Abstract
Purpose
Triple-negative breast cancer (TNBC) is the most lethal subtype of breast cancer owing to high heterogeneity, aggressive nature, and lack of treatment options, which has a substantial deleterious effect on patients' lives. HOXD antisense growth-associated long noncoding RNA (lncRNA) (HAGLR) plays tumor-promoting roles in many cancers. In this study, we aimed to explore the role of HAGLR in TNBC.
Methods
Quantitative real-time polymerase chain reaction assays were used to examine the expression of RNAs. Functional experiments were conducted to test the biological behavior of TNBC cells. Moreover, MS2-RNA immunoprecipitation, luciferase reporter, and RNA pull-down assays were conducted to verify the binding relationship between HAGLR, microRNA-143-5p (miR-143-5p), and serine- and arginine-rich splicing factor 1 (SRSF1).
Results
HAGLR was found to be highly expressed in TNBC tissues and cells, and inhibiting HAGLR suppressed cell proliferation, migration, and invasion and promoted cell apoptosis in TNBC. Meanwhile, miR-93-5p was shown to bind to HAGLR and SRSF1. In addition, SRSF1 plays an oncogenic role in TNBC. Importantly, HAGLR could activate the Wnt signaling pathway by sponging miR-93-5p to upregulate SRSF1; thus, accelerating TNBC progression.
Conclusion
HAGLR could promote the progression of TNBC through the miR-93-5p/SRSF1 axis to activate the Wnt signaling pathway.
Keywords: RNA, long noncoding; MicroRNAs; Triple negative breast neoplasms; Wnt signaling pathway
INTRODUCTION
Triple-negative breast cancer (TNBC) is characterized by a lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) [1]. Treatment of TNBC is difficult because only one of the three receptors can be targeted by chemotherapy. Recently, auxiliary radiotherapy has been found to improve the survival rate of patients with TNBC [2]. Unfortunately, radiotherapy efficiency remains low owing to the radioresistance of TNBC cells [3]. Therefore, it is necessary to explore the ongoing molecular mechanism in TNBC cells to identify possible therapies to improve the quality of life of patients with TNBC.
Long noncoding RNAs (lncRNAs) are noncoding RNA (ncRNA) transcripts that play crucial roles in regulating the function of cancer cells [4]. In addition, unusual changes in the expression of lncRNAs, which eventually promote or relieve progression, occur in various cancers [5,6,7]. HOXD antisense growth-associated lncRNA (HAGLR) is an important regulator in many cancers. For example, upregulated HAGLR accelerates the growth and invasion of non-small cell lung cancer cells [8]. In addition, HAGLR modulates epithelial-mesenchymal transition (EMT) progression in esophageal cancer by acting as a microRNA-143-5p (miR-143-5p) sponge to regulate the expression of LAMP3 [9]. Meanwhile, HAGLR hinders the progression of lung adenocarcinoma by silencing E2F1 [10]. In the present study, we investigated the relationship between HAGLR and TNBC development to detect possible targets for TNBC therapy.
Recently, a competing endogenous RNA (ceRNA) network was proposed for analyzing lncRNA function in regulating gene expression at the post-transcriptional level via sponging microRNAs (miRNAs) [11]. miRNAs are another type of ncRNA transcripts that may also be important for cancer development [12]. Of note, the significance of miRNAs in the interplay between lncRNAs and miRNA-targeted messenger RNAs (mRNAs) has been increasingly validated [13]. For example, LINC00339 competes with HOXC6 to bind to miR-377-3p, thereby contributing to TNBC progression [14]. In the present study, miR-93-5p was selected as a HAGLR-interacting miRNA. miR-93-5p may play tumor-suppressing roles in colorectal cancer [15] and may be an effective tumor inhibitor in breast cancers [16], including TNBC [17]. However, the interplay between HAGLR and miR-93-5p has not yet, to our knowledge, been explored in cancer cells, let alone the influence of their interaction on TNBC progression.
The Wnt signaling pathway is associated with various biological behaviors in cancers [18]. lncRNA MIR100HG modulates cetuximab resistance via the Wnt pathway [19]. lncRNA CRNDE improves the growth and chemosensitivity of colorectal cancer cells by modulating the Wnt pathway [20]. lncRNA H19 can modulate the Wnt pathway via Dkk4 [21]. These findings indicate that lncRNAs can regulate the activity of the Wnt signaling pathway and affect the biological function of cancer cells. In this study, we investigated the impact of HAGLR on the Wnt signaling pathway in TNBC cells.
METHODS
Clinical sample collection
Clinical tumors and matched para-tumors were surgically acquired from 42 patients with TNBC at the Affiliated Tumor Hospital of Xinjiang Medical University. These patients were treated with no other therapies, and each patient provided informed consent before the operation. The samples were stored at −80°C after immediate freezing in liquid nitrogen. The sample collection was approved by the Ethics Committee of the Affiliated Tumor Hospital of Xinjiang Medical University (2019-39).
Cell culture
The normal human breast epithelial cell line (MCF-10A) and TNBC cell lines (MDA-MB-231, MDA-MB-468, MDA-MB-436, and HCC-1937) were obtained from The Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). Dulbecco's modified Eagle medium (DMEM) was used to cultivate these cells with 10% fetal bovine serum (FBS) as the supplement. Cells were incubated at 37°C under a 5% CO2 atmosphere.
Construction of transfection plasmids
To silence HAGLR or serine- and arginine-rich splicing factor 1 (SRSF1), specific shRNAs were constructed by Genepharma (Shanghai, China), with the nonsense sequences as the negative control (sh-NC). For HAGLR or SRSF1 overexpression, pcDNA3.1 vectors acquired from Invitrogen were used to load corresponding cDNA sequences, with the empty vector as the negative control. miR-93-5p mimics/inhibitor and negative controls (NC mimics/inhibitor) were obtained from RiboBio (Guangzhou, China). The cells were then cultivated to 70% confluency. Lipofectamine 2000 reagent was then applied to transfect the indicated plasmids into 5 × 105 cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
In accordance with the manufacturer's instructions, TRIzol reagent (TaKaRa, Shiga, Japan) was used to extract total RNA from TNBC cells. RNA was then subjected to cDNA synthesis using the PrimeScript Reverse Transcriptase Kit (TaKaRa). Thereafter, qRT-PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, USA) to estimate the expression levels of genes using the SYBR Green PCR Kit (TaKaRa). The 2−ΔΔCt method was used to determine the relative expression of RNA for comparison with the NC primer group [22] with the housekeeping genes GAPDH (for HAGLR and mRNAs) and U6 (for miRNAs) as the normalized controls. All experiments were conducted in triplicate. The primer sequences are listed in Table 1.
Table 1. Primer sequences of all genes involved in this study.
| Gene | Primer sequences |
|---|---|
| HAGLR | F: TGGGGCTTCCTTTGTGTGTT |
| R: AGCGGAAGAGTAGGTCTGGT | |
| GAPDH | F: GACAGTCAGCCGCATCTTCT |
| R: GCGCCCAATACGACCAAATC | |
| U6 | F: TCCCTTCGGGGACATCCG |
| R: AATTTTGGACCATTTCTCGATTTGT | |
| miR-519d-3p | Sequence: caaagtgcctccctttagagtg |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCcactct | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFTCGGcaaagtgcctcccttt | |
| miR-182-5p | Sequence: tttggcaatggtagaactcacact |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCagtgtg | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFGCCtttggcaatggtagaact | |
| miR-106b-5p | Sequence: taaagtgctgacagtgcagat |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCatctgc | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFGCCGAtaaagtgctgacagt | |
| miR-17-5p | Sequence: actgcagtgaaggcacttgtag |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCctacaa | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFTCGAactgcagtgaaggcac | |
| miR-519c-3p | Sequence: aaagtgcatctttttagaggat |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCatcctc | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFCGGCAGGaaagtgcatcttttta | |
| miR-526b-3p | Sequence: gaaagtgcttccttttagaggc |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCgcctct | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFGCCGgaaagtgcttcctttt | |
| miR-20b-5p | Sequence: caaagtgctcatagtgcaggtag |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCctacct | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFGCCcaaagtgctcatagtgc | |
| miR-93-5p | Sequence: caaagtgctgttcgtgcaggtag |
| Reverse transcription stem loop primer: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCctacct | |
| Stem loop universal reverse primer: CTCAACTGGTGTCGTGGAFTCGcaaagtgctgttcgtgc | |
| β-catenin | F: ACGGAGGAAGGTCTGAGGAG |
| R: AGCCGCTTTTCTGTCTGGTT | |
| c-MYC | F: TGAAAAGGCCCCCAAGGTAG |
| R: CTCGTCGTTTCCGCAACAAG | |
| PAI-1 | F: CTGCCTCCAGCTACCGTTATT |
| R: ACAAAGCTTACTTGAGAAACCAC | |
| Fibronectin | F: TCATTAATGGCGGCGAGAGT |
| R: ACAAGAAGGAAGACTCAGTTAATG | |
| α-SMA | F: AGCCAAGCACTGTCAGGAATC |
| R: GTCACCCACGTAGCTGTCTT | |
| SRSF1 | F: TCTCATGAGGGAGAAACTGCC |
| R: CTTCTGCTACGGCTTCTGCT | |
| MKNK2 | F: GAGTTCCCCGACAAGGACTG |
| R: AAGTGACTGTCCCACCTCTG | |
| UBC | F: ACTCTGCACTTGGTCCTGC |
| R: GAATGCAACAACTTTATTGAAAGGA | |
| NABP1 | F: TGTGCGGTGTTCTGTCTCC |
| R: GACACTTCTGCCCTGAGTCC |
HAGLR = HOXD antisense growth-associated lncRNA; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; miR = microRNA; PAI-1 = plasminogen activator inhibitor-1; α-SMA = α-smooth muscle actin; SRSF1 = serine- and arginine-rich splicing factor 1; MKNK2 = MAPK interacting serine/threonine kinase 2; UBC = ubiquitin C; NABP1 = nucleic acid binding protein 1.
Colony formation assay
To detect colony formation ability, cells were placed in DMEM supplemented with 10% FBS. They were then incubated at 37°C and under a 5% CO2 atmosphere. After 21 days of cultivation, colonies were fixed with 4% paraformaldehyde, stained with crystal violet, and those with more than 50 cells were counted manually. The media were replenished every 3 days.
Immunofluorescence
Since Ki-67 is a reliable marker of cell proliferation and is expressed during mitosis, it is an acceptable alternative for indicating cell proliferation [23]. Here, we applied immunofluorescence staining of Ki-67 to analyze changes in cell proliferation, as previously described [24]. First, phosphate-buffered saline (PBS) was used to rinse the sections after fixing with 4% paraformaldehyde, then 0.05% Triton X was used to permeabilize them for 5 minutes. Subsequently, antibodies against Ki-67 (Ab15580; Abcam, Cambridge, USA) were used to cultivate the sections at 4°C for one night after the sections were sealed with 1% goat serum. Next, PBS and 0.01% Triton X were applied to rinse the slides. Secondary antibodies conjugated with Alexa Fluor were used to cultivate the sections. The 4′,6-diamidino-2-phenylindole (DAPI) and 90% glycerol were used to stain and mount the sections. Images were taken using a Zeiss Axio-Observer microscope (Carl Zeiss AG, Oberkochen, Germany), and the ratio of Ki-67-positive cells was calculated.
Transwell assay
Transwell chambers (Corning Incorporated, New York, USA) with or without Matrigel pre-coating were utilized to estimate cell migration and invasion. Cells were placed in the upper chamber in the medium without FBS, and the lower chamber was filled with medium containing 10% FBS. The cultivation process was carried out for 24 hours. Afterward, cells that failed to invade or migrate were eliminated using cotton swabs, while those in the bottom membrane were processed with 1% formalin and 0.1% crystal violet for fixing and staining, respectively. Thereafter, the cells were observed and photographed under a microscope.
TUNEL assay
The In Situ Cell Death Detection kit (Roche Diagnostics, Basel, Switzerland) was used to conduct the TUNEL assay. Briefly, 3% H2O2 was used to cultivate the sections at 25°C for 5 minutes. Then, the TdT labeling reaction mix was used to label the sections for 1 hour at 25°C. Afterward, 3, 3′-diaminobenzidine was used to cultivate the nuclei for 15 minutes. Using a fluorescence microscope, TUNEL-positive cells in these sections were visualized and photographed.
JC-1 assay
Recently, the JC-1 assay has been applied as a reliable method for probing the mitochondrial transmembrane potential (ΔΨm) changes that occur in the very early stage of cell apoptosis [25]. Here, we used the JC-1 assay to analyze changes in the apoptosis of TNBC cells. The variation in ΔΨm was estimated using a mitochondria staining kit (Sigma-Aldrich, St. Louis, USA), which used JC-1. Briefly, 96-well black plates were used to seed 5 × 103 cells per well. After 48 hours of treatment, JC-1 was used to cultivate the cells for 15 minutes. The excitation and emission wavelengths of the live cell bioimager were set at 490 and 530 nm, respectively, for the imagery of JC-1 monomers. The emission and excitation wavelengths were set at 590 and 525 nm, respectively, for the J-aggregates.
Subcellular fraction assay
The PARIS kit (Life Technologies, Thermo Fisher, Inc., Waltham, USA) was used to isolate the cytosolic and nuclear fractions. Briefly, re-suspension buffer was used to resuspend the cells for 15 minutes then the cells were subjected to homogenization. The cytoplasmic fraction was extracted from the supernatant after centrifugation. The pellet was then resuspended in RNase-free H2O, nuclear isolation buffer, and PBS for ice cultivation. After centrifugation, the pellet was subjected to nuclear fractionation. Finally, qRT-PCR was used to detect HAGLR expression in both fractions, with GAPDH as the cytoplasmic control and U6 as the nuclear control.
MS2-RNA immunoprecipitation (RIP) assay
This experiment was performed according to a previous protocol [26]. Briefly, pcDNA3.1-MS2 or pcDNA3.1-MS2-HAGLR was co-transfected into MDA-MB-436 and HCC-1937 cells with pMS2-GFP (Addgene, Watertown, USA) using Viafect reagent. Two days later, cells were harvested for lysis and the cell lysates were subjected to RIP experiments using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, USA) and the GFP antibody (Roche). Finally, the precipitated RNA was isolated and purified for qRT-PCR analysis.
Western blot analysis
The cell lysis buffer, which included proteinase inhibitors, was used to extract total protein from cells. SDS buffer was used to denature the proteins for 10 minutes at 95°C, and 15% polyacrylamide gels were used to isolate the proteins that were transferred to the PVDF membranes. Then, 5% non-fat milk was used to block the membranes, and the membranes were treated with primary antibodies at 4°C overnight. Subsequently, the membranes were treated with secondary antibodies. Intensive chemiluminescence (Pierce, Rockford, USA) was used to observe the bands.
Luciferase reporter assay
The pmirGLO reporter vectors were employed to carry the mutant HAGLR (or SRSF1 3′UTR) and wild-type HAGLR (or SRSF1 3′UTR). Lipofectamine 2000 was used to co-transfect MDA-MB-436 and HCC-1937 cells with miR-93-5p mimics (or NC mimics) and the above recombinant vectors. After transfection, the dual-luciferase reporter assay system (Promega, Madison, USA) was used to measure the luciferase activity. The rate of Renilla luciferase activity to firefly luciferase activity was defined as the relative luciferase activity.
Fluorescence in situ hybridization (FISH) analysis
In accordance with the protocols of Biosearch Technologies (Steinach, Germany), HAGLR or miR-93-5p conjugated with fluorescent probes were created. The cells were then hybridized with the indicated probes under non-denaturing conditions. After the completion of hybridization, samples were stained with DAPI (1:1,000) for 5 minutes. Confocal microscopy was used to observe the samples.
RNA pull-down assay
To estimate the interaction of miR-93-5p with HAGLR or SRSF1, RNA pull-down assays were conducted. Before that, miR-93-5p sequences with wild-type or mutant seed regions were synthesized and biotin-labeled (Bio-miR-93-5p-WT/Mut) with a nonsense sequence biotinylated as the negative control (Bio-NC). Then, cells were rinsed with cold PBS solution three times after they were collected, followed by cell lysis in lysis buffer containing a protease inhibitor. Thereafter, the biotin-labeled RNAs were mixed with the cell lysates overnight, and the mixtures were subsequently cultivated with M-280 streptavidin-coated magnetic beads at 25°C for 30 minutes. Next, high-salt buffer, low-salt buffer, and ice-cold lysis buffer were used to rinse the beads in succession. Then, the bound RNAs were extracted using TRIzol reagent and measured through qRT-PCR.
Statistical analysis
All assays were conducted in triplicate. Data are represented as the mean ± standard deviation. GraphPad Prism 5 software was used to conduct statistical analyses. Student's t-test was used to analyze the significance between 2 groups, and one-way analysis of variance was utilized to analyze the significance between at least three groups. Statistical significance was set at p < 0.05.
RESULTS
Inhibited HAGLR could suppress the malignant phenotypes of TNBC cells
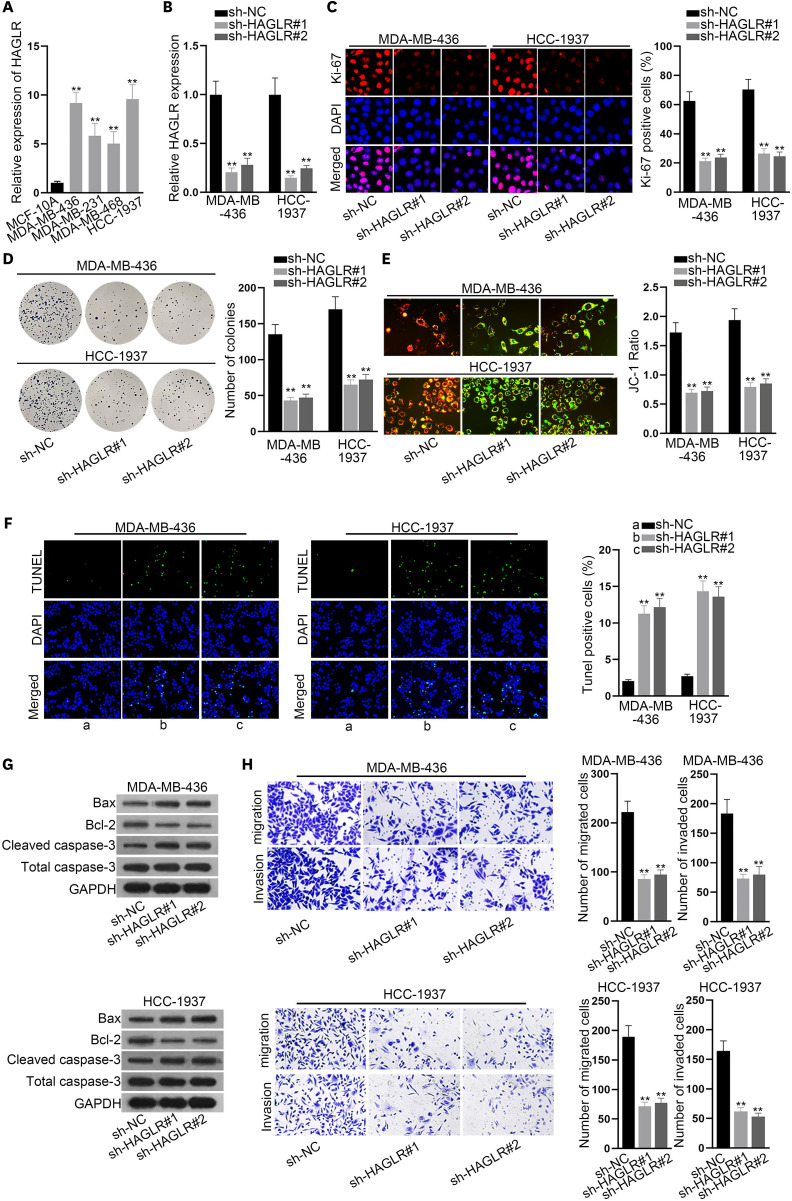
HAGLR plays an important role in many cancers. In this study, we first investigated the expression pattern of HAGLR in TNBC. Intriguingly, HAGLR expression was relatively high in 42 TNBC samples compared with adjacent non-cancerous samples (Supplementary Figure 1A). We also investigated the expression of HAGLR in a human normal breast epithelial cell line (MCF-10A) and TNBC cell lines (MDA-MB-436, MDA-MB-231, MDA-MB-468, and HCC-1937) via qRT-PCR. The results showed that HAGLR was highly expressed in TNBC cells (Figure 1A), indicating the possible role of HAGLR in TNBC. Then, we tested the inhibition efficiency of sh-HAGLR using qRT-PCR. The results showed a marked decrease in HAGLR expression in cells transfected with sh-HAGLR#1 or sh-HAGLR#2 (Figure 1B); therefore, both sh-HAGLR#1 and sh-HAGLR#2 were chosen for our functional experiments. The proliferation of TNBC cells was probed by immunofluorescence and colony formation assays. As a result, we found that the number of Ki-67-positive cells and the number of colonies in these 2 cell lines both decreased under the inhibition of HAGLR (Figure 1C and D), revealing the suppression of HAGLR inhibition on TNBC cell proliferation. In addition, JC-1 and TUNEL assays were performed to detect TNBC cell apoptosis. The JC-1 ratio decreased prominently, whereas the number of TUNEL-positive cells increased significantly in response to the absence of HAGLR (Figure 1E and F), which indicated that silencing HAGLR could promote the apoptosis of TNBC cells. Meanwhile, western blot data confirmed that the levels of pro-apoptotic proteins (Bax and cleaved caspase-3) increased, while those of anti-apoptosis protein Bcl-2 decreased due to inhibited HAGLR (Figure 1G), which further verifies the conclusion that inhibiting HAGLR promotes the apoptosis of TNBC cells. Moreover, a Transwell assay was carried out, and the results showed decreased levels of both migratory and invading cells under HAGLR deficiency (Figure 1H). In conclusion, silencing HAGLR could restrict the malignant phenotype of TNBC cells.
Figure 1. Inhibited HAGLR could suppress the malignant phenotypes of TNBC cells.
(A) HAGLR expression was tested in a human normal breast epithelial cell line and TNBC cell lines via qRT-PCR. (B) Inhibition efficacy of sh-HAGLR was tested in MDA-MB-436 and HCC-1937 cells through qRT-PCR. (C, D) Immunofluorescence and colony formation assays were implemented to detect the proliferation of MDA-MB-436 and HCC-1937 cells when HAGLR was inhibited. (E, F) JC-1 and TUNEL assays were implemented to evaluate cell apoptosis when HAGLR was inhibited. (G) Western blot was carried out to examine the changes in the levels of the apoptosis-related proteins. (H) A Transwell assay was performed to detect cell migratory and invasive capabilities when HAGLR was inhibited.
HAGLR = HOXD antisense growth-associated lncRNA; TNBC = triple-negative breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction; DAPI = 4′,6-diamidino-2-phenylindole; NC = negative control.
**p < 0.01.
miR-93-5p can bind to HAGLR
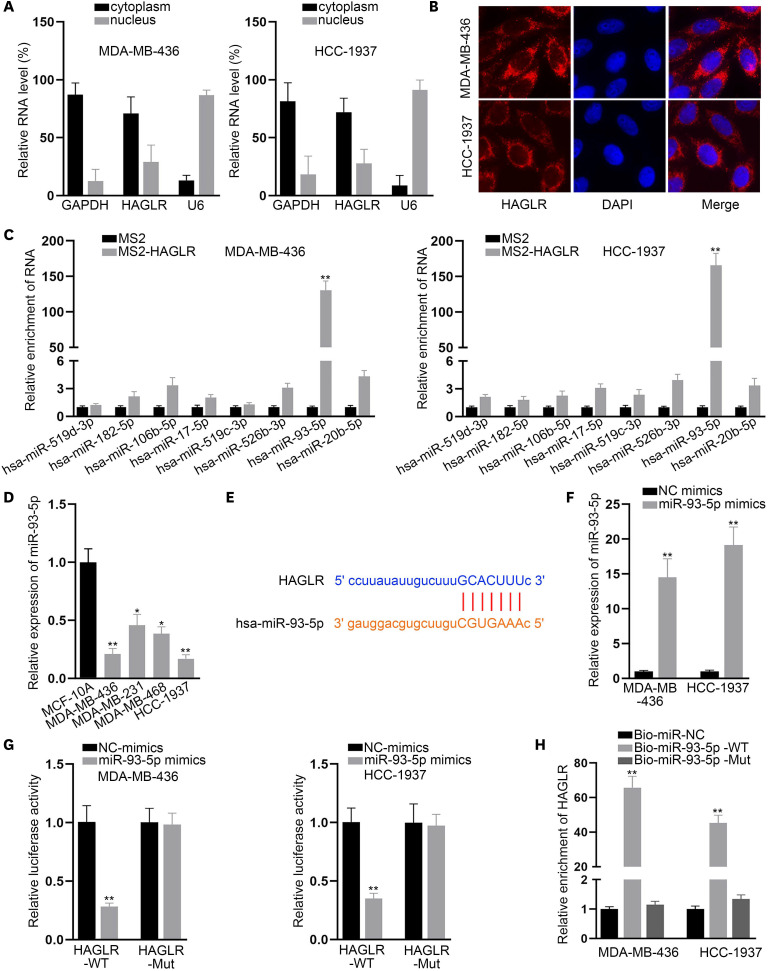
To understand the regulatory mechanism of HAGLR in TNBC, we determined the location of HAGLR in MDA-MB-436 and HCC-1937 cells. As HAGLR was mainly present in the cytoplasm, as detected by the analysis of the subcellular fraction and FISH assays (Figure 2A and B), we predicted that it might act as a miRNA sponge. Therefore, we searched ENCORI (http://starbase.sysu.edu.cn/index.php) and identified eight miRNAs that might bind to HAGLR. As revealed by the results of the MS2-RIP assay, only miR-93-5p was highly enriched by MS2-HAGLR (Figure 2C). In addition, the decreased expression of miR-93-5p in TNBC cells relative to that in MCF-10A cells was observed via qRT-PCR (Figure 2D). Moreover, FISH assay results validated that, similar to HAGLR, the main distribution of miR-93-5p was in the cytoplasm of TNBC cells (Supplementary Figure 1B). Alignment between miR-93-5p and HAGLR was observed in ENCORI (Figure 2E). After confirming the efficient overexpression of miR-93-5p in both TNBC cell lines (Figure 2F), we performed luciferase reporter and RNA pull-down assays. The results revealed that the overexpression of miR-93-5p substantially reduced the luciferase activity of HAGLR-WT, while exerting no apparent impact on that of HAGLR-Mut in either cell line (Figure 2G). Only Bio-miR-93-5p-WT pulled down HAGLR in both TNBC cell lines (Figure 2H). Based on these results, we deduced that miR-93-5p could bind to HAGLR in TNBC cells.
Figure 2. miR-93-5p could bind to HAGLR.
(A, B) Subcellular fraction and FISH assays were implemented to explore the location of HAGLR in MDA-MB-436 and HCC-1937 cells. (C). A MS2-RIP assay was conducted to detect the interaction of HAGLR with the indicated miRNAs. (D) miR-93-5p expression in TNBC cells was probed by qRT-PCR. (E). The binding site between miR-93-5p and HAGLR was presented according to ENCORI. (F) miR-93-5p overexpression was detected by qRT-PCR. (G, H) The binding relationship between miR-93-5p and HAGLR was tested by luciferase reporter assay and RNA pull-down assay.
HAGLR = HOXD antisense growth-associated lncRNA; FISH = fluorescence in situ hybridization; RIP = RNA immunoprecipitation; miR = microRNA; TNBC = triple-negative breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; DAPI = 4′,6-diamidino-2-phenylindole; NC = negative control; WT = wild-type; Mut = mutant.
*p < 0.05, **p < 0.01.
HAGLR activated the Wnt signaling pathway by upregulating SRSF1
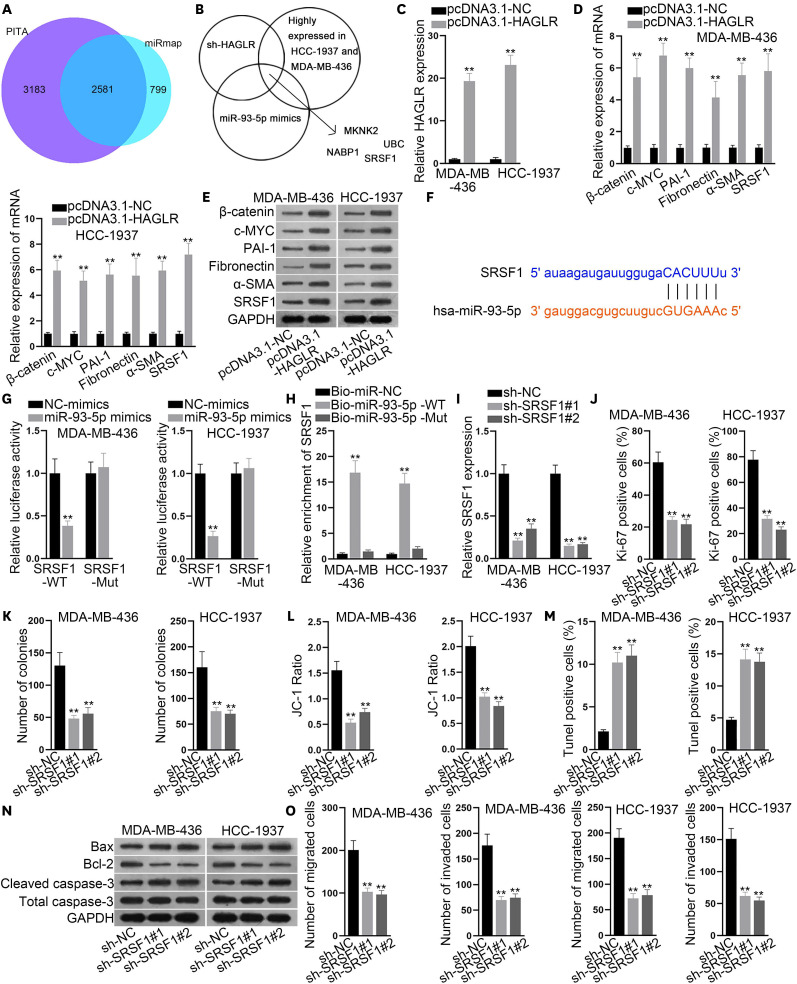
Next, we attempted to detect possible mRNAs that could bind to miR-93-5p and found 2,581 candidates under the collection of PITA and miRmap (Figure 3A). Then, qRT-PCR was performed to investigate the mRNAs whose expression could be affected by inhibited HAGLR and overexpressed miR-93-5p, and which also displayed aberrantly high expression in MDA-MB-436 and HCC-1937 cells (Figure 3B). As a result, MKNK2, UBC, NABP1, and SRSF1 were identified. However, only the expression of SRSF1 was enhanced in all four TNBC cell lines (Supplementary Figure 1C). Considering that SRSF1 could act as an activator of the Wnt signaling pathway [27], we selected SRSF1 for our subsequent experiments. Overexpression efficiency of pcDNA3.1-HAGLR was detected by qRT-PCR in MDA-MB-436 and HCC-1937 cells (Figure 3C). Then, qRT-PCR and western blotting were performed to detect the changes in the expression of SRSF1 and Wnt signaling-related molecules (β-catenin, c-MYC, PAI-1, fibronectin, and α-SMA). The results showed that the expression of these genes at both mRNA and protein levels increased significantly (Figure 3D and E), which indicated that HAGLR activated the Wnt signaling pathway by upregulating SRSF1. We then presented the binding site of miR-93-5p in SRSF1 3′UTR (Figure 3F). Results from the luciferase reporter and RNA pull-down assays confirmed the binding of miR-93-5p to SRSF1 at the predicted site (Figure 3G and H).
Figure 3. HAGLR activated the Wnt signaling pathway by upregulating SRSF1.
(A) A Venn diagram was constructed to present the possible mRNAs targeted by miR-93-5p, as predicted by 2 tools (PITA and miRmap) in ENCORI. (B) qRT-PCR was implemented to test mRNAs whose expression was affected by the inhibited HAGLR and overexpressed miR-93-5p, and which were also highly expressed in MDA-MB-436 and HCC-1937 cells. (C) Overexpression efficiency of pcDNA3.1-HAGLR was detected by qRT-PCR in MDA-MB-436 and HCC-1937 cells. (D, E) qRT-PCR and western blot were implemented to detect the expression change of genes related to the Wnt signaling pathway. (F) The binding site of miR-93-5p to SRSF1 3′UTR was presented by ENCORI. (G, H) Luciferase reporter and RNA pull-down assays were used to examine the binding of miR-93-5p to SRSF1. (I) The inhibition efficiency of sh-SRSF1 was detected by qRT-PCR in MDA-MB-436 and HCC-1937 cells. (J, K) Immunofluorescence and colony formation assays were conducted to detect the proliferation of MDA-MB-436 and HCC-1937 cells when SRSF1 was inhibited. (L-N) JC-1 assay, TUNEL assay, and western blot were implemented to detect apoptosis when SRSF1 was inhibited. (O) Transwell assays were implemented to analyze cell migratory and invasive capabilities when SRSF1 was inhibited.
HAGLR = HOXD antisense growth-associated lncRNA; SRSF1 = serine- and arginine-rich splicing factor 1; mRNA = messenger RNA; miR = microRNA; qRT-PCR = quantitative real-time polymerase chain reaction; NC = negative control; Bio = biotin-labeled; WT = wild-type; Mut = mutant.
**p < 0.01.
We also assessed the function of SRSF1 in TNBC cells. Initially, the inhibition efficiency of sh-SRSF1 was detected by qRT-PCR (Figure 3I). Immunofluorescence and colony formation assays were conducted to detect the proliferation of MDA-MB-436 and HCC-1937 cells when SRSF1 was inhibited. The results showed that the proportion of Ki-67-positive cells as well as the number of colonies decreased by the inhibition of SRSF1 (Figure 3J and K). Moreover, a JC-1 assay, a TUNEL assay, and western blotting were performed to detect apoptosis when SRSF1 was inhibited. The results also indicated that loss of SRSF1 facilitated the apoptosis of MDA-MB-436 and HCC-1937 cells (Figure 3L-N). Furthermore, the results of the Transwell assay confirmed that the inhibition of SRSF1 hindered the migration and invasion of MDA-MB-436 and HCC-1937 cells (Figure 3O). These results proved that the inhibition of SRSF1 could suppress the malignant behavior of TNBC cells.
The HAGLR/miR-93-5p/SRSF1 axis contributed to the malignancy of TNBC cells
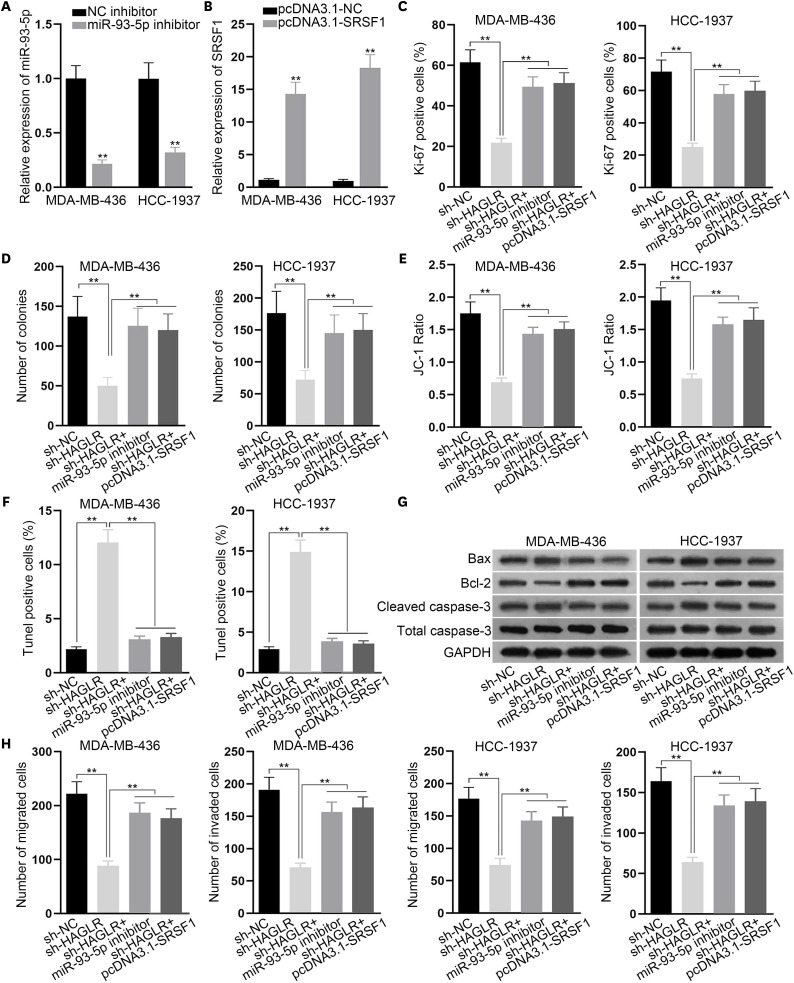
Finally, we performed rescue assays to verify the existence of the HAGLR/miR-93-5p/SRSF1 axis in TNBC cells. The knockdown efficacy of miR-93-5p inhibitor and the overexpression efficacy of pcDNA3.1-SRSF1 were tested by qRT-PCR (Figure 4A and B). Then, we designed 4 groups for the rescue experiments conducted in MDA-MB-436 and HCC-1937 cells: cells transfected with sh-NC, cells transfected with sh-HAGLR, cells co-transfected with sh-HAGLR and miR-93-5p inhibitor, and cells co-transfected with sh-HAGLR and pcDNA3.1-SRSF1. From the results of the immunofluorescence and colony formation assays, we found that the impaired proliferation caused by inhibited HAGLR was compensated by the inhibition of miR-93-5p or overexpression of SRSF1 (Figure 4C and D). In the meantime, results from the JC-1 assay, TUNEL assay, and western blot demonstrated that the accelerated apoptosis of MDA-MB-436 and HCC-1937 cells owing to the inhibition of HAGLR was recovered following the inhibition of miR-93-5p or the overexpression of SRSF1 (Figure 4E-G). Transwell assay results showed that the inhibitory effects of silencing HAGLR on cell migration and invasion were offset by miR-93-5p inhibition or SRSF1 overexpression (Figure 4H). In summary, the HAGLR/miR-93-5p/SRSF1 axis facilitated the malignant behavior of TNBC cells.
Figure 4. The HAGLR/miR-93-5p/SRSF1 axis contributed to the malignance of TNBC cells.
(A, B) The knockdown efficacy of the miR-93-5p inhibitor and the overexpression efficacy of pcDNA3.1-SRSF1 were analyzed by qRT-PCR. (C, D) Immunofluorescence and colony formation assays were implemented to investigate the proliferation of the indicated MDA-MB-436 and HCC-1937 cells. (E-G) JC-1 assay, TUNEL assay, and western blot were implemented to detect cell apoptosis under different circumstances. (H) A Transwell assay was carried out to determine the migration and invasion of the indicated MDA-MB-436 and HCC-1937 cells.
HAGLR = HOXD antisense growth-associated lncRNA; miR = microRNA; SRSF1 = serine- and arginine-rich splicing factor 1; TNBC = triple-negative breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction; NC = negative control; GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
**p < 0.01.
DISCUSSION
TNBC is difficult to cure due to the lack of the expression of ER, PR, and HER-2, as only one of the three receptors can be targeted by chemotherapy [1]. Meanwhile, auxiliary radiotherapy can improve the survival rate of patients with TNBC [2]. lncRNAs have been studied in various cancers, revealing that they can promote or relieve cancer progression [5,6]. HAGLR plays an important role in many cancers, such as non-small cell lung cancer cells [8], esophageal cancer [9] and lung adenocarcinoma [10]. Herein, we explored the function of HAGLR in TNBC cells. We first discovered that HAGLR presented abnormally high expression in TNBC tissues and cells, and this was in agreement with the findings of a recently published study [28]. Additionally, results of functional assays indicated that inhibiting HAGLR could hamper proliferative, migratory, and invasive capabilities while strengthening the apoptotic ability of TNBC cells. In other words, HAGLR was revealed to contribute to the progression of TNBC.
Considering that HAGLR could act as a miRNA sponge, as has been reported in esophageal cancer [9], we localized HAGLR in TNBC cells and observed that HAGLR primarily accumulated in the cytoplasm. Furthermore, we found that miR-93-5p could bind to HAGLR and that SRSF1 was targeted by miR-93-5p in TNBC cells. Though miR-93-5p appears to play different roles—either oncogenic or tumor-suppressive—in different cancer types [15,29], its tumor-restraining function in breast cancers, including TNBC, has already been revealed [16,17]. In addition, consistent with the findings of previous studies [30], SRSF1 was upregulated in TNBC cells and exerted a tumor-promoting function in TNBC. SRSF1 can act as an activator of the Wnt signaling pathway [27]. Here, we validated that HAGLR elevated SRSF1 levels via the ceRNA mechanism to activate the Wnt signaling pathway in TNBC cells. Moreover, we confirmed that HAGLR accelerated the malignancy of TNBC by miR-93-5p/SRSF1 signaling.
In conclusion, this study revealed that HAGLR boosted SRSF1 expression to activate the Wnt pathway by sponging miR-93-5p; thus, facilitating the development of TNBC. These findings might provide new avenues for treating TNBC, although further evidence is needed.
ACKNOWLEDGMENTS
We appreciate the support of our researchers.
Footnotes
Conflict of Interest: The authors declare that they have no competing interests.
- Conceptualization: Wu X.
- Data curation: Wu X.
- Formal analysis: Wu X.
- Funding acquisition: Zhang C.
- Investigation: Zhang C.
- Methodology: Zhang C.
- Project administration: Yang Y.
- Resources: Yang Y.
- Software: Yi L.
- Supervision: Paizula X.
- Validation: Yi L.
- Visualization: Xu W.
- Writing - original draft: Paizula X.
- Writing - review & editing: Xu W.
SUPPLEMENTARY MATERIAL
The expression of genes in TNBC tissues and cells and the distribution of miR-93-5p in TNBC cells.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Ren YQ, Fu F, Han J. MiR-27a modulates radiosensitivity of triple-negative breast cancer (TNBC) cells by targeting CDC27. Med Sci Monit. 2015;21:1297–1303. doi: 10.12659/MSM.893974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111:4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H, Zhang J, Shi J, Guo Z, He C, Ding L, et al. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016;37:11623–11631. doi: 10.1007/s13277-016-5125-8. [DOI] [PubMed] [Google Scholar]
- 5.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol. 2017;39:1010428317697574. doi: 10.1177/1010428317697574. [DOI] [PubMed] [Google Scholar]
- 9.Yang C, Shen S, Zheng X, Ye K, Sun Y, Lu Y, et al. Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge to regulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer by regulating LAMP3. FASEB J. 2019;33:10490–10504. doi: 10.1096/fj.201802543RR. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Chen Z, Zhao L, Cheng D, Song W, Zhang X. Long non-coding RNA-HAGLR suppressed tumor growth of lung adenocarcinoma through epigenetically silencing E2F1. Exp Cell Res. 2019;382:111461. doi: 10.1016/j.yexcr.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figliuzzi M, Marinari E, De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys J. 2013;104:1203–1213. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Chen T, Zhang Y, Zhang N, Li C, Li Y, et al. Long noncoding RNA Linc00339 promotes triple-negative breast cancer progression through miR-377-3p/HOXC6 signaling pathway. J Cell Physiol. 2019;234:13303–13317. doi: 10.1002/jcp.28007. [DOI] [PubMed] [Google Scholar]
- 15.Chen YL, Wang GX, Lin BA, Huang JS. MicroRNA-93-5p expression in tumor tissue and its tumor suppressor function via targeting programmed death ligand-1 in colorectal cancer. Cell Biol Int. 2020;44:1224–1236. doi: 10.1002/cbin.11323. [DOI] [PubMed] [Google Scholar]
- 16.Xiang Y, Liao XH, Yu CX, Yao A, Qin H, Li JP, et al. MiR-93-5p inhibits the EMT of breast cancer cells via targeting MKL-1 and STAT3. Exp Cell Res. 2017;357:135–144. doi: 10.1016/j.yexcr.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Shyamasundar S, Lim JP, Bay BH. miR-93 inhibits the invasive potential of triple-negative breast cancer cells in vitro via protein kinase WNK1. Int J Oncol. 2016;49:2629–2636. doi: 10.3892/ijo.2016.3761. [DOI] [PubMed] [Google Scholar]
- 18.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Xue Y, Liu J, Zhuang J, Shen L, Shen B, et al. Long noncoding RNA ASAP1-IT1 promotes cancer stemness and predicts a poor prognosis in patients with bladder cancer. Neoplasma. 2017;64:847–855. doi: 10.4149/neo_2017_606. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO, Kim KH, et al. The long noncoding RNA HOXA11 antisense induces tumor progression and stemness maintenance in cervical cancer. Oncotarget. 2016;7:83001–83016. doi: 10.18632/oncotarget.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojtowicz JM, Kee N. BrdU assay for neurogenesis in rodents. Nat Protoc. 2006;1:1399–1405. doi: 10.1038/nprot.2006.224. [DOI] [PubMed] [Google Scholar]
- 24.Wang JJ, Liu C, Shan K, Liu BH, Li XM, Zhang SJ, et al. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018;8:3408–3415. doi: 10.7150/thno.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvioli S, Ardizzoni A, Franceschi C, Cossarizza A. JC-1, but not DiOC6(3) or rhodamine 123, is a reliable fluorescent probe to assess delta psi changes in intact cells: implications for studies on mitochondrial functionality during apoptosis. FEBS Lett. 1997;411:77–82. doi: 10.1016/s0014-5793(97)00669-8. [DOI] [PubMed] [Google Scholar]
- 26.Hu YP, Jin YP, Wu XS, Yang Y, Li YS, Li HF, et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer. 2019;18:167. doi: 10.1186/s12943-019-1097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Huang B, Shi Z, Han J, Wang Y, Huangfu J, et al. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing β-catenin biosynthesis. EMBO Mol Med. 2013;5:737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahlab-Aviv S, Zohar K, Cohen Y, Peretz AR, Eliyahu T, Linial M, et al. Spliceosome-associated microRNAs signify breast cancer cells and portray potential NOVEL NUCLEAR TARGETS. Int J Mol Sci. 2020;21:8132. doi: 10.3390/ijms21218132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma DH, Li BS, Liu JJ, Xiao YF, Yong X, Wang SM, et al. miR-93-5p/IFNAR1 axis promotes gastric cancer metastasis through activating the STAT3 signaling pathway. Cancer Lett. 2017;408:23–32. doi: 10.1016/j.canlet.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, et al. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60:105–117. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression of genes in TNBC tissues and cells and the distribution of miR-93-5p in TNBC cells.