Abstract
There is a growing awareness that traits do not evolve individually but rather are organized as modular networks of covarying traits. Although the importance of multi‐trait correlation has been linked to the ability to evolve in response to new environmental conditions, the evolvability of the network itself has to date rarely been assessed experimentally. By following the evolutionary dynamics of a model bacterium adapting to plant roots, we demonstrate that the whole structure of the trait correlation network is highly dynamic. We experimentally evolved Pseudomonas protegens, a common rhizosphere dweller, on the roots of Arabidopsis thaliana. We collected bacteria at regular intervals and determined a range of traits linked to growth, stress resistance, and biotic interactions. We observed a rapid disintegration of the original trait correlation network. Ancestral populations showed a modular network, with the traits linked to resource use and stress resistance forming two largely independent modules. This network rapidly was restructured during adaptation, with a loss of the stress resistance module and the appearance of new modules out of previously disconnected traits. These results show that evolutionary dynamics can involve a deep restructuring of phenotypic trait organization, pointing to the emergence of novel life history strategies not represented in the ancestral phenotype.
Keywords: Bacteria, experimental evolution, rhizosphere, trait correlation network
Most microorganisms are typically challenged by rapidly shifting environmental conditions, making long‐term survival dependent on the ability to evolve rapidly to the new conditions. Such adaptability is of special relevance for host‐associated microorganisms that must cope with multiple challenges associated with their lifestyle (Mandel et al. 2009; Damkiaer et al. 2013). Evolution to new environmental conditions requires an optimization of a complex range of traits linked to life history (Blount et al. 2012; Folkesson et al. 2012; Meyer et al. 2012; Winstanley et al. 2016). Microbial adaptation to a new environment can for instance result in alterations of traits linked to growth, stress resistance, or secondary metabolism (Winstanley et al. 2016). To date, most studies addressing this issue have reported the evolution of patterns for specific traits of interest, but have rarely addressed evolution at the level of a complex phenotype encompassing numerous coordinated traits. We purport that single‐trait approaches overlook an important part of the evolutionary dynamics: traits may not evolve individually, rather, groups of traits may evolve in correlated patterns, a phenomenon that has been referred to as phenotypic integration (Pigliucci 2003). Traits integration can emerge from a range of biological processes including developmental, physiological, and functional interactions (Hallgrímsson and Hall 2005; Tonsor and Scheiner 2007; Herrera ). Phenotypic integration can be found across a broad range of organisms, including plants, animals, and bacteria. It has also been described with respect to different theoretical frameworks such as allometric networks (Klingenberg 2008), modular metabolic networks (Ravasz et al. 2002), or gene expression networks (Barabási and Oltvai 2004).
It remains an open debate whether phenotypic integration facilitates or constrains adaptations (Klingenberg 2008). In their natural environment, bacteria need to deal with multiple environmental constraints at once, such as resource limitation, abiotic stressors, or the presence of natural enemies. The more traits required for optimizing adaptation to a range of given stressors, the slower the evolution, a process predicted by a geometrical model and coined the “cost of complexity” (Orr 2000). A further development of this model suggested that the bundling traits into co‐regulated modules might reduce such costs, thereby speeding up the rate of evolution (Welch and Waxman 2003). A modularly structured trait covariation network allows for relatively independent evolution of each module, and ensures the robustness of adaptation (Berg 1960). However, integration patterns present in an organism will also constrain and channel its future evolutionary trajectories (Jernigan et al. 1994; Pigliucci 2003). For example, if a novel environmental change were to shift the optimum of trait A, but not the correlated trait B or shift both in opposite directions, then these bundled traits would greatly constraint the population's adaption ability, at least in the short to medium time frame. In summary, the level of phenotypic trait integration may provide new insights into the dynamics of adaptation and the evolutionary pressures driving adaptation.
Although the potential of rewiring and fine‐tuning of gene regulatory networks during adaptation has been emphasized as a core component of evolutionary dynamics (Hindré et al. 2012; Damkiaer et al. 2013), very little data are available on the real‐time evolutionary dynamics of phenotypic integration (Pigliucci 2003). Computational simulations coupled with experimental validation have revealed that phenotype integration patterns can be very dynamic and may present an essential part of the evolutionary response to new conditions (Kashtan and Alon 2005; Parter et al. 2007). This evolvable feature of phenotype integration patterns is crucial for survival under fluctuating conditions. And indeed, during long‐term adaptation, bacterial populations undergo evolutionary remodeling of global regulatory networks, as measured at the gene expression level (Damkiaer et al. 2013). Approaching evolutionary dynamics from the vantage point of trait dimensions that can emerge and vanish during adaption may open new avenues to predict evolution. We propose here to combine experimental evolution, scrutinizing parallel, evolving microbial populations, with theoretical considerations to reveal the dynamics of global phenotypic trait correlation patterns at the population level. To achieve this, we focus on a model system in which a plant root‐associated bacterial population is challenged with adapting to a new host. In this study, evolution of the root‐associated bacterium Pseudomonas protegens CHA0 in the rhizosphere of sterile Arabidopsis thaliana Col‐0 (later: Arabidopsis) was followed for six plant growth cycles. The use of a simplified two‐organism system allows one to zoom in on the importance of plant‐microbe interactions for evolution, while avoiding confounding factors emerging from interactions with soil material and other soil microorganisms.
We selected bacterial adaptation to the rhizosphere as a model system because it has far‐reaching implications for plant productivity. The rhizosphere, the space on and surrounding plant roots, represents a hotspot of plant‐microbe interactions that mediate a wide range of plant and bacterial traits. Plants deposit up to 40% of their photosynthetically fixed carbon into the rhizosphere, which renders this small zone around the roots one of the most energy‐rich habitats on Earth (Bais et al. 2006). In return, some of these enriched microorganisms can affect plant physiology, and the microbial community extends the functional repertoire of the plant (Vandenkoornhuyse et al. 2015). Understanding the dynamics of bacterial evolution on plant roots is therefore crucial to the development of more sustainable agricultural management strategies using microorganisms as a replacement for, or complement to, agrochemicals.
At the end of each cycle, bacterial populations were harvested and passed to a new, sterile plant. Sixteen bacterial colonies were isolated at random from the ancestral population and each of the five replicate evolutionary lines at the end of plant growth cycle two, four, and six. We subsequently assessed key life history traits, including ability to use different carbon sources representative of the root environment, tolerance to abiotic and biotic stressors, and the production of public goods, such as siderophores and exoproteases involved in interaction with the host. This dataset was used to build a trait correlation network for the ancestral and the evolved populations. We expected two possible outcomes.
Individual traits may evolve but retain their original trait integration patterns due to different levels of existing constraints. In that case, trait correlation networks remain identical, even if several different phenotypes emerge during the evolutionary process.
The integration pattern between traits will change, causing different traits to associate into a new correlation network. In this case, we expect that not only the expression of each trait, but also the topology of the network will change, with potential appearance or disappearance of modules of trait correlation.
Materials and Methods
EXPERIMENTAL EVOLUTION
We used green fluorescent protein (GFP)‐labeled Pseudomonas protegens (formerly Pseudomonas fluorescens) CHA0 in our study (Jousset et al. 2006). CHA0 is a reference plant‐associated strain initially isolated from a tobacco root (Stutz et al. 1986). Arabidopsis thaliana ecotype Col‐0 (Arabidopsis), a model for plant research, was used as the host. We conducted an experimental evolution assay in a gnotobiotic system with ECO2 box (http://www.eco2box.com/ov80xxl_nl.htm) for bacteria to evolve on the roots of a clonal plant as described in previous study (Li et al. 2020). Briefly, 106 cells of the ancestral CHA0 population were introduced into the rhizosphere of Arabidopsis growing in carbon‐free and Hoagland medium (Hoagland and Arnon 1950) amended silver sand. After a 4‐week growth cycle, the rhizosphere CHA0 population was harvested from the root surface and the cell density was determined with flow cytometry (BD Accuri™ C6 Plus, thresholds for FSC: 2000, SSC: 8000). Then 106 cells forming the suspension were passed to a new box with plantlet for the next growth cycle, forming an independent bacterial evolving line. In total, six growth cycles were conducted for five independent evolving lines. Sixteen colonies were retrieved from ancestral, cycle 2, 4, and 6 of each line, respectively, to represent the population at different time points. Possible contamination of the systems was checked by plating the suspension on a general‐purpose, nonselective media, Trypticase soy agar (TSA) plate, followed by the verification of fluorescent signal of GFP‐carrying colonies under UV light.
BACTERIAL LIFE HISTORY TRAITS MEASUREMENTS
For randomly picked colonies, we measured a variety of bacterial life history traits reflecting the ability of bacteria host root‐derived carbon source usage, stress resistance, and social behaviors as described in previous work (Li et al. 2020). Briefly, we applied the 96‐well microplates to measure all these traits by monitoring the optical density (OD) values of either bacterial cell density under different growth conditions or the color intensities reflecting the production of various compounds via colorimetric assays with a plate reader (SPECTROstar Nano).
The bacterial root‐derived carbon source usage ability was determined as the bacterial growth yield in modified Ornston and Stanier (OS) minimal medium (Højberg et al. 1999), which was supplemented with single carbon sources to a final concentration of 0.5g L−1. Fourteen abundant carbon sources in Arabidopsis root exudates (Chaparro et al. 2013) were measured, including alanine, arabinose, butyrolactam, fructose, galactose, glucose, glycerol, glycine, lactic acid, putrescine, serine, succinic acid, threonine, and valine. We measured bacterial growth yield in stress‐free 1 g L−1 tryptic soy broth and their relative growth, which was defined as resistance, in a variety of stresses including acidic (pH 5) and alkaline (pH 9) conditions, salt stress (2% NaCl), water potential stress (15% polyethylene glycol (PEG)‐6000), oxidative stress (0.0025% H2O2), and antibiotics tetracycline (1 μg mL−1), streptomycin (1 μg mL−1), and penicillin (5 μg mL−1). For production of public goods, we measured bacterial auxin (indole‐3‐acetic acid or IAA) with a colorimetric test (Glickmann and Dessaux 1995), proteolytic activity by the adapted assay from Smeltzer et al. (Smeltzer et al. 1993), iron‐chelating ability using a modified chrome azurol S (CAS) assay (Alexander and Zuberer 1991), biofilm formation with a modified Chrystal Violet staining assay (Moskowitz et al. 2004), and tryptophan side chain oxidase activity with a colorimetric assay (Oberhänsli et al. 1991). We further assessed CHA's antimicrobial activity by quantifying their effects on the growth of the fungal pathogen Verticillium dahliae and Fusarium oxysporum, and that of the bacterium pathogen Ralstonia solanacearum.
STATISTICAL ANALYSES
Innovation of correlation patterns
We assessed how the correlation patterns between all possible pairs of the measured traits changed in evolved populations (ti; i = 2, 4, 6 plant cycle) compared to ancestral population (t 0). We measured a total of 28 bacterial life history traits, resulting in a total of (28 × 27)/2 = 378 pairs of traits that were tested for their interaction conservativism for each replicate's evolved population and each time point (ti), respectively. We classified changes in correlation patterns into five possible categories (Fig. 1):
Figure 1.
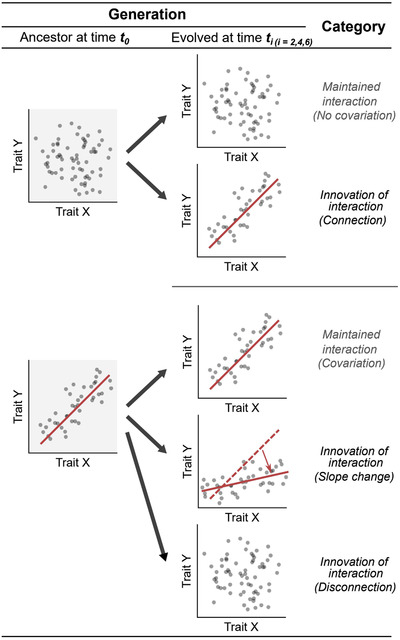
. Overview of the classification of the possible evolutionary dynamics of trait correlation patterns. Categories are based on how the correlation pattern of a pair of traits changes from ancestral to evolved populations. Solid lines represent significant correlation.
Maintained lack of interaction (no correlation): a trait pair X and Y remains independent during the whole experiment;
Maintained interaction (correlation): a trait pair X and Y correlates in the same way during the whole experiment;
Innovation of interaction (connection): a trait pair X and Y shows no correlation at t 0 but is significantly correlated at t i;
Innovation of interaction (slope change): a trait pair X and Y shows an association at t 0 and ti, but with an altered slope; and
Innovation of interaction (disconnection): a trait pair X and Y shows the association at t 0 but this association is no longer observed at ti.
Note that for category (iv), we fitted a linear model including the interaction term (X*time) to assess if the slope of the linear model changes statistically significantly over time. Here, the distinction between association and no association is made based on statistical significance (P ≤ 0.05). P‐values were corrected via the Benjamini‐Hochberg (BH) procedure to control the false discovery rate, which is considered as an appropriate correction method where the number of tests is considerably larger than the number of samples (Benjamini and Hochberg 1995). Finally, the relative proportion of each category was calculated.
Correlation network analysis
To illustrate the topology structure of the bacterial trait integration network, Pearson correlations were used for weighted undirected correlation network construction with igraph package (Messier et al. 2017; Kleyer et al. 2019). Pearson correlation test was applied for all pairwise traits to obtaining a correlation matrix at each cycle and at each line with R function corr.test(). To alleviate the inflation of Type I error and control the false discovery rate, the P‐values were adjusted with the Benjamini‐Hochberg procedure, and only statistically significant correlations (P ≤ 0.05) are included as network edges. Therefore, a network represents bacterial traits as nodes and statistically significant correlations as undirected edges. The edges are weighted according to the absolute values of correlation coefficient. The metric “modularity” is calculated with modularity() function, which described the tendency to form distinct modules in a trait network. Distinct modules were determined via “cluster_edge_betweenness” function. Each module represents a subset of traits that correlate more tightly among themselves than with others. The distinction is based on the fraction of edges within the given module minus the fraction if the edges were random. “layout.graphopt” layout has been used for network visualization.
Results
EMERGENCE OF NOVEL PATTERNS OF TRAIT CO‐VARIATION
We assessed whether bacterial phenotypic integration patterns persist or if new populations rewire the interactions between traits during host adaptation. Generalized linear models assessing the changes in correlation between all possible pairs of measured traits for each population revealed that a large proportion of traits rapidly change their relationships (Fig. 2). In all replicate lines, we observed a parallel pattern of rapid rewiring of trait correlations, with a steady decline over time of the proportion of the conserved interactions relative to the ancestral population (Fig. 2A). After two plant growth cycles, on average about 80% of the initial pairwise interactions between traits were still preserved, but this proportion dropped to about 60% by the end of the experiment. Although the same global trend was observed across all lines, distinct differences were observed between the different replicates, with retained interactions ranging from below 50% (line 4 and 5) to 78 % (line 1) after the last growth cycle.
Figure 2.
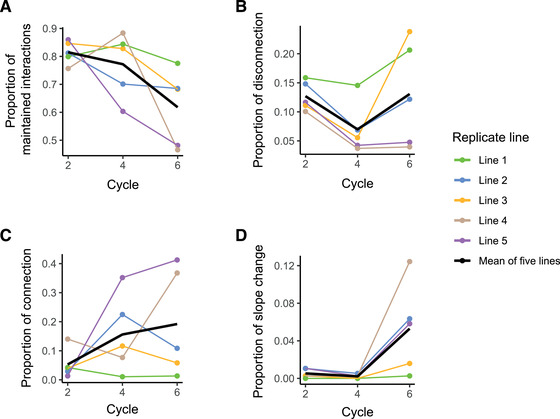
. Rapid rewiring of trait correlation networks during evolution of Pseudomonas protegens CHA0 in the rhizosphere of gnotobiotic Arabidopsis thaliana Col‐0. The systematic classification scheme for traits correlation patterns has been presented in Methods and Figure 1. (A) We defined maintained bacterial trait interactions as the fraction of the pairwise trait combinations that either remained with no correlation or kept significant association with unchanged correlation coefficient in the evolved population as compared to the ancestral population (ANOVA, nonsignificant Slope × Time interaction). (B–D) Proportion of pairs of traits in which we observed innovation, defined as significant changes of pairwise variable interactions, including disconnection (B, loss of significant correlation), connection (C, emergence of a significant correlation), and slope change (D, significant change in slope). Cycle numbers on the x‐axis represent the number of plant growth cycles, with bacteria being transferred to a new plant at the end of each growth cycle. Different colors indicate the five replicate lines, and the black line represents the average of all lines. In total, (28 × 27)/2 = 378 pairs of traits were tested for their interaction conservativism. The distinction between association and no association is made based on statistical significance. P‐values were adjusted with Benjamini‐Hochberg (BH) procedure.
We further scrutinized changes in correlations between pairs of traits to assess innovation of trait interactions in more detail. We broke down innovations of new interactions into three categories: disconnections, connections, and slope changes. In the disconnection scenario, two traits that were previously significantly correlated in the ancestral population lose their correlation with each other in the evolved population (ANOVA, P > 0.05) (Fig. 1). At an earlier evolutionary stage (plant cycle two), disconnection occurred in 10–15% of initial pairwise interactions (Fig. 2B). In lines 1 and 3, the disconnection proportion reached about 20%, whereas line 4 and 5 showed markedly different dynamics with disconnections accounting for only 5% of the initial pairwise interactions in the last cycle (Fig. 2B).
In the connection scenario, two initially uncorrelated traits became covarying in the evolved population (Fig. 1). This kind of innovation was relatively common in our study, affecting on average about 20% of initial pairwise interactions in the last cycle (Fig. 2C). Replicate lines 1 and 3 showed the lowest proportion of connection, accounting for less than 10% of initial pairwise interactions along different plant cycles, mirroring the high level of disconnection in these two lines. In contrast, the lines with the lowest disconnection proportion, lines 4 and 5, harbored the highest proportion of connection, which accounted for up to 40% of initial pairwise interactions after the last plant growth cycle.
Finally, we considered the slope change scenario, in which a pair of traits would remain significantly associated during the experiment, yet the slope of the correlation would change (ANOVA, Significant Slope × Time interaction, P < 0.05) (Fig. 1). The proportion of this type of innovation was relatively low in our study, although there was a trend of increasing slope change with time. Slope change could be found in only about 1% of initial pairwise interactions at cycle 2 and reached about 6% after six cycles (Fig. 2F). Replicate lines 1 displayed the lowest proportion of slope change, which remained at about 1% of initial pairwise interactions during the whole experiment. In contrast, replicate line 4 showed the highest proportion of slope change innovation, with this scenario accounting for approximately 12% of initial pairwise interactions.
EVOLUTION OF PHENOTYPIC TRAIT NETWORKS
To address in more detail the changes in the topology of phenotypic trait networks during bacterial evolution, we constructed trait correlation networks, respectively, for the ancestral and each of the five evolved populations at the end of each plant growth cycle (each network was based upon the phenotypic data of 16 random isolates). We described the general topology as modularity, an index reflecting the clustering of traits into discrete modules, which are weakly connected to other modules (Clauset et al. 2004).
We observed two separate modules in the ancestral population, one consisting solely of carbon usage‐related traits (later: “resource use” module) and a second one containing all traits linked to biotic and abiotic stress resistance (later: “stress resistance” module). All traits in the resource use modules were linked to growth on carbon sources representative of the plant root environment and all were positively correlated with each other (Fig. 3A). The second, stress resistance‐related module, was formed by positively correlated stress‐related traits (Fig. 3A). Most of the public goods traits, including the production of iron‐scavenging siderophores, extracellular proteases, biofilm formation, and inhibition of bacteria and fungi, were scattered around the networks, with none of them forming a separate module nor belonging to the “resource use” or “stress resistance” module.
Figure 3.
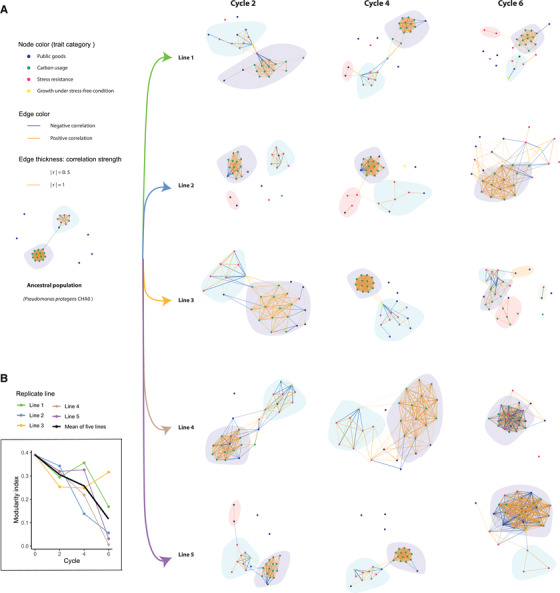
Evolutionary dynamics of the topology of Pseudomonas protegens CHA0 trait correlation networks in the rhizosphere of gnotobiotic Arabidopsis thaliana Col‐0. (A) Networks of ancestral population and evolved populations at end of each cycle (each network was based upon the phenotypic data of 16 random isolates). Each node represents one of the 28 measured phenotypic traits. Node colors represent a trait's functional category. The two colors of the edge represent positive and negative correlations, respectively. Edges are weighted according to correlation strength, the absolute values of correlation coefficient “r.” Only significant correlations at 0.05 alpha level are included. The P‐values were adjusted with Benjamini‐Hochberg (BH) procedure. Distinct modules are indicated with light background colors. (B) Modularity index. The different replicate evolutionary lines are indicated by lines of different colors; the black line shows the average modularity across all replicate lines. Cycle numbers represent the number of plant growth cycles, with bacteria being transferred to a new plant at the end of each growth cycle.
The modular structure of the ancestral network was rapidly lost during bacterial adaptation to the rhizosphere environment (Fig. 3). In cycle 2 and 4, there were still clear patterns of two separate modules in all replicate lines. However, the “stress resistance” module showed a trend of deterioration over time. After cycle 6, only line 3 still contained a separate small module consisting of a subset of stress traits. Although become disconnected in the other lines, in line 4 these stress traits merged with the “resource use” module forming one meta‐module. Unlike the dynamic change of the “stress resistance” module, the “resource use” module remained stable over the course of rhizosphere adaptation. As a result of the networks’ topological change, the modularity index shown a parallel steady decrease, dropping to zero in three‐out‐of‐five lines after cycle 6. The only exception was line 3, where we observed a splitting of the “resource use” module after cycle 6, resulting in increased modularity with separate modules formed by carbon usage‐related traits and remaining stress resistance traits. It is worth noting that, there are also new small modules appeared in most of the lines at different cycles. In contrast to the disintegration of stress resistance‐related traits, some of the public good traits, which were originally disconnected, became integrated into modules at the end of the experiment (Fig. 3). In line 4 and 5, all measured traits integrated into a meta‐module after cycle 6.
PARALLEL EVOLUTION OF FUNCTIONAL TRAITS CATEGORIES
We classified traits into a priori functional categories of carbon source usage, stress resistance, and public goods. We then further zoomed in on the changes in trait correlation for each functional group of traits. The rationale behind this classification was to shed light on the evolutionary implications for the bacteria (resource use reflects the ability to consume plant‐derived resources; stress resistance, the ability to withstand a range of abiotic stressors; the selected public goods being involved in host plant nutrition and protection against diseases) (Lugtenberg and Kamilova 2009).
The ability to use several carbon sources remained a cohesive unit, with a correlation index oscillating around 0.8 in four‐out‐of‐five replicate lines (Fig. 4). The dramatic drop in line 3 after cycle 6 is also visible in Figure 3, where the “resource use” module split. In contrast, stress resistance traits rapidly become disconnected from each other (Fig. 4), with correlation index dropping in all replicates from 0.8 to 0.4–0.6. This trend is in line with the collapse of the “stress resistance” module as seen in Figure 3.
Figure 4.
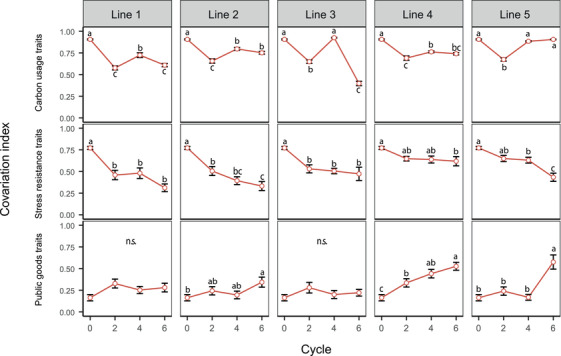
Correlation of different functional category traits of Pseudomonas protegens CHA0 populations. Correlation levels were calculated as the mean of absolute values of correlation coefficient r of each functional group of traits. Carbon usage traits consisted of the usage of 14 carbon sources representative for Arabidopsis root exudates. Stress resistance traits included two biotic and five abiotic stress resistance traits. Public goods include the production of iron‐scavenging siderophores, extracellular proteases, biofilm formation, and inhibition of bacteria and fungi. The correlation changes of each functional category over cycles were demonstrated by one‐way ANOVA, with different letters indicating significant differences and n.s. standing for not significant.
Public goods were not integrated in the initial network and did not co‐vary in the ancestral population. However, they rapidly became connected to each other over the course of the experiment, especially in lines 4 and 5, where the correlation index increased from 0.1 to an average of 0.6 within six plant growth cycles. This indicates that at the end of the experiment, bacteria produced either all or none of the measured public goods.
ADAPTATION THROUGH DISCONNECTION
As previously shown in Figure 2, several traits became disconnected during the experiment. We illustrate how disconnection gradually occurred with the evolution of the relationship between resistance to acidic and alkaline conditions along the bacterial population transfer cycles (Fig. 5; Table S1). We highlight here pH‐related stresses as an example, given the fact that the rhizosphere environment is often more acidic than the surrounding soil (Hinsinger et al. 2003) and pH has been shown to be a major driver of microbial community structure and activity in soils (Fierer and Jackson 2006; Rousk et al. 2010).
Figure 5.
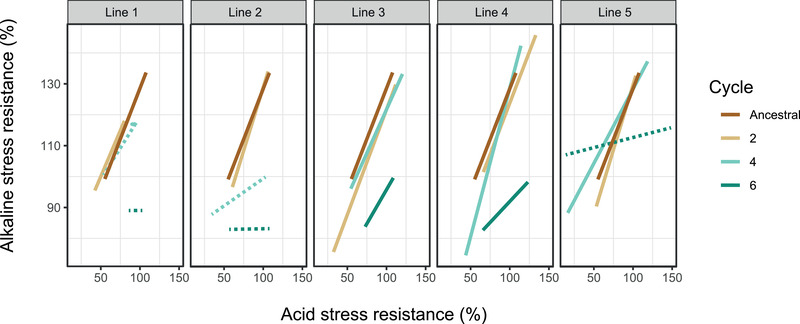
Evolutionary change of bacterial acid and alkaline stress resistance correlation patterns in the rhizosphere. The x‐axis presents the acid stress (pH 5) resistance capacity, and the y‐axis depicts the alkaline stress (pH 9) resistance capacity. Each block shows the bacterial acidic and alkaline stress resistance correlation patterns of the ancestral and all five replicates population. Different cycles are indicated by different colors, as shown in the legend. Solid lines indicate significant linear relationship, whereas dash lines indicate for no correlation. The linear regression indices are listed in Table S1.
In the ancestral population, resistance to high and low pH was significantly positively correlated, suggesting that the mechanisms involved in the resistance to these stressors were coregulated. In the early evolutionary stage (cycle 2), all lines were similar to the ancestral population (Fig. 5; Table S1), whereas at later evolutionary stages disconnection appeared: disconnection occurred after cycle 4 in lines 1 and 2, and in line 5 after cycle 6 (Fig. 5; Table S1). This parallel disconnection phenomenon indicates that, at the end of the experiment, bacteria varied in their ability to withstand acidic stress, but all had lost most of their ability to cope with alkaline stress. This fits to the acidic conditions in the rhizosphere, making resistance to acidic conditions relevant while high pH tolerance being a costly and nonrequired trait in the rhizosphere environment.
Discussion
EVOLUTION OF BACTERIAL TRAIT CORRELATION PATTERNS
Evolution has often been addressed from the perspective of single traits. More recently, various evolutionary scientists have proposed that assessing traits at the level of the whole phenotype, where the interplay between traits rather than the value of individual ones matters, may allow better understanding evolutionary dynamics (Pigliucci 2003; Hallgrímsson and Hall 2005; Tonsor and Scheiner 2007; Herrera ). Although evidence supporting for phenotypic integration has been reported for observational studies or simulations (Berg 1960; Pigliucci 2003; Kashtan and Alon 2005; Parter et al. 2007; Klingenberg 2008), to our knowledge the process of adjustment in phenotypic integration has rarely been shown experimentally. Here, we address this knowledge gap by following in real time the rapid evolution of phenotypic integration patterns during bacterial adaptation to the rhizosphere environment. A central result of this work is that traits do not evolve independently. Although one might expect several traits to evolve in a new environment, we show here that traits evolve as syndromes, and that the link between associated traits is also subject to evolution. A parallel reduction in maintained bacterial phenotypic trait interactions was observed, with about half of the initial interactions being significantly changed after evolution in the rhizosphere of Arabidopsis for 6 months (Fig. 1). We found that the way traits interconnected is very dynamic and changes within few hundreds of generations, shedding a new perspective on evolutionary processes. This is an important step toward being able to understand and further predict the complex bacterial adaptation trajectories in the changing environment. In the following paragraphs, we discuss some of the main findings and their implications for bacterial evolution.
A TRAIT NETWORK APPROACH TO MICROBIAL ADAPTATION
One of the salient features of the observed reshaping of trait correlation networks is the stability of traits within the “Resource use” module. This module, encompassing the ability to grow on the various carbon sources representative of the plant root environment (Fig. 3A), persisted during the whole experiment (Fig. 4). The carbon sources used in our study were chosen to represent the dominant Arabidopsis root exudates. This strong cohesion of main carbon source usage traits suggests that most resource acquisition traits might share a particular genetic base or conserved metabolic pathways. Even though root exudates are mainly small and soluble molecules that can be taken up directly, diverse metabolic investments are still required. The colocalization and co‐occurrence of these highly concentrated root‐derived carbon sources may have facilitated the evolutionary persistence of the “Resource use” module. In addition, the strong cohesion of main carbon source usage traits could allow bacteria to respond to carbon fluctuations more sensitively and robustly, as root exudates are mainly excreted from root tips that move through the soil, resulting in a dynamically changing resource locations (Van Bruggen et al. 2008). Also, we propose that the persistence of the resource use module during our experiment indicates that maintaining the ability to use several resources is important for competitive ability under the tested rhizosphere conditions. This is in line with the evolution of generalist mutants in Pseudomonas spp. in response to competition (Jousset et al. 2016).
In contrast, traits linked to stress resistance were strongly coupled in the ancestral population but rapidly declined over the course of the experiment (Figs. 3A and 4). Coordinated resistance to stresses allows bacteria to respond to changing environmental and ecological pressures more sensitively and robustly. This co‐regulation may be useful in a changing environment, where bacteria may expect to face at any time a range of unrelated stressors. Exposing bacteria to a stressor such as starvation, low pH, or oxidative stress triggers bacterial responses that not only protect them against the current stressor, but also other stressors that they have not yet encountered (Hecker et al. 2007; Battesti et al. 2011). However, in the present simplified experimental system containing only one plant and one bacterial species, simultaneous activation of various stress defense mechanisms may have turned to a costly feature. The disaggregation of this module (Fig. 4) may therefore be seen as a bet‐hedging strategy, in which the population diversifies into specialists each coping with a few stressors (Boles et al. 2004).
Unlike the other two functional trait categories, public goods traits did not form a separate module in the ancestral population. Rather than forming a separate module, many public goods traits merged into the overall correlation trait network over the course of the experiment. This shift was particularly evident in evolutionary lines 4 and 5 at cycle 6 (Fig. 3A). In contrast, in line 1 these traits were still kept disconnected. Interestingly, these different trajectories among replicate lines were not reflected in the trajectory of the broader topology structure of trait correlation networks. Rather, the trajectory represented a parallel evolution toward a lower modularity that was observed in all replicate lines (Fig. 3B).
It is possible that the decline of network modularity, observed in all experimental lines, is simply due to drift, with each trait getting connected or disconnected to other traits at random during the evolutionary processes. However, the following observations speak in favor of a selective process. First, the abilities to use different carbon sources remained consistently correlated to each other (Fig. 4), whereas other traits showed more temporal variability. This association of temporal dynamics with the functionality of the observed trait may be a hint for the underlying evolutionary pressures. Second, the deterioration of “stress resistance” module occurred in all replicate lines, even with similar rate of change (Figs. 4 and 5). Such a convergent patterns is unlikely to emerge as the result of pure stochastic processes. Therefore, although we do not deny the role of random drift completely, we conclude that the reduction of network modularity was resulted from a strong selective pressure breaking the ancestral patterns.
PARALLEL EVOLUTION OF TRAIT DISCONNECTION
The loss of correlation between different stress resistance traits was highly reproducible across all replicate lines (Fig. 4). Furthermore, this uncoupling process happened gradually over the course of the experiment, as illustrated by loss of alkaline stress resistance, a trait originally strongly correlated with acidity tolerance (Fig. 5). A static trait integration network in which correlation between traits remains constant would not have permitted the observed adaptation, as loss of resistance to high pH would have implied a simultaneous loss of resistance to the acidic conditions prevailing in the rhizosphere (Hinsinger et al. 2003). Thus, dynamic trait integration may provide the flexibility to evolve stress resistance strategies matching the current needs under nonfluctuating conditions.
Coordination between traits and the lack of intricacy may hinder the ability to respond in any particular direction (Hansen and Houle 2008; Singh et al. 2008). At a kingdom level, a comparison of 300 bacterial genomes covering the major phyla revealed that the metabolic network modularity index showed a parallel trend of modularity reduction from ancestors to their descendants (Kreimer et al. 2008). Further, generalist species occupying a broader range of niches harbored higher modularity scores than specialists such as endosymbionts or mammal‐specific pathogens. Another comparative genomic study revealed that bacteria evolved in variable environments have more modular metabolic networks than bacteria from more constant conditions (Parter et al. 2007). Based on these studies, we interpret the rapid decline in trait correlation network modularity in the evolved bacterial populations as a consequence of a constant selection pressure in the rhizosphere. This conclusion is, however, at odds with the general notion that the rhizosphere is a highly dynamic environment (Philippot et al. 2013). We propose as an alternative explanation that the measured trait networks may reveal the diversification process to several micro‐niches such as root tips and old roots, each selecting for specific life histories (Achouak et al. 2007).
IMPLICATION FOR THE EVOLUTION OF PLANT‐ASSOCIATED BACTERIA
Our study demonstrates the importance of a phenotype‐level approach to study adaptation. We show that evolution rapidly reshapes the level of trait integration. As modular bundling is important to increase the rate of adaptation (Orr 2000; Welch and Waxman 2003), the decline in modularity may cause a slower adaptation rate in future generations confronted with a new environment, such as bulk soil or a new host species. Further, this study sheds a new perspective regarding the adaptation of microorganisms to the rhizosphere. To date, microbial adaptation to a new host has mainly been approached using directional selection or diversification into different life history strategies along existing trade‐offs such as growth rate versus stress resistance (Ferenci 2016). By demonstrating that existing trade‐offs are malleable over time, we show that diversification processes may generate truly new phenotypes out of the boundaries set by the ancestor. This knowledge may serve as a basis for the development of new genetic improvement strategies to enhance mutualistic microorganisms with improved abilities to foster plant growth while still expressing the traits needed for survival in the rhizosphere. We anticipate that the genetics of trait network plasticity may gain in scientific relevance as microorganisms are increasingly sought to be used to replace pesticides. These insights may help to overcome previously unreliable results stemming from unreliable survival and efficiency under field conditions. Finally, the reshaping of trait co‐variance networks may be explained by constraints emerging from trait integration: The decline in modularity that is favorable in simplified and constant conditions, such as the ones prevailing in this experiment, could lead to a specialization to these conditions, which at the same time could give rise to deleterious effects when the bacteria are again faced with more complex and unpredictable conditions typical of the soil environment.
AUTHOR CONTRIBUTIONS
EL, PB, and AJ designed the experiments. EL performed the experiment. EL, MR, and AJ analyzed the data. All authors collegially wrote the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
The data and R script are deposited at https://doi.org/10.17632/wc2xk6mtjj.1.
Associate Editor: T. Chapman
Handling Editor: T. Chapman
Supporting information
Table s1. Statistic table of linear regression indices of bacterial acid and alkaline stress resistance abilities during adaptation in the rhizosphere in different replicate lines.
ACKNOWLEDGMENT
EL is funded by the China Scholarship Council (CSC) scholarship program.
Contributor Information
Erqin Li, Email: erqinli22@gmail.com.
Alexandre Jousset, Email: A.L.C.Jousset@uu.nl.
LITERATURE CITED
- Achouak, W. , Conrod S., Cohen V., and Heulin T.. 2007. Phenotypic variation of Pseudomonas brassicacearum as a plant root‐colonization strategy. Mol. Plant‐Microbe Interact. 17:872–879. [DOI] [PubMed] [Google Scholar]
- Alexander, D. B. , and Zuberer D. A.. 1991. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12:39–45. [Google Scholar]
- Bais, H. P. , Weir T. L., Perry L. G., Gilroy S., and Vivanco J. M.. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57:233–266. [DOI] [PubMed] [Google Scholar]
- Barabási, A.‐L. , and Oltvai Z. N.. 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5:101–113. [DOI] [PubMed] [Google Scholar]
- Battesti, A. , Majdalani N., and Gottesman S.. 2011. The RpoS‐mediated general stress response in Escherichia coli . Annu. Rev. Microbiol. 65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289–300. [Google Scholar]
- Berg, R. L. 1960. The ecological significance of correlation pleiades. Evolution 14:171–180. [Google Scholar]
- Blount, Z. D. , Barrick J. E., Davidson C. J., and Lenski R. E.. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles, B. R. , Thoendel M., and Singh P. K.. 2004. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. 96:1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J. M. , Badri D. V., Bakker M. G., Sugiyama A., Manter D. K., and Vivanco J. M.. 2013. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 8:e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauset, A. , Newman M. E. J., and Moore C.. 2004. Finding community structure in very large networks. Phys. Rev. E 70:066111. [DOI] [PubMed] [Google Scholar]
- Damkiaer, S. , Yang L., Molin S., and Jelsbak L.. 2013. Evolutionary remodeling of global regulatory networks during long‐term bacterial adaptation to human hosts. Proc. Natl. Acad. Sci. USA 110:7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci, T. 2016. Trade‐off mechanisms shaping the diversity of bacteria. Trends Microbiol. 24:209–223. [DOI] [PubMed] [Google Scholar]
- Fierer, N. , and Jackson R. B.. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson, A. , Jelsbak L., Yang L., Johansen H. K., Ciofu O., Høiby N., and Molin S.. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev Microbiol. 10:841–851. [DOI] [PubMed] [Google Scholar]
- Glickmann, E. , and Dessaux Y.. 1995. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61:793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson, B. , and Hall B. K.. 2005. Variation: a central concept in biology. 1st ed. Academic Press, Burlington, MA. [Google Scholar]
- Hansen, T. F. , and Houle D.. 2008. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 21:1201–1219. [DOI] [PubMed] [Google Scholar]
- Hecker, M. , Pané‐Farré J., and Uwe V.. 2007. SigB‐dependent general stress response in Bacillus subtilis and related gram‐positive bacteria. Annu. Rev. Microbiol. 61:215–236. [DOI] [PubMed] [Google Scholar]
- Hindré, T. , Knibbe C., Beslon G., and Schneider D.. 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 10:352–365. [DOI] [PubMed] [Google Scholar]
- Hinsinger, P. , Plassard C., Tang C., and Jaillard B.. 2003. Origins of root‐mediated pH changes in the rhizosphere and their responses to environmental constraints: a review. Plant Soil 248:43–59. [Google Scholar]
- Hoagland, D. R. , and Arnon D. I.. 1950. The water‐culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 347:32. [Google Scholar]
- Højberg, O. , Schnider U., Winteler H. V., Sørensen J., and Haas D.. 1999. Oxygen‐sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low‐oxygen habitats in soil. Appl. Environ. Microbiol. 65:4085–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan, R. W. , Culver D. C., and Fong D. W.. 1994. The dual role of selection and evolutionary history as reflected in genetic correlations. Evolution 48:587–596. [DOI] [PubMed] [Google Scholar]
- Jousset, A. , Eisenhauer N., Merker M., Mouquet N., and Scheu S.. 2016. High functional diversity stimulates diversification in experimental microbial communities. Sci. Adv. 2:e1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset, A. , Lara E., Wall L. G., and Valverde C.. 2006. Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl. Environ. Microbiol. 72:7083–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan, N. , and Alon U.. 2005. Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. 102:13773–13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyer, M. , Trinogga J., Cebrián‐Piqueras M. A., Trenkamp A., Fløjgaard C., Ejrnaes R., Bouma T. J., Minden V., Maier M., Mantilla‐Contreras J., et al. 2019. Trait correlation network analysis identifies biomass allocation traits and stem specific length as hub traits in herbaceous perennial plants. J. Ecol. 107:829–842. [Google Scholar]
- Klingenberg, C. P. 2008. Morphological integration and developmental modularity. Annu. Rev. Ecol. Evol. Syst. 39:115–132. [Google Scholar]
- Kreimer, A. , Borenstein E., Gophna U., and Ruppin E.. 2008. The evolution of modularity in bacterial metabolic networks. Proc. Natl. Acad. Sci. 105:6976–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E. , de Jonge R., Liu C., Jiang H., Friman V.‐P., Bakker P. H. M., and Jousset A.. 2020. Rapid evolution of bacterial mutualism in the plant rhizosphere. bioRxiv 10.1101/2020.12.07.414607. [DOI] [PMC free article] [PubMed]
- Lugtenberg, B. , and Kamilova F.. 2009. Plant growth promoting rhizobacteria. Annu. Rev. Microbiol. 63:541–556. [DOI] [PubMed] [Google Scholar]
- Mandel, M. J. , Wollenberg M. S., Stabb E. V., Visick K. L., and Ruby E. G.. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 457:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier, J. , Lechowicz M. J., McGill B. J., Violle C., and Enquist B. J.. 2017. Interspecific integration of trait dimensions at local scales: the plant phenotype as an integrated network. J. Ecol. 105:1775–1790. [Google Scholar]
- Meyer, J. R. , Dobias D. T., Weitz J. S., Barrick J. E., Quick R. T., and Lenski R. E.. 2012. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz, S. M. , Foster J. M., Emerson J., and Burns J. L.. 2004. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 42:1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhänsli, T. , Défago G., and Haas D.. 1991. Indole‐3‐acetic acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens: role of tryptophan side chain oxidase. Microbiology 137:2273–2279. [DOI] [PubMed] [Google Scholar]
- Orr, H. A. 2000. Adaptation and the cost of complexity. Evolution 54:13–20. [DOI] [PubMed] [Google Scholar]
- Parter, M. , Kashtan N., and Alon U.. 2007. Environmental variability and modularity of bacterial metabolic networks. BMC Evol. Biol. 7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot, L. , Raaijmakers J. M., Lemanceau P., and van der Putten W. H.. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11:789–799. [DOI] [PubMed] [Google Scholar]
- Pigliucci, M. 2003. Phenotypic integration: studying the ecology and evolution of complex phenotypes. Ecol. Lett. 6:265–272. [Google Scholar]
- Ravasz, E. , Somera A. L., Mongru D. A., Oltvai Z. N., and Barabási A. L.. 2002. Hierarchical organization of modularity in metabolic networks. Science 297:1551–1555. [DOI] [PubMed] [Google Scholar]
- Rousk, J. , Bååth E., Brookes P. C., Lauber C. L., Lozupone C., Caporaso J. G., Knight R., and Fierer N.. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351. [DOI] [PubMed] [Google Scholar]
- Singh, A. H. , Wolf D. M., Wang P., Arkin A. P., and Lenski R. E.. 2008. Modularity of stress response evolution. Proc. Natl. Acad. Sci. 105:7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeltzer, M. S. , Hart M. E., and Iandolo J. J.. 1993. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus . Infect. Immun. 61:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz, E. W. , Défago G., and Kern H.. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181–185. [Google Scholar]
- Tonsor, S. J. , and Scheiner S. M.. 2007. Plastic trait integration across a CO2 gradient in Arabidopsis thaliana . Am. Nat. 169:E119–E140. [DOI] [PubMed] [Google Scholar]
- Van Bruggen, A. H. C. , Semenov A. M., Zelenev V. V., Semenov A. V., Raaijmakers J. M., Sayler R. J., and De Vos O.. 2008. Wave‐like distribution patterns of Gfp‐marked Pseudomonas fluorescens along roots of wheat plants grown in two soils. Microb. Ecol. 55:466–475. [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse, P. , Quaiser A., Duhamel M., van Le A., and Dufresne A.. 2015. The importance of the microbiome of the plant holobiont. New Phytol. 206:1196–1206. [DOI] [PubMed] [Google Scholar]
- Welch, J. J. , and Waxman D.. 2003. Modularity and the cost of complexity. Evolution 57:1723–1734. [DOI] [PubMed] [Google Scholar]
- Winstanley, C. , O'Brien S., and Brockhurst M. A.. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 24:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table s1. Statistic table of linear regression indices of bacterial acid and alkaline stress resistance abilities during adaptation in the rhizosphere in different replicate lines.
Data Availability Statement
The data and R script are deposited at https://doi.org/10.17632/wc2xk6mtjj.1.


