there to have been 8 recombination events among 128 gametes giving a recombination fraction (r) of 8/128 = 0.0625 (± 0.0214) and the distance between Inc I and Del, (d) c. 6.7 (± 2.19) cM, meaning Inc I and Del lie about 6.7 cM apart. In the high quality genome sequence for A. majus (Li et al., 2019) Incolorata I (Am02g53780) and Delila (Am02g33340) are both located on chromosome 2, 20.7 Mb apart (Fig. 2a).
Fig. 2.
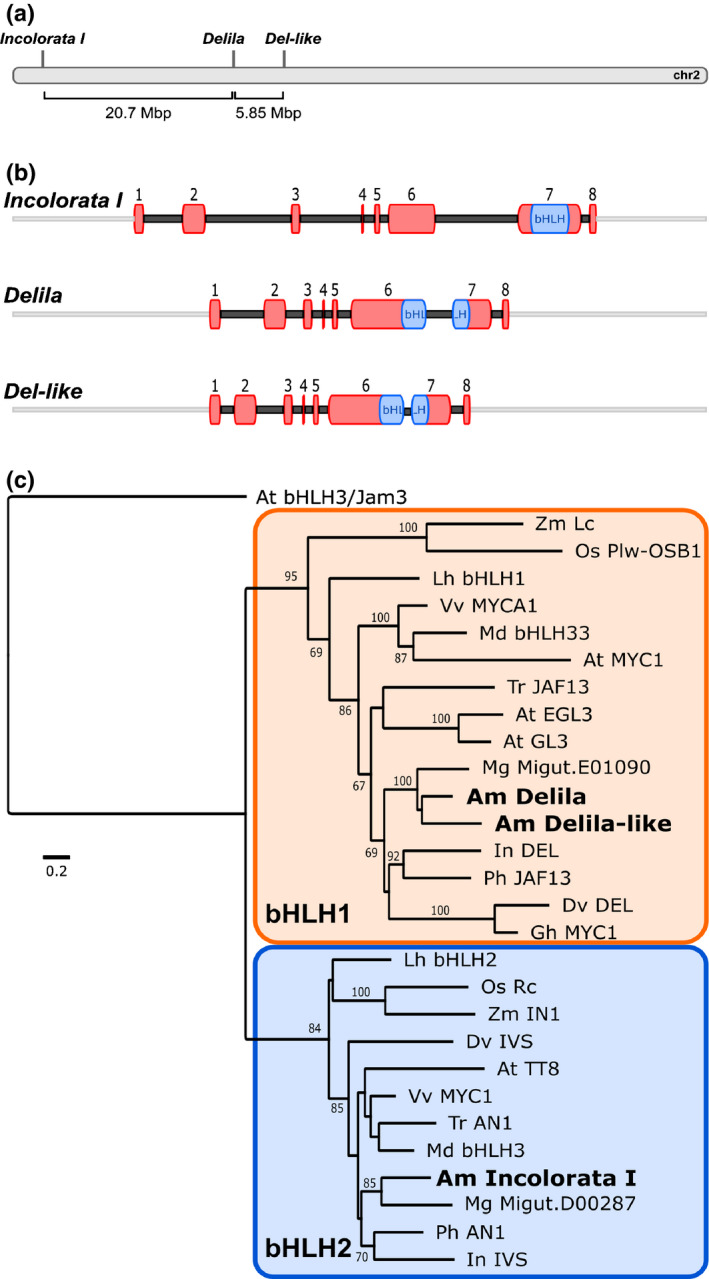
Molecular and phylogenetic analysis of Incolorata I, Delila and Del‐like bHLH transcription factors. (a) Location of Incolorata I, Delila and Delila‐like genes on Chromosome 2 of Antirrhinum majus. (b) Structure of Delila, Delila‐like and Incolorata I bHLH genes. Exons are numbered and the bHLH domain is indicated in blue. (c) Maximum likelihood phylogenetic tree of subgroup IIIf bHLH proteins, with AtbHLH3 (subgroup IIId) included as an outgroup (Heim et al., 2003). 1000 bootstrap replicates, ≥60% support indicated. Gene IDs/accession numbers of sequences used: A. majus Am Delila (AAA32663.1; Am02g33340); Delila‐like (Am02g28470); Incolorata I (Am02g53780) (A. majus genome, v.3); Arabidopsis thaliana At bHLH3/JAM3 (NP_193376); EGL3 (OAP12509); GL3 (NP_680372); TT8 (Q9FT81); AtMYC1 (NP_001154194); Dahlia variabilis DvDel (BAJ33516); IVS (BAJ33515); Gossypium hirsutum Gh: MYC1 (CAA07615); Ipomoea nil InDel (XP_019171149); IVS (XP_019197480); Lilium hybrida LhbHLH1 (BAE20057); bHLH2 (BAE20058); Malus domestica MdbHLH3 (ADL36597); bHLH33 (ABB84474); Mimulus gattatus (Erythranthe guttata) MgMigut.E01090.1; Migut.D00287.1; Oryza sativa OsPlw‐OsB1 (BAB64301); Rc (ADK36625); Petunia hybrida Ph AN1 (AAG25928); JAF13 (AAC39455); Trifolium repens TrTrAN1 (AIT76559) TrJAF13 (AIT76563); Vitis vinifera Vv MYC1 (NM_001281253); MYCA1 (NP_001267954); Zea mays Zm Lc (AAA33504); In1 (AAB03841).
Lying 5.85 Mb beyond Del on chromosome 2 is a sequence encoding a third bHLH protein, very similar to Del, which we named Delila‐like (Am02g28470) (Fig. 2a,c). The Delila‐like gene encodes a bHLH transcription factor in the bHLH‐1 clade, and its intron/exon structure (established by amplification of a cDNA), was characteristic of bHLH‐1 genes (Fig. 2b).
Phylogenetic analysis of the predicted amino acid sequences of Incolorata I, Delila and Delila‐like together with other bHLH transcription factors that regulate flavonoid biosynthesis confirmed that they represent the two bHLH clades within subgroup IIIf (Fig. 2c). The sequence similarity and the conserved exon/intron structure between Inc I and genes such as AN1 (petunia) and TT8 (Arabidopsis), with the bHLH domain encoded entirely in exon 7, placed Inc I firmly within the bHLH‐2 clade.
Confirmation of functional redundancy of Del, Del‐like and Inc I in controlling anthocyanin biosynthesis
We confirmed the functional redundancy of Inc I, Del and Del‐like in inducing anthocyanin biosynthesis by bombarding petal lobes of the inc I:del double mutant line with plasmids carrying each cDNA driven by the CaMV 35S promoter. All three genes were able to restore anthocyanin production to single cells of the petal lobes, by contrast to the CaMV 35S: GFP control (Fig. 3).
Fig. 3.
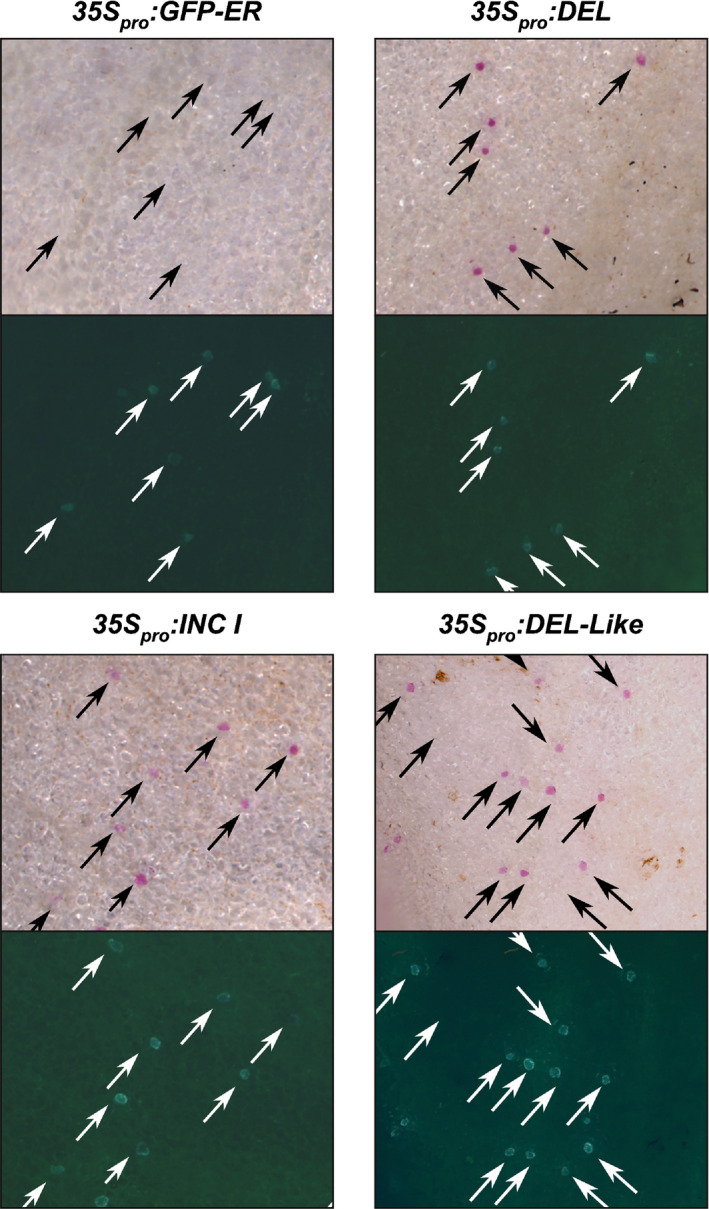
Complementation of incolorata I1 with bHLH transcription factors. Inc I1 petals of A. majus were biolistically transformed with 35Spro : Delila, 35Spro : Inc I or 35Spro : Delila‐like, co‐transformed with GFP‐ER to identify transformed cells (indicated with arrows). Red anthocyanin accumulation was visible in cells transformed with Delila, Delila‐like or Inc I, but was not detected with the GFP‐ER control.
Genetic evidence that Del acts redundantly with Inc I in inducing anthocyanin pigmentation
Crosses were performed between incolorata I (inc I1), mutabilis (inc I2) and an unstable delilarecurrens mutant (delrec). JI:602 delrec flowers have red petal lobes, but the petal tubes are ivory with red sectors, resulting from the excision of a transposable element within the Del gene, that restores Del activity (Goodrich et al., 1992). Del revertant sectors were able to complement both inc I1 and inc I 2 mutant backgrounds, restoring pigmentation in lobes (Fig. 4a). Revertant Del sectors were observed that extended throughout the tube and lobes, confirming Del to be active in both tissues and to mask the phenotype of inc I mutations (Figs 4,S4). Individuals were recovered in the F3 generation where the transposable element had excised from the Del locus in the germline of an incI/incI : delrec/del parental plant. These plants had full red flowers with coloured lobes and tubes (Fig. 4b).
Fig. 4.
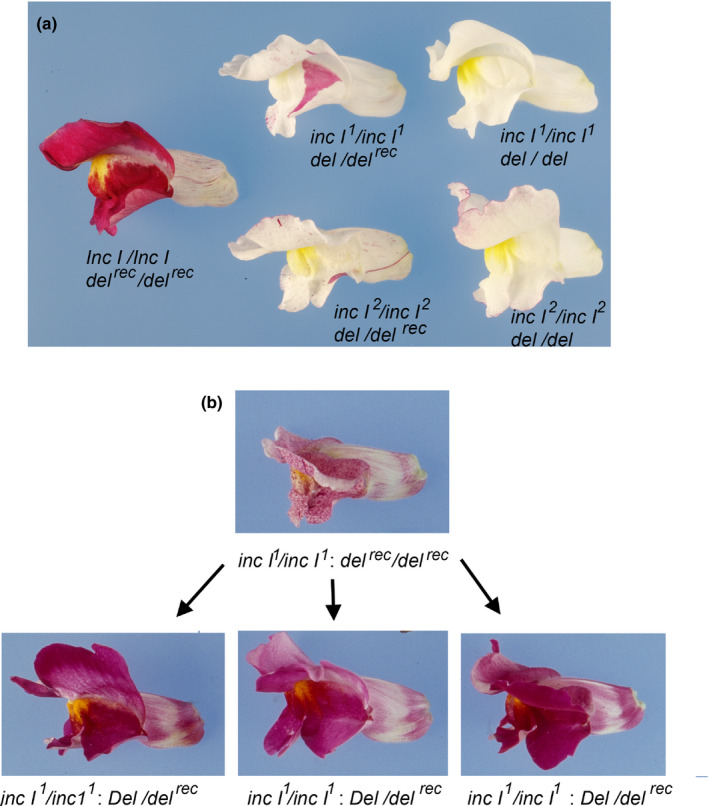
Inc I acts redundantly with Del in controlling pigmentation in petal lobes of A. majus. (a) Revertant sectors of Del arising from transposon excision from delrec complement fully loss of inc I function in lobes of inc I : delrec flowers segregating in F2 populations of inc I1 : del × Inc I : delrec and inc I2 : del × Inc I : delrec crosses. (b) Independent full red germinal revertants from an inc I1/ inc I1 : delrec/delrec parental line have full red lobes and tubes in F3 plants.
The functional redundancy between Inc I and Del was investigated further by a cross between the stock line JI:23 (Martin et al., ) and inc I1. JI23 carries a weak allele of Delila (del23) that results in slightly reduced anthocyanin pigmentation in the tube, particularly the upper region of the tube (Fig. S5). The F2 population from JI:23 Inc I/Inc I : del23/del23 × inc I1/inc I1 : del/del segregated for flower colour in 96 plants scored, with six distinct phenotypes. In addition to the parental inc I1 : del and Inc I : del23 phenotypes, four recombinant phenotypes were observed: 21 plants were del (ivory tube) of which seven were inc I1 : del; 61 plants were Inc I/Inc I or Inc I/inc I1 with del23, nine plants were inc I1/inc I1; del/del23 with a flush of pigmentation on the lobes and 5 plants were inc I1/inc I1 : del23/del23 with a dark flush on the lobes (Fig. 5). The phenotypic classes and their frequencies were consistent with segregation between del23 and del such that the phenotype of del23 with a flush of pigmentation on the lobes (9/96) was due to the inc I1/inc I1 : del23/del genotype and the phenotype of del23 with a dark flush on lobes (5/96) was due to the inc I1/inc I1 : del23/del23 genotype. These genotypes were confirmed in the F3 where the del23 with a dark flush on lobes (inc I1/inc I 1 : del23/del23) bred true, and del23 with a flush of pigmentation on the lobes segregated three del23 with a dark flush on lobes (inc I1/inc I1 : del23/del23): nine del23 with a flush of pigmentation on the lobes (inc I1/inc I1 : del23/del): three ivory flowers (inc I1/inc I1 : del/del), from a family of 15 plants. These data demonstrated that Del acts within both the tube and lobes to regulate anthocyanin pigmentation, acting redundantly with Inc I in the lobes, but predominating in the tube of the flowers.
Fig. 5.

Phenotypes of individual plants segregating in the A. majus F2 population of Inc I : del 23 x inc I1 : del cross. Top row: Floral phenotypes of the parental lines Inc I : del 23 and inc I1 : del. Second, third and fourth rows: Floral phenotypes that segregated in the F2 generation. The genotypes conferring these phenotypes are shown below each flower together with the number of individuals with that phenotype from a total F2 population of 96 individual plants (shown in parentheses).
Interestingly by comparing the flowers of Inc I/Inc I : del23/del23 with those of inc I1/inc I1 : del23/del23it was apparent that inc I activity affected anthocyanin accumulation in the tubes of the flowers as well as in the lobes, although its contribution to anthocyanin accumulation is substantially greater in the lobes than in the tubes (Fig. 5). This was confirmed by comparing the colour of Inc I/Inc I : del23/del flowers to inc I1/inc I1 : del23/del flowers (Fig. 5). By contrast, although Del operates in both the lobes and the tubes, its contribution is substantially greater in tubes than in lobes as shown by comparing the phenotypes of inc I1/inc I1 : del23/del and inc I1/inc I1 : del23/del23 (Fig. 5).
To confirm that del23 was indeed a weak allele of Del, cDNA and genomic DNA were amplified from flowers of JI:23 (Fig. S6). The genomic sequence of Del in JI:23 (del23) had 29 SNPs within the coding sequence (14 were nonsynonymous) compared with the reference genome (Goodrich et al., 1992; Li et al., 2019), additional SNPs (79) and small insertions into introns, including one near a splice acceptor site within intron five (Fig. S6b) that resembled an intron acceptor site. Sequences of 8/11 clones of the Del cDNA from this line were mis‐spliced into the insertion, resulting in mRNA sequences that encoded an insertion of an extra amino acid within a conserved region of the predicted Del protein, one transcript spliced correctly, one retained two introns and one had an entire portion of the cDNA missing (Fig. S6b). We concluded that this insertion causes mis‐splicing of the Del mRNA in a significant proportion of the transcripts in JI:23 flowers, confirming that del23 was a weak del allele.
Identification of target genes of Inc I in anthocyanin biosynthesis
To determine the target genes regulated by Inc I, transcript analyses of biosynthetic genes were conducted in dissected tubes and lobes of del or wild‐type flowers (Fig. 6a) or whole petals of wild‐type, roseadorsea (a mutant of Rosea 1 which encodes an R2R3Myb transcription factor) and inc I1 : del flowers (Fig. 6b). Transcripts of the genes encoding DFR, anthocyanidin synthase (ANS) and flavonoid: 3‐glycosyltransferase (3GT) were at trace amounts in inc I1 : del flowers and the transcript levels of flavanone 3‐hydroxylase (F3H) were substantially reduced. Transcripts of the genes encoding chalcone synthase (CHS) and chalcone isomerase (CHI), were not reduced (Fig. 6a). Similarly, transcript abundance for F3H, DFR, ANS and 3GT were severely reduced in whole petals of roseadorsea, and inc I1 : del (Fig. 6b). These data were confirmed by RNA gel blots of the inc I2 : del double mutant, mutabilis (Fig. S7) establishing that mutation of inc I causes down regulation of expression of the genes encoding F3H, DFR, ANS and 3GT (Fig. 6a; Martin et al. 1991), but not CHS or CHI. These effects were very similar to those of mutations in Del (Martin et al., 1991; Jackson et al., 1992), suggesting that the two transcription factors share the same target genes in anthocyanin biosynthesis.
Fig. 6.
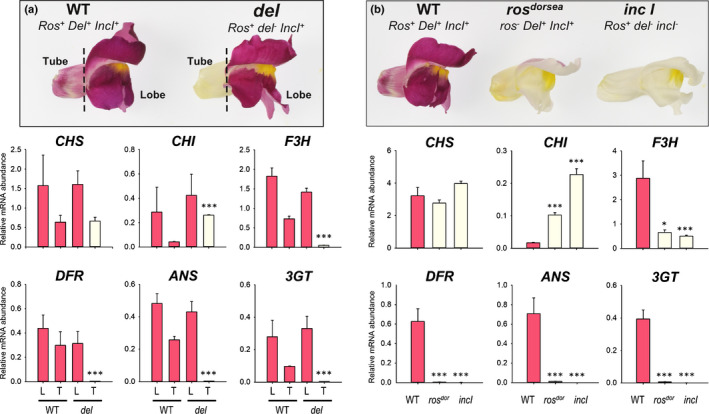
Anthocyanin biosynthesis genes are downregulated in incolorata I 1 petals of A. majus. (a) Wild‐type (WT) and delila mutant flowers were separated into tube and lobes for analysis of CHS, CHI, F3H, DFR, ANS and 3GT transcript abundance by qRT‐PCR in tube (T) and lobe (L) tissue of WT and delila mutant petals. (b) Petals from whole flowers of WT, rosdorsea and inc I1 : del were analysed for CHS, CHI, F3H, DFR, ANS and 3GT transcript levels by qRT‐PCR. Bar colour indicates pigmentation status. Means ± SEM, n = 3 biological replicates. Means that are significantly different (Student’s t‐test) from WT are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Model of MBW regulation of Pallida (DFR)
Some time ago, Almeida et al. (1989) proposed a model for bicoloured patterning of delila mutants based on analysis of mutations in the promoter of the Pallida (pal) gene encoding DFR (Coen et al., 1986; Almeida et al., 1989), which proposed that Del and a lobe‐specific transcription factor bound to the same upstream region of the DFR promoter (Box C: Fig. S8) in tubes and lobes respectively. At the time, none of the regulatory genes had been identified but, with these regulatory genes now to hand, we tested this model using some of the same DFR promoter mutations. We expressed Del, Inc I, Ros 1 and a cDNA encoding a WD repeat protein from A. majus homologous to An11 in petunia (WDR1) under the control of the CaMV35S promoter in Nicotiana benthamiana, and tested the different Antirrhinum DFR promoter sequences using the dual luciferase assay to measure promoter responsiveness.
The wild‐type DFR promoter has two starts of transcription (TS), one 66 bp upstream of the initiating ATG (TS1) and the second (TS2), 99 bp upstream of the initiating ATG (Coen et al., 1986). TS1 is the stronger initiation site in A. majus (Coen et al., 1986; Robbins et al., 1989). TS1 is preceded by a TATA box (TATA‐1: −86 to −93) and TS2 is preceded by TATA‐2 (−120 to −127; Fig. 7a). There was significant induction of luciferase driven by the wild‐type DFR promoter (p2532) by Ros 1 alone. This was possibly due to binding of Ros 1 to an AC‐box which lies between −184 and −190 nucleotides upstream of the initiating ATG codon of the DFR gene (AC‐box 1; Fig. 7a) or to a second AC‐box (AC‐box 2) lying between −157 and −162 bp upstream of the start of translation of the DFR gene. This transcriptional response to Ros 1 alone may be a feature specific to tobacco. AC‐box 1 is likely to be a Myb binding site, because its deletion following transposon excision reduced DFR gene expression and anthocyanin production to very low levels in the tube of the flower (Almeida et al., 1989). Just eight bp gene proximal to the AC‐box 1 box is G‐box 1 (CACGTG: −171 to −176), a recognised binding site for bHLH transcription factors (Fig. 7a; equivalent to Box B in Almeida et al., 1989; Goodrich et al., 1992). Displacement of these sequences (AC‐box 1 box and G‐box 1) from the downstream TATA boxes/transcriptional start sites causes complete loss of DFR expression in A. majus as shown by the insertion of Tam 3 at −169 bp upstream of the initiating ATG in the DFR promoter (Fig. S8; Carpenter et al., 1987).
Fig. 7.
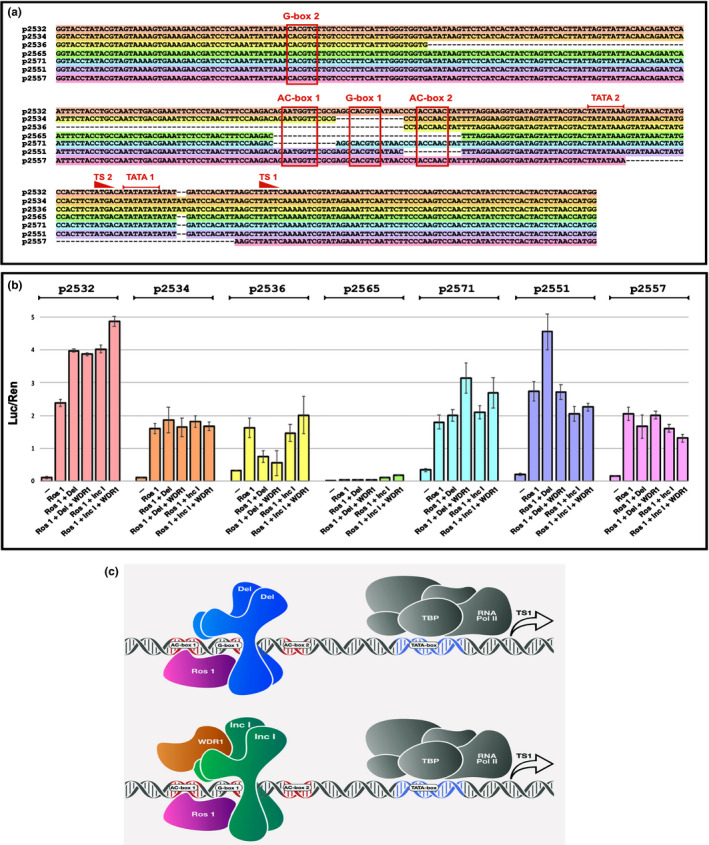
Assessment of responsiveness of the DFR promoter from A. majus to components of the MBW complex from A. majus using dual luciferase assays in N. benthamiana. (a) Structure of the wild‐type promoter of the DFR gene from A. majus (p2532) showing the gene proximal transcriptional start site (TS1) and associated TATA box (TATA 1) and the gene distal transcriptional start site (TS2) and its associated TATA box (TATA 2) (Coen et al., 1986).G‐box 1 is shown flanked on its gene distal side by AC‐box 1 and on its gene proximal side by AC‐box 2. It should be noted that AC‐box 1 reads 5′ to 3′ on the lower DNA strand whereas AC‐box 2 reads 5′ to 3′ on the upper strand. The location of a second G‐box (G‐box 2) is also shown. Below the sequence of the WT promoter (p2532; highlighted in pink) the deletions assayed using the dual luciferase assay are shown, coloured in accordance with the bars detailing the effects of these deletions on luciferase reporter activity shown in panel b). (b) Activation of luciferase by MBW components on wild‐type DFR promoter (pink) and promoters carrying deletions of the key upstream activating sequences (UAS) detailed in panel (a). All values relate luciferase to Renilla luminescence and are the results from at least five independent assays. Error bars show SEMs. (c) Model illustrating the predominant binding of MBW components to the wild‐type DFR promoter from A. majus. The bHLH proteins Del and Inc I bind to G‐box 1 probably as a dimer. The N‐termini of Del and Inc I interact with the surface exposed region of R3 of the Myb DNA binding domain of Ros1 carrying the bHLH interaction signature motif (Zimmermann et al., 2004). TBP = TATA Binding Protein For Inc I the additional binding by WDR1 is necessary for full activity.
There was substantial induction of luciferase activity when Del was included together with Ros 1 on the wild‐type DFR promoter (p2532), but no further enhancement was observed when WDR1 was also included (Fig. 7b). This suggested that Ros 1 and Del act together to induce DFR expression and the WDR1 does not enhance this interaction on the DFR promoter. In combination with Ros 1, Inc I gave similar increases in luciferase activity on the wild‐type DFR promoter but inclusion of the WDR1 protein slightly enhanced the induction of luciferase by Ros 1 and Inc I suggesting that Inc I and WDR1 interacted positively in the MBW complex to induce transcription from the DFR promoter.
Deletion of the sequences between −93 and −116 bp upstream of the initiating ATG removed the TATA‐1 box associated with TS1 (p2557). This deletion resulted in loss of induction of the DFR promoter except for the background response to Ros 1, suggesting that the MBW complex primarily regulates transcription of the DFR gene using TATA‐1 and TS1. The remaining transcriptional response to Ros 1 alone, presumably is mediated by TATA‐2 and TS2.
Assay of a mutant DFR promoter lacking 12 nucleotides immediately downstream of AC‐box 1 that included G‐box 1 known to be recognised by bHLH proteins, (p2534), abolished the induction of luciferase activity by both Del and Inc I (with or without the WDR1 protein) (Fig. 7b). These data indicated that both bHLH proteins, Del and Inc I, recognise and bind G‐box 1 to activate DFR gene expression. G‐box 1 is therefore likely bound by both bHLH proteins in the MBW complex and the Myb protein, Ros 1, likely binds to the AC‐box 1 in the wild‐type promoter especially when interacting with Del. Deletion of the DFR promoter involving loss of G‐box 1, AC‐box 1 and 200 nucleotides further upstream placed another G‐box (G‐box 2) −188 to −193 bp upstream of the initiating ATG codon of the DFR gene. Dual luciferase assays using this promoter mutation, which in A. majus results in almost complete loss of DFR expression (Almeida et al., 1989), did not restore responsiveness to Del nor to Inc I plus the WDR1 protein, over and above responsiveness to Ros 1 alone. This showed that the sequence context and proximity of the AC‐box and G‐box motifs relative to the basal transcriptional machinery assembled around the TATA box are important in defining the response of the target gene to the MBW complex.
To unravel the recognition of sequence motifs by the MBW complexes further, new mutations of the DFR promoter were created (p2551; p2565 and p2571). Removal of 12 bp downstream of G‐box 1 (loss of −154 to −165 bp), including AC‐box 2 (AACACC; −157 to −162; p2551) gave good induction of the DFR promoter in response to Ros 1 and Del, although inclusion of WDR1 in this assay inhibited luciferase activity to the levels achieved by Ros 1 alone (Fig. 7b). Interestingly, loss of AC‐box 2 eliminated responsiveness to Inc I and WDR1. We tested the effects of the MBW components on deletion of AC‐box 1 (−180 to −192; p2571; Fig. 7b). This caused large reductions in responsiveness of the DFR promoter to Del with Ros 1 and, by contrast to all other versions of the DFR promoter, the presence of WDR1 enhanced expression by the Ros 1–Del complex suggesting that Ros 1–Del binding to AC‐box 2 (in the absence of AC‐boz 1) and G‐box‐1 is enhanced by WDR1. Loss of AC‐box 1 eliminated the response to Inc I, although there was perhaps a small induction when Inc I was combined with WDR1. To confirm these results, we tested a promoter deletion that included both AC boxes and G‐box 1 (−154 to −192; p2565). This deletion completely eliminated responsiveness of the DFR promoter to the MBW complex (Fig. 7b), confirming that the MBW components work through the combined AC‐box 1, G‐box 1 and AC‐box 2 upstream activator sequence (UAS) and implying that there are structural and consequently functional differences in the way Ros 1 interacts with the bHLH‐1 and bHLH‐2 proteins on the DFR promoter. A model for how this might work is proposed in Fig. 7c.
Of course, the effects of the MBW complex on anthocyanin production reflect the net responses of all anthocyanin biosynthetic genes and the relative changes in enzyme and transporter activity on anthocyanin accumulation. To investigate the net effect of the MBW components on the regulation of anthocyanin production in N. benthamiana, we tested different combinations of regulatory proteins for induction of anthocyanins in the leaves of N. benthamiana ‘Northern Territory’ which does not suffer from reported problems with induction of anthocyanins in leaves of the more commonly used laboratory strain (Bally et al., 2015; Thole et al., 2019). Anthocyanin levels were significantly induced by Ros 1 on its own (>25 fold) and these were further elevated by inclusion of a vector expressing Del in the transient assay (>45 fold) (Fig. 8). Inclusion of a vector expressing the WDR1 protein repressed the induction of anthocyanins by Ros 1 and Del significantly, suggesting that the combination of Ros 1 and Del is negatively impacted by inclusion of the WDR1 protein on anthocyanin production. Induction of anthocyanin production by the combination of Ros 1 and Inc I was not as great as for Ros 1 and Del, but was enhanced slightly by inclusion of the WDR1 protein suggesting that the WDR protein interacts positively with Inc I to promote net anthocyanin production by the MBW complex in N. benthamiana.
Fig. 8.
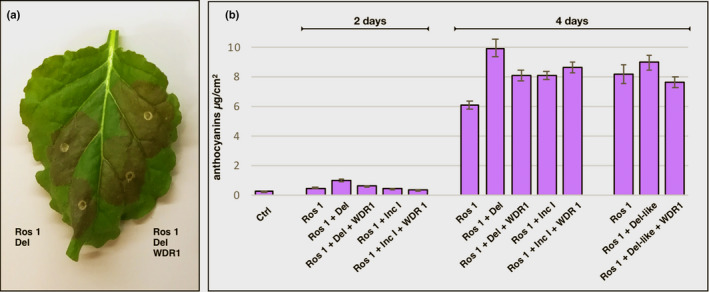
Anthocyanin production in leaves of Nicotiana benthamiana var NT in response to MBW components from A. majus. (a) Production of anthocyanin in N. benthamiana ‘NT’ leaves 4 d post inoculation. (b) Total anthocyanin production in response to MBW components of A. majus in N. benthamiana ‘NT’. Total anthocyanin production reflects the net effects of the MBW components on the activities of all anthocyanin biosynthetic enzymes and transporters. Error bars show SEMs
We also tested the functionality of Del‐like in inducing anthocyanin production in N. benthamiana. Although Del‐like did induce a small increase in anthocyanin produced in leaves, this was not significantly greater than Ros 1 alone. Inclusion of WDR1 in the inoculum reduced this small increase below levels for Ros 1 alone (Fig. 8). We concluded that the weak enhancement of anthocyanin biosynthesis by Del‐like in N. benthamiana suggested that it plays a negligible role in controlling anthocyanin biosynthesis directly in flowers of A. majus.
The regulatory network controlling expression of the components of the MBW complex
In petunia, the bHLH‐1 protein (JAF13) forms an MBW complex that regulates the expression of the An1 gene (bHLH‐2), enabling new MBW complexes to form with specific Myb proteins that target anthocyanin, proanthocyanidin and acidification pathways (Albert et al., 2014). While features of this hierarchical regulatory network have been reported in several plants (Baudry et al., 2006; Albert et al., 2014; Albert, 2015; Liu et al., 2014; Montefiori et al., 2015; Li et al., 2020), it is not known whether hierarchical regulation is universal.
Hierarchical regulation was investigated by analysing the expression of anthocyanin regulatory genes in petals (Fig. 9). Ros 1 abundance was unaffected by the del mutation and transcripts were present in both tube and lobe tissues. Inc I transcripts were detected only in the lobes of del mutants but, interestingly, Inc I was expressed in both the tube and lobes of wild‐type petals, indicating that Del promotes Inc I expression in tubes. Delila‐like transcripts were detected in the lobes and tubes of both wild‐type and del mutants albeit at relatively low levels, suggesting that Del‐like is not regulated by the MBW complex. WDR1 transcripts were reduced in the tube of del mutants compared with wild‐type, suggesting that its expression is controlled, in part, by the MBW complex.
Fig. 9.
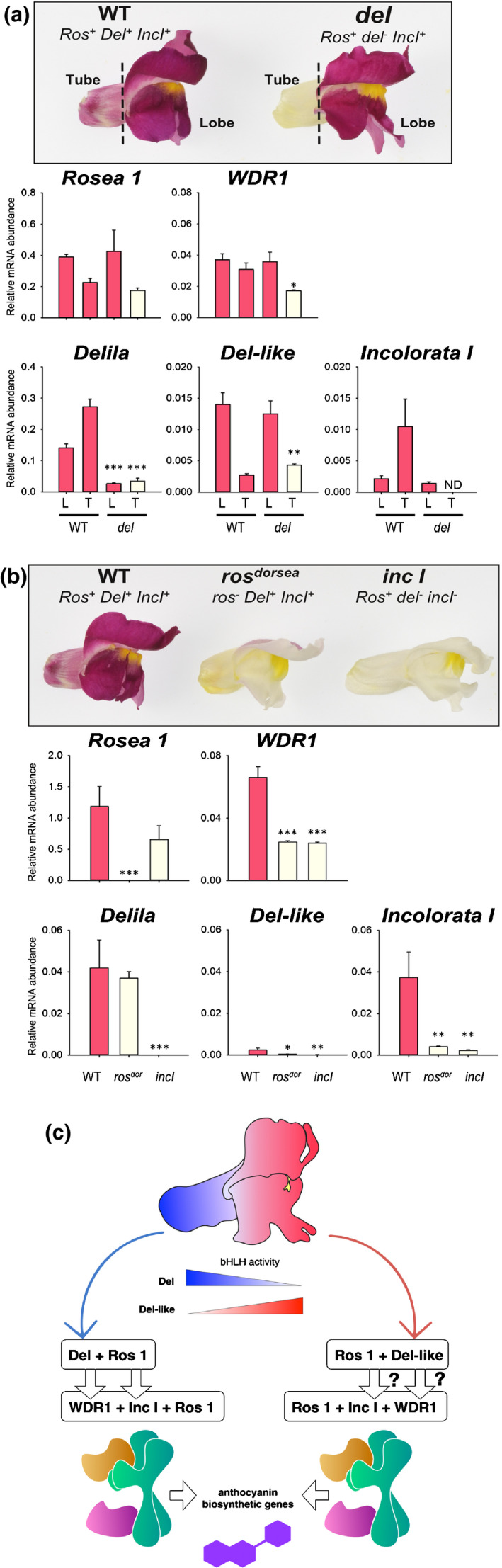
Bicolour patterning in the delila mutant arises from mis‐regulation of Incolorata I in flowers of A. majus. (a) Wild‐type (WT) and delila mutant flowers were analysed for Rosea1, WDR1, Delila, Del‐like and Incolorata I transcript levels by qRT‐PCR in tube (T) and lobe (L) tissue of WT and delila mutant petals. (b) Petals were analysed for Rosea 1, WDR1, Delila, Del‐like and Incolorata I transcript levels by qRT‐PCR. Bar colour indicates pigmentation status. Means ± SEM, n = 3 independent, biological replicates. Means that are significantly different (Student’s t‐test) from WT are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (c) Model illustrating how bicolour patterning may result from the tube‐ and lobe‐centred activities of Del and Del‐like respectively regulating expression of Inc I and WDR1 which, in turn, is necessary for full Inc I activity in the MBW complex.
Analysis of transcript levels in rosdor lines confirmed which regulatory genes were under MBW control. Inc I showed significantly lower levels of expression in rosdor compared with wild‐type flowers, confirming that it was under MBW control. Although Del‐like showed reduced expression in rosdor, the transcript levels detected were so low as to make the significance of the reduction ambiguous. Del was not impacted in its transcript levels in rosdor compared with wild‐type, confirming that Del is not regulated by the MBW complex. Ros 1 expression in inc I : del showed no significant changes in transcript levels compared with wild‐type, confirming that Ros 1 is not controlled by the MBW complex.
Discussion
We have identified Incolorata I encoding a bHLH‐2 protein involved in regulating anthocyanin biosynthesis in flowers of A. majus. Inc I has partially overlapping functions with Del, a bHLH‐1 protein. Both regulate anthocyanin biosynthesis directly and their differential activity in flowers determines bicoloured patterning of petals in del mutants. This differential activity of the two transcription factors can also explain the weak, patterned pigmentation of mutations of the Pallida gene encoding DFR, an enzyme essential for anthocyanin biosynthesis (Coen et al., 1986; Almeida et al., 1989). However, Inc I and Del are not completely functionally redundant; Del positively regulates Inc I (and WDR1) expression in flower tubes, and it is the Del‐independent expression of Inc I that gives bicoloured patterning of flowers. It is possible that Del‐independent, lobe‐specific expression of Inc I is dependent on Del‐like, which is expressed more highly in lobes than in tubes. It is unlikely that Del‐like regulates anthocyanin biosynthesis directly because inc I : del homozygotes completely lack anthocyanins in their flowers, or in any other parts of the plant. In addition, the Myb transcription factor Ros 1 showed no significant promotion of anthocyanin production in combination with Del‐like (Fig. 8) in N. benthamiana. Although Del‐like does not appear to activate the expression of anthocyanin biosynthetic genes substantially it might activate Inc I expression specifically in lobes.
Activity of Ros 1 and Del in inducing anthocyanin biosynthesis is not enhanced by the presence of the WDR1 protein and may, in fact, be inhibited by it. Activity of Ros 1 with Inc I is enhanced by the presence of WDR1, meaning that Inc I may act in an MBW complex, but that the most effective complex of Ros1 and Del may lack the WDR1 protein. Competition between Myb and WDR proteins in forming complexes with bHLH‐1 proteins has been reported by Pesch et al. (2015) in trichome formation in Arabidopsis, with the conclusion that this competition results in either an MB complex or a BW complex that regulate expression of different genes. In an extensive analysis of different subgroup IIIf bHLH proteins, Zhang et al. (2019) demonstrated that only bHLH‐1 proteins show competitive complex formation with Myb and WDRs whereas bHLH‐2 proteins always show enhanced complex formation with Myb and WDR proteins. This same difference between the bHLH proteins was shown by net anthocyanin biosynthesis in N. benthamiana, reflecting the integrated output of the MBW complex in leaves and implying that this difference also impacted expression of other genes involved in anthocyanin accumulation (Fig. 8).
Del and Inc I bind to G‐Box 1 in the DFR promoter of A. majus. Both positioning and sequence context of this G‐box are likely to be important for the binding and activity of Inc I and Del, as introduction of a G‐box displaced upstream in a different sequence context did not rescue the induction of gene expression driven by Del or Inc I from the DFR promoter lacking G‐box 1. The residual tube pigmentation observed in the equivalent mutants of A. majus (pal‐32 and pal‐33; Almeida et al., 1989) is likely to be driven by Del association with Ros 1 even where G‐box 1 has been deleted, so preventing DNA binding by bHLH transcription factors. However, binding by the bHLH protein to the target gene DNA may not be essential for inducing low level anthocyanin production in A. majus, (Fig. S8a). This ability of Del to interact with Ros 1 and promote anthocyanin production without DNA binding has been reported in tobacco (Applehagen et al., 2018), and for the B bHLH protein in regulation of the bz1 promoter in maize (Goff et al., 1992). Interestingly, the residual pigmentation observed in pal‐32 and pal‐33 is restricted predominantly to the flower tube (Fig. S8a; Coen et al., 1986; Almeida et al., 1989) suggesting that, although DNA binding may not be necessary for Del participation in the MBW complex on the DFR promoter, DNA binding is probably essential for Inc I‐WDR1 association with the MBW complex and DFR expression in the lobes in A. majus (Fig. S8b). This residual contribution of Del to induction of DFR gene expression is considerably lower than activation mediated by its binding to G‐box 1, and might vary from target gene to target gene as several of these promoter sequences in A. majus lack G‐box or even E‐box motifs (for example F3H: Martin et al., 1991) that are bound by bHLH proteins.
Bicolour patterning in flowers of A. majus results from Inc I being controlled by Del in tubes, reinforced by Del controlling WDR1 expression, which contributes positively to the activity of the Inc I‐containing MBW complex. In lobes there is Del‐independent expression of Inc I but this expression is probably dependent on an MB complex, possibly involving Del‐like (which is more highly expressed in lobes than tubes) interacting with Ros 1. Del‐like might have hierarchical activity on anthocyanin production through regulating Inc I, similar to the role of JAF13 in petunia (Albert et al., 2014) and tobacco (Montefiori et al., 2015). These findings reinforce the view that bHLH‐1 and bHLH‐2 proteins differ in their activity, even when their target pathways overlap, and that pigmentation patterning is established through hierarchical activities of transcription factors. The identification of transcription factors and mechanisms responsible for anthocyanin bicolouration in Antirrhinum provides a model for future studies to elucidate how the regulators of tissue and organ identity are linked to domain‐specific pigmentation patterns.
Author contributions
CM, PP, NWA, SMAM and KES cloned and characterised the Incolorata I gene and its mutant alleles, SMAM characterised the del23 mutant allele. CM and KES undertook RNA analysis using RNA gel blots and NWA analysed gene expression by qRT‐PCR. CNW performed biolistic complementation assays and KES cloned WDR1. EB performed dual luciferase assays in N. benthamiana, made all mutations of the DFR promoter for these assays and prepared Figs 7, 8, 9. NWA, KMD and CM drafted the manuscript before all authors contributed to its improvement and agreed on its final content. NWA and EB contributed equally to this work.
Supporting information
Dataset S1 Amino acid alignment of bHLH proteins.
Fig. S1 Differences in mutabilis phenotypes between glasshouse and field‐grown plants of A. majus.
Fig. S2 Lack of complementation between inc I1 and inc I2 of A. majus.
Fig. S3 RNA gel blot showing lack of Inc I transcript in flower lobes of the mutabilis mutant of A. majus.
Fig. S4 Sectors caused by excision of a transposable element (Tam 2) from the Del gene in inc I2 : delrec plants of A. majus.
Fig. S5 Phenotype of del23 allele of A. majus.
Fig. S6 Molecular analysis of del23 of A. majus.
Fig. S7 Impaired expression of anthocyanin biosynthetic genes in flowers of the inc I2 mutant of A. majus.
Fig. S8 Phenotypes of Pallida mutants of A. majus with deletions in their UAS controlling DFR expression caused by imprecise transposon excision and described by Almeida et al (1989).
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
CM, EB, PP and KES were supported by the Core Strategic Grant from the UK Biotechnology and Biological Sciences Research Council to the John Innes Centre, through the project ‘Understanding the control of flower development in Antirrhinum majus’ and the Institute Strategic Programmes ‘Understanding and Exploiting Plant and Microbial Secondary Metabolism’ (BB/J004596/1) and ‘Molecules from Nature’ (BB/P012523/1) from the UK Biotechnology and Biological Sciences Research Council. The Marsden Fund of New Zealand/Te Pūtea Rangahau A Marsden supported NWA, SMAM, CNW (contract PAF1501) and KES, KMD (contract CRO101). We are grateful to Prof. Peter Waterhouse (Queensland University of Technology, Brisbane, Australia) for providing the seeds of N. benthamiana cv Northern Territory. We thank Julie Ryan and Ian King at PFR and Peter Walker at JIC for care and maintenance of plants and Steve Mackay for laboratory support. We thank Andrew Davis and Peter Scott at JIC for photography.
References
- Albert NW. 2015. Subspecialization of R2R3‐MYB repressors for anthocyanin and proanthocyanidin regulation in forage legumes. Frontiers in Plant Science 6: 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26: 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Griffiths AG, Cousins GR, Verry IM, Williams WM. 2015. Anthocyanin leaf markings are regulated by a family of R2R3‐MYB genes in the genus Trifolium . New Phytologist 205: 882–893. [DOI] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. 2011. Members of an R2R3‐MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65: 771–784. [DOI] [PubMed] [Google Scholar]
- Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES. 1989. Genetic interactions underlying flower color patterns in Antirrhinum majus . Genes & Development 3: 1758–1767. [DOI] [PubMed] [Google Scholar]
- Appelhagen I, Wulff‐Vester AK, Wendell M, Hvoslef‐Eide AK, Russell J, Oertel A, Martens S, Mock HP, Martin C, Matros A. 2018. Colour bio‐factories: Towards scale‐up production of anthocyanins in plant cell cultures. Metabolic Engineering 48: 218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally J, Nakasugi K, Jia FZ, Jung HT, Ho SYW, Wong M, Paul CM, Naim F, Wood CC, Crowhurst RN et al. 2015. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nature Plants 1: 6. [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. 2006. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell‐specific accumulation of flavonoids in Arabidopsis thaliana . The Plant Journal 46: 768–779. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. 2004. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . The Plant Journal 39: 366–380. [DOI] [PubMed] [Google Scholar]
- Baur E. 1910. Vererbungs‐ und Bastardierungsversuche mit Antirrhinum . Z. Indukt. Abstammungs. Vererbungsl. 3: 34–98. [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. 2007. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiology 143: 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Moser M, Amrad A, Bapaume L, Barry C, Bliek M, Boersma M, Borghi L, Bruggmann R, Bucher M et al. 2016. Whole genome sequences of the wild parents of the garden petunia give insights into the evolution of Solanaceae genomes. Nature Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Ramadugu C, Durand‐Hulak M, Celant A, Reforgiato Recupero G, Froelicher Y, Martin C. 2019. Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Current Biology 29: 158–164. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Martin C, Coen ES. 1987. Comparison of genetic behavior of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus . Molecular & General Genetics 207: 82–89. [Google Scholar]
- Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D. 1989. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. The Plant Cell 1: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Carpenter R, Martin C. 1986. Transposable elements generate novel spatial patterns of gene‐expression in Antirrhinum majus . Cell 47: 285–296. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiology 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Greenblatt I, Kermicle JL, Hicks JB, Wessler SR. 1988. Molecular cloning of the maize R‐nj allele by transposon tagging with Ac. In: Gustafon JP, Appels R, eds. Chromosome structure and function. Boston MA, USA: Springer, 263–282. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfeld K, E Berardi A, Moser M, Bossolini E, Freitas L, Kuhlemeier C 2018. Pseudogenization and resurrection of a speciation gene. Current Biology 28: 3776–3786.e3777. [DOI] [PubMed] [Google Scholar]
- Feller A, MacHemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Feyissa DN, Lovdal T, Olsen KM, Slimestad R, Lillo C. 2009. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 230: 747–754. [DOI] [PubMed] [Google Scholar]
- Fincham JRS, Harrison BJ. 1967. Instability at the pal locus in Antirrhinum majus II. Multiple alleles produced by mutation of one original unstable allele. Heredity 22: 211–224. [Google Scholar]
- Frohman MA, Dush MK, Martin GR. 1988. Rapid production of full‐length cDNAs from rare transcripts: amplification using a single gene‐specific oligonucleotide primer. Proceedings of the National Academy of Sciences USA 85: 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. 1992. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes & Development 6: 864–875. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES. 1992. A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Haselhoff J, Siemering KR, Prasher DC, Hodge S. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA 94: 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. 2003. The basic helix‐loop‐helix transcription factor family in plants: A genome‐wide study of protein structure and functional diversity. Molecular Biology and Evolution 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Moreau C, Lin‐Wang K, Schwinn KE, Thomson SJ, Fiers MWEJ, Frew TJ, Murray SR, Hofer JMI, Jacobs JME et al. 2010. Identification of Mendel’s white flower character. PLoS ONE 5: e13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah ME, Gübitz M, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Kuhlemeier C. 2007. Single gene‐mediated shift in pollinator attraction in Petunia . The Plant Cell 19: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Roberts K, Martin C. 1992. Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinum majus . The Plant Journal 2: 425–434. [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang D, Gao Q, Luo Y, Zhang H, Ma B, Chen C, Whibley A, Zhang Y, Cao Y et al. 2019. Genome structure and evolution of Antirrhinum majus L. Nature Plants 5: 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PH, Chen BB, Zhang GY, Chen LX, Dong Q, Wen JQ, Mysore KS, Zhao J. 2016. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytologist 210: 905–921. [DOI] [PubMed] [Google Scholar]
- Li Y, Shan X, Gao R, Han T, Zhang J, Wang Y, Kimani S, Wang L, Gao X. 2020. MYB repressors and MBW activation complex collaborate to fine‐tune flower coloration in Freesia hybrida . Communications Biology 3: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnert G. 1972. Localization of some highly mutable loci of Antirrhinum majus L. by linage studies. Theoretical and Applied Genetics 42: 244–249. [DOI] [PubMed] [Google Scholar]
- Liu C, Jun JH, Dixon RA. 2014. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula . Plant Physiology 165: 1424–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. 1989. Lc a member of the maize R gene family responsible for tissue‐specific anthocyanin production encodes a protein similar to transcriptional activators and contains the Myc‐homology region. Proceedings of the National Academy of Sciences, USA 86: 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P et al. 2007. Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana . The Plant Journal 50: 678–695. [DOI] [PubMed] [Google Scholar]
- Martin C, Carpenter R, Coen ES, Gerats AGM. 1987. The control of floral pigmentation in Antirrhinum majus. In: Thomas H, Grierson D, eds. Developmental mutants in plants, SEB Seminar Series. Cambridge, UK: Cambridge University Press, 19–53. [Google Scholar]
- Martin C, Carpenter R, Sommer H, Saedler H, Coen ES. 1985. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO Journal 4: 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E. 1991. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus . The Plant Journal 1: 37–49. [DOI] [PubMed] [Google Scholar]
- Montefiori M, Brendolise C, Dare AP, Lin‐Wang K, Davies KM, Hellens RP, Allan AC. 2015. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. Journal of Experimental Botany 66: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. 2000. The TT8 gene encodes a basic helix‐loop‐helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell 12: 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KI, Choi JD, Hoshino A, Morita Y, Iida S. 2004. An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale‐colored flowers and seeds with fine spots in Ipomoea tricolor . The Plant Journal 38: 840–849. [DOI] [PubMed] [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S. 2007. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. The Plant Journal 49: 641–654. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd A. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Schultheiss I, Klopffleisch K, Uhrig JF, Koegl M, Clemen CS, Simon R, Weidtkamp‐Peters S, Hulskamp M. 2015. TRANSPARENT TESTA GLABRA1 and GLABRA1 compete for binding to GLABRA3 in Arabidopsis. Plant Physiology 168: 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. 2006. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic‐helix‐loop‐helix transcription factors of the anthocyanin pathway. The Plant Cell 18: 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. 2005. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Robbins TP, Carpenter R, Coen ES. 1989. A chromosome rearrangement suggests that donor and recipient sites are associated during Tam3 transposition in Antirrhinum majus . EMBO Journal 8: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. 2006. A small family of MYB‐regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum . The Plant Cell 18: 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. 2002. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. The Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. 2000. Anthocyanin1 of petunia encodes a basic helix‐loop‐helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell 12: 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzer P, Spelt CE, Li S, Bliek M, Federici CT, Roose ML, Koes R, Quattrocchio FM. 2019. Hyperacidification of Citrus fruits by a vacuolar proton‐pumping P‐ATPase complex. Nature Communications 10: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe H. 1966. In: Genetik und Zytologie von Antirrhinum L. sect. Antirrhinum. Jena, Germany: VEB Gustav Fischer Verlag. [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C. 2009. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiology 149: 1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Bassard JE, Ramirez‐Gonzalez R, Trick M, Afshar BG, Breitel D, Hill L, Foito A, Shepherd L, Freitag S et al. 2019. RNA‐seq, de novo transcriptome assembly and flavonoid gene analysis in 13 wild and cultivated berry fruit species with high content of phenolics. BMC Genomics 20: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheldale M. 1907. The inheritance of flower colour in Antirrhinum majus . Proceedings of the Royal Society of London, Series B: Biological Sciences 79: 288–305. [Google Scholar]
- Xu W, Grain D, Bobet S, Le Gourrierec J, Thevenin J, Kelemen Z, Lepiniec L, Dubos C. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB‐bHLH‐WDR complexes and their targets in Arabidopsis seed. New Phytolologist 202: 132–144. [DOI] [PubMed] [Google Scholar]
- Yuan YW, Sagawa JM, Frost L, Vela JP, Bradshaw HD Jr. 2014. Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytologist 204: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BP, Chopra D, Schrader A, Hulskamp M. 2019. Evolutionary comparison of competitive protein‐complex formation of MYB, bHLH, and WDR proteins in plants. Journal of Experimental Botany 70: 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BP, Hulskamp M. 2019. Evolutionary Analysis of MBW Function by Phenotypic Rescue in Arabidopsis thaliana . Frontiers in Plant Science 10: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1‐dependent pathways of Arabidopsis . Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. 2008. The TTG1‐bHLH‐MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B‐like BHLH proteins. The Plant Journal 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 Amino acid alignment of bHLH proteins.
Fig. S1 Differences in mutabilis phenotypes between glasshouse and field‐grown plants of A. majus.
Fig. S2 Lack of complementation between inc I1 and inc I2 of A. majus.
Fig. S3 RNA gel blot showing lack of Inc I transcript in flower lobes of the mutabilis mutant of A. majus.
Fig. S4 Sectors caused by excision of a transposable element (Tam 2) from the Del gene in inc I2 : delrec plants of A. majus.
Fig. S5 Phenotype of del23 allele of A. majus.
Fig. S6 Molecular analysis of del23 of A. majus.
Fig. S7 Impaired expression of anthocyanin biosynthetic genes in flowers of the inc I2 mutant of A. majus.
Fig. S8 Phenotypes of Pallida mutants of A. majus with deletions in their UAS controlling DFR expression caused by imprecise transposon excision and described by Almeida et al (1989).
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


