Summary
Mycorrhizal fungi are central to the biology of land plants. However, to what extent mycorrhizal shifts – broad evolutionary transitions in root‐associated fungal symbionts – are related to changes in plant trophic modes remains poorly understood.
We built a comprehensive DNA dataset of Orchidaceae fungal symbionts and a dated plant molecular phylogeny to test the hypothesis that shifts in orchid trophic modes follow a stepwise pattern, from autotrophy over partial mycoheterotrophy (mixotrophy) to full mycoheterotrophy, and that these shifts are accompanied by switches in fungal symbionts.
We estimate that at least 17 independent shifts from autotrophy towards full mycoheterotrophy occurred in orchids, mostly through an intermediate state of partial mycoheterotrophy. A wide range of fungal partners was inferred to occur in the roots of the common ancestor of this family, including ‘rhizoctonias’, ectomycorrhizal, and wood‐ or litter‐decaying saprotrophic fungi. Phylogenetic hypothesis tests further show that associations with ectomycorrhizal or saprotrophic fungi were most likely a prerequisite for evolutionary shifts towards full mycoheterotrophy.
We show that shifts in trophic mode often coincided with switches in fungal symbionts, suggesting that the loss of photosynthesis selects for different fungal communities in orchids. We conclude that changes in symbiotic associations and ecophysiological traits are tightly correlated throughout the diversification of orchids.
Keywords: ancestral state reconstruction, mixotrophy, mycoheterotrophy, orchid mycorrhiza, phylogenetic correlation, symbiont switching
Introduction
The history of evolution and biodiversity is fundamentally a history of the evolution of species interactions (Margulis, 1991; Thompson, 1999, 2005). Many of the major events in the diversification of life can be traced back to the appearance of novel species interactions (Szathmáry & Smith, 1995). As such, the colonization and subsequent domination of land by plants – a fundamental turning point in the evolutionary history of the earth – was probably facilitated by new interactions with mutualistic symbiotic fungi (‘mycorrhizas’) that promote plant growth by facilitating the acquisition of essential nutrients (e.g. phosphorus, nitrogen and sometimes carbon) (Pirozynski & Malloch, 1975; Selosse & Le Tacon, 1998; Field et al., 2015; Martin et al., 2017; Feijen et al., 2018). Today, the symbiosis with mycorrhizal fungi is found in over 90% of extant species of all major lineages of land plants, except for mosses, and involves 40 000–50 000 species of three different fungal phyla (Brundrett & Tedersoo, 2018; Tedersoo et al., 2020). Based on the morphology of the interaction and the identity of the interacting plants and fungi, four major mycorrhizal types can be distinguished: arbuscular mycorrhiza, ectomycorrhiza, ericoid mycorrhiza and orchid mycorrhiza (Smith & Read, 2008; van der Heijden et al., 2015; Brundrett & Tedersoo, 2018).
Interestingly, phylogenetic mapping of these four mycorrhizal types has shown that they are evolutionary‐conserved and that transitions between mycorrhizal types are relatively rare among land plants, fueling questions about what drives these switches (Feijen et al., 2018; Werner et al., 2018). Recent studies have suggested that the composition of mycorrhizal fungal communities differs between plant nutritional modes, ranging from an exclusively autotrophic to a fully mycoheterotrophic mode of life, in which photosynthesis has been replaced by the uptake of carbon from root‐associated fungi (Yagame et al., 2016; Jacquemyn & Merckx, 2019). This raises the hypothesis that shifts in trophic modes are correlated with switches in symbiotic associations in land plants.
Orchids are particularly relevant to the investigation of this hypothesis in more detail as they form unique mycorrhizas with basidiomycete and ascomycete fungi, known as orchid mycorrhizas (OrM) (van der Heijden et al., 2015; Brundrett & Tedersoo, 2018). Previous research has suggested that OrM probably evolved from an ancestor with arbuscular mycorrhizal fungi, in which the gain of OrM fungi is explained by pathogenic infection (Yukawa et al., 2009; Rasmussen & Rasmussen, 2014). The evolution of OrM has been regarded as one of the major drivers for the evolutionary success of the Orchidaceae (Dressler, 2005; Rasmussen & Rasmussen, 2014; Chase et al., 2015; Givnish et al., 2015; Jacquemyn et al., 2017a). Moreover, unlike the majority of green plants, orchids rely on carbon from OrM fungi for the germination of their dust seeds, a phenomenon known as ‘initial mycoheterotrophy’ (Leake, 1994; Rasmussen, 1995; Merckx, 2013). While most species likely become fully autotrophic as adults, several partially mycoheterotrophic species retain the ability to obtain carbon from their mycorrhizal fungi to compensate for photosynthesis (also termed mixotrophy; Selosse & Roy, 2009), and more than 250 fully mycoheterotrophic species have completely replaced their photosynthetic capacity by the uptake of fungal carbon (Jacquemyn & Merckx, 2019). Although these trophic modes have also evolved outside the Orchidaceae, no other plant family displays such a high frequency of shifts in trophic modes (Merckx, 2013; Jacquemyn & Merckx, 2019).
Traditionally, orchid mycorrhizas have been considered to consist primarily, if not only, of members of the ‘rhizoctonias’ complex, which comprises taxa from three distinct fungal families: Tulasnellaceae, Ceratobasidiaceae and Serendipitaceae (Smith & Read, 2008; Dearnaley et al., 2012), and this association has been regarded as the ancestral state of the family (Yukawa et al., 2009; Dearnaley et al., 2012; Weiß et al., 2016). However, more recently, other saprotrophic and ectomycorrhizal (ECM) fungal lineages have been found to form mycorrhizal associations with orchids (Bidartondo et al., 2004; Dearnaley et al., 2012; Ogura‐Tsujita et al., 2021). The number and composition of fungal taxa associating with a single orchid are variable and depend on the phylogenetic relatedness of the orchids (Shefferson et al., 2010; Jacquemyn et al., 2011; Martos et al., 2012), their developmental stages (Bidartondo & Read, 2008; Těšitelová et al., 2015; Waud et al., 2017), ecological conditions (Bidartondo et al., 2004; Jacquemyn et al., 2016; Duffy et al., 2019) and also trophic modes (Motomura et al., 2010; Ogura‐Tsujita et al., 2012). Autotrophic orchids generally associate with rhizoctonia fungi, while partially and fully mycoheterotrophic orchids mostly associate with ECM and wood‐ and litter‐decaying fungi, suggesting that transitions in trophic modes are probably linked to shifts in fungal lifestyles in Orchidaceae (Jacquemyn & Merckx, 2019). Detailed studies on Neottieae (Selosse et al., 2004; Selosse & Roy, 2009; Yagame et al., 2016) and Cymbidium (Ogura‐Tsujita et al., 2012) have proposed an evolutionary correlation between fungal lifestyle and trophic mode in these taxa. However, this evolutionary correlation remains to be tested in a comprehensive family‐wide phylogenetic framework.
In the past two decades, the ecophysiology of orchids and the identity of their fungal partners have been studied extensively, fueled by the development of stable isotope measurements and novel DNA sequencing methods (Gebauer & Meyer, 2003; Bidartondo et al., 2004; Martos et al., 2009; Jacquemyn et al., 2017b; Schiebold et al., 2017, 2018; Schweiger et al., 2019). In this study, we compiled a family‐wide dataset of mycorrhizal interactions and orchid trophic modes, and employed a phylogenetic framework to test the hypothesis that shifts in trophic mode in the orchid family are correlated with switches in fungal communities. Specifically, we aimed to answer the following questions: what is the evolutionary history of fungal associations and trophic modes within the family is the evolution towards mycoheterotrophy correlated with switches in fungal communities and what is the most common evolutionary scenario towards mycoheterotrophy in Orchidaceae in the context of fungal partner switches?
Materials and Methods
Orchid mycorrhiza, fungal lifestyle, and trophic mode
To compile a family‐wide dataset of orchid fungal associations, we searched for molecular data using the keywords ‘orchid mycorrhizal fungi’ in Mendeley Reference Management Desktop (before August 2019) and only retained articles that contained fungal nuclear ribosomal internal transcribed spacer (ITS) accessions generated with either Sanger or high‐throughput sequencing (HTS) techniques. By manually checking and filtering the obtained articles, c. 250 were kept as original references for the orchid mycorrhiza dataset. The final dataset contained information on 750 orchid species covering nearly all major clades of orchids (20 tribes and 39 subtribes belonging to five subfamilies) (see Supporting Information Table S1). In addition, we searched for isotope data of the orchid species included in our mycorrhizal dataset. For each orchid species, all available DNA accessions of the markers’ ITS, matK, rbcL and trnL‐F, were downloaded from the NCBI GenBank database.
Taxonomic information was assigned to each fungal sequence using Usearch v.11 (Edgar, 2010). We further categorized the sequences at the family level because half of all sequences could not be assigned to a lower taxonomic level (genus or species) by blasting against the UNITE local database (Abarenkov et al., 2010). Detailed procedures of fungal operational taxonomic unit (OTU) clustering and taxonomic assignment are described in Notes S1. After removing fungal families associated with only a single orchid species, we assigned the remaining fungal families in Basidiomycota and Ascomycota to different lifestyles based on the information provided in the original publications (Dearnaley et al., 2012; Tedersoo & Brundrett, 2017; Põlme et al., 2020) supplemented by funguild (Nguyen et al., 2016) using default parameters. We categorized the rhizoctonia families (Tulasnellaceae, Ceratobasidiaceae, and Serendipitaceae – Sebacinales ‘group B’) as ‘rhizoctonia‐like’ fungi (RHI). Families mainly comprising ECM fungi (Sebacinaceae – Sebacinales ‘group A’, Thelephoraceae, Russulaceae, and others) were classified as ECM. Families mainly containing saprotrophic fungi (e.g. Mycenaceae, Psathyrellaceae) were classified as saprotrophic (SAP). Fungal families having both ECM and SAP fungi were classified as the ECM/SAP lifestyle. Because a large number of fungal families contain members that are pathogens, endophytes, or belong to unknown ecological guilds, we adopted a conservative approach in all further analyses by restricting the number of fungal families to the 17 families that are known to contain putative orchid mycorrhizal fungi (Table 12.1 in Dearnaley et al., 2012).
Based on morphological descriptions and stable isotope signatures (13C and 15N), we assigned orchid species to three trophic modes: autotrophy (AU), partial mycoheterotrophy (PMH), and full mycoheterotrophy (MH). Due to the inconsistency in 13C and 15N signatures of several orchids, either a strict or more relaxed definition of PMH was adopted. A detailed description of the criteria to assign an orchid to one of the trophic modes can be found in Notes S1.
Orchid phylogeny reconstruction and divergence time estimation
To reconstruct a time‐calibrated phylogeny of Orchidaceae, we performed a relaxed molecular clock analysis with Beast v.2.5 (Bouckaert et al., 2019) using backbone trees from Chase et al., (2015), Chomicki et al., (2015) and Givnish et al., (2015). Sequence alignment, phylogenetic reconstruction, and time calibration are described in detail in the Notes S1.
Phylogenetic signal
Pagel’s lambda (λ) (Pagel, 1999) was used to investigate whether the distribution of the character states of trophic mode and fungal lifestyle showed some degree of phylogenetic signal on the orchid phylogeny. The λ value was estimated using ‘fitDiscrete’ function in the R package geiger (Pennell et al., 2014).
Ancestral state estimations
Ancestral state reconstruction was used to trace the evolutionary history of trophic modes and symbiotic associations represented by fungal lifestyles over the reconstructed orchid phylogeny. Ancestral state reconstructions were performed with the ‘make.simmap’ function in the R package phytools (Revell, 2012) using stochastic character mapping (Bollback, 2006). An initial model with independent rates of state transitions (‘All Rates Different’, ARD) was applied among symbiotic associations and trophic modes, respectively. Because MH is associated with rampant plastid gene loss (Graham et al., 2017) and is thus likely irreversible (Merckx, 2013), we specified the transition rates from MH to PMH and AU as zero in the state transition rate matrix (Q matrix). The ancestral states of trophic mode were inferred based on both a relaxed and a strict definition of PMH. In addition, analyses of ancestral state reconstruction were also performed using ‘MultiState’ Markov chain Monte Carlo (MCMC) analysis implemented in bayestraits v.3, adopting the same model for each discrete trait as with stochastic character mapping. We ran each of our analyses for 1010 000 iterations with the first 10 000 generations as burn‐in. Reversible‐jump MCMC analyses (Green, 1995; Pagel & Meade, 2006) were applied to reduce model complexity and over‐parameterization.
Hypothesis tests
Discrete Independent and Dependent models in bayestraits detected correlated trait evolution between symbiotic association and trophic mode (Notes S1). However, with this approach, the path of trait evolution was not yet evident. Therefore, we combined the two traits into a coupled multistate character and performed explicit hypothesis tests to verify plausible evolutionary scenarios towards MH. By setting constraints on the Q matrix, we tested which state was best supported to be the intermediate state that enabled the evolution of mycoheterotrophy (Notes S1). To compare models, we estimated the marginal likelihood of the free model where no constraints were set to the Q matrix. One constrained model disallowing specific transitions between states in the Q matrix that best fitted the data will result in the marginal likelihood that differs most significantly from the less constrained free model. We ran each ‘MultiState’ analysis in triplicate for 106 generations and calculated the average marginal likelihood using a stepping stone sampler. Finally, we compared the constrained models with the free model by Bayesian information criterion (BIC), and the model with the lowest BIC value was selected.
Results
Mycorrhizal interactions in Orchidaceae
Adopting a strict definition of PMH, a total of 455 AU, 27 PMH, and 37 MH orchid species were used for phylogenetic reconstruction (Table S2). Under a relaxed definition of PMH, there were 414 AU, 69 PMH species, and 37 MH species (Table S2). After removing fungal families that associated with a single orchid species, a total of 68 fungal families in Basidiomycota and Ascomycota were identified based on ITS sequences generated from both Sanger and HTS techniques (Fig. S1). Autotrophic orchids (455 species) were associated with the highest diversity of fungal taxa (66 fungal families), and shared 67% and 50% of their fungal families with PMH and MH orchids, respectively (Fig. S2). The number of fungal families detected in our dataset showed small differences between Sanger sequencing and HTS techniques (see details in Notes S1; Fig. S3). Seventeen of all detected fungal families that have previously been identified as putative orchid mycorrhiza fungi (Dearnaley et al., 2012) were mapped on the orchid phylogeny (Figs 1, S4), including three rhizoctonia families (Tulasnellaceae, Ceratobasidiaceae and Serendipitaceae), seven families mainly comprising ECM fungi (Sebacinaceae, Thelephoraceae, Russulaceae, Tuberaceae, Clavulinaceae, Hymenogastraceae, Inocybaceae), and seven families containing SAP and/or ECM fungi (Pezizaceae, Pyronemataceae, Hymenochaetaceae, Marasmiaceae, Psathyrellaceae, Mycenaceae, Physalacriaceae). The three rhizoctonia families were present in the roots of 454 (87.5%) of 519 orchid species, followed by major ECM families (e.g. Sebacinaceae, Thelephoraceae, and Russulaceae) (Figs 1, S4).
Fig. 1.
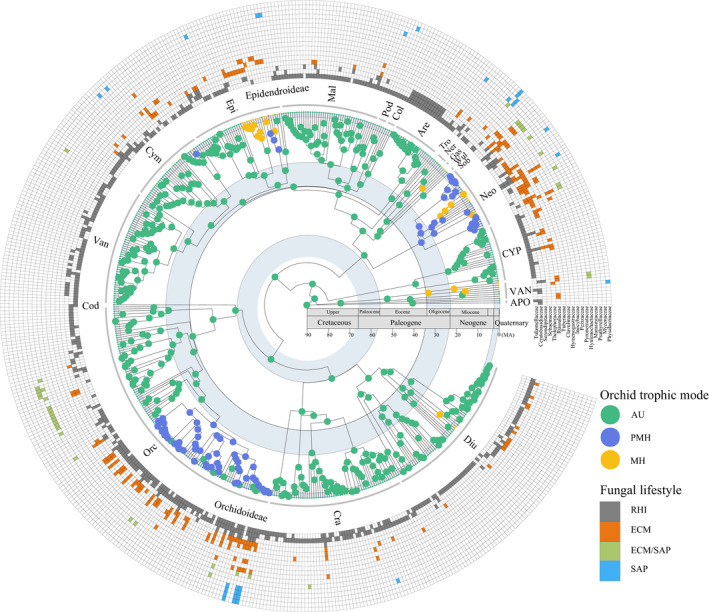
Orchid phylogeny, ancestral state reconstruction of trophic mode and symbiotic association. The orchid chronogram is annotated with subfamilies and tribes represented by three‐letter initials (see the full name in Supporting Information Table S2); the highest posterior probability state of trophic modes is mapped on all internal nodes of the tree based on ancestral state reconstruction (Fig. S5). AU, autotrophy; PMH, partial mycoheterotrophy; MH, full mycoheterotrophy. The geological time scales are visualized by circles from upper Cretaceous (c. 90–66 million yr ago (Ma)), via Paleocene (c. 66–23 Ma), and Neogene (c. 23–2.58 Ma), to Quaternary (from 2.58 Ma to the present). The outside matrix represents the presence or absence of 17 fungal families containing putative orchid mycorrhizal fungi (Dearnaley et al., 2012). Fungal families are ranked by lifestyles and the number of orchid species they associate with. The lifestyles (‘rhizoctonia‐like’ fungi (RHI), ectomycorrhizal (ECM), ECM/saprotrophic (ECM/SAP), and SAP) of each fungal family are visualized using different colors. Visualization was generated using the R package ggtree (Yu, 2020).
Ancestral states of trophic mode and symbiotic association
Both stochastic character mapping and ‘MultiState’ MCMC analysis inferred that the common ancestor of orchids was most likely autotrophic at adult stage (Figs 2a,b, S5, S6). From the ancestral state of AU, there have been 42 or 28 transitions to PMH, and 17 or 18 shifts to MH by using the relaxed and strict definition of PMH, respectively (Figs S5, S6). Few transitions were detected directly from AU to MH using both definitions of PMH (Figs S5, S6). Shifts to mycoheterotrophy mainly occurred in the subfamilies Vanilloideae (Vanilleae), Orchidoideae (Orchideae, Cranichideae, Diurideae), and Epidendroideae (Neottieae, Gastrodieae, Wullschlaegelieae, Epidendreae, Cymbidieae) (Figs S5, S6). By tracing transitions in trophic modes through geological time, we showed a general trend of transitions from AU towards PMH and MH, and transitions to MH were predated by transitions to PMH (Fig. S7a). Full mycoheterotrophy appeared only recently in the evolutionary history of Orchidaceae at the start of the Oligocene (c. 35 million years ago; Fig. S7a).
Fig. 2.
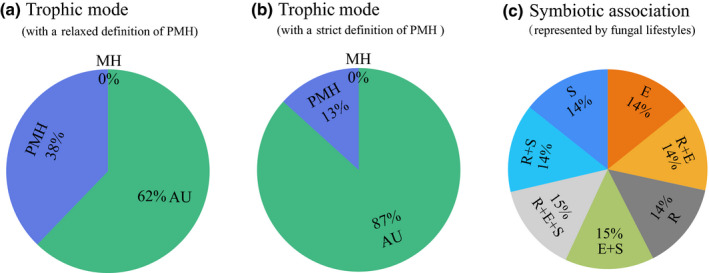
Ancestral state reconstruction of trophic mode and symbiotic association using bayestraits ‘MultiState’ analysis. (a) The posterior probability of trophic modes of the common ancestor of orchids under a relaxed definition of partial mycoheterotrophy (Supporting Information Notes S1). AU, autotrophy; PMH, partial mycoheterotrophy; MH, full mycoheterotrophy. (b) The posterior probability of trophic modes of the common ancestor of orchids under a strict definition of PMH (Notes S1). (c) The posterior probability of symbiotic associations of the common ancestor of orchids. Symbiotic associations are represented by the lifestyles of fungal families that one orchid associates with. R, ‘rhizoctonias’; E, ectomycorrhizal fungi; S, saprotrophic fungi.
Ancestral state reconstruction using both methods of stochastic character mapping and ‘MultiState’ MCMC analysis inferred that each symbiotic association has a comparatively equal probability of the ancestral state (Figs 2c, 3), indicating that the ancestor of orchids was probably associated with a broad range of fungi. Most transitions started from the combined state R + E + S to a variety of symbiotic associations, mainly including ECM and SAP fungi (Fig. 3c). The majority of transitions to ECM and wood‐ or litter‐decaying SAP fungi involved 34 orchid genera belonging to nine tribes (Fig. 3), of which six genera do not contain PMH or MH species (Table S2). By trancing transitions in symbiotic associations through geological time, we inferred a general trend of shifts from a fungal community comprising diverse fungal lifestyles in the ancestor of orchids towards more specialized fungal lifestyles among extant lineages of Orchidaceae (Fig. S7b). A large proportion of evolutionary shifts towards an exclusive association with rhizoctonia‐like fungi was observed, whereas only a small proportion of transitions towards ECM and wood‐ or litter‐decaying SAP fungi was detected (Fig. S7b).
Fig. 3.
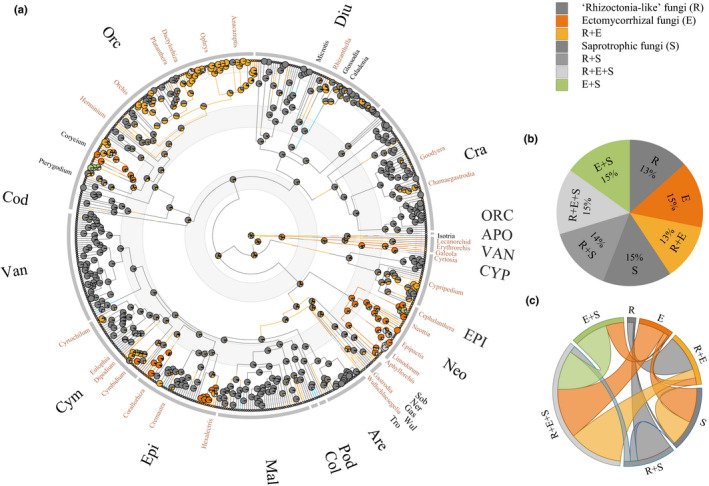
Ancestral state reconstruction of symbiotic association in Orchidaceae. (a) Likelihood pie reconstruction of each symbiotic association for all nodes. The symbiotic association used for ancestral state estimation is based on the lifestyles of 17 fungal families containing putative orchid mycorrhizal fungi (Dearnaley et al., 2012). The orchid chronogram is annotated with subfamilies and tribes represented by three‐letter initials (see the full name in Supporting Information Table S2), and genera where shifts to ectomycorrhizal or saprotrophic fungi occur. The genera comprising mycoheterotrophic species are colored in brown. The geological time scales are visualized by circles from upper Cretaceous (c. 90–66 million yr ago (Ma)), via Paleocene (c. 66–23 Ma), and Neogene (c. 23–2.58 Ma), to Quaternary (from 2.58 Ma to the present). (b) The likelihood of each symbiotic association for the root. R, ‘rhizoctonias’, E, ectomycorrhizal fungi, S, saprotrophic fungi. (c) The transitions between symbiotic associations. The band size for each symbiotic association represents the proportion of transitions among all transitions; the width of the ribbons represents the proportion of transitions starting from that state.
Phylogenetic signals and trait correlations
We detected a robust phylogenetic signal for both trophic mode and fungal lifestyle (Table S3). A strong correlation between trophic mode and symbiotic association was inferred by both the Discrete Independent and Dependent models, as supported by Bayes factor scores (Table S4). Furthermore, our ‘MultiState’ analyses of trophic mode and symbiotic association showed that the fourth model is favored with the lowest BIC compared with the other three models, which sets constraints on the forward transition from autotrophic states (states 1, 2 and 3) to mycoheterotrophic states (states 4 and 5) (Table 1). This model assumes that a forward transition from AU to MH is associated with an obligate intermediate transition of symbiotic association – a combination of rhizoctonias and ECM or SAP fungi (Notes S1). By setting further constraints on the reverse direction of transitions, our results showed that the sixth model best fitted our data (Table 1). This model assumes that the reverse transition from the MH to the AU state only occurs from the intermediate symbiotic association with a combination of rhizoctonias and ECM or SAP fungi (Notes S1).
Table 1.
Model tests of possible evolutionary scenarios of the coupled character combining trophic mode and symbiotic association using ‘MultiState’ analyses implemented in bayestraits.
| Models | No. of parameters | Loge L | BIC |
|---|---|---|---|
| The free model | 20 | −487.99 | 1101.01 |
| Model 1 | 16 | −499.07 | 1098.17 |
| Model 2 | 16 | −486.45 | 1072.93 |
| Model 3 | 16 | −484.67 | 1069.37 |
| Model 4 | 15 | −485.3 | 1064.37 |
| Model 5 | 12 | −481.95 | 1038.92 |
| Model 6 | 11 | −479.63 | 1028.03 |
BIC, Bayesian information criterion.
Trophic mode (autotrophy and full mycoheterotrophy) and symbiotic association represented by fungal lifestyle (‘rhizoctonias’ (R), ectomycorrhizal fungi (E) and saprotrophic fungi (S)) in our dataset were converted into a five‐state character: (1) AU‐R; (2) AU‐ES; (3) AU‐RES; (4) MH‐RES; and (5) MH‐ES. We developed plausible models to test different evolutionary paths towards full mycoheterotrophy contributed by symbiotic shifts (see the diagram of models in Supporting Information Notes S1). The BIC is used to compare models, and the model with the lowest BIC values is selected.
Discussion
Evolutionary shifts in symbiotic associations
Our results suggest that the common ancestor of orchids probably did not associate with fungi of a single lifestyle, but rather with a fungal community of multiple lifestyles (Figs 2c, 3). Although rhizoctonias were most likely part of this community, as previously suggested by Yukawa et al. (2009), our analyses indicate the possibility of a much wider partner breadth in the ancestors of orchids (Figs 2c, 3). Our results further show that the largest proportion of evolutionary shifts in symbiotic associations occurred from the combined state of rhizoctonias, ECM, and SAP fungi (R + E + S) towards more specialized associations (Fig. 3c). Similarly, a dual symbiotic association was also recently inferred as a prerequisite for shifts between major mycorrhizas among land plants (Werner et al., 2018). These studies collaboratively support the overall hypothesis that the evolution of symbiotic shifts follows a stepwise process, in which fungal partners in a later stage of a mycorrhizal association have been latently present in the fungal community of the ancestor (Selosse et al., 2010; van der Heijden et al., 2015; Jacquemyn & Merckx, 2019; Suetsugu & Matsubayashi, 2021).
Support for this so‐called ‘waiting room’ hypothesis (Selosse et al., 2010, 2018; van der Heijden et al., 2015) has been mounting. For instance, several ECM fungi colonizing orchid roots are also endophytes in surrounding plants or even in orchid tissues, such as Sebacinaceae (Selosse et al., 2009; Oliveira et al., 2014; Weiß et al., 2016) and Tuberaceae (Gryndler et al., 2014; Schneider‐Maunoury et al., 2018). Likewise, some groups of free‐living SAP fungi have been suggested to transit to a mycorrhizal association with orchid species through an endophytic state, such as Mycenaceae (Martos et al., 2009; Ogura‐Tsujita et al., 2009) and Psathyrellaceae (Yamato et al., 2005; Yagame et al., 2007; Ogura‐Tsujita & Yukawa, 2008). These findings suggest that the ability to associate with non‐mycorrhizal fungi is probably a predisposition for tight mycorrhizal associations in Orchidaceae (e.g. Shubin et al., 2009). However, due to the lack of microscope observations or germination tests, we cannot be sure that all fungal taxa recorded in this study are truly mycorrhizal, and thus some might reside in orchid roots as endophytes. Considering that a large number of fungal associates of orchids was recorded in this study (Figs S1, S2), previous studies, especially those that have restrictions in primer sets and sequencing methods, may have overlooked a wide range of fungal associates that reside in orchid roots (Selosse et al., 2010, 2018).
Evolutionary shifts in trophic modes
Our analyses further show that from the ancestral autotrophic state (Figs 1, 2a,b), shifts from AU or PMH to MH have occurred at least 17 times across the orchid family (Figs S5, S6), accounting for a large fraction of the estimated total of 40 of these shifts across land plants (Merckx & Freudenstein, 2010; Merckx, 2013; Jacquemyn & Merckx, 2019). The occurrence of mycoheterotrophy in orchids exhibits phylogenetic conservatism and is mainly confined to nine tribes in the subfamilies Vanilloideae, Orchidoideae, and Epidendroideae (Figs S5, S6). In addition, a recent study has shown that in the early divergent subfamily Apostasioideae a photosynthetic orchid species is potentially mycoheterotrophic due to its enriched 13C and 15N signatures (Suetsugu & Matsubayashi, 2021). As the 13C and 15N signatures of only few species have been investigated, and because these isotopes cannot detect low degrees of PMH in orchids, especially for those associated with rhizoctonias (Gebauer et al., 2016; Schiebold et al., 2018; Schweiger et al., 2019), the number of shifts to mycoheterotrophy may have been underestimated here.
Transitions to MH occurred dominantly through an intermediate stage of PMH (Figs S5, 6), which is in line with previous phylogenetic studies on narrower phylogenetic scales (Selosse & Roy, 2009; Motomura et al., 2010; Ogura‐Tsujita et al., 2012; Yagame et al., 2016; Jacquemyn & Merckx, 2019). Furthermore, stable isotope studies have confirmed that 13C and 15N signatures of PMH orchids are on a dynamic continuum between AU and MH (Jacquemyn et al., 2017b; Schiebold et al., 2018; May et al., 2020). These findings corroborate the hypothesis that transitions to MH are accomplished by a gradual increase in the level of mycoheterotrophy, rather than by abrupt shifts of AU to MH. This gradual transition has also been suggested in the genus Pyrola in the family Ericaceae, where a similar full range of trophic modes has been observed (Tedersoo et al., 2007; Zimmer et al., 2007; Hynson et al., 2009).
Correlation between symbiotic switches and trophic mode shifts
Ancestral state reconstructions show that symbiotic switches often co‐occur with shifts in trophic modes (Figs 1, 3, S4–S6). Discrete Independent and Dependent models further revealed a strong correlation between transitions in trophic modes and fungal partners across the orchid phylogeny (Table S4). Such correlation has been suggested based on observations in genus‐level analyses (Bidartondo et al., 2004; Selosse et al., 2004; Motomura et al., 2010; Ogura‐Tsujita et al., 2012; Jacquemyn et al., 2016, 2017b; Yagame et al., 2016). Fully mycoheterotrophic orchids primarily associate with either ECM or wood and litter‐decaying SAP fungi, while rhizoctonias dominate the associations of their autotrophic relatives (Motomura et al., 2010; Ogura‐Tsujita et al., 2012; Yagame et al., 2016), suggesting that full or partial loss of photosynthesis selects for different mycorrhizal communities. Therefore, the evolutionary transition from AU over PMH to MH was accompanied by a shift in fungal partners from rhizoctonias to ECM or SAP fungi in the orchid family (Figs 1, 3; Table 1). The latter fungal groups are often detected alongside rhizoctonia‐like fungi in the roots of autotrophic orchids, and here we infer that the association with a broad range of fungal partners was the ancestral state in orchids (Fig. 3). The symbiotic shift to ECM or wood‐ and litter‐decaying saprotrophic fungi is therefore likely an essential predisposition for the evolution of mycoheterotrophy (Table 1). The trophic mode shift to mycoheterotrophy seems thus to go in parallel with increased importance of ECM and SAP fungi while gradually discarding the rhizoctonias that were associated with the autotrophic ancestors.
In arbuscular and ECM systems, several experimental studies have shown that plants can reward greater nutrient‐providing symbionts with increased carbon supplies, and thus ‘choose’ optimal fungal partners from the local environment (Bever et al., 2009; Kiers et al., 2011; Bogar et al., 2019). An increased level of mycoheterotrophy requires an increased dependence on fungal carbon, ultimately replacing photosynthesis in MH plants. Because carbon is the primary resource that non‐photosynthetic mycoheterotrophic plants receive from their mycorrhizal interactions, Taylor & Bruns (1997) hypothesized that the driving force for orchids to change their fungal partners is to obtain a more stable and higher amount of carbon and/or nitrogen. It has been suggested that ECM fungi, which are tightly associated with forest trees, or SAP fungi living on dead wood or decaying leaves are more beneficial partners for mycoheterotrophic orchids in deeply shaded forest habitats than rhizoctonias, which may have limited SAP capabilities (Roberts, 1999). Recent genomic studies have shown that multiple lineages of ECM fungi exhibit a reduced capability to acquire C from soil organic matter and plant cell walls, which is elucidated by rampant loss of genes encoding lignocellulose‐degrading enzymes present in their SAP ancestors (Miyauchi et al., 2020). Interestingly, ECM fungi have evolved novel and species‐specific genes, which may contribute to their tight symbiotic associations with forest trees (Kohler et al., 2015; Hess et al., 2018; Miyauchi et al., 2020), and in this way provide more stable nutrient supplies to nearby orchids species in the tripartite network (Merckx, 2013).
Author contributions
VSFTM initiated and supervised the project. DW compiled the data and performed the analyses with input from HJ, SIFG, and RAV All authors contributed to writing of the manuscript.
Supporting information
Fig. S1 The number of orchid species associated with each fungal family.
Fig. S2 Fungal family composition among orchid species with different trophic modes.
Fig. S3 A comparison between Sanger sequencing and high‐throughput sequencing (HTS) techniques.
Fig. S4 Orchid phylogeny, ancestral states of trophic mode, and symbiotic association.
Fig. S5 Ancestral state reconstruction of the trophic mode under a relaxed definition of partial mycoheterotrophy (PMH) using stochastic character mapping.
Fig. S6 Ancestral state reconstruction of the trophic mode under a strict definition of partial mycoheterotrophy (PMH) using stochastic character mapping.
Fig. S7 Changes of trophic mode and symbiotic association through time.
Notes S1 Additional methodological details and results.
Table S1 Orchid mycorrhiza dataset.
Table S2 The list of orchid species for phylogenetic reconstruction and trait analyses.
Table S3 Phylogenetic signals of symbiotic association and trophic mode using Pagel’s lambda.
Table S4 Phylogenetic correlations between orchid trophic mode and symbiotic association using the Discrete Independent and Dependent models implemented in bayestraits.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank the editor and the four reviewers who provided constructive comments on an earlier version of this manuscript. Funding was provided by the China Scholarship Council (Grant no. 201804910634) and the Ecology Fund of the Royal Netherlands Academy of Arts and Sciences (KNAWWF/807/19039).
References
- Abarenkov K, Henrik Nilsson R, Larsson K‐H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T et al. 2010. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. 2009. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecology Letters 12: 13–21. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Burghardt B, Gebauer G, Bruns TD, Read DJ. 2004. Changing partners in the dark: isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proceedings of the Royal Society of London B: Biological Sciences 271: 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Read DJ. 2008. Fungal specificity bottlenecks during orchid germination and development. Molecular Ecology 17: 3707–3716. [DOI] [PubMed] [Google Scholar]
- Bogar L, Peay K, Kornfeld A, Huggins J, Hortal S, Anderson I, Kennedy P. 2019. Plant‐mediated partner discrimination in ectomycorrhizal mutualisms. Mycorrhiza 29: 97–111. [DOI] [PubMed] [Google Scholar]
- Bollback JP. 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido‐Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC, Tedersoo L. 2018. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytologist 220: 1108–1115. [DOI] [PubMed] [Google Scholar]
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Van Den Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Botanical Journal of the Linnean Society 177: 151–174. [Google Scholar]
- Chomicki G, Bidel LPR, Ming F, Coiro M, Zhang X, Wang Y, Baissac Y, Jay‐Allemand C, Renner SS. 2015. The velamen protects photosynthetic orchid roots against UV‐B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytologist 205: 1330–1341. [DOI] [PubMed] [Google Scholar]
- Dearnaley JDW, Martos F, Selosse M‐A. 2012. Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects. In: Esser K, ed. Fungal associations. Berlin, Germany: Springer, 207–230. [Google Scholar]
- Dressler RL. 2005. How many orchid species? Selbyana 96: 1997–2009. [Google Scholar]
- Duffy KJ, Waud M, Schatz B, Petanidou T, Jacquemyn H. 2019. Latitudinal variation in mycorrhizal diversity associated with a European orchid. Journal of Biogeography 46: 968–980. [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Feijen FAA, Vos RA, Nuytinck J, Merckx VSFT. 2018. Evolutionary dynamics of mycorrhizal symbiosis in land plant diversification. Scientific Reports 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. 2015. Symbiotic options for the conquest of land. Trends in Ecology & Evolution 30: 477–486. [DOI] [PubMed] [Google Scholar]
- Gebauer G, Meyer M. 2003. 15N and 13C natural abundance of autotrophic and myco‐heterotrophic orchids provides insight into nitrogen and carbon gain from fungal association. New Phytologist 160: 209–223. [DOI] [PubMed] [Google Scholar]
- Gebauer G, Preiss K, Gebauer AC. 2016. Partial mycoheterotrophy is more widespread among orchids than previously assumed. New Phytologist 211: 11–15. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens‐Mack J et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proceedings of the Royal Society of London. Series B: Biological Sciences 282: 20151553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SW, Lam VK, Merckx VS. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytologist 214: 48–55. [DOI] [PubMed] [Google Scholar]
- Green PJ. 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82: 711–732. [Google Scholar]
- Gryndler M, Černá L, Bukovská P, Hršelová H, Jansa J. 2014. Tuber aestivum association with non‐host roots. Mycorrhiza 24: 603–610. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Martin FM, Selosse MA, Sanders IR. 2015. Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytologist 205: 1406–1423. [DOI] [PubMed] [Google Scholar]
- Hess J, Skrede I, Chaib De Mares M, Hainaut M, Henrissat B, Pringle A. 2018. Rapid divergence of genome architectures following the origin of an ectomycorrhizal symbiosis in the genus Amanita . Molecular Biology and Evolution 35: 2786–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynson NA, Preiss K, Gebauer G, Bruns TD. 2009. Isotopic evidence of full and partial myco‐heterotrophy in the plant tribe Pyroleae (Ericaceae). New Phytologist 182: 719–726. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Duffy KJ, Selosse M‐A. 2017a. Biogeography of orchid mycorrhizas. In: Tedersoo L, ed. Biogeography of mycorrhizal symbiosis. Tartu, Estonia: Springer, 159–177. [Google Scholar]
- Jacquemyn H, Merckx V, Brys R, Tyteca D, Cammue BPA, Honnay O, Lievens B. 2011. Analysis of network architecture reveals phylogenetic constraints on mycorrhizal specificity in the genus Orchis (Orchidaceae). New Phytologist 192: 518–528. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Merckx VSFT. 2019. Mycorrhizal symbioses and the evolution of trophic modes in plants. Journal of Ecology 107: 1567–1581. [Google Scholar]
- Jacquemyn H, Waud M, Lievens B, Brys R. 2016. Differences in mycorrhizal communities between Epipactis palustris, E. helleborine and its presumed sister species E. neerlandica . Annals of Botany 118: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn H, Waud M, Brys R, Lallemand F, Courty P‐E, Robionek A, Selosse M‐A. 2017b. Mycorrhizal associations and trophic modes in coexisting orchids: an ecological continuum between auto‐ and mixotrophy. Frontiers in Plant Science 8: 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A et al. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A et al. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nature Genetics 47: 410–415. [DOI] [PubMed] [Google Scholar]
- Leake JR. 1994. The biology of myco‐heterotrophic (saprophytic) plants. New Phytologist 127: 171–216. [DOI] [PubMed] [Google Scholar]
- Margulis L. 1991. Symbiogenesis and symbionticism. In: Margulis L, Fester R, eds. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge, MA, USA: MIT Press, 1–14. [PubMed] [Google Scholar]
- Martin FM, Uroz S, Barker DG. 2017. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 356: eaad4501. [DOI] [PubMed] [Google Scholar]
- Martos F, Dulormne M, Pailler T, Bonfante P, Faccio A, Fournel J, Dubois MP, Selosse MA. 2009. Independent recruitment of saprotrophic fungi as mycorrhizal partners by tropical achlorophyllous orchids. New Phytologist 184: 668–681. [DOI] [PubMed] [Google Scholar]
- Martos F, Munoz F, Pailler T, Kottke I, Gonneau C, Selosse MA. 2012. The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Molecular Ecology 21: 5098–5109. [DOI] [PubMed] [Google Scholar]
- May M, Jąkalski M, Novotná A, Dietel J, Ayasse M, Lallemand F, Figura T, Minasiewicz J, Selosse M‐A. 2020. Three‐year pot culture of Epipactis helleborine reveals autotrophic survival, without mycorrhizal networks, in a mixotrophic species. Mycorrhiza 30: 51–61. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT. 2013. Mycoheterotrophy: an introduction. In: Merckx VSFT, ed. Mycoheterotrophy: the biology of plants living on fungi. New York, NY, USA: Springer, 1–17. [Google Scholar]
- Merckx V, Freudenstein JV. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytologist 185: 605–609. [DOI] [PubMed] [Google Scholar]
- Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez‐García M, Morin E, Andreopoulos B, Barry KW, Bonito G et al. 2020. Large‐scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nature Communications 11: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura H, Selosse MA, Martos F, Kagawa A, Yukawa T. 2010. Mycoheterotrophy evolved from mixotrophic ancestors: Evidence in Cymbidium (Orchidaceae). Annals of Botany 106: 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. 2016. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology 20: 241–248. [Google Scholar]
- Ogura‐Tsujita Y, Gebauer G, Hashimoto T, Umata H, Yukawa T. 2009. Evidence for novel and specialized mycorrhizal parasitism: the orchid Gastrodia confusa gains carbon from saprotrophic Mycena . Proceedings of the Royal Society of London. Series B: Biological Sciences 276: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura‐Tsujita Y, Yokoyama J, Miyoshi K, Yukawa T. 2012. Shifts in mycorrhizal fungi during the evolution of autotrophy to mycoheterotrophy in Cymbidium (Orchidaceae). American Journal of Botany 99: 1158–1176. [DOI] [PubMed] [Google Scholar]
- Ogura‐Tsujita Y, Yukawa T. 2008. High mycorrhizal specificity in a widespread mycoheterotrophic plant, Eulophia zollingeri (Orchidaceae). American Journal of Botany 95: 93–97. [DOI] [PubMed] [Google Scholar]
- Ogura‐Tsujita Y, Yukawa T, Kinoshita A. 2021. Evolutionary histories and mycorrhizal associations of mycoheterotrophic plants dependent on saprotrophic fungi. Journal of Plant Research 134: 19–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira SF, Bocayuva MF, Veloso TGR, Bazzolli DMS, da Silva CC, Pereira OL, Kasuya MCM. 2014. Endophytic and mycorrhizal fungi associated with roots of endangered native orchids from the Atlantic forest, Brazil. Mycorrhiza 24: 55–64. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401: 877–884. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible‐jump Markov chain Monte Carlo. American Naturalist 167: 808–825. [DOI] [PubMed] [Google Scholar]
- Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30: 2216–2218. [DOI] [PubMed] [Google Scholar]
- Pirozynski KA, Malloch DW. 1975. The origin of land plants: a matter of mycotrophism. Biosystems 6: 153–164. [DOI] [PubMed] [Google Scholar]
- Põlme S, Abarenkov K, Henrik Nilsson R, Lindahl BD, Clemmensen KE, Kauserud H, Nguyen N, Kjøller R, Bates ST, Baldrian P et al. 2020. FungalTraits: a user‐friendly traits database of fungi and fungus‐like stramenopiles. Fungal Diversity 105: 1–16. [Google Scholar]
- Rasmussen HN. 1995. Terrestrial orchids: from seed to mycotrophic plant. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Rasmussen HN, Rasmussen FN. 2014. Seedling mycorrhiza: a discussion of origin and evolution in Orchidaceae. Botanical Journal of the Linnean Society 175: 313–327. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Roberts P. 1999. Rhizoctonia‐forming fungi: a taxonomic guid. London, UK: Royal Botanic Gardens, Kew. [Google Scholar]
- Schiebold JMI, Bidartondo MI, Karasch P, Gravendeel B, Gebauer G. 2017. You are what you get from your fungi: Nitrogen stable isotope patterns in Epipactis species. Annals of Botany 119: 1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebold JMI, Bidartondo MI, Lenhard F, Makiola A, Gebauer G. 2018. Exploiting mycorrhizas in broad daylight: Partial mycoheterotrophy is a common nutritional strategy in meadow orchids. Journal of Ecology 106: 168–178. [Google Scholar]
- Schneider‐Maunoury L, Leclercq S, Clément C, Covès H, Lambourdière J, Sauve M, Richard F, Selosse M‐A, Taschen E. 2018. Is Tuber melanosporum colonizing the roots of herbaceous, non‐ectomycorrhizal plants? Fungal Ecology 31: 59–68. [Google Scholar]
- Schweiger JMI, Kemnade C, Bidartondo MI, Gebauer G. 2019. Light limitation and partial mycoheterotrophy in rhizoctonia‐associated orchids. Oecologia 189: 375–383. [DOI] [PubMed] [Google Scholar]
- Selosse M‐A, Dubois M‐P, Alvarez N. 2009. Do Sebacinales commonly associate with plant roots as endophytes? Mycological Research 113: 1062–1069. [DOI] [PubMed] [Google Scholar]
- Selosse M‐A, Faccio A, Scappaticci G, Bonfante P. 2004. Chlorophyllous and achlorophyllous specimens of Epipactis microphylla (Neottieae, Orchidaceae) are associated with ectomycorrhizal septomycetes, including truffles. Microbial Ecology 47: 416–426. [DOI] [PubMed] [Google Scholar]
- Selosse M‐A, Le Tacon F. 1998. The land flora: a phototroph‐fungus partnership? Trends in Ecology & Evolution 13: 15–20. [DOI] [PubMed] [Google Scholar]
- Selosse MA, Martos F, Perry BA, Padamsee M, Roy M, Pailler T. 2010. Saprotrophic fungal mycorrhizal symbionts in achlorophyllous orchids: finding treasures among the ‘molecular scraps’? Plant Signaling and Behavior 5: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse M‐A, Roy M. 2009. Green plants that feed on fungi: Facts and questions about mixotrophy. Trends in Plant Science 14: 64–70. [DOI] [PubMed] [Google Scholar]
- Selosse MA, Schneider‐Maunoury L, Martos F. 2018. Time to re‐think fungal ecology? Fungal ecological niches are often prejudged. New Phytologist 217: 968–972. [DOI] [PubMed] [Google Scholar]
- Shefferson RP, Cowden CC, McCormick MK, Yukawa T, Ogura‐Tsujita Y, Hashimoto T. 2010. Evolution of host breadth in broad interactions: Mycorrhizal specificity in East Asian and North American rattlesnake plantains (Goodyera spp.) and their fungal hosts. Molecular Ecology 19: 3008–3017. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457: 818–823. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis. Cambridge, UK: Academic Press. [Google Scholar]
- Suetsugu K, Matsubayashi J. 2021. Evidence for mycorrhizal cheating in Apostasia nipponica, an early‐diverging member of the Orchidaceae. New Phytologist 229: 2302–2310. [DOI] [PubMed] [Google Scholar]
- Szathmáry E, Smith JM. 1995. The major evolutionary transitions. Nature 374: 227–232. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Bruns TD. 1997. Independent, specialized invasions of ectomycorrhizal mutualism by two nonphotosynthetic orchids. Proceedings of the National Academy of Sciences, USA 94: 4510–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Zobel M. 2020. How mycorrhizal associations drive plant population and community biology. Science 367: eaba1223. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Brundrett MC. 2017. Evolution of ectomycorrhizal symbiosis in plants. In: Tedersoo L, ed. Biogeography of mycorrhizal symbiosis. New York, NY, USA: Springer, 407–467. [Google Scholar]
- Tedersoo L, Pellet P, Koljalg U, Selosse MA. 2007. Parallel evolutionary paths to mycoheterotrophy in understorey Ericaceae and Orchidaceae: ecological evidence for mixotrophy in Pyroleae. Oecologia 151: 206–217. [DOI] [PubMed] [Google Scholar]
- Těšitelová T, Kotilínek M, Jersáková J, Joly F‐X, Košnar J, Tatarenko I, Selosse M‐A. 2015. Two widespread green Neottia species (Orchidaceae) show mycorrhizal preference for Sebacinales in various habitats and ontogenetic stages. Molecular Ecology 24: 1122–1134. [DOI] [PubMed] [Google Scholar]
- Thompson JN. 1999. The evolution of species interactions. Science 284: 2116–2118. [DOI] [PubMed] [Google Scholar]
- Thompson J. 2005. The geographic mosaic of coevolution. Chicago, IL, USA: University of Chicago Press. [Google Scholar]
- Waud M, Brys R, Van Landuyt W, Lievens B, Jacquemyn H. 2017. Mycorrhizal specificity does not limit the distribution of an endangered orchid species. Molecular Ecology 26: 1687–1701. [DOI] [PubMed] [Google Scholar]
- Weiß M, Waller F, Zuccaro A, Selosse M. 2016. Sebacinales – one thousand and one interactions with land plants. New Phytologist 211: 20–40. [DOI] [PubMed] [Google Scholar]
- Werner GDA, Cornelissen JHC, Cornwell WK, Soudzilovskaia NA, Kattge J, West SA, Kiers ET. 2018. Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proceedings of the National Academy of Sciences, USA 115: 5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagame T, Ogura‐Tsujita Y, Kinoshita A, Iwase K, Yukawa T. 2016. Fungal partner shifts during the evolution of mycoheterotrophy in Neottia . American Journal of Botany 103: 1630–1641. [DOI] [PubMed] [Google Scholar]
- Yagame T, Yamato M, Mii M, Suzuki A, Iwase K. 2007. Developmental processes of achlorophyllous orchid, Epipogium roseum: from seed germination to flowering under symbiotic cultivation with mycorrhizal fungus. Journal of Plant Research 120: 229–236. [DOI] [PubMed] [Google Scholar]
- Yamato M, Iwase K, Yagame T, Suzuki A. 2005. Isolation and identification of mycorrhizal fungi associating with an achlorophyllous plant, Epipogium roseum (Orchidaceae). Mycoscience 46: 73–77. [Google Scholar]
- Yu G. 2020. Using ggtree to visualize data on tree‐like structures. Current Protocols in Bioinformatics 69: e96. [DOI] [PubMed] [Google Scholar]
- Yukawa T, Ogura‐Tsujita Y, Shefferson RP, Yokoyama J. 2009. Mycorrhizal diversity in Apostasia (Orchidaceae) indicates the origin and evolution of orchid mycorrhiza. American Journal of Botany 96: 1997–2009. [DOI] [PubMed] [Google Scholar]
- Zimmer K, Hynson NA, Gebauer G, Allen EB, Allen MF, Read DJ. 2007. Wide geographical and ecological distribution of nitrogen and carbon gains from fungi in pyroloids and monotropoids (Ericaceae) and in orchids. New Phytologist 175: 166–175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The number of orchid species associated with each fungal family.
Fig. S2 Fungal family composition among orchid species with different trophic modes.
Fig. S3 A comparison between Sanger sequencing and high‐throughput sequencing (HTS) techniques.
Fig. S4 Orchid phylogeny, ancestral states of trophic mode, and symbiotic association.
Fig. S5 Ancestral state reconstruction of the trophic mode under a relaxed definition of partial mycoheterotrophy (PMH) using stochastic character mapping.
Fig. S6 Ancestral state reconstruction of the trophic mode under a strict definition of partial mycoheterotrophy (PMH) using stochastic character mapping.
Fig. S7 Changes of trophic mode and symbiotic association through time.
Notes S1 Additional methodological details and results.
Table S1 Orchid mycorrhiza dataset.
Table S2 The list of orchid species for phylogenetic reconstruction and trait analyses.
Table S3 Phylogenetic signals of symbiotic association and trophic mode using Pagel’s lambda.
Table S4 Phylogenetic correlations between orchid trophic mode and symbiotic association using the Discrete Independent and Dependent models implemented in bayestraits.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


