Abstract
The novel coronavirus disease, COVID‐19, has grown into a global pandemic and a major public health threat since its breakout in December 2019. To date, no specific therapeutic drug or vaccine for treating COVID‐19 and SARS has been FDA approved. Previous studies suggest that berberine, an isoquinoline alkaloid, has shown various biological activities that may help against COVID‐19 and SARS, including antiviral, anti‐allergy and inflammation, hepatoprotection against drug‐ and infection‐induced liver injury, as well as reducing oxidative stress. In particular, berberine has a wide range of antiviral activities such as anti‐influenza, anti‐hepatitis C, anti‐cytomegalovirus, and anti‐alphavirus. As an ingredient recommended in guidelines issued by the China National Health Commission for COVID‐19 to be combined with other therapy, berberine is a promising orally administered therapeutic candidate against SARS‐CoV and SARS‐CoV‐2. The current study comprehensively evaluates the potential therapeutic mechanisms of berberine in preventing and treating COVID‐19 and SARS using computational modeling, including target mining, gene ontology enrichment, pathway analyses, protein‐protein interaction analysis, and in silico molecular docking. An orally available immunotherapeutic‐berberine nanomedicine, named NIT‐X, has been developed by our group and has shown significantly increased oral bioavailability of berberine, increased IFN‐γ production by CD8+ T cells, and inhibition of mast cell histamine release in vivo, suggesting a protective immune response. We further validated the inhibition of replication of SARS‐CoV‐2 in lung epithelial cells line in vitro (Calu3 cells) by berberine. Moreover, the expression of targets including ACE2, TMPRSS2, IL‐1α, IL‐8, IL‐6, and CCL‐2 in SARS‐CoV‐2 infected Calu3 cells were significantly suppressed by NIT‐X. By supporting protective immunity while inhibiting pro‐inflammatory cytokines; inhibiting viral infection and replication; inducing apoptosis; and protecting against tissue damage, berberine is a promising candidate in preventing and treating COVID‐19 and SARS. Given the high oral bioavailability and safety of berberine nanomedicine, the current study may lead to the development of berberine as an orally, active therapeutic against COVID‐19 and SARS.
Keywords: anti‐viral, apoptosis, berberine, computational modeling, COVID‐19 and SARS
Abbreviations
- 3CLpro3
chymotrypsin‐like protease
- ACE2
angiotensin‐converting enzyme 2
- AGE‐RAGE
advanced glycation endproducts‐receptor for advanced glycation endproducts
- Akt
AKT serine/threonine kinase
- BAX
BCL2 associated X, apoptosis regulator
- BCL2
BCL2 apoptosis regulator
- BCL2L1
BCL2 Like 1
- BID
BH3 interacting domain death agonist
- CASP
critical assessment of structure prediction
- CCL2
C‐C motif chemokine ligand 2
- CCND1
cyclin D1
- CDK4
cyclin‐dependent kinase 4
- CDKN1A
cyclin‐dependent kinase inhibitor 1A
- CHUK
component of inhibitor of nuclear factor kappa B kinase complex
- CoV
coronavirus
- COVID‐19
coronavirus disease 2019
- COX‐2
cyclooxygenase‐2
- C‐T‐P‐D
compound‐target‐pathway‐disease
- CXCL2
C‐X‐C motif chemokine ligand 2
- EGFR
epidermal growth Factor Receptor
- ERK
extracellular signal‐regulated kinase
- FDR
false discovery rate
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- GO
gene ontology
- IFN
interferon
- IKBA
nuclear factor of kappa light polypeptide gene enhancer in B‐cells inhibitor, alpha
- IL
interleukin
- JUN
Jun proto‐oncogene, AP‐1 transcription factor subunit
- MAPKs
mitogen‐activated protein kinases
- MERS‐CoV
middle east respiratory syndrome‐related coronavirus
- MMP
matrix metallopeptidase
- MYC
MYC proto‐oncogene, BHLH transcription factor
- NADPH
nicotinamide adenine dinucleotide phosphate
- NFκB1
nuclear factor kappa B subunit 1
- NFκB1A
NF‐κB inhibitor alpha
- NF‐κB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- NLRP3
NOD‐like receptor family pyrin domain containing 3
- NOD
nucleotide binding oligomerization domain
- PGE2
prostaglandin E2
- Plpro
papain‐like protease
- PPI
protein‐protein interaction
- PTGS2
prostaglandin‐endoperoxide synthase 2
- RdRp
RNA‐dependent RNA polymerase
- ROS
reactive oxygen species
- SARS
severe acute respiratory syndrome
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus‐2
- TCM
traditional Chinese medicine
- TMPRSS2
transmembrane serine protease 2
- TNF
tumor necrosis factor
- TP53
tumor protein P53
- VEGFA
vascular endothelial growth factor A
1. INTRODUCTION
Coronavirus disease‐19 (COVID‐19) is an infectious disease caused by a newly discovered coronavirus SARS‐CoV2 that has reached global pandemic status and become a major global health threat. As of December 1 2020, there have been 63,751,931 confirmed cases and 1,477,976 deaths worldwide. 1 The United States hit record‐high daily COVID‐19 cases in November, 2020. Since the pandemic started in March, 2020, the nation has surpassed 12 million cases and more than 266,000 Americans have died. United States could see “a surge upon a surge” of COVID‐19 cases this winter. 2 During just two full days at the end of November, 2020, the country saw over 360,000 new COVID‐19 cases nationwide, in addition to over 2,700 new deaths. In 2003, a zoonotic coronavirus outbreak of SARS‐CoV had resulted in severe SARS with fatality rates of 10%. 3 , 4 , 5 , 6 The SARS‐CoV‐2 genome shares approximately 70%‐80% sequence similarity to SARS‐CoV, and causes similar clinical symptoms. 7 , 8 Key clinical features of COVID‐19 and SARS include fever, chills, muscle pain, headache, sore throat, new loss of taste or smell, cough, shortness of breath, gastrointestinal problems in mild to moderate cases, and more serious disease involving pneumonia, acute respiratory distress syndrome, cardiovascular and hepatic failure with high morbidity. 7 , 8 Individuals with pre‐existing conditions like cardiovascular disease, hypertension, asthma, and diabetes, 9 and elderly patients are at a higher risk to become infected with severe symptoms. 10 To date, no specific therapeutic drug or vaccine COVID‐19 and SARS is available, resulting in an urgent need for broad‐spectrum therapeutics for COVID‐19 and other CoV infections.
Traditional Chinese medicine (TCM) has been highly recommended by the government of China to treat COVID‐19 patients. 11 Natural products from TCM remain a rich source for the development of novel therapeutic agents for the treatment of COVID‐19 and SARS. Berberine, a natural isoquinoline alkaloid, is a medicinally valuable natural compound with published anti‐inflammatory, antiviral, antibacterial, anticancer, and antiparasitic activities, and having beneficial effects in hypertension, diabetes, and neurodegenerative conditions 12 , 13 , 14 Existing literature suggests that berberine has protective effects against drug‐ 15 or infection‐induced 16 liver, heart, and neuronal cell damage. 12 , 13 , 14 Moreover, berberine has shown a wide range of antiviral activities 17 such as anti‐influenza, 18 anti‐hepatitis C, 19 anti‐cytomegalovirus, 20 and anti‐alphaviruses. 21 Berberine is also an ingredient recommended in guidelines issued by China National Health Commission for COVID‐19 to be combined with other therapies. 8 In 2014, we discovered the IgE‐lowering property of berberine, 22 , 23 showing excellent efficacy in treating allergic diseases. Moreover, to overcome berberine's low biovaliability, 24 , 25 we developed an oral immunotherapeutic berberine nanomedicine (NIT‐X), for treating food allergy. 26 Our previous studies showed that a once‐a‐day oral NIT‐X (2 mg/mouse) for 4‐weeks resulted in 98%‐100% reduction in IgE and 100% protection against anaphylaxis in peanut‐allergic mice, which is associated with suppressing histamine release by mast cells, and induction of IFN‐γ by CD8+ T cells (Srivastava et al manuscript in preparation, 2020).
Pro‐inflammatory cytokines like IL‐6, IL‐1α/β, TNF‐α, IL‐8, and MCP‐1 (CCL2) promote severity of disease and tissue damage in COVID‐19. 27 The major cause of mortality is associated with hyper‐inflammation resulting in a cytokine storm. Moreover, recent meta‐analysis showed that a high IL‐6/IFN‐γ ratio was associated with severe COVID‐19 cases. 28 SARS‐CoV‐2 infection may primarily affect T lymphocytes, particularly CD4+ and CD8+ T cells, resulting in decreased IFN‐γ production 29 and adding IFN‐γ to type I IFNs as a synergistic combination therapy for COVID‐19 has been suggested. 30 Chronic inflammation and allergy share several molecular targets with COVID‐19. For example, the NFκB signaling pathway—one of two signals required for allergy‐prone IgE production 31 —and NFκB driven inflammatory cytokine production (TNF‐a, IL‐6, IL‐1β, IL‐8, etc) are involved in recalcitrant asthma. 32 , 33 Impaired IFN‐γ leads to susceptibility to hyperreactivity 34 and was associated with severe COVID‐19 cases in young people. 35 In addition, mast cell activation, the key mechanism involved in anaphylaxis, has been suggested to be involved in hyper‐inflammation in COVID‐19 patients and anti‐IgE and mast cell mediator antagonists appear to be helpful for COVID‐19 patients. 36 , 37 , 38 , 39
Early response of IFNs is critical for combatting SARS‐CoV‐2. 29 , 30 Berberine has been shown to increase the production of INF‐γ, and inhibit Th2 responses, 40 indicating its potential role in protective immunity for infectious diseases. 41 Previously, we also found that in vivo treatment with berberine/NIT‐X resulted in significant elevation of IFN‐γ and CD8+ T cells in peanut‐restimulated splenocytes in a murine model. Furthermore, berberine was shown to mitigate inflammatory cytokine production by downregulation of MAP Kinase and ERK and downregulation of pro‐inflammatory transcription factors NFκB and AP‐1. 12 , 42 , 43 , 44 The hallmarks of SARS and COVID‐19 disease are unchecked viral replication and serious multiorgan tissue damage. 45 , 46 , 47 , 48 Berberine/NIT‐X might alleviate tissue damage by reducing inflammation‐induced death signals (RAGE, NLRP3/Caspase). 49 , 50 With excellent efficacy in treating allergy, we hypothesized that berberine/NIT‐X would likewise be effective against COVID‐19 and SARS by supporting protective immunity 41 while inhibiting pro‐inflammatory cytokines and viral replication, inducing apoptosis, and protecting against tissue damage.
The current study seeks to evaluate the potential therapeutic mechanisms underlying the efficacy of berberine/NIT‐X in preventing and treating COVID‐19 and SARS using computational modeling—target mining, gene ontology enrichment, pathway, and protein‐protein interaction analyses, and in silico molecular docking. The entire workflow of the study is shown in Figure 1. In addition, the inhibition of replication of SARS‐CoV‐2 by berberine/NIT‐X has been validated in vitro. The suppression of ACE2, TMPRSS2, IL‐1α, IL‐8, IL‐6, and CCL‐2 by berberine/NIT‐X were further validated. The results of these analyses will contribute to a molecular‐level understanding of how berberine functions in the prevention and treatment of COVID‐19 and SARS, providing both the rationale and tools needed to further validate its efficacy. Given the discovery of the high oral bioavailable ability of berberine nanomedicine (NIT‐X), 26 the current study may lead to a development of berberine/NIT‐X as an orally active therapeutic against COVID‐19 and SARS.
FIGURE 1.
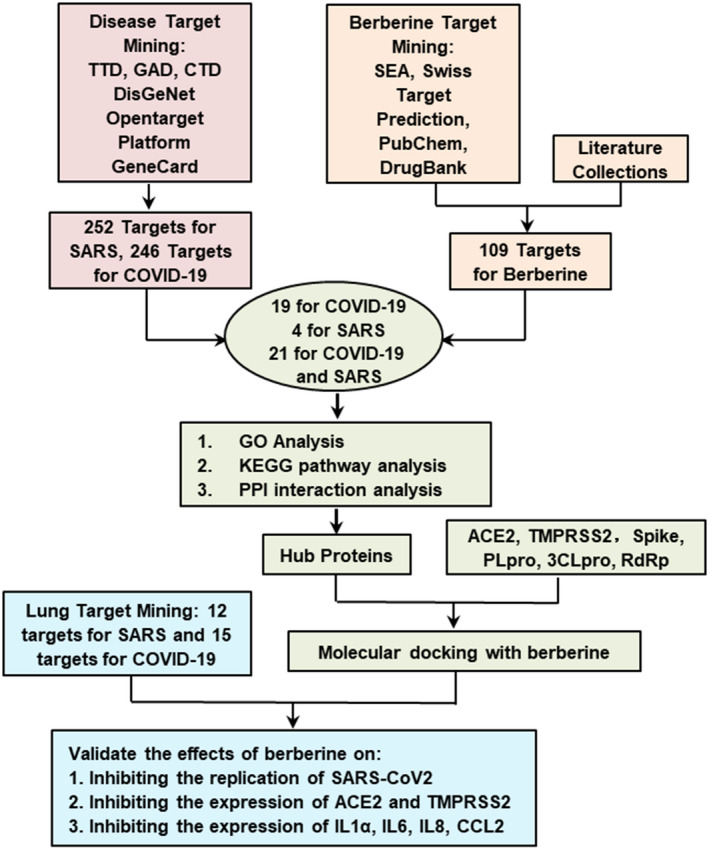
The workflow of computational modeling used for analysis of berberine as a promising candidate against COVID‐19 and SARS. TTD: therapeutic target database; GAD: genetic association database; SEA: similarity ensemble approach; PPI: protein‐protein interactions; ACE2: angiotensin‐converting enzyme 2; TMPRSS2: transmembrane serine protease 2; PLpro: Papain‐like Protease; 3CLpro: coronavirus main proteinase; RdRp: RNA‐dependent RNA polymerase. First, a total of 254 genes for SARS and 247 genes for COVID‐19 were selected as possible targets for berberine. Separately, 109 inflammatory and biological targets of berberine were collected based on literatures and following published databases: hitpick, swiss target prediction, SEA, pubchem, and drugbank. Mining berberine targets onto identified disease targets uncovers that berberine might potentially regulate 21 targets for both COVID‐19 and SARS, 4 targets for SARS specifically, and 19 targets for COVID‐19 specifically. With biological targets of berberine for the prevention and treatment of COVID‐19 and SARS established, GO, KEGG pathway, and PPI analysis were conducted to uncover the mechanistic details of berberine regulation in key pathways. Moreover, hub host proteins are determined for further molecular docking analysis. Combining crucial virus proteins and vital host receptor proteins in virus infection and duplication process, molecular docking was further applied to investigate the possible binding modes of berberine to these targets. Based on the lung target mining and molecular docking results, the inhibition of berberine on ACE2, TMPRSS2, IL1α, IL6, IL8, and CCL2 were further validated
2. MATERIALS AND METHODS
2.1. Target mining
Biological targets of berberine were identified from literature reports 51 , 52 and published databases including Similarity Ensemble Approach, 56 , 57 PubChem, 58 , 59 and DrugBank. 60 , 61 The relevant human genes associated with COVID‐19 and SARS were selected as drug targets from various databases including Therapeutic Target Database, 62 , 63 Genetic Association Database, 64 , 65 GeneCards, 66 , 67 Open Targets Platform, 68 , 69 and Comparative Toxicogenomics Database. 70 , 71 The lung specific targets were collected from Open Targets Platform 68 , 69 and literature. 72 To ensure the predominance of targets, only the top 100 genes in each database were considered. Selected targets were finally mapped to UniProt Database 73 , 74 for normalization. Next, the shared targets of berberine with COVID‐19 and SARS were obtained and these were considered to be potential regulated targets of berberine for the prevention and treatment of COVID‐19 and SARS.
2.2. Gene ontology (GO), pathway, and protein‐protein interaction (PPI) analysis
Target enrichment gene ontology, pathway, and protein‐protein interaction (PPI) analyses provided a molecular‐level mechanistic insight into biological function. GO was introduced by mapping potential targets to the DAVID database. 75 , 76 The GO biological process terms with a false discovery rate of (FDR) < 0.01 were selected. Pathways were obtained by mapping targets to KOBAS 3.0 77 , 78 and the significant pathways with FDR < 0.01 were selected. Potential targets were mapped to String database, obtaining their interaction. The protein interactions were further used to construct the PPI network using Cytoscape (v3.2.1).
2.3. Compound‐target‐pathway‐disease (C‐T‐P‐D) network construction and analysis
With obtained targets and significant pathways, C‐T‐P‐D biological networks were constructed using Cytoscape (v3.2.1). The C‐T‐P‐D network, containing berberine, its related targets for COVID‐19 and SARS, and significant principal pathways provided general information about pharmacological mechanisms of berberine for the prevention and treatment of COVID‐19 and SARS at a molecular level. The properties of C‐T‐P‐D networks were validated by NetworkAnalyzer, 79 a plugin of Cytoscape.
2.4. Molecular docking analysis
The hub 10 proteins in C‐T‐P‐D network were selected as highly promising targets of berberine. Moreover, human receptors and virus proteins involved in viral infection and replication processes were all considered as potential targets of berberine. Molecular docking was performed on berberine with obtained targets by AutoDock Vina 80 to further explore their binding modes. Protein crystal structures including NFκB1 (PDB:1NFK), CHUK (PDB:5EBZ), 81 MAPK3 (PDB:4QTB), 82 MAPK1 (PDB:4O6E), 83 CASP3 (PDB:1PAU), IL6 (PDB: 1ALU), 84 MAPK8 (PDB:2H96), 85 BAX (PDB:4S0O), 86 and TNF (PDB:2AZ5), 87 ACE2 (PDB:1R4L), 88 3CLpro (PDB:6LU7), 89 Spike (PDB: 6VW1), PLpro (PDB: 6W9C), and RdRp (PDB:6M71) 90 with excellent resolution were downloaded from RCSB protein data bank (www.rcsb.org/). 91 Protein structures of NFκB1A were built by homology modeling. 92 Structure of TMPRSS2 was obtained from a reported model. 93 The structure of berberine was directly downloaded from PubChem (pubchem.ncbi.nlm.nih.gov/) 59 , 94 without further optimization. Proteins and berberine were prepared by AutoDockTools (v1.5.6). 95 The molecular graphics were prepared by PyMOL system 96 (http://www.pymol.org) and Discovery Studio. 97 Generally, all hydrogens and Gasteriger charges were added to each molecule. Docking areas and AutoGrid parameters were set based on the binding pockets of proteins.
2.5. Cell culture
Human epithelial cells, Calu‐3 (American Type Culture Collection; Rockville, MD), were used to evaluate the effect of NIT‐X on SARS‐CoV‐2 viral replication, ACE2, TMPSS2, and cytokine and chemokine expression. Cells were cultured at 37°C under 5% CO2 in complete EMEM medium supplemented with 20% FBS, and 1% penicillin‐streptomycin. Cells were seeded at an initial concentration of 5 × 105 cells/mL and medium was changed every 3 days.
2.6. Cell infection and real time polymerase chain reaction (PCR)
Calu‐3 cells were seeded in six well plates with 5 × 105 cells per well. After 24 hours, cells were incubated in media containing 20 µg/mL, 40 µg/mL berberine/NIT‐X, or DMSO (v/v 1:1000). After 3 days incubation, cells were infected with SARS‐CoV‐2 (MOI = 0.005) or mock (‐) for 1 hour and grown in indicated media for 24 hours. At 1‐day post infection (1 dpi), cells were harvested and total RNA in mock or infected cells were extracted using Trizol. A 200 ng of total RNA was used for cDNA synthesis. Real Time‐PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) with SARS‐Cov‐2 Nucleocapsid (N) proteins, ACE, TMPSS2, IL‐6, IL‐8, IL‐1α, CCL2, and HPRT primers. The primer sequences are listed as in Table S1. Data were normalized to HPRT and presented as 2‐ΔCT.
2.7. Cell viability assay
Cell viability by CCK‐8 assay of berberine/NIT‐X on Calu3 cells was performed using a commercial kit (Dojindo, Rockville, MD). A 6.5 × 103/well Calu3 cells were seeded on 96 well plate and preincubated for 24 hours. Next, berberine/NIT‐X at 20 µg/mL and 40 µg/mL were added in corresponding wells and cells were incubated for another 24 hours. A 10 µL of CCK‐8 solution was then added into each well and incubated for 1 hour. The plate was read at 450 nm. Cell viability was analyzed by comparing the OD value.
3. RESULTS AND DISCUSSION
3.1. Target mining identifies the shared biological targets between berberine, COVID‐19, and SARS
The Venn diagram in Figure 2 shows that 252 genes for SARS and 246 genes for COVID‐19 were selected, totaling 336 genes. Of these, 155 genes were shared between COVID‐19 and SARS, supporting the rationale to develop therapies for both diseases. Moreover, 109 biological targets of berberine were collected from the literature, 51 , 52 and published databases. Among them, 21 shared targets for berberine with both COVID‐19 and SARS; 19 shared targets for berberine and COVID‐19; and four shared targets for berberine and SARS were discovered, yielding 44 shared targets between berberine and COVID‐19 and SARS, which were finally selected as the main targets of berberine in preventing and treating COVID‐19 and SARS.
FIGURE 2.
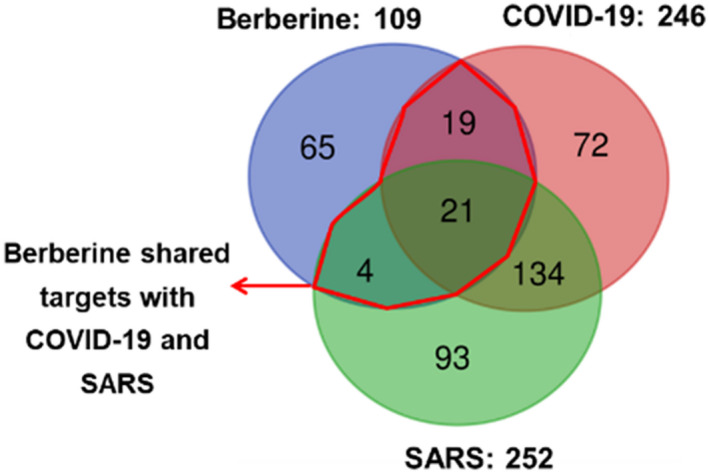
Venn diagram showing shared targets between berberine, COVID‐19, and SARS. Among them, 21 shared targets are identified for berberine with both COVID‐19 and SARS, 19 shared targets for berberine and COVID, and 4 shared targets for berberine and SARS
3.2. Gene ontology (GO) reveals potential regulation of berberine in apoptosis, proliferation, and host systematic reactions
GO biological process terms in DAVID database were obtained with the identified targets as an enriched gene‐set. The top 15 biological process GO terms with FDR < 0.01 were ranked by enrichment score (‐logFDR) in Figure 3A. The host acts against pathogenic microbes by inducing both innate and adaptive immune responses. 98 Most significant GO biological process terms are closely associated with the change in states or activities of cells encountering viral infection, such as proliferation, differentiation, secretion, gene expression, and apoptosis. Among them, inducing apoptosis in host cells at early‐stage infection by viruses has been considered as a self‐defense mechanism. 99 , 100 In infected cells, the viruses are likely to be destroyed along with phagocytized and digested processes of apoptotic cells. Cell apoptosis has been observed in a wide range of human viral infections, 101 for instance, cardiocyte apoptosis during both active and chronic viral myocarditis, 102 hepatocyte apoptosis during hepatitis B virus 103 and hepatitis C virus infections, 104 and apoptosis during influenza virus infection. 105 Berberine has been demonstrated to induce biphasic cell death in treating hepatitis C virus‐induced hepatocellular carcinoma—first triggering apoptosis in early‐stage at 24 hours post‐berberine treatment that then progressing to necrotic cell death at 48 hours posttreatment. 106 Apoptosis induced by berberine through mitochondria/caspases pathway in cancer cells has been widely investigated, 107 , 108 , 109 which may be a vital regulated process of berberine to prevent viral proliferation in early infection. Based on GO analysis results, regulated apoptosis by berberine includes “negative regulation of apoptotic process,” “apoptotic process,” “extrinsic apoptotic signaling pathway in absence of ligand,” “activation of cysteine‐type endopeptidase activity involved in apoptotic process,” and “regulation of apoptotic process.” In addition, most viral infections including SARS‐CoV infection may induce a transient state of immune suppression and cell proliferation inhibition. 110 , 111 Cell proliferation, especially immune cell proliferation, plays a crucial role in combatting viral infection. However, in hyperinflammation caused by COVID‐19 and SARS, immunosuppression is likely to be beneficial. 112 Berberine has been reported to cause G0/G1 113 and G2/M 114 , 115 cell arrest leading to inhibition of cell proliferation in different cell lines. The proliferation‐related process, including “positive regulation of cell proliferation” and “cell proliferation,” displays the potential efficacy of berberine in regulating cell proliferation and balancing the immune response to resist COVID‐19 and SARS. Moreover, the comprehensive cell activities include “response to drug,” “cellular response to organic cyclic compound,” “response to estradiol,” “cellular response to mechanical stimulus,” “response to toxic substance,” and “cellular response to DNA damage stimulus,” standing for systemic regulation of berberine for host systematic reaction after a viral stimulus.
FIGURE 3.
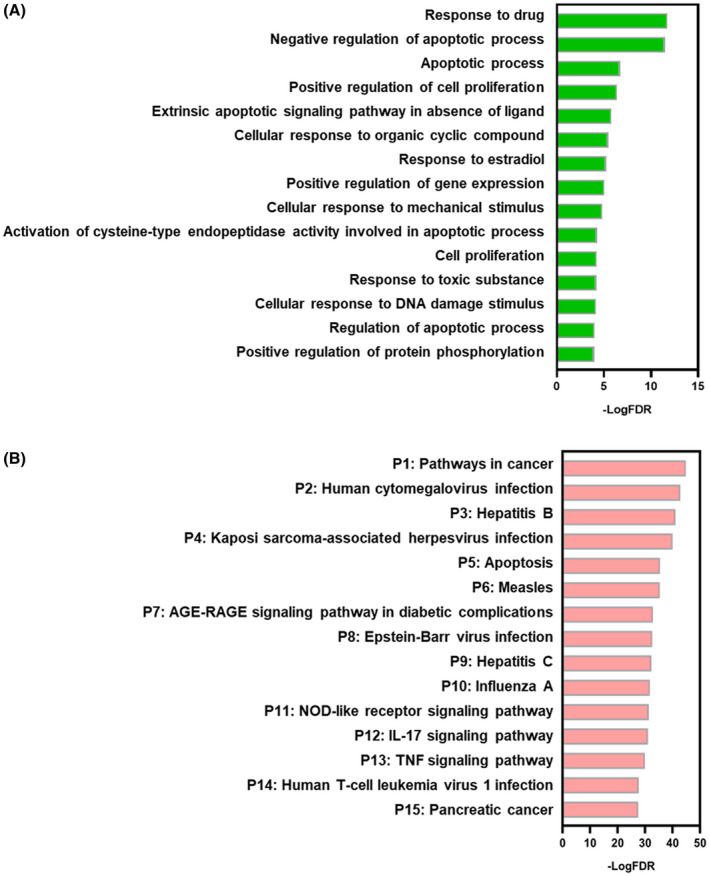
Gene ontology (GO) biological process (BP) analyses and pathway analyses of the targets. A, GO BP analyses; B, Pathway analysis; Y‐axis: top 15 biological processes (A) and top 15 pathway (B) relevant to the enriched targets; X‐axis: significance of each term ranked with –log (false discovery rate) (FDR)
3.3. Pathway analysis reveals complex signal transduction regulated by berberine
Regulated Kegg pathways were obtained with these identified targets as an enriched gene‐set. The top 15 significant pathways with FDR < 0.01 were ranked by enrichment score (‐logFDR) in Figure 3B. The involved genes in each pathway are listed in Figure S1. The pathway results are consistent with the main points deduced from GO analysis. Most pathways (53%) are related to host immune responses to viral infection including “Human cytomegalovirus infection (P2),” “Hepatitis B (P3),” “Kaposi sarcoma‐associated herpesvirus infection (P4),” “Measles (P6),” “Epstein‐Barr virus infection (P8),” “Hepatitis C (P9),” “Influenza A (P10),” and “Human T‐cell leukemia virus 1 infection (P14).” During viral infection, both innate and adaptive immune reactions are activated to regulate various cell activities through signaling transduction. 98 Moreover, “NOD‐like receptor signaling pathway (P11)” is an immune‐related pathway, which is responsible for detecting various pathogens and generating innate immune response. 116 Especially, NLRP3 activation is well recognized as a trigger for CoV inflammatory cascade and tissue damage. 117 , 118 , 119 Berberine has been reported to alleviate influenza virus‐induced inflammatory lesions by restricting NLRP3 inflammasome activation through decreasing ROS generation. 49 NLRP3 may, therefore, be a high priority target of berberine against CoVs in the regulation of “NOD‐like receptor signaling pathway (P11).” “Apoptosis (P5)” further emphasizes the regulation of apoptosis by berberine, which is an innate immune response to viral infection. The cytokine‐related pathways include “IL‐17 signaling pathway (P12)” and “TNF signaling pathway (P13),” which play a crucial role in modulating immune pathophysiology of viral infection. 120 , 121 It is reported that the severity of COVID‐19 and SARS positively correlates with levels of Th17 cell‐related pro‐inflammatory cytokines including IL‐17, IL‐6, IL‐1, TNF, and IFN‐γ. 122 And berberine has been found to regulate differentiation and amelioration of Th1 and Th2 cell to impact the corresponding cytokines, 123 indicating IL‐17 and TNF might be the main cytokines regulated by berberine to control the course of infection. In addition, the “AGE‐RAGE signaling pathway in diabetic complications (P7)” is related to inflammatory regulation. AGE‐RAGE signaling elicits activation of multiple intracellular signaling pathways involving NADPH oxidase, protein kinase C, and MAPKs, then, resulting in NF‐κB activation. 124 Much evidence 125 shows that pulmonary tissues express a remarkably high basal level of RAGE, which is a key molecule in the onset and sustainment of the inflammatory response in many disease pathologies. Berberine has been discovered to regulate AGEs‐RAGE signaling pathway in mesangial cells exerting renoprotective effects during diabetic nephropathy, 50 which might indicate its possible role in regulation of AGEs‐RAGE signaling during SARS‐CoV and SARS‐CoV‐2 infection. 126 In addition, “Pathway in cancer (P1)” and “Pancreatic cancer (P15)” stand for complex pathways involving various activities including inflammatory process, metabolic regulation, cell proliferation, and cell apoptosis, part of which may be regulated by berberine to resist against the virus.
3.4. Compound‐target‐pathway‐disease (C‐T‐P‐D) network construction to select the crucial proteins
C‐T‐P‐D network (Figure 4) containing berberine, selected targets, top 15 pathways, and COVID‐19 and SARS as diseases, provides general information about the potential pharmacological mechanisms of berberine for prevention and treatment of COVID and SARS at the molecular level. We believe that the frequency of targets appearing in the top 15 pathways imply their influence and importance. Node sizes from large to small, and color from red to green, are proportional to degree value, displaying their importance from high to low in the network. The encoded proteins of NFκB1 (p50 and p105), CHUK (IKK), NFκB1A (IKBA), and TNF (TNF‐α) all play crucial roles in NF‐κB signaling pathway, which participates in multiple aspects of innate and adaptive immune functions and serves as a pivotal mediator of inflammatory responses. 127 The activation of NF‐κB can not only induce the expression of various pro‐inflammatory cytokines and chemokines 127 , 128 such as TNF‐α, IL‐8, IL‐6, IL‐1β, and COX‐2, but also regulate the survival, activation and differentiation of innate immune cells and inflammatory T cells, 129 , 130 causing hyper‐inflammation and severe tissue damage during viral infection. It has been demonstrated that treatment with NF‐κB inhibitors did not affect virus titers but reduced expression of cytokines such as TNF, CCL2, and CXCL2; alleviated lung pathology in both SARS‐CoV‐infected cultured cells and mice; and significantly increased mouse survival. 131 In addition, NF‐κB signaling participated in most of the top 15 pathways, further indicating its pivotal role. Berberine has been found to suppress the activation of NF‐κB through inhibition of various inflammatory agents, such as direct inhibition of IκB kinase (IKK) activation and inhibition of PPARγ activation, which may contribute in part to the inhibition of NF‐κB. 132 Thus, proteins in NF‐κB signaling transduction might be promising main targets of berberine to suppress the hyper‐inflammation during SARS‐CoV and SARS‐CoV2 infection. Similarly, MAPKs are members of all top 15 pathways. MAPK3, MAPK1, and MAPK8, signal transducers responding to extracellular stimulation by cytokines, growth factors, viral infection, and stress can regulate cell differentiation, proliferation, survival, and apoptosis. 133 , 134 , 135 It is reported that SARS‐CoV causes the activation of physiological intracellular signaling cascades leading to the phosphorylation and activation of p38 MAPK signaling pathway. 135 Moreover, a subset of the licensed kinase inhibitors targeting the ERK/MAPK pathway has been demonstrated to significantly inhibit MERS‐CoV propagation in vitro regardless of whether they were added before, or after viral infection. 101 Downregulated expression of MAPKs was detected in recovered COVID‐19 patients, indicating the MAPK signal pathway may be one sign of patient recovery. 136 Berberine has been found to hamper influenza A and enterovirus 71 replication through inhibition of MAPK/ERK signaling pathway, 137 , 138 thus, inhibition of the ERK signaling pathway is one of the primary potential mechanisms of action for berberine suppression of SARS‐CoV and SARS‐CoV‐2 replication. 139 Apoptosis‐related genes including CASP3, CASP8, BAX, BID, BCL2, and BCL2L1 can initiate apoptotic signaling (P5) via the extrinsic pathway or intrinsic pathway. The importance of apoptotic regulation for COVID‐19 and SARS treatment has been discussed in both GO and pathway analysis. CASP3, CASP8, and BAX are known targets of berberine in regulating apoptosis 109 , 140 , 141 and may in fact be the critical targets in inducing apoptosis of cells infected by SARS‐CoV and SARS‐CoV‐2. Moreover, the inhibition of pro‐inflammatory cytokines (eg, TNF‐α, IL‐1α/β, and IL‐6) by berberine would prevent infected cells from pyroptosis and reduce the tissue damage in late stages. Cytokines and chemokines, including IL‐6, TNF‐α, IFN‐β, IL‐8, IL‐1α/β, CCL2, MMP9, and MMP2, have all been confirmed to be closely associated with pathogenesis of COVID‐19 and SARS. 142 , 143 , 144 Among them, it has been reported that the high levels of IL‐6 are activated by the viral nucleocapsid SARS‐CoV N protein, causing lung lesions in SARS patients. 145 The increased expression of IL‐6 and IL‐8 in serum is expected to predict the severity of COVID‐19 pneumonia and the prognosis of patients in the clinic 146 and IL‐1α/β could mediate the inflammation of the lungs, fever, and fibrosis thus causing respiratory complications in the infected host. 146 Berberine has been shown to significantly suppress TNF‐α and IL‐6 expression induced by HIV protease inhibitors even at low concentrations. 147 The production of IL‐1α/β and TNF‐α have been reported to be suppressed by berberine via the inhibition of IκB degradation in human lung cells. 44 , 148 Berberine can regulate the production or effects of inflammatory cytokines directly or indirectly, 149 , 150 which make it a promising agent in the prevention and treatment of COVID‐19 and SARS. Moreover, the regulation of cell proliferation by berberine has been mentioned previously. The targets related with cell proliferation and cell cycle include CDK4, TP53, CCND1, CDKN1A, and MYC and it is known that berberine can up‐regulate varicocele‐induced CDK4 and CCND1 expression reduction in rat testicles. 151 Berberine may also balance cell proliferation and apoptosis during viral infection via signaling regulation. In conclusion, these genes are all promising targets on which berberine may act to regulate immune responses, inflammatory processes, and cell activities against COVID‐19 and SARS infection. The top 10 targets in the network were chosen for further molecular docking analysis.
FIGURE 4.
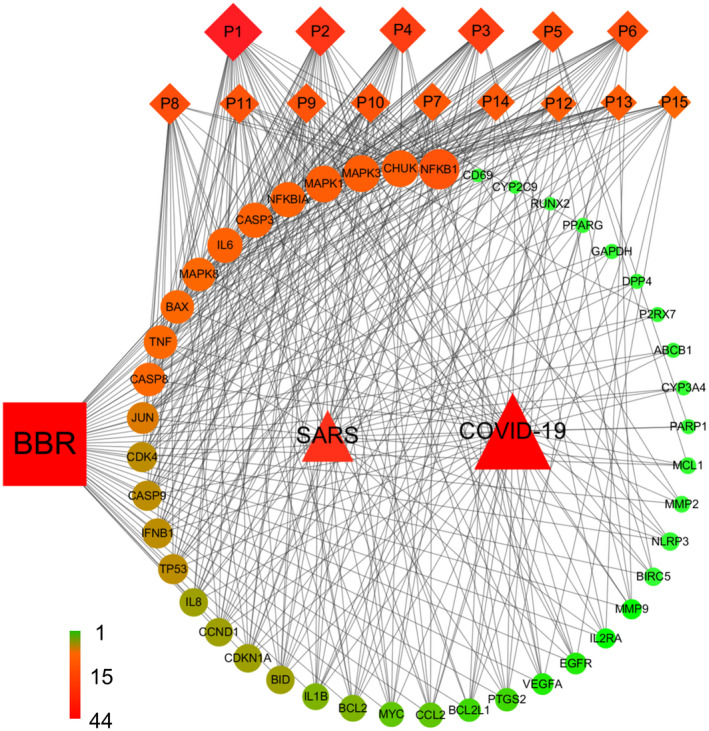
Compound‐Target‐Pathway‐Disease (C‐T‐P‐D) network of the berberine for COVID‐19 and SARS treatment. Squares, circles, diamonds, and triangles represent berberine, common targets, pathways and diseases, respectively. Node size and node color (ie, from green (lowest) to red (highest) indicate a measure of degree. Black lines represent interaction between nodes
3.5. Protein‐Protein interaction (PPI) network construction to confirm the vital function of proteins
The PPI network (Figure 5) was constructed by mapping potential targets to the String database. 152 The size of the node from large to small is proportional to its degree value in the network. It is well known that protein‐protein interactions are critical to a wide range of biological processes, including cell‐to‐cell interaction and metabolic and developmental control. 153 Deeper understanding of such complex relationships among disease‐related proteins provides new opportunities to investigate the molecular mechanisms of diseases. 154 Recently, PPI has become a reliable tool to evaluate protein functions in the network and determine hub proteins in the regulation of diseases. As expected, TNF (TNF‐α) occupied the central position in the network, indicating its close associations with the other proteins. TNF‐α belongs to the TNF superfamily of cytokines, which regulates dozens of pathways related to cell proliferation, differentiation, survival, and death. 155 Following, IL‐6, NFκB1, CASPs, and MAPKs have previously all been selected as pivotal targets of berberine, and will not be discussed further here. As with the C‐T‐P‐D network, proteins such as TP53 and JUN hold a moderate rank in the PPI network; however, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) seems to display more connections in the PPI network. Besides association with cellular glycolysis, GAPDH can modulate cellular signaling pathways in response to oxidative stress, and participates in cell death/dysfunction processes in the nucleus. 156 In particular, GAPDH has been demonstrated to bind with telomeres and protect telomeric DNA from rapid degradation, 157 with telomere length possibly playing an important role in maintaining the rapid replicative response of leukomonocytes in the face of SARS‐CoV and SARS‐CoV‐2 infection. 158 Berberine has been proven to regulate the level of GAPDH in adipocyte differentiation, 159 indicating that berberine may affect telomere status through regulating GAPDH during SARS‐CoV and SARS‐CoV‐2 infection. Moreover, it's worth noting that targets including VEGFA, CCND1, EGFR, and PTGS2 play a critical role in signal transmissions in PPI. Vascular Endothelial Growth Factor (VEGF) is considered to be the most potent vascular permeability inducer and recent evidence 77 has revealed higher VEGF levels in COVID‐19 patients compared with healthy controls. The rise of VEGF levels may be caused by hypoxia, severe inflammation, and upregulation of the infected respiratory tract epithelium itself. 160 , 161 CCND1 is related to cell proliferation and apoptosis, and this has been fully discussed earlier. Activation of EFGR triggers the signaling of MAPK, Akt, and JNK pathways, resulting in a range of outcomes, such as inhibition of apoptosis, increase in cell proliferation and migration, activation of the inflammatory response, and increase of IL‐8 production. 162 EFGR signaling has been shown to mediate pulmonary fibrosis in the hyperactive host response to SARS‐CoV infection. And the inhibition of EGFR signaling may prevent an excessive fibrotic response to SARS‐CoV and other respiratory viral infections. 163 It has been reported that berberine could suppress the constitutive activation of EGFR in tumor cells to inhibit growth and induce apoptosis. 164 Thus, EGFR may be an upstream target of berberine to prevent lung fibrosis during SARS‐CoV and SARS‐CoV‐2 infection. Cyclooxygenases (COXs/PTGSs) play a significant role in many different viral infections with respect to replication and pathogenesis, 165 such as herpesviruses, 166 bovine leukemia virus, 167 and rotavirus. 168 Moreover, structural proteins from the SARS‐CoV were shown to induce the expression of COX‐2 in vitro 169 and elevated levels of PGE2 were found in the blood of SARS‐CoV‐infected individuals 170 caused by increased COX‐2 expression, suggesting a role for COXs and PGs in CoV pathogenesis. Both in vitro and in vivo studies have reported that berberine decreased the expression of both TNF‐α and COX‐2 in a hepatotoxicity rat model induced by cyclophosphamide. 171 The inhibition of COX‐2 expression 172 by berberine further increases the possibility for berberine to become a promising candidate in the prevention and treatment of COVID‐19 and SARS.
FIGURE 5.
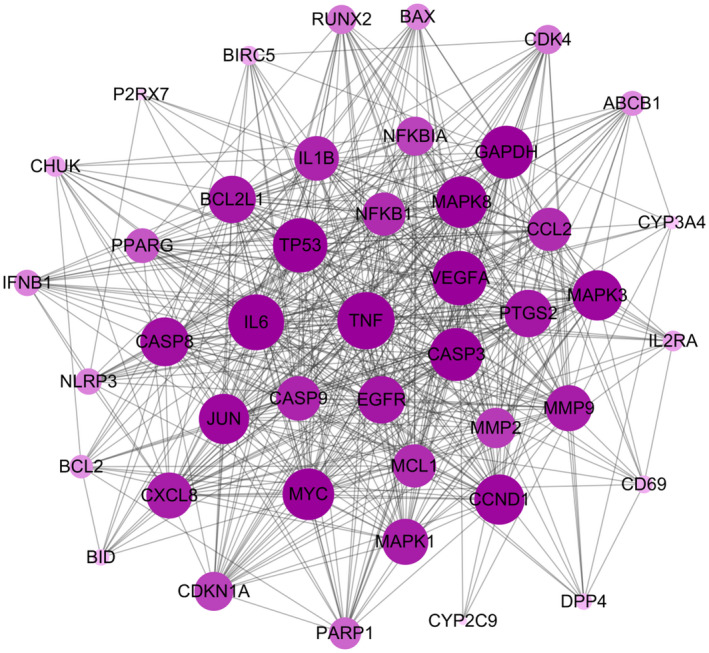
Protein‐protein interactions. Circles represent the targets. Black lines represent the interaction between nodes. Node size is proportional to its degree in network
3.6. Molecular docking analysis predicts the binding modes between berberine and its crucial targets
Berberine targets can be categorized into three categories: host immune‐related proteins, host receptors, and virus proteins. During SARS‐CoV2 infection, the immune system is activated by stimulating lymphocytes to release cytokines including IL‐6, IL‐8, IL‐7, IL‐2, IL‐1, IFN‐γ, and TNF‐α, which may cause hyperinflammation. 122 In addition, the cytokine storm has been considered to be one of the major causes of acute respiratory distress syndrome (ARDS) and multiple organ failure in COVID‐19 and SARS. 144 Regulating immune‐related proteins is therefore likely to be beneficial for the treatment of COVID‐19 and SARS. The top 10 vital proteins in the C‐T‐P‐D network were chosen as host immune‐related proteins for molecular docking analysis, including NFκB1, CHUK, MAPK3, MAPK1, NFκB1A, CASP3, IL6, MAPK8, BAX, and TNF, all of which have been shown to be strongly associated with the pathogenesis of COVID‐19 and SARS (and most of which are equally prominent in PPI analysis). Blocking the viral entrance process itself is another strategy to inhibit viral infection and ACE2 has been proven to be a cellular entry receptor of COVID‐19 and SARS. 173 In addition, TMPRSS2 helps the CoV spike (S) glycoprotein, a key target for the development of vaccines, therapies, and diagnostics, prime and fuse with the cellular membrane, which is crucial for SARS‐CoV and SARS‐CoV‐2 infection and spread throughout host cells. 174 Thus, ACE2 175 and TMPRSS2 were chosen as host receptor, which are vital host targets during the viral infection process. In addition, the S protein helps SARS‐CoV and SARS‐CoV‐2 gain entry into host cells by fusing the viral membrane with the host cell membrane. 176 , 177 RNA‐dependent RNA polymerase (RdRp), a key part of the CoV replication machinery, is involved in processing protein production during infection. 90 RdRp has been considered one of the main drug targets against SARS‐CoV and SARS‐CoV‐2. Moreover, PLpro and 3CLpro cysteine proteases can process polyproteins of virus, which are essential for maturation and infectivity of SARS‐CoV and SARS‐CoV‐2. 178 Thus, spike (S), RdRp, PLpro, and 3CLpro from SARS‐CoV‐2 179 have been selected as the viral targets for the molecular docking to predict the binding modes of the viral proteins with berberine.
We assume that berberine regulates these targets by suppressing their gene expression or blocking binding sites at the protein level directly. Thus, molecular docking was used to calculate the binding energy and evaluate binding favorability (Table 1). The docking results show that berberine would be a promising inhibitor for each of the selected targets with moderate to strong binding affinities (−6.4‐‐9.8 Kcal/mol), which is supported by data in the literature indicating that free binding energy greater than −5.5 kcal/mol is an indication that the compound is inactive. 180 For host immune‐related proteins, the best results were obtained for the MAPK3‐berberine and MAPK8‐berberine complexes, with free binding energies of −8.9 kcal/mol and −8.6 kcal/mol, respectively. TNF, MAPK1, and BAX showed a moderate binding affinity with berberine, with binding energies of −8.2 kcal/mol, −8.2 kcal/mol, and −8.1 kcal/mol, respectively. The NFκB1‐berberine and CHUK‐berberine complexes displayed modest binding affinities of −7.3 kcal/mol in each case. By contrast, berberine exhibited weaker binding to NFκB1A, CASP3, and IL6. For host infection‐related proteins, berberine showed the highest potential to inhibit the ACE2 receptor with a binding energy of −9.8 kcal/mol. Interestingly, berberine bound to the contact interface of the Spike‐ACE2 complex, indicating that berberine may be an inhibitor of ACE2 enzyme activities rather than an inhibitor of ACE2‐driven viral infections. 93 The binding energy of TMPRSS2‐berberine was −6.7 kcal/mol, which was weaker compared with ACE2‐berberine, but still notable. Berberine may also be a potential inhibitor of 3CLpro, RdRp, PLpro, and Spike, based on the modest binding energy of those complexes. It is worth mentioning that berberine has been evaluated as a potential inhibitor of 3CLpro with the lowest binding energy compared to other natural products. 181
TABLE 1.
Molecular docking results between relevant proteins with berberine
| Protein type | Gene name | Affinity (Kcal/mol) |
|---|---|---|
| Immune‐related proteins | MAPK3 | −8.9 |
| MAPK8 | −8.6 | |
| TNF | −8.2 | |
| MAPK1 | −8.2 | |
| BAX | −8.1 | |
| NFκB1 | −7.3 | |
| CHUK | −7.3 | |
| IL6 | −6.9 | |
| NFκB1A | −6.4 | |
| CASP3 | −6.4 | |
| Host receptor | ACE2 | −9.8 |
| TMPRSS2 | −6.7 | |
| Viral proteins | 3CLpro | −6.7 |
| RdRp | −6.6 | |
| PLpro | −6.6 | |
| Spike | −6.5 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The optimal binding modes of each studied berberine complex (bold shown in Table 1, ie, MAPK3‐berberine, TNF‐berberine, BAX‐berberine, NFκB1‐berberine, CHUK‐berberine, ACE2‐berberine, TMPRSS2‐berberine, and 3CLpro‐berberine) are demonstrated in Figure 6 MAPKs showed a better binding affinity with berberine in host immune‐related proteins. Considering the similar structures of MAPKs, only complex MAPK3‐berberine was selected and illustrated in Figure 6A, where berberine fitted well into the binding cavity of MAPK3 as a result of hydrophobic interactions with TYR53, VAL56, and LEU173. For TNF‐berberine (Figure 6B), a hydrogen bond between berberine and TYR151, along with π‐π stacking between berberine and TYR59, significantly contributed to the stability of the complex. For complex BAX‐berberine (Figure 6C), the hydrogen bonds between berberine with GLN32 and ALA35 stabilized the left structure of berberine, and hydrophobic bonds between berberine with ALA46 and ILE133 further stabilized the right structure of berberine.
FIGURE 6.
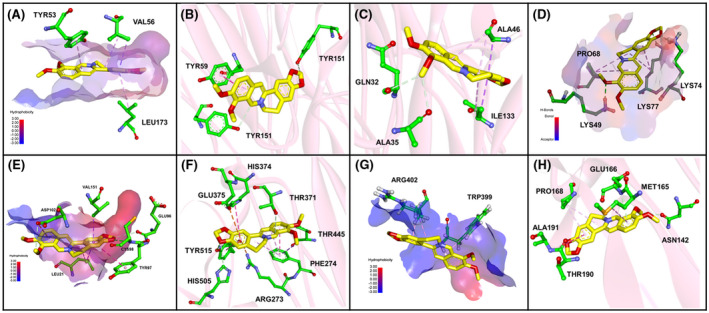
Binding explorations of complex MAPK3‐berberine, TNF‐Berberine, BAX‐berberine, BAX‐berberine, NFκB1‐berberine, CHUK‐berberine, ACE2‐berberine, TMPRSS2 ‐berberine, and 3CLpro‐berberine. Predicted lowest‐energy binding mode of berberine with the following proteins: A, MAPK3; B, TNF; C, BAX; D, NFκB1; E, CHUK; F, ACE2; G,TMPRSS2; H, 3CLpro. For berberine, the C, O, and N are highlighted in yellow, red, and blue, respectively. For residues of proteins, the green, red, and blue stand for C, O, and N, respectively. The green, purple, and orange lines stand for hydrogen binding, hydrophobic interaction, and anion‐π interaction between berberine and residues, respectively
For NFκB1‐berberine (Figure 6D), hydrogen bonds with LYS49, LYS74, and LYS77 facilitated the interaction between the small molecule and its target. With respect to CHUK (Figure 6E), berberine could be successfully docked to the target, with hydrogen bonds between the berberine and CYS98, GLU96, and ASP102 further increasing the stability of the complex. For ACE2‐berberine, hydrogen bonds (TYR515, HIS505, THR371, and THR445), π‐π stacking (HIS374, TYR515, and PHE274), and anion‐π interactions (ARG273 and GLU375) formed between protein residues and berberine contributed to stability of the complex and were clearly illustrated in Figure 6F. For TMPRSS2‐berberine (Figure 6G), the berberine was attached to the hydrophobic surface of the protein. For the 3CL pro‐berberine complex, key amino acid residues (ASN142, MET165, GLU166, PRO168, ALA191, and THR190) bound the berberine molecule tightly through hydrogen bonds and hydrophobic interactions (Figure 6H). More detailed two‐dimensional interactions were illustrated in Figure S2.
To explain the binding significance of the berberine structure, protein ACE2 was selected to conduct molecular docking with two derivatives of berberine. Analog1 (5,6‐dihydroisoquinolino[3,2‐a]isoquinolin‐7‐ium) only retains the main skeleton without the methoxy substituent (Figure S3), while analog2 (7,8‐dimethoxy‐1,2,3,4‐tetrahydropyrido[1,2‐b]isoquinolin‐5‐ium) modifies the polycyclic isoquinoline‐based skeleton by removing one conjugated benzene ring. The docking score of ACE2‐analog1 and ACE2‐analog2 are −9.3 kcal/mol and −7.4 kcal/mol, respectively. Compared to berberine, analog1 still displays most anion‐π stacking and π‐π stacking interactions with key protein residues, but loses the hydrogen bonds between the methoxy groups and the protein. While the binding affinity of ACE2‐analog1 decreases slightly compared to ACE2‐berberine, the binding affinity of ACE2‐analog2 declines significantly. This implies that the anion‐π stacking and π‐π stacking interactions formed by the conjugated skeleton of berberine and residues from ACE2 are critical features in a tightly binding complex between the small molecule and ACE2.
3.7. Berberine/NIT‐X inhibits SARS‐Cov‐2 replication, ACE2, and TMPSS2 gene expression in SARS CoV‐2‐infected human lung epithelial cell line
To provide experimental support for the hypothesis that berberine/NIT‐X could be an effective inhibitor SARS‐Cov‐2, the replication level of SARS‐CoV‐2 in infected calu‐2 cells was evaluated by detecting SARS‐CoV‐2 N protein gene expression using qPCR. We observed that 1 day post infection, there was a marked increase in SARS‐CoV‐2 N protein gene expression in SARS‐CoV‐2 infected cells compared with mock infected Calu‐3 cells (Figure 7A, P < .01). Berberine/NIT‐X treatment at both 20 μg/mL and 40 μg/mL significantly inhibited the viral levels (Figure 7A, P < .01 vs. untreated). ACE2 and TMPSS2 levels were further analyzed and an increasing trend was observed for both genes when compared with noninfected cells. We next determined whether berberine/NIT‐X inhibits viral infection (entry) by suppression of ACE2 and TMPSS2 expression, with data showing that berberine/NIT‐X inhibited the ACE2 and TMPSS2 levels at both 20 μg/mL and 40 μg/mL (Figure 7B,C, P < .05 vs. un‐treated). No cell toxicity was observed at these two concentrations (Figure 7D). The data further validate our hypothesis that berberine inhibits viral infection by regulating host receptor ACE2 and TMPSS2.
FIGURE 7.
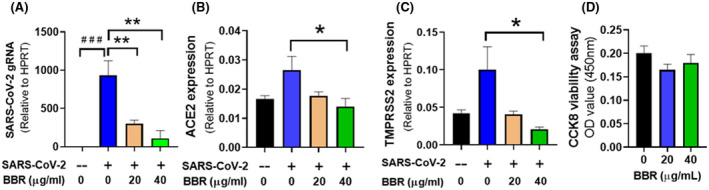
Effect of BBR (berberine/NIT‐X) on ACE2, TMPRSS2 expression in infected Calu‐3 cells. Calu‐3 cells were seeded in six well plates with 5 × 105 cells per well. After 24 hours, cells were incubated in media containing 20 µg/mL, 40 µg/mL BBR/NIT‐X or DMSO (v/v 1:1000). After 3 days incubation, cells were infected with SARS‐CoV‐2 (MOI = 0.005) or mock (‐) for 1hr and grown in indicated media for 24 hours. One day post infection, total RNA were for cDNA synthesis. Real Time‐PCR was performed with the primer for SARS‐CoV‐2 (A) gRNA of SARS‐CoV2, (B) ACE2, and (C) TMPRSS2. Data were normalized to HPRT and presented as 2‐ΔCT. Data represent two sets of qPCR with six readouts. D, Cytotoxicity was performed using commercial CCK8 toxicity kit, *P < .05 versus infected, but not treated. # P < .05 versus uninfected/untreated
3.8. Berberine/NIT‐X inhibits inflammatory cytokine and chemokine gene expression by SARS‐CoV‐2 infected human lung epithelial cells line
We also investigated the effect of berberine/NIT‐X on the expression of different cytokines and chemokine in infected Calu3 cells; specific lung gene expression differentiation of SARS and COVID‐19 were illustrated in (Figure S4). There are 12 and 15 pharmacological targets for SARS and COVID‐19, respectively, all of which are potential berberine targets. Among them, eight shared targets (IL1α, IL6, IL8, CCL2, PTGS2, NLRP3, VEGFA, and ERBB2) were displayed in the overlap area. IL‐1α, IL‐8, IL‐6, and CCL2 are shared genes that were selected as targets of berberine for further validation. The data showed that the IL‐1α, IL‐8, and IL‐6 gene expression were significantly increased in virus infected, nontreated cells. After the treatment of berberine/NIT‐X at 20 μg/mL, the expression of IL‐1α, IL‐8, and IL‐6 were all significantly decreased (Figure 8A‐C, *P < .05; **P < .01). No significant difference was observed in CCL‐2 levels in untreated infected cells versus untreated noninfected cells; however, under the treatment of berberine/NIT‐X, CCL‐2 expression was significantly reduced (Figure 8D, **P < .01). These data demonstrate that berberine/NIT‐X effectively suppresses the expression of pro‐inflammatory cytokines and chemokines including IL‐1α, IL‐8, IL‐6, and CCL2, which reduce the risk of cytokine storm and pneumonia in COVID‐19. The data further support our hypothesis that berberine can inhibit various pro‐inflammatory cytokines and protect against tissue damage during viral infection.
FIGURE 8.
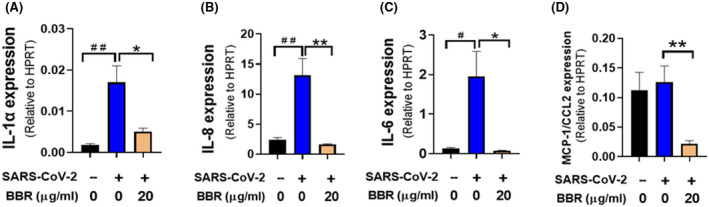
Effect of BBR (berberine/NIT‐X) cytokine expression in infected Calu‐3 cells. Calu‐3 cells were cultured, treated, infected and qPCR were performed. *P < .05,**P < .01 versus infected, but not treated. Data represent two sets of qPCR with six readouts. # P < .05 versus uninfected/untreated
4. CONCLUSION
We have identified potential therapeutic targets of berberine against both SARS and COVID‐19 using computational modeling. The most prominent targets for berberine relevant to host immune response include NF‐κB and MAPKs, which are important proteins regulating the cytokine storm, and CASPs and BAX, which are relevant targets in preventing tissue damage via suppressing cell death signaling pathways. Besides balancing host immune responses, our molecular docking analysis identifies berberine as a potential antagonist of host receptor for viral entry, such as ACE2 and TMPSS2, and may inhibit virus proteins. Furthermore, as the first step to validate our computational modeling results, we for the first time demonstrate that berberine significantly reduced viral replication, suppressed viral entry host receptor ACE2 and TMPSS2, and decreased inflammatory markers including IL‐6, IL‐8, IL‐1α, and CCL2 in SARS‐CoV‐2 infected lung epithelial cells. Given that berberine/NIT‐X exhibits high oral bioavailability and has previously been shown to have in vivo immunomodulatory effects and suppression of hyper‐mast cells activation, berberine/NIT‐X has the potential to become a promising, orally active therapeutic against COVID‐19 and SARS. However, direct evidence of berberine antagonist to ACE2, TMPRSS2 protein, and binding activities with these receptors and other targets have not been elucidated in this study and will be further investigated in our future research.
Berberine/NIT‐X may exhibit a number of possible clinical prospective applications as follows: (1) Given that berberine improves the Th1 immune response at the early stage of infection, then, inhibits inflammatory responses triggered by viruses at the late stage, it is possible that berberine can be both a preventive and treatment option for individuals at a higher risk of viral infection such as immune‐compromised patients. (2) Our computational modeling showed that berberine targets a cancer pathway, an AGE‐RAGE signaling pathway in diabetic complications, and a number of other viral infection‐related immune response pathways. Therefore, berberine/NIT‐X may have be potentially useful for patients with preexisting condition such as cancer, diabetes, and patients with other viral infection including influenza, EBV, CMV, HBV, etc. (3) Clinically, a high dosage of berberine may cause gastrointestinal side‐effects. berberine/NIT‐X boosts oral bioavailability and reduces the dosage of oral administration by 6 times, and thereby may evade the side‐effects caused by high dosage. (4) Berberine might regulate host immune responses, inhibit host viral receptors, and block virus proteins to prevent and treat SARS and COVID‐19. Therefore, the co‐treatment with immune supplements such as ascorbic acid, antiviral drugs such as remdesivir, or antibodies from convalescent plasma might be interesting avenues to enhance the effects of berberine. Recently, strong synergistic in vitro antiviral activities between remdesivir and berberine have been reported, 182 which further supports our hypothesis.
Taken together, our current study may lead to berberine/NIT‐X as an orally active therapeutic candidate in the prevention and treatment of COVID‐19, SARS, and other viral infections. In vitro and in vivo studies are currently in progress to investigate, confirm, and expand the knowledge into the molecular alterations induced by berberine in preventing and treating COVID‐19 and SARS.
CONFLICT OF INTEREST
ZZW, YL, WH, RT, ANW, JG, and MM have no financial conflict of interests to disclose. XML received research support from the National Institutes of Health (NIH)/National Center for Complementary and Alternative Medicine (NCCAM); Food Allergy Research and Education (FARE) and Winston Wolkoff Integrative Medicine Fund for Allergies and Wellness; received consulting fees from Food Allergy Research and Education (FARE), Johnson & Joh nson Pharmaceutical Research & Development, LLC, Bayer Global Health LLC; Received grant from Henan University of Chinese Medicine & New York Medica College for TCM Immunopharmacology and Integrative Medicine; received royalties from UpToDate; received travel expenses from the National Center for Complementary and Alternative Medicine (NCCAM) and FARE; received practice compensation from the Integrative Health and Acupuncture PC; US Times Technology Inc is managed by the related party; Share patents (PCT/US14/857772, PCT/US14/68396, PCT/US2014/012306, PCT/US2005/008417, and (PCT/US2017/056822 (Pending), is a member of Herbs Springs, LLC, General Nutraceutical Technology LLC, and Health Freedom LLC. NY shares US patent PCT/US14/68396 and is a member of Health Freedom LLC. KS share patent (PCT/US2017/056822 (Pending), Salary support by General Nutraceutical Technology LLC.
AUTHOR CONTRIBUTIONS
Z.‐Z. Wang and X.‐M. Li designed this study. Z.‐Z. Wang, K. Li, N. Yang, and K. Srivastava performed the data collection, analysis, target validation, and manuscript preparation. A.R. Maskey helped prepare manuscript. A.A. Toutov, W. Huang, M. Miao, N. Yang, K. Srivastava, R. Tiwari, J. Geleibter, and X.‐M. Li contributed to the manuscript preparation/revision and provided guidance. All authors read and approved the final manuscript.
Supporting information
Fig S1‐S4
TableS1
ACKNOWLEDGMENTS
This research was founded by Henan University of Chinese Medicine, visiting fellowship grant to ZZW (CTIM‐01‐2019), Scientific Research Foundation (KYQD 2019‐05), and Henan Province Scientific and Technological Project (NO. 202102310472). Study of Integrative Medicine for Immunology and Wellness, New York Medical College.
Wang Z‐Z, Li K, Maskey AR, et al. A small molecule compound berberine as an orally active therapeutic candidate against COVID‐19 and SARS: A computational and mechanistic study. The FASEB Journal. 2021;35:e21360. 10.1096/fj.202001792R
Zhen‐Zhen Wang and Kun Li contributed equally to this manuscript.
REFERENCES
- 1. COVID‐19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) . https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed December 1, 2020
- 2.Dr. Fauci: US may see ‘surge upon surge’ of virus this winter. https://www.clickondetroit.com/health/2020/11/29/dr‐fauci‐us‐may‐see‐surge‐upon‐surge‐of‐virus‐this‐winter/. Accessed December 1, 2020
- 3. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1986‐1994. [DOI] [PubMed] [Google Scholar]
- 5. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 6. de Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS‐CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790‐7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hui DS, Wong PC, Wang C. SARS: clinical features and diagnosis. Respirology. 2003;8(s1):S20‐S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berberine. Altern Med Rev. 2000;5(2):175‐177. [PubMed] [Google Scholar]
- 9. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cases of Coronavirus Disease (COVID‐19) in the U.S. https://www.cdc.gov/coronavirus/2019‐ncov/cases‐updates/cases‐in‐us.html. Accessed December 1, 2020
- 11. Yang Y, Islam MS, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019‐new coronavirus (SARS‐CoV‐2): a review and perspective. Int J Biol Sci. 2020;16(10):1708‐1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu L, Zheng X, Wang Y, et al. Berberine protects acute liver failure in mice through inhibiting inflammation and mitochondria‐dependent apoptosis. Eur J Pharmacol. 2018;819:161‐168. [DOI] [PubMed] [Google Scholar]
- 13. Wen C, Huang C, Yang M, et al. The secretion from bone marrow mesenchymal stem cells pretreated with berberine rescues neurons with oxidative damage through activation of the Keap1‐Nrf2‐HO‐1 signaling pathway. Neurotox Res. 2020;38(1):59‐73. [DOI] [PubMed] [Google Scholar]
- 14. Chen X, Guo H, Li Q, et al. Protective effect of berberine on aconiteinduced myocardial injury and the associated mechanisms. Mol Med Rep. 2018;18(5):4468‐4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hao G, Yu Y, Gu B, Xing Y, Xue M. Protective effects of berberine against doxorubicin‐induced cardiotoxicity in rats by inhibiting metabolism of doxorubicin. Xenobiotica. 2015;45(11):1024‐1029. [DOI] [PubMed] [Google Scholar]
- 16. Yan F, Li F, Liu J, et al. The formulae and biologically active ingredients of Chinese herbal medicines for the treatment of atopic dermatitis. Biomed Pharmacother. 2020;127:110142. [DOI] [PubMed] [Google Scholar]
- 17. Warowicka A, Nawrot R, Goździcka‐Józefiak A. Antiviral activity of berberine. Arch Virol. 2020;165(9):1935‐1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan YQ, Fu YJ, Wu S, et al. Anti‐influenza activity of berberine improves prognosis by reducing viral replication in mice. Phytother Res. 2018;32(12):2560‐2567. [DOI] [PubMed] [Google Scholar]
- 19. Hung TC, Jassey A, Liu CH, et al. Berberine inhibits hepatitis C virus entry by targeting the viral E2 glycoprotein. Phytomedicine. 2019;53:62‐69. [DOI] [PubMed] [Google Scholar]
- 20. Hayashi K, Minoda K, Nagaoka Y, Hayashi T, Uesato S. Antiviral activity of berberine and related compounds against human cytomegalovirus. Bioorg Med Chem Lett. 2007;17(6):1562‐1564. [DOI] [PubMed] [Google Scholar]
- 21. Luganini A, Mercorelli B, Messa L, Palu G, Gribaudo G, Loregian A. The isoquinoline alkaloid berberine inhibits human cytomegalovirus replication by interfering with the viral Immediate Early‐2 (IE2) protein transactivating activity. Antiviral Res. 2019;164:52‐60. [DOI] [PubMed] [Google Scholar]
- 22. Yang N, Wang J, Liu C, et al. Berberine and limonin suppress IgE production by human B cells and peripheral blood mononuclear cells from food‐allergic patients. Ann Allergy Asthma Immunol. 2014;113(5):556‐564.e4. [DOI] [PubMed] [Google Scholar]
- 23. Yang N, Srivastava K, Song Y, et al. Berberine as a chemical and pharmacokinetic marker of the butanol‐extracted Food Allergy Herbal Formula‐2. Int Immunopharmacol. 2017;45:120‐127. [DOI] [PubMed] [Google Scholar]
- 24. Liu CS, Zheng YR, Zhang YF, Long XY. Research progress on berberine with a special focus on its oral bioavailability. Fitoterapia. 2016;109:274‐282. [DOI] [PubMed] [Google Scholar]
- 25. Mirhadi E, Rezaee M, Malaekeh‐Nikouei B. Nano strategies for berberine delivery, a natural alkaloid of Berberis. Biomed Pharmacother. 2018;104:465‐473. [DOI] [PubMed] [Google Scholar]
- 26. Parker S, Li X‐M, Liu C, Srivastava K, Yu H. Treatment of immunological disease using berberine nanoparticles. 2017, PCT/US2017/056822.
- 27.Correction to Lancet Infect Dis 2020; https://doi.org/10.1016/S1473‐3099(20)30144‐4. Lancet Infect Dis. 2020. [DOI] [PMC free article] [PubMed]
- 28. Lagunas‐Rangel FA, and Chavez‐Valencia V. High IL‐6/IFN‐gamma ratio could be associated with severe disease in COVID‐19 patients. J Med Virol. 2020;92(no.,), 1789–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shahabi nezhad F, Mosaddeghi P, Negahdaripour M, et al. Therapeutic approaches for COVID‐19 based on the dynamics of interferon‐mediated immune responses. Preprints. 2020.
- 31. Laurencikiene J, Deveikaite V, Severinson E. HS1,2 enhancer regulation of germline epsilon and gamma2b promoters in murine B lymphocytes: evidence for specific promoter‐enhancer interactions. J Immunol. 2001;167(6):3257‐3265. [DOI] [PubMed] [Google Scholar]
- 32. Hansbro PM, Kim RY, Starkey MR, et al. Mechanisms and treatments for severe, steroid‐resistant allergic airway disease and asthma. Immunol Rev. 2017;278(1):41‐62. [DOI] [PubMed] [Google Scholar]
- 33. Hinks TS, Brown T, Lau LC, et al. Multidimensional endotyping in patients with severe asthma reveals inflammatory heterogeneity in matrix metalloproteinases and chitinase 3‐like protein 1. J Allergy Clin Immunol. 2016;138(1):61‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thio CL, Lai AC, Chi PY, Webster G, Chang YJ. Toll‐like receptor 9‐dependent interferon production prevents group 2 innate lymphoid cell‐driven airway hyperreactivity. J Allergy Clin Immunol. 2019;144(3):682‐697.e9. [DOI] [PubMed] [Google Scholar]
- 35. van der Made CI, Simons A, Schuurs‐Hoeijmakers J, et al. . Presence of genetic variants among young men with severe COVID‐19. JAMA. 2020;324(7):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Afrin LB, Weinstock LB, Molderings GJ. Covid‐19 hyperinflammation and post‐Covid‐19 illness may be rooted in mast cell activation syndrome. Int J Infect Dis. 2020;100:327‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conti P, Caraffa A, Tetè G, et al. Mast cells activated by SARS‐CoV‐2 release histamine which increases IL‐1 levels causing cytokine storm and inflammatory reaction in COVID‐19. J Biol Regul Homeost Agents. 2020;34:1629‐1632. [DOI] [PubMed] [Google Scholar]
- 38. Criado PR, Pagliari C, Criado RFJ, Marques GF, Belda W Jr. What the physicians should know about mast cells, dendritic cells, urticaria, and omalizumab during COVID‐19 or asymptomatic infections due to SARS‐CoV‐2? Dermatol Ther. 2020;33:e14068. [DOI] [PubMed] [Google Scholar]
- 39. Malone RW, Tisdall P, Fremont‐Smith P, et al. COVID‐19: famotidine, histamine, mast cells, and mechanisms. Res Sq. 2020. https://www.researchsquare.com/article/rs‐30934/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim TS, Kang BY, Cho D, Kim SH. Induction of interleukin‐12 production in mouse macrophages by berberine, a benzodioxoloquinolizine alkaloid, deviates CD4+ T cells from a Th2 to a Th1 response. Immunology. 2003;109(3):407‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahmoud MA, Ghareeb DA, Sahyoun HA, Elshehawy AA, Elsayed MM. In vivo interrelationship between insulin resistance and interferon gamma production: protective and therapeutic effect of berberine. Evid Based Complement Alternat Med. 2016;2016:2039897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou H, Mineshita S. The effect of berberine chloride on experimental colitis in rats in vivo and in vitro. J Pharmacol Exp Ther. 2000;294(3):822‐829. [PubMed] [Google Scholar]
- 43. Fukuda K, Hibiya Y, Mutoh M, Koshiji M, Akao S, Fujiwara H. Inhibition of activator protein 1 activity by berberine in human hepatoma cells. Planta Med. 1999;65(4):381‐383. [DOI] [PubMed] [Google Scholar]
- 44. Jeong HW, Hsu KC, Lee JW, et al. Berberine suppresses proinflammatory responses through AMPK activation in macrophages. Am J Physiol Endocrinol Metab. 2009;296(4):E955‐E964. [DOI] [PubMed] [Google Scholar]
- 45. Su H, Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID‐19 in China. Kidney Int. 2020;98(1):219‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun J, Aghemo A, Forner A, Valenti L. COVID‐19 and liver disease. Liver Int. 2020;40(6):1278‐1281. [DOI] [PubMed] [Google Scholar]
- 48. Yan Y, Yang Y, Wang F, et al. Clinical characteristics and outcomes of patients with severe covid‐19 with diabetes. BMJ Open Diabetes Res Care. 2020;8(1):e001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou H, Feng L, Xu F, et al. Berberine inhibits palmitate‐induced NLRP3 inflammasome activation by triggering autophagy in macrophages: a new mechanism linking berberine to insulin resistance improvement. Biomed Pharmacother. 2017;89:864‐874. [DOI] [PubMed] [Google Scholar]
- 50. Qiu YY, Tang LQ, Wei W. Berberine exerts renoprotective effects by regulating the AGEs‐RAGE signaling pathway in mesangial cells during diabetic nephropathy. Mol Cell Endocrinol. 2017;443:89‐105. [DOI] [PubMed] [Google Scholar]
- 51. Chen XW, Di YM, Zhang J, Zhou ZW, Li CG, Zhou SF. Interaction of herbal compounds with biological targets: a case study with berberine. Sci World J. 2012;2012:708292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu H, You L, Wu J, et al. Berberine suppresses influenza virus‐triggered NLRP3 inflammasome activation in macrophages by inducing mitophagy and decreasing mitochondrial ROS. J Leukoc Biol. 2020;108(1):253‐266. [DOI] [PubMed] [Google Scholar]
- 53. HitPick . mips.helmholtz‐muenchen.de/hitpick/. Accessed April 10, 2020.
- 54. Swiss Target Prediction . swisstargetprediction.ch/. Accessed April 10, 2020.
- 55. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357‐W364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Similarity Ensemble Approach . sea.bkslab.org/. Accessed April 10, 2020.
- 57. Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197‐206. [DOI] [PubMed] [Google Scholar]
- 58. PubChem . pubchem.ncbi.nlm.nih.gov/. Accessed April 10, 2020.
- 59. Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102‐D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. DrugBank . drugbank.ca. Accessed April 10, 2020.
- 61. Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901‐D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Therapeutic target database . db.idrblab.net/ttd/. Accessed January 30, 2020.
- 63. Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031‐D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Genetic association database . geneticassociationdb.nih.gov/.
- 65. Piñero J, Ramírez‐Anguita JM, Saüch‐Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845‐D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. GeneCards . genecards.org/. Accessed April 10, 2020.
- 67. Safran M, Solomon I, Shmueli O, et al. GeneCards 2002: towards a complete, object‐oriented, human gene compendium. Bioinformatics. 2002;18(11):1542‐1543. [DOI] [PubMed] [Google Scholar]
- 68. Open Targets Platform . targetvalidation.org/. Accessed April 10, 2020.
- 69. Carvalho‐Silva D, Pierleoni A, Pignatelli M, et al. Open targets platform: new developments and updates two years on. Nucleic Acids Res. 2019;47(D1):D1056‐D1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Comparative Toxicogenomics Database . ctdbase.org/. Accessed April 10, 2020.
- 71. Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948‐D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID‐19. Nat Commun. 2020;11(1):5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. UniProt Database . uniprot.org/. Accessed April 10, 2020.
- 74. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506‐D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. DAVID . david.ncifcrf.gov/. Accessed April 10, 2020.
- 77. Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(suppl_2):W316‐W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. KOBAS 3.0 . kobas.cbi.pku.edu.cn. Accessed April 10, 2020.
- 79. Assenov Y, Ramírez F, Schelhorn S‐E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2007;24(2):282‐284. [DOI] [PubMed] [Google Scholar]
- 80. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Polley S, Passos DO, Huang DB, et al. Structural basis for the activation of IKK1/α. Cell Rep. 2016;17(8):1907‐1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chaikuad A, Tacconi EM, Zimmer J, et al. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat Chem Biol. 2014;10(10):853‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Blake JF, Gaudino JJ, De Meese J, et al. Discovery of 5,6,7,8‐tetrahydropyrido[3,4‐d]pyrimidine inhibitors of Erk2. Bioorg Med Chem Lett. 2014;24(12):2635‐2639. [DOI] [PubMed] [Google Scholar]
- 84. Somers W, Stahl M, Seehra JS. 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. EMBO J. 1997;16(5):989‐997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhao H, Serby MD, Xin Z, et al. Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c‐Jun NH2‐terminal kinase inhibitors. J Med Chem. 2006;49(15):4455‐4458. [DOI] [PubMed] [Google Scholar]
- 86. Garner TP, Reyna DE, Priyadarshi A, et al. An autoinhibited dimeric form of BAX regulates the BAX activation pathway. Mol Cell. 2016;63(3):485‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. He MM, Smith AS, Oslob JD, et al. Small‐molecule inhibition of TNF‐alpha. Science. 2005;310(5750):1022‐1025. [DOI] [PubMed] [Google Scholar]
- 88. Towler P, Staker B, Prasad SG, et al. ACE2 X‐ray structures reveal a large hinge‐bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279(17):17996‐18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jin Z, Du X, Xu Y, et al. Structure of M(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature. 2020;582(7811):289‐293. [DOI] [PubMed] [Google Scholar]
- 90. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368(6492):779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yazdi S, Durdagi S, Naumann M, Stein M. Structural modeling of the N‐terminal signal‐receiving domain of IκBα. Front Mol Biosci. 2015;2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Müller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF‐kappa B p50 homodimer bound to DNA. Nature. 1995;373(6512):311‐317. [DOI] [PubMed] [Google Scholar]
- 95. Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17(1):57‐61. [PubMed] [Google Scholar]
- 96. DeLano WL. Pymol: an open‐source molecular graphics tool. CCP4 Newsletter on Protein Crystallography. 2002;40:82‐92. [Google Scholar]
- 97. DassaultSystèmesBIOVIAD . Discovery Studio, 2020; 2020.
- 98. Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S3‐S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7(4):160‐165. [DOI] [PubMed] [Google Scholar]
- 100. Clarke P, Tyler KL. Apoptosis in animal models of virus‐induced disease. Nat Rev Microbiol. 2009;7(2):144‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kindrachuk J, Ork B, Hart BJ, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59(2):1088‐1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Alter P, Jobmann M, Meyer E, Pankuweit S, Maisch B. Apoptosis in myocarditis and dilated cardiomyopathy: does enterovirus genome persistence protect from apoptosis? An endomyocardial biopsy study. Cardiovasc Pathol. 2001;10(5):229‐234. [DOI] [PubMed] [Google Scholar]
- 103. Elizalde MM, Sevic I, González López Ledesma MM, Campos RH, Barbini L, Flichman DM. Human hepatocytes apoptosis induced by replication of hepatitis B virus subgenotypes F1b and F4: role of basal core promoter and preCore mutations. Virology. 2018;513:160‐167. [DOI] [PubMed] [Google Scholar]
- 104. Walsh MJ, Vanags DM, Clouston AD, et al. Steatosis and liver cell apoptosis in chronic hepatitis C: a mechanism for increased liver injury. Hepatology. 2004;39(5):1230‐1238. [DOI] [PubMed] [Google Scholar]
- 105. Fujikura D, Miyazaki T. Programmed cell death in the pathogenesis of influenza. Int J Mol Sci. 2018;19(7):2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tai CJ, Jassey A, Liu CH, Richardson CD, Wong SH, Lin LT. Targeting autophagy augments BBR‐mediated cell death in human hepatoma cells harboring hepatitis C virus RNA. Cells. 2020;9(4).908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hwang JM, Kuo HC, Tseng TH, Liu JY, Chu CY. Berberine induces apoptosis through a mitochondria/caspases pathway in human hepatoma cells. Arch Toxicol. 2006;80(2):62‐73. [DOI] [PubMed] [Google Scholar]
- 108. Yip NK, Ho WS. Berberine induces apoptosis via the mitochondrial pathway in liver cancer cells. Oncol Rep. 2013;30(3):1107‐1112. [DOI] [PubMed] [Google Scholar]
- 109. Lin C‐C, Yang J‐L, Lu C‐C, Chung J‐G. Berberine induces cell cycle arrest and apoptosis in human HSC‐3 oral cancer cells. FASEB J. 2007;21(6):A1189. [PubMed] [Google Scholar]
- 110. Marshall HD, Urban SL, Welsh RM. Virus‐induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J Virol. 2011;85(12):5929‐5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mizutani T, Fukushi S, Iizuka D, et al. Inhibition of cell proliferation by SARS‐CoV infection in Vero E6 cells. FEMS Immunol Med Microbiol. 2006;46(2):236‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Li F, Dong X, Lin P, Jiang J. Regulation of Akt/FoxO3a/Skp2 axis is critically involved in berberine‐induced cell cycle arrest in hepatocellular carcinoma cells. Int J Mol Sci. 2018;19(2).327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yang IW, Chou CC, Yung BY. Dose‐dependent effects of berberine on cell cycle pause and apoptosis in Balb/c 3T3 cells. Naunyn Schmiedebergs Arch Pharmacol. 1996;354(2):102‐108. [DOI] [PubMed] [Google Scholar]
- 115. Lin CC, Lin SY, Chung JG, Lin JP, Chen GW, Kao ST. Down‐regulation of cyclin B1 and up‐regulation of Wee1 by berberine promotes entry of leukemia cells into the G2/M‐phase of the cell cycle. Anticancer Res. 2006;26(2a):1097‐1104. [PubMed] [Google Scholar]
- 116. Wen H, Miao EA, Ting JP. Mechanisms of NOD‐like receptor‐associated inflammasome activation. Immunity. 2013;39(3):432‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yue Y, Nabar NR, Shi CS, et al. SARS‐coronavirus open reading frame‐3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 2019;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Nieto‐Torres JL, Verdia‐Baguena C, Jimenez‐Guardeno JM, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ryzhakov G, Lai CC, Blazek K, To KW, Hussell T, Udalova I. IL‐17 boosts proinflammatory outcome of antiviral response in human cells. J Immunol. 2011;187(10):5357‐5362. [DOI] [PubMed] [Google Scholar]
- 121. Kimura H, Yoshizumi M, Ishii H, Oishi K, Ryo A. Cytokine production and signaling pathways in respiratory virus infection. Front Microbiol. 2013;4:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lingeswaran M, Goyal T, Ghosh R, et al. Inflammation, Immunity and Immunogenetics in COVID‐19: A Narrative Review. Indian J Clin Biochem. 2020;35(3):260‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Qin X, Guo BT, Wan B, et al. Regulation of Th1 and Th17 cell differentiation and amelioration of experimental autoimmune encephalomyelitis by natural product compound berberine. J Immunol. 2010;185(3):1855‐1863. [DOI] [PubMed] [Google Scholar]
- 124. Kierdorf K, Fritz G. RAGE regulation and signaling in inflammation and beyond. J Leukoc Biol. 2013;94(1):55‐68. [DOI] [PubMed] [Google Scholar]
- 125. Oczypok EA, Perkins TN, Oury TD. All the “RAGE” in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr Respir Rev. 2017;23:40‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rojas A, Gonzalez I, Morales MA. SARS‐CoV‐2‐mediated inflammatory response in lungs: should we look at RAGE? Inflamm Res. 2020;69(7):641‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Liu T, Zhang L, Joo D, Sun S. NF‐kB signaling in inflammation. Signal Transduct Target Ther. 2017;2:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Front Immunol. 2014;5:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tak PP, Firestein GS. NF‐kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107(1):7‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lawrence T. The nuclear factor NF‐kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1(6):a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. DeDiego ML, Nieto‐Torres JL, Regla‐Nava JA, et al. Inhibition of NF‐κB‐mediated inflammation in severe acute respiratory syndrome coronavirus‐infected mice increases survival. J Virol. 2014;88(2):913‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Li Z, Geng YN, Jiang JD, Kong WJ. Antioxidant and anti‐inflammatory activities of berberine in the treatment of diabetes mellitus. Evid Based Complement Alternat Med. 2014;2014:289264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Coskun M, Olsen J, Seidelin JB, Nielsen OH. MAP kinases in inflammatory bowel disease. Clin Chim Acta. 2011;412(7–8):513‐520. [DOI] [PubMed] [Google Scholar]
- 134. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK‐activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus‐infected cells. Biochem Biophys Res Commun. 2004;319(4):1228‐1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Huang L, Shi Y, Gong B, et al. Blood single cell immune profiling reveals the interferon‐MAPK pathway mediated adaptive immune response for COVID‐19. medRxiv 2020, 2020.03.15.20033472.
- 137. Botwina P, Owczarek K, Rajfur Z, et al. Berberine hampers influenza A replication through inhibition of MAPK/ERK pathway. Viruses. 2020;12(3):344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wang H, Li K, Ma L, et al. Berberine inhibits enterovirus 71 replication by downregulating the MEK/ERK signaling pathway and autophagy. Virol J. 2017;14(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cai Y, Liu Y, Zhang X. Suppression of coronavirus replication by inhibition of the MEK signaling pathway. J Virol. 2007;81(2):446‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mantena SK, Sharma SD, Katiyar SK. Berberine, a natural product, induces G1‐phase cell cycle arrest and caspase‐3‐dependent apoptosis in human prostate carcinoma cells. Mol Cancer Ther. 2006;5(2):296‐308. [DOI] [PubMed] [Google Scholar]
- 141. Ho YT, Lu CC, Yang JS, et al. Berberine induced apoptosis via promoting the expression of caspase‐8, ‐9 and ‐3, apoptosis‐inducing factor and endonuclease G in SCC‐4 human tongue squamous carcinoma cancer cells. Anticancer Res. 2009;29(10):4063‐4070. [PubMed] [Google Scholar]
- 142. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID‐19: an overview of the involvement of the chemokine/chemokine‐receptor system. Cytokine Growth Factor Rev. 2020;53:25‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus‐infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79(12):7819‐7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID‐19. J Infect. 2020;80(6):607‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhang X, Wu K, Wang D, et al. Nucleocapsid protein of SARS‐CoV activates interleukin‐6 expression through cellular transcription factor NF‐kappaB. Virology. 2007;365(2):324‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Russell B, Moss C, George G, et al. Associations between immune‐suppressive and stimulating drugs and novel COVID‐19‐a systematic review of current evidence. Ecancermedicalscience. 2020;14:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zha W, Liang G, Xiao J, et al. Berberine inhibits HIV protease inhibitor‐induced inflammatory response by modulating ER stress signaling pathways in murine macrophages. PLoS ONE. 2010;5(2):e9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lee CH, Chen JC, Hsiang CY, Wu SL, Wu HC, Ho TY. Berberine suppresses inflammatory agents‐induced interleukin‐1beta and tumor necrosis factor‐alpha productions via the inhibition of IkappaB degradation in human lung cells. Pharmacol Res. 2007;56(3):193‐201. [DOI] [PubMed] [Google Scholar]
- 149. Wang Q, Qi J, Hu R, Chen Y, Kijlstra A, Yang P. Effect of berberine on proinflammatory cytokine production by ARPE‐19 cells following stimulation with tumor necrosis factor‐α. Invest Ophthalmol Vis Sci. 2012;53(4):2395‐2402. [DOI] [PubMed] [Google Scholar]
- 150. Liu YF, Wen CY, Chen Z, Wang Y, Huang Y, Tu SH. Effects of berberine on NLRP3 and IL‐1β expressions in monocytic THP‐1 cells with monosodium urate crystals‐induced inflammation. Biomed Res Int. 2016;2016:2503703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Amin M, Razi M, Sarrafzadeh‐Rezaei F, Shalizar Jalali A, Najafi G. Berberine inhibits experimental varicocele‐induced cell cycle arrest via regulating cyclin D1, cdk4 and p21 proteins expression in rat testicles. Andrologia. 2018;50(4):e12984. [DOI] [PubMed] [Google Scholar]
- 152. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607‐D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein‐protein interaction detection: methods and analysis. Int J Proteomics. 2014;2014:147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466‐2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mahdavi Sharif P, Jabbari P, Razi S, Keshavarz‐Fathi M, Rezaei N. Importance of TNF‐alpha and its alterations in the development of cancers. Cytokine. 2020;130:155066. [DOI] [PubMed] [Google Scholar]
- 156. Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23(2):317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sundararaj KP, Wood RE, Ponnusamy S, et al. Rapid shortening of telomere length in response to ceramide involves the inhibition of telomere binding activity of nuclear glyceraldehyde‐3‐phosphate dehydrogenase. J Biol Chem. 2004;279(7):6152‐6162. [DOI] [PubMed] [Google Scholar]
- 158. Aviv A. Telomeres and COVID‐19. Faseb j. 2020;34(6):7247‐7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhang J, Tang H, Zhang Y, et al. Identification of suitable reference genes for quantitative RT‐PCR during 3T3‐L1 adipocyte differentiation. Int J Mol Med. 2014;33(5):1209‐1218. [DOI] [PubMed] [Google Scholar]
- 160. Zhang W, Zhao Y, Zhang F, et al. The use of anti‐inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID‐19): the perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID‐19 treatment. Rev Panam Salud Publica. 2020;44:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127‐137. [DOI] [PubMed] [Google Scholar]
- 163. Venkataraman T, Frieman MB. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus‐induced pulmonary fibrosis. Antiviral Res. 2017;143:142‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wang J, Yang S, Cai X, et al. Berberine inhibits EGFR signaling and enhances the antitumor effects of EGFR inhibitors in gastric cancer. Oncotarget. 2016;7(46):76076‐76086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Raaben M, Einerhand AW, Taminiau LJ, et al. Cyclooxygenase activity is important for efficient replication of mouse hepatitis virus at an early stage of infection. Virol J. 2007;4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Rott D, Zhu J, Burnett MS, et al. Effects of MF‐tricyclic, a selective cyclooxygenase‐2 inhibitor, on atherosclerosis progression and susceptibility to cytomegalovirus replication in apolipoprotein‐E knockout mice. J Am Coll Cardiol. 2003;41(10):1812‐1819. [DOI] [PubMed] [Google Scholar]
- 167. Pyeon D, Diaz FJ, Splitter GA. Prostaglandin E(2) increases bovine leukemia virus tax and pol mRNA levels via cyclooxygenase 2: regulation by interleukin‐2, interleukin‐10, and bovine leukemia virus. J Virol. 2000;74(12):5740‐5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Rossen JW, Bouma J, Raatgeep RH, Büller HA, Einerhand AW. Inhibition of cyclooxygenase activity reduces rotavirus infection at a postbinding step. J Virol. 2004;78(18):9721‐9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Liu M, Yang Y, Gu C, et al. Spike protein of SARS‐CoV stimulates cyclooxygenase‐2 expression via both calcium‐dependent and calcium‐independent protein kinase C pathways. FASEB J. 2007;21(7):1586‐1596. [DOI] [PubMed] [Google Scholar]
- 170. Lee CH, Chen RF, Liu JW, et al. Altered p38 mitogen‐activated protein kinase expression in different leukocytes with increment of immunosuppressive mediators in patients with severe acute respiratory syndrome. J Immunol. 2004;172(12):7841‐7847. [DOI] [PubMed] [Google Scholar]
- 171. Germoush MO, Mahmoud AM. Berberine mitigates cyclophosphamide‐induced hepatotoxicity by modulating antioxidant status and inflammatory cytokines. J Cancer Res Clin Oncol. 2014;140(7):1103‐1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Yu C‐S, Kuo H‐M, Chung J‐G. The role of cyclooxygenase‐2 in berberine induced apoptosis and inhibited cell migration of human gastric adenocarcinoma RF‐1 and RF‐48 cell lines. FASEB J. 2006;20(5):A1131. [Google Scholar]
- 173. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yan T, Xiao R, Lin G. Angiotensin‐converting enzyme 2 in severe acute respiratory syndrome coronavirus and SARS‐CoV‐2: a double‐edged sword? FASEB J. 2020;34(5):6017‐6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Xiu S, Dick A, Ju H, et al. Inhibitors of SARS‐CoV‐2 entry: current and future opportunities. J Med Chem. 2020;63(21):12256‐12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lindner HA, Fotouhi‐Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R. The papain‐like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79(24):15199‐15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Gil C, Ginex T, Maestro I, et al. COVID‐19: drug targets and potential treatments. J Med Chem. 2020;63(21):12359‐12386. [DOI] [PubMed] [Google Scholar]
- 180. Eleftheriou P, Amanatidou D, Petrou A, Geronikaki A. In silico evaluation of the effectivity of approved protease inhibitors against the main protease of the novel SARS‐CoV‐2 virus. Molecules. 2020;25(11):2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Narkhede RR, Pise AV, Cheke RS, Shinde SD. Recognition of natural products as potential inhibitors of COVID‐19 main protease (Mpro): in‐silico evidences. Nat Prod Bioprospect. 2020;10(5):297‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Pizzorno A, Padey B, Dubois J, et al. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS‐CoV‐2. Antiviral Res. 2020;181:104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S4
TableS1


