Abstract
Objective
Infection with the novel coronavirus SARS–CoV‐2 triggers severe illness with high mortality in a subgroup of patients. Such a critical course of COVID‐19 is thought to be associated with the development of cytokine storm, a condition seen in macrophage activation syndrome (MAS) and secondary hemophagocytic lymphohistiocytosis (HLH). However, specific data demonstrating a clear association of cytokine storm with severe COVID‐19 are still lacking. The aim of this study was to directly address whether immune activation in COVID‐19 does indeed mimic the conditions found in these classic cytokine storm syndromes.
Methods
Levels of 22 biomarkers were quantified in serum samples from patients with COVID‐19 (n = 30 patients, n = 83 longitudinal samples in total), patients with secondary HLH/MAS (n = 50), and healthy controls (n = 9). Measurements were performed using bead array assays and single‐marker enzyme‐linked immunosorbent assay. Serum biomarker levels were assessed for correlations with disease outcome.
Results
In patients with secondary HLH/MAS, we observed pronounced activation of the interleukin‐18 (IL‐18)–interferon‐γ axis, increased serum levels of IL‐1 receptor antagonist, intercellular adhesion molecule 1, and IL‐8, and strongly reduced levels of soluble Fas ligand in the course of SARS–CoV‐2 infection. These observations appeared to discriminate immune dysregulation in critical COVID‐19 from the well‐recognized characteristics of other cytokine storm syndromes.
Conclusion
Serum biomarker profiles clearly separate COVID‐19 from MAS or secondary HLH in terms of distinguishing the severe systemic hyperinflammation that occurs following SARS–CoV‐2 infection. These findings could be useful in determining the efficacy of drugs targeting key molecules and pathways specifically associated with systemic cytokine storm conditions in the treatment of COVID‐19.
INTRODUCTION
The novel coronavirus SARS–CoV‐2 has been infecting ever increasing numbers of people around the globe. While the infection results in mild‐to‐moderate symptoms in most individuals, it triggers a severe illness with high mortality in a subgroup of patients.
Early in the pandemic, it was proposed that a severe (fatal) course of COVID‐19 correlated with the presence of hyperinflammation, as is seen in classic cytokine storm syndromes (1), including secondary hemophagocytic lymphohistiocytosis (HLH). Secondary HLH may occur in the context of, for example, infection, malignancy, metabolic disease, trauma, or rheumatic disease (in the latter case, referred to as macrophage activation syndrome [MAS]). MAS is frequently associated with adult‐onset Still’s disease (AOSD) and systemic juvenile idiopathic arthritis in children, but it has also been seen in Kawasaki disease and other rheumatic conditions. Current data suggest that there is a strong clinical and immunophenotypic overlap between secondary HLH and MAS (2).
Key molecules or pathways that drive HLH/MAS, such as interleukin‐1β (IL‐1β), IL‐6, IL‐18, interferon‐γ (IFNγ), or JAK/STAT, can be targeted by state‐of‐the‐art therapies, and ever since the proposal regarding an overlap of (critical) COVID‐19 with classic cytokine storm conditions was put forward (1, 3), those types of conditions have been considered therapeutic targets in COVID‐19 or have already been studied in respective clinical trials (ClinicalTrials.gov identifiers: NCT04372186, NCT04317092, NCT04324021, and NCT04338958). Yet, at the same time, studies addressing the relevance of cytokine storm conditions in COVID‐19 are frequently limited to discussions focused on the effects of IL‐6 (4) and tend to draw conclusions based on comparisons with many different critical clinical conditions or even with healthy controls. However, to draw such conclusions, we believe it is necessary to investigate scenarios of severe immunologic disease, classified, as a group, as “cytokine storm conditions” on the basis of clinical and laboratory criteria. Therefore, in this study, we set out to directly compare the cytokine signatures in patients with secondary HLH and patients with MAS to the cytokine signatures observed in patients with COVID‐19, with the aim of identifying serum biomarkers that could clearly separate the different entities.
PATIENTS AND METHODS
Study subjects and samples
Serum samples from COVID‐19 patients (n = 30 patients, n = 83 longitudinal samples) were collected at the Department of Gastroenterology, Hepatology, Endocrinology and Clinical Infectiology of the University Hospital Muenster in Germany from March until May 2020. Samples were collected at the time of hospital admission and throughout the disease course. All serum samples from patients with COVID‐19 were collected during the first wave of the COVID‐19 pandemic in Germany, and none of the enrolled COVID‐19 patients had received immunosuppressive or biologic therapies or (experimental) antiviral treatment. However, in cases of bacterial or fungal superinfection, patients did receive anti‐infection drugs.
Disease severity was defined as critical (presence of acute respiratory distress syndrome [ARDS] and/or deceased), severe (requiring oxygen supplementation), or moderate (ARDS not present and oxygen supplementation not required). ARDS was diagnosed according to the Berlin definition (i.e., presence of ground‐glass opacities bilaterally on chest radiograph, and exclusion of other causes of respiratory failure) (5). COVID‐19 patients were categorized according to the comorbidity designated as their worst condition over the course of hospitalization.
For comparison, serum samples from adult patients with secondary HLH (n = 20) and patients with AOSD‐MAS (n = 17), which were collected in the course of previous studies (6, 7, 8), were used. In addition, serum samples were collected from pediatric/adolescent patients with secondary HLH (n = 4), pediatric/adolescent patients with MAS (n = 9), and healthy control subjects (n = 9) at the University Children’s Hospital Muenster in Germany. Samples from patients with secondary HLH and patients with MAS were collected during a state of active disease. Disease classification is further detailed in the Supplementary Methods and Supplementary Table 1 (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract).
All study subjects or the childrens’ caregivers provided their written informed consent. The study was approved by ethics committees in previously reported studies (6, 7, 8) as well as the local ethics committee of the University Hospital Muenster (approval nos. 2020‐210‐s‐S and 2015‐670‐f‐S).
Quantification of serum markers
For quantification of biomarkers in the serum of all subjects, we used multiplex assays to measure IL‐1β, interleukin‐1 receptor antagonist (IL‐1Ra), IL‐4, IL‐6, IL‐8, IL‐10, IL‐18, tumor necrosis factor, IFNα, IFNβ, IFNγ, monocyte chemoattractant protein 2 (MCP‐2; CCL8), MCP‐3 (CCL7), CXCL9, CXCL10, macrophage colony‐stimulating factor, leucine‐rich α2‐glycoprotein 1, soluble FasL (sFasL), intercellular adhesion molecule 1 (ICAM‐1), vascular cell adhesion molecule 1 (VCAM‐1), and galectin 3. Specific reagents (all purchased from R&D Systems) and the sera were prepared according to the manufacturer’s instructions. Data acquisition and analysis were performed on a MAGPIX instrument (Merck Millipore) using xPONENT version 4.2 software (Luminex). Concentrations of S100A12 in the subjects’ sera were quantified by sandwich enzyme‐linked immunosorbent assay using in‐house monoclonal antibodies.
Data analysis
Serum marker data were assessed using unsupervised clustering analysis, including correlation distance and ward.D linkage in the pheatmap R package and RStudio platforms (RStudio Team 2015 and RStudio: Integrated Development for R; http://www.rstudio.com/). Principal components analysis (PCA) of the serum marker expression data was performed using the ggfortify and autoplot R packages and RStudio software. Multiple serum analytes were assessed for correlations with severe COVID‐19 using Spearman’s rank correlation analyses, with the data plotted using the corrplot R package and RStudio or GraphPad Prism software (version 8.0 for Mac OS X; GraphPad Software).
Data on individual serum markers were analyzed for normality distribution with the D’Agostino‐Pearson normality test, using GraphPad Prism software. The majority of the data did not pass this test, and therefore the nonparametric data were tested using a Kruskal‐Wallis test followed by Dunn’s test for multiple comparisons (GraphPad Prism version 8.0). Receiver operating characteristic (ROC) curve analyses were performed using GraphPad Prism software.
RESULTS
Serum marker profiling of COVID‐19 compared to classic cytokine storm syndromes
In our cohort of COVID‐19 patients who were hospitalized during the first wave of SARS–COV‐2 infections (n = 30), 17 patients had critical disease, of whom 7 died (Table 1). Six patients presented with severe disease, and 7 were classified as having moderate disease.
Table 1.
Characteristics of the patients with COVID‐19 compared to patients with secondary HLH/MAS and healthy controls*
| Patients with COVID‐19 |
Patients with secondary HLH (n = 22)/MAS (n = 28) (total n = 50) |
Healthy controls (n = 9) |
||||
|---|---|---|---|---|---|---|
|
Total (n = 30) |
Critical disease (n = 17) |
Severe disease (n = 6) |
Moderate disease (n = 7) |
|||
| Sex, no. (%) | ||||||
| Male | 28 (93) | 16 (94) | 6 (100) | 6 (86) | 24 (48) | 4 (44) |
| Female | 2 (7) | 1 (6) | 0 (0) | 1 (14) | 26 (52) | 5 (55) |
| Age, median (range) years | 57 (30–81) | 60 (49–76) | 53 (49–73) | 54 (30–81) | 48 (1.5–86.5) | 28 (7–55) |
| BMI, median (IQR) kg/m2 | 25 (23–29) | 27 (24.5–30.5) | 23 (22.8–25.3) | 23 (22–26) | ND | ND |
| Medical history, no. (%) | ||||||
| Cardiovascular insufficiency | 4 (13) | 3 (18) | 0 (0) | 1 (14) | 0 (0) | 0 (0) |
| Respiratory insufficiency | 1 (3) | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| COPD | 1 (3) | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Kidney insufficiency | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Metastatic neoplasm | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Diabetes | 1 (3) | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Hematologic malignancy | 3 (10) | 2 (12) | 0 (0) | 1 (14) | 0 (0) | 0 (0) |
| SAPS II score, median (IQR) | – | 55 (34.5–73) | 22 (18.3–24.5) | 15 (13–28) | ND | ND |
| Leukocytes, median (IQR) × 109/liter | 6.41 (4.23–8.40) | 7.51 (4.99–7.32) | 5.71 (4.89–7.32) | 4.41 (3.32–6.80) | 9.6 (2.94–13.53) | ND |
| Creatinine, median (IQR) mg/dl | 0.95 (0.78–1.53) | 1.4 (0.75–1.70) | 0.8 (0.70–0.93) | 1.00 (0.80–1.00) | 1.17 (0.43–2.1) | ND |
| CRP, median (IQR) mg/dl | 8.3 (3.3–16.9) | 14.2 (6.9–25.5) | 7.3 (4.3–11) | 1.6 (1.3–3.4) | 12.2 (0.8–19.1) | ND |
| Ferritin, median (IQR) µg/liter | 811 (582–1,363) | 1,084 (720–2,024) | 811 (608–1,426) | 596 (437–706) | 3,897 (1,792–10,787) | ND |
HLH = hemophagocytic lymphohistiocytosis; MAS = macrophage activation syndrome; BMI = body mass index; IQR = interquartile range; ND = not determined; COPD = chronic obstructive pulmonary disease; SAPS II = Simplified Acute Physiology Score II; CRP = C‐reactive protein.
Unsupervised hierarchical clustering analysis (Figure 1A) and PCA analysis (Figures 1B–D) of early serum (i.e., first blood sample following hospitalization) marker profiles were carried out in the serum from patients with COVID‐19 compared to the serum from patients with secondary HLH/MAS (n = 50) and healthy controls (n = 9) (Table 1; see also Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). The results revealed distinct groupings of patients based on marker profiles. PCA discriminated patients with critical COVID‐19 from those with severe or moderate COVID‐19, and the latter 2 groups of COVID‐19 patients clustered with healthy controls (Figure 1B). Patients with secondary HLH/MAS clustered separately from those with COVID‐19 (Figures 1C and D), particularly in the comparison of patients with secondary HLH or MAS and patients with COVID‐19 individually (Figures 1A and D).
Figure 1.
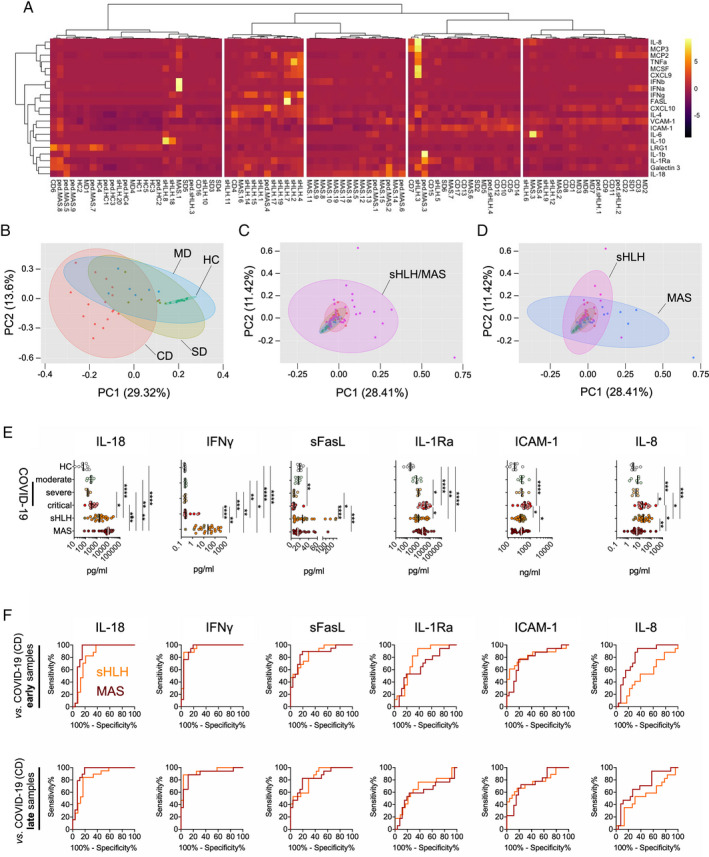
Serum biomarker profiles in patients with COVID‐19 compared to patients with secondary hemophagocytic lymphohistiocytosis (sHLH)/macrophage activation syndrome (MAS). A, Heatmap from unsupervised clustering analysis using correlation distance and ward.D linkage showing biomarker levels in the first serum sample obtained following hospitalization from patients with critical COVID‐19 (CD) (presence of acute respiratory distress syndrome [ARDS] and/or deceased; n = 17), those with severe disease (SD) (requiring oxygen supplementation; n = 6), or those with moderate disease (MD) (ARDS not present and oxygen supplementation not required; n = 7) in relation to measurements of serum biomarker levels in patients with active secondary HLH (adult, n = 18; pediatric, n = 4) and patients with MAS (adult‐onset Still’s disease, n = 17; systemic lupus erythematosus [SLE], n = 2; systemic juvenile idiopathic arthritis, n = 8; juvenile SLE, n = 1) as well as healthy controls (HC) (n = 9). Color coding indicates the row Z score for expression levels in each sample. B–D, Principal components (PC) analyses of the serum samples described in A, analyzing biomarker profiles in serum from patients with COVID‐19 according to disease severity compared to healthy controls (B) and patients with secondary HLH/MAS (C) and from patients with secondary HLH compared to patients with MAS (D). E, Individual biomarkers showing differential expression in patients with COVID‐19 according to disease severity compared to patients with secondary HLH, patients with MAS, and healthy controls. Results are shown as scatterplots, in which symbols represent individual samples, and vertical lines show the median. * = P < 0.05; ** = P < 0.01; *** = P < 0.001; **** = P < 0.0001, by Kruskal‐Wallis test followed by Dunn’s test for multiple comparisons. F, Receiver operating characteristic curve analyses of individual serum biomarkers (corresponding to those in E) for the differentiation of patients with critical COVID‐19 from patients with secondary HLH or MAS. Results are shown according to the time of sample collection from patients with COVID‐19: early = first serum sample obtained following hospitalization; late = later in disease progression. IL‐8 = interleukin‐8; MCP‐3 = monocyte chemoattractant protein 3; TNFA = tumor necrosis factor; MCSF = macrophage colony‐stimulating factor; IFNb = interferon‐β; VCAM‐1 = vascular cell adhesion molecule 1; ICAM‐1 = intercellular adhesion molecule 1; LRG‐1 = leucine‐rich α2‐glycoprotein 1; sFasL = soluble FasL; IL‐1Ra = IL‐1 receptor antagonist.
In patients whose COVID‐19 developed with a critical course, the majority of assessed biomarker levels were elevated to a range similar to that seen in patients with secondary HLH or MAS (see Supplementary Figure 1A, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). No differences in serum levels of IFNα, IFNβ, and MCP‐2 were noted (Supplementary Figure 1B). In contrast, patients with secondary HLH/MAS could be separated from patients with critical and/or severe COVID‐19 based on 6 of the serum markers assessed (Figure 1E). Levels of IL‐18 and IFNγ were markedly elevated in those with secondary HLH and those with MAS, while the ratio of IL‐18 to CXCL9 discriminated only those with MAS from those with critical COVID‐19 (see Supplementary Figure 1C).
Serum concentrations of IL‐1Ra and IL‐8 were significantly increased in patients with critical COVID‐19 compared to those with secondary HLH and those with MAS, respectively. Furthermore, serum levels of soluble ICAM‐1 were increased in patients with critical COVID‐19 compared to those with secondary HLH and those with MAS. In contrast to these elevations in serum markers, the serum levels of sFasL were markedly decreased in patients with COVID‐19 in comparison to patients with secondary HLH and those with MAS (Figure 1E).
Unlike the included study patients with COVID‐19, some patients with secondary HLH and patients with MAS had received immunosuppressive medications (see Supplementary Table 1 [http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract]). However, when samples from immunosuppressant‐treated patients with secondary HLH or MAS were removed from the data set, the previously recorded significant differences in serum marker levels remained unchanged (Figure 1E and Supplementary Figure 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract).
Separation of critical COVID‐19 from classic cytokine storm syndromes based on selected serum biomarkers, irrespective of disease severity
Over the course of the disease, levels of inflammation biomarkers in the serum from patients with COVID‐19 varied with respect to the time point of sampling from first manifestation of symptoms (see Supplementary Figure 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). When the serum marker levels in the first available sample following hospitalization (median 12.5 days, interquartile range [IQR] 11–21 days since first symptoms) were compared to those in the last available sample (median 31 days, IQR 21–36 days since first symptoms), we noted that the biomarker concentrations in some patients with critical COVID‐19 had escalated during the disease course, whereas in other patients with COVID‐19, the levels were approaching those seen in healthy controls (see Supplementary Figure 4, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). However, none of these changes reached the level of significance.
When we compared samples collected early in the disease course to samples collected late in the disease course, the underlying serum marker signatures still clearly distinguished patients with secondary HLH from patients with critical COVID‐19, regardless of the time point of sample collection (see Supplementary Figure 5, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). ROC curve analyses of specifically IL‐18, IFNγ, sFasL, and ICAM‐1 serum levels collected at different time points during the course of critical COVID‐19 revealed an almost identical performance in terms of separating critical COVID‐19 from secondary HLH and MAS (Figure 1F and Supplementary Table 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). In contrast, IL‐1Ra and IL‐8 serum levels quantified in samples collected late in the course of critical COVID‐19 revealed less power in separating critical COVID‐19 from either secondary HLH (with IL‐1Ra) or MAS (with IL‐8), compared to the respective serum concentrations of these markers in samples collected early in the disease course (Figure 1F and Supplementary Table 2).
When we tested the identified parameters for their power in differentiating secondary HLH or MAS from critical COVID‐19 in samples collected within defined time frames following the onset of the first symptoms, our findings confirmed a universal strong differentiation of critical COVID‐19 from both secondary HLH and MAS based on the serum levels of IL‐18, IFNγ, sFasL, and ICAM‐1 (see Supplementary Figure 6, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract). Of all of the tested markers, the serum levels of IFNγ were the best at separating secondary HLH and MAS from critical COVID‐19 (Figure 1F and Supplementary Table 2).
Dysregulation of the IL‐18–IFNγ axis in classic cytokine storm syndromes when compared to COVID‐19
IL‐18 and IFNγ have a central role in viral defense (9), but also in the pathogenesis of hyperinflammation as observed in patients with secondary HLH/MAS (2). Importantly, our serum biomarker analyses revealed a pronounced differential expression of these cytokines in patients with SARS–COV2–induced inflammation as compared to that in patients with secondary HLH/MAS. In multiple correlation analyses, we noted a prevalence of positive associations of both IL‐18 and IFNγ with almost all of the quantified serum markers in patients with secondary HLH/MAS (Figure 2A), whereas we did not observe these associations in patients with critical COVID‐19 (Figure 2B); similar correlation patterns were observed for many of the other blood biomarkers assessed. When we further analyzed associations of IL‐18 serum levels with serum levels of IFNγ or the IFNγ signaling surrogates CXCL9 and CXCL10, as well as with the serum ferritin and thrombocyte counts (as has been previously established to confirm a role of IFNγ in MAS pathogenesis [10]), we noted a poor correlation of these parameters in patients with critical COVID‐19 (Figures 2A and B). Although the serum ferritin levels and blood thrombocyte counts did not differ significantly between patients with critical COVID‐19 and patients with secondary HLH/MAS (see Supplementary Figure 7, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41763/abstract), correlations of the serum ferritin levels and blood thrombocyte counts with other investigated parameters were strikingly different in patients with COVID‐19 compared to patients with secondary HLH/MAS (Figures 2C and D versus Figures 2E and F).
Figure 2.
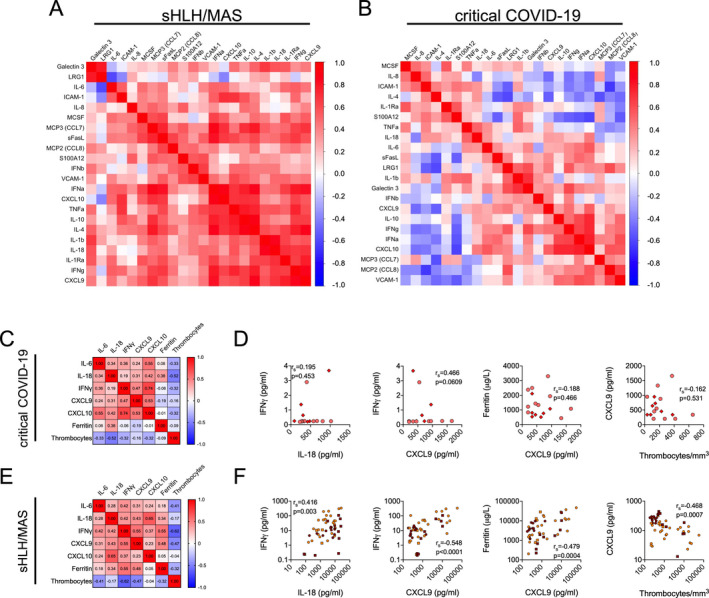
Dysregulation of the IL‐18–IFNγ axis in patients with classic cytokine storm syndromes as compared to patients with COVID‐19. A and B, Hierarchical clustering analyses showing multiple correlations by Spearman’s rank correlation test of serum biomarker levels in patients with active secondary HLH/MAS (n = 50) (A) and patients with critical COVID‐19 (n = 17) (B). Positive associations are depicted in red; negative associations are depicted in blue. C and E, Hierarchical clustering analyses of Spearman’s rank correlations between serum levels of IL‐6, IL‐18, IFNγ, and IFNγ signaling surrogates CXCL9 and CXCL10, as well as serum ferritin and thrombocyte cell counts, in patients with critical COVID‐19 (n = 17) (C) and patients with secondary HLH/MAS (n = 50) (E). D and F, Correlations of expression levels between the same serum biomarkers as indicated in C and E. In D, circles represent patients with critical COVID‐19, and diamonds represent patients who are deceased (n = 7). In F, orange circles represent patients with secondary HLH (n = 22), dark red circles represent patients with MAS (n = 28), and squares represent pediatric/adolescent patients with secondary HLH (n = 4) or MAS (n = 9). See Figure 1 for definitions.
DISCUSSION
The initial proposal of cytokine storm as a relevant element of (critical) COVID‐19 pathogenesis (1) intrigued physicians and researchers, particularly in the field of rheumatology, as such conditions are seen and investigated on a regular basis in patients with rheumatic autoinflammatory diseases (3). However, while the scientific discussion on the relevance and impact of cytokine storm following SARS–COV‐2 infection is still ongoing (11), to our knowledge there are yet no data that explicitly compare the immunology in COVID‐19 with that in classic, inflammation‐induced cytokine storm conditions as defined by clinical and laboratory criteria. Therefore, in this study, we analyzed serum biomarker signatures in patients with COVID‐19 as compared to patients with secondary HLH or MAS (as classic cytokine storm syndromes), and found that the IL‐18–IFNγ axis, as well as the serum levels of sFasL and ICAM‐1, could clearly differentiate patients with SARS–CoV‐2–induced immune dysregulation.
In our patient cohort, the quantified serum inflammation biomarkers in patients with COVID‐19 increased with disease severity and could indicate disease outcome at an early time point in the course of the disease, which supports previous data (12). In patients with COVID‐19 with a critical course, the majority of assessed biomarker levels, including IL‐6, were elevated to a range similar to that seen in patients with secondary HLH or MAS. Importantly, none of the enrolled COVID‐19 patients received immunosuppressive or biologic therapies or (experimental) antiviral treatment, which may have confounded our results.
In contrast to the many quantified parameters, the IFNγ axis, including IL‐18 as an IFNγ‐inducing factor (9) as well as IFNγ itself, appeared to be dysregulated in patients with secondary HLH or MAS, which echoes previous data (10). While a reduction in the expression of IFNγ in the serum of patients with COVID‐19 was already reported in an earlier study (13), we herein showed that IFNγ, as well as IL‐18, could significantly distinguish COVID‐19 from hyperferritinemic cytokine storm conditions.
The IL‐18–IFNγ axis appeared to be strongly dysregulated in patients with secondary HLH or MAS, and the corresponding serum levels of these cytokines could be found in substantially increased concentration ranges as compared to those seen in patients with COVID‐19; a similar pattern was seen for the serum concentrations of sFasL, except that the levels of sFasL were strongly decreased in patients with critical COVID‐19 compared to both healthy controls and patients with secondary HLH or MAS. Decreasing sFasL levels according to the level of COVID‐19 disease activity, as has been similarly reported very recently (14), may indicate a selective SARS–CoV‐2–induced immunosuppressive effect rather than general overactivation and hyperinflammation (15). Furthermore, these data could point to an evasive strategy resulting from apoptosis, as has been previously reported for HIV on the level of FasL expression (16).
In contrast to IL‐18, IFNγ, and sFasL, the serum concentrations of ICAM‐1 were significantly elevated in patients with COVID‐19 compared to patients with secondary HLH or MAS. Earlier reports of increased soluble ICAM‐1 levels in the serum of patients with COVID‐19 suggest that excessive endothelial activation and barrier dysfunction is occurring (17). Within our data set, we observed similar changes in soluble VCAM‐1 levels, albeit those remained below the level of significance.
Similar to ICAM‐1, IL‐8 and IL‐1Ra serum levels were significantly increased in patients with severely critical COVID‐19 but not in patients with MAS or secondary HLH. Elevated serum levels of these markers can indicate general inflammatory activity. However, with respect to the specific clinical presentation of critical COVID‐19, increased IL‐8 serum concentrations may indeed reflect the pathologic features of ARDS. In patients with ARDS, IL‐8 has been shown to enable both neutrophil influx and survival in lung tissue (18). Correspondingly, the therapeutic efficacy of IL‐8 blockade is currently being tested in patients with COVID‐19 (ClinicalTrials.gov identifier: NCT04347226).
While our analyses suggest that we may identify particular axes of inflammation to contrast COVID‐19 from inflammation‐ or infection‐induced cytokine storm as in MAS or secondary HLH, we are well aware of 3 limitations of our study. First, the study is descriptive and limited to a rather small number of patients. Second, even though we were able to significantly extend our findings beyond those previously reported with regard to associations with IL‐6 (4), our serum marker panel still comprises comparably few analytes, but covers those with reported relevance in classic cytokine storm conditions (2). Third, we carried out our comparisons of serum marker signatures between clinical conditions with a predominant lung involvement (COVID‐19) and those with systemic pathology (secondary HLH/MAS).
Yet, despite these limitations, we believe our data provide important insights into the proposed overlap between SARS–CoV‐2–induced immune dysregulation and classic cytokine storm conditions (3), and raise questions regarding the significance of systemic hyperinflammation in COVID‐19 (19). Furthermore, our analyses may raise doubt about the efficacy of clinical trials targeting key molecules and pathways associated with secondary HLH and/or MAS in the treatment of COVID‐19.
Therapeutic blockade of IFNγ, which appears to be a promising therapeutic option in the treatment of HLH (20) and potentially also MAS (21), may be less effective in COVID‐19 (ClinicalTrials.gov identifier: NCT04324021) as the overall activation of the IL‐18–IFNγ axis seems far less pronounced in the context of SARS–CoV‐2 infection. In contrast to IL‐18 and IFNγ, the serum levels of IL‐1Ra in patients with COVID‐19 are substantially elevated. This observation may point to a limited utility of therapeutic IL‐1 blockade in patients with COVID‐19 (22, 23), since high endogenous levels of IL‐1Ra have been reported to indicate a rather poor response to treatment with drugs neutralizing IL‐1β or IL‐1 signaling (24). However, elevated circulating concentrations of IL‐1Ra usually reflect an IL‐1 signature, and the correct timing of IL‐1 blockade in COVID‐19 may be critical and likely complicates the interpretation of the present data (22). Thus, early intervention upon the development of acute hyperinflammatory respiratory failure in patients with COVID‐19 can have a therapeutic effect (25, 26, 27). Furthermore, albeit at a different level compared to that in patients with secondary HLH/MAS, the IL‐18–IFNγ axis is certainly active in patients with critical COVID‐19, and targeting this and IL‐1 simultaneously may constitute a rescue treatment for extremely ill patients (28). A corresponding randomized controlled trial is ongoing. Indeed, our data may further support the use of combined medications directed against different targets or the use of medications with broader immunoregulatory effects, such as glucocorticoids or dexamethasone (29), and may suggest strategies to bypass low sFasL expression or block IL‐8 signaling in the treatment of patients with COVID‐19 (ClinicalTrials.gov identifier: NCT04347226).
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Kessel had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Kessel, Foell, Tepasse.
Acquisition of data
Kessel, Vollenberg, Masjosthusmann, Hinze, Wittkowski, Schmidt, Tepasse.
Analysis and interpretation of data
Kessel, Vollenberg, Masjosthusmann, Hinze, Wittkowski, Debaugnies, Nagant, Corazza, Vély, Kaplanski, Girard‐Guyonvarc’h, Gabay, Schmidt, Foell, Tepasse.
Supporting information
Supplementary Material
Supported by the German Research Foundation (DFG) (grant FO 354/14‐1) and the European Union Horizon 2020 Programme (grant 779295 [ImmunAID]).
Drs. Kessel and Vollenberg contributed equally to this work. Drs. Foell and Tepasse contributed equally to this work.
Dr. Kessel has received consulting fees from Novartis (less than $10,000). Dr. Hinze has received consulting fees, speaking fees, and/or honoraria from Novartis (less than $10,000). Dr. Wittkowski has received consulting fees, speaking fees, and/or honoraria from Novartis (less than $10,000). Dr. Gabay has received consulting fees from AB2 Bio (less than $10,000), research grants from AB2 Bio, and owns stock or stock options in AB2 Bio. Dr. Foell has received consulting fees, speaking fees, and/or honoraria from Chugai‐Roche, Novartis, and Sobi (less than $10,000 each) and research support from Novartis, Pfizer, and Sobi. No other disclosures relevant to this article were reported.
References
- 1. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, et al. COVID‐19: consider cytokine storm syndromes and immunosuppression [letter]. Lancet 2020;395:1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crayne CB, Albeituni S, Nichols KE, Cron RQ. The immunology of macrophage activation syndrome [review]. Front Immunol 2019;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henderson LA, Canna SW, Schulert GS, Volpi S, Lee PY, Kernan KF, et al. On the alert for cytokine storm: immunopathology in COVID‐19. Arthritis Rheumatol 2020;72:1059–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID‐19? [editorial]. JAMA Intern Med 2020;180:1152–4. [DOI] [PubMed] [Google Scholar]
- 5. Force AD, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA 2012;307:2526–33. [DOI] [PubMed] [Google Scholar]
- 6. Carvelli J, Piperoglou C, Farnarier C, Vely F, Mazodier K, Audonnet S, et al. Functional and genetic testing in adults with HLH reveals an inflammatory profile rather than a cytotoxicity defect. Blood 2020;136:542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Debaugnies F, Mahadeb B, Nagant C, Meuleman N, De Bels D, Wolff F, et al. Biomarkers for early diagnosis of hemophagocytic lymphohistiocytosis in critically ill patients. J Clin Immunol 2021;41:658–65. [DOI] [PubMed] [Google Scholar]
- 8. Girard C, Rech J, Brown M, Allali D, Roux‐Lombard P, Spertini F, et al. Elevated serum levels of free interleukin‐18 in adult‐onset Still's disease. Rheumatology (Oxford) 2016;55:2237–47. [DOI] [PubMed] [Google Scholar]
- 9. Dinarello CA. Immunological and inflammatory functions of the interleukin‐1 family [review]. Annu Rev Immunol 2009;27:519–50. [DOI] [PubMed] [Google Scholar]
- 10. Bracaglia C, de Graaf K, Marafon DP, Guilhot F, Ferlin W, Prencipe G, et al. Elevated circulating levels of interferon‐γ and interferon‐γ‐induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann Rheum Dis 2017;76:166–72. [DOI] [PubMed] [Google Scholar]
- 11. Nigrovic PA. COVID‐19 cytokine storm: what is in a name? [editorial]. Ann Rheum Dis 2021;80:3–5. [DOI] [PubMed] [Google Scholar]
- 12. Del Valle DM, Kim‐Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, et al. An inflammatory cytokine signature predicts COVID‐19 severity and survival. Nat Med 2020;26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science 2020;369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abers MS, Delmonte OM, Ricotta EE, Fintzi J, Fink DL, de Jesus AA, et al. An immune‐based biomarker signature is associated with mortality in COVID‐19 patients. JCI Insight 2021;6:e144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Remy KE, Mazer M, Striker DA, Ellebedy AH, Walton AH, Unsinger J, et al. Severe immunosuppression and not a cytokine storm characterizes COVID‐19 infections. JCI Insight 2020;5:e140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sieg S, Smith D, Yildirim Z, Kaplan D. Fas ligand deficiency in HIV disease. Proc Natl Acad Sci U S A 1997;94:5860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin Y, Ji W, Yang H, Chen S, Zhang W, Duan G. Endothelial activation and dysfunction in COVID‐19: from basic mechanisms to potential therapeutic approaches [review]. Signal Transduct Target Ther 2020;5:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodman RB, Strieter RM, Martin DP, Steinberg KP, Milberg JA, Maunder RJ, et al. Inflammatory cytokines in patients with persistence of the acute respiratory distress syndrome. Am J Respir Crit Care Med 1996;154:602–11. [DOI] [PubMed] [Google Scholar]
- 19. Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, et al. Cytokine elevation in severe and critical COVID‐19: a rapid systematic review, meta‐analysis, and comparison with other inflammatory syndromes. Lancet Respir Med 2020;8:1233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Locatelli F, Jordan MB, Allen C, Cesaro S, Rizzari C, Rao A, et al. Emapalumab in children with primary hemophagocytic lymphohistiocytosis. N Engl J Med 2020;382:1811–22. [DOI] [PubMed] [Google Scholar]
- 21. De Benedetti F, Brogan P, Bracaglia C, Pardeo M, Marucci G, Sacco E, et al. Emapalumab (anti‐interferon‐gamma monoclonal antibody) in patients with macrophage activation syndrome (MAS) complicating systemic juvenile idiopathic arthritis (sJIA). Arthritis Rheumatol 2020;72 Suppl 4. URL: https://acrabstracts.org/abstract/emapalumab‐anti‐interferon‐gamma‐monoclonal‐antibody‐in‐patients‐with‐macrophage‐activation‐syndrome‐mas‐complicating‐systemic‐juvenile‐idiopathic‐arthritis‐sjia/. [Google Scholar]
- 22. The CORIMUNO‐19 Collaborative Group . Effect of anakinra versus usual care in adults in hospital with COVID‐19 and mild‐to‐moderate pneumonia (CORIMUNO‐ANA‐1): a randomised controlled trial. Lancet Respir Med 2021;9:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kooistra EJ, Waalders NJ, Grondman I, Janssen NA, de Nooijer AH, Netea MG, et al. Anakinra treatment in critically ill COVID‐19 patients: a prospective cohort study. Crit Care 2020;24:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arthur VL, Shuldiner E, Remmers EF, Hinks A, Grom AA, Foell D, et al. IL1RN variation influences both disease susceptibility and response to recombinant human interleukin‐1 receptor antagonist therapy in systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2018;70:1319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cauchois R, Koubi M, Delarbre D, Manet C, Carvelli J, Blasco VB, et al. Early IL‐1 receptor blockade in severe inflammatory respiratory failure complicating COVID‐19. Proc Natl Acad Sci U S A 2020;117:18951–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavalli G, De Luca G, Campochiaro C, Della‐Torre E, Ripa M, Canetti D, et al. Interleukin‐1 blockade with high‐dose anakinra in patients with COVID‐19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol 2020;2:e325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pontali E, Volpi S, Antonucci G, Castellaneta M, Buzzi D, Tricerri F, et al. Safety and efficacy of early high‐dose IV anakinra in severe COVID‐19 lung disease [letter]. J Allergy Clin Immunol 2020;146:213–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaplanski G, Bontemps D, Esnault P, Blasco V, Carvelli J, Delarbre D, et al. Combined anakinra and ruxolitinib treatment to rescue extremely ill COVID‐19 patients: a pilot study [letter]. Autoimmun Rev 2021;20:102726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, et al. Effect of dexamethasone on days alive and ventilator‐free in patients with moderate or severe acute respiratory distress syndrome and COVID‐19: the CoDEX Randomized Clinical Trial. JAMA 2020;324:1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


