Abstract
Background
In biological cells, promoters drive gene expression by specific binding of RNA polymerase. They determine the starting position, timing and level of gene expression. Therefore, rational fine-tuning of promoters to regulate the expression levels of target genes for optimizing biosynthetic pathways in metabolic engineering has recently become an active area of research.
Results
In this study, we systematically detected and characterized the common promoter elements in the unconventional yeast Yarrowia lipolytica, and constructed an artificial hybrid promoter library that covers a wide range of promoter strength. The results indicate that the hybrid promoter strength can be fine-tuned by promoter elements, namely, upstream activation sequences (UAS), TATA box and core promoter. Notably, the UASs of Saccharomyces cerevisiae promoters were reported for the first time to be functionally transferred to Y. lipolytica. Subsequently, using the production of a versatile platform chemical isoamyl alcohol as a test study, the hybrid promoter library was applied to optimize the biosynthesis pathway expression in Y. lipolytica. By expressing the key pathway gene, ScARO10, with the promoter library, 1.1–30.3 folds increase in the isoamyl alcohol titer over that of the control strain Y. lipolytica Po1g KU70∆ was achieved. Interestingly, the highest titer increase was attained with a weak promoter PUAS1B4-EXPm to express ScARO10. These results suggest that our hybrid promoter library can be a powerful toolkit for identifying optimum promoters for expressing metabolic pathways in Y. lipolytica.
Conclusion
We envision that this promoter engineering strategy and the rationally engineered promoters constructed in this study could also be extended to other non-model fungi for strain improvement.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-021-02002-z.
Keywords: Y. lipolytica, Metabolic engineering, Hybrid promoter, Isoamyl alcohol, Synthetic promoter
Background
Promoters are one of the most important components in synthetic biology, and well-controlled promoters are very critical for regulating gene expression in eukaryotes. The activity of a promoter is co-regulated by various elements. In yeast, the common promoter elements usually include upstream activation sequences (UAS), TATA box and core promoter [1, 2]. By rational modification of these elements, the activity of promoters can be fine-tuned.
At the beginning of transcription, regulatory signals are transmitted from the UAS to the core promoter, the site where the transcription factors and the RNA polymerase II assembles to form the transcription preinitiation complex (PIC) [3]. The core promoter significantly contributes to the regulation of gene expression and is also the key factor determining the promoter strength. Although core promoters were initially thought to be invariant, researchers have found that they exhibit great structural and functional diversity [4, 5]. TATA box, the recognition site of the transcription factor TATA binding protein (TBP), is one of the first kind of functional elements identified to regulate the promoter strength of the core promoter and typically located 40–120 bp upstream of the transcription start site (TSS) [3, 6]. Mutations in the TATA box usually alter the promoter strength [1, 7–10]. The UAS, which is usually located at the 5′ end of the promoter [3, 11], is also known to have an impact on the strength of the promoter by varying its copy number. By analysing the Yarrowia lipolytica alkaline extracellular protease 2 (XPR2) gene promoter, PXPR2, Madzak et al. identified UAS1B to be the most significant functional element that activates the promoter [12]. Subsequently, evaluation of a hybrid promoter library consisting of a minimal PLEU fragment and different copy numbers of the UAS1B indicates that enhancement in promoter strength is correlated to increased copy number of UAS1B [12]. When present in a promoter, distinct types of UAS can cooperate to control transcription. For example, by combining different UAS elements (UASTEF and UAS1B) in Y. lipolytica, the expression level of a constructed promoter was sevenfold higher than the wild-type promoter [2]. Taken together, by exploring the synergy between various promoter elements and understanding the working mechanism of the promoter, promoters with stronger activity and a wider range of expression levels can be constructed.
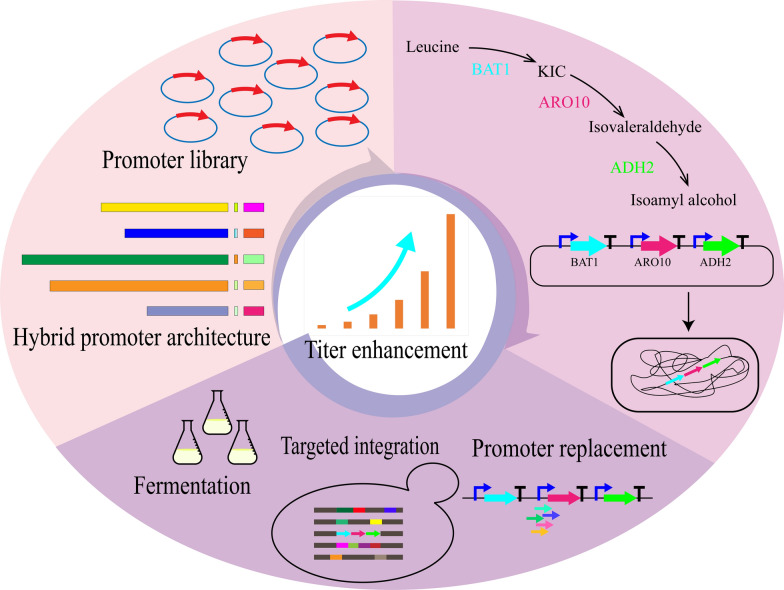
In this study, to explore the mechanism of synergy between various elements in Y. lipolytica, the promoter elements were characterized and rearranged. Consequently, a library of hybrid promoters that enables stable expression and covers a wide range of promoter strength was constructed. Subsequently, we employed the hybrid promoters for promoter engineering of the pathway genes in isoamyl alcohol biosynthesis. Isoamyl alcohol, an important platform chemical, is widely applied in the production of biofuels, fragrances, medicines and fine chemicals [13] and has been produced in recent years by metabolic engineering of diverse microbial cells, such as Escherichia coli, Corynebacterium glutamicum and Aspergillus oryzae [14–16]. While there is a report on improving the production of isoamyl alcohol in Y. lipolytica by metabolic engineering, a native promoter was employed [17]. Therefore, we used this pathway as a testbed and demonstrated the efficacy of our promoter library for optimizing metabolic pathways by significantly improving the isoamyl alcohol titer (Fig. 1). The outcome of this work shows that promoter engineering is an effective strategy for facilitating metabolic engineering efforts to biosynthesize valuable chemicals and our hybrid promoter library is a powerful toolkit for future metabolic engineering work in Y. lipolytica.
Fig. 1.
Strategy of promoter engineering using isoamyl alcohol production as the test study. An artificial hybrid promoter library that covers a wide range of promoter strength was constructed, and applied to optimize the isoamyl alcohol synthesis pathway in Y. lipolytica
Results and discussion
Characterization of native promoters as a basis for the construction of hybrid promoters for Yarrowia lipolytica
The strengths of different native promoters are known to vary greatly in microbes. We systematically characterized the expression of different GFPs to determine the gene that may function as an ideal reporter in Y. lipolytica Po1g KU70∆, which was the host strain for this work. The results show that hrGFPO was the most suitable reporter gene, as its fluorescence was high and consistent. Thus, hrGFPO was utilized for subsequent promoter characterization (Additional file 1: Figures S1, S2).
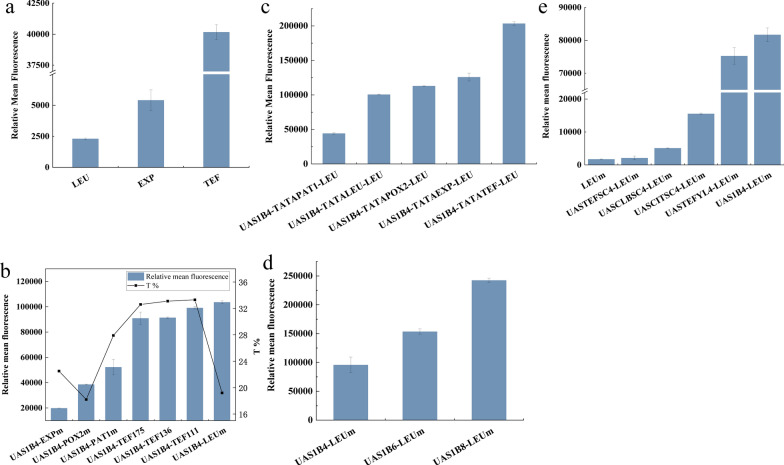
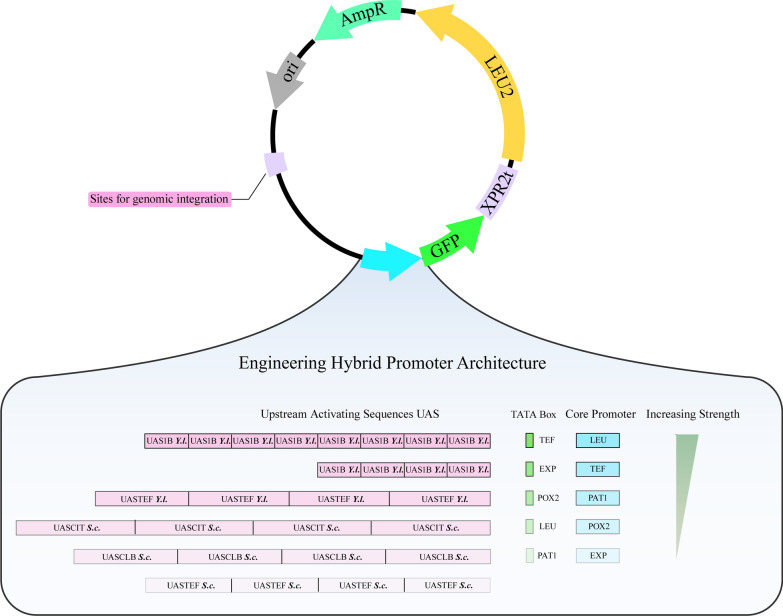
To form a basis for our hybrid promoter library, we sought to use the hrGFPO reporter gene to evaluate the promoter strengths of several commonly used native Y. lipolytica promoters: β-isopropylmalate dehydrogenase (LEU2) gene promoter PLEU, export protein (EXP1) gene promoter PEXP and translation elongation factor-1α (TEF1) gene promoter PTEF. Based on the results of our experiments (Fig. 2a), the relative fluorescence intensities of the corresponding strains from high to low are PTEF > PEXP > PLEU, whereby the strength of PTEF is about an order stronger than both PEXP and PLEU. Subsequently, these promoters were dissected into the various promoter elements, i.e., UAS, TATA box and core promoter. Based on the structures of these native promoters, other known promoter elements were added to build hybrid promoters. In most previous studies on the construction on hybrid promoters, the focus was mainly on the utilization of UAS but few studies explored varying the other promoter elements. Thus, in this study, we investigated the mixing of promoter constituent elements and examined the influence of various combinations on the strengths of the resulting hybrid promoters in Y. lipolytica (Table 1, Fig. 3).
Fig. 2.
Fluorescence strength of the promoters. a Characterization of the native promoters PLEU, PEXP and PTEF. b Characterization of different core promoters, and the relationship between T content upstream of TSS and relative mean fluorescence. Bars represent relative mean fluorescence and lines represent percentage of T content upstream of TSS. c Characterization of different TATA boxes. d Relationship between the copy number of UAS and relative mean fluorescence. e Characterization of different UASs from S. cerevisiae and Y. lipolytica. At least three biological replicates were measured by flow cytometry, and a cell count of 10,000 was analyzed. The error bars represent standard deviation
Table 1.
List of promoters used in this study
| Promoters | UAS type | TATA box | Core promoter | Strength | References |
|---|---|---|---|---|---|
| LEU | + | [40] | |||
| EXP | + | [31] | |||
| TEF | + + | [31] | |||
| LEUm | LEU | LEU | + | [12] | |
| UASTEFSC4-LEUm | UASTEFSC4 | LEU | LEU | + | This study |
| UASCLBSC4-LEUm | UASCLBSC4 | LEU | LEU | + | This study |
| UASCITSC4-LEUm | UASCITSC4 | LEU | LEU | + + | This study |
| UAS1B4-EXPm | UAS1B4 | EXP | EXP | + + | This study |
| UAS1B4-POX2m | UAS1B4 | POX2 | POX2 | + + | This study |
| UAS1B4-TATAPAT1-LEU | UAS1B4 | PAT1 | LEU | + + | This study |
| UAS1B4-PAT1m | UAS1B4 | PAT1 | PAT1 | + + + | This study |
| UASTEFYL4-LEUm | UASTEFYL4 | LEU | LEU | + + + | This study |
| UAS1B4-TEF175 | UAS1B4 | TEF | TEF175 | + + + | This study |
| UAS1B4-TEF136 | UAS1B4 | TEF | TEF136 | + + + | This study |
| UAS1B4-TEF111 | UAS1B4 | TEF | TEF111 | + + + | This study |
| UAS1B4-LEUm | UAS1B4 | LEU | LEU | + + + + | [20] |
| UAS1B4-TATAPOX2-LEU | UAS1B4 | POX2 | LEU | + + + + | This study |
| UAS1B4-TATAEXP-LEU | UAS1B4 | EXP | LEU | + + + + | This study |
| UAS1B6-LEUm | UAS1B6 | LEU | LEU | + + + + + | This study |
| UAS1B4-TATATEF-LEU | UAS1B4 | TEF | LEU | + + + + + | This study |
| UAS1B8-LEUm | UAS1B8 | LEU | LEU | + + + + + | [26] |
+ Means the range of relative mean fluorescence is 0–10,000; + + means the range of relative mean fluorescence is 10,000–50,000; + + + means the range of relative mean fluorescence is 50,000–100,000; + + + + means the range of relative mean fluorescence is 100000–150,000; + + + + + means the range of relative mean fluorescence is > 150,000
Fig. 3.
Strategy of engineering hybrid promoter architecture. Different promoter elements (UAS, TATA box and core promoter) were tested and ligated to the upstream of the reporter gene hrGFPO to characterize promoter strength in this study
Characterization of features in core promoters that influence promoter strength
The core promoter, first identified in the mammalian gene regulatory region, plays a key role in regulating the initiation of gene transcription and is defined as ‘the smallest DNA element for transcription’ [3]. In yeast systems, many studies have proved that the regulation mechanism of the core promoter has a very complex impact on the activity and strength of the promoter, and thus modulate gene expression. For example, in S. cerevisiae, the T content in the core promoter upstream of the TSS has a pronounced influence on the promoter activity. When the gene expression was high, the T content upstream of the TSS was abundant, and the A content downstream of TSS was rich [18]. Thus, we hypothesized that a similar trend exists in Y. lipolytica. Therefore, a series of endogenous core promoters of different lengths and which contain a TATA box, namely, LEU, TEF, EXP, POX2 and PAT1, were selected to calculate the content of T upstream of the TSS and verify the functions of the core promoters in Y. lipolytica. To confirm the function of the core promoters, the UAS1B4 elements which activate gene transcription were linked to the upstream of the core promoter to express the hrGFPO reporter gene for characterizing the promoter strengths by fluorescence. The results show that the promoter strengths of the hybrid promoters that we constructed in general increased with the T content upstream of the TSS (Fig. 2b), implying that the activity of the promoter was related to the upstream T content of the TSS [1, 18]. Nevertheless, two exceptions, LEUm and POX2m, suggest that other factors may be involved in determining the promoters’ activity, e.g., co-regulation by known elements such as UAS and TATA box studied in this study.
We further investigated the effect of promoter truncation on the activity of the TEF core promoter by analyzing the promoters TEF111, TEF136 [1] and TEF175, which are truncated PTEF from the 3′ terminal with lengths of 111, 136 and 175 bp, respectively (Additional file 1: Figure S3). The results indicate that the hybrid promoter strength increases with decreasing length of the core promoter in the promoter PTEF for Y. lipolytica (Fig. 2b). These data suggest that expression level of genes can be regulated largely by both the types and length of core promoters. While there appears to be a relationship between T content and promoter strength in Y. lipolytica, further studies are required to elucidate the specific relationship between the base content and promoter strength.
Modulating the promoter strength by varying the TATA box
Functional elements of the core promoter including TATA box, initiator element, downstream promoter element, TFIIB recognition element and motif ten element have been identified [19, 20]. The sequence lengths of these functional elements are short, the specificities are low and the combinations in various promoters are different. All these functional elements, except the TATA box, are clearly nonconserved in yeast [21, 22]. The TATA box, which is the binding site of the TATA binding protein (TBP), is the first element identified in the core promoter. Previous studies have shown that the TATA box has a significant effect on promoter strength [1]. Therefore, a series of TATA boxes (Table 2) were selected to study their specific performance in promoters in Y. lipolytica. PUAS1B4+LEU, which has the highest activity among the hybrid promoters constructed in the previous section, was selected as the control for engineering. First, we selected several TATA boxes to replace TATA LEU by site-directed mutagenesis. The expression of hrGFPO under the promoter variants was evaluated by fluorescence and it was shown that different TATA boxes significantly affected the promoter strength (Fig. 2c). Notably, the fluorescence intensity of the strain with the hybrid promoter containing TATA TEF was more than twice that of the control strain with TATA LEU. Therefore, the results validate the important role of TATA box in influencing the strength of a promoter and provide a theoretical basis for future promoter engineering studies.
Table 2.
TATA box tested in this study
| TATA box | Sequence |
|---|---|
| LEU | TATATATA |
| TEF | TATAAAA |
| EXP | ATTATATATAA |
| PAT1 | TATATACC |
| POX2 | GTATACTTATATA |
Construction of promoters with various UAS elements from Yarrowia lipolytica and Saccharomyces cerevisiae
The process of transcriptional regulation begins with the recognition of specific sequences by transcription factors (TFs), such as the recognition of UASs by transcriptional activators and upstream repression sequences (URSs) by repressors. Many studies have shown that UAS has a strong influence on transcriptional regulation. Several UASs have been identified in S. cerevisiae, such as UASTEF [23], UASCLB [24] and UASCIT [25]. However, only a few UASs have been identified in Y. lipolytica, among which the UAS1B is the most well-studied. In previous studies, it has been demonstrated that the copy number of UAS has a significant impact on hybrid promoter strength as well [2, 12]. Four tandem UAS1B of PXPR2 and one PLEU core promoter have been combined to construct the strong constitutive promoter PUAS1B4+LEUm [26]. We increased the copy number of UAS and verified that the copy number of UAS is proportional to the hybrid promoter strength (Fig. 2d), which corroborates with published data [26]. In addition, while it has been shown that synthetic terminators can be effectively transferred between S. cerevisiae and Y. lipolytica [27], there is no research on the transferability of promoter elements across diverse yeast species. Therefore, different UASs (UASCIT S.c., UASCLB S.c., UASTEF S.c. and UASTEF Y.l.) [2, 23–25] from S. cerevisiae and Y. lipolytica were used to replace UAS1B4 in PUAS1B4+LEUm at the same copy number to explore the influence of UAS types and origin on hybrid promoter activity. By expressing the hrGFPO gene under the hybrid promoters with different UASs, the activities of the promoters were proved to be significantly affected by the variation in UAS. The relative fluorescence intensity from the GFP expressed from the promoters containing various UASs, from high to low, is UAS1B > UASTEF Y.l. > UASCIT S.c. > UASCLB S.c. > UASTEF S.c. (Fig. 2e). These results demonstrate for the first time that UAS from S. cerevisiae are functional in Y. lipolytica.
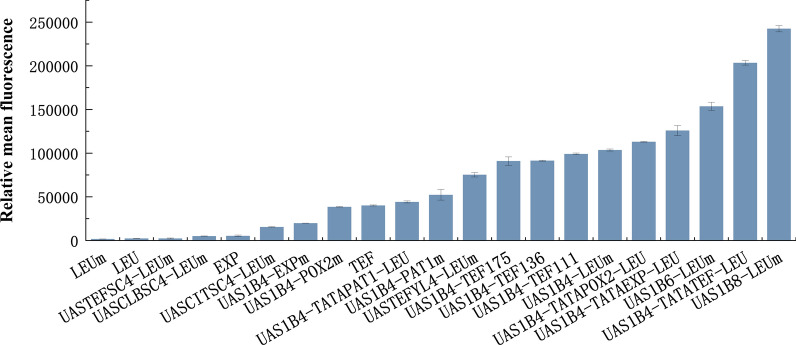
Taken together, we have constructed a library of hybrid promoters with different promoter strengths using various combinations of UASs, TATA boxes and core promoters, as summarized in Fig. 4 and Table 1. As a testbed to demonstrate the application of our hybrid promoter library, we aimed to optimize a biosynthesis pathway, i.e., isoamyl alcohol production, by promoter engineering using our hybrid promoters to regulate gene expression and improve the production level of the target compound.
Fig. 4.
Fluorescence of the promoter library constructed in this study. The hrGFPO was used as a reporter gene for the promoter library constructed, and the fluorescence were detected by the BD Accuri C6 flow cytometer (BD Biosciences) using 488-nm excitation wavelength and FL1 channel. The error bars represent standard deviation
Construction of the isoamyl alcohol overexpression pathway in Yarrowia lipolytica
As an important platform chemical, isoamyl alcohol is a promising biofuel and biochemical with a huge market demand. However, the titer of isoamyl alcohol in Y. lipolytica is quite low natively at a mere 0.37 mg/L (Fig. 5). Thus, the production titer of isoamyl alcohol has much room for improvement and the biosynthesis pathway serves as a suitable testbed for optimization by promoter engineering using our hybrid promoter library.
Fig. 5.
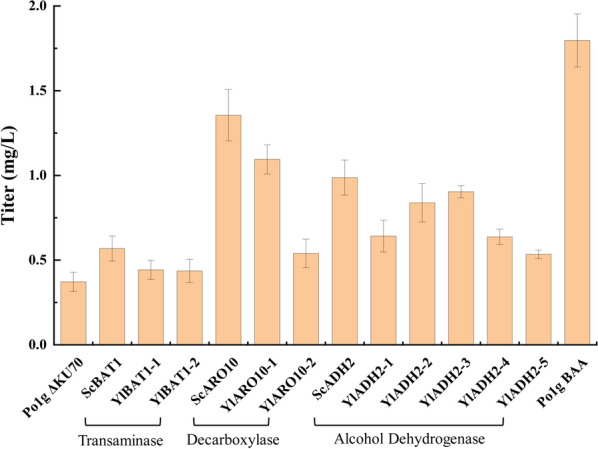
Production of isoamyl alcohol in engineered Y. lipolytica. The cultures were grown in 40 mL YPD medium with an initial OD600 of 0.1 and 10% of n-dodecane in a 250 mL shake flask at 225 rpm and 28 °C for 3 days. The organic phase was analyzed by GC/MS. ScARO10 is the key gene in the heterologous isoamyl alcohol production pathway. The isoamyl alcohol titer of Po1g ScARO10, which expressed ScARO10 under PUAS1B4-LEUm, was 1.36 mg/L, which was 2.7-fold higher than the control strain Po1g KU70Δ. The strain Po1g BAA, which co-expressed ScBAT1, ScARO10 and ScADH2 under PUAS1B4-LEUm, achieved an isoamyl alcohol titer of 1.8 mg/L, which was 3.9-fold that of control strain Po1g KU70Δ. The error bars represent standard deviation
In yeast, isoamyl alcohol is generally produced through the Ehrlich pathway, which usually involves three reaction steps: transamination, decarboxylation and reduction. Twelve genes encoding transaminases (ScBAT1, YlBAT1-1 and YlBAT1-2), decarboxylases (ScARO10, YlARO10-1 and YlARO10-2) and alcohol dehydrogenases (ScADH2, YlADH2-1, YlADH2-2, YlADH2-3, YlADH2-4 and YlADH2-5) were selected and individually overexpressed to determine the key genes of isoamyl alcohol biosynthesis in the Ehrlich pathway. For this purpose, twelve strains overexpressing native and heterologous genes in the Ehrlich pathway were constructed. All genes were individually integrated into the genome of Y. lipolytica Po1g KU70Δ and driven by the constitutive promoter PUAS1B4+LEUm. After 3 days of cultivation, individual overexpression of the pathway genes enhanced the isoamyl alcohol titer in the engineered strains compared to that of the control strain Po1g KU70Δ (Fig. 5). The results show that among the three evaluated classes of enzymes in the Ehrlich pathway, the strains overexpressing decarboxylase genes resulted in the most significant increase in isoamyl alcohol production. Among them, the highest isoamyl alcohol production was obtained by the ScARO10-overexpressed strain, which reached 1.36 mg/L. The strains that overexpressed the transaminase gene ScBAT1 and the dehydrogenase gene ScADH2 also increased the production of isoamyl alcohol moderately. The results indicate that the decarboxylase encoded by ScARO10 is the key limiting step in the Ehrlich pathway, which improved isoamyl alcohol biosynthesis in Y. lipolytica upon overexpression (Fig. 5). To further improve the yield of isoamyl alcohol, the genes ScBAT1, ScARO10 and ScADH2 were chosen for overexpression to construct the strain Po1g BAA. After 3 days of cultivation, the titer of isoamyl alcohol reached 1.8 mg/L, which was 3.9-fold higher than that of the control strain Po1g KU70Δ (Fig. 5). Thus, strain Po1g BAA was selected for subsequent engineering by promoter replacement with our hybrid promoter library.
Application of the hybrid promoter library to improve the isoamyl alcohol biosynthesis pathway
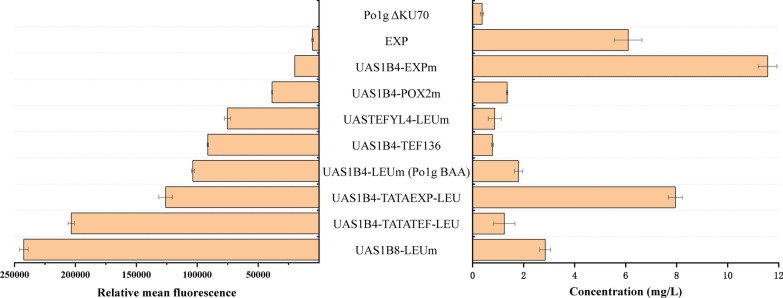
In metabolic engineering, studies have shown that the yield of the target product can be increased by replacing promoters of pathway genes with stronger ones [28, 29]. Therefore, to demonstrate the application of our promoter library for optimizing metabolic pathways, we employed our hybrid promoters in the heterologous isoamyl alcohol pathway of Po1g BAA. Nine representative promoters were selected from our hybrid promoter library to cover a wide spectrum of strengths for optimizing the expression of ScARO10, a gene that can overcome a major bottleneck in the isoamyl alcohol pathway when overexpressed. These constructed strains were cultured for 3 days, and the titer of the isoamyl alcohol was quantified (Fig. 6). It can be seen from the results that the isoamyl alcohol titers do not correlate with the strengths of the promoters used. For example, strain Po1g BA + PUAS1B4+EXPm + ARO10 with a low-activity promoter had the highest isoamyl alcohol titer of 11.57 mg/L, which was about 30.3-fold higher than that of Po1g KU70Δ and 5.4-fold that of Po1g BAA. This result is consistent with the opinion of Dulermo et al. that stronger promoters do not necessarily increase the expression level and/or function of a protein [30]. In addition, we found that although the activity of PEXP was low, several strains containing PEXP elements (PEXP, PUAS1B4-EXPm, PUAS1B4+TATAEXP-LEUm) had higher titers of isoamyl alcohol, suggesting that the elements of the PEXP have greater beneficial effects to the expression of the ARO10 gene. It is unclear why the expression of ARO10 benefitted from the elements of PEXP, which drives the expression of a necessary gene YALI0C12034g that is highly homologous to an S. cerevisiae encoding a non-classical export protein 2 [31]. It is possible that transcriptional/post-transcriptional regulation or protein stability and modification plays a regulatory role in the activity of decarboxylases (ARO10) but more studies are needed to better understand the mechanism between the PEXP and gene expression which resulted in the improved production titer. Nevertheless, our results indicate that different promoters can affect the titer of isomayl alcohol and demonstrate the capability of using our hybrid promoter library for pathway optimization. Attempts to further improve isoamyl alcohol titer may be achieved by applying the hybrid promoter library to other key genes in the pathway. Further understanding of the relationship between promoter elements, genes and gene expression will offer valuable insights to facilitate high production of the target compounds.
Fig. 6.
Comparison of partial promoter strength with isoamyl alcohol titer. Several promoters that cover a range of strengths (left) were selected to replace PUAS1B4-LEUm for overexpressing ScARO10 and the titers of isomayl alcohol were measured (right). The strain with PUAS1B4-EXPm achieved the highest isoamyl alcohol titer of 11.57 mg/L, which was approximately 30.3-fold higher than that of Po1g KU70Δ and 5.4-fold that of Po1g BAA. The error bars represent standard deviation
Conclusions
Promoters are one of the most essential components of synthetic biology for determining protein expression. Compared to prokaryotes, the regulatory mechanism of the promoter structure in eukaryotes is extremely complex [1, 32]. Increasing the promoter strength is a common method to improve gene transcription and protein expression level. However, recent studies have revealed that not all strong promoters can achieve the highest protein expression and activity [30]. We explored the structure and functional characteristics of the promoters in Y. lipolytica, and subsequently constructed a series of promoters which are stable and efficient in this study. This is the first time that the T content upstream of the TSS has been shown to positively correlate with the hybrid promoter strength in Y. lipolytica, although it is worth noting that some core promoter elements, such as POX2m and LEUm, did not conform to the trend. Therefore, the relationship between the T content upstream of the TSS and the promoter strength in Y. lipolytica needs to be further studied. Next, the effects of different UAS elements from S. cerevisiae and Y. lipolytica on promoter strength were investigated and we discovered for the first time that UAS elements can be transferred between yeast species. However, the functions of the UASs from S. cerevisiae were weaker than the native ones from Y. lipolytica (Fig. 2e), and the underlying molecular mechanisms need to be investigated further in future work. Nevertheless, these findings lay the groundwork for the development of hybrid promoters which can be effectively transferred across diverse yeast species.
To demonstrate application of our hybrid promoter library, the isoamyl alcohol production pathway was constructed to serve as a testbed by co-expressing multiple genes from S. cerevisiae and Y. lipolytica. ScARO10, the key gene of the isoamyl alcohol pathway, was selected as the test gene for expression under various hybrid promoters from our library to optimize the enzyme’s expression and activity for enhancing isoamyl alcohol production. Consequently, the titer of the isoamyl alcohol was 30.3-fold higher than the control strain. Although the titer of isoamyl alcohol from Y. lipolytica achieved here is lower compared with other studies [17, 33, 34], it is the first time that promoter engineering has been applied for the biosynthesis of isoamyl alcohol to provide an advanced solution for the production of biofuels and alcohols. Regulation of expression by promoters involves numerous factors, such as temperature, pH and substrate [1, 35]. In the future, we will further study the mechanisms of promoters to construct hybrid promoters with stronger activity and a wider range of expression levels for optimum expression of biosynthesis pathway genes to achieve high-level production of value-added chemicals.
Materials and methods
Strains and media
Escherichia coli strain DH5α was used for all cloning and plasmid propagation, and DH5α was grown at 37 °C in Luria Bertani (LB), and supplemented with ampicillin to final concentration of 100 μg/mL for plasmid propagation. Yarrowia lipolytica strain Po1g KU70Δ, a leucine auxotroph devoid of any secreted protease activity, was used as the base strain in this study. The original plasmid pYLEX1, a pBR322-based monocopy integrative vector [12, 36], is produced by Yeastern Biotech Co., Ltd. Y. lipolytica Po1g KU70Δ was fitted with an integrated pBR322 docking platform. The vector pYLEX1 digested with Spe I or Not I will be inserted to the pBR322 locus of the Po1g KU70Δ strain. After transformation, the positive Y. lipolytica KU70 Δ transformants were selected on YNB-leu plates and subsequently confirmed by genomic DNA PCR analysis. Yarrowia lipolytica Po1g KU70 Δ containing plasmid was routinely cultivated at 28 °C and 225 rpm with YPD media consisting of 20 g/L glucose, 20 g/L peptone, and 10 g/L yeast extract. In this study, PCR primers were synthesized by Genewiz (Jiangsu, China) and are listed in Additional file 1: Table S1, plasmids are listed in Additional file 1: Table S2 and strains used are listed in Additional file 1: Table S3.
Chemicals and enzymes
All restriction enzymes were purchased from New England Biolabs (Beijing, China), 2× Phanta® max master mix, 2× Rapid Taq master mix, ClonExpress® II one step cloning kit, FastPure® Plasmid Mini Kit and FastPure® Gel DNA Extraction Mini Kit were purchased from Vazyme Biotech Co., Ltd. (Nanjing, China), peptone and yeast extract were purchased from Thermo Scientific Oxoid Microbiology Products (Basingstoke, England), isoamyl alcohol and n-dodecane were purchased from Aladdin® (Shanghai, China).
Plasmid construction of promoter library
The GFPuv gene was preserved in this laboratory, and cloned into pYLEX1 with primers GFPuv-F/GFPuv-R (Additional file 1: Table S1) yield plasmid pYLGFPuv (Additional file 1: Table S2). The hrGFP gene and hrGFPO gene were synthesized and cloned into pYLEX1 to yield plasmids pYLhrGFP and pYLhrGFPO (Additional file 1: Table S2), respectively, by Genewiz (Jiangsu, China). The UASCIT S.c.4, UASCLB S.c.4, UASTEF S.c.4, UASTEF Y.l.4, UAS1B6 and UAS1B8 motifs were synthesized and cloned into plasmids pYLhrGFPO to replace UAS1B4 to yield plasmids pYLPUASCITSC4-LEUm + hrGFPO, pYLPUASCLBSC4-LEUm + hrGFPO, pYLPUASTEFSC4-LEUm + hrGFPO, pYLPUASTEFYL4-LEUm + hrGFPO, pYLPUAS1B6-LEUm + hrGFPO and pYLPUAS1B8-LEUm + hrGFPO (Additional file 1: Table S2), respectively, by Genewiz (Jiangsu, China). Three endogenous promoters PLEU, PTEF and PEXP were cloned into vector pYLhrGFPO with primers PLEU-F/LEU-hrGFPO-R, PTEF-F/TEF-hrGFPO-R and PEXP-F/EXP-hrGFPO-R (Additional file 1: Table S1) yield plasmids pYLPLEU + hrGFPO, pYLPTEF + hrGFPO and pYLPEXP + hrGFPO (Additional file 1: Table S2), respectively. The core promoters were amplified by primer pairs PAT1m-F/PAT1-hrGFPO-R, POX2m-F/POX2-hrGFPO-R, EXPm-F/EXP-hrGFPO-R, TEFm111-F/TEF-hrGFPO-R, TEFm136-F/TEF-hrGFPO-R and TEFm175-F/TEF-hrGFPO-R (Additional file 1: Table S1), and then replace the core promoter LEU in PUAS1B4-LEU. These promoters were ligated to pYLhrGFPO in place of the PUAS1B4-LEU to yield plasmids pYLPUAS1B4-PAT1m + hrGFPO, pYLPUAS1B4-POX2m + hrGFPO, pYLPUAS1B4-EXP1m + hrGFPO, pYLPUAS1B4-TEF111 + hrGFPO, pYLPUAS1B4-TEF136 + hrGFPO and pYLPUAS1B4-TEF175 + hrGFPO (Additional file 1: Table S2), respectively. The TATA box LEU in PUAS1B4-LEU was replaced by TATA box TEF, EXP, PAT1 and POX2 using primer pairs TATA TEF-F/LEU-hrGFPO-R, TATA EXP-F/LEU-hrGFPO-R, TATA PAT1-F/LEU-hrGFPO-R and TATA POX2-F/LEU-hrGFPO-R (Additional file 1: Table S1). These hybrid promoters were ligated to pYLhrGFPO in place of the PUAS1B4-LEU to yield plasmids pYLPUAS1B4-TATATEF-LEU + hrGFPO, pYLPUAS1B4-TATAEXP-LEU + hrGFPO, pYLPUAS1B4-TATAPAT1-LEU + hrGFPO and pYLPUAS1B4-TATAPOX2-LEU + hrGFPO (Additional file 1: Table S2), respectively.
All plasmids, linearized by Not I or Spe I, were transformed into competent cells of Y. lipolytica strains using the lithium acetate method [37].
Plasmid construction of exogenous isoamyl alcohol pathway
The transaminase gene (BAT1, GenBank ID: 856615), decarboxylase gene (ARO10, GenBank ID: 851987) and alcohol dehydrogenase gene (ADH2, GenBank ID: 855349) from S. cerevisiae S288C were codon-optimized and synthesized and cloned into pYLEX1 to yield plasmids pYLSCBAT1, pYLSCARO10 and pYLSCADH2 (Additional file 1: Table S2), respectively, by Genewiz (Jiangsu, China). In Y. lipolytica, the homologous sequences that YlBAT1-1 and YlBAT1-2 of ScBAT1 were cloned into pYLEX1 with primers YLBAT1-1-F/YLBAT1-1-R and YLBAT1-2-F/YLBAT1-2-R (Additional file 1: Table S1) to yield plasmids pYLYLBAT1-1 and pYLYLBAT1-2 (Additional file 1: Table S2), respectively. The homologous sequences that YlARO10-1 and YlARO10-2 of ScARO10 were cloned into pYLEX1 with primers YLARO10-1-F/YLARO10-1-R and YLARO10-2-F/YLARO10-2-R (Additional file 1: Table S1) to yield plasmids pYLYLARO10-1 and pYLYLARO10-2 (Additional file 1: Table S2), respectively. The homologous sequences that YlADH2-1, YlADH2-2, YlADH2-3, YlADH2-4 and YlADH2-5 of ScADH2 were cloned into pYLEX1 with primers YLADH2-1-F/YLADH2-1-R, YLADH2-2-F/YLADH2-2-R, YLADH2-3-F/YLADH2-3-R, YLADH2-4-F/YLADH2-4-R and YLADH2-5-F/YLADH2-5-R (Additional file 1: Table S1) to yield plasmids pYLYLADH2-1, pYLYLADH2-2, pYLYLADH2-3, pYLYLADH2-4 and pYLYLADH2-5 (Additional file 1: Table S2), respectively.
The expression cassettes of ScARO10 and ScADH2 were cloned into pYLSCBAT1 with primers BDH-ADH2-F/BDH-ADH2-R and BDH-ARO10-F/ BDH-ARO10-R (Additional file 1: Table S1) to yield plasmid pYLBAA (Additional file 1: Table S2). All plasmids, linearized by Not I or Spe I, were transformed into competent cells of Y. lipolytica strains using the lithium acetate method [37].
Expressing the isoamyl alcohol synthesis pathway using the promoter library
Several promoters from the promoter library were used to express the ARO10 gene which is the key gene in the isoamyl alcohol pathway. The promoters PEXP and PUAS1B4+EXPm were amplified by primers BDH-ARO10-F/PEXP-ARO10-R (Additional file 1: Table S1), and then ligated to ScARO10 in pYLBAA to yield plasmid pYLBA + PEXP + ARO10 and pYLBA + PUAS1B4-EXPm + ARO10 (Additional file 1: Table S2), respectively. The promoters PUAS1B4-POX2m and PUAS1B4-TEF136 were amplified by primers BDH-ARO10-F/POX2-ARO10-R and BDH-ARO10-F/PTEF-ARO10-R (Additional file 1: Table S1), and then ligated to ScARO10 in pYLBAA to yield plasmid pYLBA + PUAS1B4-POX2m + ARO10 and pYLBA + PUAS1B4-TEF136 + ARO10 (Additional file 1: Table S2), respectively. The promoters PUASTEFLY4-LEUm, PUAS1B4-TATAEXP-LEU, PUAS1B4-TATATEF-LEU and PUAS1B8- LEUm were amplified by primers BDH-ARO10-F/PLEU-ARO10-R (Additional file 1: Table S1), and then ligated to ScARO10 in pYLBAA to yield plasmids pYLBA + PUASTEFLY4-LEUm + ARO10, pYLBA + PUAS1B4-TATAEXP-LEU + ARO10, pYLBA + PUAS1B4-TATATEF-LEU + ARO10 and pYLBA + PUAS1B8-LEUm + ARO10 (Additional file 1: Table S2), respectively.
All plasmids, linearized by Not I or Spe I, were transformed into competent cells of Y. lipolytica strains using the lithium acetate method [38].
Yeast strain construction
The competent cell scheme and transformation method are referred to Pang et al.[39]. After selection, the following engineered Y. lipolytica strains were generated: Po1g PUAS1B4-LEUm + GFPuv, Po1g PUAS1B4-LEUm + hrGFP, Po1g PUAS1B4-LEUm + hrGFPO, Po1g PUAS1B6-LEUm + hrGFPO, Po1g PUAS1B8-LEUm + hrGFPO, Po1g PLEUm + hrGFPO, Po1g PLEU + hrGFPO, Po1g PEXP + hrGFPO, Po1g PTEF + hrGFPO, Po1g PUAS1B4-EXPm + hrGFPO, Po1g PUAS1B4-POX2m + hrGFPO, Po1g PUAS1B4-PAT1m + hrGFPO, Po1g PUAS1B4-TEF111 + hrGFPO, Po1g PUAS1B4-TEF136 + hrGFPO, Po1g PUAS1B4-TEF175 + hrGFPO, Po1g PUAS1B4-TATAPAT1-LEUm + hrGFPO, Po1g PUAS1B4-TATAPOX2-LEUm + hrGFPO, Po1g PUAS1B4-TATAEXP-LEUm + hrGFPO, Po1g PUAS1B4-TATATEF-LEUm + hrGFPO, Po1g PUASTEFSC4-LEUm + hrGFPO, Po1g PUASCITSC4-LEUm + hrGFPO, Po1g PUASCLBSC4-LEUm + hrGFPO, Po1g PUASTEFYL4-LEUm + hrGFPO, Po1g ScBAT1, Po1g YlBAT1-1, Po1g YlBAT1-2, Po1g ScARO10, Po1g YlARO10-1, Po1g YlARO10-2, Po1g ScADH2, Po1g YlADH2-1, Po1g YlADH2-2, Po1g YlADH2-3, Po1g YlADH2-4, Po1g YlADH2-5, Po1g BAA, Po1g BA + PEXP + ARO10, Po1g BA + PUAS1B4-EXPm + ARO10, Po1g BA + PUAS1B4-POX2m + ARO10, Po1g BA + PUASTEFYL4-LEUm + ARO10, Po1g BA + PUAS1B4-TEF136 + ARO10, Po1g BA + PUAS1B4-LEUm + ARO10, Po1g BA + PUAS1B4-TATAEXP-LEU + ARO10, Po1g BA + PUAS1B4-TATATEF-LEU + ARO10, Po1g BA + PUAS1B8-LEUm + ARO10 (Additional file 1: Table S3).
The Y. lipolytica strains Po1g PUAS1B4-LEUm + GFPuv, Po1g PUAS1B4-LEUm + hrGFP, Po1g PUAS1B4-LEUm + hrGFPO, Po1g PUASTEFYL4-LEUm + hrGFPO, Po1g ScBAT1, Po1g YlBAT1-1, Po1g YlBAT1-2, Po1g ScARO10, Po1g YlARO10-1, Po1g YlARO10-2, Po1g ScADH2, Po1g YlADH2-1, Po1g YlADH2-2, Po1g YlADH2-3, Po1g YlADH2-4 and Po1g YlADH2-5 were verified by the primers pYL-F/pYL-R (Additional file 1: Table S1). The Y. lipolytica strain Po1g BAA was verified by the primers ADH2-CX-4–1/BAT1-YZ-R (Additional file 1: Table S1). The Y. lipolytica strains Po1g PUAS1B6-LEUm + hrGFPO, Po1g PUAS1B8-LEUm + hrGFPO, Po1g PLEUm + hrGFPO, Po1g PLEU + hrGFPO, Po1g PEXP + hrGFPO, Po1g PTEF + hrGFPO, Po1g PUAS1B4-EXPm + hrGFPO, Po1g PUAS1B4-POX2m + hrGFPO, Po1g PUAS1B4-PAT1m + hrGFPO, Po1g PUAS1B4-TEF111 + hrGFPO, Po1g PUAS1B4-TEF136 + hrGFPO, Po1g PUAS1B4-TEF175 + hrGFPO, Po1g PUAS1B4-TATAPAT1-LEUm + hrGFPO, Po1g PUAS1B4-TATAPOX2-LEUm + hrGFPO, Po1g PUAS1B4-TATAEXP-LEUm + hrGFPO, Po1g PUAS1B4-TATATEF-LEUm + hrGFPO, Po1g PUASTEFSC4-LEUm + hrGFPO, Po1g PUASCITSC4-LEUm + hrGFPO and Po1g PUASCLBSC4-LEUm + hrGFPO were verified by the primers CX-2/P-CX-R (Additional file 1: Table S1). The Y. lipolytica strains Po1g BA + PEXP + ARO10, Po1g BA + PUAS1B4-EXPm + ARO10, Po1g BA + PUAS1B4-POX2m + ARO10, Po1g BA + PUASTEFYL4-LEUm + ARO10, Po1g BA + PUAS1B4-TEF136 + ARO10, Po1g BA + PUAS1B4-LEUm + ARO10, Po1g BA + PUAS1B4-TATAEXP-LEU + ARO10, Po1g BA + PUAS1B4-TATATEF-LEU + ARO10 and Po1g BA + PUAS1B8-LEUm + ARO10 were verified by the primers ADH2-CX-F/A-CX-R (Additional file 1: Table S1).
Flow cytometry
The green fluorescent proteins GFPuv, hrGFP and hrGFPO were selected as reporter proteins. At least three biological replicates were measured by flow cytometry. The colonies of transformants were selected from plates and grew in 5 mL of fresh YPD medium in tubes for 24 h. After that, the seed culture was diluted to OD600 0.1 in 250 mL flasks containing 40 mL YPD medium. The cultures were cultivated with shaking at 225 rpm and 28 °C for 3 days. Before flow cytometric analysis, the cultures were centrifuged at 12,000 rpm for 1 min, and washed in 0.1 M phosphate-buffered saline (PBS), then resuspended in PBS. A cell count of 10,000 was analyzed with a BD Accuri C6 flow cytometer (BD Biosciences) using 488-nm excitation wavelength and FL1 channel for fluorescence detection. The CFlow software was used to analyze the data and compute mean fluorescence values. The background fluorescence was normalized when calculating the relative mean fluorescence.
GC/MS analysis of isoamyl alcohol produced in the engineered Yarrowia lipolytica strains
The engineered Y. lipolytica transformants were selected from plate and prepared in 5 mL of fresh YPD medium in tube for 24 h. The seed culture solution was inoculated to 250 mL flasks containing 40 mL of YPD medium, starting from OD600 0.1. The cultures were shaken at 225 rpm and 28 °C for 3 days. To extract isoamyl alcohol from the cultures, 10% n-dodecane was added to the cultures, and the mixture was vortexed for 3 min, then centrifuged at 7500 rpm for 5 min. The organic phase of 1ul was detected by GC/MS using an Agilent 7890B GC with an 5977B MSD equipped with a HP-5MS column (60 m × 0.25 mm × 0.25 μm, Agilent, Santa Clara, CA, USA). GC oven temperature was initially held at 60 °C for 2 min, and then ramped to 140 °C at a rate of 5 °C/min. It was then subsequently ramped at 10 °C/min to 280 °C and held for 5 min. The split ratio was 10:1. Helium was used as the carrier gas, with an inlet pressure of 13.8 psi. The injector was maintained at 280 °C and the ion source temperature was set to 230 °C. Final data analysis was achieved using MassHunter Workstation Software (Agilent, Santa Clara, CA, USA).
Supplementary Information
Additional file 1: Figure S1. Screening of reporter genes. Figure S2. Characterization of different green fluorescent proteins expressed under the promoter PUAS1B4-LEUm. Figure S3. Schematic of theTEF core promoter. Figure S4. Time course of fluorescence by Po1g PUAS1B4-LEUm + hrGFPO. Table S1. Primers used in PCR. Table S2. Plasmids used in this study. Table S3. Strains used in this study.
Acknowledgements
This work was supported by the Natural Science Foundation of Tianjin, China (17JCYBJC40800), the Research Foundation of Tianjin Municipal Education Commission, China (2017ZD03), the Innovative Research Team of Tianjin Municipal Education Commission, China (TD13-5013), Tianjin Municipal Science and Technology Project (18PTSYJC00140, 19PTSYJC00060), Startup Fund for ‘Haihe Young Scholars’ of Tianjin University of Science and Technology, the Thousand Young Talents Program of Tianjin, China.
Abbreviations
- GC/MS
Gas chromatography/mass spectrometry
- OD600
Optical density at 600 nm
- LB medium
0.5% Yeast extract, 1% tryptone and 1% NaCl
- YPD medium
1% Yeast extract, 2% peptone and 2% glucose
- YNB plate
2% Glucose, 0.67% yeast nitrogen base without amino acids and 2% agar
- PCR
Polymerase chain reaction
- YlBAT1-1
YALI0_D01265g
- YlBAT1-2
YALI0_F19910g
- YlARO10-1
YALI0_D06930g
- YlARO10-2
YALI0_E07325g
- YlADH2-1
YALI0_A16379g
- YlADH2-2
YALI0_D25630g
- YlADH2-3
YALI0_E17787g
- YlADH2-4
YALI0_A15147g
- YlADH2-5
YALI0_E07766g
Authors’ contributions
AQY, JLF, DGX and CYZ conceived and designed the study. YZ, SQL, ZHL, BXZ and SHW performed plasmid and strain construction, and fermentation experiments. AQY, JLF, DGX, CYZ revised the manuscript. All authors read and approved the final manuscript.
Funding
The Natural Science Foundation of Tianjin, China (17JCYBJC40800), the Research Foundation of Tianjin Municipal Education Commission, China (2017ZD03), the Innovative Research Team of Tianjin Municipal Education Commission, China (TD13-5013), Tianjin Municipal Science and Technology Project (18PTSYJC00140, 19PTSYJC00060), Startup Fund for “Haihe Young Scholars” of Tianjin University of Science and Technology, the Thousand Young Talents Program of Tianjin, China.
Availability of data and materials
All relevant data generated or analyzed during this study were included in this published article.
Declarations
Ethics approval and consent to participate
This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors give consent to publish the research in Biotechnology for Biofuels.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Zhaoa and Shiqi Liu contributed equally to this work
Contributor Information
Yu Zhao, Email: zhaoyu@mail.tust.edu.cn.
Shiqi Liu, Email: liushiqi@mail.tust.edu.cn.
Zhihui Lu, Email: zhihui1997@mail.tust.edu.cn.
Baixiang Zhao, Email: zhaobaixiang@mail.tust.edu.cn.
Shuhui Wang, Email: wsh150697@mail.tust.edu.cn.
Cuiying Zhang, Email: Cyzhangcy@tust.edu.cn.
Dongguang Xiao, Email: Xiao99@tust.edu.cn.
Jee Loon Foo, Email: jeeloon.foo@nus.edu.sg.
Aiqun Yu, Email: yuaiqun@tust.edu.cn.
References
- 1.Shabbir Hussain M, Gambill L, Smith S, Blenner MA. Engineering promoter architecture in oleaginous yeast Yarrowia lipolytica. ACS Synth Biol. 2016;5(3):213–223. doi: 10.1021/acssynbio.5b00100. [DOI] [PubMed] [Google Scholar]
- 2.Blazeck J, Reed B, Garg R, Gerstner R, Pan A, Agarwala V, Alper HS. Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Appl Microbiol Biotechnol. 2013;97(3):3037–3052. doi: 10.1007/s00253-012-4421-5. [DOI] [PubMed] [Google Scholar]
- 3.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189(3):705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16(16):2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 5.Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15(19):2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- 7.Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 8.Grosveld GC, Shewmaker CK, Jat P, Flavell RA. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- 9.Hu SL, Manley JL. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci USA. 1981;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasylyk B, Derbyshire R, Guy A, Molko D, Roget A, Téoule R, Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci USA. 1980;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fantino E, Marguet D, Lauquin GJ. Downstream activating sequence within the coding region of a yeast gene: specific binding in vitro of RAP1 protein. Mol Gen Genet. 1992;236(1):65–75. doi: 10.1007/BF00279644. [DOI] [PubMed] [Google Scholar]
- 12.Madzak C, Tréton B, Blanchin-Roland S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J Mol Microbiol Biotechnol. 2000;2(2):207–216. [PubMed] [Google Scholar]
- 13.Jang YS, Kim B, Shin JH, Choi YJ, Choi S, Song CW, Lee J, Park HG, Lee SY. Bio-based production of C2–C6 platform chemicals. Biotechnol Bioeng. 2012;109(10):2437–2459. doi: 10.1002/bit.24599. [DOI] [PubMed] [Google Scholar]
- 14.Connor MR, Liao JC. Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol. 2008;74(18):5769–5775. doi: 10.1128/AEM.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogt M, Brüsseler C, Ooyen JV, Bott M, Marienhagen J. Production of 2-methyl-1-butanol and 3-methyl-1-butanol in engineered Corynebacterium glutamicum. Metab Eng. 2016;38:436–445. doi: 10.1016/j.ymben.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita N, Motoyoshi T, Nishimura A. Molecular cloning of the isoamyl alcohol oxidase-encoding gene (mreA) from Aspergillus oryzae. J Biosci Bioeng. 2000;89(6):522–527. doi: 10.1016/S1389-1723(00)80050-X. [DOI] [PubMed] [Google Scholar]
- 17.Celinska E, Borkowska M, Bialas W, Kubiak M, Korpys P, Archacka M, Ledesma-Amaro R, Nicaud JM. Genetic engineering of Ehrlich pathway modulates production of higher alcohols in engineered Yarrowia lipolytica. FEMS Yeast Res. 2019 doi: 10.1093/femsyr/foy122. [DOI] [PubMed] [Google Scholar]
- 18.Lubliner S, Keren L, Segal E. Sequence features of yeast and human core promoters that are predictive of maximal promoter activity. Nucleic Acids Res. 2013;41(11):5569–5581. doi: 10.1093/nar/gkt256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2009;339(2):225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 21.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116(5):699–709. doi: 10.1016/S0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 22.Sugihara F, Kasahara K, Kokubo T. Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39(1):59–75. doi: 10.1093/nar/gkq741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazeck J, Garg R, Reed B, Alper HS. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng. 2012;109(11):2884–2895. doi: 10.1002/bit.24552. [DOI] [PubMed] [Google Scholar]
- 24.Van Slyke C, Grayhack EJ. The essential transcription factor Reb1p interacts with the CLB2 UAS outside of the G2/M control region. Nucleic Acids Res. 2003;31(15):4597–4607. doi: 10.1093/nar/gkg638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenkrantz M, Kell CS, Pennell EA, Webster M, Devenish LJ. Distinct upstream activation regions for glucose-repressed and derepressed expression of the yeast citrate synthase gene CIT1. Curr Genet. 1994;25(3):185–195. doi: 10.1007/BF00357161. [DOI] [PubMed] [Google Scholar]
- 26.Blazeck J, Liu LQ, Redden H, Alper HS. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77(22):7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curran KA, Morse NJ, Markham KA, Wagman AM, Gupta A, Alper HS. Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synth Biol. 2015;4(7):824–832. doi: 10.1021/sb5003357. [DOI] [PubMed] [Google Scholar]
- 28.Hong SP, Seip J, Walters-Pollak D, Rupert R, Jackson R, Xue Z, Zhu Q. Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter. Yeast. 2012;29(2):59–72. doi: 10.1002/yea.1917. [DOI] [PubMed] [Google Scholar]
- 29.Xiong X, Chen S. Expanding toolbox for genes expression of Yarrowia lipolytica to include novel inducible, repressible, and hybrid promoters. ACS Synth Biol. 2020;9(8):2208–2213. doi: 10.1021/acssynbio.0c00243. [DOI] [PubMed] [Google Scholar]
- 30.Dulermo R, Brunel F, Dulermo T, Ledesma-Amaro R, Vion J, Trassaert M, Thomas S, Nicaud JM, Leplat C. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb Cell Fact. 2017;16(1):31. doi: 10.1186/s12934-017-0647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damude HG, Gillies PJ, Macool DJ, Picataggio SK, Pollak DWM, Ragghianti JJ, Xue Z. High eicosapentaenoic acid producing strains of Yarrowia lipolytica. United States patent US 8518674 B2. 2013.
- 32.Portela RM, Vogl T, Kniely C, Fischer JE, Oliveira R, Glieder A. Synthetic core promoters as universal parts for fine-tuningexpression in different yeast species. ACS Synth Biol. 2017;6(3):471–484. doi: 10.1021/acssynbio.6b00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dombrink-Kurtzman MA. A gene having sequence homology to isoamyl alcohol oxidase is transcribed during patulin production in Penicillium griseofulvum. Curr Microbiol. 2008;56(3):224–228. doi: 10.1007/s00284-007-9061-0. [DOI] [PubMed] [Google Scholar]
- 34.Yuan J, Chen X, Mishra P, Ching CB. Metabolically engineered Saccharomyces cerevisiae for enhanced isoamyl alcohol production. Appl Microbiol Biotechnol. 2017;101(1):465–474. doi: 10.1007/s00253-016-7970-1. [DOI] [PubMed] [Google Scholar]
- 35.Madzak C. Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol. 2015;99(11):4559–4577. doi: 10.1007/s00253-015-6624-z. [DOI] [PubMed] [Google Scholar]
- 36.Barth G. Yarrowia lipolytica: biotechnological applications. Berlin: Springer; 2013. [Google Scholar]
- 37.Yu AQ, Pratomo N, Ng TK, Ling H, Cho HS, Leong SS, Chang MW. Genetic engineering of an unconventional yeast for renewable biofuel and biochemical production. J Vis Exp. 2016;20(115):54371. doi: 10.3791/54371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Zhu K, Miao L, Rong LX, Zhao Y, Li SL, Ma LJ, Li JX, Zhang CY, Xiao DG, Foo JL, Yu A. Simultaneous improvement of limonene production and tolerance in Yarrowia lipolytica through tolerance engineering and evolutionary engineering. ACS Synth Biol. 2021;10(4):884–896. doi: 10.1021/acssynbio.1c00052. [DOI] [PubMed] [Google Scholar]
- 39.Pang YR, Zhao YK, Li SL, Zhao Y, Li J, Hu ZH, Zhang CY, Xiao DG, Yu AQ. Engineering the oleaginous yeast Yarrowia lipolytica to produce limonene from waste cooking oil. Biotechnol Biofuels. 2019;12:241. doi: 10.1186/s13068-019-1580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xuan JW, Fournier P, Declerck N, Chasles M, Gaillardin C. Overlapping reading frames at the LYS5 locus in the yeast Yarrowia lipolytica. Mol Cell Biol. 1990;10(9):4795–4806. doi: 10.1128/mcb.10.9.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Screening of reporter genes. Figure S2. Characterization of different green fluorescent proteins expressed under the promoter PUAS1B4-LEUm. Figure S3. Schematic of theTEF core promoter. Figure S4. Time course of fluorescence by Po1g PUAS1B4-LEUm + hrGFPO. Table S1. Primers used in PCR. Table S2. Plasmids used in this study. Table S3. Strains used in this study.
Data Availability Statement
All relevant data generated or analyzed during this study were included in this published article.