Abstract
Background
First vaccines for prevention of Coronavirus disease 2019 (COVID‐19) are becoming available but there is a huge and unmet need for specific forms of treatment. In this study we aimed to evaluate the anti‐SARS‐CoV‐2 effect of siRNA both in vitro and in vivo.
Methods
To identify the most effective molecule out of a panel of 15 in silico designed siRNAs, an in vitro screening system based on vectors expressing SARS‐CoV‐2 genes fused with the firefly luciferase reporter gene and SARS‐CoV‐2‐infected cells was used. The most potent siRNA, siR‐7, was modified by Locked nucleic acids (LNAs) to obtain siR‐7‐EM with increased stability and was formulated with the peptide dendrimer KK‐46 for enhancing cellular uptake to allow topical application by inhalation of the final formulation – siR‐7‐EM/KK‐46. Using the Syrian Hamster model for SARS‐CoV‐2 infection the antiviral capacity of siR‐7‐EM/KK‐46 complex was evaluated.
Results
We identified the siRNA, siR‐7, targeting SARS‐CoV‐2 RNA‐dependent RNA polymerase (RdRp) as the most efficient siRNA inhibiting viral replication in vitro. Moreover, we showed that LNA‐modification and complexation with the designed peptide dendrimer enhanced the antiviral capacity of siR‐7 in vitro. We demonstrated significant reduction of virus titer and lung inflammation in animals exposed to inhalation of siR‐7‐EM/KK‐46 in vivo.
Conclusions
Thus, we developed a therapeutic strategy for COVID‐19 based on inhalation of a modified siRNA‐peptide dendrimer formulation. The developed medication is intended for inhalation treatment of COVID‐19 patients.
Keywords: COVID‐19, LNA, peptide dendrimers, SARS‐CoV‐2, siRNA
siR‐7, targeting SARS‐CoV‐2 RdRp inhibits viral replication in vitro. LNA‐modification and complexation with peptide dendrimer KK‐46 enhanced the antiviral capacity of siR‐7 in vitro. Inhalation exposure of SARS‐CoV‐2‐infected Syrian Hamsters with siR‐7‐EM/KK‐46 reduces the virus titer and lung inflammation.
Abbreviations: GFP, green fluorescent protein; LNA, locked nucleic acid; LUC, luciferase; RISC, RNA‐induced silencing complex; RdRp, RNA‐dependent RNA polymerase; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; siRNA, small interfering RNA; siR‐7‐EM, LNA‐modified small interfering RNA‐7

Abbreviations
- GFP
green fluorescent protein
- LNA
locked nucleic acid
- LUC
luciferase
- RISC
RNA‐induced silencing complex
- RdRp
RNA‐dependent RNA polymerase
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- siRNA
small interfering RNA
- siR‐7‐EM
LNA‐modified small interfering RNA ‐7
1. INTRODUCTION
In December 2019, an outbreak of severe acute respiratory infections was reported in the Chinese city of Wuhan. In the meantime, the disease named COVID‐19 (CoronaVirus Disease‐19) has flooded the globe and became the deadliest respiratory disease pandemic since 1918 when the Spanish influenza pandemic killed more than 50 million people. 1 , 2 The culprit of the current COVID‐19 pandemic outbreak is the novel coronavirus (CoV) which was named as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) by the International Committee on Taxonomy of Viruses (ICTV). SARS‐CoV‐2 such as SARS‐CoV and MERS‐CoV which all can cause severe respiratory illness belong to the closely related ß‐corona viruses 3 and have originated as human pathogens by animal‐to‐human‐host switching. 4 The two SARS viruses are derived from viruses enzootic in bats for which a rich reservoir exists in Asian countries and in particular in China. 5 As of March 3, 2021, there are >115 million confirmed cases and >2.5 million deaths related to COVID‐19 according to the Johns Hopkins Coronavirus Resource Center (https://coronavirus.jhu.edu/map.html). Currently, multiple vaccine candidates have entered into clinical trials, some of them have already been approved by health authorities and recently vaccination has been started in several countries. 6 , 7 , 8 , 9
However, in addition to prophylactic vaccines SARS‐CoV‐2‐specific therapies for treatment of infected patients are urgently needed. Accordingly, several antiviral drugs, among them remdesivir, hydroxychloroquine, lopinavir and interferons, have been evaluated for treatment of COVID‐19 but so far with limited clinical success. 10 , 11 Convalescent plasma treatment is another possibility for treatment of severe COVID‐19 12 and achieved FDA approval for treatment of critically ill patients but there are limitations to its production. 13 In addition, first monoclonal SARS‐CoV‐2‐specific antibodies have been developed, showed first promising results in experimental animal models 14 and reduced viral loads in patients. 15 Therefore, a huge unmet need for SARS‐CoV‐2‐specific treatment strategies remains.
The SARS‐CoV‐2 viral genome is a single‐stranded positive RNA (+ssRNA) of almost 30 kb, encoding at least five open reading frames (ORFs). The first ORF (ORF1a/b) occupies about 70% of the entire genome and encodes 16 nonstructural proteins (nsp1‐16). The remaining 30% of the genome encodes four major structural proteins necessary for virion assembly: spike (S), membrane (M), envelope (E), and nucleocapsid (N). 16 , 17 , 18
Gene expression in general and viral gene expression in infectious diseases can be suppressed through the mechanism of RNA interference (RNAi) for therapeutic purposes. 19 , 20 , 21 , 22 , 23 The RNAi approach is based on negative regulation of gene expression at the post‐transcriptional level and therefore highly specific. 24 , 25 Hence, the main advantage of the RNAi strategy over most other therapeutic approaches is its specificity but the technology has also limitations among them how to enhance specific targeting of and efficient transportation into infected cells.
In this study we screened a panel of SARS‐CoV‐2‐specific siRNAs for their potential to silence SARS‐CoV‐2 gene expression. The most potent silencing siRNA was then modified to enhance siRNA stability and formulated with a novel non‐toxic peptide dendrimer KK‐46 as vehicle for efficient siRNA delivery into the target cells. Finally, we show in an in vivo model of SARS‐CoV‐2 infection in Syrian hamsters 26 that topical treatment by inhalation of the modified siRNA‐peptide dendrimer formulation has the potential to reduce viral replication and to ameliorate SARS‐CoV‐2‐induced lung inflammation.
2. MATERIALS AND METHODS
2.1. Plasmid constructs and design of siRNA
The primary screening of siRNAs silencing activity was performed in vitro using vectors expressing viral genes fused with the firefly luciferase reporter gene. Two vectors expressing full‐size genes and gene fragments of SARS‐CoV‐2 (NCBI RefSeq NC_045512.2) RdRp RNA‐dependent RNA polymerase (2796 bp) and nucleocapsid protein N (1260 bp) were designed (pRdRp‐full and pVAX‐N‐IRES‐LUC, respectively) (Figure 1). Plasmid pVAX‐1 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) ensuring stable protein expression in eukaryotic cells, was used as expression vector. The expression plasmids containing virus‐specific inserts were synthesized as three bicistronic expression plasmids containing IRES fragment (Takara Bio USA, Mountain View, California, USA) (Figure 1a,b). The vectors allow mRNA transcription of target (RdRp or N) and reporter (Luc) genes under a single CMV promoter followed by translation of the target and reporter proteins. A single RNA‐transcript assures comparable expression of both genes.
FIGURE 1.
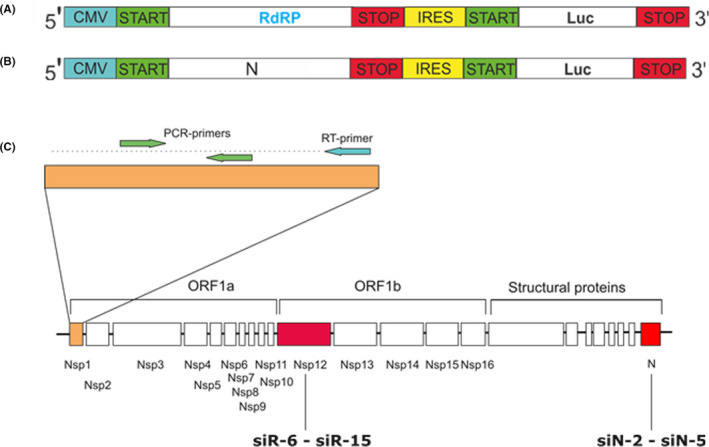
Plasmid constructs and design of siRNAs. Schematic representation of the bicistronic expression plasmid coding genes of firefly luciferase (Luc) and full size RdRp (pRdRp‐full) (A) or N (pVAX‐N‐IRES‐LUC) (B) genes of SARS‐CoV‐2. (C) Positions of the siRNA targeting SARS‐CoV‐2 RdRp (siR‐6‐siR‐15) and N (siN‐2‐siN‐5) genes. The RT‐PCR–amplified region at the most upstream region of ORF1 (NSP1‐leader protein) is marked
In total 13 siRNA duplexes targeting RdRp and N proteins of SARS‐CoV‐2 (Figure 1c) were designed in silico. Sequences of siRNAs are described in Table S1.siRNA against firefly luciferase (siLuc) 27 and GFP were used as controls.
To increase stability, we incorporated LNA (locked nucleic acid)‐modifications to the 3’ ends of the siR‐7 sense (S) and antisense sequences (aS) as follow (LNA; symbols of bases, +G and+T), 5’‐3’:
S +Ggaaggaaguucuguugaa+T+Tt
aS uucaacagaacuuccuucc+T+Tt
2.2. Co‐transfection of HEp‐2 cells with plasmid constructs and siRNAs
HEp‐2 cells (ATCC® Number: CCL‐23™) were cultivated with 10% fetal bovine serum (FBS) (HyClone, US) and 1% penicillin/streptomycin (PS; PanEco, Russia) in Dulbecco's modified Eagle's medium (DMEM; PanEco, Russia). The cells were seeded into 24‐well plates at a concentration of 105 cells/well in 0.5 mL of media and incubated at 37°C with 5% CO2 for 12–24 h. The cells were consistently co‐transfected with each of the plasmid constructs (0.25 μg/well) pRdRp‐full, and pVAX‐N‐IRES‐LUC/Lipofectamine 3000 (0.5 μL/well) complexes and then in 30 min with each of the 13 different siRNAs (0.75 μg/well) with Lipofectamine 3000 (1.5 μL/well). In both cases complexes with Lipofectamine 3000 were made in serum‐free Opti‐MEM media (Gibco) according to the manufacturer's protocol. After four hours, media with complexes were replaced with fresh DMEM. Then, after 24 h cells were harvested in Cell Culture Lysis Reagent (CCLR) and assayed for firefly luciferase activity with the Luciferase Assay System (Promega, Madison, Wisconsin) according to the manufacturer's instructions. Cells transfected with siLuc and siGFP were used as a positive and negative control, respectively.
2.3. Cytotoxicity assay
Vero cells were seeded into 96‐well cell culture plates (NUNC, Denmark) at 2 × 105 cells/mL with 5% FBS, allowed to attach and grow for 24 h, and treated with 2‐fold dilutions of KK‐46 (29–2500 μg/mL) in FBS‐free culture media for 24 h. The cytotoxicity of KK‐46 was assessed using “CellTiter96® Non‐Radioactive Cell Proliferation Assay” kit (Promega, USA) in accordance with the manufacturer's instructions. The absorbance was recorded at 570 nm wavelength (with reference wavelength 700 nm).
2.4. Spectrophotometry and luminescence measurements
All measurements were performed with the Varioskan®Flash 2.4 spectral scanning multimode reader (Thermo Fisher Scientific, USA).
2.5. Synthesis, purification and characterization of the peptide dendrimer KK‐46
The design of the cationic dendrimeric peptide KK‐46 for intracellular delivery of siRNA was based on in silico calculation of the molecular properties (Chemsketch software, Dock Prep software, PyMOL software) to achieve an optimal range of positive charge and amphiphilicity for cell penetration and RNA‐binding. The positive charge was attributable to the use of arginine and histidine residues for the N‐terminal ends of the peptide branches. Details about KK‐46 synthesis, purification, transfection efficacy and cytotoxicity are summarized in the Supplement (Figure S1‐S3). The peptide was tested for its ability to inhibit the binding of RBD to ACE2 using a molecular interaction assay. 28
2.6. Transfection of HEp‐2 cells with pGL‐3/KK‐46 complexes
The HEp‐2 cells were propagated as described above. Cells were transfected with the pGL3 Luciferase Reporter Vector (Promega) (0.5 μg/well) in complexes with peptide dendrimer KK‐46. In these experiments several weight ratios of KK‐46 and vector (5:1,12.5:1, 20:1, 25:1, 50:1, 100:1 µg KK‐46: µg pGL3) were used to transfect the cells. Commercial Lipofectamine 3000 was used as positive control. The transfection protocol and assays were performed as described above.
2.7. Viral Stocks
The SARS‐CoV‐2 strain PMVL‐1 (GISAID Number EPI_ISL_421275) was propagated and titrated in Vero E6 cells (ATCC® Number: CRL‐1586 TM) to determine TCID50/mL (50% tissue culture infectivity dose). This virus was used for experiments in vitro. The SARS‐CoV‐2 strain B was grown in Vero C1008 cells (ATCC® CRL‐1586™) and used for in vivo experiments. SARS‐CoV‐2 strain B was titrated on Vero C1008 cells by plaque assay to determine the number of plaque forming units (PFU) as described. 29 Viral stocks were used at 1.0 x 105 TCID50/mL and 1.0x106 PFU/mL for strain PMVL‐1 and strain B of SARS‐CoV‐2, respectively. Experiments with SARS‐CoV‐2 have been conducted in labs certified as biosafety level 3.
2.8. Transfection of antiviral siRNA/siLNA with Lipofectamine 3000 or peptide dendrimer KK‐46
Vero E6 cells (ATCC® Number: CRL‐1586 TM) were cultivated with 5% of FBS (HyClone, Logan, UT, USA) and 1% PS in DMEM (PanEco, Moscow, Russia). The cells were seeded into 24‐well plates at a concentration of 105 cells/well in 0.5 mL of medium and incubated at 37°C, 5% CO2 for 12–24 h.
The medium was replaced with FBS‐free Opti‐MEM (Gibco, USA). Aliquots of 100 μL containing 0.5 μg of specific or non‐specific siRNAs, 1.5 μL of Lipofectamine3000 (Invitrogen) in Opti‐MEM were added to VeroE6 cells. After four hours, complexes were removed, and cells were infected with SARS‐CoV‐2 at a multiplicity of infection (MOI) of 0.0001 (in 0.5 mL of media) and incubated for 1 h at 37°C, 5% CO2. The dose was chosen in titration experiments. Infection with low MOI 0.0001 allowed to achieve virus‐specific cytotoxicity and at the same time this was detectable by qRT‐PCR measurement of viral load. Cells were washed and non‐adherent virus was removed and re‐suspended in fresh medium. Cell culture supernatants and RNA lysates were harvested at times indicated and stored at −80°C.
The same procedure was used for transfection with siRNA/KK‐46 complexes. The only exception was the amount of the components. Three increasing concentrations of complexes 21, 42 and 84 μg/per well which included the sums of KK‐46+siRNA/siLNA, μg: 20+1, 40+2 and 80+4, respectively (20:1 weight ratio) were used. Unspecific siRNA targeting the gene of firefly luciferase (siLuc) and SARS‐CoV‐2 infected cells were used as negative controls.
2.9. RNA extraction and quantitative RT‐PCR
Total RNA was extracted from cell culture supernatants and cell lysates using the Extract RNA Reagent (Eurogen, Moscow, Russia) following the manufacturer's instructions. Amplification and quantification of SARS‐CoV‐2 RNA were carried out by using a one‐step RT‐qPCR technique. To perform one‐step RT‐qPCR the reaction mixture containing (for one reaction) 5 pmol of each primer, 3 pmol of probe, 0.025 mM of each dNTP (Eurogen, Moscow, Russia), 5 μl of 10× in‐house one‐step RT‐PCR buffer (100 mM Tris‐HCl (pH 8.3 at 25°C), 150 mM KCl, 10 mM MgCl2, 8 mM DTT), 0.25 μl of in‐house M‐MLV reverse (100units), 0.25 μl of in‐house Taq polymerase (5 units) and 10 μl of RNA (0.5 μg) was used. The total volume of one reaction mixture was 25 μl. The primers and probes were designed to target the gene coding NSP1 (Leader Protein) of SARS‐CoV‐2, the oligonucleotides were as follows: forward primer – 5’‐ GTA CGT GGC TTT GGA GAC TC −3’, reverse primer – 5’‐ ACT AAG CCA CAA GTG CCA TC −3’, probe 5’‐Cy5‐ AGG AGG TCT TAT CAG AGG CAC GTC A ‐BHQ2‐3’. Amplification was performed using a Real‐time CFX96 Touch instrument (Bio‐Rad, USA). The conditions of the one‐step RT‐qPCR reaction were as follows: 50°C for 15 min, 95°C for 5 min, followed by 45 cycles of 95°C for 10 s and 55°C for 1 min. The number of copies of viral RNA was calculated using a standard curve generated by amplification of a plasmid DNA template containing the amplified fragment. More details on qPCR‐RT and standard curve can be found in Supplementary materials (Figure S4).
2.10. Serum stability
Duplexes of siRNA (siR‐7) and siLNA (siR‐7‐EM) (3 μg) were incubated at 37°C in 50% FBS (Invitrogen) diluted in phosphate buffered saline for 264 h. Aliquots of 10 μg were withdrawn at indicated time points and stored at −20°C. Samples were then subjected to electrophoresis in a 1.5% agarose gel, stained with Ethidium Bromide and bands corresponding to full‐length siRNAs were quantified by gel densitometry using Gel Doc XR+software (BioRad, USA) and compared to the sample collected immediately after dissolving the siRNA in the FBS solution.
2.11. Experiments in vivo
Experiments with animals were carried out in accordance with the EU Directive 2010/63/EU for animal experiments. Ethics approval 09–2020 from 17.09.2020 was obtained from the local research ethics committee of the NRC Institute of Immunology FMBA. The Syrian hamsters (females, 4–5 weeks of age, 40–60 g weight) were used to assess the antiviral activity of siR‐7‐EM(siLNA)/KK‐46 complexes in vivo. A schematic representation of treatment protocols is shown in Figure S5.
In the first set of in vivo experiments we aimed to find the optimal concentration of the siR‐7‐EM/KK‐46 complexes. Six groups of animals (N = 10) were formed. Five out of the six groups were intranasally infected with 105 PFU/animal of SARS‐CoV‐2 strain B on day 0. On the same day (one hour after infection) and day 1 infected animals were exposed to a siR‐7‐EM/KK‐46 aerosol at three increasing doses: 0.7, 1.96 or 5.6 mg/kg. The hamsters were anesthetized and placed in an inhalation exposure chamber. Aerosols were formed using a conventional Xiaomi Andon VP‐M3A Micro Mesh Nebulizer. A virus‐only control group included animals infected with SARS‐CoV‐2. Group “Intact” did not receive any treatment and served as a negative control. Five animals from each group were sacrificed at day 2 after infection, lungs were removed. Macroscopic evaluation and scoring of the histopathology lesions of the lung were performed. Lung tissues were freshly fixed in 10% buffered formaldehyde, embedded in paraffin wax, and stained with haematoxylin‐eosin (H&E). The histopathological changes were graded according to a modified semiquantitative scoring system by a blinded investigator (none, 0; mild, 1.0; moderate, 2.0; or severe, 3.0). The right lobe of the lung was homogenated and the viral titer was assessed by plaque assay to determine the number of PFU as described. 29 Five animals left in each group were exposed to the siR‐7‐EM/KK‐46 aerosol at days 3, 4, 5 and were sacrificed at day 6 after infection. Lungs were removed and processed as described above.
The second series of in vivo experiments were performed according to the same scheme with two exceptions. First, we exposed anesthetized animals to siR‐7‐EM/KK‐46 aerosol twice a day with two hours interval to yield a daily dose of the antiviral complexes of 0.35, 0.7 and 2.0 mg/kg, respectively. Second, the control group of animals received orally Favipiravir (within 1 hour after infection, a dose of 1.2 mg/animal was administered twice a day, and then daily for 6 days post infection 0.4 mg/animal were given twice a day). According to the manufacturer's instruction Favipiravir was administered perorally. The pills were resuspended in a 1% starch solution (w/v, dissolved in distilled boiled water) and given to Hamsters without prior anesthesia. The efficacy of Favipiravir was assessed as described for siR‐7‐EM/KK‐46.
2.12. Statistical analysis
The Shapiro–Wilk test for normality was performed. Data were presented as means +s.e.m for parametric analyses and as medians +s.d. for nonparametric analyses, as indicated in the figure legends. Statistical analyses were performed using Prism version 8 (GraphPad Software). For normally distributed data we performed ordinary one‐way ANOVA or one‐way repeated measures ANOVA with Tukey's or Dunnetts's post hoc tests, respectively, for multiple comparisons to determine differences between groups, or if not normally distributed the Kruskal–Wallis test followed by post hoc testing (if the Kruskal–Wallis was significant) using un‐paired Mann‐Whitney U tests was performed, as mentioned in the respective figure legends. Data were accepted as significantly different when p < 0.05.
3. RESULTS
3.1. Screening of siRNAs for silencing of SARS‐CoV‐2 genes by bioluminescence assay
To determine the silencing activity of the designed anti‐SARS‐CoV‐2 siRNAs in vitro, we constructed plasmids pRdRp‐full and pVAX‐N‐IRES‐LUC simultaneously expressing SARS‐CoV‐2 genes RdRp (RNA dependent RNA polymerase) and N (nucleocapsid), respectively, and the reporter gene of firefly luciferase. Hep‐2 cells were co‐transfected with vectors pRdRp‐full, and pVAX‐N‐IRES‐LUC and 13 different siRNA variants to evaluate their knockdown activity. siRNA against luciferase (siLuc) and GFP (siGFP) were used as a positive and negative control, respectively. As indicated in Table 1 and Figure 2 the siRNAs siN‐4, siR‐7 and siR‐11 which significantly decreased the level of luminescence for more than 80% were chosen as the most potent SARS‐CoV‐2 silencing siRNAs and were further studied. Of note, we found no significant difference in luciferase expression between cells transfected with plasmid only or after co‐transfection with plasmid and non‐specific siGFP (Table 1).
TABLE 1.
The suppression of luciferase activity by siRNA in Hep‐2 cells consecutively transfected with plasmid coding SARS‐CoV‐2 genes fused with firefly luciferase gene and specific or nonspecific siRNAs
| № | siRNA name | The suppression of luciferase activity (%, N=4, Mean ±SD) in comparison with: | ||
|---|---|---|---|---|
| plasmid | nonspecific siGFP | |||
| 1 | siLuc | pRdRp‐full | 82.9 ± 6.2 | 79.3 ± 10.2 |
| 1a | siLuc | pVAX‐N‐IRES‐LUC | 75.2 ± 10.9 | 72.4 ± 9.1 |
| 2 | siN−2 | pVAX‐N‐IRES‐LUC | 42.3 ± 27.4 | 36.4 ± 20.7 |
| 3 | siN−3 | pVAX‐N‐IRES‐LUC | 64.6 ± 17.3 | 60.8 ± 14.7 |
| 4 | siN−4 | pVAX‐N‐IRES‐LUC | 81.2 ± 9.2 | 78.8 ± 10.05 |
| 5 | siN−5 | pVAX‐N‐IRES‐LUC | 64.7 ± 13.4 | 60.4 ± 6.1 |
| 6 | siR−7 | pRdRp‐full | 91.3 ± 3.2 | 89.2 ± 4.9 |
| 7 | siR−8 | pRdRp‐full | 47.2 ± 28.3 | 56.6 ± 22.01 |
| 8 | siR−9 | pRdRp‐full | 67.6 ± 7.9 | 61.7 ± 14.6 |
| 9 | siR−10 | pRdRp‐full | 70.6 ± 19.3 | 64.6 ± 22.9 |
| 10 | siR−11 | pRdRp‐full | 84.5 ± 8.6 | 81.3 ± 10.6 |
| 11 | siR−12 | pRdRp‐full | 56.8 ± 23.5 | 48.01 ± 28.9 |
| 12 | siR−13 | pRdRp‐full | 77.9 ± 8.8 | 73.4 ± 12.2 |
| 13 | siR−14 | pRdRp‐full | 47.3 ± 22.4 | 36.3 ± 31.06 |
| 14 | siR−15 | pRdRp‐full | 71.1 ± 11.3 | 65.03 ± 17.4 |
FIGURE 2.
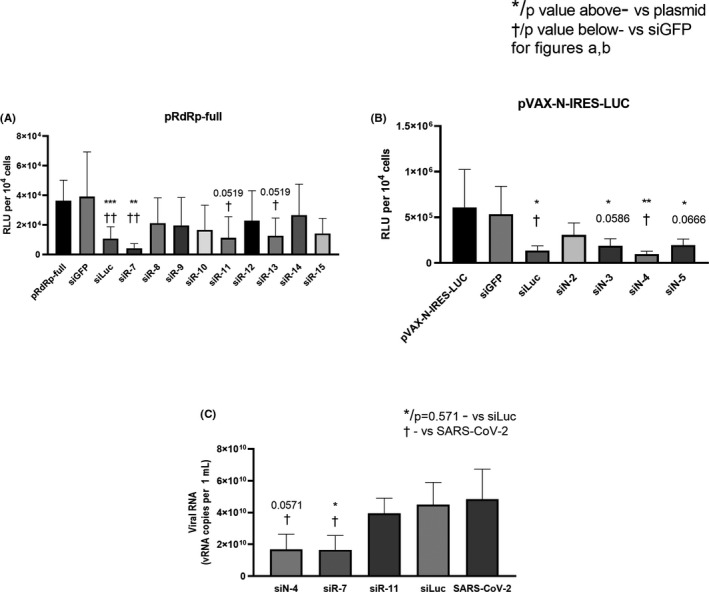
Properties of designed siRNA. (A, B) Inhibition of gene expression with synthetic siRNA. Hep‐2 cells were transfected with each of the plasmids coding SARS‐CoV‐2 genes fused with firefly luciferase gene (pRdRp‐full) (A) or pVAX‐N‐IRES‐LUC (B) followed by transfection with SARS‐CoV‐2‐specific or control siRNA. siLuc and siGFP were used as positive and negative controls, respectively. LipofectamineTM 3000 was used as vehicle for both pDNA and siRNA. After 24 h cells were harvested and luciferase activity was determined. Data are expressed as relative light units (RLU) per 104 cells. Footnotes: */† or adjusted p value above/below represents the difference compared to cells transfected with plasmid only/non‐specific siGFP, respectively. *†p < 0.05, **††p < 0.01, ***p < 0.001. (D) Inhibition of SARS‐CoV‐2 reproduction with synthetic siRNA. Vero E6 cells were transfected with siRNA/ LipofectamineTM 3000 complexes. Media with complexes were removed four hours after transfection and cells were infected with SARS‐CoV‐2 at MOI 0.0001. Viral load was determined by qRT‐PCR. The results are expressed as number of viral RNA copies per mL. Footnotes: * or p = 0.571 represent differences with cells infected with SARS‐CoV‐2 and treated with non‐specific siLuc. † represent differences with SARS‐CoV‐2 only infected cells.*†p < 0.05. Differences between multiple groups were estimated using a Kruskal–Wallis test followed by post‐hoc testing (if the Kruskal–Wallis was significant) using un‐paired Mann–Whitney U tests. Bars show means of four (A, B) and five (C) independent experiments ± SDs
3.2. Antiviral effect of siRNA in vitro
Next, we investigated the antiviral effects of the three selected siRNAs (siN‐4, siR‐7 and siR‐11) in vitro. To evaluate their antiviral effect, we transfected Vero E6 cells with complexes of siRNA/Lipofectamine 3000 and four hours after transfection cells were infected with SARS‐CoV‐2 at MOI 0.0001. After 48 hours of infection cells were lysed and viral load was determined by qRT‐PCR. We found a significant difference in vRNA concentration between cells treated with specific siR‐7, compared to both unspecific siLuc‐transfected and infected cells (p < 0.05) (Figure 2c). In addition, a significant reduction of vRNA in cells treated with siN‐4 (p < 0.05) compared to SARS‐CoV‐2 infected cells and a trend towards significance (p = 0.0571) to reduce the amount of vRNA compared to unspecific siLuc‐transfected cells were observed. According to these results the siR‐7 targeting SARS‐CoV‐2 RdRp gene was chosen as best candidate for further analysis.
3.3. Modified siR‐7‐EM shows increased nuclease resistance
Introduction of LNA into classic antisense oligos has been shown to increase their stability. 30 Therefore, we designed LNA‐modified siR‐7‐EM to test if these modifications could improve the properties of siR‐7 for in vivo application. Stability of unmodified siR‐7 and LNA‐modified siR‐7‐EM was evaluated in vitro by incubation in 50% mouse serum for 264 h at 37°C. As shown in Figure 3, unmodified siR‐7 was degraded after 48 h of incubation (up to 50%) and only 10% were left after 264 h of incubation. In contrast, LNA‐modified siR‐7‐EM remained largely undegraded even after 264 h of incubation (around 70% of initial amount remained intact). Moreover, we found that the half‐life (t1/2) of siR‐7 and siR‐7‐EM were 72.4 and 256.8 h, respectively (Figure 3a). These experiments showed that incorporation of LNA molecules in siR‐7 contributes to increased half‐life and thus may have a positive influence on overall biostability.
FIGURE 3.
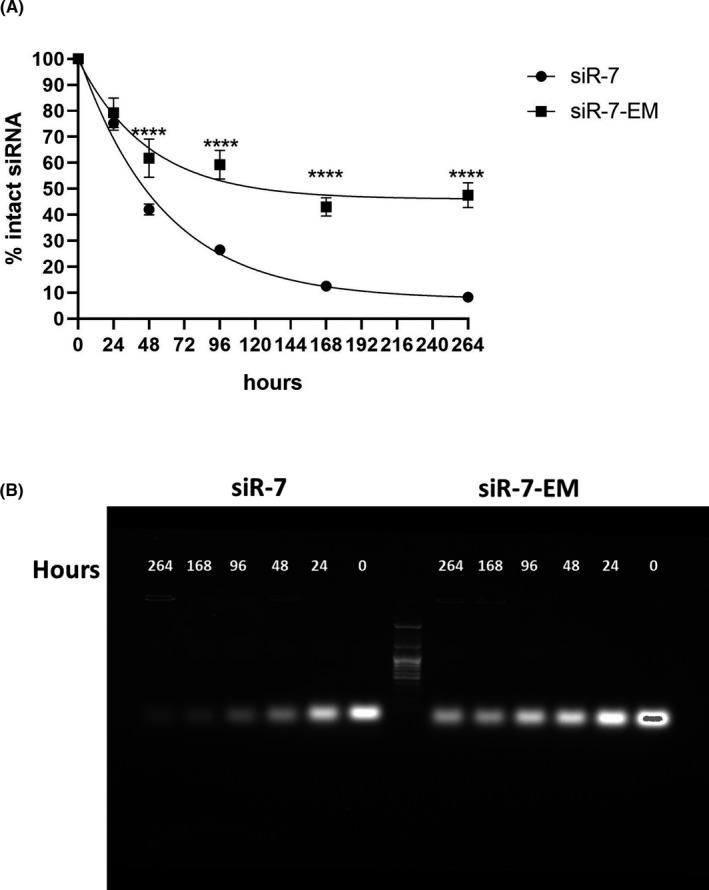
Modified siRNA have increased resistance to nuclease degradation. A, The stability of unmodified siR‐7 (circles) and modified siR‐7‐EM (squares) in 50% mouse serum are compared over a period of 264 h at 37°C (N = 4). siRNA quantities at various time points were calculated by dividing the total counts of full‐length siRNA by the input starting material. B, For this purpose, aliquots of each sample (10 μg siRNA per lane) were analyzed by 1.5% agarose gel electrophoresis. Differences between multiple groups were analyzed using repeated measures one‐way ANOVA with Dunnetts's post hoc test. Bars show medians of four independent experiments+SEM. **** p < 0.0001
3.4. siR‐7‐EM/peptide dendrimer KK‐46 complexes exhibit strong anti‐SARS‐CoV‐2 activity in vitro
Modifications of siRNA may influence different stages of RNA‐induced silencing complex (RISC) actions. 31 , 32 To examine whether the LNA‐modification may influence the knockdown activity and to evaluate KK‐46 transfection activity with siRNA, Vero E6 cells were transfected with unmodified siR‐7 or LNA‐ modified siR‐7‐EM/ KK‐46 complexes and infected with SARS‐CoV‐2. Three increasing concentrations of complex 21, 42 and 84 μg/per well which included sum of siRNA/siLNA+KK‐46, μg: 1+20, 2+40 and 4+80 were used, respectively. As shown in Figure 4, up to a two‐log reduction in vRNA levels in cell lysates (Figure 4a; p < 0.01) and supernatants (Figure 4b; p < 0.01, p < 0.05), were achieved after 48 h using either 42 or 84 μg of both siR‐7 and siR‐7‐EM /KK‐46 complexes. Complexes containing unspecified siLuc (targeting gene of firefly luciferase) with SARS‐CoV‐2 infected cells were used as negative control showing no significant reduction of vRNA levels as compared to SARS‐CoV‐2‐infected cells (Figure 4). Significant reduction of vRNA in both cell lysate and supernatants compared to unspecific siLuc and infectious control was observed at a dose of 42 μg (p < 0.01, p < 0.05). The target‐specificity of the siR‐7 or siR‐7‐EM /KK‐46 complexes was reflected by the use of an untargeted siLuc control, which showed no significant off‐target effects on the vRNA levels neither in cell lysates nor in supernatants. We found no significant difference in antiviral effects of unmodified siR‐7 or LNA‐ modified siR‐7‐EM but complexes with siR‐7‐EM seemed to be more effective than siR‐7 complexes which was also reflected by the fact that only the 21 µg dose of siR‐7‐EM gave significant results compared to siLuc, p < 0.05. Probably this is a result of the enhanced stability of siLNA as the viral load in cells lysate and supernatants was determined by qRT‐PCR 48 h after infection when there was already a difference in stability between unmodified and modified siRNA (Figure 3).
FIGURE 4.
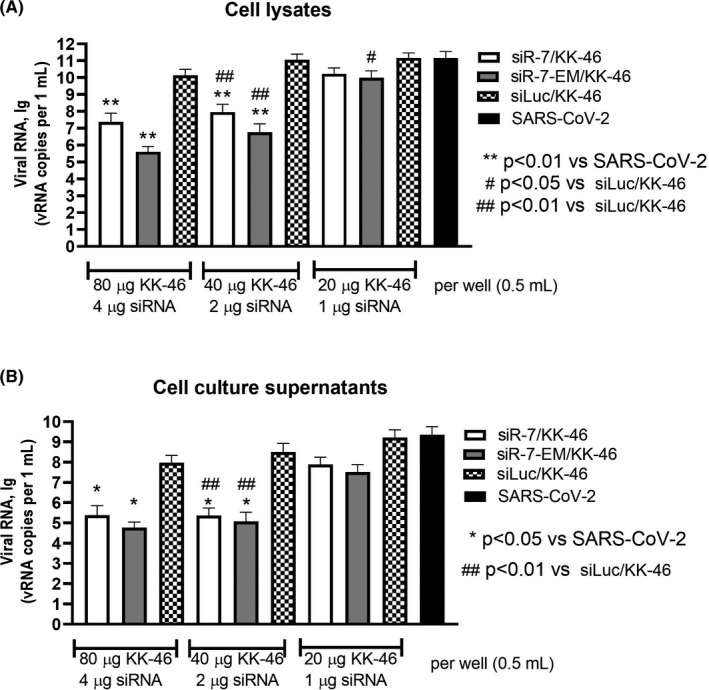
Inhibition of SARS‐CoV‐2 reproduction with unmodified or LNA‐modified siR‐7/peptide dendrimer KK‐46 complexes in vitro. Vero E6 cells were transfected with unmodified siR‐7 or LNA‐modified siR‐7‐EM‐peptide dendrimer KK‐46 complexes at three concentrations (x‐axes). Media with complexes were removed four hours after transfection and cells were infected with SARS‐CoV‐2 at MOI 0.0001. After 48 hours supernatants and cells were harvested. Viral load was determined by qRT‐PCR in cells lysates (A) and supernatants (B). The results are expressed as viral RNA copies per mL. Differences between multiple groups were estimated using a Kruskal–Wallis test followed by post‐hoc testing (if the Kruskal–Wallis was significant) using un‐paired Mann‐Whitney U tests. Bars show medians of five independent experiments +SDs. Footnotes: # represent differences with cells infected with SARS‐CoV‐2 and treated with non‐specific siLuc.*represent differences with SARS‐CoV‐2 only infected cells. *#p < 0.05, **##p < 0.01
3.5. siR‐7‐EM / KK‐46 complexes show strong anti‐SARS‐CoV‐2 effects in vivo
In order to investigate the therapeutic potential of the siR‐7‐EM/KK‐46 complex in vivo, female Syrian hamsters were infected with SARS‐CoV‐2 at dose of 105 PFU/animal and then given a single inhalation exposure of 0.7, 1.96 and 5.6 mg/kg of siR‐7‐EM/KK‐46 per day for 6 days. This schedule was thought as an early topical post‐exposure treatment. Half of the animals were sacrificed two and six days post infection, respectively. We found that progeny virus production was significantly and dose‐dependently decreased. Up to a 1.5–3 times reduction, in the lungs of the siR‐7‐EM/KK‐46‐treated animals on day two (5.0 ± 0.5, 4.7 ± 0.2, 4.5 ± 0.3 in log10 PFU/mL for 0.7, 1.96 and 5.6 mg/kg of siR‐7‐EM/KK‐46, respectively), compared to infected and untreated animals was found (6.1 ± 0.2), p < 0.01 (Figure 5a). Similar results were obtained for the other half of animals which were analyzed on day six. Again, treatment with siR‐7‐EM/KK‐46 significantly and dose‐dependently decreased viral loads in the lungs (2.2 ± 0.2, 2.0 ± 0.2, 1.1 ± 0.1 in log10 PFU/mL for 0.7, 1.96 and 5.6 mg/kg of siR‐7‐EM/KK‐46, respectively) as compared with the infected and untreated animals (3.5 ± 0.2), (p < 0.01).
FIGURE 5.
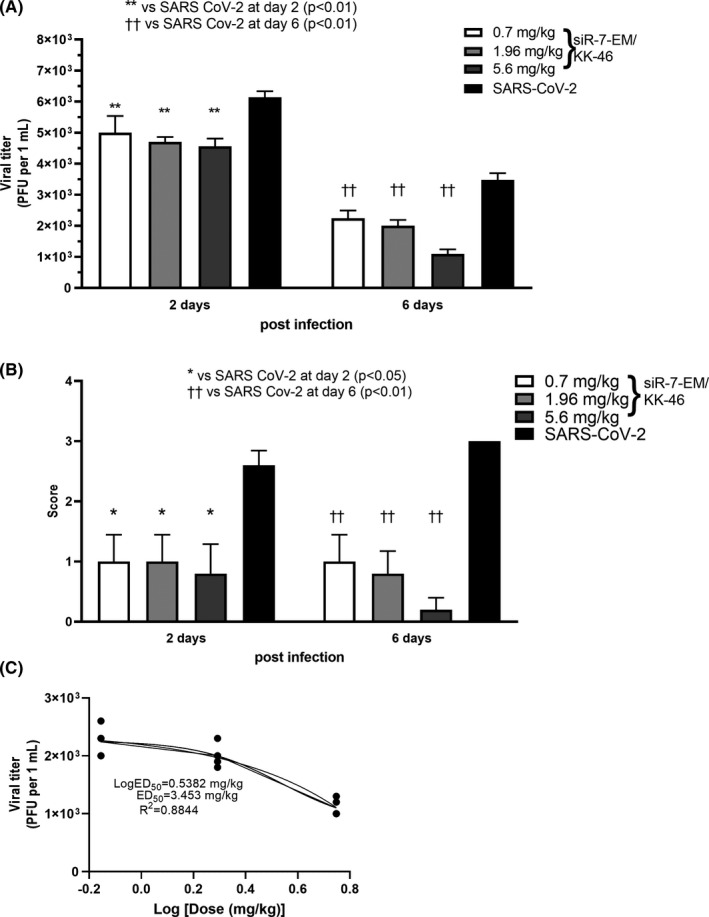
Dose‐dependent inhibition of SARS‐CoV‐2 reproduction with modified siR‐7‐EM‐peptide dendrimer KK‐46 complexes in Syrian hamsters after repeated inhalation exposure. Syrian hamsters were infected with SARS‐CoV‐2 with a dose of 105 PFU/animal and treated with three doses of the modified siR‐7‐EM‐peptide dendrimer KK‐46 complexes (35, 98 and 289 μg siR‐7‐EM/animal, 0.7, 1.96 and 5.6 mg KK‐46/kg, respectively). Inhalations with complexes were repeated daily for six days. Two and six days after infection animals were sacrificed and determination of viral titer (A) and macroscopic evaluation plus scoring of histopathology (B) in the lung were performed. The results are expressed as plaque forming units (PFU) per mL (A) or scores (B) obtained for histological analysis of pathologic alterations in hamsters’ lung. (C) Dose‐response curve of SARS‐CoV‐2 levels in the lungs at day six after infection, where the ED50 estimation is ∼ 3.453 mg/kg. Differences between multiple groups were estimated using a Kruskal–Wallis test followed by post‐hoc testing (if the Kruskal–Wallis was significant) using un‐paired Mann‐Whitney U tests. Bars show medians of one experiment (five animals per group) + SDs. Data are provided for one experiment representative of two independent experiments with five hamsters /group. Footnotes: */† represent differences with hamsters infected with SARS‐CoV‐2 and assessed at day two/six after infection, respectively. **††p < 0.01, *p < 0.05
Histological analysis (Figure 5b, S6a) revealed significantly (p < 0.05 and p < 0.01, at day two and six, respectively) decreased lung inflammation as assessed by macroscopic indicators of disease in animals exposed to 0.7, 1.96 and 5.6 mg/kg of siR‐7‐EM/KK‐46 as compared to SARS‐CoV‐2 infected, untreated animals group (Figure 5b). Taken together these results suggested that inhalation of siR‐7‐EM/KK‐46 at a dose of 3.5 mg/kg significantly and dose‐dependently attenuates pathological effects in vivo by hindering viral replication.
The estimated effective dose 50 (ED50) of siR‐7‐EM/KK‐46 assessed at day 6 post infection was ∼3.453 mg/kg/day (R2 = 0.88; Figure 5c).
To further investigate alternative dosing schemes of siR‐7‐EM/KK‐46, a multiple dose study with repeated low doses of siR‐7‐EM/KK‐46 was conducted. In this second set of in vivo experiments Syrian hamsters were infected with SARS‐CoV‐2 and treated with 0.175, 0.35, and 1.0 mg/kg (yielding daily doses of 0.35, 0.7, and 2.0 mg/kg, respectively) of siR‐7‐EM/KK‐46 inhalation exposure twice a day with two hours interval for 6 days. The SARS‐CoV‐2 viral titers were found to be significantly reduced by the siR‐7‐EM/KK‐46 compared with the untreated virus‐infected animals (p < 0.01 for all dose levels) at days two and six (Figure 6a). In particular, viral titers were reduced a 1.3 (26) and 2.5 (54) times (%) on day two and six, respectively, in the lungs of the siR‐7‐EM/KK‐46‐treated animals on day two (5.1 ± 0.1, 4.9 ± 0.2, 4.9 ± 0.1 in log10 PFU/mL for 0.35, 0.7 and 2.0 mg/kg, of siR‐7‐EM/ KK‐46, respectively), compared with results for the untreated, virus‐infected animals (6.7 ± 0.03), p < 0.01. Also, on day six viral titers in the siR‐7‐EM/KK‐46‐treated animals were significantly reduced (1.7 ± 0.1, 1.6± 0.2, 1.3 ± 0.1 in log10 PFU/mL for 0.35, 0.7 and 2.0 mg/kg of siR‐7‐EM/KK‐46, respectively) as compared with the untreated, virus‐infected animals (3.3 ± 0.1), (p < 0.01). Oral administration of Favipiravir reduced the viral titer approximately 1.2‐times on day two (5.4 ± 0.1) compared to SARS‐CoV‐2–infected and untreated animals (p < 0.01).
FIGURE 6.
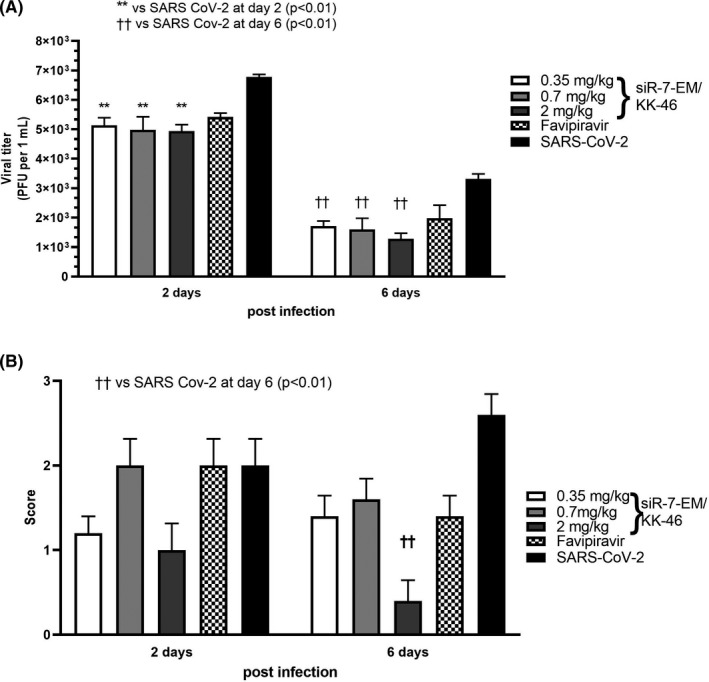
Effect of repeated low dose modified siR‐7‐EM‐peptide dendrimer KK‐46 complexes in Syrian hamsters. Syrian hamsters were infected with SARS‐CoV‐2 at a dose of 105 PFU/animal and exposed to different doses (0.175, 0.35 and 1.0 mg/kg of siR‐7‐EM/KK‐46 aerosol) twice a day with two hours interval. Two and six days after infection animals were sacrificed and viral titers (A) and macroscopic evaluation and scoring of the histopathology lesions (B) in the lung were analyzed. Orally administrated Favipiravir (1 h after infection, a dose of 1.2 mg/animal was administered twice a day, and then daily during 6 days after infection 0.4 mg/animal twice a day) served as positive control. The results are expressed as plaque forming units (PFU) per mL (A) or scores (B) obtained for histological analysis of pathologic lung alterations. Differences between multiple groups were estimated using a Kruskal–Wallis test followed by post‐hoc testing (if the Kruskal–Wallis was significant) using un‐paired Mann–Whitney U tests. Bars show medians of one experiment (five animals per group) + SDs. Data are from one experiment representative of two independent experiments with five hamsters/group are shown. Footnotes: */† represents difference with hamsters infected with SARS‐CoV‐2 and assessed at day two/six after infection, respectively. **††p < 0.01
Again, we performed scoring of lung pathology and found significantly (p < 0.01) decreased lung inflammation in animals exposed to 2 mg/kg of siR‐7‐EM/KK‐46 aerosol compared with SARS‐CoV‐2 infected group at day six but not in the animals treated with lower doses of siR‐7‐EM/KK‐46 or with Favipiravir as compared to the infected but untreated animals (Figure S6b). However, this significant reduction was only found after 6 days suggesting that the dose should be 2 mg/kg or higher.
4. DISCUSSION
The novel human coronavirus disease, COVID‐19, caused by SARS‐CoV‐2 has become the first coronavirus pandemic in history. 33 Numerous clinical trials and ongoing research aimed to identify therapeutic strategies based on existing and repurposed as well as novel antiviral drugs for COVID‐19. 34 Furthermore, active vaccination and passive immunization with convalescent plasma and recombinant SARS‐CoV‐2‐specific antibodies has reached clinical application. However, a huge unmet need for virus‐specific treatment remains. Here we present data showing that topical application by inhalation of SARS‐CoV‐2‐specific modified siRNA with enhanced stability formulated with a peptide dendrimer facilitating siRNA transfer into infected cells has the potential to treat SARS‐CoV‐2‐induced lung inflammation using the Syrian hamster model. In fact, small interfering RNA–mediated gene silencing technology (siRNA) holds promise for drug development. Recently two RNA interference (RNAi)‐based drugs GIVLAARI® for acute hepatic porphyria and ONPATTRO™ for the treatment of the polyneuropathy of hereditary transthyretin‐mediated amyloidosis 35 were approved in both the European Union and the United States. Several anti‐HBV, HCV, HIV, Zaire ebolavirus (ZEBOV), and RSV (respiratory syncytial virus) siRNA‐based formulations have been evaluated in clinical trials. 36 Furthermore, it has been shown that siRNAs could inhibit the replication of SARS‐CoV both in vitro 37 , 38 , 39 , 40 and in vivo 41 , 42 and in silico exploration of potential siRNA targets in the SARS‐CoV‐2 genome has been reported. 43
Research in COVID‐19 is rapidly expanding. First approaches for siRNA treatment have been described 44 but to the best of our knowledge our study as summarized in Figure S7 is the first which aimed not only to evaluate the antiviral efficiency of siRNAs targeting the SARS‐CoV‐2 genome but also sought to overcome possible bottlenecks of a therapeutic SARS‐CoV‐2 siRNA approach. Designing siRNAs with antiviral activity is a challenging task. 45 First, sequences of siRNAs must be identified which are highly specific minimizing potential off‐target effects and are highly effective in silencing. Second, effective and safe forms of delivery must be developed to introduce the siRNAs into infected cells. Finally, it is important to prevent degradation of siRNAs by nucleases and to avoid unwanted inflammation/immunostimulation.
In order to identify SARS‐CoV‐2‐specific siRNAs which are highly effective in silencing siRNA sequences targeting N and RdRP genes of SARS‐CoV‐2 were designed in silico 46 and tested in vitro to choose the siRNAs which are most powerful in silencing. In order to reduce working with infectious virus the first screening of potential antiviral siRNAs was performed using plasmids containing SARS‐CoV‐2 genes fused with the firefly luciferase gene, which allows estimating the silencing potency of the siRNAs by assessing the redaction of the luciferase activity in the treated cells. Specific siRNAs targeting the gene of firefly luciferase and GFP were used as positive and negative control, respectively. According to this screening assay 3 out of 13 tested siRNAs (siN‐4, siR‐7 and siR‐11) targeting SARS‐CoV‐2 genes N and RdRp, respectively, were found to be most effective. The specificity of the screening assay was demonstrated by the fact that siGFP (negative control had no effects). Next, we evaluated the specific antiviral activity of the chosen siRNA molecules in vitro using Vero E6 cells infected with SARS‐CoV‐2. These experiments identified siR‐7 as best molecule because it significantly decreases the number of viral RNA (vRNA) in infected cells as compared with cells only infected with SARS‐CoV‐2 as well as in infected cells which had been transfected with a SARS‐CoV‐2‐unrelated siRNA (i.e., siLuc) (Figure 2c). These results are important because they showed that siR‐7 targeting of the RdRp gene was specific and not due to a non‐specific effect of the transfection of cells with siRNA per se.
In initial experiments using cultivated virus‐infected cells, commercial Lipofectamine 3000 was used as vehicle for siRNA. However, it is not recommended for in vivo use. Although it has been reported that local delivery of unmodified naked siRNA to the lung can be successful 47 , 48 , 49 we decided to further modify siR‐7 to enhance its entrance into cells, to render it resistant to nuclease and to decrease off‐target effects. Therefore, we designed a novel formulation KK‐46 based on peptide dendrimers (PD) to achieve safe and efficient siRNA delivery into the lung. Dendrimers are branched three‐dimensional structures containing a central core surrounded by peripheral positively charged groups which promote their binding and condensation to nucleic acid molecules. 50 PDs contains mostly unnatural ε‐amide bonds on lysine residues, which increase resistance against proteolytic digestion. 51 , 52 Moreover, PDs are less toxic than linear peptides comprising the same combinations of amino acids. 53 Several types of dendrimers have been explored for siRNA delivery and gave promising results. 54 Based on our previous results 52 and in silico calculation of the molecular properties for achieving positive charge and amphiphilicity for cell penetration and RNA‐binding we designed the cationic PD KK‐46. The positive charge was attributable to the use of arginine and histidine residues for the N‐terminal ends of the peptide branches. A dendrimeric lysine core and hydrophobic amino acid residues contributed to the increase of amphiphilicity of the peptide. To determine the optimal concentration for in vivo use of KK‐46 we transfected Hep‐2 cells with pGL3 Luciferase Reporter Vectors /KK‐46 complexes at different concentrations and found that the optimal concentration of KK46 at 20–25 μg/mL for transfection was significantly lower than its IC50 = 418.9 μg/mL as evaluated by MTT testing of Vero cells (Figure S3). As DNA and RNA possess a similar structure in terms of nucleic acid framework and their electronegative nature, 54 we calculated a 20:1 ratio of KK‐46:siRNA for further experiments.
In addition to identifying KK‐46 as a possible pharmacologically acceptable vehicle for in vivo use we also tried to optimize the siR‐7 molecule itself. It has been shown that potential off‐target effects and unwanted immunostimulatory effects of siRNAs as well as their biodegradation may be reduced by incorporation of Locked nucleic acids (LNAs) in the siRNA sequence. LNA nucleotides contain a methylene linkage connecting the 2’ oxygen and 4’ carbon of the ribose ring that leads to a reduction of the conformational flexibility of the ribose. 55 siRNA molecules have the potential to induce inflammatory response by effects on the innate immune system through activation of Toll‐like receptors. 56 LNA incorporation has been shown to inhibit such immune stimulation. In particular, LNA modification at the 3′ end of the siRNA passenger strand, strongly inhibited IFNα induction without affecting knockdown activity. 57 Moreover LNA‐incorporation can increase endo‐ and exo‐nuclease resistance and sequence‐related off‐target effects. 58 We therefore incorporated LNA‐modification to the 3’ ends of siR‐7 sense and antisense sequences to improve it stability and functionality and found that this significantly increases the half‐life compared with unmodified siRNA (Figure 3). Next, we investigated whether the LNA‐modification may have a negative influence on the knockdown efficacy. For this purpose, we transfected Vero E6 cells with unmodified siR‐7 or LNA‐modified siR‐7‐EM/peptide dendrimer complex at three increasing concentrations of siRNA/siLNA and KK‐46 (Figure 4). We found that LNA‐modification did not affect specific virus gene silencing which is in accordance with a previous study. 58 It has been demonstrated that there is a sequence‐ and target‐independent effect by siRNA via toll‐like receptor 3 (TLR3). 59 However, the experiments carried out in SARS‐CoV‐2‐infected Vero cells expressing TLR3 in Figure 4 with the control siRNA siLuc demonstrate that there is no such TLR3‐mediated non‐specific effect. We thus could establish a formulation of LNA‐modified siR‐7‐EM targeted to RdRp gene of SARS‐CoV‐2 in complex with the KK‐46 peptide dendrimer which revealed strong and specific antiviral activity in vitro.
Topically applied siRNAs have been shown to be highly effective for inhibition of herpes simplex virus (HSV) and RSV in animal models. 60 , 61 We therefore envisaged topical treatment in the lung by inhalation for the treatment of COVID‐19. Furthermore, the lung is a major target for the disease due to high expression of ACE2 and long‐term lung damage is a major complication of COVID‐19. 62 Moreover, nebulizers or inhalers are available to generate aerosols for drug delivery by inhalation. 63
For the in vivo studies we used the model based on Syrian hamsters which has been shown to be a suitable small animal model for COVID‐19 64 and was also used to evaluate the effects of vaccination and passive immunization strategies with monoclonal antibodies 14 , 65 which afterwards also showed promising results in clinical trials. As the half‐life of siR‐7‐EM/KK‐46 after inhalation was only short (i.e., 23 min) we performed daily treatment of SARS‐CoV‐2‐infected animal with two applications of different doses of siR‐7‐EM/KK‐46 (0.7, 1.96 or 5.6 mg/kg daily) and evaluated the effects after two and six days. We found a dose‐dependent effect of treatment in terms of a significant reduction of viral load and most importantly, reduced lung inflammation on days 2 and day 6 as compared to non‐treated infected animals. In addition, we conducted additional studies evaluating lower doses which confirmed that treatment by daily twice inhalation of siR‐7‐EM/KK‐46 reduced viral load and lung inflammation. Collectively the in vivo experiments indicated 3.453 mg/kg/day as the optimal dose for treatment in the Syrian Hamster model.
The effect of treatment siR‐7‐EM/KK‐46 may be estimated by comparing our results with those obtained by passive immunization with the monoclonal SARS‐CoV‐2‐specific antibodies REGN10987 and REGN10933 which were used in a prophylactic and treatment setting in the Syrian Hamster model. 14 In the latter study prophylactic treatment reduced viral load, albeit not in a significant manner, and significantly reduced lung inflammation as we observed for early therapeutic application of siR‐7‐EM/KK‐46. The REGN cocktail of SARS‐CoV‐2 antibodies, which has demonstrated similar effects as siR‐7‐EM/KK‐46 in the Syrian Hamster model, could also reduce viral load in a clinical trial performed in COVID‐19 patients 15 and encourages us to further evaluate topical treatment by inhalation of siR‐7‐EM/KK‐46 in COVID‐19 patients to further explore the clinical utility of silencing SARS‐CoV‐2 by siRNA technology for specific treatment of COVID‐19. The main limitation which we were faced was that the siR‐7‐EM/KK‐46 composition is manufactured in lyophilized form. It is intended to administer the drug immediately after reconstitution in buffer. Therefore, it will be necessary to educate patients for the preparation of the solution and the proper use of inhalation devices. It is therefore likely that the siR‐7‐EM/KK‐46 composition will be applied mainly in specialized clinics.
In fact, preclinical evaluation of siR‐7‐EM/KK‐46 medication is now finished and a permission for clinical trial has been received from Ministry of Health of Russian Federation. In case of the successful completion of clinical trials it is planned to use the developed drug for therapy of COVID‐19 by inhalation.
CONFLICT OF INTEREST
Rudolf Valenta has received research grants from the Austrian Science Fund (FWF), HVD Biotech, Vienna, Austria, Worg Pharmaceuticals, Hangzhou, China and Viravaxx, Vienna, Austria and serves as a consultant for Viravaxx. Veronica Skvortsova currently serves as head of the Federal Medico‐biological Agency of Russia (FMBA Russia). Musa Khaitov, Alexandra Nikonova, Ksenia Kozhikova, Ilya Kofiadi, Igor Shilovskiy, Valeriy Smirnov, Ivan Kozlov, Sergey Andreev, Olesya Koloskova and Ilya Sergeev are authors on a patent application related to this study. The other authors do not have any conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing of the manuscript and have approved the final version for publication. Conceptualization, Methodology: Khaitov M., Valenta R., Nikonova A., Shilovskiy I., Kofiadi I., Kozhikhova K., Smirnov V., Koloskova O., Andreev S., Martynov A., Berzin I., Khaitov R., Skvortsova V.; Investigation: Vishnyakova L., Nikolsky A., Kovchina V., Barvinskaya E., Yumashev K., Maerle A., Kozlov I., Shatilov A., Timofeeva A., Kuznetsova N., Vasina D., Nikiforova M., Rybalkin S., Sergeev I., Trofimov D., Gushchin V., Kovalchuk A., Borisevich S., P. Gattinger; Formal analysis: Nikonova A., Shilovskiy I.; Writing ‐ Original Draft: Khaitov M, Nikonova A.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
This study was funded by Federal Medico‐biological Agency of Russia. We are grateful to Prof. Andrew Fire, Stanford University for having critically read our manuscript.
REFERENCES
- 1. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 2020;75(7):1730‐1741. [DOI] [PubMed] [Google Scholar]
- 2. Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy 2020;75(7):1564‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang D, Comish P, Kang R. The hallmarks of COVID‐19 disease. PLoS Pathog 2020;16(5):e1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morens DM, Folkers GK, Fauci AS. Emerging infections: a perpetual challenge. Lancet Infect Dis 2008;8(11):710‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morens DM, Breman JG, Calisher CH, et al. The origin of COVID‐19 and why it matters. Am J Trop Med Hyg 2020;103(3):955‐959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramasamy MN, Minassian AM, Ewer KJ, et al. Safety and immunogenicity of ChAdOx1 nCoV‐19 vaccine administered in a prime‐boost regimen in young and old adults (COV002): a single‐blind, randomised, controlled, phase 2/3 trial. Lancet 2021;396(10267):1979‐1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Logunov DY, Dolzhikova IV, Zubkova OV, et al. Safety and immunogenicity of an rAd26 and rAd5 vector‐based heterologous prime‐boost COVID‐19 vaccine in two formulations: two open, non‐randomised phase 1/2 studies from Russia. Lancet 2020;396(10255):887‐897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mulligan MJ, Lyke KE, Kitchin N, et al. Phase I/II study of COVID‐19 RNA vaccine BNT162b1 in adults. Nature 2020;586(7830):589‐593. [DOI] [PubMed] [Google Scholar]
- 9. Krammer F. SARS‐CoV‐2 vaccines in development. Nature 2020;586(7830):516‐527. [DOI] [PubMed] [Google Scholar]
- 10. Pan H, Peto R, Henao‐Restrepo AM, et al. Repurposed Antiviral Drugs for Covid‐19 ‐ Interim WHO solidarity trial results. N Engl J Med 2020;384:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 ‐ final report. N Engl J Med 2020;383(19):1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu STH, Lin HM, Baine I, et al. Convalescent plasma treatment of severe COVID‐19: a propensity score‐matched control study. Nat Med 2020;26(11):1708‐1713. [DOI] [PubMed] [Google Scholar]
- 13. Tanne JH. Covid‐19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ 2020;368:m1256. [DOI] [PubMed] [Google Scholar]
- 14. Baum A, Ajithdoss D, Copin R, et al. REGN‐COV2 antibodies prevent and treat SARS‐CoV‐2 infection in rhesus macaques and hamsters. Science 2020;370(6520):1110‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN‐COV2, a neutralizing antibody cocktail, in outpatients with Covid‐19. N Engl J Med. 2021;384(3):238‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrison AG, Lin T, Wang P. Mechanisms of SARS‐CoV‐2 transmission and pathogenesis. Trends Immunol 2020;41(12):1100‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thi Nhu Thao T, Labroussaa F, Ebert N, et al. Rapid reconstruction of SARS‐CoV‐2 using a synthetic genomics platform. Nature 2020;582(7813):561‐565. [DOI] [PubMed] [Google Scholar]
- 18. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19(3):155‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet 2002;3(10):737‐747. [DOI] [PubMed] [Google Scholar]
- 20. Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV‐1 infection in human T cells by lentiviral‐mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A 2003;100(1):183‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carmichael GG. Medicine: silencing viruses with RNA. Nature 2002;418(6896):379‐380. [DOI] [PubMed] [Google Scholar]
- 22. Jacque JM, Triques K, Stevenson M. Modulation of HIV‐1 replication by RNA interference. Nature 2002;418(6896):435‐438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahlquist P. RNA‐dependent RNA polymerases, viruses, and RNA silencing. Science 2002;296(5571):1270‐1273. [DOI] [PubMed] [Google Scholar]
- 24. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans. Nature 1998;391(6669):806‐811. [DOI] [PubMed] [Google Scholar]
- 25. Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 2003;67(4):657‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imai M, Iwatsuki‐Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS‐CoV‐2 infection and countermeasure development. Proc Natl Acad Sci U S A 2020;117(28):16587‐16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Faizuloev E, Marova A, Nikonova A, Volkova I, Gorshkova M, Izumrudov V. Water‐soluble N‐[(2‐hydroxy‐3‐trimethylammonium)propyl]chitosan chloride as a nucleic acids vector for cell transfection. Carbohydr Polym 2012;89(4):1088‐1094. [DOI] [PubMed] [Google Scholar]
- 28. Gattinger P, Borochova K, Dorofeeva Y, et al. Antibodies in serum of convalescent patients following mild COVID‐19 do not always prevent virus‐receptor binding. Allergy. 2021;76(3):878‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borisevich SV, Syromyatnikova SI, Khamitov RA, Markov VI, Maximov VA, Pistsov MN. Inventor composition of agar coating for titration of coronavirus‐the causative agent of severe acute respiratory syndrome by plaque forming units. Russia. 2008:RU2325436. [Google Scholar]
- 30. Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid‐modified small interfering RNA in vitro and in vivo. Mol Cancer Ther 2007;6(3):833‐843. [DOI] [PubMed] [Google Scholar]
- 31. Prakash TP, Allerson CR, Dande P, et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J Med Chem 2005;48(13):4247‐4253. [DOI] [PubMed] [Google Scholar]
- 32. Piasecka J, Lenartowicz E, Soszynska‐Jozwiak M, Szutkowska B, Kierzek R, Kierzek E. RNA secondary structure motifs of the influenza a virus as targets for siRNA‐mediated RNA interference. Mol Ther Nucleic Acids 2020;19:627‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu YC, Kuo RL, Shih SR. COVID‐19: The first documented coronavirus pandemic in history. Biomed J 2020;43(4):328‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jomah S, Asdaq SMB, Al‐Yamani MJ. Clinical efficacy of antivirals against novel coronavirus (COVID‐19): A review. J Infect Public Health 2020;13(9):1187‐1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu B, Weng Y, Xia XH, Liang XJ, Huang Y. Clinical advances of siRNA therapeutics. J Gene Med 2019;21(7):e3097. [DOI] [PubMed] [Google Scholar]
- 36. Levanova A, Poranen MM. RNA Interference as a prospective tool for the control of human viral infections. Front Microbiol 2018;9:2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li T, Zhang Y, Fu L, et al. siRNA targeting the leader sequence of SARS‐CoV inhibits virus replication. Gene Ther 2005;12(9):751‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu A, Zhang H, Zhang X, et al. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA‐dependent RNA polymerase. Virology 2004;324(1):84‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Li T, Fu L, et al. Silencing SARS‐CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett 2004;560(1–3):141‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He ML, Zheng B, Peng Y, et al. Inhibition of SARS‐associated coronavirus infection and replication by RNA interference. JAMA 2003;290(20):2665‐2666. [DOI] [PubMed] [Google Scholar]
- 41. Tang Q, Li B, Woodle M, Lu PY. Application of siRNA against SARS in the rhesus macaque model. Methods Mol Biol 2008;442:139‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li BJ, Tang Q, Cheng D, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med 2005;11(9):944‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen W, Feng P, Liu K, Wu M, Lin H. Computational identification of small interfering RNA targets in SARS‐CoV‐2. Virol Sin 2020;35(3):359‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berber B, Aydin C, Kocabas F, et al. Gene editing and RNAi approaches for COVID‐19 diagnostics and therapeutics. Gene Ther 2020;14:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rothe D, Wade EJ, Kurreck J. Design of small interfering RNAs for antiviral applications. Methods Mol Biol 2011;721:267‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khaitov MR, Shilovskiy IP, Kofiadi IA, Sergeev IV, et al. The siRNA‐based drug for inhibition of the SARS‐CoV‐2 replication. Russia. 2020:N2733361. [Google Scholar]
- 47. Lam JK, Liang W, Chan HK. Pulmonary delivery of therapeutic siRNA. Adv Drug Deliv Rev 2012;64(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khaitov MR, Shilovskiy IP, Nikonova AA, et al. Small interfering RNAs targeted to interleukin‐4 and respiratory syncytial virus reduce airway inflammation in a mouse model of virus‐induced asthma exacerbation. Hum Gene Ther 2014;25(7):642‐650. [DOI] [PubMed] [Google Scholar]
- 49. Nikonova A, Shilovskiy I, Galitskaya M, et al. Respiratory syncytial virus upregulates IL‐33 expression in mouse model of virus‐induced inflammation exacerbation in OVA‐sensitized mice and in asthmatic subjects. Cytokine. 2021;138:155349. [DOI] [PubMed] [Google Scholar]
- 50. Santos A, Veiga F, Figueiras A. Dendrimers as pharmaceutical excipients: synthesis, properties. toxicity and biomedical applications. Materials (Basel) 2019;13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luo K, Li C, Wang G, et al. Peptide dendrimers as efficient and biocompatible gene delivery vectors: Synthesis and in vitro characterization. J Control Release 2011;155(1):77‐87. [DOI] [PubMed] [Google Scholar]
- 52. Kozhikhova KV, Andreev SM, Shilovskiy IP, et al. A novel peptide dendrimer LTP efficiently facilitates transfection of mammalian cells. Org Biomol Chem 2018;16(43):8181‐8190. [DOI] [PubMed] [Google Scholar]
- 53. Eggimann GA, Blattes E, Buschor S, et al. Designed cell penetrating peptide dendrimers efficiently internalize cargo into cells. Chem Commun (Camb) 2014;50(55):7254‐7257. [DOI] [PubMed] [Google Scholar]
- 54. Wu J, Huang W, He Z. Dendrimers as carriers for siRNA delivery and gene silencing: a review. ScientificWorldJournal 2013;2013:630654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Braasch DA, Corey DR. Locked nucleic acid (LNA): fine‐tuning the recognition of DNA and RNA. Chem Biol 2001;8(1):1‐7. [DOI] [PubMed] [Google Scholar]
- 56. Meng Z, Lu M. RNA interference‐induced innate immunity, off‐target effect, or immune adjuvant? Front Immunol 2017;8:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jackson AL, Linsley PS. Recognizing and avoiding siRNA off‐target effects for target identification and therapeutic application. Nat Rev Drug Discov 2010;9(1):57‐67. [DOI] [PubMed] [Google Scholar]
- 58. Elmen J, Thonberg H, Ljungberg K, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 2005;33(1):439‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kleinman ME, Yamada K, Takeda A, et al. Sequence‐ and target‐independent angiogenesis suppression by siRNA via TLR3. Nature 2008;452(7187):591‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paavilainen H, Lehtinen J, Romanovskaya A, et al. Topical treatment of herpes simplex virus infection with enzymatically created siRNA swarm. Antivir Ther 2017;22(7):631‐637. [DOI] [PubMed] [Google Scholar]
- 61. Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med 2005;11(1):50‐55. [DOI] [PubMed] [Google Scholar]
- 62. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Front Med 2020;14(2):185‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Youngren‐Ortiz SR, Gandhi NS, Espana‐Serrano L, Chougule MB. Aerosol Delivery of siRNA to the Lungs. Part 1: Rationale for Gene Delivery Systems. Kona 2016;33:63‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Munoz‐Fontela C, Dowling WE, Funnell SGP, et al. Animal models for COVID‐19. Nature 2020;586(7830):509‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tostanoski LH, Wegmann F, Martinot AJ, et al. Ad26 vaccine protects against SARS‐CoV‐2 severe clinical disease in hamsters. Nat Med 2020;26(11):1694‐1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


